Highlights
-
•
A fully automated segmentation of new or enlarged multiple sclerosis (MS) lesions.
-
•
3D convolutional neural network (CNN) with U-net-like encoder-decoder architecture.
-
•
Simultaneous processing of baseline and follow-up scan of the same patient.
-
•
Trained on 3253 patient data from over 103 different MR scanners.
-
•
Fast (<1min), robust algorithm with segmentation results in inter-rater variability.
Keywords: Multiple sclerosis, Lesion activity, Convolutional neural network, U-net, Lesion segmentation
Abstract
The quantification of new or enlarged lesions from follow-up MRI scans is an important surrogate of clinical disease activity in patients with multiple sclerosis (MS). Not only is manual segmentation time consuming, but inter-rater variability is high. Currently, only a few fully automated methods are available. We address this gap in the field by employing a 3D convolutional neural network (CNN) with encoder-decoder architecture for fully automatic longitudinal lesion segmentation.
Input data consist of two fluid attenuated inversion recovery (FLAIR) images (baseline and follow-up) per patient. Each image is entered into the encoder and the feature maps are concatenated and then fed into the decoder. The output is a 3D mask indicating new or enlarged lesions (compared to the baseline scan). The proposed method was trained on 1809 single point and 1444 longitudinal patient data sets and then validated on 185 independent longitudinal data sets from two different scanners. From the two validation data sets, manual segmentations were available from three experienced raters, respectively. The performance of the proposed method was compared to the open source Lesion Segmentation Toolbox (LST), which is a current state-of-art longitudinal lesion segmentation method.
The mean lesion-wise inter-rater sensitivity was 62%, while the mean inter-rater number of false positive (FP) findings was 0.41 lesions per case. The two validated algorithms showed a mean sensitivity of 60% (CNN), 46% (LST) and a mean FP of 0.48 (CNN), 1.86 (LST) per case. Sensitivity and number of FP were not significantly different (p < 0.05) between the CNN and manual raters.
New or enlarged lesions counted by the CNN algorithm appeared to be comparable with manual expert ratings. The proposed algorithm seems to outperform currently available approaches, particularly LST. The high inter-rater variability in case of manual segmentation indicates the complexity of identifying new or enlarged lesions. An automated CNN-based approach can quickly provide an independent and deterministic assessment of new or enlarged lesions from baseline to follow-up scans with acceptable reliability.
1. Introduction
Over the last decade, magnetic resonance imaging (MRI) has become a key tool in the diagnosis and disease monitoring of multiple sclerosis (MS) (Polman et al., 2011). New or enlarged MS lesions (also referred to as lesion activity) from T2-weighted MRI follow-up scans are the most important surrogate marker of clinical disease activity in MS patients (Fahrbach et al., 2013). While, new or enlarged lesion have traditionally served as a primary endpoint in phase II studies, T2 lesion evolution has recently been suggested as a new primary end point in MS phase III clinical trials by experts in the field (Sormaniet al., 2013, Sormani et al., 2014). Fluid attenuated inversion recovery (FLAIR) sequences have begun to complement or even replace standard T2-weighted imaging, since FLAIR suppresses not only the signal originating from cerebrospinal fluid but also blood flow effects, thereby improving the detection of white matter, and possibly even grey matter lesions (Gramsch et al., 2015).
Manual segmentation of lesion activity from follow-up MRI scans is time consuming and inter-rater variability is high (Egger et al., 2017). Numerous automated methods are currently available for cross-sectional detection and quantification of MS lesions (Schmidtet al., 2012, Shieeet al., 2010, Van Leemputet al., 2001, Lao et al., 2008, Griffanti et al., 2016, Cabezaset al., 2014, Salemet al., 2017, Rouraet al., 2015). However, only a few fully automated algorithms have been provided for the longitudinal quantification of lesion activity from repeated MRI scans so far. Among other methods, image differences (Battagliniet al., 2014, Ganileret al., 2014) and deformation fields (Boscet al., 2003, Cabezaset al., 2016, Salemet al., 2017) have been used to detect new lesions. Also, intensity-based approaches using local context between scans have been proposed (Lesjak et al., 2016). Overall, methods for detection of lesion growth have largely relied on classic image processing methods so far (Cheng et al., 2018, Schmidt et al., 2019). Elliott et al. (2013) used classical learning methods – employing a Bayesian classifier and a random-forest based lesion-level classification.
Latest developments in deep-learning based methods have led to an increase in the number of available automated segmentation tools, with convolutional neural networks (CNNs) gaining popularity as a promising method (Akkuset al., 2017, Danelakiset al., 2018, Litjens et al., 2017, Valverdeet al., 2017, Chen et al., 2018, Isenseeet al., 2018, Kamnitsaset al., 2017). Currently for the segmentation purposes, the most commonly used networks follow a U-net- like architecture (Ronneberger et al., 2015, Broschet al., 2016) with an encoder-decoder structure and long-range connections between the encoder and decoder. The network usually uses (a portion of) an MRI scan as input and produces a segmentation mask indicating lesioned tissue as an output (Danelakis et al., 2018).
As of yet, deep learning methods have only considered lesion segmentation from a single MRI volume. The most intuitive approach to derive the lesion activity between two scans is to subtract lesion maps from two independent segmentation masks of both scans (baseline and follow-up) (Jainet al., 2016, Ganileret al., 2014, Köhler et al., 2019). However, this approach is associated with high variability and inconsistency (García-Lorenzo et al., 2013). Others have tried to explicitly incorporate information from both MRI volumes, e. g., intensity-based approaches considering local context between volumes have shown promising results (Lesjak et al., 2016). Overall, longitudinal methods have relied primarily on classical image processing methods until today (Cheng et al., 2018).
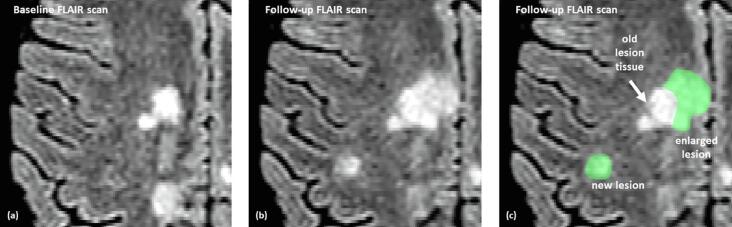
This study utilizes a fully convolutional neural network approach to segment new or enlarged lesions using two MRI scans from patients with a confirmed diagnosis of MS, acquired at different time points. A baseline (BL) and follow-up (FU) FLAIR volume was provided as input into the network, while the output produced a segmentation mask indicating new or enlarged lesions. Fig. 1 shows an example of the targeted lesion segmentation in green.
Fig. 1.
