Abstract
Monitoring of pathogen resistance profiles is necessary to guide empirical antibiotic therapy before culture and sensitivity results become available. The aim of this study was to describe current antibiotic resistance patterns of five most frequent causative uropathogens in a Department of Urology of a tertiary referral centre in Central Europe over a period of nine years. The Hospital Department of Clinical Microbiology database was used to extract data on all positive urine samples from inpatients in the Department of Urology between 2011 and 2019. Numbers of susceptible and resistant isolates per year were calculated for five most frequent uropathogens: Escherichia coli, Enterococcus spp., Klebsiella spp., Pseudomonas aeruginosa, and Proteus spp. Antimicrobial agents selected for the survey included: ampicillin, amoxicillin/clavulanic acid, piperacillin/tazobactam; cefuroxime, cefotaxime, ceftazidime and cefepime; ciprofloxacin and ofloxacin; gentamicin and amikacin; ertapenem, meropenem and imipenem; trimethoprim-sulfamethoxazole (co-trimoxazole), nitrofurantoin, colistin, and vancomycin. High resistance rates of Gram-negative uropathogens were demonstrated to most common antimicrobials, with statistically significant increasing or decreasing trends in some cases. No carbapenem-resistant Enterobacteriaceae were isolated. Vancomycin-resistant Enterococcus spp. strains were rare in our population.
Keywords: antibiotics, Enterobateriaceae, resistance, urinary tract infection, E. coli, Klebsiella, Proteus, Pseudomonas, Enterococcus
1. Introduction
Less than a century after Sir Alexander Flemming’s discovery of Penicillinum notatum, the growing antimicrobial resistance (AMR) of most clinically relevant bacteria to antimicrobial agents represents a major challenge to the public health. AMR is caused by a mixture of factors, from improper antibiotics use in health care, to their unrestrained use in agriculture [1]. AMR has become a widespread phenomenon (albeit with important spatial and temporal differences) occurring in all parts of the world, including Europe [2,3,4], the Americas [5,6,7], Africa [8], and Australasia [9,10,11]. In the United States, more than 2.8 million people acquire an antibiotic-resistant infection every year, resulting in more than 35,000 deaths; the corresponding figure for Europe is 33,000 annually [12]. Besides the associated morbidity and mortality, infections by multi-drug-resistant pathogens increase the pressure on heath care systems, too: An episode of a carbapenem-resistant Enterobacteriaceae urinary tract infection has been estimated to cost the health care system 66,000 USD [13].
Urinary tract infections (UTIs) belong among the most common types of infectious disease, accounting for approximately 150–250 million cases globally per year [14]. They are usually caused by the host’s endogenous microbial flora and Gram-negative enteric rods, such as E. coli, Klebsiella, Proteus etc., are their most frequent etiological agents [15]. Approximately 50% of women acquire a UTI at least once in their lifetime and recurrences are common [15]. UTIs are therefore an economic problem, too: The costs of treatment of adult female UTIs were estimated at 58 million euro in France alone in 2012 [16]. A complicated UTI (one associated with structural or functional abnormalities of the urinary tract, immunocompromised host, virulent microorganism, etc.) [15] may cost the health care system between 4028 and 7740 euro [17].
Antibiotic consumption is a primary driver for AMR, a fact documented on a hospital, regional, and country level [18,19]. A north-to-south gradient in AMR exists in Europe [20], with southern countries having higher rates of antibiotic-resistant UTIs. Perhaps not coincidentally, all four top antibiotic consumers (Greece, Cyprus, France, and Italy) belong among southern European states [21].
Despite the existence of international guidelines, research shows improper antibiotic prescribing is commonplace [22]. According to a survey from the United States, 30% of primary care antibiotic prescriptions were classified as inadequate [23]. Chardavoyne et al. reported appropriate antibiotic treatment in 68% of adult cystitis cases and 46% of pyelonephritis cases [24]. Treatment in the absence of infection is common [24] and although not recommended in international guidelines for lower UTI management, fluoroquinolones are frequently prescribed [25].
In clinical practice of the past, the problem of AMR used to be overcome by the advent of new more potent antimicrobials. This is no longer the case. Many pharmaceutical companies have withdrawn from the field, leaving the antibiotic pipeline virtually empty. There are multiple reasons for this that are out of the scope of this paper (high cost of antibiotic development, availability of inexpensive generic antimicrobials, antibiotic stewardship reducing overall antibiotic use, and regulated use of new antibiotics to maintain their efficiency) [26]. The few antimicrobials under development are mostly derivatives of the major existing antibiotic classes (particularly beta-lactams, beta-lactamase inhibitors, and tetracyclines) and all exhibit some degree of pre-existing cross-resistance [27]. Antimicrobial agents with non-traditional therapeutic targets (virulence factors inactivators, inhibitors of adhesion and biofilm formation, monoclonal antibodies against bacterial exotoxins, quorum-sensing and other communication channels disrupting agents or those countering immune evasion) are being developed but not yet ready for clinical use [28]. The same holds true for alternative substances with antimicrobial effect, such as zinc-oxide nano-particles [29], cell-free probiotic suspensions [30], essential oils [31], or phage therapy [32].
As stated above, flaws in antibiotic prescribing are among the causes of increasing AMR. The knowledge of local and regional antimicrobial susceptibility patterns is one of the ways to improve antibiotic prescription and at least in part to counter the development of AMR. The aim of this study was to describe the antibiotic resistance patterns of the five most frequent causative uropathogens in a Department of Urology of a tertiary referral centre in Central Europe over a period of nine years.
