Summary
Satellite cells are main muscle stem cells that could provide myonuclei for myofiber growth and synaptic-specific gene expression during the early postnatal development. Here, we observed that splicing factor SRSF1 is highly expressed in myoblasts and its expression is closely related with satellite cell activation and proliferation. By genetic deletion of SRSF1 in myogenic progenitors, we found that SRSF1 is critical for satellite cell proliferation in vitro and in vivo. Most notably we also observed that SRSF1 is required for the functional neuromuscular junction (NMJ) formation, as SRSF1-deficient mice fail to form mature pretzel-like NMJs, which leads to muscle weakness and premature death in mice. Finally, we demonstrated that SRSF1 contributes to muscle innervation and muscle development likely by regulating a restricted set of tissue-specific alternative splicing events. Thus, our data define a unique role for SRSF1 in postnatal skeletal muscle growth and function in mice.
Keywords: splicing factor SRSF1, satellite cell proliferation, neuromuscular junction maturation, postnatal myogenesis, alternative splicing regulation
Highlights
-
•
SRSF1 is highly expressed in activated satellite cells
-
•
Loss of SRSF1 dramatically impairs satellite cell proliferation in vitro and in vivo
-
•
SRSF1 is also required for the functional neuromuscular junction formation in mice
-
•
SRSF1-deficient mice display muscle weakness and die prematurely
In this article, Feng and colleagues show that the splicing factor SRSF1 is highly expressed in activated satellite cells, and its expression is closely related with satellite cell activation and proliferation. SRSF1 is also required for functional neuromuscular junction (NMJ) formation in mice. SRSF1-deficient mice display severe NMJ disorders, muscle weakness, and premature death.
Introduction
Skeletal muscle formation is a precisely coordinated process, which involves highly complicated regulatory mechanisms. After a period of progenitor proliferation, differentiation and fusion to form myofibers during embryonic development, postnatal development is strongly influenced by satellite cell activity, innervation, and electrical activity (White, et al., 2010; Dhawan and Rando, 2005).
Satellite cells are located at the periphery of muscle fibers, which are main muscle stem cells (Mauro, 1961). They are most abundant within 3 weeks after birth and quickly decline after that to adult levels in mice (Bachman, et al., 2018). During early postnatal development, satellite cells undergo extensive proliferation and most of their progeny (myoblasts) fuse with existing myofibers, leading to a continuous increase in the numbers of myonuclei in the developing fibers (Bachman, et al., 2018). Expression of PAX7 is indispensable for satellite cell survival during the postnatal period, as inactivation of PAX7 led to decreased numbers of satellite cells and hypotrophic muscles in mice (Oustanina, et al., 2004; Seale, et al., 2000). MyoD and Myf5 are master transcription factors that can induce a unique gene expression profile for myogenesis in activated satellite cells (Ustanina, et al., 2007; Megeney, et al., 1996). Combined disruption of MyoD and Myf5 genes leads to depletion of satellite cells and a complete loss of skeletal muscle (Rudnicki, et al., 1993). Despite a lot of progress, the precise mechanism underlying satellite cell proliferation during postnatal muscle growth still remains elusive.
In addition to satellite cells, development of skeletal muscle relies heavily on the synapse between the motor neuron terminals and endplates on the muscle fibers, which is known as NMJ. During embryonic development, the acetylcholine receptors (AChRs) become clustered in the middle region of the muscle fibers, where motoneuron terminals will innervate and overlap AChRs. Within the first few weeks after birth, multiple innervations of muscle fibers are reduced until each muscle fiber becomes innervated by only one motor axon; fetal AChRs composed of α2ßγδ subunits are gradually replaced by adult-type AChRs, where the ε-subunit replaces the fetal γ-subunit, and AChR clusters undergo plaque-to-pretzel transition as the postsynaptic membrane invaginates to form junctional folds (Li, et al., 2018; Liu and Chakkalakal, 2018).
