Abstract
Extracellular vesicles (EVs) are produced by all domains of life. In fungi, these structures were first described in Cryptococcus neoformans and, since then, they were characterized in several pathogenic and non-pathogenic fungal species. Cryptococcal EVs participate in the export of virulence factors that directly impact the Cryptococcus–host interaction. Our knowledge of the biogenesis and pathogenic roles of Cryptococcus EVs is still limited, but recent methodological and scientific advances have improved our understanding of how cryptococcal EVs participate in both physiological and pathogenic events. In this review, we will discuss the importance of cryptococcal EVs, including early historical studies suggesting their existence in Cryptococcus, their putative mechanisms of biogenesis, methods of isolation, and possible roles in the interaction with host cells.
Keywords: extracellular vesicles, biogenesis, Cryptococcus spp., extracellular vesicle analysis
1. Vesicular Export: A General System of Extracellular Delivery of Biological Structures
Extracellular vesicles (EVs) are vehicles exporting molecules from cells to the extracellular milieu and this kind of transport has been observed in organisms from all domains of life [1]. EVs are round-shaped, bilayered lipid membranes loaded with a diverse nature of molecular classes, including proteins [2], lipids [3,4], glycans [4,5], nucleic acids [6], and pigments [7,8].
In eukaryotes, EVs can be classified according to their mechanism of biogenesis. Apoptotic bodies are extracellular vesicles larger than 1 𝜇m that are released when the producing cells undergo apoptosis [9]. Microvesicles or ectosomes, which range from 100 nm to 1 𝜇m, are originated by shedding at the plasma membrane level [10]. Exosomes range from 30 to 200 nm and they result from the fusion of multivesicular bodies (MVBs) with the plasma membrane [11].
EV-like particles have been observed in fungi since the advent of high-resolution techniques of electron microscopy. In Cryptococcus neoformans, EV-like structures were first observed in 1973 using freeze-etching electron microscopy [12]. Two types of periplasmic, round-shaped structures ranging from 100 to 150 nm and 200 to 600 nm in diameter were at that time called paramural bodies [12,13]. These particles had properties that were all compatible with the presently known EVs, including dimensions, morphology, and bilayered membranes. In 2000, vesicle-like compartments containing glucosylceramide were detected at the cell wall of C. neoformans [14]. In 2007, EVs were isolated from C. neoformans culture supernatants [4]. Compositional and morphological studies showed that cryptococcal EVs corresponded to structures composed of several molecules participating in both fungal metabolism and pathogenicity [4,7]. After the seminal studies with C. neoformans, EVs were described in several fungal genera, including Candida [3,15], Histoplasma [3], Saccharomyces [3], Sporothrix [3,16], Paracoccidioides [17], Malassezia [18], Alternaria [19], Rhizopus [20], Trichophytum [21], Pichia [22], Aspergillus [23,24], Fusarium [25], Trichoderma [26], and Exophiala [27].
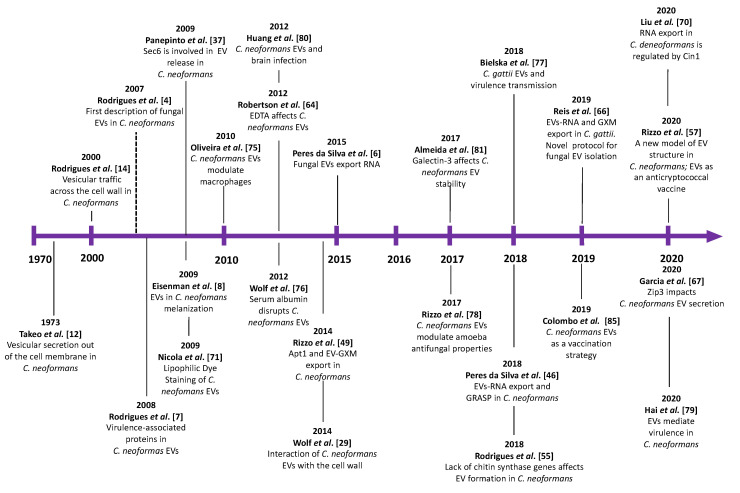
The knowledge of EVs in fungi is in frank expansion [28]. This review will focus on cryptococcal EVs, and we will discuss the main findings in this field, from the early description of EV-like particles in the 1970s to current times (Figure 1).
Figure 1.
Main discoveries related to cryptococcal extracellular vesicles (EVs). The timeline herein illustrated for Cryptococcus findings was based on that described for all fungal EVs [28].
2. EV Biogenesis and Secretory Pathways in Cryptococcus
Several studies indicated that fungal EVs are produced and released under the coordination of multiple mechanisms. EVs may be formed at different cellular sites [29], with the possible participation of both post-Golgi conventional secretion and unconventional secretory pathways [30]. Several comprehensive reviews on the regulation of secretory pathways in eukaryotes are available in the literature [31,32,33], and the details of these processes are not in the scope of this manuscript.
The conventional secretory pathway in eukaryotes results in the fusion of post-Golgi vesicles with the plasma membrane, and subsequent release of luminal molecules to the extracellular milieu [34,35]. This general process requires to a large extent the participation of members of the SEC gene family, which regulates the traffic from the endoplasmic reticulum (ER) to the Golgi, and then to the cell surface [34,36]. The role of the SEC6 gene in EV formation was evaluated in Cryptococcus [37]. The disruption of SEC6 resulted in the negative detection of EVs in C. neoformans. However, the functional connection between SEC genes and EV formation has not been established in Cryptococcus [37].
Exosome formation requires the maturation of endosomes into MVBs [11]. The later compartments can be targeted to the cell surface, allowing fusion with the plasma membrane and consequent release of luminal MVB vesicles in the outer space [30,38]. MVB formation requires the functionality of the endosomal sorting complex required for transport (ESCRT). The ESCRT pathway is very complex, and its functionality demands a series of finely regulated events [39,40]. Briefly, it initiates with the phosphorylation of phosphatidylinositol 3-phosphate at the endosomal membrane by the phosphatidylinositol 3-kinase Vps34, resulting in the formation of the ESCRT-0 subcomplex. ESCRT-0 formation will consequently regulate the formation of the ESCRT-I, -II, and -III subcomplexes that will finally result in MVB formation and EV release [39,40].
