Single-molecule imaging and tracking reveal how rRNA transcription is regulated by phase separation in the nucleolus.
Abstract
The nucleolus is a nuclear body with multiphase liquid droplets for ribosomal RNA (rRNA) transcription. How rRNA transcription is regulated in the droplets remains unclear. Here, using single-molecule tracking of RNA polymerase I (Pol I) and chromatin-bound upstream binding factor (UBF), we reveal suppression of transcription with phase separation. For transcription, active Pol I formed small clusters/condensates that constrained rDNA chromatin in the nucleolus fibrillar center (FC). Treatment with a transcription inhibitor induced Pol I to dissociate from rDNA chromatin and to move like a liquid within the nucleolar cap that transformed from the FC. Expression of a Pol I mutant associated with a craniofacial disorder inhibited transcription by competing with wild-type Pol I clusters and transforming the FC into the nucleolar cap. The cap droplet excluded an initiation factor, ensuring robust silencing. Our findings suggest a mechanism of rRNA transcription suppression via phase separation of intranucleolar molecules governed by Pol I.
INTRODUCTION
A subset of macromolecules self-organize into liquid droplets in a process termed phase separation, which allows specific molecules to form a concentrate while others are excluded (1, 2). According to this phase separation principle, cells organize liquid-like compartments such as droplets, allowing the regulation of complex biochemical reactions in space and time.
The first membraneless compartment to be found was the nucleolus, where ribosomes, or translation machinery, are synthesized (2–6). The nucleolus is thought to be composed of immiscible multiphase droplets in three layers (Fig. 1A). The inner layer is the fibrillar center (FC), which contains RNA polymerase I (Pol I) machinery responsible for the transcription of ribosomal RNA genes (rDNA). The intermediate and outer layers are the dense fibrillar component (DFC; fibrillarin rich) and granular component [GC; nucleophosmin (NPM1) rich], respectively.
Fig. 1. Generation of cell lines expressing HaloTag-RPA194 or HaloTag-UBF and single-molecule imaging of HaloTag-RPA194 and HaloTag-UBF.
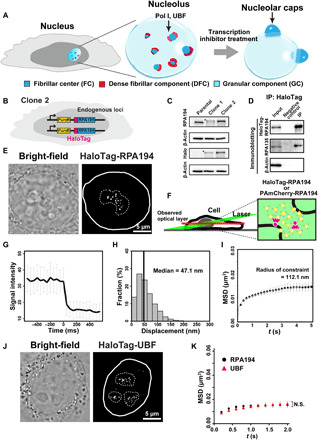
(A) Diagram of nucleolar reorganization in response to transcription inhibition. (B) Genome-edited HeLa cell line used for labeling Pol I molecules. (C) Immunoblotting of the parental HeLa cells, clones 1 (monoallelic tagging) and 2 (biallelic tagging) cell lysates with antibodies of RPA194, HaloTag, and β-actin. Asterisks indicate the position of HaloTag-RPA194. (D) Coimmunoprecipitation of RPA194-RPA135 in the Pol I complex from the clone 2 cell lysate. (E) Bright-field cell nucleus image (left) and oblique illumination microscopy image of HaloTag-RPA194 molecules (right) in a live HeLa cell. The nucleus and nucleolar area are indicated by white and dashed lines, respectively. (F) Left: Diagram of oblique illumination. Right: Sparse fluorescence labeling (red). (G) Single-step photobleaching of HaloTag-RPA194 dots. The vertical and horizontal axes represent the fluorescence intensities of the HaloTag-RPA194 dots and the tracking time series (the photobleaching point is set as time = 0; n = 35 cells), respectively. (H) Displacement (movement) distribution of HaloTag-RPA194 over 50 ms (n = 24 cells). (I) MSD plots of HaloTag-RPA194 molecules from 0.05 to 5 s (n = 24 cells). The data were fitted to a subdiffusive curve (gray line). Error bars indicate the 95% CI computed via bootstrap resampling. (J) Localization of TMR-HaloTag-UBF in the nucleolus. (K) MSD plots of HaloTag-UBF (red, n = 24 cells) and HaloTag-RPA194 (black, n = 24 cells) molecules with 95% CIs. N.S., not significant; P = 0.34 via bootstrapping for HaloTag-UBF versus HaloTag-RPA194.
While the DFC and GC droplet compartments of nucleoli can be recapitulated by purified fibrillarin and NPM1, which separate into two distinct phases in a droplet in vitro (5, 7), the nature of the FC remains unclear. According to observations of FC behavior in living cells, FC is likely organized by phase separation (8–10). Furthermore, it is well established that when RNA Pol I transcription is perturbed by treatment with actinomycin D (AMD) or other DNA intercalators (11, 12), the FC components segregate to the nucleolar periphery, where they coalesce to form large bodies called nucleolar caps. This transformation process resembles a liquid droplet fusion event (Fig. 1A) (8, 13).
Because Pol I is the major molecule present in the FC (14–16) and is involved in various cellular activities including ribosome biogenesis (14, 17, 18), maintenance of genome integrity (19), and aging (20), spatial and temporal regulation of Pol I behavior is a critical issue in cell biology. Fluorescence recovery after photobleaching (FRAP) analysis has revealed the rapid turnover of nucleolar proteins including Pol I and shown that ~10% of Pol I has longer retention times and is correlated with transcription activity in living cells (21–23). Consistent with these findings, a recent study based on super-resolution microscopy and spatiotemporal clustering analysis found that transcriptionally active nucleolar centers appeared as stable clusters of Pol I, whereas prolonged Pol I inhibition (24 hours) made the clusters dynamic (24). However, the distinctive behavior of nucleolar proteins, including Pol I, within multiphase droplets and the relationship of these behaviors within droplets with rRNA synthesis remain unclear, as we lack direct measurements of individual molecule movements in these tiny spaces.
To address these questions, we performed single-molecule tracking of the largest subunit of Pol I, POLR1A (RPA194), and rDNA chromatin bound protein, upstream binding factor (UBF), in living human cells (25–29). Active Pol I molecules were relatively stable and formed small clusters that constrained rDNA chromatin. Treatment with a transcription inhibitor resulted in encasement of Pol I in the large nucleolar cap, caused Pol I to move freely like a liquid, and released the chromatin constraint. On the other hand, accumulation in the FC of a Pol I variant with an RPA194 mutation, which was identified in a patient with cardioskeletal malformation syndrome (30), caused the wild-type (WT) Pol I clusters to become unstable and rRNA transcription to decrease because the mutant Pol I bound stably to chromatin. An initiation factor was segregated from the cap, which could stably suppress transcription within the cap. Our results revealed transcription suppression of rDNA with phase separation of Pol I and other nucleolar proteins.
RESULTS
Generation of genome-edited cell lines to label Pol I
To study the dynamic behaviors of nucleolar molecules in living human cells, we first labeled the large catalytic subunit of Pol I, RPA194, in HeLa cells (Fig. 1B)(31). Using CRISPR-Cas9 genome editing, we introduced a sequence encoding a HaloTag at the initiation site of the endogenous Pol I gene locus (Fig. 1B and fig. S1, A and B). Once proper insertion of the tag sequence into the RPA194 gene had been verified using polymerase chain reaction (PCR), we isolated clones 1 and 2 with mono- and biallelic tagging, respectively (fig. S1C). Immunoblotting (Fig. 1C) showed that the entire RPA194 protein fraction in clone 2 was fused with the HaloTag, which was expressed at a level similar to that of endogenous RPA194 in the parental HeLa cell line. Further analysis via immunoprecipitation of HaloTag-RPA194 using HaloTag-PEG-biotin ligand and streptavidin beads confirmed the interaction between HaloTag-RPA194 and another large subunit, RPA135, in the Pol I complex (Fig. 1D). HaloTag-RPA194 was fluorescently labeled with an excess amount of the HaloTag ligand tetramethylrhodamine (TMR) dye (fig. S2A) and exhibited the expected colocalization with the FC protein UBF (fig. S2B). Quantitative measurement of rRNA transcription showed that clone 2, with biallelic tagging, produced similar amounts of pre-rRNA as did the parental HeLa cell line (fig. S2C), indicating that the expressed HaloTag-RPA194 was functional in rRNA transcription. We used clone 2 for further analysis.
