Abstract
Epigenetic regulators play a critical role in normal and malignant hematopoiesis. Deregulation, including epigenetic deregulation, of the HOXA gene cluster drives transformation of about 50% of acute myeloid leukemia (AML). We recently showed that the histone 3 lysine 9 methyltransferase SETDB1 negatively regulates the expression of the proleukemic genes Hoxa9 and its cofactor Meis1 through deposition of promoter H3K9 trimethylation in MLL-AF9 leukemia cells. Here, we investigated the biological impact of altered SETDB1 expression and changes in H3K9 methylation on AML. We demonstrate that SETDB1 expression is correlated to disease status and overall survival in AML patients. We recapitulated these findings in mice, where high expression of SETDB1 delayed MLL-AF9 mediated disease progression by promoting differentiation of leukemia cells. We also explored the biological impact of treating normal and malignant hematopoietic cells with an H3K9 methyltransferase inhibitor, UNC0638. While myeloid leukemia cells demonstrate cytotoxicity to UNC0638 treatment, normal bone marrow cells exhibit an expansion of cKit+ hematopoietic stem and progenitor cells. Consistent with these data, we show that bone marrow treated with UNC0638 is more amenable to transformation by MLL-AF9. Next generation sequencing of leukemia cells shows that high expression of SETDB1 induces repressive changes to the promoter epigenome and downregulation of genes linked with AML, including Dock1 and the MLL-AF9 target genes Hoxa9, Six1, and others. These data reveal novel targets of SETDB1 in leukemia that point to a role for SETDB1 in negatively regulating pro-leukemic target genes and suppressing AML.
Introduction
Epigenetic deregulation has emerged as an important contributor to oncogenesis and disease progression in a variety of malignancies, including leukemia.1 Deep sequencing has revealed that genes encoding epigenetic modifying proteins are mutated in over 70% of acute myeloid leukemia (AML) patients.2 H3K9me2/3 marks large regions of condensed transcriptionally inactive chromatin, such as pericentric heterochromatin.3 H3K9me2/3 also plays a functional role in the dynamic repression of genes in euchromatic regions of the genome.4 Two families of proteins are associated with deposition of H3K9 methylation: the SUV39 family and PRDM family. The SUV39 family of H3K9 methyltransferases consists of SUV39H1/2, EHMT1/2, and SETDB1/2.4 Our lab and others have previously demonstrated that members of the SUV39 family of H3K9 methyltransferases bind to the polymerase associated factor complex (PAF1c).5,6 SETDB1, G9a (EHMT2), and GLP (EHMT1), were identified in a proteomics study exploring the interactome of the PAF1c in AML.5 The PAF1c is an epigenetic regulator complex that physically associates with RNA polymerase II (RNAPII) and both positively and negatively regulates gene transcription.7–11 In AML, the PAF1c is critical for the regulation of a proleukemic HOXA gene program in AML cells through the recruitment of MLL and MLL-fusion proteins to the HOXA locus via direct physical interactions.12–15 Hoxa9 and its co-factor MEIS1 are upregulated in about 50% of AML and associated with a poor patient prognosis.16 Given our recent data linking H3K9 methyltransferases with Hoxa9 and Meis1 repression along with altered H3K9me3 in AML patients compared to CD34+ cells,17 it is important to understand the epigenetic and biological impact of H3K9 methyltransferases on AML.
SETDB1 is a H3K9 mono/di/tri-methyltransferase involved in heterochromatin regulation and euchromatic gene silencing.18 SETDB1 was shown to bind gene loci associated with development in mouse embryonic stem (ES) cells, such as the Hoxd cluster of genes.19 SETDB1 has been implicated as an oncogene in melanoma, breast cancer, liver cancer, and lung cancer.4 Importantly, Ceol et al. reported amplification of SETDB1 in melanoma results in aberrant binding and regulation of the HOXA locus.20 In contrast to these oncogenic roles for SETDB1, Avgustinova and colleagues report that depletion of the H3K9 methyltransferase G9a in squamous tumors leads to a delayed, but more aggressive phenotype due to expanded cancer progenitor pools with increased genomic instability. 21 In the hematopoietic system, the methyltransferase activity of G9a is required for leukemogenesis due to a physical interaction with HOXA9.22 Importantly, loss of G9a has no effect on hematopoietic stem cells.22,23 SETDB1, however, is required for both hematopoietic stem and progenitor cell (HSPC) maintenance and leukemic stem cells.23 Further, Cuellar and colleagues show that SETDB1 mediated silencing of endogenous retroviral elements is required for the growth of AML cell lines.24 Together, these studies suggest that therapeutic targeting of SETDB1 may benefit AML patients. However, we recently demonstrated that SETDB1 negatively regulates the expression of the pro-leukemic Hoxa9 and Meis1 genes in MLL-AF9 transformed AML cells through association with the PAF1c, which localizes to Hoxa and Meis1 loci. The PAF1c-SETDB1 interaction mediates promoter H3K9me3 and repression of Hoxa9 and Meis1 expression.5 Further, SETDB1 expression is inversely correlated with HOXA9 and MEIS1 expression in AML patient samples.5 These data imply a more complex role for H3K9 methylation in AML similar to skin tumors whereby H3K9 methyltransferases display both oncogenic and suppressive roles.20,21 Thus, further investigation into the role of H3K9 methyltransferases in AML is required.
Here we show that AML patients with higher expression of SETDB1 display a better prognosis, consistent with repression of HOXA9 and MEIS1 expression. SETDB1 overexpression induces cellular differentiation and delays disease onset in a mouse model of AML, recapitulating AML patient survival. We also investigated the utility of inhibiting H3K9 methyltransferases in AML cells and HSPC, demonstrating that inhibition of H3K9 methylation in HSPC leads to retention of self-renewal capacity in HSPC and more efficient transformation by the MLL-AF9 fusion protein. Finally, we show that SETDB1 regulates gene expression by inducing changes in the epigenetic landscape and chromatin accessibility at gene targets critical to leukemogenesis.
Methods
Patient sample data
Data for patient gene expression relative to normal hematopoietic cells were mined from BloodPool on BloodSpot database.25 Bloodspot assigns AML patient samples to a closest normal hematopoietic counterpart using transcriptomic profiles.26 AML patient RNA sequencing (RNA-seq) and survival data were mined from The Cancer Genome Atlas.2
Cell line generation
Cell lines were generated from C57Bl/6 (Taconic Farms) mouse bone marrow or from SETDB1floxed27 mice. Platinum-E viral packaging cells were transfected with the indicated constructs: MSCVneo-FLAG-MLL-AF9 (MA9), MSCVneo-FLAG-E2A-HLF (EHF), MSCVhygro-FLAG-EHMT2 (G9a) (Ge lab; Addgene #41721), MSCVpuro-HA-SETDB1 or empty vector (EV) controls. Cells were spinfected with viral supernatants and 5 ug/mL polybrene (Millipore), selected with 1 mg/mL G418 (Invitrogen) and 1 ug/mL puromycin (Invitrogen) or 200 ug/mL hygromycin (Invitrogen) and cultured in IMDM with 15% stem cell fetal bovine serum (Millipore), 1% penicillin/ streptomycin (Invitrogen), 10 ng/mL interleukin-3 (IL-3) and 100 ng/mL stem cell factor (R&D).
Mouse modeling
Primary MLL-AF9 mouse leukemia cells were retrovirally transduced with MSCVpuro-HA-SETDB1 or EV and selected in 2 ug/mL puromycin for 4 days. 100,000 cells were injected intravenously into lethally irradiated (950 rads) C57Bl/6 mice. Mice were monitored for survival, moribund mice were euthanized, and bone marrow, spleen, and liver were harvested. Animal studies were approved by the University of Michigan’s Committee on Use and Care of Animals and Unit for Laboratory Medicine.
