Abstract
Resistance to the last-line polymyxins is increasingly reported in multidrug-resistant Gram-negative pathogens, including Acinetobacter baumannii which develops resistance via either lipid A modification or even lipopolysaccharides (LPS) loss in the outer membrane (OM). Considering these two different mechanisms, quantitative membrane lipidomics data were utilized to develop three OM models representing polymyxin-susceptible and -resistant A. baumannii strains. Through all-atom molecular simulations with enhanced sampling techniques, the effect of lipid A-pEtN modification and LPS loss on the action of colistin (i.e. polymyxin E) was examined for the first time, with a focus on the dynamics and energetics of colistin penetration into these OMs. Lipid A-pEtN modification improved the OM stability, impeding the penetration of colistin into the OM; this differed from the current literature that lipid A-pEtN modification confers resistance by diminishing the initial interaction with polymyxins. In contrast, LPS deficiency significantly reduced the negative charges on the OM surface, diminishing the binding of colistin. Moreover, both lipid A-pEtN modification and LPS loss also constituted colistin resistance through disturbing the conformational transitions of the colistin molecule. Collectively, atomic-scale interactions between polymyxins and different bacterial OMs are very different and the findings may facilitate the discovery of new-generation polymyxins against the Gram-negative ‘superbugs’.
Keywords: Antimicrobial resistance, polymyxin, colistin, Acinetobacter baumannii, molecular dynamics
Graphical Abstract
Polymyxins are a last-line therapy against MDR Gram-negative pathogens. Unfortunately, resistance to polymyxins can emerge via the remodeling of their outer membranes. Here, we employed all-atom molecular dynamics simulations to elucidate the mechanisms of polymyxin resistance in A. baumannii due to lipid A modification and loss of lipopolysaccharide.
Multidrug-resistant (MDR) Gram-negative bacteria such as Acinetobacter baumannii present a significant medical challenge due to the resistance to most and in some cases all antibiotics currently available in the clinic.[1] The World Health Organization has listed MDR A. baumannii as a “critical” priority for global human health requiring research and development of new antibiotics.[2] Due to a lack of new antibiotic development in recent years, the polymyxins (i.e. polymyxin B and colistin) are increasingly used as a last-line therapy to treat infections caused by these life-threatening ‘superbugs’.[3-5] Alarmingly, reports of polymyxin resistance in Gram-negative bacteria have increased rapidly in the last decade.[6, 7] Therefore, there is an urgent need to investigate the mechanism of polymyxin resistance and develop novel polymyxin antibiotics to cope with this severe situation.
Polymyxin resistance in Gram-negative bacteria is primarily resulted from modifications of the lipopolysaccharide (LPS) molecule that is the major component of the outer layer of the bacterial outer membrane (OM).[6, 8] In most polymyxin-resistant strains, cationic moieties such as 4-amino-4-deoxy-L-arabinose (L-Ara4N), phosphoethanolamine (pEtN) or galactosamine are added to the lipid A or core components of LPS, which is believed to diminish the electrostatic interactions with polymyxins.[8] Expression of the majority of genes in the LPS modification pathway is modulated by a variety of two-component systems (TCSs) such as PhoPQ and PmrAB.[8, 9] Stimuli including high Fe3+, low pH and antibiotics are known to induce the expression of several genes (e.g. eptA and arnT) which in turn leads to the translation of enzyme machineries that mediate the modification of LPS molecules with the aforementioned cationic moieties. Several mutations in the genes encoding these TCSs result in constitutive upregulation of the LPS modification pathway and thus polymyxin resistance[6, 10] Furthermore, plasmid-borne mcr genes encode a pEtN transferase that chemically modifies lipid A molecules and causes horizontal and cross-dissemination of polymyxin resistance.[11] Alternatively, the loss of LPS has been identified as another critical mechanism of polymyxin resistance in A. baumannii wherein mutations in lpxACD genes responsible for the synthesis of the lipid A component of LPS.[12] Notably, our recent quantitative membrane lipidomics study discovered that loss of LPS caused significant changes in the composition of different phospholipids in the OM of A. baumannii.[13] The initial interaction of the polymyxins with the OM of Gram-negative bacteria is largely dependent on the electrostatic interactions between the positively charged L-α-γ-diaminobutyric (Dab) residues of polymyxins and negatively charged phosphate groups of lipid A in the bacterial OM.[3] The aforementioned LPS structural alternations diminish the negative charges on the OM surface, thus inhibiting the binding of polymyxins and protecting the bacteria from the antimicrobial action of polymyxins.[3]
Although the genetic mechanisms of polymyxin resistance are well studied, it is still unclear how modifications and loss of LPS impact the action of polymyxins differently at the atomic level. A previous molecular dynamics (MD) simulation study examining the impact of lipid A modifications on the bacterial OM structure found the addition of hydroxyl, L-Ara4N or extra palmitoyl groups to lipid A molecule increased the lateral interactions between neighboring LPS molecules, thus strengthening the OM barrier.[14] Another simulation study revealed that polymyxin B bound to the bacterial OM, causing membrane curvature in susceptible but not in resistant LPS chemotypes; notably, the polymyxin molecule showed different conformations in susceptible and resistant OMs.[15] Unfortunately, due to the limitations of conventional MD simulations, the effect of OM remodeling due to LPS modification and deficiency on the penetration of polymyxins into the OM has not been investigated in previous studies. Therefore, advanced simulation techniques are required to investigate the penetration of polymyxins into different bacterial OMs and the relationship between OM remodeling and polymyxin resistance needs to be addressed.
In the present study, all-atom MD simulations with enhanced sampling techniques were employed to investigate the different thermodynamics governing the penetration of colistin into three different bacterial OMs. As A. baumannii can develop polymyxin resistance via lipid A-pEtN modification or LPS loss, our quantitative membrane lipidomics results were utilized to developed three representative OM models of A. baumannii, namely a lipid A OM, a lipid A-pEtN modified OM and an LPS-deficient OM.[13] In summary, our novel findings highlight that lipid A-pEtN modification and LPS loss exert different impacts on the OM structural integrity, and confer resistance via different mechanisms and at different stages of colistin action on the bacterial OM.
