Abstract
Resistance to platinum-based drugs, such as cisplatin (CDDP), has been one of the major factors adversely affecting the clinical prognosis of patients with advanced non-small cell lung cancer (NSCLC). While it has been demonstrated that dysregulation of microRNAs (miRNAs) may contribute to cisplatin resistance in NSCLC, the underlying mechanisms remain largely unclear. In the present study, the effect of exosomal miR-1273a on cisplatin sensitivity of NSCLC was investigated. Microarray analysis was conducted to analyze the miRNA expression profiles in exosomes isolated from A549 cells treated with or without CDDP, and miR-1273a was found to be the most prominently downregulated miRNA in CDDP-treated exosomes. Overexpression of miR-1273a significantly increased the cytotoxicity of CDDP and induced apoptosis in A549 cells. Syndecan binding protein (SDCBP) was predicted to be a direct target of miR-1273a by bioinformatics and was found to be downregulated by miR-1273a in A549 cells. Furthermore, decreased plasma exosomal miR-1273a and increased plasma SDCBP levels were found to be associated with worse therapeutic outcomes of patients with advanced NSCLC receiving platinum-based chemotherapy. These findings suggest that miR-1273a is closely associated with the development of cisplatin resistance and may serve as a potential prognostic biomarker and therapeutic target for NSCLC.
Keywords: non-small cell lung cancer, exosomes, miR-1273a, cisplatin
Introduction
Lung cancer is the most common malignant tumor worldwide, and its morbidity and mortality rank first among all cancers (1). The most common histological subtype is non-small cell lung cancer (NSCLC), accounting for more than 80% of all lung cancer cases (2). Unfortunately, the majority of patients with NSCLC are at an advanced stage at the time of initial diagnosis, and have missed the opportunity to undergo surgery, which results in a poor prognosis (3). Although platinum-based chemotherapy is currently the standard first-line treatment for these patients, some NSCLC patients are not sensitive to platinum-based chemotherapy, with an overall response rate of only 30% (4,5). In addition, most patients with NSCLC eventually develop resistance to the chemotherapy, thus greatly limiting the clinical application of chemotherapeutic drugs and leading to a worse prognosis. Therefore, it is crucial to explore the molecular mechanism underlying the low response rate of NSCLC to platinum-based chemotherapy.
Exosomes are nanoscale membrane vesicles secreted by cells into the extracellular space to mediate intercellular communication (6). Therefore, exosomes are not only important carriers of bioactive substances, but also crucial mediators for information exchange between cells (7,8). It has been demonstrated that exosomes can transmit biological information by transporting coated contents, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and proteins, and participate in the malignant processes of NSCLC through various pathways (9,10). miRNAs are a family of small non-coding RNAs that can alter gene expression post-transcriptionally, thereby regulating tumor cell proliferation, migration, invasion and metastasis, angiogenesis and apoptosis as well as genomic instability (11,12). Since miRNAs in exosomes have the ability to regulate gene expression, are stable in the extracellular environment and display strong resistance to degradation, they may be widely used as diagnostic biomarkers of cancers and other diseases (13). More importantly, secreted exosomal miRNAs may be taken up by recipient cells, and then regulate a variety of biological processes such as proliferation, differentiation and apoptosis by inhibiting the expression of target genes (14–16).
In recent years, miRNAs in exosomes have attracted considerable attention as the changes in the type and amount of miRNAs in exosomes are associated with the resistance of NSCLC cells to chemotherapeutic drugs (17). A recent study demonstrated that miRNAs were differentially expressed in the exosomes of cisplatin (CDDP)-resistant and CDDP-sensitive NSCLC cells. In CDDP-resistant NSCLC cell lines, the amount of miR-100-5p was significantly decreased in exosomes and was functionally involved in CDDP resistance of NSCLC (18). It should be noted that platinum stimulation may also change the expression profiles of miRNAs, many of which are considered to be responsible for drug resistance (19). However, few studies have focused on the expression of miRNAs in exosomes stimulated by platinum drugs and the effect of these miRNAs on the sensitivity of NSCLC to platinum drugs. In the present study, microarray analysis was performed to examine the differential expression profiles of exosomal miRNAs in the NSCLC cell line A549 with or without CDDP treatment, and miR-1273a was identified as a candidate miRNA involved in the sensitivity of A549 cells to CDDP. In addition, the expression of miR-1273a in plasma exosomes of patients with advanced NSCLC before and after CDDP chemotherapy was further evaluated to confirm the clinical significance.
Materials and methods
Cell culture and human specimens
A549 cells were purchased from the American Type Culture Collection (ATCC) and grown in RPMI-1640 medium supplemented with 10% FBS (Biological Industries) and 100 U/ml penicillin-streptomycin at 37°C in an atmosphere of 95% air and 5% CO2. Plasma samples were collected from 49 patients with advanced NSCLC at Daping Hospital, Chongqing, China from January to December 2018 and stored at −80°C until use. The average age of the patients was 57.0±9.8, 32 (65.3%) were male and 17 (34.7%) were female patients. All patients received standard first-line platinum-based chemotherapy (Table S1). To evaluate therapeutic outcomes, the patients were divided into responder [complete response (CR) + partial response (PR)] and non-responder [stable disease (SD) + progressive disease (PD)] groups according to the Response Evaluation Criteria In Solid Tumors (version 1.1) (20). The present study was conducted in accordance with the principles outlined in the Declaration of Helsinki, and was approved by the Ethics Committee of Daping Hospital (ratification no 2018113).