In fungi, the importance of the ESCRT complex for EV formation was suggested in strains where distinct genes regulating ESCRT functions were disrupted. In Candida albicans, the deletion of several genes related to the ESCRT complex resulted in a significant decrease in EV production [41]. In C. neoformans, a mutant strain lacking expression of Vps27, a component of the ESCRT-0 subcomplex, manifested abnormal vesicle traffic and release, resulting in an accumulation of MVBs in the cytosol [42]. The deletion of other genes belonging to the ESCRT pathway led to significant defects in the delivery of virulence factors associated with EVs [42,43,44,45]. The cryptococcal mutant strains vps27Δ (ESCRT-0), vps23Δ (ESCRT-I), and snf7Δ (ESCRT-III) had attenuated virulence in a mice model of cryptococcosis [42,43,44]. Although the impact of gene deletion on EV production was studied only in the vps27Δ strain [42], the attenuation of virulence in ESCRT mutants suggests important connections between the unconventional secretory pathway and the pathogenesis of Cryptococcus.
Other regulators of unconventional secretion were linked to EV formation in C. neoformans. The Golgi reassembly and stacking protein (GRASP), for instance, regulates EV cargo and dimensions in Cryptococcus [46]. In C. neoformans, a graspΔ mutant strain produced EVs with dimensions that significantly differed from those produced by wild-type cells [46]. This strain also manifested attenuated virulence [47] and a different RNA composition [46]. Autophagy regulators, which also participate in the formation of EVs in other eukaryotes, also participate in the formation of cryptococcal EVs. An atg7Δ strain manifested hypovirulence [48] and EVs produced by this strain had a slightly different RNA composition, in comparison with wild-type cells [46]. Similarly, the flippase Apt1, which plays an essential role in membrane architecture and, consequently, in secretory mechanisms [49,50], was required for correct EV formation and virulence in C. neoformans [49]. Together, these results strongly suggest that EV formation, virulence, and unconventional secretion are connected in C. neoformans.
3. Cell Wall Passage
In fungi, exported particles and molecules have to overcome the cell wall to reach the outer environment [4,29,51]. EVs supposedly use three putative mechanisms to cross the cell wall. First, EV accumulation in the periplasmic space would create a turgor pressure shoving the vesicles to pass across the naturally existing pores of the cell wall. Second, EVs could catalyze their passage across the cell wall using glycan hydrolases, including β-glucosidases and endochitinases. Third, EVs could use pore channels to reach the outer environment, by getting deformed to adapt to the pore morphology, and moving out through cytoskeleton-dependent mechanisms [3,13,51,52].
In C. neoformans, EVs were found to be released collectively or individually [29], but the exact mechanism explaining how they cross the cell wall has not been characterized. Microscopic analyses demonstrated vesicles near to damaged areas in the cell wall, but a clear association between cell wall breakage and vesicle passage has not been established [29]. Indeed, intra-wall vesicles in apparently intact regions were found in C. neoformans [4]. Microscopic observations also revealed that melanization in C. neoformans is associated with the accumulation of vesicle-like structures in the periplasmic space [29,53]. During this process, a significant reduction in the porosity of the cell wall was observed, and vesicles were observed crossing the cell wall directly [29,53]. In summary, there has been no evidence so far that cryptococcal vesicles use pore channels to reach the extracellular space, reinforcing the hypotheses of pressure-induced release and/or vesicle-mediated cell wall hydrolysis. The latter hypothesis has been recently validated in bacteria. In Bacillus subtilis, EV formation was demonstrated to be a result of endolysins that degraded bacterial peptidoglycan and generated cell wall holes, which finally facilitated EV release [54].
Cell wall porosity can directly impact the efficacy of EV export through the fungal wall. Therefore, the composition of the cell wall might affect EV release. In this sense, C. neoformans mutants lacking each of the eight putative chitin synthase genes (CHS1-8) had their ability to produce EVs analyzed in a recent study. The C. neoformans mutants indeed manifested variable cell wall defects, but the analysis of EV production was puzzling, since the pattern of EV detection in the chsΔ mutants was highly variable [55]. For instance, it was initially predicted that disruption of CHS3, a gene encoding a class IV synthase mainly responsible for chitin synthesis in C. neoformans, would be more efficient in releasing vesicles, based on its previously suggested enhanced cell wall porosity [56]. However, this mutant was the one with the lowest efficacy in EV release. Other mutants (chs4Δ and chs5Δ) with no apparent cell wall alterations produced high amounts of EVs. Therefore, the differences observed in the EV analysis were not a consequence of altered cell wall porosity, although the possibility that the mutant strains simply had different abilities to produce EVs could not be ruled out. These results efficiently illustrate the need for a better understanding of how EVs traverse the fungal cell wall. In this sense, a recent study demonstrated that the vast majority of cryptococcal EVs are decorated with mannoproteins [57], suggesting that vesicle composition is directly affected by the presence of cell wall components. These results formed the basis for the proposal of a novel structural model of cryptococcal EVs, in which the outer vesicular layer is composed of the capsular polysaccharide glucuronoxylomannan (GXM), with the lipid bilayer carrying a fibrillar, protein coat enriched with mannoproteins [57].
4. Bioactive Components of Cryptococcal EVs
The first virulence-associated component characterized in cryptococcal EVs was GXM [4,7,58], the main component of the polysaccharide capsule [59,60,61]. It is now known that approximately 70% of cryptococcal EVs are coated with GXM [57]. In contrast to most of the microbial polysaccharides, GXM is synthesized intracellularly, in the Golgi [58]. In C. neoformans, disruption of the SAV1 gene, which encodes a homolog of the Sec4/Rab8 subfamily GTPases that conservatively regulates exocytosis in yeast, resulted in an accumulation of vesicles loaded with GXM in the cytosol [58]. Additionally, the treatment of C. neoformans cells with brefeldin A, an inhibitor of the Golgi-derived transport, inhibited capsule formation [62]. Finally, deletion of the gene encoding GRASP resulted in aberrant Golgi morphology and reduced GXM secretion, with a negative impact on capsule size and attenuation of virulence in in vitro and in vivo models [47]. Together, these results point to the participation of the Golgi in GXM synthesis and export to the cell surface. The extracellular stage of GXM traffic, however, was not studied until cryptococcal EVs were first characterized. Since GXM is a major extracellular component in the Cryptococcus genus, the above-mentioned results implied the existence of mechanisms of trans-cell wall export.
The deletion of genes related to EV export through the ESCRT complex directly impacted the Cryptococcus capsule. Disruption of VPS34, VPS27, HSE1, VPS23, VPS22, VPS25, VPS20, and SNF7 genes led to a significant decrease in capsule size [42,44,45,63]. These results could be related to the observation of EVs altered in size distribution and reduced capsule dimensions in the C. neoformans vps27Δ strain [42]. In this sense, capsular growth was correlated with EV detection. We observed that induction of capsule growth in vitro was accompanied by an increase in the detection of EVs carrying GXM [4]. Robertson et al. (2012) found that the treatment of C. neoformans cells with EDTA resulted in a remarkable reduction in EV detection, and a significant reduction in capsular diameter [64]. On the other hand, a C. neoformans mutant strain lacking a putative G1/S cyclin (Cln1) displayed an abnormal increase in capsule size, and a significantly increased production of EVs [65].
