Abstract
Polychlorinated biphenyls (PCBs) are ubiquitously detected and have been linked to metabolic diseases. Gut microbiome is recognized as a critical regulator of disease susceptibility; however, little is known how PCBs and gut microbiome interact to modulate hepatic xenobiotic and intermediary metabolism. We hypothesized the gut microbiome regulates PCB-mediated changes in the metabolic fingerprints and hepatic transcriptome. Ninety-day-old female conventional and germ-free mice were orally exposed to the Fox River Mixture (synthetic PCB mixture, 6 or 30 mg/kg) or corn oil (vehicle control, 10 ml/kg), once daily for 3 consecutive days. RNA-seq was conducted in liver, and endogenous metabolites were measured in liver and serum by LC-MS. Prototypical target genes of aryl hydrocarbon receptor, pregnane X receptor, and constitutive androstane receptor were more readily upregulated by PCBs in conventional conditions, indicating PCBs, to the hepatic transcriptome, act partly through the gut microbiome. In a gut microbiome-dependent manner, xenobiotic, and steroid metabolism pathways were upregulated, whereas response to misfolded proteins-related pathways was downregulated by PCBs. At the high PCB dose, NADP, and arginine appear to interact with drug-metabolizing enzymes (ie, Cyp1–3 family), which are highly correlated with Ruminiclostridium and Roseburia, providing a novel explanation of gut-liver interaction from PCB-exposure. Utilizing the Library of Integrated Network-based Cellular Signatures L1000 database, therapeutics targeting anti-inflammatory and endoplasmic reticulum stress pathways are predicted to be remedies that can mitigate PCB toxicity. Our findings demonstrate that habitation of the gut microbiota drives PCB-mediated hepatic responses. Our study adds knowledge of physiological response differences from PCB exposure and considerations for further investigations for gut microbiome-dependent therapeutics.
Keywords: bioinformatics, cytochrome P450, environmental chemicals, hepatotoxicity, metabolomics, molecular mechanisms, RNA-seq, toxicogenomics, exposure, environmental, biotransformation
Polychlorinated biphenyls (PCBs) were widely used as electric coolants and insulators due to their chemical stability and fire resistance and were banned from production in the United States in 1979 due to their toxicity (Korrick and Sagiv, 2008). PCBs bioaccumulate and are environmentally persistent with an estimated 150 million pounds of PCBs dispersed in the environment (Korrick and Sagiv, 2008). PCBs can be detected in food, soil, water, human tissues, and serum (Boesen et al., 2019; Ellsworth et al., 2015; Grimm et al., 2015; Jahnke and Hornbuckle, 2019; Jin et al., 2019; Stremy et al., 2019; Weber et al., 2018). There are several known adverse health effects of PCB exposure; epidemiological and animal models have demonstrated dysregulation in neurodevelopment (Klocke and Lein, 2020; Schug et al., 2015; Zhang et al., 2017), interference of thyroid hormone signaling (Li et al., 2018), perturbation of the gut microbiome (Cheng et al., 2018; Petriello et al., 2018), and development of type II diabetes (Dzierlenga et al., 2019; Meek et al., 2019; Rahman et al., 2019; Tornevi et al., 2019) and cancer (Georgiadis et al., 2019; Ghosh et al., 2018; He et al., 2017; Leng et al., 2016).
Comprised of an estimated 38 trillion bacteria in the gut, millions of microbial genes functionally exist in the human body (Kho and Lal, 2018; Sender et al., 2016; Yang et al., 2009). Altered gut microbiome composition may indicate altered microbial function that affects the host, and gut dysbiosis may serve as a biomarker for toxic exposure and diseases. The gut microbiome contributes to xenobiotic biotransformation via reactions including reduction and hydrolysis reactions. In addition, gut microbiome may also indirectly modify xenobiotic biotransformation activities of the host via microbial metabolites, which can enter circulation and remotely regulate host signaling in target organs (Fu and Cui, 2017; Klaassen and Cui, 2015; Spanogiannopoulos et al., 2016). In addition to xenobiotic biotransformation, gut microbiota modulates host intermediary metabolism by producing distinct microbial metabolites such as secondary bile acids and short-chain fatty acids, and modifying host metabolites such as amino acids, carbohydrates, and nucleotides (den Besten et al., 2013; LeBlanc et al., 2017; Prawitt et al., 2011, Scoville et al., 2019). It is increasingly recognized that these intermediary metabolites are essential signaling molecules in complex human diseases. Quantifying altered endogenous metabolites in biological systems can lead to detailed physiologically relevant biochemical status during diseases and xenobiotic exposure (Ramirez et al., 2013). Integrating microbial and metabolomic changes from toxic exposures can provide additional insights to understanding the mechanisms of toxicity or diseases related to dysbiosis.
The liver is a critical organ in xenobiotic metabolism and nutrient homeostasis. The individual steps in liver function are delicately regulated by the interplay of hormones, cellular signaling pathways, and nuclear receptors (NRs) and other transcription factors (Bechmann et al., 2012). Through the biliary tract, portal vein, and systemic mediators, the gut and liver extensively communicate with one another. It has been increasingly recognized that the gut microbiome, through microbial metabolites, mediate liver functions and play a role in intermediary metabolism modification and progression of toxicant-induced liver diseases (Kolodziejczyk et al., 2019; Scoville et al., 2019; Zhang et al., 2020). Using RNA-seq, transcriptomic signatures following toxic exposure can reveal the perturbed biological pathways, which serves as an initial step in predicting the mechanisms of toxification. Furthermore, through comparing the transcriptomic signatures following toxicant exposure and xenobiotic-induced transcriptomic cellular responses, one may predict the potential therapeutic remedies that may mitigate the toxicity of these chemicals.
Previously, using the same PCB-exposed mice, our group has showed that PCBs produce dysbiosis and modulates bile acid metabolism in a gut microbiome-dependent manner (Cheng et al., 2018). However, the global effect of PCB exposure and gut microbiome interactions on the hepatic transcriptome has not been characterized. Therefore, we tested our hypothesis that the gut microbiome is a critical regulator of the interface between xenobiotic biotransformation and intermediary metabolic fingerprints following PCB exposure.
MATERIALS AND METHODS
Chemical preparation
The Fox River PCB mixture was prepared from technical PCB mixtures as described previously (Cheng et al., 2018). Briefly, Aroclor 1242, 1248, 1254, and 1260 were prepared in pesticide grade acetone (50 mg/ml acetone) (purchased from Fisher Scientific, Far Lawn, New Jersey) and mixed at a ratio of 35:35:15:15, respectively, by weight. The acetone was then evaporated and the solute PCB mixture was used in the study upon successful congener-specific analysis.
Animals and chemical exposure
The experimental design is summarized in Figure 1A. All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines (https://aaalac.org/resources/theguide.cfm). All experiments were approved by Institutional Animal Care and Use Committee (IACUC) at the University of Washington. Three-month-old conventional (CV) SPF-free female C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, Maine) and were acclimated to the animal facility at the University of Washington at 74 degrees Fahrenheit, 26% humidity, and 12-h light and dark cycle for 1 week prior to the experiment. Germ-free (GF) breeders in C57BL/6 background were purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill). Pups born from GF mice were raised in the Gnotobiotic Animal Core (GNAC) Facility at the University of Washington. All mice had ad libitum access to standard laboratory autoclaved rodent diet (LabDiet No. 5010 after weaning and LabDiet No. 5021 for breeding; LabDiet, St Louis, Missouri) and water (nonacidified autoclaved water). Animals were housed with autoclaved bedding (autoclaved Enrich-N’Pure; Andersons, Maumee, Ohio). Animals were monitored daily by the University of Washington Department of Comparative Medicine technicians.
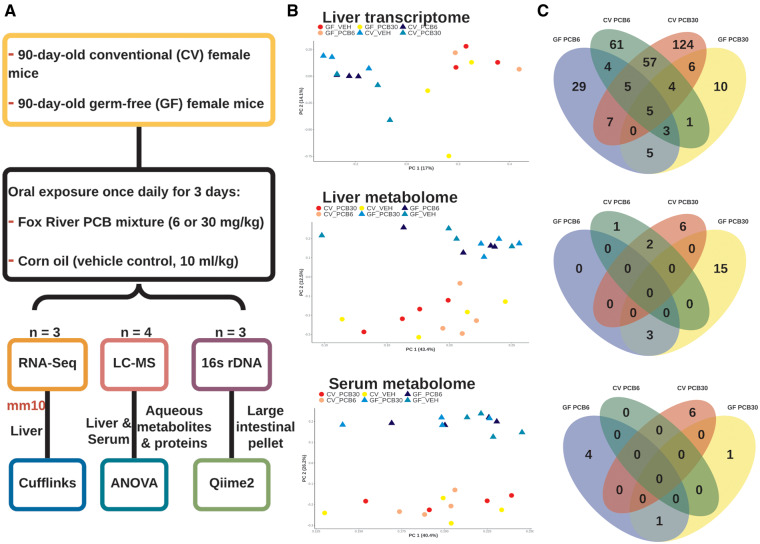
Figure 1.
Experimental design of the study and summary of data. A, 90-day-old female conventional (CV) and germ-free (GF) mice were orally exposed to the Fox River polychlorinated biphenyl (PCB) mixture (6 or 30 mg/kg) or corn oil (vehicle control, 10 ml/kg). Twenty-four hours the final dosing, organs were collected. RNA-seq, 16s rDNA-seq, LC-MS were performed on the liver, large intestinal pellets, and liver and serum, respectively. B, Principal component analysis of the liver transcriptome and liver and serum metabolome. Individual points on the principal components scale are labeled by enterotype and exposure. C, Venn diagrams of differentially expressed genes of the liver and differentially abundant metabolites in the liver and serum.
Female mice were used in this study to compare results and as a follow-up with a previous publication on PCB-mediated developmental neurotoxicity, which used female mice (Kania-Korwel et al., 2008a,b,c, 2010; Wu et al., 2013, 2015). Mice were exposed to corn oil (vehicle control; 10 ml/kg body weight), or Fox River mixture low dose (6 mg/kg body weight) or high dose (30 mg/kg body weight), once daily between 8 and 10 Am for 3 consecutive days. Doses of PCBs were chosen to compare results from a previous study (Cheng et al., 2018). Similar doses of PCBs have been used to identify the induction of hepatic xenobiotic metabolism-related expression, PCB disposition, and developmental neurotoxicity (Kania-Korwel et al., 2008c, 2012; Poon et al., 2013; Sable et al., 2011; Yang et al., 2009). The higher doses of PCBs used in this study was to detect alterations in the gut microbiome in a short time to connect to the hepatic responses to add to future mechanistic studies. These PCB doses are also below the dose of another human health relevant PCB mixture that have been found in contaminated food, namely PCB-138, PCB-153, and PCB-180 (150 μmol/kg or 43 mg/kg), which has been shown to result in a PCB plasma level of 5 μM (Choi et al., 2010), and is comparable with PCB plasma levels in an acutely exposed human population (Jensen, 1989; Wassermann et al., 1979).
Tissues were harvested 24 h after the final dose. Whole blood was collected via cardiac puncture and transferred to a MiniCollect serum separator (Greiner Bio-One, Kremsmunster, Austria), and was kept on ice for at least 30 min to allow sufficient time for coagulation. Serum was collected by centrifugation at 4000 rpm (1503 × g) at 4°C for 20 min and was stored at −80°C until use. Large intestinal contents was flushed out in 15 ml of ice-cold phosphate-buffered saline (PBS) that contains 0.1% dithiothreitol (DTT) (Sigma Aldrich, St Louis, Missouri), and immediately frozen on dry ice Microbial DNA was extracted using a with a method as previously (Cheng et al., 2018). All tissues were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
RNA isolation
Total RNA was extracted from frozen livers using RNA-Bee reagent (Tel-Test, Inc, Friendswood, Texas) following the manufacturer’s protocol. RNA concentrations were quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts) at 260 nm. The integrity of total RNA samples was evaluated by agarose gel electrophoresis with visualization of 18S and 28S rRNA bands under UV light, and confirmed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, California). Samples with RNA integrity numbers above 8.0 were used for RNA-seq in triplicates.
RNA sequencing
The cDNA library was constructed using a ribosomal depletion method, and reads were sequenced using a 75 bp paired end sequencing per the Illumina manufacturer’s protocol. Data were analyzed in triplicates FASTQ files were demultiplexed and concatenated for each sample. Quality control the FASTQ files was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed July 3, 2020). Sequenced reads from the FASTQ files were then mapped to the mouse reference genome (National Center for Biotechnology Information [NCBI GRCm38/mm10]) using HISAT2 version 2.1 (Kim et al., 2019). The sequencing alignment/map (SAM) files were converted to binary alignment/map (BAM) format using SAMtools version 1.8 (Li et al., 2009) and were analyzed by Cufflinks version 2.2.1 to estimate the transcript abundance (Trapnell et al., 2012) using Gencode mouse version 22 (vM22) gene transfer format (GTF). The abundance was expressed as fragments per kilobase of transcript per million mapped reads (FPKM) and was converted to transcripts per million (TPM). Genes were considered expressed if the TPM of each gene were greater than the total sample number and if the variance was greater than 1. Differential expression analysis was performed using Cuffdiff (Trapnell et al., 2012). The differentially expressed genes were defined as false discovery rate (FDR) Benjamini-Hochberg adjusted p value < .05 in the chemical-exposed groups compared with the vehicle-exposed control group. Differentially expressed genes were also categorized and matched with genes in categories of interest (xenobiotic biotransformation, epigenetic modifiers, NRs, oxidative stress, and inflammation). Gene categories were generated based on literature. If the absolute value of a gene was greater than 1.5, it was considered as an up- or downregulated gene. Dysregulation was defined as genes that were either up- or downregulated. Venn diagrams were plotted for differentially regulated genes for exposure group and enterotype using the R package VennDiagram (Chen and Boutros, 2011). Hierarchical clustering was performed using R package ComplexHeatmap (Gu et al., 2016). Lists of differentially up- and downregulated genes were used as input for gene ontology (GO) enrichment using the R package topGO (https://bioconductor.org/packages/release/bioc/vignettes/topGO/inst/doc/topGO.pdf) for all groups with the list of genes in the unfiltered expression table was used as the background. Box and whiskers plots were created using ggplot2 (Wickham, 2016, http://ggplot2.org). The raw and analyzed RNA-seq data are available Dryad: https://doi.org/10.5061/dryad.wwpzgmsgf.
