Abstract
The fast dynamics occurring in natural processes increases the difficulty of creating biomaterials capable of mimicking Nature. Within synthetic biomaterials, water-soluble supramolecular polymers show great potential in mimicking the dynamic behavior of these natural processes. In particular, benzene-1,3,5-tricaboxamide (BTA)-based supramolecular polymers have shown to be highly dynamic through the exchange of monomers within and between fibers, but their suitability as biomaterials has not been yet explored. Herein we systematically study the interactions of BTA supramolecular polymers bearing either tetraethylene glycol or mannose units at the periphery with different biological entities. When BTA fibers were incubated with bovine serum albumin (BSA), the protein conformation was only affected by the fibers containing tetraethylene glycol at the periphery (BTA-OEG4). Coarse-grained molecular simulations showed that BSA interacted with BTA-OEG4 fibers rather than with BTA-OEG4 monomers that are present in solution or that may exchange out of the fibers. Microscopy studies revealed that, in the presence of BSA, BTA-OEG4 retained their fiber conformation although their length was slightly shortened. When further incubated with fetal bovine serum (FBS), both long and short fibers were visualized in solution. Nevertheless, in the hydrogel state, the rheological properties were remarkably preserved. Further studies on the cellular compatibility of all the BTA assemblies and mixtures thereof were performed in four different cell lines. A low cytotoxic effect at most concentrations was observed, confirming the suitability of utilizing functional BTA supramolecular polymers as dynamic biomaterials.
Introduction
Nature is the source of inspiration for the fabrication of fascinating materials.1,2 Although the field of biomaterials has made huge progress in recent years, mimicking Nature remains an extremely challenging task. This is mainly due to the highly dynamic character of natural systems, which is complex to simulate artificially or synthetically. Noncovalent recognition motifs occurring through hydrogen bonding, π–π stacking, metal chelation, van der Waals, and hydrophobic interactions allow natural systems to continuously evolve, reshape and reorganize in time and space to fulfill specific functions. Thus, biomaterials assembled through these noncovalent interactions have emerged as promising candidates to mimic the unique dynamics of Nature. Supramolecular chemistry offers the essential tools to reach this ambitious goal. As a result, water-compatible supramolecular polymers have been extensively studied as building blocks for the fabrication of dynamic biomaterials.3,4
A plethora of supramolecular polymers assemble into one-dimensional (1D) fibers in water and buffers,5 making them extremely powerful to mimic the abundant number of fibrillary components in natural systems.6 In particular, host–guest interactions have been used to assemble 1D supramolecular polymers by exploiting their hydrophobic character.7,8 Nevertheless, achieving complex modular host–guest biomaterials is challenging due to synthetic limitations. Alternatively, π-conjugates have been chosen as modular supramolecular building blocks, keeping the hydrophobic interactions as driving forces for the assembly. In this case, direct coupling to specific biofunctional moieties is feasible, which facilitates the synthetic challenge.9,10
A further example of fiber-like materials is represented by nature-inspired peptide assemblies that fully mimic 1D fibrillary native entities, containing natural amino acids and thus having intrinsic biofunctionality. Among them, peptide amphiphiles (PA), made of peptide sequences covalently bound onto hydrophobic chains, have shown promising properties as biofunctional materials.11−14 These PA fibers have been used in a number of exciting applications, while at the same time, the fundamental understanding of their structure–property relationship has been crucial to progress.15
In order to achieve broader modularity and expand the bioapplications of supramolecular polymers, ureido-pyrimidinone (UPy)-based 1D materials have been developed. UPy moieties dimerize via quadruple hydrogen bonding, and the addition of urea groups allows for lateral stacking into fibers held together by a combination of hydrogen bonding, hydrophobic interactions and π–π stacking.16,17 In dilute form, UPy fibers have been enriched with cationic moieties to allow intracellular delivery of siRNA,18 while the incorporation of specific peptides enabled growth factor stabilization.19
Another example that involves hydrogen bonding and hydrophobic effects is benzene-1,3,5-tricaboxamide (BTA)-based supramolecular polymers. The increased understanding gathered from fundamental studies on their assembly in water renders this class of materials highly promising for bioapplications.20−23 BTAs are known to form μm-long 1D fibers via intermolecular 3-fold hydrogen bonding between the amides and hydrophobic effects24,25 and they have been proven to be highly dynamic.26−29 Additionally, in analogy to covalent polymers, copolymerization has been successfully achieved,30,31 thus, allowing the modular insertion of multiple functionalities. Peptides, charges, carbohydrates, and DNA have been used to decorate BTA fibers, inducing cellular recognition, allowing intracellular delivery, mimicking the glycocalyx, and recruiting proteins.23,31−35 Furthermore, hydrogels were recently obtained by increasing the concentration of fibers.34,36 The dynamic and self-healing characters of these hydrogels make them extremely powerful tools to mimic the extracellular matrix. Nevertheless, their application as biomaterials in, for example, tissue engineering is still limited, also due to the lack of detailed knowledge on how these supramolecular materials behave in physiological environments and, especially, on how they interact with biologically relevant species, that is, proteins and cells.
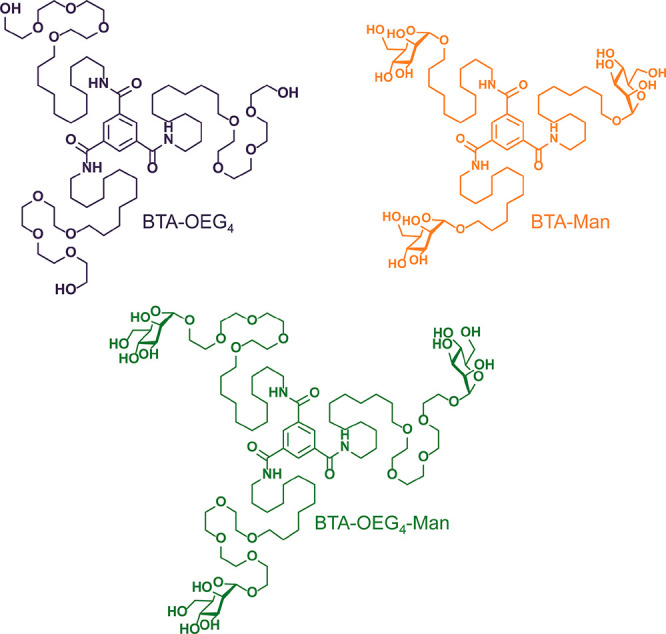
In this study, we assess the potential of water-compatible BTA derivatives as biomaterials both in the fiber and in the hydrogel state. By applying a fully realistic approach, the stability of different BTA homopolymers (BTA-OEG4, BTA-OEG4-Man, and BTA-Man; Scheme 1) and copolymers (BTA-OEG4-Man/BTA-OEG4 and BTA-Man/BTA-OEG4) upon incubation with physiological concentrations of bovine serum albumin (BSA) was investigated. Remarkably, the BTAs retained their fiber-like structure in protein-rich media, as proven by complementary techniques. Furthermore, several cell lines showed high viability, even upon exposure to high BTA concentrations, providing unprecedented insight into the applicability of BTA-based supramolecular polymers as biomaterials.
Scheme 1. Chemical Structure of the Water-Compatible BTA Monomers Selected for This Study.
Experimental Section
Materials
Dulbecco’s Modified Eagle Medium (DMEM), Eagle’s Minimum Essential Medium (MEM), penicillin–streptomycin, trypsin, and fetal bovine serum (FBS) were purchased from Gibco (Life Technologies). Phosphate buffer saline (PBS), bovine serum albumin (BSA), urea, 10× DMEM, 10× MEM, and human recombinant insulin were purchased from Sigma-Aldrich. (4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) solution (CellTiter 96Aqueous One Solution Assay) was purchased from Promega. Cy5-BSA was obtained from ProteinMods.