The content of cryptococcal EVs has been also linked to capsule formation. Deletion of the C. gattii encoding a putative scramblase (Aim25) resulted in an increased capsule size [66]. No differences in the amount of EVs were observed in WT and mutant strains. However, an enrichment of a population of larger EVs with a significantly increased GXM concentration was detected in the mutant. Interestingly, the acapsular strain cap67Δ was more efficient in incorporating GXM from EVs obtained from the aim25Δ strain than the WT strain [66]. The importance of membrane regulators on the proper EV formation and GXM export was also suggested in studies of the Apt1 flippase in C. neoformans. Mutant strains produced EVs with lower concentration of GXM and had smaller capsules in vivo [49,50]. More recently, it has been suggested that ZIP3, a cryptococcal regulator of manganese homeostasis, also participates in EV formation, as concluded from the observation of a higher concentration of GXM in culture supernatants of zip3Δ mutants and a high production of EVs, with an enrichment of an EV population with higher dimensions [67]. Together, these studies suggest the existence of connections between EV production and export of the most important capsule component of Cryptococcus spp.
Laccase, the enzyme catalyzing melanin synthesis, is another major virulence factor of Cryptococcus associated with EVs [4,7]. In vitro, C. neoformans EVs incubated with the melanin precursor L-3,4-dihydroxyphenylalanine (L-DOPA) became melanized [8]. It has been further proposed that melanin can be synthesized inside vesicles [68]. Melanin synthesis inside vesicles could protect Cryptococcus cells from the toxic compounds produced during melanin polymerization [68,69].
The C. neoformans mutant strains vps34Δ, vps27Δ, and hseIΔ, all showing functional defects in the ESCRT-0 complex, failed to export laccase to the cell wall [42], which might suggest an association between exosome formation and melanization in Cryptococcus. These results might be related to those observed with a C. neoformans sec6Δ mutant. Sec6 is a protein involved in the polarized fusion of exocytic vesicles with the plasma membrane, and its disruption in Cryptococcus resulted in an increased formation of MVB-like structures, affecting the transport of laccase to the cell wall [37]. A similar interpretation can apply to urease, another EV-linked virulence factor of cryptococci [7]. Interruption of the ESCRT pathway by disruption of the VPS27 gene in C. neoformans resulted in reduced urease activity in vitro [42], and the same phenotype was also observed in the C. neoformans sec6Δ mutant [37]. These studies reinforce the notion that both conventional and unconventional secretory pathways participate in the release of cryptococcal EV-associated virulence molecules.
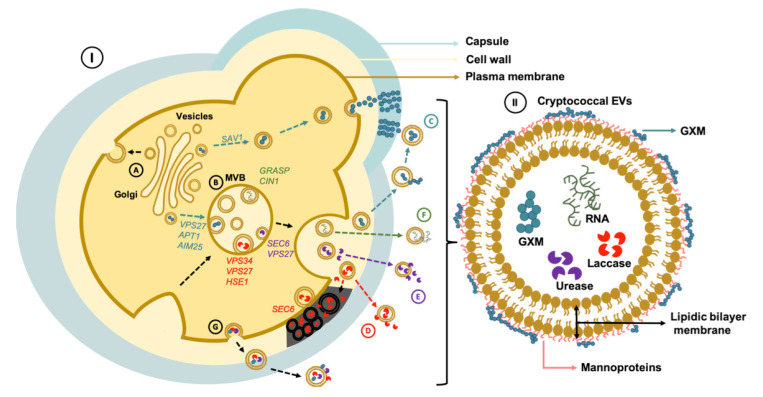
The diversity of molecules inside the Cryptococcus EVs is not restricted to virulence factors. Several RNA subclasses were described in cryptococcal EVs [6,66,70,71]. The first evidence of the presence of RNA in Cryptococcus EVs was provided by Nicola et al. (2009) using an RNA-selective nucleic acid dye to stain vesicular structures [71]. Different subclasses of RNA were further described in cryptococcal EVs, including, small nuclear RNA, ribosomal RNA, transfer RNA, microRNA, long noncoding RNA, and messenger RNA [6,70,72]. Recently, Liu et al. (2020) showed that Cin1, a multidomain adaptor protein that regulates cryptococcal growth, intracellular transport, and the production of several virulence factors [73], also plays an important role in regulating RNA export in C. deneoformans [70]. RNA export in C. neoformans EVs relies on the unconventional secretory pathway. Disruption of GRASP in C. neoformans leads to a significant change in the RNA cargo in EVs when compared to the WT strain [46]. Since disruption of GRASP also resulted in decreased GXM export, these results reinforce the notion that EVs and unconventional secretory mechanisms are connected in Cryptococcus. Figure 2 illustrates the importance of vesicles and EV cargo in physiology and virulence of Cryptococcus.
Figure 2.
EV export and transport of virulence traits in Cryptococcus spp. (I) Vesicle-mediated export of cryptococcal virulence factors are impacted by both conventional (A) and unconventional (B) secretory pathways. (C) Transport of GXM, a major virulence factor that is transported in vesicles, is impacted by both pathways, as illustrated by the need of the SAV1 gene (conventional secretion), and the possible involvement of the VPS27 gene (unconventional secretion) for polysaccharide export. GXM-containing EVs in the outer space might operate as a polysaccharide source for capsule construction. At least two proteins related to membrane architecture and EV formation (Aim25 and Apt1) participate in GXM export. (D) Laccase export is affected by the expression of genes operating in the ESCRT complex (VPS34, VPS27, and HSEI). SEC6, a component of the secretory pathway, also participate in vesicle-mediated melanization. (E) Urease is another virulence factor exported in EVs and its activity is also influenced by genes participating in conventional (SEC6) and unconventional (VPS27) secretory pathways. (F) RNA is also exported in EVs (H) through mechanisms that require CIN1 and GRASP. (G) EVs carrying virulence factors of Cryptococcus may also originate directly from the plasma membrane. However, the molecular mechanisms behind this process are still unknown. (II) Illustration of a cryptococcal EV.
The participation of cryptococcal EVs and their components in fungal virulence suggests that targeting proteins participating in the secretory machinery could lead to the development of novel chemotherapies. Pharmacological inhibitors of EV formation in fungi have not been characterized so far. However, in other eukaryotes, compounds reported to inhibit EV formation (microvesicles or exosomes) were characterized [74]. If these molecules can also affect EV formation in Cryptococcus and other fungi, they could interfere with their pathogenic potential.