Potential therapeutics were inferred by Enrichr (https://amp.pharm.mssm.edu/Enrichr/) using the Library of Integrated Network-based Cellular Signatures (LINCS) L1000 database as a reference, which is a National Institutes of Health-driven database catalog containing around 28 000 drug and small molecule-induced gene expressions at different doses and time points (Subramanian et al., 2017; Wang et al., 2016). The Enrichr analysis using the LINCS database is a computational prediction through comparing the gene expression signatures from published datasets and the dataset from this study. Specifically, genes that were upregulated by PCBs in a gut microbiome-dependent manner were queried against the chemicals that downregulated the same genes; whereas genes that were downregulated by PCBs in a gut microbiome-dependent manner were queried against the chemicals that downregulated the same genes. The fundamental assumption of this analysis is that chemicals that reverse the PCB-mediated gene expression signatures may serve as candidate therapies to mitigate PCB toxicity in liver. The top 15 hits were shown, with a FDR below 0.05.
Serum and liver preparation for aqueous metabolite measurement
Frozen serum samples stored at −80°C were thawed overnight under 4°C. 50 μl of each serum sample was placed in a 2 ml Eppendorf tube and 250 μl of MeOH was added for the initial step for protein precipitation and metabolite extraction. Each liver sample (10 mg) was homogenized in 200 μl water in an Eppendorf tube using a Bullet Blender homogenizer (Next Advance, Averill Park, New York). An 800 μl of MeOH was added, and after vortexing for 10 s, the samples were stored on dry ice for 30 min. The samples were then sonicated in an ice bath for 10 min. Each serum and liver sample was then vortexed for 10 s and stored at −20°C for 30 min, followed by centrifugation at 14 000 rpm for 10 min at 4°C. The supernatants (200 μl for serum and 800 μl for liver) were collected into a new Eppendorf vial and dried under vacuum using an Eppendorf Vacufuge (Eppendorf, Hauppauge, New York). The dried samples were reconstituted in 200 μl of 40% PBS/60% ACN. Liver and serum metabolomics were performed in quadruplicates.
The targeted LC-MS/MS method used here was modeled after that developed and used in a growing number of studies (Carroll et al., 2015; Shi et al., 2019; Zhu et al., 2014). Briefly, all LC-MS/MS experiments were performed on an Agilent 1290 UPLC-6490 QQQ-MS (Santa Clara, California) system. Each sample was injected twice for analysis, with 10 µl using the negative ionization mode and 4 µl using the positive ionization mode. Both chromatographic separations were performed using the hydrophilic interaction chromatography mode on a Waters XBridge BEH Amide column (150 × 2.1 mm, 2.5-µm particle size, Waters Corporation, Milford, Massachusetts). The flow rate was set to 0.3 ml/min, autosampler temperature was kept at 4°C, and the column compartment was set at 40°C. The mobile phase was composed of Solvents A (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% H2O/5% ACN) and B (10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% ACN/5% H2O). After the initial 1 min isocratic elution of 90% B, the percentage of Solvent B decreased to 40% at t = 11 min. The composition of Solvent B was maintained at 40% for 4 min (t = 15 min). The percentage of B gradually went back to 90%, and the next injection was prepared.
The mass spectrometer is equipped with an electrospray ionization source. Targeted data acquisition was performed in multiple-reaction-monitoring (MRM) mode. We monitored 118 and 160 MRM transitions in negative and positive mode, respectively (278 transitions in total). The whole LC-MS system was controlled by Agilent MassHunter Workstation software (Santa Clara, California). The extracted MRM peaks were integrated using Agilent MassHunter Quantitative Data Analysis (Santa Clara, California).
Data were analyzed in quadruplicates. Relative abundance values for each metabolite in liver and serum were compared using 1-way analysis of variance followed by Tukey’s post hoc test using base R (R Core Team, 2017). Venn Diagrams and heat maps were built using differentially abundant metabolites in liver and serum using packages VennDiagram (Chen and Boutros, 2011). Gene-metabolite network analysis was performed using differentially abundant metabolites and differentially regulated genes per each exposure group and enterotype using Network explorer function in MetaboAnalyst (Chong et al., 2018). Spearman correlation for differentially abundant metabolites and differentially regulated genes were plotted using ComplexHeatmap in R (Gu et al., 2016) for GF groups. Box and whiskers plots were created using ggplot2 (Wickham, 2016).
16s rDNA sequencing and data analysis
For CV and GF mice exposed to PCBs, in triplicates, the V4-amplified rDNA sequencing FASTQ files were obtained as described previously (Cheng et al., 2018, Dryad: https://doi.org/10.5061/dryad.wwpzgmsgf). Bacterial abundance table for CV and conventionalized (exGF mice introduced to bacteria by living in CV cage conditions for 2 months) was obtained as previously described (Selwyn et al., 2016). The paired-end sequence reads, in triplicates, were merged, demultiplexed, quality-checked, and chimera-filtered using QIIME 2 (Callahan et al., 2016; Hall and Beiko, 2018; Mandal et al., 2015; Vazquez-Baeza et al., 2013, 2017). Community composition and (Bokulich et al., 2013) the Silva 99 version 132 reference (Quast et al., 2012) using QIIME2 (Hall and Beiko, 2018). The OTU table annotated with classified bacteria information was read into R for further analysis (R Core Team, 2017). Negative binomial differential abundance testing was performed using the R package DESeq2 (Love et al., 2014) following pipelines in phyloseq (McMurdie and Holmes, 2013). Differentially abundant bacteria and correlation tables of differentially abundant bacteria (relative abundance of OTU) and metabolites, and differentially regulated hepatic genes (Spearman correlation) were plotted using ComplexHeatmap (Gu et al., 2016).
RESULTS
Effect of PCB Exposure on the Hepatic Transcriptome as Well as Hepatic and Serum Metabolome in CV and GF Mice
Principal component analysis was performed on the Z-normalized expression and abundance matrix and was labeled by enterotype and exposure group (Figure 1B). Clusters on normalized expression levels on the principal component space showed that enterotype (ie, presence or absence of the gut microbiome) was the primary factor in grouping the transcriptomic and metabolomic signatures within the same exposure. Although the coordinates of GF mice exposed to the PCB high dose were distinct in the first principal component from the other GF exposure groups, overall, no clear clusters were formed among PCB-exposed groups within the same enterotype.
Venn diagrams were used to visualize the commonly and differentially regulated genes and abundant metabolites between PCB-exposed group and the vehicle-expose group of the same dose and enterotype (Figure 1C). Regarding the liver transcriptome, exposure to PCBs had a greater effect on the CV mice, having more differentially regulated genes than the GF mice (Figure 1C top panel). Regarding the effect of the 2 PCB doses, the number of differentially regulated genes was higher in the high PCB dose group than the low PCB dose group in livers of CV mice; however, in livers of the GF mice, the low PCB dose differentially regulated more genes than the high PCB dose. The majority of genes were uniquely regulated by PCBs in different enterotype and PCB dose groups, with only 5 genes commonly differentially regulated, namely Cyp1a2, Arntl, G6pc, and Sult2a1 (Supplementary Table 1A–D). In the liver transcriptome of CV mice, most genes were commonly regulated between the low and high PCB doses (ie, Cyp2a5, Mt1, Mt2, Slc16a1). Interestingly, the bile acid metabolizing enzyme encoding Cyp8b1 was differentially regulated by both the PCB doses only in GF conditions (Supplementary Table 7).
Regarding the liver metabolome of amino acids, carbohydrates, and nucleotides, in general, GF mice had more differentially regulated metabolites following PCB exposure than CV mice that were exposed to the same dose of PCBs. The differentially regulated metabolites were not shared between the 2 enterotypes or the 2 PCB doses (Figure 1C middle panel, Supplementary Figure 2A and Table 7B). Specifically, the high PCB dose-exposed GF mice had the most differentially regulated metabolites (18), followed by the high PCB dose-exposed CV mice (8), whereas the PCB low dose-exposed CV and GF mice both had 3 differentially regulated metabolites. Aminoisobutyric acid and dimethylglycine for commonly regulated by PCB exposure in the livers of CV mice, as well as raffinose, glucose-6 phosphate (G6P) and fructose-6 phosphate (F6P) for in the livers of GF mice (Figure 1C middle panel, Supplementary Figure 2A and Table 7A).
The serum metabolome, similar to the liver metabolome, showed no commonly regulated metabolites in CV and GF mice (Figure 1C bottom panel, Supplementary Figure 2B and Table 7C). In CV mice, the low PCB dose did not alter any serum metabolites, whereas the high PCB dose altered 6 serum metabolites (levulinic acid, 6-methyl-Dl-tryptophan, serine, asparagine, proline, and urocanic acid). In GF mice, the low PCB dose uniquely altered 4 serum metabolites, namely mucic acid, mannose, norvaline, and cytosine and the high PCB dose uniquely altered 1 serum metabolite, 2-hydroxyphenylacetic acid. Glucuronic acid was commonly regulated in serum of low and high PCB dose-exposed GF mice.
PCB-mediated Functional Changes Predicted From the Transcriptomic Response in Livers of CV and GF Groups
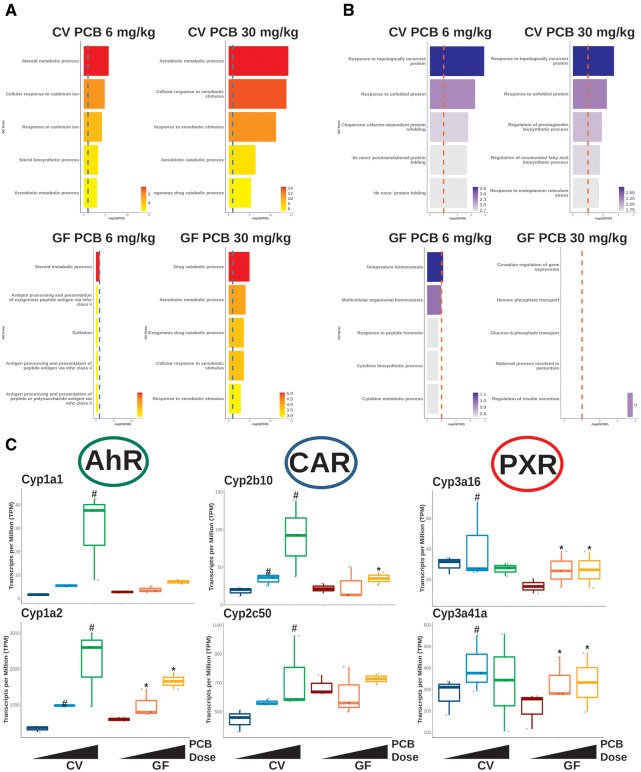
As shown in Figures 2A and 2B, GO enrichment was performed to determine the transcriptional change of biological functions using up- and downregulated genes. For both the low and high PCB doses, up- and downregulated GO terms were more highly enriched in livers of the CV than GF mice, indicating that the presence of the gut microbiome facilitates PCB-mediated effects. As shown in Figure 2A, upregulated GO terms were mostly comprised of steroid or xenobiotic metabolism-related pathways, and there appeared to be similarities in the predicted changes in pathways between the low PCB dose exposure. For both CV (Figure 2A top) and GF (Figure 2A bottom) mice, the enrichment significance (−log10p value) followed a dose-response pattern. For the downregulated GO terms, responses to misfolded protein were the top enriched terms, and this enrichment disappeared in the GF group (Figure 2B), suggesting that the gut microbiome may promote PCB-induced endoplasmic reticulum (ER)-stress (Cybulsky, 2017; Gardner et al., 2013; Rashid et al., 2015). For the GF group exposed to the low dose PCBs (Figure 2A bottom left), temperature homeostasis was the only significantly downregulated enriched term due to the downregulation in heat shock protein encoding genes and no significantly downregulated GO terms were found for GF mice exposed to the PCB high dose (Figure 2A bottom right).
Figure 2.
Gene ontology (GO) enrichment of differentially regulated genes and gene expression of prototypical target genes of xenobiotic-sensing transcription factors. A, Top 5 downregulated GO enrichment terms for conventional (CV) (top) and germ-free (GF) (bottom) mice. The red dotted lines represents −log10(FDR) = 0.1. B, Box plots of differentially regulated prototypical target genes of the transcription factors, aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and pragnane X receptor (PXR). The pound signs and asterisks represent differential expression in the conventional and GF groups, respectively.
Effect of PCB Exposure on the Hepatic Expression of Xenobiotic-sensing Transcription Factor Target Genes in CV and GF Mice
As shown in Figure 2C, the prototypical target genes of the major xenobiotic-sensing transcription factors aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and pregnane X receptor (PXR), were more readily upregulated by PCBs in livers of the CV mice as compared with the GF mice (Figure 2C). Specifically, the mRNA transcripts of Cyp1a1 and Cyp1a2, which are the prototypical target genes of AhR (Flaveny et al., 2010; Lo and Matthews, 2012), were more upregulated by PCBs in livers of CV mice than in GF mice; this such trend was more prominent at the high PCB dose. Similarly, the mRNAs of the CAR prototypical target genes in the Cyp2 family (ie, Cyp2b10, Cyp2c50) (Cui and Klaassen, 2016; Kiyosawa et al., 2008; Maglich et al., 2003) had higher PCB-mediated fold induction in livers of CV mice, and the induction change was also more prominent at the high PCB dose. The mRNAs of the prototypical target genes for PXR in the Cyp3 family (Cui and Klaassen, 2016; Kiyosawa et al., 2008; Maglich et al., 2003), including Cyp3a16 and Cyp3a41a, were more induced by the low PCB dose in CV mice than in GF mice, whereas the high PCB dose did not alter these Cyp3a gene isoforms in CV livers, but moderately upregulated these genes in GF livers.
Taken together, our data at the mRNA level showed that the presence of the gut microbiome is necessary for the PCB-mediated upregulation of the prototypical target genes of major xenobiotic-sensing transcription factors.
