Abstract
Honey bees are essential pollinators threatened by colony losses linked to the spread of parasites and pathogens. Here we report a new approach for manipulating bee gene expression and protecting bee health. We engineered a symbiotic bee gut bacterium, Snodgrassella alvi, to induce eukaryotic RNA interference (RNAi) immune responses. We show that engineered S. alvi can stably re-colonize bees and produce double-stranded RNA to activate RNAi and repress host gene expression, thereby altering bee physiology, behavior, and growth. We use this approach to improve bee survival following a viral challenge and show that engineered S. alvi can kill parasitic Varroa mites by triggering the mite RNAi response. This symbiont-mediated RNAi approach is a tool for bee functional genomics and potentially for safeguarding bee health.
One Sentence Summary:
Engineered gut bacteria induce immune responses to control honey bee gene expression and protect bees against pathogens and parasites.
Honey bees (Apis mellifera) are dominant crop pollinators worldwide and a model organism for studying development, behavior, and learning. Recently, high honey bee colony mortality (1), attributed largely to synergistic interactions between parasitic mites (Varroa destructor) and RNA viruses (2), has become a critical problem for agriculture and the maintenance of natural biodiversity. Despite their importance, studies of honey bee biology are limited by bees’ unusual social structure and reproductive biology. New genetic tools and methods for deterring pathogens are vital for understanding and protecting honey bees.
Honey bees possess the molecular machinery for RNA-interference (RNAi) (3), a eukaryotic antiviral immune system in which double-stranded RNA (dsRNA) triggers degradation of other RNAs with similar sequences. RNAi can be induced by feeding or injecting dsRNA, and this has been used to knock down expression of bee genes and to impair replication of RNA viruses including Deformed Wing Virus (DWV) (4–8). dsRNA administered to bees is transmitted to their eukaryotic parasites and can induce parasite RNAi responses. This approach has been used to suppress Varroa (9) and Nosema (10) by using dsRNAs that silence essential parasite genes. However, using dsRNA for sustained manipulation of bee gene expression or control of bee pests has proven difficult. Even administration of dsRNA to individual bees yields patchy and transient gene knockdown (11), and dsRNA can have off-target effects (12–14). There are even greater obstacles to using dsRNA to defend entire hives located in the field against pathogens, as dsRNA is expensive to produce and degrades rapidly in the environment.
Here we describe successful efforts to engineer Snodgrassella alvi wkB2, a symbiotic bacterium found in bee guts, to continuously produce dsRNA to manipulate host gene expression and protect bees against pathogens and parasites.
S. alvi is a core member of the conserved gut microbiota of honey bees (15). To test whether engineered S. alvi robustly colonizes bees, we inoculated bees en masse with S. alvi transformed with a plasmid expressing GFP and then monitored bacterial colonization (Fig. 1). Even at a dose of 500 CFU engineered S. alvi establishes within worker bees, grows to ~5.0 × 107 CFU after five days (Fig. 1A), and persists stably throughout the lifespan of bees reared in the lab (Fig. 1B). Most engineered S. alvi cells remained functional throughout our 15-day experiments, although some bees contained cells that lost fluorescence at the final timepoint (Fig. 1C). We also confirmed that 11 days after colonization engineered S. alvi was found along the gut wall with the same localization as the wild-type strain (Fig. 1D–F) (15).
Fig. 1. Engineered S. alvi colonizes and functions in bee guts.