BTA-OEG4, BTA-OEG4-Man, BTA-Man, and BTA-Cy3 were synthesized and assembled into 1D fibers as previously reported.24,26,34 BTA-OEG4 and BTA-OEG4-Man samples were assembled by weighing the solid material into a glass vial, adding Milli-Q (MQ) water to obtain the desired concentration, stirring the sample at 90 °C for 15 min, and vortexing the sample for 15 s immediately afterward. The samples were then left to equilibrate overnight at room temperature before being used for any measurements. The samples in MEM in DMEM were prepared by first preparing a 1 mM sample of BTA-OEG4 in MQ water following the standard heating–cooling method. After equilibration for 8 h, the samples were diluted to 250 μM with medium (including 10% FBS) and allowed to equilibrate overnight at 4 °C. BTA-Man was dissolved in methanol to obtain a concentrated stock solution, which was subsequently injected into MQ water. After overnight equilibration, dialysis against MQ water was performed to remove the methanol content. Co-assembled samples were prepared by allowing the individual stocks to cool down to room temperature for 10 min after heating the stocks for 10 min at 80 °C. The 1:1 mixture was prepared by pipetting the correct amounts together. The samples were vortexed for 10 s, heated for another 10 min at 80 °C, vortexed for 10 s, and allowed to equilibrate at room temperature overnight before measuring. Hydrogels were prepared using the same procedure as in the assembly but with an additional step of equilibration in an ice bath after vortexing for 15 s.
Tryptophan Fluorescence Emission
BSA (20 mg/mL in PBS) was incubated overnight at 37 °C in a 96-well-plate with different concentrations (1.6 to 100 μM in PBS) of BTA-OEG4 and the copolymers of BTA-OEG4-Man/BTA-OEG4 and BTA-Man/BTA-OEG4. Tryptophan fluorescence emission was measured with a TECAN SPARK Multimode microplate-reader in the wavelength range of 320 to 500 nm after excitation at 280 nm. The experiments were performed in triplicate, and the average of the spectra was calculated. BSA alone was measured at negative control and BSA incubated with urea (9 M) was considered as positive control.
Förster Resonance Energy Transfer (FRET)
Cy5-BSA (1 μM in PBS) was incubated overnight at 37 °C in a 96-well plate with different concentrations (1–20 μM in PBS) of 5% BTA-Cy3, 95% BTA-OEG4, (Cy3- BTA-OEG4), 5% BTA-Cy3, 95% BTA-OEG4-Man/BTA-OEG4 (Cy3-BTA-OEG4-Man/BTA-OEG4), and 5% BTA-Cy3, 95% BTA-Man/BTA-OEG4 (Cy3-BTA-Man/BTA-OEG4). FRET was measured with a TECAN SPARK Multimode microplate reader in the wavelength range of 560 to 800 nm after excitation at 520 nm. Cy5/Cy3 ratio was calculated after subtraction of the Cy5-BSA fluorescence and normalized for the Cy3 amount in each sample.
Cryogenic Transmission Electron Microscopy (CryoTEM)
Imaging was performed on samples with a BTA concentration of 250 μM incubated overnight with 20 mg/mL BSA in PBS. Vitrified films were prepared in a “Vitrobot” instrument (PC controlled vitrification robot, patent applied, Frederik et al. 2002, patent licensed to FEI, Vitrobot Mark III) at 22 °C and at a relative humidity of 100%. In the preparation chamber of the “Vitrobot”, 3 μL samples were applied on Lacey grids (LC200-Cu, Electron Microscopy Sciences), which were surface plasma treated just prior to use (Cressington 208 carbon coater operating at 5 mA for 40 s). Excess sample was removed by blotting using filter paper for 4 s at −3 mm, and the thin film thus formed was plunged (acceleration about 3 g) into liquid ethane just above its freezing point. Vitrified films were transferred into the vacuum of a CryoTITAN equipped with a field emission gun that was operated at 300 kV, a postcolumn Gatan energy filter, and a 2048 × 2048 Gatan CCD camera. Virtrified films were observed in the CryoTITAN microscope at temperatures below −170 °C. Micrographs were taken at low dose conditions, starting at a magnification of 6500 with a defocus setting of −40 μm and at a magnification of 24000 with a defocus setting of −10 μm.
Total Internal Reflection Fluorescence Microscopy (TIRF)
TIRF images were acquired with a Nikon N-STORM system. Cy3 was excited using a 561 nm laser. Fluorescence was collected by means of a Nikon ×100, 1.4NA oil immersion objective and passed through a quad-band-pass dichroic filter (97335 Nikon). Images were recorded with an EMCCD camera (ixon3, Andor, pixel size 0.17 μm). BTA fibers were prepared at a total BTA concentration of 20 μM with Cy3-BTA-OEG4 and incubated overnight with 20 mg/mL of BSA or 10% of FBS in PBS at 37 °C. The samples were diluted to 2.5 μM total BTA and flown in a chamber between a glass microscope coverslip (Menzel-Gläser, no. 1, 21 × 26 mm) and a glass slide that were separated by double-sided tape.
CG-MD Simulations of BTA-OEG4-BSA Interaction–BSA Destabilization
A coarse-grained (CG) description of the system was chosen by building the molecular model based on the MARTINI force-field,37,38 as it is suited to study the interactions between supramolecular polymers and biomolecules.39,40 The CG model of the BSA protein dimer was based on the PDB structure 4f5s,41 and the topology and interaction parameters were constructed using the martinize.py tool (github.com/cgmartini/martinize.py), employing the MARTINI force-field extension for proteins.42,43 The limited accuracy of MARTINI in treating the interactions among residues in the protein structure was overcome by using an elastic network potential,44,45 which introduces a set of restraints (harmonic forces) among these residues to preserve the correct native protein structure. Elastic network bonds were imposed between those residues that were less than 0.9 nm apart in the native structure using a standard force constant of 500 kJ mol–1 nm–2. Since the network potential could hamper the ability of studying eventual perturbations to the protein structure, BSA was simulated with and without the elastic network.
A previously developed and optimized CG model46 based on the MARTINI force field and compatible with BSA was employed for BTA-OEG4. A BTA-OEG4 fiber model was preliminarily equilibrated in explicit MARTINI water. This model was used to study the interactions of a BTA-OEG4 fiber with BSA compared with interaction of dispersed/disassembled monomers.
The BSA + BTA-OEG4 monomers system contained the protein dimer, 100 BTA-OEG4 monomers, and 34 Na+ ions to neutralize the protein charge. The BSA + BTA-OEG4 fiber model contained the same number of proteins, BTA-OEG4 monomers (arranged into a pre-equilibrated fiber), and neutralizing Na+ ions as the BSA + BTA-OEG4 monomer system. Both CG models were solvated in standard MARTINI CG water (W) beads.
We conducted 2 μs of molecular dynamics (CG-MD) simulations starting from model configurations where the BTA-OEG4 monomers or the BTA-OEG4 fiber were placed in proximity to the BSA dimer. During these runs the interaction between BSA and the BTA-OEG4 monomers or fiber reached the equilibrium in the CG-MD regime. From this point, the simulation box size was resized in order to have the same number of water beads in the two models. An additional 0.5 μs of CG-MD simulations were conducted to calculate the energies of the two systems and compare the two cases of BSA interacting with BTA-OEG4 monomers versus with a BTA-OEG4 fiber.