Quantitative PCR (qPCR)
RNA was harvested using the Qiagen RNeasy mini plus kit. cDNA synthesis was performed using the SuperScript III kit (Invitrogen). qPCR was performed using fast SYBR-green mastermix (Thermo Fisher). Primers sequences are listed in the Online Supplementary Table S2.
Sequencing libraries preparation
MA9+ empty vector (EV) and MA9+SETDB1 cells were harvested for next generation sequencing library preparation for RNAseq, ChIP-seq, and ATAC-seq. ChIP-seq antibodies were validated using Epicypher histone peptide arrays (Online Supplementary Figure S6). Library preparation details are in the Online Supplementary Materials and Methods.
Data availability
Sequencing data is available via the Gene Expression Omnibus, accession GSE136850.
Results
SETDB1 expression is correlated with AML patient prognosis
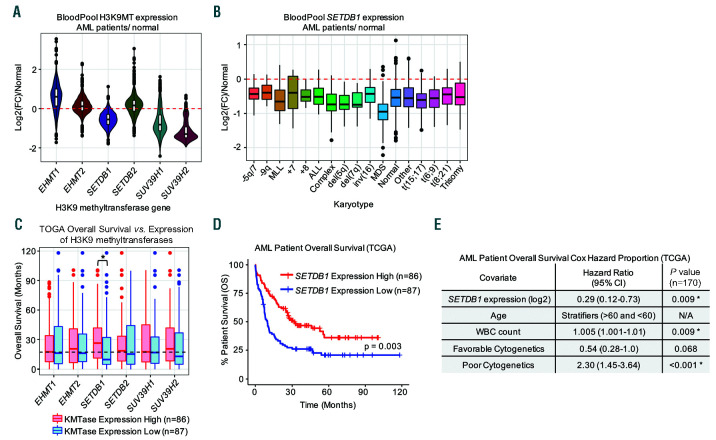
Given our data linking H3K9 methyltransferases with HOXA9 and MEIS1 expression, we investigated their expression in AML patient sample data. Interestingly, SETDB1, SUV39H1, and SUV39H2 exhibit lower expression in AML patient samples when compared with normal hematopoietic cells, with median expression levels that are 66%, 56%, and 41% relative to their nearest normal hematopoietic counterparts, respectively (Figure 1A).25,26 The downregulation of these genes in AML samples was consistent regardless of AML karyotype (Figure 1B and Online Supplementary Figure S1A-B).25,26 We next tested whether H3K9 methyltransferase gene expression significantly correlated with patient survival using publicly available RNA-seq data2 and found that only SETDB1 expression significantly correlated with patient survival (Figure 1C). Median survival for patients with SETDB1 expression above the median was 26.3 months and 9.5 months in patients with SETDB1 expression below the median (Figure 1D).2 Univariate and multivariate analyses reveal that higher expression of SETDB1 is associated with a higher overall survival rate with a P<0.003 and a lower expected hazard ratio of 0.29 per two-fold change in expression (Figure 1D-E).2
Figure 1.
SETDB1 expression is low in acute myeloid leukemia (AML) and correlated with AML patient survival. (A) BloodPool microarray gene expression data mined from Bloodspot shows AML patient gene expression relative to the nearest normal hematopoietic counterpart for the indicated genes encoding H3K9 methyltransferases (n=1991) in a violin plot. Max probe was used for all genes. (B) BloodPool data for AML patients’ SETDB1 expression relative to nearest normal counterpart divided by karyotype (n=1,991) shown in a boxplot. Max probe was used for SETDB1 expression. (C) TCGA RNA-seq patient sample data is divided by “high” expression (above median) and “low” expression (below median) for the indicated gene encoding an H3K9 methyltransferase. Boxplot shows overall survival for each stratified gene (n=173). (D) Kaplan Meier curve showing overall survival of AML patients stratified by SETDB1 expression above (“high”) or below (“low”) median (n=173). (E) Multivariate analysis using Cox Hazard Proportion analysis to assess the hazard ratio associated with changing levels of SETDB1 expression. SETDB1 expression is expressed in log2 (RSEM), so the hazard ratio is associated with a two-fold change in SETDB1 expression. Age did not satisfy the model and was stratified by patients <60 years old and patients >60 years old (n=173). Statistics: log-rank followed by multiple testing correction using Benjamani-Hochberg false discovery rate (FDR) (C); log-rank (D); Cox Proportional Hazard Model (E); n: biological replicates; *: P/padj<0.05; RNA-seq: RNA sequencing.
SETDB1 or G9A expression reduces AML growth and colony formation
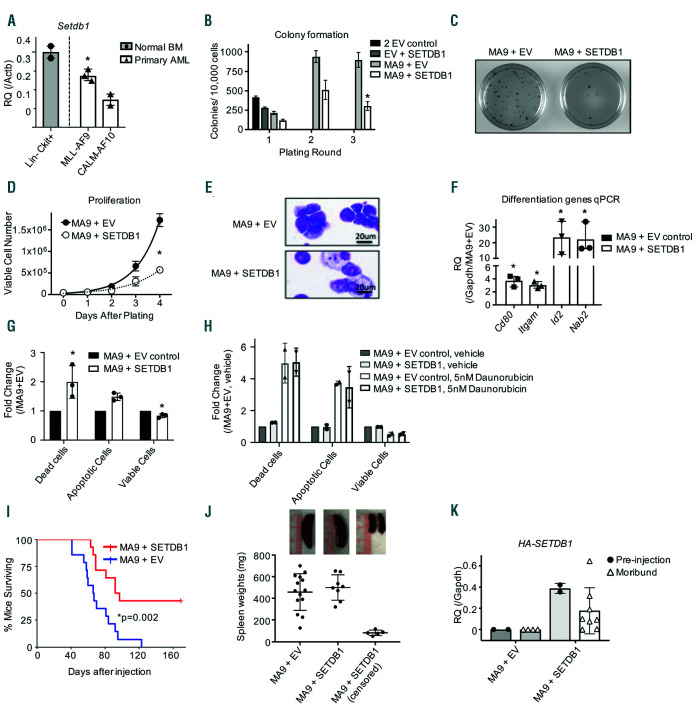
We next explored the biological effects of SETDB1 expression on the transformation and growth of AML cells. First, we explored whether there is a difference in Setdb1 expression in mouse AML relative to normal HSPC by isolating lineage negative (Lin–) cKit+ cells from mouse bone marrow. qPCR demonstrates that Setdb1 expression is reduced in murine MLL-AF9 and CALM-AF10 leukemias compared to normal HSPC, consistent with the patient sample data (Figure 2A). We next performed colony replating assays where Lin– mouse bone marrow cells were retrovirally co-transduced with the MLL-AF9 fusion oncogene with and without SETDB1 and plated in semi-solid methylcellulose. Overexpression of human SETDB1 significantly reduced MLL-AF9 mediated colony formation (Figure 2B-C) while not affecting the expression of the exogenous MLL-AF9 (Online Supplementary Figure S2A).24 Colony formation driven by a separate leukemic fusion protein, E2A-HLF, was also reduced in the presence of SETDB1, suggesting a general effect on AML transformation (Online Supplementary Figure S2B-C). Ex vivo proliferation assays demonstrate that overexpression of SETDB1 in MLL-AF9 or E2A-HLF transformed AML cells leads to a significant reduction in cellular proliferation (Figure 2D and Online Supplementary Figure S2D). Interestingly, we observed a strong selective pressure to reduce exogenous SETDB1 expression in cultured MLLAF9+ SETDB1 cells (Online Supplementary Figure S2E) resulting in rescue of MLL-AF9 cellular proliferation (Online Supplementary Figure S2F). MLL-AF9 cells that overexpress SETDB1 undergo morphological changes consis-tent with differentiation (Figure 2E). Genes associated with hematopoietic differentiation, including Id2, Cd80, Nab2, and Itgam have significantly increased expression upon overexpression of SETDB1 (Figure 2F). Additionally, overexpression of SETDB1 in MLL-AF9 cells leads to increased apoptosis (Figure 2G and Online Supplementary Figure S2G). We next examined whether SETDB1 expression is associated with reduced chemoresistance. MA9+SETDB1 do not exhibit reduced apoptosis after treatment with Daunorubicin at their IC50 (Figure 2H and Online Supplementary Figure S2H-I), suggesting SETDB1 expression is not associated with increased Daunorubicin sensitivity. Overexpression of another H3K9 methyltransferase, human G9A, also reduces colony formation and proliferation of MLL-AF9 cells (Online Supplementary Figure S3A-C) and induces morphological changes consistent with differentiation (Online Supplementary Figure S3D). These data demonstrate that expression of multiple H3K9 methyltransferases reduces AML cell proliferation and colony forming potential and induces AML differentiation.
