Abstract
The changes of microbiota in lungs could change interleukin-17a (IL-17a) expression by altering microRNAs (miRNAs) profile, thus contributing to the pathogenesis of chronic obstructive pulmonary disease (COPD). In this study, we aimed to study molecular mechanisms’ underlying effect of microbiota imbalance on COPD deterioration. Real-time polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) were performed to analyze expression of miRNAs and IL-17a mRNA. ELISA was used to evaluate abundance of IL-17a in plasma, peripheral blood monocyte, and sputum of COPD mice and patients. Luciferase assay was performed to explore underlying molecular mechanisms. The expression of miR-122, miR-30a, and miR-99b were remarkably decreased in COPD mice, while the expression of IL-17a was notably increased in plasma, peripheral blood monocytes, and lung tissues of COPD mice. The levels of Lactobacillus/Moraxella and IL-17a expression were significantly enhanced in sputum of exacerbated COPD patients, along with notably decreased expression of miR-122 and miR-30a. Luciferase assay confirmed that miR-122 and miR-30a played an inhibitory role in IL-17a expression. We identified miR-122 and miR-30a as differentially expressed miRNAs in sputum and plasma of COPD patients in exacerbation-month12 group. Furthermore, downregulated miR-122 and miR-30a expression associated with microbiota imbalance may contribute to COPD deterioration by enhancing IL-17a production.
Keywords: COPD, microbiota, miRNA, IL-17a, lactobacillus
Graphical Abstract
miR-122 and miR-30a were differentially expressed miRNAs in sputum and plasma of COPD patients in the exacerbation-month12 group. Furthermore, downregulated miR-122 and miR-30a expression associated with microbiota imbalance may contribute to COPD deterioration by enhancing IL-17a production.
Introduction
As a frequently diagnosed lung disease related to smoking, chronic obstructive pulmonary disease (COPD) has been defined as the long term obstruction in the airflow.1 COPD is featured by long-term inflammation, tissue remodeling, and fibrosis in small airways, as well as the destruction and impairment of parenchymal tissues in the lungs.2
Microbiota are an important part of human body and can contain very complex ecosystems made of viruses, protozoa, and bacteria, as well as fungi, all of which can live under different conditions in different parts of the body, including the skin, the gut, the oral cavity, and the respiratory airways, as well as the vagina.3 In particular, the microbiota living in the respiratory airways were shown to play a key role in inducing the IL-17a reactions in local tissues. The absence of microbiota decreased the production of IL-17a, while the intranasal transfer of fluid enriched with microbiota isolated from mice with chronic pulmonary inflammation enhanced the production of IL-17A in the lungs of antibiotic treated or axenic recipients. Moreover, the long-term activation of IL-17a activities in the body can consequently lead to the onset of chronic lung diseases such as COPD.4 In fact, the signaling pathway of IL-17a can impact adaptive and innate immunity to regulate the neogenesis of lymphocytes, as well as the proliferation of auto-reactive B/T cells.4
As a type of short non-coding RNA transcripts with a length of less than 24 nucleotides, microRNAs (miRNAs) can act as key regulators in the expression of their target genes.5 In addition, many miRNAs can interact with the 3′ untranslated region (3′ UTR) of their target mRNA transcripts to result in the degradation of these target mRNA transcripts or the suppression of their translation.5 In addition, an abnormal level of miRNA expression was found in multiple types of human malignancies including lung, renal, breast, ovarian, and cervical cancers.6 Several past studies showed that the expression of different miRNAs in patients with renal cancer is different from that in normal tissue samples.7
In addition, as one of the most enriched miRNAs in human body, miR-122 was shown to be highly expressed in the liver, making it important in the normal operation of many liver processes.8 In addition, the absence of miR-122 expression in the liver can suppress the generation of hepatic phenotypes, as well as blocking the invasion of liver cancer cells.9 On the other hand, an increased expression level of miR-122 in the liver can predict a poor prognosis of T cell lymphoma while increasing the resistance of T cell lymphoma cells to chemotherapy.10 As a result, the expression level of miR-122 might be used as a biomarker to predict the prognosis of T cell lymphoma.
The family of miR-30 includes 6 miRNAs, i.e., miR-30c-1, miR-30c-2, miR-30a, miR-30b, miR-30d, and miR-30e.11 According to the evidence reported in the literature, the dysregulation in the expression of miR-30a can contribute to the onset of multiple human malignancies, such as lung cancer, gastric cancer, breast cancer, thyroid cancer, and colon cancer.12, 13, 14, 15 In fact, miR-30a can play a role of tumor suppressor by blocking the process of tumorigenesis in the above human malignancies via binding to genes involved in tumorigenesis.12,16 As an example, the presence of miR-30a can suppress the growth of colon cancer cells via targeting the activity of insulin receptor substrate.12,16
As one of the most extensively investigated proteins in the family of IL-17,17 IL-17a, has been discovered to play critical roles in the induction of host immune response against a wide range of microbes.18 The changes of microbiota in lungs could influence the profile of miRNAs and the expression of IL-17a, a factor functionally involved in the pathogenesis of COPD.19 In this study, we tested the profiles of candidate miRNAs selected by miRNA library screening for CODP-regulating miRNAs and IL-17a in smoke-induced COPD in mice and human subjects.