Single Pol I imaging and tracking in living cells
We fluorescently labeled a small number of RPA194 with a low concentration of the HaloTag ligand TMR. Using oblique illumination microscopy, which can illuminate a limited area within the cell (Fig. 1F) (27, 28, 32, 33), we detected clear HaloTag-RPA194-TMR dots (Fig. 1E and left, movie S1). Single-step photobleaching of these dots suggested that each dot represented a single Pol I molecule (Fig. 1G; for multiple molecules, see fig. S2D). Although some Pol I molecules diffused rapidly in the nucleus, most remained in the dense area visible in bright-field images, which corresponded to the nucleolus (Fig. 1E and left, movie S1). Simultaneous expression of enhanced green fluorescent protein (EGFP)–fibrillarin showed that in terms of localization, Pol I remained adjacent to the DFC in living cells (Fig. 1A and fig. S2E). We tracked Pol I movements in the nucleoli at 50 ms per frame (20 Hz) for 5 s (27, 28). The median Pol I displacement over 50 ms was 47.1 nm (Fig. 1H). When plotted, the mean square displacements (MSDs) of Pol I fitted to a subdiffusive curve (Fig. 1I), suggesting that Pol I movement in the FC was somehow constrained. With a tracking time of 5 s, the curve approached a plateau (p), which was proportional to the square of the radius of constraint [Rc; p = 6/5 × Rc2; (34)]. The diameter of the constrained space of Pol I was estimated at approximately 224.2 nm, which is consistent with the diameter of the FC in HeLa cells measured via electron microscopy (~270 nm) (35, 36), validating our imaging analysis. These results indicate that Pol I movement is restricted and that the molecules are relatively stable in the FC. Furthermore, the observed Pol I dynamics in the FC were unaffected by inhibition of posttranscriptional rRNA maturation in the DFC (fig. S3, A and B) (37) and GC (fig. S3, C and D) (38).
Pol I constrains rDNA chromatin motion
We next analyzed the behavior of rDNA chromatin in the FC by tracking UBF, which binds extensively across transcriptionally active rDNA repeats (39, 40). We established a cell line expressing HaloTag-UBF from endogenous loci in a similar manner to HaloTag-RPA194 and performed single-molecule imaging of HaloTag-UBF (Fig. 1J). The MSD plot showed that UBF movement is highly restricted and appears very similar to that of Pol I (Fig. 1K). As active Pol I is known to form clusters on active rDNA in the FC (24), as shown in fig. S3E, clustered active Pol I appears to constrain rDNA chromatin, which is consistent with observations of RNA Pol II (27, 28, 41) and the transcription factory model of RNA transcription (42, 43).
Transcription inhibitor treatment drastically changes the movements of RNA Pol I and UBF
We explored effects of transcription inhibition on the behaviors of Pol I and rDNA chromatin. We monitored Pol I and UBF movements by selectively inhibiting Pol I transcription initiation using the drug CX-5461, which prevents the selective factor-1 (SL-1) complex from binding to the rDNA promoter (fig. S4A) (44). Treatment with CX-5461 for 2 hours decreased transcription to <20% (Fig. 2A), and conventional microscopy images showed that the treatment quickly transforms the FC, including the molecules Pol I and UBF, into larger condensates around the nucleolar periphery (Figs. 1A and 2B and fig. S4, B to D); these condensates are referred to as nucleolar caps (11). In the nucleolar caps, Pol I was much more dynamic than in the FC (right, movie S1). Displacement distribution and MSD analyses showed that following CX-5461 treatment, Pol I dynamics were significantly greater than those in the untreated control (Fig. 2, C and D). The Rc of Pol I motion after inhibition was 193.7 nm, which is highly similar to the reported size of the nucleolar cap (12).
Fig. 2. Dynamics of Pol I and UBF with and without transcription inhibitor.
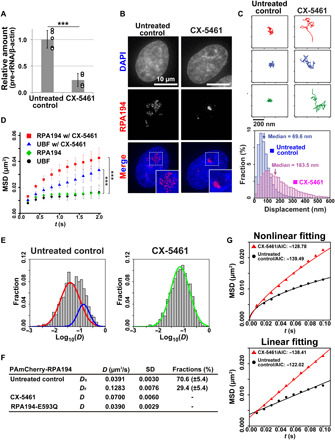
(A) 47S pre-rRNA levels in the cells treated with (right) or without (left) CX-5461. Error bars represent the SD calculated from four to five independent experiments. ***P < 0.001, two-sided unpaired Student’s t test. (B) Localizations of fluorescently labeled HaloTag-RPA194 in clone 2 HeLa cells with (right) or without (left) CX-5461. (C) Top: Three representative trajectories of HaloTag-RPA194 dots in untreated (left column) and CX-5461-treated (right column) cells over 2 s. Bottom: Displacement distributions of HaloTag-RPA194 molecules over 2 s in CX-5461–treated (pink) and untreated control (blue) cells. Significant differences were determined using the Wilcoxon rank sum test (P < 0.01). (D) MSD plots of HaloTag-RPA194 molecules without (green, n = 24 cells) and with CX-5461 treatment (red, n = 23 cells) and HaloTag-UBF molecules without (black, n = 29 cells) and with CX-5461 treatment (blue, n = 29 cells) with 95% CIs. ***P < 0.0001 via bootstrapping for HaloTag-RPA194 in untreated control versus CX-5461–treated cells (P = 1.0 × 10−6) and ***P < 0.001 via bootstrapping for HaloTag-UBF in untreated control versus CX-5461–treated cells (P = 2.2 × 10−4). (E) D distributions of PAmCherry-RPA194 in untreated control (left, n = 20 cells) and CX-5461–treated cells (right, n = 20 cells) with a logarithmic scale. The D distributions of untreated control and CX-5461 treatment were fitted with a mixture of two slow (red) and fast (blue) Gaussian distributions and a single Gaussian distribution (green), respectively. (F) Summary table of the D of PAmCherry-RPA194. DS is the diffusion coefficient of the slow fraction, and DF is that of the fast fraction in untreated cells. (G) Fitting of the MSD of PAmCherry-RPA194 to a subdiffusion (top) or free-diffusion model (bottom). The AIC (48) for each fit is shown.
Following 2-hour treatment with CX-5461, UBF movement in the nucleolar caps increased (Fig. 2D), while UBF continued to bind stably to rDNA under CX-5461 treatment in a previous study (45). The MSD plots of UBF and Pol I indicate that transcription inhibition causes the dynamics of UBF-bound rDNA chromatin to become lower than those of Pol I (Fig. 2D). A log-log plot of UBF MSD data (fig. S4E) revealed that the MSD of UBF under inhibited transcription conditions was proportional to Δt~0.5. This property represents the dynamics of a Rouse polymer (46), an ideal chain consisting of beads connected by harmonic springs, which is driven by thermal fluctuations. Consistent with this concept, the local motion of UBF-bound chromatin in the nucleolar cap did not require adenosine 5′-triphosphate (ATP), as ATP depletion via treatment with sodium azide and 2-deoxyglucose did not significantly affect UBF movement under transcriptional inhibition within 1.0 s (fig. S4, F and G).
We then investigated how much of the Pol I population in the FC was bound to rDNA chromatin and how this population changed upon transcription inhibitor treatment. We, therefore, labeled Pol I with photoactivatable mCherry (PAmCherry) (27, 47). This type of labeling allowed us to label many more Pol I molecules with shorter bleaching time than we might have with HaloTag labeling. We performed single-molecule imaging at a higher frame rate (100 Hz; 10 ms per frame) and calculated the diffusion coefficients (D) of individual Pol I molecules (Fig. 2E). As expected, the D distribution for Pol I molecules fitted a two-species model with slow (DS) and fast (DF) components, which presumably correspond to rDNA-bound Pol I and freely mobile Pol I in the FC, respectively (left, Fig. 2E). Upon inhibitor treatment, the slow Pol I fraction disappeared and the fast fraction became prominent (right, Fig. 2E), leading to an increase in MSD, whereas 70.6 and 29.4% of Pol I molecules were slow and fast in the untreated control, respectively (Fig. 2F). DF was greater than D under inhibition, presumably because the nucleolar caps, in which high levels of materials such as RNA accumulated, might have higher density and provide a more viscous environment than the FC, which slows the movement of the molecules.
Liquid like movement of inactive Pol I molecules in the nucleolar caps
Pol I molecules that are engaged in transcription are constrained by binding with rDNA chromatin (fig. S4H). We inferred that Pol I molecules dissociate from rDNA upon transcription inhibition and move like a liquid in the nucleolar caps. To test this hypothesis, we statistically analyzed PAmCherry-RPA194 tracking data by fitting them to a free-diffusion or subdiffusion model (Fig. 2G). According to the Akaike information criterion (AIC) (48), the MSD of untreated cells fitted a subdiffusive curve better (AIC, −139.4), whereas that of CX-5461–treated cells fitted a linear curve better (AIC, −138.4) (Fig. 2G). Together, these findings suggest that transformation of the FCs into nucleolar caps upon transcription inhibition caused Pol I to be released from rDNA chromatin and to acquire liquid-like behavior through further phase separation (fig. S4H). This conclusion supports the concept of the classical nucleolar cap as a distinct droplet in the nucleolus. Consistently, this liquid-like Pol I movement in the nucleolar cap did not require ATP, as depletion of ATP upon treatment with sodium azide and 2-deoxyglucose did not change Pol I movement significantly upon transcriptional inhibition (fig. S4I).
Mutant Pol I associated with cranioskeletal malformation syndrome
To investigate the phase separation during rRNA transcription suppression in a more physiological context, we focused on a mutant Pol I, an RPA194 variant causing cranioskeletal malformation syndrome (Fig. 3A, c.1777G > C [p. Glu593Gln], [E593Q]) (30). In the patient, one mutant allele of the RPA194 gene, a de novo mutation near the active site for Mg2+ binding, was sufficient to cause a major deficit in cranioskeletal development (30), although how the Pol I mutation affects its behavior and activity remains unknown. We first transiently expressed RPA194-E593Q fused with HaloTag at the N terminus (Fig. 3B). The mutant Pol I exhibited slightly faster movement than normal Pol I (Fig. 3C), but slower than WT Pol I with inhibitor treatment (Fig. 3C).