Figure 2.
Overexpression of SETDB1 delays acute myeloid leukemia growth. (A) Quantitative PCR (qPCR) using primers for mSetdb1 to determine expression levels in primary MLL-AF9 (n=3) or CALM-AF10 (n=2) acute myeloid leukemia (AML) cells compared to Lin-cKit+ mouse bone marrow hematopoietic stem and progenitor cell (HSPC) (n=2 pools of 5 mice each). (B) Mouse Lin- bone marrow was retrovirally transduced with the indicated plasmid vectors and plated in methylcellulose. Colonies were counted after 7 days and re-plated, for a total of three rounds. Shown is one representative experiment of n=4, error bars are standard deviations of the technical replicates. (C) Representative Iodonitrotetrazolium chloride (INT) staining of colony assay plates for MA9 cells with or without SETDB1 overexpression. Images were taken on a BioRad ChemiDoc XRS+ at 1X magnification at the same time with the same contrast. (D) Lin- bone marrow cells were retrovirally transduced with MA9 in the presence or absence of SETDB1 overexpression, selected for 2 weeks, then proliferation was monitored by viable cell count daily. Shown is one representative experiment of n=4. Error bars are standard deviations of technical replicates. (E) Representative cytospin and Hema3 stained MA9 cells in the presence or absence of SETDB1 overexpression (n=3). (F) qPCR measurement of genes associated with differentiation in MA9 cells in the presence or absence of SETDB1 overexpression. Data are normalized to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) relative to MA9+EV gene expression (delta delta ct), n=3. Error bars are standard deviation of delta delta ct. (G-H) Quantification of fluorescence-activated cell sorting (FACS) analysis to determine the relative amount of live, dead, and dying cells in (G) resting AML cells or (H) AML cells treated with Daunorubicin. Cells were stained with FITC-Annexin V and DAPI and run on an LSRII flow cytometer. Cells are defined as follows: AnnexinV+,DAPI+ = dead; AnnexinV-,DAPI- = alive; AnnexinV+,DAPI- = apoptotic. Shown are relative quantifications comparing the SETDB1 overexpression cells to control. Error bars indicate standard deviations. (G) n=3, (H) n=2. (I) Survival for MLL-AF9 AML mouse model. Primary MA9 cells were transduced with SETDB1 or EV control and selected for 4 days prior to tail vein injections in sublethally irradiated mice. Shown is the Kaplan Meier survival curve; MA9+EV n=14; MA9+SETDB1 n=8 (six censored mice did not develop AML). (J) Spleen weights of moribund or censored mice from MLL-AF9 AML mouse model. Error bars represent standard deviations. Above are representative images of spleens from euthanized mice. (K) qPCR using primers specific for the HA-SETDB1 exogenous construct from the AML mouse model to measure expression of the plasmid. RNA was harvested before injection or from moribund mice (n=2, 4 MA9+EV preinjection, moribund mice; n=2, 8 MA9+SETDB1 pre-injection, moribund mice). Error bars represent standard deviations. Statistics: significance was determined by: two-sample t-test comparing relative expression (Actb or Gapdh) of AML primary cells to Lin-cKit+ for Setdb1 expression(A) or MA9+SETDB1 to MA9+EV for four different genes’ expression (F); generalized linear modeling followed by ANOVA where each MA9+SETDB1 replicate was paired to the MA9+EV control from the same biological replicate. Main effect is reported if there are no significant interactions (see statistical analysis in the Online Supplementary Materials and Methods). (B/D/G/H); Log-rank test (I). MA9: MLL-AF9; EV: empty vector control; n: biological replicates; *: P<0.05: Actb: beta actin; Gapdh: glyceraldehyde 3-phosphate dehydrogenase.
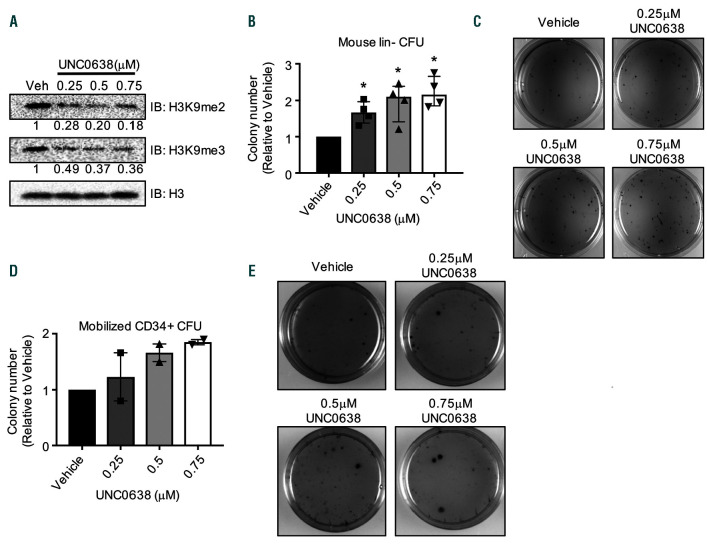
Figure 3.
H3K9 methyltransferase inhibitor UNC0638 enhances hematopoietic stem and progenitor cell colony formation capacity. (A-B) Lin- mouse bone marrow was isolated and treated in culture for 4 days with the indicated dose of UNC0638. (A) Cells pretreated with UNC0638 were harvested, lysed in SDS loading buffer, and run on SDS-PAGE. Shown is a Western blot probed for the indicated antibodies. Below the H3K9me2/3 Western blots are numbers indicating the band densities normalized to the Western blot probing for total H3 and normalized to vehicle control. (C) Cells pretreated with UNC0638 were plated in methylcellulose and colonies were counted after 7 days, n=4. Colony numbers are shown relative to the vehicle/non-silencing control for each replicate, bar graphs represent the mean and error bars represent the standard deviation of these normalized values. (C) INT stained representative colonies from (B). (D) Isolated human CD34+ cells were treated for 4 days with the indicated doses of UNC0638 and plated in methylcellulose. Colonies were counted after 14 days, n=2. Colony numbers are shown relative to the vehicle/ non-silencing control for each replicate, bargraphs represent the mean and error bars represent the standard deviation of these normalized values. (E) INT stained representative colonies from (D). Statistics: significance was determined by generalized linear modeling followed by ANOVA where each treated group was paired to the vehicle treatment from the same biological replicate. Main effect is reported if there are no significant interactions (B/D) (See statistical analysis in Online Supplementary Materials and Methods); n: biological replicates; *: P<0.05: INT: 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride; HSPC: hematopoietic stem and progenitor cell.
SETDB1 expression delays MLL-AF9 mediated AML
To examine the effects of SETDB1 in vivo, we transplanted primary mouse MLL-AF9 AML cells retrovirally transduced with or without SETDB1 into sublethally irradiated syngeneic recipient mice and monitored survival. Consistent with AML patient data, overexpression of SETDB1 significantly delays MLL-AF9 mediated leukemogenesis in vivo (Figure 2I). All moribund mice from both the control MLL-AF9 group and MLL-AF9+SETDB1 group exhibited splenomegaly, leukemic infiltration in the liver (Figure 2J and Online Supplementary Figure S3E) and similar MLL-AF9 expression levels (Online Supplementary Figure S3F). We measured expression of exogenous SETDB1 in MLL-AF9 leukemic cells before injection and in bone marrow of moribund mice and observed a decrease in exogenous SETDB1 expression in 6 of 8 mice (Figure 2K). These data demonstrate that SETDB1 expression suppresses MLL-AF9 mediated leukemic progression in vivo.
