Abstract
Hereditary hemochromatosis (HH), an iron-overload disease, is a prevalent genetic disorder. As excess iron causes a multitude of metabolic disturbances, we postulated that iron overload in HH disrupts colonic homeostasis and colon–microbiome interaction and exacerbates the development and progression of colonic inflammation and colon cancer. To test this hypothesis, we examined the progression and severity of colitis and colon cancer in a mouse model of HH (Hfe−/−), and evaluated the potential contributing factors. We found that experimentally induced colitis and colon cancer progressed more robustly in Hfe−/− mice than in wild-type mice. The underlying causes were multifactorial. Hfe−/− colons were leakier with lower proliferation capacity of crypt cells, which impaired wound healing and amplified inflammation-driven tissue injury. The host/microflora axis was also disrupted. Sequencing of fecal 16S RNA revealed profound changes in the colonic microbiome in Hfe−/− mice in favor of the pathogenic bacteria belonging to phyla Proteobacteria and TM7. There was an increased number of bacteria adhered onto the mucosal surface of the colonic epithelium in Hfe−/− mice than in wild-type mice. Furthermore, the expression of innate antimicrobial peptides, the first-line of defense against bacteria, was lower in Hfe−/− mouse colon than in wild-type mouse colon; the release of pro-inflammatory cytokines upon inflammatory stimuli was also greater in Hfe−/− mouse colon than in wild-type mouse colon. These data provide evidence that excess iron accumulation in colonic tissue as happens in HH promotes colitis and colon cancer, accompanied with bacterial dysbiosis and loss of function of the intestinal/colonic barrier.
Keywords: colitis, colon cancer, dysbiosis, hemochromatosis, HFE-null mouse, iron/heme overload
Introduction
Colorectal cancer (CRC) is one of the most frequent cancers worldwide [1]. Free iron as well as heme have been suspected to play a role in the development of colitis-associated CRC because it facilitates Fenton reaction to generate hydroxyl radical (Fe2+ + H2O2 → Fe3+ + OH− + OH•), which causes oxidative damage [2]. However, details of molecular/biochemical mechanisms by which excessive iron drives CRC remain poorly understood. This is of great concern given the fact that 1 in 250 people of Caucasian and Hispanic origin are at risk for iron overload due to hereditary hemochromatosis (HH) [3], a disease often underdiagnosed/misdiagnosed [4]. Furthermore, as HH symptoms appear only at 50–60 years of age despite being a genetic disease, most patients are treated for symptoms without being diagnosed for HH [5]. As free iron is an oxidant with the potential to cause damage to colon, do individuals with HH have an increased risk for colitis and CRC? Even though directly relevant to millions of people with this disorder, there have been no systematic investigations to address this question.
HH is characterized by increased iron absorption in the proximal intestine due to two recessive mutations (C282Y mostly in Caucasians; H63D mostly in Hispanics) in the major histocompatibility class-I-like gene HFE [6]. HFE is a component of the iron-sensing complex that, when systemic iron is high, stimulates the production of hepcidin [7], a hepatic hormone that inhibits iron entry into the blood from the intestine and also from macrophages that recirculate iron from hemoglobin degradation [8]. Mutations in HFE reduce hepcidin and cause systemic and organ iron overload [5]. The primary site of iron deposition in HH is liver, and unequivocal evidence links hepatocellular carcinoma to excessive tissue iron in HH [9]. Iron overload occurs in other tissues as well in HH, though at a lower magnitude than in liver, but the potential connection of this iron accumulation to cancers in other tissues has not been investigated. HH patients do exhibit oxidative damage and histological abnormalities in the colon indicative of inflammation [10], a finding reproducible in Hfe−/− mice [11], an animal model of HH [12]. There is also an increased incidence of HFE mutations in patients with CRC [13–17]. Another factor that potentially links HH to cancer is HIF-2α, an important regulator of cellular response to hypoxia [18]. There is a direct biological connection between iron and oxygen availability to tissues because of the obligatory role of iron in hemoglobin, the oxygen transport protein. In fact, activation of HIF-2α potentiates CRC progression by affecting iron homeostasis via the up-regulation of the divalent metal iron transporter, which mediates the first step in the intestinal absorption of dietary iron [19]. Based on these data, we hypothesized that iron overload as occurs in HH disrupts colonic homeostasis and exacerbates the progression of colonic inflammation and CRC. To test this hypothesis, we used Hfe−/− mouse, which phenocopies the disease in humans [12], and investigated the impact of the disease on the development and severity of experimentally induced colitis and CRC. As colonic bacteria play an obligatory role in colitis and CRC, we also evaluated the impact of HH on the host-microbiome ecosystem.
Materials and methods
Animals
Hfe−/− mice and ApcMin/+ mice, both on C57BL/6 background, were purchased from Jackson Laboratory and were maintained at our animal facility. ApcMin/+ mice on Hfe−/− background were generated in our laboratory. Animals were maintained on the laboratory rodent diet. There is no information available on the heme content of this diet, but it is unlikely to contain heme at any significant level because the diet is devoid of animal products. The iron content in the diet is 0.024% (w/w). Only male mice of ages more than 4 months were used in this study. All experimental procedures were approved by the TTUHSC Institutional Animal Care and Use Committee (IACUC — protocol number 18005) and the Institutional Review Board (IRB). For tissue collection, mice were killed by cervical dislocation under CO2 anesthesia in accordance with the guidelines from the American Veterinary Medical Association.
Measurement of the free iron and heme
The content of free iron (Fe2+/Fe3+) in the colon was measured by a colorimetry-based Iron Assay Kit (Abcam). Tissue sections of the colon were also subjected to Perls Prussian stain protocol to detect iron (Fe3+) by immunohistochemistry. Heme content in the colon was measured by a colorimetry-based Hemin Assay Kit (Sigma).
Experimental colitis
Mice were administered 2.5% dextran sulfate sodium (DSS), molecular mass 36–50 kDa, (MP Biochemicals, Santa Ana, CA, U.S.A.) in drinking water for 7 days followed by 7 days of drinking water only (Supplementary Figure S1A). Diarrhea was scored as 0 for normal stool, 1 for soft but formed pellet, 2 for very soft pellet, 3 for diarrhea (no pellet) or 4 for dysenteric diarrhea. Rectal bleeding was recorded as 0 for no bleeding, 2 for the presence of occult blood in stool or 4 for gross macroscopic bleeding.
Measurement of gut epithelial barrier permeability
Gut epithelial barrier function was evaluated using fluorescent permeability-tracer FITC-dextran, molecular mass 4 kD (Sigma, St. Louis, MO, U.S.A.). Mice received an oral gavage of FITC-dextran at a dose of 50 mg per 100 g of body weight. The serum was collected 6 h after administration. The presence of FITC-dextran in mouse serum was measured by fluorescence spectrometry, and its concentration was determined from standard curve generated by serial dilution of known concentrations of FITC-dextran in control serum that was run in parallel.
Experimental colitis-associated CRC
Mice received a single intraperitoneal injection of azoxymethane (AOM) (Sigma) in phosphate-buffered saline (PBS) at a dose of 10 mg/kg body weight. Five days later, mice received 2.5% DSS in drinking water for 5 days followed by 5 days of drinking water only. The cycle was repeated two more times. The daily body index was recorded as described above. Mice were killed on day 70, counted from the day on which the first exposure to DSS was initiated (Supplementary Figure S1B).
Protein isolation and Western blot
Tissues and cells were lysed in Pierce™ RIPA buffer (Thermo Fisher Scientific, Waltham, MA, U.S.A.) supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, U.S.A.). Homogenates were centrifuged, and supernatants were used for protein measurement via Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A.). Western blot samples were prepared in Laemmli Sample Buffer (Bio-Rad Laboratories, Hercules, CA, U.S.A.). They were loaded onto a 10% SDS–PAGE gel and transferred onto a PVDF membrane (Bio-Rad Laboratories, Hercules, CA, U.S.A.). The membrane was blocked, and antibodies diluted in 5% nonfat dry milk (Bio-Rad Laboratories, Hercules, CA, U.S.A.) or in 5% bovine serum albumin (Irvine Scientific, Santa Ana, CA, U.S.A.) were used. Protein bands were visualized using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and developed on the autoradiography film (Santa Cruz, Dallas, TX, U.S.A.). Anti-PCNA (D3H8P XP® #13110) was purchased from Cell Signaling Technology (Danvers, MA, U.S.A.). Horseradish peroxidase-conjugated goat anti-rabbit (#1706515) was purchased from Bio-Rad Laboratories (Hercules, CA, U.S.A.). For quantification of protein levels by the densitometric analysis, the experiment was carried out in triplicate and the data were collected from the resultant three Western blots.