Example of input data for one patient: each set consists of a baseline (a) and a follow-up (b) FLAIR scan. The ideal segmentation (green, (c)) indicates new or enlarged lesions in the follow-up scan in contrast to the baseline scan. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A U-net-like encoder-decoder architecture was used, that combines the output of the encoder for both scans (BL, FU), before feeding those features into the decoder. The proposed algorithm was evaluated on 185 independent pairs of MRI follow-up data from two different clinical studies. For validation, its performance was compared with the lesion segmentation toolbox (LST) (http://www.applied-statistics.de/lst.htm, Schmidt et al. (2012, 2019)), which is available under the Statistical Parametric Mapping (SPM12, https://www.fil.ion.ucl.ac.uk/spm/software/spm12/, Friston et al. (2007)) software package.
The purpose of our study was to introduce our deep learning methods for the detection of new or enlarged lesions and compare it with a current state-of-the-art non-deep-learning method.
2. Material and methods
2.1. Data
For the training and evaluation of the proposed method, several data sets were available.
2.1.1. Training data
2.1.1.1. Routine data 1 (Rou1), 1 time point
1809 2D and 3D single time point FLAIR images were acquired in clinical routine on 156 different MRI scanners. The mean patient age was 45.75 (±15.12) years. MRIs were sent to jung diagnostics GmbH for image analysis. The images were anonymized and the ground truth for the MS lesion segmentation were annotated semi-automatically during the manual quality control process of jung diagnostics GmbH.
2.1.1.2. Routine data 2 (Rou2), longitudinal
1444 predominantly MS patient data sets with follow-up FLAIR image pairs (BL and FU) were available (acquired on 103 different MRI scanners with 18 different models; excluded scanner models are Philips Ingenia and SIEMENS Verio). The mean patient age was 43.22 (±12.46) years and the mean follow-up time was 1.16 (±0.60) years. The data originated from clinical routine and were sent to the company jung diagnostics GmbH for image analysis. The images were anonymized and the ground truth for the new or enlarged lesions (BL/FU) annotated semi-automatically during the manual quality control process of jung diagnostics GmbH.
2.1.1.3. Philips Ingenia (PhIng), longitudinal
130 predominantly MS patient data sets with follow-up FLAIR image pairs (BL and FU) were acquired on 13 different 3.0 T Philips Ingenia scanners. The mean patient age was 40.44 (±12.05) years and the mean follow-up time was 0.98 (±0.38) years. The data originated from clinical routine and were sent to the company jung diagnostics GmbH for image analysis. The images were anonymized and the ground truth for the new or enlarged lesions (BL/FU) annotated semi-automatically during the manual quality control process of jung diagnostics GmbH.
2.1.2. Evaluation data
2.1.2.1. Zurich data (ZURICH), longitudinal
This data set composed of 89 patient data is part of an observational study on MS heterogeneity carried out at the University Hospital of Zurich, Switzerland. All images were acquired with a 3.0 T Philips Ingenia Scanner (Philips, Eindhoven, the Netherlands). The mean age of the patients was 34.22 (±8.72) years with a mean follow-up time of 2.24 (±1.17) years. The MRI data were annotated by three independent raters. The ZURICH data is not included in the PhIng data set.
2.1.2.2. Dresden data (DRESDEN), longitudinal
For 32 patients 4 scans at 4 time points were performed on a 3 T Siemens Verio scanner. The subjects with a mean age of 39.64 (±10.78) years were scanned at University Hospital Carl Gustav Carus at Technische Universität Dresden, Germany. The mean follow-up time between the 3 scan pairs (between time point 1 and 2, time point 2 and 3 and time point 3 and 4) was 1.01 (±0.08) years. For the resulting 96 scan pairs (32 patients × 3 scan pairs) three different annotations were available from three independent raters. Additionally, the scan pairs between time point 1 and time point 4 with a mean follow-up time of 3.02 (±0.12) were annotated by the three raters.
The routine data (Rou2) do not contain the scanner types Philips Ingenia and SIEMENS Verio since these scanners are used for the evaluation data. Furthermore, the training data set PhIng is exclusively acquired on the Philips Ingenia scanner (but not the same machine as used for the ZURICH data set). Table 1 summarizes the described data sets. A more detailed description of all involved scanner models and applied protocols of the training and evaluation data is presented in Table 1 of the supplementary.
Table 1.
Summary of the available evaluation (ZURICH and DRESDEN) and training (Rou2 and PhIng) data with number of cases (BL, FU pairs) with and without new or enlarged lesions, mean new or enlarged lesion count per case, mean lesion size and mean time between the two scans (BL and FU). For the DRESDEN data the values for an additional evaluation between time point one and four are also given (DRESDEN 1-4).
| Sets | #cases | #new/enl. lesions | #new/enl. Lesion per case | lesion size | follow-up time | |
|---|---|---|---|---|---|---|
| Training sets: | ||||||
| Rou2 (without SIEM. | all cases | 1444 | 2447 | 1.69 (±4.59) | 1.16 (±0.60) yr. | |
| Verio, Ph. Ingenia) | cases with > 1 lesion | 614 | 2447 | 3.98 (±6.36) | 0.18 (±0.54) ml | |
| PhIng (Ph. Ingenia) | all cases | 130 | 288 | 2.21 (±5.60) | 0.98 (±0.38) yr. | |
| cases with > 1 lesion | 55 | 288 | 5.23 (±7.68) | 0.19 (±0.50) ml | ||
| Evaluation sets: | ||||||
| ZURICH | all cases | 89 | 209 | 2.34 (±4.44) | 2.24 (±1.17) yr. | |
| (Ph. Ingenia) | cases with > 1 lesion | 51 | 209 | 4.09 (±5.23) | 0.13 (±0.14) ml | |
| DRESDEN | all cases | 96 | 157 | 1.67 (±2.49) | 1.01 (±0.08) yr. | |
| (SIEM. Verio) | cases with > 1 lesion | 53 | 157 | 2.96 (±2.69) | 0.13 (±0.18) ml | |
| DRESDEN 1–4 | all cases | 32 | 108 | 3.36 (±3.74) | 3.02 (±0.12) yr. | |
| (SIEM. Verio) | cases with > 1 lesion | 26 | 108 | 4.13 (±3.75) | 0.12 (±0.19) ml | |
2.2. Manual annotation of data
For the MR scans with only one available time point (data set Rou1), a semi-automatic threshold-based method for the lesion segmentation was employed. Subsequently, the results were checked manually by experienced raters. For the new or enlarged lesions, all FLAIR data sets with two time points were segmented by one to three experienced raters. For diagnostic purposes, the number of new or enlarged lesions in the FU scan was evaluated, therefore, the BL scan was co-registered to the FU scan rigidly using SPM12, to get the resulting masks in the FU space. The annotated ground truth for all the images follows the scheme detailed in Fig. 1: only regions with new or enlarged lesion tissue was marked. The ground truth was encoded as a 1/0-mask, where new appearing lesion tissue was marked with 1-values and healthy (without lesions) brain tissue as well as old lesion tissue (already present in the BL scan) were not segmented (0-values) (see Fig. 1).