Fig. 3. Mutant Pol I with an RPA194 variant (RPA194-E593Q) that causes a craniofacial disorder.
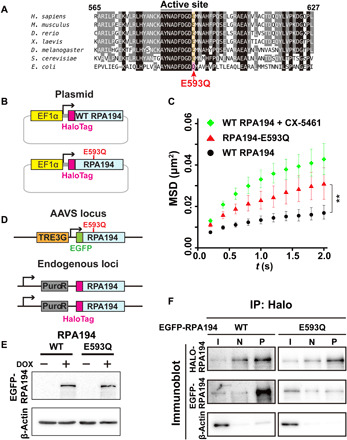
(A) Amino acid sequence comparison of RPA194 including the active site for Mg2+ binding among Homo sapiens, Mus musculus, Danio rerio, Xenopus laevis, Drosophila melanogaster, Saccharomyces cerevisiae, and Escherichia coli. In the RPA194 variant from the patient, the conserved glutamic acid residue (E) at position 593 (highlighted orange) was substituted with glutamine (Q). (B) Diagram of plasmids expressing HaloTag-WT RPA194 and HaloTag-RPA194-E593Q used for single-molecule imaging. (C) MSD plots of HaloTag-RPA194 molecules after expressing HaloTag-RPA194 (WT, black, n = 18 cells) or HaloTag-RPA194-E593Q (E593Q, red, n = 17 cells), with 95% CIs. For comparison, MSD data for WT HaloTag-RPA194 with CX-5461 treatment are reproduced from Fig. 2D (red). **P < 0.01 via bootstrapping for the WT RPA194 versus RPA194-E593Q (P = 5.2 × 10−3). (D) Diagram of the clone 2 cell line (Fig. 1B) expressing EGFP-RPA194-E593Q (or EGFP-RPA194) from an exogenous AAVS locus. (E) Expression of EGFP-RPA194-E593Q induced by doxycycline (DOX; 2 μg/ml) for 24 hours. (F) Coimmunoprecipitation of endogenous HaloTag-RPA194 with EGFP-RPA194 or EGFP-RPA194-E593Q. I, input; N, negative control precipitant (HaloTag-ligand minus); P, precipitant (HaloTag-ligand plus).
Mutant Pol I compromises the stable clustering of WT Pol I
To investigate the behavior of mutant Pol I and its effects on other nucleolar components, we generated a stable cell line with a tetracycline-inducible system consisting of RPA194-E593Q fused with EGFP at the safe-harbor AAVS locus using CRISPR-Cas9 (Fig. 3, D and E). In these cells, two versions of RPA194 are expressed, HaloTag-RPA194 (WT) and EGFP-RPA194-E593Q (mutant); the system allowed us to distinguish these two types. Immunoblotting showed that the expression of EGFP-RPA194 induced by doxycycline increased approximately twofold compared with endogenous HaloTag-RPA194 (fig. S5, A to C). When binding of the two largest subunits in the mutant Pol I complex, RPA194-E593Q and RPA135, was tested via immunoprecipitation (fig. S5D), mutant Pol I exhibited impaired association ability compared with the WT (Fig. 3F).
We then examined the localization of mutant Pol I. Upon induction, the mutant RPA194 was segregated at the nucleolar periphery in most cells (EGFP-RPA194-E593Q; Fig. 4, A and C), in a manner reminiscent of its localization in the nucleolar cap following transcription inhibitor treatment. Mutant Pol I condensate formation was dependent on the amount of RPA194-E593Q expressed (fig. S5E). By contrast, WT RPA194 expression did not affect Pol I localization (WT RPA194; Fig. 4A). Not only the mutant but also the WT Pol I was localized to the condensates (HaloTag-WT RPA194; Fig. 4A), suggesting that mutant Pol I compromised the stable Pol I cluster. Extensive immunofluorescence observations (fig. S6) of other subunits of Pol I (RPA43 and RPA34) and other FC components [UBF and treacle ribosome biogenesis factor 1 (TCOF1)] confirmed that they were all colocalized with the RPA194-E593Q condensates. We then monitored the Pol I population’s behavior using time-lapse live-cell imaging following induction of the mutant RPA194 (Fig. 4B) and found that Pol I foci often fused with one another to form large condensates like droplets. Collectively, mutant Pol I transformed the FC into the nucleolar cap structure, which is a hallmark of the inactive status of rRNA transcription.
Fig. 4. Expression of mutant Pol I with RPA194-E593Q induces nucleolar cap structure formation.
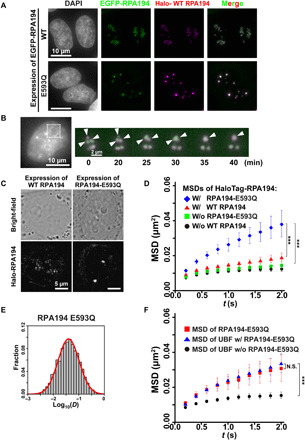
(A) Localizations of HaloTag-RPA194 (magenta) and conditionally expressed EGFP-RPA194 (top, green) or EGFP-RPA194-E593Q (bottom, green) in cells fixed with formaldehyde. Note that HaloTag-RPA194 was fluorescently labeled with an excess amount of the HaloTag ligand TMR. (B) Time-lapse images of RPA194 foci in a cell expressing HaloTag-RPA194 (magenta) and EGFP-RPA194-E593Q (green). HaloTag-RPA194 was fluorescently labeled with an excess amount of the HaloTag ligand TMR. Left: Nucleolar regions were identified using Hoechst 33342 DNA staining in the same live cell. Right: Enlarged time-lapse images of the boxed region on the left. Arrows indicate individual RPA194 foci fusing. (C) Live-cell images of the localization of EGFP-RPA194. (D) MSD plots of HaloTag-RPA194 molecules before (black) and after induction of EGFP-RPA194 (WT, red), or before (green) and after induction of EGFP-RPA194-E593Q (E593Q, blue) with 95% CIs. For each condition, n = 23 to 26 cells. ***P < 0.0001 via bootstrapping for WT expression (red) versus E593Q expression (blue) (P = 6.8 × 10−5) and no expression of E593Q (green) versus E593Q expression (blue) (P = 8.0 × 10−6). (E) D distribution of PAmCherry-RPA194-E593Q (n = 30 cells) with a logarithmic scale (for details, see Materials and Methods), which was fitted to a single Gaussian distribution (red). (F) MSD plots of HaloTag-UBF molecules before (black) and after induction of EGFP-RPA194 (blue) with 95% CIs. For each condition, n = 40 to 45 cells. For comparison, the MSD plot of HaloTag-RPA194-E593Q (red) was reproduced from Fig. 3C. ***P < 0.0001 via bootstrapping for UBF (black) versus UBF with RPA194-E593Q (blue) (P = 1.0 × 10−6). P = 0.29 for UBF with RPA194-E593Q versus RPA194-E593Q.
We inferred that the WT Pol I molecules also behave in a dynamic manner in the nucleolar cap following expression of RPA194-E593Q. Single-molecule imaging of endogenous HaloTag-RPA194 demonstrated that following induction of RPA194-E593Q, WT Pol I moved faster than those without induction, whereas ectopic expression of WT RPA194 had little effect (Fig. 4D). These results suggest that upon mutant Pol I accumulation, WT Pol I clusters become unstable and dissociate from rDNA chromatin, leading to liquid-like behavior (fig. S7A).
Mutant Pol I can stably bind to UBF-bound rDNA chromatin and competitively inhibit WT Pol I binding
To further explore the behavior of mutant Pol I, we performed single-molecule imaging of mutant Pol I at a higher frame rate (100 Hz; 10 ms per frame) based on transient expression of PAmCherry-RPA194-E593Q (Fig. 4E). The D distribution obtained for mutant Pol I molecules could be well fitted with a single component model, similar to the slow DS of Pol I (left, Fig. 2E), which presumably corresponds to rDNA-bound Pol I in the FC. This finding suggests that the mutant form stably binds to rDNA.
Furthermore, we studied UBF behavior in the nucleolar caps based on the expression of RPA194-E593Q (Fig. 4F). The MSD plots showed that UBF movement was quite similar to that of RPA194-E593Q (Fig. 4F). Together, our findings suggest that the RPA194-E593Q mutant can bind stably to UBF-bound rDNA chromatin and competitively inhibit WT Pol I-binding (fig. S7A).
Mutant pol I represses rRNA transcription
Next, we confirmed that the ectopic expression of mutant Pol I results in a defect in rRNA transcription by measuring 5-ethynyl uridine (5-EU) incorporation into nascent rRNA in the nucleolus (fig. S7B) (49). This induction reduced the level of 5-EU in the nucleolus to 16.3% (Fig. 5A and fig. S7B). Consistently, the 47S pre-rRNA transcript was decreased to 30% in cells expressing RPA194-E593Q compared with control cells (Fig. 5B). These reduction rates may be underestimated; in some cells, mutant Pol I was not expressed at high levels upon induction and rRNA transcription was not suppressed. Nonetheless, the obtained results indicate that the mutant Pol I complex with the RPA194 variant is defective in its transcription ability. Accumulation of the mutant form severely perturbed rRNA transcription by preventing WT Pol I from binding to rDNA (fig. S7A).
