Abstract
Background
The effect of metformin on gut microbiota has been reported, but whether metformin can suppress colorectal cancer (CRC) by affecting gut microbiota composition and rescue F. nucleatum-induced tumourigenicity remains unclear.
Methods
To identify microbiota associated with both CRC occurrence and metformin treatment, first, we reanalyzed the gut microbiome of our previous data on two human cohorts of normal and CRC individuals. Subsequently, we summarized microbiota altered by metformin from published literatures. Several taxa, including Fusobacterium, were associated with both CRC occurrence and metformin treatment. We investigated the effect of metformin on APCMin/+ mice given with or without F. nucleatum. 16S rRNA gene sequencing was performed.
Findings
We summarized 131 genera altered by metformin from 18 published literatures. Five genera reported to be changed by metformin, including Bacteroides, Streptococcus, Achromobacter, Alistipes and Fusobacterium, were associated with CRC in both of our human cohorts. Metformin relieved the symptoms caused by F. nucleatum administration in APCMin/+ mice, and showed promise in suppressing intestinal tumour formation and rescuing F. nucleatum-induced tumourigenicity. Administration of F. nucleatum and/or metformin had effect on gut microbiome structure, composition and functions of APCMin/+ mice.
Interpretation
This study pioneers in predicting critical CRC-associated taxa contributing to the antitumour effect of metformin, and correlating gut microbiome with the antitumour effect of metformin in experimental animals. We presented a basis for future investigations into metformin's potential effect on suppressing F. nucleatum-induced tumor formation in vivo.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31701250).
Keywords: Colorectal cancer, Metformin, Microbiome, Fusobacterium nucleatum
Abbreviations: AMPK, AMP-activated protein kinase; ANOVA, analysis of variance; AUROC, area under receiver operating curve; CRC, colorectal cancer; H&E, Hematoxylin and Eosin; IV, indicator value; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, Kyoto Encyclopedia of Genes and Genomes orthologs; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis; MFV, metformin score; OTU, operational taxonomic units; PCoA, principal coordinate analysis; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; ROC, receiver operator characteristic; SPF, specific pathogen-free
Research in context.
Evidence before this study
Accumulating evidences from experimental animals and patients with CRC have demonstrated that specific bacteria like Fusobacterium nucleatum (F. nucleatum) may promote colorectal carcinogenesis. Metformin, a first-line antidiabetic drug, has been widely used in clinic for more than 60 years with minimal side-effects and low cost. Recent studies suggest that metformin also acts through pathways in the gut, and intestinal microbial changes might be responsible for the therapeutic effect of metformin. Apart from the effect on gut microbiota, metformin was also effective for chemoprevention of multiple cancers including CRC in animal experiments and clinical trials. However, it is still not known whether metformin can suppress CRC by affecting gut microbiota composition.
Added value of this study
In this study, we discovered that Fusobacterium was enriched in CRC samples and could also be altered by metformin. Our results showed that metformin relieved the symptoms caused by F. nucleatum administration in APCMin/+ mice, and showed promise in suppressing intestinal tumour formation and rescuing F. nucleatum-induced tumourigenicity. We found that gut microbiota was altered when comparing APCMin/+ mice treated with or without metformin, and APCMin/+ mice given F. nucleatum with and without metformin, which indicated that modulation of the gut microbiota might contribute to the antitumour effects of metformin. Our work revealed the structural, compositional, and functional dysbiosis of the gut microbiome after F. nucleatum and/or metformin administration. In addition, we demonstrated correlations among taxa and involvement of gut microbiome in functional activities. To the best of our knowledge, this study pioneers in implicating that altered gut microbiota may mediate some of metformin's antitumour effects using both analyses of published data and experimental methods.
Implications of all the available evidence
This study proposed the possibility that F. nucleatum might be a site of metformin treatment action in CRC. We intend to undertake follow-up studies to investigate the association between the antitumour effect and other promising microbial therapeutic targets of metformin predicted in this study.
Alt-text: Unlabelled box
1. Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause for cancer-related deaths worldwide [1]. With the advancement in metagenome sequencing, recent studies have demonstrated that an altered microbiome environment in the gut is associated with colorectal tumourigenesis [2], [3], [4]. Accumulating evidences from experimental animals and patients with CRC have demonstrated that specific bacteria like enterotoxigenic Bacteroides fragilis, Fusobacterium nucleatum (F. nucleatum), and Peptostreptococcus anaerobius, may promote colorectal carcinogenesis [5]. We have previously demonstrated that F. nucleatum accumulated in colorectal adenomas and colorectal carcinomas, and played a vital role in colorectal tumour initiation and development [6,7]. We also showed that CRC patients with high amount of F. nucleatum had shorter recurrence free survival (RFS) and shorter five-year recurrence survival than the F. nucleatum low group [7]. In The Cancer Genome Atlas (TCGA) cohort, poorer overall survival was observed in correlation with tumour Fusobacterium load in patients with cecum and ascending colon tumours [2]. Yang et al. demonstrated that F. nucleatum administration reduced the survival of ApcMin/+ mice [8].Other studies showed that F. nucleatum had good performance in diagnosing CRC [9,10]. Most studies investigating F. nucleatum concentrated on its performance in CRC diagnosis or its effect on intestinal cancer progression, and few of them regarded F. nucleatum as a site of treatment action [10].
Metformin, a first-line antidiabetic drug, has been widely used in clinic for more than 60 years with minimal side-effects and low cost. It is generally acknowledged that metformin suppresses hepatic gluconeogenesis through the activation of AMP-activated protein kinase (AMPK)-dependent and AMPK-independent pathways [11]. Recent studies suggest that intestinal microbial changes might be responsible for the therapeutic effect of metformin in the gut [12], [13], [14]. However, the major changes of metformin in the gut microbiota are not always consistent among different studies. A multi-nation type II diabetes (T2D) metagenomic dataset built by Forslund et al. reported a significant decrease in Intestinbacter and a significant elevation in Escherichia in metformin treated T2D individuals from a Swedish cohort. The Danish cohort showed similar results, while the Chinese cohort did not exhibit an increase in Escherichia [15]. In two different animal models, metformin was positively associated with the genus Bacteroides [16,17]. Apart from the effect on gut microbiota, metformin was also effective for chemoprevention of multiple cancers including CRC in animal experiments and clinical trials [11,[18], [19], [20]]. A multicentre, double-blind, placebo-controlled, randomised phase 3 trial reported that low-dose metformin reduced the prevalence and number of metachronous adenomas or polyps after polypectomy, indicating that metformin has a potential role in the chemoprevention of colorectal cancer [11]. Additionally, it has been demonstrated that in metastatic colorectal cancer (mCRC), metformin-only diabetes patients had ameliorative overall survival (OS). Other antidiabetic drug users had no improved outcome compared with mCRC patients without diabetes or diabetic mCRC patients without hypoglycemic treatment [21]. However, it is still not known whether metformin can suppress CRC by affecting gut microbiota composition.
We hypothesized that metformin could suppress colorectal carcinogenesis by modulation of gut microbiota. First, we reanalyzed our previous data on gut microbiome of normal and CRC individuals to identify CRC-associated taxa. Subsequently, we summarized the bacteria altered by metformin after reviewing 18 literatures. We observed that Fusobacterium was enriched in CRC samples and could be altered by metformin treatment. We assumed that metformin could exert its antitumour effect by modulating F. nucleatum. 28 C57BL/6-APCMin/+ mice were randomly divided into 4 groups: negative control (NC), F. nucleatum (FN), F. nucleatum plus metformin (FM), and metformin (MET). 16S ribosomal RNA (rRNA) gene sequencing was performed in 4 groups of APCMin/+ mice given F. nucleatum and/or metformin. We analyzed the structure, composition, and functions of the gut microbiome after F. nucleatum and/or metformin administration. In addition, we demonstrated correlations among taxa and involvement of gut microbiome in functional activities.
2. Materials and methods
2.1. Human cohorts
The data was originally published by Yu et al. [6]. Patients who had undergone colonoscopy or colorectal carcinoma surgery in the Shanghai Jiao-Tong University School of Medicine, Renji Hospital were recruited. Fecal samples and left colonic tissues were collected. The protocol had the approval of the Ethics Committee of the Shanghai Jiao-Tong University School of Medicine, Renji Hospital. Informed consent was obtained from all the subjects. An independent data and safety committee monitored the trial and reviewed the results.
2.2. Mice
4- to 5-week-old C57BL/6-APCMin/+ mice, used extensively for intestinal cancer studies, were purchased from Shanghai Model Organisms Center and housed in specific pathogen-free (SPF) and barrier conditions with controlled temperature (20–26 °C) and humidity (40–70%). After 1-week period of adaptive breeding, all mice were randomly divided into 4 groups: negative control (NC), F. nucleatum (FN), F. nucleatum plus metformin (FM), and metformin (MET). F. nucleatum was administrated at 109 colony forming units (CFU) suspended with PBS twice per week via gavage. Metformin (Sangon Biotech, China) was fed at a dose of 250 mg/kg by oral gavage every day for 12 weeks. PBS was acted as the control treatment. All mice were sacrificed at 18 weeks. The whole intestinal tract was removed and longitudinally opened to estimate tumour progression. Tumour numbers and sizes (diameter) were carefully recorded. The mouse experiment was conducted according to guidelines approved by the Institutional Animal Care and Use Committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University.
2.3. Bacterial strain, culture condition and in vitro bacterial growth experiments
Fusobacterium nucleatum strain (ATCC 25586) was obtained from American Type Culture Collection. F. nucleatum was cultured overnight at 37°C in an anaerobic glove box (DG250, UK) with 80% N2, 10% H2 and 10% CO2 in brain heart infusion (BHI) broth supplemented with hemin, Vitamin K1, K2HPO4, and L-Cysteine.
F. nucleatum was streaked onto BHI agar plates supplemented with hemin, Vitamin K1, K2HPO4, and L-Cysteine in the presence or absence of 10 mM metformin in a 37 °C anaerobic glove box for 2days. For growth curve, F. nucleatum was cultured in BHI broth supplemented with hemin, Vitamin K1, K2HPO4, and L-Cysteine in an anaerobic glove box in the presence or absence of 10 mM metformin and OD600 value was measured.
2.4. H&E staining and immunohistochemistry
Intestinal tissues of the colorectum removed from the abdomen was slit open and then rolled up longitudinally, with the mucosa outwards, using a wooden stick. Next, the proportions were carefully placed in 10% formalin solution immediately. Then, the tissues were embedded into paraffin, blocked, cut into 4–5 μm long pieces, and were deparaffinized in xylene, which was followed by Hematoxylin and Eosin (H&E) staining.
For immunohistochemistry, paraffin-embedded tissues were used to analyze the expression of Ki-67. All sections were deparaffinized and hydrated using graded concentrations of ethanol to deionized water. Tissue sections were subjected to quenching of endogenous peroxidase and antigen retrieval using microwaving in low pH citrate buffer. Anti-Ki-67 (CST, 12202s, dilution 1:400) was then applied to the tissues and incubated overnight at 4 °C. Next, bound antibody was visualized with DAB chromogen (MXB), and later followed by incubating with hematoxylin. The proportion of Ki-67 positive cells was evaluated using ImmunoRatio in ImageJ, determined by counting immunostain-positive cells as a percentage to the total number of nuclei in the field.
2.5. F. nucleatum quantification in tissue sample
F. nucleatum quantification was performed as described previously [7]. DNA from CRC tissue were extracted using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The amounts of F. nucleatum was determined by qPCR. Relative abundance was calculated by 2−ΔCt method. Universal 16s was used as reference gene. The following primer sets were used:
Fusobacterium nucleatum-F, 5’-CAACCATTACTTTAACTCTACCATGTTCA-3’,
Fusobacterium nucleatum-R, 5’-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3’ [7];
Universal 16S-F, 5’-GGTGAATACGTTCCCGG-3’,
Universal 16S-R, 5’-TACGGCTACCTTGTTACGACTT-3’.
2.6. Fecal sample collection and DNA extraction
Fresh stool samples were collected carefully from each mouse into sterile tubes before sacrifice and immediately stored at −80°C until extraction. Genomic DNA was extracted from fecal samples by QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. The concentration and purification of bacterial DNA was measured using Nanodrop 2000 (Thermo Scientific, USA).