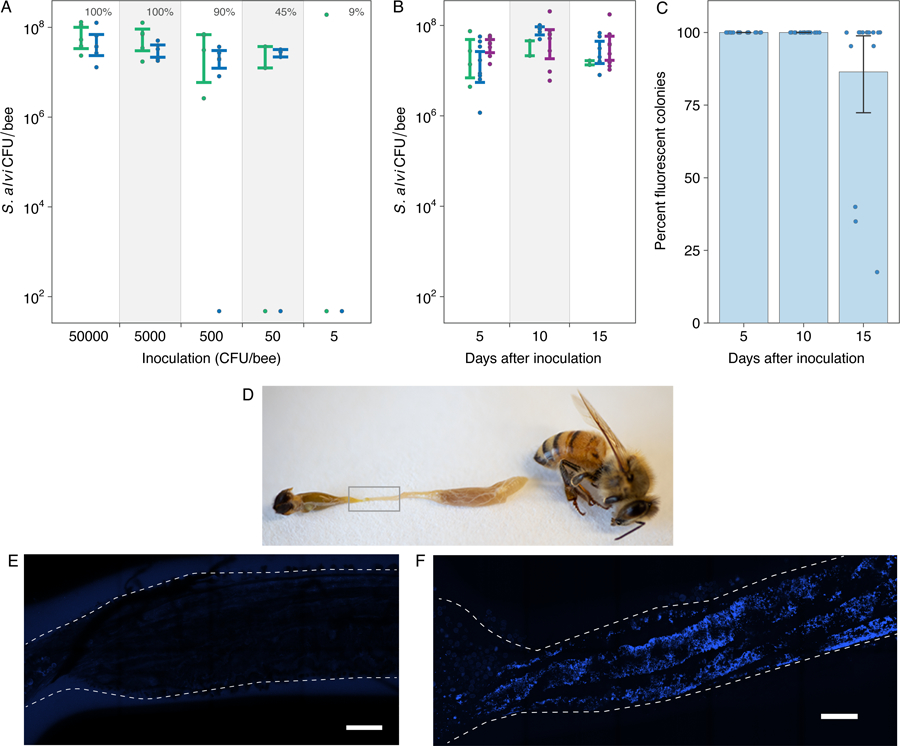
(A) Colonization of newly emerged honey bees by different inoculum sizes. The percentage of bees colonized in each treatment is annotated above the inoculation dose. N = 53 bees from 2 hives. (B) Stability of S. alvi colonization over time. N = 48 bees from 3 hives. Colors in (A) and (B) correspond to different source hives. (C) Stability of GFP expression by engineered S. alvi over time. (D) Photograph of dissected bee. S. alvi resides in the ileum (gray box). (E-F) Ilea of bees 11 days after colonization with non-fluorescent (E) or fluorescent (F) S. alvi. E2-Crimson fluorescence from engineered S. alvi is blue. Scale bars are 150 µm. Error bars in (A)–(C) are 95% bootstrap confidence intervals.
To test whether S. alvi could deliver dsRNA in situ, we designed a modular platform to assemble plasmids that produce dsRNA from an inverted arrangement of two promoters (Fig. S1). First, we assessed whether S. alvi produced dsRNA during colonization and whether there was a general bee immune response to symbiont production of dsRNA. We inoculated bees with S. alvi wkB2 transformed with either a plasmid that expresses no dsRNA (pNR) or a plasmid that expressed dsRNA corresponding to the GFP coding sequence (pDS-GFP). At 5, 10, and 15 days after inoculation, we sampled and dissected bees to measure RNA levels in different body regions. We detected GFP RNA in the head, gut, and hemolymph of bees colonized with dsRNA-producing bacteria at all sampling times (Fig. S2). The presence of GFP RNA in the hemolymphs and heads of bees, where no bacteria reside, suggests that RNA is transported throughout their bodies, as previously reported (8). We also detected upregulation and differential expression of immune pathway genes in the bees colonized with S. alvi bearing the pDS-GFP plasmid, and for some genes this upregulation correlates with the amount of dsRNA produced in the gut (Fig. S2). The upregulated genes included DDX52 and DHX33, which encode RNA helicases previously implicated in the bee immune response to dsRNA (8). Other upregulated genes include cact1 and cact2 (in abdomens) which remain upregulated for the entire 15-day trial; cact1 and cact2 were previously shown to be upregulated following injection of dsRNA, but only for a few hours following injection (8). RNAi components dicer and argonaute were not consistently upregulated, but dicer expression in abdomens did increase 5 – 10 days after colonization, as was observed for dicer shortly after dsRNA injection (8). Thus, engineered S. alvi persistently produces dsRNA in situ, and the bee host responds by activating immune pathway genes.