Fig. 5. Accumulation of RPA194-E593Q in the nucleolus inhibits rRNA transcription.
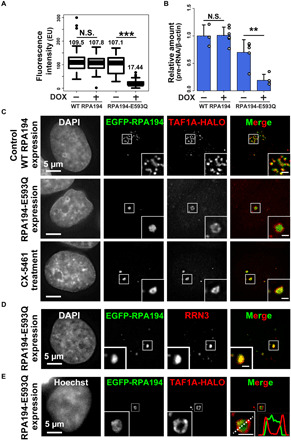
(A) rRNA synthesis was monitored by incorporating EU. Boxplot of fluorescence intensity of EU conjugated with Alexa Fluor 594 in cells before and after DOX induction of RPA194 (WT) or RPA194-E593Q (E593Q). Median values are indicated. ***P < 0.001, Wilcoxon rank sum test. (B) 47S pre-rRNA levels before and after DOX induction of RPA194 or RPA194-E593Q. Error bars represent SD calculated from three to five independent experiments. **P < 0.01, Student’s t test. (C) Localization of the TAF1A-HaloTag (red) in cells transiently expressing EGFP-WT RPA194 (first row) or EGFP-RPA194-E593Q (second row). Third row: WT RPA194 localization in a CX-5461–treated cell. Insets show enlarged images of regions indicated in the box. Scale bars, 1 μm. (D) Localization of RRN3 in cells expressing EGFP-RPA194-E593Q. Insets show enlarged images of the regions indicated in the box. Scale bar, 1 μm. (E) Localizations of the TAF1A-HaloTag and transiently expressed EGFP-RPA194-E593Q in a living HeLa cell. The merged image has line plots of measurements along the dotted line.
We then explored how mutant Pol I expression stably silenced rDNA chromatin in the nucleolar cap droplets, where WT Pol I diffuses freely around UBF-bound rDNA chromatin. To address this topic, we investigated the locations of specific transcription initiation factors (fig. S7C). We found that upon mutant Pol I expression, TAF1A, which is a TATA box–associated factor (TAF) (fig. S7C), segregated from the cap droplets, whereas it was localized in the nucleolar cap with transcription inhibitor treatment (Fig. 5C and fig. S7D). By contrast, RRN3, which forms a bridge between Pol I and TAFs (fig. S7C), was colocalized with the cap droplets with mutant Pol I expression (Fig. 5D). Segregation of TAF1A from the cap droplets was also observed in living cells expressing mutant Pol I (Fig. 5E). We found that the disorder tendency exhibited by the TAF1A polypeptide was much lower than those of primary FC components (fig. S8A) that we previously identified using mass spectrometry analysis (16), suggesting that differences in the disorder tendency of FC proteins drive the exclusion of SL-1 from cap droplets, which is induced by mutant Pol I.
Although cap droplet formation with inhibitor treatment is often accompanied by DNA damage (50, 51), DNA damage is not a prerequisite to the transformation of Pol I clusters on the mutant accumulation pathway because γ-H2AX foci, which are DNA damage indicators, were not detectable in mutant-expressing cells, in contrast with observations from CX-5461 treatment (fig. S8, B and C). Together, these findings suggest that the exclusion of an essential factor stably silenced transcription within the droplet cap and that silencing by mutant Pol I accumulation is a distinct mechanism from that of the inhibitor treatment (fig. S7A).
Pol I is a modulator of phase separation in the nucleolus
Transcription inhibitor treatment and expression of the mutant form transform the FC into nucleolar caps through further phase separation processes (figs. S4H and S7A). As both methods of transcription inhibition release active Pol I clusters from rDNA chromatin, we tested whether Pol I could act as a modulator of phase separation in the nucleolus. To test this idea, we exploited the rapid depletion of Pol I by the auxin-inducible degron (AID) system (52) to minimize the risk of various indirect effects from Pol I depletion. Using CRISPR-Cas9–based genome editing, we introduced a cassette encoding mini-AID (mAID) and the fluorescent protein mClover (mAID + mClover) at the initiation site of the endogenous RPA194 gene locus (Fig. 6A) in human colon adenocarcinoma HCT116 cells expressing OsTIR1, which is involved in the induced degradation process (for details of the establishment of this cell line, see Materials and Methods) (52). In the established cells, RPA194 was depleted within 3 hours after auxin addition (+auxin; Fig. 6B). In accordance with previous observations (figs. S4D and S6B), rapid depletion of RPA194 transformed the FCs into nucleolar caps (Fig. 6C), supporting our conclusion that Pol I is a modulator of phase separation in the nucleolus. Pol I and/or synthesized RNAs might drive the nucleolar phase separation.
Fig. 6. Rapid depletion of Pol I and diagram of two distinct suppression mechanisms of rRNA transcription.
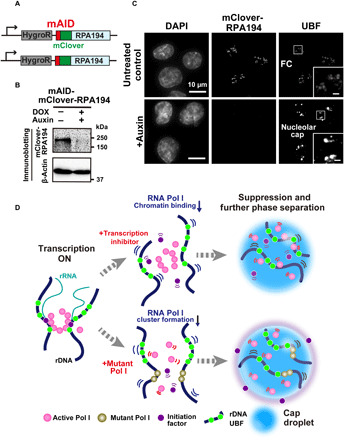
(A) Diagram of a genome-edited HCT116 line expressing mAID-mClover-RPA194 from the endogenous RPA194 gene locus. (B) Rapid Pol I degradation in HCT116 cells after auxin addition. (C) Localization of UBF in the cells before (first row, untreated control) and after rapid depletion of Pol I (second row, +auxin). Insets show enlarged images of the regions indicated with boxes. Scale bars, 1 μm. (D) Left: Active Pol I molecules form a stable cluster/condensate to transcribe rRNA genes (rDNA), constraining rDNA chromatin. Top: Once transcription is inhibited by a drug, the Pol I cluster/condensate detaches from chromatin, thus releasing the chromatin constraint. Pol I behaves like a liquid in the nucleolar cap through further phase separation. Bottom: Mutant Pol I stably binds to rDNA chromatin and inhibits WT Pol I cluster/condensate formation. The whole Pol I population becomes mobile in the nucleolar cap. An initiation factor is excluded from the cap, ensuring robust transcription suppression within the cap. Figure 6D consists of illustrations reproduced from figs. S3E, S4H, and S7A. Note that this model is highly simplified.
DISCUSSION
The nucleolus is thought to be composed of three subcompartments with liquid-like properties—the FC, DFC, and GC (Fig. 1A) (4, 5). The DFC and GC droplet compartments in nucleoli can be recapitulated by purified components in vitro (5, 7). Recent studies have suggested that the phase separation process of the nucleolus is connected to ribosome biogenesis and that this process drives the initial processing of pre-rRNA in the DFC (53) and facilitates selective exclusion of fully assembled ribonucleoprotein complexes from the nucleolus (54). Moreover, the phase-separated GC compartment serves as a storage vessel for a subset of misfolded proteins under stress conditions, such as heat shock (55). However, the nature of the FC remains unclear.
Our single-molecule imaging study of Pol I and UBF revealed that these molecules formed stable clusters in the FC for rRNA transcription and that upon transcription inhibitor treatment, Pol I acquired liquid-like behavior that coincided with nucleolar-cap droplet formation (Fig. 6D). Although the dynamic turnover of nucleolar components has been previously investigated using FRAP analysis (21, 22), recent studies have highlighted the difficulties of validating liquid-like properties in macromolecular condensates using FRAP (2, 56). Our study eliminated these difficulties by directly measuring the movement of molecules that remained within the FC; as a result, we found that approximately 70% of Pol I in the FC was bound to rDNA chromatin (Figs. 2F and 6D). Considering that approximately 10% of Pol I molecules are engaged in transcription elongation with rDNA (22), most active Pol I molecules are involved in cluster/condensate formation through self-association, together with other FC components, and constrain UBF-bound rDNA chromatin (Fig. 6D). This finding is consistent with the observation in our recent report that active Pol II globally constrains chromatin (27, 28, 41), suggesting that chromatin constraint by clusters/condensates of RNA polymerases may be a general mechanism for efficient transcription.
CX-5461 treatment and mutant Pol I expression revealed two distinct pathways for the suppression of rDNA transcription based on multilayered phase separation in the nucleolus (Fig. 6D). Pol I is a modulator of the phase separation process (Fig. 6C). CX-5461 inhibitor treatment released active Pol I clusters from rDNA chromatin, causing their movement to resemble that of a liquid in the nucleolar cap, which forms through transformation of the FC (top, Fig. 6D), presumably because CX-5461 treatment prevents the SL-1 complex from binding the rDNA promoter and further recruiting Pol I (fig. S7C) (44). Considering that transcription inhibitor treatment often causes DNA damage (50, 51), the repair process may be efficiently enhanced by the flexibility of rDNA chromatin in the cap. Consistently, it is well known that the nucleolar cap is also induced by ataxia-telangiectasia mutated (ATM) kinase–dependent signaling pathways with double-strand breaks in rRNA genes (57, 58).