Differentially Abundant Bacteria From Exposure to PCBs
V4-amplified 16s rDNA sequencing was conducted to study the gut microbiota changes in the large intestinal pellets of CV mice in response to exposure to PCBs as we described before (Cheng et al., 2018). Overall, as seen in Figure 3B, the PCB low dose had a larger effect on bacteria abundance than the PCB high dose. The low PCB dose uniquely downregulated Ruminococcaceae, Muribaculaceae, Lachnospiraceae ASF356, and 2 taxa in Lachnospiraceae UCG-001, whereas it uniquely upregulated Akkermansia muciniphila, Erysipelatoclostridium, Ruminiclostridium 6, Enterohabdus, and Clostridiales bacterium CIEAF 016. The high PCB dose uniquely downregulated Dubosiella newyorkensis, Anaerotruncus, Acetatifactor, Turicibacter, and Muribaculaceae, whereas it uniquely upregulated Ruminococcaceae, Clostridium sp. Clone-44, Lachnospiraceae GCA-900066575, and Lachnospiraceae A2. Clostridium scindens, Ruminococcaceae UCG-013, Oscillibacter, Roseburia, Family XIII UCG-001, Clostridium sp. Culture-1 were upregulated by both the low and the high PCB doses. There were no bacteria that were commonly downregulated by the low and the high PCB dose, or regulated in opposite directions by the 2 PCB doses.
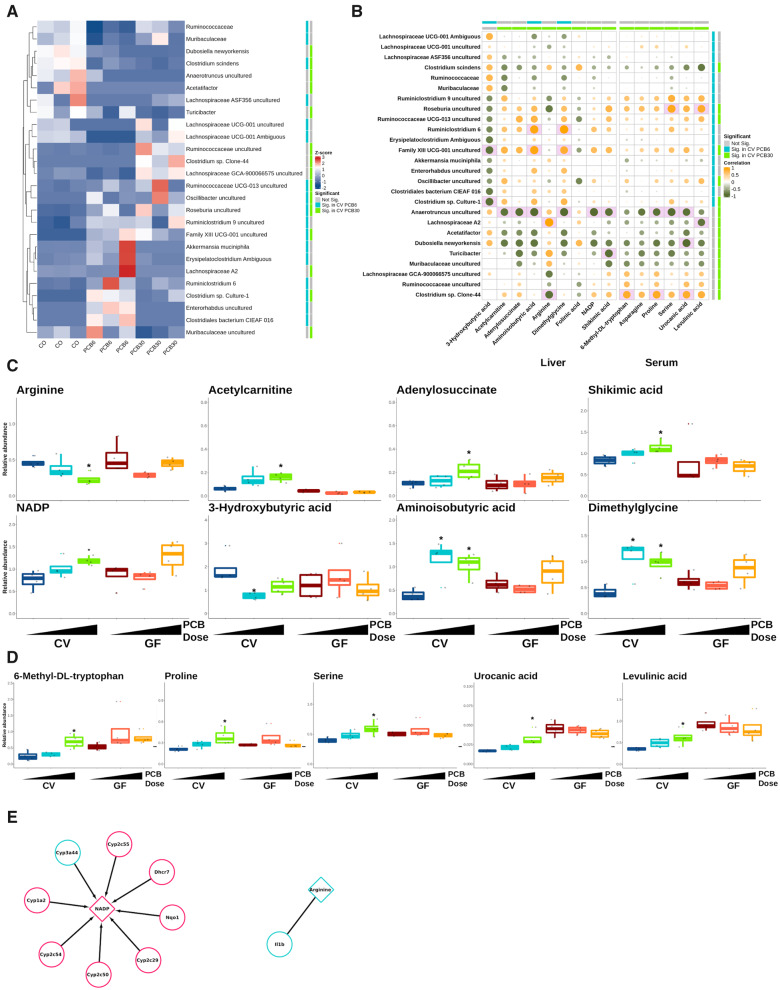
Figure 3.
Differentially abundant bacteria and correlation with differentially abundant liver and serum metabolites in conventional mice. A, Scaled relative abundance of bacteria for conventional mice. Differential abundance (Significant) information is provided on the colored bar (ie, blue shows differential expression for conventional mice exposed to polychlorinated biphenyl [PCB] 6 mg/kg and gray indicates genes that are not differentially regulated). B, Spearman correlation for differentially abundant bacteria (rows) and liver and serum metabolites (columns). Green-orange color scale represents the direction of correlation and sizes of circles show strength of correlation. Differential abundant bacteria and metabolites are shown as blue and green colored bars, representing significance for mice exposed to PCB 6 and 30 mg/kg, respectively. Pink filled box indicates significant correlation at FDR < 0.1 with absolute Spearman’s ρ > .8. Box plots of differentially abundant metabolites in liver (C) and serum (D) for conventional mice exposed to PCBs. Data are shown as relative abundance. The asterisks represent differential abundance in conventional groups exposed to PCBs. E, Differentially abundant metabolites are shown as triangles and differentially regulated genes are represented in circles. Colors indicate metabolite abundance and gene expression relative to the control (blue, lower abundance or expression; red, higher abundance or expression). Abbreviations: CV, conventional; GF, germ free.
Comparison Between CV and Conventionalized Mice
Supplementary Figure 3 shows the microbiome diversity for the top 15 most abundant bacteria in the colon at the family level for CV (n = 3) and conventionalized mice (GF mice bred in CV conditions for 2 weeks for microbiome colonization, n = 2). The top family included Clostridiaceae, Erysipelotrichaceae, Lachnospiraceae, Lactobacillaceae, Ruminococcaceae, S24-7, Staphylococcaceae, and an unidentified family in the Clostridiales order.
Metabolite Changes in Response to PCBs for CV and GF Groups
Endogenous metabolites in liver (Figs. 3C and 4A, and Supplementary Figure 2A) and serum (Figs. 3D and 4B, and Supplementary Figure 2B) were measured and analyzed to investigate the effect of the gut microbiome on PCB-mediated changes on essential endogenous metabolites for energy metabolism. In general, the PCB-mediated differences in relative abundances for metabolites were greater in liver than serum in both CV and GF mice. Dysregulated liver metabolites for the CV group exposed to PCBs were mainly from the amino acid metabolism pathways, including acetylcarnitine, adenylosuccinate, 3-hydroxybutyric acid, aminoisobutyric acid, and dimethylglycine (Figure 3C). Specifically, in livers of CV mice exposed to the PCB high dose, arginine was downregulated by the high PCB dose in CV mice and acetylcarnitine was upregulated in CV PCB high, dose-dependently. Adenylosuccinate, which is involved in nucleotide metabolism, and shikimic acid, which is a metabolite from bacteria, were upregulated by PCB exposure dose-dependently in the CV mice and were significant in the PCB high dose. The cofactor, NADP (upregulated by the high PCB dose in livers of CV mice), dose-dependently increased with exposure to PCBs in CV mice. The abundances of 2 liver metabolites from butyric acid metabolism, namely 3-hydroxybutyric acid (downregulated in CV PCB low) and aminoisobutyric acid (upregulated in CV mice in either doses) were altered only in CV mice. Dimethylglycine, a derivative of glycine, was upregulated by PCBs only in livers of CV mice (Figure 3C).
In serum, as shown in Figure 3D, amino acid metabolism-related serum metabolites, namely, 6-methyl-Dl-tryptophan, serine, asparagine, proline, and urocanic acid were upregulated in CV mice exposed to the PCB high dose. Levulinic acid, a metabolite from cellulose degradation, was upregulated from PCB high dose in CV mice in serum. Asparagine was upregulated in CV mice exposed to the PCB high dose (Supplementary Figure 2B). No metabolites were dysregulated from PCB low exposure in the CV group.
To establish the regulatory relationships between differentially expressed genes and differentially abundant metabolites (Figs. 3C and 3D and 5A and 5B), gene-metabolite integrated analysis on MetaboAnalyst was used as described previously (Chong et al., 2018). Links between significantly altered hepatic gene expression and liver metabolites by PCB exposure were only found in CV mice exposed to the PCB high dose (Figure 3E). The upregulation of many P450s and other phase-I xenobiotic biotransformation enzymes (except for Cyp3a44), through the utilization of NADPH, induced the abundance of NADP. In addition, an association between arginine abundance and the proinflammatory cytokine Il1b gene expression was observed (El-Sayed et al., 2019; Fultang et al., 2019; Pulugulla et al., 2018; Weldy et al., 2012).
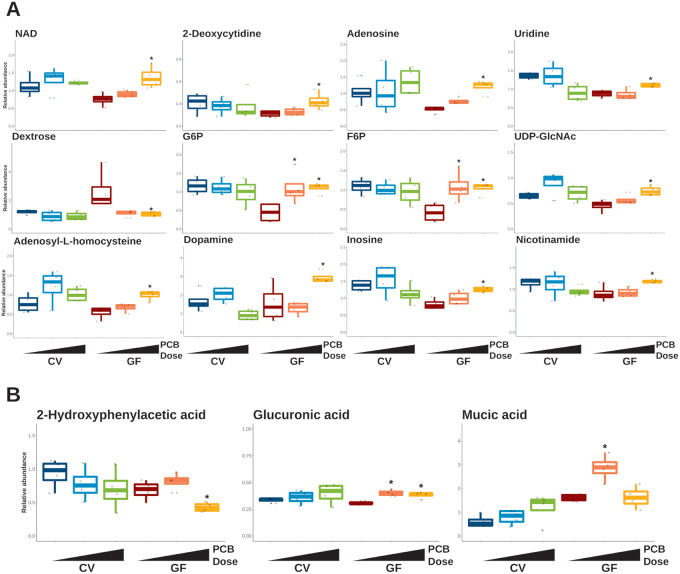
In GF mice, altered hepatic metabolites were mostly from carbohydrate metabolism or nucleotide metabolism (Figure 4A). Uridine, adenosine, and 2-deoxycytidine were upregulated in GF mice exposed to the PCB high dose in the liver. Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), a nucleotide sugar and a coenzyme involved in cell maintenance regulation (Love and Hanover, 2005), was upregulated in the livers of GF group exposed to the PCB high dose. The relative abundance for dextrose (also known as d-glucose) in the liver was lower for both PCB doses in GF mice, but only downregulated in the GF PCB high group. The relative abundance of the hepatic G6P and F6P were upregulated for both PCB doses in the GF group. In addition, NAD, an essential cofactor, was significantly higher for PCB high in the GF group. Adenosyl-l-homocysteine, dopamine, inosine, and nicotinamide were upregulated in GF mice exposed to the PCB high dose (Figure 4A). Guanosine triphosphate, 2,3-dihydroxybenzoic acid, 4-methylvaleric acid, and allantoin were upregulated by GF mice exposed to the PCB high dose and raffinose was downregulated by GF mice exposed to either PCB dose (Supplementary Figure 2).
Figure 4.
Selected liver and serum metabolites differentially abundant in germ-free (GF) mice exposed to polychlorinated biphenyls (PCBs). Box plots of differentially abundant metabolites in liver (A) and serum (B). Data are shown as relative abundance. The asterisks represent differential abundance in GF groups exposed to PCBs. Abbreviation: CV, conventional.
In serum, cytosine was upregulated by the low PCB dose in GF mice (Supplementary Figure 2A). Carbohydrate metabolism-related metabolites were upregulated only in GF mice, including mucic acid and mannose (upregulated in GF PCB low) and glucuronic acid (upregulated in GF PCB low or high) (Figure 4B and Supplementary Figure 2B). Norvaline (isomer of valine) was upregulated in GF mice exposed to the low PCB dose and 2-hydroxyphenylacetic acid, a metabolite of phenylalanine, was downregulated in GF mice exposed to the PCB high dose (Figure 4B and Supplementary Figure 2B).
Highly Correlated Changes Between Differentially Abundant Bacteria and Metabolites From Exposure to PCBs
Spearman’s correlation analysis was conducted to determine the association between gut dysbiosis and differentially regulated metabolites by PCB exposure in CV mice (Figure 3B and Supplementary Table 5A and 5B). Only the highly correlated relationships with an absolute value of ρ above .8 with FDR-adjusted p value < .1 were considered a significant correlation (indicated by the pink background in the cell).
In the liver, Family XIII UCG-001 (−0.81) and Clostridium sp. Culture-1 (−0.83) were both negatively associated with 3-hydroxybutyric acid. In addition, Family XIII UCG-001 was positively correlated with aminoisobutyric acid (0.88) and dimethylglycine (0.81). Clostridium sp. Culture-1 was negatively associated with the hepatic 3-hydroxybutyric acid (−0.83). Lachnospiraceae A2 was positively associated with arginine (0.90), which was negatively associated with Clostridium sp. Clone-44 (−0.82). Interestingly, Anaerotruncus was negatively associated with the majority (6 out of 9 in CV mice) of dysregulated liver metabolites, namely acetylcarnitine (−0.80), adenylosuccinate (−0.84), aminoisobutyric acid (−0.85), dimethylglycine (−0.86), NADP (−0.85), and shikimic acid (−0.82). In addition, Turicibacter was negatively associated with shikimic acid (−0.84).
As seen in Figure 5B and Supplementary Table 5A, in serum, Roseburia was positively associated with serine (0.87). Roseburia was positively correlated and Lachnospiraceae A2 was negatively correlated with levulinic acid (0.83 and −0.80, respectively). Clostridium sp. Clone-44 was positively associated with 6-methyl-Dl-tryptophan (0.82), and proline (0.84). Furthermore, Clostridium sp. Clone-44 was positively associated and D. newyorkensis was negatively associated with serum urocanic acid (0.82 and −0.86, respectively). Anaerotruncus was negatively associated with proline (−0.82), serine (−0.86), and urocanic acid (−0.82).
Figure 5.
