Abstract
The basic leucine zipper transcription factor ATF5 is over-expressed in many tumor types and interference with its expression or function inhibits cancer cell survival. As a potential therapeutic approach to exploit these findings, we created dominant-negative (DN) ATF5 forms lacking DNA-binding ability that retain the ATF5 leucine zipper, and thus associate with and sequester ATF5’s requisite leucine-zipper binding partners. Preclinical studies with DN-ATF5, including a cell-penetrating form, show in vitro and in vivo efficacy in compromising cancer cell survival. However, DN-ATF5’s targets, and particularly those required for tumor cell survival have been unknown. We report that cells lacking ATF5 succumb to DN-ATF5, indicating that ATF5 itself is not DN-ATF5’s obligate target. Unbiased pulldown assays coupled with mass spectrometry and immunoblotting revealed that DN-ATF5 associates in cells with the basic leucine zipper proteins CEBPB and CEBPD and coiled-coil protein CCDC6. Consistent DN-ATF5 affecting tumor cell survival by suppressing CEBPB and CEBPD function, DN-ATF5 interferes with CEBPB and CEBPD transcriptional activity, while CEBPB or CEBPD knockdown promotes apoptotic death of multiple cancer cells lines, but not of normal astrocytes. We propose a two-pronged mechanism by which DN-ATF5 kills tumor cells. One is by inhibiting heterodimer formation between ATF5 and CEBPB and CDBPD, thus suppressing ATF5-dependent transcription. The other is by blocking formation of transcriptionally active CEBPB and CEBPD homodimers as well as heterodimers with partners in addition to ATF5.
Implications:
This study indicates that the potential cancer therapeutic DN-ATF5 acts by associating with and blocking the transcriptional activities of CEBPB and CEBPD.
Introduction
Dominant-negative (DN) proteins are mutated forms that lack the activities of their wild-type counterparts, but that retain the capacity to associate with the latters’ substrates or binding partners (1). Consequently, over-expressed DN proteins can disrupt the activities of their normal counterparts as well as that of their interacting partners. For this reason, a variety of strategies employing dominant-negative proteins have been proposed as possible cancer therapies (2).
One particularly promising use of the DN approach is for the basic leucine zipper (bZip) transcription factor ATF5 (3). Based on findings that ATF5 over-expression promotes cell survival and blocks differentiation and cell-cycle exit of neuroprogenitor cells, we created DN forms of this protein to facilitate studies of its functions (4–6). This was initially achieved following the approach described by Vinson and colleagues (7,8) for generation of DN forms of bZip transcription factors in which the ATF5 leucine zipper was left intact to permit association with obligatory binding partners and the DNA binding domain was mutated both to abolish transcriptional regulatory activity and to extend the leucine zipper. The N-terminus of the full-length ATF5 protein was also truncated to enhance stability. Experiments with DN-ATF5 led to the surprising observation that it promoted massive apoptotic death of glioblastoma cells in vitro and in animal models without affecting survival of normal, non-transformed cells (6,9). The idea of targeting cancer cells with DN-ATF5 has been further supported by reports of ATF5 over-expression in a variety of tumor types (3, 6,10–12), negative correlations between ATF5 expression and cancer patient survival (11–13), as well as observations of ATF5-dependent tumor cell aggressiveness (14), invasiveness (15) and therapeutic resistance (15,16). In line with these findings, interference with ATF5 expression or function with si/sh-RNAs or DN-ATF5 constructs interferes with growth and survival of a variety of cancer cell types in vitro and in vivo (6, 9–11,17,18).
To exploit these observations for potential therapeutic purposes, we designed a cell penetrating (CP) form of DN-ATF5 in which further truncations were made at the N and C termini (13,19) and in which the remaining portion (which includes the intact leucine zipper) was fused to a cell penetrating “penetratin” peptide derived from the Drosophila Antennapedia Homeodomain protein (20). CP-DN-ATF5, which is produced as either recombinant or synthetic peptides, rapidly enters cells and passes into various tissues including the brain when delivered subcutaneously or intraperitoneally (13,19). CP-DN-ATF5 peptide causes death of a wide variety of tumor cell types in culture and in mouse models and appears to have no evident side effects or adverse effects on normal cells (13,19). CP-DN forms of ATF5 thus appear to have potential therapeutic benefit.
The efficacy of DN-ATF5 in selectively killing tumor cells has led to the important and hitherto unresolved issue of its molecular targets. In addition to providing mechanistic insight, identifying DN-ATF5’s primary targets could uncover the most appropriate malignancies for its use, rationale combination therapies, and potential side effects. It could also open additional therapeutic strategies targeted to its binding partners.
DN proteins can function to interfere with activity by associating with homomeric and/or heteromeric binding partners. In the case of ATF5, early reports indicating homomeric dimerization (21) led to expectations that a DN form would act by sequestering endogenous ATF5, thereby interfering with its activity. Consistent with this, ATF5 knockdown also kills tumor cells (6,11,18). On the other hand, DN-ATF5 could also function to deprive cellular ATF5 of heteromeric partners that are required for its activity. With respect to these possibilities, resonance energy transfer assays carried out with isolated bZIP domains of a number transcription factors failed to establish a tight homodimerization of the ATF5 bZIP domain, but did suggest a limited number of possible heteromeric interactions (22).
Given the uncertainty about DN-ATF5’s relevant partners in promoting tumor cell death, in this study we used an unbiased approach to identify its cellular targets. This revealed association with the leucine zipper transcription factors CEBPB and CEBPD and the coiled-coil domain protein CCDC6. We further assessed the roles of these identified targets in maintaining tumor cell survival and found that knockdown of CEBPB or CEBPD phenocopied tumor cell death promoted by DN-ATF5.
Materials and Methods
Cells culture and transfection
HAP1 WT, ATF5KO-10 (HZGHC005144c003) AND ATF5KO-1 (HZGHC006497c012) cells were purchased from Horizon Discovery and were authenticated by the supplier, including sequencing to verify the indicated deletions. The deletions were also verified by sequencing after purchase. The lines were grown in IMDM supplemented with 10% FBS. GBM 12 cells were kindly supplied by Dr. Jann Sarkaria (Mayo Clinic, Rochester MN). T98G, LN229, PC3, U87 and MDA-MB-468 cells were purchased from and authenticated by the ATCC. All were grown in DMEM supplemented with 10% FBS and 100U/ml Penicillin-Streptomycin. Human astrocytes and astrocyte growth medium were purchased from Sciencell Research Laboratories and the cells were authenticated by the supplier. All cell lines were used within 10 passages from thawing or purchase and were not otherwise authenticated nor tested for mycoplasma unless indicated. siCEBPB-1, siCEBPB-2, siCEBPD-1, siCEBPD-2, siATF5-1, siATF5-2, siSurvivin (silencer Select siRNAs s2891, s2892, s2894, s2895, s22424, s22425, s21458, respectively) and siCTR (4390843) were from Ambion and were transfected into cells using Oligofectamine™ (Invitrogen) following the supplier’s protocol. All plasmids were transfected using Lipofectamine™ 3000 (Invitrogen) following the supplier’s protocol. Unless otherwise specified, for experiments concerning cell number, survival, or protein/mRNA expression, cells were seeded into 48-well (approximately 1x104 cells/well) or 12-well (approximately 6x105 cells/well) plates. The plates were precoated with a poly-L-lysine (0.1 μg/μl) solution for 2 hours and then air-dried for 10 minutes. CP-DN-ATF5 was either purchased from CS Bio or was the generous gift (as ST-36) of Sapience Therapeutics; ST-47 was the generous gift of Sapience Therapeutics. The peptides were dissolved at 1 mM in a stock buffer of 20 mM Tris (pH 8) and 150 mM NaCl and stored as aliquots at −80C.
Cell counts
For apoptotic cell counting, cultures were fixed with 4% PFA for 10 minutes and then stained with Hoechst 33342 nuclear dye. Total cells and cells with apoptotic nuclei were scored in 3 random fields (totaling 120-250 cells) using a Zeiss epifluorescence microscope equipped with a digital camera and Axiovision software. To determine total cell numbers, cultures were exposed to a lysis buffer (13) and intact nuclei were quantified with a hemocytometer.
Western immunoblot and antibodies
Cells were homogenized in cell lysis buffer (#9803, Cell Signaling Technology) with protease inhibitor mixture (#11836170001, Roche). Protein samples were prepared in x2 loading buffer (#161-0737, BioRad) with β-ME following the manufacturer’s instructions. Protein was loaded on SDS-polyacrylamide gels, resolved by electrophoresis, transferred onto PVDF membranes (Bio-Rad) and probed with the following antisera/antibodies: anti-CEBPB (MABF769, Millipore), anti-CEBPD (PA5-3466, Invitrogen), anti-phospho-CEBPB (Thr235) (#3084, Cell Signaling Technology), anti-FLAG (#8146, Cell Signaling Technology), mouse anti-actin (#3700, Cell Signaling Technology), anti-GFP (#sc-9996, Santa Cruz), anti-Survivin (#2808, Cell signaling), anti-CCDC6 (#13717, Proteintech), anti-CEBPG (ab74045, Abcam). Images were obtained using a KwikQuant Imager or Li-COR Odyssey. Band intensities were determined using ImageJ and normalized to actin as loading control.
Silver stain
Samples were electrophoretically resolved on NuPAGE Bis-Tris protein gels (Invitrogen), which were fixed (50% ethanol, 5% acetic acid) for 2 hours, washed with 50% ethanol 3X for 10 minutes, then washed with ddH2O for 2x 10 minutes and incubated with sensitizer (0.025% sodium dithionite) for 1 minute and washed with ddH2O for 2 minutes. The gels were then incubated with staining solution (0.1% AgNO3, 0.08% formalin) for 30 minutes, washed with ddH2O for 1 minute, treated with developer solution (2% sodium carbonate, 0.04% formalin) for 2-3 minutes, and then terminated by addition of stop solution (5% acetic acid).
Flow cytometry
Cells were dissociated with trypsin, washed twice with cold PBS and resuspended in 1x Binding buffer (BD flow cytometry kit #556547) at a concentration of approximately 1 x 106 cells/ml. 100 μl of the solution were mixed with 5 μl of FITC Annexin V and 5 μl PI (#556547, BD) for 15 minutes at room temperature in the dark. The cell solution was then added to 400 μl of 1x Binding Buffer and analyzed on a BD FACScalibur flow cytometer following the manufacturer’s instructions.