Expression of the mutant Pol I, which is associated with a craniofacial disorder, leads to another form of transcription suppression (bottom, Fig. 6D). After mutant Pol I, which exhibits stable rDNA binding with low self-association, accumulated in the nucleolus, the WT Pol I clusters became unstable, and the whole Pol I population exhibited rapid movement in the nucleolar cap (bottom, Fig. 6D). The initiation factor TAF1A was segregated from the cap, ensuring robust suppression of transcription. Although the detailed segregation mechanism remains unknown, differences in the disorder of FC proteins may drive the exclusion of SL-1 from cap droplets induced by mutant Pol I (fig. S8A). Upon inhibitor treatment, SL-1 complex modification or other factors may overcome the low protein disorder tendency to maintain the complex within the nucleolar cap droplet.
Transcription inhibition induced Pol I to behave like a liquid in the nucleolar cap and released the chromatin constraint. Both the local motion of UBF-bound chromatin and the liquid-like Pol I movement did not require ATP (fig. S4, G and I), suggesting that both types of movement in the nucleolar cap are driven by thermal fluctuations. UBF-bound chromatin behaves like a Rouse polymer (46), which is an ideal chain consisting of beads connected by harmonic springs; these dynamics are likely sufficient to create specific cis- or trans-chromatin contacts over distances of less than ~400 nm (46). Therefore, the state of rDNA chromatin in the nucleolar cap may be sufficient for rapid and efficient recruitment of repair machinery and subsequent homologous recombination to repair spontaneously damaged regions (58) or regions damaged by inhibitor treatment (50, 51). This rDNA chromatin status would also facilitate the restarting of rRNA gene transcription and subsequent nucleolus reorganization.
This finding is consistent with the recent observation of stable clusters of active Pol I (24) and reports that active Pol II, mediator, and other transcription factors form dynamic clusters/condensates, presumably through a phase separation process (25, 26, 59–63). Within the clusters/condensates, RNA polymerases and other transcription factors may be concentrated together to promote functional interactions between factors, leading to highly efficient transcription initiation reactions and subsequent entry into elongation. Because the mutant form of Pol I, which exhibits stable rDNA binding and low self-association, compromises stable clustering/condensation of the WT and represses rRNA transcription, our findings also suggest that RNA polymerase cluster/condensate formation is important during the transcription process. However, to our knowledge, transcription inhibition–dependent Pol II droplets have not been observed. Liquid droplet formation by RNA polymerases, which is associated with transcription suppression, may be a system that is unique to Pol I.
We also describe here a potential mechanism underlying a craniofacial disorder. When primary progenitors of the craniofacial skeleton—i.e., neural crest cells—require abundant rRNA (64), they may increase levels of Pol I machinery components, thus increasing the production of the mutant RPA194. This process causes the failure of ribosome biogenesis up-regulation in a tissue-specific manner, followed by severe growth and differentiation defects. Some heterozygous missense mutations in the genetic codes of other subunits of Pol I, such as POLR1C, and POLR1D, have been identified; these mutations cause a craniofacial development disorder called Treacher Collins syndrome (65). An allelic imbalance in the expression of these genes may also induce further phase separation of nucleolar components and perturbation of ribosome synthesis.
MATERIALS AND METHODS
Cell culture
HeLa S3 cells (66) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; D5796-500ML; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; FB-1061/500, Biosera) at 37°C under 5% CO2.
Plasmid construction
Construction of the pX330 CRISPR-Cas9 plasmid (#42230, Addgene) expressing guide RNA for each target site and the donor plasmid was performed as follows. Gene-specific guide RNA sequences for RPA194, UBF, TCOF1, RPA34, RPA43, TAF1A, and AAVS1 were designed using the CRISPR design website and inserted into the pX330 Bbs I cloning site, as previously described (67). The guide RNA sequences were as follows: RPA194 gRNA, 5′-TGGGGTGTCGGAATTCAAAA-3′; UBF gRNA, 5′-TTCCAGGTCTGTGGGGCAGT-3′; RPA43 gRNA, 5′-ATTCCTAGGGGCGTGGTCG-3′; RPA34 gRNA, 5′-TCAACCCGCACCCTCACCGC-3′; TCOF11 gRNA, 5′-AAGTAGCTCCCGCCGCTTCC-3′; TAF1-A gRNA, 5′-TAGCTACACAGTAAGTAGCT-3′; and AAVS1 gRNA, 5′-GGCCACTAGGGACAGGAT-3′. For donor plasmids (fig. S1B), the left and right homologous arms were PCR amplified using KOD FX (KFX-101, Toyobo) from HeLa S3 genomic DNA, which was isolated using a Wizard Genomic DNA Purification kit (A1120, Promega). The HaloTag sequence was PCR amplified from the pFC14A HaloTag CMV Flexi Vector (G965A, Promega). The homologous arms, antibiotic resistance open reading frame, and HaloTag fragment were joined using standard overlapping PCR and inserted between the Eco RI and Sal I sites of the pGEM-T (Easy) vector (A1360, Promega) using In-Fusion (639648, Clontech). The primers used to amplify the homologous arms of the RPA194, UBF, RPA43, RPA34, TCOF1, TAF1A, and AAVS1 loci are shown in table S2. The HaloTag coding sequence of pGEM-HaloTag-RPA194 was replaced with the PAmCherry fragment from pPAmCherry-N1 (632584, Clontech) to generate pGEM-PAmCherry-RPA194.
To clone full-length RPA194, total RNA was isolated from human RPE-1 cells (CRL-4000, American Type Culture Collection) using an RNeasy Mini kit (74104; Qiagen), and first-strand cDNA was synthesized using a SuperScript III First-Strand Synthesis System (18080-400; Thermo Fisher Scientific) with oligo(dT). The coding region of RPA194 was amplified from first-strand cDNA using the following primers: 5′-GATCTGGTGGCGGCGGTTCAATGTTGATCTCCAAGAACATGC-3′ and 5′-GCCACTGTGCTGGATCTATCTCAGAGGCTGCTTGAG-3′. The HaloTag fragment was amplified from pGEM-T easy-HaloTag-RPA194 using the following primers: 5′-TGGAATTCTGCAGATGCCACCATGGGATCCGAAATCGGTAC-3′ and 5′-CGCCGCCACCAGATCCACCTCCACCAGATCCACCTCCACCACCGGAAATCTCCAGAGTAG-3′. The amplified fragment was inserted into pEF1-FRT digested with Eco RV to generate pEF1-FRT-HaloTag-RPA194. For the mutant RPA194 (RPA194-E593Q), site-directed mutagenesis was performed to substitute G for C at 1777 base pairs (bp) downstream of the start codon using PCR with the following primers: 5′-AGATGAATGCCCATTTCCCC-3′ and 5′-GGTCTCCATCAAAGTCGGCA-3′ (the underlined letter represents the mutation site).
To generate the inducible vector of RPA194-E593Q (pAAVS1-EGFP-RPA194-E593Q), RPA194-E593Q fragments were amplified from pEF1-FRT-HaloTag-RPA194-E593Q using the following primers: 5′-GCAGTCGACGGTACCATGTTGATCTCCAAGAACATGC-3′ and 5′-GGTACCGTCGACTGCAGAATTC-3′. The EGFP fragment was amplified from pEGFP-C1-fibrillarin (#26673, Addgene) using the following primers: 5′-GTAAACTTAAGGTTAATTAACGCCACCATGGTGAGCAAGGGCGAGGAG-3′ and 5′-GGTACCGTCGACTGCAGAATTC-3′. The amplified RPA194-E593Q and EGFP fragments were joined together using standard overlapping PCR and inserted into the Pac I and Age I sites of pAAVS1-NDi-CRISPRi (#73498, Addgene) (68) using In-Fusion.
Construction of pEF1-EGFP-fibrillarin-FRT was performed as follows. First, pEGFP-C1-fibrillarin (Addgene, #26673) was digested with Bam HI and Age I to excise the EGFP-fibrillarin fragment. This fragment was then blunt ended using a DNA Blunting Kit (6025, TaKaRa) and inserted into the Eco RV site of a pEF5/FRT/V5-DEST Gateway vector (V602020, Invitrogen) to obtain pEF1-EGFP-fibrillarin-FRT.
Isolation of stable cell lines
To establish HeLa S3 cells that stably express HaloTag-RPA194 from the endogenous locus, we used the CRISPR-Cas9 system (67, 69). HeLa S3 cells at 25% confluence in a 6-cm dish were cotransfected with 500 ng each of pX330 (with a target gene-specific guide sequence inserted) and an antibiotic resistance donor plasmid using Effectene Transfection reagent (301425, Qiagen). At 24 hours after transfection, the medium was changed, and at 48 hours after transfection, transformants in the 6-cm dish were transferred to a 10-cm dish and selected using puromycin (1 μg/ml; P8833-25MG, Sigma-Aldrich) or G418 (800 μg/ml; ALX- 380-013-G001, Enzo Life Sciences). The medium with antibiotics was changed every 3 days until untransfected cells had all died (after 2 weeks of selection). To establish HeLa S3 cells that stably express EGFP-fibrillarin, we used an Flp-In system (K601002, Invitrogen) as previously described.