Results
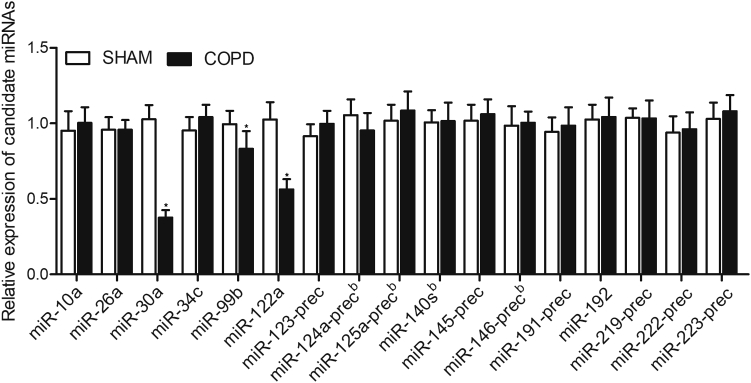
Downregulation of miR-122a, miR-30a, and miR-99b in COPD Mice
A smoke-induced COPD mouse model was established. We evaluated the expression of 17 candidate miRNAs in COPD mice and SHAM controls using qPCR. As shown in Figure 1, the expression of miR-30a, miR-99b, and miR-122a was significantly decreased in COPD mice. However, no obvious difference was observed in the expression of miR-10a, miR-26a, miR-34c, miR-123-prec, miR-124a-prec, miR-125a-prec, miR-140 s, miR-145-prec, miR-146-prec, miR-191-prec, miR-192, miR-219-prec, miR-222-prec, and miR-223-prec between COPD mice and the control mice.
Figure 1.
Expression of miRNAs
Real-time PCR observed that the expression of miR-122, miR-30a, and miR-99b was significantly repressed in the smoke-induced chronic obstructive pulmonary disease (COPD) mice, and no difference was observed for the expression of miR-10a, miR-26a, miR-34c, miR-123-prec, miR-124a-prec, miR-125a-prec, miR-140 s, miR-145-prec, miR-146-prec, miR-191-prec, miR-192, miR-219-prec, miR-222-prec, and miR-223-prec. ∗p < 0.05, versus Sham group.
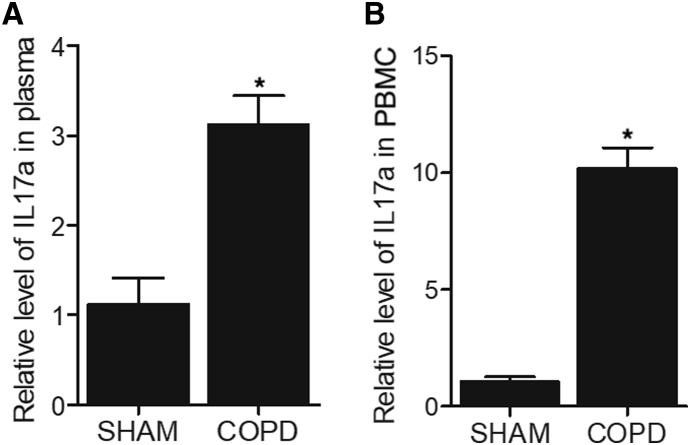
IL-17a Expression Was Enhanced in the Plasma and Peripheral Blood Monocytes in COPD Mice
ELISA was performed to analyze the expression of IL-17a in the plasma and peripheral blood monocytes of COPD mice. The expression of IL-17a in the plasma was remarkably elevated in the plasma of COPD mice (Figure 2A). Moreover, a more dramatic increase was observed in the expression of IL-17a in the peripheral blood monocytes of COPD mice (Figure 2B).
Figure 2.
ELISA Assay Indicated that the Expression of IL-17a Was Elevated in the Plasma and Peripheral Blood Monocytes of COPD Mice
(A) The relative level of IL-17a in plasma was increased in smoke-induced COPD mice. (B) The relative level of IL-17a in the peripheral blood monocytes (PBCs) was dramatically enhanced in smoke-induced COPD mice. ∗p < 0.05, versus Sham group.
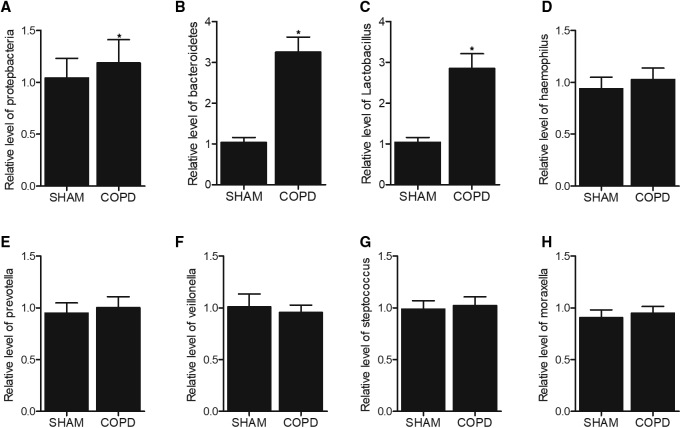
The Levels of Bacteroidetes and Lactobacillus Were Increased in the BAL of COPD Mice
The bronchoalveolar lavage (BAL) samples were collected and subjected to microbiota examination. As shown in Figure 3, we found that the levels of Bacteroidetes (Figure 3B) and Lactobacillus (Figure 3C) were obviously elevated in the BAL of COPD mice, whereas no obvious changes were detected for Proteobacteria (Figure 3A), Haemophilus (Figure 3D), Prevotella (Figure 3E), Veillonella (Figure 3F), Streptococcus (Figure 3G), and Moraxella (Figure 3H).
Figure 3.
The Levels of Bacteroidetes and Lactobacillus Were Increased in the Bronchoalveolar Lavage (BAL) of COPD Mice
(A) The level of Proteobacteria remained unchanged in the BAL of COPD mice. (B) The level of Bacteroidetes was increased in the BAL of COPD mice. (C) The level of Lactobacillus was elevated in the BAL of COPD mice. (D) The level of Haemophilus remained unchanged in the BAL of COPD mice. (E) The level of Prevotella remained unchanged in the BAL of COPD mice. (F) The level of Veillonella remained unchanged in the BAL of COPD mice. (G) The level of Streptococcus remained unchanged in the BAL of COPD mice. (H) The level of Moraxella remained unchanged in the BAL of COPD mice. ∗p < 0.05, versus Sham group.