Pulldown assays
Cells (approximately 5x105) were transfected with plasmids, then harvested 3 days later in 450 μl of lysis buffer (20 mM Tris, 200 mM NaCl, 0.2% Triton X100, pH7.4 containing proteasome inhibitor (#11836170001, Roche)), and centrifuged at 12,000 rpm in a microfuge for 15 minutes. The cleared lysates were incubated with 12 μl/sample of Anti-FLAG M2 Magnetic beads (M8823, Sigma-Aldrich) in a tube rotator for 2-3 hours at 4 degrees. For competition with CP-DN-ATF5 peptides, stock solution was added to the lysates to reach the indicated final concentrations during incubation with the beads. Controls received equal volumes of 1 mM BSA in the same stock buffer. After incubation, magnetic beads were collected in a magnetic separator, washed 3x with cell lysis buffer, and FLAG-fusion protein complex was eluted with 40 μl of 3x FLAG peptide (F4799, Millipore). For GFP pulldowns, GFP-Trap Magnetic Agarose (gtma-20, Chromotek) was employed following the manufacturer’s instructions and bound protein was released by boiling in sample buffer for 10 minutes.
Mass spectroscopic analysis
In-gel digestion.
The eluates were separated by 4-12% gradient SDS-PAGE and stained with Simply Blue Protein Stain (Thermo Scientific). In-gel digestion (of the entire gel) was performed as described earlier (23), with minor modifications. The gels were washed with 1:1 acetonitrile:100 mM ammonium bicarbonate (v/v) for 30 minutes, dehydrated with 100% acetonitrile for 10 minutes, dried in a speed-vac for 10 minutes without heat and treated with 5 mM DTT for 30 minutes at 56 °C in an air thermostat and then alkylated with 11 mM iodoacetamide for 30 minutes at room temperature in the dark. Gels were then washed with 100 mM ammonium bicarbonate and 100% acetonitrile for 10 minutes, excess acetonitrile was removed by drying in a speed-vac for 10 minutes without heat, rehydrated in a solution of 25 ng/μl of trypsin in 50 mM ammonium bicarbonate for 30 minutes on ice, and then trypsin digestions was performed overnight at 37 °C. Digested peptides were collected and further extracted in buffer (1:2 5% formic acid/acetonitrile (v/v)) with high-speed shaking. Supernatants were dried in a speed-vac, and peptides were dissolved in a solution containing 3% acetonitrile and 0.1% formic acid and desalted with C18 disk-packed stage-tips.
LC-MS/MS analysis:
Desalted peptides were injected onto an EASY-Spray PepMap RSLC C18 25 cm x 75 μm column (Thermo Scientific), coupled to an Orbitrap Fusion™ Tribrid™ mass spectrometer (Thermo Scientific). Peptides were eluted with a non-linear 70-minute gradient of 2-30% buffer B (0.1% (v/v) formic acid, 100% acetonitrile) at a flow rate of 300 nL/minute. The column temperature was maintained at 50 °C. Survey scans of peptide precursors were performed from 400 to 1500 m/z at 120K FWHM resolution (at 200 m/z) with a 2 x 105 ion count target and a maximum injection time of 50 ms. The instrument was set to run in top speed mode with 3 second cycles for the survey and the MS/MS scans. After a survey scan, tandem MS was performed on the most abundant precursors exhibiting a charge state from 2 to 6 of greater than 5 x 103 intensity by isolating them in the quadrupole at 1.6 Th. CID fragmentation was applied with 35% collision energy and resulting fragments were detected using the rapid scan rate in the ion trap. The AGC target for MS/MS was set to 1 x 104 and the maximum injection time limited to 35 ms. The dynamic exclusion was set to 60 second with a 10 ppm mass tolerance around the precursor and its isotopes. Monoisotopic precursor selection was enabled.
Data analysis:
Raw mass spectrometric data were processed and searched using the Sequest HT search engine within the Proteome Discoverer 2.2 (PD2.2, Thermo) with the Swiss-Prot human database. The default search settings used for protein identification in PD2.2 searching software were as follows: two mis-cleavages for full trypsin with fixed carbamidomethyl modification of cysteine and oxidation of methionine, acetylation on the N-terminal were used as variable modifications. Identified peptides were filtered for a maximum 1% false discovery rate using the Percolator algorithm in PD2.2. The PD2.2 output combined folder was uploaded in Scaffold (Proteome Software) for data visualization. Spectral counting and LFQ intensities were used for analysis to compare the samples.
Plasmids
pLe-GFP and pLe-GFP-Flag-DN (GFP-FLAG fusion ATF5 DN) were generated as previous described (4). 3xFLAG-DN-ATF5, 3XFLAG-DN-ATF5-MUT and 3xFLAG-DN-ATF5- were cloned into pCMV-1A backbones. To generate these constructs, DNA optimized for human codon usage with a 5’- BamHI site and a 3’-XhoI site were synthesized as gBlock fragments (Integrated DNA Technologies Inc) encoding the wild-type or mutant DN-ATF5 sequences. The DN-ATF5 sequences are as follows: DN-ATF5, LEQRAEELARENEELLEKEAEELEQENAELEGECQGLEARNRELRERAESVEREIQYVKDLLIEVYKARSQRTRSA; DN-ATF5-MUT, GEQRAEEGARENEEGGEKEAEEGEQENAEGEGECQGGEARNREGRERAESVEREIQYVKDGGIEVYKARSQRTRSA; DN-ATF5-TRUNC, LEGECQGLEARNRELRERAESV in which the italics represent the enhanced leucine zipper/mutated DNA binding domain, the bolded sequences the ATF5 leucine zipper and the underlined amino acids the L to G mutations. The fragments were subcloned into the BamH1 and XhoI site of pCMV-3Tag-1A (Agilent Technologies Inc) plasmid for in-frame N-terminal 3xFlag-tagged expression of DN-ATF5 or mutant DN-ATF5. CMV-FLAG LIP (24) was a gift from Joan Massague (Addgene plasmid # 15737; http://n2t.net/addgene:15737 ; RRID:Addgene_15737).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were carried out using SimpleChIP® Enzymatic Chromatin IP Kit (#9003, Cell Signaling) according to the supplier’s instructions. 1.5 X 107 T98G cells with or without CP-ATF5-DN treatment for 24 hours were cross-linked with formaldehyde and approximately 4 X 106 cells were used for each immunoprecipitation. Antibodies used were normal rabbit IgG (#2729, Cell Signaling), anti-CEBPB (ab32358, Abcam), and anti-CEBPD (ab198230, Abcam). Immunoprecipitation employed ChIP-Grade Protein G Magnetic Beads (#9005, Cell Signaling) and DNA purified from the immunoprecipitates was analyzed by qPCR using previously described primers for human IL6 and IL8 (25).
CEBP-B DNA Binding activity
T98G cells (approximately 2x106) were treated with or without 100 μM CP-ATF5-DN for 24 hours followed by isolation of nuclear extracts. The extracts (normalized for protein concentration) were assessed for CEBPB DNA binding activity using ELISA kit LS-F816. Extraction and binding activity were carried out using the supplier’s instructions.
Luciferase reporter assay
The luciferase reporter assay was carried out as previously described (26) using plasmids pmIL-6 FL which drives luciferase from the full length wild type mouse Il6 promoter and pmIL-6 mut CEBP which drives luciferase from the same promoter in which the CEBP binding site is mutated (gifts from Gail Bishop; Addgene plasmid # 61286; http://n2t.net/addgene:61286;RRID:Addgene_61286 and Addgene plasmid # 61291; http://n2t.net/addgene:61291; RRID:Addgene_61291). Approximately 1.2x104 T98G cells/well were seeded into coated 48-well dishes and transfected with pmIL-6 mut C/EBP or pmIL-6 for 24 hours. The cells were then treated with or without 100 μM CP-ATF5-DN for 24 hours. After washing, the cells were harvested in 40 μl of lysis reagent (# E1531, Promega) and 20 μl were mixed with 100 μl of luciferase assay reagent (#E1500, Promega) and the signal was recorded with a SpectraMax i3X plate reader (Molecular Devices).
qPCR
Cells were lysed and total RNA was purified using TRI regent (Molecular Research Center) following the manufacturer’s protocol. 1 μg of mRNA was used for the synthesis of cDNA using the First-strand cDNA synthesis kit (Origene). qPCR was performed using FastStart SYBR Green Master Mix (Roche) using the following primer pairs, with values normalized to 18S ribosomal RNA.
SURVIVIN Forward primer: 5’-CCACTGAGAACGAGCCAGACTT-3’
SURVIVIN Reverse primer: 5’-GTATTACAGGCGTAAGCCACCG-3’
18S ribosomal RNA Forward primer: 5’-AGTCCCTGCCCTTTGTACACA-3’
18S ribosomal RNA Reverse primer: 5’-GATCCGAGGGCCTCACTAAAC-3’
BCL2 Forward primer: 5’-TACTTAAAAAATACAACATCACAG-3’
BCL2 Reverse primer: 5’-GGAACACTTGATTCTGGTG-3’
IL6 Forward primer 1: 5’-CCAGAGCTGTGCAGATGAGTA-3’
IL6 Reverse primer 1: 5’-TGGGTCAGGGGTGGTTATTG-3’
IL6 Forward primer 2: 5’-CCAGGAGCCCAGCTATGAAC-3’
IL6 Reverse primer 2: 5’-CCCAGGGAGAAGGCAACTG-3’
CEBPB Forward primer: 5’-GCCGCCGCCTGCCTTTAAATC-3’
CEBPB Reverse primer: 5’-AGCCAAGCAGTCCGCCTCGTAG-3’
CEBPD Forward primer: 5’-CCATGTACGACGACGAGAG-3’
CEBPD Reverse primer: 5’-TGTGATTGCTGTTGAAGAGG-3’
IL8 Forward primer: 5’-CTCTTGGCAGCCTTCCTGATT-3’
IL8 Reverse primer: 5’-TATGCACTGACATCTAAGTTCTTTAGCA-3’
ATF5 Forward primer: 5’-CTGGCTCCCTATGAGGTCCTTG-3’
ATF5 Reverse primer: 5’-GAGCTGTGAAATCAACTCGCTC-3’
MCL1 Forward primer: 5’-GCTGCATCGAACCATTAGCA-3’
MCL1 Reverse primer: 5’-ATGCCAAACCAGCTCCTACT-3’
Statistical analysis
Experiments were carried out with ≥ 3 independent times each with ≥3 independent cultures unless otherwise indicated. Data are plotted either as individual data points with mean or are given as mean ± SEM as indicated. P values were calculated using a two-tailed Student’s t test.