In all CG-MD runs, the boundaries of the simulation box were treated with Periodic Boundary Conditions to reduce finite size effects. To prevent the BSA from rotating and directly interacting with its periodic images, a restraining potential was imposed, acting on two residues (Asp363 and Lys312 of first and second BSA monomers, respectively). As verified by the comparison of two trajectories with and without the restraining potential, the effect of these restrains was negligible. The leapfrog algorithm was used to integrate the equations of motion, with a standard CG time step of 20 fs. The temperature of the systems was set at 293 K by means of the v-rescale thermostat method,47 with a coupling time of 2 ps, while the pressure was kept at 1 bar by means of Parrinello–Rahman barostat,48 with a time constant of 6 ps.
CG-MetaD Simulations–Mechanism of BSA–BTA Interaction
Metadynamics (MetaD)49 was employed as previously described29 to efficiently explore the configurations and exchange of a BTA-OEG4 monomer across the interface between the fiber and the BSA dimer. The applied bias acted along two collective variables A and B, relative to a preselected monomer M, initially interacting with both the fiber and the protein surface. A was defined as the minimum distance between the M core atoms (the benzene ring of BTA-OEG4) and the backbone of the fiber. B was defined as Cfiber – Cprotein, where Cfiber was the number of contacts between the core atoms of M and the backbone of the fiber (the other BTA cores) and Cprotein was the number of contacts between the core of M and the BSA atoms. The bias was constructed by depositing every 1000 CG-MD steps Gaussian kernels of height 0.2 kJ/mol and width of 0.05 (in the A direction) and 0.25 (in the B direction). All MD and MetaD simulations and analyses were performed with GROMACS 2018,50,51 equipped with PLUMED 2.5.52
Rheology
Rheological measurements were performed on a TA Instruments DHR-3 rheometer (TA Instruments). A hydrogel was deposited onto the rheometer stage. A 20 mm stainless-steel cone plate was used, and the gap height was set to 56 μm for measurement. The temperature was controlled strictly at 37 °C using a water trap for all the measurements, and each measurement was repeated multiple times for reliable data. We monitored the storage modulus G′ and loss modulus G″ under an applied strain of 0.1% to 1000% at a frequency of 1 rad s–1 for the strain sweep.
Cell Culture
HEK293, RAW264.7, and MDA-MB-231 cells were cultured in a medium composed as follows: DMEM, 10% FBS and 1% penicillin-streptomycin. MCF7 cells were cultured in a medium composed as follows: MEM, 10% FBS, 0.01 mg/mL human recombinant insulin, and 1% penicillin–streptomycin. The cell culture was performed under 5% CO2 and at 37 °C. Detachment of the cells from the culture flask was performed by trypsination for HEK293, MDA-MB-231, and MCF7 and by scraping for RAW264.7.
Cytotoxicity
For the determination of the cytotoxicity of BTAs, cells were seeded in a 96-well cell culture plate at a density of 5 × 103 cells per well for HEK293 and MDA-MB-231, 10 × 103 cells per well for MCF7 and 25 × 103 cells per well for RAW264.7 in complete media (100 μL) and allowed to grow for 24 h at 37 °C in a humidified atmosphere with 5% of CO2. Subsequently, the medium was aspirated, and the cells were washed twice with PBS before adding different concentrations of BTAs (from 0.8 to 150 μM). The final concentration of BTAs was obtained by diluting the samples with 10× DMEM and 10× MEM. The samples were incubated for 24 h at 37 °C in a humidified atmosphere with 5% CO2. Control cells were treated with equivalent volumes of the corresponding medium. Afterward, 20 μL of MTS solution (CellTiter 96Aqueous One Solution Assay) was added to each well of a 96-well plate containing cells. The cells were then incubated for 3 h at 37 °C in a humidified atmosphere with 5% CO2, and the absorbance was read at 490 nm. The experiments were run in triplicate. The average of the triplicate was normalized based on the controls.
Results and Discussion
Interaction of BTA Fibers with BSA Probed by Fluorescence Spectroscopy
Since protein adsorption onto materials is depicted as the first event occurring in any biological system,53 the interaction of BTA fibers with proteins was first investigated by using bovine serum albumin (BSA) as model protein. The BSA quaternary structure shields two tryptophans in hydrophobic pockets, allowing them to emit fluorescence. Upon protein unfolding, these amino acids get exposed to the surrounding aqueous environment, and their fluorescence quenches.54−56 By spectroscopically monitoring this process, interactions between BSA and BTA fibers were investigated.
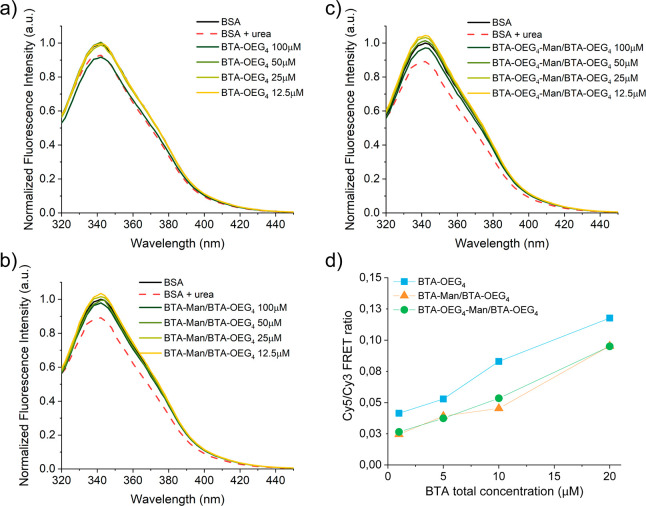
Different amounts of BTA-OEG4 fibers, assembled through the standard heating–cooling procedure, were incubated overnight at 37 °C with 20 mg/mL (303 μM) BSA in PBS, which corresponds to its physiological concentration, and the changes in tryptophan fluorescence emission were followed. The fluorescence quenching was overall low and only at the highest concentration of BTA-OEG4, when the ratio BTA-OEG4/BSA was close to 1:3, the fluorescence emission spectra overlapped with the one containing urea, which is a known protein denaturing agent (Figure 1a). In order to investigate the mechanism of this interaction, the same experiment was repeated with more hydrophilic BTA-based supramolecular copolymers. In particular, a physiological concentration of BSA was exposed to different amounts of 1:1 copolymers composed of BTA-OEG4-Man and BTA-OEG4 (BTA-OEG4-Man/BTA-OEG4) and BTA-Man and BTA-OEG4 (BTA-Man/BTA-OEG4). The tryptophan fluorescence showed almost no quenching by these two copolymers (Figure 1b,c), indicating a minor interaction between BSA and BTA copolymers bearing mannose moieties at the periphery. Remarkably, the role of mannose in reducing the BTA interaction with proteins was further confirmed by lower Forster Resonance Energy Transfer (FRET). Both Cy3-labeled BTA-OEG4-Man/BTA-OEG4 (Cy3-BTA-OEG4-Man/BTA-OEG4) and Cy3-labeled BTA-Man/BTA-OEG4 (Cy3-BTA-Man/BTA-OEG4) showed lower FRET when incubated overnight with Cy5-BSA than Cy3-labeled BTA-OEG4 (Cy3-BTA-OEG4) incubated with Cy5-BSA (Figure 1d). These experiments also showed that higher BTA concentrations induced an increase in interactions, corroborating the tryptophan fluorescence emission results.
Figure 1.
BSA tryptophan fluorescence emission showed interaction between BTA-OEG4 and physiological concentration of BSA at 37 °C (a). The introduction of mannose moieties at the fiber periphery reduced this interaction (b, c). FRET experiments (d) showed enhanced interaction between Cy3-labeled BTA fibers and Cy5-BSA increasing the fibers concentrations.
Since experimentally BSA showed a somewhat stronger interaction with BTA-OEG4 fibers compared to the two mannose copolymers, the interaction and stability of BTA-OEG4 fibers was further unraveled in the presence of BSA.