The annotation of new or enlarged lesions was performed with a tool, which was developed by MEVIS specifically for this task (MEVIS, Fraunhofer Institute for Digital Medicine, Lübeck, Germany). The co-registered FLAIR images (BL and FU) are loaded simultaneously into the viewer and then displayed side by side. All image interactions like scrolling or adjusting window-level-setting are coupled between the image pair. To further facilitate the comparison between BL and FU scan the user can toggle in one of the viewers between the BL and FU image. This way changes in the images can be detected easily. Newly appearing lesion tissue are then annotated by an interactive thresholding tool. With a brush tool the user draws an arbitrary region of interest. Within this region a threshold is applied. The user then interactively adjusts the threshold in order to optimally fit the resulting segmentation to the boundary of the lesion. These annotations do not distinguish between newly appearing lesions (left annotation in Fig. 1(c)) and newly appearing lesion tissue which is connected to an already existing lesion (right annotation in Fig. 1(c)). The latter would be considered as an enlarged lesion. However, in the evaluation of the algorithm in the next section we do not distinguish between new and enlarged lesions.
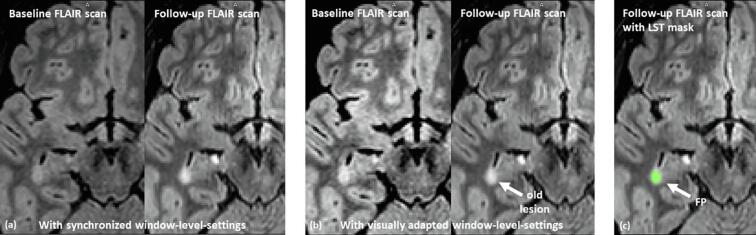
New lesions were only annotated if the diameter exceeded 3 mm, which corresponds to a lesion volume of 0.01 ml (rounded). This minimal lesion size was recently suggested by Rovira et al. (2015) for defining a new or enlarged lesion. In addition, new appearing lesion tissue (connected to an existing lesion) was only annotated if the increase in size and shape was pronounced and not explainable by variations of the image acquisition such as different angulations or changes in image contrast (see Fig. 7 (a) as an example).
Fig. 7.
Example of problematic signal value changes between the BL and FU FLAIR scans: the images in (a) visualize the BL and FU scan with the same level-windows settings for both scans. This shows that the contrast and the signal value distribution between the two scans changed. If the window-level-setting are adjusted (b) to create a similar visual impression between the images, it becomes obvious that the lesion in this image slide has already be present in the BL scan. The CNN-based method correctly identified this lesion as old lesion tissue while the LST-method (c) detected a false positive (FP) new lesion. In the whole MR scan LST found 13 FP while our CNN method found no FP.
2.3. Convolutional neural network
Since the voxel spacing and slice thickness of the FLAIR scans were very heterogeneous, all co-registered scans (and their segmentation masks) were re-sampled to an isotropic 3D volume with spacing of 1 mm × 1 mm × 1 mm. The signal values of each volume were standardized individually to have zero mean and unit variance.
In our preparatory work, we focused on the topic of MS lesion segmentation at a single point in time. For this work, experiments were carried out to determine the optimal patch size for the problem of lesion segmentation. When comparing 643 voxels, 963 voxels, 1283 voxels and 1603 voxel patch-sizes, the method showed best results for 1283 voxels. Therefore, we decided to also use a patch-wise approach with a patch-size of 128 × 128 × 128 voxels (=128 mm × 128 mm × 128 mm) for the longitudinal lesion segmentation. In addition, results from lesion segmentation based on only FLAIR data were compared to results based on coupled FLAIR and T1 data. The evaluation showed that FLAIR images contain sufficient information and additional use of T1 data did not bring any further advantages.
2.3.1. Architecture
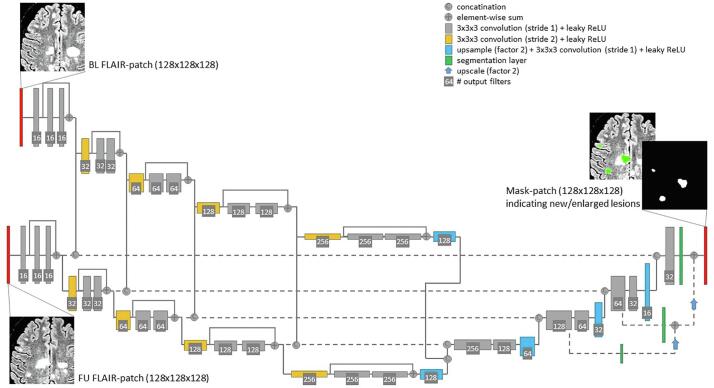
The employed network follows a fully convolutional encoder-decoder (U-net-like) architecture with 3D convolutions with 3 × 3 × 3 kernel size. Residual blocks are used (He et al., 2015) in the encoder. In addition, deep supervision (Dou et al., 2017) is employed by including additional segmentation layers at several stages in the decoder. Dou et al. (2017) showed that deep supervision increased the stability of the training process for 3D volume-to-volume learning, since 3D convolutional neural networks suffer from the optimization problem of gradients vanishing or exploding. A preliminary experiment confirmed that without deep supervision the convergence behaviour of the loss function is worse than with deep supervision and was hence included in the architecture. Fig. 2 visualizes the proposed architecture.
Fig. 2.
The proposed network: a fully convolutional encoder-decoder architecture with 3D convolutions, residual-block-connections and four reductions of the feature map size. The two input images (BL and FU FLAIR-patch) are fed into the same encoder path – both visualized paths have shared weights. After each residual-block the feature maps for each input are concatenated and fed into the decoder, respectively. As an output a segmentation mask is predicted indicating new and enlarged lesions.
The encoder reduces the spatial feature map size four times (using convolution with stride 2) and doubles the feature map number with each reduction. Starting with 16 feature maps of size 128 × 128 × 128 in the first layer, this leads to 256 maps of size 8 × 8 × 8 in the last encoder layer. The two input volumes (BL, FU) are fed into the same encoder-path independently (in Fig. 2 the two encoder paths have shared weights). Both feature maps are concatenated before being fed into the decoder.
The decoder uses convolution layers, followed by nearest-neighbour up-sampling and deep supervision in three layers (Dou et al., 2017). For the long-range connections between encoder and decoder a feature concatenation is employed. Furthermore, after each scaling-level of the encoder the feature maps of the BL and FU scan are concatenated before going into the long-range connections.
2.3.2. Parameters
As the activation function a leaky ReLU (Maas et al., 2013) is used in each layer. Due to the rather large patches of 128 × 128 × 128 voxels the batch size is 1. Therefore, instance normalization, a special case of group normalization, is used instead of batch normalization. The model is trained using the Adam optimizer (“Adaptive Moment Estimation”, a version of stochastic gradient descent (Kingma and Ba, 2014)) with a cross-entropy loss function and batch size of 1 for Ne = 200 epochs. A starting learning rate of α0 = 10−4 is used with an exponential decay of α = α0(1 – e/Ne)0.9 with e as current epoch.