Demographic, Clinicopathological, and Genotypic Parameters of the Participants Recruited in This Study
A group of smoke-related COPD patients were recruited for our research and divided into 4 groups according to the status of their disease at 12 months after diagnosis: (1) stable-month0 (n = 22); (2) stable-month12 (n = 22); (3) exacerbation-month0 (n = 16); and (4) exacerbation-month12 (n = 16). Then, sputum and peripheral blood samples were collected from all subjects. The demographic and clinicopathological characteristics of the participants in stable-month0 and exacerbation-month0 groups, such as their age, gender, body mass index, number of current smokers, COPD medication, and COPD status (mild, moderate, severe, and very severe), were summarized in Table 1. Student’s t test was utilized to perform the statistical comparison, which revealed no obvious difference in all parameters between the above two groups.
Table 1.
The Demographic and Clinicopathological Characteristics of the Participants in Stable-Month0 and Exacerbation-Month0 Groups
| Characteristics | Stable-Month0 (n = 22) | Exacerbation-Month0 (n = 16) | p Value |
|---|---|---|---|
| Age (years) at enrollment | 62.4 ± 5.7 | 63.8 ± 4.5 | 0.776 |
| Female sex, n (%) | 10 (45.5) | 6 (37.5) | 0.675 |
| Body mass index at enrollment | 25.7 ± 4.1 | 24.9 ± 5.1 | 0.738 |
| Current smokers | 8 (36.4) | 7 (43.8) | 0.280 |
| Medication for COPD | 22 (100) | 16 (100) | |
| Inhaled corticosteroids | 20 (90.9) | 15 (93.8) | 0.392 |
| COPD status, GOLD stage | 0.273 | ||
| Mild | 0 (0) | 0 (0) | |
| Moderate | 8 (36.4) | 7 (43.8) | |
| Severe | 9 (40.9) | 7 (43.8) | |
| Very severe | 5 (22.7) | 2 (12.4) |
GOLD, Global Initiative for Chronic Obstructive Lung Disease.
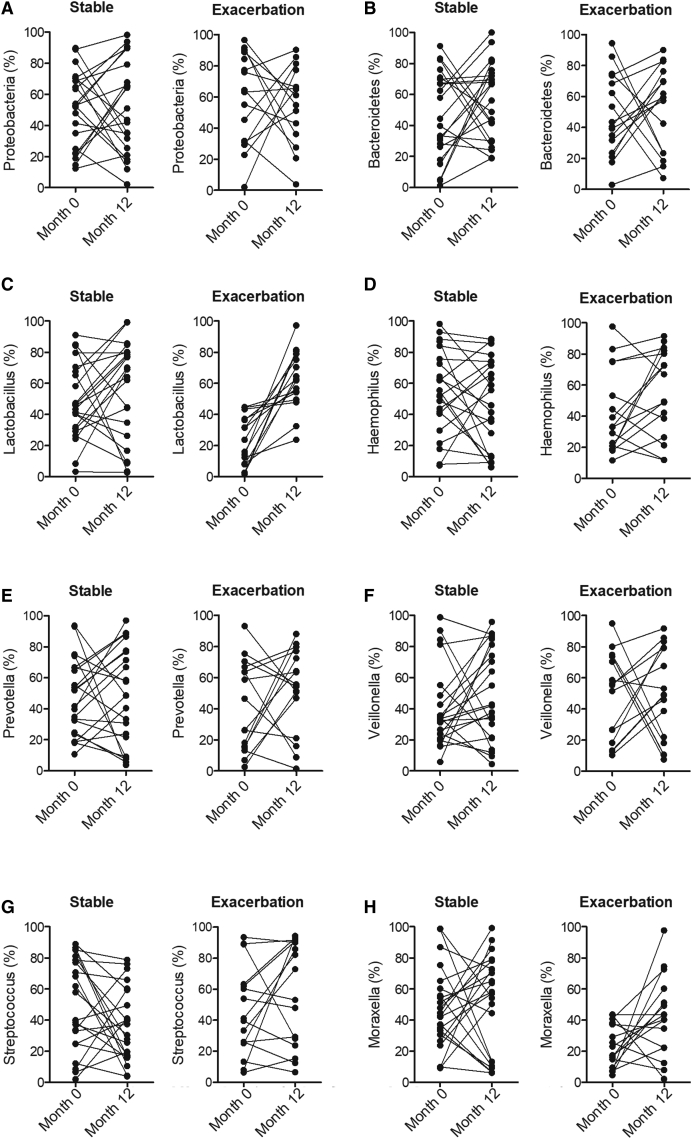
Lactobacillus and Moraxella Levels Were Increased in the Sputum of Exacerbated COPD Patients
A group of smoke-related COPD patients were recruited and divided into 4 groups according to their status of the disease at 12 months after diagnosis: (1) stable-month0 (n = 22); (2) stable-month12 (n = 22); (3) exacerbation-month0 (n = 16); and (4) exacerbation-month12 (n = 16). The sputum samples were collected from all patients at the corresponding time points. The levels for bacteria including Proteobacteria (Figure 4A), Bacteroidetes (Figure 4B), Lactobacillus (Figure 4C), Haemophilus (Figure 4D), Prevotella (Figure 4E), Veillonella (Figure 4F), Streptococcus (Figure 4G), and Moraxella (Figure 4H) were examined in the sputum samples of COPD patients. As shown in Figure 4, for the stable patients, no differences in respect to the levels of all bacteria investigated were observed (Figure 4). However, for the exacerbation patients, the levels of Lactobacillus (Figure 4C) and Moraxella (Figure 4H) were obviously increased in the month12 compared with month0, although no significant difference was observed for other bacteria (Figure 4).
Figure 4.
The Levels of Lactobacillus and Moraxella Were Increased in the Exacerbation-Month12 Group.
(A) No obvious difference was detected in the levels of Proteobacteria between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (B) No obvious difference was detected in the levels of Bacteroidetes between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (C) The level of Lactobacillus was remarkably increased in the exacerbation-month12 group. (D) No obvious difference was detected in the levels of Haemophilus between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (E) No obvious difference was detected in the levels of Prevotella between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (F) No obvious difference was detected in the levels of Veillonella between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (G) No obvious difference was detected in the levels of Streptococcus between the stable-month0 and stable-month12 groups or between the exacerbation-month0 and exacerbation-month12 groups. (H) The level of Moraxella was remarkably increased in the exacerbation-month12 group.