Quantification of colon bacterial load
DNA from colon tissue (∼100 mg) was isolated as described previously for the fecal DNA isolation [20,21]. Quantitative polymerase chain reaction using group-specific PCR primers were used to quantify the relative abundance of different groups in the total gut microbiota [20,21]. The sequences of PCR primers are provided in Supplementary Table S1.
Colony formation unit (CFU) for adherent bacteria
Colons excised from wild-type mice and Hfe−/− mice were thoroughly washed in PBS to remove feces. Tissues were homogenized in PBS using 0.1 mm diameter zirconium beads (Sigma) in Beadbeater (BioSpec, Bartlesville, OK, U.S.A.) and plated on agar plates.
Library preparation and 16S rRNA gene sequencing
Sequencing library preparation and sequencing were performed at the Center for Biotechnology and Genomics, Texas Tech University, Lubbock, TX, using the Illumina 16S-metagenomics library prep protocol. Paired-end sequencing was performed on a MiSeq using a 600 cycle reagent cartridge. The preparation of the samples for high throughput sequencing involved using a two-step PCR approach according to Illumina protocol. The amplification of the variable region V3 and V4 of bacterial rRNA gene was done using universal bacterial primer set 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) containing Illumina adaptors as described by Klindworth et al. [22]. The concentration of the DNA was determined for each sample based on the average of triplicates of concentration (ng/µl) and average library size of 615 bp. Sequencing was done using MiSeq Reagent Kit v3 (600 cycle kit) (Illumina) and the cartridge and reagents were handled according to manufacturer's instructions.
Processing of sequencing data
Sequencing reads in fastq format were obtained by unzipping the fastq.gz files by gunzip command on Linux server. Processing of the demultiplexed samples was done using quantitative insights into microbial ecology (QIIME, version 1.8.0) pipeline. Assembling of the paired end reads of each sample was done using PEAR software. This was followed by quality trimming the assembled sequences to remove poor quality reads using split_libraries_fastq.py script with default parameters [23]. Sequences that were not at least 75% of the expected amplicon length together with ambiguous base calls were removed from the data pool. The 16S rRNA gene sequence were clustered based on 97% similarity of the reads and OTUs (operational taxonomic units) were recognized against the subset Greengenes database (http://greengenes.lbl.gov) using UCLUST algorithm. We aligned the sequences on PyNast aligner [24] and allocated representative sequences from each OUT to taxonomy based on Greengenes 16S rRNA reference sequences.
Isolation of colonic mucosa
The colon was cut open, washed with the ice-cold PBS and was placed on an ice-cold glass surface, with the lumen side facing up. The mucosa was mechanically detached and collected by scraping with a glass slide. Samples were centrifuged and pellet was used for further analysis.
Real-time quantitative PCR
Total RNA was isolated from tissues using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, U.S.A.). Two micrograms of RNA was used for cDNA synthesis via SuperScript III First-Strand Synthesis kit (Thermo Fisher). Relative mRNA levels were detected by a SYBR Green (Bio-rad) detection system using StepOne Plus Real-Time PCR system (Applied Biosystems). Gene expression was normalized to internal control (Hypoxanthine-guanine phosphoribosyl transferase, HPRT or glyceraldehyde-3-phophate dehydrogenase, GAPDH) and was quantified using 2ΔΔCt method. Sequences of PCR primers are given in Supplementary Table S1.
Organ culture and ELISA
Colons from control and DSS-treated wild-type mice and Hfe−/− mice were cut into 1–2 cm pieces and cultured in medium containing penicillin/streptomycin for 24 h. Supernatants were collected, and cytokines measured by ELISA using respective antibodies (Biolegend, San Diego, CA, U.S.A.).
Patient-derived xenografts (PDX)
Colon adenocarcinoma PDX samples were obtained from the TTUHSC School of Medicine Cancer Center (Lubbock, TX, U.S.A.). The protocol for the collection and use of these patient-derived materials was in accordance with the World Medical Association Declaration of Helsinki. All participating subjects, from whom the tissues were collected, provided written informed consent. RNA was isolated from the tissues and used for cDNA synthesis. Commercially available RNA from the normal human colon with mucosal lining pooled from five males (TaKaRa Bio, Kusatsu, Shiga, Japan) served as the control.
Data mining
The data for the expression of HFE and SLC40A1 mRNA in CRC tissues were obtained from the cancer genome atlas (TCGA) and gene expression omnibus (GEO) databases. For TCGA analysis, we queried TCGA for colon cancer patients. ‘Level 3 of Exp-Gene’ files from COAD Data Matrix Datasets were downloaded and used to extract data for the expression levels of HFE mRNA and SLC40A1 mRNA. For GEO data analysis, we downloaded and analyzed data that is publicly available from the GEO: 82 samples from GSE9348, 585 samples from GSE39582, 96 samples from GSE33113 and 240 samples from GSE41258.
RNA sequencing
RNA was extracted from colonic epithelial cells. Sequencing library preparation and sequencing were performed using Illumina library prep protocol. All the sequences for data analysis were uploaded into the NCBI-Bio Project data archive. Differentially expressed genes were determined using log2(fold change).
Immunohistochemistry and histopathology
Histology and immunohistochemical staining were performed on 5-µm thick formalin-fixed, paraffin-embedded colon sections. Colon sections stained with hematoxylin and eosin (H&E), Periodic acid–Schiff (PAS) stain, and Gram stain. Briefly, sections were deparaffinized, followed by antigen retrieval using the antigen retrieval solution (Agilent Technologies, Santa Clara, CA, U.S.A.). Endogenous peroxidase activity was quenched with 3% H2O2 in PBS. Sections were stained using specific primary antibody (Ki-67, Cell Signaling, Danvers, MA, U.S.A.) and VECTASTAIN ABC Kit and diaminobenzidine (Vector Laboratories, Burlingame, CA, U.S.A.). Counterstaining was performed with hematoxylin and stained sections were visualized by Leica DM550B microscope. Ki-67 immunostaining was quantified by counting Ki-67-positive cells in at least 20 crypts per mouse.
Nuclei characterization using Image J (FIJI)
For each mouse, a 63X objective focus as seen through Leica DM 2000 LED microscope was captured as 2592 × 1944 px TIFF images by a LEICA MC 170 HD camera. FIJI accessioned these TIFFs; from each image were excised by free hand the 50 most aberrant epithelial nuclei to a new image, yielding 100 HFE and 100 WT carcinoma nuclei. From each image-bearing 50 nuclei, nuclear areas in square pixels and circularity, a measure of irregularity of outline, were calculated. The HSB color field has, as one of its three parameters, brightness; on brightness images; for each nucleus, the standard deviation was divided by the mean brightness to obtain a brightness c.v. as a measure of chromatin variability.
Statistical analysis
The data shown are means ± SEM. Statistical significance was calculated using Student's t-test with two-tailed analysis. A P-value <0.05 was used to detect statistically significant differences.