Next, we tested whether symbiont-produced dsRNA can be used to silence specific host genes. Insulin/insulin-like signaling (IIS) controls bee feeding behavior and development, including the transition of worker bees from nurses to foragers (16). We built a dsRNA plasmid targeting the insulin receptor InR1 (pDS-InR1) (Fig. 2A, Fig. S3), transformed this plasmid into S. alvi, and assayed its effects on bees. Compared to the pDS-GFP off-target control, we saw significantly lower expression of InR1 over multiple days and in all tested body regions (Fig. 2B). In contrast, previous studies found that direct injections of dsRNA into honey bee brains cause only transient (<1 day) knockdown (17). Bees colonized by bacteria harboring the pDS-InR1 plasmid showed increased sensitivity to low concentrations of sucrose (Fig. 2C), and gained more weight over time in each of two independent trials (Fig. 2D, Fig S4). InR1-suppressing bacteria led to significantly heavier bees at 10 and 15 days after colonization, likely a product of increased feeding behavior. Thus, symbiont-mediated RNAi systemically silences bee genes, and can lead to persistent behavioral and physiological changes.
Fig. 2. Symbiont-mediated RNAi reduces expression of a specific host gene and alters feeding behavior and physiology.
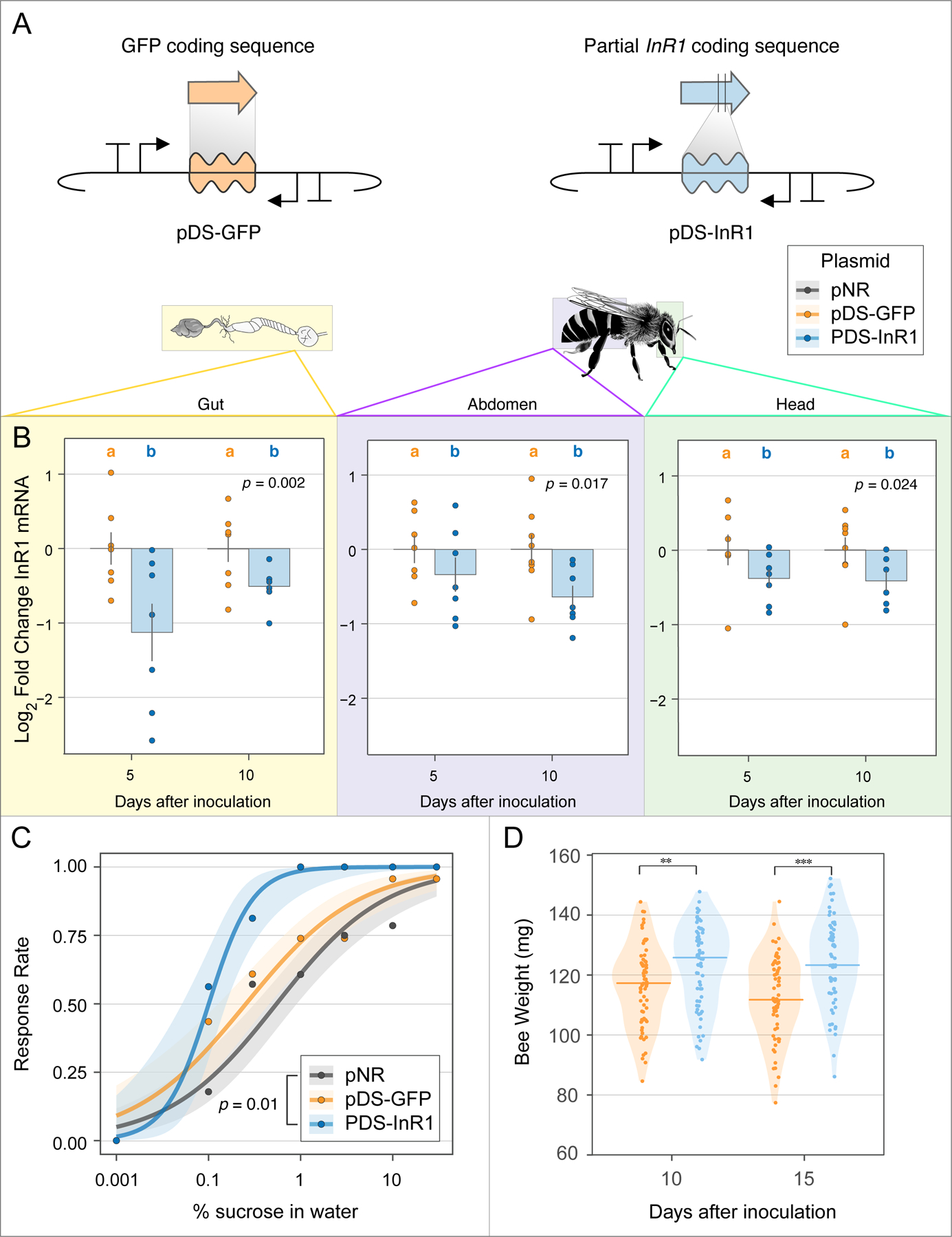
(A) Plasmid design for off-target dsRNA control (pDS-GFP) and InR1 knockdown plasmid (pDS-InR1). (B) Bees colonized with engineered S. alvi expressing InR1 dsRNA (pDS-InR1 plasmid) show reduced expression of InR1 throughout bee body regions for 10 days, as compared to bees colonized with off-target dsRNA control (pDS-GFP). Total N = 29 bees from one hive. (C) pDS-InR1 plasmid increases host feeding activity (sucrose sensitivity response) measured 5 days after inoculation. Curves are a binomial-family generalized