Results
HAP1 cells survive and proliferate without ATF5 expression but are responsive to DN-ATF5.
To assess whether ATF5 is the obligatory target of DN-ATF5, we acquired HAP1 chronic myelogenous leukemia cell lines in which expression of endogenous ATF5 protein was compromised by either of two different deletions (1 and 10 BP). These deletions in the coding region disrupt expression of the full length protein including the DNA binding and leucine zipper domains (Supplementary Fig. S1A). Both ATF5-compromised lines (designated ATF5KO-1 and ATF5KO-10 showed growth rates and background apoptotic levels similar to that of the WT cells, though the knockout lines appeared to be somewhat more rounded and less adhesive to the culture dish substrate (see Fig. 1A, for example). To further verify the independence of HAP1 cells on ATF5 for growth and survival, we used two different siRNAs to knock down ATF5 (Supplementary Fig. S1B and Fig. 1B). ATF5 knockdown compromises survival of a number of cancer cell lines (6,11), including T98G glioblastoma cells that we used as a positive control (Supplementary Fig. S1B and Fig. 1B). Consistent with the ability of WT and ATF5KO cells to endure ATF5 knockout, siRNA-promoted ATF5 knockdown had no effect on their growth/survival (Fig. 1B), but did affect that of T98G cells. As an additional positive control, an siRNA directed against BIRC5 (encoding the survivin anti-apoptotic protein) effectively reduced survivin expression and cell numbers in HAP1 cells cultures as well as in T98G cultures (Supplementary Fig. S1C, Fig 1B). Together, these findings support the idea that HAP1 cells do not require ATF5 for survival.
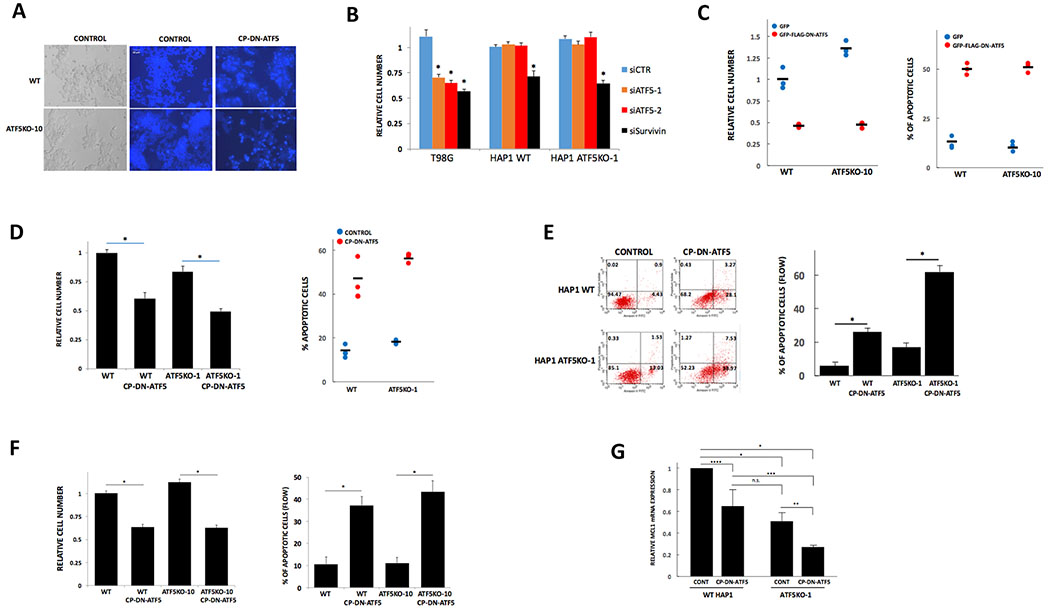
Figure 1.
HAP1 cells lacking active ATF5 respond to DN-ATF5, indicating that ATF5 is not the obligate target for DN-ATF5. A, Photomicrographs of WT and ATF5KO-10 HAP1 cells in bright field (left) or stained with Hoechst 33342 under control conditions (center) or after exposure to 100 μM CP-DN-ATF5 for 3 days in (right). B, ATF5 knockdown does not affect survival of HAP1 or HAP1 ATF5KO-1 cells, but does kill T98G cells. In contrast survivin knockdown compromises survival of all 3 cell types. Cultures were transfected with indicated siRNAs and 3 days later were evaluated for relative cell number. Values are means ± SEM. For T98G cells, data are from 5 independent experiments, total N=19-20 per condition. For HAP1 WT cells, data are from 5 independent experiments, total N=16 per condition. For HAP1 ATF5KO-1 cells, data are from two independent experiments, total N=10 per condition. *P<0.001 compared with siCTR for each cell type. C, FLAG-GFP-DN-ATF5 transfection reduces survival of WT and ATF5KO-1 HAP1 cells. Replicate cultures (N=3 per condition) were transfected as indicated and assessed 3 days later for relative numbers of transfected (GFP+) cells (left panel) and for proportion of transfected cells with apoptotic nuclei (right panel). D, CP-DN-ATF5 treatment reduces survival of WT and ATF5KO-1 HAP1 cells. Replicate cultures as indicated were treated without or with 100 μM CP-DN-ATF5 and assessed 3 days later for total cell number (left panel) and proportion of cells with apoptotic nuclei (right panel). For left panel, values represent means ± SEM for 3 independent experiments, each carried out in triplicate. *P<0.001. Right panel data are from one experiment carried out in replicate cultures. E, CP-DN-ATF5 treatment reduces survival of WT and ATF5KO-1 HAP1 cells as determined by flow cytometry. Left panel shows example of flow cytometric analysis of cell death in HAP1 WT and HAP1 ATF5KO-1 cultures treated with or without 100 μM CP-DN-ATF5 for 3 days. Right panel shows quantification of apoptotic cells with values representing means ± SEM for 2 independent experiments, each carried in triplicate (total N=6). *P<0.001. F, CP-DN-ATF5 treatment reduces survival of WT and ATF5KO-10 cells. Replicate cultures of WT and ATF5KO-10 cells as indicated were treated without or with 100 μM CP-DN-ATF5 and assessed 3 days later for cell number (left panel) or for % of apoptotic cells by flow cytometry. Values for cell numbers represent means ± SEM for 3 independent experiments, each carried out in triplicate. *P<0.001Values for % apoptosis represent means ± SEM for 4 independent experiments, each carried with 2-3 replicates (total N=10). *P<0.001. G, MCL1 mRNA levels are reduced in ATF5KO-1 cells compared to WT HAP1 cells and CP-DN-ATF5 lowers MCL1 mRNA levels in both cell types. Indicated cells were treated with or without 100 μM CP-DN-ATF5 for 3 days and assessed for MCL1 mRNA levels. Data represent mean values ±SEM and are from 9 independent determinations. *P<0.001; **P=0.01; ***P=0.02; ****P=0.03.
We reasoned that if HAP1 cells do not require ATF5 for survival, they should be unresponsive to DN-ATF5-promoted killing if ATF5 is the only target of this construct. On the other hand, if DN-ATF5 targets other factors required for maintenance, then both WT and ATF5KO cells should be vulnerable to the DN protein. To distinguish between these possibilities, WT and ATF5KO-10 HAP1 cells were transfected with GFP-FLAG-DN-ATF5 or GFP alone (as control) and the cultures were assessed 3 days later for numbers of surviving GFP+ cells and for percentage of GFP+ cells with apoptotic nuclei (Fig. 1C). In each instance, transfection with DN-ATF5 elicited robust cell death similar to that previously seen with multiple other cancer cell types (6,10). To confirm and extend these findings, we also treated WT, ATF5KO-1 and ATF5KO-10 cells with the cell-penetrating DN-ATF5 peptide, CP-DN-ATF5 (13,19). This too had similar effects on WT and both ATF5KO lines in reducing cell numbers and triggering apoptosis as assessed by monitoring cell nuclei and by FACS analysis of PI-Annexin-V stained cells (Fig. 1D–F). Taken together, these observations thus indicate that DN-ATF5 promotes cancer cell death by engaging targets other than ATF5.
MCL1 has been described as a direct ATF5 target (11), and consistent with the loss of ATF5 activity, expression of MCL1 transcripts was substantially diminished in ATF5KO cells compared with WT cells (Fig. 1G). Moreover, consistent with the idea that DN-ATF5 acts via a mechanism independent of ATF5 expression, both cell types responded to CP-DN-ATF5 treatment by showing reduced expression of MCL1 transcripts (Fig. 1G).
Affinity pulldown/mass spectroscopic analysis reveals CEBPB, CEBPD and CCDC6, but not ATF5, as binding partners for DN-ATF5.
To identify potential binding partners for DN-ATF5, we transfected PC3 prostate tumor cells with constructs expressing either GFP or GFP-FLAG-DN-ATF5 protein. PC3 cells were chosen for their high transfection efficiency and sensitivity to DN-ATF5 (13). Two days later (when the death response to DN-ATF5 is still minimal), the cells were harvested in buffer containing 0.4% Triton X100, sonicated and the pre-cleared supernatants were incubated with anti-FLAG M2 magnetic beads. GFP-FLAG-DN-ATF5 and its potential binding partners were released from the beads by 3x-FLAG peptide and subjected to SDS-PAGE and mass spectrometry proteomic analysis. Silver-staining and FLAG immunoblotting verified both the presence of GFP-FLAG-DN-ATF5 in the eluate as well as absence of most proteins in the input lysate (Fig. 2A).