2. Results
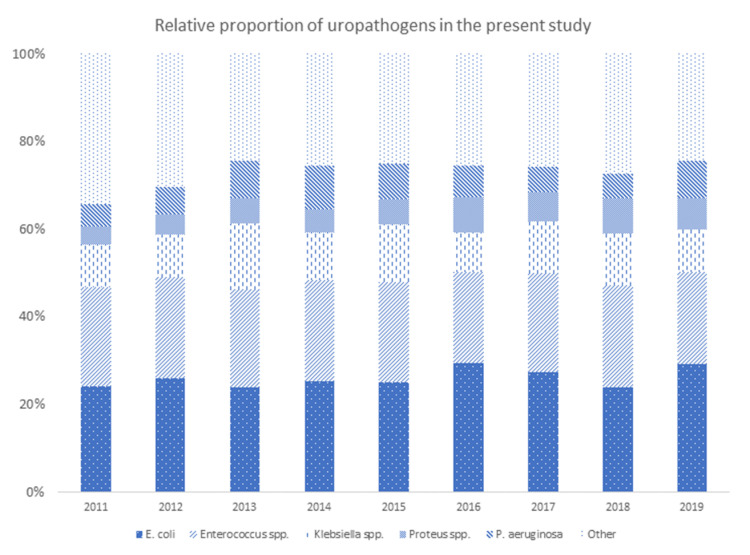
From a total of 25,593 urine cultures from inpatients reported between 1 January 2011 and 31 December 2019, 6897 (26.9%) samples yielded a positive result. Spontaneous mid-stream urine samples represented 49.3% of all positive urine cultures; 33.8% were catheterised urine specimens; 8.9% originated from nephrostomy tubes; and suprapubic catheter, ureteroileostomy, ureteric catheter, and “not specified” represented 1.3%, 4.6%, 1.6%, and 0.4% of all positive urine samples, respectively. E. coli was the most frequent bacteria isolated (26.0%), followed by Enterococcus spp. (22.4%), Klebsiella spp. (11.3%), P. aeruginosa (7.3%), and Proteus spp. (6.2%). The relative prevalence of these microorganisms changed somewhat over time, with E. coli and Proteus spp. showing an increase in prevalence (p = 0.049 and p < 0.0001, respectively) (Figure 1). Table 1 outlines the cumulative resistance rates for individual uropathogens and antibiotics, respectively, for the entire study period.
Figure 1.
Spectrum of uropathogens during the study period (relative proportions).
Table 1.
Cumulative resistance rates for individual uropathogens and antibiotics, respectively, 2011–2019. N, total number of isolates; n, number of resistant isolates; NT, not tested. Percentages represent the rate of resistant isolates.
| Uropathogen | Antimicrobial Agent | |||||||
|---|---|---|---|---|---|---|---|---|
| Ampicillin | Amoxicillin/clavulanic acid | Piperacillin/tazobactam | ||||||
| n/N | n/N | n/N | ||||||
| E. coli | 641/1149 | 55.8% | 163/1138 | 14.3% | 50/358 | 14.0% | ||
| Klebsiella spp. | 522/522 | 100.0% | 191/492 | 38.8% | 129/245 | 52.7% | ||
| P. aeruginosa | NT | NT | 55/180 | 30.6% | ||||
| Proteus spp. | 138/241 | 57.3% | 30/239 | 12.6% | 1/113 | 0.9% | ||
| Enterococcus spp. | 80/756 | 10.6% | NT | NT | ||||
| Cefuroxime | Cefotaxime | Ceftazidime | Cefepime | |||||
| n/N | n/N | n/N | n/N | |||||
| E. coli | 143/1143 | 12.5% | 108/1135 | 9.5% | 100/1131 | 8.8% | 86/367 | 23.4% |
| Klebsiella spp. | 219/523 | 41.9% | 173/505 | 34.3% | 180/503 | 35.8% | 151/260 | 58.1% |
| P. aeruginosa | NT | NT | 47/252 | 18.7% | 56/178 | 31.5% | ||
| Proteus spp. | 38/242 | 15.7% | 14/240 | 5.8% | 7/240 | 2.9% | 8/109 | 7.3% |
| Enterococcus spp. | NT | NT | NT | NT | ||||
| Ciprofloxacin | Ofloxin | |||||||
| n/N | n/N | |||||||
| E. coli | 161/382 | 42.1% | 267/1015 | 26.3% | ||||
| Klebsiella spp. | 191/278 | 68.7% | 183/460 | 39.8% | ||||
| P. aeruginosa | 99/260 | 38.1% | 12/12 | 100.0% | ||||
| Proteus spp. | 55/113 | 48.7% | 107/210 | 51.0% | ||||
| Enterococcus spp. | NT | NT | ||||||
| Gentamicin | Amikacin | |||||||
| n/N | n/N | |||||||
| E. coli | 82/1149 | 7.1% | 5/381 | 1.3% | ||||
| Klebsiella spp. | 170/521 | 32.6% | 7/278 | 2.5% | ||||
| P. aeruginosa | 81/258 | 31.4% | 24/258 | 9.3% | ||||
| Proteus spp. | 70/242 | 28.9% | 0/112 | 0.0% | ||||
| Enterococcus spp. | NT | NT | ||||||
| Ertapenem | Meropenem | Imipenem | ||||||
| n/N | n/N | n/N | ||||||
| E. coli | 1/377 | 0.3% | 0/388 | 0.0% | 0/385 | 0.0% | ||
| Klebsiella spp. | 9/276 | 3.3% | 0/283 | 0.0% | 0/282 | 0.0% | ||
| P. aeruginosa | NT | 54/170 | 31.8% | 21/135 | 15.6% | |||
| Proteus spp. | 0/112 | 0.0% | 0/114 | 0.0% | 1/114 | 0.9% | ||
| Enterococcus spp. | NT | NT | NT | |||||
| Vancomycin | Colistin | Nitrofurantoin | Cotrimoxazole | |||||
| n/N | n/N | n/N | n/N | |||||
| E. coli | NT | 1/123 | 0.8% | 47/973 | 4.8% | 398/1150 | 34.6% | |
| Klebsiella spp. | NT | 2/100 | 2.0% | 144/313 | 46.0% | 248/522 | 47.5% | |
| P. aeruginosa | NT | 0/260 | 0.0% | NT | NT | |||
| Proteus spp. | NT | NT | 173/173 | 100.0% | 162/242 | 66.9% | ||
| Enterococcus spp. | 8/796 | 1.0% | NT | 34/711 | 4.8% | NT | ||
2.1. Penicillin Derivatives
