Abstract
Bacteria of the family Chlamydiaceae are globally disseminated and able to infect many bird species. So far, 11 species of Chlamydia have been detected in wild birds, and several studies found chlamydial strains classified as genetically intermediate between Chlamydia (C.) psittaci and C. abortus. Recently, a group of these intermediate strains was shown to form a separate species, i.e., C. buteonis. In the present study, 1128 samples from 341 raptors of 16 bird species and 253 corvids representing six species were examined using a stepwise diagnostic approach. Chlamydiaceae DNA was detected in 23.7% of the corvids and 5.9% of the raptors. In corvids, the most frequently detected Chlamydia species was C. psittaci of outer membrane protein A (ompA) genotype 1V, which is known to have a host preference for corvids. The most frequently detected ompA genotype in raptors was M56. Furthermore, one of the raptors harbored C. psittaci 1V, and two others carried genotype A. C. buteonis was not detected in the bird population investigated, so it remains unknown whether this species occurs in Switzerland. The infection rate of Chlamydiaceae in corvids was high compared to rates reported in other wild bird species, but neither Chlamydiaceae-positive corvids nor raptors showed overt signs of disease. Since the Chlamydiaceae of both, raptors and crows were identified as C. psittaci and all C. psittaci genotypes are considered to be zoonotic, it can be suggested that raptors and crows pose a potential hazard to the health of their handlers.
Keywords: Chlamydiaceae, raptors, crows, Switzerland, C.psittaci/C.abortus intermediates
1. Introduction
Microorganisms of the family Chlamydiaceae are Gram-negative, obligate intracellular bacteria characterized by their unique biphasic lifecycle [1]. The family Chlamydiaceae currently comprises a single genus, Chlamydia, including 14 characterized species [2,3,4]. Chlamydiaceae are globally disseminated and have a broad host range including mammals, birds, reptiles, and amphibians [2]. Chlamydia (C). psittaci, the best-known chlamydial species associated with birds, has been reported to infect more than 460 avian species comprising at least 30 orders [5]. Wild birds serve as an important reservoir not only for C. psittaci but also for several other chlamydial species. To date, eleven Chlamydia species have been detected in birds [3,6,7,8,9,10,11].
Avian chlamydiosis caused by C. psittaci is a notifiable disease in Switzerland and other countries. Between 2010 and 2019, 46 cases were reported to the Federal Food Safety and Veterinary Office, of which 35 cases occurred in domestic and eleven in wild birds [12]. The clinical signs in infected birds can be variable, depending on the virulence of the strain, the susceptibility of the host species, and the immune status of the individual [1,13]. Shedding of the bacteria occurs in both diseased birds and asymptomatic carriers and can be intermittently activated by stressful events like migration, breeding or illness [14].
The zoonotic risk associated with C. psittaci and C. abortus infections is well-known for other chlamydial species harbored by birds; zoonotic transmission is suspected (e.g., C. gallinacea [15]) or unknown (e.g., C. pecorum [2], C. buteonis [3], C. avium [16]).
There are few studies on infection rates of Chlamydiaceae in birds in Switzerland but no study concerning raptors and crows. One study focusing on C. psittaci in pigeons, songbirds, and waterfowl found infection rates of 14.3%, 0.4%, and 4.3%, respectively [17]. Mattmann et al. (2019) investigated Chlamydiaceae infection rates in pigeons from different geographical areas in Switzerland and found a total infection rate of 16.9% [18].
In some European countries, however, the infection rates of Chlamydiaceae in raptors have been investigated. In Sweden, one study reported a C. psittaci infection rate of 1.3% in peregrine falcons (Falco peregrinus) and white-tailed sea eagles (Haliaeetus albicilla) using real-time PCR (qPCR) [19]. Gerbermann and Korbel (1993) reported a C. psittaci infection rate of 13.2% in raptors from southern Germany by antigen ELISA, whereas in eastern Germany Schettler et al. (2003) found 74.4% of the sampled raptors to be positive for C. psittaci using nested PCR [20,21]. Data on Chlamydiaceae in corvids from Europe appears to be even scarcer. One study from Poland found an infection rate of 13.4% based on qPCR, while in Italy an infection rate of 28.9% has been reported in corvids using nested PCR [8,10].
Several studies investigating wild birds found chlamydial species that could not be classified but were identified as genetic intermediates between C. psittaci and C. abortus [22,23,24,25,26,27,28,29,30]. One of these intermediates had initially been detected in a red-tailed hawk (Buteo jamaicensis) in the 1990s [31]. At that time, the organism was identified as C. psittaci. The genome of this isolate was later re-evaluated and recently classified as the new species C. buteonis, together with a new isolate found in a red-shouldered hawk (Buteo lineatus) [3,32]. The clinical importance of C. buteonis is still unknown as few studies have focused on clinical signs associated with chlamydial infections in raptors. However, both the red-tailed hawk and the red-shouldered hawk from which C. buteonis was isolated showed clinical signs of avian chlamydiosis, respiratory distress, and diarrhea in the first, and conjunctivitis in the latter.
The aims of the present study were (i) the collection of data on the infection rates of Chlamydiaceae in raptors and corvids in Switzerland also related to a potential health hazard to humans and (ii) the characterization of the chlamydial species involved, with particular interest in the aforementioned, so far not fully characterized “intermediates” and the new species C. buteonis in view of the limited information available for these organisms.
2. Results
2.1. Chlamydiaceae 23S rRNA qPCR
2.1.1. Species
Results of qPCR testing using an assay targeting the 23S ribosomal RNA (rRNA) gene of Chlamydiaceae (“Chlamydiaceae 23S rRNA qPCR”) for the different bird species are presented in Table 1. In total, 119 samples (10.5%) from 80 birds (13.5%) were positive for Chlamydiaceae. In corvids, Chlamydiaceae were detected in 60/253 animals (23.7%), while 20/341 raptors (5.9%) were positive. The odds ratio showed that the odds of Chlamydiaceae infection was five times higher in corvids than in raptors (OR = 4.99 (95% confidence interval (CI): 2.92–8.53), p < 0.01). Chlamydiaceae were detected in representatives of six raptor species, namely in 13/142 common buzzards (Buteo buteo), 3/32 Eurasian sparrowhawks (Accipiter nisus), 1/23 red kites (Milvus milvus), 1/66 common kestrels (Falco tinnunculus), 1/17 long-eared owls (Asio otus), and 1/17 barn owls (Tyto alba). In corvids, 59/207 carrion crows (Corvus corone) and 1/3 rooks (Corvus frugilegus) were positive for Chlamydiaceae.
Table 1.
Total number and percentage of raptors and corvids positive for Chlamydiaceae per species and number and percentage of chlamydial species identified.
| Species Name | Chlamydiaceae qPCR Pos. (%) | Final Classification | ||||||
|---|---|---|---|---|---|---|---|---|
|
C. Abortus/ C. Psittaci (%) |
C. Psittaci M56 (%) | C. Psittaci A (%) | C. Psittaci 6N (%) | C. Psittaci 1V (%) | C. Psittaci D (%) | Not Further Identified (%) | ||
| Bearded vulture | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Black kite | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Common buzzard | 13 (9.2%) | 0 | 5 (38.5%) | 1 (7.7%) | 0 | 1 (7.7%) | 0 | 6 (46.2%) |
| Eurasian sparrowhawk | 3 (9.4%) | 0 | 0 | 1 (33.3%) | 0 | 0 | 0 | 2 (66.7%) |
| European honey-buzzard | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Golden eagle | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Montagu’s harrier | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Northern goshawk | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Red kite | 1 (4.3%) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100%) |
| Accipitridae subtotal | 17 (7.9%) | 0 | 5 (29.4%) | 2 (11.8%) | 0 | 1 (5.9%) | 0 | 9 (52.9%) |
| Common kestrel | 1 (1.5%) | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 0 |
| Eurasian hobby | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Peregrine falcon | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Falconidae subtotal | 1 (1.4%) | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 0 |
| Eurasian eagle-owl | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Long-eared owl | 1 (5.9%) | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 0 |
| Tawny owl | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Strigidae subtotal | 1 (2.6%) | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 0 |
| Barn owl | 1 (5.9%) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100%) |
| Tytonidae subtotal | 1 (5.9%) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100%) |
| Carrion crow | 59 (28.5%) | 23 (39.0%) | 0 | 0 | 1 (1.7%) | 21 (35.6%) | 3 (5.1%) | 11 (18.6%) |
| Eurasian jay | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eurasian magpie | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hooded crow | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rook | 1 (33.3%) | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 0 |
| Western jackdaw | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Corvidae subtotal | 60 (23.7%) | 23 (38.3%) | 0 | 0 | 1 (1.7%) | 22 (36.7%) | 3 (5.0%) | 11 (18.3%) |
2.1.2. Geographical Origin
Chlamydiaceae-positive birds were detected in nine Swiss cantons as shown in Table 2. There was a strong trend towards higher rates of Chlamydiaceae positivity in the cantons Zug (52.9%) and Zurich (24.2%) compared to the other cantons tested. The lowest infection rate of the cantons of which at least one bird was positive was found in Lucerne (1.7%).
Table 2.