H3K9 methylation impairs HSPC colony formation and suppresses leukemic transformation
We examined how loss of Setdb1 affects normal HSPC and AML growth and transformation. First, using inducible CreER-mediated knock-out of Setdb1, we confirmed that Setdb1 is required for MLL-AF9 cell growth23 and demonstrated that Setdb1 is also required for the growth of E2A-HLF leukemic cells (Online Supplementary Figure S4A-B). We next sought to determine the effect of reduced expression but not deletion of Setdb1 on AML cell growth. However, heterozygous deletion of Setdb1 in MLL-AF9-Setdb1fl/+-CreER cells did not reduce Setdb1 protein (Online Supplementary Figure S4C). Therefore, we utilized short hairpin RNA (shRNA) mediated knockdown of Setdb1 using a doxycycline inducible MLL-AF9 Tet-on cell line in competitive growth assays. Interestingly, our results were inconclusive where two shRNA constructs with efficient knockdown displayed negative selection, while two less efficient shRNA showed a positive selection compared to MLL-AF9-Tet-on cells relative to the untreated control (Online Supplementary Figure S4D-E). This may suggest Setdb1 is maintained within a narrow expression window in AML cells.
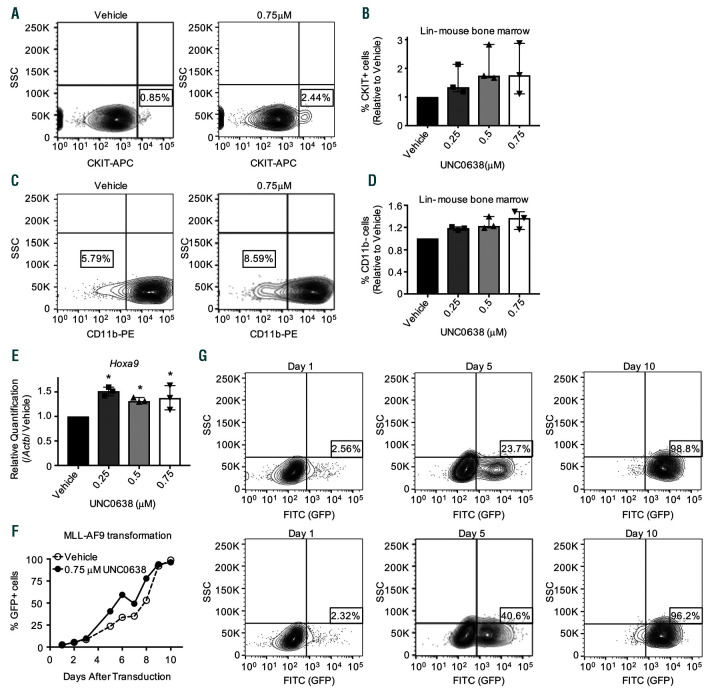
Figure 4.
H3K9 methyltransferase inhibitor UNC0638 preserves primitive hematopoietic cells amenable to MLL-AF9 transformation. (A-G) Lin- mouse bone marrow was treated with the indicated doses of UNC0638 for 4 days. (A-D) Cells were stained with anti-CKIT conjugated to APC fluorophore (A-B) or anti-Cd11b conjugated to PE fluorophore (C-D). Flow cytometry was performed to analyze cKit+ or Cd11b– populations. (A/C) are representative flow plots for 0.75 μM UNC0638 treatments. B/D show the biological replicates for cKit+ populations (B) or Cd11b– populations (D) of treated cells relative to vehicle (n=3). Bar graphs represent the median and error bars show the range of these normalized values. (E) RNA was harvested after 4 days of treatment with UNC0638 and quantitative PCR (qPCR) was used to determine changes in Hoxa9 expression. Plotted are the biological replicates relative to vehicle. Bar graphs represent the median and error bars show the range of these normalized values. (F) After treatment with UNC0638, cells were spinfected with MigR1-MLL-AF9, which also expresses a green fluorescent protein (GFP) reporter. Cells were monitored for GFP expression by flow cytometry until 100% GFP was achieved. (G) Representative flow plots from different time points during the GFP monitoring experiment. For F-G: shown is one representative experiment of n=4. Statistics: significance was determined by two-sample t-test comparing relative expression (Gapdh) of treated cells compared to vehicle cells (E). Significance was determined by generalized linear modeling followed by ANOVA where each treated group was paired to the vehicle treatment from the same biological replicate. Main effect is reported if there are no significant interactions(C-D) (see statistical analysis in the Online Supplementary Materials and Methods); n: biological replicates; *: P<0.05; Gapdh: glyceraldehyde 3-phosphate dehydrogenase.
Figure 5.
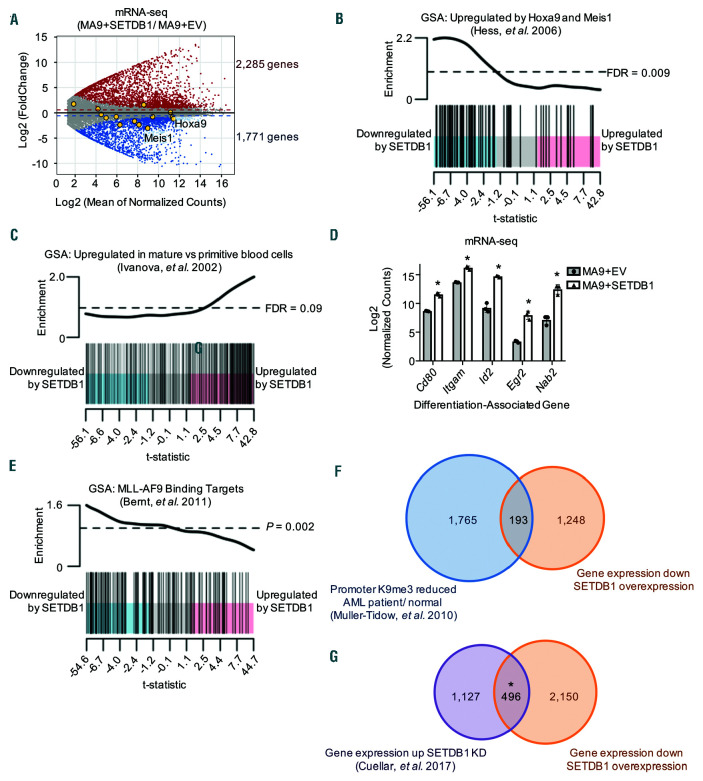
SETDB1 overexpression downregulates oncogenic gene programs and upregulates differentiation gene programs in acute myeloid leukemia. (A) MA plot shows fold changes of genes in MA9+SETDB1/ MA9+EV versus the average expression of those genes in all samples. Red highlighted genes are significantly upregulated; blue highlighted genes are significantly downregulated; yellow highlighted genes are the Hoxa cluster of genes and the Hoxa9 cofactor Meis1 (n=3/ condition). (B) Gene set analysis (GSA) using CAMERA shows that genes that are upregulated by Hoxa9 and Meis1 are downregulated by SETDB1. (C) GSA analysis using CAMERA shows that genes that are upregulated in mature blood cells relative to primitive blood cells are upregulated by SETDB1. (D) DESeq2-normalized RNA-seq counts of genes associated with differentiation (n=3/ condition). Error bars represent the range of counts. (E) GSA analysis using ROAST shows that genes that are directly bound by MLL-AF9 are downregulated by SETDB1. (F) Overlap analysis of genes that are downregulated by SETDB1 in mouse MA9 cells and genes that have reduced promoter H3K9me3 in acute myeloid leukemia (AML) patient samples compared to normal human CD34+ cells. (G) Overlap analysis of genes that are downregulated by SETDB1 in mouse MA9 cells and genes that are upregulated upon SETDB1 knockdown by Crispr-Cas9 in human THP-1 cells. For both data sets in G, FDR of 0.1 was used as a cutoff for consistency with the previously published THP-1 dataset. Statistics: unless otherwise noted, significant gene expression changes are defined by DESeq2 algorithm with fold change >1.5 and padj<0.05 (A/D/F); false discovery rate (FDR) is calculated by CAMERA using Benjamani-Hochberg correction (B-C); Hypergeometric tests (F-G). For RNA-seq, n = 3 for each cell condition. EV: empty vector control; n: biological replicates; *: P/padj<0.05.