linear model (GLM) fit to the response data for N = 67 bees from two hives. (D) pDS-InR1 plasmid significantly increases bee weight, measured 10 and 15 days post inoculation (Mann-Whitney U test). Total N = 135 bees from one hive. See Fig. S4 for additional trial. Error bars and shading represent standard error. **, *** indicate p < 0.01, 0.001, respectively.
Next, we tested whether symbiont-produced dsRNA can protect bees against a common viral pathogen. We designed three dsRNA-producing plasmids targeting different sections of the DWV genome (pDS-DWV1–3) (Fig. S5), and then initially assessed whether S. alvi with these plasmids could help bees resist DWV infection (Fig. S6). We orally inoculated bees with DWV and 48 hours later assessed viral replication in the hemolymph using qPCR. DWV levels were lower in bees colonized by S. alvi with any dsRNA-producing plasmid (Fig. S6A, S7). Dicer was upregulated in bees inoculated with pDS-DWV1 or pDS-DWV2 that were exposed to virus (Fig. S6B), and pDS-DWV2 significantly increased the survival of bees injected with purified virus (Fig. S6C).
To validate these initial findings, we performed a larger experiment to assess whether dsRNA-producing bacteria improved survival following DWV injection. This procedure mimics the natural route of DWV transmission via Varroa mites feeding on bees (2). We injected cohorts of seven-day-old bees with DWV and monitored their survival over ten days (Fig. 3). After DWV injection, bees with bacteria bearing pNR died rapidly. Likewise, pDS-GFP provided no significant protection. In contrast, pDS-DWV2 significantly improved survival of virus-injected bees. Thus, symbiont-mediated RNAi can protect honey bees from DWV by reducing viral proliferation and increasing bee survival.
Fig. 3. Symbiont-produced RNAi can improve honey bee survival after viral injection.