E. coli and Proteus spp. resistance rates for ampicillin exceeded 50%, precluding its empirical use for the treatment of UTIs. Enterococcus spp. resistance to ampicillin more than doubled, from 7.2% to 16.3% (p for trend 0.013). Resistance to amoxicillin/clavulanic acid was above 10% for E. coli and Proteus spp. in most study years and ranged between 20.4% and 58.9% for Klebsiella spp. with a significant increasing trend (p = 0.041). Resistance rates for all Gram-negatives in the study decreased significantly for piperacillin/tazobactam during the study period (Supplementary Material Table S1). Supplementary Material Figure S1a–e show statistically significant trends in antimicrobial resistance for each of the five uropathogens covered in this survey.
2.2. Cephalosporines
Among cephalosporines, cefuroxime resistance rates fluctuated above 10% and 15% for E. coli and Proteus spp., respectively. Proteus spp. showed a significant decrease in resistance to cefuroxime (p = 0.012). Cefotaxime and ceftazidime had a slightly more favourable resistance profile for E. coli and Proteus spp. (<10% in most of the study period) than cefuroxime. Over 30% of the Klebsiella spp. strains tested were resistant to cefuroxime, cefotaxime, and ceftazidime. Cefepime resistance rates were above 25%, 50%, and 30% for E. coli, Klebsiella spp., and P. aeruginosa, respectively, in most study years despite a significant decreasing trend (Supplementary Material Table S2).
2.3. Fluoroquinolones
Ciprofloxacin susceptibility was tested on Gram-negatives only; resistance rates fluctuated above 40% but showed a consistently decreasing trends for all of them except Proteus spp. (Supplementary Material Table S3). Resistance to ofloxacin exceeded 25% in most examined years for E. coli, 35% for Klebsiella spp., and 40% for Proteus spp., with an increasing trend for the latter two.
2.4. Aminoglycosides
The E. coli resistance rate to gentamicin did not exceed 10% (with one year’s exception) but was above 30% for all other Gram-negatives surveyed. Amikacin showed a better resistance profile with >95% susceptible isolates of E. coli, Klebsiella spp., and Proteus spp.
The P. aeruginosa resistance rate against amikacin was 9% compared to 31% for gentamicin. Both exhibited decreasing trends in resistance (p = 0.046 and p = 0.006 for gentamicin and amikacin, respectively; see Supplementary Material Table S4 for details).
2.5. Carbapenems
E. coli, Klebsiella spp., and Proteus spp. isolates were all susceptible to ertapenem, meropenem, and imipenem except one E. coli and nine Klebsiella spp. strains resistant to ertapenem during the entire study period. P. aeruginosa showed a >20% resistance rate for both meropenem and imipenem in most study years despite a decreasing trend in the case of meropenem (p = 0.002) (Supplementary Material Table S5).
2.6. Co-Trimoxazole and Nitrofurantoin
Co-trimoxazole resistance for all Gram-negatives far exceeded 30% without any demonstrable trend. E. coli and the only Gram-positive pathogen in the survey, Enterococcus spp., were resistant to nitrofurantoin in less than 10%, and often less than 5% of strains. Klebsiella spp. resistance exceeded 50% and was on the increase up to 83% in 2019 (p for trend 0.002) (Supplementary Material Table S6).
2.7. Vancomycin and Colistin
Enterococcus resistance to vancomycin remained low despite an increasing trend (p = 0.018). All P. aeruginosa isolates were susceptible to colistin. There were three individual isolates of E. coli and Klebsiella spp. displaying colistin resistance (Supplementary Material Table S7).
3. Discussion
In the present study, we report the AMR rates for five major causative agents of UTIs. These data, together with other papers in the literature, complement the picture of AMR worldwide and its trends. The resistance rates of Gram-negative uropathogens to a majority of commonly used antimicrobials in the present study were high, a fact to be expected given the epidemiological situation in the world today [2,3,4,5,6,7,8,9,10,11]. With increasing individual mobility and international travel easier than ever, a global spread of multi-drug-resistant bacterial strains seems an inevitable reality [13].