To test the effect of Setdb1 knock-down on HSPC colony formation we transduced Lin– bone marrow with shRNA targeting Setdb1. Our results show a trend for increased HSPC colony formation without changes in Hoxa9 or Meis1 expression following Setdb1 knockdown (Online Supplementary Figure S4F, data not shown). Because inhibition of H3K9 methyltransferases has been proposed as a therapeutic option to treat AML,22,23 we explored the effects of chemical inhibition of H3K9 methylation in normal and malignant hematopoietic cells. Without a selective SETDB1 small molecule inhibitor, we utilized the G9a inhibitor UNC0638, which addresses the function of H3K9 methyltransferases more broadly. Lin– mouse bone marrow cells treated with UNC0638 exhibit a reduction in both H3K9me2 and H3K9me3 (Figure 3A).28,29 Given our data demonstrating Hoxa9 and Meis1 are repressed by H3K9 methylation,5 we tested the effect of UNC0638 on HSPC self-renewal. Previous reports have demonstrated that mouse Lin–Ska1+cKit+ cells (LSK) are preserved in culture following treatment with UNC0638.30 To explore this further, Lin– cells were treated with increasing doses of UNC0638 for five days prior to plating in semi-solid methylcellulose in the presence of SCF and IL-3. Interestingly, treatment with UNC0638 significantly increased colony formation capacity of Lin– cells in a dosedependent manner (Figure 3B-C). Further, human CD34+ cells isolated from mobilized peripheral blood and treated with increasing doses of UNC0638 demonstrate increased colony formation capacity (Figure 3D-E), consistent with reports that UNC0638 preserves CD34+ cells in culture.31 Thus, chemical or genetic inhibition of H3K9 methyltransferases preserves self-renewal capacity of bone marrow cells.
We also examined the effects of UNC0638 chemical inhibition of H3K9 methylation on AML cells. Consistent with previous studies, treatment with UNC0638 results in reduced cellular proliferation of MLL-AF9 cells (Online Supplementary Figure S5A).22 Since UNC0638 preserves self-renewal of HSPC (Figure 3), we asked whether inhibition of H3K9me alters MLL-AF9 mediated transformation of bone marrow cells. First, we found that UNC0638 treatment of Lin– bone marrow cells preserve more primitive cKit+ and Cd11b– populations (Figure 4A-D and Online Supplementary Figure S5B-C). Lin– cells display significantly increased Hoxa9 expression in response to UNC0638 treatment (Figure 4E). Lin–cKit+ (LK) and related primitive hematopoietic cell populations are more amenable to transformation than more differentiated subtypes.32 To explore whether this expansion of LK cells results in greater AML transformation capacity, we pretreated Lin– bone marrow cells with UNC0638 for four days then retrovirally transduced them with MigR1-MLL-AF9 and monitored green fluorescent protein (GFP) by flow cytometry. We observed a more rapid expansion of GFP+ MLLAF9 cells following treatment with UNC0638 compared to vehicle, with a 1.4-1.7 fold increase in GFP+ cells before both populations reached 100% GFP positivity (Figure 4FG and Online Supplementary Figure S5D). One of four replicates demonstrated a more rapid expansion of vehicle treated cells; however, GFP+ cells were increased two-fold in vehicle treated cells at day 1 suggesting this is attributable to differences in transduction rates. Our combined data suggests that H3K9 methyltransferases can suppress leukemic transformation and may point to a narrow window of H3K9 methylation that is optimal for leukemic transformation and cell growth.
SETDB1 regulates oncogenic gene programs in AML
To explore the gene programs regulated by SETDB1 in AML, we performed RNA-sequencing experiments on MLL-AF9 cells overexpressing SETDB1. 2,285 genes are upregulated and 1,771 genes are downregulated by SETDB1 overexpression with a fold-change of 1.5 or more and an false discovery rate (FDR) of <0.05 (Figure 5A and Online Supplementary Table S4). Notably, many genes found in the Hoxa cluster were significantly downregulated, including Hoxa3, Hoxa5, Hoxa6, Hoxa9, and the Hoxa9 cofactor Meis1; while only one Hoxa gene was upregulated, the long non-coding RNA Hoxa11os (Figure 5A, highlighted genes). In fact, gene programs that are upregulated by forced expression of HOXA9 and MEIS1 in mouse cells are significantly downregulated by SETDB1 overexpression using gene set analysis33 (Figure 5B). We show significant upregulation of genes that exhibit increased expression in mature blood cells compared to HSPC, consistent with the differentiation observed upon SETDB1 overexpression in AML34,35 (Figure 5C-D and Figure 2F-G). Because SETDB1 binds the PAF1c, which is required for localization of MLL fusion proteins,5 we asked how SETDB1 expression affects direct targets of MLL-AF9. Interestingly, genes bound by regulation of MLL-AF9 were significantly downregulated upon SETDB1 overexpression, 36 suggesting H3K9me3 regulation MLL-AF9 gene programs in leukemic cells (Figure 5E). 193 genes downregulated by SETDB1 overexpression are reported to have reduced promoter H3K9 methylation in AML relative to normal CD34+ cells,17 suggesting SETDB1 may be responsible for regulating a subset of these genes, including Kit, Cbl, Ptpn11, Six1, and other genes that are important in AML (Figure 5F and Online Supplementary Table S4). There is significant overlap between genes downregulated by SETDB1 and genes upregulated by Crispr/Cas9 mediated knockdown of SETDB1 in human THP-1 AML cells harboring an MLL-AF9 fusion (Figure 5E and Online Supplementary Table S4).24 This suggests SETDB1 regulates conserved pro-leukemic gene programs in leukemic cells including direct MLL fusion targets.