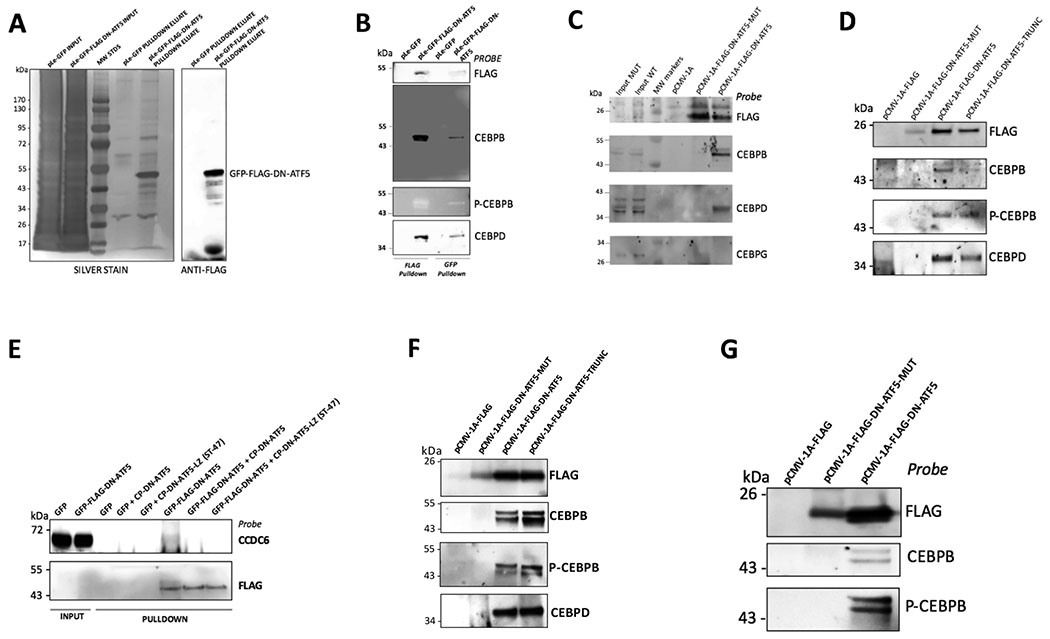
Figure 2.
Bead pulldown experiments confirm that DN-ATF5 associates with CEBPB, CEBPD and CCDC6 in cells. A, Silver stain of SDS-PAGE for input and FLAG-bead pulldown material obtained from PC3 cells transfected with GFP or GFP-FLAG-DN-ATF5 (left lanes) and a Western blot of the pulldown material probed with anti-FLAG (right lanes). B, Pulldown with FLAG- or GFP-beads followed by Western immunoblotting confirms association of CEBPB, phospho-CEBPB (Thr235) (P-CEBPB) and CEBPD with GFP-FLAG-DN-ATF5 in PC3 cells. Cultures transfected with GFP or GFP-FLAG-DN-ATF5 and lysates for the pulldown were prepared 2 days later. The blot was probed with antisera to the indicated proteins. C, FLAG-DN-ATF5 associates with CEBPB and CEBPD in T98G cells and this requires the intact ATF5 leucine zipper. T98G cells were transfected with FLAG-DN-ATF5 or FLAG-DN-ATF5-mut in which the leucines in the zipper domain were mutated to glycine residues. Lysates prepared two days later were subjected to pulldown with FLAG beads and the eluates probed by Western blotting for the indicated proteins. D, Association of CEBPB, P-CEBPB and CEBPD with FLAG-DN-ATF5 in T98G cells requires the intact ATF5 leucine zipper, but not the mutated DNA binding domain. T98G cells were transfected with the indicated constructs (including FLAG-DN-ATF5-TRUNC in which the mutated DNA binding domain was deleted) and two days later lysates were subjected to pulldown experiments with FLAG beads and the eluates probed as indicated by Western immunoblotting. E, GFP-FLAG-DN-ATF5 associates with CCDC6 in T98G cells as determined by FLAG-bead pulldown assay and this association is competed off by CP-DN-ATF5 and a CP-DN-ATF5 peptide [CP-DN-ATF5-LZ (ST-47)] containing the ATF5 leucine domain but not the mutated DNA binding domain. T98G cells were transfected with the indicated constructs and 2 days later lysates were subjected to pulldown with FLAG beads in the presence or absence of 100 μM CP-DN-ATF5 or 40 μM ST-47. Following the pulldown procedure, 3x-FLAG eluates were subjected to Western immunoblotting and probed for the indicated epitopes. F, FLAG-DN-ATF5 associates with CEBPB, P-CEBPB and CEBPD in LN229 cells and the association requires the intact ATF5 leucine zipper, but not the modified DNA binding domain. LN229 cells were transfected with the indicated constructs and two days later lysates were subjected to pulldown experiments with FLAG beads and the eluates probed as indicated by Western immunoblotting. G, FLAG-DN-ATF5 associates with CEBPB and P-CEBPB in U87 glioblastoma cells and this requires the intact ATF5 leucine zipper. U87 cells were transfected with the indicated constructs and two days later lysates were subjected to pulldown experiments with FLAG beads and the eluates probed as indicated by Western immunoblotting.
Filtering of spectral count data from two independent mass spectrometry experiments using results from the GFP-only transfected cells and the CRAPome data base (27) returned three robust, consistent “hits”, the bZIP transcription factors CEBPB and CEBPD and the coiled-coil domain protein CCDC6 (Table 1). This was also confirmed in one experiment by comparing LFQ intensities for control and GFP-FLAG-DN-ATF5 transfected cells (Table1). Although there were abundant signals associated with ATF5 protein for samples transfected with GFP-FLAG-DN-ATF5, none of these corresponded to sequences outside of the bZIP domain and so were assigned to the DN-ATF5 construct. Additional abundant species were identified in the pulldown material, but these failed to show specificity or reproducibility (see Table 1 for examples).
Table 1. Pulldown-mass spectrometric analysis reveals CEBPB, CEBPD and CCDC6 as binding partners for DN-ATF5.
PC3 cells were transfected with GFP-FLAG-DN-ATF5 or GFP and two days later cell lysates were prepared, incubated with anti-FLAG a beads and the beads eluted with 3x-FLAG peptide. The eluates were subjected to mass spectrometry. Data from two independent experiments are presented. Table includes examples of 5 abundantly recovered proteins in the eluates
| PROTEIN ID | EXPT 1 SPECTRAL COUNTS GFP | EXPT 1 SPECTRAL COUNTS DN-ATF5 | EXPT 2 SPECTRAL COUNTS GFP | EXPT 2 SPECTRAL COUNTS DN-ATF5 | EXPT 2 LFQ INTENSITY DN-ATF5/GFP | ||
|---|---|---|---|---|---|---|---|
| DN-ATF5 | 0 | 411 | 22 | 415 | 88.1 | ||
| CEBPB | 0 | 20 | 7 | 23 | 11.1 | ||
| CEBPD | 0 | 3 | 1 | 4 | 10.7 | ||
| CCDC6 | 0 | 9 | 0 | 19 | 68.1 | ||
| EIF4B | 35 | 50 | 50 | 20 | 0.65 | ||
| STK38 | 38 | 40 | 75 | 36 | 0.52 | ||
| PPM1B | 18 | 36 | 15 | 6 | 0.79 | ||
| FLNA | 6 | 20 | 47 | 17 | 0.54 | ||
| PLEC | 5 | 39 | 35 | 15 | 0.79 |
Bead pull-down experiments confirm association between DN-ATF5 and CEBPB, CEBPD and CCDC6.
To confirm association between DN-ATF5 and CEBPB, CEBPD and CCDC6, we carried out pulldown studies as above using anti-FLAG beads with lysates from GFP-FLAG-DN-ATF5 or FLAG-DN-ATF5 transfected PC3 and T98G cells and subjected the 3x-FLAG-released materials to immunoblotting with probes for all 3 proteins. Species corresponding to CEBPB, CEBPD and CCDC6 were detected for DN-ATF5, but not for GFP-only pulldowns (Fig. 2 B–E). CEBPB is reported to require phosphorylation at Thr235 to bind DNA (28). Probing the blots revealed that p(Thr235)-CEBPB also interacts with DN-ATF5 (Fig. 2 B,D). We additionally observed pulldown of CEBPB, pCEBPB and CEBPD in LN229 glioblastoma cells and of CEBPB and pCEBPB in U87 glioblastoma cells (which did not express detectable levels of CEBPD) (Fig. 2 F,G). In contrast, we did not detect pulldown of another CEBP family member, CEBPG in T98G cells (Fig. 2C). To confirm our findings, we also took advantage of the GFP tag on GFP-FLAG-DN-ATF5 to carry out parallel pulldown studies with beads coated with anti-GFP. These too identified CEBPB, pCEBPB and CEBPD as DN-ATF5 binding partners (Fig. 2B).
Three CEBPB isoforms have been described, LAP1, LAP2 and LIP, that are the products of alternative translation of CEBPB transcripts (29). Different functional activities have been ascribed to each isoform (29). Based on its electrophoretic mobility, the form described above in our pull-down experiments corresponds to LAP1. All 3 isoforms contain the CEBPB leucine zipper domain, so all would be anticipated to associate with DN-ATF5. Because we were unable to unequivocally identify endogenous LAP2 and LIP in our WB analyses, we sought to confirm interaction of DN-ATF5 with LIP, for which an expression vector is available. Such interaction was detected when FLAG-LIP and GFP-FLAG-DN-ATF5 were co-expressed in 293T cells and subjected to pulldown-WB analysis with anti-GFP beads and probed with anti-FLAG and anti-GFP (Supplementary Fig. 2).
The ATF5 leucine zipper is required for interaction of DN-ATF5 with CEBPB and CEBPD.
We next assessed whether association between DN-ATF5 and its identified targets is dependent on the ATF5 leucine zipper domain as anticipated. To achieve this, we carried out parallel pulldown assays in T98G, LN229 and U87 cells with a DN-ATF5 mutant in which each leucine residue in the zipper domain was replaced with glycine (FLAG-DN-ATF5-mut). In a past study, this mutant form failed to promote tumor cell death (13). In contrast to the wild-type construct, the form with the mutated leucine zipper failed to pull down CEBPB, pCEBPB or CEBPD (Fig. 2C,D,F,G). These observations confirm that the ATF5 leucine zipper is required for interaction of DN-ATF5 with these targets and serve as an additional control to exclude their non-specific association with either DN-ATF5 or anti-FLAG beads.
As originally designed, DN-ATF5 contains an extended leucine zipper generated by mutation of the basic DNA binding domain (4). To determine whether this domain is also required for interaction with CEBPB and CEBPD, we produced and tested an additional mutant construct in which the extended leucine zipper was deleted and the native leucine zipper was retained (FLAG-DN-ATF5-TRUNC). This construct also associated with CEBPB, pCEBPB and CEBPD in pulldown assays with T98 and LN229 cells (Fig. 2D,F), thus indicating that the extended leucine zipper is dispensable for interaction with these proteins in cells.