Chlamydiaceae infection rates of raptors and corvids per canton.
| Greater Region | Swiss Canton | Number of Birds | Chlamydiaceae Positive (%) |
|---|---|---|---|
| Lake Geneva | Geneva | 15 | 0 |
| Valais | 11 | 0 | |
| Espace Mittelland | Bern | 56 | 5 (8.9%) |
| Fribourg | 18 | 1 (5.6%) | |
| Solothurn | 13 | 0 | |
| Northwestern Switzerland | Aargau | 31 | 1 (3.2%) |
| Basel District | 4 | 0 | |
| Zurich | Zurich | 132 | 32 (24.2%) |
| Eastern Switzerland | Glarus | 1 | 0 |
| Grisons | 10 | 0 | |
| Schaffhausen | 9 | 1 (11.1%) | |
| St. Gallen | 6 | 0 | |
| Thurgau | 14 | 1 (7.1%) | |
| Central Switzerland | Lucerne | 115 | 2 (1.7%) |
| Nidwalden | 3 | 0 | |
| Obwalden | 5 | 1 (20.0%) | |
| Uri | 1 | 0 | |
| Zug | 51 | 27 (52.9%) | |
| Ticino | Ticino | 6 | 0 |
| Unknown | Unknown | 93 | 9 (9.7%) |
2.1.3. Swab Type
Chlamydiaceae were detected in 13.9% of the choanal (n = 72), 8.8% of the cloacal (n = 46), and 1.1% of the fecal (n = 1) swabs (Table 3). Paired choanal and cloacal swabs were available from 79 birds that tested positive for Chlamydiaceae in at least one site. Regarding these 79 birds, Chlamydiaceae were detected in both swabs in 39 (49.4%) birds, in choanal swabs only in 33 (41.8%) birds and in cloacal swabs only in 7 (8.9%) birds. Based on chi-squared test, successful detection of Chlamydiaceae-positive birds was significantly higher (p < 0.01) with choanal swabs, which detected 72/79 (91.1%) of the cases, compared to cloacal swabs, which only detected 46/79 cases (58.2%). No appropriate comparison with fecal swabs was possible due to the limited number of birds where all three swab types were available.
Table 3.
Number of swabs per sampling site from raptors and corvids positive for Chlamydiaceae.
| Choanal Swabs Positive/Total (%) | Cloacal Swabs Positive/Total (%) | Fecal Swabs Positive/Total (%) | |
|---|---|---|---|
| Raptors | 15/299 (5.0%) | 16/304 (5.3%) | 0/42 (0%) |
| Corvids | 57/220 (25.9%) | 30/216 (13.9%) | 1/47 (2.1%) |
| Total | 72/519 (13.9%) | 46/520 (8.8%) | 1/89 (1.1%) |
2.2. C. Psittaci qPCR and C. Buteonis qPCR
Of the 119 Chlamydiaceae-positive samples, all were negative in C. buteonis species-specific qPCR, and two were positive in C. psittaci-specific qPCR. Both positives originated from raptors, one from a Eurasian sparrowhawk (Nr. 268C), the other one from a common buzzard (Nr. 683C). Both specimens were choanal swabs, and in both animals, the cloacal swab was negative using Chlamydiaceae 23S rRNA qPCR.
2.3. 16S rRNA Conventional PCR and Sequencing
Partial sequences of the 16S ribosomal RNA (278 bp) were successfully obtained from 74 samples of 55 individuals that met the requirements of being negative by both previously described species-specific qPCRs and having a mean cycle quantification (Cq) value <35 in the Chlamydiaceae-specific 23S rRNA qPCR, i.e., eight samples from six raptors and 66 samples from 49 crows (Table 4). Seven samples (Nr. 14C, 311K, 556C, 556K, 669K, 671C, 671K) from five raptors were identified as C. psittaci M56 (accession number: CP003795.1). The remaining sample (Nr. 566C), a choanal swab from a common buzzard, showed 100% sequence identity with two C. abortus strains, C. abortus 15-58d44 (LS974600.1) and C. abortus 15-58d/44 (KX870502.1), and with three C. psittaci isolates, C. psittaci nier_A97 (KX603686.1), C. psittaci nier_A101 (KX603687.1), and C. psittaci nier_A113 (KX603688.1). The 16S rRNA sequences obtained from all the 66 samples from corvids also showed the highest similarity with the sequences of these five strains with identities ranging between 95.6% and 100%. The amplified sequence was identical in these five strains.
Table 4.
Sequence length, sequence quality, first hit by nucleotide identity when compared against the NCBI database and accession number of partial 16S rRNA sequences generated in this study from eight samples from six raptors and 66 samples from 49 corvids from Switzerland.
| Sample Nr. | Species Name (English) | Sequence Length (bp) | Sequence Quality (%) | First Hit | Nucleotide Identity (%) | Accession Number |
|---|---|---|---|---|---|---|
| Raptors | ||||||
| 14C | Common buzzard | 245 | 26.1 | C. psittaci M56 | 99.59 | MT423441 |
| 311K | Common kestrel | 269 | 80.3 | C. psittaci M56 | 100 | MT423442 |
| 556C | Common buzzard | 239 | 30.1 | C. psittaci M56 | 100 | MT423443 |
| 556K | Common buzzard | 278 | 83.1 | C. psittaci M56 | 100 | MT423444 |
| 566C | Common buzzard | 278 | 80.9 | C. abortus 15-58d44 | 100 | MT423446 |
| 669K | Common buzzard | 271 | 83.0 | C. psittaci M56 | 100 | MT423448 |
| 671C | Common buzzard | 260 | 32.7 | C. psittaci M56 | 98.85 | MT423449 |
| 671K | Common buzzard | 269 | 77.3 | C. psittaci M56 | 100 | MT423450 |
| Corvids | ||||||
| 565C | Carrion crow | 253 | 82.2 | C. abortus 15-58d44 | 100 | MT423445 |
| 621C | Rook | 274 | 75.2 | C. abortus 15-58d44 | 100 | MT423447 |
| 686C | Carrion crow | 271 | 40.6 | C. abortus 15-58d44 | 99.63 | MT423451 |
| 688C | Carrion crow | 252 | 80.2 | C. abortus 15-58d44 | 100 | MT423452 |
| 688K | Carrion crow | 279 | 73.8 | C. abortus 15-58d44 | 100 | MT423453 |
| 689C | Carrion crow | 271 | 79.3 | C. abortus 15-58d44 | 100 | MT423454 |
| 689K | Carrion crow | 249 | 25.7 | C. abortus 15-58d44 | 95.58 | MT423455 |
| 690C | Carrion crow | 253 | 80.6 | C. abortus 15-58d44 | 100 | MT423456 |
| 696C | Carrion crow | 276 | 78.3 | C. abortus 15-58d44 | 100 | MT423457 |
| 702C | Carrion crow | 275 | 80.7 | C. abortus 15-58d44 | 100 | MT423458 |
| 705C | Carrion crow | 278 | 71.6 | C. abortus 15-58d44 | 100 | MT423459 |
| 706C | Carrion crow | 256 | 80.1 | C. abortus 15-58d44 | 100 | MT423460 |
| 711C | Carrion crow | 260 | 85.4 | C. abortus 15-58d44 | 100 | MT423461 |
| 716K | Carrion crow | 272 | 84.2 | C. abortus 15-58d44 | 100 | MT423462 |
| 721C | Carrion crow | 253 | 81.4 | C. abortus 15-58d44 | 100 | MT423463 |
| 725C | Carrion crow | 267 | 81.3 | C. abortus 15-58d44 | 100 | MT423464 |
| 735C | Carrion crow | 267 | 76.8 | C. abortus 15-58d44 | 100 | MT423465 |
| 736C | Carrion crow | 253 | 80.6 | C. abortus 15-58d44 | 100 | MT423466 |
| 736K | Carrion crow | 271 | 76.8 | C. abortus 15-58d44 | 100 | MT423467 |
| 737C | Carrion crow | 275 | 77.5 | C. abortus 15-58d44 | 100 | MT423468 |
| 737K | Carrion crow | 315 | 63.8 | C. abortus 15-58d44 | 96.96 | MT423469 |
| 740C | Carrion crow | 260 | 82.7 | C. abortus 15-58d44 | 100 | MT423470 |
| 740K | Carrion crow | 260 | 73.5 | C. abortus 15-58d44 | 100 | MT423471 |
| 744C | Carrion crow | 278 | 77.3 | C. abortus 15-58d44 | 100 | MT423472 |
| 746C | Carrion crow | 266 | 84.6 | C. abortus 15-58d44 | 100 | MT423473 |
| 750C | Carrion crow | 276 | 76.4 | C. abortus 15-58d44 | 100 | MT423474 |
| 750K | Carrion crow | 250 | 21.2 | C. abortus 15-58d44 | 98.40 | MT423475 |
| 751C | Carrion crow | 269 | 85.9 | C. abortus 15-58d44 | 100 | MT423476 |
| 751K | Carrion crow | 226 | 32.7 | C. abortus 15-58d44 | 100 | MT423477 |
| 752C | Carrion crow | 278 | 62.2 | C. abortus 15-58d44 | 100 | MT423478 |
| 752K | Carrion crow | 253 | 51.0 | C. abortus 15-58d44 | 100 | MT423479 |
| 753C | Carrion crow | 274 | 75.9 | C. abortus 15-58d44 | 100 | MT423480 |
| 754C | Carrion crow | 267 | 82.8 | C. abortus 15-58d44 | 100 | MT423481 |
| 756C | Carrion crow | 278 | 75.5 | C. abortus 15-58d44 | 100 | MT423482 |
| 756K | Carrion crow | 240 | 28.3 | C. abortus 15-58d44 | 99.17 | MT423483 |