2.3.3. Data augmentation
For the purpose of data augmentation, the input patches of 128 × 128 × 128 voxels are randomly cropped from the entire scan volumes. The patches are flipped randomly around both axes of the axial plane. Furthermore, random noise and a random “bias field” are added, to handle inhomogeneous signal distributions between the two scans. To deal with a non-perfect rigid alignment between BL and FU scan, we randomly translate one of the scans by 0, 1 or 2 mm (randomly chosen) in each dimension.
2.4. Training and pre-training
2.4.1. Pre-training of encoder path with single time point data
The task of finding new or enlarged lesions is related to finding lesions in FLAIR scans at a single time point. Similar image features (convolution filters of the encoder) should be of interest for segmentation algorithms in both tasks. Therefore, the encoder was pre-trained with single time point images (routine data set with 1 time point (Rou1) – one FLAIR image as input and one segmentation mask indicating lesion load at one time point as ground truth/output). Another reason for the pre-training was the large amount of single time point data available (1809 scans). Since these were acquired on many different MR scanners, this data set also increased the variability of the training data, which is very important due to the heterogeneity of the FLAIR data. For the purpose of pre-training, a U-net-like network was trained similarly to the proposed network. This pre-training network included the same structure for the encoder, however, only one encoder path was utilized and therefore, did not involve concatenations between encoder-outputs in each scaling level.
This model was trained for 300 epochs using the 1809 2D and 3D single time point FLAIR images (Rou1, see Section 2.1).
2.4.2. Training with longitudinal data
To train the decoder, the weights and parameters of the encoder were initialized with the values of the pre-trained net. Then the entire net with the proposed architecture (Fig. 2) was trained on 1444 images pairs of BL and FU FLAIR images, acquired during clinical routine (Rou2). The training and evaluation data sets are described in Table 1. Since all of the data were acquired in clinical routine or in MS specific studies, almost all the images contained MS lesions. However, new or enlarged lesions were identified by the raters in only 614 cases of the total available training data. Because this means an unbalanced distribution of positive and negative mask values, each epoch alternated between two training data sets: one set with all the images and one set with only image pairs that include at least one new or enlarged lesion.
The net was trained for 200 epochs, whereby in the first 100 epochs only the parameters of the decoder were updated while the encoder parameters were left fix and in the last 100 epochs all parameters (including the pre-trained encoder) were updated.
To determine the benefit of pre-training and of training on a large heterogeneous data set versus a training on a specific MR scanner (in this case Philips Ingenia (PhIng)), three different variants with different training data were compared in total. To evaluate the following three variants, the ZURICH data, acquired on a Philips Ingenia MR scanner, were employed. See Table 1 for the description of the training data sets. The three different training variants were as follows:
PhIng_PhIng - All parameters (encoder and decoder) are trained for 200 epochs on the 130 training data acquired on Philips Ingenia MR scanner.
Rou1_PhIng - Same as PhIng_PhIng, whereby the encoder was pre-trained on the routine data with one time point (Rou1), as described in Section 2.4.1.
Rou1_Rou2 - The encoder was pre-trained on routine data with one time point (Rou1), as described in Section 2.4.1. Then the decoder was trained on 1444 routine data sets with BL and FU images (Rou2). The training was done entirely without the evaluation data (ZURICH and DRESDEN) and the MR scanners (Philips Ingenia, SIEMENS Verio), respectively. This method was described above.
3. Evaluation
For the evaluation of the nets, 27 (3 × 3 × 3) evenly distributed overlapping crops of 128 × 128 × 128 mm3 were taken from both rigidly aligned scans. For each crop, the predicted segmentation mask was computed and merged to the entire volume by taking the mean mask values of the overlapping regions. Due to the binary cross entropy loss function used, the resulting masks contain values between 0 and 1. Those predicted masks were thresholded with a value of 0.4 (determined in preliminary tests by analysing a ROC curve comparing sensitivity and false positive rate of the different thresholds) and compared to the given ground truth segmentation in the following.
3.1. Compared method: Lesion segmentation toolbox (LST)
To compare with our current method, the given 185 evaluation cases (BL and FU for each case) were evaluated with the longitudinal lesion segmentation pipeline of the LST (Schmidt et al., 2012; Schmidt et al., 2019). LST is an open source toolbox which is freely available under the Statistical Parametric Mapping (SPM12) software package.
As an initial step, the longitudinal pipeline of the LST requires that lesion segmentation is performed separately for each time point. The resulting lesion maps are then compared between the time points and the algorithm decides whether changes in lesion maps are significant or if they are due to potential natural variations of the FLAIR signal. Lesion change labels are produced as the final product from this pipeline. Lesion decrease, no change, and lesion increase are labelled by the numbers 1, 2, and 3, respectively. Since we were primarily interested in the evaluation of new or enlarged lesions, only label 3 was considered as a marked region. LST provides two different algorithms for (single time point) lesion segmentation. The lesion growth algorithm (LGA, Schmidt et al. (2012)) requires a T1 image in addition to the FLAIR image. The lesion prediction algorithm (LPA) requires a FLAIR image only. Since the CNN pipeline proposed in this paper uses the FLAIR image only, we used the LPA in the longitudinal pipeline of the LST.
3.2. Metrics and statistical analysis
To investigate the performance of our algorithm, we compared lesion segmentation masks indicating new or enlarged lesions, employing lesion-wise metrics. For clinical routine, the number of new or enlarged lesions is an important parameter. Therefore, the lesion-wise sensitivity (lesion-wise true positive/number of lesions), lesion-wise false positive count (FP) as well as lesion-wise false positive rate (FPR: FP/number of lesions) were assessed. A cluster of voxels was defined as a lesion if voxels were inter-connected (over a 3D 26-voxel-neighborhood). A lesion was defined as true positive (TP) if it had an overlap with a lesion in the second map/ground truth map. These metrics are best suited for determining if the segmentation methods were able to detect the correct number of lesions. To evaluate the resulting overlap of the annotated lesion, the voxel-wise Dice coefficient of correctly identified lesions per image pair was reported:
| (2 × true_positive_voxels)/(2 × true_positive_voxels + false_positive_voxels + false_negative_voxels). |
here, only the voxels of overlapping lesions in the compared maps were considered.
A lesion was defined as relevant if its size exceeds 0.01 ml. This is the minimal lesion size recently suggested by Rovira et al. (2015) for defining a new or enlarged lesion. Therefore, all available ground truth and computed segmentation maps were thresholded by size of connected components, to remove noise and lesions with a size < 0.01 ml.
All following values are given as means and standard deviations over the evaluated cases. Since a number of ground truth masks were available for each case, the metrics comparing the results to all available ground truth segmentations were first averaged for each case. Subsequently, the mean values for the defined data sets are reported. For the inter-rater variability, all rater masks were evaluated against each other and averaged.
Since the chosen metrics were not normally distributed, the Wilcoxon test was used for comparisons of the results.