Relative expression of differentially regulated genes grouped by functional categories and correlation of differentially abundant bacteria and differentially regulated genes in conventional mice. A, Scaled expression of genes in epigenetic modification (Epi.), inflammation and oxidative stress, and nuclear receptor (NR) categories for conventional and germ-free (GF) mice. Differential regulation (Significant) information is provided on the colored bar (ie, blue shows differential expression for conventional mice exposed to polychlorinated biphenyl [PCB] 6 mg/kg and gray indicates genes that are not differentially regulated). B, Scaled expression of genes in phase-I, -II, and transporter categories for conventional and GF mice. Differential regulation (Significant) information is provided on the colored bar (ie, blue shows differential expression for conventional mice exposed to PCB 6 mg/kg and gray indicates genes that are not differentially regulated). Spearman correlation for differentially abundant bacteria (rows) and differentially regulated genes (columns) in phase-I and -II (C), and transporters, inflammation and oxidative stress, and NR categories (D). Green-orange color scale represents the direction of correlation and sizes of circles show strength of association. Differential abundant bacteria and metabolites are shown as blue and green colored bars, representing significance for mice exposed to PCB 6 and 30 mg/kg, respectively. Pink filled box indicates significant correlation at FDR < 0.1 with absolute spearman’s ρ > .8. Abbreviation: CV, conventional.
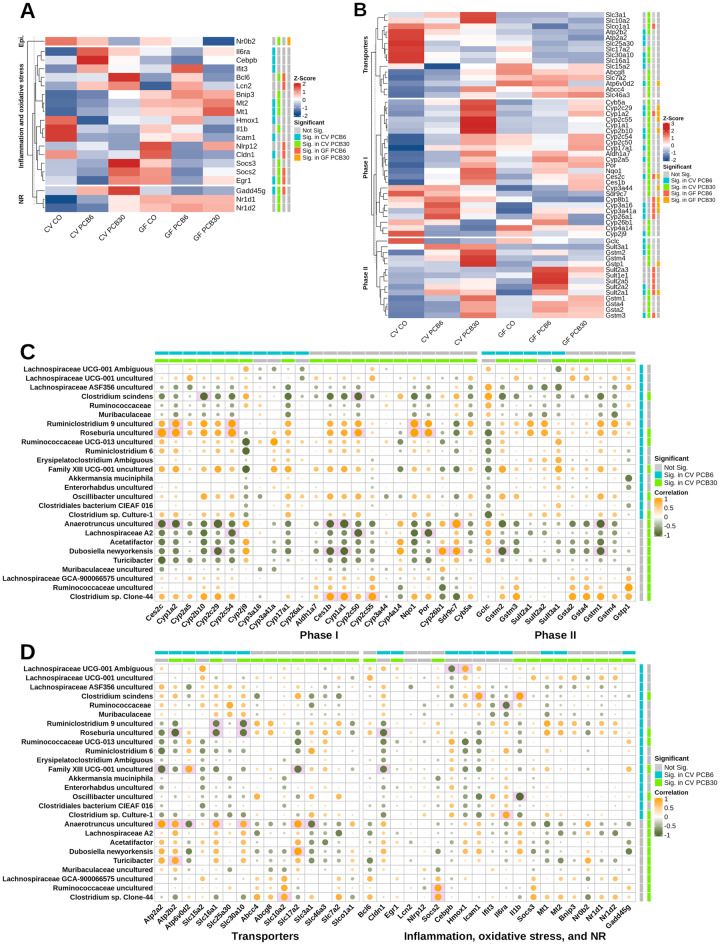
Transcriptomic Response Involved in Critical Pathways in Livers of CV and GF Groups Following PCB Exposure
Specific examples of important hepatic pathways are presented in Figures 5A and 5B. Dysregulated genes were grouped into categories to determine the regulation of the following pathways: epigenetic modification factors (Epi.), inflammation and oxidative stress, NRs, phase-I and -II drug metabolizing enzymes, and transporters. In general, unique expression patterns were observed comparing each enterotype (Figs. 5A and 5B). The mRNA of Nr0b2 (also known as SHP) was downregulated by the high PCB dose in both CV and GF mice; however, the degree of decrease was greater for the GF group (Figure 5A). None of the gene categories shown in Figure 3C were commonly regulated by both enterotypes and doses, and no genes in these categories were uniquely regulated for GF mice. Most of the genes were dysregulated for the CV group exposed to PCBs for both doses in the inflammation and oxidative stress, and NR category, which included Il6ra, Mt1, Mt2, Il1b, Nr1d1, and Nr1d2 (Figure 5A).
For genes that are important in xenobiotic biotransformation (Figure 5B), similar to the gene expression pattern shown in Figure 3C, gene induction by PCBs occurred more readily in CV mice than in GF mice. Specifically, as shown in Figure 4D top cluster, genes that encode transporters in liver were more regulated by PCBs of CV mice than the GF mice. In the CV group exposed to PCBs, manganese efflux transporter (Slc30a10) and proton-coupled monocarboxylate transporter (Slc16a1) were commonly downregulated in livers of CV mice by either dose of PCBs. The membrane glycoprotein transporter involved in neutral amino acid transport (Slc3a1) and sodium/bile acid cotransporter (Slc10a2 [Ntcp]) were upregulated by the PCB high dose in livers of the CV mice. Sodium-dependent phosphate transporter (Slc17a2) and proton-coupled peptide transporter (Slc15a2) were uniquely downregulated by PCB high dose and PCB low dose in CV mice, respectively. ATP-binding cassette subfamily G, member 8 (Abcg8 [sterolin]), which is important for sterol regulation and excretion, cationic amino acid transporter 2 (Slc7a2), multidrug resistance-associated protein 4 (Abcc4 [Mrp4]), and lysosomal catabolite transporter (Slc46a3) were upregulated in CV mice exposed to the PCB high dose. Sodium-independent organic anion transporter Slco1a1 (Oatp1a1) was downregulated in CV mice exposed to the PCB high dose and GF mice exposed to PCB low dose. The lysosomal proton pump Atp6v0d2 was upregulated for CV mice exposed to either PCB dose, however was downregulated in GF mice exposed to the PCB high dose.
For genes that encode phase-I enzymes (Figure 5B middle cluster), in general, genes in the Cyp1, 2, and 3 families are known to be important for drug metabolism and Cyp4 families are involved in fatty acid metabolism. Except for Cyp1a2 (prototypical target gene for AhR), Cyp2c29, and Cyp2b10 (prototypical target gene for CAR), most of the genes in the Cyp1 and 2 families were uniquely differentially regulated in the CV group, including Cyp1a1, Cyp2c54, Cyp17a1, and Cyp2a5. Other phase-I drug metabolizing enzymes, such as aldehyde dehydrogenase (Aldh1a7), P450 oxidoreductase (Por), NAD(P)H dehydrogenase [quinone] (Nqo1), carboxylesterase (Ces1b and Ces2c), and short chain dehydrogenase/reductase (Sdr9c7) followed the same trend as Cyp1 and 2 families, being more upregulated by PCBs in livers of CV mice than in livers of GF mice. Cyp26b1 (involved in retinoic acid metabolism) and Cyp4a14 were downregulated in CV mice exposed to the PCB high dose and Cyp2j9 (involved in arachidonic acid metabolism) was downregulated for CV mice exposed to either PCB doses. Genes in the Cyp3 family (Cyp3a16 and Cyp3a41a) were upregulated in CV mice exposed to the low dose PCBs and either dose for GF mice. Cyp26a1 was upregulated for mice exposed to the PCB low dose for both enterotypes.
Interestingly, as seen in the bottom cluster on Figure 5B, for the phase-II conjugation enzymes, genes in the glutathione-S-transferase (Gst) category were uniquely upregulated by CV mice, with the exception of Gstm3 (upregulated by CV mice in either PCB dose and GF mice exposed to PCB low dose), whereas genes in the sulfotransferase (Sult) 1 and 2 categories, except the Sult2a1 mRNA (upregulated by either enterotype or dose) were upregulated by GF mice. The Gstp1 mRNA was upregulated by the PCB high dose in either enterotype. For the upregulated phase-II enzymes that are shared between enterotypes, baseline relative expression (mice exposed to CO) and the degree of induction were also different, such that the Gstm3 mRNA was lower, whereas the Gstp1 and Sult2a1 mRNAs were higher in CV than GF mice. In summary, the dysregulated genes show that the PCB-mediated liver transcriptome is regulated in part by the gut microbiome.
Correlation of Differentially Regulated Intestinal Bacteria and Expression of Hepatic Xenobiotic-biotransformation-related Genes Following PCB Exposure
To investigate to what extent the gut microbiome regulates xenobiotic biotransformation-related functions upon responses to PCB toxic exposure, we focused on the potential relationship between differentially regulated genes and differentially abundant bacteria as examined using Spearman’s correlation (Figs. 5C and 5D, and Supplementary Table 5C and 5D). In the phase-I category (Figure 5C left panel), C. scindens was negatively associated with Cyp2b10 and Cyp2c50 (−0.85 for both genes). Ruminiclostridium 9 was positively associated with Cyp1a2, Cyp2c54, and Nqo1 (0.82, 0.83, and 0.85, respectively). Roseburia was positively associated with Ces2c, Cyp1a2, Cyp2c54, Cyp2c50, Nqo1, and Por (0.93, 0.87, 0.88, 0.82, 0.9, and 0.85, respectively). Anaerotruncus was negatively associated with Ces2c, Cyp1a2, Cyp2c29, Ces1b, and Cyp1a1 (−9.84, −0.86, −0.91, −0.83, and −0.91, respectively), and positively associated with Sdr9c7 (0.91). Lachnospiraceae A2 was negatively associated with Cyp2c54, Cyp2c50, and Por (−0.81, −0.85, and −0.84, respectively). Dubosiella newyorkensis was negatively associated with Cyp2c29, Ces1b, Cyp1a1 (−0.85 for all 3 genes), but positively associated with Cyp26b1 and Sdr9c7 (0.81 for both genes). Clostridium sp. Clone-44 was positively associated with Ces1b, Cyp1a1, and Cyp2c55 (0.82, 0.82, and 0.84, respectively).
In the Phase-II category (Figure 5C right panel), D. newyorkensis was negatively associated with Gstm2 (−0.83) and Gstm1 (−0.81). In addition, Anaerotruncus was negatively associated with Gstm1 (−0.84).
For transporters (Figure 5D left panel), Roseburia was negatively associated with Atp2b2 (−0.93) and Roseburia and Ruminiclostridium 9 were negatively associated with Slc16a1 (−0.87 and −0.82, respectively) and Slc30a10 (0.87 for both bacteria), however, Anaerotruncus was associated with both genes (0.86 and 0.80, respectively), in addition to Atp2a2, Atp2b2, Atp6v0d2, Slc17a2, and Slc3a1 (0.91, 0.84, −0.82, 0.89, and −0.86, respectively). Family XIII UCG-001 was associated positively with Atp6v0d2 (0.85), and negatively with Slc17a2, Atp2a2 (−0.88 and −0.90, respectively). Dubosiella newyorkensis was positively associated with Slc17a2. Turicibacter was positively associated with Atp2b2 (0.85) and Clostridium sp. Clone-44 was positively associated with Slc10a2 (0.82).
In the immune response, oxidative stress, and NRs category (Figure 5D right panel), Roseburia and Family XIII UCG-001 were negatively associated with Cldn1 (−0.88 and −0.85, respectively). Ruminococcaceae and Clostridium sp. Clone-44 were positively association with Socs2 (0.87 and 0.84, respectively). Lachnospiraceae UCG-001 Ambiguous was associated negatively with Cebpb (0.83) and positively with Hmox1 (0.81). Clostridium scindens was positively associated with Icam1 (0.87) and Il1b (0.9). Oscillibacter uncultured was negatively associated with Il1b (−0.95) and Clostridium sp. Culture-1 was negatively associated with Il6ra (−0.87).
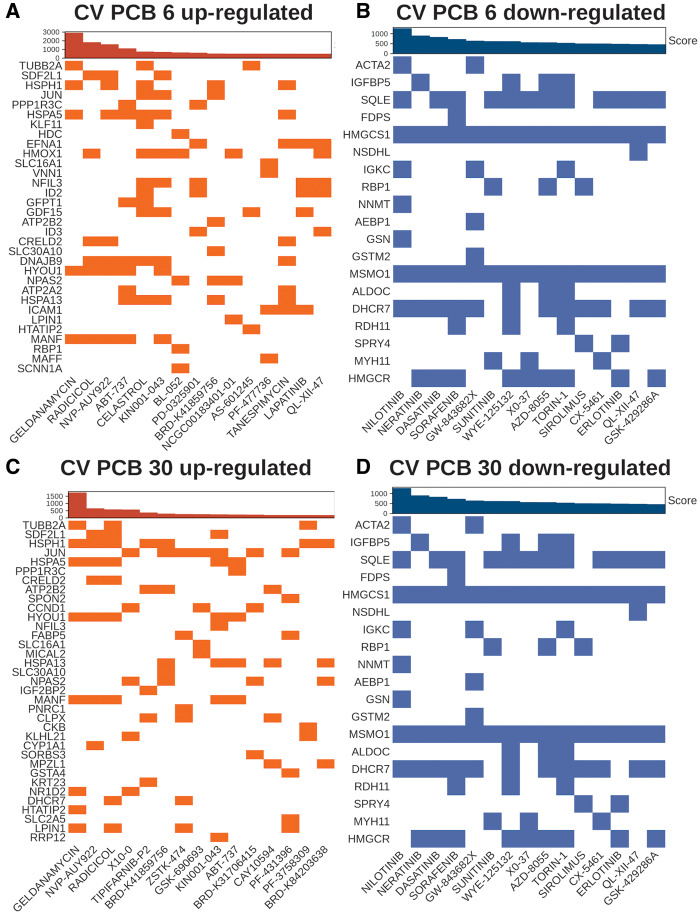
Potential Therapeutics to Target Gut Microbiome-dependent PCB-induced Toxicity
To find potential therapeutics against the gut microbiome-dependent PCB effects, we performed an inverse correlation analysis between dysregulated genes from PCB low or high dose exposure and drugs and small molecule response were done using the LINCS L1000 database (Figure 6). For example, upregulated genes unique to CV mice exposed to the PCB low dose compared with GF PCB low were associated with target genes that were downregulated from drugs and other small molecule exposure to human cell lines (inverse association). The top drugs ranked by the combined score in the upregulated signatures using downregulated genes from PCB exposure in both doses included geldanamycin, radicicol, and NVP-AUY922, which are antitumor compounds that potently inhibit HSP90 function to regulate cell and growth, and angiogenesis (He et al., 2013; Jensen et al., 2008; Schulte et al., 1998) (Figs. 6A and 6C). In addition, as seen in Figure 6A, tanespimycin, another drug that inhibits HSP90, was observed in the top drug list for the CV PCB low dose (Yang et al., 2009). Other top compounds for downregulated gene expression signatures for CV PCB low included ABT-737 (Bcl-2 and Bcl-XL inhibitor), celastrol (sirtuin 1 inhibitor), PD-0325901 (MEK inhibitor), AS-601245 (JNK inhibitor), PF-477736 (cell cycle checkpoint kinase 1 [chk1] inhibitor), lapatinib (epidermal growth factor receptor [EGFR] inhibitor), QL-XII-47 (BTK kinase inhibitor), as well as KIN001-043, NRD-K41859756, and NCGC00183401-01, which the mechanism of actions are not defined (Bryant et al., 2014; Cerbone et al., 2012; de Wispelaere et al., 2017; Huang et al., 2009; Medina and Goodin, 2008; Parrondo et al., 2013; Venkatesha and Moudgil, 2016). Drugs targeting the gut microbiome-dependent downregulated genes from PCB high exposure included QS11 (GTPase activating protein of ADP-ribosylation factors [ARFGAPs] inhibitor), tipifarnib (farnesyltransferase inhibitor), ZSTK-474 (IFNg and IL-17 proliferation inhibitor), GSK690692 (pan-AKT inhibitor), CAY10594 (phospholipase D2 inhibitor), as well as PF-43758309, BRD-K84203638, and BRD-K31706415 (unknown mechanisms) (Kong and Yamori, 2010; Lee et al., 2019; Levy et al., 2009; Reid and Beese, 2004; Zhang et al., 2007).