E. coli resistance to most antimicrobials surveyed in our study approached or exceeded 30% in the case of ampicillin, both fluoroquinolones, cotrimoxazole, and was above 10% for amoxicillin/clavulanate, piperacillin/tazobactam, cefuroxime, and cefepime. Third-generation cephalosporines proved reasonably efficient as well as gentamicin and nitrofurantoin. Resistance rates approached 0% for amikacin, carbapenems, and colistin. In a Hungarian study by Magyar et al., similar ranges of resistance for penicillins, cefuroxime, cefotaxim, aminoglycosides, carbapenems, co-trimoxazole, and nitrofurantoin were reported [33]. Our isolates were, however, twice as resistant to ciprofloxacin and five times more resistant to cefepime. Our data show twice higher resistance rates for ciprofloxacin and cefuroxime than an older survey (1999–2009) from Ireland [4].
During the study period, 8.7% of E. coli strains were reported to be producers of extended-spectrum beta-lactamase (ESBL; yearly incidence 5.4% to 12.3%, no significant trend observed). This compares to 5.4% and 10.5% of ESBL producers in a recent British and Moroccan study, respectively [34,35]. Some authors report excellent rates of susceptibility to nitrofurantoin and fosfomycin among ESBL-producing E. coli [2,34,35,36,37]; these antimicrobials should be used for the treatment of uncomplicated UTIs only. Most inpatients will, however, be classified as having a complicated UTI, and their therapeutic options will be limited to reserve antibiotics, such as carbapenems or amikacin [34].
Alarming rates of 30% to 90% resistance were demonstrated for Klebsiella spp. against all penicillin derivatives, cephalosporins, fluoroquinolones, co-trimoxazole, nitrofurantoin, and gentamicin. Amikacin and carbapenems seem the only suitable antimicrobials that should be considered for empirical treatment of complicated UTIs caused by Klebsiella spp. in our patient population. Resistance rates reported in the Hungarian study are essentially similar [33]. Fajfr et al. reported comparable resistance rates for cefuroxime and penicillin/beta-lactamase inhibitor combinations as we do and “only” 47% of Klebsiella strains were resistant to ciprofloxacin [37]. This may be because their study included outpatients whose bacterial isolates are presumed to be generally more susceptible.
Klebsiella spp. is the genus with the highest proportion of ESBL-producing isolates [35]. In fact, 33.9% of Klebsiella spp. strains in our study were ESBL producers (between 15.4% and 52.3% in the study period; no significant trend demonstrated). This compares to 18.6% of ESBL-producing Klebsiella isolates from inpatients in a British report [34] and 25.8% within a mixed population of in- and outpatients in a Moroccan study [35]. Intriguingly, a study of urine cultures from 14 urology services in the Netherlands covering the decade 1998–2009 reported a much more favourable Klebsiella spp. antimicrobial susceptibility pattern with resistance rates one order of magnitude lower (piperacillin/tazobactam, cefepime, cefotaxime, ciprofloxacin, gentamicin) than our and other authors’ data [38].
Proteus spp. is a typical nosocomial pathogen that has been isolated particularly from patients with complicated UTIs. Proteae (a tribe including Proteus, Morganella spp., and Providentia spp.) are intrinsically resistant to nitrofurantoin, colistin, and have decreased susceptibility to imipenem [39]. In our patient population, Proteus spp. retained susceptibility to piperacillin/tazobactam, third- and fourth-generation cephalosporins, amikacin, and carbapenems. Resistance rates for all other antibiotics make them unsuitable for empirical treatment of Proteus-caused UTIs.
P. aeruginosa belongs to the so-called ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.) [11] that have been associated with serious health care-associated infections worldwide and frequently display a multi-drug-resistant phenotype. It is intrinsically resistant to many antibiotics due to complementary mechanisms, including low outer membrane permeability, AmpC-beta-lactamase production, and the production of several efflux systems. In addition, it can acquire other resistance determinants, such as beta-lactamases and carbapenemases [40], and its survival is enhanced by biofilm formation [41]. In the present study, P. aeruginosa displayed resistance rates exceeding 30% for most antimicrobials, including heavy-weight agents, such as piperacillin/tazobactam, cefepime, or meropenem. No isolate was resistant to colistin and a relatively reasonable resistance fluctuating around 10% was demonstrated to amikacin. Magyar et al. reported P. aeruginosa resistance rates roughly 50% lower for most antibiotics where a comparison was possible; good susceptibility to amikacin and colistin was also noted. Furthermore, their group reported a statistically significant decrease in P. aeruginosa resistance to ciprofloxacin (from 38% to 13%, p = 0.025), a trend corroborated by our data (p = 0.009).
Against our own expectations, the data show a statistically significant decrease in resistance to ciprofloxacin for E. coli, Klebsiella spp., and P. aeruginosa in the nine years covered by our study. Nonetheless, their resistance rates remain too high to justify their use for empirical treatment of UTIs. An explanation of this unexpected finding may be a reasonable antibiotic prescribing policy in the department: Intravenous fluoroquinolones are seldom prescribed (never as first-line treatment) and they have not been used for preoperative prophylaxis in the department in the past 15 years. Of note, a warning has recently been issued by the European Medicines Agency discouraging the use of fluoroquinolones [42], and although not strictly forbidden, their administration and prescription should happen on a discernible benefit-to-harm ratio for the patient [43].
Enterococcus spp. resistance to ampicillin increased significantly from 7.2% to 16.3% in the study period (p = 0.013). The resistance rate to nitrofurantoin remained below 5–10% and only several cases of vancomycin-resistant enterococcus (VRE) were detected, mostly in recent years (p for trend 0.018). This compares favourably with a British study, where 40% of Enterococcus spp. isolates showed resistance to nitrofurantoin and a prevalence of VRE was 9.8% [44]. Magyar et al. [33] reported a much lower resistance rate to ampicillin, which may be explained by the inclusion of both E. faecalis and E. faecium in our study, the latter being often ampicillin resistant.