Results
Hfe−/− mouse colons accumulate iron and heme
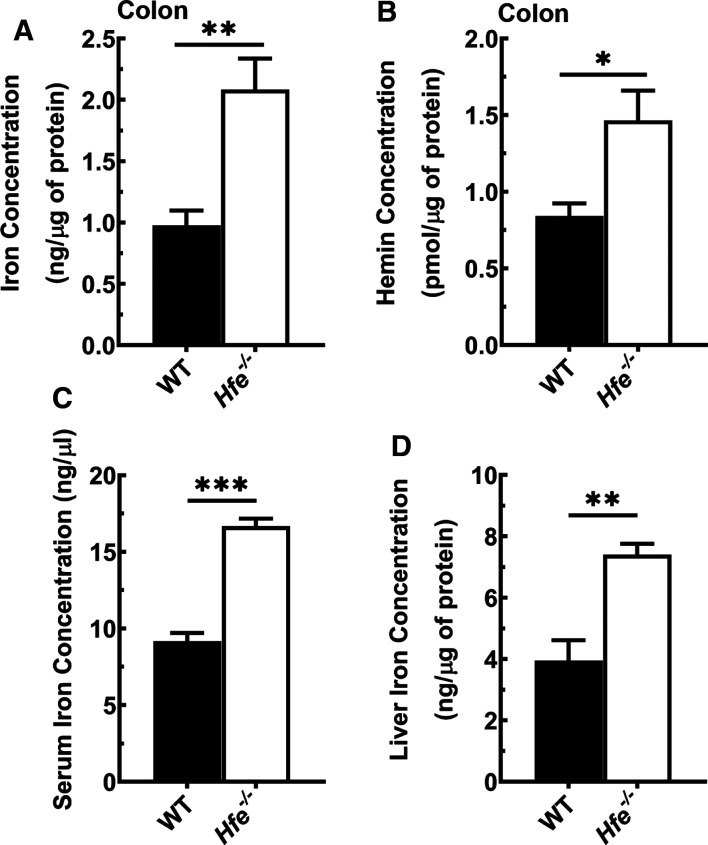
There are no published reports on iron status in the colon in HH. Therefore, we first evaluated the iron status in the colon in wild-type mice and Hfe−/− mice. We found a 2-fold increase in free iron and heme in Hfe−/− mouse colon compared with wild-type mouse colon (Figure 1A,B). This was in parallel with systemic and hepatic iron accumulation in Hfe−/− mice (Figure 1C,D). We also performed Perls staining in tissue sections of the colon to detect iron, but the technique yielded very weak staining, most likely due to the lower sensitivity of the method compared with the colorimetry-based assay, thus precluding any meaningful comparison between wild-type mouse colon and Hfe−/− mouse colon for iron content by immunohistochemistry. As wild-type mice and Hfe−/− mice were fed the same diet, the increase in free iron and heme in Hfe−/− mouse colon is most likely due to the systemic iron overload that occurs in this mouse line. All mammalian cells are capable of heme synthesis, and excess iron in Hfe−/− mouse colon likely feeds into heme synthesis to some extent, thus increasing the cellular heme content.
Figure 1. Iron overload in Hfe−/− mouse colon, serum, and liver.
(A) Total iron (Fe+2 and Fe+3) concentration in wild-type mouse colon and Hfe−/− mouse colon. (B) Heme concentration in wild-type mouse colon and Hfe−/− mouse colon. (C) Total iron (Fe+2 and Fe+3) concentration in serum of wild-type mice and Hfe−/− mice. (D) Total iron (Fe+2 and Fe+3) concentration in liver of wild-type mice and Hfe−/− mice. Data show mean values (±SEM) for three mice per group. *P < 0.05; **P < 0.01; ***P < 0.001.
Hfe−/− mouse colon is more permeable and is more susceptible to the experimental colitis
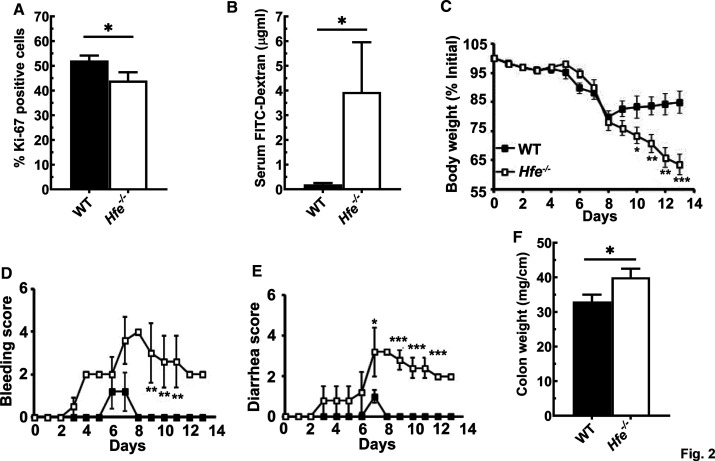
We first assessed the proliferation capacity of the Hfe−/− mouse crypts, by identifying the fraction of the Ki-67-positive cells. The colonic monolayer in Hfe−/− mice consisted of ∼40% of cells positive for Ki-67, which was ∼10% less than the fraction found in wild-type mouse colon (Figure 2A and Supplementary Figure S2A). The significantly reduced proliferative index of the Hfe−/− mouse crypts manifested with the increase in the intestinal permeability. FITC-dextran is a fluorescent compound that can enter the circulation solely through intestinal gaps; when FITC-dextran was administered orally, fluorescence was ∼4-fold higher in Hfe−/− mouse serum than in wild-type mouse serum (Figure 2B). Furthermore, the histological examination showed substantial damage in the Hfe−/− mouse colonic mucosa under basal conditions (Supplementary Figure S2B); there was a noticeable decrease in the villus height and an increased disruption of villus integrity in Hfe−/− mouse colon compared with wild-type mouse colon. Therefore, the Hfe−/− mice manifested with a leaky, injured gut and an impairment in the colonocyte turnover. To examine if this renders them more prone to the development of colonic inflammation, we compared the development and progression of DSS-induced colitis between wild-type mice and Hfe−/− mice. In this model of acute colitis, the severity of inflammation was markedly greater in H./e−/− mice than in wild-type mice. This difference was seen in all indices examined (weight loss, rectal bleeding, diarrhea, and colon weight) (Figure 2C–F). At the end of the DSS treatment, wild-type mice successfully improved their body weight, recovered stool consistency, and stopped rectal bleeding, which indicated that their colons healed from the inflammation. On the contrary, Hfe−/− mice continued to lose body weight and have severe rectal bleeding and diarrhea, indicating that they suffered from the impaired wound healing capacity. Furthermore, the histological analyses confirmed that during DSS administration, the Hfe−/− mice had higher colonic mucosal injury with frequent ulcerations and a markedly increased loss of villus integrity compared with wild-type mice (Supplementary Figure S2B). Overall, the magnitude of colonic inflammation in Hfe−/− mice was greater than in wild-type mice.
Figure 2. Hfe−/− mice manifest with the leaky gut and are more susceptible to the development of experimental colitis.
(A) Quantification of the Ki-67 positive cells present in wild type and Hfe−/−mouse crypts. Data show mean values (±SEM) for three mice per group. (B) The permeability of the wild type and Hfe−/− mouse intestinal tracts was monitored by the accumulation of the FITC-Dextran in the serum upon its administration by oral gavage. Data are represented as mean values (±SEM) for five mice per group. (C–F) The degree of inflammation of the experimental colitis in Hfe−/− mice (□) and wild type (▪) mice was assessed by measuring daily body index that consisted of observing body weight change, rectal bleeding, diarrhea and upon the sacrifice, colon weight. Data show mean values (±SEM) for five mice per group. *P < 0.05; **P < 0.01; ***P < 0.001.
Hfe−/− mice are more susceptible to colorectal cancer
We then compared the development of colitis-associated CRC between wild-type mice and Hfe−/− mice. This experimental model involved the exposure to the colon-specific carcinogen AOM in the presence of chronic colitis. We found a significant increase in number and size of adenomas in distal colon in Hfe−/− mice compared with wild-type mice (Figure 3A). To examine if HH accelerates genetically driven colon cancer, we used ApcMin/+ mice, which develop adenomatous polyps spontaneously in small intestine and colon. We compared the number of polyps in this mouse when raised on Hfe+/+ and Hfe−/− backgrounds. There was a significant increase in the number of polyps on Hfe−/− background, both in small intestine and colon. The difference was detectable with macroscopic examination of the mucosal surface of the colons for counting the visible polyps (Figure 3B), and substantiated further with histologic analysis (Supplementary Figure S3A). Histopathologic examination of the polyps identified significant changes in the morphology of nuclei in tumor cells (Supplementary Figure S3B,C). On Hfe−/− background, the nuclei were smaller and less bright, and the nucleoli were smaller. Furthermore, we examined the difference in the proliferation capacity of the adjacent normal and colonic polyps in ApcMin/+/Hfe+/+ mice and ApcMin/+/Hfe−/− mice by observing the protein levels of proliferation marker PCNA. We found that, deletion of Hfe increased the proliferation capacity, even in the normal adjacent colon tissue due to the mutant ApcMin (Figure 3C). This stimulatory effect of Hfe deletion on the proliferation marker PCNA was not readily apparent in polyps because of the robust basal proliferation already induced by the mutant Apc under these conditions.
Figure 3. Hfe−/− mice are more susceptible to the development of CRC driven by inflammation and ApcMin mutation.