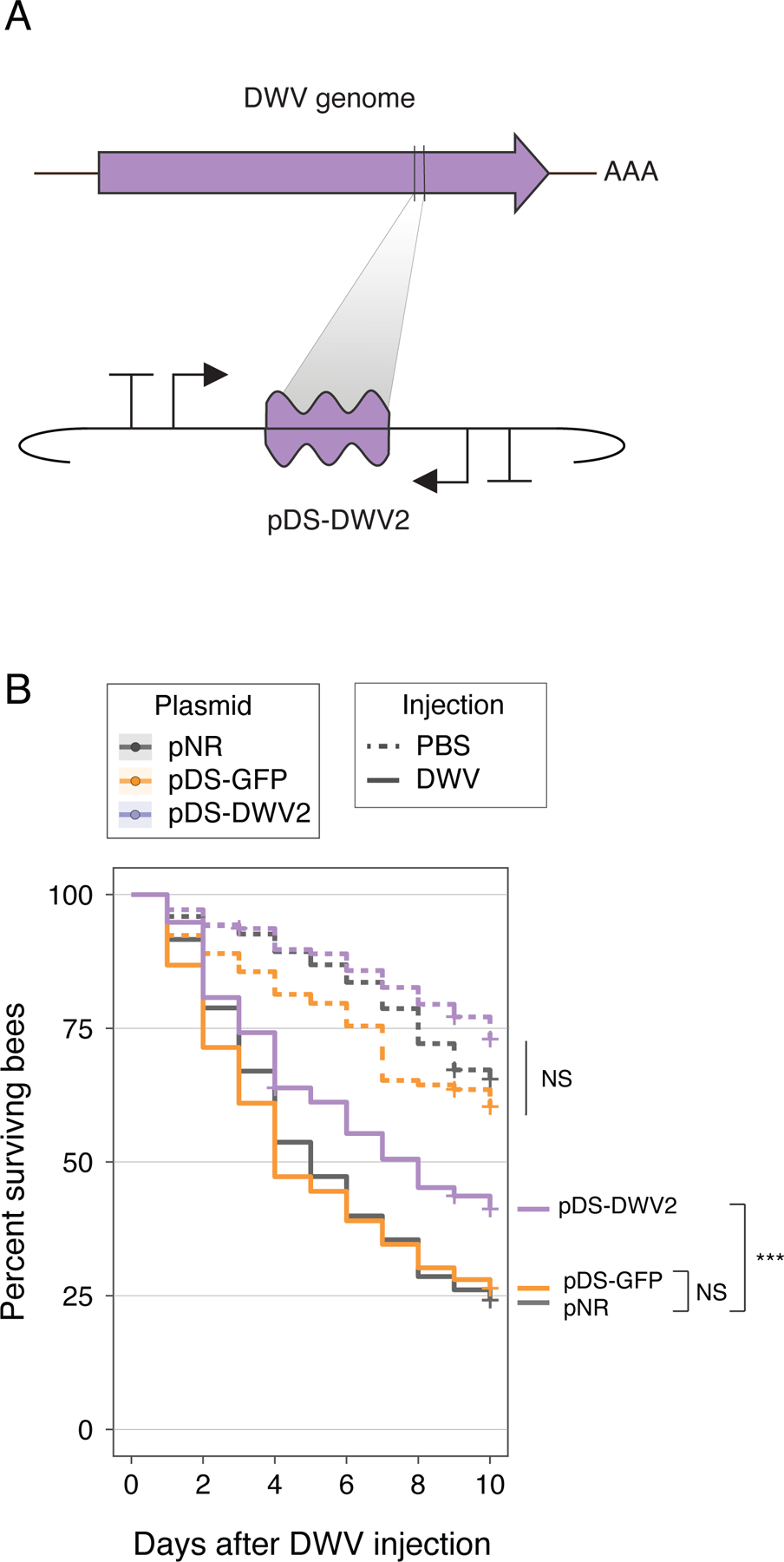
(A) Design of the DWV knockdown construct, pDS-DWV2. (B) Survival curve of bees monitored for 10 days after injection with DWV or PBS control. Bees inoculated with pNR, pDS-GFP, or pDS-DWV2 and then injected with PBS showed no significant change in survival (dotted lines). When injected with DWV, bees inoculated with pDS-DWV2 showed increased survival compared to bees inoculated with pNR (no dsRNA control) or pDS-GFP (off-target dsRNA control) (***, p < 0.001, Wald test). Total N = 980 bees, sourced from three separate hives.