Results
Colistin A penetrates and disorganizes the bacterial OM
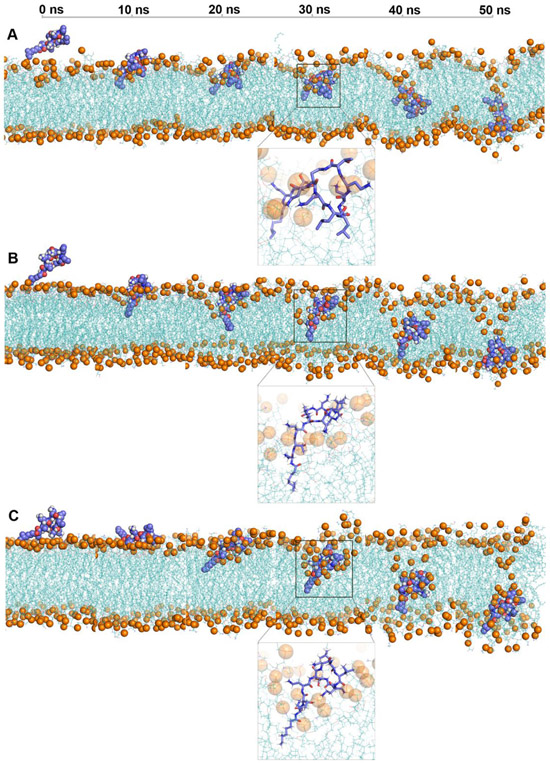
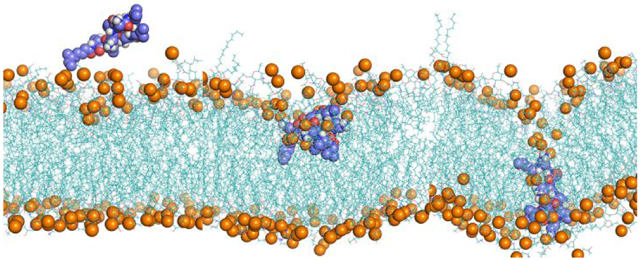
Figure 1 shows structural snapshots of colistin A penetration in the lipid A, lipid A-pEtN and LPS-deficient model OM systems from the steered simulations trajectories. The entire trajectory space covered the translocation of the colistin molecule through both the OM headgroup and the hydrophobic regions; that together constitute the major fortification layers of the bacterial OM.[16] In all three OM systems, the fatty acyl tail of colistin passed through the OM headgroup first, followed by the gradual penetration of its cyclic head. Notably, the colistin molecule formed a unique folded conformation in the lipid A OM system when it traversed the OM headgroup region (Figure 1A), which is consistent with previous NMR studies.[17-18] Within this unique conformation, the fatty acyl tail and d-Leu6-l-Leu7 of colistin A adopted an inward orientation and were embedded in the hydrophobic layer of the OM, while the Dab side chains retained interactions with the OM lipid headgroups. The details of the folded conformation of colistin in lipid A OM was analyzed in the following section. In contrast, this folded conformation was absent in lipid A-pEtN and LPS-deficient systems wherein the colistin molecule maintained an extended conformation during the penetration trajectory (Figure 1B and 1C).
Figure 1.
Snapshots of colistin penetration into the outer membrane (OM) of A. baumannii, including (A) Lipid A OM; (B) Lipid A-pEtN OM; (C) LPS-deficient OM. Colistin A: blue spheres or sticks in amplified figures; OM: cyan lines. The lipid phosphate atoms are shown with orange spheres to indicate the lipid headgroups. Water molecules and ions are not shown for clarity.
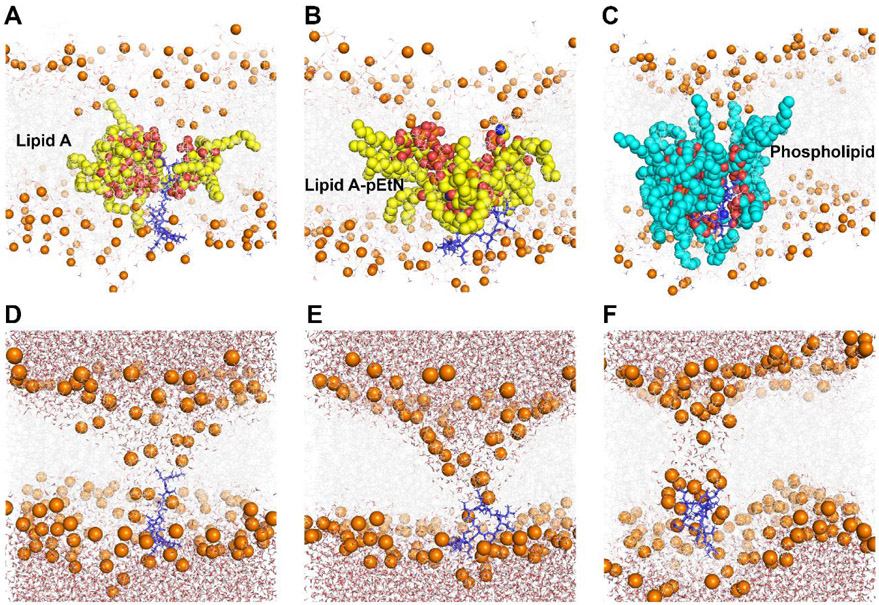
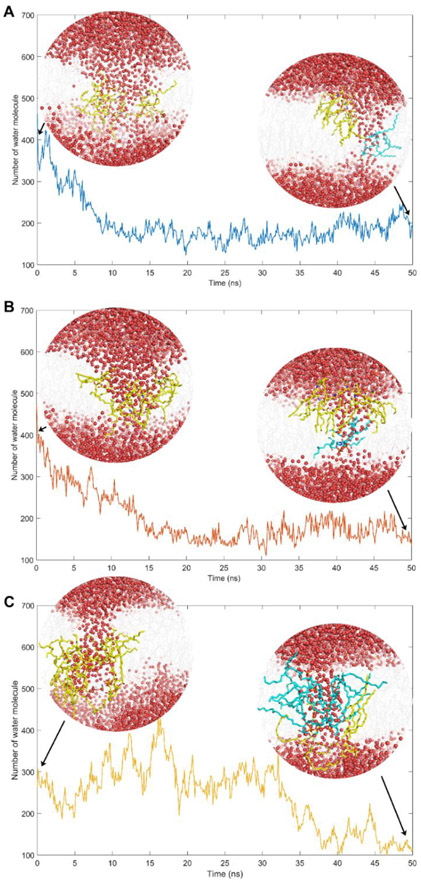
Colistin penetration induced significant disorganization of the OMs, especially their outer leaflets (Figure 2). The inserted colistin molecule occupied a major portion in the hydrophobic core of the OMs,[19-20] inducing negative membrane curvature. Subsequently, colistin insertion disrupted the intramolecular hydrogen bonds between adjacent lipid molecules (Figure S1), causing multiple lipid molecules originally in the outer leaflet to enter the OM hydrophobic region. To elaborate, 3 or 4 lipid A molecules, 4 lipid A-pEtN molecules and 11 or 12 phospholipid molecules were affected in the lipid A, lipid A-pEtN and LPS-deficient systems, respectively (Figure 2A-C). The polar headgroups of these relocated lipid A, lipid A-pEtN and phospholipid molecules surrounded the colistin molecule, while their hydrocarbon tails protruded into the hydrophobic layer of the OMs. This special structural re-arrangement mediated the formation of a hydrophilic channel within the OMs, through which a large number of water molecules moved into the OM hydrophobic regions (Figure 2D-F). Specifically, the average volume of the water pores per colistin molecule was 5.48 nm3, 6.52 nm3 and 2.85 nm3 in the lipid A, lipid A-pEtN and LPS-deficient OM systems, respectively (Figure S2). These results revealed the different disorganizing effects of colistin penetration on different bacterial OMs.