Isolation of exosomes
When the cells had grown to reach a confluence of ~70%, the culture medium was replaced with fresh RPMI-1640 containing 10% Exosome-Depleted FBS (VivaCell). After 48 h of incubation, the supernatant was collected and the exosomes were extracted according to the following steps. The supernatant was centrifuged at 10,000 × g for 30 min at 4°C and filtered by a 0.22-mm filter. Subsequently, the exosomes were precipitated twice by ultracentrifugation at 100,000 × g for 70 min at 4°C. The isolated exosomes were added into the culture medium of the cells for further analysis. To isolate exosomes from patient plasma, plasma samples were centrifugated at 10,000 × g for 20 min at 4°C, and the plasma exosomes were isolated by the Total Exosome Isolation Kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The collected exosomes were resuspended in sterile PBS and stored at −80°C until use.
Transmission electron microscopy (TEM) and nanoparticle-tracking analysis (NTA)
Exosome sample were added to copper grids at room temperature for 5 min, stained with 2% uranyl acetate solution for 1 min, dried for 20 min at room temperature, and then observed using TEM (JEOL, Ltd.). NTA was performed by a nanoparticle tracing assay (ZetaVIEW S/N 17–310; Particle Metrix GmbH) and analyzed using ZetaVIEW 8.04.02 software (Particle Merix GmbH).
Western blotting
Cells and exosomes were lysed in RIPA Lysis Buffer (Beyotime Institute of Biotechnology). A total of 20 µg protein was then separated by 10% SDS-PAGE and then transferred onto a PVDF membrane according to methods described previously (21). Antibodies against CD63 (dilution 1:500; cat. no. ab59478), CD9 (dilution 1:500; cat. no. ab58989), Alix (dilution 1:500; cat. no. ab117600), Calnexin (dilution 1:500; cat. no. ab92573) and β-actin (dilution 1:500; cat. no. ab8226) were purchased from Abcam. Antibody against syndecan binding protein (SDCBP; dilution 1:1,000; cat. no. 22399-1-AP) was purchased from Proteintech (Sanying Biotechnology, Inc.). The band intensity of the western blot images was quantified using Image Lab software (version 1.2.0.12, Bio-Rad Laboratories, Inc.).
Cellular uptake of exosomes
Exosomes were stained with PKH26 and incubated with A549 cells at 37°C for 4 h, and then the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. The nuclei were stained with DAPI. For the control group, PKH26 was added to an equal amount of PBS and then used to treat A549 cells. The fluorescence-labelled exosomes were observed under a confocal microscope (PerkinElmer, Inc.) at ×400 magnification.
Analysis of cell viability and apoptosis
For cell viability analysis, the A549 cells were seeded onto 96-well plates at a density of 5,000 cells per well and incubated for 12 h. The cells were then treated with the indicated concentration of CDDP for an additional 48 h, and the cell viability was determined using Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology), according to the manufacturer's instructions. For apoptosis analysis, the cells were seeded onto 6-well plates at a density of 2×105 cells per well and cultured for 24 h. Subsequently, the cells were seeded into 6-well plates overnight and then treated with 2 µg/ml CDDP. After 36 h of CDDP treatment, the apoptotic cells were detected with the Annexin V-FITC kit (CalbioChem) and quantified using flow cytometry (Gallios, Beckman Coulter).
Microarray analysis of exosomal RNA
Microarray analysis was performed to detect differentially expressed miRNAs using Affymetrix GeneChip miRNA 4.0 array (Affymetrix, Thermo Fisher Scientific, Inc.). GeneChip® Scanner 3000 7G (Affymetrix, Thermo Fisher Scientific, Inc.) was used to scan the arrays. CEL files obtained from the Affymetrix GeneChip were analyzed by the Gminix-Cloud Biotechnology Information (GCBI) platform (http://www.gcbi.com.cn) using Affymetrix default analysis settings and global scaling as the normalization method. SAM method was used to analyze the difference (22). According to the filter condition |Fold Change|>2, the final difference results were obtained.