A conditional AID mutant was generated in an HCT116 cell line expressing OsTIR1 (52). We transfected a CRISPR-Cas9 plasmid targeting the first methionine site of the RPA194 gene (POLR1A, CCTGGAGGATGTTGATCTCCAAGAACATG), along with a donor harboring a Hygro-P2A-mAID-mClover cassette flanked by a 700-bp homology arm using Effectene transfection reagent. After selection with hygromycin (100 μg/ml; 10687010; Thermo Fisher Scientific), colonies were isolated for further analysis through immunoblotting.
PCR genotyping of HaloTag tagging
Genomic DNA was isolated using proteinase K and phenol (Molecular Cloning 6.1) and genotyped using KOD FX (KFX-101, Toyobo) and the following primers: dhRPA194-a3 5′-AAAGAGCGAGCAGAGAGTCG-3′ and dhRPA194-a4 5′-CGCCTGAACTGACACTTGAA-3′. The positions of the primers along the genomic sequence are shown in fig. S1B.
Single-molecule live-cell imaging
For single-molecule imaging of HaloTag- or PAmCherry-fused proteins in living cells, all cell types were plated onto glass-bottomed dishes (3970-035, Iwaki) treated with poly-lysine (P1524-500MG; Sigma-Aldrich). Before HaloTag-fused protein imaging, cells were incubated in a medium containing 100 pM TMR-HaloTag ligand (8251, Promega) for 30 min, washed three times with 1× Hanks’ balanced salt solution (HBSS; H1387, Sigma-Aldrich), and incubated with DMEM containing 10% FBS, without phenol red. To maintain constant cell culture conditions (37°C, 5% CO2) and humidity during imaging, we used a live-cell chamber (INU-TIZ-F1, Tokai Hit) and GM-8000 digital gas mixer (Tokai Hit).
Single-molecule live-cell imaging was performed using an inverted Nikon Eclipse Ti microscope with a 100-mW sapphire 561-nm laser (Coherent) and sCMOS ORCA-Flash 4.0 camera (Hamamatsu Photonics). Fluorescently labeled Halo with TMR or PAmCherry in living cells was excited with a 561-nm laser through an objective lens [100× PlanApo total internal reflection fluorescence (TIRF), numerical aperture 1.49, Nikon] and detected at 575 to 710 nm. An oblique illumination system with a TIRF unit (Nikon) was used to excite Halo-TMR or PAmCherry molecules within a limited thin area of the cell’s nucleus and reduce background noise. Movies of 600 sequential frames were acquired using MetaMorph software (Molecular Devices) at 50 ms per frame under continuous illumination.
Data analysis for single-molecule live-cell imaging and tracking
The methods used for image processing, single-molecule tracking, and single-nucleosome movement analysis have been described previously (27). Briefly, sequential images were converted into 8-bit grayscale images, and background noise signals were subtracted using rolling ball background subtraction (50) in Fiji software (70). The nucleolar regions in the images were extracted manually. Following this step, the centroid of each fluorescent dot in each image was determined, and its trajectory was then tracked with u-track using MATLAB (MathWorks) (71). The numbers of trajectories obtained in each experiment are listed in table S1. The MSD was originally calculated in two dimensions. To obtain three-dimensional (3D) values, we multiplied 2D values by 1.5 (4 to 6 Dt). Histograms, MSD plots, and trajectories of the molecules were prepared using R software (R Development Core Team). Error bars for each data point [representing the average MSD(t)] were calculated as 95% confidence intervals (CIs) via bootstrap resampling of the population. Statistical analyses of single-nucleosome MSD values among transcription conditions were performed using the bootstrap resampling method.
Regarding the single-molecule imaging and tracking of UBF, which binds extensively across transcriptionally active rDNA repeats (39, 40), we considered the free fraction of UBF to be negligible. According to reported FRAP data for UBF (21), only ~3.8% of UBF molecules on rDNA can dissociate during our 0.5-s tracking period and ~14.3% over 2 s of tracking in the untreated condition. Upon transcription inhibition, only ~5.4% could dissociate within 2 s of tracking.
To generate super-resolution images of Pol I (fig. S2E), molecule positions were mapped using the R package base (65 nm per pixel), and Gaussian blur (sigma = 1 pixel) was added using ImageJ software (National Institutes of Health) for smoother rendering.
Data analysis for determining D
To determine the D values of Pol I molecules, we plotted MSD (R2 > 0.8) against time (40 ms) using PAmCherry-RPA194 tracking data (three independent experiments, n = 20 cells per experiment) and then calculated D from the slope of the plot using the Einstein relation (MSD = 6 Dt). Tracking data for PAmCherry-RPA194 molecules, which photobleach rapidly, were used for D measurement, as acquiring fluorescence protein data over a shorter period reduces the bias toward slow-moving molecules.
Model-fitting analysis
To test the fit of models for each MSD plot, we calculated the AIC score (48) using R software following linear [MSD(t) = a × t + b] or nonlinear regression [MSD(t) = a × tb] analyses of PAmCherry-RPA194 MSD data collected from 0 to 0.1 s.
Immunoblotting
A small proportion of cells were reserved for cell counting. Cells were lysed in Laemmli sample buffer supplemented with 10% 2-mercaptoethanol (133-1457, Wako) and incubated at 95°C for 5 min to denature proteins. We then performed 10% tris-glycine sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis to separate the proteins. Proteins in the gel were transferred to an Immobilon-P membrane (IPVH00010, Merck) and blocked with phosphate-buffered saline (PBS) with Tween 20 containing 5% nonfat milk (190-12865, Wako) for 30 min at room temperature. Proteins were detected using anti-RPA194 (1:1000 dilution; sc48385, Santa Cruz Biotechnology), anti–β-actin (1:50,000 dilution; A5441, Sigma-Aldrich), anti-HaloTag (1:1000 dilution; G9211, Promega), and anti-GFP (1:2000 dilution; 632381, TaKaRa) antibodies.
Immunostaining
Cells were grown on coverslips coated with poly-lysine, washed twice in PBS, and fixed with 1.85% formaldehyde (063-04815, Wako) in PBS. For blocking, sections were treated with 5% normal goat serum (NGS; 143-06561, Wako) for 30 min. The fixed cells were stained with anti-RPA194 (1:500 dilution; sc48385, Santa Cruz Biotechnology), anti-UBF (1:250 dilution; sc13125, Santa Cruz Biotechnology), anti-fibrillarin (1:250 dilution; ab4566, Abcam), anti-NPM1 (1:1000 dilution; B0556, Sigma-Aldrich), anti–γ-H2AX (1:1000 dilution; B0556, Abcam), and anti-RRN3 (1:300 dilution; HPA049837, ATLAS) antibodies in PBS containing 1% NGS and incubated with the following mouse and/or goat secondary antibodies in PBS containing 1% NGS for 1 hour: goat anti-mouse immunoglobulin G (IgG) Alexa Fluor 488 (1:1000 dilution; A11029, Thermo Fisher Scientific), goat anti-mouse IgG Alexa Fluor 594 (1:1000 dilution; A11032, Thermo Fisher Scientific), goat anti-mouse IgG Alexa Fluor 647 (1:1000 dilution; A21235, Thermo Fisher Scientific), goat anti-rabbit IgG Alexa Fluor 594 (1:1000 dilution; A11037, Thermo Fisher Scientific), and goat anti-rabbit IgG Alexa Fluor 647 (1:1000 dilution; A21245, Thermo Fisher Scientific). After washing three times with PBS for 5 min and counterstaining with DAPI (4′,6-diamidino-2-phenylindole) (1 μg/ml; 10236276001, Roche), coverslips (c018001, Matsunami) were mounted in PPDI [20 mM Hepes (pH 7.4), 1 mM MgCl2, 100 mM KCl, 78% glycerol, and paraphenylene diamine (1 mg/ml); 695106-1G, Sigma-Aldrich) and sealed with nail polish. To stain HaloTag-RPA194 and TAF1A-HaloTag, cells were incubated in a medium containing 0.1 and 0.2 μM TMR-HaloTag ligand for 24 hours, respectively. Section images were recorded using a DeltaVision Elite microscope (Applied Precision) or a DeltaVision Ultra microscope (Cytiva) and deconvolved using the DeltaVision Softworx software to eliminate out-of-focus blur to obtain clearer pictures. Deconvolved images were projected using the Quick Projection tool in the Softworx software to obtain the maximum intensity of focus signals.
For live-cell imaging of TAF1A-HaloTag following transient expression of RPA194-E593Q, cells were incubated in medium containing 5 μM TMR-HaloTag ligand for 60 min and then washed as described above. The nucleolar region was identified using Hoechst 33342 DNA staining in the same live cell.