(A) Number and size of AOM/DSS polyps in wild-type mice and Hfe−/− mice. Data show mean values (±SEM) for eight mice per group. (B) Number of polyps in colon and small intestine in ApcMin/+ mice on Hfe+/+ and Hfe−/− backgrounds. Data show mean values (±SEM) for five mice on Hfe+/+ background and seven mice on Hfe−/−background. (C) Western blot analysis of PCNA protein levels in colon polyps and normal, healthy adjacent tissue of ApcMin/+ on Hfe+/+ and Hfe−/− backgrounds. Western blot band intensities were estimated using ImageJ software and the band intensities that correspond to the PCNA protein were normalized to the respective β-actin levels. Data are means ± SEM. *P < 0.05; ***P < 0.001.
Expression of ferroptosis markers and p53 in Hfe−/− mouse colon
Ferroptosis is a non-apoptotic form of cell death that is dependent on iron (Fe2+) and p53 [25]. Fe2+ participates in Fenton reaction to generate hydroxyl radicals from H2O2, and the hydroxyl radicals promote oxidation of polyunsaturated fatty acids in biological membranes (lipid peroxidation). This process can be prevented by glutathione, which detoxifies H2O2 by converting it to H2O. The levels of glutathione in cells are, however, dependent on the activity of the cystine/glutamate exchanger (SLC7A11) [26]. The expression of this transporter is repressed by the tumor suppressor p53 [25]. Therefore, we examined the expression of ferroptosis markers and p53 in normal colon and in polyps in both models of CRC. We found increased expression of p53 and Slc7a11 in polyps compared with normal colon; this was evident in ApcMin/+ mice on both Hfe+/+ and Hfe−/− backgrounds (Figure 4A). There was, however, no detectable change in the expression of glutathione peroxidase-4 (Gpx4), another enzyme involved in the glutathione-mediated protection of the cells from ferroptosis. The increase in the expression of p53 in polyps corroborated with the corresponding increase in the expression of two of the three targets of p53 examined in the study, namely cyclin D1 and Bax (Figure 4B). These data suggest that chronic exposure to excess iron as occurs in HH may not lead to ferroptosis in contrast with what happens in acute exposure to iron. We repeated the same experiments with wild-type polyps and Hfe−/− polyps in the AOM/DSS model of CRC; we found no changes in any of these parameters (data not shown).
Figure 4. Changes in p53 and ferroptosis markers in normal colon and in polyps in ApcMin/+ mice on Hfe+/+ and Hfe−/− backgrounds.
(A) qPCR analysis of mRNA levels for the tumor suppressor p53, cystine/glutamate exchanger Slc7a11, and glutathione peroxidase 4 (Gpx4) with GAPDH mRNA as an internal control. (B) qPCR analysis of mRNA levels for p53 targets. ns, not significant; **P < 0.01; ***P < 0.001.
HIF-2α and DMT1 in wild-type mouse colon and Hfe−/− mouse colon
As there is evidence from published reports that HIF-2α is an important determinant of iron homeostasis with a tumor-promoting role [19], we examined the levels of mRNA and protein for this protein in wild-type mouse colon and in Hfe−/− mouse colon. We found no difference in either HIF-2α mRNA or protein between wild type and Hfe−/− colon (Figure 5A). With regard to DMT1 expression, we found no impact of Hfe deletion on colonic expression even though we detected a decrease in expression the in duodenum (Figure 5B).
Figure 5. Expression of HIF-2α and DMT1 in wild type and Hfe−/− mouse colon.
(A) HIF-2α expression was monitored in colon at the protein level by Western blot with β-actin as an internal control. (B) DMT1 (divalent metal ion transporter 1) expression was monitored in colon as well as in the duodenum at the mRNA level by qPCR with GAPDH mRNA as an internal control. ns, not significant; *P < 0.05.
Adherence of bacteria onto luminal surface of Hfe−/− mouse colon
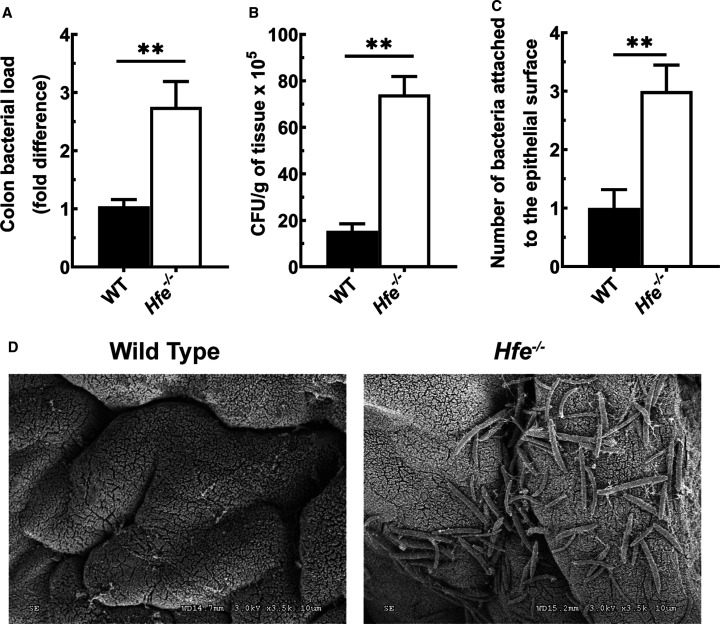
Next, we investigated if the bacteria adheres to the mucosal surface of the colonic epithelium and if there is any difference between wild-type mice and Hfe−/− mice in this phenomenon. Using the bacterial group-specific primers and quantitative PCR, we found an increase in the total colon bacterial load in Hfe−/− mice compared with wild-type mice (Figure 6A). Furthermore, we performed the CFU assay using homogenized colon tissues from wild-type mice and Hfe−/− mice. We observed a significant increase in the total CFU counts in the Hfe−/− mice, as well as the enhanced abundance in the Gram-positive and Gram-negative bacteria (Figure 6B and Supplementary Figure S4A). The Gram-negative group included different coliforms; the Gram-positive group included Staphylococci, Lactobacilli, Enterococci, and Streptococci. Furthermore, the staining for Gram-negative bacteria confirmed the increase in the number of bacteria attached to the epithelial surface of the Hfe−/− mouse colon; this number was minimal in the wild-type mouse colon (Figure 6C and Supplementary Figure S4B). The penetrance of the bacteria was not influenced, however; the PAS staining did not show any difference between wild-type mice and Hfe−/− mice (Supplementary Figure S4C). Finally, the bacterial adherence in Hfe−/− mouse colon was confirmed by transmission electron microscopy (Figure 6D).
Figure 6. Increased adherence of bacteria to the mucosal surface of Hfe−/− mouse colon.
(A) qPCR analysis of colon bacterial load in wild-type mice and Hfe−/− mice. Data show mean values (±SEM) for six mice per group. (B) The colony formation units (CFU) counts of total bacteria recovered from wild-type mouse colon and Hfe−/− mouse colon. Bacterial load is expressed as the number of CFU per gram of colon tissue. Data show mean values (±SEM) for three mice per group. (C) Number of bacteria attached to the colonic epithelial cells surface, as assessed by staining for Gram-negative bacteria. Data show mean values (±SEM) for five mice per group. (D) Visualization of bacterial adherence onto Hfe−/− mouse colon lumen by utilizing scanning electron microscopy. **P < 0.01.