Consequences of the Interactions between BSA and BTAs Probed by Microscopy
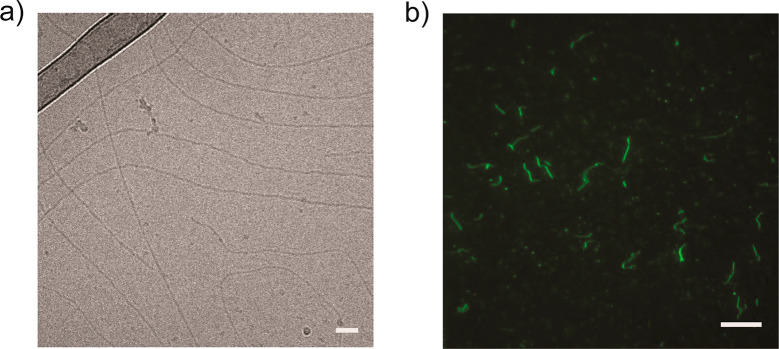
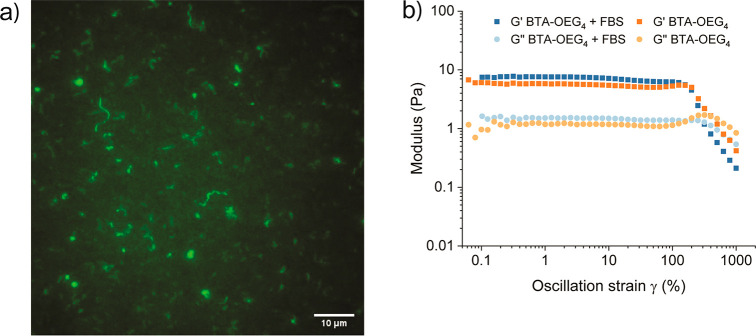
In order to assess whether BTA-OEG4 assemblies were retaining a fiber-like morphology upon incubation with BSA, cryogenic transmission electron microscopy (cryoTEM) and total internal reflection fluorescence microscopy (TIRF) were measured. For cryoTEM imaging, BTA-OEG4 fibers were incubated overnight with BSA at physiological protein concentration in PBS. Even though fluorescence spectroscopy on tryptophan exposure and FRET experiments suggested an interaction of BSA with BTA-OEG4 fibers, long 1D assemblies were visualized (Figure 2a). Nevertheless, some fibers showed chain ends, suggesting that the interaction with BSA partially affects the BTA-OEG4 microstructure by shortening the fiber length, as seen by cryoTEM imaging of BTA-OEG4 with and without BSA at the same magnification (6.5k) and BTA concentration (250 μM; Figure S4). The BTA-OEG4 fiber-like structure preservation upon incubation with BSA was further confirmed by TIRF microscopy, which showed the presence of micrometer-long fibers (Figure 2b). In some cases, those were slightly shorter compared to Cy3-BTA-OEG4 not incubated with BSA (Figure S5). This result showed the correlation with cryoTEM results and suggested that even though the fibers retained their morphology, their length was partially affected due to the interaction with BSA.
Figure 2.
(a) CryoTEM of BTA-OEG4 incubated with BSA (scale bar: 50 nm) and (b) TIRF imaging of Cy3-BTA-OEG4 incubated with BSA (scale bar: 10 μm).
Molecular Simulations Elucidating the Interaction of BTAs with BSA
Two mechanisms were initially hypothesized for the BTA-OEG4/BSA interaction: either BTA-OEG4 monomers were sequestered by BSA and trapped into the protein hydrophobic pockets (i.e., monomer exchange out of the fibers into the protein), or BSA was destabilized by BTA-OEG4 fibers (i.e., interaction of BSA with BTA-OEG4 fibers, rather than monomers). In the first case, the BSA/BTA-OEG4 interaction would be governed by the dynamics of the monomer exchange out of the BTA-OEG4 fibers, which determines how many disassembled monomers were present in the solution. In the second case, the destabilization would involve a macromolecular interaction between BSA and the whole BTA-OEG4 fibers.
In order to shed light onto the most probable BSA/BTA-OEG4 interaction mechanism, we employed coarse-grained molecular dynamics (CG-MD) simulations. In particular, two in silico experiments were designed to assess the effect on the BSA in the two types of interactions, that is, BSA interaction with disassembled BTA-OEG4 monomers in solution vs BSA interaction with assembled BTA-OEG4 fiber. We thus simulated two cases where one BSA dimer interacts with 100 initially disassembled BTA monomers versus one pre-equilibrated BTA fiber composed of 100 assembled monomers in aqueous solution. The effect of the interaction with BTA-OEG4 on the BSA structure in both cases was then compared.
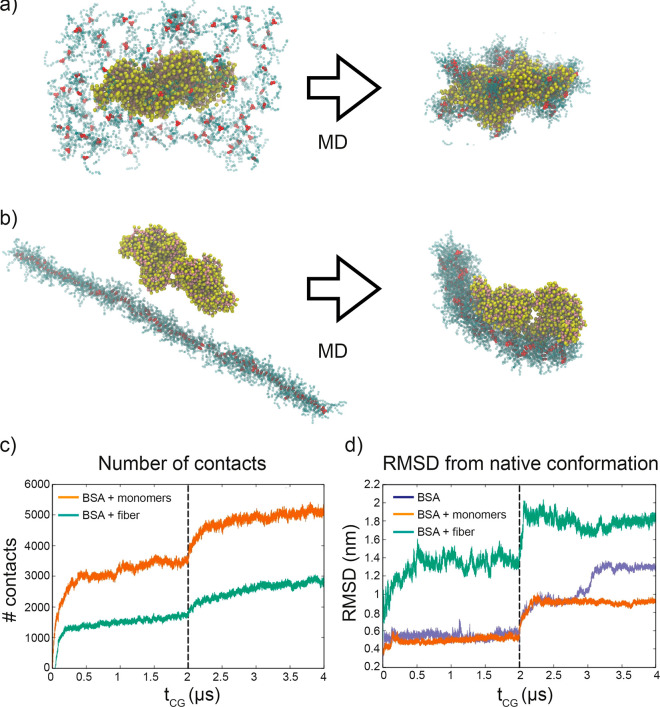
In general, both CG-MD simulations showed that the protein had a strong interaction with BTA-OEG4. In the first case, the monomers tended to distribute across the BSA surface, targeting specific hydrophobic patches (Figure 3a). In the second situation, the BTA-OEG4 fiber strongly attached to the protein surface and bent surrounding it (Figure 3b). Analysis of the CG-MD trajectories indicated that the number of contacts between BSA and BTA-OEG4 was higher in the case of free/disassembled BTA-OEG4 monomers in solution (Figure 3c). Nevertheless, the BSA 3D structure deviated more from its native conformation when interacting with the BTA-OEG4 fiber (Figure 3d).
Figure 3.
CG modeling of BSA interaction with BTA-OEG4. (a) BSA dimer (yellow and pink beads) interacting with 100 disassembled BTA-OEG4 monomers (red and blue beads) and (b) BSA dimer interacting with the BTA-OEG4 fiber composed of 100 monomers. The snapshots on the left show the initial configuration, while those on the right are taken after 2 μs of CG-MD. VMD was used for visualization and rendering of the systems.57 (c) Evolution of the number of contacts between BSA and BTA-OEG4 in the two systems (a) and (b). (d) Evolution of root-mean-square displacement (RMSD) of BSA conformation from the native structure, for the two systems (a) and (b) and for a control system with BSA in water. The first 2 μs of CG-MD in (c) and (d) employed an elastic network potential, which was removed after 2 μs (vertical dashed line).