2.7. 16S ribosomal RNA gene sequencing analysis
For APCMin/+ mice, the V3–V4 hypervariable region of the bacteria's 16S ribosomal RNA (rRNA) gene was amplified by PCR with barcode-indexed primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), using FastPfu Polymerase. Amplicons were further purified by gel extraction (AxyPrep DNA GelExtraction Kit, Axygen Biosciences, Union City, California, USA) and quantified using QuantiFluor-ST (Promega, USA). Purified amplicons from PCR were pooled in equimolar concentrations and paired-end sequencing was performed based on an Illumina MiSeq platform (Illumina, San Diego, USA). Using the Quantitative Insights into Microbial Ecology (QIIME2, v2019.7.0), 16S rRNA gene sequences were demultiplexed and quality filtered. Operational taxonomic units (OTUs) were picked at 97% similarity cut-off, and the identified taxonomy was then aligned using SILVA (Release 132). Only OTUs and genera present in at least 25% of samples were remained for subsequent analysis. Additionally, rarefaction was performed on the OTU table to prevent methodological artefacts arising from varying sequencing depth. α-Diversity was measured by community richness from the rarefied OTU table by using R package vegan. β-Diversity was estimated by computing unweighted Unifrac, weighted Unifrac, Euclidean and Euclidean binary distances respectively. Significance of clustering was determined by Adonis test with 999 permutations. Then results of β-Diversity was visualized with principal coordinate analysis (PCoA). Functional contributions of the microbial communities were predicted based on OTU using R package Tax4Fun [22] with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [23]. The Tax4Fun tool is based on SILVA-labeled OTU abundances. We used UProC model for computation of functional reference profile in terms of KEGG orthologs (KOs) of bacterial origins [24]. Differences in bacterial relative abundances were determined by Mann–Whitney U test [25].
Three 16S rRNA sequencing datasets were respectively accessed through the BioProject (accession number PRJNA325931) at the NCBI, the European Nucleotide Archive (ENA) (accession number ERP117168), and the Sequence Read Archive (SRA) (accession number SRP099828). These sequences were demultiplexed and quality filtered using QIIME2 (v2019.7.0). Operational taxonomic units (OTUs) were delineated at the frequently used 97% similarity threshold. The OTUs were assigned to taxa by matching to the Greengenes database (Release 13.8). The R package DESeq2 was used to calculate genera that differed significantly between metformin untreated groups and metformin treated groups [26]. The community function was predicted from the OTU table using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)(27) with KEGG database [23].
2.8. Literature selection and data extraction
To identify gut microbiota significantly altered after metformin treatment, PubMed searches were performed with combinations of the terms “microbiome” and “metformin”. Full texts of all articles were screened. Articles with 16S rRNA gene or metagenomic raw sequences, processed sequences or differential analysis results of control group and metformin treatment group were remained for further evaluation. Author/Year, DOI, model, sample group, metformin dosage, subject age, weight/BMI, gender, sample used, sequencing method, region amplified, platform, reference database, source of data and calculation of statistical significance were extracted from the included studies. The altered genera in each article were recorded. Unclassified genera changed after metformin treatment were excluded.
2.9. Receiver Operator Characteristic (ROC) analysis
The analyses were carried out to distinguish metformin-untreated and metformin-treated patients or rodents by bacterial genera. To evaluate the effect of metformin on multiple genera, we defined and computed the metformin score (MFV) for each human or rodent on the basis of the frequently altered genera identified by literature screening. For each human or rodent, MFV equals to the sum of abundances of the genus more frequent in metformin-untreated humans or rodents subtracted by the sum of the abundances of genus more frequent in metformin-treated humans or rodents. MFV was used to generate ROC curve. By using the pROC package of R software [28], area under receiver operating curve (AUROC) was calculated. Pairwise comparison of ROC curves was performed using the bootstrap percentile method.
2.10. Statistical analysis
Statistical analyses were performed with R (V.3.5.1), GraphPad Prism or SPSS version 26.0. 2*2 factorial analysis of variance (ANOVA) was used to determine interactions between F. nucleatum and metformin. Differences in diversity, KEGG pathways abundances, KOs abundances were determined by Mann–Whitney U test [25]. We conducted linear discriminant analysis effect size (LEfSe) analysis to identify taxa differentially abundant between cases and controls [29]. This method first uses the non-parametric factorial Kruskal–Wallis sum-rank test or Wilcoxon sum-rank test to detect features with significant differential abundance and then applies linear discriminant analysis (LDA) to calculation of the effect size of each feature. Indicator genera specific to a given group were identified based on the normalized abundances of genera using the R package indicspecies, and the significant indicator value (IV) index was calculated by the 9999-permutation test [30]. Larger IV indicates greater specificity of taxa and P < 0.05 was considered statistically significant. The abundances of genera in the samples were standard normalized using the Rhea pipeline [31]. Spearman's correlation coefficient and significance were calculated using the R package Hmisc. Correlation network within genera was constructed using R package igraph, then visualized with Cytoscape (ver. 3.7.2).
3. Results
3.1. Alterations of gut microbiome in colorectal carcinoma
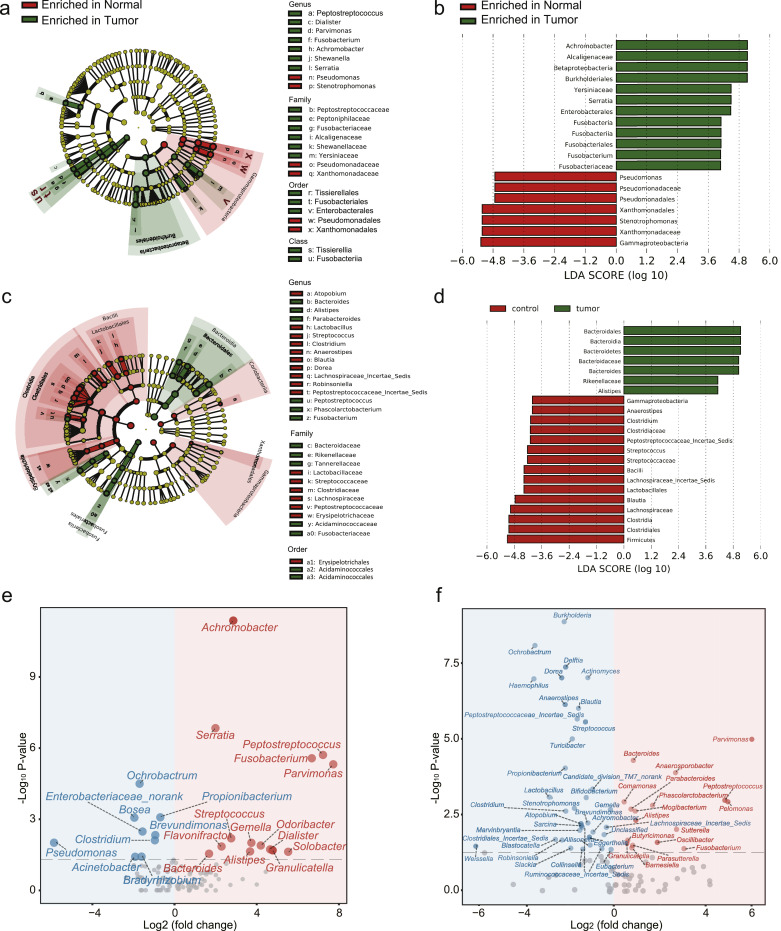
To examine alterations of gut microbiome in CRC, we re-analyzed our previous data [6]. 16S rRNA gene sequencing data using a Roche 454 GS FLX obtained from the mucosal samples of 32 normal Chinese individuals and 33 CRC Chinese patients (Fig. 1(a), Cohort 1), and 16S rRNA gene sequencing data using a Roche 454 GS FLX obtained from the fecal samples of 51 normal Chinese individuals and 42 CRC Chinese patients (Fig. 1(c), Cohort 2) were respectively analyzed. Only genera with a mean of relative abundance greater than 0.01% were included in the analysis. We used the LEfSe algorithm [29] and comparative analysis based on Wilcoxon sum-rank test to identify microbiota associated with CRC occurrence. Results of LEfSe analysis showed that Achromobacter, Serratia and Fusobacterium were more enriched in CRC patients of Cohort 1 (Fig. 1(b)), and Bacteroides and Alistipes were more enriched in CRC patients of Cohort 2 (Fig. 1(d)). Results of comparative analysis showed that Parvimonas, Pelomonas, Peptostreptococcus and Fusobacterium were the most significantly changed genera in Cohort 1 (Fig. 1(e)). Peptostreptococcus, Parvimonas and Fusobacterium were the most significantly changed genera in Cohort 2 (Fig. 1(f)).
Fig. 1.
16S rRNA sequencing analysis of the gut microbiome in Cohort 1 (mucosal) and Cohort 2 (fecal). (a) A cladogram representation of data in Cohort 1. (Green) Tumour-enriched taxa; (red) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size. (b) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in (a). Tumour-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in normal tissue have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. (c) A cladogram representation of data in Cohort 2. (Green) Tumour-enriched taxa; (red) taxa enriched in normal tissue. The brightness of each dot is proportional to its effect size. (d) LDA coupled with effect size measurements identifies the significant abundance of data in (c). Tumour-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in normal tissue have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. (e) Volcano plot of significantly (Wilcoxon test, P < 0.05) altered genera between normal and CRC samples in Cohort 1. Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue. (f) Volcano plot of significantly (Wilcoxon test, P < 0.05) altered genera between normal and CRC samples in Cohort 2. Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue.
3.2. Alterations of gut microbiome by metformin treatment
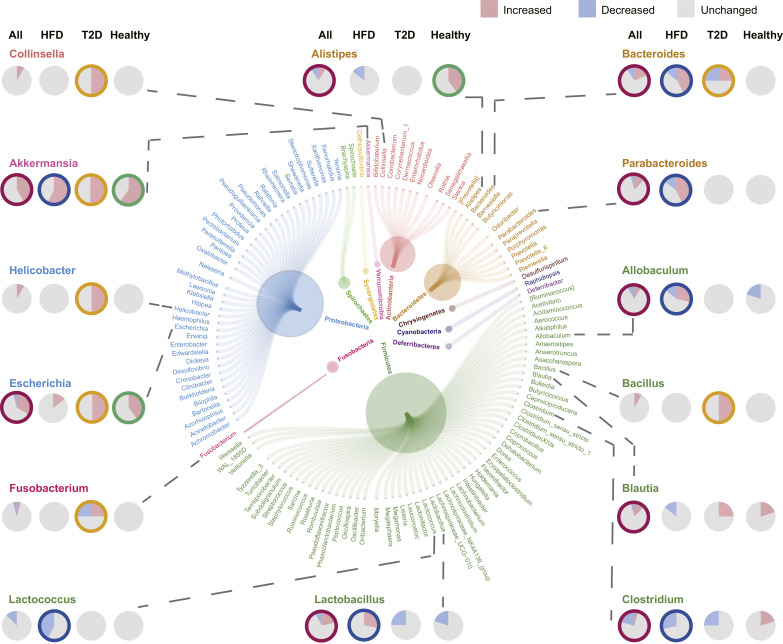
To identify microbiota correlated with metformin treatment, we summarized taxa significantly altered by metformin from 18 literatures. Raw sequences from 3 articles [13,32,33] were processed and differential analyses between metformin-treated and metformin-untreated humans or rodents were performed. Notably, de la Cuesta-Zuluaga et.al [13] reported a cohort with 28 diabetes patients, of which 14 were taking metformin. According to the patient information provided by the authors in the supplementary materials, two diabetes patients without metformin were taking other anti-diabetes drugs (MI-236 taking glibenclamide and MI-315 taking insulin). To avoid confounding factors, these two patients were excluded from the following analyses. Differential analysis was performed based on the OTU table achieved from supplementary files of 1 literature [17]. 7 articles [14,16,[34], [35], [36], [37], [38]] provided results of differential analysis in tables or supplementary tables. Significantly changed microbiota were collected from the figures of 7 articles [12,[39], [40], [41], [42], [43], [44]]. Characteristics of included studies were presented in Supplementary Table. S1. Genera altered by metformin in 18 articles involving 22 metformin treatment models were presented in Fig. 2 and Supplementary Table. S2. In total, 131 genera altered by metformin were achieved from literatures. Akkermansia was significantly more abundant in 10 models and was a genus most frequently increased by metformin treatment. Other frequently altered genera (altered in no less than 4 models) among all models were Alistipes, Allobaculum, Bacteroides, Blautia, Clostridium, Escherichia, Lactobacillus and Parabacteroides. Frequently altered genera (altered in no less than 2 models) of T2D models included Akkermansia, Bacillus, Collinsella, Escherichia, Helicobacter, Bacteroides, and Fusobacterium. Frequently altered genera of HFD models (altered in no less than 2 models) included Akkermansia, Clostridium, Lactobacillus, Lactococcus, Allobaculum, Bacteroides and Parabacteroides. Frequently altered genera (altered in no less than 2 models) of normal healthy models included Akkermansia, Alistipes and Escherichia.