| 759C | Carrion crow | 278 | 72.7 | C. abortus 15-58d44 | 100 | MT423484 |
| 760C | Carrion crow | 279 | 68.1 | C. abortus 15-58d44 | 100 | MT423485 |
| 760K | Carrion crow | 250 | 53.2 | C. abortus 15-58d44 | 100 | MT423486 |
| 761C | Carrion crow | 256 | 81.3 | C. abortus 15-58d44 | 100 | MT423487 |
| 764C | Carrion crow | 276 | 71.7 | C. abortus 15-58d44 | 100 | MT423488 |
| 765C | Carrion crow | 278 | 78.8 | C. abortus 15-58d44 | 100 | MT423489 |
| 769C | Carrion crow | 276 | 81.2 | C. abortus 15-58d44 | 100 | MT423490 |
| 770C | Carrion crow | 256 | 75.4 | C. abortus 15-58d44 | 100 | MT423491 |
| 772C | Carrion crow | 267 | 82.8 | C. abortus 15-58d44 | 100 | MT423492 |
| 772K | Carrion crow | 270 | 80.4 | C. abortus 15-58d44 | 100 | MT423493 |
| 773C | Carrion crow | 269 | 76.6 | C. abortus 15-58d44 | 100 | MT423494 |
| 774C | Carrion crow | 277 | 75.1 | C. abortus 15-58d44 | 100 | MT423495 |
| 797C | Carrion crow | 275 | 77.1 | C. abortus 15-58d44 | 100 | MT423496 |
| 797K | Carrion crow | 265 | 64.2 | C. abortus 15-58d44 | 100 | MT423497 |
| 798C | Carrion crow | 277 | 75.8 | C. abortus 15-58d44 | 100 | MT423498 |
| 798K | Carrion crow | 271 | 81.2 | C. abortus 15-58d44 | 100 | MT423499 |
| 814C | Carrion crow | 266 | 82.7 | C. abortus 15-58d44 | 100 | MT423500 |
| 814K | Carrion crow | 278 | 74.8 | C. abortus 15-58d44 | 100 | MT423501 |
| 826C | Carrion crow | 266 | 77.8 | C. abortus 15-58d44 | 100 | MT423502 |
| 846C | Carrion crow | 276 | 71.7 | C. abortus 15-58d44 | 100 | MT423503 |
| 847C | Carrion crow | 238 | 43.3 | C. abortus 15-58d44 | 99.58 | MT423504 |
| 848C | Carrion crow | 267 | 82.8 | C. abortus 15-58d44 | 100 | MT423505 |
| 850C | Carrion crow | 256 | 75.4 | C. abortus 15-58d44 | 100 | MT423506 |
| 850K | Carrion crow | 278 | 65.5 | C. abortus 15-58d44 | 100 | MT423507 |
| 851C | Carrion crow | 264 | 82.6 | C. abortus 15-58d44 | 100 | MT423508 |
| 856C | Carrion crow | 267 | 80.1 | C. abortus 15-58d44 | 100 | MT423509 |
| 858C | Carrion crow | 255 | 74.9 | C. abortus 15-58d44 | 100 | MT423510 |
| 858K | Carrion crow | 271 | 77.1 | C. abortus 15-58d44 | 100 | MT423511 |
| 861C | Carrion crow | 276 | 80.1 | C. abortus 15-58d44 | 100 | MT423512 |
| 861K | Carrion crow | 270 | 40.0 | C. abortus 15-58d44 | 99.63 | MT423513 |
| 972C | Carrion crow | 260 | 83.8 | C. abortus 15-58d44 | 100 | MT423514 |
The ten samples selected for 16S rRNA (1481 bp) conventional PCR originated from one Eurasian sparrowhawk (Nr. 268C), one common kestrel (Nr. 311K), one rook (Nr. 621C), two common buzzards (Nr. 556K, 566C), and five carrion crows (Nr. 565C, 746C, 769C, 814C, 972C). The results were very similar to those of the partial 16S rRNA PCR (Table 5). Two strains found in a common buzzard (Nr. 556K) and a common kestrel (Nr. 311K) showed high nucleotide identity with C. psittaci M56 with identities of 99.1% and 100%, respectively. The strains found in the five carrion crows (Nr. 565C, 746C, 769C, 814C, 972C) one rook (Nr. 621C), and one common buzzard (Nr. 566C) again showed high sequence similarity with the five aforementioned C. psittaci and C. abortus strains with identities ranging from 99.6% to 100%. The strain detected in the Eurasian sparrowhawk (Nr. 268C) showed high sequence identity (98%) with several C. psittaci and C. abortus strains, including C. psittaci Ful127 (CP033059.1), C. abortus 84/2334 (CP031646.1), C. psittaci GIMC 2005 (CP024451.1), and C. psittaci WC (CP003796.1).
Table 5.
Sequence length, sequence quality, first hit by nucleotide identity when compared against the NCBI database and accession number of 16S rRNA (1481 bp) sequences generated in this study from four raptors and six corvids from Switzerland.
| Sample Nr. | Species Name (English) | Sequence Length (bp) | Sequence Quality (%) | First Hit | Nucleotide Identity (%) | Accession Number |
|---|---|---|---|---|---|---|
| Raptors | ||||||
| 268C | Eurasian sparrowhawk | 1000 | 86.1 | C. psittaci Ful127 | 97.99 | MT430892 |
| 311K | Common kestrel | 921 | 78.8 | C. psittaci M56 | 99.57 | MT429304 |
| 556K | Common buzzard | 1395 | 95.8 | C. psittaci M56 | 100 | MT430893 |
| 566C | Common buzzard | 1147 | 94.9 | C. psittaci nier_A113 | 100 | MT430895 |
| Corvids | ||||||
| 565C | Carrion crow | 996 | 98.4 | C. psittaci nier_A113 | 99.90 | MT430894 |
| 621C | Rook | 1357 | 91.4 | C. psittaci nier_A113 | 100 | MT430896 |
| 746C | Carrion crow | 1218 | 95.7 | C. psittaci nier_A113 | 99.92 | MT430897 |
| 769C | Carrion crow | 1370 | 95.5 | C. psittaci nier_A113 | 99.85 | MT430898 |
| 814C | Carrion crow | 1379 | 93.8 | C. psittaci nier_A97 | 99.93 | MT430899 |
| 972C | Carrion crow | 1071 | 93.8 | C. psittaci nier_A113 | 99.72 | MT430900 |
2.4. Outer Membrane Protein A (ompA) Genotyping
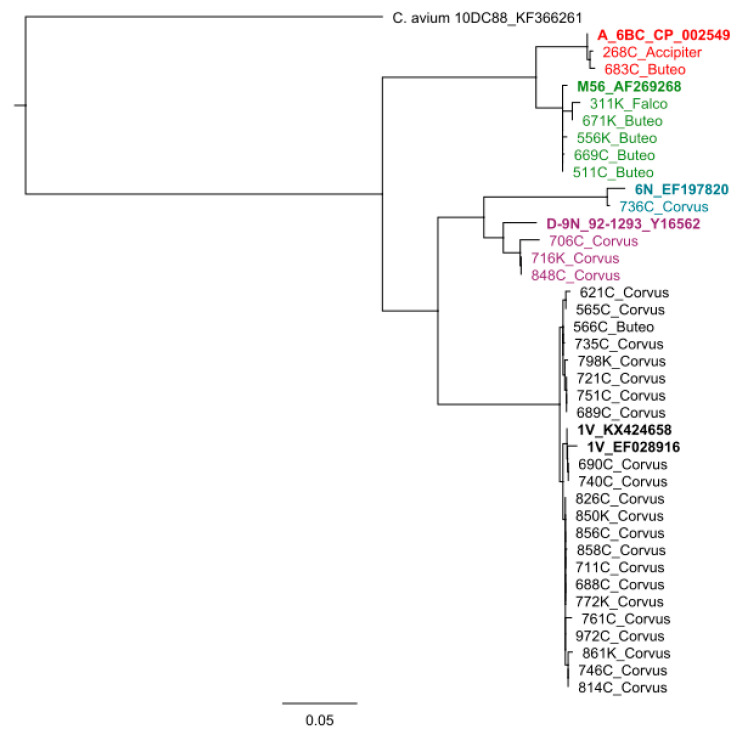
Amplification and sequencing of the ompA gene was successful in both qPCR-positive samples for C. psittaci, as well as 33 selected samples that were positive for Chlamydiaceae, but negative in both species-specific qPCRs (Supplementary Table S1). The 33 samples were selected based on the mean Cq value in the Chlamydiaceae 23S rRNA qPCR, on host species, and geographical location. They originated from five common buzzards (Nr. 511C, 556K, 566C, 669C, 671K), one common kestrel (Nr. 311K), one long-eared owl (877K), one rook (621C), and 25 carrion crows (565C, 688C, 689C, 690C, 706C, 711C, 716K, 721C, 735C, 736C, 740C, 746C, 751C, 752C, 761C, 772K, 798K, 814C, 826C, 848C, 850K, 856C, 858C, 861K, 972C). Both organisms detected in the Eurasian sparrowhawk (Nr. 268C) and the common buzzard (Nr. 683C) positive by C. psittaci qPCR shared the highest ompA sequence identity with the strain C. psittaci Ful127 (CP033059.1) with identities of 99.9% and 99.5%, respectively. This strain had been detected in Northern fulmars (Fulmarus glacialis) from the Faroe Islands and belongs to ompA genotype A [33]. The ompA sequence of four common buzzards (Nr. 511C, 556K, 669C, 671K), the common kestrel (Nr. 311K), and the long-eared owl (Nr. 877K) shared the highest nucleotide identity with C. psittaci M56 (LS974600.1) with identities ranging from 97.2% to 100%. The remaining common buzzard (Nr. 566C), as well as 22 corvids (Nr. 565C, 621C, 688C, 689C, 690C, 711C, 721C, 735C, 740C, 746C, 751C, 752C, 761C, 772K, 798K, 814C, 826C, 850K, 856C, 858C, 861K, 972C) harbored a chlamydial species that shared the highest ompA sequence identity with C. abortus strain 15-58d/44 (KX870484.1), C. psittaci isolate 15-58D/43 (KX424658.1), and C. abortus strain 15-58d44 (LS974600.1) with identities ranging from 99.2% to 100%. All three strains are classified within the ompA genotype 1V. The ompA sequence of the sample of one carrion crow (Nr. 736C) shared the highest sequence similarity with C. psittaci isolate nier_A113 (KX603696.1), C. psittaci isolate nier_A97 (KX603693.1), and C. psittaci isolate 6N (EF197829.1), all belonging to the ompA genotype 6N with identities of 100%, 99.8%, and 98.4%, respectively. Furthermore, chlamydial organisms sharing the highest ompA sequence similarity with C. psittaci NJ1 (CP003798.1), belonging to ompA genotype D, were detected in three carrion crows (Nr. 706C, 716K, 848C). Identities ranged from 96.8% to 97.1%. Results of the ompA sequencing are shown in Table 6. In Figure 1, an ompA based Neighbor Joining dendrogram is shown. Two obtained ompA sequences (752C, 877K) were not included in the dendrogram due to poor sequence quality.