Gut microflora dysbiosis in Hfe−/− mouse colon
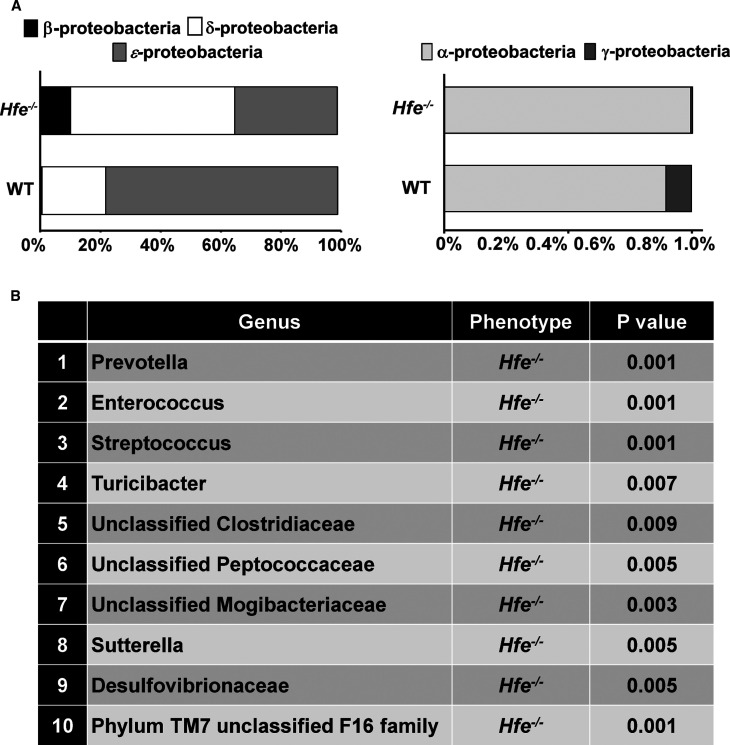
As deletion of Hfe had significant impact on colitis and colon cancer, along with abnormal bacterial attachment onto the colonic mucosal surface, we asked if Hfe−/− mice had altered colonic microbiota. To address this question, we performed 16S metagenomic sequencing using fecal DNA from wild-type mice and Hfe−/− mice. The relative taxonomic distribution and prevalence of bacteria, determined at the phylum level, in the two groups of mice are presented in Supplementary Figure S5. We observed a significant reduction in Firmicutes and expansion in Bacteroidetes in Hfe−/− mice compared with wild-type mice. Furthermore, the Proteobacteria and TM7 phyla, which are associated with pathologic conditions in colon [27,28], were more prevalent in Hfe−/− mice than in wild-type mice (Supplementary Figure S5). The diversity of the Proteobacteria phylum was also altered in Hfe−/− mice; α-proteobacteria, β-proteobacteria and δ-proteobacteria were more abundant in Hfe−/− mice than in wild-type mice (Figure 7A). Species enrichment analysis revealed that, among the enriched genera, the pro-inflammatory bacteria Desulfovibrionaceae, Turicibacter, Prevotella, and Sutterella were expanded in Hfe−/− mice (Figure 7B).
Figure 7. Colonic bacterial dysbiosis in Hfe−/− mice.
Fecal DNA was extracted from wild-type mice and Hfe−/− mice and 16S metagenomic sequencing was performed to determine the bacterial composition in two animal groups. (A) The distribution of five Proteobacteria classes in the wild type and Hfe−/− mouse fecal material. (B) Species enrichment analysis in the wild type and Hfe−/− mouse fecal material. Data show mean values for six mice per group of the gut microflora components whose changes were statistically significant.
Impaired innate immune response in Hfe−/− mouse colon
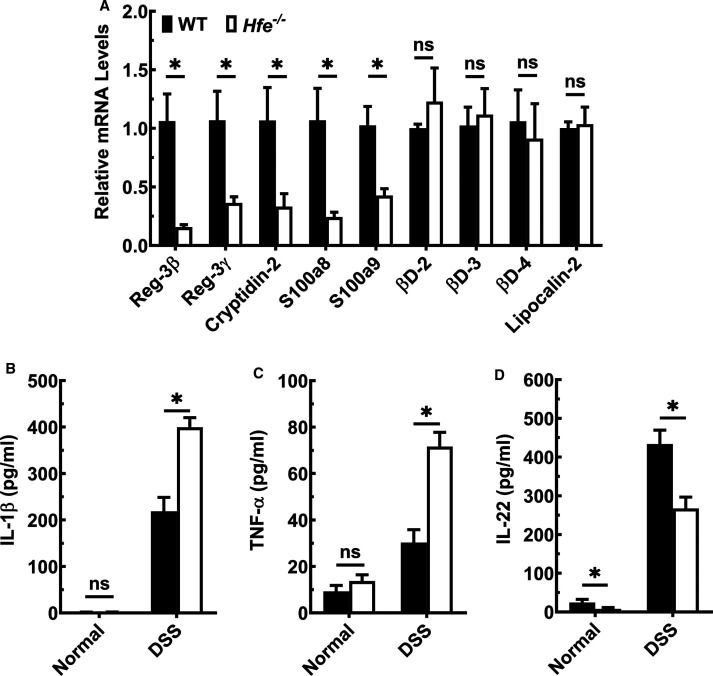
We then analyzed the expression of genes that determine innate mucosal antibacterial response. As shown in Figure 8A, qPCR revealed that Hfe−/− mouse colonic epithelium had lower expression of C-type lectins RegIIIβ and RegIIIγ than wild-type mice. The decrease was also evident in mRNA levels of Paneth cell-derived defensin, cryptidin-2, and calgranulin A (S100A8) and calgranulin B (S100A9), whose activity is directed towards Gram-positive and Gram-negative bacteria. The expression of β-defensins, however, was comparable between the two groups. To address the colonic immune response, we cultured control and DSS-exposed colons from Hfe−/− mice and wild-type mice ex vivo and quantified selected cytokines in the culture media (Figure 8B–D). There was no difference in the secretion of IL-β and TNF-α between the two genotypes under basal conditions. However, upon DSS exposure, the secretion of these pro-inflammatory cytokines was greater in control mice, but the increase was comparatively higher in Hfe−/− mice. In contrast, the secretion of IL-22, a cytokine that responds to inflammation by promoting cell proliferation and tissue repair, was decreased in Hfe−/− mice under basal conditions as well as upon DSS exposure. Furthermore, we evaluated the transcriptomic profiles of colonic epithelium from wild type and Hfe−/− mice by RNAseq. We found that there were 45 up-regulated and 23 down-regulated genes (Supplementary Figure S6A,B). Using Ingenuity Pathway Analysis, we observed significant differences in the actin cytoskeleton, integrin-linked kinase and integrin signaling between wild-type mice and Hfe−/− mice (Supplementary Figures S6C and S7–S9).
Figure 8. Down-regulation of the antimicrobial response and up-regulation of pro-inflammatory immune system in Hfe−/− mouse colon.
(A) qPCR analysis of the antimicrobial peptides (AMPs) mRNA in colonic epithelial cells of wild-type mice and Hfe−/− mice. Data show mean values (±SEM) for three mice per group. (B–D) The amount of secreted IL-1β, TNF-α, and IL-22 on basal levels and upon DSS challenge quantified by ELISA, in wild type (closed bars) and Hfe−/− (open bars) mice. Data show mean values (±SEM) for three mice per group. *P < 0.05.
Expression of Hfe and Slc40a1 (ferroportin) in colon cancer
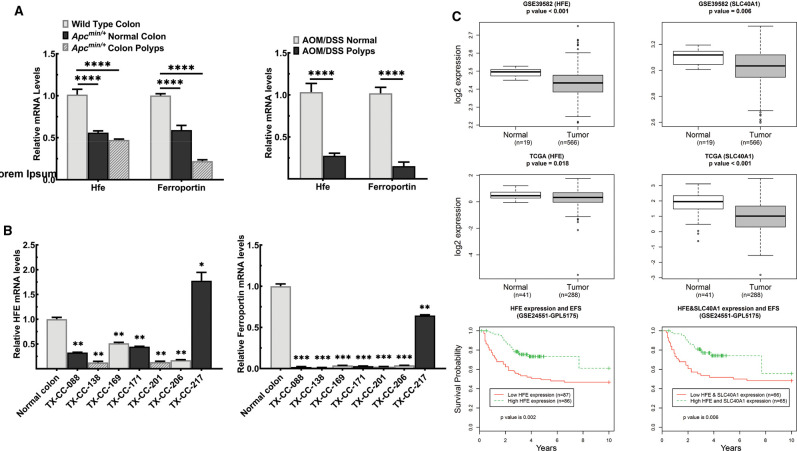
Because there is an increasing evidence that excess iron/heme drives carcinogenesis in multiple tissues [29–31], we examined if the expression levels of HFE and ferroportin, a transporter that exports iron from the cell, in colon cancer. Decreased expression of HFE disrupts iron homeostasis, leading to excessive iron accumulation in cells; the same is true with ferroportin as decreased expression of this transporter suppresses iron export from cells leading to increased iron accumulation. We first examined their expression levels in the polyps and healthy adjacent tissues isolated from the spontaneous mouse model of colon cancer (ApcMin/+mouse). When compared with the healthy colon tissue in wild-type mice, we observed that the ApcMin mutation resulted in a significant down-regulation of Hfe and ferroportin (Figure 9A). The down-regulation in the colonic polyps was also seen in the experimental model of colitis-driven colon cancer (AOM/DSS model) (Figure 9A). Furthermore, when compared with healthy, unmatched colon samples, HFE and Ferroportin mRNA levels were markedly decreased in seven independent colon cancer PDX (Figure 9B). In addition, we analyzed the publicly available databases for HFE expression in colon cancer (Figure 9C). The expression levels were significantly lower in tumors than in normal tissues. When the survival probability of the patients, stratified as low HFE expressors and high HFE expressors, was analyzed, low HFE expression was associated with reduced survival. The same was true for ferroportin. The expression was markedly lower in colon cancer than in normal colon. As was the case with HFE, colon cancer patients with lower expression of ferroportin had decreased survival probability than those with higher expression.