Immunoprecipitation
For immunoprecipitating RPA135 with HaloTag-fused RPA194, cells were resuspended in radioimmunoprecipitation assay buffer [25 mM tris-HCl (pH 7.6), 300 mM NaCl, 1% NP-40 (11754599001, Roche), 1% sodium deoxycholate (192-08312, Wako), 0.1% SDS, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride (PMS; P7626-1G, Sigma-Aldrich), and protease inhibitor cocktail (11836153001, Sigma-Aldrich)] and incubated for 5 min on ice. After centrifugation at 13,000g for 10 min at 4°C, cell extracts were used for subsequent immunoprecipitation procedures. For immunoprecipitating EGFP-RPA194 with HaloTag-fused RPA194, cells were first harvested for nuclear extraction. Cell pellets were swollen in hypotonic buffer [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.8), 1.5 mM MgCl2, 10 mM KCl, 0.2 mM PMSF, and 0.5 mM dithiothreitol (DTT)] and Dounce homogenized using five passes of a tight pestle. After centrifugation at 1500g for 5 min at 4°C, the nuclei were resuspended in a salt buffer [20 mM HEPS (pH 7.8), 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 125 mM KCl, 0.2 mM PMSF, 0.5 mM DTT, and protease inhibitor cocktail] and incubated at 4°C on a rotator. After centrifugation at 13,000g for 15 min, the supernatant was collected as nuclear extracts. For immunoprecipitating EGFP-fused RPA194, we used the GFP-Trap (gtma-10, ChromoTek) following the manufacturer’s instructions for cell extract preparation.
For immunoprecipitating HaloTag-fused RPA194, cell extracts or nuclear extracts were split equally into two tubes. The contents of one tube was reacted with 500 pmol HaloTag-PEG-biotin ligand (G859A, Promega), and that of the other was mixed with 500 pmol d-biotin (B-20656, Thermo Fisher Scientific) to examine nonspecific binding of proteins on the beads (negative control) overnight at 4°C on a rotator. MyONE C1 magnetic beads (DB65002, Thermo Fisher Scientific) were added into both tubes. After washing five times, the beads were boiled in Laemmli buffer to obtain the binding proteins. We subjected 5% of the input fraction (Input), pull-down fraction without ligand (negative control), and pull-down fraction immunoprecipitant (IP) to immunoblotting with antibodies against HaloTag-RPA194, RPA135, and β-actin.
EU labeling
EU incorporation was performed using Click-iT RNA Alexa Fluor 594 imaging kits (C10330, Thermo Fisher Scientific) according to the manufacturer’s instructions. Section images were recorded using a DeltaVision microscope, and nondeconvolved pictures were used for the quantitative analysis of EU incorporation.
Quantification of pre-rRNA using quantitative PCR
Total RNA was isolated with TRIzol reagent (10296-010, Thermo Fisher Scientific) and reverse transcribed using a SuperScript III First-Strand Synthesis system (18080-400, Thermo Fisher Scientific) with random primers. First-strand complementary DNAs (cDNAs) were analyzed with SYBR Premix Ex Taq II (RR820A, TaKaRa) using a Thermal Cycler Dice Real-Time System TP800 (TaKaRa) for quantitative PCR (qPCR). Standard curves for relative quantification with each primer set were obtained following fivefold dilution of the input sample. Pre-rRNA (47S pre-rRNA) and β-actin were detected using the following primer pairs: 5′-CCTTCCCCAGGCGTCCCTCG-3′ and 5′-GGCAGCGCTACCATAACGGA-3′; and 5′-TGCGTCTGGACCTGGCTGGC-3′ and 5′-GCCTCAGGGCAGCGGAACCG-3′, respectively. Pre-rRNA levels were normalized to β-actin mRNA levels.
RNA interference
Transfection of siRNA was performed using Lipofectamine RNAiMAX (13778-075, Thermo Fisher Scientific) according to the manufacturer’s instructions. The small interfering RNA (siRNA) oligos for fibrillarin (#M-011269-00-0005, GE Healthcare) and the negative control from the siGenome siRNA library (#D-001206-14-05, GE Healthcare) were used. Cells were transfected with 20 nM siRNA and incubated for 60 hours before imaging.
Chemical treatments and perturbations
To inhibit RNA Pol I transcription, cells were treated with 0.5 μM CX-5461(M66052-2 s; Xcess Biosciences Inc.) for 2 hours. To inhibit pre-rRNA processing, cells were treated with 75 μM roscovitine (R7772-1MG, Sigma-Aldrich) for 2 hours. To rapidly degrade mAID-mClover-RPA194, cells were incubated in medium supplemented with doxycycline (1 μg/ml) for 24 hours and then treated with 500 μM indole-3-acetic acid (19119-61, Nacalai), a natural auxin, in the presence of doxycycline for 3 hours. After treatment, the cells were imaged or chemically fixed.
ATP depletion and intracellular ATP measurement based on luciferase activity
Aliquots of 0.5 × 105 cells were seeded into a 12-well culture plate (IWAKI). For ATP reduction, the cells were incubated on coverslips with 10 mM sodium azide and 50 mM 2-deoxyglucose in HBSS (Gibco) for 10 min. To measure ATP, Cell ATP Assay Reagent (300-15363, Toyo B-Net Co. Ltd.) was used according to the manufacturer’s instructions. Bioluminescence was measured using a Lumat LB 9507 tube luminometer (EG&G Berthold). A standard plot of ATP concentration versus bioluminescence intensity verified that our measured ATP concentrations fell within a linear range. Both the reaction and measurement were performed at 23°C in the dark. The incubation time was 5 min from the addition of assay reagent to measurement.
Supplementary Material
Acknowledgments
We thank S. Tamura for the illustrations and contribution to the ATP experiment; S. Hirose, M. Sasai, S. Iida, Y. Hiromi, M. Kurusu, and H. Seino for the critical review of the manuscript; and Y. Murayama for contribution to the biochemical experiments. We also thank members of the Maeshima laboratory for the discussions and support. Funding: This work was supported by the Japan Society for the Promotion of Science KAKENHI grants (15K18580, 15H01361, and 18K06187 to S.I.; 16H04746 and 19H05273 to K.M.), a Japan Science and Technology Agency CREST grant (JPMJCR15G2 to K.M.), and the Takeda Science Foundation (to K.M.). Author contributions: S.I. and K.M. designed the project. S.I. and H.O. performed all of the experiments. S.I. and R.I. analyzed the single-molecule tracking data. S.I. and K.M. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/42/eabb5953/DC1
REFERENCES AND NOTES
- 1.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 2.McSwiggen D. T., Mir M., Darzacq X., Tjian R., Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 33, 1619–1634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pederson T., The nucleolus. Cold Spring Harb. Perspect. Biol. 3, a000638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brangwynne C. P., Mitchison T. J., Hyman A. A., Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., Brangwynne C. P., Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitrea D. M., Cika J. A., Stanley C. B., Nourse A., Onuchic P. L., Banerjee P. R., Phillips A. H., Park C.-G., Deniz A. A., Kriwacki R. W., Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitrea D. M., Cika J. A., Guy C. S., Ban D., Banerjee P. R., Stanley C. B., Nourse A., Deniz A. A., Kriwacki R. W., Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangan H., Gailín M. Ó., McStay B., Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 284, 3977–3985 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Falahati H., Pelham-Webb B., Blythe S., Wieschaus E., Nucleation by rRNA dictates the precision of nucleolus assembly. Curr. Biol. 26, 277–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falahati H., Wieschaus E., Independent active and thermodynamic processes govern the nucleolus assembly in vivo. Proc. Natl. Acad. Sci. U.S.A. 114, 1335–1340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds R. C., Montgomery P. O., Hughes B., Nucleolar “Caps” produced by actinomycin D. Cancer Res. 24, 1269–1277 (1964). [PubMed] [Google Scholar]
- 12.Jordan E. G., McGovern J. H., The quantitative relationship of the fibrillar centres and other nucleolar components to changes in growth conditions, serum deprivation and low doses of actinomycin D in cultured diploid human fibroblasts (strain MRC-5). J. Cell Sci. 52, 373–389 (1981). [DOI] [PubMed] [Google Scholar]
- 13.Tchelidze P., Benassarou A., Kaplan H., O’Donohue M.-F., Lucas L., Terryn C., Rusishvili L., Mosidze G., Lalun N., Ploton D., Nucleolar sub-compartments in motion during rRNA synthesis inhibition: Contraction of nucleolar condensed chromatin and gathering of fibrillar centers are concomitant. PLOS ONE 12, e0187977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell J., Zomerdijk J. C., RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30, 87–96 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen J. S., Lam Y. W., Leung A. K., Ong S.-E., Lyon C. E., Lamond A. I., Mann M., Nucleolar proteome dynamics. Nature 433, 77–83 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ide S., Dejardin J., End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nat. Commun. 6, 6674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grummt I., Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Moss T., At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 14, 210–217 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ide S., Miyazaki T., Maki H., Kobayashi T., Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327, 693–696 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi Y., Horiuchi T., Kobayashi T., Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17, 1497–1506 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D., Huang S., Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L. I., Phair R. D., Misteli T., A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Gorski S. A., Snyder S. K., John S., Grummt I., Misteli T., Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol. Cell 30, 486–497 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews J. O., Conway W., Cho W.-K., Narayanan A., Spille J.