Transfection experiment
When the cells reached 50% confluence, 20 nM miR-1273a mimic or inhibitors or negative control (Guangzhou RiboBio Co., Ltd.) were mixed with 10 µl of HiPerfect transfection reagent (Qiagen GmbH), and then transfected into A549 cells according to the HiPerfect transfection reagent operating instructions. The sequence of miR-1273a mimic sense was GGGCGACAAAGCAAGACUCUUUCUU and antisense was AAGAAAGAGUUUUGCUUUGTCGCCC. The sequence of miR-1273a inhibitor was CAGUACUUUUGUGUAGUACAA. The sequence of mimics-control was as follows: Sense, UUUGUACUACACAAAAGUACUG and antisense, CAGUACUUUUGUGUAGUACAAA. The sequence of inhibitor-control was CAGUACUUUUGUGUAGUACAA. After 48 h of transfection, the cells were used for the subsequent experiment.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from exosomes or cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized from 200 ng total RNA and amplified by RT-qPCR using TB Green PremixExTaq II (Takara Biotechnology Co., Ltd.). The thermocycling condition for miRNA consisted of 95°C for 20 sec followed by 40 cycles of 10 sec at 95°C, 20 sec at 60°C and 10 sec at 70°C; for mRNA: 95°C for 5 min followed by 39 cycles of 10 sec at 95°C, 15 sec at 60°C and 20 sec at 72°C. Cel-miR-39 was used as an external control in exosomes, and U6 was used as an internal reference in cells. β-actin was used as the endogenous control to normalize the expression of SDCBP. All the primers were synthesized by Guangzhou RiboBio Co., Ltd. The relative expression level of miRNAs and mRNAs were calculated by the 2−ΔΔCt method (23). The expression level of miRNAs in plasma exosomes was calculated using the log2 (2−ΔCtx1010) formula, as previously described (24).
Quantitation of SDCBP by ELISA
Plasma was collected from NSCLC patients and assessed by ELISA using the SDCBP ELISA kit (catalog no. MK4079A, Meike Biotechnology Co., Ltd.), according to the manufacturer's instructions.
Statistical analysis
Statistical analyses were performed by GraphPad Prism software 7.0 (GraphPad Software, Inc.) or statistical package SPSS (IBM SPSS Statistics for Windows, version 19.0). Parametric data are expressed as the means ± standard deviation (SD). Difference analyses between different cell groups were compared with Student's t-test. Difference analyses between different patient groups were compared with Mann-Whitney U test. The expression levels of miR-1273a before and after chemotherapy were compared with Wilcoxon paired non-parametric test. Correlation was analyzed using Pearson correlation. P<0.05 was considered to indicate statistically significant differences.
Results
Identification of isolated exosomes from CDDP-treated A549 cells
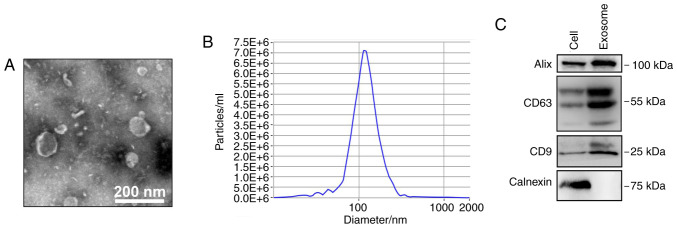
A549 cells were treated with 2 µg/ml CDDP for 48 h based on dose-response evaluation (Appendix S1 and Fig. S1), and the exosomes were then isolated from the cell culture medium. To ensure successful isolation of exosomes from A549 cells, the collected exosomes were observed by TEM and analyzed by nanoparticle tracking and western blotting. The expected size range of exosomes and bilayer membrane-shaped morphology were readily observed using TEM (Fig. 1A). NTA demonstrated that the mean particle diameter was 30–150 nm (Fig. 1B). Western blotting demonstrated that the exosomes were positive for the exosomal markers CD9, CD63 and ALIX, but negative for calnexin (an endoplasmic reticulum protein) (Fig. 1C).
Figure 1.
Characterization of exosomes from CDDP-treated A549 cells. (A) Representative transmission electron microscopy micrograph of isolated exosomes from A549 cells (scale bar, 200 nm). (B) Nanoparticle tracking analysis of the size distribution of isolated exosomes. (C) Western blotting analysis of the exosomal markers ALIX, CD63, and CD9 and the non-exosomal marker calnexin in A549 cells and exosomes isolated from A549 cells. CDDP, cisplatin.
Exosomes secreted from CDDP-treated cells reduce the sensitivity of A549 cells to CDDP
Before examining the effects of exosomes isolated from CDDP-treated NSCLC cells on NSCLC sensitivity to CDDP treatment, it was investigated whether exosomes can be taken up by A549 cells. Compared with control cells, following incubation with PKH26-stained exosomes for 4 h, a large number of PKH26 fluorescence-labeled exosomes appeared inside the A549 cells (Fig. 2A), suggesting that the cells can take up exosomes. In order to investigate the effects of exosomes secreted by CDDP-treated cells on the sensitivity of NSCLC cells to cisplatin treatment, A549 cells were co-cultured with PBS (used as control), EXOCDDP (exosomes isolated from the culture medium of CDDP-treated cells) for 48 h. Compared with the control group, the cell viability of the EXOCDDP group was significantly enhanced following treatment with various concentrations of CDDP (Fig. 2B). Consistently with these findings, the apoptotic rate of the EXOCDDP group was significantly reduced compared with that of the control group (25.03±1.24 vs. 10.23±2.76%, respectively; P<0.01; Fig. 2C). These results suggest that the transmission of exosomes derived from CDDP-stimulated cells may reduce the sensitivity of A549 cells to CDDP.