Fig. 2.
Microbial genera most frequently found to be altered by metformin. Genera (outer circle) with the same color are classified in the same phylogenetic phylum level (inner circle). A certain genus was increased, reduced or unchanged by metformin treatment in certain models. Pie charts show the distribution of models reporting that a certain genus was increased (red), decreased (blue) or unchanged (grey). Pie charts in the first column show the distributions of all metformin models included. Pie charts in the second, third and fourth column show the distributions of HFD models, T2D models and models of healthy humans or rodents, respectively. Pie chart with colored circular edge indicates that the corresponding genus is considered as a biomarker for the corresponding model (all metformin treatment models, red; HFD models, blue; T2D models, yellow; models for healthy humans or rodents, green). See Supplementary Table. S2 for detailed information.
3.3. Key microbial biomarkers of metformin treatment
To identify key taxa associated with the beneficial effect of metformin, we evaluated the performance of discriminating metformin-treated and metformin-untreated humans or rodents with the frequently altered genera summarized above. These frequently altered genera were considered as bacterial biomarkers. For example, the 9 frequently altered genera (altered in no less than 4 models) of all models, including Akkermansia, Alistipes, Allobaculum, Bacteroides, Blautia, Clostridium, Escherichia, Lactobacillus and Parabacteroides, were considered as biomarkers for discrimination of all different metformin models. The 7 frequently altered genera (altered in no less than 2 models) of T2D models, including Akkermansia, Bacillus, Collinsella, Escherichia, Helicobacter, Bacteroides and Fusobacterium, were considered as biomarkers particularly for discriminating metformin-treated and metformin-untreated T2D humans or rodents. We defined and calculated the metformin value (MFV) to evaluate the performance of biomarkers combinations. Detailed definitions of biomarkers combinations (MFV1-all, MFV2-T2D, MFV3-T2D, MFV4-HFD, MFV5-HFD, MFV6-Healthy) were shown in Supplementary Table. S3. ROC curves were generated to assess the performance of single biomarkers and biomarkers combinations in 3 articles [13,32,33] with raw 16S rRNA gene sequences (Supplementary Fig. S1(a)–(f)). Performances of biomarkers particularly for T2D, obese and healthy humans or rodents were assessed alone or combined in three datasets published by de la Cuesta-Zuluaga et al. [13], Adeshirlarijaney et al. [33] and Ma et al. [32], respectively. Notably, not all bacterial biomarkers were detected in the 3 datasets. Therefore, we were unable to evaluate the performance of some genera in discriminating metformin-treated and metformin-untreated humans or rodents.
De la Cuesta-Zuluaga et al. [13] reported a cohort involving diabetes patients taking or not taking metformin. In the evaluation of single biomarker, Akkermansia presented the best performance in discriminating metformin-treated and metformin-untreated T2D individuals with an area under receiver operating curve (AUROC) of 0.737 (95% confidence interval, 0.541 0.933; bootstrap percentile method, P = 0.046, Supplementary Fig. S1(a)). When evaluating the performance of biomarkers combinations, MFV3-T2D gave the highest AUROC of 0.851 (95% confidence interval, 0.701–1.000; bootstrap percentile method, P = 0.003, Supplementary Fig. S1(b)). However, no statistical significance of performance was found between Akkermansia and MFV1-all (bootstrap percentile method, P = 0.29), Akkermansia and MFV2-T2D (bootstrap percentile method, P =0.27), Akkermansia and MFV3-T2D (bootstrap percentile method, P = 0.15). Adeshirlarijaney et al. [33] fed male C57BL/6 mice with a high-fat diet (HFD) and injected HFD-fed mice intraperitoneally with normal saline (NS) or metformin. Allobaculum presented the highest AUROC of 1 (95% confidence interval, 1.000–1.000; bootstrap percentile method, P = 0.000, Supplementary Fig. S1(c)), followed by Lactococcus with an AUROC of 0.944 (95% confidence interval, 0.844–1.000; bootstrap percentile method, P = 0.001, Supplementary Fig. S1(c)). Akkermansia also showed a high AUROC of 0.789 (95% confidence interval, 0.576–1.000; bootstrap percentile method, P = 0.034, Supplementary Fig. S1(c)). No significance was detected between the performance of Akkermansia and Lactococcus (bootstrap percentile method, P = 0.245). When comparing the performance of Akkermansia and Allobaculum, P-value was very close to 0.05 (bootstrap percentile method, P = 0.049). Ma et al. [32] separated 19 C57BL/6 healthy mice into two groups: 9 were controls and 10 were treated with metformin. Results showed that Akkermansia exhibited the best performance in discriminating metformin-treated and metformin-untreated healthy rodents with an AUROC of 1 (95% confidence interval, 1.000-1.000; bootstrap percentile method, P = 0.050, Supplementary Fig. S1(e)).
Subsequently, we predicted functional characterizations in 3 datasets [13,32,33] using PICRUSt [27] with KEGG database [23]. Overlapping significantly (Wilcoxon test, P < 0.05) altered pathways in 3 datasets were plotted in Supplementary Fig. S1(g). Pathways involved in genetic information processing, cellular processes and signaling, lipid metabolism, glycan biosynthesis and metabolism, biosynthesis of some secondary metabolites were highly associated with metformin treatment. These results were consistent with current published studies. The association between metformin and lipid metabolism has been widely reported [12,34,42,45]. For example, metformin could improve glucose homeostasis in obese mice [12]. In addition, it has been reported that in newly diagnosed T2D patients naively treated with metformin for 3 days, bile acid glycoursodeoxycholic acid (GUDCA) was increased in the gut [46].
3.4. Prediction of microbiota critical for the antitumour effect of metformin
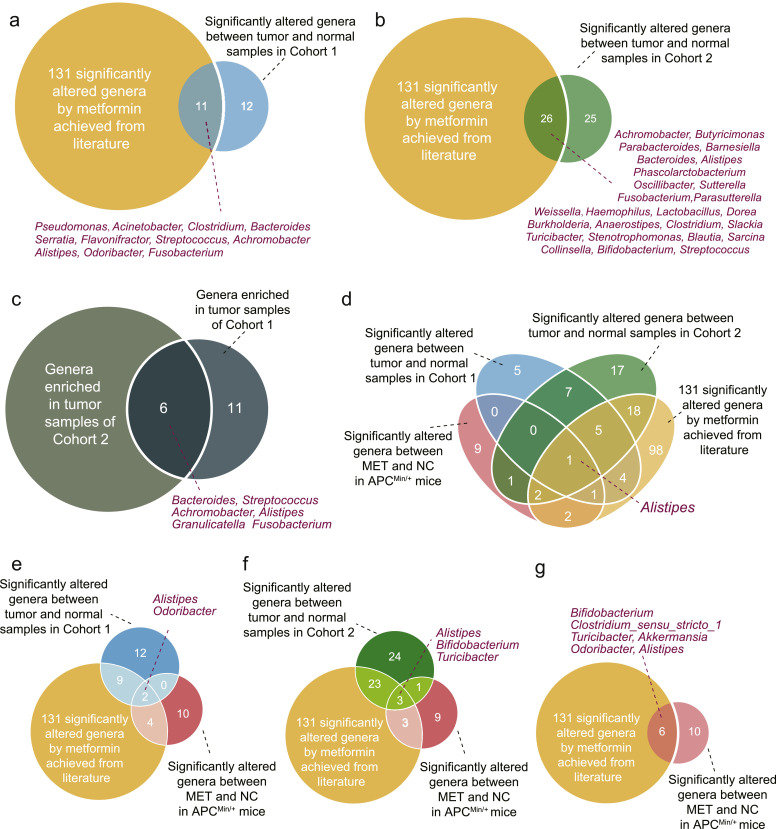
To predict potential microbiota critical for the antitumour effect of metformin, we intersected the significantly altered genera between normal and CRC samples in Cohort 1 and Cohort 2 with the 131 genera obtained from literatures that were altered by metformin. 11 and 26 overlapping genera were identified for Cohort 1 and Cohort 2 respectively (Fig. 6(a) and (b)). Bacteroides, Streptococcus, Achromobacter, Alistipes, Granulicatella and Fusobacterium were enriched in CRC patients from both Cohort 1 and Cohort 2 (Fig. 6(c)). Notably, results of LEfse analysis also showed that Fusobacterium enriched in CRC samples (Fig. 1(b)). Our previous work demonstrated that F. nucleatum colonization in the intestine prompted colorectal tumourigenesis [6]. Also, F. nucleatum was associated with CRC reoccurrence and patient outcome [7]. An abundant number of researches have revealed the significant role of F. nucleatum in CRC diagnosis [10]. It has been reported that Fusobacterium could be altered by metformin (Supplementary Table. S2). Moreover, a research published in 2018 reported that metformin would inhibit F. nucleatum at a colon concentration of 1.5 mM [47]. Therefore, we hypothesized that metformin exerted its antitumour effect by modulating F. nucleatum.
Fig. 6.
Venn diagrams showing the overlap of genera. (a) Venn diagram shows the intersection between significantly altered genera in normal and CRC samples in Cohort 1, and 131 significantly altered genera by metformin achieved from literature. (b) Venn diagram shows the intersection between significantly altered genera in normal and CRC samples in Cohort 2, and 131 significantly altered genera by metformin achieved from literature. (c) Venn diagram shows the intersection between genera enriched in tumour samples of Cohort 1 and Cohort 2. (d) Venn diagram shows the intersection among significantly altered genera in normal and CRC samples in Cohort 1 and Cohort 2, 131 significantly altered genera by metformin achieved from literature, and the significantly altered genera in MET and NC of APCMin/+ mice. (e) Venn diagram shows the intersection among significantly altered genera in normal and CRC samples in Cohort 1, 131 significantly altered genera by metformin achieved from literature, and the significantly altered genera in MET and NC of APCMin/+ mice. (f) Venn diagram shows the intersection among significantly altered genera in normal and CRC samples in Cohort 2, 131 significantly altered genera by metformin achieved from literature, and the significantly altered genera in MET and NC of APCMin/+ mice. (g) Venn diagram shows the intersection between 131 significantly altered genera by metformin achieved from literature, and the significantly altered genera in MET and NC of APCMin/+ mice. NC: normal control group; MET: metformin group.