The main limitation of the present study is the impossibility to differentiate community- and hospital-acquired infections as the dates of urine cultures could not be linked to admission and discharge dates of each inpatient episode. It might also be insightful to discriminate urine samples representing asymptomatic bacteriuria as opposed to a clinical UTI, but the nature of our retrospective data would not allow for this. Unfortunately, this trait is common to most other AMR reports in the literature.
The present survey fulfils the recommendation of the European Association of Urology guidelines [45] to monitor local resistance patterns of common uropathogens so that empirical antibiotic treatment can be tailored to the local epidemiological situation. Ideally, these collected data should be gathered in a broader survey, such as the global prevalence of infections in urology (GPIU) [46] study, that allows for comparisons between countries and institutions and plays an important role in the monitoring of global resistance trends.
Our data suggest only a handful of options exist for the treatment of patients with a complicated UTI. In everyday practice, however, we do keep in mind the threat of AMR and avoid the empirical use of carbapenems and piperacilin/tazobactam despite their favourable resistance profiles seen on our data. Empirical treatment of newly admitted UTI patients starts with a third-generation cephalosporine +/− gentamicin. Higher-class antimicrobials are reserved for culture-proven non-susceptibility to standard antibiotic regimens and only rarely used without a culture in clinically deteriorating patients.
4. Materials and Methods
The Department of Clinical Microbiology electronic database was searched and all urinary cultures between January 2011 and December 2019 were reviewed. These included midstream urine cultures, urine sampled during theatre procedures, from indwelling catheters, suprapubic catheters, nephrostomy tubes, and urine samples from uretero-ileostomies. Only inpatient samples were considered for analysis. Duplicates were excluded, allowing only one isolate of a given pathogen per patient per year. Prevalence of major uropathogenic organisms and their antimicrobial susceptibility patterns were analysed.
Antimicrobial resistance patterns for five most common uropathogens are reported here: Escherichia coli (E. coli), including subspecies E. coli haemolytica, Enterococcus spp. (including both E. faecalis and E. faecium), Klebsiella spp., Pseudomonas aeruginosa (P. aeruginosa), and Proteus spp. (including P. mirabilis, P. vulgaris, and other species).
Culture Methods and Susceptibility Testing
Urine samples were processed by a semi-quantitative dilution method. Uncentrifuged urine was inoculated with a 0.01-mL loop on blood and UriSelect chromogenic agar (Bio-Rad, Berkeley, CA, USA). Diluted urine 1:10 in saline was inoculated with a 0.01-mL and 0.001-mL loop on Columbia blood agar (Bio-Rad, Berkeley, CA, USA) and UriSelect chromogenic agar. Agar plates were incubated at 37 °C for 20–24 h. Detected pathogens in significant amounts were identified according to phenotypical characteristics (growth properties, biochemical tests) and using the semi-automatic system MIKROLATEST ID (Erba-Lachema, Brno, Czech Republic). Antibiotic susceptibility testing was performed by the disc diffusion method on Mueller-Hinton agar plates (Bio-Rad, Berkeley, CA, USA) and the MIC dilution method (TRIOS MIC, Prague, Czech Republic until 2017, then MICRO-LA-TEST ATB (MIC) Erba-Lachema, Brno, Czech Republic). Results were evaluated according to EUCAST MIC breakpoint tables. Intermediate results (newly termed “susceptible, increased exposure”) were excluded from analysis.
The following antimicrobial agents were selected for the survey: ampicillin, amoxicillin/clavulanate and piperacillin/tazobactam to include an unprotected penicillin, a penicillin-beta-lactamase inhibitor combination and an antipseudomonad penicillin with a beta-lactamase inhibitor; cefuroxime, cefotaxime (2nd and 3rd generation cephalosporine, respectively), ceftazidime and cefepime (3rd generation with antipseudomonad coverage and 4th generation, respectively); ciprofloxacin and ofloxacin to represent fluoroquinolones, gentamicin and amikacin for aminoglycosides; the carbamenems ertapenem, meropenem, and imipenem; and trimethoprim-sulfamethoxazole (co-trimoxazole), nitrofurantoin, colistin, and vancomycin.