Figure 2.
Exosomes secreted from CDDP-treated cells reduce the sensitivity of A549 cells to CDDP. (A) Exosomes labelled by fluorescent PKH26 were visualized by laser confocal microscopy (scale bar, 32 nm). (B) A549 cells pre-treated with PBS or EXOCDDP were further incubated with various concentrations of CDDP for 48 h. Cell viability was determined by the Cell Counting Kit-8 assay (*P<0.05, **P<0.01 vs. the A549+PBS group). (C) Cell apoptosis rates were analyzed by flow cytometry (**P<0.01 vs. the A549+PBS group). EXOCDDP, exosomes isolated from the culture medium of CDDP-treated cells; CDDP, cisplatin.
Changes in microRNA expression profiles in exosomes derived from CDDP-treated A549 cells
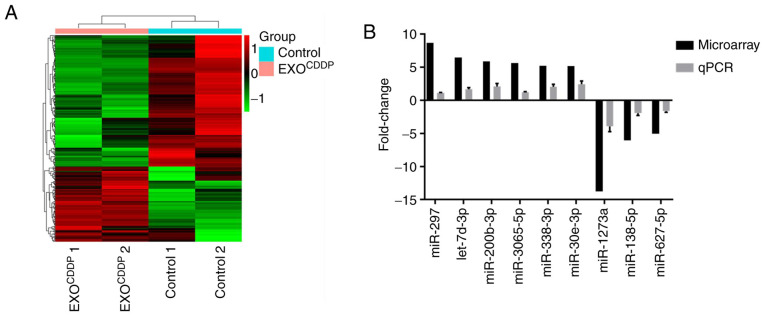
Numerous studies have demonstrated that miRNAs play an important role in exosome-mediated chemoresistance. Therefore, miRNAs may be involved in EXOCDDP-mediated CDDP resistance. Microarray analysis was performed to examine the expression profiles of miRNAs in EXOCDDP. Exosomes derived from A549 cells (EXO) were used as control. Compared with the control group, a total of 276 miRNAs in EXOCDDP were altered >2-fold (Fig. 3A). In particular, there was a >5-fold difference in the expression of 9 miRNAs between the two groups. Among those, the expression of miR-297, let-7d-3p, miR-200b-3p, miR-3065-5p, miR-338-3p and miR-30e-3p was upregulated, whereas the expression of miR-1273a, miR-138-5p, and miR-627-5p was downregulated (Table I). Next, the expression levels of the above mentioned 9 miRNAs were further confirmed by RT-qPCR analysis. As shown in Fig. 3B, compared with control exosomes, the RT-qPCR results on the changes in the 9 miRNAs in EXOCDDP were consistent with the microarray data. Since the expression difference of miR-1273a was the most prominent among these miRNAs, miR-1273a was selected for further experiments.
Figure 3.
Expression profiles of microRNAs in the EXOCDDP and control groups. (A) Hierarchical cluster analysis of exosomal miRNA expression profiles in the EXOCDDP and control groups. (B) The fold-changes in the expression of 9 miRNAs were determined and compared by reverse transcription-quantitative PCR and microarray analyses. EXOCDDP, exosomes isolated from the culture medium of CDDP-treated cells; CDDP, cisplatin.
Table I.
Top 9 miRNAs with differential expression (fold change >5) between the EXO and EXOCDDP groups.
| Upregulation | Fold | Rank | Downregulation | Fold | Rank |
|---|---|---|---|---|---|
| hsa-miR-297 | 8.69 | 1 | hsa-miR-1273a | 13.76 | 1 |
| hsa-let-7d-3p | 6.49 | 2 | hsa-miR-138-5p | 6.05 | 2 |
| hsa-miR-200b-3p | 5.90 | 3 | hsa-miR-627-5p | 5.03 | 3 |
| hsa-miR-3065-5p | 5.65 | 4 | |||
| hsa-miR-338-3p | 5.21 | 5 | |||
| hsa-miR-30e-3p | 5.20 | 6 |
EXO, exosomes derived from A549 cells; EXOCDDP, exosomes isolated from the culture medium of CDDP-treated cells; CDDP, cisplatin.
Overexpression of miR-1273a enhances the sensitivity of A549 cells to CDDP
Given the decreased expression of miR-1273a in EXOCDDP, it was hypothesized that miR-1273a acts as a tumor suppressor. To explore whether miR-1273a affects CDDP sensitivity, A549 cells were transfected with miR-1273a mimic or mimic negative control (NC) (Fig. 4A), followed by treatment with different concentrations of CDDP. The CCK-8 assay demonstrated that overexpression of miR-1273a significantly increased the toxicity of CDDP at different concentrations (Fig. 4B). Consistently, the apoptosis analysis revealed that overexpression of miR-1273a significantly enhanced CDDP-induced apoptosis compared with the control group (25.28±1.23 vs. 38.75±3.01%, respectively; P<0.01; Fig. 4C). Furthermore, it was also observed that miR-1273a in exosomes can enhance the sensitivity of A549 cells to cisplatin by exosomal transmission (Appendix S1 and Fig. S2). These results indicate that miR-1273a may enhance CDDP sensitivity in A549 cells.