-H., Jayanth N., Inoue T., Mullen S., Thaler J., Cissé I. I., qSR: A quantitative super-resolution analysis tool reveals the cell-cycle dependent organization of RNA Polymerase I in live human cells. Sci. Rep. 8, 7424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cisse I. I., Izeddin I., Causse S. Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., Darzacq X., Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cissé I. I., Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozaki T., Imai R., Tanbo M., Nagashima R., Tamura S., Tani T., Joti Y., Tomita M., Hibino K., Kanemaki M. T., Wendt K. S., Okada Y., Nagai T., Maeshima K., Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67, 282–293.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Nagashima R., Hibino K., Ashwin S. S., Babokhov M., Fujishiro S., Imai R., Nozaki T., Tamura S., Tani T., Kimura H., Shribak M., Kanemaki M. T., Sasai M., Maeshima K., Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwin S. S., Nozaki T., Maeshima K., Sasai M., Organization of fast and slow chromatin revealed by single-nucleosome dynamics. Proc. Natl. Acad. Sci. U.S.A. 116, 19939–19944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver K. N., Watt K. E. N., Hufnagel R. B., Navajas Acedo J., Linscott L. L., Sund K. L., Bender P. L., König R., Lourenco C. M., Hehr U., Hopkin R. J., Lohmann D. R., Trainor P. A., Wieczorek D., Saal H. M., Acrofacial dysostosis, cincinnati type, a mandibulofacial dysostosis syndrome with limb anomalies, is caused by POLR1A dysfunction. Am. J. Hum. Genet. 96, 765–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seither P., Coy J. F., Pouska A., Grummt I., Molecular cloning and characterization of the cDNA encoding the largest subunit of mouse RNA polymerase I. Mol. Gen. Genet. 255, 180–186 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Hihara S., Pack C.-G., Kaizu K., Tani T., Hanafusa T., Nozaki T., Takemoto S., Yoshimi T., Yokota H., Imamoto N., Sako Y., Kinjo M., Takahashi K., Nagai T., Maeshima K., Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2, 1645–1656 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga M., Imamoto N., Sakata-Sogawa K., Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Dion V., Gasser S. M., Chromatin movement in the maintenance of genome stability. Cell 152, 1355–1364 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Hozák P., Cook P. R., Schöfer C., Mosgöller W., Wachtler F., Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 107, 639–648 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Cheutin T., O’Donohue M.-F., Beorchia A., Vandelaer M., Kaplan H., Deféver B., Ploton D., Thiry M., Three-dimensional organization of active rRNA genes within the nucleolus. J. Cell Sci. 115, 3297–3307 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C., Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72, 443–457 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Sirri V., Hernandez-Verdun D., Roussel P., Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 156, 969–981 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bazett-Jones D. P., Leblanc B., Herfort M., Moss T., Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264, 1134–1137 (1994). [DOI] [PubMed] [Google Scholar]
- 40.O’Sullivan A. C., Sullivan G. J., McStay B., UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22, 657–668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babokhov M., Hibino K., Itoh Y., Maeshima K., Local chromatin motion and transcription. J. Mol. Biol. 432, 694–700 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Cook P. R., The organization of replication and transcription. Science 284, 1790–1795 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Edelman L. B., Fraser P., Transcription factories: Genetic programming in three dimensions. Curr. Opin. Genet. Dev. 22, 110–114 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Drygin D., Lin A., Bliesath J., Ho C. B., O’Brien S. E., Proffitt C., Omori M., Haddach M., Schwaebe M. K., Siddiqui-Jain A., Streiner N., Quin J. E., Sanij E., Bywater M. J., Hannan R. D., Ryckman D., Anderes K., Rice W. G., Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71, 1418–1430 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Quin J., Chan K. T., Devlin J. R., Cameron D. P., Diesch J., Cullinane C., Ahern J., Khot A., Hein N., George A. J., Hannan K. M., Poortinga G., Sheppard K. E., Khanna K. K., Johnstone R. W., Drygin D., McArthur G. A., Pearson R. B., Sanij E., Hannan R. D., Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget 7, 49800–49818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hajjoul H., Mathon J., Ranchon H., Goiffon I., Mozziconacci J., Albert B., Carrivain P., Victor J.-M., Gadal O., Bystricky K., Bancaud A., High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome Res. 23, 1829–1838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subach F. V., Patterson G. H., Manley S., Gillette J. M., Lippincott-Schwartz J., Verkhusha V. V., Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Y. Sakamoto, M. Ishiguro, G. Kitagawa, Akaike Information Criterion Statistics (KTK Scientific Publishers; D. Reidel; Sold and distributed in the U.S.A. and Canada by Kluwer Academic Publishers, Tokyo Dordrecht; Boston Hingham, MA, 1986). [Google Scholar]
- 49.Jao C. Y., Salic A., Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 105, 15779–15784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trask D. K., Muller M. T., Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc. Natl. Acad. Sci. U.S.A. 85, 1417–1421 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N. J., Santos N. D., Silvester J., Wei V., Garcia J., Kabeer F., Lai D., Soriano P., Banáth J., Chiu D. S., Yap D., Le D. D., Ye F. B., Zhang A., Thu K., Soong J., Lin S.-C., Tsai A. H. C., Osako T., Algara T., Saunders D. N., Wong J., Xian J., Bally M. B., Brenton J. D., Brown G. W., Shah S. P., Cescon D., Mak T. W., Caldas C., Stirling P. C., Hieter P., Balasubramanian S., Aparicio S., CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 8, 14432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natsume T., Kiyomitsu T., Saga Y., Kanemaki M. T., Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Yao R.-W., Xu G., Wang Y., Shan L., Luan P.-F., Wang Y., Wu M., Yang L.-Z., Xing Y.-H., Yang L., Chen L.-L., Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell 76, 767–783.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Zhu L., Richardson T. M., Wacheul L., Wei M.-T., Feric M., Whitney G., Lafontaine D. L. J., Brangwynne C. P., Controlling the material properties and rRNA processing function of the nucleolus using light. Proc. Natl. Acad. Sci. U.S.A. 116, 17330–17335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frottin F., Schueder F., Tiwary S., Gupta R., Körner R., Schlichthaerle T., Cox J., Jungmann R., Hartl F. U., Hipp M. S., The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Taylor N. O., Wei M.-T., Stone H. A., Brangwynne C. P., Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J. 117, 1285–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruhlak M., Crouch E. E., Orlov M., Montaño C., Gorski S. A., Nussenzweig A., Misteli T., Phair R. D., Casellas R., The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734 (2007). [DOI] [PubMed] [Google Scholar]
- 58.van Sluis M., McStay B., A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 29, 1151–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho W. K., Jayanth N., English B. P., Inoue T., Andrews J. O., Conway W., Grimm J. B., Spille J. H., Lavis L. D., Lionnet T., Cissé I. I., RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife 5, e13617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A. S., Yu T., Marie-Nelly H., McSwiggen D. T., Kokic G., Dailey G. M., Cramer P., Darzacq X., Zweckstetter M., RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Boija A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R., Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabari B. R., Dall’Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross A. P., Zarbalis K. S., The emerging roles of ribosome biogenesis in craniofacial development. Front. Physiol. 5, 26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dauwerse J. G., Dixon J., Seland S., Ruivenkamp C. A., van Haeringen A., Hoefsloot L. H., Peters D. J., Boers A. C., Daumer-Haas C., Maiwald R., Zweier C., Kerr B., Cobo A. M., Toral J. F., Hoogeboom A. J., Lohmann D. R., Hehr U., Dixon M. J., Breuning M. H., Wieczorek D., Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 43, 20–22 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Maeshima K., Yahata K., Sasaki Y., Nakatomi R., Tachibana T., Hashikawa T., Imamoto F., Imamoto N., Cell-cycle-dependent dynamics of nuclear pores: Pore-free islands and lamins. J. Cell Sci. 119, 4442–4451 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mandegar M. A., Huebsch N., Frolov E. B., Shin E., Truong A., Olvera M. P., Chan A. H., Miyaoka Y., Holmes K., Spencer C. I., Judge L. M., Gordon D. E., Eskildsen T. V., Villalta J. E., Horlbeck M. A., Gilbert L. A., Krogan N. J., Sheikh S. P., Weissman J. S., Qi L. S., So P.-L., Conklin B. R., CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park A., Won S. T., Pentecost M., Bartkowski W., Lee B., CRISPR/Cas9 allows efficient and complete knock-in of a destabilization domain-tagged essential protein in a human cell line, allowing rapid knockdown of protein function. PLOS ONE 9, e95101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S. L., Danuser G., Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dosztányi Z., Csizmok V., Tompa P., Simon I., IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Yahata K., Maeshima K., Sone T., Ando T., Okabe M., Imamoto N., Imamoto F., cHS4 insulator-mediated alleviation of promoter interference during cell-based expression of tandemly associated transgenes. J. Mol. Biol. 374, 580–590 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/42/eabb5953/DC1