Since CG models can be intrinsically limited by the approximations in the CG scheme and by the use of the elastic network applied on the protein model, we cross-checked these comparative simulations in presence and in absence of an elastic network potential (preserving the BSA structure). These additional simulations confirmed the trends observed in the data shown in Figure 3d for both cases. This demonstrated that the BSA/fiber interaction entailed a strong effect on the protein secondary and tertiary structure. This phenomenon, which can be compatible with protein denaturation, is experimentally corroborated by the similar effect of urea and BTA-OEG4 seen in the fluorescence spectroscopy. On the other hand, although the BSA interaction with free monomers was observed in the CG-MD (Figure 3c: number of contacts with monomers was even higher than those with the BTA-OEG4 fiber), the effect on the BSA’s 3D structure was considerably smaller. Altogether, this indicated that the interaction with a full fiber had a stronger destabilization effect on the BSA structure than the interaction with the same number of individual BTA-OEG4 monomers. Since the concentration of free monomers when the fibers are present is very low,29 BSA proteins interacted more likely with BTA-OEG4 fibers than with disassembled BTA-OEG4 monomers. Thus, these CG-MD simulations suggested that the tryptophan fluorescence quenching and the shortening of the fibers observed experimentally were caused by the interaction of BSA with the BTA-OEG4 fiber rather than with the monomers. This was further confirmed by thermodynamic data extracted from the CG-MD simulations, which showed a more favorable interaction energy for BSA with BTA-OEG4 fibers compared with the same number of disassembled BTA-OEG4 monomers (with a difference in interaction energy of ∼2.2 kcal/mol per monomer, see Table S1).
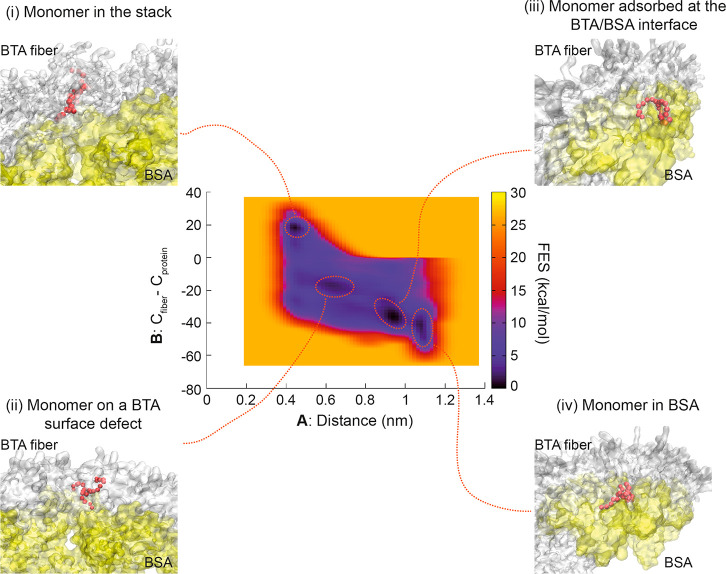
Additional evidence for stronger interaction of BSA with BTA-OEG4 fibers rather than with BTA-OEG4 monomers was obtained by CG metadynamics (CG-MetaD) simulations. Starting from a BTA-OEG4/BSA bound state (i.e., BTA-OEG4 fiber bent and bound to the BSA surface), a CG-MetaD simulation was performed where one BTA-OEG4 monomer at the interface was biased to exchange/diffuse from the BTA-OEG4 fiber within the protein and back. During the CG-MetaD run, the monomer crossed multiple times back and forth from the fiber into the protein. From this CG-MetaD simulation, we computed the free energy surface (FES) of the monomer exchange between fiber and protein. The FES showed that a BTA-OEG4 monomer could exist in different states with favorable free energy (see Figure 4, dark free energy minima). In the first case, the monomer could be stacked into the BTA-OEG4 fiber backbone having negligible interaction with BSA (Figure 4i). Another possibility could see the monomer stacked onto a defect of the BTA-OEG4 fiber backbone at the interface between BSA and the BTA-OEG4 fiber and interacting with both (Figure 4ii). The monomer could also be placed at the interface between BSA and the BTA-OEG4 fiber but adsorbed on the fiber surface (Figure 4iii). In this case, the monomer interaction was stronger with BSA than with the fiber. Finally, the monomer could be sequestered by the protein, having no interaction with the fiber (Figure 4iv). Remarkably, the FES demonstrated that the global free energy minimum was achieved when the exchanging monomer was at the interface, interacting more with BSA (Figure 4iii). The shape of the FES suggested that, once a BSA/BTA-fiber binding was completed, the diffusion of monomers into the protein was quite likely. From the global minimum, the monomer could indeed diffuse inside the BSA structure by crossing a free energy barrier of just ∼5 kcal/mol, which is half of the free energy (∼10 kcal/mol) required to exchange a monomer out from the fiber into water.29 This means that BTA-OEG4 monomers were more favorably exchanged directly from the fiber into the BSA structure via a direct surface/interface contact, rather than through a multistep mechanism involving monomer exchange from fiber into water and from water into BSA. This further supported the hypothesis that the conformational change of BSA observed from the tryptophan fluorescence quenching experiment was also likely due to the interaction of BSA with BTA-OEG4 fibers rather than with exchanged BTA-OEG4 monomers present in solution.
Figure 4.
Free-energy surface (FES) for the event of monomer exchange from BTA-OEG4 fiber to BSA. In the center, the FES is represented as a function of the two variables A and B (defined in the methods section). Snapshots from the main free-energy minima are reported: (i) The monomer (red beads) is part of the backbone of the fiber; (ii) The monomer is in a defect configuration, interacting with the protein; (iii) The monomer is adsorbed at the interface between BSA and BTA-OEG4 fiber; (iv) The monomer detaches from the fiber and is sequestered inside the protein. VMD was used for visualization and rendering of the system.57
Stability of BTA-OEG4 Fibers and Hydrogels in FBS and Cell Media
Since the interaction between BTA-OEG4 and BSA did not disrupt the fiber morphology, the stability of the BTA fibers was further assessed in the presence of fetal bovine serum (FBS) as a more complex biological medium. For this purpose, Cy3-BTA-OEG4 fibers were incubated overnight with FBS, and TIRF microscopy was measured (Figure 5a). Compared to Cy3-BTA-OEG4 alone (Figure S5) and Cy3-BTA-OEG4 incubated with BSA (Figure 2b), the presence of FBS induced heterogeneity between fibers, as micrometer long assemblies could be visualized, together with smaller aggregates. This indicated that FBS was capable of disrupting some of the fibers into smaller assemblies, but also that long micrometer fibers remained present. The stability of BTA-OEG4 fibers was further assessed by overnight incubation in DMEM and MEM cell media containing 10% FBS. CryoTEM imaging of these samples showed the presence of 1D supramolecular polymers with visible chain ends in both media that were shorter in length when dispersed in MEM than in DMEM (Figure S6). These results show that even in complex cell culture media, the fiber morphology was retained and only shortened due to interactions with proteins.
Figure 5.
(a) TIRF imaging of BTA-OEG4 after overnight exposure to FBS. (b) Strain-dependent oscillatory rheology with a fixed angular frequency of 1 rad/s of a 2 wt % BTA-OEG4 hydrogel with and without the addition of FBS.
All the above measurements were done at relatively low concentrations. As a next step, we investigated how the presence of FBS affects the material properties of BTA-OEG4. At a concentration of 2 wt %, BTA-OEG4 forms a hydrogel. This hydrogel was exposed to FBS by adding 100 μL of pure FBS on top of the gel and incubating the system overnight. To assess how far FBS has a negative effect on the hydrogel, the mechanical properties of the hydrogels were investigated by rheological measurements (Figure 5b). Remarkably, both hydrogels showed the same linear behavior and a similar gel-to-sol transition upon 300% strain, indicating that, upon FBS incubation, the hydrogel retained its viscoelastic properties. Furthermore, no significant difference of G′ was observed upon addition of the FBS, indicating that the strength of the hydrogel was hardly affected by the presence of FBS. Even though TIRF imaging showed that FBS affected the length of the BTA fibers, hydrogels exposed to FBS maintained their mechanical properties. This suggests that the interaction of FBS with the fibers is not strong enough to inhibit fiber entanglement as the driving force for the hydrogel formation. These results showed that, although interactions exist at the microscopic scale between serum proteins and BTA-based fibers, they did not have a large impact on the material properties; thus, showing that BTA-based supramolecular biomaterials can be used for bioapplications.