3.5. Metformin could suppress colonic tumourigenicity in APCMin/+ mice
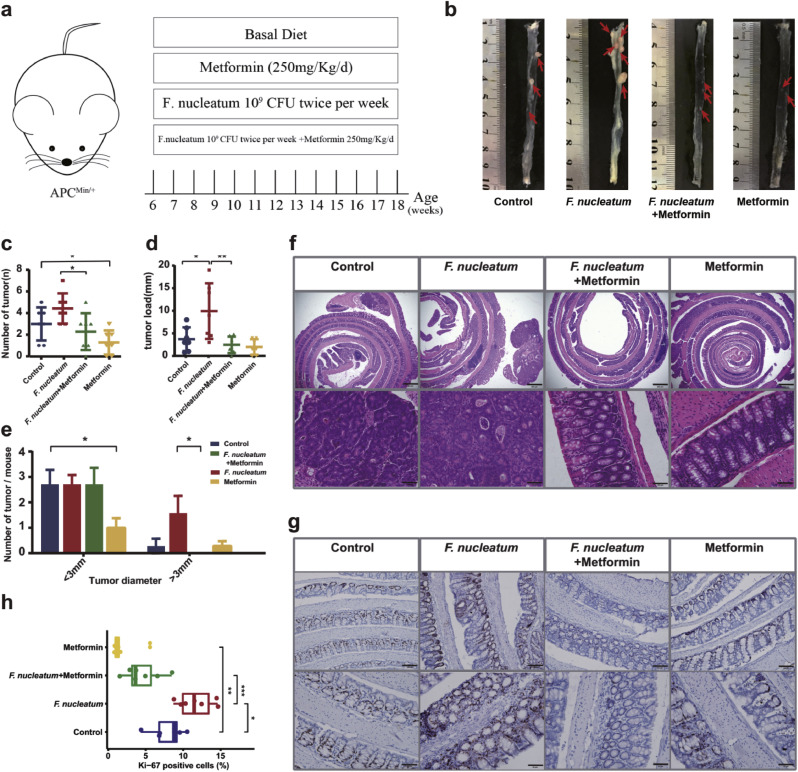
To confirm our hypothesis, first, we cultured F. nucleatum in the presence or absence of 10mM metformin to investigate whether metformin can suppress the growth of F. nucleatum in vitro. Interestingly, metformin inhibited the growth of F. nucleatum both in broth tube (Supplementary Fig. S2(a)) and on agar plate (Supplementary Fig. S2(b)) under anaerobic conditions. To investigate whether metformin can suppress intestinal tumour formation and rescue F. nucleatum-induced tumourigenicity, 6-week-old APCMin/+ mice were given 109 CFU F. nucleatum and/or 250 mg/kg/d metformin [20], which was roughly equivalent to 2000 mg per day in human (Fig. 3(a)). Although the amount of metformin we used in this study was higher than that used in diabetes patients (30–50 mg/kg), previous reports investigating the antidiabetic and antitumour effects of metformin in the mouse model used a higher amount of metformin (250–350 mg/kg) because of the different drug sensitivity between rodent and human [20]. Therefore, we used metformin at the dose of 250 mg/kg in this study. Although metformin is associated with modest weight loss, we did not obverse significant body weight alteration by metformin during the period (Supplementary Fig. S2(c)). 12 weeks after treatment, 2 groups of mice exposed to metformin showed lower incidence of adenoma (Fig. 3(b) and (c)). In detail, NC developed 3.00 ± 0.58 colon tumour per mouse while metformin reduced this number to 1.29 ± 0.42 per mouse (Fig. 3(c)). Additionally, a decrease in tumour size (the number of tumours smaller than 3 mm; Fig. 3(e)) were identified in MET compared with NC. Consistently, histopathological examinations showed that metformin attenuated the colonic neoplasia in APCMin/+ mice (Fig. 3(f)). Metformin-treated APCMin/+ tumours showed decreased cell proliferation, as evidenced by lower proportion of Ki-67+ cells compared with tumours from NC group (Fig. 3(g) and (h)). What is more, we surprisingly found that metformin could reverse the F. nucleatum-induced tumourigenicity. After F. nucleatum introduction, 6 APCMin/+ mice presented serious bloody diarrhea while none of the mice in FM developed this complication. FM showed a decrease in tumour number (2.29 ± 0.64 versus 4.43 ± 0.53, Wilcoxon test, P = 0.0244, Fig. 3(c)) compared with FN. We further discovered that the colorectum showed decrease in tumour load (2.47 ± 0.69 versus 9.9 ± 2.3, Wilcoxon test, P = 0.0097, Fig. 3(d)), and tumour size (the number of tumours larger than 3 mm; Fig. 3(e)) when comparing FM with FN. Metformin also alleviated F. nucleatum-induced neoplasia histologically (Fig. 3(f)). A higher percentage of Ki-67+ cells was discovered in FN compared with NC. Metformin could attenuate F. nucleatum-induced cell proliferation, as evidenced by lower proportion of Ki-67+ cells in FM compared with FN (Fig. 3(g) and (h)). Further factorial analyses showed interactions between metformin and F. nucleatum for tumour load (2*2 factorial ANOVA, P = 0.04). In addition, the small intestine demonstrated similar results (Supplementary Fig. S2(d)–(g)).
Fig. 3.
Metformin suppresses colonic tumourigenicity in APCMin/+ mice. (a) Experimental design for mice study. 4- to 5-week-old APCMin/+ mice were divided into 4 groups and were administrated with metformin and/or F. nucleatum for 12 weeks (n=7 mice/group). (b) Representative images of colons. The red arrows indicated the positive location. (c) Numbers of colon tumour from APCMin/+ mice. (d) Colon tumour load. (e) Comparison of colon tumour size below 3 mm and over 3 mm in diameter. (f) Representative images of the colon stained with hematoxylin and eosin. Scale bar = 500 μm (top panels); Scale bar = 100 μm (bottom panels). (g) Immunohistochemistry showing Ki-67+ cells in colon tumours. Scale bar = 100 μm (top panels); Scale bar = 50 μm (bottom panels). (h) Box plot showing Ki-67+ cells in colon tumours. Significance between every two groups was calculated using unpaired two-tailed Student's t test. Data are expressed as the means ± SE. *** P < 0.001, ** P < 0.01, * P < 0.05.
3.6. Effect of metformin and F. nucleatum on microbial communities
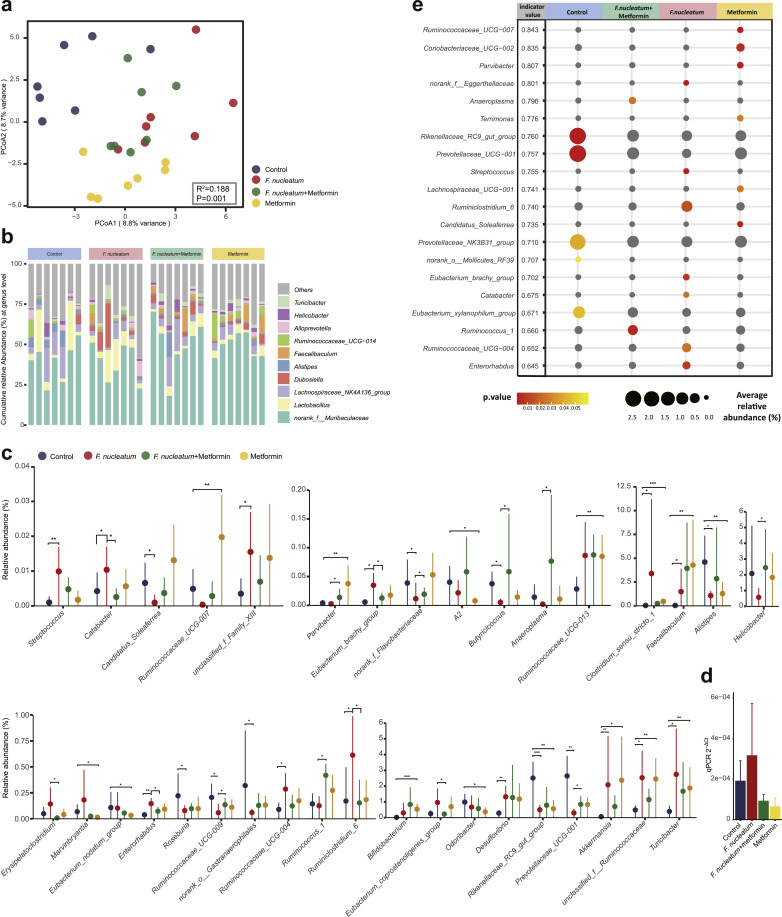
To explore the effect of metformin on gut microbiota, 16S rRNA gene sequencing was performed in 4 groups of APCMin/+ mice. In total, 1,377,375 high-quality sequences were generated from 28 samples, with an average length of 420.05bp per sequence, yielding 634, 602, 583 and 628 OTUs at a 97% identity cut-off in NC, FN, FM and MET respectively (Supplementary Fig. S3(a)). PCoA based on Euclidean binary distances (Adonis P = 0.001, R2 = 0.188) at the OTU level revealed the most significant separation among groups (Fig. 4(a)) compared with PCoA on weighted Unifrac and unweighted Unifrac distances (Adonis weighted Unifrac: R2 = 0.207, P = 0.002, Supplementary Fig. S3(b); unweighted Unifrac: R2 = 0.307, P = 0.001, Supplementary Fig. S3(c)). In addition, PCoA on weighted Unifrac distances showed a clearer separation than unweighted Unifrac distances. PCoA on Euclidean distances demonstrated an unclear separation between groups (R=0.122, P=0.003, data not shown). Results of PCoA indicated a more significant difference in evenness than richness of microbiota communities among groups. No significant change in alpha diversity was detected by pairwise comparison (Supplementary Fig. S3(d)).
Fig. 4.
Microbial community structure and composition in APCMin/+ mice. (a) Principal coordinate analysis (PCoA) on Euclidean binary distances is shown for NC (blue), FN (red), FM (green) and MET (yellow). Significance of clustering was determined using the Adonis test. (b) Microbial compositional profiling of NC, FN, FM and MET at genus level. (c) Relative abundance of genera that are significantly altered when comparing NC with MET, NC with FN, FN with FM, respectively. The plots represent the median relative abundance of genera (filled circles) and the interquartile range of the distribution (whiskers). The Wilcoxon rank-sum test was used for statistical comparisons. ∗∗∗ P < 0.001, ∗∗ P < 0.01, ∗ P < 0.05. (d) The abundance of F. nucleatum in the colon tissue of APCMin/+ mice. No statistical significance was detected by Student's t test between groups. n = 5–6 mice per group. Data in the bar plots are presented as the means ± SEM. (e) Indicator genera characterizing the NC, FN, FM, MET bacterial communities. Indicator values (IV) plotted next to the name of taxa range from zero to one, with larger values denoting greater specificity. The average relative abundance (circle size) and P-values (circle color) were plotted. Circles colored in grey indicate insignificant taxa. NC: normal control group; FN: F. nucleatum group; FM: F. nucleatum + metformin group; MET: metformin group.
Compositional profiling revealed that the phyla Firmicutes and Bacteroidetes exhibited the highest proportions in all APCMin/+ mice (Supplementary Fig. S3(e)). Muribaculaceae was the most abundant family in APCMin/+ mice but its abundance was not significantly different among groups (Supplementary Fig. S3(f)). Turicibacter, Helicobacter, Alloprevotella, Ruminococcaceae_UCG−014, Faecalibaculum, Alistipes, Dubosiella, Lachnospiraceae_NK4A136_group and Lactobacillus constituted the vast majority of genera in all samples (Fig. 4(b)).
We compared the relative abundances of genera in APCMin/+ mice (Fig. 4(c)). Introduction of F. nucleatum elevated the abundances of Catabacter, Eubacterium_brachy_group, Enterohabdus, Ruminiclostridium_6, Prevotellaceae_UCG-001, which were reversed by metformin. Conversely, the abundances of Butyricicoccus, norank_Flavobacteriaceae, Ruminococcaceae_UCG-009, Prevotellaceae_UCG-001 were depleted by F. nucleatum, but were elevated by metformin treatment. Treatment with metformin in APCMin/+ mice increased the relative abundances of Ruminococcaceae_UCG-007, Parvibacter, Ruminococcaceae_UCG-013, Clostridum_sensu_stricto_1, Faecalibaculum, Bifidobacterium, Akkermansia, unclassified_Ruminococcaceae and Turicibacter while decreased A2, Alistipes, Marvinbryantia, Eubacterium_nodatum_group, Odoribacter and Rikenellaceae_RC9_gut_group. Unexpectedly, F. nucleatum was not detected by 16S rRNA gene sequencing in any group, even in APCMin/+ mice given F. nucleatum. Nevertheless, microbiota composition has been altered by introduction of F. nucleatum. We then performed qPCR to detect the abundance of F. nucleatum in tumour tissue of APCMin/+ mice. Although no statistical significance was detected among groups, FN group showed high abundance of F. nucleatum, and metformin seemed to reverse this change (Fig. 4(d)).
We then used the LEfSe method to identify the key communities among 4 groups. The relative abundance of altered genera (P < 0.05) was showed by the LEfSe taxonomy cladogram (Fig. S3(g)). A linear discriminant analysis (LDA) score higher than 3 suggested a higher relative abundance in the corresponding group than in the other 3 groups (P < 0.05) (Fig. S3(h)). Metformin showed selective enrichment in Faecalibaculum, Akkermansia, Anaerofilum, Dechloromonas, Bdellovibrio, Blastocatellaceae, OPB56 and Ruminococcaceae. Clostridium_sensu_stricto_1, Ruminococcaceae, Desulfovibrio, Eggerthellaceae, Streptococcus, Ruminiclostridium_6 and Staphylococcus were more enriched in FN.