NMJ formation involves a complex exchange of signals between motoneurons and skeletal muscle fibers. Studies in the past two decades have brought significant progress in identifying NMJ components and assembly mechanisms (Li, et al., 2018; Liu and Chakkalakal, 2018). The Agrin/Lrp4/Musk/Dok7 pathway is the best characterized to be implicated in NMJ development, in which the muscle-specific kinase Musk is a master organizer (Okada, et al., 2006; Weatherbee, et al., 2006; DeChiara, et al., 1996; Glass, et al., 1996). Mutations in Musk, Lrp4, agrin, and Dok7 have been identified in congenital neuromuscular disorders termed as congenital myasthenic syndromes (CMS), although the majority of CMS cases are due to mutations in AChR subunits (Engel, et al., 2015). Some mutations in CMS are due to mutations affecting pre-mRNA splicing (Ohno, et al., 2003), and analysis of splicing defects has disclosed unprecedented roles of RNA-binding proteins in maintaining NMJ integrity (Ohno, et al., 2017.
In higher eukaryotes, the majority of genes express primary transcripts that undergo alternative splicing (AS) regulation in the nucleus to generate different functional mRNAs (Black, 2003; Chen and Manley, 2009). The serine/arginine-rich (SR) proteins are well-known splicing factors that play critical roles in AS regulation. We have observed that SR proteins can activate inclusion of alternative exons to generate splicing variants that significantly stimulate cell growth in different cell lines (Chen, et al., 2017; Luo, et al., 2017; Zhou, et al., 2014a). Moreover, knockdown of SR genes in the mouse results in multi-organ dysfunction accompanied by unusual AS changes. Targeted inactivation of SRSF1 affects splicing transition of the CaMKIId pre-mRNA, causing severe excitation-contraction coupling defects in mice (Xu, et al., 2005). SRSF2 and SRSF3 have been shown to regulate liver-specific AS events, and their expression is essential for liver homeostasis and hepatic metabolism (Cheng, et al., 2016; Sen, et al., 2013). These findings have demonstrated that SR proteins and their regulated AS events play critical roles in cell proliferation and tissue development.
In this study, we observed that SRSF1 was highly expressed in satellite cells, and loss of SRSF1 significantly impaired satellite cell proliferation, causing the cell number to decrease sharply after birth in mice. Meanwhile, we also observed disassembly of AChRs and lack of motor neuron terminals on the muscle fiber, indicating severe NMJ disorders. As a result, SRSF1-deficient mice failed to thrive and all died before they reached maturity.
Results
SRSF1 Is Mainly Expressed in Activated Satellite Cells
Analysis of published microarray data have revealed that activated satellite cells possess a unique transcriptional profile that distinguishes them from the quiescent satellite cells or non-muscle satellite cells (Figure S1). Interestingly, we found that several genes encoding splicing factors could be induced to be highly expressed upon satellite cell activation. We decided to focus on the SRSF1 gene, as SRSF1 was reported to be essential for the development of heart muscle in mice (Xu, et al., 2005) and also it could promote the proliferation of smooth muscle cells (Xie, et al., 2017).
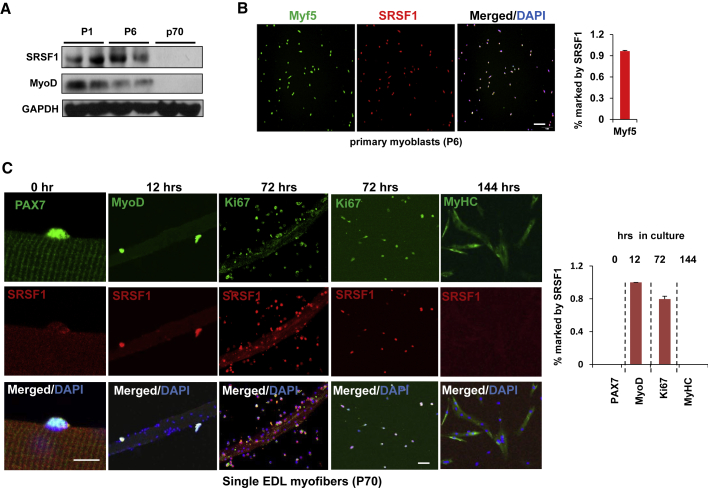
Next we wanted to examine whether SRSF1 is expressed in activated satellite cells. We first performed western blot analysis using neonatal skeletal muscle isolated from mice at 1 day and 6 days after birth (P1 and P6), when satellite cells are highly proliferative, and adult skeletal muscle lacking appreciable numbers of proliferating satellite cells was used as a control. As shown in Figure 1A, SRSF1 was only examined in neonatal but not adult skeletal muscle, and its expression pattern is similar to that observed for MyoD in skeletal muscle. Then we purified primary satellite cells from the P6 mice, using the fluorescence-activated cell sorting (FACS) technique. After confirming the satellite cell identity of FACS-isolated cells (Figure S2), we observed that SRSF1 was detected in approximately 90% of satellite cells, and co-localized with Myf5 (Figure 1B). The results suggested that SRSF1 was highly expressed in proliferating satellite cells.