Cytotoxicity Studies of BTA Fibers Using Different Cell Lines
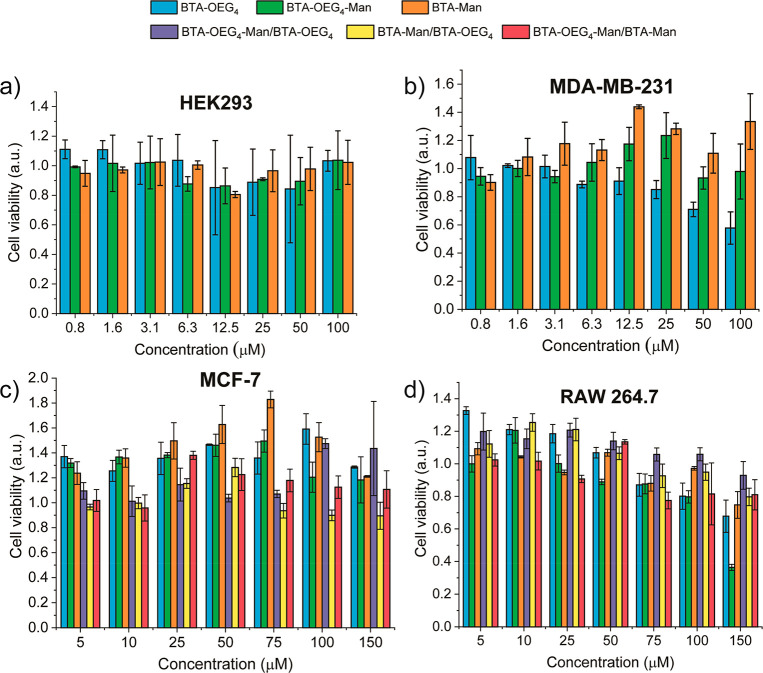
Once the stability of the BTA fibers in the presence of serum proteins was assessed and cell culture media, the cytotoxic effect of different BTA assemblies was evaluated in different cell lines. First, the cytotoxicity of pure BTA-OEG4, BTA-OEG4-Man, and BTA-Man was assessed. Whereas BTA-OEG4 and BTA-Man form micrometer long 1D fibers, BTA-OEG4-Man forms small micelles instead of 1D fibers.31 The human embryonic kidney cell line HEK293 was chosen as a common human cell line that does not express mannose receptor on the cell membrane and the breast cancer cell line MDA-MB-231 was chosen as human cell line expressing mannose receptor. The three homoassemblies were incubated for 24 h with the two different cell lines at increasing concentrations of BTAs and the cytotoxicity was assessed using the MTS viability assay. The homoassemblies showed no cytotoxic effect when incubated with HEK293 cells at any of the concentrations tested (Figure 6a). Interestingly, the mannose receptor expressing cell line MDA-MB-231 showed a decrease in cell viability to 60% and 70% when incubated with the highest concentrations of BTA-OEG4 (100 and 50 μM, respectively), which did not contain mannose (Figure 6b). The two mannose-containing homoassemblies of BTA-OEG4-Man and BTA-Man did not show any cytotoxic effect at any of the concentrations tested.
Figure 6.
HEK293 (a), MDA-MB-231 (b), MCF-7 (c), and RAW 264.7 (d) viability after 24 h incubation with increasing concentrations of BTA homo- and coassemblies.
To further assess whether the coassemblies of each of the monomers could affect the cell viability, all the combinations of the three monomers were coassembled (BTA-OEG4-Man/BTA-OEG4, BTA-Man/BTA-OEG4, BTA-OEG4-Man/BTA-Man) and the cytotoxicity was tested. Since only the mannose receptor expressing cell line showed a slight decrease in cell viability upon exposure to a high concentration of BTA-OEG4, two additional cell lines containing mannose receptor were chosen for the cytotoxicity assay, that is, the human breast cancer cell line MCF-7 and the murine macrophages RAW264.7. All the materials were incubated at different concentrations with the cells for 24 h and the cell viability was assessed using the MTS assay. In contrast to the viability trend shown with MDA-MB-231 breast cancer cells, none of the BTAs showed a cytotoxic effect against MCF-7 breast cancer cells at any of the concentrations tested (Figure 6c). The three BTA homopolymers incubated with macrophages RAW264.7 showed a slight decrease in cell viability at the highest concentration of materials. This phenomenon was particularly pronounced in the case of BTA-OEG4-Man homopolymers that showed only 40% of cell viability (Figure 6d). Since it is known that spherical nanoparticles are internalized by macrophages to a higher extent compared to other more elongated morphologies,58 this could be attributed to the mannose-displaying micelle-like morphology of BTA-OEG4-Man. Nevertheless, this cytotoxic effect was improved by coassembly of the BTA monomers with each other, leading to negligible cytotoxicity even at the highest concentrations. Hence, in summary, the BTA assemblies did not show a general cytotoxic trend at any of the concentrations studied.
Conclusion
In order to assess the suitability of water-soluble BTA supramolecular polymers as biomaterials, three functional BTA materials bearing tetraethylene glycol, mannose, or both tetraethylene glycol and mannose at the periphery (BTA-OEG4, BTA-Man, and BTA-OEG4-Man, respectively) were studied in the presence of different biological entities (BSA, FBS, and cells). By following the tryptophan fluorescence of BSA as well as FRET between labeled BTAs and BSA, it could be concluded that only BTA-OEG4 interacted with BSA. CG-MD simulations corroborated this interaction and showed that the BSA conformation was affected by the interaction with BTA-OEG4 fibers rather than with BTA-OEG4 monomers. Additional CG-MetaD simulations showed that the exchange of monomers from the BTA-OEG4 fibers to BSA occurred most likely via a direct contact between the fiber and the BSA surface without the need of a monomer exchange to water as an intermediate step. Gratifyingly, microscopy imaging of BTA-OEG4 in the presence of BSA and FBS showed that the fiber conformation was mostly preserved. Only the length was affected and in the case of FBS, the samples were more polydisperse, but still containing long fibers. Moreover, upon incubation with FBS, BTA-OEG4 hydrogels retained the viscoelastic properties of the materials showing their stability in protein-rich media. However, when a specific binding of fibers to certain proteins should be limited, the oligoethyleneglycol groups should be avoided and carbohydrate units are preferred. BTA fiber’s biocompatibility was further assessed in four different cell lines. Some BTA materials showed a slight cytotoxic effect in specific cell lines (BTA-OEG4 with MDA-MD-231 and BTA-OEG4-Man with RAW264.7), but this phenomenon only happened when incubated at the highest concentrations (100 and 150 μM). No cytotoxicity was observed in any of the other cases. The stability of the functional BTA fibers, as well as their compatibility with different cell lines, make these polymers excellent candidates for biomaterials development. Furthermore, their direct accessibility to modularity paves the way for the fabrication of tailor-made materials for selected bioapplications.