We obtained the indicator taxa based on genus level profiles of the 4 groups (Fig. 4(e)). Higher indicator values (IV) suggested better performances in signature taxa. Parvibacter, Coriobacteriaceae_UCG−002, Ruminococcaceae_UCG−007 and Terrimonas were highly specific for metformin treated APCMin/+ mice, with IVs of 0.843, 0.835, 0.807, 0.776, respectively. Prevotellaceae_UCG−001 and Rikenellaceae_RC9_gut_group were highly specific for metformin untreated APCMin/+ mice with IVs of 0.760 and 0.757, respectively. We then evaluated the performance of indicator taxa in discriminating NC and MET using ROC curves. Parvibacter exhibited the highest AUROC of 0.98. MFV was used to evaluate the performance of all indicators of MET and NC. MFV-indicator presented an AUROC of 1 (Supplementary Fig. S4(a)). Then, the performances in discriminating APCMin/+ mice fed with and without metformin were evaluated using biomarkers that were obtained from literatures for all metformin models (Supplementary Fig. S4(b)). Akkermansia gave the highest AUROC of 0.878, which was higher than all biomarkers combined. We then compared the performance of Akkermansia and Parvibacter, and no significance was identified between the two biomarkers (Supplementary Fig. S4(c)).
3.7. Functional shifts of microbiome by metformin and F. nucleatum
Functional characterizations were explored in this study to disclose the effect of metformin on APCMin/+ mice with or without F. nucleatum introduction. A total of 5943 KO functional categories were yielded. Pairwise comparisons of the functional differences were quantified using a two-tailed Wilcoxon test, and P < 0.05 was considered statistically significant (Supplementary Table S4). The significantly altered KOs were filtered with the criterion of abs(logFC) ≥0.2 and visualized in Supplementary Fig. S5(a)–(c). Through this filter, the greatest variations in KO functional categories were discovered when comparing MET with NC. In pairwise comparisons, 1649 KOs were significantly enriched in MET and 94 were significantly enriched in NC (Supplementary Fig. S5(a)). 804 KOs were significantly enriched in FN and 195 KOs were significantly enriched in NC (Supplementary Fig. S5(b)). 152 KOs were significantly enriched in FM and 262 KOs were significantly enriched in FN (Supplementary Fig. S5(c)).
We further explored KEGG pathways in which KO functional categories were involved. Pairwise comparisons were performed with a two-tailed Wilcoxon test, and P < 0.05 was considered statistically significant. Representative altered pathways with the highest relative abundances were visualized in Supplementary Fig. S5(d)–(f).
3.8. Interactions of microbial taxa altered by metformin and/or F. nucleatum
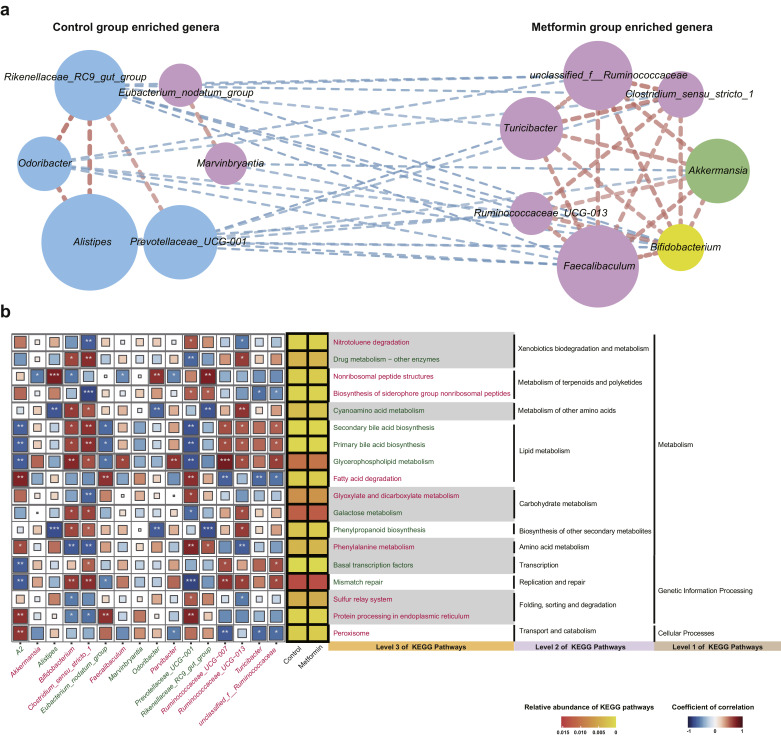
To investigate whether metformin would promote the interactions among gut bacteria, we inferred pairwise taxonomic correlations in significantly altered genera in APCMin/+ mice. Spearman's correlation coefficients among genera were calculated, and correlations with absolute coefficients ≥ 0.3 and P < 0.05 were retained. Results showed that metformin increased positive taxonomic correlations in APCMin/+ mice either given without (Fig. 5(a)) or with F. nucleatum (Supplementary Fig. S7(a)). F. nucleatum administration could also change the interactions among gut bacteria (Supplementary Fig. S6(a)). To provide possible explanations for the relationships between taxa, we examined the correlations between taxa and significantly altered KEGG pathways to obtain an overview of how significantly altered taxa act during dysfunction (Fig. 5(b), Supplementary Fig. S6(b), Supplementary Fig. S7(b)). Only KEGG pathways with mean relative abundances over 0.03% and genera with a sum of relative abundance greater than 0.5% were included in the analysis. Spearman's correlation coefficients were calculated between KEGG pathways and taxa, and P < 0.05 was considered significant. Results showed that positively correlated genera usually had synergy in functional pathways while negatively correlated genera often presented opposite associations with specific functional pathways. Moreover, within group interactions were often positive while between groups relationships were often negative. Members of the same phylum were more likely to form strong relationships with one another.
Fig. 5.
Interactions of gut microbiota altered by metformin. (a) Correlation networks of significantly altered genera between NC and MET based on the Wilcoxon rank-sum test. Spearman’ s absolute correlation coefficients ≥ 0.3 and P-values < 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level. (b) Associations between significantly altered genera and significantly altered KEGG pathways in NC and MET. Spearman’ s correlation coefficients are plotted by the color of squares. The sizes of squares correspond to the significance of the P-values (the larger the square, the smaller the P-value). ∗∗∗ P < 0.001, ∗∗ P < 0.01, ∗ P < 0.05. Average relative abundance of KEGG pathways is plotted by heatmap next to visualization of correlation matrix. The names of genera and pathways enriched in MET were colored in red. The names of genera and pathways enriched in NC were colored in green. NC: normal control group; MET: metformin group.
In the correlation network of NC and MET, the strongest relation was found between Odoribacter and Rikenellaceae_RC9_gut_group. The two genera belonging to the same phylum Bacteroidetes were positively correlated and synergistically involved in phenylpropanoid biosynthesis, cyanoamino acid metabolism and nonribosomal peptide structures (Fig. 5). Strong relationships were also formed between Alistipes and Rikenellaceae_RC9_gut_group, Alistipes and Odoribacter, Clostridium_sensu_stricto_1 and unclassified_f__Ruminococcaceae, Faecalibaculum and Rikenellaceae_RC9_gut_group, Prevotellaceae_UCG-001 and Turicibacter. Alistipes and Rikenellaceae_RC9_gut_group belonging to the same phylum Bacteroidetes were positively correlated, exerting synergistic influence on legionellosis, phenylpropanoid biosynthesis, cyanoamino acid metabolism and nonribosomal peptide structures. Alistipes and Odoribacter belonging to the same phylum Bacteroidetes showed strong within group positive association and both were negatively involved in phenylalanine metabolism, cyanoamino acid metabolism, and positively involved in nonribosomal peptide structures. Clostridium_sensu_stricto_1 and unclassified_f__Ruminococcaceae belonging to the same phylum Firmicutes were positively correlated and coordinated in mismatch repair, legionellosis, lipid metabolism and biosynthesis of siderophore group nonribosomal peptides. A negative relation was formed between Faecalibaculum and Rikenellaceae_RC9_gut_group, but Faecalibaculum seemed to have relatively weak correlations with functional pathways despite its high relative abundance. A negative relation was formed between Prevotellaceae_UCG-001 and Turicibacter, and they demonstrated opposite impact on fatty acid degradation and biosynthesis of siderophore group nonribosomal peptides.
In the correlation network of NC and FN, Faecalibaculum and Ruminococcaceae_UCG-009 exhibited the strongest relation. Alistipes and Rikenellaceae_RC9_gut_group, Faecalibaculum and Rikenellaceae_RC9_gut_group, unclassified_f__Ruminococcaceae and Ruminococcaceae_UCG-004, Alistipes and Ruminococcaceae_UCG-009, Prevotellaceae_UCG-001 and Desulfovibrio also showed strong relations. Alistipes and Rikenellaceae_RC9_gut_group both belonged to the phylum Bacteroidetes and showed strong associations with multiple functional pathways (Supplementary Fig. S6). The two genera had synergy and almost the same significance in various pathways including cell growth and death, transport and catabolism, folding, sorting and degradation, amino acid metabolism, biosynthesis of other secondary metabolites, energy metabolism, glycan biosynthesis and metabolism, lipid metabolism, environmental adaptation, xenobiotics biodegradation and metabolism, metabolism of terpenoids and polyketides, cyanoamino acid metabolism. This suggested that Alistipes and Rikenellaceae_RC9_gut_group might coordinate intensively in resisting the functional shifts caused by F. nucleatum introduction. Prevotellaceae_UCG-001 and Desulfovibrio were negatively correlated and showed significant opposite associations with several pathways, including nitrotoluene degradation, drug metabolism-other enzymes, galactose metabolism, phenylalanine metabolism, mismatch repair, homologous recombination and sulfur relay system.
The correlation network and functional characterizations of FN and FM greatly differed from those of NC and MET, suggesting a certain effect of F. nucleatum on microbiome composition and metabolic dysfunction. Erysipelatoclostridium and Eubacterium_coprostanoligenes_group were both members of Firmicutes, and had the strongest correlation. Helicobacter was abundantly enriched in FM, and had significant negative correlation with insulin signaling pathway. Ruminococcus_1 was significantly involved in multiple pathways, including N−Glycan biosynthesis, phosphonate and phosphinate metabolism, pyruvate metabolism, butanoate metabolism, valine, leucine and isoleucine degradation (Supplementary Fig. S7).
Although F. nucleatum was quantified by qPCR using colon tumour tissue in APCMin/+ mice, it was not detected by 16S rRNA gene sequencing. Therefore, we were unable to explore the interaction between F. nucleatum and other bacteria during metformin treatment using 16S rRNA gene sequencing data of APCMin/+ mice. To provide more information for future investigations into the taxonomic correlations of Fusobacterium under metformin treatment, we predicted potential bacteria co-occurred and interacted with Fusobacterium by examination of the correlation between Fusobacterium and other genera in CRC patients. Only genera with a sum of relative abundance greater than 0.5% were included in the analysis. Correlations with absolute coefficients ≥ 0.3 and P < 0.05 were retained (Supplementary Fig. S8(a) and Supplementary Fig. S8(b)). However, none of the genera highly correlated with F. nucleatum in Cohort 1 and Cohort 2 was significantly altered when comparing FN with NC or FN with FM. Correlation networks were constructed in Cohort 1 and Cohort 2 (Supplementary Fig. S8(c) and Supplementary Fig. S8(d)).
To further explore bacteria co-varied with Fusobacterium, we classified CRC patients of both cohorts into Fusobacterium-high and Fusobacterium-low groups based on the median relative abundance of Fusobacterium. We conducted LEfse analysis (Supplementary Fig. S9(a)–(d)) as well as differential analysis based on the genus level (Supplementary Fig. S9(e) and (f)). LEfse analysis showed that Streptococcus was significantly enriched in Fusobacterium-high group of Cohort 2 (Supplementary Fig. S9(c) and (d)) and was also elevated in FN compared with NC (Fig. 4(c)). Results of differential analysis showed that Streptococcus was significantly elevated in F. nucleatum-high patients compared with F. nucleatum-low patients of Cohort 1 (Supplementary Fig. S9(e)) and was also significantly increased in FN compared with NC (Fig. 4(c)). A study [2] reported significantly enriched genera in F. nucleatum “High” (>1% relative abundance) colon adenocarcinoma (COAD) from The Cancer Genome Atlas (TCGA cohort) [48]. We observed that Desulfovibrio was significantly enriched in both F. nucleatum “High” TCGA patients and FN (Fig. 4(c)). We inferred from above results that Streptococcus and Desulfovibrio were vital bacteria co-varied with F. nucleatum.