Figure 9. HFE and SLC40A1 expression is down-regulated in the colon cancer and low expression is associated with poor patient survival.
(A) qPCR analysis of Hfe and Slc40a1 mRNA levels in polyps and healthy, adjacent tissue obtained from two mouse models of colon cancer: the genetically driven model of colon cancer elicited by the Min mutation in the Apc gene and the experimental model of colitis-associated colon cancer elicited by a combination of azoxymethane and dextran sodium sulfate (AOM/DSS). Data show mean values (±SEM) for four mice per group for ApcMin/+ cohort and five mice per group for AOM/DSS cohort. (B) qPCR analysis of HFE and SLC40A1 mRNA in the normal, healthy colon and in the colon cancer PDX samples. Data show mean values (±SEM) for three measurements. (C) HFE and SLC40A1 mRNA levels were assessed with publicly available information from the gene expression omnibus databases (GSE39582), and the cancer genome atlas (TCGA). Colon cancer patient survival with regard to low and high HFE and SLC40A1 expression. Bounds of the boxes denote interquartile range, solid lines denote median, whickers denote 95% and 5% confidence limits, respectively. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
Here, we tested the hypothesis that chronic exposure to excess iron/heme as occurs in HH promotes colonic inflammation and colon cancer. This study was prompted by intriguing observations in published reports which showed that patients with HH exhibit histologic abnormalities in colon indicative of inflammation [10] and that Hfe−/− mice show evidence of oxidative damage in colon [11]. In addition to colitis, patients with HH also have increased infiltration of lymphocytes and neutrophils in the intestinal tract [10]. There is also an increased incidence of mutations in the hemochromatosis gene HFE in colon cancer [13–17]. Furthermore, iron-rich diets exacerbate colitis [32,33] and there is a strong connection between excessive dietary heme intake and colon cancer [34–38]. Even though these epidemiological observations suggest a potential link between excessive iron and inflammation/carcinogenesis in colon, there is no direct experimental evidence for such a link.
We undertook the present study to examine systematically the impact of chronic exposure to excess iron/heme on colonic inflammation and colon cancer. We have now established, using the Hfe−/− mouse as a model of hemochromatosis, that this iron-overload disease markedly enhances the development and progression of colitis and colon cancer in experimental model systems. This study reveals that the HH represents a substantial risk for higher incidence and more aggressive forms of colitis and CRC. This study also shows that HH causes dysbiosis and disrupts the host-bacteria cross-talk. The expression of antimicrobial peptides is decreased in Hfe−/− mouse colon compared with normal mouse colon, probably contributing to dysbiosis. When challenged with DSS, the expression of pro-inflammatory cytokines IL-1β and TNF-α is elevated, whereas the expression of IL-22, a beneficial cytokine under conditions of inflammation, is decreased in Hfe−/− mouse colons compared with normal mouse colons. This cytokine profile supports the observations that experimentally induced colonic inflammation is exacerbated in Hfe−/− mice. Our findings expanded the work of Buhnik-Rosenblau et al. [39], which also noted dysbiosis in Hfe−/− mice. We found a marked increase in adherent bacteria on the mucosal surface of colon in Hfe−/− mice compared with WT mice. Kortman et al. [40] found that iron overload selects for pathogenic bacteria at the expense of commensals. Ijssennagger et al. [41,42] found that heme iron increases Gram-negative bacteria and increases the sulfate-reducing bacteria Desulfovibrionaceae. In our study, we found that Hfe−/− mice manifested a leaky gut and impairment in crypt cell proliferation even under basal conditions; the bacterial dysbiosis, clearly evident in Hfe−/− mouse colon, is very likely to be involved in the pathogenesis of this decrease in intestinal/colonic barrier function and consequently in the increased risk for colonic inflammation observed in response to iron/heme overload.
Multiple mechanisms contribute to the increased severity in experimentally induced colonic inflammation and colon cancer seen in Hfe−/− mice. The most obvious factor is the iron/heme-induced oxidative damage to the tissue. Free iron leads to the generation of hydroxyl radical via Fenton reaction using endogenously produced H2O2. Oxidation of membrane lipids by iron-induced reactive free radicals is detrimental to cells. This phenomenon, known as lipid peroxidation, is detrimental to the survival of cells. Under normal physiologic conditions, the intestinal and colonic mucosal cells are highly proliferative, a feature essential for the maintenance of the barrier function and for the elimination of unhealthy cells damaged by exogenous chemicals and materials introduced into the intestinal tract via diet. This process also offers protection against carcinogenesis because altered cells that are prone to cancer are routinely eliminated by the rapid cellular renewal process. Of note is the fact that the entire epithelial cellular layer in the intestinal tract is replaced with new cells every 4–5 days. Accumulation of excessive iron and heme as occurs in hemochromatosis disrupts this important function, thus producing a pro-inflammatory environment that is conducive for colitis and colon cancer.
Suppression of the signaling pathways associated with the tumor suppressor p53 is another mechanism likely to be associated with the increased risk of colitis and colon cancer observed in Hfe−/− mice. Recent studies have uncovered that p53 is a heme-binding protein and that the p53-heme complex is a substrate for proteasomal degradation [25]. Chronic exposure to excess iron leads to decreased p53 protein levels in cells. We have reported recently that p53 protein levels are significantly decreased in Hfe−/− mouse colon and that the phenomenon is associated with decreased transcriptional activity of this protein [43]. As p53 is either mutated or decreased in most cancers, including colon cancer, the decrease in p53 protein levels in colon in response to excessive iron/heme certainly contributes to the observed potentiation of colitis and colon cancer in Hfe−/− mice. This phenomenon is also related to cancer initiation via epithelial-to-mesenchymal transition where p53 plays an integral role. Recently, we provided experimental evidence for the promotion of epithelial-to-mesenchymal transition in pancreatic ductal cells upon chronic exposure to excess iron [44]. The decrease in the p53 protein levels in Hfe−/− colon is a post-translational process caused by enhanced protein degradation. There is no change in p53 mRNA levels due to iron overload. Interestingly, the levels of p53 mRNA are increased in ApcMin/+ polyps, and this change is independent of the Hfe status (Hfe+/+ or Hfe−/−). However, as we did not see any alteration in p53 mRNA in polyps from the DSS/AOM colon cancer model, the observed phenomenon appears to be specific to ApcMin/+ mutation.
We also have to consider the iron-induced cell death process, known as ferroptosis [45,46], in the context of the findings reported in the present study. As iron initiates this cell death process, one might think that excess iron is detrimental to cancer cells. Surprisingly, this is not true; excess iron is actually a tumor promoter. Cancer cells accumulate iron by reprogramming iron-regulatory pathways to promote their growth [30,31]. The data presented in this paper as well as those in publicly available databases show that the expression of HFE is markedly down-regulated in colon cancer. Suppression of HFE function as an iron-sensing mechanism would lead to the accumulation of iron in cells. The same is true with the iron exporter ferroportin, which is also down-regulated in colon cancer. Decreased expression of this transporter in cancer cells ensures retention of iron inside the cells. How are the cancer cells able to accumulate iron and use it to promote their growth without risking ferroptosis? The iron/heme-induced promotion of proteasomal degradation of p53 provides an answer to this puzzle. This tumor suppressor protein is a negative regulator of the cystine transporter SLC7A11 [47], which promotes the synthesis of glutathione inside the cells by providing cysteine via the influx of extracellular cystine [19,48]. Glutathione detoxifies H2O2, thus suppressing Fenton reaction and hence decreasing the generation of hydroxyl radical and consequent ferroptosis. This could be the mechanism by which chronic exposure to excess iron as occurs in hemochromatosis promotes cancer without risking the cancer cells to ferroptosis. In fact, one of our earlier studies showed that primary retinal pigment epithelial cells isolated from Hfe−/− mice expressed higher levels of SLC7A11 demonstrable at the mRNA and functional level [49]. This phenomenon was also associated with decreased senescence and other cell biological features that are known to be the hallmarks of cancer cells. Up-regulation of SLC7A11 seems to be a common phenomenon in multiple cancers as this transporter protects the cancer cells from oxidative stress and lipid peroxidation. In the present study, the mRNA levels for Slc7a11 are increased markedly in polyps associated with ApcMin/+ mutation irrespective of whether the polyps were from Hfe+/+ mice or Hfe−/− mice. The protection caused by enhanced expression of SLC7A11 is related to increased synthesis of glutathione. As there was no change in the expression of glutathione peroxidase (Gpx4) in both models of colon cancer used in the present study, increased availability of glutathione for the enzyme Gpx4 seems to be the primary driver of protection against oxidative stress in cancer cells. The changes observed in the mRNA levels for the apoptosis-related protein Bax or for the cell cycle-associated proteins cyclin D1 and p21 in the polyps seem to depend on the etiology of carcinogenesis as we noticed the changes only in ApcMin/+ polyps but not in polyps from the DSS/AOM cancer model.