Acknowledgments
G.M. and S.V.A. acknowledge the funding received by the Gravitation Program “Materials Driven Regeneration”, funded by The Netherlands Organization for Scientific Research (024.003.013). G.M. acknowledges the funding received by the Swiss National Science Foundation (SNSF “Early PostDoc Mobility” P2EZP2-178435). G.M.P. acknowledges the funding received by the Swiss National Science Foundation (SNSF grant number 200021_175735) and by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 818776-DYNAPOL). The authors also acknowledge the computational resources provided by the Swiss National Supercomputing Center (CSCS) and by CINECA. The authors acknowledge the ICMS Animation Studio for providing the artwork.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.0c00904.
CD spectra of BTA fiber incubated with BSA, cryoTEM images of BTA-OEG4 fibers incubated with and without BSA, TIRF imaging of Cy3-BTA-OEG4 in PBS, and cryoTEM images of BTA-OEG4 fibers in cell culture media (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Author Contributions
† These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Huebsch N.; Mooney D. J. Inspiration and application in the evolution of biomaterials. Nature 2009, 462 (7272), 426–432. 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart A. R. Biologically Inspired Dynamic Material Systems. Angew. Chem., Int. Ed. 2015, 54 (11), 3400–3416. 10.1002/anie.201410139. [DOI] [PubMed] [Google Scholar]
- Goor O. J. G. M.; Hendrikse S. I. S.; Dankers P. Y. W.; Meijer E. W. From supramolecular polymers to multi-component biomaterials. Chem. Soc. Rev. 2017, 46 (21), 6621–6637. 10.1039/C7CS00564D. [DOI] [PubMed] [Google Scholar]
- Webber M. J.; Appel E. A.; Meijer E. W.; Langer R. Supramolecular biomaterials. Nat. Mater. 2016, 15 (1), 13–26. 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- Krieg E.; Bastings M. M. C.; Besenius P.; Rybtchinski B. Supramolecular Polymers in Aqueous Media. Chem. Rev. 2016, 116 (4), 2414–2477. 10.1021/acs.chemrev.5b00369. [DOI] [PubMed] [Google Scholar]
- Frantz C.; Stewart K. M.; Weaver V. M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123 (24), 4195–4200. 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yang H.; Wang Z.; Zhang X. Cucurbit[8]uril-Based Supramolecular Polymers. Chem. - Asian J. 2013, 8 (8), 1626–1632. 10.1002/asia.201300151. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Ma P. X. Host–guest interactions mediated nano-assemblies using cyclodextrin-containing hydrophilic polymers and their biomedical applications. Nano Today 2010, 5 (4), 337–350. 10.1016/j.nantod.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choisnet T.; Canevet D.; Sallé M.; Nicol E.; Niepceron F.; Jestin J.; Colombani O. Robust supramolecular nanocylinders of naphthalene diimide in water. Chem. Commun. 2019, 55 (64), 9519–9522. 10.1039/C9CC04723A. [DOI] [PubMed] [Google Scholar]
- Görl D.; Zhang X.; Stepanenko V.; Würthner F. Supramolecular block copolymers by kinetically controlled co-self-assembly of planar and core-twisted perylene bisimides. Nat. Commun. 2015, 6 (1), 7009. 10.1038/ncomms8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajangam K.; Behanna H. A.; Hui M. J.; Han X.; Hulvat J. F.; Lomasney J. W.; Stupp S. I. Heparin Binding Nanostructures to Promote Growth of Blood Vessels. Nano Lett. 2006, 6 (9), 2086–2090. 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- Mata A.; Geng Y.; Henrikson K. J.; Aparicio C.; Stock S. R.; Satcher R. L.; Stupp S. I. Bone regeneration mediated by biomimetic mineralization of a nanofiber matrix. Biomaterials 2010, 31 (23), 6004–6012. 10.1016/j.biomaterials.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartgerink J. D.; Beniash E.; Stupp S. I. Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science 2001, 294 (5547), 1684–1688. 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- Hartgerink J. D.; Beniash E.; Stupp S. I. Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (8), 5133–5138. 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp S. I.; Clemons T. D.; Carrow J. K.; Sai H.; Palmer L. C. Supramolecular and Hybrid Bonding Polymers. Isr. J. Chem. 2020, 60 (1–2), 124–131. 10.1002/ijch.202000005. [DOI] [Google Scholar]
- Dankers P. Y. W.; Hermans T. M.; Baughman T. W.; Kamikawa Y.; Kieltyka R. E.; Bastings M. M. C.; Janssen H. M.; Sommerdijk N. A. J. M.; Larsen A.; van Luyn M. J. A.; Bosman A. W.; Popa E. R.; Fytas G.; Meijer E. W. Hierarchical Formation of Supramolecular Transient Networks in Water: A Modular Injectable Delivery System. Adv. Mater. 2012, 24 (20), 2703–2709. 10.1002/adma.201104072. [DOI] [PubMed] [Google Scholar]
- Hendrikse S. I. S.; Wijnands S. P. W.; Lafleur R. P. M.; Pouderoijen M. J.; Janssen H. M.; Dankers P. Y. W.; Meijer E. W. Controlling and tuning the dynamic nature of supramolecular polymers in aqueous solutions. Chem. Commun. 2017, 53 (14), 2279–2282. 10.1039/C6CC10046E. [DOI] [PubMed] [Google Scholar]
- Bakker M. H.; Kieltyka R. E.; Albertazzi L.; Dankers P. Y. W. Modular supramolecular ureidopyrimidinone polymer carriers for intracellular delivery. RSC Adv. 2016, 6 (112), 110600–110603. 10.1039/C6RA22490C. [DOI] [Google Scholar]
- Hendrikse S. I. S.; Spaans S.; Meijer E. W.; Dankers P. Y. W. Supramolecular Platform Stabilizing Growth Factors. Biomacromolecules 2018, 19 (7), 2610–2617. 10.1021/acs.biomac.8b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straßburger D.; Stergiou N.; Urschbach M.; Yurugi H.; Spitzer D.; Schollmeyer D.; Schmitt E.; Besenius P. Mannose-Decorated Multicomponent Supramolecular Polymers Trigger Effective Uptake into Antigen-Presenting Cells. ChemBioChem 2018, 19 (9), 912–916. 10.1002/cbic.201800114. [DOI] [PubMed] [Google Scholar]
- Alemán García M. Á.; Magdalena Estirado E.; Milroy L.-G.; Brunsveld L. Dual-Input Regulation and Positional Control in Hybrid Oligonucleotide/Discotic Supramolecular Wires. Angew. Chem., Int. Ed. 2018, 57 (18), 4976–4980. 10.1002/anie.201800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. K.; Brunsveld L. A Supramolecular Polymer as a Self-Assembling Polyvalent Scaffold. Angew. Chem., Int. Ed. 2009, 48 (16), 2921–2924. 10.1002/anie.200900143. [DOI] [PubMed] [Google Scholar]
- Bakker M. H.; Lee C. C.; Meijer E. W.; Dankers P. Y. W.; Albertazzi L. Multicomponent Supramolecular Polymers as a Modular Platform for Intracellular Delivery. ACS Nano 2016, 10 (2), 1845–1852. 10.1021/acsnano.5b05383. [DOI] [PubMed] [Google Scholar]
- Leenders C. M. A.; Albertazzi L.; Mes T.; Koenigs M. M. E.; Palmans A. R. A.; Meijer E. W. Supramolecular polymerization in water harnessing both hydrophobic effects and hydrogen bond formation. Chem. Commun. 2013, 49 (19), 1963–1965. 10.1039/c3cc38949a. [DOI] [PubMed] [Google Scholar]
- Lafleur R. P. M.; Lou X.; Pavan G. M.; Palmans A. R. A.; Meijer E. W. Consequences of a cosolvent on the structure and molecular dynamics of supramolecular polymers in water. Chem. Sci. 2018, 9 (29), 6199–6209. 10.1039/C8SC02257G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi L.; van der Zwaag D.; Leenders C. M. A.; Fitzner R.; van der Hofstad R. W.; Meijer E. W. Probing Exchange Pathways in One-Dimensional Aggregates with Super-Resolution Microscopy. Science 2014, 344 (6183), 491–495. 10.1126/science.1250945. [DOI] [PubMed] [Google Scholar]