The Cochrane-Armitage test was used to assess the statistical significance of trends. Statistical analyses were performed in XLSTAT 2020.1.3 (Addinsoft, New York, NY, USA). Alpha level of 0.05 was considered significant. Ethical approval was waived for this retrospective analysis of anonymised data.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/9/630/s1. Table S1. Resistance rates against penicillin derivatives. The table shows resistance rates against ampicillin, amoxicillin/clavulanic acid and piperacillin/tazobactam 2011–2019. Klebsiella spp. is inherently resistant to ampicillin. N, total number of isolates; n, number of resistant isolates; NS, trend not significant. Percentages represent the rate of resistant isolates * Note small number of isolates in some or all years when interpreting statistical significance. †) Statistical significance of trend is retained when only 4 consecutive years with N > 30 are included in analysis. Table S2. Resistance rates against cephalosporines. The table shows resistance rates against cefuroxime, cefotaxime, ceftazidime and cefepime 2011–2019. * Note small number of isolates in some or all years when interpreting statistical significance. ‡) Statistical significance of trend is lost when only 4 consecutive years with N > 30 are included in analysis. Table S3. Resistance rates against fluoroquinolones. The table shows resistance rates against ciprofloxacin and ofloxacin 2011–2019. * Note small number of isolates in some or all years when interpreting statistical significance. †) Statistical significance of trend is retained when only 4 consecutive years with N > 30 are included in analysis. Table S4. Resistance rates against aminoglycosides. The table shows resistance rates against gentamicin and amikacin 2011–2019. * Note small number of isolates in some or all years when interpreting statistical significance. Table S5. Resistance rates against carbapenems. The table shows resistance rates against ertapenem, meropenem and imipenem 2011–2019. * Note small number of isolates in some or all years when interpreting statistical significance. Table S6. Resistance rates against cotrimoxazol and nitrofurantoin. The table shows resistance rates against cotrimoxazol and nitrofurantoin 2011–2019. Proteus spp. is inherently resistant to nitrofurantoin. * Note small number of isolates in some or all years when interpreting statistical significance. ‡) Statistical significance of trend is lost when only 5 consecutive years with N > 30 are included in analysis. Table S7. Resistance rates against vancomycin and colistin. The table shows resistance rates against vancomycin and colistin 2011–2019. Figure S1a–e: Statistically significant trends in antimicrobial resistance 2011–2019 for E. coli, Klebsiella spp., Proteus spp., P. aeruginosa and Enterococcus spp., respectively. The figures should be interpreted along with the Supplementary Material Tables S1–S7. AMP ampicillin, AMC amoxicillin/clavulanic acid, PPT piperacillin/tazobactam, CRX cefuroxime, CPM cefepime, CIP ciprofloxacin, OFL ofloxacin, GEN gentamicin, AMI amikacin, MER meropenem, NTF nitrofurantoin, VAN vancomycin.
Author Contributions
J.H.: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Visualization, Writing—original draft, Writing—review and editing. P.C.: Resources, Validation, Writing—review and editing. R.Z.: Data curation, Supervision, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Manyi-Loh C., Mamphweli S., Meyer E., Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules. 2018;23:795. doi: 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonkat G., Müller G., Braissant O., Frei R., Tschudin-Suter S., Rieken M., Wyler S., Gasser T.C., Bachmann A., Widmer A.F. Increasing prevalence of ciprofloxacin resistance in extended-spectrum-beta-lactamase-producing Escherichia coli urinary isolates. World J. Urol. 2013;31:1427–1432. doi: 10.1007/s00345-013-1031-5. [DOI] [PubMed] [Google Scholar]
- 3.Iacchini S., Sabbatucci M., Gagliotti C., Rossolini G.M., Moro M.L., Iannazzo S., D’Ancona F., Pezzotti P., Pantosti A. Bloodstream infections due to carbapenemas eproducing Enterobacteriaceae in Italy: Results from nationwide surveillance, 2014 to 2017. Eurosurveillance. 2019;24:1800159. doi: 10.2807/1560-7917.ES.2019.24.5.1800159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen I.M., Manecksha R.P., Mccullagh E., Ahmad S., O’Kelly F., Flynn R.J., McDermott T., Murphy P., Grainger R., Fennell J.P., et al. The changing pattern of antimicrobial resistance within 42 033 Escherichia coli isolates from nosocomial, community and urology patient-specific urinary tract infections, Dublin, 1999–2009. BJUI Int. 2011;109:1198–1206. doi: 10.1111/j.1464-410X.2011.10528.x. [DOI] [PubMed] [Google Scholar]
- 5.Lob S.H., Nicolle L.E., Hoban D.J., Kazmierczak K.M., Badal R.E., Sahm D.F. Susceptibility patterns and ESBL rates of Escherichia coli from urinary tract infections in Canada and the United States, SMART 2010–2014. Diagn. Microbiol. Infect. Dis. 2016;85:459–465. doi: 10.1016/j.diagmicrobio.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Guyomard-Rabenirina S., The Laboratory Working Group. Malespine J., Ducat C., Sadikalay S., Falord M., Harrois D., Richard V., Dozois C., Breurec S., et al. Temporal trends and risks factors for antimicrobial resistant Enterobacteriaceae urinary isolates from outpatients in Guadeloupe. BMC Microbiol. 2016;16:121. doi: 10.1186/s12866-016-0749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sierra-Díaz E., Hernández-ríos C.J., Bravo-cuellar A. Antibiotic resistance: Microbiological profile of urinary tract infections in Mexico. Cir. Cir. 2019;87:176–182. doi: 10.24875/CIRU.18000494. [DOI] [PubMed] [Google Scholar]