Figure 2.
Permeabilizing effect of colistin on three different outer membranes (OMs) of A. baumannii. (A, D) lipid A OM system; (B, E) lipid A-pEtN OM system; (C, F) LPS-deficient OM system. Colistin A: blue sticks; OM: grey lines; Phosphate atom: orange spheres; collapsed lipid A/lipid A-pEtN molecules: yellow spheres; collapsed phospholipid molecules: cyan spheres; Water molecules: sticks with red oxygen and white hydrogen atoms.
Free energy profiles of colistin penetration into the bacterial OM
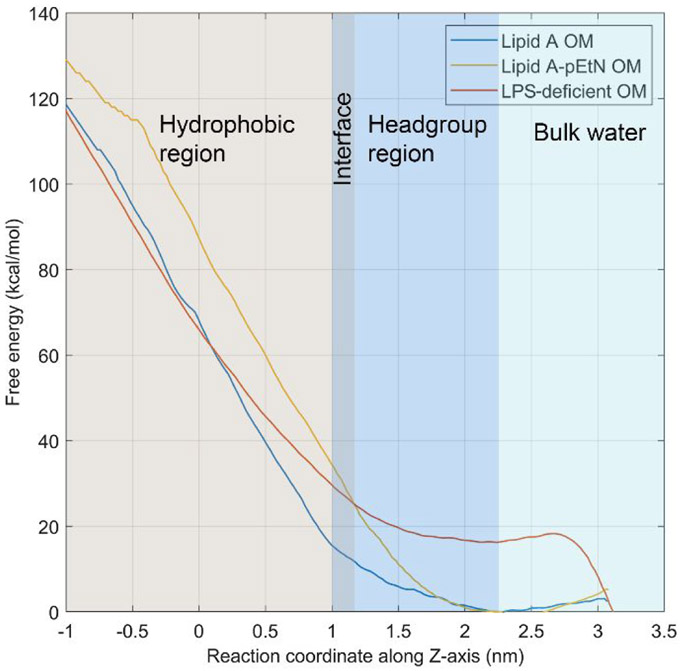
A series of umbrella sampling simulations were performed to characterize the free energy profiles of colistin penetration into the bacterial OMs. It was clear that the changing trends of the free energy were significantly different in the lipid A, lipid A-pEtN and LPS-deficient systems (Figure 3). As the colistin molecule bound to the OM headgroup regions (reaction coordinate from 2.3 to 3.1 nm), the free energy slightly decreased in the lipid A and lipid A-pEtN systems but sharply increased in the LPS-deficient system. The free energy barriers for the binding process (ΔGbind) were −3.0 kcal mol−1 (energy-favorable), −5.3 kcal mol−1 (energy-favorable) and 16.3 kcal mol−1 (energy-unfavorable) in the lipid A, lipid A-pEtN and LPS-deficient systems, respectively. As the colistin molecule moved to the OM hydrophobic center (reaction coordinate from 0 to 2.3 nm), the free energy steadily increased in all three systems. However, the free energy barrier of the translocation process (ΔGtranslocate) was higher in the lipid A-pEtN system (87.4 kcal mol−1) than the lipid A (68.5 kcal mol−1) and LPS-deficient systems (50.4 kcal mol−1). The distinct free energy profiles in different OM systems indicate that lipid A-pEtN modification and LPS deficiency conferred resistance at different stages of colistin action on the OM of A. baumannii. The lipid A-pEtN modification primarily inhibited the translocation of colistin through the OM hydrophobic layer, whereas LPS deficiency significantly reduced the binding of colistin to the OM without LPS.
Figure 3.
Free energy profiles of colistin A penetration into the outer membrane. The free energy minimum is shifted to zero in the profiles.
Effects on the OM properties due to lipid A-pEtN modification and LPS deficiency
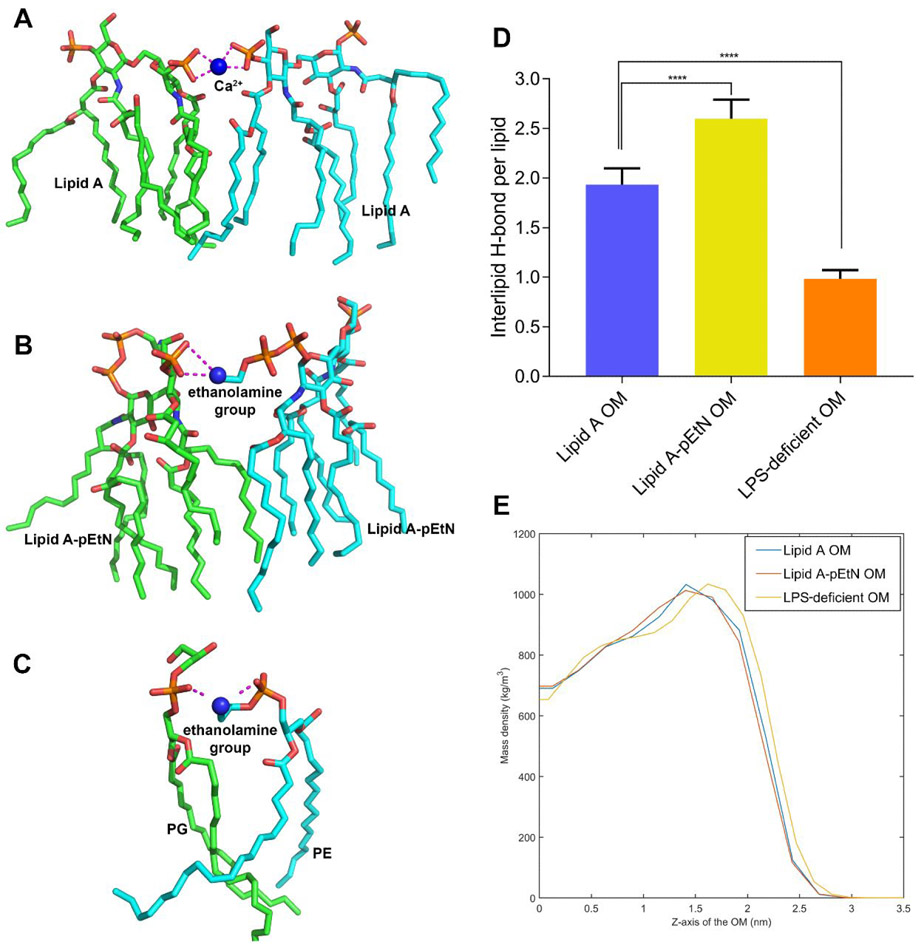
Owing to the addition of the cationic pEtN or the complete removal of LPS molecules (PE dominated after LPS loss), the number of negatively charged phosphate groups on the OM surface was decreased, compared to the wild-type lipid A OM. Quantitatively, the density of the negative charge on the OM surface was −1.77, −1.37 and −0.63 e/nm2 in the lipid A, lipid A-pEtN and LPS-deficient systems, respectively. The significantly reduced charge density on the LPS-deficient OM surface accounted for the energy-unfavorable binding of the colistin molecule to the LPS-deficient OM (Figure 3). Notably, the change in the charge density also decreased the binding of Ca2+ to the lipid A-pEtN and LPS-deficient OM surface (Table S1). Divalent cations generally form bridging electrostatic interactions between adjacent lipid A phosphate groups (Figure 4A) and contribute to the lamellar structure of the bacterial OM.[21] However, in the lipid A-pEtN and LPS-deficient OMs, the ethanolamine group of pEtN and PE replaced Ca2+ as the main cross-linking agent between neighboring lipid molecules (Figure 4B and 4C). These OM changes most likely render the bacteria cell less susceptible to polymyxins, particularly given the well-known competitive displacement mechanism of polymyxins.[3, 15]
Figure 4.