Figure 1.
Expression of SRSF1 Was Closely Related to Satellite Cell Activation and Proliferation
(A) Skeletal muscle protein samples were harvested from P1, P6, and P70 C57BL/6 mice and western blots were performed using the indicated antibodies (n = 3).
(B) Primary satellite cells were isolated from C57BL/6 mice at the age of P6. Cells were cultured in growth medium and stained with Myf5 (green) or SRSF1 (red). Right panels show merged images with DAPI (blue). Percentage of Myf5+ cells also marked with SRSF1 staining was shown on the right histogram. Scale bar, 100 μm. Data were shown as mean ± SEM in an independent experiment (n = 3).
(C) Extensor digitorum longus (EDL) myofibers were prepared from adult mice and cultured in vitro. Immunostaining was performed using antibodies against PAX7 (green), MyoD (green), MyHc (green), Ki67 (green), or SRSF1 (red) at indicated time. Bottom panels show merged images with DAPI (blue). Fraction of PAX7+, MyoD+, ki67+, or MyHc+ nuclei that are also marked by SRSF1 staining was shown in histogram on the right panel. Scale bars, 20 μm (0 h), 20 μm (12 and 72 h), or 50 μm (144 h), respectively. Data were shown as mean ± SEM in an independent experiment (n = 3).
In mature muscle, satellite cells are typically quiescent but retain the ability to proliferate and differentiate, as demonstrated by rapid activation and proliferation in single fiber cultures (Danoviz and Yablonka-Reuveni, 2012). Next, we decided to perform immunostaining on single myofibers obtained from extensor digitorum longus (EDL) muscles of adult C57BL/6 mice. As shown in Figure 1C, SRSF1 was not expressed in satellite cells attached on the freshly isolated single fiber that expressed PAX7 (0 h). After 12-h culture when satellite cells were activated to produce MyoD, expression of SRSF1 was detected and fully co-localized with MyoD. Over the next 48 h, SRSF1 was still expressed and co-localized with the proliferation marker Ki67 in progeny myoblasts (72 h). However, expression of SRSF1 significantly diminished after myoblasts were differentiated into myotubes marked by MyHc expression (144 h). These data strongly indicated that expression of SRSF1 was closely associated with satellite cell activation and proliferation.
Inactivation of SRSF1 in Skeletal Muscle Resulted in Premature Death in Mice
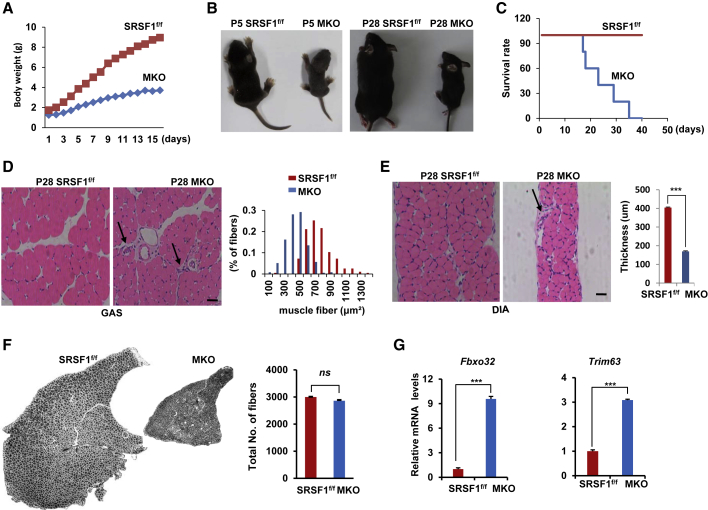
As SRSF1 was expressed in highly proliferative satellite cells, next we wanted to investigate the physiological consequences of satellite cells lacking SRSF1. We therefore inactivated SRSF1 selectively in skeletal muscle by crossing SRSF1f/f mice (Xu, et al., 2005) with MyoDcre mice (Chen, et al., 2005) (Figure S3A). Knockout (MKO) mice appeared normal at birth, but shortly after birth they showed significant differences in body weight when compared with heterozygous-Cre mice and SRSF1f/f mice. In the first few weeks, we found that MKO animals showed severe deficits in weight gain when compared with control littermates (Figure 2A). The body weight of MKO mice at P5 was 50% reduced in comparison with control littermates. Such weight difference increased with time, and MKO animals were only 20% the weight of control littermates at P28 (Figure 2B). Meanwhile, the levels of serum glucose and TG decreased significantly in the mutant mice (Figure S3B). To rule out the weight loss of MKO mice due to inability to compete for milk, only MKO mice and a few control mice were allowed to have maternal care shortly after birth. Under meticulous care, we observed that MKO mice still showed underweight phenotypes and most died around the time of weaning, with none surviving past 5 weeks (Figure 2C). Besides severe weight gain deficiency, MKO mice also showed abnormal movement. For example, they usually had droopy eyelids, and became much less active than their control littermates. Then we prepared protein samples from hindlimb muscles and diaphragm (DIA) muscles from control and MKO mice at P8, or from gastrocnemius (GAS) muscles, vastus muscles, DIA muscles, and tibialis anterior (TA) muscles from mice at P28. Western blot analysis confirmed that levels of SRSF1 proteins were greatly decreased in various skeletal muscles of MKO mice (Figure S3C). Together, these findings demonstrated that inactivation of SRSF1 in skeletal muscle resulted in premature death in mice.