5. Impact of EVs during Cryptococcus Infection of Host Cells
EVs can interfere with the outcome of the interaction of cryptococci with infected cells. Murine macrophages RAW 264.7 and J774 can incorporate C. neoformans EVs [75,76]. Similarly, C. gattii EVs were incorporated by J774 macrophages [77]. The uptake of EVs by mouse macrophages is very efficient, as concluded from the incorporation of C. gattii EVs in only five minutes [77]. Actin polymerization inhibitors blocked EV uptake, suggesting the participation of cytoskeleton plasticity [77].
Exposure to cryptococcal EVs resulted in alterations of phagocyte functionality. The treatment of RAW 264.7 macrophages with C. neoformans EVs resulted in increased phagocytosis of non-opsonized C. neoformans [75]. A more prominent increase in the phagocytosis levels was observed when the macrophages were stimulated with EVs produced by a C. neoformans acapsular strain, which indicates that changes in vesicular composition differentially impact their functions [75]. In the same study, C. neoformans EVs were demonstrated to affect cytokine production by RAW 264.7 macrophages. Stimulation of the macrophages with EVs led to increased production of TNF-α, TGF-β, and IL-10. Once again, differences were found between stimulation of the phagocytes with EVs from acapsular or encapsulated C. neoformans strains. EVs from the acapsular strain led to an increase in the production of TNF-α, which induced antifungal activity. On the other hand, EVs from the encapsulated C. neoformans strain led to a significant increase in the production of TGF-β and IL-10, which are known to be positively modulated by GXM [75]. C. neoformans EVs also modulated nitric oxide (NO) production. Curiously, the stimulation of NO production was significantly less effective when the macrophages were treated with EVs isolated from the acapsular C. neoformans strain cap67Δ [75]. These results might be related to the ability of the EVs to modulate fungal killing by host phagocytes. Accordingly, environmental phagocytes are also affected by Cryptococcus EVs. Rizzo et al. (2017) observed stimulation of Acanthamoeba castellanii with EVs resulted in a significantly increased survival of phagocytized C. neoformans [78].
Besides influencing the performance of phagocytes, cryptococcal EVs also modulated important features of the Cryptococcus physiology during macrophage infection. C. gattii EVs obtained from a virulent strain were used to treat macrophages infected with a non-virulent C. gattii isolate, which resulted in the accumulation of the vesicles in the phagosomes [77]. Inside the phagosomes, the EVs from the pathogenic C. gattii strain stimulated the intracellular replication of the non-pathogenic isolate. Negative results were observed when EVs from the non-pathogenic strain or produced by an acapsular mutant were tested [77]. Similarly, Hai et al. (2020) demonstrated that culture filtrates from a high virulent strain induced an increased virulence in a hypovirulent strain [79]. This effect was only observed under conditions of EV availability. These results indicate that cryptococcal EVs are vehicles operating in the transfer of virulence traits between distinct Cryptococcus strains and demonstrate an important function of the vesicles in cell-to-cell communication processes.
Cryptococcus EVs were also suggested to positively impact both adhesion and invasion of the blood–brain barrier (BBB) by fungal cells [80]. In a mice model, C. neoformans EVs induced an enhanced fungal burden in the brain and the cerebrospinal fluid in a dose-dependent manner, with an accumulation of structures that could correspond to EVs surrounding the brain lesions on infected mice [80]. More recently, additional modulatory effects on the host’s immune mechanisms were demonstrated. The mammalian β-galactoside-binding protein Galectin-3 (Gal-3) recognized EVs and promoted vesicle disruption, resulting in decreased levels of interaction of the fungi with macrophages in vitro, reduced recovery of intact EVs, and a diminished uptake of EVs by macrophages [81].
6. Cryptococcal EVs: Vaccine Candidates?
The ability of cryptococcal vesicles to modulate the host’s immunological functions might result in vaccinal potential. Since licensed antifungal vaccines are still not available [82], information on how fungal EVs activate the immune response could be greatly impactful. The vaccinal potential of fungal EVs was first suggested in the Candida model [83,84], and similar observations were recently described in Cryptococcus. In an immunization model of Galleria mellonella with vesicular structures enriched in sterolglycosides (SGs) and GXM, EV administration resulted in the protection of the invertebrate host against a lethal challenge with C. neoformans [85], revealing a potential vaccination strategy for cryptococcosis using sgl1∆ EVs [85]. The vaccinal potential of cryptococcal EVs was recently confirmed in a murine model of cryptococcosis. Mice immunized with EVs obtained from an acapsular C. neoformans mutant strain induced a strong antibody response and significantly prolonged survival of mice upon a lethal challenge with C. neoformans [57]. Importantly, the immunological mechanisms associated with this protection are still unknown, but cryptococcal EVs were recognized by antibodies produced by infected mice [7,57]. Figure 3 presents an overview of the role of EVs during the interaction of Cryptococcus with host cells, including their vaccinal potential.
Figure 3.
Cryptococcus EVs are biologically active and modulate the activity of immune cells. Cryptococcal EVs containing diverse compounds leave the fungal cell and are incorporated by host phagocytes, where they have a number of modulatory activities. (A) EVs can reach the intracellular space, where their cargo modulates the host response, as illustrated by the increased production of TGF-β and IL-10, and decreased release of TNF-α and nitric oxide (NO). A possible outcome of EV exposure is a defective antifungal response by the macrophages, favoring cryptococcal intracellular replication. (B) Galectin-3 (Gal-3) can interact with EVs and disrupt them, leading to failure in the delivery of concentrated virulence factors into host cells and tissues. (C) Cryptococcal EVs represent a potential vaccine strategy to cryptococcosis. EVs from an acapsular C. neoformans mutant or enriched with sterylglucosides (SGs) favor the survival of mice and G. mellonella after a lethal challenge with C. neoformans, respectively. (D) Cryptococcus EVs also play a role in cell-to-cell communication, turning hypovirulent C. gattii cells into a virulent population.
7. Facilitated Methods for the Analysis of Cryptococcal Vesicles
The generation of knowledge on the functions and mechanisms of the biogenesis of cryptococcal vesicles has been continuously affected by methodological limitations. Empirically, it is known in the field that Cryptococcus EVs are produced in low yields, in comparison to other models. Indeed, our laboratory experience shows that other yeast genera, including Saccharomyces and Candida, are more efficient producers of EVs. Therefore, the perception that improved methods of EV analysis were necessary for the Cryptococcus model has been clear for years.