Table 6.
Identified outer membrane protein A (ompA) genotype of Chlamydiaceae detected in nine raptors and 26 crows from various Swiss cantons.
| Sample Nr. | Species Name (English) | Canton of Origin | Year of Sampling | Mean Cq Value Chlamydiaceae qPCR | OmpA Genotype | Accession Number |
|---|---|---|---|---|---|---|
| Raptors | ||||||
| 268C | Eurasian sparrowhawk | Unknown | 2018 | 26.3 | A | MT450242 |
| 311K | Common kestrel | Unknown | 2018 | 26.8 | M56 | MT450243 |
| 511C | Common buzzard | Unknown | 2019 | 38.3 | M56 | MT450244 |
| 556K | Common buzzard | Zurich | 2019 | 14.1 | M56 | MT450245 |
| 566C | Common buzzard | Obwalden | 2019 | 29.8 | 1V | MT450247 |
| 669C | Common buzzard | Unknown | 2019 | 27.9 | M56 | MT450249 |
| 671K | Common buzzard | Thurgau | 2019 | 27.1 | M56 | MT450250 |
| 683C | Common buzzard | Unknown | 2019 | 33.3 | A | MT450251 |
| 877K | Long-eared owl | Bern | 2019 | 31.3 | M56 | MT450275 |
| Corvids | ||||||
| 565C | Carrion crow | Aargau | 2019 | 27.9 | 1V | MT450246 |
| 621C | Rook | Unknown | 2019 | 23.9 | 1V | MT450248 |
| 688C | Carrion crow | Zurich | 2019 | 28.2 | 1V | MT450252 |
| 689C | Carrion crow | Zurich | 2019 | 26.8 | 1V | MT450253 |
| 690C | Carrion crow | Zurich | 2019 | 29.4 | 1V | MT450254 |
| 706C | Carrion crow | Zurich | 2019 | 30.2 | D | MT450255 |
| 711C | Carrion crow | Bern | 2019 | 31.6 | 1V | MT450256 |
| 716K | Carrion crow | Bern | 2019 | 30.6 | D | MT450257 |
| 721C | Carrion crow | Zurich | 2019 | 29.9 | 1V | MT450258 |
| 735C | Carrion crow | Zug | 2019 | 28.4 | 1V | MT450259 |
| 736C | Carrion crow | Zug | 2019 | 28.7 | 6N | MT450260 |
| 740C | Carrion crow | Zug | 2019 | 27.7 | 1V | MT450261 |
| 746C | Carrion crow | Zug | 2019 | 24.9 | 1V | MT450262 |
| 751C | Carrion crow | Zug | 2019 | 20.2 | 1V | MT450263 |
| 752C | Carrion crow | Zug | 2019 | 30.4 | 1V | MT450264 |
| 761C | Carrion crow | Zug | 2019 | 29.7 | 1V | MT450265 |
| 772K | Carrion crow | Zug | 2019 | 26.8 | 1V | MT450266 |
| 798K | Carrion crow | Zurich | 2019 | 26.7 | 1V | MT450267 |
| 814C | Carrion crow | Zurich | 2019 | 25.6 | 1V | MT450268 |
| 826C | Carrion crow | Zurich | 2019 | 29.3 | 1V | MT450269 |
| 848C | Carrion crow | Zurich | 2019 | 26.6 | D | MT450270 |
| 850K | Carrion crow | Zurich | 2019 | 27.9 | 1V | MT450271 |
| 856C | Carrion crow | Zurich | 2019 | 29.3 | 1V | MT450272 |
| 858C | Carrion crow | Zurich | 2019 | 27.8 | 1V | MT450273 |
| 861K | Carrion crow | Unknown | 2019 | 23.6 | 1V | MT450274 |
| 972C | Carrion crow | Fribourg | 2019 | 27.4 | 1V | MT450276 |
Figure 1.
Outer membrane protein A (ompA) based Neighbor Joining dendrogram of Chlamydiaceae from raptors and corvids from Switzerland. Representative sequences from various C. psittaci genotypes are included in boldface. Designation of study isolates correspond to Table 6. Samples in the same color belong to the same ompA genotype.
3. Discussion
3.1. Corvids
The Chlamydiaceae infection rate found in this study (23.7%) is in accordance with the findings of Di Francesco et al. (2015), who detected Chlamydiaceae in 28.9% (n = 22) of the 76 corvids sampled [8]. This study suggests, that C. psittaci of ompA genotype 1V is widespread in the Swiss crow population. Genotypes 1V and 6N are considered to be intermediates between C. psittaci and C. abortus. [10,34,35]. Genotype D, which was detected in three carrion crows, has a known host preference for turkeys [36].
As all corvid samples were negative in the species-specific qPCR for the recently described species C. buteonis, it remains unknown whether this species is able to infect corvids or not.
3.2. Raptors
The Chlamydiaceae infection rate in raptors (5.9%) was towards the lower end of the wide range of infection rates (1.3–74.4%) reported in European raptors [19,20,21] and is in agreement with the findings of Konicek et al. (2016) [37]. Two studies performed in the neighboring country of Germany reported higher infection rates of 13.2% and 74.4% in the southern and eastern part of the country based on antigen ELISA and nested PCR, respectively [20,21]. However, the significance of the 74.4% has to be put in perspective as the authors only tested a small number of birds (n = 39) for Chlamydiaceae [19].
Regarding the three orders of raptors, no significant differences in the Chlamydiaceae infection rate could be observed, as reported earlier [19,30]. Although members of the Accipitriformes (7.9%) showed a considerably higher infection rate than members of the Falconiformes (1.4%), this difference proved statistically not significant (p = 0.050).
C. psittaci M56, which was identified in seven raptors, is considered a mammalian strain with a host preference for muskrats and hares [38]; none of these samples were identified as C. psittaci-positive using the C. psittaci-specific qPCR by Pantchev et al. (2009). This lack of coverage had already been noticed by other researchers (Sachse K., personal communication). These raptors presumably got infected through their prey, as this strain is usually found in mammals, and it was detected only in raptors in this study. The mean Cq value in the Chlamydiaceae qPCR in these birds was 29.1 with values ranging from 14.1–38.3, thus suggesting that the bacteria in samples with low Cq values may have been actively replicating, and the positive result was not only due to residual bacteria from infected prey. Further, ompA genotyping revealed that one common buzzard harbored C. psittaci genotype 1V, a C. psittaci/C. abortus intermediate that has a host preference for corvids [10,34,39]. As all samples were found negative for C. buteonis using species-specific qPCR, it remains unknown whether this recently described chlamydial species does occur in raptors in Switzerland.
3.3. Geographical Distribution
Due to a geographically uneven distribution of samples, owed to the nature of sample collection, it was decided to forego a statistical analysis of differences in infection rate between the cantons. However, the finding that birds from the canton of Zurich showed a higher infection rate compared to birds from Lucerne is supported by previous studies [17,18]. Zweifel et al. (2009) reported a Chlamydiaceae infection rate of 3.3% in feral pigeons from Lucerne and 41.7% in feral pigeons from Zurich [17]. Mattmann et al. (2019) found an infection rate of 17.4% in pigeons from Lucerne and 27.5% in pigeons from Zurich [18]. Mattmann et al. (2019) explained the difference between the infection rates of these two areas with the fact that culling of pigeons, as performed in Zurich, may lead to an increased contact rate of individual pigeons due to frequent restructuring of the population, and therefore, the transmission of pathogens might be increased [18]. In Lucerne on the other hand, different population management programs, including city lofts, are in use.
3.4. Swab Types
Swabs of the choana detected significantly more Chlamydiaceae-positive birds than cloacal swabs, which is in accordance with studies on farmed chickens, ducks, geese, pigeons, turkeys, and cockatiels [9,40,41]. One study investigating the pathogenicity of different C. psittaci strains in chickens found that the overall pharyngeal excretion was slightly higher than the cloacal excretion and that the intensity of excretion varies depending on the C. psittaci strain involved [42]. These findings suggest that the respiratory tract plays a major role in the infection and transmission of chlamydiae [43]. Although the present study highlights that choanal swabs have a higher sensitivity, some birds were negative in the choanal but positive in the cloacal swab as the site of shedding depends on the stage of infection. Thus, for clinical sampling, it can be suggested to use both choanal and cloacal swabs for detection of Chlamydiaceae, since both are relatively easily accessible.