Finally, we tested whether symbiont-produced dsRNA can protect bees against Varroa mites. When Varroa parasitize bees they feed on fat bodies (18) and ingest dsRNA present in that tissue, triggering their own RNAi response. Using mite RNAi to target essential mite genes results in mite death or lowered reproduction (8). We designed a dsRNA-producing plasmid with 14 concatenated sequences from essential genes previously shown to kill Varroa (pDS-VAR) (Fig. 4A, Fig. S8) (8). We inoculated bees with S. alvi bearing pNR, pDS-GFP, or pDS-VAR, and then introduced adult Varroa mites five days later and monitored mite survival for 10 days. Mites that fed on bees colonized with pDS-VAR bacteria died more quickly than mites fed on control bees (Fig. 4B).
Fig. 4. Symbiont-produced RNAi kills Varroa mites feeding on honey bees.
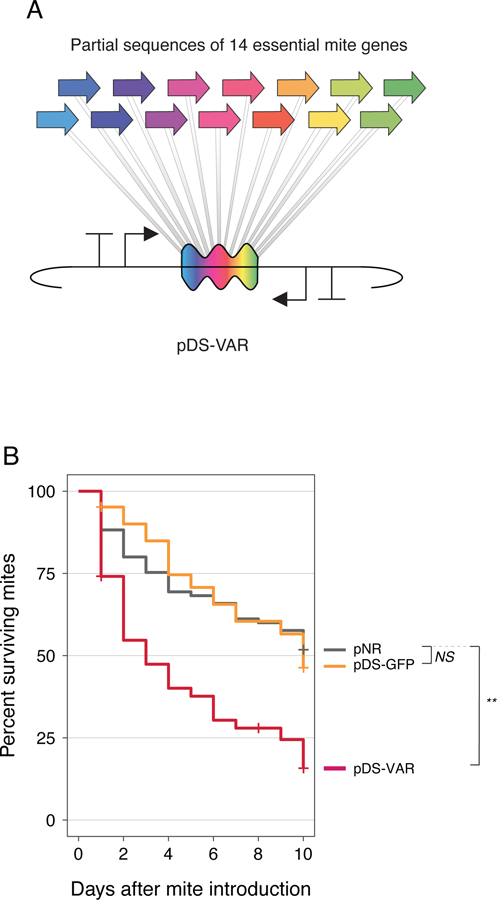
(A) Design of pDS-VAR plasmid targeting essential Varroa genes. (B) Survival curves for Varroa mites fed on bees colonized with engineered S. alvi. Total N = 253 mites. All mites came from a single infested hive. Bees were sourced from three separate hives (**, p < 0.01, Wald test).
Determining whether engineered symbiotic bacteria can improve whole hive health will require further testing. Inoculating bees with dsRNA-producing strains alone had no effect on their survival (Fig. S9). Ongoing within-hive transmission could increase the effectiveness of this treatment by promoting the persistence and spread of engineered strains to new bees. The natural transmission routes of S. alvi and other bee gut symbionts is through direct social contact within hives (15), and engineered S. alvi strains are transferred between co-housed bees in the lab (Fig. S10), suggesting that within-hive transmission is likely. Less is known about between-hive transmission of the bee gut microbiota. Using this approach outside of the laboratory would require understanding these processes and potentially adding biocontainment safeguards.