Figure 6.
Potential therapeutics from gut microbiome-dependent polychlorinated biphenyl (PCB) exposure. Drugs and small molecule (columns) and their respective target genes (rows) in human cell lines. Top bar plot represents the combined score using the adjusted p value and odds ratio. Top 15 inversely correlated target signatures with the differentially regulated genes from PCB exposure are shown. Upregulated target genes from exposure to drugs and small molecules (downregulated from PCB low and or high exposure) are highlighted in orange (A and B, respectively). Downregulated target genes from exposure to drugs and small compounds (upregulated from PCB low or high exposure) are highlighted in blue (C and D, respectively). Abbreviation: CV, conventional.
Downregulated drug response signatures using upregulated genes from PCB exposure were the same for both PCB doses (Figs. 6B and 6D). Nilotinib, a transduction inhibitor targeting BCR-ABL oncogene, c-kit (CD117), and platelet-derived growth factor receptor (PDGF), was among the top potential therapeutics to counter-regulate upregulated genes related to actin function (ie, actin alpha2 [Acta2], Gelsolin [Gsn]), xenobiotic biotransformation (ie, nicotinamide N-methyltransferase [Nnmt]), immune response (ie, immunoglobulin kappa constant [Igkc]), and lipid/steroid metabolism (ie, squalene epoxidase [Sqle], hydromethylglutaryl-CoA synthase [Hmgcs1], methylsterol monooxygenase 1 [Msmo1], 7-dehydrocholesterol reductase [Dhcr7]) from PCB low and high exposure (Blay and von Mehren, 2011) (Figs. 6B and 6D). In addition, neratinib (human epidermal growth factor inhibitor 2 [Her2] and EGFR inhibitor), dasatinib (dual BCR-ABL and Src family tyrosine kinase inhibitor), sorafenib (Raf kinase, PDGF, VEGFR, and c-kit inhibitor), GW-843682X (ATP-competitive polo-like kinase 1 and 3[PLK1 and 3] inhibitor), sunitinib (multireceptor tyrosine kinase inhibitor), WYE-125132, AZD-8055, torin 1, and sirolimus (ATP-competitive mTOR inhibitor), RAF265 (B-raf kinase and VEGFR2 phosphorylation inhibitor), CX-5461 (p53 stabilizer), QL-XII-47 (selective BTK inhibitor), and GSK-429286A (selective Rho-kinase inhibitor) were inversely correlated with the upregulated gene expression signatures from PCB exposure at both doses (Chresta et al., 2010; Dumont and Su, 1995; Huang et al., 2017; Mena et al., 2010; Schade et al., 2008; Segovia-Mendoza et al., 2015; Thompson et al., 2017).
DISCUSSION
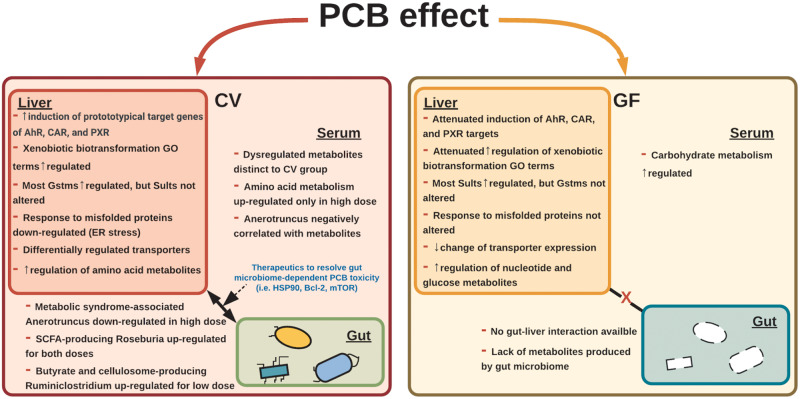
This study demonstrates that the gut microbiome is a necessary component in mediating PCB-induced effects on the liver transcriptome and metabolome of liver and serum (Figure 7). The presence of gut microbiome drives PCB-mediated activation of xenobiotic-sensing transcription factors AhR, CAR, and PXR, evidenced by a more prominent increase in the expression of their prototypical target genes, as well as a more upregulated GO involved in xenobiotic biotransformation. In addition, the presence of gut microbiome lead to the downregulation of the GO terms involved in response to misfolded proteins, which indicates the contribution of gut microbiome in PCB-mediated ER stress. Regarding intermediary metabolism, the presence of gut microbiome promotes the PCB-mediated upregulation of amino acid metabolites, but suppresses the metabolites involved in nucleotide metabolism. In particular, the genus Anaerotruncus, as well as the short-chain fatty acid-producing bacteria Roseburia and Ruminiclostridium, was highly associated with hepatic xenobiotic biotransformation enzymes and metabolites of liver and serum. Taken together, our findings demonstrate that the habitation of the gut microbiota drives PCBs-mediated toxic responses including bioactivation of xenobiotics, ER stress, and dysregulation of intermediary metabolism.
Figure 7.
Summary of study findings. From polychlorinated biphenyl (PCB) exposure, in livers of conventional (CV) mice, higher induction of prototypical target genes of aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and pragnane X receptor (PXR) were observed, compared with the germ-free (GF) mice. In CV mice, xenobiotic biotransformation-related gene ontology (GO) terms were more enriched and ER stress-related GO terms were uniquely downregulated. Most Gstms were upregulated in CV mice, whereas most Sults were upregulated in GF mice exposed to PCBs. In addition, CV mice exposed to PCBs had differentially regulated transporters; however, less change in transporter expression was observed in GF mice. In the liver and serum, amino acid metabolism-related metabolites were preferably upregulated in CV mice. Conversely, nucleotide and carbohydrate metabolism-related metabolites were upregulated in GF mice. Short-chain fatty acid-producing bacteria, such as Roseburia (PCB both doses) and Ruminiclostridium (PCB low dose) were upregulated and Anaerotruncus was downregulated in the PCB high dose following PCB exposure and were correlated with metabolites, such as aminoisobutyric acid, dimethylglycine, and arginine, and genes important in xenobiotic biotransformation, such as Cyp1a1, Cyp1a2, and Atp2a2, showing the possible role of these bacteria in mediating PCB-induced toxicity. Therapeutics that target critical pathways, such as mTOR and Bcl-2, have the potential to resolve signatures from PCB exposure.
Gut Microbiome-mediated Transcriptomic Responses From PCB Exposure
The gut microbiome mediates the activation of transcription factors and their targets (Bjorkholm et al., 2009; Richards et al., 2019), which is possibly due to the metabolite interaction with the biochemical environment (Venkatesh et al., 2014). The PCB-mediated upregulation of the prototypical target genes of AhR, CAR, and PXR act partly through the gut microbiome, indicating that the PCB may increase microbial metabolites that can further activate these host receptors, or the gut microbiome may convert PCBs to more potent ligands for these host receptors. In contrast to PCBs, we have previously shown that the absence of gut microbiota potentiated the liver to polybrominated diphenyl ether congeners (PBDE-47 and 99)-mediated upregulation of the prototypical target genes of these host receptors (Li et al., 2017a), suggesting that the 2 classes of persistent environmental toxicants (PCBs and PBDEs) may modify the gut-liver axis through 2 distinct mechanisms.
To note, the xenobiotic-sensing transcription factor genes themselves were not altered in livers of GF mice. Previous studies have shown that the activity of host xenobiotic-sensing transcription factors, such as AhR, PXR, and CAR can alter the composition of the gut microbiome (Dempsey et al., 2019; Hudson et al., 2017). To the best of our knowledge, the microbial AhR and CAR receptors have not been known in the literature. Even if these microbial receptors do exist, it is unlikely they will contribute to the upregulation of host hepatic AhR- and CAR-target genes, because these receptors are expected to interact with portions of the PCB mixtures. This study provides novel findings that the absence of the gut microbiome greatly attenuated the expression of the prototypical targets, which indicates that the effect of PCBs in host transcription factor activation acts partly through the gut microbiome, likely due to the microbial biotransformation of PCBs or host-derived PCB metabolites into more potent ligands for theses host receptors. The relative abundance for the most abundant bacteria for the conventionalized exGF mice resemble those of the CV mice (Supplementary Figure 3). This result indicates that recolonization of the gut microbiome reaches a similar microbial abundance level as the CV mice. In addition, previous studies have shown that the conventionalization of GF mice have restored xenobiotic metabolism signatures to similar levels of those of the CV mice and conventionalization of GF mice led to the induction of host xenobiotic-sensing transcription factors, indicating that the gut microbiome, through bacterial metabolites, can regulate the activation of these receptors (Claus et al., 2011; Selwyn et al., 2016). Therefore, the attenuation effects are likely due to the lack of gut microbiome. Although CV and GF mice have different physiology, colonization of healthy bacteria to PCB-exposed GF mice may result in responses similar to the CV mice.
PCBs are well-known to activate host transcription factors, including AhR, PXR, CAR, and PPARα (Wahlang et al., 2014), which are known to upregulate their prototypical target genes following a dose-response curve, and this may subsequently lead to changes in endogenous metabolites in a dose-dependent manner. In this study, although the hepatic transcriptome and metabolome tended to have a dose-response relationship from exposure to the 2 different PCB doses, the effect of PCBs on the gut microbiome is not monotonic, possibly involving different regulatory mechanisms. It has been shown that the PCB low dose tended to lower the richness of the gut microbiota to a greater extent than the PCB high dose (Cheng et al., 2018). It is possible that the PCB high dose may overwhelm the system and involve less specific regulatory pathways.
The gut microbiome appears to have fundamental roles in PCB-mediated upregulation of P450 activities, because the 2 genes that encode essential enzymes that sustain P450 reactions, namely Cyb5b (regulates P450 activity through P450-dependent monooxygenase system) and Por (transfers electrons from NADPH to P450s) were upregulated by PCBs in a gut microbiome-dependent manner; thus, PCB-exposed GF mice may have less efficiency and decreased hepatic P450 activity than PCB-exposed CV mice. In addition, the gut microbiome-dependent upregulation of Gsts may be due to Epi. from the gut microbial metabolites, such as butyrate, succinate, and acetate (Fellows et al., 2018; Hullar and Fu, 2014; Xu et al., 2018). The lack of Gst upregulation in PCB-exposed GF mice may send a stress signal to upregulate other phase-II detoxification pathways such as sulfonation, evidenced by a PCB-mediated upregulation of multiple Sults only in livers of GF mice.
Differences in PCB-induced Metabolism Changes Between CV and GF Mice
ER stress has been shown to be one of the key mechanisms of PCB-mediated hepatotoxicity (Ruan et al., 2020; Xu et al., 2015), but little is known to what extent gut microbiome contributes to this mechanism. This study is the first to show that the presence of the gut microbiome is linked to the PCB-mediated downregulation of pathways related to response to misfolded proteins, which is a hallmark for ER stress. Oxidative damage and impaired amino acid metabolism are known to cause ER stress, which involves Ca2+ signaling (Bahar et al., 2016; Chong et al., 2017; Harding et al., 2003). Altered amino acid metabolites were found primarily in the livers of PCB-exposed CV mice, including acetylcarnitine, adenylosuccinate, 3-hydroxybutyric acid, aminoisobutyric acid, and dimethylglycine (Figure 3C). Furthermore, numerous liver genes related to oxidative stress were upregulated in CV mice exposed to PCBs, such as Arntl, Atp2a2, Jun, Mt1, Mt2, and Cebpb (Han et al., 2008; Kujiraoka et al., 2013; Moloney and Cotter, 2018; Wilking et al., 2013). These results suggest that oxidative damage and altered amino acid metabolism from PCB exposure possibly drive hepatic ER stress as suggested by downregulation GO terms in CV mice (Figure 2B), downregulation of heat shock proteins, as well as Ca2+ transporters involved in intrinsic apoptotic signaling pathways in response to ER stress (Concilli et al., 2016; Xu et al., 2018; Yang et al., 2015) (Figs. 2B and 5B, and Supplementary Table 1A and 1B).