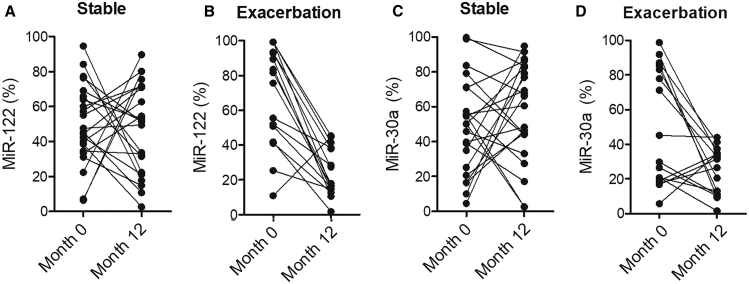
The Expression of miR-122 and miR-30a Was Elevated in the Sputum and Plasma of Exacerbated COPD Patients
The expression of miR-122 (Figure 5A) and miR-30a (Figure 5B) was decreased in the sputum and plasma of COPD patients under different conditions. No obvious changes were detected for miR-122 and miR-30a expression between COPD patients in stable-month0 and stable-month12 groups. However, the expression of miR-122 and miR-30a was notably decreased in the sputum and plasma of COPD patients in the exacerbation-month12 group compared to the exacerbation-month0 group (Figure 5).
Figure 5.
The Expression of miR-122 and miR-30a Was Significantly Downregulated in the Sputum and Plasma of COPD Patients in the Exacerbation-Month12 Group
(A) The expression of miR-122 was notably inhibited in the sputum and plasma of COPD patients in the exacerbation-month12 group. (B) The expression of miR-30a was notably inhibited in the sputum and plasma of COPD patients in the exacerbation-month12 group.
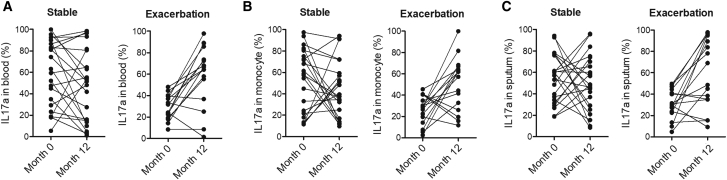
IL-17a Expression Was Enhanced in the Plasma and Peripheral Blood Monocytes of Exacerbated COPD Patients
Plasma, peripheral blood monocytes, and sputum samples were collected from COPD patients in different groups. Then, ELISA was performed to measure the expression of IL-17a. The expression of IL-17a remained unchanged between the stable-month0 and stable-month12 groups (Figure 6). In contrary, IL-17a expression was elevated in the plasma (Figure 6A), peripheral blood monocytes (Figure 6B), and sputum (Figure 6C) of COPD patients in the exacerbation-month12 group.
Figure 6.
The Expression of IL-17a Was Obviously Upregulated in the Plasma, Peripheral Blood Monocytes, and Sputum of COPD Patients in the Exacerbation-Month12 Group
(A) The expression of IL-17a was enhanced in the plasma of COPD patients in the exacerbation-month12 group. (B) The expression of IL-17a was enhanced in the peripheral blood monocytes of COPD patients in the exacerbation-month12 group. (C) The expression of IL-17a was enhanced in the sputum of COPD patients in the exacerbation-month12 group.
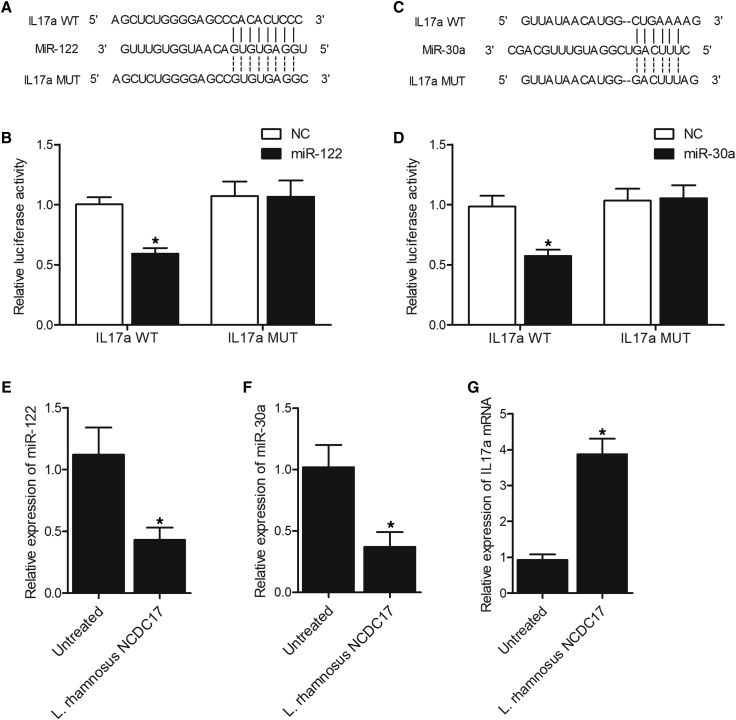
miR-122 and miR-30a Suppressed the Expression of IL-17a in PASMCS Cells
Sequence analysis showed that the 3′ UTR of IL-17a contained candidate binding sites for miR-122 (Figure 7A) and miR-30a (Figure 7C). Luciferase vectors containing wild-type and mutant IL-17a 3′ UTR were constructed and co-transfected into pulmonary arteray smooth muscle cells (PASMCS) cells with miR-122 and miR-30a. The luciferase activity of wild-type IL-17a vector was remarkably repressed by miR-122 (Figure 7B) and miR-30a (Figure 7D). Moreover, we treated PASMCS cells with L. rhamnosus NCDC17 and analyzed the expression of miR-122, miR-30a, and IL-17a mRNA using qPCR. The expression of miR-122 (Figure 7E) and miR-30a (Figure 7F) was remarkably inhibited in PASMCS cells treated with L. rhamnosus NCDC17. On the contrary, the expression of IL-17a mRNA (Figure 7G) was notably increased in PASMCS cells treated with L. rhamnosus NCDC17.