3.9. Correlation between gut microbiota and the antitumour effect of metformin
To reveal the relationship between gut microbiota and the antitumour effect of metformin, we intersected the significantly altered genera in MET and NC, significantly altered genera in normal samples and CRC samples in Cohort 1 and Cohort 2, 131 significantly altered genera by metformin achieved from literature (Fig. 6). Only 1 genus, Alistipes, was overlapped when intersecting results from these 4 sections (Fig. 6(d)). Alistipes was more enriched in CRC patients, and decreased after metformin treatment in APCMin/+ mice. In Cohort 1, 2 genera, Alistipes and Odoribacter, were significantly increased in CRC patients (Fig. 6(e)) while decreased after metformin treatment in APCMin/+ mice (Fig. 4(c)). In Cohort 2, Alistipes was significantly elevated in CRC patients (Fig. 6f) while significantly decreased after metformin treatment in APCMin/+ mice (Fig. 4(c)). Bifidobacterium and Turicibacter significantly reduced in CRC patients (Fig. 6(f)) while significantly increased after metformin treatment in APCMin/+ mice (Fig. 4(c)). In addition, Bifidobacterium, Turicibacter and Odoribacter were among the 131 genera altered by metformin achieved from literature (Fig. 6(e) and (f)). These results suggested that Alistipes, Bifidobacterium, Turicibacter and Odoribacter might played critical roles in the suppression of colonic tumourigenicity by metformin.
6 overlapping genera, Bifidobacterium, Clostridium_sensu_stricto_1, Turicibacter, Akkermansia, Odoribacter, Alistipes, were identified when intersecting the significantly altered genera in MET and NC, with 131 significantly altered genera by metformin achieved from literature (Fig. 6(g)). This indicated the effect of metformin on these taxa had been reported previously.
4. Discussion
In this study, we reanalyzed our previous data on gut microbiome of normal and CRC individuals. Subsequently, we summarized bacteria altered by metformin after reviewing 18 literatures. We discovered that Fusobacterium was enriched in CRC samples and could also be altered by metformin. We hypothesized that metformin exerted its antitumour effect by modulating F. nucleatum. To confirm this hypothesis, we conducted experiments on APCMin/+ mice given F. nucleatum and/or metformin. Results showed that metformin relieved the symptoms caused by F. nucleatum administration in APCMin/+ mice, and showed promise in suppressing intestinal tumour formation and rescuing F. nucleatum-induced tumourigenicity. Additionally, metformin had interactions with F. nucleatum. Our findings that gut microbiota were significantly altered when comparing NC with MET, and FN with FM indicated that modulation of the gut microbiota might contribute to the antitumour effects of metformin. Our work revealed the structural, compositional, and functional dysbiosis of the gut microbiome after F. nucleatum and/or metformin administration. In addition, we demonstrated correlations among taxa and involvement of gut microbiome in functional activities. To the best of our knowledge, this study pioneers in implicating that altered gut microbiota may mediate some of metformin's antitumour effects using both analyses of published data and experimental methods.
Literature screening of previous investigations into the effect of metformin on gut microbiota was performed to explore taxa frequently altered by metformin. We found that several bacteria frequently altered by metformin, including Bacteroides [5], Alistipes [49], Escherichia [5], Blautia [50], Clostridium [50], Lactobacillus [50] and Fusobacterium [5], had significant change when comparing microbiota of normal and CRC samples. We also discovered that Bacteroides, Streptococcus, Achromobacter, Alistipes and Fusobacterium could be altered by metformin according to published literatures, and they enriched in CRC samples of Cohort 1 and Cohort 2. Besides F. nucleatum, several strains belonging to these overlapping genera showed associations with colorectal tumourigenesis. For example, Clostridium symbiosum, a strain of the Clostridium genus, has been tested as a marker for early CRC detection [51]. Enterotoxigenic Bacteroides fragilis (ETBF), a species widely reported to be associated with CRC occurrence, is a strain of Bacteroides [52,53]. It has been proposed that ETBF had direct effect on β-Catenin activation and MAPK signalling activation [5]. Colibactin produced by some Escherichia coli strains is thought to play a role in colorectal carcinogenesis [54]. Streptococcus bovis, a strain of Streptococcus, has long been associated with CRC [55]. Whether metformin could inhibit these CRC-associated species in vitro and in vivo requires further investigations.
The reasons that we decided to inoculate mice with F. nucleatum instead of other CRC-associated bacteria mainly have three respects. First, a research published in 2018 screened the impact of more than 1000 non-antibiotic drugs including metformin on gut bacteria [47]. The authors found that metformin would inhibit F. nucleatum at a colon concentration of 1.5 mM, which is considered a safe dose for humans as metformin concentrates approximately 150-fold higher than in plasma [56]. Second, before the start of our animal experiments, we have performed growth curve of both F. nucleatum and ETBF with and without metformin. Interestingly, growth of ETBF was not inhibited by metformin but growth of F. nucleatum was inhibited by metformin. The growth curve of ETBF was not presented in this study. Third, our lab has been focusing on the association between F. nucleatum and colorectal carcinoma. Our team has conducted basic researches on the vital role of F. nucleatum in colorectal tumour initiation, development, and patient outcome [7]. We have reported the contribution of F. nucleatum to the occurrence of colorectal adenomas and colorectal carcinomas [6]. We are well experienced in conducting experiments on F. nucleatum. The association between CRC and F. nucleatum has also been reported in an abundant number of researches [2,[57], [58], [59]]. Based on a comprehensive consideration of the great significance of F. nucleatum in colorectal carcinogenesis and the feasibility of experiments, we aimed to explore the effect of metformin on F. nucleatum. We not only revealed the effect of metformin on rescuing F. nucleatum-induced tumourigenicity, but also showed an antagonistic effect of metformin on F. nucleatum according to factorial analysis, confirming our hypothesis that metformin could exert its antitumour effect by interacting with F. nucleatum. We intend to undertake further studies to investigate the mechanism of metformin and F. nucleatum interactions, and to examine the effect of metformin on other taxa.
16S rRNA sequencing analysis showed that Alistipes, Bifidobacterium, Turicibacter and Odoribacter significantly altered when comparing NC and MET, and changed in an opposite direction between normal and CRC samples in our cohorts. Notably, Alistipes was the only one genus that enriched in both Cohort 1 and Cohort 2, and has been previously reported to be reduced by metformin [12,36]. Previous study has also reported that Alistipes was augmented in CRC [60]. Taken together, evidences from our experiment, published literatures and analyses of CRC cohorts collectively indicated that modulation of gut microbiota highly associated with CRC may contribute to the antitumour effect of metformin.
In the comparison of microbiota changes between APCMin/+ mice treated with and without metformin, Faecalibaculum, Akkermansia, and Bifidobacterium were three beneficial genera with the most significant increase in feces from APCMin/+ mice administered metformin. It has recently been demonstrated that Faecalibaculum rodentium protected the host from intestinal tumour growth [61]. Akkermansia not only showed a good performance in discriminating metformin-treated and metformin-untreated humans or rodents in published literatures, but also showed good performance in discriminating APCMin/+ mice treated with and without metformin in this study. The contribution of Akkermansia to the therapeutic effect of metformin has been reported in a number of studies [12,13]. Wu et al. showed that the growth of A. muciniphila but not of E. coli could be directly promoted by metformin in pure cultures [14]. The authors have also observed a metformin-induced increase in Bifidobacterium. The health-promoting effects of Bifidobacterium is widely acknowledged. Some Bifidobacterium strains are considered as probiotic microorganisms and have been included as bioactive ingredients in functional foods [62]. Consistent with previous studies, we found that Bifidobacterium was diminished in CRC [60], and increased after metformin treatment [14]. As is shown by the results of current study and previous publications regarding the effect of metformin on gut microbiota, it seems that the increase in probiotics was more pronounced than the decrease in pathobionts. As the effect of metformin on gut protective bacteria has been reported [12,46] but few researches investigated how pathobionts reacted to metformin, we inoculated mice with a pathogenic CRC-associated bacterium, F. nucleatum, aiming to obtain a comprehensive view of the influence of metformin on microbiota. We intend to carry out follow-up researches investigating metformin's effect on both harmful and beneficial bacteria, and the interactions between multiple bacteria.
Although F. nucleatum was not detected by 16S rRNA gene sequencing in APCMin/+ mice administrated F. nucleatum, we managed to detect F. nucleatum in the tumour tissue. As metformin significantly reduced tumour number and tumour load in mice given F. nucleatum and could also inhibit the growth of F. nucleatum in vitro, we speculated that statistical significance of F. nucleatum abundance between groups would be identified with a larger sample size.
There are several merits of this work. First, through analyses of previous CRC cohorts and data collection from literatures, we predicted critical CRC-associated taxa correlating with the beneficial effects of metformin, providing new directions for future investigations into the interactions between gut microbiota and metformin in tumours. We showed that metformin might have the potential to suppress F. nucleatum-induced tumourigenesis in vivo. In addition, we proposed the possibility that certain taxa can be used to discriminate metformin-treated and metformin-untreated individuals, and may ultimately act as therapeutic targets of metformin. Second, according to the literature search in this study, most investigations into the effect of metformin on gut microbiome were conducted on experimental animals or patients with metabolic disorders such as obesity and T2D. To our knowledge, this is the first study correlating gut microbiome with the antitumour effect of metformin in experimental animals. Our work illustrated the effect of metformin on gut microbiome structure, composition and function, which may be conducive to future investigations into tumour suppression by interactions between metformin and gut microbiota. Third, current work revealed the significance of multiple taxa in the therapeutic effect of metformin, indicating that this effect may be attributed to the collective contribution of various bacteria rather than a single bacterium. The mechanisms of direct or indirect effect of metformin on gut microbiota need further validations in future researches.
Limitation existed in this work. First, different taxonomic classifications, including SILVA, RDP and Greengenes Database, were used for read assignment to taxonomic units in the 18 literatures included (Supplementary Table S1). Greengenes is the smallest taxonomy of the three taxonomic classifications, and it shares 80% taxa in genus ranks with either SILVA or RDP [63], indicating that the Greengenes taxonomy is mostly contained in the other two taxonomies. In this study, genera associated with both CRC and metformin were overlapped taxa of the three taxonomic classifications. Therefore, a small fraction of taxa unique to either Greengenes, SILVA or RDP might be omitted, preventing us from making a comprehensive prediction of bacteria involving in both CRC occurrence and metformin treatment. Second, data analyzed in this study were all based on 16S rRNA gene sequencing that rarely identify (annotate) microbes at species or strain levels. However, deep shotgun sequencing was unaffordable for us to identify specific strains and microbial genes associated with metformin in CRC. All analyses performed in this study was based on genus level. Further researches focusing on the genomics, transcriptome, proteome or metabolome of metformin treatment are needed. Third, considering that the sample size of our animal experiment was small and multiple comparisons with corrections might diminish statistical power and interfere with potential meaningful interpretation, statistical differences were determined using P values without correction for multiple hypothesis testing. Thus, the probability of type I errors occurring within this study might be inflated and our study might be underpowered to draw a definite conclusion. Human or animal studies with larger sample size in the future could give a more unequivocally answer on this subject.
In summary, through data collection from literatures and analyses of previous CRC cohorts, we observed that Bacteroides, Streptococcus, Achromobacter, Alistipes and Fusobacterium might be critical for the suppression of colonic tumourigenicity by metformin. We examined the antitumour effect of metformin on F. nucleatum and demonstrated that metformin showed promise for suppressing intestinal tumour formation and rescuing F. nucleatum-induced tumourigenicity in APCMin/+ mice. Administration of F. nucleatum and/or metformin had effect on gut microbiome structure, composition, and functions of APCMin/+ mice. We presented a basis for future investigations into metformin's potential effect on suppressing F. nucleatum-induced tumor formation in vivo. We intend to undertake follow-up studies to investigate the association between the antitumour effect and other promising microbial therapeutic targets of metformin predicted in this study, including Odoribacter, Bifidobacterium, Turicibacter, Bacteroides, Streptococcus, Alistipes, Achromobacter, Akkermansia, Blautia, Clostridium, Escherichia, Lactobacillus, etc.