Regarding the microbial signatures, the PCB low dose upregulated Ruminiclostridium 6 and Family XIII UCG-013, and both of them were positively associated with dimethylglycine and aminoisobutyric acid (Figure 3B). The abundance of the genus Ruminiclostridium is increased by immune response and oxidative damage, which PCB is known to produce inflammation and oxidative stress (Petriello et al., 2018; Phillips et al., 2018; Sarkar et al., 2019). The Family XIII UCG-013 was negatively correlated with Atp2a2 (sarco/ER Ca2+-ATPase [SERCA2]). Atp2a2 has been shown to be downregulated in oxidative damage and ER stress (Kujiraoka et al., 2013). Dimethylglycine is generated from homocysteine metabolism, which is related to protein degradation and ER stress. Therefore, the upregulation of Ruminiclostridium 6 and dimethylglycine may reflect PCB-mediated oxidative damage, ER stress, and inflammation. Aminoisobutyric acid is one of the end products of pyrimidine metabolism (van Kuilenburg et al., 2006). Multiple species of Ruminiclostridium, such as Ruminiclostridium cellulolyticum, have enzymes that contribute to generating aminoisobutyric acid, such as pyrimidine-nucleoside phosphorylase (DumitracHe et al., 2017). Aminoisobutyric acid concentration has been suggested to be protective against metabolic disorder (Roberts et al., 2014). In turn, the upregulated oxidative stress elements may have upregulated aminoisobutyric acid, as a compensatory response, to alleviate the PCB responses in liver. In summary, the oxidative damage and immune response pathways as regulated by the PCB low dose, associated with upregulated Ruminiclostridium 6 and Family XIII UCG-013, may have contribute to the gut microbiome-dependent ER stress, and modulating amino acid metabolites may have a therapeutic value in alleviating adverse health effects from PCB exposures.
The absence of gut microbiome potentiated PCB-mediated dysregulation of metabolites involved in carbohydrate metabolism and nucleotide metabolism (Figure 4 and Supplementary Figure 2). Specifically, hepatic G6P and F6P were upregulated in GF mice exposed to both PCB doses, whereas d-glucose was downregulated. This may indicate that the glucose utilization in the liver is impaired in GF mice exposed to PCBs. It is known that the lack of gut microbiome impairs liver regeneration (Liu et al., 2015; Wu et al., 2015), suggesting the importance of gut microbiome in providing the energy to the host liver.
Clostridium sp. Clone-44 was positively associated with genes involved in inflammation and oxidative stress (Ces1b, Cyp1a1, Cyp2c55, Slc10a2, and Socs2), but was negatively associated with arginine levels (Figs. 3 and 5). Arginine can reduce inflammatory response and oxidative stress by inhibiting cytokine secretion and inducing glutathione synthesis (Hnia et al., 2008; Liang et al., 2018; Wu et al., 2016). Multiple species in the Clostridium genus have been shown to metabolize arginine (Mitruka and Costilow, 1967; Neumann-Schaal et al., 2019). The gut microbiome-dependent decrease in arginine may be due to increased abundance of Clostridium sp. Clone-44, which may explain the upregulated pathways involved in inflammation and oxidative stress. Arachidonic acid is suggested to interact with arginine by sharing the nitric oxide pathway. Milner and Perkins (1978) showed that rats deficient in arginine had increased levels of arachidonic acid. In CV mice exposed to the high dose PCB, more arachidonic acid metabolizing P450s were upregulated, such as Cyp1a1 and Cyp2c55, which metabolizes arachidonic acid to eicosanoids, suggesting a rapid inflammation-mediating transition, compared with the GF state.
Anaerotruncus, a butyrate producer, is a normal gut inhabitant in mice with enhanced growth in the presence of mucin (Wang et al., 2019; Wlodarska et al., 2017). Following PCB exposure, Anaerotruncus was negatively associated with amino acid metabolites, such as acetylcarnitine, adenylosuccinate, aminoisobutyric acid, and dimethylglycine in liver, as well as proline, serine, and urocanic acid in serum (Figs. 5A and 5B). Anaerotruncus was also negatively associated with expression of Ces2c, Cyp1a2, Cyp2c29, Ces1b, Cyp1a1, and positively associated with Atp1a2, Atp2b2, Slc16a1, Slc30a10, and Slc17a2 (Figs. 6C and 6D). It has been reported that Anaerotruncus has the capabilities to metabolize fatty acids and its abundance was negatively correlated with xenobiotic exposure, such as dipeptidyl peptidase 4 inhibitors, which reduces blood glucose levels (Olivares et al., 2018; Visconti et al., 2019; Yan et al., 2016; Zhang et al., 2017). Anaerotruncus has been linked to glucose intolerance, gut permeability, muscle lipid content, and type 2 diabetes in humans, cats, and rats, which suggests that Anaerotruncus may have a protective role in oxidative damage, and the decrease in the abundance of Anaerotruncus may have contributed to PCB-induced toxicities (Everard et al., 2011; Kieler et al., 2019; Li et al., 2017b).
Potential Therapeutics to Limit Impact of Exposure to PCBs via Gut Microbiome-dependent Signatures
Results from the inverse relationships between transcriptomic signatures from the LINCS L1000 database and the gut microbiome-dependent gene expression signatures following PCB exposure highlighted the potential remedy to mitigate gut microbiome-potentiated PCB toxicity. The majority of the drugs and other small compounds that were identified to have opposite gene expression signatures as compared with PCB-exposed conditions are anticancer drugs. These results may indicate that the gut microbiome is contributes to the tumorigenesis mechanism of PCBs. Geldanamycin, which inhibits the function of HSP90 and regulates cell cycle, growth, and apoptosis, was the top inversely correlated drug that upregulates the genes that make up the downregulated signatures, such as Hyou1 and Hspa5, from PCB exposure. In addition, other top drugs and small molecules that could restore expression of downregulated genes included ABT-737, celastrol, and KIN001-043, which may reverse the ER damage and oxidative stress signatures, such as Hyou1, Hspa5, Hspa13, and Atp2b2, which are potentially due to and closely related to changes in altered bacteria, such as Ruminiclostridium, Roseburia, and Anaerotruncus (Figs. 3B, 5C, and 5D and 6A and 6C). Similarly, the top inversely correlated drug that could inhibit the gut microbiome-dependent upregulated genes from PCB exposure was shown to be nilotinib, which may normalize the levels of upregulated lipid and sterol metabolism- and actin function-related genes (Figs. 6B and 6D). Interestingly, compounds inhibiting the mTOR pathway (ie, WYE-125132, AZD-8055, torin 1, and sirolimus) were repeatedly observed (Figs. 6B and 6D). It has been shown that PCB-95 activated mTOR in primary rat hippocampal neurons and developing zebrafish (Frank et al., 2017; Keil et al., 2018). Activation of mTOR pathway in the liver has been linked to liver tumor, NASH, and liver damage from autophagy in both mouse models and humans (Okuno et al., 2018; Saxton and Sabatini, 2017). PCB-mediated inhibition of mTOR signaling pathway may reduce the risk of liver cancer and metabolic disease development.
This study includes limitations, such as the not having information on all of the dysregulated bacteria at the species level and metabolites in specific pathways, and a small sample size for the transcriptomic responses and 16s bacteria changes. Species and strain levels of dysregulated bacteria through deeper sequencing methods, such as from using shotgun metagenomics, and anaerobic culturing-coupled sequencing may give more mechanistic evidence of how bacteria regulate and respond with the gut and liver during PCB exposure. However, with the given breadth of data, we showed that the higher induction of xenobiotic biotransformation, immune response and oxidative stress-related mRNA signatures, and ER stress-mediated damage is unique to mice with a normal gut microbiome. Future investigations should focus on the specific metabolic capacities of the dysregulated microbiome to study the mechanisms of biochemical perturbation from PCB exposure. Furthermore, despite the limitations, our study identifies the gut microbiome as a key player in biochemical regulation. To delineate whether the transcription factor signaling pathway is modified by the presence of the gut microbiome, an in vitro experiment using primary hepatocytes from CV and GF mice can be planned. However, the in vitro hepatocytes loses many in vivo features within hours after plating, including downregulation of many transporters as well as drug-metabolizing enzymes; in addition, the in vitro system lacks the gut-liver axis and is not exposed to gut-derived microbial metabolites. The gut-liver axis is the key focus of this study and can only be assessed in vivo. When cells are isolated from an organism, the resulting responses are likely not directly comparable from the introduction of an artificial environment to cells, which may not function as equivalent. Furthermore, using isolated cells exposed to PCBs would drastically alter the mode of exposure, which the exposed cells would lack intestinal biotransformation pathways. In order for this approach to be comparable with in vivo results, a cell battery consisting of multiple bio-compartments, including enterocytes colonized with key microbiota and liver, should be used.
For people with severe dysbiotic states (eg, from antibiotics treatment), our study adds knowledge of potential differences in physiological responses from ubiquitous environmental contaminants, such as PCBs and potential considerations for further investigations regarding therapeutics.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the members of Dr Cui’s laboratory for their help in tissue collection and manuscript revision.
FUNDING
National Institutes of Health (R01 ES025708, R01 ES030197, R01 GM111381, R01 ES031098, T32 ES007032, ES013661, ES005605); University of Washington Center for Exposures, Diseases, Genomics, and Environment (NIH P30 ES0007033); and University of Washington (Sheldon Murphy Endowment).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Bahar E., Kim H., Yoon H. (2016). ER stress-mediated signaling: Action potential and Ca(2+) as key players. Int. J. Mol. Sci. 17, 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann, L. P., Hannivoort, R. A., Gerken, G., Hotamisligil, G. S., Trauner, M., and Canbay, A. (2012). The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol 56, 952–964. [DOI] [PubMed] [Google Scholar]
- Bjorkholm B., Bok C. M., Lundin A., Rafter J., Hibberd M. L., Pettersson S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay J. Y., von Mehren M. (2011). Nilotinib: A novel, selective tyrosine kinase inhibitor. Semin. Oncol. 38, S3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesen, A. C., Martinez, A., and Hornbuckle, K. C. (2020). Air-water PCB fluxes from southwestern Lake Michigan revisited. Environ Sci Pollut Res Int. 27, 8826–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., Mills D. A., Caporaso J. G. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C., Scriven K., Massey A. J. (2014). Inhibition of the checkpoint kinase Chk1 induces DNA damage and cell death in human Leukemia and Lymphoma cells. Mol. Cancer 13, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P. A., Diolaiti D., McFerrin L., Gu H., Djukovic D., Du J., Cheng P. F., Anderson S., Ulrich M., Hurley J. B., et al. (2015). Deregulated Myc requires mondoA/MIx for metabolic reprogramming and tumorigenesis. Cancer Cell 27, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbone A., Toaldo C., Pizzimenti S., Pettazzoni P., Dianzani C., Minelli R., Ciamporcero E., Roma G., Dianzani M. U., Canaparo R., et al. (2012). AS601245, an anti-inflammatory JNK inhibitor, and Clofibrate have a synergistic effect in inducing cell responses and in affecting the gene expression profile in CaCo-2 colon cancer cells. PPAR Res. 2012, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Boutros P. C. (2011). VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. L., Li X., Lehmler H. J., Phillips B., Shen D., Cui J. Y. (2018). Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol. Sci. 166, 269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. J., Seelbach M. J., Pu H., Eum S. Y., Chen L., Zhang B., Hennig B., Toborek M. (2010). Polychlorinated biphenyls disrupt intestinal integrity via NADPH oxidase-induced alterations of tight junction protein expression. Environ. Health Perspect. 118, 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., Xia J. (2018). MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong W. C., Shastri M. D., Eri R. (2017). Endoplasmic reticulum stress and oxidative stress: A vicious nexus implicated in bowel disease pathophysiology. Int. J. Mol. Sci. 18, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta C. M., Davies B. R., Hickson I., Harding T., Cosulich S., Critchlow S. E., Vincent J. P., Ellston R., Jones D., Sini P., et al. (2010). AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298. [DOI] [PubMed] [Google Scholar]
- Claus S. P., Ellero S. L., Berger B., Krause L., Bruttin A., Molina J., Paris A., Want E. J., de Waziers I., Cloarec O., et al. (2011). Colonization-induced host-gut microbial metabolic interaction. mBio 2, e00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concilli M., Iacobacci S., Chesi G., Carissimo A., Polishchuk R. (2016). A systems biology approach reveals new endoplasmic reticulum-associated targets for the correction of the ATP7B mutant causing Wilson disease. Metallomics 8, 920–930. [DOI] [PubMed] [Google Scholar]
- Cui J. Y., Klaassen C. D. (2016). RNA-seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim. Biophys. Acta 1859, 1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulsky A. V. (2017). Endoplasmic reticulum stress, the unfolded protein response and autophagy in kidney diseases. Nat. Rev. Nephrol. 13, 681–696. [DOI] [PubMed] [Google Scholar]
- Dempsey J. L., Wang D., Siginir G., Fei Q., Raftery D., Gu H., Yue Cui J. (2019). Pharmacological activation of PXR and CAR downregulates distinct bile acid-metabolizing intestinal bacteria and alters bile acid homeostasis. Toxicol. Sci. 168, 40–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., van Eunen K., Groen A. K., Venema K., Reijngoud D. J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wispelaere M., Carocci M., Liang Y., Liu Q., Sun E., Vetter M. L., Wang J., Gray N. S., Yang P. L. (2017). Discovery of host-targeted covalent inhibitors of dengue virus. Antiviral Res. 139, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrache A., Klingeman D. M., Natzke J., Rodriguez M. Jr, Giannone R. J., Hettich R. L., Davison B. H., Brown S. D. (2017). Specialized activities and expression differences for Clostridium thermocellum biofilm and planktonic cells. Sci. Rep. 7, 43583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont F. J., Su Q. (1995). Mechanism of action of the immunosuppressant rapamycin. Life Sci. 58, 373–395. [DOI] [PubMed] [Google Scholar]
- Dzierlenga, M. W., Yoon, M., Wania, F., Ward, P. L., Armitage, J. M., Wood, S. A., Clewell, H. J., and Longnecker, M. P. (2019). Quantitative bias analysis of the association of type 2 diabetes mellitus with 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB-153). Environ Int. 125, 291–299. [DOI] [PubMed] [Google Scholar]
- Ellsworth, R. E., Mamula, K. A., Costantino, N. S., Deyarmin, B., Kostyniak, P. J., Chi, L. H., Shriver, C. D., and Ellsworth, D. L. (2015). Abundance and distribution of polychlorinated biphenyls (PCBs) in breast tissue. Environ Res. 138, 291–297. [DOI] [PubMed] [Google Scholar]
- El-Sayed R. M., Ahmed H. I., Abd El-Lateef A. E. S., Ali A. A. (2019). Apoptosis perturbations and expression of regulatory inflammatory factors in cisplatin-depleted rat livers under l-arginine protection. Can. J. Physiol. Pharmacol. 97, 359–369. [DOI] [PubMed] [Google Scholar]