Bacterial dysbiosis observed in Hfe−/− mice would also be a likely contributing factor to the increased risk of colitis and colon cancer in response to excess iron. Even though exact molecular mechanisms linking bacterial dysbiosis to colonic diseases are lacking for the most part, the connection has been well established. We also found marked differences in the transcriptomic profile in Hfe−/− mouse colonic epithelium compared with wild-type mouse colonic epithelium. The biochemical pathways related to cytoskeletal network and integrin signaling are obviously altered in the response of Hfe deletion, but it is difficult to predict whether these alterations represent primary changes or secondary to oxidative stress and changes in p53 signaling. It is interesting to note that a recent study has documented an active role for gut microbiota in iron homeostasis in the host, implicating specific bacterial metabolites in the regulatory process [50]. Additional studies are needed to tease out the functional significance of the transcriptomic changes with regard to increased propensity of Hfe−/− colon to inflammation and carcinogenesis.
Even though we placed emphasis in the present study on chronic exposure to excess iron/heme as the pathologic factor in the initiation and progression of colitis and colon cancer in Hfe−/− mice, other iron/heme-independent pathways in this process cannot be ignored. HFE strongly interacts with β2-microglobulin, which functions as a chaperone to promote the trafficking of HFE to the plasma membrane, and the deletion of Hfe in mice leads to increased degradation of this chaperone. β2-microglobulin has a broad spectrum of biological functions, particularly in the immune system [51]. Therefore, the decreased levels of this regulatory protein in Hfe−/− mice might influence the pathways involved in inflammation and carcinogenesis. Another possibility is the signaling via the iron-storage protein ferritin. The cellular levels of this protein are elevated in Hfe−/− mice because of the iron overload. In addition to serving as the iron-binding protein, ferritin also has a signaling function [52]. We do not know if these changes in ferritin levels due to the deletion of Hfe contribute in any way to the observations made in the present study.
Our findings have important implications for patients with HH. Chronic exposure to excessive iron/heme as occurs in HH increases the risk for colitis and colon cancer. The enhanced risk of cancer could also be true for other organs as well. This study suggests that strategies to reduce iron accumulation in HH such as regular phlebotomy and/or iron chelation therapy, coupled with avoidance of iron/heme-rich food but increased consumption of appropriate probiotics and prebiotics, could protect colon from inflammation and carcinogenesis. As iron accumulation to toxic levels takes several decades in HH, it might be desirable to take such preventive measures as early as possible. This requires information as to whether or not a given individual carries HH mutations, thus making a case for recommending genetic screening in susceptible populations.
Abbreviations
- AOM
azoxymethane
- APC
adenomatous polyposis coli
- CFU
colony-forming unit
- CRC
colorectal cancer
- DSS
dextran sulfate sodium
- FITC
fluorescein isothiocyanate
- HFE
high Fe or histocompatibility class I-like gene involved in Fe regulation
- HH
hemochromatosis
- PDX
patient-derived xenograft
- SLC40A1
solute carrier gene family 40, subfamily A, member 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Welch Endowed Chair in Biochemistry, Grant no. BI-0028, at Texas Tech University Health Sciences Center.
Open Access
Open access of this article was enabled by the participation of Texas Tech University Health Sciences Center in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society.
Author Contribution
S.S. and B.R. performed all the experiments related to colitis and colon cancer in mice and in human samples, analyzed the publicly available expression data for colon cancer, and contributed to the preparation of the manuscript; N.M., A.N.H., and J.C.-H. performed the experiments related to the characterization of fecal and adherent bacteria; M.S.W. and A.G.N. did the morphological analysis of the colonic polyps; K.R.K. did the 16S rRNA gene sequencing; V.G. designed the experiments and wrote the manuscript.
Supplementary Material
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chua A.C., Klopcic B., Lawrance I.C., Olynyk J.K. and Trinder D. (2010) Iron: an emerging factor in colorectal carcinogenesis. World J. Gastroenterol. 16, 663–672 10.3748/wjg.v16.i6.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merryweather-Clarke A.T., Pointon J.J., Shearman J.D. and Robson K.J.H. (1997) Global prevalence of putative haemochromatosis mutations. J. Med. Genet. 34, 275–278 10.1136/jmg.34.4.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherfane C.E., Hollenbeck R.D., Go J. and Brown K.E. (2013) Hereditary hemochromatosis: missed diagnosis or misdiagnosis? Am. J. Med. 126, 1010–1015 10.1016/j.amjmed.2013.07.013 [DOI] [PubMed] [Google Scholar]
- 5.Pietrangelo A. (2010) Hereditary hemochromatosis: pathogenesis, diagnosis, and treatment. Gastroenterology 139, 393–408 10.1053/j.gastro.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 6.Feder J.N., Gnirke A., Thomas W., Tsuchihashi Z., Ruddy D.A., Basava A. et al. (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat. Genet. 13, 399–408 10.1038/ng0896-399 [DOI] [PubMed] [Google Scholar]
- 7.Wu X., Wang Y., Wu Q., Cheng W., Liu W., Zhao Y. et al. (2014) HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood 124, 1355–1343 10.1182/blood.V124.21.1355.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeth E., Tuttle M.S., Powelson J., Vaughn M.B., Donovan A., McVey Ward D. et al. (2004) Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093 10.1126/science.1104742 [DOI] [PubMed] [Google Scholar]
- 9.Kowdley K.V. (2004) Iron, hemochromatosis, and hepatocellular carcinoma. Gastroenterology 127, S79–S86 10.1016/j.gastro.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 10.Zhou W.X., Wu X.R., Bennett A.E. and Shen B. (2014) Endoscopic and histologic abnormalities of gastrointestinal tract in patients with hereditary hemochromatosis. J. Clin. Gastroenterol. 48, 336–342 10.1097/MCG.0b013e3182a9be10 [DOI] [PubMed] [Google Scholar]
- 11.Stevens R.G., Morris J.E., Cordis G.A., Anderson L.E., Rosenberg D.W. and Sasser L.B. (2003) Oxidative damage in colon and mammary tissue of the HFE-knockout mouse. Free Radic. Biol. Med. 34, 1212–1216 10.1016/S0891-5849(03)00072-8 [DOI] [PubMed] [Google Scholar]
- 12.Zhou X.Y., Tomatsu S., Fleming R.E., Parkkila S., Waheed A., Jiang J. et al. (1998) HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc. Natl. Acad. Sci. U.S.A. 95, 2492–2497 10.1073/pnas.95.5.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne N.J., Gurrin L.C., Allen K.J., Constantine C.C., Delatycki M.B., McLaren C.E. et al. (2010) HFE c282y homozygotes are at increased risk of breast and colorectal cancer. Hepatology 51, 1311–1318 10.1002/hep.23448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaheen N.J., Silverman L.M., Keku T., Lawrence L.B., Rohlfs E.M., Martin C.F. et al. (2003) Association between hemochromatosis (HFE) gene mutation carrier status and the risk of colon cancer. J. Natl. Cancer Inst. 95, 154–159 10.1093/jnci/95.2.154 [DOI] [PubMed] [Google Scholar]
- 15.Chen A.T., Ma J., Tranah G.J., Giovannucci E.L., Rifai N., Hunter D.J. et al. (2005) Hemochromatosis gene mutations, body iron stores, dietary iron, and risk of colorectal adenoma in women. J. Natl. Cancer Inst. 97, 917–926 10.1093/jnci/dji165 [DOI] [PubMed] [Google Scholar]
- 16.Chen W., Zhao H., Li T. and Yao H. (2013) HFE gene C282Y variant is associated with colorectal cancer in Caucasians: a meta-analysis. Tumor Biol. 34, 2255–2259 10.1007/s13277-013-0766-3 [DOI] [PubMed] [Google Scholar]