In Section 2.1 the two evaluation sets with 185 cases are described. The data included some cases without any new or enlarged lesions. Table 1 summarizes the mean lesion count per case and the mean lesion size the for all given evaluation sets. Because the sensitivity, the FPR, and the Dice coefficient are not defined if no new or enlarged lesion is given in an image, those metrics were only computed for the 104 cases including new or enlarged lesions. The FP count is reported for all 185 cases.
3.3. Results
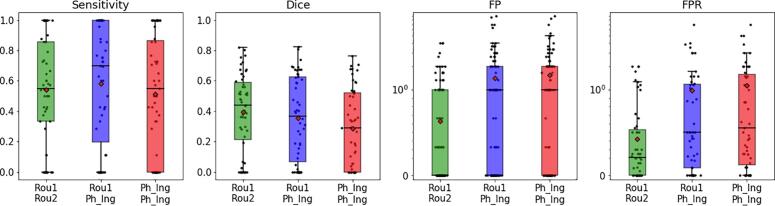
In Section 2.4.2 the three training variants on different data sets (to evaluate the effect of the pre-training using single time point data and the effect of a heterogeneous training set) are described. Fig. 3 summarizes the results of the comparison of the performance on the 89 ZURICH data sets: the network with pre-trained encoder (Rou1_PhIng – blue boxes) performed better than the net trained only on the longitudinal data of the evaluation set (PhIng_PhIng – red boxes) (similar sensitivity and similar Dice coefficient but significantly lower FP count and FPR – p-values < 0.05).
Fig. 3.
The boxplots summarize the comparison between the three variants of the trained networks: sensitivity, Dice coefficient, FP count (visualized using logarithmic scale) and FPR (log scale) for all 89 ZURICH data sets were compared between the nets trained on the routine data (Rou1_Rou2, green), trained on the Philips Ingenia data with pre-trained encoder (Rou1_PhIng, blue) and trained completely on the Philips Ingenia data (PhIng_PhIng, red). See Section 2.4.2 for the description of the variants. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the performance increased (significantly for FP count and FPR with p-values < 0.05 for Wilcoxon test) when the network was trained on a higher number of routine data (Rou1_Rou2 – green boxes), whereby the evaluation sets (ZURICH and DRESDEN) as well as the MR scanners used for the evaluation data (Philips Ingenia and SIEMENS Verio) were not included at all in the training. Therefore, in the following analysis only the values of the Rou1_Rou2 network were compared with the inter-rater performance and the LST algorithm.
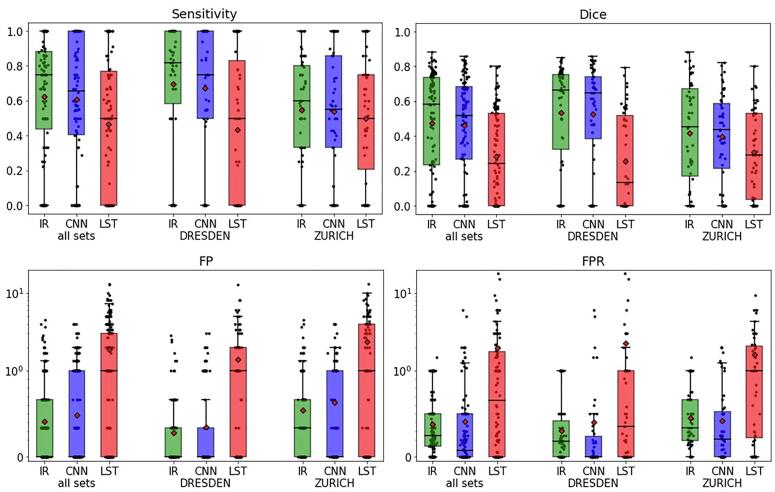
Results of the comparative performance between manual rating (inter-rater, IR), the proposed CNN-based method and LST are shown in Fig. 4 and Table 2: for all 185 evaluation data sets, the CNN outperformed LST. CNN showed a higher sensitivity (0.60 for CNN and 0.46 for LST), a higher Dice coefficient (0.45 for CNN and 0.28 for LST) and lower a FP count (0.48 for CNN and 1.86 for LST) compared to LST. For all applied measures the CNN was not significantly (p > 0.05) different from the inter-rater performance (sensitivity 0.62, Dice 0.47, FP count 0.41).
Fig. 4.
The boxplots summarize the segmentation results for the evaluation data sets: sensitivity, Dice coefficient, FP count (visualized using logarithmic scale) and FPR (log scale) were compared with inter-rater values (IR, green), the proposed CNN based algorithm (blue) and LST (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Summary of the comparison of lesion segmentations for new or enlarged lesion between raters (inter-rater variability (IR)), as well as between the proposed CNN method and LST. The results are given as mean and standard deviation for each of the two data sets (ZURICH and DRESDEN) and both data sets). The numbers marked with an asterisk indicate values which are significantly (p < 0.05 for Wilcoxon test) different from the inter-rater-variability (IR).
| Sets | sensitivity | Dice coefficient | FP count | FPR | |
|---|---|---|---|---|---|
| all sets | Inter-rater (IR) | 0.62 (±0.34) | 0.47 (±0.29) | 0.41 (±0.73) | 0.37 (±0.35) |
| CNN vs. raters | 0.60 (±0.36) | 0.45 (±0.28) | 0.48 (±0.82) | 0.41 (±0.88) | |
| LST vs. raters | 0.46 (±0.38)* | 0.28 (±0.26)* | 1.86 (±2.48)* | 1.92 (±4.54)* | |
| ZURICH | Inter-rater (IR) | 0.54 (±0.31) | 0.41 (±0.28) | 0.54 (±0.87) | 0.45 (±0.33) |
| CNN vs. raters | 0.54 (±0.34) | 0.39 (±0.25) | 0.63 (±0.94) | 0.42 (±0.56) | |
| LST vs. raters | 0.49 (±0.34) | 0.30 (±0.25)* | 2.35 (±2.70)* | 1.61 (±1.97)* | |
| DRESDEN | inter-rater (IR) | 0.69 (±0.35) | 0.53 (±0.29) | 0.28 (±0.54) | 0.30 (±0.35) |
| CNN vs. raters | 0.67 (±0.37) | 0.52 (±0.29) | 0.34 (±0.66) | 0.40 (±1.11) | |
| LST vs. raters | 0.43 (±0.41)* | 0.25 (±0.27)* | 1.40 (±2.14)* | 2.24 (±6.09)* |
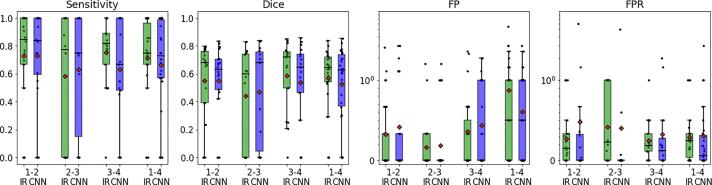
Fig. 5 shows an additional analysis concerning the time between the two compared MR scans (BL, FU). Since the DRESDEN data contain four scans at four time points for each patient, we compared the performance for scan pairs with approximately one year between BL and FU scan (between time point one and two (1–2), two and three (2–3) and three and four (3–4)) and for pairs with approximately three years between BL and FU scan (time point one and four (1–4)). The results show that the CNN-based algorithm (blue boxes) performs independently of the time between scans and is within the inter-rater variability (green boxes in Fig. 5).