SETDB1 regulates the epigenome to affect changes in chromatin accessibility and gene expression
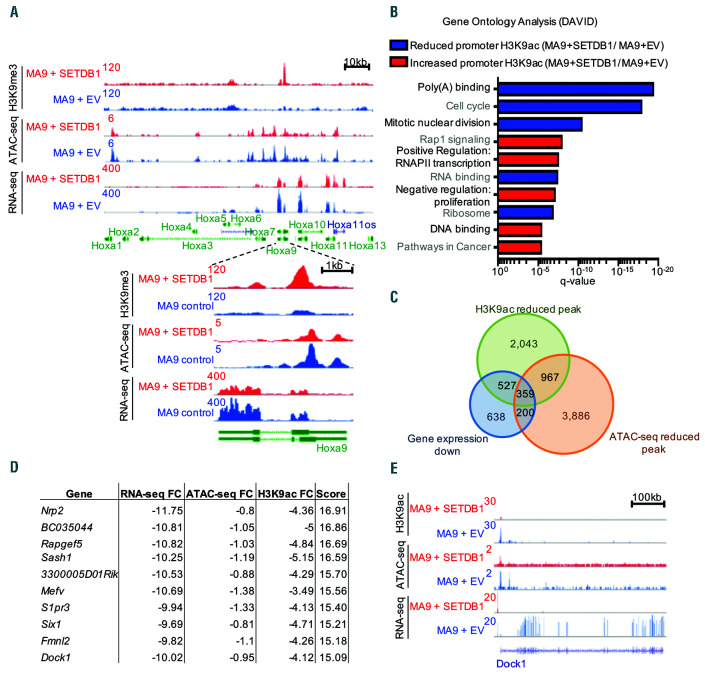
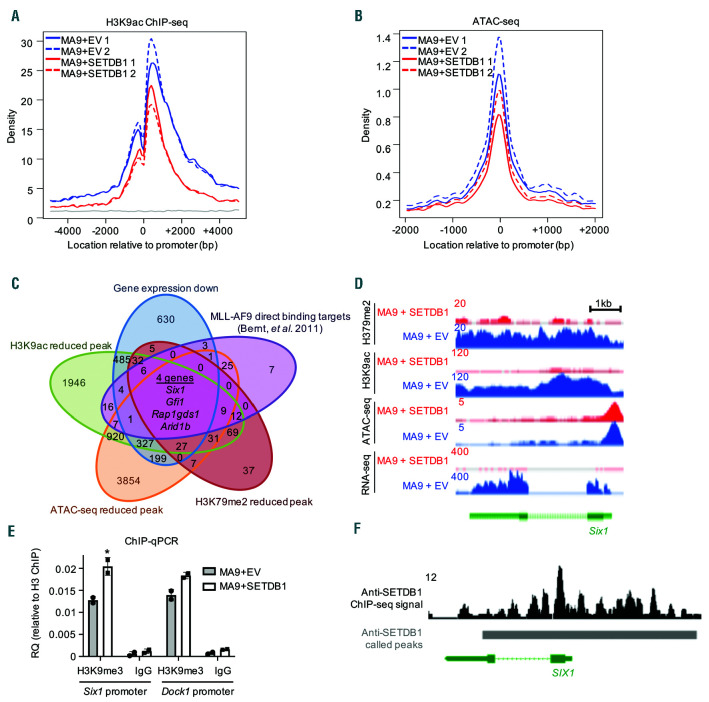
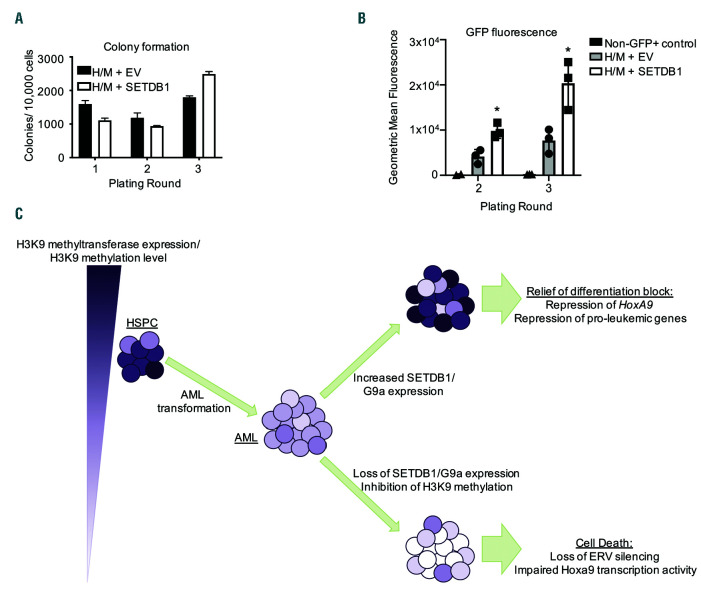
We have shown that overexpression of SETDB1 in MLL-AF9 cells leads to global increases in H3K9me3.5 To understand the specific epigenomic changes induced by SETDB1 in AML, we performed ChIP-seq for H3K9me3 in MLL-AF9 and MLL-AF9+SETDB1 leukemic cells. We also performed ATAC-seq to assess changes in chromatin accessibility. We first explored differences at the HOXA locus due to its importance in a large subset of AML, including MLL leukemias.5,37 Overexpression of SETDB1 reduced chromatin accessibility and increased H3K9me3 at posterior Hoxa genes, which results in reduced transcription of Hoxa9 (Figure 6A). We sought to define whole genome epigenetic regulation mediated by SETDB1 but observed only 552 consensus H3K9me3 peaks (Online Supplementary Figure S7A and Online Supplementary Table S5), which included regions enriched for H3K9me3, such as the zinc finger protein cluster on chromosome 7 (Online Supplementary Figure S7B). Repetitive elements may consume H3K9me3 sequencing depth making it difficult to map with stringency. We circumvented this by performing ChIP-Seq for H3K9ac, which is mutually exclusive of H3K9me3 and associated with gene activation, in contrast to H3K9me3.38 We performed H3K9ac ChIP-seq on MLLAF9 and MLL-AF9+SETDB1 cells and saw changes at over 6,000 promoter regions, including both increased and decreased H3K9ac signal (Online Supplementary Figure S7C and Online Supplementary Table S5). Gene ontology analysis reveals that genes with decreased promoter H3K9ac in MLL-AF9+SETDB1 cells were associated with cell cycle and RNA binding, whereas genes with increased promoter H3K9ac were associated with signaling pathways and negative regulation of proliferation (Figure 6B). Overlapping downregulated genes with genes that exhibit reduced promoter ATAC-seq and H3K9ac ChIP-seq signal in MLL-AF9+SETDB1 cells and scoring by their combined fold changes reveals several interesting targets including Six1 and Mefv, which are implicated as biomarkers in AML patients39,40 (Figure 6C-D and Online Supplementary Table S5). We also observed significant loss of promoter H3K9ac and chromatin accessibility and reduced gene expression of Dock1 (Figure 6E), which is a prognostic marker of AML that displays changes in DNA methylation in AML patient samples relative to normal HSPC.41,42 We identified several direct binding targets of MLL-AF9 that underwent epigenetic remodeling and expression changes with overexpression of SETDB1. Thus, we asked whether SETDB1 impacts H3K79me2, which is deposited by DOT1L and associated with MLL-fusion proteins,36 using ChIP-seq for H3K79me2. We found a marked decrease of H3K9ac and ATAC-seq signal at H3K79me2 peaks in MLL-AF9+SETDB1 cells suggesting a role for H3K9 modifications in regulating genes marked with H3K79me2 (Figure 7A-B). To further explore the role of SETDB1 in regulating MLL-AF9 targets, we performed an overlap analysis for genes that, upon SETDB1 overexpression, are downregulated, lose promoter H3K9ac, compact chromatin, lose gene body H3K79me2 signal, and are bound by MLL-AF9.36 This defined a target list of genes that may be coregulated by SETDB1 and MLL-AF9. Included in this group is Gfi1, which has been shown to affect AML cell growth;43 Rap1gds1, a nucleotide exchange factor; Arid1b, a member of the SWI/SNF complex; and Six1, which promotes formation of leukemic stem cells40 (Figure 7C). Six1 is of particular interest given its role in promoting leukemogenesis and the striking reductions observed in H3K9ac, H3K79me2, ATAC-seq signal, and gene expression at this locus (Figure 7D). We investigated H3K9me3 levels at specific loci by performing ChIP-qPCR at the promoter of Six1 and Dock1. We detected increased H3K9me3 in MLL-AF9 cells overexpressing SETDB1 (Figure 7E). Additionally, ENCODE data from human K562 cells shows SETDB1 binding at SIX1 (Figure 7F), GFI1 (Figure S7D), RAP1GDS1, and ARID1B (data not shown), but not DOCK1 (data not shown), suggesting we identified both direct and indirect targets. To determine whether SETDB1 expression inhibits cell growth primarily through Hoxa9 repression, we asked whether HOXA9/MEIS1 driven colony formation is affected by SETDB1 overexpression. A modest but insignificant decrease in colony formation is observed in HOXA9/MEIS1 transformed cells following SETDB1 overexpression (Figure 8A). However, SETDB1 overexpression is accompanied by significant upregulation of exogenous Meis1 (Figure 8B and Online Supplementary Figure S8A-B) that may account for the modest effects. Thus, Hoxa9, Meis1, and their downstream targets are likely affected by SETDB1 to influence AML cell growth. Taken together, our data demonstrate profound effects of SETDB1 on the epigenome and transcriptome that affects genes critical for AML.