3.5. Public Health Concerns
The Chlamydiaceae of raptors and crows in this study were all identified as C. psittaci, belonging to the ompA genotypes M56, A and 1V (raptors), and ompA genotypes 1V, 6N and D (crows). C. psittaci is the best characterized zoonotic species of the family Chlamydiaceae, and all genotypes are considered zoonotic, including C. psittaci M56 [44,45,46,47,48]. The reported case numbers of human psittacosis indicate that the disease is most likely underdiagnosed, due to lack of disease awareness among the general public and physicians [49,50,51,52].
Sporadic outbreaks of psittacosis are regularly reported in the literature. In 2002, there was a psittacosis outbreak in the Blue Mountains, New South Wales, Australia, with 95 suspected cases with community-acquired pneumonia [53]. From January to April 2013, 25 individuals from southern Sweden were diagnosed with psittacosis [54]. Wild birds were thought to be the source of infection in both outbreaks [53,55]. C. psittaci Ful127 is thought to be the responsible agent for a psittacosis outbreak with 174 human cases on the Faroe Islands in the 1930s [33]. Humans contracted C. psittaci while catching juvenile fulmars and preparing them for cooking [56].
These examples show that C. psittaci outbreaks are still a possible threat to human health. Humans with an increased risk for psittacosis include those coming into close contact with birds on a regular basis (e.g., workers in a zoo or in pet shops, veterinarians, veterinary assistants, and pet bird owners) [57,58,59,60,61,62]. These individuals should take appropriate safety and hygiene measures when handling wild birds. A study showed that bird handlers applying simple measures like wearing protective gloves and washing their hands after handling birds were less likely to get infected by C. psittaci [63].
4. Materials and Methods
4.1. Samples
Sampling was performed between April 2018 and January 2020. A total of 1128 samples were collected from 594 birds representing 22 species belonging to four orders (Supplementary Table S2). In detail, 483 samples were collected from 253 corvids of six species and 645 samples from 341 raptors representing 16 species. Samples consisted of dry choanal (n = 519), cloacal (n = 520), and fecal (n = 89) swabs. Choanal and cloacal swabs were obtained from deceased birds (n = 528), whereas from living birds (n = 66) only fresh fecal material was sampled with swabs after defecation (Table 7). Twenty-three birds died or were euthanized during treatment; therefore, all three swab types were available from these birds. Paired choanal and cloacal swabs were available from 511 birds. For sampling, dry swabs (FLOQSwab®, Copan Flock Technologies, Brescia, Italy) were used and stored in cryovials at −80 °C until further processing.
Table 7.
Number of raptors and corvids with sample types obtained in this study per bird species.
| Order | Family | Species Name (Latin) | Species Name (English) | Number of Birds | Number of Choanal Swabs | Number of Cloacal Swabs | Number of Fecal Swabs |
|---|---|---|---|---|---|---|---|
| Accipitriformes | Accipitridae | Gypaetus barbatus | Bearded vulture | 1 | 1 | 1 | 0 |
| Milvus migrans | Black kite | 6 | 4 | 4 | 3 | ||
| Buteo buteo | Common buzzard | 142 | 127 | 128 | 14 | ||
| Accipiter nisus | Eurasian sparrowhawk | 32 | 32 | 32 | 1 | ||
| Pernis apivorus | European honey-buzzard | 1 | 1 | 1 | 0 | ||
| Aquila chrysaetos | Golden eagle | 6 | 3 | 6 | 0 | ||
| Circus pygargus | Montagu’s harrier | 1 | 1 | 1 | 0 | ||
| Accipiter gentilis | Northern goshawk | 2 | 2 | 2 | 0 | ||
| Milvus milvus | Red kite | 23 | 20 | 21 | 2 | ||
| Falconiformes | Falconidae | Falco tinnunculus | Common kestrel | 66 | 54 | 55 | 12 |
| Falco subbuteo | Eurasian hobby | 4 | 3 | 3 | 1 | ||
| Falco peregrinus | Peregrine falcon | 1 | 1 | 1 | 0 | ||
| Passeriformes | Corvidae | Corvus corone | Carrion crow | 207 | 190 | 187 | 19 |
| Garrulus glandarius | Eurasian jay | 9 | 9 | 8 | 1 | ||
| Pica pica | Eurasian magpie | 30 | 16 | 16 | 22 | ||
| Corvus cornix | Hooded crow | 1 | 1 | 1 | 0 | ||
| Corvus frugilegus | Rook | 3 | 3 | 3 | 2 | ||
| Corvus monedula | Western jackdaw | 3 | 1 | 1 | 3 | ||
| Strigiformes | Strigidae | Bubo bubo | Eurasian eagle-owl | 4 | 4 | 4 | 0 |
| Asio otus | Long-eared owl | 17 | 15 | 14 | 2 | ||
| Strix aluco | Tawny owl | 18 | 14 | 14 | 6 | ||
| Tytonidae | Tyto alba | Barn owl | 17 | 17 | 17 | 1 | |
| Total | 594 | 519 | 520 | 89 |
Dead birds or their samples were obtained from the bird rehabilitation center of the Swiss Ornithological Institute in Sempach, Lucerne, the Wildlife Rehabilitation Center Landshut, Utzenstorf, Berne, the Clinic for Zoo Animals, Exotic Pets and Wildlife, Vetsuisse Faculty, University of Zurich, the Berg am Irchel Bird of Prey Sanctuary, as well as from gamekeepers and local hunters of various cantons. In total, sampled birds originated from 19 Swiss cantons. Carcasses of birds of prey and Corvidae were found dead or were euthanized due to incurable trauma or disease. In addition, carcasses of corvids shot in the scope of cantonal population control programs to reduce the number of birds were available. All living birds were inpatients either at the bird rehabilitation center of the Swiss Ornithological Institute or at the Wildlife Rehabilitation Center Landshut, Utzenstorf, canton of Berne. For all species, the canton of origin and date of sampling were noted if available.
4.2. DNA Extraction
DNA of the choanal and cloacal swabs was extracted using a commercial kit (Genomic DNA from tissue, NucleoSpin® Tissue from Macherey-Nagel, Düren, Germany) according to manufacturer’s instructions. For each extraction lot, a negative control was prepared by using “Buffer T1” instead of the sample. DNA of the fecal samples was extracted with the NucleoSpin® Stool kit (Macherey-Nagel, Düren, Germany) according to the company recommendations. Quality (260/280 value) and quantity of extracted DNA was measured using a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The extracted DNA was stored at −20 °C until further use.
4.3. Chlamydiaceae 23S rRNA qPCR
All samples (n = 1128) were analyzed with a 23S rRNA based Chlamydiaceae family-specific real-time PCR as described previously, modified to include an internal amplification control (eGFP) to control for inhibition [64,65,66]. The cycle conditions were 95 °C for 20 s, followed by 45 cycles of 95 °C for 3 s, and 60 °C for 30 s. Detailed information about all primers and probes used in this study are listed in Table 8. All samples were tested in duplicates. The cycle threshold was set at 0.1 in each run, and a sevenfold dilution series of C. abortus was included as a standard curve in each run. Molecular grade water was used as a negative control. Samples were interpreted as positive if the mean Cq value was <38. Samples with questionable results with Cq values >38 were retested in duplicates. Samples with inhibited amplification were retested undiluted and tenfold diluted, both in duplicates. Samples repeatedly showing a Cq value >38 were considered as positive.
Table 8.
Detailed information about the primers and probes used in this study for the detection of Chlamydiaceae in raptors and corvids, including their final concentration in the PCR reagent mix. eGFP = enhanced green fluorescent protein (used as internal amplification control), ompA = outer membrane protein A; qPCR = real-time PCR.