Figure 7.
Luciferase Assay Indicated that miR-122 and miR-30a Inhibited the Expression of IL-17a by Binding to the 3′ UTR of IL-17a.
(A) Sequence analysis indicated a potential binding site of miR-122 in the 3′ UTR of IL-17a (wild-type [WT], mutant [MUT]). (B) The luciferase activity of WT IL-17a was inhibited by miR-122 in PASMCS cells (∗p < 0.05, versus negative control [NC] group). (C) Sequence analysis indicated a potential binding site of miR-30a in the 3′ UTR of IL-17a. (D) The luciferase activity of WT IL-17a was inhibited by miR-30a in PASMCS cells (∗p < 0.05, versus NC group). (E) The expression of miR-122 was inhibited in PASMCS cells treated with L. rhamnosus NCDC17 (∗p < 0.05, versus untreated group). (F) The expression of miR-30a was repressed in PASMCS cells treated with L. rhamnosus NCDC17 (∗p < 0.05, versus untreated group). (G) The expression of IL-17a mRNA was increased in PASMCS cells treated with L. rhamnosus NCDC17. ∗p < 0.05, versus untreated group. GOLD, Global Initiative for Chronic Obstructive Lung Disease.
Discussion
Some past studies tried to analyze the microbiota profile in lung tissues collected from late stage COPD patients.20,21 The results of above studies showed the presence of different types of microbiota in different locations of a same patient, suggesting that both the lower and upper lobes of the lungs may undergo certain pathologic changes during the progression of COPD, as well as upon receiving certain types of treatment.20 In this study, we used a smoke-induced COPD mouse model and performed qPCR to evaluate the expression of 14 candidate miRNAs. We found that the expression of miR-122a, miR-30a, and miR-99b was significantly decreased in the COPD mice. In addition, we evaluated the expression of IL-17a in plasma, peripheral blood monocyte, and lung tissue of COPD mice, which showed a remarkable increased in IL-17a expression. Furthermore, we analyzed the levels of several bacteria in the BAL of COPD mice, and the levels of Lactobacillus and Bacteroidetes were notably enhanced in the BAL of COPD mice.
As a family of diversified, as well as heterogeneous, bacterial population with the capability to produce lactic acid, Lactobacillales contain Lactobacillus, Pediococcus, Leuconostoc, Granulicatella, Facklamia, and Streptococcus. These bacteria can generate lactic acid by utilizing carbohydrates in the body.22 In addition, Lactobacillus can affect the expression level of certain cytokines and miRNAs such as miR-375, miR-122-5p, miR-193a-5p, and miR-3525, as well as miR-215-5p in chickens infected by Salmonella Typhimurium.23 In addition, Lactobacillus was shown to be present in the fecal samples collected from healthy centenarians and was used to alleviate the severity of food induced allergies in mice by promoting the production of transforming growth factor β (TGF-β) in regulatory T cells (T-regs).24 Furthermore, the administration of an oral dose of Lactobacillus in rats could alleviate the symptoms of AHR and reduce the severity of inflammations in the airways, as well as reduce the response of IL-17 induced by the exposure to PM2.5 and OVA.25
A past study has shown that the increased level of miR-122 in humans and animals could elevate the severity of apoptosis of hepatic cells during liver injuries.26 On the other hand, few studies have studied the function of miR-122 in diseases including heart infarction and COPD, although some recent studies have shown the correlation between the altered expression level of serum miR-122 and the progression of sepsis.27, 28, 29 In addition, miR-30a plays a regulatory role in the transduction of IL-17 signals by negatively regulating the activation of MAPK/NF-κB pathways, so as to reduce the synthesis of chemokines, as well as inflammatory cytokines in the body. Moreover, miR-30a can also reduce the stability of IL-17 mRNA, thus reducing the intensity of IL-17 signals. Therefore, a reduced level of miR-30a expression might increase the expression of IL-17 to facilitate the onset of autoimmune disorders.30 In this study, we performed luciferase assay to explore the regulatory role of miR-122 and miR-30a in the expression of IL-17a. We found that the expression of IL-17a was inhibited by miR-122 and miR-30a.
IL-17a is an important inflammatory cytokine expressed by many types of cells, such as epithelial cells, mesenchymal cells, and macrophages. IL-17a is involved in the onset of many autoimmune diseases including multiple sclerosis and rheumatoid arthritis.31 In this study, we enrolled smoking-related COPD patients and collected their sputum and peripheral blood samples. We found that the levels of Lactobacillus and Moraxella in the sputum were obviously enhanced in the exacerbation-month12 group. In addition, we carried out qPCR and ELISA to analyze the expression of miR-122, miR-30a, and IL-17a in the sputum and peripheral blood of COPD patients. The expression of miR-122 and miR-30a was differently inhibited between the exacerbation-month12 group and the exacerbation-month0 group. The expression of IL-17a was remarkably elevated in the exacerbation-month12 group. Most of IL-17a is secreted by memory T cells during the induction of inflammatory responses.32,33 In the lungs, IL-17a plays a key role in the onset of COPD by stimulating the growth of fibroblasts and endothelial cells in the airways. IL-17a can also promote the secretion of IL-6 and IL-8 to impair lung tissues in the onset of COPD and chronic airway inflammation.34
Meanwhile, the results and conclusions drawn from our study are limited. Although the miRNA library screening for COPD-regulated miRNAs were applied, the sample size of subjects recruited for the study was small. And the cohort mainly is restricted to patients whose nationality was Chinese. Therefore, in our future study, an appropriate sample size with varied nationality is preferred.
In conclusion, we tested the expression of several miRNAs in the subjects with stable and exacerbated COPD to identify miR-122 and miR-30a as differentially expressed miRNAs. Furthermore, downregulated miR-122 and miR-30a expression associated with microbiota imbalance may contribute to the deterioration of COPD by enhancing the production of IL-17a.