- Everard A., Lazarevic V., Derrien M., Girard M., Muccioli G. G., Neyrinck A. M., Possemiers S., Van Holle A., Francois P., de Vos W. M., et al. (2011). Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60, 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows R., Denizot J., Stellato C., Cuomo A., Jain P., Stoyanova E., Balazsi S., Hajnady Z., Liebert A., Kazakevych J., et al. (2018). Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaveny C. A., Murray I. A., Perdew G. H. (2010). Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol. Sci. 114, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. F., Miller G. W., Connon R. E., Geist J., Lein P. J. (2017). Transcriptomic profiling of mTOR and ryanodine receptor signaling molecules in developing zebrafish in the absence and presence of PCB 95. PeerJ 5, e4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z. D., and Cui, J. Y. (2017). Remote Sensing Between Liver and Intestine: Importance of Microbial Metabolites. Curr. Pharmacol. Rep. 3, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultang L., Gamble L. D., Gneo L., Berry A. M., Egan S. A., De Bie F., Yogev O., Eden G. L., Booth S., Brownhill S., et al. (2019). Macrophage-derived IL1beta and TNFalpha regulate arginine metabolism in neuroblastoma. Cancer Res. 79, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B. M., Pincus D., Gotthardt K., Gallagher C. M., Walter P. (2013). Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis P., Gavriil M., Rantakokko P., Ladoukakis E., Botsivali M., Kelly R. S., Bergdahl I. A., Kiviranta H., Vermeulen R. C. H., Spaeth F., et al. (2019). DNA methylation profiling implicates exposure to PCBs in the pathogenesis of B-cell chronic lymphocytic leukemia. Environ. Int. 126, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Loffredo C. A., Mitra P. S., Trnovec T., Palkovicova Murinova L., Sovcikova E., Hoffman E. P., Makambi K. H., Dutta S. K. (2018). PCB exposure and potential future cancer incidence in Slovak children: An assessment from molecular finger printing by Ingenuity Pathway Analysis (IPA(R)) derived from experimental and epidemiological investigations. Environ. Sci. Pollut. Res. Int. 25, 16493–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, F. A., He, X., Teesch, L. M., Lehmler, H. J., Robertson, L. W., and Duffel, M. W. (2015). Tissue Distribution, Metabolism, and Excretion of 3,3′-Dichloro-4′-sulfooxy-biphenyl in the Rat. Environ Sci Technol. 49, 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Hall M., Beiko R. G. (2018). 16S rRNA gene analysis with QIIME2. Methods Mol. Biol. 1849, 113–129. [DOI] [PubMed] [Google Scholar]
- Han E. S., Muller F. L., Perez V. I., Qi W., Liang H., Xi L., Fu C., Doyle E., Hickey M., Cornell J., et al. (2008). The in vivo gene expression signature of oxidative stress. Physiol. Genomics 34, 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- He Y., Li Y., Zhang S., Perry B., Zhao T., Wang Y., Sun C. (2013). Radicicol, a heat shock protein 90 inhibitor, inhibits differentiation and adipogenesis in 3T3-L1 preadipocytes. Biochem. Biophys. Res. Commun. 436, 169–174. [DOI] [PubMed] [Google Scholar]
- He Y., Peng L., Huang Y., Peng X., Zheng S., Liu C., Wu K. (2017). Association of breast adipose tissue levels of polychlorinated biphenyls and breast cancer development in women from Chaoshan, China. Environ. Sci. Pollut. Res. Int. 24, 4778–4790. [DOI] [PubMed] [Google Scholar]
- Hnia K., Gayraud J., Hugon G., Ramonatxo M., De La Porte S., Matecki S., Mornet D. (2008). l-Arginine decreases inflammation and modulates the nuclear factor-kappaB/matrix metalloproteinase cascade in mdx muscle fibers. Am. J. Pathol. 172, 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Chen J., Lu H., Zhou M., Chai Z., Hu Y. (2017). Two mTOR inhibitors, rapamycin and Torin 1, differentially regulate iron-induced generation of mitochondrial ROS. Biometals 30, 975–980. [DOI] [PubMed] [Google Scholar]
- Huang W., Yang A. H., Matsumoto D., Collette W., Marroquin L., Ko M., Aguirre S., Younis H. S. (2009). PD0325901, a mitogen-activated protein kinase kinase inhibitor, produces ocular toxicity in a rabbit animal model of retinal vein occlusion. J. Ocul. Pharmacol. Ther. 25, 519–530. [DOI] [PubMed] [Google Scholar]
- Hudson G. M., Flannigan K. L., Erickson S. L., Vicentini F. A., Zamponi A., Hirota C. L., Alston L., Altier C., Ghosh S., Rioux K. P., et al. (2017). Constitutive androstane receptor regulates the intestinal mucosal response to injury. Br. J. Pharmacol. 174, 1857–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullar M. A., Fu B. C. (2014). Diet, the gut microbiome, and epigenetics. Cancer J. 20, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke, J. C., and Hornbuckle, K. C. (2019). PCB Emissions from Paint Colorants. Environ Sci Technol. 53, 5187–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A. A. (1989). Background levels in humans In Halogenated Biphenyls, Terphenyls, Naphthalenes, Dibenzodioxins and Related Products (Kimbrough and R. D., Jensen A. A., Eds.), pp. 345–364. Elsevier Science Publishers, New York, NY. [Google Scholar]
- Jensen M. R., Schoepfer J., Radimerski T., Massey A., Guy C. T., Brueggen J., Quadt C., Buckler A., Cozens R., Drysdale M. J., et al. (2008). NVP-AUY922: A small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 10, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Dai, W., Li, Y., Hu, X., Zhu, J., Wu, P., Wang, W., and Zhang, Q. (2019). Semi-volatile organic compounds in tap water from Hangzhou, China: Influence of pipe material and implication for human exposure. Sci Total Environ. 677, 671–678. [DOI] [PubMed] [Google Scholar]
- Kania-Korwel I., Barnhart C. D., Stamou M., Truong K. M., El-Komy M. H., Lein P. J., Veng-Pedersen P., Lehmler H. J. (2012). 2,2′,3,5′,6-Pentachlorobiphenyl (PCB 95) and its hydroxylated metabolites are enantiomerically enriched in female mice. Environ. Sci. Technol. 46, 11393–11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., El-Komy M. H., Veng-Pedersen P., Lehmler H. J. (2010). Clearance of polychlorinated biphenyl atropisomers is enantioselective in female C57Bl/6 mice. Environ. Sci. Technol. 44, 2828–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008. a). Dose-dependent enantiomeric enrichment of 2,2′,3,3′,6,6′-hexachlorobiphenyl in female mice. Environ. Toxicol. Chem. 27, 299–305. [DOI] [PubMed] [Google Scholar]
- Kania-Korwel I., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008. b). Influence of dietary fat on the enantioselective disposition of 2,2′,3,3′,6,6′-hexachlorobiphenyl (PCB 136) in female mice. Food Chem. Toxicol. 46, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Xie W., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008. c). Enantiomeric enrichment of 2,2′,3,3′,6,6′-hexachlorobiphenyl (PCB 136) in mice after induction of CYP enzymes. Arch. Environ. Contam. Toxicol. 55, 510–517. [DOI] [PubMed] [Google Scholar]
- Keil K. P., Miller G. W., Chen H., Sethi S., Schmuck M. R., Dhakal K., Kim J. W., Lein P. J. (2018). PCB 95 promotes dendritic growth in primary rat hippocampal neurons via mTOR-dependent mechanisms. Arch. Toxicol. 92, 3163–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho Z. Y., Lal S. K. (2018). The human gut microbiome—A potential controller of wellness and disease. Front. Microbiol. 9, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieler I. N., Osto M., Hugentobler L., Puetz L., Gilbert M. T. P., Hansen T., Pedersen O., Reusch C. E., Zini E., Lutz T. A., et al. (2019). Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria. Sci. Rep. 9, 4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa N., Kwekel J. C., Burgoon L. D., Dere E., Williams K. J., Tashiro C., Chittim B., Zacharewski T. R. (2008). Species-specific regulation of PXR/CAR/ER-target genes in the mouse and rat liver elicited by o, p′-DDT. BMC Genomics 9, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. D., Cui J. Y. (2015). Review: Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab. Dispos. 43, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocke, C., and Lein, P. J. (2020). Evidence Implicating Non-Dioxin-Like Congeners as the Key Mediators of Polychlorinated Biphenyl (PCB) Developmental Neurotoxicity. Int J Mol Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk, A. A., Zheng, D., Shibolet, O., and Elinav, E. (2019). The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D. X., Yamori T. (2010). ZSTK474, a novel phosphatidylinositol 3-kinase inhibitor identified using the JFCR39 drug discovery system. Acta Pharmacol. Sin. 31, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrick, S. A. and Sagiv, S. K. (2008). Polychlorinated biphenyls, organochlorine pesticides and neurodevelopment. Curr Opin Pediatr. 20, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujiraoka T., Satoh Y., Ayaori M., Shiraishi Y., Arai-Nakaya Y., Hakuno D., Yada H., Kuwada N., Endo S., Isoda K., et al. (2013). Hepatic extracellular signal-regulated kinase 2 suppresses endoplasmic reticulum stress and protects from oxidative stress and endothelial dysfunction. J. Am. Heart Assoc. 2, e000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. G., Chain F., Martin R., Bermudez-Humaran L. G., Courau S., Langella P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 16, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Bae G. H., Kim Y. S., Kim H. S., Lee M., Ghim J., Zabel B. A., Ryu S. H., Bae Y. S. (2019). A phospholipase D2 inhibitor, CAY10594, ameliorates acetaminophen-induced acute liver injury by regulating the phosphorylated-GSK-3beta/JNK axis. Sci. Rep. 9, 7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng L., Li J., Luo X. M., Kim J. Y., Li Y. M., Guo X. M., Chen X., Yang Q. Y., Li G., Tang N. J. (2016). Polychlorinated biphenyls and breast cancer: A congener-specific meta-analysis. Environ. Int. 88, 133–141. [DOI] [PubMed] [Google Scholar]
- Levy D. S., Kahana J. A., Kumar R. (2009). AKT inhibitor, GSK690693, induces growth inhibition and apoptosis in acute lymphoblastic leukemia cell lines. Blood 113, 1723–1729. [DOI] [PubMed] [Google Scholar]
- Li C. Y., Lee S., Cade S., Kuo L. J., Schultz I. R., Bhatt D. K., Prasad B., Bammler T. K., Cui J. Y. (2017. a). Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals polybrominated diphenyl ethers. Drug Metab. Dispos. 45, 1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Qi T., Huang Z. S., Ying Y., Zhang Y., Wang B., Ye L., Zhang B., Chen D. L., Chen J. (2017. b). Relationship between gut microbiota and type 2 diabetic erectile dysfunction in Sprague-Dawley rats. J. Huazhong Univ. Sci. Technol. Med. Sci. 37, 523–530. [DOI] [PubMed] [Google Scholar]
- Li, Z. M., Hernandez-Moreno, D., Main, K. M., Skakkebaek, N. E., Kiviranta, H., Toppari, J., Feldt-Rasmussen, U., Shen, H., Schramm, K. W., and De Angelis, M. (2018). Association of In Utero Persistent Organic Pollutant Exposure With Placental Thyroid Hormones. Endocrinology 159, 3473–3481. [DOI] [PubMed] [Google Scholar]
- Liang M., Wang Z., Li H., Cai L., Pan J., He H., Wu Q., Tang Y., Ma J., Yang L. (2018). l-Arginine induces antioxidant response to prevent oxidative stress via stimulation of glutathione synthesis and activation of Nrf2 pathway. Food Chem. Toxicol. 115, 315–328. [DOI] [PubMed] [Google Scholar]
- Liu H. X., Keane R., Sheng L., Wan Y. J. (2015). Implications of microbiota and bile acid in liver injury and regeneration. J. Hepatol. 63, 1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R., Matthews J. (2012). High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol. Sci. 130, 349–361. [DOI] [PubMed] [Google Scholar]
- Love D. C., Hanover J. A. (2005). The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE 2005, re13. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich J. M., Parks D. J., Moore L. B., Collins J. L., Goodwin B., Billin A. N., Stoltz C. A., Kliewer S. A., Lambert M. H., Willson T. M., et al. (2003). Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J. Biol. Chem. 278, 17277–17283. [DOI] [PubMed] [Google Scholar]
- Mandal S., Van Treuren W., White R. A., Eggesbo M., Knight R., Peddada S. D. (2015). Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 26, 27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina P. J., Goodin S. (2008). Lapatinib: A dual inhibitor of human epidermal growth factor receptor tyrosine kinases. Clin. Ther. 30, 1426–1447. [DOI] [PubMed] [Google Scholar]
- Meek, E. C., Jones, D. D., Crow, J. A., Wills, R. W., Cooke, W. H., 3rd, and Chambers, J. E. (2019). Association of serum levels of p,p'- Dichlorodiphenyldichloroethylene (DDE) with type 2 diabetes in African American and Caucasian adult men from agricultural (Delta) and non-agricultural (non-Delta) regions of Mississippi. J Toxicol Environ Health A 82, 387–400. [DOI] [PubMed] [Google Scholar]
- Mena A. C., Pulido E. G., Guillen-Ponce C. (2010). Understanding the molecular-based mechanism of action of the tyrosine kinase inhibitor: Sunitinib. Anticancer Drugs 21(Suppl. 1), S3–11. [DOI] [PubMed] [Google Scholar]
- Milner J. A., Perkins E. G. (1978). Liver lipid alterations in rats fed arginine deficient diets. Lipids 13, 563–565. [DOI] [PubMed] [Google Scholar]
- Mitruka B. M., Costilow R. N. (1967). Arginine and ornithine catabolism by Clostridium botulinum. J. Bacteriol. 93, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney J. N., Cotter T. G. (2018). ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 80, 50–64. [DOI] [PubMed] [Google Scholar]
- Neumann-Schaal M., Jahn D., Schmidt-Hohagen K. (2019). Metabolism the difficile way: The key to the success of the pathogen Clostridioides difficile. Front. Microbiol. 10, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno T., Kakehashi A., Ishii N., Fujioka M., Gi M., Wanibuchi H. (2018). mTOR activation in liver tumors is associated with metabolic syndrome and non-alcoholic steatohepatitis in both mouse models and humans. Cancers (Basel) 10, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M., Neyrinck A. M., Potgens S. A., Beaumont M., Salazar N., Cani P. D., Bindels L. B., Delzenne N. M. (2018). The DPP-4 inhibitor vildagliptin impacts the gut microbiota and prevents disruption of intestinal homeostasis induced by a Western diet in mice. Diabetologia 61, 1838–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrondo R., de Las Pozas A., Reiner T., Perez-Stable C. (2013). ABT-737, a small molecule Bcl-2/Bcl-xL antagonist, increases antimitotic-mediated apoptosis in human prostate cancer cells. PeerJ 1, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriello M. C., Brandon J. A., Hoffman J., Wang C., Tripathi H., Abdel-Latif A., Ye X., Li X., Yang L., Lee E., et al. (2018). Dioxin-like PCB 126 increases systemic inflammation and accelerates atherosclerosis in lean LDL receptor-deficient mice. Toxicol. Sci. 162, 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. C., Dheer R., Santaolalla R., Davies J. M., Burgueno J., Lang J. K., Toborek M., Abreu M. T. (2018). Intestinal exposure to PCB 153 induces inflammation via the ATM/NEMO pathway. Toxicol. Appl. Pharmacol. 339, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E., Monaikul S., Kostyniak P. J., Chi L. H., Schantz S. L., Sable H. J. (2013). Developmental exposure to polychlorinated biphenyls reduces amphetamine behavioral sensitization in Long-Evans rats. Neurotoxicol. Teratol. 38, 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J., Caron S., Staels B. (2011). Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr. Diabetes Rep. 11, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulugulla S. H., Workman R., Rutter N. W., Yang Z., Adamik J., Lupish B., Macar D. A., El Abdouni S., Esposito E. X., Galson D. L., et al. (2018). A combined computational and experimental approach reveals the structure of a C/EBPbeta-Spi1 interaction required for IL1B gene transcription. J. Biol. Chem. 293, 19942–19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F. O. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. L., Zhang, C., Smarr, M. M., Lee, S., Honda, M., Kannan, K., Tekola-Ayele, F., and Buck Louis, G. M. (2019). Persistent organic pollutants and gestational diabetes: A multi-center prospective cohort study of healthy US women. Environ Int. 124, 249–258. [DOI] [PubMed] [Google Scholar]
- Ramirez, T., Daneshian, M., Kamp, H., Bois, F. Y., Clench, M. R., Coen, M., Donley, B., Fischer, S. M., Ekman, D. R., Fabian, E., Guillou, C., Heuer, J., Hogberg, H. T., Jungnickel, H., Keun, H. C., Krennrich, G., Krupp, E., Luch, A., Noor, F., Peter, E., Riefke, B., Seymour, M., Skinner, N., Smirnova, L., Verheij, E., Wagner, S., Hartung, T., van Ravenzwaay, B., and Leist, M. (2013). Metabolomics in toxicology and preclinical research. ALTEX 30, 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid H. O., Yadav R. K., Kim H. R., Chae H. J. (2015). ER stress: Autophagy induction, inhibition and selection. Autophagy 11, 1956–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/.