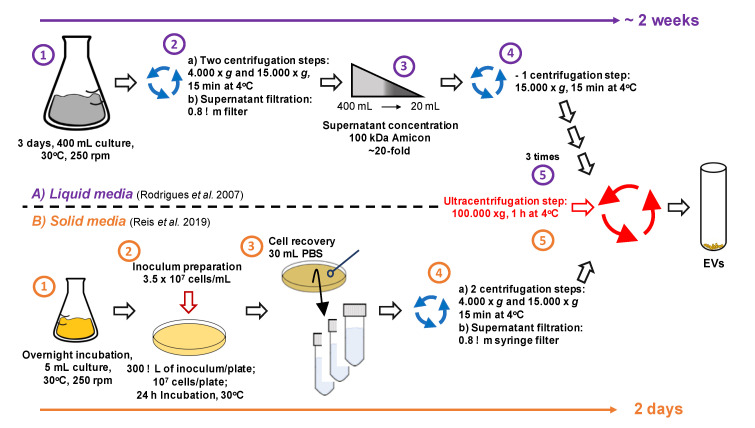
EV analysis in fungi and other eukaryotes has historically included isolation of membrane structures from the supernatants of liquid cultures by ultracentrifugation methods, followed by particle analysis by a combination of microscopic and physical methods [4]. Fungal EVs have been analyzed according to these protocols for more than a decade. Although this model has been helpful to address several questions, it must be highlighted that fungal cells are rarely distributed in liquid matrices both in the environment and during infection. The isolation of cryptococcal EVs from liquid media can take up to two weeks, with very low yields of EV isolation. This was the basis for the design of a novel protocol of isolation of EVs from the Cryptococcus genus. We hypothesized that EVs could be recovered from cultures obtained in solid media since there was no evidence in the literature suggesting that EVs were exclusively produced in liquid matrices.
Cultivation of C. neoformans and C. gattii in regular agar plates followed by suspension of yeast cells in PBS for further centrifugation steps resulted in facilitated detection of typical EVs [66]. However, since this study and earlier articles used diverse methods for EV quantification, a reliable comparison between the yields of the different methods is still not available. Of note, the solid medium protocol successfully allowed EV detection independently on the medium used, and all fungal species tested, including Candida albicans, Histoplasma capsulatum, and Saccharomyces cerevisiae, gave positive results. The protocol was shown to be highly reproducible, and fast: from the recovery of fungal cells to the analysis of ultracentrifugation pellets, the time estimated was of 5 h. Isolated EVs were reliably detected by ELISA targeting GXM, nanoparticle tracking analysis, and transmission electron microscopy. Our most recent unpublished results indicate that the facilitated EV isolation method allows efficient analysis of samples obtained from multiple isolates, separation of vesicles by gradient centrifugation, and a small molecule composition. We anticipate that, in this new scenario, it will be possible to experimentally address currently complicated questions related to vesicle fractionation, diversity, and biogenesis. The most recent methods of EV isolation from cryptococcal cultures are summarized in Figure 4.
Figure 4.
Protocols to isolate cryptococcal EVs. The upper panel (A) summarizes EV isolation from liquid cultures. The lower panel (B) summarizes the protocol used for EV isolation from agar plates. For details, please visit the references mentioned in panel (A) [4] and (B) [66].
8. Gaps, Unanswered Questions, and Perspectives
Despite the recent progress in the field of fungal EVs, in particularly in the Cryptococcus model, it is unquestionable that several gaps and questions remain open. For instance, most studies performed so far were based on single, standard strains of C. neoformans rather than C. gattii, which limits our knowledge on the compositional diversity of cryptococcal EVs. Considering that EV composition is a major determinant of their functions, studies on the diversity in the production of EVs by different cryptococcal species and strains are necessary. Similarly, it is still unknown whether the production of EVs changes at the various life-stages of Cryptococcus.
Novel methods for EV separation are similarly necessary. All studies performed with Cryptococcus so far used centrifugation protocols that resulted in the coisolation of diverse EV populations, as recently illustrated in early [7] and recent [57] studies. This limitation directly impacts, for instance, immunological studies, since these studies are testing mixed EV populations that can manifest divergent immunological functions. Therefore, methods separating EVs based on their biogenesis and/or physical chemical properties are required for refining the functional studies, and they likely improve the knowledge on their vaccinal potential.
Finally, as previously mentioned in this manuscript and several others, we do know that cryptococcal EVs have different cellular origins, but we still do not know where exactly they come from. The identification of genes regulating EV formation and/or pharmacological inhibitors of EV release in Cryptococcus will likely open new venues of investigation, with the potential to change the way we understand the functions of cryptococcal EVs.
Acknowledgments
Authors’ findings that were reviewed and discussed in the manuscript originated from projects that were supported by grants from the Brazilian Ministry of Health (grant 440015/2018-9), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; grants 405520/2018-2 and 301304/2017-3), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Finance code 001), and Fiocruz (grants VPPCB-007-FIO-18 and VPPIS-001-FIO18). JR is supported by the CAPES-COFECUB Franco-Brazilian Research Exchange Program (88887.357947/2019-00). We also acknowledge support from the Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças de Populações Negligenciadas (INCT-IDPN). MLR is currently on leave from the position of Associate Professor in the Federal University of Rio de Janeiro. We are grateful to Leonardo Nimrichter and Jorge J. Jó Bastos Ferreira for helpful insights over a decade of studies on cryptococcal EVs.