Effects of lipid A-pEtN modification and LPS deficiency on the outer membrane properties. (A-C) The cross-linking electrostatic interactions between the neighboring lipid molecules mediated by Ca2+ or ethanolamine group are shown with purple dashed lines. (D) The number of inter-lipid hydrogen bonds in different systems. P values are used to indicate significant difference (P < 0.0001). (E) The mass density of the outer leaflet of the different outer membranes. Z = 0 indicates the membrane center.
Consistent with a recent report,[14] the addition of pEtN groups strengthened the lateral interactions between neighboring lipid A molecules in the OM. The intermolecular hydrogen bonds between the headgroups were increased significantly (Figure 4D), while the hydrophobic packing between lipid carbon tails was also slightly improved as indicated by the increase in membrane density within the hydrocarbon tail region (Figure 4E). Interestingly, the loss of LPS destabilized the OM with decreased intermolecular hydrogen bonding and looser packing between neighboring phospholipid molecules (Figure 4D and 4E).
Effects on the conformational dynamics of the colistin molecule due to lipid A-pEtN modification and LPS deficiency
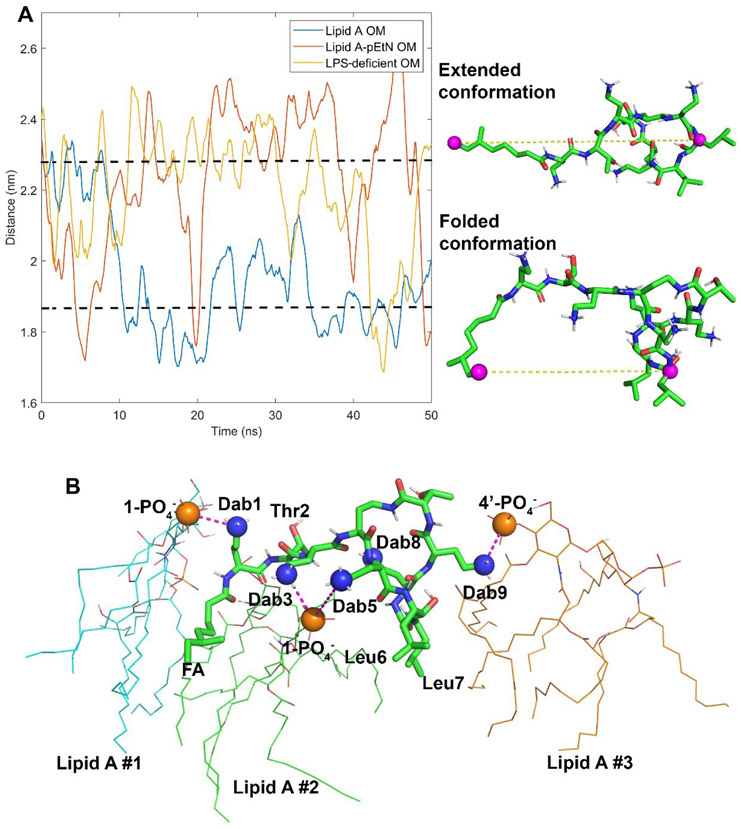
Conformational differences in the colistin molecule were evident during penetration into the different OMs (Figure 1A). To characterize the conformational ensembles of the colistin molecule in the three different OM systems, the distance between the last carbon atom of the fatty acyl tail and the Cα of d-Leu6 was analyzed (Figure 5A). The colistin molecule changed from an extended conformation to a folded conformation within 20-ns simulations in the wild-type lipid A system. Moreover, the folded conformation was retained during the subsequent simulation trajectory. In contrast, the colistin molecule formed the folded conformation transiently in the lipid A-pEtN system (only at approximately 5 ns, 20 ns, 40 ns, and 50 ns), while folding in the LPS-deficient system only occurred for the simulation period from 42 ns to 45 ns.
Figure 5.
Conformational dynamics of the colistin molecule during outer membrane (OM) penetration. (A) The last carbon atom of fatty acyl tail and the α-carbon atom of d-Leu6 are shown in the colistin structure with purple spheres. The distances between these two atoms are used to describe the conformational dynamics of the colistin molecule in different OMs. (B) The interaction model of the colistin molecule with the lipid A OM. The electrostatic interactions between the side chains of the Dab residues and the lipid A phosphate groups are shown with purple dashed lines.
The interactions between the colistin molecule and the wild-type lipid A OM were investigated. As shown in Figure 5B, the colistin molecule interacted with three lipid A molecules as follows: l-Dab1 formed electrostatic interaction with the 1-PO4− group of the left lipid A molecule; l-Dab3 and l-Dab5 bound to the 1-PO4− of the middle lipid A molecule; and l-Dab9 attached to the 4′-PO4− of the right lipid A molecule. Meanwhile, the hydrophobic residues of colistin (fatty acyl tail, d-Leu6 and l-Leu7) inserted into the gaps between neighboring lipid A molecules and formed hydrophobic interactions with their hydrocarbon tails. The overall interaction pattern was reproducible in two other different simulation replicates despite some minor differences (i.e. the colistin molecule bound to four lipid A molecules in the second simulation replicate and l-Dab8 electrostatically interacted with the 4′-PO4− of lipid A molecule in two simulation replicates; Figure S3). In the case of both the pEtN modified and LPS-deficient OM systems, the interactions between the colistin Dab residues and the OM were inevitably perturbed and the conformational transitions of the colistin molecule were markedly different (Figure 5A).