Fig. 5.
The boxplots summarize an analysis on the DRESDEN data comparing different scan intervals between BL and FU scans. Therefore, the 4 scans for the 32 patients were paired as follows: time point one and two (1–2), two and three (2–3) and three and four (3–4) as well as time point one and four (1–4)). The mean time interval between the first three pairings is 1.01 (±0.08) years and between the first and the last (1–4) is 3.02 (±0.12) years.
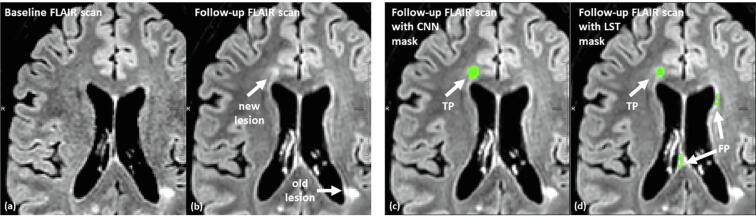
Fig. 6, Fig. 7 visualize the problem of the FP lesions in LST. Fig. 6 shows a representative case, where the CNN-based method outperformed the LST, which detected two false positive lesions in the MR slide. Fig. 7 visualizes another representative case where LST detected 13 FP lesions and the CNN-based algorithm found no FP lesions.
Fig. 6.
Segmentation results for the CNN-based method and the LST method: Between BL (a) and FU (b) scan one new lesion occurred in the visualized MR slice. The CNN (c) identified the lesion correctly (true positive (TP)) and LST (d) reported further false positive (FP) new/enlarged lesions in addition to the correctly detected lesion.
4. Discussion
The quantification of new or enlarged MS lesions from T2-weighted MRI follow-up scans is an important surrogate of clinical disease activity in MS. A fully automated CNN-based deep learning method is proposed here as a tool to identify MS lesion activity between two consecutive MRI scans. In contrast to conventional methods, in which the two scans are first processed independently, and then combined, our algorithm considers the two scans (BL, FU) simultaneously in order to make a decision based on the combined information. This mimics how lesion activity is assessed by radiologists.
Firstly, in many MS clinical drug trials new or enlarged lesions are used as a composite secondary end point. Furthermore, common risk stratification algorithms like Rio score (Sormani et al., 2013) or the modified Rio Score do not distinguish between new appearing and enlarged lesions. Thus clinically, both new and enlarged lesions are used as surrogate markers for disease activity. Additionally, in many cases it is not easy to distinguish between new and enlarged lesions. If a lesion appears in the proximity of an existing lesion and is connected to an already existing lesion by some voxels, it is unclear if that lesion is new or enlarged. This may introduce inter-rater variability without any additional clinical benefit. In this study it was therefore decided not to distinguish between new or enlarged lesions.
The segmentation of MS lesions in MRI FLAIR scans is challenging due to the highly heterogeneous nature of FLAIR protocols between radiology centres as well as between scans of individual patients. To investigate the impact of acquisition heterogeneity two independent data sets acquired on two independent MR scanner types were analysed (evaluation data as defined in Section 2.1.2): 89 image pairs from ZURICH acquired with a Philips Ingenia scanner and 96 image pairs from DRESDEN acquired with SIEMENS Verio scanner. To evaluate the performance of the proposed algorithm with respect to scanner and/or protocol independence, the training was done with two different data sets: First the algorithm was trained on 130 routine data sets acquired with a single scanner, a Philips Ingenia (DRESDEN), and evaluated using 89 image pairs also acquired with a Philips Ingenia scanner (ZURICH). We then trained the network on 1444 routine data sets from 103 different scanners excluding the evaluation data. The analysis showed that the network trained on more heterogeneous data was able to segment the evaluation data of an unknown scanner. To further accommodate for this high variability of the FLAIR scans, additional single time point data sets were used for a pre-training of parts of the network. The results suggest that the problem of lesion segmentation can be solved independently of the scanner and protocol, despite the high heterogeneity of the MR scans, if the amount of data in the training data is chosen to be correspondingly heterogeneous.
To evaluate the performance of our CNN-based algorithm, we compared the results to the manual inter-rater performance, as well as to a current state-of-the-art non-deep-learning method (LST) for detecting lesion activity (Schmidt et al., 2019). As described in the method section the LST provides two different algorithms for (single time point) lesion segmentation: the lesion growth algorithm (LGA) and the lesion prediction algorithm (LPA). Since the CNN pipeline proposed in this paper uses the FLAIR image only, we used the LPA in the longitudinal pipeline of the LST. However, the LGA was deployed in the recent study by Schmidt et al., 2019. In order to provide a fair comparison between the tools also the performance of the LGA was tested as an additional experiment, which is not shown in the results section. For all evaluation data sets also a corresponding 3D-T1 image was available. The longitudinal LST pipeline using the LGA was executed for a range of kappa values (0.001, 0.1, 0.2, 0.3, and 0.4). As for the other algorithms the FP and sensitivity was computed for the DRESDEN and the ZURICH data. For all kappa values the LPA outperformed the LGA. We therefore decided to focus on the LPA algorithm in this paper.
To derive the inter-rater performance two to three manual segmentations were available for 185 evaluation data sets, provided by different experienced raters. The rather low inter-rater performance (e.g. sensitivity of 0.62) signifies the complexity and uncertainty of identifying new or enlarged lesions. The small values of the Dice coefficients are due to the small lesion volumes, which is on average only 0.13 ml. Only 2% of all lesions in the evaluation data are between 1.0 and 1.66 ml in volume. An additional experiment with the DRESDEN data was performed, in which the segmentation results of data with different follow-up times were compared. The CNN-based algorithm performed independent on the time between BL and FU scan.
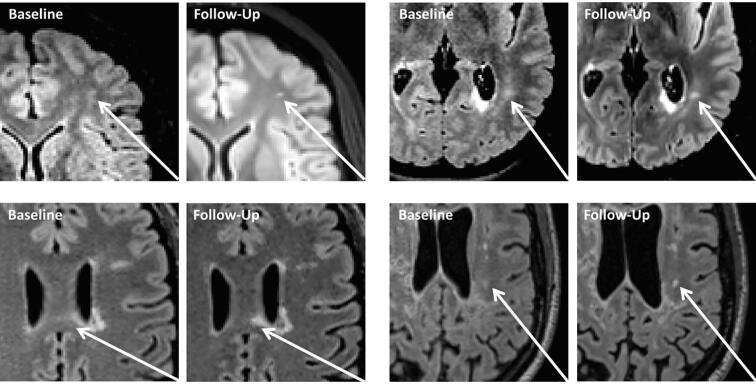
Fig. 8 visualizes four representative examples, where raters graded differently. Some of the evaluated data suffered from fluctuating signal values between the BL and FU scan. Such fluctuations are common in clinical routine due to different MR scanners or different scanner/protocol settings, providing a realistic evaluation setting. FLAIR is not a quantitative MRI sequence, meaning that a signal intensity in a given image voxel is arbitrary and cannot be compared between scans and patients. The proposed CNN algorithm showed a significantly better false positive rate than the compared LST. A visual evaluation (Fig. 7) indicated, that LST is more sensitive to signal changes between the two scans due to MR parameter differences.