Author Contributions
H.C.d.O. and R.F.C. contributed with writing to all sections and prepared Figure 2, Figure 3 and Figure 4. F.C.G.R. wrote Section 7. J.R. wrote about the timeline of scientific discoveries in the field and prepared Figure 1. M.L.R. wrote Section 8 and was responsible for editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The authors received no specific funding for this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Woith E., Fuhrmann G., Melzig M.F. Extracellular Vesicles—Connecting Kingdoms. Int. J. Mol. Sci. 2019;20:5695. doi: 10.3390/ijms20225695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues M.L., Nakayasu E.S., Almeida I.C., Nimrichter L. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J. Proteom. 2014;97:177–186. doi: 10.1016/j.jprot.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albuquerque P.C., Nakayasu E.S., Rodrigues M.L., Frases S., Casadevall A., Zancope-Oliveira R.M., Almeida I.C., Nosanchuk J.D. Vesicular transport in Histoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 2008;10:1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues M.L., Nimrichter L., Oliveira D.L., Frases S., Miranda K., Zaragoza O., Alvarez M., Nakouzi A., Feldmesser M., Casadevall A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albuquerque P.C., Cordero R.J.B., Fonseca F.L., Peres da Silva R., Ramos C.L., Miranda K.R., Casadevall A., Puccia R., Nosanchuk J.D., Nimrichter L., et al. A Paracoccidioides brasiliensis glycan shares serologic and functional properties with cryptococcal glucuronoxylomannan. Fungal Genet. Biol. 2012;49:943–954. doi: 10.1016/j.fgb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peres da Silva R., Puccia R., Rodrigues M.L., Oliveira D.L., Joffe L.S., César G.V., Nimrichter L., Goldenberg S., Alves L.R. Extracellular vesicle-mediated export of fungal RNA. Sci. Rep. 2015;5:7763. doi: 10.1038/srep07763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues M.L., Nakayasu E.S., Oliveira D.L., Nimrichter L., Nosanchuk J.D., Almeida I.C., Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenman H.C., Frases S., Nicola A.M., Rodrigues M.L., Casadevall A. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology. 2009;155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caruso S., Poon I.K.H. Apoptotic cell-derived extracellular vesicles: More than just debris. Front. Immunol. 2018;9:1486. doi: 10.3389/fimmu.2018.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocucci E., Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Raposo G., Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeo K., Uesaka I., Uehira K., Nishiura M. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J. Bacteriol. 1973;113:1442–1448. doi: 10.1128/JB.113.3.1442-1448.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown L., Wolf J.M., Prados-Rosales R., Casadevall A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues M.L., Travassos L.R., Miranda K.R., Franzen A.J., Rozental S., de Souza W., Alviano C.S., Barreto-Bergter E. Human Antibodies against a Purified Glucosylceramide from Cryptococcus neoformans Inhibit Cell Budding and Fungal Growth. Infect. Immun. 2000;68:7049–7060. doi: 10.1128/IAI.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson J., Mihalik R., Soll D.R. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J. Bacteriol. 1990;172:224–235. doi: 10.1128/JB.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M.A.K., de Almeida J.R.F., Jannuzzi G.P., Cronemberger-Andrade A., Torrecilhas A.C.T., Moretti N.S., da Cunha J.P.C., de Almeida S.R., Ferreira K.S. Extracellular Vesicles from Sporothrix brasiliensis Are an Important Virulence Factor That Induce an Increase in Fungal Burden in Experimental Sporotrichosis. Front. Microbiol. 2018;9:2286. doi: 10.3389/fmicb.2018.02286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallejo M.C., Matsuo A.L., Ganiko L., Medeiros L.C., Miranda K., Silva L.S., Freymüller-Haapalainen E., Sinigaglia-Coimbra R., Almeida I.C., Puccia R. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic α-Galactosyl epitopes. Eukaryot. Cell. 2011;10:343–351. doi: 10.1128/EC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehrmann U., Qazi K.R., Johansson C., Hultenby K., Karlsson M., Lundeberg L., Gabrielsson S., Scheynius A. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses-novel mechanisms for host-microbe interactions in atopic eczema. PLoS ONE. 2011;6:e21480. doi: 10.1371/journal.pone.0021480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva B.M., Prados-Rosales R., Espadas-Moreno J., Wolf J.M., Luque-Garcia J.L., Goncalves T., Casadevall A. Characterization of Alternaria infectoria extracellular vesicles. Med. Mycol. 2014;52:202–210. doi: 10.1093/mmy/myt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M., Bruni G.O., Taylor C.M., Zhang Z., Wang P. Comparative genome-wide analysis of extracellular small RNAs from the mucormycosis pathogen Rhizopus delemar. Sci. Rep. 2018;8:5243. doi: 10.1038/s41598-018-23611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bitencourt T.A., Rezende C.P., Quaresemin N.R., Moreno P., Hatanaka O., Rossi A., Martinez-Rossi N.M., Almeida F. Extracellular Vesicles from the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Front. Immunol. 2018;9:2343. doi: 10.3389/fimmu.2018.02343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone F., Bellani L., Muccifora S., Giorgetti L., Bongioanni P., Simili M., Maserti B., Del Carratore R. Analysis of extracellular vesicles produced in the biofilm by the dimorphic yeast Pichia fermentans. J. Cell. Physiol. 2018;233:2759–2767. doi: 10.1002/jcp.25885. [DOI] [PubMed] [Google Scholar]
- 23.Souza J.A.M., Baltazar L.M., Carregal V.M., Gouveia-Eufrasio L., de Oliveira A.G., Dias W.G., Campos Rocha M., Rocha de Miranda K., Malavazi I., Santos D.A., et al. Characterization of Aspergillus fumigatus Extracellular Vesicles and Their Effects on Macrophages and Neutrophils Functions. Front. Microbiol. 2019;10:2008. doi: 10.3389/fmicb.2019.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo J., Chaze T., Miranda K., Roberson R.W., Gorgette O., Nimrichter L., Matondo M., Latgé J.-P., Beauvais A., Rodrigues M.L. Characterization of Extracellular Vesicles Produced by Aspergillus fumigatus Protoplasts. mSphere. 2020;5:e00476-20. doi: 10.1128/mSphere.00476-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bleackley M.R., Samuel M., Garcia-Ceron D., McKenna J.A., Lowe R.G.T., Pathan M., Zhao K., Ang C.S., Mathivanan S., Anderson M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2019;10:1610. doi: 10.3389/fpls.2019.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Paula R.G., Antoniêto A.C.C., Nogueira K.M.V., Ribeiro L.F.C., Rocha M.C., Malavazi I., Almeida F., Silva R.N. Extracellular vesicles carry cellulases in the industrial fungus Trichoderma reesei. Biotechnol. Biofuels. 2019;12:146. doi: 10.1186/s13068-019-1487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavrin T., Konte T., Kostanjšek R., Sitar S., Sepčič K., Prpar Mihevc S., Žagar E., Župunski V., Lenassi M., Rogelj B., et al. The Neurotropic Black Yeast Exophiala dermatitidis Induces Neurocytotoxicity in Neuroblastoma Cells and Progressive Cell Death. Cells. 2020;9:963. doi: 10.3390/cells9040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzo J., Rodrigues M.L., Janbon G. Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives. Front. Cell. Infect. Microbiol. 2020;10:346. doi: 10.3389/fcimb.2020.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf J.M., Espadas-Moreno J., Luque-Garcia J.L., Casadevall A. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot. Cell. 2014;13:1484–1493. doi: 10.1128/EC.00111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues M.L., Nosanchuk J.D., Schrank A., Vainstein M.H., Casadevall A., Nimrichter L. Vesicular transport systems in fungi. Future Microbiol. 2011;6:1371–1381. doi: 10.2217/fmb.11.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viotti C. Unconventional Protein Secretion. Humana Press; New York, NY, USA: 2016. ER to golgi-dependent protein secretion: The conventional pathway. [DOI] [PubMed] [Google Scholar]