Materials and Methods
Animal Model Establishment and Treatment
A smoke-induced COPD mouse model was established by using male C57BL/6J mice (n = 20) weighing from 20∼23 g. After 7 days of environmental adaptation, the mice were divided into 2 groups, i.e., (1) SHAM (N = 10) and (2) COPD (N = 10), and then treated differently to establish a COPD mouse model. In brief, one group of mice was placed in an environment free of smoke and was used as the SHAM group. The other group of mice was administered with 50 μL/mouse of saline containing 7.5 μg of LPS (Sigma Aldrich, St. Louis, MO, USA). The administration of LPS was given via two doses of intratracheal instillation administered on D1 and D14, respectively. During the model establishment, all mice were housed in fume boxes with a dimension of 60 cm × 55 cm × 100 cm and then exposed to smoke generated by burning nine cigarettes equipped with a filter tip. The smoke exposure was carried out two times a day and 6 days each week and such smoke exposure was conducted for 3 consecutive months to establish a chronic model of COPD. After this time, mice were anesthetized with chloral hydrate (40 mg/kg, intraperitoneally). All animal procedures were approved by the institutional ethics committee and were in strict compliance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH).
Subjects
A group of smoke-related COPD patients were divided into 4 groups according to the status of their disease at 12 months after diagnosis: (1) stable-month0 (n = 22); (2) stable-month12 (n = 22); (3) exacerbation-month0 (n = 16); and (4) exacerbation-month12 (n = 16). Then, sputum and peripheral blood samples were collected from all subjects. The demographic and clinicopathological characteristics of all participants, such as their age, gender, body mass index, number of current smokers, COPD medication, and COPD status (mild, moderate, severe, and very severe), FEV1, FVC, and FEV1/FVC, were collected, summarized, and compared among different groups. To confirm the diagnosis of COPD, all participating patients should have long-term impaired airflow featured by a ratio of forced expiratory volume in 1 s [FEV1] to forced vital capacity [FVC] < 0.7. For the patients in the exacerbated groups, they should show signs of exacerbation for at least one time in the 6 months prior to their participation in the study. The human research ethics committees of our institute have approved this research and all methods were performed in accordance with the last vision of the Declaration of Helsinki.
Sample Collection and Processing
Sputum samples of each patient (including exacerbated patients) were taken via airway inductions under stable conditions prior to the application of any antibiotics. The sputum samples were collected via airway inductions, and the sputum samples were then nebulized with different concentrations of sodium chloride (5.0%, 4.0%, 3.0%, and 0.9%). All sputum samples were processed following the procedure described below: 2 h after the sample collection, the mucus content in each sputum sample was separated and transferred into a tube containing PBS. 0.2% DTT solution (EMD Millipore, Etobicoke, ON, Canada) was then added into the samples before the samples were stored frozen in a −80°C freeze for further analysis.
Processing of Sputum Samples
The sputum samples with their mucous content were solubilized by using the 0.1% DTT solution and then diluted 4 times by using EDTA. In the next step, the diluted sputum samples were filtered by using a filter membrane. The cells separated from each sputum sample was centrifuged and then stained by using a Wright’s-Giemsa staining kit (Diff-Quik, Thermo Fisher Scientific, St. Louis, MO, USA). The results of Wright’s-Giemsa staining were evaluated underneath a light microscope by using a hemacytometer to identify the presence of neutrophils, macrophages, and epithelial cells in each sample.
RNA Isolation and Real-Time PCR
In this study, the expression level of multiple miRNAs, such as miR-10a, miR-26a, miR-30a, miR-34c, miR-99b, miR-122a, miR-123-prec, miR-124a miR-125a-precb, miR-140 sb, miR-145-prec, miR-146-precb, miR-192, miR-219-prec, miR-222-prec, miR-223-prec, and miR-122, as well as the mRNA expression level of IL-17a was analyzed using real-time PCR. The total RNA content in each plasma sample collected from the mice models was isolated by using a Trizol kit assay kit (Invitrogen, Carlsbad, CA, USA) following the standard protocol provided by the kit manufacturer.35,36 In the next step, the isolated RNA was subjected to real-time PCR to determine the relative expression of miR-10a, miR-26a, miR-30a, miR-34c, miR-99b, miR-122a, miR-123-prec, miR-124a-precb, miR-125a-precb, miR-140 sb, miR-145-prec, miR-146-precb, miR-191-prec, miR-192, miR-219-prec, miR-222-prec, miR-223-prec, miR-122, and IL-17a mRNA by the calculation provided by the 2−ΔΔCt method. The qPCR data for miRNAs were normalized to RNU48, while the qPCR data for IL-17a mRNA was normalized to GAPDH. The primers used in this study were demonstrated in Table S1.
Isolation of Peripheral Blood Monocytes from Human Subjects
Peripheral blood monocytes were isolated from the peripheral blood samples collected from each subject by using a Lymphocytes Separation assay kit (Dakewei, Beijing, China) following the standard protocol provided by the assay kit manufacturer.
Cell Culture and Transfection
To study the effect of Lactobacillus rhamnosus NCDC17 treatment on the expression of IL-17a, as well as miR-122 and miR-30a, primary PASMCs were isolated and cultured as previously described.37,38 The cells were cultured in a modified DMEM medium supplemented with 10% FBS and appropriate antibiotics. When the cell confluency reached 70%, the cells were divided into two groups: (1) untreated group and (2) L. rhamnosus NCDC17 group. Then, the cells were treated accordingly for 48 h before they were harvested to measure the expression of IL-17a, as well as miR-122 and miR-30a.
ELISA
The expression of IL-17a in collected plasma samples was measured using a commercial ELISA assay kit (eBioscience, San Diego, CA, USA) following the standard protocol provided by the assay kit manufacturer.
Detection of Microbiota Abundance in BAL and Sputum Samples
We have used a specific algorithm based on bacteria 16S rRNA following previously described instructions39 to calculate and detect the genera of microbiota in BAL and SPUTUM samples collected from study subjects.