| Method | Target | Final Concentration | Primer & Probe | Sequence (5′–3′) | Amplicon Size (Base Pairs) | Annealing Temperature (°C) | References |
|---|---|---|---|---|---|---|---|
|
Chlamydiaceae 23S rRNA qPCR |
23S rRNA | 500 nM | Ch23S-F Ch23S-R |
CTGAAACCAGTAGCTTATAAGCGGT ACCTCGCCGTTTAACTTAACTCC |
111 | 60 | Ehricht et al. (2006) [64] |
| 200 nM | Ch23S-P | FAM-CTCATCATGCAAAAGGCACGCCG-TAMRA | |||||
| Internal amplification control |
eGFP | 200 nM | eGFP-1-F | GACCACTACCAGCAGAACAC | 177 | Hoffmann et al. (2006) [65] PCR modified by Blumer et al. (2011) [66] |
|
| eGFP-10-R | CTTGTACAGCTCGTCCATGC | ||||||
| eGFP-HEX | HEX-AGCACCCAGTCCGCCCTGAGCA-BHQ1 | ||||||
|
C. psittaci ompA qPCR |
ompA | 900 nM | CppsOMP1-F | CACTATGTGGGAAGGTGCTTCA | 76 | 60 | Pantchev et al. (2009) [67] |
| CppsOMP1-R | CTGCGCGGATGCTAATGG | ||||||
| 200 nM | CppsOMP1-S | FAM-CGCTACTTGGTGTGAC-TAMRA | |||||
| Internal amplification control |
eGFP | 900 nM | eGFP-1-F | GACCACTACCAGCAGAACAC | 132 | Hoffmann et al. (2005) [68] | |
| eGFP-2-R | GAACTCCAGCAGGACCATG | ||||||
| 200 nM | eGFP-HEX | HEX-AGCACCCAGTCCGCCCTGAGCA-BHQ1 | |||||
| 16S rRNA PCR (partial) |
16S rRNA | 300 nM | 16S IGF (short) 16S IGR (short) |
GATGAGGCATGCAAGTCGAACG CCAGTGTTGGCGGTCAATCTCTC |
278 | 65 | Blumer et al. (2007) [70], Modified from Everett et al. (1999) [75] |
| 16S rRNA PCR (near-full length) |
16S rRNA | 300 nM | 16S-IGF 16S-B1 |
CGGCGTGGATGAGGCAT TACGGYTACCTTGTTACGACTT |
1481 | 57.5 | Everett et al. (1999) [69] Hosokawa et al. (2006) [72] |
| C. buteonis oxaA qPCR |
oxaA | 600 nM | RSHA-F | ATTTCCAACACGCACTGCAT | 80 | 60 | Laroucau et al. (2019) [3] |
| RSHA-R | TGGGACTAGGTGTTCTCCCT | ||||||
| 200 nM | RSHA-P | FAM-GGACAACATGCCTAGATGAAGA-TAMRA | |||||
| Internal amplification control |
eGFP | 400 nM | eGFP-1-F | GACCACTACCAGCAGAACAC | 132 | Hoffmann et al. (2005) [68] | |
| eGFP-2-R | GAACTCCAGCAGGACCATG | ||||||
| 200 nM | eGFP-HEX | HEX-AGCACCCAGTCCGCCCTGAGCA-BHQ1 | |||||
| ompA typing PCR | ompA | 200 nM | ompA F (CTU) | ATGAAAAAACTCTTGAAATCGG | 1212 | 49 | Sachse et al. (2008) [73] |
| ompA rev | TCCTTAGAATCTGAATTGAGC |
4.4. C. Psittaci OmpA qPCR
All Chlamydiaceae-positive samples were subsequently tested with the C. psittaci-specific qPCR according to the protocol as described by Pantchev et al. (2009) including an internal amplification control [67,68]. The reaction mix contained 4 μL (<150 ng/μL) sample template, 1 μL eGFP template, 1x TaqMan Universal PCR MasterMix (Thermo Fisher Scientific, Waltham, MA, USA), 900 nM of the primers CppsOMP1-F and CppsOMP1-R, 200 nM probe CppsOMP1-S, 900 nM of the primers eGFP-1-F and eGFP 2-R, and 200 nM probe eGFP-HEX in a final volume of 25 μL. A negative control (aqua bidest.) and a positive control (synthesized oligonucleotide of the ompA gene of a C. psittaci field isolate “T0592/03, amazon parrot” (National Reference Centre for poultry and Rabbit Disease, University of Zurich); synthesized by Microsynth) were used in duplicates in each run [18].
4.5. C. Buteonis OxaA qPCR
The C. buteonis-specific qPCR was performed as previously described in all Chlamydiaceae-positive samples, modified to include an internal amplification control [3,68]. The reaction mix contained 4 μL sample template, 1 μL eGFP template, 12.5 μL TaqMan Universal PCR MasterMix (Thermo Fisher Scientific, Waltham, MA, USA), 600 nM of the primers RSHA-F and RSHA-R, 200 nM probe RSHA-P, 400 nM of the primers eGFP-1-F and eGFP-2-R, and 200 nM probe eGFP-HEX in a final volume of 25 μL. A negative control (aqua bidest.) and a positive control (DNA of C. buteonis RSHA, kindly provided by Karine Laroucau, ANSES, Maison-Alfort, France) were used in duplicates in each run.
4.6. 16S rRNA PCR and Sequencing
Samples negative by both previously described species-specific qPCRs and fulfilling the requirement of a mean Cq value <35 in the Chlamydiaceae 23S rRNA qPCR were subjected to the 16S rRNA conventional PCR as previously described [69], using the modified primers 16S IGF (short) and 16S IGR (short) [70] to amplify a partial sequence of 278 bp. Per sample, a 50 μL reaction mix was prepared, containing 5 μL (<150 ng/μL) sample template, 25 μL Red Taq Ready Mix (Merck KGaA, Darmstadt, Germany), and 300 nM of both the forward (16S IGF) and the reverse (16S IGR) primer. Cycling conditions were 95 °C for 5 min, followed by 40 cycles of 95 °C for 60 s, 65 °C for 60 s, 72 °C for 90 s, and a final extension of 72 °C for 10 min. 16S rRNA sequences generated in this study are available in GenBank under accession numbers MT423441–MT423514.
Ten samples were selected based on the result of the 16S (partial) sequencing, host species, geographical location, and mean Cq value in the Chlamydiaceae 23S rRNA qPCR and subjected to the near-full length 16S rRNA conventional PCR to amplify a sequence of 1481 bp [71]. The reaction mix was identical to the reaction mix described above, but instead of 16S IGF (short) and 16S IGR (short), the forward and reverse primers 16S-IGF [69] and 16S-B1 [72] were used, respectively. Cycling conditions were identical to those described above, with the only difference that the annealing temperature was set at 57.5 °C instead of 65 °C. The nearly complete 16S rRNA gene sequences are available in GenBank under accession numbers MT429304 and MT430892–MT430900.
Products from all conventional PCRs were purified using the QIAquick® PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Purified amplicons were Sanger sequenced by Microsynth. The obtained sequences were assembled and analyzed using the Geneious Prime software (version 2019.2.3, https://www.geneious.com) and compared against the NCBI database using the BLASTn tool (NCBI, https://blast.ncbi.nlm.nih.gov/).
4.7. OmpA Genotyping PCR
Per sample, a reaction mix with a final volume of 50 μL containing 25 μL REDTaq ReadyMix (Merck KGaA, Darmstadt, Germany), 200 nM of the primers ompA F (CTU) and ompA rev [73], and 3 μL sample template with a DNA concentration of 25 ng/μL was prepared. Cycling conditions were 10 min at 95 °C, followed by 35 cycles of 95 °C for 30 s, 49 °C for 30 s, 72 °C for 60 s, and a final elongation at 72 °C for 7 min [73]. If amplification resulted in weak bands, a modified cycling protocol with 40 cycles of 95 °C for 60 s, 49°C for 60 s, 72 °C for 90 s was used [18]. The ompA sequences obtained in this study are available in GenBank under accession numbers MT450242–MT450276. Analysis of ompA nucleotide sequences was conducted using Geneious version 10.2 (Biomatters Ltd., available from https://www.geneious.com). Multiple sequence alignments were handled using MAFFT v7.450 [74] using the Auto algorithm and scoring matrix: 200PAM/k = 2. Phylogenetic trees were reconstructed using RAxML v8 [75] with nucleotide model GTR GAMMA and the Rapid hill climbing algorithm.
4.8. Statistical Analysis
Statistical analyses were carried out using SPSS version 26 software. For differences in detection rate of Chlamydiaceae from different swab sites, the chi-squared test was performed. The value of p < 0.05 was considered statistically significant.
4.9. Ethical Statement
All animal housing and sampling were conducted in strict accordance to the Swiss law of animal welfare. None of the birds were killed for this study. The birds of which choanal and cloacal swabs were taken were euthanized due to incurable trauma or disease prior to sampling.
Acknowledgments
We would like to thank Brigitte Sigrist from the NRGK and Barbara Prähauser and Theresa Pesch from the Institute of Veterinary Pathology for their excellent technical assistance. We would also like to thank Prisca Mattmann from the Swiss Ornithological Institute; Jean-Michel Hatt from the Clinic for Zoo Animals, Exotic Pets and Wildlife, University of Zurich; Ulrike Cyrus-Eulenberger from the Wildlife Rehabilitation Center Landshut; Andreas Lischke from the Berg am Irchel Bird of Prey Sanctuary; as well as Federico Tettamanti, Sandro Stoller, Gottlieb Dändliker, Hannes Jenni, This Schenkel, Hugo Schober, Elmar Bürgy, Sven Wirthner, Nicolas Zürcher, and Stephan Liersch for their contribution in sample collection. Many thanks also to Karine Laroucau for providing C. buteonis DNA as a positive control for the C. buteonis qPCR.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/9/724/s1, Table S1: Sequence length, sequence quality, first hit by nucleotide identity when compared against the NCBI database and accession number of outer membrane protein A (ompA) sequences generated in this study from nine raptors and 26 corvids from Switzerland, Table S2: Details on origin and analysis results of all swab samples collected and processed in the frame of the present study.