Figure 4.
Overexpression of miR-1273a enhances the CDDP sensitivity of A549 cells. (A) After A549 cells were transfected with miR-1273a mimic or negative control, the relative expression of miR-1273a was determined (**P<0.01 vs. the NC group). (B) A549 cells transfected with miR-1273a mimic or negative control were exposed to the indicated concentrations of CDDP for 48 h, and cell viability was determined by the Cell Counting Kit-8 assay (*P<0.05, **P<0.01 vs. the A549 NC group). (C) Apoptosis rates in cells transfected with miR-1273a mimic were analyzed by flow cytometry (**P<0.01 vs. the the A549 NC group). CDDP, cisplatin.
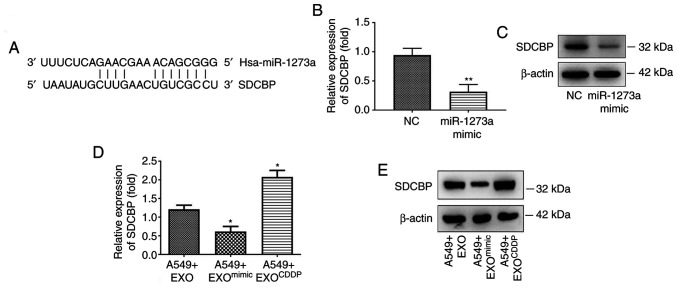
SDCBP may be one of the downstream targets of miR-1273a
To explore the mechanisms underlying the regulatory role of miR-1273a in cell sensitivity to CDDP, it was next attempted to identify the target gene of miR-1273a. TargetScan (www.targetscan.org) and microRNA.org (www.microrna.org) were first used to identified the 3′-untranslated region (UTR) of SDCBP which was identified as the potential target binding region of miR-1273a (Fig. 5A). The expression of SDCBP was detected by both RT-qPCR and western blot analyses after A549 cells were transfected with miR-1273a mimic or mimic negative control. As shown in Fig. 5B and C, RT-qPCR and western blotting analyses revealed that overexpression of miR-1273a significantly decreased the expression of SDCBP at both the mRNA and protein levels. Furthermore, according to our findings (Fig. S2B), miR-1273a was significantly downregulated in EXOCDDP and upregulated in EXOmimic (exosomes from miR-1273a mimic transfected cells). To verify whether miR-1273a could regulate SDCBP expression through exosome-mediated delivery, A549 cells were treated with EXO, EXOCDDP or EXOmimic for 48 h, and the expression of SDCBP in cells was detected by RT-qPCR and western blotting analyses. Compared with EXO-treated cells, SDCBP expression was significantly increased in the EXOCDDP-treated cells but decreased in EXOmimic-treated cells (Fig. 5D and E). These results indicated that SDCBP may be a promising downstream target of miR-1273a.
Figure 5.
SDCBP may be a downstream target of miR-1273a. (A) Prediction of miR-1273a binding site in the 3′-untranslated region of SDCBP by TargetScan. (B and C) After transfecting cells with miR-1273a mimics, the expression of SDCBP was detected by reverse transcription-quantitative PCR and western blot analyses, respectively (**P<0.01 vs. the NC group). (D and E) After treating cells with EXO, EXOmimic, or EXOCDDP, the expression of SDCBP was detected by reverse transcription-quantitative PCR and western blot analyses, respectively (*P<0.05 vs. the A549+EXO group). SDCBP, syndecan binding protein; EXO, exosomes derived from A549 cells; EXOCDDP, exosomes isolated from the culture medium of CDDP-treated cells; EXOmimic, exosomes from miR-1273a mimic-transfected cells; CDDP, cisplatin.
Exosomal miR-1273a in plasma is associated with the therapeutic effect of platinum-based chemotherapy in NSCLC
Exosomes are known to contain a variety of proteins and RNAs from tumor cells (25). Several studies have reported that exosomal miRNAs are secreted into the plasma, and these miRNAs may thus be used as potential biomarkers for the diagnosis of different types of cancer (26). To further confirm whether exosomal miR-1273a in the plasma reflects the therapeutic effect of platinum-based chemotherapy in NSCLC, exosomal miR-1273a was measured in the plasma in 49 patients with advanced NSCLC receiving platinum-based chemotherapy. Exosomes isolated from the patient plasma samples displayed typical oval-shaped vesicles on TEM examination, with a diameter of 30–150 nm (Fig. 6A). Furthermore, the exosomal markers CD9 and CD63 in isolated exosomes from 4 random plasma samples were detected by western blotting (Fig. 6B), and the appropriate particle size distribution of isolated exosomes was confirmed by NTA (Fig. 6C).
Figure 6.