The degree of protection of bees that we observed in our experiments could likely be improved by further optimizing this symbiont-mediated RNAi delivery system. The specific dsRNA sequence chosen will affect the efficacy of targeted RNAi knockdown, as has been shown for suppression of DWV by oral delivery of RNAi (19). Engineering S. alvi to deliver more dsRNA to bees (e.g., by reducing RNase III activity) could also improve efficacy (20). The deleterious effects of Varroa mites and viruses are interdependent, in part because Varroa vector viruses (2); both types of pests could be targeted at the same time by symbiont-mediated RNAi, which might lead to synergistic improvements in bee health.
We have demonstrated that microbiome engineering can increase resistance to pathogens, a strategy proposed for humans (21) and honey bees (22, 23). Insect-associated microbes have been engineered to interfere with mosquito transmission of malaria (24) and to kill crop pests (25), but not to improve pollinator health. Our experiments imply movement of symbiont-produced dsRNA from the gut lumen into bee cells, but do not identify the mechanism of transfer. Possibly, lysis of S. alvi cells releases dsRNA to be taken up through the same route as orally administered dsRNA. Alternatively, symbiont-mediated dsRNA delivery may co-opt an uncharacterized interaction of S. alvi with its bee host, such as outer membrane vesicle production (26) or direct RNA export (27). Symbiont-mediated RNAi provides a new tool to study bee biology and to improve resilience against current and future challenges to honey bee health.
Supplementary Material
Acknowledgments:
We thank Kim Hammond for laboratory support and work on figures and Howard Ochman for useful discussion.
Funding: DARPA BRICS HR0011-15-C-0095, DARPA HR0011-16-2-0019, and US NIH award R01GM108477
Footnotes
Competing interests: Authors SPL, JEB, and NAM have filed a patent application (62/529,754) on the commercial use of engineered gut bacteria to improve honey bee health.
Data and materials availability: All data are available in the main text or the supplementary materials. Materials will be shared as requested.
References and Notes:
- 1.Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE, Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol 25, 345–353 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Nazzi F, Brown SP, Annoscia D, Del Piccolo F, Di Prisco G, Varricchio P, Della Vedova G, Cattonaro F, Caprio E, Pennacchio F, Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog 8, e1002735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brutscher LM, Flenniken ML, RNAi and antiviral defense in the honey bee. J. Immunol. Res 2015, 941897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amdam GV, Norberg K, Page RE Jr., Erber J, Scheiner R, Downregulation of vitellogenin gene activity increases the gustatory responsiveness of honey bee workers (Apis mellifera). Behav. Brain Res 169, 201–205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai SD, Eu YJ, Whyard S, Currie RW, Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol. Bio 21, 446–455 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, Sela I, IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol. Bio 18, 55–60 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Hunter W, Ellis J, vanEngelsdorp D, Hayes J, Westervelt D, Glick E, Williams M, Sela I, Maori E, Pettis J, Cox-Foster D, Paldi N, Large-scale field application of RNAi technology reducing Israeli Acute Paralysis Virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog 6, e1001160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brutscher LM, Daughenbaugh KF, Flenniken ML, Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep 7, 6448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbian Y, Maori E, Kalev H, Shafir S, Sela I, Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog 8, e1003035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paldi N, Glick E, Oliva M, Zilberberg Y, Aubin L, Pettis J, Chen Y, Evans JD, Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol 76, 5960–5964 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, Zhu KY, Douglas AE, Towards the elements of successful insect RNAi. J. Insect Physiol 59, 1212–1221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mogren CL, Lundgren JG, In silico identification of off-target pesiticidal dsRNA binding in honey bees (Apis mellifera). PeerJ 5:e4131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarosch A, Moritz RF, RNA interference in honeybees: off-target effects caused by dsRNA. Apidologie 43, 128–138 (2012). [Google Scholar]