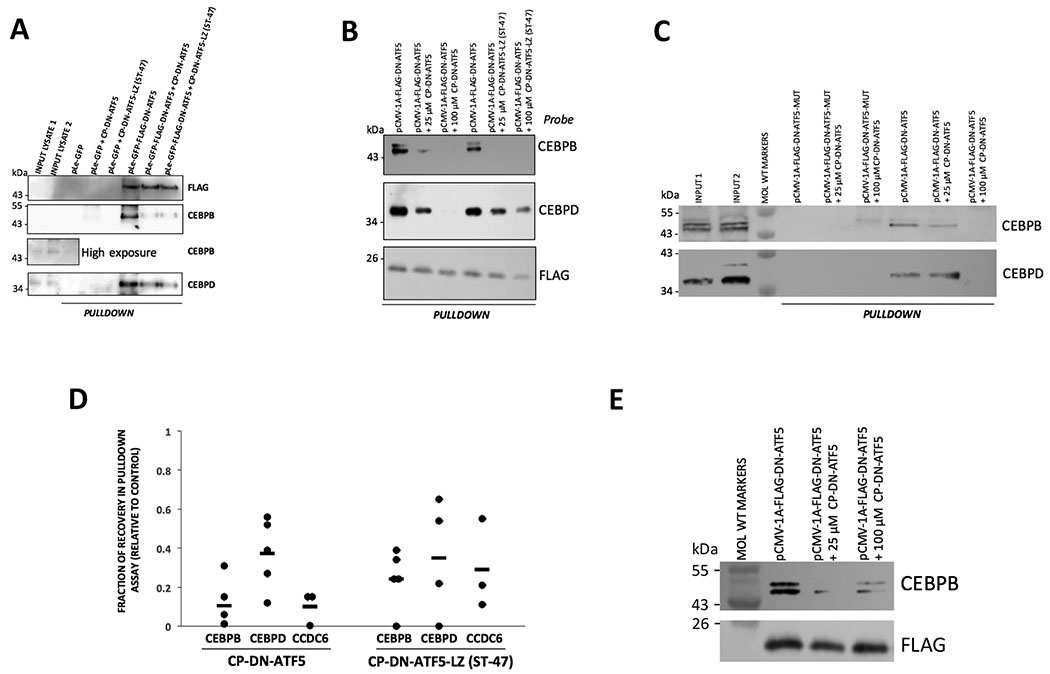
CP-DN-ATF5 competes for association with CEBPB, CEBPD and CCDC6.
We have generated a cell-penetrating form of DN-ATF5 (CP-DN-ATF5) that, like transfected DN-ATF5, selectively kills tumor cells in vitro and in vivo (13,19). Because this form, which is synthetic, lacks a tag and cannot be used for conventional pulldown assays, we asked whether it would compete with GFP-FLAG-DN-ATF5 or FLAG-DN-ATF5 for association with CEBPB, CEBPD and CCDC6. Accordingly, pulldown experiments with T98G cells as described above were carried out in which CP-DN-ATF5 was added or not to the lysates during incubation with anti-FLAG beads. After extensive washing, the material remaining on the beads was released with 3x-FLAG and probed by immunoblotting. The results revealed that 100 μM CP-DN-ATF5 successfully competed for association with CEBPB, CEBPD and CCDC6 (Figs. 2E, 3A–D). Probing with anti-FLAG for FLAG-DN-ATF5/GFP-FLAG-DN-ATF5 in the eluates indicated that this effect was not due to reduction by CP-DN-ATF5 on the association of FLAG-DN-ATF5 with the anti-FLAG beads (Figs. 2E, 3A,B). 25 μM CP-DN-ATF5 also successfully competed for association with CEBPB and CEBPD (Fig 3B–D). To affirm that the intact ATF5 leucine zipper is sufficient for association with CEBPB, CEBPD and CCDC6, we also carried out competition experiments with a synthetic peptide of the penetratin domain fused to the ATF5 leucine zipper sequence (CP-DN-ATF5-LZ; ST-47). AT 25-100 μM, this too competed with FLAG-DN-ATF5 for binding to the three target proteins (Figs. 2E, 3A,B,D). Finally, we confirmed that CP-DN-ATF5 also competes for association with CEBPB in lysates from LN229 cells (Fig. 3E).
Figure 3.
CP-DN-ATF5 peptides compete with FLAG-GFP-DN-ATF5 and FLAG-DN-ATF5 for association with CEBPB, CEBPD and CCDC6. A,B, CP-DN-ATF5 and CP-DN-ATF5-LZ (ST-47) compete with GFP-FLAG-DN-ATF5 for association with CEBPB and CEBPD. T98G cells were transfected with the indicated constructs and 2 days later lysates were subjected to pulldown with FLAG beads in the presence or absence of 100 (A,B) or 25 (B) μM CP-DN-ATF5 or 25 (B) or 40 (A) μM ST-47. Following the pulldown procedure, 3x-FLAG eluates were subjected to Western immunoblotting and probed for the indicated epitopes. C, CP-DN-ATF5 competes with FLAG-DN-ATF5 for association with CEBPB and CEBPD. T98G cells were transfected with the indicated constructs and 2 days later lysates were subjected to pulldown with FLAG beads in the presence or absence of 25 or 100 μM CP-DN-ATF5. Following the pulldown procedure, 3x-FLAG eluates were subjected to Western immunoblotting and probed for CEBPB and CEBPD. D, CP-DN-ATF5 and CP-DN-ATF5-LZ (ST-47) compete with GFP-FLAG-DN-ATF5 for association with CEBPB, CEBPD and CCDC6. T98G cells were transfected with the indicated constructs and 2 days later lysates were subjected to pulldown with FLAG beads in the presence or absence of 100 μM CP-DN-ATF5 or 100 μM ST-47. Following the pulldown procedure, 3x-FLAG eluates were subjected to Western immunoblotting and probed for the indicated epitopes. The band intensities of the blots were then determined and relative intensities for the indicated proteins were normalized relative to that of parallel control samples not treated with the peptides. Data represent 3-5 independent experiments. E, CP-DN-ATF5 competes with FLAG-DN-ATF5 for association with CEBPB in LN229 cells. LN22 cells were transfected with FLAG-DN-ATF5 and 2 days later lysates were subjected to pulldown with FLAG beads in the presence or absence of 25 or 100 μM CP-DN-ATF5. Following the pulldown procedure, 3x-FLAG eluates were subjected to Western immunoblotting and probed for CEBPB and FLAG (FLAG-DN-ATF5).
CP-DN-ATF5 and regulation of CEBPB and CEBPD expression.
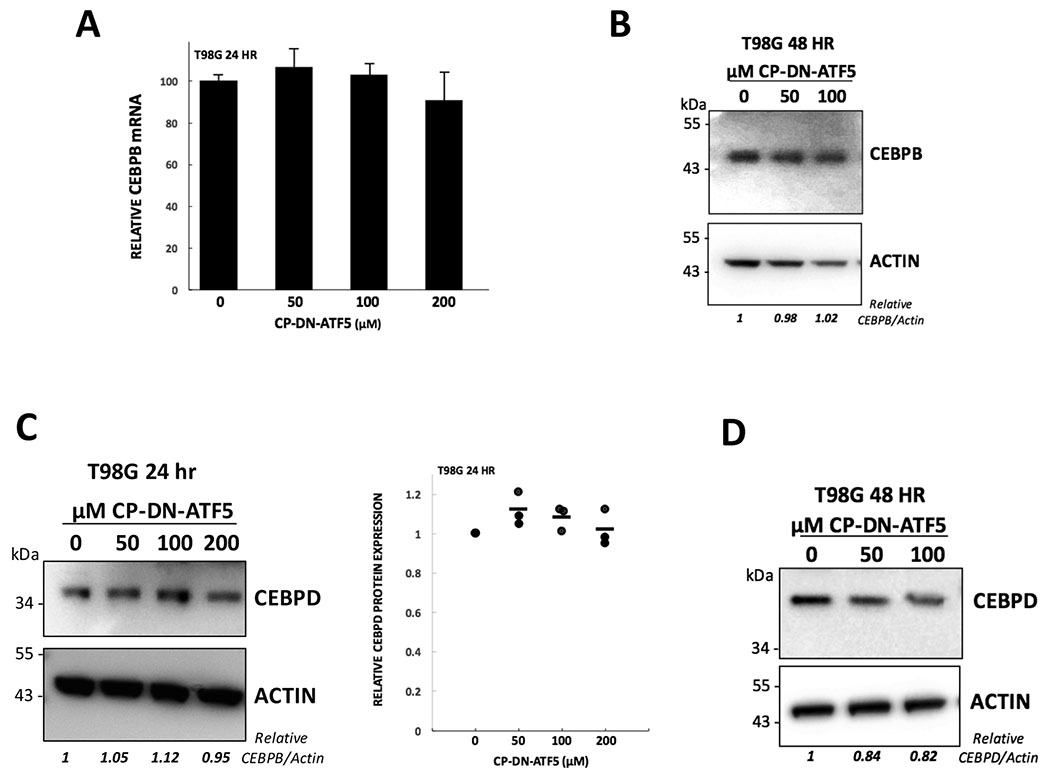
We next asked whether CP-DN-ATF5 might affect CEBPB and CEBPD signaling by altering their expression levels. qPCR carried out with T98G cells revealed no significant changes in CEBPB mRNA levels after 24 hours of treatment (Fig. 4A) and in two experiments with LN229 cells at 100 μM CP-DN-ATF5, CEBPB mRNA levels were 84±10% (mean ± SEM) of those in controls (n=2 independent experiments, each in triplicate). In addition, immunoblots performed on T98G, U87 and LN229 cells at 24-48 hours after exposure to CP-DN-ATF5 also showed no major effects on CEBPB protein levels (Fig. 4B–D; Supplementary Fig. S3A–C). Additionally, we also found no major changes in CEBPD protein levels in T98G cells at 24 and 48 hours after CP-DN-ATF5 treatment (Fig. 4C,D).
Figure 4.