The levels of exosomal miR-1273a in the plasma are correlated with the therapeutic efficacy of platinum-based chemotherapy in NSCLC. (A) Morphology of the isolated exosomes from patients was determined by transmission electron microscopy (scale bar, 200 nm). (B) The expression of exosomal markers CD63 and CD9 was detected by western blotting. (C) Nanoparticle tracking analysis of exosomes derived from NSCLC patients. (D) Expression of plasma exosomal miR-1273a in the non-responder and responder groups before and after chemotherapy (**P<0.01; ns, not significant). (E) Expression of plasma exosomal miR-1273a in the responder and non-responder groups after chemotherapy (*P<0.05). (F) Expression of plasma SDCBP in the responder and non-responder groups after chemotherapy (*P<0.05). (G) Pearson's correlation analysis revealed an association between the expression of miR-1273a and SDCBP (SDCBP expression values are log2 transformed). SDCBP, syndecan binding protein; NSCLC, non-small cell lung cancer.
Next, we examined the miR-1273a and SDCBP expression levels in NSCLC patients based on the therapeutic outcomes. As shown in Fig. 6D, the level of plasma exosomal miR-1273a in the non-responder group was significantly reduced after chemotherapy, whereas the level of plasma exosomal miR-1273a in the responder groups did not differ significantly before and after chemotherapy. These results indicate that decreased exosomal miR-1273a during chemotherapy may be associated with poor outcome of cisplatin therapy in patients with NSCLC. The expression of miR-1273a and SDCBP in NSCLC patients after chemotherapy was next examined. The level of exosomal miR-1273a in the non-responder group was significantly lower compared with that in the responder group, whereas the expression of SDCBP was significantly higher in the responder group (Fig. 6E and F). Furthermore, a negative correlation between plasma exosomal miR-1273a and plasma SDCBP expression was identified (Fig. 6G). Based on the results mentioned above, it may be inferred that decreased exosomal miR-1273a may reduce the therapeutic effect of platinum-based agents by regulating SDCBP expression.
Discussion
Platinum-based chemotherapy has been the major treatment method for patients with advanced NSCLC for more than 30 years (27). However, resistance to platinum during the course of therapy greatly limits the therapeutic efficacy (28). Previous research has demonstrated that exosomes derived from CDDP-resistant NSCLC cells exhibit a differential miRNA expression profile compared with CDDP-sensitive cells. Futhermore, exosomal miR-100-5p and miR-425-3p were found to be involved in the development of cisplatin resistance in NSCLC (18,29). However, cisplatin resistance may occur rapidly in the early stages of chemotherapy in several NSCLC cases (30). Therefore, unlike previous studies, the present study mainly focused on the role of exosomal miRNAs in CDDP resistance under condition of short-term CDDP stimulation.
The present study demonstrated that exosomes secreted by CDDP-stimulated cells can reduce cell sensitivity to CDDP, which was consistent with the findings of Xiao et al (31). Next, our microarray analysis revealed that the level of exosomal miR-1273a exhibited the most notable change under CDDP treatment; therefore, it was hypothesized that the change in exosomal miR-1273a level is associated with CDDP resistance of A549 cells under CDDP stimulation. Of note, although most studies have shown that increased miRNA levels in exosomes promote the malignant behavior of tumors, several studies have found that decreased exosomal miRNAs may also mediate tumor progression (32). Therefore, the decreased miR-1273a levels in exosomes may still make sense under CDDP treatment. A recent study demonstrated that miR-1273a overexpression abrogated the oncogenic function by rescuing the expression of activator protein-1, interferon regulatory factor-4, CDX-2 and Zic-1 in colon cancer cells (33), highlighting its potential role as a tumor suppressor miRNA. Consistently, the present study revealed that miR-1273a increased the sensitivity of NSCLC cells to CDDP and an increase in the amount of miR-1273a in exosomes promoted cell apoptosis. However, drug resistance is likely the combined result of several factors. As our microarray data also revealed other miRNA changes, additional miRNAs that may affect the sensitivity of NSCLC cells to CDDP must be further investigated.
A large number of studies have demonstrated that microRNAs delivered by exosomes maintain their biological activity after reaching the recepient cells, and then play a role by inhibiting their target genes (34). To this end, exosomes with high expression of miR-1273a were constructed, and it was observed that the drug sensitivity of cells was also increased after co-incubation with these miR-1273a-rich exosomes (Appendix S1 and Fig. S2). Furthermore, it was hypothesized that SDCBP may be a downstream target of miR-1273a, based on the bioinformatics prediction of its binding site and its oncogenic role in cell survival, stemness and chemoresistance, in various types of human cancer (35,36). It was next confirmed that miR-1273a overexpression inhibited the expression of SDCBP at both the mRNA and protein levels. Furthermore, co-incubation of cells with exosomes containing miR-1273a at different expression levels may also result in changes in the SDCBP expression in receptor cells. However, the effect of SDCBP on cisplatin sensitivity in NSCLC must be thoroughly explored in future studies.