Author contributions
Xiaowen Huang searched the literature. Hua Xiong, Jingyuan Fang, Jilin Wang, Tiantian Sun designed the experiment and supervised the study. Xialu Hong performed the experiments. Xiaowen Huang, Xialu Hong, Yanan Yu and TaChung Yu collected the data. Xiaowen Huang and Xialu Hong analyzed and interpreted the data. Xiaowen Huang and Xialu Hong drafted the manuscript.
Declaration of Competing Interest
None declared.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31701250). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The corresponding authors confirm to have full access to all the data and have final responsibility for the decision to submit for publication. The authors declare no conflict of interest.
Footnotes
Xiaowen Huang and Xialu Hong are Co-first authors.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.103037.
Contributor Information
Jingyuan Fang, Email: JY.Fang@sjtu.edu.cn.
Hua Xiong, Email: huaxong88@126.com.
Appendix. Supplementary materials
Supplementary Fig. S1. Characteristics of bacterial biomarkers for discriminating metformin-treated and metformin-untreated humans or rodents. ROC curves evaluating the performance of discriminating metformin-treated and metformin-untreated humans or rodents using single bacterial biomarkers for a particular metformin treatment model, generated from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al. (a), Adeshirlarijaney et\elsamp #x00A0;al. (c), and Ma et\elsamp #x00A0;al. (e) respectively. ROC curves evaluating the performance of discriminating metformin-treated and metformin-untreated humans or rodents using bacterial biomarkers combinations for a particular metformin treatment model, generated from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al. (b), Adeshirlarijaney et\elsamp #x00A0;al. (d), and Ma et\elsamp #x00A0;al. (f) respectively. See Supplementary Table. S3 for detailed definitions of bacterial biomarkers combinations. (g) Heatmap showing differences between mean abundance of overlapped KEGG pathways predicted from metformin-treated and metformin-untreated humans or rodents in from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al., Adeshirlarijaney et\elsamp #x00A0;al., and Ma et\elsamp #x00A0;al. respectively. A positive difference between means indicates higher abundance of a KEGG pathway in metformin-treated humans or rodents.
Supplementary Fig. S2. Metformin suppresses tumourigenicity in APCMin/\elsamp #x002B; mice. (a) Growth curve of F. nucleatum with or without metformin in a laboratory culture. n\elsamp #x2009;\elsamp #x003D;\elsamp #x2009;3 replicates/treatment. (b) Colony counts on agarose plates of F. nucleatum with or without metformin in a laboratory culture. n\elsamp #x2009;\elsamp #x003D;\elsamp #x2009;3 replicates/treatment. Statistical significance was detected by Student\elsamp #x0027;s t test. Data in the bar plots are presented as the means \elsamp #x00B1; SEM. (c) Mean weights of APCMin/\elsamp #x002B; mice in each group. No statistical difference in body weight was observed between groups. (d) Representative images of small intestine. The red arrows indicated the positive location. (e) Numbers of small intestine tumour. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test. (f) Small intestine tumour load. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test. (g) Comparison of small intestine tumour size below 1.5 mm and over 1.5 mm in diameter. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test.
Supplementary Fig. S3. 16S rRNA sequencing analysis of the gut microbiome in APCMin/\elsamp #x002B; mice. (a) Venn diagram showing overlapped operational taxonomic unit (OTU) among 4 groups. Principal coordinate analysis (PCoA) on weighted Unifrac (b) and unweighted Unifrac distances (c) is shown for NC (blue), FN (red), FM (green) and MET (yellow). Significance of clustering was determined using the Adonis test. (d) No significant difference is identified for alpha-diversity (Chao1, richness, Ace index, Shannon index) between NC and MET, NC and FN, FN and FM. Significance between every two groups was calculated using the Wilcoxon rank-sum test. (e) Microbial compositional profiling of NC, FN, FM and MET at phylum level. (f) Microbial compositional profiling of NC, FN, FM and MET at family level. (g) A cladogram representation of data in NC, FN, FM and MET. (Blue) Taxa enriched in NC; (red) taxa enriched in FN; (green) taxa enriched in FM; (yellow) taxa enriched in MET. The brightness of each dot is proportional to its effect size. The prefixes \elsamp #x2018;p\elsamp #x2019;, \elsamp #x2018;c\elsamp #x2019;, \elsamp #x2018;o\elsamp #x2019;, \elsamp #x2018;f\elsamp #x2019; and \elsamp #x2018;g\elsamp #x2019; represent the level of phylum, class, order, family, and genus. (h) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in G. (Blue) Taxa enriched in NC; (red) taxa enriched in FN; (green) taxa enriched in FM; (yellow) taxa enriched in MET. Only taxa meeting an LDA significant threshold of 3 are shown. NC: normal control group; FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group; MET: metformin group.
Supplementary Fig. S4. Assessment of performance in discriminating NC and MET using bacterial biomarkers in APCMin/\elsamp #x002B; mice. (a) ROC curves of bacterial indicators. (b) ROC curves of bacterial biomarkers for all metformin treatment models. (c) ROC curves comparing the performance of biomarkers with the highest AUROC in (a) and (b). NC: normal control group; MET: metformin group.
Supplementary Fig. S5. Significantly (Wilcoxon test, P \elsamp #x003C; 0.05, abs(logFC) \elsamp #x2265; 0.2) different KO functional categories between NC and MET (a), NC and FN (b), FN and FM (c). Representative significantly (Wilcoxon test, P \elsamp #x003C; 0.05) enriched KEGG pathways with the highest relative abundances between NC and MET (d), NC and FN (e), FN and FM (f). NC: normal control group; FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group; MET: metformin group.
Supplementary Fig. S6. Interactions of gut microbiota altered by F. nucleatum. (a) Correlation networks of significantly altered genera between NC and FN based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level. (b) Associations between significantly altered genera and significantly altered KEGG pathways in NC and FN. Spearman\elsamp #x2019; s correlation coefficients are plotted by the color of squares. The sizes of squares correspond to the significance of the P-values (the larger the square, the smaller the P-value). \elsamp #x2217;\elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.001, \elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.01, \elsamp #x2217; P \elsamp #x003C; 0.05. Average relative abundance of KEGG pathways is plotted by heatmap next to visualization of correlation matrix. The names of genera and pathways enriched in FN were colored in red. The names of genera and pathways enriched in NC were colored in green. NC: normal control group; FN: F. nucleatum group.
Supplementary Fig. S7. Interactions of gut microbiota altered by metformin and F. nucleatum. (a) Correlation networks of significantly altered genera between FM and FN based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level. (b) Associations between significantly altered genera and significantly altered KEGG pathways in FM and FN. Spearman\elsamp #x2019; s correlation coefficients are plotted by the color of squares. The sizes of squares correspond to the significance of the P-values (the larger the square, the smaller the P-value). \elsamp #x2217;\elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.001, \elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.01, \elsamp #x2217; P \elsamp #x003C; 0.05. Average relative abundance of KEGG pathways is plotted by heatmap next to visualization of correlation matrix. The names of genera and pathways enriched in FM were colored in red. The names of genera and pathways enriched in FN were colored in green. FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group.
Supplementary Fig. S8. Interactions between Fusobacterium and other taxa associated with CRC. Correlations between taxa and Fusobacterium in CRC patients of Cohort 1 (a) and Cohort 2 (b). Correlations with absolute coefficients \elsamp #x2265; 0.3 and P \elsamp #x003C; 0.05 are plotted. P-values are plotted by sizes of the circles. Correlation networks of significantly altered genera between normal and tumour samples of Cohort 1 (c) and Cohort 2 (d) based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level.
Supplementary Fig. S9. Alteration in gut microbiome between Fusobacterium-high and Fusobacterium-low patients. (a) Cladograms representation of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 1. (Green) Taxa enriched in Fusobacterium-low patients; (red) taxa enriched in Fusobacterium-high patients. The brightness of each dot is proportional to its effect size. (b) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 1. Taxa enriched in Fusobacterium-high patients are indicated with a positive LDA score (red), and taxa enriched in Fusobacterium-low patients have a negative score (green). Only taxa meeting an LDA significant threshold of 4 are shown. (c) Cladograms representation of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 2. (Green) Taxa enriched in Fusobacterium-high patients; (red) taxa enriched in Fusobacterium-low patients. The brightness of each dot is proportional to its effect size. (d) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 2. Taxa enriched in Fusobacterium-high patients are indicated with a positive LDA score (green), and taxa enriched in Fusobacterium-low patients have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. Volcano plot of significantly (Wilcoxon test, P \elsamp #x003C; 0.05) altered genera between normal and CRC samples in Cohort 1 (e) and Cohort 2 (f). Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue. Differential relative abundances of genera were evaluated using the two-tailed Wilcoxon test.
Supplementary Table. S1. Study characteristics of included literatures.
Supplementary Table. S2. Significantly altered taxa by metformin achieved from literatures.
Supplementary Table. S3. Definitions of bacterial biomarker combinations for discriminating metformin-treated and metformin-untreated humans or rodents.
Supplementary Table. S4. Differentially abundant KO categories.