Vector Construction, Mutagenesis, and Luciferase Assay
A combination of TargetScan (http://www.targetscan.org/) and the miRNA database miRBase (http://www.mirbase.org/) were utilized to predict the candidate binding sites for miR-122 and miR-30a in the 3′ UTR of IL-17a mRNA. To confirm the regulatory relationship between IL-17a and miR-122, miR-125b, and miR-30a, we cloned the 3′ UTR of IL-17a containing the binding sites for miR-122, miR-125b, and miR-30a into pcDNA vectors (Promega, Madison, WI, USA) to generate a wild-type luciferase vector of IL-17a 3′ UTR.40,41 At the same time, we used a Quick Change mutagenesis assay kit (Stratagene, San Diego, CA, USA) following the standard protocol provided by the assay kit manufacturer to induce site-directed mutations in the miR-122, miR-125b, and miR-30a binding sites of IL-17a 3′ UTR, respectively, to generate various mutant vectors of IL-17a 3′ UTR. In the next step, PASMCS cells were co-transfected with wild-type or mutant luciferase vectors of IL-17a 3′ UTR in conjunction with miR-122, miR-125b, or miR-30a mimics using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the standard protocol provided by the assay kit manufacturer. The transfected cells were harvested 48 h later and the luciferase activity of transfected cells was measured.
Statistical Analysis
In general, each experiment was done in triplicates for each condition. SPSS 19.0 statistical software was used in the statistical analysis. All experimental results were represented by means ± standard error of the mean (SEM), which were calculated by Microsoft Excel. The measurement data were displayed as x ± s. For the inter-group comparisons, an unpaired two-tailed Student’s t test was applied. For the multi-group comparisons, a one-way ANOVA followed by Tukey’s test as the post hoc test was applied. The level of statistical significance was set to p <0.05.
Author Contributions
R.W. designed the project. K.Z. carried out most of the experiments, analyzed the data, and wrote the manuscript. S.Z. and A.X. acquired, analyzed, and interpreted data and drafted the manuscript. L.S., M.L., H.J., and B.Z. were responsible for the concept, analyzed and interpreted data, and performed critical revision of the manuscript. D.Z. and G.F. helped to design and coordinate the experiment.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the fund for Natural Science Foundation of China(grant number 81970051), Excellent Top Talent Cultivation Project of Anhui Higher Education Institutions (gxyqZD2017030), the fund from Reserve Candidate for Anhui Province Academic and Technical Leader and scientific research fund from Anhui medical university. The authors thank the Center for Scientific Research of Anhui Medical University for valuable help in our experiment.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.017.
Contributor Information
Daxiong Zeng, Email: zengdxsz@126.com.
Guanghe Fei, Email: guanghefei@hotmail.com.
Ran Wang, Email: ranwangtjmu@hotmail.com.
Supplemental Information
References
- 1.Hogg J.C., Chu F., Utokaparch S., Woods R., Elliott W.M., Buzatu L., Cherniack R.M., Rogers R.M., Sciurba F.C., Coxson H.O., Paré P.D. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 2.McDonough J.E., Yuan R., Suzuki M., Seyednejad N., Elliott W.M., Sanchez P.G., Wright A.C., Gefter W.B., Litzky L., Coxson H.O. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N. Engl. J. Med. 2011;365:1567–1575. doi: 10.1056/NEJMoa1106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sender R., Fuchs S., Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadava K., Pattaroni C., Sichelstiel A.K., Trompette A., Gollwitzer E.S., Salami O., von Garnier C., Nicod L.P., Marsland B.J. Microbiota Promotes Chronic Pulmonary Inflammation by Enhancing IL-17A and Autoantibodies. Am. J. Respir. Crit. Care Med. 2016;193:975–987. doi: 10.1164/rccm.201504-0779OC. [DOI] [PubMed] [Google Scholar]
- 5.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Chow T.F., Mankaruos M., Scorilas A., Youssef Y., Girgis A., Mossad S., Metias S., Rofael Y., Honey R.J., Stewart R. The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J. Urol. 2010;183:743–751. doi: 10.1016/j.juro.2009.09.086. [DOI] [PubMed] [Google Scholar]
- 8.Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 9.Coulouarn C., Factor V.M., Andersen J.B., Durkin M.E., Thorgeirsson S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manfè V., Biskup E., Rosbjerg A., Kamstrup M., Skov A.G., Lerche C.M., Lauenborg B.T., Odum N., Gniadecki R. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS ONE. 2012;7:e29541. doi: 10.1371/journal.pone.0029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q., Tang Q., Qin D., Yu L., Huang R., Lv G., Zou Z., Jiang X.C., Zou C., Liu W. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol. Cell. Biol. 2015;35:988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Visone R., Pallante P., Vecchione A., Cirombella R., Ferracin M., Ferraro A., Volinia S., Coluzzi S., Leone V., Borbone E. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2016;35:5214. doi: 10.1038/onc.2016.139. [DOI] [PubMed] [Google Scholar]