CP-DN-ATF5 does not have major effects on expression of CEBPB or CEBPD. A, CP-DN-ATF5 treatment for 24 hours does not significantly affect CEBPB mRNA levels in T98G cells. Values are means ± SEM for 3 independent experiments each carried out in triplicate. B, CP-DN-ATF5 treatment for 48 hours does not significantly affect CEBPB protein levels in T98G cells. Replicate cultures were treated with 0, 50 or 100 μM CP-DN-ATF5 for 48 hours and relative (to ACTIN) levels of CEBPB protein were determined by Western immunoblotting and evaluation of relative band intensities. C, CP-DN-ATF5 treatment for 24 hours has relatively little effect on CEBPD expression in T98G cells as determined by Western immunoblotting. Left panel shows representative blot; right panel, data show quantification of Western immunoblotting in 3 independent experiments. D, CP-DN-ATF5 treatment for 48 hours has relatively little effect on CEBPD expression in T98G cells as determined by Western immunoblotting.
CP-DN-ATF5 decreases DNA binding and transcriptional regulatory activity of CEBPB and CEBPD.
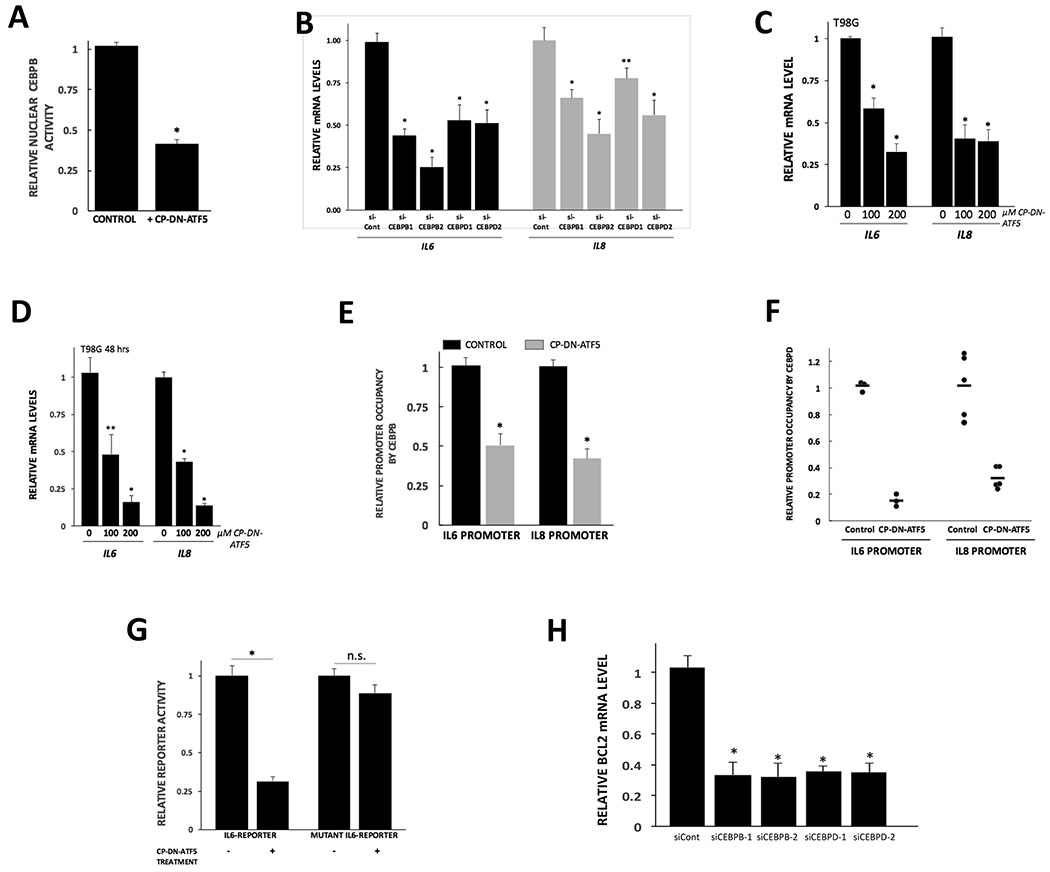
As a dominant-negative transcription factor, DN-ATF5 would be anticipated to form inactive complexes with its partners and thus ultimately interfere with their capacity to associate with and regulate their transcriptional targets. To test whether DN-ATF5 suppresses the transcriptional capacities of CEBPB and CEBPD, we again employed CP-DN-ATF5. We first assessed whether CP-DN-ATF5 reduces the levels of active CEBPB in cell nuclei that are available to bind a CEBP consensus binding site on a synthetic dsDNA oligonucleotide. This revealed that 24 hours treatment of T98G cells with 100 μM CP-DN-ATF5 reduced such levels by approximately 60% (Fig. 5A).
Figure 5.
CP-DN-ATF5 interferes with CEBPB and CEBPD transcriptional activity. A, CP-DN-ATF5 treatment diminishes the DNA binding activity of nuclear CEBPB in T98G cells. Cultures were treated without (control) or with 100 μM CP-DN-ATF5 for 24 hours and then nuclear extracts were assessed for CEBPB binding activity to a CEBP consensus binding site on a synthetic dsDNA oligonucleotide as described in Methods. Activity was normalized to protein content in the extracts. Values represent mean ± SEM for 3 independent experiments performed in duplicate or triplicate for a total N=7. *P<0.001 compared with control. B, siRNA-mediated knockdown of CEBPB and CEBPD reduces expression of IL6 and IL8 mRNAs in T98G cells. Cultures were transfected with indicated siRNAs and 3 days later were assessed for relative levels of IL6 and IL8 mRNA. Values represent means ± SEM. For si-Control, N=13 for 3 independent experiments; for siCEBPB1, N=9(IL6) and 8(IL8) for 3 independent experiments; for siCEBPB2 and siCEBPD1, N=6 for 3 independent experiments; for siCEBPD2, N=9 for 3 independent experiments. *P<0.001, **P=0.003 compared with corresponding control. C,D, CP-DN-ATF5 treatment reduces the expression of IL6 and IL8 mRNAs in T98G cells. Replicate cultures were exposed to the indicated concentrations of CP-DN-ATF5 for 24 (C) or 48 (D) hours and then assessed for relative levels of IL6 and IL8 transcripts. Values represent means ± SEM and were derived from 3 independent experiments carried out in triplicate. *P<0.001 compared with corresponding control, **P=0.01 compared with corresponding control. E, CP-DN-ATF5 treatment reduces the occupancy by CEBPB of the IL6 and IL8 promoters. T98G cells were treated without (control) or with 100 μM CP-DN-ATF5 for 24 hours and then chromatin immunoprecipitation (ChIP) assays were carried out as described in methods. Values are normalized to those of corresponding control cultures and are given as means ± SEM. Data represent 3 independent experiments carried out in triplicate. *P<0.001 compared with corresponding control. F, CP-DN-ATF5 treatment reduces the occupancy by CEBPD of the IL6 and IL8 promoters. T98G cells were treated without (control) or with 100 μM CP-DN-ATF5 for 24 hours and then ChIP assays were carried out. Values are normalized to those of corresponding control cultures. G, CP-DN-ATF5 reduces IL6 reporter activity in T98G cells. Replicate cultures were transfected with either an IL6-promoter-luciferase reporter or an IL6 reporter mutated at the CEBP binding site. 2 days later the cultures were treated without or with 100 μM CP-DN-ATF5 for 24 hours and then assessed for reporter activity as described in methods. Values represent means ± SEM and represent 3 independent experiments carried out in triplicate. *P<0.001. H, Knockdown of CEBPB and CEBPD reduces expression of BCL2 in T98G cells. Replicate cultures were transfected with the indicated siRNAs and assessed 4 days later for BCL2 mRNA levels. Values represent means ± SEM. Data are from 3 independent experiments. Total N values = 11 (siCont), 8 (siCEBPB1, siCEBPB2, siCEBPD1) and 6 (siCEBPD2). *P<0.001 compared with siCont.
IL6 and IL8 are two well-characterized direct targets of CEBPB and CEBPD which bind to defined sites in the IL6 and IL8 promoters (25,30,31). To confirm that IL6 and IL8 are CEBPB and CEBPD targets in T98G cells, we knocked down both CEBPB and CEBPD with siRNAs (Supplementary Fig. S4A,B,C). This resulted in significant decreases in IL6 and IL8 transcripts (Fig. 5B). Moreover, consistent with disruption of CEBPB and CEBPD transcriptional activation by DN-ATF5, we found that CP-DN-ATF5 (100 μM, 24-48 hours) substantially reduced the levels of IL6 and IL8 transcripts in T98G cells (Fig. 5C,D) as well as in LN229 cells (Supplementary Fig. S5). We additionally observed that CP-DN-ATF5 diminished IL6 and IL8 transcripts in both WT and ATF5KO-10 HAP1 cells, thus indicating that ATF5 expression is not required for DN-ATF5 to interfere with CEBPB/CEBPD activity (Supplementary Fig. S5).
To determine whether DN-ATF5 interferes with association of CEBPB and CEBPD with the IL6 and IL8 promoters, we next carried out chromatin immunoprecipitation (ChIP) and qPCR assays with T98G cells treated without or with CP-DN-ATF5 (24 hours, 100 μM). These revealed a substantial decrease in occupancy of the IL6 and IL8 promoters by CEBPB and CEBPD in cells treated with CP-DN-ATF5 compared with controls (Fig. 5E,F).
Finally, we examined the effects of CP-DN-ATF5 on activity of a luciferase reporter driven by the IL6 promoter which contains a CEBP consensus binding site for CEBPB and CEBPD. Treatment of T98G cells with CP-DN-ATF5 (100 μM) for 24 hours reduced reporter activity by about 70% (Fig. 5G). In contrast, there was no significant effect on a reporter in which the CEBP binding site was mutated (Fig. 5G). Taken together, these findings indicate that DN-ATF5 significantly reduces the DNA binding and transcriptional activities of CEBPB and CEBPD in intact cells.
Knockdown of CEBPB or CEBPD reduces expression of BCL2.
BCL2 has been established as a direct anti-apoptotic target of ATF5 (17) and treatment with CP-DN-ATF5 reduces BCL2 levels in multiple tumor cell lines (13). It is also reported that BCL2 is a direct target of CEBPB in multiple myeloma cells (32). We reasoned that if ATF5 interacts with CEBPB and CEBPD to regulate BCL2, then CEBPB and/or CEBPD knockdown should also affect BCL2 expression. As shown in Fig. 5H, siRNA-mediated knockdown of either CEBPB or CEBPD in T98G cells reduces BCL2 mRNA levels by about 70%.
Knockdown of CEBPB and CEBPD promotes cancer cell death.