Reference
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 4.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15(2):109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 6.Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6(31):32013–32026. doi: 10.18632/oncotarget.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2019 doi: 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higurashi T, Hosono K, Takahashi H, Komiya Y, Umezawa S, Sakai E. Metformin for chemoprevention of metachronous colorectal adenoma or polyps in post-polypectomy patients without diabetes: a multicentre double-blind, placebo-controlled, randomised phase 3 trial. Lancet Oncol. 2016;17(4):475–483. doi: 10.1016/S1470-2045(15)00565-3. [DOI] [PubMed] [Google Scholar]
- 12.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 13.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the Gut. Diabetes Care. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 15.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405. doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao K, Yang R, Zhang J, Wang Z, Jia C, Zhang F. Effects of Qijian mixture on type 2 diabetes assessed by metabonomics, gut microbiota and network pharmacology. Pharmacol Res. 2018;130:93–109. doi: 10.1016/j.phrs.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Fujimori T, Kato K, Fujihara S, Iwama H, Yamashita T, Kobayashi K. Antitumor effect of metformin on cholangiocarcinoma: In vitro and in vivo studies. Oncol Rep. 2015;34(6):2987–2996. doi: 10.3892/or.2015.4284. [DOI] [PubMed] [Google Scholar]
- 19.Kato K, Gong J, Iwama H, Kitanaka A, Tani J, Miyoshi H. The antidiabetic drug metformin inhibits gastric cancer cell proliferation in vitro and in vivo. Mol Cancer Therap. 2012;11(3):549–560. doi: 10.1158/1535-7163.MCT-11-0594. [DOI] [PubMed] [Google Scholar]
- 20.Tomimoto A, Endo H, Sugiyama M, Fujisawa T, Hosono K, Takahashi H. Metformin suppresses intestinal polyp growth in ApcMin/+ mice. Cancer Sci. 2008;99(11):2136–2141. doi: 10.1111/j.1349-7006.2008.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie J, Xia L, Xiang W, He W, Yin H, Wang F. Metformin selectively inhibits metastatic colorectal cancer with the KRAS mutation by intracellular accumulation through silencing MATE1. Proc Nat Acad Sci USA. 2020 doi: 10.1073/pnas.1918845117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asshauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2015;31(9):1382–1388. doi: 10.1093/bioinformatics/btu843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119(10):1674–1684. [Google Scholar]
- 31.Lagkouvardos I, Fischer S, Kumar N, Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ. 2017;5:e2836. doi: 10.7717/peerj.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma W, Chen J, Meng Y, Yang J, Cui Q, Zhou Y. Metformin alters gut microbiota of healthy mice: implication for its potential role in gut microbiota homeostasis. Front Microbiol. 2018;9:1336. doi: 10.3389/fmicb.2018.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adeshirlarijaney A, Zou J, Tran HQ, Chassaing B, Gewirtz AT. Amelioration of metabolic syndrome by metformin associates with reduced indices of low-grade inflammation independently of the gut microbiota. Am J Physiol Endocrinol Metabol. 2019;317(6):E1121–E1E30. doi: 10.1152/ajpendo.00245.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji S, Wang L, Li L. Effect of metformin on short-term high-fat diet-induced weight gain and anxiety-like behavior and the gut microbiota. Front Endocrinol. 2019;10:704. doi: 10.3389/fendo.2019.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ. Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metabol. 2018;27(1):101–117.e5. doi: 10.1016/j.cmet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 36.Tong X, Xu J, Lian F, Yu X, Zhao Y, Xu L. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. mBio. 2018;9(3) doi: 10.1128/mBio.02392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryrup T, Thomsen CW, Kern T, Allin KH, Brandslund I, Jorgensen NR. Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia. 2019;62(6):1024–1035. doi: 10.1007/s00125-019-4848-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elbere I, Kalnina I, Silamikelis I, Konrade I, Zaharenko L, Sekace K. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PloS One. 2018;13(9) doi: 10.1371/journal.pone.0204317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol. 2014;80(19):5935–5943. doi: 10.1128/AEM.01357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Feng R, Yang M, Qian C, Wang Z, Liu W. Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats. BMJ Open Diabetes Res Care. 2019;7(1) doi: 10.1136/bmjdrc-2019-000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F, Wang M, Yang J, Xu Q, Liang C, Chen B. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine. 2019;66(3):485–493. doi: 10.1007/s12020-019-02041-5. [DOI] [PubMed] [Google Scholar]
- 42.Ejtahed HS, Tito RY, Siadat SD, Hasani-Ranjbar S, Hoseini-Tavassol Z, Rymenans L. Metformin induces weight loss associated with gut microbiota alteration in non-diabetic obese women: a randomized double-blind clinical trial. Eur J Endocrinol. 2018 doi: 10.1530/EJE-18-0826. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, Liang L, Yu G, Li Q. Pumpkin polysaccharide modifies the gut microbiota during alleviation of type 2 diabetes in rats. Int J Biol Macromol. 2018;115:711–717. doi: 10.1016/j.ijbiomac.2018.04.127. [DOI] [PubMed] [Google Scholar]
- 44.Lee H, Lee Y, Kim J, An J, Lee S, Kong H. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes. 2018;9(2):155–165. doi: 10.1080/19490976.2017.1405209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67(3):534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 46.Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abu-Ghazaleh N, Chua WJ, Gopalan V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. 2020 doi: 10.1111/jgh.15042. [DOI] [PubMed] [Google Scholar]
- 51.Xie YH, Gao QY, Cai GX, Sun XM, Sun XM, Zou TH. Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120(Pt 11):1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 54.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical importance of streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2011;53(9):870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 56.Paleari L, Burhenne J, Weiss J, Foersch S, Roth W, Parodi A. High accumulation of metformin in colonic tissue of subjects with diabetes or the metabolic syndrome. Gastroenterology. 2018;154(5):1543–1545. doi: 10.1053/j.gastro.2017.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haruki K, Kosumi K, Hamada T, Twombly TS, Vayrynen JP, Kim SA. Association of autophagy status with amount of Fusobacterium nucleatum in colorectal cancer. J Pathol. 2019 doi: 10.1002/path.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF, Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Espanola Enfermedades Digest: Org Of Soc Espanol Patol Digest. 2015;107(11):659–671. doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 61.Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol. 2020;5(3):511–524. doi: 10.1038/s41564-019-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their health-promoting effects. Microbiol Spectrum. 2017;5(3) doi: 10.1128/microbiolspec.bad-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balvociute M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT – how do these taxonomies compare? BMC Genom. 2017;18(Suppl 2):114. doi: 10.1186/s12864-017-3501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Characteristics of bacterial biomarkers for discriminating metformin-treated and metformin-untreated humans or rodents. ROC curves evaluating the performance of discriminating metformin-treated and metformin-untreated humans or rodents using single bacterial biomarkers for a particular metformin treatment model, generated from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al. (a), Adeshirlarijaney et\elsamp #x00A0;al. (c), and Ma et\elsamp #x00A0;al. (e) respectively. ROC curves evaluating the performance of discriminating metformin-treated and metformin-untreated humans or rodents using bacterial biomarkers combinations for a particular metformin treatment model, generated from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al. (b), Adeshirlarijaney et\elsamp #x00A0;al. (d), and Ma et\elsamp #x00A0;al. (f) respectively. See Supplementary Table. S3 for detailed definitions of bacterial biomarkers combinations. (g) Heatmap showing differences between mean abundance of overlapped KEGG pathways predicted from metformin-treated and metformin-untreated humans or rodents in from three datasets published by de la Cuesta-Zuluaga et\elsamp #x00A0;al., Adeshirlarijaney et\elsamp #x00A0;al., and Ma et\elsamp #x00A0;al. respectively. A positive difference between means indicates higher abundance of a KEGG pathway in metformin-treated humans or rodents.
Supplementary Fig. S2. Metformin suppresses tumourigenicity in APCMin/\elsamp #x002B; mice. (a) Growth curve of F. nucleatum with or without metformin in a laboratory culture. n\elsamp #x2009;\elsamp #x003D;\elsamp #x2009;3 replicates/treatment. (b) Colony counts on agarose plates of F. nucleatum with or without metformin in a laboratory culture. n\elsamp #x2009;\elsamp #x003D;\elsamp #x2009;3 replicates/treatment. Statistical significance was detected by Student\elsamp #x0027;s t test. Data in the bar plots are presented as the means \elsamp #x00B1; SEM. (c) Mean weights of APCMin/\elsamp #x002B; mice in each group. No statistical difference in body weight was observed between groups. (d) Representative images of small intestine. The red arrows indicated the positive location. (e) Numbers of small intestine tumour. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test. (f) Small intestine tumour load. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test. (g) Comparison of small intestine tumour size below 1.5 mm and over 1.5 mm in diameter. Significance between every two groups was calculated using unpaired two-tailed Student\elsamp #x0027;s t test.
Supplementary Fig. S3. 16S rRNA sequencing analysis of the gut microbiome in APCMin/\elsamp #x002B; mice. (a) Venn diagram showing overlapped operational taxonomic unit (OTU) among 4 groups. Principal coordinate analysis (PCoA) on weighted Unifrac (b) and unweighted Unifrac distances (c) is shown for NC (blue), FN (red), FM (green) and MET (yellow). Significance of clustering was determined using the Adonis test. (d) No significant difference is identified for alpha-diversity (Chao1, richness, Ace index, Shannon index) between NC and MET, NC and FN, FN and FM. Significance between every two groups was calculated using the Wilcoxon rank-sum test. (e) Microbial compositional profiling of NC, FN, FM and MET at phylum level. (f) Microbial compositional profiling of NC, FN, FM and MET at family level. (g) A cladogram representation of data in NC, FN, FM and MET. (Blue) Taxa enriched in NC; (red) taxa enriched in FN; (green) taxa enriched in FM; (yellow) taxa enriched in MET. The brightness of each dot is proportional to its effect size. The prefixes \elsamp #x2018;p\elsamp #x2019;, \elsamp #x2018;c\elsamp #x2019;, \elsamp #x2018;o\elsamp #x2019;, \elsamp #x2018;f\elsamp #x2019; and \elsamp #x2018;g\elsamp #x2019; represent the level of phylum, class, order, family, and genus. (h) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in G. (Blue) Taxa enriched in NC; (red) taxa enriched in FN; (green) taxa enriched in FM; (yellow) taxa enriched in MET. Only taxa meeting an LDA significant threshold of 3 are shown. NC: normal control group; FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group; MET: metformin group.
Supplementary Fig. S4. Assessment of performance in discriminating NC and MET using bacterial biomarkers in APCMin/\elsamp #x002B; mice. (a) ROC curves of bacterial indicators. (b) ROC curves of bacterial biomarkers for all metformin treatment models. (c) ROC curves comparing the performance of biomarkers with the highest AUROC in (a) and (b). NC: normal control group; MET: metformin group.
Supplementary Fig. S5. Significantly (Wilcoxon test, P \elsamp #x003C; 0.05, abs(logFC) \elsamp #x2265; 0.2) different KO functional categories between NC and MET (a), NC and FN (b), FN and FM (c). Representative significantly (Wilcoxon test, P \elsamp #x003C; 0.05) enriched KEGG pathways with the highest relative abundances between NC and MET (d), NC and FN (e), FN and FM (f). NC: normal control group; FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group; MET: metformin group.
Supplementary Fig. S6. Interactions of gut microbiota altered by F. nucleatum. (a) Correlation networks of significantly altered genera between NC and FN based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level. (b) Associations between significantly altered genera and significantly altered KEGG pathways in NC and FN. Spearman\elsamp #x2019; s correlation coefficients are plotted by the color of squares. The sizes of squares correspond to the significance of the P-values (the larger the square, the smaller the P-value). \elsamp #x2217;\elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.001, \elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.01, \elsamp #x2217; P \elsamp #x003C; 0.05. Average relative abundance of KEGG pathways is plotted by heatmap next to visualization of correlation matrix. The names of genera and pathways enriched in FN were colored in red. The names of genera and pathways enriched in NC were colored in green. NC: normal control group; FN: F. nucleatum group.
Supplementary Fig. S7. Interactions of gut microbiota altered by metformin and F. nucleatum. (a) Correlation networks of significantly altered genera between FM and FN based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level. (b) Associations between significantly altered genera and significantly altered KEGG pathways in FM and FN. Spearman\elsamp #x2019; s correlation coefficients are plotted by the color of squares. The sizes of squares correspond to the significance of the P-values (the larger the square, the smaller the P-value). \elsamp #x2217;\elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.001, \elsamp #x2217;\elsamp #x2217; P \elsamp #x003C; 0.01, \elsamp #x2217; P \elsamp #x003C; 0.05. Average relative abundance of KEGG pathways is plotted by heatmap next to visualization of correlation matrix. The names of genera and pathways enriched in FM were colored in red. The names of genera and pathways enriched in FN were colored in green. FN: F. nucleatum group; FM: F. nucleatum\elsamp #x00A0;\elsamp #x002B;\elsamp #x00A0;metformin group.
Supplementary Fig. S8. Interactions between Fusobacterium and other taxa associated with CRC. Correlations between taxa and Fusobacterium in CRC patients of Cohort 1 (a) and Cohort 2 (b). Correlations with absolute coefficients \elsamp #x2265; 0.3 and P \elsamp #x003C; 0.05 are plotted. P-values are plotted by sizes of the circles. Correlation networks of significantly altered genera between normal and tumour samples of Cohort 1 (c) and Cohort 2 (d) based on the Wilcoxon rank-sum test. Spearman\elsamp #x2019; s absolute correlation coefficients \elsamp #x2265; 0.3 and P-values \elsamp #x003C; 0.05 were retained. The edge width is proportional to the correlation strength. The node size is proportional to the mean abundance of the genera. Nodes with the same color are classified in the same phylogenetic phylum level.
Supplementary Fig. S9. Alteration in gut microbiome between Fusobacterium-high and Fusobacterium-low patients. (a) Cladograms representation of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 1. (Green) Taxa enriched in Fusobacterium-low patients; (red) taxa enriched in Fusobacterium-high patients. The brightness of each dot is proportional to its effect size. (b) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 1. Taxa enriched in Fusobacterium-high patients are indicated with a positive LDA score (red), and taxa enriched in Fusobacterium-low patients have a negative score (green). Only taxa meeting an LDA significant threshold of 4 are shown. (c) Cladograms representation of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 2. (Green) Taxa enriched in Fusobacterium-high patients; (red) taxa enriched in Fusobacterium-low patients. The brightness of each dot is proportional to its effect size. (d) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the significant abundance of data in Fusobacterium-high and Fusobacterium-low patients in Cohort 2. Taxa enriched in Fusobacterium-high patients are indicated with a positive LDA score (green), and taxa enriched in Fusobacterium-low patients have a negative score (red). Only taxa meeting an LDA significant threshold of 4 are shown. Volcano plot of significantly (Wilcoxon test, P \elsamp #x003C; 0.05) altered genera between normal and CRC samples in Cohort 1 (e) and Cohort 2 (f). Tumour-enriched genera are colored in red and genera enriched in normal samples are colored in blue. Differential relative abundances of genera were evaluated using the two-tailed Wilcoxon test.
Supplementary Table. S1. Study characteristics of included literatures.
Supplementary Table. S2. Significantly altered taxa by metformin achieved from literatures.
Supplementary Table. S3. Definitions of bacterial biomarker combinations for discriminating metformin-treated and metformin-untreated humans or rodents.
Supplementary Table. S4. Differentially abundant KO categories.