- 14.Li X., Zhang Y., Zhang Y., Ding J., Wu K., Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C.W., Wang H.W., Chang C.W., Chu H.W., Chen C.Y., Yu J.C., Chao J.I., Liu H.F., Ding S.L., Shen C.Y. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res. Treat. 2012;134:1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 16.Duchman K.R., Gao Y., Miller B.J. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015;39:593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Weaver C.T., Hatton R.D., Mangan P.R., Harrington L.E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 18.Chen K., Kolls J.K. Interluekin-17A (IL17A) Gene. 2017;614:8–14. doi: 10.1016/j.gene.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richmond B.W., Brucker R.M., Han W., Du R.H., Zhang Y., Cheng D.S., Gleaves L., Abdolrasulnia R., Polosukhina D., Clark P.E. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat. Commun. 2016;7:11240. doi: 10.1038/ncomms11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erb-Downward J.R., Thompson D.L., Han M.K., Freeman C.M., McCloskey L., Schmidt L.A., Young V.B., Toews G.B., Curtis J.L., Sundaram B. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sze M.A., Dimitriu P.A., Hayashi S., Elliott W.M., McDonough J.E., Gosselink J.V., Cooper J., Sin D.D., Mohn W.W., Hogg J.C. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012;185:1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre M., Collins M.D. Lactic acid bacteria and human clinical infection. J. Appl. Bacteriol. 1993;75:95–107. doi: 10.1111/j.1365-2672.1993.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q., Tong C., Ma S., Zhou L., Zhao L., Zhao X. Involvement of MicroRNAs in Probiotics-Induced Reduction of the Cecal Inflammation by Salmonella Typhimurium. Front. Immunol. 2017;8:704. doi: 10.3389/fimmu.2017.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Ren F., Zhang H., Jiang L., Hao Y., Luo X. Induction of Regulatory Dendritic Cells by Lactobacillus paracasei L9 Prevents Allergic Sensitization to Bovine β-Lactoglobulin in Mice. J. Microbiol. Biotechnol. 2015;25:1687–1696. doi: 10.4014/jmb.1503.03022. [DOI] [PubMed] [Google Scholar]
- 25.Iseppi R., Messi P., Camellini S., Sabia C. Bacteriocin activity of Lactobacillus brevis and Lactobacillus paracasei ssp. paracasei. J. Med. Microbiol. 2019;68:1359–1366. doi: 10.1099/jmm.0.001045. [DOI] [PubMed] [Google Scholar]
- 26.Trebicka J., Anadol E., Elfimova N., Strack I., Roggendorf M., Viazov S., Wedemeyer I., Drebber U., Rockstroh J., Sauerbruch T. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013;58:234–239. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 27.D’Alessandra Y., Devanna P., Limana F., Straino S., Di Carlo A., Brambilla P.G., Rubino M., Carena M.C., Spazzafumo L., De Simone M. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Su Y., Xu F., Kong J., Yu H., Qian B. Circulating microRNAs in relation to EGFR status and survival of lung adenocarcinoma in female non-smokers. PLoS ONE. 2013;8:e81408. doi: 10.1371/journal.pone.0081408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Zhang P., Chen W., Feng D., Jia Y., Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLoS ONE. 2012;7:e38885. doi: 10.1371/journal.pone.0038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan Q., Zhou Z., Ding S., He J. The miR-30a Negatively Regulates IL-17-Mediated Signal Transduction by Targeting Traf3ip2. J. Interferon Cytokine Res. 2015;35:917–923. doi: 10.1089/jir.2014.0146. [DOI] [PubMed] [Google Scholar]
- 31.Gaffen S.L., Jain R., Garg A.V., Cua D.J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildebrand J.M., Luo Z., Manske M.K., Price-Troska T., Ziesmer S.C., Lin W., Hostager B.S., Slager S.L., Witzig T.E., Ansell S.M. A BAFF-R mutation associated with non-Hodgkin lymphoma alters TRAF recruitment and reveals new insights into BAFF-R signaling. J. Exp. Med. 2010;207:2569–2579. doi: 10.1084/jem.20100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacNee W. Update in chronic obstructive pulmonary disease 2007. Am. J. Respir. Crit. Care Med. 2008;177:820–829. doi: 10.1164/rccm.200801-167UP. [DOI] [PubMed] [Google Scholar]
- 34.Bozinovski S., Vlahos R. Multifaceted Role for IL-17A in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2015;191:1213–1214. doi: 10.1164/rccm.201504-0714ED. [DOI] [PubMed] [Google Scholar]
- 35.Zhou S., Zhu K., Du Y., Jiang H., Li M., Wu P., Xu A., Ding X., Sun L., Cao C. Estrogen administration reduces the risk of pulmonary arterial hypertension by modulating the miR-133a signaling pathways in rats. Gene Ther. 2020;27:113–126. doi: 10.1038/s41434-019-0103-6. [DOI] [PubMed] [Google Scholar]
- 36.Wang R., Zhou S., Wu P., Li M., Ding X., Sun L., Xu X., Zhou X., Zhou L., Cao C., Fei G. Identifying Involvement of H19-miR-675-3p-IGF1R and H19-miR-200a-PDCD4 in Treating Pulmonary Hypertension with Melatonin. Mol. Ther. Nucleic Acids. 2018;13:44–54. doi: 10.1016/j.omtn.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou S., Sun L., Cao C., Wu P., Li M., Sun G., Fei G., Ding X., Wang R. Hypoxia-induced microRNA-26b inhibition contributes to hypoxic pulmonary hypertension via CTGF. J. Cell. Biochem. 2018;119:1942–1952. doi: 10.1002/jcb.26355. [DOI] [PubMed] [Google Scholar]
- 38.Zhou S., Liu Y., Li M., Wu P., Sun G., Fei G., Xu X., Zhou X., Zhou L., Wang R. Combined Effects of PVT1 and MiR-146a Single Nucleotide Polymorphism on the Lung Function of Smokers with Chronic Obstructive Pulmonary Disease. Int. J. Biol. Sci. 2018;14:1153–1162. doi: 10.7150/ijbs.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan D.A., Willger S.D., Dolben E.L., Hampton T.H., Stanton B.A., Morrison H.G., Sogin M.L., Czum J., Ashare A. Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease. PLoS ONE. 2016;11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding X., Zhou S., Li M., Cao C., Wu P., Sun L., Fei G., Wang R. Upregulation of SRF Is Associated With Hypoxic Pulmonary Hypertension by Promoting Viability of Smooth Muscle Cells via Increasing Expression of Bcl-2. J. Cell. Biochem. 2017;118:2731–2738. doi: 10.1002/jcb.25922. [DOI] [PubMed] [Google Scholar]
- 41.Wang R., Li M., Zhou S., Zeng D., Xu X., Xu R., Sun G. Effect of a single nucleotide polymorphism in miR-146a on COX-2 protein expression and lung function in smokers with chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2015;10:463–473. doi: 10.2147/COPD.S74345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.