Numerous studies have confirmed that exosomal miRNAs may be used as biomarkers for early cancer detection and monitoring compared with traditional blood-based cancer markers (37–39). The present study also demonstrated a significant reduction in exosomal miR-1273a in the plasma of patients who were not sensitive to platinum-based chemotherapy. In agreement with our in vitro results, there was a corresponding increase in SDCBP levels in the plasma of non-responders. Currently, the detection of exosomal miR-1273a in the plasma appears to be an effective and convenient method for evaluating the efficacy of chemotherapy. Due to the unique biological characteristics of exosomes, such as stability, biocompatibility, permeability, low toxicity and low immunogenicity, they may be used as a chemotherapeutic drug delivery system (40,41). Thus, plasma exosomal miR-1273a may also be used in the future as a promising therapeutic molecule for NSCLC patients who are not sensitive to chemotherapy.
In conclusion, it was herein demonstrated that decreased exosomal miR-1273a reduced the sensitivity of NSCLC to CDDP. In addition to cell-based evidence, it was further observed that the expression of miRNA-1273a in plasma exosomes was significantly decreased in patients who were not sensitive to chemotherapy. These effects may be associated with the regulatory effect of miR-1273a on SDCBP. Therefore, for NSCLC patients who are not sensitive to chemotherapy, exosomal miR-1273a may prove to be a useful biomarker and feasible treatment strategy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (no. 81672312).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
Conception and design of the study were carried out by DW, ML and ND. Administrative and experimental support were carried out by YY and XD. Provision of study materials and recruitment of patients and samples were achieved by YY and YP. Collection and assembly of data were carried out by XZ and XD. Data analysis and interpretation were performed by XZ and CX. Writing of the manuscript was conducted by XZ. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients signed informed consent forms, and this study was authorized by the Ethics Committees of Daping Hospital (Chongqing, China) (ratification no. 2018113). This study was conducted in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lam KC, Mok TS. Targeted therapy: An evolving world of lung cancer. Respirology. 2011;16:13–21. doi: 10.1111/j.1440-1843.2010.01821.x. [DOI] [PubMed] [Google Scholar]
- 3.Mack PC, Redman MW, Chansky K, Williamson SK, Farneth NC, Lara PN, Jr, Franklin WA, Le QT, Crowley JJ, Gandara DR, SWOG Lower osteopontin plasma levels are associated with superior outcomes in advanced non-small-cell lung cancer patients receiving platinum-based chemotherapy: SWOG study S0003. J Clin Oncol. 2008;26:4771–4776. doi: 10.1200/JCO.2008.17.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Brahmer JR, Juergens RA, Borghaei H, Gettinger S, Chow LQ, Gerber DE, Laurie SA, Goldman JW, et al. Nivolumab in combination with platinum-based doublet chemotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:2969–2979. doi: 10.1200/JCO.2016.66.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Chen Y, Feng H, Zhang B, Wang H. Prognostic and predictive value of HURP in nonsmall cell lung cancer. Oncol Rep. 2018;39:1682–1692. doi: 10.3892/or.2018.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.H Rashed M, Bayraktar E, K Helal G, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C. Exosomes: From garbage bins to promising therapeutic targets. Int J Mol Sci. 2017;18:538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig AK, Giebel B. Exosomes: Small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44:11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Park S, Kim H, Choi YJ, Kim SY, Sung KJ, Sung YH, Choi CM, Yun M, Yi YS, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020;475:2–13. doi: 10.1016/j.canlet.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, Varming K, Jørgensen MM, Sorensen BS. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. 2016;11:1701–1710. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 11.Del Vescovo V, Denti MA. microRNA and lung cancer. Adv Exp Med Biol. 2015;889:153–177. doi: 10.1007/978-3-319-23730-5_9. [DOI] [PubMed] [Google Scholar]
- 12.Czarnecka KH, Szmyd B, Baranska M, Kaszkowiak M, Kordiak J, Antczak A, Pastuszak-Lewandoska D, Brzeziańska-Lasota E. A strong decrease in TIMP3 expression mediated by the presence of miR-17 and 20a enables extracellular matrix remodeling in the NSCLC lesion surroundings. Front Oncol. 2019;9:1372. doi: 10.3389/fonc.2019.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu G, Drescher KM, Chen XM. Exosomal miRNAs: Biological properties and therapeutic potential. Front Genet. 2012;3:56. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshii S, Hayashi Y, Iijima H, Inoue T, Kimura K, Sakatani A, Nagai K, Fujinaga T, Hiyama S, Kodama T, et al. Exosomal microRNAs derived from colon cancer cells promote tumor progression by suppressing fibroblast TP53 expression. Cancer Sci. 2019;110:2396–2407. doi: 10.1111/cas.14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berrout J, Kyriakopoulou E, Moparthi L, Hogea AS, Berrout L, Ivan C, Lorger M, Boyle J, Peers C, Muench S, et al. TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p. Nat Commun. 2017;8:947. doi: 10.1038/s41467-017-00983-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, Liu Y. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34:1003–1010. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and ‘exosomal shuttle microRNA’ in tumorigenesis and drug resistance. Cancer Lett. 2015;356:339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, Shen B, Liu S, Yan D, Feng J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721–3733. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drayton RM. The role of microRNA in the response to cisplatin treatment. Biochem Soc Trans. 2012;40:821–825. doi: 10.1042/BST20120055. [DOI] [PubMed] [Google Scholar]