- 17.Asberg A., Thorstensen K., Irgens W.O., Romundstad P.R. and Hveem K. (2013) Cancer risk in HFE C282Y homozygotes: results from the HUNT2 study. Scand. J. Gastroenterol. 48, 189–195 10.3109/00365521.2012.752028 [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishnan S.K. and Shah Y.M. (2016) Role of intestinal HIF-2α in health and disease. Annu. Rev. Physiol. 78, 301–325 10.1146/annurev-physiol-021115-105202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue X., Taylor M., Anderson E., Hao C., Qu A., Greenson J.K. et al. (2012) Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 72, 2285–2293 10.1158/0008-5472.CAN-11-3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh N., Gurav A., Sivaprakasam S., Brady E., Padia R., Shi H. et al. (2014) Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivaprakasam S., Ganapathy P.K., Sikder M.O.F., Elmassry M., Ramachandran S., Kottapalli K.R. et al. (2019) Deficiency of dietary fiber in Slc5a8-null mice promotes bacterial dysbiosis and alters colonic epithelial transcriptome towards pro-inflammatory milieu. Can. J. Gastroenterol. Hepatol. 2019, 2543082 10.1155/2019/2543082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M. et al. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R. et al. (2013) Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 10, 57–59 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S.J., Ou Y., Jiang L. and Gu W. (2015) Ferroptosis: a missing puzzle piece in the p53 blueprint? Mol. Cell. Oncol. 3, e1046581 10.1080/23723556.2015.1046581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewerenz J., Hewett S.J., Huang Y., Lambros M., Gout P.W., Kalivas P.W. et al. (2013) The cystine/glutamate antiporter system xc- in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox. Signal. 18, 522–555 10.1089/ars.2011.4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuehbacher T., Rehman A., Lepage P., Hellmig S., Folsch U.R., Schreiber S. et al. (2008) Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57, 1569–1576 10.1099/jmm.0.47719-0 [DOI] [PubMed] [Google Scholar]
- 28.Jakobsson H.E., Rodriguez-Pineiro A.M., Schutte A., Ermund A., Boysen P., Bemark M. et al. (2015) The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao L. and Feng Y. (2013) Intakes of heme iron and zinc and colorectal cancer incidence: a meta-analysis of prospective studies. Cancer Causes Control 24, 1175–1183 10.1007/s10552-013-0197-x [DOI] [PubMed] [Google Scholar]
- 30.Torti S.V., Manz D.H., Paul B.T., Blanchette-Farra N. and Torti F.M. (2018) Iron and cancer. Annu. Rev. Nutr. 38, 97–125 10.1146/annurev-nutr-082117-051732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown R.A.M., Richardson K.L., Kabir T.D., Trinder D., Ganss R. and Leedman P.J. (2020) Altered iron metabolism and impact in cancer biology, metastasis, and immunology. Front. Oncol. 10, 476 10.3389/fonc.2020.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uritski R., Barshack I., Bilkis I., Ghebremeskel K. and Reifen R. (2004) Dietary iron affects inflammatory status in a rat model of colitis. J. Nutr. 134, 2251–2255 10.1093/jn/134.9.2251 [DOI] [PubMed] [Google Scholar]
- 33.Seril D.N., Liao J., West A.B. and Yang G.Y. (2006) High-iron diet: foe or feat in ulcerative colitis and unlcerative colitis-associated carcinogenesis. J. Clin. Gastroenterol. 40, 391–397 10.1097/00004836-200605000-00006 [DOI] [PubMed] [Google Scholar]
- 34.Larsson S.C. and Wolk A. (2006) Meat consumption and risk of colorectal cancer: a meta-analysis of prospective studies. Int. J. Cancer 119, 2657–2664 10.1002/ijc.22170 [DOI] [PubMed] [Google Scholar]
- 35.Cross A.J., Leitzmann M.F., Gail M.H., Hollenbeck A.R., Schatzkin A. and Sinha R. (2007) A prospective study of red and processed meat intake in relation to cancer risk. PLoS Med. 4, e325 10.1371/journal.pmed.0040325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bastide N.M., Pierre F.H. and Corpet D.E. (2011) Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved. Cancer Prev. Res. 4, 177–184 10.1158/1940-6207.CAPR-10-0113 [DOI] [PubMed] [Google Scholar]
- 37.Chan D.S., Lau R., Aune D., Viera R., Greenwood D.C., Kampman E. et al. (2011) Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS One 6, e20456 10.1371/journal.pone.0020456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raza M., Chakraborty S., Choudhury M., Ghosh P.C. and Nag A. (2014) Cellular iron homeostasis and therapeutic implications of iron chelators in cancer. Curr. Pharm. Biotechnol. 15, 1125–1140 10.2174/138920101512141202111915 [DOI] [PubMed] [Google Scholar]
- 39.Buhnik-Rosenblau K., Moshe-Belizowski S., Danin-Poleg Y. and Meyron-Holtz E.G. (2012) Genetic modification of iron metabolism in mice affects the gut microbiota. Biometals 25, 883–892 10.1007/s10534-012-9555-5 [DOI] [PubMed] [Google Scholar]
- 40.Kortman G.A.M., Boleij A., Swinkels D.W. and Tjalsma H. (2012) Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 7, e29968 10.1371/journal.pone.0029968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.IJssennagger N., Derrien M., van Doorn G.M., Rijnierse A., van den Bogert B., Müller M. et al. (2012) Dietary heme alters microbiota andmucosa of mouse colon without functional changes in host-microbe cross-talk. PLoS One 7, e49868 10.1371/journal.pone.0049868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ijssennagger N., Belzer C., Hooiveld G.J., Dekker J., van Mil S.W., Müller M. et al. (2015) Gut microbiota facilitates dietary heme-inducedepithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. U.S.A. 112, 10038–10043 10.1073/pnas.1507645112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ristic B., Sivaprakasam S., Narayanan M. and Ganapathy V. (2020) Hereditary hemochromatosis disrupts uric acid homeostasis and causes hyperuricemia via altered expression/activity of xanthine oxidase and ABCG2. Biochem. J. 477, 1499–1513 10.1042/BCJ20190873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhutia Y.D., Ogura J., Grippo P.J., Torres C., Sato T., Wachtel M. et al. (2020) Chronic exposure to excess iron promotes EMT and cancer via p53 loss in pancreatic cancer. Asian J. Pharm. Sci. 15, 237–251 10.1016/j.ajps.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. et al. (2017) Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayir H., Anthonymuthu T.S., Tyurina Y.Y., Patel S.J., Amoscato A.A., Lamade A.M. et al. (2020) Achieving life through death: redox biology of lipid peroxidation in ferroptosis. Cell. Chem. Biol. 27, 387–408 10.1016/j.chembiol.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H. et al. (2015) Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhutia Y.D., Babu E., Ramachandran S. and Ganapathy V. (2015) Amino acid transporters in cancer and their relevance to ‘glutamine addiction’: novel targets for the design of a new class of anticancer drugs. Cancer Res. 75, 1782–1788 10.1158/0008-5472.CAN-14-3745 [DOI] [PubMed] [Google Scholar]
- 49.Gnanaprakasam J.P., Veeranan-Karmegam R., Coothankandaswamy V., Reddy S.K., Martin P.M., Thangaraju M. et al. (2013) Loss of Hfe leads to progression of tumor phenotype in primary retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 54, 63–71 10.1167/iovs.12-10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das N.K., Schwartz A.J., Barthel G., Inohara N., Liu Q., Sankar A. et al. (2020) Microbial metabolite signaling is required for systemic iron homeostasis. Cell Metab. 31, 115–130.e6 10.1016/j.cmet.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Argyropoulos C.P., Chen S.S., Ng Y.H., Roumelioti M.E., Shaffi K., Singh P.P. et al. (2017) Rediscovering beta-2 microglobulin as a biomarker across the spectrum of kidney diseases. Front. Med. 4, 73 10.3389/fmed.2017.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Recalcati S., Invernizzi P., Arosio P. and Cairo G. (2008) New functions for an iron storage protein: The role of ferritin in immunity and autoimmunity. J. Autoimmun. 30, 84–89 10.1016/j.jaut.2007.11.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.