- Reid T. S., Beese L. S. (2004). Crystal structures of the anticancer clinical candidates R115777 (Tipifarnib) and BMS-214662 complexed with protein farnesyltransferase suggest a mechanism of FTI selectivity. Biochemistry 43, 6877–6884. [DOI] [PubMed] [Google Scholar]
- Richards A. L., Muehlbauer A. L., Alazizi A., Burns M. B., Findley A., Messina F., Gould T. J., Cascardo C., Pique-Regi R., Blekhman R., et al. (2019). Gut microbiota has a widespread and modifiable effect on host gene regulation. mSystems 4, e00323–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. D., Bostrom P., O’Sullivan J. F., Schinzel R. T., Lewis G. D., Dejam A., Lee Y. K., Palma M. J., Calhoun S., Georgiadi A., et al. (2014). beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 19, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Guo J., Huang Y., Mao Y., Yang Z., Zuo Z. (2020). Adolescent exposure to environmental level of PCBs (Aroclor 1254) induces non-alcoholic fatty liver disease in male mice. Environ. Res. 181, 108909. [DOI] [PubMed] [Google Scholar]
- Sable H. J., Monaikul S., Poon E., Eubig P. A., Schantz S. L. (2011). Discriminative stimulus effects of cocaine and amphetamine in rats following developmental exposure to polychlorinated biphenyls (PCBs). Neurotoxicol. Teratol. 33, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kimono D., Albadrani M., Seth R. K., Busbee P., Alghetaa H., Porter D. E., Scott G. I., Brooks B., Nagarkatti M., et al. (2019). Environmental microcystin targets the microbiome and increases the risk of intestinal inflammatory pathology via NOX2 in underlying murine model of Nonalcoholic Fatty Liver Disease. Sci. Rep. 9, 8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A., Sabatini D. M. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade A. E., Schieven G. L., Townsend R., Jankowska A. M., Susulic V., Zhang R., Szpurka H., Maciejewski J. P. (2008). Dasatinib, a small-molecule protein tyrosine kinase inhibitor, inhibits T-cell activation and proliferation. Blood 111, 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, T. T., Blawas, A. M., Gray, K., Heindel, J. J., and Lawler, C. P. (2015). Elucidating the links between endocrine disruptors and neurodevelopment. Endocrinology 156, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T. W., Akinaga S., Soga S., Sullivan W., Stensgard B., Toft D., Neckers L. M. (1998). Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville D. K., Li C. Y., Wang D., Dempsey J. L., Raftery D., Mani S., Gu H., Cui J. Y. (2019). Polybrominated diphenyl ethers and gut microbiome modulate metabolic syndrome-related aqueous metabolites in mice. Drug Metab. Dispos. 47, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia-Mendoza M., Gonzalez-Gonzalez M. E., Barrera D., Diaz L., Garcia-Becerra R. (2015). Efficacy and mechanism of action of the tyrosine kinase inhibitors gefitinib, lapatinib and neratinib in the treatment of HER2-positive breast cancer: Preclinical and clinical evidence. Am. J. Cancer Res. 5, 2531–2561. [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cheng S. L., Klaassen C. D., Cui J. Y. (2016). Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab. Dispos. 44, 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Jan J., Hardesty J. E., Falkner K. C., Prough R. A., Balamurugan A. N., Mokshagundam S. P., Chari S. T., Cave M. C. (2019). Polychlorinated biphenyl exposures differentially regulate hepatic metabolism and pancreatic function: Implications for nonalcoholic steatohepatitis and diabetes. Toxicol. Appl. Pharmacol. 363, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Wang S., Jasbi P., Turner C., Hrovat J., Wei Y., Liu J., Gu H. (2019). Database-assisted globally optimized targeted mass spectrometry (dGOT-MS): Broad and reliable metabolomics analysis with enhanced identification. Anal. Chem. 91, 13737–13745. [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos, P., Bess, E. N., Carmody, R. N., and Turnbaugh, P. J. (2016). The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremy, M., Sutova, Z., Murinova, L. P., Richterova, D., Wimmerova, S., Conka, K., Drobna, B., Fabelova, L., Jureckova, D., Jusko, T. A., Tihanyi, J., and Trnovec, T. (2019). The spatial distribution of congener-specific human PCB concentrations in a PCB-polluted region. Sci Total Environ. 651, 2292–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Narayan R., Corsello S. M., Peck D. D., Natoli T. E., Lu X., Gould J., Davis J. F., Tubelli A. A., Asiedu J. K., et al. (2017). A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. M., Nguyen Q. H., Singh M., Pavesic M. W., Nesterenko I., Nelson L. J., Liao A. C., Razorenova O. V. (2017). Rho-associated kinase 1 inhibition is synthetically lethal with von Hippel-Lindau deficiency in clear cell renal cell carcinoma. Oncogene 36, 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornevi, A., Sommar, J., Rantakokko, P., Akesson, A., Donat-Vargas, C., Kiviranta, H., Rolandsson, O., Rylander, L., Wennberg, M., and Bergdahl, I. A. (2019). Chlorinated persistent organic pollutants and type 2 diabetes - A population-based study with pre- and post- diagnostic plasma samples. Environ Res. 174, 35–45. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg A. B., Stroomer A. E., Abeling N. G., van Gennip A. H. (2006). A pivotal role for beta-aminoisobutyric acid and oxidative stress in dihydropyrimidine dehydrogenase deficiency? Nucleosides Nucleotides Nucleic Acids 25, 1103–1106. [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y., Gonzalez A., Smarr L., McDonald D., Morton J. T., Navas-Molina J. A., Knight R. (2017). Bringing the dynamic microbiome to life with animations. Cell Host Microbe 21, 7–10. [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. (2013). EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A. P., Qiu Z., Maher L., Redinbo M. R., Phillips R. S., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesha S. H., Moudgil K. D. (2016). Celastrol and its role in controlling chronic diseases. Adv. Exp. Med. Biol. 928, 267–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti A., Le Roy C. I., Rosa F., Rossi N., Martin T. C., Mohney R. P., Li W., de Rinaldis E., Bell J. T., Venter J. C., et al. (2019). Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10, 4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B., Falkner K. C., Clair H. B., Al-Eryani L., Prough R. A., States J. C., Coslo D. M., Omiecinski C. J., Cave M. C. (2014). Human receptor activation by Aroclor 1260, a polychlorinated biphenyl mixture. Toxicol. Sci. 140, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lang T., Shen J., Dai J., Tian L., Wang X. (2019). Core gut bacteria analysis of healthy mice. Front. Microbiol. 10, 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Clark N. R., Ma’ayan A. (2016). Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics 32, 2338–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann M., Wassermann D., Cucos S., Miller H. J. (1979). World PCBs map: Storage and effects in man and his biologic environment in the 1970s. Ann. NY Acad. Sci. 320, 69–124. [DOI] [PubMed] [Google Scholar]
- Weber, R., Herold, C., Hollert, H., Kamphues, J., Blepp, M., and Ballschmiter, K. (2018). Reviewing the relevance of dioxin and PCB sources for food from animal origin and the need for their inventory, control and management. Environ Sci Eur. 30, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldy C. S., Luttrell I. P., White C. C., Morgan-Stevenson V., Bammler T. K., Beyer R. P., Afsharinejad Z., Kim F., Chitaley K., Kavanagh T. J. (2012). Glutathione (GSH) and the GSH synthesis gene Gclm modulate vascular reactivity in mice. Free Radic. Biol. Med. 53, 1264–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York, NY.. [Google Scholar]
- Wilking M., Ndiaye M., Mukhtar H., Ahmad N. (2013). Circadian rhythm connections to oxidative stress: Implications for human health. Antioxid. Redox Signal. 19, 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Luo C., Kolde R., d’Hennezel E., Annand J. W., Heim C. E., Krastel P., Schmitt E. K., Omar A. S., Creasey E. A., et al. (2017). Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 22, 25–37.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Wang C., Ding L., Shen Y., Cui H., Wang M., Wang H. (2016). Arginine relieves the inflammatory response and enhances the casein expression in bovine mammary epithelial cells induced by lipopolysaccharide. Mediators Inflamm. 2016, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Barnhart C., Lein P. J., Lehmler H. J. (2015). Hepatic metabolism affects the atroposelective disposition of 2,2′,3,3′,6,6′-hexachlorobiphenyl (PCB 136) in mice. Environ. Sci. Technol. 49, 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Duffel M., Lehmler H. J. (2013). Oxidation of polychlorinated biphenyls by liver tissue slices from phenobarbital-pretreated mice is congener-specific and atroposelective. Chem. Res. Toxicol. 26, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Sun R., Chen Y., Zheng X., Bai L., Lian Z., Wei H., Tian Z. (2015). Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology 62, 253–264. [DOI] [PubMed] [Google Scholar]
- Xu D., Su C., Song X., Shi Q., Fu J., Hu L., Xia X., Song E., Song Y. (2015). Polychlorinated biphenyl quinone induces endoplasmic reticulum stress, unfolded protein response, and calcium release. Chem. Res. Toxicol. 28, 1326–1337. [DOI] [PubMed] [Google Scholar]
- Xu J. Y., Xu Z., Liu X., Tan M., Ye B. C. (2018). Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol. Cell. Proteomics 17, 1156–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Feng B., Li P., Tang Z., Wang L. (2016). Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: An animal study. J. Diabetes Res. 2016, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Kim K. H., Phimister A., Bachstetter A. D., Ward T. R., Stackman R. W., Mervis R. F., Wisniewski A. B., Klein S. L., Kodavanti P. R., et al. (2009). Developmental exposure to polychlorinated biphenyls interferes with experience-dependent dendritic plasticity and ryanodine receptor expression in weanling rats. Environ. Health Perspect. 117, 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wei J., Wu Y., Wang Z., Guo Y., Lee P., Li X. (2015). Metformin induces ER stress-dependent apoptosis through miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis 4, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Xie L., Li Y., Wei C. (2009). More than 9,000,000 unique genes in human gut bacterial community: Estimating gene numbers inside a human body. PLoS One 4, e6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Major M. B., Takanashi S., Camp N. D., Nishiya N., Peters E. C., Ginsberg M. H., Jian X., Randazzo P. A., Schultz P. G., et al. (2007). Small-molecule synergist of the Wnt/beta-catenin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 7444–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xiao X., Li M., Yu M., Ping F., Zheng J., Wang T., Wang X. (2017). Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS One 12, e0184735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Djukovic D., Deng L., Gu H., Himmati F., Chiorean E. G., Raftery D. (2014). Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 13, 4120–4130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.