Fig. 8.
Four Examples of BL/FU image pairs with “maybe” new/enlarged lesions, in which the opinion of the raters diverged. In each example one of the raters marked a new or enlarged lesion in the follow-up scan, whereby the other rater interpreted the lesion as already existing in the baseline scan.
Classical methods based on image differences or deformation-based approaches require a certain consistency between the two scans (BL and FU). In order to be able to catch possible deviations from this consistency, probability models and regularization methods (e. g. for image registration) are used. In our experience, however, the inconsistency in MRI data in routine clinical operation – outside the highly standardized conditions of clinical trials – is very high. As a consequence, “manual” designed adjustments are not feasible for the level of heterogeneity we aim to address with our algorithm. Furthermore, most of the previously proposed methods (including LST) consider both scans individually (segmentation of lesion load in BL and in FU) and then invest effort in combining the two resulting lesion maps. The approach of the proposed CNN algorithm, however, is that both scans are considered simultaneously and the combination of signal values of BL and FU is done before lesion segmentation. Therefore, no rules are necessary to combine the masks. Rather, the network is trained to identify the correct combination of (very heterogeneous) image features that indicate new or enlarged lesions.
LST is time consuming as compared to the proposed CNN algorithm. The mean computation time of LST was around 30 min. For cases with a high number of lesions the computation time increases to over 2 or even 3 h. The mean computation time of the described CNN-based method was under 1 min (GPU: NVIDIA GeForce RTX2080Ti, 11 GB GDDR6, 14 Gbps, 4352 cores) – independent on lesion count or image resolution. An automated CNN-based approach can quickly (<1 min) provide an independent and deterministic assessment of lesions from baseline and follow-up scans to support disease and therapy monitoring in MS as a rapid alternative to manual segmentation.
CRediT authorship contribution statement
Julia Krüger: Conceptualization, Methodology, Software, Writing - original draft, Visualization, Formal analysis. Roland Opfer: Software, Writing - original draft. Nils Gessert: Software. Ann-Christin Ostwaldt: Data curation. Praveena Manogaran: Writing - review & editing. Hagen H. Kitzler: Data curation. Alexander Schlaefer: Supervision. Sven Schippling: Supervision, Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was funded by Hamburgische Investitions- und Förderbank (IFB Hamburg), Nr.: 51084589. Praveena Manogaran has received travel grants from Merck Sereno. Sven Schippling reports compensation for consulting, serving on scientific advisory boards, speaking, or other activities from Biogen, Celgene, Merck, Sanofi and TEVA. Hagen H. Kitzler has received travel grants, speaker's honoraria, financial research support, and consultancy fees from Bayer, Biogen Idec, Novartis, Siemens and TEVA. He served on advisory boards for Biogen, Novartis and Ixico. Sven Schippling is currently an employee of Hoffmann La Roche pharmaceutical Research and Early Development.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102445.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Akkus Z., Galimzianova A., Hoogi A., Rubin D.L., Erickson B.J. Deep learning for brain mri segmentation: State of the art and future directions. J. Digit. Imaging. 2017;30(4):449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini M., Rossi F., Grove R.A., Stromillo M.L., Whitcher B., Matthews P.M., De Stefano N. Automated identification of brain new lesions in multiple sclerosis using subtraction images. J. Magn. Reson. Imaging. 2014;39(6):1543–1549. doi: 10.1002/jmri.24293. [DOI] [PubMed] [Google Scholar]
- Bosc M., Heitz F., Armspach J., Namer I., Gounot D., Rumbach L. Automatic change detection in multimodal serial mri: Application to multiple sclerosis lesion evolution. NeuroImage. 2003;20:643–656. doi: 10.1016/S1053-8119(03)00406-3. [DOI] [PubMed] [Google Scholar]
- Brosch T., Tang L.Y.W., Yoo Y., Li D.K.B., Traboulsee A., Tam R. Deep 3d convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging. 2016;35(5):1229–1239. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- Cabezas M., Corral J., Oliver A., Díez Y., Tintoré M., Auger C., Montalban X., Lladó X., Pareto D., Rovira À. American Journal of Neuroradiology. 2016;37(10):1816–1823. doi: 10.3174/ajnr.A4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas M., Oliver A., Roura E., Freixenet J., Vilanova J.C., Ramió-Torrentà L., Rovira A., Llado X. Computer methods and programs in biomedicine. 2014. Automatic multiple sclerosis lesion detection in brain mri by flair thresholding; p. 115. [DOI] [PubMed] [Google Scholar]
- Chen H., Dou Q., Yu L., Qin J., Heng P.-A. Voxresnet: Deep voxelwise residual networks for brain segmentation from 3d mr images. NeuroImage. 2018;170:446–455. doi: 10.1016/j.neuroimage.2017.04.041. Segmenting the Brain. [DOI] [PubMed] [Google Scholar]
- Cheng, M., Galimzianova, A., Lesjak, i., Spiclin, Z., B. Lock, C., and Rubin, D. (2018). A Multi-scale Multiple Sclerosis Lesion Change Detection in a Multi-sequence MRI: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings, pages 353–360.