- 32.Rabouille C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Owji H., Nezafat N., Negahdaripour M., Hajiebrahimi A., Ghasemi Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018;97:422–441. doi: 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Schekman R. Lasker Basic Medical Research Award. SEC mutants and the secretory apparatus. Nat. Med. 2002;8:1055–1058. doi: 10.1038/nm769. [DOI] [PubMed] [Google Scholar]
- 35.Lee M.C.S., Miller E.A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 36.Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 37.Panepinto J., Komperda K., Frases S., Park Y.D., Djordjevic J.T., Casadevall A., Williamson P.R. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 2009;71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira D.L., Nakayasu E.S., Joffe L.S., Guimaraes A.J., Sobreira T.J., Nosanchuk J.D., Cordero R.J., Frases S., Casadevall A., Almeida I.C., et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE. 2010;5:e11113. doi: 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henne W.M., Buchkovich N.J., Emr S.D. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Williams R.L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 41.Zarnowski R., Sanchez H., Covelli A.S., Dominguez E., Jaromin A., Bernhardt J., Mitchell K.F., Heiss C., Azadi P., Mitchell A., et al. Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018;16:e2006872. doi: 10.1371/journal.pbio.2006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park Y.D., Chen S.H., Camacho E., Casadevall A., Williamson P.R. Role of the ESCRT pathway in Laccase Trafficking and Virulence of Cryptococcus neoformans. Infect. Immun. 2020;88:e00954-19. doi: 10.1128/IAI.00954-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godinho R.M., Crestani J., Kmetzsch L., Araujo Gde S., Frases S., Staats C.C., Schrank A., Vainstein M.H., Rodrigues M.L. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci. Rep. 2014;4:6198. doi: 10.1038/srep06198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu G., Caza M., Cadieux B., Chan V., Liu V., Kronstad J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 2013;81:292–302. doi: 10.1128/IAI.01037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu G., Caza M., Cadieux B., Bakkeren E., Do E., Jung W.H., Kronstad J.W. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol. Microbiol. 2015;96:973–992. doi: 10.1111/mmi.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peres da Silva R., Martins S.T., Rizzo J., Dos Reis F.C.G., Joffe L.S., Vainstein M., Kmetzsch L., Oliveira D.L., Puccia R., Goldenberg S., et al. Golgi Reassembly and Stacking Protein (GRASP) Participates in Vesicle-Mediated RNA Export in Cryptococcus neoformans. Genes. 2018;9:400. doi: 10.3390/genes9080400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kmetzsch L., Joffe L.S., Staats C.C., de Oliveira D.L., Fonseca F.L., Cordero R.J., Casadevall A., Nimrichter L., Schrank A., Vainstein M.H., et al. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol. Microbiol. 2011;81:206–218. doi: 10.1111/j.1365-2958.2011.07686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveira D.L., Fonseca F.L., Zamith-Miranda D., Nimrichter L., Rodrigues J., Pereira M.D., Reuwsaat J.C., Schrank A., Staats C., Kmetzsch L., et al. The putative autophagy regulator Atg7 affects the physiology and pathogenic mechanisms of Cryptococcus neoformans. Future Microbiol. 2016;11:1405–1419. doi: 10.2217/fmb-2016-0090. [DOI] [PubMed] [Google Scholar]
- 49.Rizzo J., Oliveira D.L., Joffe L.S., Hu G., Gazos-Lopes F., Fonseca F.L., Almeida I.C., Frases S., Kronstad J.W., Rodrigues M.L. Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot. Cell. 2014;13:715–726. doi: 10.1128/EC.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzo J., Colombo A.C., Zamith-Miranda D., Silva V.K.A., Allegood J.C., Casadevall A., Del Poeta M., Nosanchuk J.D., Kronstad J.W., Rodrigues M.L. The putative flippase Apt1 is required for intracellular membrane architecture and biosynthesis of polysaccharide and lipids in Cryptococcus neoformans. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:532–541. doi: 10.1016/j.bbamcr.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues M.L., Godinho R.M., Zamith-Miranda D., Nimrichter L. Traveling into Outer Space: Unanswered Questions about Fungal Extracellular Vesicles. PLoS Pathog. 2015;11:e1005240. doi: 10.1371/journal.ppat.1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamith-Miranda D., Nimrichter L., Rodrigues M.L., Nosanchuk J.D. Fungal extracellular vesicles: Modulating host-pathogen interactions by both the fungus and the host. Microbes Infect. 2018;20:501–504. doi: 10.1016/j.micinf.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobson E.S., Ikeda R. Effect of melanization upon porosity of the cryptococcal cell wall. Med. Mycol. 2005;43:327–333. doi: 10.1080/13693780412331271081. [DOI] [PubMed] [Google Scholar]
- 54.Toyofuku M., Cárcamo-Oyarce G., Yamamoto T., Eisenstein F., Hsiao C.-C., Kurosawa M., Gademann K., Pilhofer M., Nomura N., Eberl L. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 2017;8:481. doi: 10.1038/s41467-017-00492-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigues J., Ramos C.L., Frases S., Godinho R.M.d.C., Fonseca F.L., Rodrigues M.L. Lack of chitin synthase genes impacts capsular architecture and cellular physiology in Cryptococcus neoformans. Cell Surf. 2018;2:14–23. doi: 10.1016/j.tcsw.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banks I.R., Specht C.A., Donlin M.J., Gerik K.J., Levitz S.M., Lodge J.K. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzo J., Wong S.S.W., Gazi A.D., Moyrand F., Chaze T., Commere P.-H., Matondo M., Novault S., Pehau-Arnaudet G., Reis F., et al. New insights into Cryptococcus extracellular vesicles suggest a new structural model and an antifungal vaccine strategy. bioRxiv. 2020 doi: 10.1101/2020.08.17.253716. [DOI] [Google Scholar]