Author Contributions
S.A., B.R.V., and N.B. designed the study; S.S. performed the experiments; S.S., H.M., and K.S. analyzed the data; S.S. wrote the original draft; S.A., B.R.V., N.B., H.M., and K.S. reviewed and edited the original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sachse K., Laroucau K., Vanrompay D. Avian chlamydiosis. Curr. Clin. Micro. Rpt. 2015;2:10–21. doi: 10.1007/s40588-014-0010-y. [DOI] [Google Scholar]
- 2.Cheong H.C., Lee C.Y.Q., Cheok Y.Y., Tan G.M.Y., Looi C.Y., Wong W.F. Chlamydiaceae: Diseases in primary hosts and zoonosis. Microorganisms. 2019;7:146. doi: 10.3390/microorganisms7050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laroucau K., Vorimore F., Aaziz R., Solmonson L., Hsia R.C., Bavoil P.M., Fach P., Hölzer M., Wuenschmann A., Sachse K. Chlamydia buteonis, a new Chlamydia species isolated from a red-shouldered hawk. Syst. Appl. Microbiol. 2019;42:125997. doi: 10.1016/j.syapm.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Sachse K., Bavoil P.M., Kaltenboeck B., Stephens S.S., Kuo C.C., Rosselló-Móra R., Horn M. Emendation of the family Chlamydiaceae: Proposal of a single genus, Chlamydia, to include all currently recognized species. Syst. Appl. Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Kaleta E.F., Taday E.M. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 2003;32:435–461. doi: 10.1080/03079450310001593613. [DOI] [PubMed] [Google Scholar]
- 6.Sachse K., Kuehlewind S., Ruettger A., Schubert E., Rohde G. More than classical Chlamydia psittaci in urban pigeons. Vet. Microbiol. 2012;157:476–480. doi: 10.1016/j.vetmic.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Vorimore F., Hsia R.C., Huot-Creasy H., Bastian S., Deruyter L., Passet A., Sachse K., Bavoil P., Myers G., Laroucau K. Isolation of a new Chlamydia species from the feral sacred ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS ONE. 2013;8:e74823. doi: 10.1371/journal.pone.0074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Francesco A., Donati M., Laroucau K., Balboni A., Galuppi R., Merialdi G., Salvatore D., Renzi M. Chlamydiae in corvids. Vet. Rec. 2015;177:466. doi: 10.1136/vr.103218. [DOI] [PubMed] [Google Scholar]
- 9.Guo W., Li J., Kaltenboeck B., Gong J., Fan W., Wang C. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus) Sci. Rep. 2016;6:19638. doi: 10.1038/srep19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymańska-Czerwińska M., Mitura A., Niemczuk K., Zaręba K., Jodełko A., Pluta A., Scharf S., Vitek B., Aaziz R., Vorimore F., et al. Dissemination and genetic diversity of chlamydial agents in Polish wildfowl: Isolation and molecular characterisation of avian Chlamydia abortus strains. PLoS ONE. 2017;12:e0174599. doi: 10.1371/journal.pone.0174599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokes H.S., Martens J.M., Chamings A., Walder K., Berg M.L., Segal Y., Bennet A. Identification of Chlamydia gallinacea in a parrot and in free-range chickens in Australia. Aust. Vet. J. 2019;97:398–400. doi: 10.1111/avj.12856. [DOI] [PubMed] [Google Scholar]
- 12.Bundesamt für Lebensmittelsicherheit und Veterinärwesen (BLV) Informationssystem Seuchenmeldungen InfoSM. [(accessed on 3 April 2020)]; Available online: https://www.infosm.blv.admin.ch.
- 13.Borel N., Polkinghorne A., Pospischil A. A review on chlamydial diseases in animals: Still a challenge for pathologists? Vet. Pathol. 2018;55:374–390. doi: 10.1177/0300985817751218. [DOI] [PubMed] [Google Scholar]
- 14.Knittler M.R., Sachse K. Chlamydia psittaci: Update on an underestimated zoonotic agent. Pathog. Dis. 2015;73:1–15. doi: 10.1093/femspd/ftu007. [DOI] [PubMed] [Google Scholar]
- 15.Laroucau K., Vorimore F., Aaziz R., Berndt A., Schubert E., Sachse K. Isolation of a new chlamydial agent from infected domestic poultry coincided with cases of atypical pneumonia among slaughterhouse workers in France. Infect. Genet. Evol. 2009;9:1240–1247. doi: 10.1016/j.meegid.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Creasy H.H., Weidmann M., Myers G., Vorimore F., Vicari N., et al. Evidence of the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst. Appl. Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Zweifel D., Hoop R., Sachse K., Pospischil A., Borel N. Prevalence of Chlamydophila psittaci in wild birds—Potential risk for domestic poultry, pet birds, and public health? Eur. J. Wildlife Res. 2009;55:575–581. doi: 10.1007/s10344-009-0275-2. [DOI] [Google Scholar]
- 18.Mattmann P., Marti H., Borel N., Jelocnik M., Albini S., Vogler B.R. Chlamydiaceae in wild, feral and domestic pigeons in Switzerland and insight into population dynamics by Chlamydia psittaci multilocus sequence typing. PLoS ONE. 2019;14:e0226088. doi: 10.1371/journal.pone.0226088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blomqvist M., Christerson L., Waldenström J., Lindberg P., Helander B., Gunnarsson G., Herrmann B., Olsen B. Chlamydia psittaci in birds of prey, Sweden. Infect. Ecol. Epidemiol. 2012;2 doi: 10.3402/iee.v2i0.8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbermann H., Korbel R. The occurrence of Chlamydia psittaci infections in raptors from wildlife preserves. Tierarztl. Prax. 1993;21:217–224. [PubMed] [Google Scholar]
- 21.Schettler E., Fickel J., Hotzel H., Sachse K., Streich W.J., Wittstatt U., Frölich K. Newcastle disease virus and Chlamydia psittaci in free-living raptors from eastern Germany. J. Wildl. Dis. 2003;39:57–63. doi: 10.7589/0090-3558-39.1.57. [DOI] [PubMed] [Google Scholar]
- 22.Fukushi H., Hirai K. Immunochemical diversity of the major outer membrane protein of avian and mammalian Chlamydia psittaci. J. Clin. Microbiol. 1988;26:675–680. doi: 10.1128/JCM.26.4.675-680.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanrompay D., Butaye P., Sayada C., Ducatelle R., Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res. Microbiol. 1997;148:327–333. doi: 10.1016/S0923-2508(97)81588-4. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann B., Rahman R., Bergström S., Bonnedahl J., Olsen B. Chlamydophila abortus in a brown skua (Catharacta antarctica lonnbergi) from a subantarctic island. Appl. Environ. Microbiol. 2000;66:3654–3656. doi: 10.1128/AEM.66.8.3654-3656.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Loock M., Vanrompay D., Herrmann B., vander Stappen J., Volckaert G., Goddeeris B.M., Everett K.D.E. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int. J. Syst. Evol. Microbiol. 2003;53:761–770. doi: 10.1099/ijs.0.02329-0. [DOI] [PubMed] [Google Scholar]
- 26.Madani S.A., Peighambari S.M. PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathol. 2013;42:38–44. doi: 10.1080/03079457.2012.757288. [DOI] [PubMed] [Google Scholar]
- 27.Aaziz R., Gourlay P., Vorimore F., Sachse K., Siarkou V.I., Laroucau K. Chlamydiaceae in North Atlantic seabirds admitted to a wildlife rescue center in western France. Appl. Environ. Microbiol. 2015;81:4581–4590. doi: 10.1128/AEM.00778-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krawiec M., Piasecki T., Wieliczko A. Prevalence of Chlamydia psittaci and other Chlamydia species in wild birds in Poland. Vector Borne Zoonotic Dis. 2015;15:652–655. doi: 10.1089/vbz.2015.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luján-Vega C., Hawkins M.G., Johnson C.K., Briggs C., Vennum C., Bloom P.H., Hull J.M., Cray C., Pesti D., Johnson L., et al. Atypical Chlamydiaceae in wild populations of hawks (Buteo spp.) in California. J. Zoo. Wildl. Med. 2018;49:108–115. doi: 10.1638/2017-0053R.1. [DOI] [PubMed] [Google Scholar]
- 30.Liu S.Y., Li K.P., Hsieh M.K., Chang P.C., Shien J.H., Ou S.C. Prevalence and genotyping of Chlamydia psittaci from domestic waterfowl, companion birds, and wild birds in Taiwan. Vector Borne Zoonotic Dis. 2019;19:666–673. doi: 10.1089/vbz.2018.2403. [DOI] [PubMed] [Google Scholar]
- 31.Mirandé L.A., Howerth E.W., Poston R.P. Chlamydiosis in a red-tailed hawk (Buteo jamaicensis) J. Wildl. Dis. 1992;28:284–287. doi: 10.7589/0090-3558-28.2.284. [DOI] [PubMed] [Google Scholar]
- 32.Joseph S.J., Marti H., Didelot X., Castillo-Ramirez S., Read T.D., Dean D. Chlamydiaceae genomics reveals interspecies admixture and the recent evolution of Chlamydia abortus infecting lower mammalian species and humans. Genome Biol. Evol. 2015;7:3070–3084. doi: 10.1093/gbe/evv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrmann B., Persson H., Jensen J.K., Joensen H.D., Klint M., Olsen B. Chlamydophila psittaci in Fulmars, the Faroe Islands. Emerg. Infect. Dis. 2006;12:330–332. doi: 10.3201/eid1202.050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong J., An I., Oem J.K., Wang S.J., Kim Y., Shin J.H., Woo C., Kim Y., Jo S.D., Son K., et al. Molecular prevalence and genotyping of Chlamydia spp. in wild birds from South Korea. J. Vet. Med. Sci. 2017;79:1204–1209. doi: 10.1292/jvms.16-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yatsentyuk S.P., Obukhov I.L. Molecular genetic characterization of avian Chlamydophila psittaci isolates. Russ. J. Genet. 2007;43:1454–1460. doi: 10.1134/S1022795407110026. [DOI] [PubMed] [Google Scholar]