Spontaneous recovery of the OM structure
The OM disorganization caused by colistin A recovered spontaneously within the late umbrella sampling windows (Figure 6). The headgroups of the majority of the collapsed lipid A and lipid A-pEtN molecules moved back to the outer leaflet of the OM within 20-ns in the lipid A and lipid A-pEtN systems, respectively (Movie S1 and S2). Notably, only one lipid A or lipid A-pEtN molecule was still trapped with the colistin molecule in the OM hydrophobic region. Concomitantly, the number of the water molecules within the OM rapidly decreased by approximately 56.5% and 68.1% (compared to the maximum number of water molecules during the insertion) in lipid A and lipid A-pEtN systems, respectively, indicating OM recovery also involves the partial closure of the water pore (Figure 6A and 6B). However, in the LPS-deficient system, most of the collapsed phospholipid molecules remained in the OM hydrophobic center after 50-ns simulations and several phospholipid molecules flipped to the inner leaflet of the OM (Movie S3). Notably, in the LPS-deficient system the water pore persisted for a much longer duration (35 ns), compared to that in the lipid A (10 ns) and lipid A-pEtN (15 ns) systems (Figure 6C). The slower recovery process in the LPS-deficient system might be related to the significantly weakened interactions between the neighboring phospholipid molecules in the absence of lipid A molecules (Figure 4D and 4E).
Figure 6.
Spontaneous recovery of the outer membranes (OMs) in (A) lipid A, (B) lipid A-pEtN and (C) LPS-deficient systems. The time-evolution of the number of water molecules within the OM is plotted. The snapshots of the simulation system in the initial and final configurations are presented. Water molecules: red spheres; OM: grey lines; collapsed lipids in the initial configuration: yellow sticks; ‘Trapped’ lipids in the final configuration: cyan sticks.
Discussion
Understanding the mechanisms of polymyxin resistance in Gram-negative pathogens is an important step in the development of new-generation polymyxin antibiotics. Here, we report the first all-atom simulation study on the interactions of colistin A with three representative OMs of A. baumannii. Previous simulation studies employed simplified bacterial OMs with a symmetric LPS bilayer or an asymmetric LPS bilayer with model lipid A chemotype and phospholipid compositions.[14-15, 22-23] In the present study, the OM compositions were based on the recent results of quantitative membrane lipidomics in A. baumannii.[13] Our experimentally based OM models incorporated membrane heterogeneity and allowed for much improved precision when characterizing the interactions with colistin.[24] For example, Santos et al. explored the interaction between polymyxin B and the OM with different LPS chemotypes.[15] Unfortunately, the limitations of conventional MD simulations meant that the penetration of polymyxin B into the OM was not observed in their study. In the present study only one polymyxin molecule was examined in our steered MD simulations in order to elucidate the structure-activity relationship. Furthermore, only one drug molecule can be examined in steered MD simulations as the harmonic potential can only be applied to the center of mass of a single drug molecule.[25-26] Our all-atom simulations and enhanced sampling techniques revealed that lipid A-pEtN modification and LPS deficiency conferred resistance at different stages of colistin action on the A. baumannii OM. Our results provide the first comprehensive, atomic-scale mechanistic understanding of polymyxin resistance due to LPS modification and deficiency.
Our MD simulations revealed that lipid A-pEtN modification and LPS deficiency changed the OM properties in several different ways. Both the addition of the cationic pEtN moieties and the loss of LPS molecules reduced the overall negative charge on the OM surface which are the major electrostatic binding targets for the cationic Dab residues of polymyxins. This was especially so in the LPS-deficient system where the dramatic reduction of negative charges resulted in a significant loss of binding affinity with colistin, as reflected by the unfavorable free energy change (16.3 kcal mol−1) (Figure 3). In contrast to the traditional view that lipid A modification via addition of positive-charged moieties diminishes the initial interaction of polymyxin with the bacterial OM,[9] our results revealed that pEtN modification of lipid A did not cause a major change in the binding affinity of colistin to the OM (−5.3 kcal mol−1 for lipid A-pEtN OM; −3.0 kcal mol−1 for wild-type lipid A OM) (Figure 3). This was because one phosphate group of lipid A was modified with the pEtN moiety in our constructed lipid A-pEtN OM model consistent with lipid A profiling data of polymyxin-resistant A. baumannii strains;[27] in addition, only one colistin molecule was employed in our all-atom simulation systems. However, we should note that lipid A-pEtN OM retained approximately 77.4% negative charges on its surface relative to the wild-type lipid A OM. The free phosphate group on the other side of lipid A can still provide sufficient binding sites for the colistin molecules, Additionally, fewer divalent cations (e.g. Ca2+ and Mg2+) bound to the lipid A-pEtN and LPS-deficient OMs (Table S1). These divalent cations serve as bridging agents between the neighboring lipid A molecules and stabilize the bacterial OM.[21, 28] Interestingly, in both lipid A-pEtN and LPS-deficient systems, the cross-linking role of Ca2+ was largely replaced by the cationic ethanolamine groups of pEtN and PE, respectively (Figure 4B and 4C). These modifications inhibit the initial action of polymyxins on the bacterial OM and render the bacterial cells less susceptible to polymyxins.[14-15]
In addition to the changes in OM properties discussed above, both lipid A-pEtN modification and LPS deficiency affected the structural stability of the OM. Modification of lipid A with a pEtN group improved the structural integrity of the OM by increasing the number of hydrogen bonds in the headgroup region and creating tighter hydrophobic contacts in the hydrocarbon layer (Figure 4D and 4E). These changes thus require colistin to disrupt a greater number of bonds when interacting with the OM, sharply increasing the free energy required (by ~20 kcal mol−1) when traversing the OM compared to the wild-type lipid A OM (Z < 1 nm) (Figure 3). This is consistent with the work of Rice and Wereszczynski that examined several other types of lipid A modifications in Salmonella enterica including the addition of l-Ara4N, hydroxyl and extra palmitoyl groups.[14] These modifications also strengthened the lateral interactions between neighboring LPS molecules and, therefore, likely confer polymyxin resistance through a similar mechanism to that observed with the addition of pEtN group. Our results also support the role of PmrAB and PhoPQ in Gram-negative bacteria which regulate lipid A modifications to cope with environmental stimuli (e.g. low Mg2+ and high Fe3+ concentration, low pH, antimicrobial peptides).[8, 29] Interestingly, the loss of LPS molecules deteriorated the structural integrity of the OM (Figure 4D and 4E). This decreased the barrier of colistin A penetration into the LPS-deficient OM, reflective of the much lower ΔGtranslocate 50.4 kcal mol−1 (over 2.5 - 0 nm of the reaction coordinate along Z-axis in Figure 3) than that in the lipid A (68.5 kcal mol−1) and lipid A-pEtN (87.4 kcal mol−1) OMs; this low ΔGtranslocate value 50.4 kcal mol−1 is comparable to those reported with other peptides in previous studies obtained with simplified membrane models.[25-26] Besides, unlike the lipid A and lipid A-pEtN OMs whose disorganized structure recovered rapidly (Figure 6A and 6B), the weakened intermolecular interaction made the LPS-deficient OM more difficult to recover spontaneously, resulting in an increased OM permeability (Figure 6). The OM vulnerability explains why LPS-deficient A. baumannii strains become more susceptible to other antibiotics such as rifampicin, azithromycin and vancomycin.[30-31] These results further indicate that LPS deficiency confers polymyxin resistance primarily via decreasing the binding affinity of polymyxins with the OM, rather than strengthening the OM structure. Notably, experimental results revealed that the polymyxin resistance level conferred by LPS deficiency (minimum inhibitory concentration [MIC] = 128 μg mL−1) is usually much higher than that caused by lipid A-pEtN modification (MIC = 8-16 μg mL−1) in A. baumannii,[32] which supports that inhibition of the binding with polymyxins in LPS-deficient OM is more prominent in conferring polymyxin resistance than the suppression of polymyxin penetration in lipid A-pEtN OM.