- 14.Nunes FMF, Aleixo AC, Barchuk AR, Bomtorin AD, Grozinger CM, Simoes ZLP, Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assay. Insects 4, 90–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong WK, Moran NA, Gut microbial communities of social bees. Nat. Rev. Microbiol 14, 374–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ament SA, Corona M, Pollock HS, Robinson GE, Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl. Acad. Sci. U. S. A 105, 4226–4231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X, Wang Y, Sinakevitch I, Lei H, Smith BH, Comparison of RNAi knockdown effect of tyramine receptor 1 induced by dsRNA and siRNA in brains of the honey bee, Apis mellifera. J. Insect Physiol 111, 47–52 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Ramsey SD, Ochoa R, Bauchan G, Gulbronson C, Mowery JD, Cohen A, Lim D, Joklik J, Cicero JM, Ellis JD, Hawthorne D, vanEngelsdorp D, Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. U. S. A 116, 1792–1801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryabov EV, Childers AK, Lopez D, Grubbs K, Posada-Florez F, Weaver D, Girten W, vanEngelsdorp D, Chen Y, Evans JD, Dynamic evolution in the key honey bee pathogen deformed wing virus: novel insights into virulence and competition using reverse genetics. PLoS Biol 17, e3000502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmons L, Court DL, Fire A, Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Chua KJ, Kwok WC, Aggarwal N, Sun T, Chang MW, Designer probiotics for the prevention and treatment of human diseases. Curr. Opin. Chem. Biol 40, 8–16 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Rangberg A, Diep DB, Rudi K, Amdam GV, Paratransgenesis: an approach to improve colony health and molecular insight in honey bees (Apis mellifera)? Integr. Comp. Biol 52, 89–99 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Rangberg A, Mathiesen G, Amdam GV, Diep DB, The paratransgenic potential of Lactobacillus kunkeei in the honey bee Apis mellifera. Benef. Microbes 6, 513–523 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Dos-Santos ALA, Huang W, Liu KC, Oshaghi MA, Wei G, Agre P, Jacobs-Lorena M, Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitten MMA, Facey PD, Del Sol R, Fernandez-Martinez LT, Evans MC, Mitchell JJ, Bodger OG, Dyson PJ, Symbiont-mediated RNA interference in insects. Proc. R. Soc. B 283, 20160042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E, Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol 26, 401–410 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, Wilmes P, The extracellular RNA complement of Escherichia coli. MicrobiologyOpen 4, 252–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrieres L, Hemery G, Nham T, Guerout A, Mazel D, Beloin C, Ghigo J, Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol 192, 6418–6427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong WK, Moran NA, Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol 63, 2008–2018 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Leonard SP, Perutka J, Powell JE, Geng P, Richhart DD, Byrom M, Kar S, Davies BW, Ellington AD, Moran NA, Barrick JE, Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol 7, 1279–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Demares FJ, Crous KL, Pirk CWW, Nicolson S, Human H, Sucrose sensitivity of honey bees is differently affected by dietary protein and a neonicotinoid pesticide. PLoS One 11, e0156584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Miranda JR, bailey L, Ball BV, Blanchard P, Budge GE, Chejanovsky N, Chen Y, Gautheir L, Genersch E, de Graaf DC, Ribiere M, Ryabov E, De Smet L, van der Steen JJM, Standard methods for virus research in Apis mellifera. J. Apic. Res 52, 1–56 (2013) [Google Scholar]
- 34.Lee ME, DeLoache WC, Cervantes B, Dueber JE, A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol 4, 975–986 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Raymann K, Shaffer Z, Moran NA, Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 15, e2001861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen PR, Hammer K, The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol 64, 82–87 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, Frazier M, Frazier J, Cox-Foster D, Chen Y, Underwood R, Tarpy DR, Pettis JS, Colony collapse disorder: a descriptive study. PLoS One 4, e6481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