Figure 6.
SETDB1 regulates the epigenetic landscape of acute myeloid leukemia oncogenes and biomarkers. (A) Sequencing tracks showing H3K9me3 ChIP-seq (top), ATAC-seq (middle), and RNA-seq (bottom) signals for MA9+EV (blue) or MA9+SETDB1 (red) cells. Shown here is the entire Hoxa cluster of genes and a closer view of Hoxa9 specifically. (B) Gene ontology analysis using DAVID to query biological process, molecular function, and Kegg pathway gene sets that are overrepresented in the following groups: H3K9ac ChIP-seq peaks that have significantly reduced (blue) or increased (red) signal intensity in MA9+SETDB1 compared to MA9+EV. (C) Overlap analysis of genes that are downregulated by SETDB1, genes where SETDB1 drives reduced promoter H3K9ac, and genes where SETDB1 drives reduced promoter ATAC-seq signal. (D) Top ten scoring genes when fold changes for the three datasets in C are summed. (E) Sequencing tracks showing H3K9ac ChIP-seq (top), ATAC-seq (middle), and RNA-seq (bottom) signals for MA9+EV (blue) or MA9+SETDB1 (red) cells. Shown here is the locus for Dock1. Statistics: differentially bound regions are defined by DiffBind false discovery rate (FDR) <0.05 (B/C/D); Significant gene expression changes are defined by DESeq2 algorithm with fold change >1.5 and padj<0.05 (C). For all ChIP-seq and ATAC-seq studies, n = 2 for each cell condition. AML: acute myeloid leukemia; EV: empty vector control; n: biological replicates; *: P/padj<0.05; RNA-seq: RNA sequencing; ChIP-seq: chromatin immunoprecipitation followed by sequencing; ATAC-seq: assay for transposaseaccessible chromatin coupled with next-generation sequencing.
Figure 7.
SETDB1 regulates the epigenetic landscape of a subset of MLL-AF9 target genes. (A) Signal track for H3K9ac ChIP-seq centered around promoters of genes that have reduced H3K79me2 in MA9+SETDB1 relative to MA9+EV cells. (B) Signal track for ATAC-seq centered around promoters of genes that have reduced H3K79me2 in MA9+SETDB1 relative to MA9+EV cells. (C) Overlap analysis of genes that are downregulated by SETDB1 in acute myeloid leukemia (AML), have reduced H3K9ac ChIP-seq, ATAC-seq, or H3K79me2 ChIP-seq upon SETDB1 overexpression in AML, or MLL-AF9 direct binding targets (Bernt et al. 2011). (D) Sequencing tracks showing H3K79me2 ChIP-seq (top), H3K9ac ChIP-seq (second from top), ATAC-seq (third from top), and RNA-seq (bottom) signals for MA9+EV (blue) or MA9+SETDB1 (red) cells. Shown here is the Six1 gene locus. (E) ChIP-qPCR: anti-H3K9me3 or anti- immunoglobulinG (anti-IgG) immunoprecipitated DNA were subjected to quatitative PCR (qPCR) using primers for the promoter region of Six1 and Dock1. Shown is relative quantification compared to total anti-H3 immunoprecipitated DNA. n=2. (F) Anti-SETDB1 ChIP-seq track from the ENCODE project showing the SIX1 locus in K562 leukemia cells. Solid bars represent called peaks as reported by ENCODE. Statistics: differentially bound regions are defined by DiffBind false discovery rate (FDR) <0.05 (A/B/C); Significant gene expression changes are defined by DESeq2 algorithm with fold change >1.5 and padj<0.05 (C). For all ChIP-seq and ATAC-seq studies, n=2 for each cell condition. EV: empty vector control; n: biological replicates; *: P/padj<0.05. ChIP-seq: chromatin immunoprecipitation followed by sequencing; ATAC-seq: assay for transposase-accessible chromatin coupled with next-generation sequencing.
Figure 8.
SETDB1 affects Hoxa9, Meis1 and downstream targets. (A) Mouse Lin- bone marrow was retrovirally transduced with the indicated plasmid vectors and plated in methylcellulose. Colonies were counted after 7 days and re-plated, for a total of three rounds. Shown is one representative experiment of n=3, error bars are standard deviations of the technical replicates. (B) Flow cytometry showing differences in mean fluorescence intensities in HOXA9/MEIS1 transformed cells that are overexpressing SETDB1 or empty vector (EV) control. (C) Working model for the proposed role of SETDB1/H3K9 methylation in acute myeloid leukemia (AML) initiation and maintenance. AML initiates from hematopoietic stem and progenitor cell (HSPC) with variable H3K9 methylation and maintain lower SETDB1 expression. After establishment of AML, inhibition of H3K9 methyltransferases leads to loss of retroviral silencing and cell death. Stabilization of SETDB1 or G9a leads to repressed Hox gene expression and relief of blocked differentiation. Statistics: generalized linear modeling followed by ANOVA where each MA9+SETDB1 replicate was paired to the MA9+EV control from the same biological replicate. Main effect is reported if there are no significant interactions (A) (See statistical analysis in the Online Supplementary Materials and Methods).
Discussion
Here we demonstrate that SETDB1 and H3K9 methylation suppresses AML disease progression in vivo through the repression of pro-leukemic genes including direct MLL-fusion protein targets. We found that AML patient samples exhibit lower SETDB1 expression compared to normal hematopoietic cells and that higher SETDB1 expression correlates strongly with better overall AML patient survival (Figure 1). We recapitulated these findings in mice where forced expression of SETDB1 in MLL-AF9 driven AML induces differentiation of AML cells and increases disease latency (Figure 2). These data suggest SETDB1 suppresses AML cell growth and self-renewal by relieving the block in differentiation.
We attribute the phenotypes in AML cells to altered H3K9 methylation. We altered H3K9 methylation levels genetically (SETDB1 and G9a) and through small molecule inhibition (UNC0638). Similar to our results with SETDB1, manipulation of G9a suggests H3K9 methylation can suppress AML progression by promoting differentiation (Figures 3-4, and Online Supplementary Figure S3). Thus, H3K9 methylation may have a more general effect on AML initiation and progression. Interestingly, Lehnertz and colleagues reported G9a overexpression accelerated Hoxa9/Meis1 mediated leukemia in vivo. We found Hoxa9/Meis1 mediated transformation in vitro was inhibited by SETDB1, but to a lesser extent than MLL-AF9 (Figure 8). Unique experimental strategies or functions for SETDB1 and G9a may account for these differences.22 Consistent with a role for H3K9 methylation in suppressing hematopoietic transformation, deletion of the H3K9 methyltransferase, SUV39H1 (and to a lesser degree SUV39H2), leads to the development of B-cell lymphomas in mice.44 Additionally, SETDB2 resides in a region of chromosome 13 that is commonly deleted in chronic lymphocytic leukemia (CLL).45 Thus, H3K9 methylation is likely exquisitely regulated in hematopoietic cells and performs context dependent functions that require further investigation to fully understand its role in AML.