- 20.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Pan D, Chen Q, Zhu LJ, Ou J, Wabitsch M, Wang YX. Transcription factor Hlx controls a systematic switch from white to brown fat through Prdm16-mediated co-activation. Nat Commun. 2017;8:68. doi: 10.1038/s41467-017-00098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Huang YH, Liang KH, Chien RN, Hu TH, Lin KH, Hsu CW, Lin CL, Pan TL, Ke PY, Yeh CT. A Circulating MicroRNA signature capable of assessing the risk of hepatocellular carcinoma in cirrhotic patients. Sci Rep. 2017;7:523. doi: 10.1038/s41598-017-00631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HG, Grizzle WE. Exosomes and cancer: A newly described pathway of immune suppression. Clin Cancer Res. 2011;17:959–964. doi: 10.1158/1078-0432.CCR-10-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Z, Yu J, Lin J, Liu Y, Liao Y. Exosome-specific tumor diagnosis via biomedical analysis of exosome-containing microRNA biomarkers. Analyst. 2019;144:5856–5865. doi: 10.1039/C9AN00777F. [DOI] [PubMed] [Google Scholar]
- 27.Zarogoulidis K, Zarogoulidis P, Darwiche K, Boutsikou E, Machairiotis N, Tsakiridis K, Katsikogiannis N, Kougioumtzi I, Karapantzos I, Huang H, Spyratos D. Treatment of non-small cell lung cancer (NSCLC) J Thorac Dis. 2013;5(Suppl 4):S389–S396. doi: 10.3978/j.issn.2072-1439.2013.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, O'Flaherty JD, Fennell DA, Richard D, O'Leary JJ, O'Byrne KJ. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One. 2013;8:e54193. doi: 10.1371/journal.pone.0054193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M, Xue W, Wang Y, Li W, Shu Y, et al. Exosomal transfer of cisplatin-induced miR-425-3p confers cisplatin resistance In NSCLC through activating autophagy. Int J Nanomedicine. 2019;14:8121–8132. doi: 10.2147/IJN.S221383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara PN, Jr, Gandara DR, Longmate J, Gumerlock PH, Lau DH, Edelman MJ, Gandour-Edwards R, Mack PC, Israel V, Raschko J, et al. Activity of high-dose toremifene plus cisplatin in platinum-treated non-small-cell lung cancer: A phase II California cancer consortium trial. Cancer Chemother Pharmacol. 2001;48:22–28. doi: 10.1007/s002800100293. [DOI] [PubMed] [Google Scholar]
- 31.Xiao X, Yu S, Li S, Wu J, Ma R, Cao H, Zhu Y, Feng J. Exosomes: Decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25:5300–5309. doi: 10.3748/wjg.v25.i35.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooijmans SA, Schiffelers RM, Zarovni N, Vago R. Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment. Pharmacol Res. 2016;111:487–500. doi: 10.1016/j.phrs.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Jana S, Sengupta S, Biswas S, Chatterjee A, Roy H, Bhattacharyya A. miR-216b suppresses breast cancer growth and metastasis by targeting SDCBP. Biochem Biophys Res Commun. 2017;482:126–133. doi: 10.1016/j.bbrc.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Talukdar S, Das SK, Pradhan AK, Emdad L, Windle JJ, Sarkar D, Fisher PB. MDA-9/syntenin (SDCBP) is a critical regulator of chemoresistance, survival and stemness in prostate cancer stem cells. Cancers (Basel) 2019;12:53. doi: 10.3390/cancers12010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, Zhang X. Circulating MicroRNAs in cancer: Potential and challenge. Front Genet. 2019;10:626. doi: 10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang C, Hopfner F, Katsikoudi A, Hein R, Catli C, Evetts S, Huang Y, Wang H, Ryder JW, Kuhlenbaeumer G, et al. Serum neuronal exosomes predict and differentiate Parkinson's disease from atypical parkinsonism. J Neurol Neurosurg Psychiatry. 2020;91:720–729. doi: 10.1136/jnnp-2019-322588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Dou J, Cao J, Wang Y, Gao Q, Zeng Q, Liu W, Liu B, Cui Z, Teng L, et al. Uncovering the potential differentially expressed miRNAs as diagnostic biomarkers for hepatocellular carcinoma based on machine learning in the cancer genome atlas database. Oncol Rep. 2020;43:1771–1784. doi: 10.3892/or.2020.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pullan JE, Confeld MI, Osborn JK, Kim J, Sarkar K, Mallik S. Exosomes as drug carriers for cancer therapy. Mol Pharm. 2019;16:1789–1798. doi: 10.1021/acs.molpharmaceut.9b00104. [DOI] [PubMed] [Google Scholar]
- 41.Fan Z, Xiao K, Lin J, Liao Y, Huang X. Functionalized DNA enables programming exosomes/vesicles for tumor imaging and therapy. Small. 2019;15:e1903761. doi: 10.1002/smll.201903761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.