- Danelakis A., Theoharis T., Verganelakis D.A. Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput. Med. Imaging Graph. 2018;70:83–100. doi: 10.1016/j.compmedimag.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Dou Q., Yu L., Chen H., Jin Y., Yang X., Qin J., Heng P.-A. 3d deeply supervised network for automated segmentation of volumetric medical images. Med. Image Anal. 2017;41 doi: 10.1016/j.media.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Egger C., Opfer R., Wang C., Kepp T., Sormani M.P., Spies L., Barnett M., Schippling S. MRI flair lesion segmentation in multiple sclerosis: Does automated segmentation hold up with manual annotation? NeuroImage: Clinical. 2017;13:264–270. doi: 10.1016/j.nicl.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C., Collins L., Arnold D., Arbel T. IEEE transactions on medical imaging. 2013. Temporally consistent probabilistic detection of new multiple sclerosis lesions in brain mri; p. 32. [DOI] [PubMed] [Google Scholar]
- Fahrbach K., Huelin R., Martin A.L., Kim E., Dastani H.B., Rao S., Malhotra M. Relating relapse and t2 lesion changes to disability progression in multiple sclerosis: a systematic literature review and regression analysis. BMC Neurol. 2013;13(1):180. doi: 10.1186/1471-2377-13-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K., Ashburner J., Kiebel S., Nichols T., Penny W., editors. Statistical Parametric Mapping – The Analysis of Functional Brain Image. Academic Press; London: 2007. [Google Scholar]
- Ganiler O., Oliver A., Diez Y., Freixenet J., Vilanova J.C., Beltran B., Ramió-Torrentà L., Rovira À., Lladó X. A subtraction pipeline for automatic detection of new appearing multiple sclerosis lesions in longitudinal studies. Neuroradiology. 2014;56(5):363–374. doi: 10.1007/s00234-014-1343-1. [DOI] [PubMed] [Google Scholar]
- García-Lorenzo D., Francis S., Narayanan S., Arnold D.L., Collins D.L. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 2013;17(1):1–18. doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Gramsch C., Nensa F., Kastrup O., Maderwald S., Deuschl C., Ringelstein A., Schelhorn J., Forsting M., Schlamann M. Diagnostic value of 3d fluid attenuated inversion recovery sequence in multiple sclerosis. Acta Radiol. 2015;56(5):622–627. doi: 10.1177/0284185114534413. PMID: 24867222. [DOI] [PubMed] [Google Scholar]
- Griffanti L., Zamboni G., Khan A., Li L., Bonifacio G., Sundaresan V., Schulz U.G., Kuker W., Battaglini M., Rothwell P.M., Jenkinson M. NeuroImage. 2016. Bianca (brain intensity abnormality classification algorithm): A new tool for automated segmentation of white matter hyperintensities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Zhang, X., Ren, S., and Sun, J. (2015). Deep residual learning for image recognition. CoRR, abs/1512.03385.
- Isensee F., Kickingereder P., Wick W., Bendszus M., Maier-Hein K.H. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. Lect. Notes Comput. Sci. 2018:287–297. [Google Scholar]
- Jain S., Ribbens A., Sima D.M., Cambron M., De Keyser J., Wang C., Barnett M.H., Van Huffel S., Maes F., Smeets D. Two time point ms lesion segmentation in brain mri: An expectation-maximization framework. Front. Neurosci. 2016;10:576. doi: 10.3389/fnins.2016.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnitsas K., Ledig C., Newcombe V.F., Simpson J.P., Kane A.D., Menon D.K., Rueckert D., Glocker B. Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med. Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Köhler C., Wahl H., Ziemssen T., Linn J., Kitzler H.H. Exploring individual multiple sclerosis lesion volume change over time: Development of an algorithm for the analyses of longitudinal quantitative mri measures. NeuroImage: Clinical. 2019;21:101623. doi: 10.1016/j.nicl.2018.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D., Ba J. International Conference on Learning Representations. 2014. Adam: A method for stochastic optimization. [Google Scholar]
- Lao Z., Shen D., Liu D., Jawad A., Melhem E.R., Launer L.J., Bryan R.N., Davatzikos C. Computer-assisted segmentation of white matter lesions in 3d mr images using support vector machine. Acad. Radiol. 2008;15(3):300–313. doi: 10.1016/j.acra.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesjak Z., Pernus F., Likar B., Spiclin Z. Validation of white-matter lesion change detection methods on a novel publicly available mri image database. Neuroinformatics. 2016;14:03–420. doi: 10.1007/s12021-016-9301-1. [DOI] [PubMed] [Google Scholar]
- Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Medical Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Maas A.L., Hannun A.Y., Ng A.Y. Speech and Language Processing. 2013. Rectifier nonlinearities improve neural network acoustic models. In in ICML Workshop on Deep Learning for Audio. [Google Scholar]
- Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M., Fujihara K., Havrdova E., Hutchinson M., Kappos L., Lublin F.D., Montalban X., O’Connor P., Sandberg-Wollheim M., Thompson A.J., Waubant E., Weinshenker B., Wolinsky J.S. Diagnostic criteria for multiple sclerosis: 2010 revisions to the mcdonald criteria. Ann. Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer International Publishing; Cham: 2015. pp. 234–241. [Google Scholar]
- Roura E., Oliver A., Cabezas M., Valverde S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira A., Lladó X. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015;57:1031–1043. doi: 10.1007/s00234-015-1552-2. [DOI] [PubMed] [Google Scholar]
- Rovira À., Wattjes M.P., Tintoré M., Tur C., Yousry T., Sormani M.P., Stefano N.D., Filippi M., Auger C., Rocca M.A., Barkhof F., Fazekas F., Kappos L., Polman C., Miller D., Montalban X. Evidence-based guidelines: Magnims consensus guidelines on the use of mri in multiple sclerosis—clinical implementation in the diagnostic process. Nat. Rev. Neurol. 2015;11:471–482. doi: 10.1038/nrneurol.2015.106. [DOI] [PubMed] [Google Scholar]
- Salem M., Cabezas M., Valverde S., Pareto D., Oliver A., Salvi J., Rovira L., Llado X. A supervised framework with intensity subtraction and deformation field features for the detection of new t2-w lesions in multiple sclerosis. NeuroImage: Clinical. 2017;17C:607–615. doi: 10.1016/j.nicl.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P., Gaser C., Arsic M., Buck D., Förschler A., Berthele A., Hoshi M., Ilg R., Schmid V., Zimmer C., Hemmer B., Mühlau M. An automated tool for detection of flair-hyperintense white-matter lesions in multiple sclerosis. NeuroImage. 2012;59:3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Schmidt P., Pongratz V., Küster P., Meier D., Wuerfel J., Lukas C., Bellenberg B., Zipp F., Groppa S., Sämann P.G., Weber F., Gaser C., Franke T., Bussas M., Kirschke J., Zimmer C., Hemmer B., Mühlau M. Automated segmentation of changes in flair-hyperintense white matter lesions in multiple sclerosis on serial magnetic resonance imaging. NeuroImage: Clinical. 2019;23:101849. doi: 10.1016/j.nicl.2019.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiee N., Bazin P., Ozturk A., Reich D., Calabresi P., Pham D. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani M., Rio J., Tintorè M., Signori A., Li D., Cornelisse P., Stubinski B., Stromillo M., Montalban X., Stefano N.D. Scoring treatment response in patients with relapsing multiple sclerosis. Multiple Sclerosis J. 2013;19(5):605–612. doi: 10.1177/1352458512460605. PMID: 23012253. [DOI] [PubMed] [Google Scholar]
- Sormani M.P., Arnold D.L., Stefano N.D. Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann. Neurol. 2014;75(1):43–49. doi: 10.1002/ana.24018. [DOI] [PubMed] [Google Scholar]
- Valverde S., Cabezas M., Roura E., González-Villà S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira Àlex, Oliver A., Lladó X. Improving automated multiple sclerosis lesion segmentation with a cascaded 3d convolutional neural network approach. NeuroImage. 2017;155:159–168. doi: 10.1016/j.neuroimage.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Van Leemput K., Maes F., Vandermeulen D., Colchester A., Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Trans. Med. Imaging. 2001;20:677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.