- 58.Yoneda A., Doering T.L. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell. 2006;17:5131–5140. doi: 10.1091/mbc.e06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fromtling R.A., Shadomy H.J., Jacobson E.S. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia. 1982;79:23–29. doi: 10.1007/BF00636177. [DOI] [PubMed] [Google Scholar]
- 60.Chang Y.C., Cherniak R., Kozel T.R., Granger D.L., Morris L.C., Weinhold L.C., Kwon-Chung K.J. Structure and biological activities of acapsular Cryptococcus neoformans 602 complemented with the CAP64 gene. Infect. Immun. 1997;65:1584–1592. doi: 10.1128/IAI.65.5.1584-1592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang Y.C., Kwon-Chung K.J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994;14:4912–4919. doi: 10.1128/MCB.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu G., Steen B.R., Lian T., Sham A.P., Tam N., Tangen K.L., Kronstad J.W. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 2007;3:e42. doi: 10.1371/journal.ppat.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chun C.D., Madhani H.D. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS ONE. 2010;5:e12503. doi: 10.1371/journal.pone.0012503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robertson E.J., Wolf J.M., Casadevall A. EDTA inhibits biofilm formation, extracellular vesicular secretion, and shedding of the capsular polysaccharide glucuronoxylomannan by Cryptococcus neoformans. Appl. Environ. Microbiol. 2012;78:7977–7984. doi: 10.1128/AEM.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Rodas R., Cordero R.J., Trevijano-Contador N., Janbon G., Moyrand F., Casadevall A., Zaragoza O. Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. MBio. 2014;5:e00945-14. doi: 10.1128/mBio.00945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reis F.C.G., Borges B.S., Jozefowicz L.J., Sena B.A.G., Garcia A.W.A., Medeiros L.C., Martins S.T., Honorato L., Schrank A., Vainstein M.H., et al. A Novel Protocol for the Isolation of Fungal Extracellular Vesicles Reveals the Participation of a Putative Scramblase in Polysaccharide Export and Capsule Construction in Cryptococcus gattii. mSphere. 2019;4:e00080-19. doi: 10.1128/mSphere.00080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia A.W.A., Kinskovski U.P., Diehl C., Reuwsaat J.C.V., Motta de Souza H., Pinto H.B., Trentin D.d.S., de Oliveira H.C., Rodrigues M.L., Becker E.M., et al. Participation of Zip3, a ZIP domain-containing protein, in stress response and virulence in Cryptococcus gattii. Fungal Genet. Biol. 2020;144:103438. doi: 10.1016/j.fgb.2020.103438. [DOI] [PubMed] [Google Scholar]
- 68.Eisenman H.C., Casadevall A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perez-Dulzaides R., Camacho E., Cordero R.J.B., Casadevall A. Cell-wall dyes interfere with Cryptococcus neoformans melanin deposition. Microbiology. 2018;164:1012–1022. doi: 10.1099/mic.0.000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M., Zhang Z., Ding C., Wang T., Kelly B., Wang P. Transcriptomic Analysis of Extracellular RNA Governed by the Endocytic Adaptor Protein Cin1 of Cryptococcus deneoformans. Front. Cell. Infect. Microbiol. 2020;10:256. doi: 10.3389/fcimb.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicola A.M., Frases S., Casadevall A. Lipophilic Dye Staining of Cryptococcus neoformans Extracellular Vesicles and Capsule. Eukaryot. Cell. 2009;8:1373–1380. doi: 10.1128/EC.00044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Toledo Martins S., Szwarc P., Goldenberg S., Alves L.R. Extracellular Vesicles in Fungi: Composition and Functions. Curr. Top. Microbiol. Immunol. 2019;422:45–59. doi: 10.1007/82_2018_141. [DOI] [PubMed] [Google Scholar]
- 73.Shen G., Whittington A., Song K., Wang P. Pleiotropic function of intersectin homologue Cin1 in Cryptococcus neoformans. Mol. Microbiol. 2010;76:662–676. doi: 10.1111/j.1365-2958.2010.07121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Catalano M., O’Driscoll L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles. 2020;9:1703244. doi: 10.1080/20013078.2019.1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oliveira D.L., Freire-de-Lima C.G., Nosanchuk J.D., Casadevall A., Rodrigues M.L., Nimrichter L. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect. Immun. 2010;78:1601–1609. doi: 10.1128/IAI.01171-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolf J.M., Rivera J., Casadevall A. Serum albumin disrupts Cryptococcus neoformans and Bacillus anthracis extracellular vesicles. Cell. Microbiol. 2012;14:762–773. doi: 10.1111/j.1462-5822.2012.01757.x. [DOI] [PubMed] [Google Scholar]
- 77.Bielska E., Sisquella M.A., Aldeieg M., Birch C., O’Donoghue E.J., May R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 2018;9:1556. doi: 10.1038/s41467-018-03991-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rizzo J., Albuquerque P.C., Wolf J.M., Nascimento R., Pereira M.D., Nosanchuk J.D., Rodrigues M.L. Analysis of multiple components involved in the interaction between Cryptococcus neoformans and Acanthamoeba castellanii. Fungal Biol. 2017;121:602–614. doi: 10.1016/j.funbio.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 79.Hai T.P., Tuan T.L., Van Anh D., Mai T.N., Huong L.N.P., Thwaites G.E., Johnson E., Chau N.V.V., Baker S., Asthon P.M., et al. The virulence of the Cryptococcus neoformans VNIa-5 lineage is highly plastic and associated with isolate background. bioRxiv. 2020 doi: 10.1101/2020.02.24.962134. [DOI] [Google Scholar]
- 80.Huang S.H., Wu C.H., Chang Y.C., Kwon-Chung K.J., Brown R.J., Jong A. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS ONE. 2012;7:e48570. doi: 10.1371/journal.pone.0048570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Almeida F., Wolf J.M., da Silva T.A., DeLeon-Rodriguez C.M., Rezende C.P., Pessoni A.M., Fernandes F.F., Silva-Rocha R., Martinez R., Rodrigues M.L., et al. Galectin-3 impacts Cryptococcus neoformans infection through direct antifungal effects. Nat. Commun. 2017;8:1968. doi: 10.1038/s41467-017-02126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodrigues M.L., Nosanchuk J.D. Fungal diseases as neglected pathogens: A wake-up call to public health officials. PLoS Negl. Trop. Dis. 2020;14:e0007964. doi: 10.1371/journal.pntd.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vargas G., Rocha J.D.B., Oliveira D.L., Albuquerque P.C., Frases S., Santos S.S., Nosanchuk J.D., Gomes A.M.O., Medeiros L.C.A.S., Miranda K., et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell. Microbiol. 2015;17:389–407. doi: 10.1111/cmi.12374. [DOI] [PubMed] [Google Scholar]
- 84.Vargas G., Honorato L., Guimarães A.J., Rodrigues M.L., Reis F.C.G., Vale A.M., Ray A., Nosanchuk J.D., Nimrichter L. Protective Effect of Fungal Extracellular Vesicles against Murine Candidiasis. Cell. Microbiol. 2020;22:e13238. doi: 10.1111/cmi.13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colombo A.C., Rella A., Normile T., Joffe L.S., Tavares P.M., de S Araújo G.R., Frases S., Orner E.P., Farnoud A.M., Fries B.C., et al. Cryptococcus neoformans Glucuronoxylomannan and Sterylglucoside Are Required for Host Protection in an Animal Vaccination Model. MBio. 2019;10:e02909-18. doi: 10.1128/mBio.02909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]