- 8.Okeke I.N., Laxminarayan R., Bhutta Z.A., Duse A.G., Jenkins P., O’Brien T.F., Pablos-Mendez A., Klugman K.P. AMR Resistance in developing countries. Lancet Infect. Dis. 2005;5:481–493. doi: 10.1016/S1473-3099(05)70189-4. [DOI] [PubMed] [Google Scholar]
- 9.Fasugba O., Mitchell B.G., Mnatzaganian G., Das A., Collignon P., Gardner A. Five-Year Antimicrobial Resistance Patterns of Urinary Escherichia coli at an Australian Tertiary Hospital: Time Series Analyses of Prevalence Data. PLoS ONE. 2016;11:e0164306. doi: 10.1371/journal.pone.0164306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyun M., Noh C.I., Ryu S.Y., Kim H.A. Changing trends in clinical characteristics and antibiotic susceptibility of Klebsiella pneumoniae bacteremia. Korean J. Intern. Med. 2018;33:595–603. doi: 10.3904/kjim.2015.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlowsky J.A., Hoban D.J., Hackel M.A., Lob S.H., Sahm D.F. Antimicrobial susceptibility of Gram-negative ESKAPE pathogens isolated from hospitalized patients with intra-abdominal and urinary tract infections in Asia-Pacific countries: SMART 2013–2015. J. Med. Microbiol. 2017;66:61–69. doi: 10.1099/jmm.0.000421. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization . 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline. WHO; Geneva, Switzerland: 2019. [(accessed on 24 April 2020)]. Available online: https://www.who.int/medicines/areas/rational_use/antibacterial_agents_clinical_development/en/ [Google Scholar]
- 13.Frost I., Van Boeckel T.P., Pires J., Craig J., Laxminarayan R. Global geographic trends in antimicrobial resistance: The role of international travel. J. Travel Med. 2019;26:taz036. doi: 10.1093/jtm/taz036. [DOI] [PubMed] [Google Scholar]
- 14.Zowawi H.M., Harris P.N.A., Roberts M.J., Tambyah P.A., Schembri M.A., Pezzani M.D., Williamson D.A., Paterson D. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat. Rev. Urol. 2015;12:570–580. doi: 10.1038/nrurol.2015.199. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer A., Schaeffer E. Infections of the urinary tract. In: Wein A., Kavoussi L., editors. Campbell-Walsh Urology. 10th ed. Elsevier Saunders; Philadelphia, PA, USA: 2012. pp. 258–260. [Google Scholar]
- 16.François M., Hanslik T., Dervaux B., Le Strat Y., Souty C., Vaux S., Maugat S., Rondet C., Sarazin M., Heym B., et al. The economic burden of urinary tract infections in women visiting general practices in France: A cross-sectional survey. BMC Health Serv. Res. 2016;16:365. doi: 10.1186/s12913-016-1620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallejo-Torres L., Pujol M., Shaw E., Wiegand I., Vigo J.M., Stoddart M., Grier S., Gibbs J., Vank C., Cuperus N., et al. Cost of hospitalised patients due to complicated urinary tract infections: A retrospective observational study in countries with high prevalence of multidrug-resistant Gram-negative bacteria: The COMBACTE-MAGNET, RESCUING study. BMJ Open. 2018;8:e020251. doi: 10.1136/bmjopen-2017-020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stapleton P.J., Lundon D.J., McWade R., Scanlon N., Hannan M.M., O’Kelly F., Lynch M. Antibiotic resistance patterns of Escherichia coli urinary isolates and comparison with antibiotic consumption data over 10 years, 2005–2014. Ir. J. Med. Sci. 2017;186:733–741. doi: 10.1007/s11845-016-1538-z. [DOI] [PubMed] [Google Scholar]
- 19.Klein E.Y., Van Boeckel T.P., Martinez E., Pant S., Gandra S., Levin S.A., Goossens H., Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA. 2018;115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandil H., Cramp E., Vaghela T. Trends in Antibiotic Resistance in Urologic Practice. Eur. Urol. Focus. 2016;2:363–373. doi: 10.1016/j.euf.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Adriaenssens N., Coenen S., Versporten A., Muller A., Minalu G., Faes C., Vankerckhoven V., Aerts M., Hens N., Molenberghs G., et al. European Surveillance of Antimicrobial Consumption (ESAC): Outpatient antibiotic use in Europe (1997–2009) J. Antimicrob. Chemother. 2011;66:3–12. doi: 10.1093/jac/dkr190. [DOI] [PubMed] [Google Scholar]
- 22.Durkin M.J., Keller M., Butler A.M., Kwon J.H., Dubberke E.R., Miller A.C., Polgreen P.M., Olsen M.A. An assessment of inappropriate antibiotic use and guideline adherence for uncomplicated urinary tract infections. Open Forum Infect. Dis. 2018;5:ofy198. doi: 10.1093/ofid/ofy198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming-Dutra K., Hersh A.L., Shapiro D.J. Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010–2011. JAMA. 2016;315:1865–1873. doi: 10.1001/jama.2016.4151. [DOI] [PubMed] [Google Scholar]
- 24.Chardavoyne P.C., Kasmire K.E. Appropriateness of Antibiotic Prescriptions for Urinary Tract Infections. West. J. Emerg. Med. 2020;21:633–639. doi: 10.5811/westjem.2020.1.45944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai T., Palagin I., Brunelli R., Cipelli R., Pellini E., Truzzi J.C., Van Bruwaene S. Office-based approach to urinary tract infections in 50,000 patients: Results from the REWIND study. Int. J. Antimicrob. Agents. 2020;56:105966. doi: 10.1016/j.ijantimicag.2020.105966. [DOI] [PubMed] [Google Scholar]
- 26.Luepke K.H., Luepke K.H., Boucher H., Russo R.L., Bonney M.W., Hunt T.D., Mohr J.F. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacotherapy. 2017;37:71–84. doi: 10.1002/phar.1868. [DOI] [PubMed] [Google Scholar]