Our study also discovered that lipid A-pEtN modification and LPS deficiency changed the interaction pattern between colistin and the OM. In the wild-type lipid A OM, the colistin molecule adopted a folded conformation when it started to enter the hydrophobic layer of the OM (Figure 1). With this folded conformation, its hydrophobic segments and positively charged Dab residues interacted separately with the carbon tails and phosphate groups of multiple lipid A molecules, respectively (Figure 5B). A similar folded conformation has been reported in previous NMR studies, although only one lipid A molecule was employed to characterize the interaction with the polymyxin molecule.[17-18] Notably, the free energy increased slowly during the initial penetration period (1 nm < Z < 2.3 nm) compared to the following penetration (Z < 1 nm) (Figure 3), indicating that the special structural folding of the colistin molecule is likely an important intermediate to facilitate the insertion into the OM. In our OM-based polymyxin-lipid A interaction model, l-Dab1, l-Dab3 and l-Dab5 of the colistin molecule preferred to electrostatically interact with the 1-phosphate groups of multiple lipid A molecules; while l-Dab8 and l-Dab9 primarily bound to the lipid A 4’-phosphate groups (Figure 5B and S3). Interestingly, the pEtN transferase expressed in A. baumannii selectively transfers the pEtN moiety to the 1-phosphate group of lipid A.[27] This means it is very likely that the lipid A-pEtN modification primarily interferes with the electrostatic interactions from the l-Dab1, l-Dab3 and l-Dab5 residues. Presumably, the loss of LPS molecules diminished the specific binding targets of colistin (i.e. the phosphate groups of lipid A) in the LPS-deficient OM, which inevitably affects the initial interaction with all colistin Dab residues. These effects caused by lipid A-pEtN modification and LPS deficiency disturb the conformational transition of the colistin molecule and eventually make bacterial cells less susceptible to colistin. The distance between the hydrophobic segments of colistin fluctuated around 1.86 nm in the lipid A containing OM, while it was larger than 2.28 nm in the lipid A-pEtN and LPS-deficient OMs. The compacted structural folding further enhances the penetration of colistin A into the lipid A OM. New-generation polymyxins are needed for the treatment of infections caused by polymyxin-resistant bacteria. Our MD results demonstrated that the pEtN modification of lipid A and LPS loss significantly decreased the binding between the phosphate groups of lipid A and the positively charged Dab residues (in particular Dab1 and Dab5 of polymyxins). Therefore, increasing the hydrophobicity of the polymyxin molecule (e.g. positions 6 and 7) can enhance the hydrophobic interactions with the bacterial OM, thereby improving the activity against polymyxin-resistant bacteria. This approach has been validated by the activity of several novel polymyxin analogues (e.g. FADDI-002 and FADDI-019 with a fatty acyl chain at position 6 or 7) against polymyxin-resistant A. baumannii.[17, 32]
Conclusions
To the best of our knowledge, this is the first systematic all-atom simulation study examining the thermodynamics governing the penetration of the colistin into three different OMs of A. baumannii. We have shown for the first time that lipid A-pEtN modification and LPS deficiency confer polymyxin resistance at different stages of colistin action on the bacterial OM. Specifically, modification of lipid A with pEtN primarily inhibits the penetration of polymyxins through the OM, whereas LPS loss significantly attenuates the binding affinity of polymyxins to the OM. All-atom characterization of the interactions between polymyxin molecules and different bacterial OMs provides valuable information on the relationship between OM remodeling and polymyxin resistance at the membrane level. These results will facilitate the discovery of new-generation polymyxins to treat infections caused by life-threatening Gram-negative ‘superbugs’.
Methods
System construction
Unlike previous MD simulation studies in which the bacterial membranes were often mimicked by model membrane compositions[14-15, 22-23] our quantitative membrane lipidomics results in A. baumannii were employed to set the lipid compositions of three representative OM models (a wild-type lipid A OM, a lipid A-pEtN modified OM and an LPS-deficient OM).[13] We employed hepta-acylated lipid A for the development of the OM models, as it is predominant in the outer membrane of A. baumannii.[13] In addition to differences in lipid A structure, the molar ratios of phospholipids are also changed between these different OMs based on the compositional analysis from experimental lipidomics data.[13] Hence, our MD models simulated ‘real’ outer membranes of A. baumannii. Polymyxins bind to the bacterial OM primarily through the electrostatic and hydrophobic interactions with lipid A;[3, 33] therefore, lipid A and lipid A-pEtN were used to construct the OM models. The detailed information of lipid components employed in the OM constructs is listed in Table S2. The structures, topology parameters and area per lipid molecule of all types of lipids, including lipid A, lipid A-pEtN, phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and cardiolipin (CL) were from the CHARMM-GUI server based on all-atom CHARMM36 force field.[34] Both the asymmetric lipid A and lipid A-pEtN OMs and symmetric LPS-deficient OM were built in a tetragonal box using CHARMM-GUI membrane builder. CHARMM-GUI employed idealized surface area per lipid to ensure similar surface areas between the outer and inner leaflets of the membrane. Previous studies revealed that the differential stress between the outer and inner leaflets of the asymmetric membrane may increase the membrane rigidity and affect the interaction with small peptides.[35-37] In the present study, TIP3P water molecules and counter-ions were introduced to hydrate and neutralize the simulation systems, respectively.
The structure of colistin A (i.e. polymyxin E1) was generated with Chem3D (Figure S4). Energy minimization using the MM2 force field was performed to relieve potential intramolecular steric clashes.[38] The topology parameters of colistin were generated in the SwissParam server by combining bonded parameters extracted from the Merck Molecular Force Field (MMFF) and non-bonded terms from the CHARMM force field.[39] To construct the simulation systems containing colistin molecule, the internal Gromacs tool gmx insert-molecules was used to place a colistin molecule above the OM surface by replacing water molecules.[40] Additionally, a certain number of counter-ions were removed to neutralize the charges in the system before simulations. Three control simulation systems were also conducted to characterize the membrane properties of lipid A, lipid A-pEtN and LPS-deficient OMs in the absence of colistin. The details of all simulation systems are summarized in Table S3.