Mechanistically, we found that SETDB1 is linked with altered H3K9 methylation and acetylation, decreased chromatin accessibility and transcriptional repression of critical AML oncogenes (Figures 5-7). These genes included several that have been implicated in myelodysplastic syndromes (MDS) and AML.39 We show SETDB1 regulates Dock1 expression, which is correlated with leukemic stem cell gene signatures and a poor prognosis in AML patients.41,46 We also observed that SETDB1 represses genes associated with AML, such as Hoxa9 and Six1, which are direct targets of MLL-AF9.16,37,40 Interestingly, Six1 was recently shown to be important for leukemic stem cell maintenance where depletion of Six1 leads to increased disease latency.47 This suggests repression of Six1 may contribute to SETDB1 mediated extension of leukemic disease latency. These data point to SETDB1 negatively regulating a pro-leukemic gene program, many of which are potential therapeutic targets. Thus, understanding the mechanisms regulating SETDB1 at the transcriptional and post-translational level may be a valuable therapeutic approach for AML. For example, miRNA29 is a critical mediator of SETDB1 expression.48 Another potential mediator of H3K9 methylation is the PAF1c. We showed SETDB1 binds to the PAF1c and mediates promoter H3K9me3 of the Hoxa9 and Meis1 loci.5 Further, we and others identified G9a and SUV39H1 as interacting partners of the PAF1c.5,6 Interestingly, SETDB1, G9a, GLP and SUV39H1 form a complex that directs H3K9 methylation to euchromatic gene promoters.49 Thus, the PAF1c may recruit H3K9 methyltransferases to specific targets to mediate gene repression. The PAF1c is a critical regulator of transcription of several pro-leukemic genes in AML cells through direct physical interaction with wild-type MLL and MLL-fusion proteins.13,15 It will be interesting to consider the biochemical interplay between H3K9 methyltransferases and MLL-fusion proteins with the PAF1c.
Previous studies have demonstrated that SETDB1 and G9a are required for AML initiation and progression.22–24 Our current data demonstrating that SETDB1 suppresses AML growth may suggest AML cells maintain a narrow SETDB1 expression level. We show increased SETDB1 expression induces differentiation of AML cells through H3K9me3 and repression of self-renewal genes. Conversely, loss of SETDB1 is detrimental to leukemic cells due to derepression of endogenous retroviral elements (ERV) and inhibition of HOXA9 transcriptional activity22,24 (Figure 8C). Given the essential role for SETDB1 in leukemia, small molecule inhibition of H3K9 methyltransferases has been proposed as a therapeutic option.22,23 However, a recent study shows depletion of G9a increased cancer progenitor cell populations that initiate a delayed but more aggressive disease state.21 Thus, it is critical to fully understand the effects of chemically inhibiting of H3K9 methylation as a treatment for AML. Further investigation into the roles of SETDB1, G9a and more generally H3K9 methylation levels will likely shed light on the precise role of these methyltransferases in normal and malignant hematopoiesis and determine the value of these epigenetic modifiers as therapeutic targets.
Acknowledgments
The authors thank Dr. Jianyong Shou for a SETDB1 expression construct and Dr. Schahram Akbarian for Setdb1fl/fl mice. We also thank Dr. Russell Ryan, Dr. Emmalee Adelman, Dr. Maria Figueroa, and Dr. Sami Malek for helpful discussion.
Funding Statement
Funding: This work was supported by NIH grants R01-HL-136420 (AGM), T32CA140044 (JR) and P30CA046592 (University of Michigan Flow Cytometry Core).
References
- 1.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Jia ST, Jia S. New insights into the regulation of heterochromatin. Trends Genet. 2016;32(5):284-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao VK, Pal A, Taneja R. A drive in SUVs: from development to disease. Epigenetics. 2017;12(3):177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ropa J, Saha N, Chen Z, et al. PAF1 complex interactions with SETDB1 mediate promoter H3K9 methylation and transcriptional repression of Hoxa9 and Meis1 in acute myeloid leukemia. Oncotarget. 2018;9(31):22123-22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y-J, Han J-W, Youn H-D, Cho E-J. The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010; 38(2):382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799(5-6):379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Oss SB, Cucinotta CE, Arndt KM. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci. 2017;42(10):788-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen FX, Xie P, Collings CK, et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 2017;357(6357):1294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krogan NJ, Dover J, Wood A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003; 11(3):721-729. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, Yang W, Ni T, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350(6266):1383-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serio J, Ropa J, Chen W, et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene. 2018;37(4):450-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muntean AG, Chen W, Jones M, Granowicz EM, Maillard I, Hess JL. MLL fusion protein-driven AML is selectively inhibited by targeted disruption of the MLL-PAFc interaction. Blood. 2013; 122(11):1914-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muntean AG, Tan J, Sitwala K, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins CT, Hess JL. Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene. 2016;35(9):1090-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller-Tidow C, Klein H-U, Hascher A, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010; 116(18): 3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23(21):2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471 (7339):513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avgustinova A, Symeonidi A, Castellanos A, et al. Loss of G9a preserves mutation patterns but increases chromatin accessibility, genomic instability and aggressiveness in skin tumours. Nat Cell Biol. 2018;20(12):1400-1409. [DOI] [PubMed] [Google Scholar]
- 22.Lehnertz B, Pabst C, Su L, et al. The methyltransferase G9a regulates HoxA9- dependent transcription in AML. Genes Dev. 2014;28(4):317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koide S, Oshima M, Takubo K, et al. Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood. 2016;128(5):638-649. [DOI] [PubMed] [Google Scholar]
- 24.Cuellar TL, Herzner A-M, Zhang X, et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol. 2017; 216(11):3535-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagger FO, Sasivarevic D, Sohi SH, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapin N, Bagger FO, Jendholm J, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood. 2014;123(6):894-904. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y, Loh Y-HE, Rajarajan P, et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet. 2017;49(8):1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Lee H-M, Xiong Y, et al. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader-Willi syndrome. Nat Med. 2017;23(2):213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vedadi M, Barsyte-Lovejoy D, Liu F, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7(8):566-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ugarte F, Sousae R, Cinquin B, et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep. 2015;5(5):728-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Skutt-Kakaria K, Davison J, et al. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev. 2012;26(22):2499-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess JL, Bittner CB, Zeisig DT, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108(1): 297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298(5593):601-604. [DOI] [PubMed] [Google Scholar]
- 35.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755-766. [DOI] [PubMed] [Google Scholar]
- 36.Bernt KM, Zhu N, Sinha AU, et al. MLLrearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghare SS, Joshi-Barve S, Moghe A, et al. Coordinated histone H3 methylation and acetylation regulate physiologic and pathologic fas ligand gene expression in human CD4+ T cells. J Immunol. 2014;193(1):412-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Celik S, Tangi F, Oktenli C. Increased frequency of Mediterranean fever gene variants in multiple myeloma. Oncol Lett. 2014;8(4):1735-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q-F, Wu G, Mi S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117(25):6895-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S-H, Chiu Y-C, Li Y-H, et al. High expression of dedicator of cytokinesis 1 (DOCK1) confers poor prognosis in acute myeloid leukemia. Oncotarget. 2017; 8(42):72250-72259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung N, Dai B, Gentles AJ, Majeti R, Feinberg AP. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun. 2015;6:8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volpe G, Walton DS, Grainger DE, et al. Prognostic significance of high GFI1 expression in AML of normal karyotype and its association with a FLT3-ITD signature. Sci Rep. 2017;7(1):11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters AH, O’Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001; 107(3):323-337. [DOI] [PubMed] [Google Scholar]
- 45.Falandry C, Fourel G, Galy V, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010; 285(26):20234-20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Li Y, Zhang L, Fang X, Zhen C, Wang X. Prognostic importance of DOCK1 transcript levels, and biologic insights from DOCK1 -associated gene and microrna expression signatures in de novo acute myeloid leukemia. Blood. 2017;130(Suppl 1):3952. [Google Scholar]
- 47.Chu Y, Chen Y, Li M, et al. Six1 regulates leukemia stem cell maintenance in acute myeloid leukemia. Cancer Sci. 2019; 110(7):2200-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong C-M, Wei L, Law C-T, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474-487. [DOI] [PubMed] [Google Scholar]
- 49.Fritsch L, Robin P, Mathieu JRR, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37(1):46-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data is available via the Gene Expression Omnibus, accession GSE136850.