- 27.Theuretzbacher U., Bush K., Harbarth S., Paul M., Rex J.H., Tacconelli E., Thwaites G.E. Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 2020;18:286–298. doi: 10.1038/s41579-020-0340-0. [DOI] [PubMed] [Google Scholar]
- 28.Theuretzbacher U., Piddock L.J.V. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe. 2019;26:61–72. doi: 10.1016/j.chom.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Tiwari V., Mishra N., Gadani K., Solanki P.S., Shah N.A., Tiwari M. Mechanism of anti-bacterial activity of zinc oxide nanoparticle against Carbapenem-Resistant Acinetobacter baumannii. Front. Microbiol. 2018;9:1218. doi: 10.3389/fmicb.2018.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelhamid A.G., Esaam A., Hazaa M.M. Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm. J. 2018;26:603–607. doi: 10.1016/j.jsps.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iseppi R., Di Cerbo A., Aloisi P., Manelli M., Pellesi V., Provenzano C., Camellini S., Messi P., Sabia C. In vitro activity of essential oils against planktonic and biofilm cells of extended-spectrum β-lactamase (ESBL)/carbapenamase-producing gram-negative bacteria involved in human nosocomial infections. Antibiotics. 2020;9:272. doi: 10.3390/antibiotics9050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sybesma W., Zbinden R., Chanishvili N., Kutateladze M., Chkhotua A., Ujmajuridze A., Mehnert U., Kessler T.M. Bacteriophages as Potential Treatment for Urinary Tract Infections. Front. Microbiol. 2016;7:465. doi: 10.3389/fmicb.2016.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magyar A., Koves B., Nagy K., Dobák A., Arthanareeswaran V.K.A., Bálint P., Wagenlehner F., Tenke P. Spectrum and antibiotic resistance of uropathogens between 2004 and 2015 in a tertiary care hospital in Hungary. J. Med. Microbiol. 2017;66:788–797. doi: 10.1099/jmm.0.000498. [DOI] [PubMed] [Google Scholar]
- 34.Toner L., Papa N., Aliyu S.H., Dev H., Lawrentschuk N., Al-Hayek S. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in hospital urinary tract infections: Incidence and antibiotic susceptibility profile over 9 years. World J. Urol. 2016;34:1031–1037. doi: 10.1007/s00345-015-1718-x. [DOI] [PubMed] [Google Scholar]
- 35.Sbiti M., Lahmadi K., Louzi L. Profil épidémiologique des entérobactéries uropathogènes productrices de bêta-lactamases à spectre élargi. Pan Afr. Med. J. 2017;28:29. doi: 10.11604/pamj.2017.28.29.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho Y.H., Jung S.I., Chung H.S., Yu H.S., Hwang E.C., Kim S.-O., Kang T.W., Kwon D.D., Park K. Antimicrobial susceptibilities of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in health care-associated urinary tract infection: Focus on susceptibility to fosfomycin. Int. Urol. Nephrol. 2015;47:1059–1066. doi: 10.1007/s11255-015-1018-9. [DOI] [PubMed] [Google Scholar]
- 37.Fajfr M., Louda M., Paterová P., Ryskova L., Pacovský J., Košina J., Žemličková H., Brodak M. The susceptibility to fosfomycin of Gram-negative bacteria isolates from urinary tract infection in the Czech Republic: Data from a unicentric study. BMC Urol. 2017;17:33. doi: 10.1186/s12894-017-0222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Donk C.F.M., Beisser P.S., Hoogkamp-Korstanje J.A.A., Bruggeman C.A., Stobberingh E.E. A 12 year (1998–2009) antibiotic resistance surveillance of Klebsiella pneumoniae collected from intensive care and urology patients in 14 Dutch hospitals. J. Antimicrob. Chemother. 2011;66:855–858. doi: 10.1093/jac/dkq538. [DOI] [PubMed] [Google Scholar]
- 39.Gajdács M., Urbán E. Comparative Epidemiology and Resistance Trends of Proteae in Urinary Tract Infections of Inpatients and Outpatients: A 10-Year Retrospective Study. Antibiotics. 2019;8:91. doi: 10.3390/antibiotics8030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupo A., Haenni M., Madec J.-Y. Antimicrobial Resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.ARBA-0007-2017. [DOI] [PubMed] [Google Scholar]
- 41.Tolker-Nielsen T.I.M., Brinch U.C., Ragas P.C., Andersen J.B.O., Jacobsen C.S., Molin S. Development and Dynamics of Pseudomonas sp. Biofilms. J. Bacteriol. 2000;182:6482–6489. doi: 10.1128/JB.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EMEA Disabling and Potentially Permanent Side Effects Lead to Suspension or Restrictions of Quinolone and Fluoroquinolone Antibiotics. [(accessed on 12 April 2020)]; Available online: https://www.ema.europa.eu/en/news/disabling-potentially-permanent-side-effects-lead-suspension-restrictions-quinolone-fluoroquinolone.
- 43.Bonkat G., Wagenlehner F. In the Line of Fire: Should Urologists Stop Prescribing Fluoroquinolones as Default ? Eur. Urol. 2019;75:205–207. doi: 10.1016/j.eururo.2018.10.057. [DOI] [PubMed] [Google Scholar]
- 44.Toner L., Papa N., Aliyu S.H., Dev H., Lawrentschuk N., Al-Hayek S. Vancomycin resistant enterococci in urine cultures: Antibiotic susceptibility trends over a decade at a tertiary hospital in the United Kingdom. Investig. Clin. Urol. 2016;57:129–134. doi: 10.4111/icu.2016.57.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonkat G., Bartoletti R., Bruyère F., Cai T., Geerlings S., Köves B., Schubert S., Wagenlehner F. Urological Infections. [(accessed on 17 September 2020)];2020 Available online: https://uroweb.org/guideline/urological-infections/
- 46.Wagenlehner F.M., Tandoğdu Z., Bartoletti R., Cai T., Cek M., Kulchavenya E.V., Koves B., Naber K., Perepanova T.S., Tenke P., et al. The global prevalence of infections in urology study: A long-term, worldwide surveillance study on urological infections. Pathogens. 2016;5:10. doi: 10.3390/pathogens5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.