Steered MD simulations
Steered simulation technique was used to examine the penetration of colistin into different bacterial OM models.[25-26,41] Adaptive force was applied between the centers of mass (COMs) of both the colistin molecule and the membrane along the Z axis. The initial force constant of the harmonic potential was 500 kJ mol−1 nm−2 and changed adaptively based on the interaction between the colistin molecule and the OM. The distance between the COMs was changed at a rate of 1×10−4 nm ps−1. Accordingly, 50-ns simulations were sufficient to achieve the OM penetration of colistin and track its conformational transition as previous NMR studies revealed.[17-18] The motion of the colistin molecule on the XY plane was not restrained. For each system, three independent simulations were performed.
Umbrella sampling
The structural snapshots of the simulation system generated from the steered simulation trajectories were used to construct a series of umbrella sampling simulation windows in which the COM of the colistin molecule was harmonically restrained at various fixed positions at Z axis.[25-26,41]. Due to LPS loss, the lipid components are dramatically changed in the LPS-deficient OM.[13] The longer carbon tails of the phospholipids (16 carbon atoms for phospholipids versus 12 or 14 carbon atoms for lipid A/lipid A-pEtN OM) slightly increased the thickness of the LPS-deficient OM; therefore, there were 24, 24, and 28 simulation windows constructed for the lipid A, lipid A-pEtN and LPS-deficient OM systems, respectively. Specifically, the distance interval between the configurations in neighboring windows was 0.2 nm. Each window was simulated for 50 ns to enhance the sampling of the free energy barrier region during the penetration of colistin into the OMs; hence, a total of 1.2 μs or 1.4 μs simulations were performed to obtain a single Potential of Mean Force (PMF, free energy profile). In each OM system, PMFs calculated using 10-ns time blocks and the overlap of position distributions of the colistin A molecule for different simulation windows were used to check the convergence of free-energy calculation (Figure S5). The WHAM integration algorithm was used to calculate the PMF.[42] In the PMFs, the reaction coordinates were set based on the positions relative to the OM center at the Z axis. In particular, Z > 0 nm and Z < 0 nm indicated the outer and inner leaflet of the OM, respectively, with Z = 0 nm corresponding to the OM center.
Simulation protocol
All molecular simulations were conducted using GROMACS 5.1.2 with the CHARMM36 all-atom force field.[40, 43-44] Unlike the coarse-grained approach, all-atom MD simulations were conducted in the present study to precisely elucidate the atomic interactions between colistin and bacterial OM. Before a production run, energy minimization was performed using the steepest descent algorithm with the force tolerance of 1,000 kJ mol−1 nm−1. Six equilibration cycles were conducted by gradually turning off the position restraints on lipid molecules. The surface area of the OMs was monitored to ensure that the system reached the equilibrium (Figure S6). The values of surface area per lipid of the lipid A and lipid A-pEtN molecules in the OM models were 1.78 and 1.82 nm2, respectively, which were similar to that (~1.75 nm2) reported in the literature.[45] Periodic boundary conditions were considered and all simulations were conducted at constant temperature and pressure. The temperature and pressure were maintained at 313 K using the Nose-Hoover method and 1 bar using the semi-isotropic pressure coupling approach with Parrinello-Rhaman barostat, respectively.[46-48] The Particle Mesh Ewald method was applied to treat long-range electrostatic interactions with a short-range cutoff of 1.2 nm;[49] the shifted Lennard-Jones potential algorithm was employed to calculate the Van der Waals interactions with a general cutoff of 1.2 nm and a shifting cutoff of 1.0 nm. All bonds involving hydrogen atoms were constrained using the LINCS method[50] and the time step in production simulations was 2 femtoseconds (fs).
Supplementary Material
Acknowledgements
This research was supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI132154). XKJ is recipient of the 2019 Faculty Bridging Fellowship, Monash University. JL is an Australia National Health Medical Research Council (NHMRC) Principal Research Fellow. The simulations were performed on the HPC Cloud Platform (National Key Research and Development Project, 2016YFB0201702) at Shandong University (China), and the supercomputer M3 at eResearch, Monash University (Australia). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Supporting Information
Ca2+ binding; Components of different out membranes; MD simulation systems; hydrogen bonding analysis; water pore analysis; interaction of colistin A with lipid A OM in simulation replicates; structure of colistin A; convergence of umbrella sampling simulations; surface area of the OM; movies of OM structural dynamics.
References
- [1].MacLean RC, San Millan A The evolution of antibiotic resistance. Science 2019, 365, 1082–1083. [DOI] [PubMed] [Google Scholar]
- [2].Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis 2018, 18, 318–327. [DOI] [PubMed] [Google Scholar]
- [3].Velkov T, Thompson PE, Nation RL, Li J Structure-activity relationships of polymyxin antibiotics. J. Med. Chem 2010, 53, 1898–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Landman D, Georgescu C, Martin DA, Quale J Polymyxins revisited. Clin. Microbiol. Rev 2008, 21, 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Velkov T, Roberts KD, Thompson PE, Li J Polymyxins: a new hope in combating Gram-negative superbugs? Future Med. Chem 2016, 8, 1017–1025. [DOI] [PubMed] [Google Scholar]
- [6].Srinivas P, Rivard K Polymyxin Resistance in Gram-negative Pathogens. Curr. Infect. Dis. Rep 2017, 19, 38. [DOI] [PubMed] [Google Scholar]
- [7].Baron S, Hadjadj L, Rolain JM, Olaitan AO Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [DOI] [PubMed] [Google Scholar]
- [8].Jeannot K, Bolard A, Plesiat P Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [DOI] [PubMed] [Google Scholar]
- [9].Zavascki AP, Goldani LZ, Li J, Nation RL Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother 2007, 60, 1206–1215. [DOI] [PubMed] [Google Scholar]
- [10].Trimble MJ, Mlynárčik P, Kolář M, Hancock RE Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med 2016, 6, a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis 2016, 16, 161–168. [DOI] [PubMed] [Google Scholar]
- [12].Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu Y, Lu J, Han M, Jiang X, Azad MAK, Patil NA, Lin Y, Zhao J, Hu Y, Yu HH, Chen K, Boyce JD, Dunstan RA, Lithgow T, Barlow CK, Li W, Schneider-Futschik EK, Wang J, Gong B, Sommer B, Creek DJ, Fu J, Wang L, Schreiber F, Velkov T, Li J Polymyxins bind to the cell surface of unculturable Acinetobacter baumannii and cause unique dependent resistance. Adv. Sci 2020, doi: 10.1002/advs.202000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rice A, Wereszczynski J Atomistic scale effects of lipopolysaccharide modifications on bacterial outer membrane defenses. Biophys. J 2018, 114, 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Santos DES, Pol-Fachin L, Lins RD, Soares TA Polymyxin binding to the bacterial outer membrane reveals cation displacement and increasing membrane curvature in susceptible but not in resistant lipopolysaccharide chemotypes. J. Chem. Inf. Model 2017, 57, 2181–2193. [DOI] [PubMed] [Google Scholar]