Our studies have established that DN-ATF5 causes selective death of cancer cells both in vitro and in vivo. If the main targets of DN-ATF5 are CEBPB, CEBPD and CCDC6, then it follows that alternative means of reducing levels of these proteins should also trigger death of tumor cells. Indeed, past studies have reported instances in which loss of these proteins affects cancer cell growth and/or survival (31–33, 35–39). To extend such findings to the present experimental conditions, we transfected cancer cell lines with siRNAs targeting CEBPB, CEBPD or CCDC6 (Supplementary Figs. S4A–C, S6). Knockdown of CEBPB or CEBPD reduced cell numbers and enhanced apoptotic death of T98G cells (Fig. 6A–C). CEBPB and CEBPD knockdown also promoted death of LN229 and GBM22 cells and of MDA-MB-468 breast cancer cells (Fig. 6D–F). In contrast, knockdown of CEBPB or CEBPD in cultured normal astrocytes (Fig. 6G), despite also effectively interfering with expression of IL6 (Fig. 6G), did not affect their survival (Fig. 6H), as previously found for DN-ATF5 (6). Finally, targeting CCDC6 in T98G cells (Supplementary Fig. S6) was without effect on cell death (Fig. 6I). These findings thus suggest that loss of CEBPB or CEBPD activity in multiple tumor cell lines, like treatment with DN-ATF5, is sufficient to cause cell death and could thus account for DN-ATF5’s anti-tumor activity.
Figure 6.
Knockdown of CEBPB and CEBPD affects the survival of cancer cell lines, but not of normal astrocytes. A, Knockdown of CEBPB and CEBPD affects morphology and survival of T98G cells. Replicate cultures were transfected with the indicated siRNAs and photomicrographs were taken 4 days later. Scale bar =100 μm. B,C, CEBPB and CEBPD knockdown reduces cell numbers (B) and promotes apoptosis (C) in cultures of T98G cells. Replicate cultures were transfected with the indicated siRNAs and assessed 4 days later for relative cell number. Values are expressed as means ± SEM and represent data from 4 independent experiments, each in triplicate. *P<0.001 compared with siCont. D, CEBPB and CEBPD knockdown promotes apoptotic death in cultures of LN229 cells. Replicate cultures were transfected with the indicated siRNAs and assessed 4 days later for proportion of cells with apoptotic nuclei. Values are expressed as means ± SEM and represent data from 3 independent experiments, each in triplicate. *P<0.001 compared with siCont. E, CEBPB and CEBPD knockdown promotes apoptotic death in cultures of GBM 12 cells. Replicate cultures were transfected with the indicated siRNAs and assessed 4 days later for proportion of cells with apoptotic nuclei. Values are expressed as means ± SEM and represent data from 2 independent experiments, each in triplicate. *P<0.001 compared with siCont. F, CEBPB and CEBPD knockdown promotes apoptotic death in cultures of MDA-MB-468 cells. Replicate cultures were transfected with the indicated siRNAs and assessed 4 days later for proportion of cells with apoptotic nuclei. Values are expressed as means ± SEM and represent data from 2 independent experiments, each in triplicate. *P<0.001 compared with siCont. G, Knockdown efficacy of siCEBPB and siCEBPD in cultured human astrocytes. Replicate cultures were transfected with indicated siRNAs and assessed 4 days later for expression of CEBPB, CEBPD or IL6 mRNA. 2-4 cultures were assessed per point, each in triplicate. H, Knockdown of CEBPB and CEBPD does not affect astrocyte survival. Cultures of normal human astrocytes were transfected with the indicated siRNAs and 4 days later assessed for total cell numbers. I, Knockdown of CCDC6 does not affect survival of T98G cells. Cultures were transfected with the indicated siRNAs and 4 days later were assessed for cell numbers. Values are for replicate cultures in one experiment.
Discussion
We have developed a cell penetrating form of dominant negative ATF5 that shows promise in pre-clinical studies as a novel anti-tumor agent both as mono- and combination therapies (13,19). To date, this peptide has shown remarkably little toxicity in mice (13,19). Despite this potential therapeutic promise, the mechanism by which DN-ATF5 affects tumor cell survival has been unknown. Initial reports that ATF5 can form homodimers suggested that a dominant-negative form might directly interfere with the function of the corresponding endogenous protein (21). However, an indication that this might not be so was suggested by studies that analyzed interactions of purified bZIP domains and that did not detect ATF5-ATF5 homodimers (22). The later findings are consistent with our failure to detect interaction of GFP-FLAG-DN-ATF5 with endogenous ATF5 in pulldown studies. Our findings with HAP1 and HAP1-ATF5KO cells further support the idea that ATF5 is not the critical binding target of DN-ATF5. These cells survive and replicate when ATF5 is silenced or when ATF5 is absent, yet succumb when transfected with DN-ATF5 or treated with CP-DN-ATF5.
We used an unbiased pulldown/mass spectroscopic screen in PC3 prostate cancer cells to detect DN-ATF5 binding partners. This line is sensitive to DN-ATF5 treatment and thus should contain relevant DN-ATF5 targets (13). To confirm the expression of multiple basic leucine zipper proteins in PC3 cells that might be potential DN-ATF5 targets, we searched the ARCHS database (40) for the following families (number per family in parentheses): ATF(8), BACH(2), BATF(3), CEBP(6), CREB(11), DBP(1), FOS(4), HLF(1), JUN(4), MAF(7), NFE(4), NFIL3(1), NRL(1), TEF(1), AND XBP1(1). Transcripts for all proteins were well-expressed with the exceptions of CEBPE, BATF and MAFB. It is notable that of the 52 basic leucine zipper proteins noted above that are expressed in PC3 cells, only CEBPB and CEBPD were detected as DN-ATF5 partners in our screen. Nevertheless, we cannot rule out associations that were not stable enough to endure the conditions of our experiments or with proteins not expressed in PC3 cells.
A complicating feature of CEBPB biology is that it exists in 3 isoforms reported to have distinct activities (29). Although we could only reliably detect the larger LAP1 isoform in our pulldown studies with endogenous proteins, over-expression experiments also supported interaction of DN-ATF5 with the smaller LIP1 isoform. All 3 isoforms retain the same CEBPB leucine zipper domain and given that association of DN-ATF5 with its partners appears to rely on leucine zipper:leucine zipper interactions, our observations are consistent with the likelihood that all 3 isoforms are bound by DN-ATF5. The siRNAs in our knockdown studies are also anticipated to deplete all 3 isoforms. Because the 3 CEBPB isoforms appear be differentially regulated and to have distinctive activities, it will be important in future to determine whether or not they are differentially affected by DN-ATF5 treatment.
In addition to CEBPB and CEBPD, we found that DN-ATF5 associates with CCDC6, a coiled-coil domain protein. Oncogenic fusion proteins of CCDC6 with signaling receptors such as RET have been found in several tumor types (41). This oncogenicity appears due to enforced dimerization via the CCDC6 coiled-coil domain and consequent constituent receptor activation. Analysis of the CCDC6 coiled-coil domain by motif-scan (https://myhits.isb-sib.ch/cgi-bin/motif_scan) reveals a putative bZIP domain that could in principle mediate association of CCDC6 with DN-ATF5. Despite this interaction, CCDC6 knockdown in at least one cell line failed to phenocopy the death-promoting effects of DN-ATF5 or of CEBPB or CEBPD knockdown. Such findings suggest that interference with CCDC6 function does not play a required role in DN-ATF5’s overall effect on tumor cell survival. It remains to be seen whether DN-ATF5 might be effective in affecting the growth and survival of tumor cells expressing an oncogenic CCDC6-fusion protein.
Our findings indicate that the leucine zipper domain of ATF5 is both necessary and sufficient for interactions of DN-ATF5 with CEBPB, CEBPD and CCDC6. In support of this conclusion, transfected DN-ATF5 with a leucine zipper mutated to replace the leucine residues, failed to associate with the three targets. In contrast, a transfected DN-ATF5 in which the modified DNA binding domain was deleted, interacted with CEBPB or CEBPD. Additionally, a form of CP-DN-ATF5 which contains only the ATF5 leucine zipper (ST-47) competed with GFP-FLAG-DN-ATF5 for association with CEBPB, CEBPD and CCDC6.
In addition to binding CEBPB and CEBPD, our data indicate that CP-DN-ATF5 interferes with their transcriptional activity. We found that CP-DN-ATF5 treatment decreased the levels of nuclear CEBPB capable of binding to a CEBPB DNA binding sequence, that it diminished CEBPB and CEBPD occupancy of the promoters for CEBPB/CEBPD targets IL6 and IL8, and that it suppressed the activity of a CEBPB/CEBPD-driven luciferase reporter. Moreover, like depletion of CEBPB and CEBPD, CP-DN-ATF5 also reduced IL6 and IL8 expression.
In addition to being markers for interference with CEBPB and CEBPD activity, IL6 and IL8 have been variously implicated as promoters of cancer initiation, invasiveness, cell-survival, metastasis, angiogenesis, therapeutic resistance and immunoevasion (42,43). Moreover, these interleukins are synthesized by tumor cells themselves as well as in the environmental niche and may act both in autocrine and paracrine fashions (42,43). Inhibition of IL6 and IL8 expression via its effects of CEBPB and CEBPD signaling may therefore represent one of the means by which DN-ATF5 affects tumor growth and survival.
The concentrations of CP-DN-ATF5 that interfered with the activities of endogenous CEBPB and CEBPD in living cells were equivalent to those that promote cancer cell death. This raises the question as to whether such effects might be a consequence of the death process. Tumor cell death initiated by CP-DN-ATF5 is not detectable prior to at least 24-48 hours (13). Yet, the changes in CEBPB and CEBPD activity (though not levels) were seen within 24 hours of CP-DN-ATF5 treatment. These observations support the idea that interference with CEBPB and CEBPD signaling by DN-ATF5 is a cause, rather than response to tumor cell death. In support, we found that knockdown of either CEBPB or CEBPD in a variety of tumor cell lines caused apoptosis. Such findings are consistent with reports that interference with CEBPB or CEBPD levels/activity increases apoptotic susceptibility (26,32,37,44,45) as well as the ample evidence that both proteins play roles in the generation, growth, survival, metastasis and therapeutic resistance of multiple neoplasm types (32,34,36–39,45–50). Taken together, our findings suggest that DN-ATF5 affects tumor cell growth and survival both by a two-pronged mechanism. One is by inhibiting the formation of active heterodimers between ATF5 and CEBPB and CDBPD, thus depriving cells of ATF5-dependent transcription. The other is via interference with the capacities of CEBPB and CEBPD to form transcriptionally active homo- and heterodimers with partners in addition to ATF5. It is likely that the pro-apoptotic effects of interfering with ATF5, CEBPB and CEBPD activities reflects dysregulation of multiple genes. BCL2 has emerged as one such gene with reported regulation by both ATF5 and CEBPB (17,32).