- 36.Dickx V., Geens T., Deschuyffeleer T., Tyberghien L., Harkinezhad T., Beeckman D.S.A., Braeckman L., Vanrompay D. Chlamydophila psittaci zoonotic risk assessment in a chicken and turkey slaughterhouse. J. Clin. Microbiol. 2010;48:3244–4250. doi: 10.1128/JCM.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konicek C., Vodrážka P., Barták P., Knotek Z., Hess C., Račka K., Hess M., Troxler S. Detection of zoonotic pathogens in wild birds in the cross-border region Austria—Czech Republic. J. Wildl. Dis. 2016;52:850–861. doi: 10.7589/2016-02-038. [DOI] [PubMed] [Google Scholar]
- 38.Pannekoek Y., Dickx V., Beeckman D.S.A., Jolley K.A., Keijzers W.C., Vretou E., Maiden M.C.J., Vanrompay D., van der Ende A. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS ONE. 2010;5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sachse K., Ruettger A. Rapid microarray-based genotyping of Chlamydia spp. strains from clinical tissue samples. Methods Mol. Biol. 2015;1247:391–400. doi: 10.1007/978-1-4939-2004-4_28. [DOI] [PubMed] [Google Scholar]
- 40.Andersen A.A. Comparison of pharyngeal, fecal, and cloacal samples for the isolation of Chlamydia psittaci from experimentally infected cockatiels and turkeys. J. Vet. Diagn. Investig. 1996;8:448–450. doi: 10.1177/104063879600800407. [DOI] [PubMed] [Google Scholar]
- 41.Čechová L., Halánová M., Babinská I., Danišová O., Bartkovský M., Marcinčák S., Marcinčáková D., Valenčáková A., Čisláková L. Chlamydiosis in farmed chickens in Slovakia and zoonotic risk for humans. Ann. Agric. Environ. Med. 2018;25:320–325. doi: 10.26444/aaem/82948. [DOI] [PubMed] [Google Scholar]
- 42.Yin L., Lagae S., Kalmar I., Borel N., Pospischil A., Vanrompay D. Pathogenicity of low and highly virulent Chlamydia psittaci isolates for specific-pathogen-free chickens. Avian Dis. 2013;57:242–247. doi: 10.1637/10439-102612-Reg.1. [DOI] [PubMed] [Google Scholar]
- 43.Van Buuren C.E., Dorrestein G.M., van Dijk J.E. Chlamydia psittaci infections in birds: A review on the pathogenesis and histopathological features. Vet. Q. 1994;16:38–41. doi: 10.1080/01652176.1994.9694414. [DOI] [PubMed] [Google Scholar]
- 44.Beeckman D.S., Vanrompay D.C. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin. Microbiol. Infect. 2009;15:11–17. doi: 10.1111/j.1469-0691.2008.02669.x. [DOI] [PubMed] [Google Scholar]
- 45.Bavoil P., Kaltenboeck B., Greub G. In Chlamydia veritas. Pathog. Dis. 2013;67:89–90. doi: 10.1111/2049-632X.12026. [DOI] [PubMed] [Google Scholar]
- 46.Heddema E.R., van Hannen E.J., Bongaerts M., Dijkstra F., Ten Hove R.J., de Wever B., Vanrompay D. Typing of Chlamydia psittaci to monitor epidemiology of psittacosis and aid disease control in the Netherlands, 2008 to 2013. Euro Surveill. 2015;20:21026. doi: 10.2807/1560-7917.ES2015.20.5.21026. [DOI] [PubMed] [Google Scholar]
- 47.Carlier L., Kempf M., Aaziz R., Jolivet-Gougeon A., Laroucau K. A severe case of pneumopathy in a duck breeder due to Chlamydia psittaci diagnosed by 16S rDNA sequencing. JMM Case Rep. 2014:1. doi: 10.1099/jmmcr.0.001537. [DOI] [Google Scholar]
- 48.Radomski N., Einenkel R., Müller A., Knittler M.R. Chlamydia-host cell interaction not only from a bird’s eye view: Some lessons from Chlamydia psittaci. FEBS Lett. 2016;590:3920–3940. doi: 10.1002/1873-3468.12295. [DOI] [PubMed] [Google Scholar]
- 49.Spoorenberg S.M., Bos W.J., van Hannen E.J., Dijkstra F., Heddema E.R., van Velzen-Blad H., Heijligenberg R., Grutters J.C., de Jongh B.M., Ovidius Study Group Chlamydia psittaci: A relevant cause of community-acquired pneumonia in two Dutch hospitals. Neth. J. Med. 2016;74:75–81. [PubMed] [Google Scholar]
- 50.Hogerwerf L., de Gier B., Baan B., van der Hoek W. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: A systematic review and meta-analysis. Epidemiol. Infect. 2017;145:3096–3105. doi: 10.1017/S0950268817002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Gier B., Hogerwerf L., Dijkstra F., van der Hoek W. Disease burden of psittacosis in the Netherlands. Epidemiol. Infect. 2018;146:303–305. doi: 10.1017/S0950268817003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybarczyk J., Versteele C., Lernout T., Vanrompay D. Human psittacosis: A review with emphasis on surveillance in Belgium. Acta Clin. Belg. 2020;75:42–48. doi: 10.1080/17843286.2019.1590889. [DOI] [PubMed] [Google Scholar]
- 53.Telfer B.L., Moberley S.A., Hort K.P., Branley J.M., Dwyer D.E., Muscatello D.J., Correll P.K., England J., McAnulty J.M. Probable psittacosis outbreak linked to wild birds. Emerg. Infect. Dis. 2005;11:391–397. doi: 10.3201/eid1103.040601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rehn M., Ringberg H., Runehagen A., Herrmann B., Olsen B., Petersson A.C., Hjertqvist M., Kühlmann-Berenzon S., Wallensten A. Unusual increase of psittacosis in southern Sweden linked to wild bird exposure, January to April 2013. Euro Surveill. 2013;18:20478. [PubMed] [Google Scholar]
- 55.Chereau F., Rehn M., Pini A., Kühlmann-Berenzon S., Ydring E., Ringberg H., Runehagen A., Ockborn G., Dotevall L., Wallensten A. Wild and domestic bird faeces likely source of psittacosis transmission-a case-control study in Sweden, 2014-2016. Zoonoses Public Health. 2018;65:790–797. doi: 10.1111/zph.12492. [DOI] [PubMed] [Google Scholar]
- 56.Haagen E., Maurer G. Ueber eine auf den Menschen übertragbare Viruskrankheit bei Sturmvögeln und ihre Beziehung zur Psittakose. Zentralblatt für Bakteriologie, Parasitenkunde und Infektionskrankheiten. Erste Abt. Orig. 1938;143:81–88. [Google Scholar]
- 57.Filstein M.R., Ley A.B., Vernon M.S., Gaffney K.A., Glickman L.T. Epidemic of psittacosis in college of veterinary medicine. J. Am. Vet. Med. Assoc. 1981;179:569–572. [PubMed] [Google Scholar]
- 58.Schlossberg D., Delgado J., Moore M.M., Wishner A., Mohn J. An epidemic of avian and human psittacosis. Arch. Intern. Med. 1993;153:2594–2596. doi: 10.1001/archinte.1993.00410220106012. [DOI] [PubMed] [Google Scholar]
- 59.Davies A., Collins T. Respiratory Chlamydia: The management of an outbreak. Public Health. 1995;109:207–211. doi: 10.1016/S0033-3506(05)80054-X. [DOI] [PubMed] [Google Scholar]
- 60.Gosbell I.B., Ross A.D., Turner I.B. Chlamydia psittaci infection and reinfection in a veterinarian. Aust. Vet. J. 1999;77:511–513. doi: 10.1111/j.1751-0813.1999.tb12121.x. [DOI] [PubMed] [Google Scholar]
- 61.Saito T., Ohnishi J., Mori Y., Iinuma Y., Ichiyama S., Kohi F. Infection by Chlamydophila avium in an elderly couple working in a pet shop. J. Clin. Microbiol. 2005;43:3011–3013. doi: 10.1128/JCM.43.6.3011-3013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raso T.F., Carrasco A.O., Silva J.C., Marvulo M.F., Pinto A.A. Seroprevalence of antibodies to Chlamydophila psittaci in zoo workers in Brazil. Zoonoses Public Health. 2010;57:411–416. doi: 10.1111/j.1863-2378.2009.01237.x. [DOI] [PubMed] [Google Scholar]
- 63.Tolba H.M.N., Abou Elez R.M.M., Elsohaby I. Risk factors associated with Chlamydia psittaci infections in psittacine birds and bird handlers. J. Appl. Microbiol. 2019;126:402–410. doi: 10.1111/jam.14136. [DOI] [PubMed] [Google Scholar]
- 64.Ehricht R., Slickers P., Goellner S., Hotzel H., Sachse K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell. Probes. 2006;20:60–63. doi: 10.1016/j.mcp.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Hoffmann B., Depner K., Schirrmeier H., Beer M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods. 2006;136:200–209. doi: 10.1016/j.jviromet.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 66.Blumer S., Greub G., Waldvogel A., Hässig M., Thoma R., Tschuor A., Pospischil A., Borel N. Waddlia, Parachlamydia and Chlamydiaceae in bovine abortion. Vet. Microbiol. 2011;152:385–393. doi: 10.1016/j.vetmic.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 67.Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet. J. 2009;181:145–150. doi: 10.1016/j.tvjl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 68.Hoffmann B., Beer M., Schelp C., Schirrmeier H., Depner K. Validation of a real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J. Virol. Methods. 2005;130:36–44. doi: 10.1016/j.jviromet.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 69.Everett K.D., Bush R.M., Andersen A.A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 70.Blumer C., Zimmermann D.R., Weilenmann R., Vaughan L., Pospischil A. Chlamydiae in free-ranging and captive frogs in Switzerland. Vet. Pathol. 2007;44:144–150. doi: 10.1354/vp.44-2-144. [DOI] [PubMed] [Google Scholar]
- 71.Taylor-Brown A., Rüegg S., Polkinghorne A., Borel N. Characterisation of Chlamydia pneumoniae and other novel chlamydial infections in captive snakes. Vet. Microbiol. 2015;178:88–93. doi: 10.1016/j.vetmic.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 72.Hosokawa T., Kikuchi Y., Nikoh N., Shimada M., Fukatsu T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006;4:e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sachse K., Laroucau K., Hotzel H., Schubert E., Ehricht R., Slickers P. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008;8:63. doi: 10.1186/1471-2180-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.