- [16].Nikaido H, Nakae T The outer membrane of Gram-negative bacteria. Adv. Microb. Physiol 1980, 20, 163–250. [DOI] [PubMed] [Google Scholar]
- [17].Velkov T, Roberts KD, Nation RL, Wang J, Thompson PE, Li J Teaching 'old' polymyxins new tricks: new-generation lipopeptides targeting gram-negative 'superbugs'. ACS Chem. Biol 2014, 9, 1172–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mares J, Kumaran S, Gobbo M, Zerbe O Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. J. Biol. Chem 2009, 284, 11498–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Laederach A, Andreotti AH, Fulton DB Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry 2002, 41, 12359–12368. [DOI] [PubMed] [Google Scholar]
- [20].Vineeth Kumar TV, Sanil G A review of the mechanism of action of amphibian antimicrobial peptides focusing on peptide-membrane interaction and membrane curvature. Curr. Protein Pept. Sci 2017, 18, 1263–1272. [DOI] [PubMed] [Google Scholar]
- [21].Pontes FJ, Rusu VH, Soares TA, Lins RD The effect of temperature, cations, and number of acyl chains on the lamellar to non-lamellar transition in lipid-A membranes: a microscopic view. J. Chem. Theory Comput 2012, 8, 3830–3838. [DOI] [PubMed] [Google Scholar]
- [22].Jefferies D, Hsu PC, Khalid S Through the lipopolysaccharide glass: A potent antimicrobial peptide induces phase changes in membranes. Biochemistry 2017, 56, 1672–1679. [DOI] [PubMed] [Google Scholar]
- [23].Fu L, Wan M, Zhang S, Gao L, Fang W Polymyxin B loosens lipopolysaccharide bilayer but stiffens phospholipid bilayer. Biophys. J 2019, 118, 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Khondker A, Dhaliwal AK, Saem S, Mahmood A, Fradin C, Moran-Mirabal J, Rheinstädter MC Membrane charge and lipid packing determine polymyxin-induced membrane damage. Commun. Biol 2019, 2, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yesylevskyy S, Marrink SJ, Mark AE Alternative mechanisms for the interaction of the cell-penetrating peptides penetratin and the TAT peptide with lipid bilayers. Biophys. J 2009, 97, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao J, Zhao C, Liang G, Zhang M, Zheng J Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J. Chem. Inf. Model 2013, 53, 3280–3296. [DOI] [PubMed] [Google Scholar]
- [27].Raetz CR, Reynolds CM, Trent MS, Bishop RE Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem 2007, 76, 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pink D, Truelstrup Hansen L, Gill TA, Quinn BE, Jericho M, Beveridge T Divalent calcium ions inhibit the penetration of protamine through the polysaccharide brush of the outer membrane of Gram-negative bacteria. Langmuir 2003, 19, 8852–8858. [Google Scholar]
- [29].Olaitan AO, Morand S, Rolain JM Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol 2014, 5, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].García-Quintanilla M, Carretero-Ledesma M, Moreno-Martínez P, Martín-Peña R, Pachón J, McConnell MJ Lipopolysaccharide loss produces partial colistin dependence and collateral sensitivity to azithromycin, rifampicin and vancomycin in Acinetobacter baumannii Int. J. Antimicrob. Agents 2015, 46, 696–702. [DOI] [PubMed] [Google Scholar]
- [31].Li J, Nation RL, Owen RJ, Wong S, Spelman D, Franklin C Antibiograms of multidrug-resistant clinical Acinetobacter baumannii: promising therapeutic options for treatment of infection with colistin-resistant strains. Clin. Infect. Dis 2007, 45, 594–598. [DOI] [PubMed] [Google Scholar]
- [32].Velkov T, Gallardo-Godoy A, Swarbrick JD, Blaskovich MAT, Elliott AG, Han M, Thompson PE, Roberts KD, Huang JX, Becker B, Butler MS, Lash LH, Henriques ST, Nation RL, Sivanesan S, Sani MA, Separovic F, Mertens H, Bulach D, Seemann T, Owen J, Li J, Cooper MA Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative Bacteria. Cell Chem. Biol 2018, 25, 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Raetz CR, Whitfield C Lipopolysaccharide endotoxins. Annu. Rev. Biochem 2002, 77, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee J, Patel DS, Ståhle J, Park S-J, Kern NR, Kim S, Lee J, Cheng X, Valvano MA, Holst O CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput 2018, 15, 775–786. [DOI] [PubMed] [Google Scholar]
- [35].Doktorova M, Weinstein H Accurate in silico modeling of asymmetric bilayers based on biophysical principles. Biophys. J 2018, 115, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hossein A, Deserno M Spontaneous curvature, differential stress, and bending modulus of asymmetric lipid membranes. Biophys. J 2020, 118, 624–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Park S, Beaven AH, Klauda JB, Im W How tolerant are membrane simulations with mismatch in area per lipid between leaflets? J Chem Theory Comput. 2015, 77, 3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Allinger N MM2: A hydrocarbon force field utilizing V, and V2 torsional terms. J. Am. Chem. Soc 1997, 99, 8127–8134. [Google Scholar]
- [39].Zoete V, Cuendet MA, Grosdidier A, Michielin O SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem 2011, 32, 2359–2368. [DOI] [PubMed] [Google Scholar]
- [40].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ GROMACS: fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- [41].Sun D, Forsman J, Lund M, Woodward CE Effect of arginine-rich cell penetrating peptides on membrane pore formation and life-times: A molecular simulation study. Phys. Chem. Chem. Phys 2014, 16, 20785–20795. [DOI] [PubMed] [Google Scholar]
- [42].Hub JS, De Groot BL, Van Der Spoel D g_wham: A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput 2010, 6, 3713–3720. [Google Scholar]
- [43].Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr, Pastor RW Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Botan A, Favela-Rosales F, Fuchs PF, Javanainen M, Kanduč M, Kulig W, Lamberg A, Loison C, Lyubartsev A, Miettinen MS Toward atomistic resolution structure of phosphatidylcholine headgroup and glycerol backbone at different ambient conditions. J. Phys. Chem. B 2015, 119, 15075–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim S, Patel DS, Park S, Slusky J, Klauda JB, Widmalm G, Im W Bilayer properties of lipid A from various Gram-negative bacteria. Biophys. J. 2016, 111, 1750–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Parrinello M, Rahman A Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys 1981, 52, 7182–7190. [Google Scholar]
- [47].Nosé S A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys 1984, 52, 255–268. [Google Scholar]
- [48].Hoover WG Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695. [DOI] [PubMed] [Google Scholar]
- [49].Darden T, York D, Pedersen L Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys 1993, 98, 10089–10092. [Google Scholar]
- [50].Hess B, Bekker H, Berendsen HJ, Fraaije JG LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem 1997, 18, 1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.