Our observations that DN-ATF5 interferes with CEBPB and CEBPD signaling and that interference with such signaling promotes tumor cell death support the prospect that additional means of targeting CEBPB and CEBPD may be effective anti-tumor therapies. For example, DN forms of CEBPB and CEBPD may have potent anti-tumor effects by associating not only with ATF5, CEBPB and CEBPD, but also with their binding partners.
Supplementary Material
Acknowledgements
We thank Ms. Chang Shu for her outstanding technical assistance, Dr. Rajesh Kumar Soni, Proteomics Shared Resource, Herbert Irving Comprehensive Cancer Center, Columbia University Medical Center for his help in carrying out and interpreting mass spectroscopic analysis, and Sapience Therapeutics for their generous provision of ST-47 and CP-DN-ATF5 (ST-36). This work was supported in part by grants from the NIH-NINDS (1R01NS083795; JMA and LAG, JMA PI; J. Angelastro, L. Greene), Sapience Therapeutics (LAG PI; L. Greene, X. Sun), the Thompson Family Foundation Initiative at Columbia University (LAG, PI, L. Greene, X. Sun) and the Emerson Collective (LAG PI, Q. Zhou, P. Jefferson).
Abbreviations:
- DN
dominant-negative
- CP
cell-penetrating
- bZIP
basic leucine zipper
- WB
Western immunoblot
- ChIP
chromatin immunoprecipitation
Footnotes
Disclosure and Potential Conflicts of Interest
Columbia University holds equity in Sapience Therapeutics which has licensed DN-ATF5 technology from Columbia University. LAG and JMA are listed as co-inventors on a patent entitled “Methods for promoting apoptosis and treating tumor cells by inhibiting the expression or function of the transcription factor ATF5” which is owned by Columbia University and on patent application “Compositions and Methods for Inhibiting Tumor Cells by Inhibiting the Transcription Factor ATF5,” which is owned by Columbia University and the University of California, Davis. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Herskowitz I Functional inactivation of genes by dominant negative mutations. Nature 1987;329:219–22. [DOI] [PubMed] [Google Scholar]
- 2.Smith LM, Birrer MJ. Use of transcription factors as agents and targets for drug development. Oncology (Williston Park) 1996;10:1532–8; discussion 41-2. [PubMed] [Google Scholar]
- 3.Angelastro JM. Targeting ATF5 in Cancer. Trends Cancer 2017;3:471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angelastro JM, Ignatova TN, Kukekov VG, Steindler DA, Stengren GB, Mendelsohn C, et al. Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J Neurosci 2003;23:4590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelastro JM, Mason JL, Ignatova TN, Kukekov VG, Stengren GB, Goldman JE, et al. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J Neurosci 2005;25:3889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelastro JM, Canoll PD, Kuo J, Weicker M, Costa A, Bruce JN, et al. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene 2006;25:907–16. [DOI] [PubMed] [Google Scholar]
- 7.Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev 1993;7:1047–58. [DOI] [PubMed] [Google Scholar]
- 8.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J 1995;14:5329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arias A, Lame MW, Santarelli L, Hen R, Greene LA, Angelastro JM. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene 2012;31:739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer 2007;120:1883–90. [DOI] [PubMed] [Google Scholar]
- 11.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, et al. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med 2010;16:671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldheim J, Kessler AF, Schmitt D, Wilczek L, Linsenmann T, Dahlmann M, et al. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. Onco Targets Ther 2018;11:8673–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpel-Massler G, Horst BA, Shu C, Chau L, Tsujiuchi T, Bruce JN, et al. A Synthetic Cell-Penetrating Dominant-Negative ATF5 Peptide Exerts Anticancer Activity against a Broad Spectrum of Treatment-Resistant Cancers. Clin Cancer Res 2016;22:4698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nukuda A, Endoh H, Yasuda M, Mizutani T, Kawabata K, Haga H. Role of ATF5 in the invasive potential of diverse human cancer cell lines. Biochem Biophys Res Commun 2016;474:509–14. [DOI] [PubMed] [Google Scholar]
- 15.Ishihara S, Yasuda M, Ishizu A, Ishikawa M, Shirato H, Haga H. Activating transcription factor 5 enhances radioresistance and malignancy in cancer cells. Oncotarget 2015;6:4602–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rousseau J, Gagne V, Labuda M, Beaubois C, Sinnett D, Laverdiere C, et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood 2011;118:5883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dluzen D, Li G, Tacelosky D, Moreau M, Liu DX. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J Biol Chem 2011;286:7705–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Shmuel S, Rashed R, Rostoker R, Isakov E, Shen-Orr Z, LeRoith D. Activating Transcription Factor-5 Knockdown Reduces Aggressiveness of Mammary Tumor Cells and Attenuates Mammary Tumor Growth. Front Endocrinol (Lausanne) 2017;8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cates CC et al. Regression/eradication of gliomas in mice by a systemically-deliverable ATF5 dominant-negative peptide. Oncotarget 7, 12718–12730, doi: 10.18632/oncotarget.7212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupont E, Prochiantz A, Joliot A. Penetratin Story: An Overview. Methods Mol Biol 2015;1324:29–37. [DOI] [PubMed] [Google Scholar]
- 21.Peters CS, Liang X, Li S, Kannan S, Peng Y, Taub R, et al. ATF-7, a novel bZIP protein, interacts with the PRL-1 protein-tyrosine phosphatase. J Biol Chem 2001;276:13718–26. [DOI] [PubMed] [Google Scholar]
- 22.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 2003;300:2097–101. [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 2006;1:2856–60. [DOI] [PubMed] [Google Scholar]
- 24.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell 2006;10:203–14. [DOI] [PubMed] [Google Scholar]
- 25.Flanagan KC, Alspach E, Pazolli E, Parajuli S, Ren Q, Arthur LL, et al. c-Myb and C/EBPbeta regulate OPN and other senescence-associated secretory phenotype factors. Oncotarget 2018;9:21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baccam M, Woo SY, Vinson C, Bishop GA. CD40-mediated transcriptional regulation of the IL-6 gene in B lymphocytes: involvement of NF-kappa B, AP-1, and C/EBP. J Immunol 2003;170:3099–108. [DOI] [PubMed] [Google Scholar]
- 27.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 2013;10:730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A 1993;90:2207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med 2009;11:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hungness ES, Luo GJ, Pritts TA, Sun X, Robb BW, Hershko D, et al. Transcription factors C/EBP-beta and -delta regulate IL-6 production in IL-1beta-stimulated human enterocytes. J Cell Physiol 2002;192:64–70. [DOI] [PubMed] [Google Scholar]
- 31.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 2008;133:1019–31. [DOI] [PubMed] [Google Scholar]
- 32.Pal R, Janz M, Galson DL, Gries M, Li S, Johrens K, et al. C/EBPbeta regulates transcription factors critical for proliferation and survival of multiple myeloma cells. Blood 2009;114:3890–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanasopoulou A, Stravopodis DJ, Dimas KS, Schwaller J, Anastasiadou E. Loss of CCDC6 affects cell cycle through impaired intra-S-phase checkpoint control. PLoS One 2012;7:e31007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessells J, Yakar S, Johnson PF. Critical prosurvival roles for C/EBP beta and insulin-like growth factor I in macrophage tumor cells. Mol Cell Biol 2004;24:3238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YH, Wu WJ, Wang WJ, Huang HY, Li WM, Yeh BW, et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget 2015;6:31069–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilar-Morante D, Cortes-Canteli M, Sanz-Sancristobal M, Santos A, Perez-Castillo A. Decreased CCAAT/enhancer binding protein beta expression inhibits the growth of glioblastoma cells. Neuroscience 2011;176:110–9. [DOI] [PubMed] [Google Scholar]
- 37.Cao J, Wang M, Wang T. CCAAT enhancer binding protein beta has a crucial role in regulating breast cancer cell growth via activating the TGF-beta-Smad3 signaling pathway. Exp Ther Med 2017;14:1554–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010;463:318–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A 2002;99:207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun 2018;9:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerrato A, Merolla F, Morra F, Celetti A. CCDC6: the identity of a protein known to be partner in fusion. Int J Cancer 2018;142:1300–8. [DOI] [PubMed] [Google Scholar]
- 42.Alfaro C, Sanmamed MF, Rodriguez-Ruiz ME, Teijeira A, Onate C, Gonzalez A, et al. Interleukin-8 in cancer pathogenesis, treatment and follow-up. Cancer Treat Rev 2017;60:24–31. [DOI] [PubMed] [Google Scholar]
- 43.Kumari N, Dwarakanath BS, Das A, Bhatt AN. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol 2016;37:11553–72. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee S, Aykin-Burns N, Krager KJ, Shah SK, Melnyk SB, Hauer-Jensen M, et al. Loss of C/EBPdelta enhances IR-induced cell death by promoting oxidative stress and mitochondrial dysfunction. Free Radic Biol Med 2016;99:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R, et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J 2010;29:4106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balamurugan K, Mendoza-Villanueva D, Sharan S, Summers GH, Dobrolecki LE, Lewis MT, et al. C/EBPdelta links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tregnago C, Manara E, Zampini M, Bisio V, Borga C, Bresolin S, et al. CREB engages C/EBPdelta to initiate leukemogenesis. Leukemia 2016;30:1887–96. [DOI] [PubMed] [Google Scholar]
- 48.Liu D, Zhang XX, Li MC, Cao CH, Wan DY, Xi BX, et al. C/EBPbeta enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat Commun 2018;9:1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salotti J, Sakchaisri K, Tourtellotte WG, Johnson PF. An Arf-Egr-C/EBPbeta pathway linked to ras-induced senescence and cancer. Mol Cell Biol 2015;35:866–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim MH, Minton AZ, Agrawal V. C/EBPbeta regulates metastatic gene expression and confers TNF-alpha resistance to prostate cancer cells. Prostate 2009;69:1435–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.