- Lou X.; Lafleur R. P. M.; Leenders C. M. A.; Schoenmakers S. M. C.; Matsumoto N. M.; Baker M. B.; van Dongen J. L. J.; Palmans A. R. A.; Meijer E. W. Dynamic diversity of synthetic supramolecular polymers in water as revealed by hydrogen/deuterium exchange. Nat. Commun. 2017, 8 (1), 15420. 10.1038/ncomms15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparotto P.; Bochicchio D.; Ceriotti M.; Pavan G. M. Identifying and Tracking Defects in Dynamic Supramolecular Polymers. J. Phys. Chem. B 2020, 124 (3), 589–599. 10.1021/acs.jpcb.9b11015. [DOI] [PubMed] [Google Scholar]
- Bochicchio D.; Salvalaglio M.; Pavan G. M. Into the Dynamics of a Supramolecular Polymer at Submolecular Resolution. Nat. Commun. 2017, 8 (1), 147. 10.1038/s41467-017-00189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota B. N. S.; Lou X.; Bochicchio D.; Paffen T. F. E.; Lafleur R. P. M.; van Dongen J. L. J.; Ehrmann S.; Haag R.; Pavan G. M.; Palmans A. R. A.; Meijer E. W. Supramolecular Copolymerization as a Strategy to Control the Stability of Self-Assembled Nanofibers. Angew. Chem., Int. Ed. 2018, 57 (23), 6843–6847. 10.1002/anie.201802238. [DOI] [PubMed] [Google Scholar]
- Leenders C. M. A.; Jansen G.; Frissen M. M. M.; Lafleur R. P. M.; Voets I. K.; Palmans A. R. A.; Meijer E. W. Monosaccharides as Versatile Units for Water-Soluble Supramolecular Polymers. Chem. - Eur. J. 2016, 22 (13), 4608–4615. 10.1002/chem.201504762. [DOI] [PubMed] [Google Scholar]
- Baker M. B.; Gosens R. P. J.; Albertazzi L.; Matsumoto N. M.; Palmans A. R. A.; Meijer E. W. Exposing Differences in Monomer Exchange Rates of Multicomponent Supramolecular Polymers in Water. ChemBioChem 2016, 17 (3), 207–213. 10.1002/cbic.201500606. [DOI] [PubMed] [Google Scholar]
- Schoenmakers S. M. C.; Leenders C. M. A.; Lafleur R. P. M.; Lou X.; Meijer E. W.; Pavan G. M.; Palmans A. R. A. Impact of the water-compatible periphery on the dynamic and structural properties of benzene-1,3,5-tricarboxamide based amphiphiles. Chem. Commun. 2018, 54 (79), 11128–11131. 10.1039/C8CC04818E. [DOI] [PubMed] [Google Scholar]
- Hendrikse S. I. S.; Su L.; Hogervorst T. P.; Lafleur R. P. M.; Lou X.; van der Marel G. A.; Codee J. D. C.; Meijer E. W. Elucidating the Ordering in Self-Assembled Glycocalyx Mimicking Supramolecular Copolymers in Water. J. Am. Chem. Soc. 2019, 141 (35), 13877–13886. 10.1021/jacs.9b06607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnands S. P. W.; Engelen W.; Lafleur R. P. M.; Meijer E. W.; Merkx M. Controlling protein activity by dynamic recruitment on a supramolecular polymer platform. Nat. Commun. 2018, 9 (1), 65. 10.1038/s41467-017-02559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenders C. M. A.; Mes T.; Baker M. B.; Koenigs M. M. E.; Besenius P.; Palmans A. R. A.; Meijer E. W. From supramolecular polymers to hydrogel materials. Mater. Horiz. 2014, 1 (1), 116–120. 10.1039/C3MH00103B. [DOI] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; de Vries A. H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111 (27), 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. Perspective on the Martini model. Chem. Soc. Rev. 2013, 42 (16), 6801–6822. 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Torchi A.; Bochicchio D.; Pavan G. M. How the Dynamics of a Supramolecular Polymer Determines Its Dynamic Adaptivity and Stimuli-Responsiveness: Structure–Dynamics–Property Relationships From Coarse-Grained Simulations. J. Phys. Chem. B 2018, 122 (14), 4169–4178. 10.1021/acs.jpcb.8b00428. [DOI] [PubMed] [Google Scholar]
- Bochicchio D.; Pavan G. M. Molecular modelling of supramolecular polymers. Adv. Phys.-X 2018, 3 (1), 1436408. 10.1080/23746149.2018.1436408. [DOI] [Google Scholar]
- Bujacz A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68 (10), 1278–1289. 10.1107/S0907444912027047. [DOI] [PubMed] [Google Scholar]
- de Jong D. H.; Singh G.; Bennett W. F. D.; Arnarez C.; Wassenaar T. A.; Schäfer L. V.; Periole X.; Tieleman D. P.; Marrink S. J. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput. 2013, 9 (1), 687–697. 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- Monticelli L.; Kandasamy S. K.; Periole X.; Larson R. G.; Tieleman D. P.; Marrink S.-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008, 4 (5), 819–834. 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Periole X.; Cavalli M.; Marrink S.-J.; Ceruso M. A. Combining an Elastic Network With a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory Comput. 2009, 5 (9), 2531–2543. 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- Tirion M. M. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys. Rev. Lett. 1996, 77 (9), 1905–1908. 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- Bochicchio D.; Pavan G. M. From Cooperative Self-Assembly to Water-Soluble Supramolecular Polymers Using Coarse-Grained Simulations. ACS Nano 2017, 11 (1), 1000–1011. 10.1021/acsnano.6b07628. [DOI] [PubMed] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126 (1), 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52 (12), 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Laio A.; Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (20), 12562–12566. 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H. J. C.; van der Spoel D.; van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91 (1), 43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Tribello G. A.; Bonomi M.; Branduardi D.; Camilloni C.; Bussi G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 2014, 185 (2), 604–613. 10.1016/j.cpc.2013.09.018. [DOI] [Google Scholar]
- Othman Z.; Cillero Pastor B.; van Rijt S.; Habibovic P. Understanding interactions between biomaterials and biological systems using proteomics. Biomaterials 2018, 167, 191–204. 10.1016/j.biomaterials.2018.03.020. [DOI] [PubMed] [Google Scholar]
- Varlan A.; Hillebrand M. Bovine and human serum albumin interactions with 3-carboxyphenoxathiin studied by fluorescence and circular dichroism spectroscopy. Molecules 2010, 15 (6), 3905–3919. 10.3390/molecules15063905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z.; Zhou B.; Liu Y.-X.; Zhou C.-X.; Ding X.-L.; Liu Y. Fluorescence Study on the Interaction of Bovine Serum Albumin with P-Aminoazobenzene. J. Fluoresc. 2008, 18 (1), 109–118. 10.1007/s10895-007-0247-4. [DOI] [PubMed] [Google Scholar]
- Das N. K.; Pawar L.; Kumar N.; Mukherjee S. Quenching interaction of BSA with DTAB is dynamic in nature: A spectroscopic insight. Chem. Phys. Lett. 2015, 635, 50–55. 10.1016/j.cplett.2015.06.033. [DOI] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14 (1), 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Li Z.; Sun L.; Zhang Y.; Dove A. P.; O’Reilly R. K.; Chen G. Shape Effect of Glyco-Nanoparticles on Macrophage Cellular Uptake and Immune Response. ACS Macro Lett. 2016, 5 (9), 1059–1064. 10.1021/acsmacrolett.6b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.