Figure 2.
Targeting SRSF1 Gene in Skeletal Muscle Resulted in Premature Death in Mice
(A) Comparison of body weight between SRSF1f/f mice and MKO mice (n = 9) at the indicated days after birth.
(B) Macroscopic view of SRSF1f/f mice and MKO mice at P5 or P28, respectively.
(C) Survival analysis of SRSF1f/f mice and MKO mice treated with meticulous care (n = 20).
(D) H&E staining was performed with GAS sections isolated from control and MKO mice at P28. Comparison of CSA (cross-section area) between control and MKO animals was shown in histogram on the right. Scale bar, 50 μm.
(E) H&E staining was performed with DIA sections isolated from control and MKO mice at P28. Quantification of DIA thickness between control and MKO animals is shown on the right histogram. Scale bar, 50 μm. Error bars depicted mean ± SEM (n = 3).
(F) TA muscles from P30 control and MKO mice were isolated, cross-sectioned, and then stained with laminin a2 antibodies. Comparison of total TA fiber number between control and MKO mice was shown on the right (n = 3).
(G) Levels of two atrophy marker genes were measured by qRT-PCR using total RNA isolated from the vastus muscles from control and MKO mice at P28. All results were normalized to 36B4 (n = 3).
Severe Muscle Atrophy Was Observed in MKO Mice
Next we performed histological analysis of SRSF1f/f and MKO mice at P28. Transverse sections from GAS muscles revealed a clear reduction in muscle cross-sectional area and a strong shift of myofiber diameters toward smaller fiber sizes in MKO mice compared with controls (Figure 2D), and notably thinner DIA muscles were even evident in MKO mice than in control animals (Figure 2E). Then we isolated single myofibers from EDL muscles of MKO or control mice at P23 and stained them with DAPI. As shown in Figure S3D, the mutant fibers were nearly half thinner than control fibers. And the number of myonuclei per fiber was also significantly reduced in MKO mice.
To determine whether reduction in muscle mass is due to fiber cross-sectional area and/or fiber number, we prepared whole cross-sections of TA muscles from P30 MKO and control mice, and stained for laminin to mark the myofibers. The whole TA section from MKO mice showed a clear reduction in muscle area; however, the total number of fibers in the TA muscles did not differ significantly between control and MKO mice (Figure 2F). qPCR further verified increased levels of two well-known atrophy makers Fbxo32 and Trim63 in the mutant muscles at P28 (Figure 2G). All the data indicated that MKO mice suffered from severe muscle atrophy.
In addition, we also observed that a few areas in Figures 2D and 2E (indicated by arrows) displayed signs of fibrosis in MKO animals, but no signs of necrosis or apoptosis. We then stained sections with anti-CD68 antibodies for inflammation, or with antibodies for fibronectin and counterstained with DAPI (Figure S4). Only a few positive signals were visible for CD68 staining in MKO GAS muscles. There was no difference for fibronectin staining in most areas for both GAS and DIA muscles, with a small amount of region staining enhanced in MKO muscles. Taken together, these findings indicated that SRSF1-deficient muscles were free from gross pathology.
Satellite Cell Proliferation Was Severely Impaired in MKO Mice
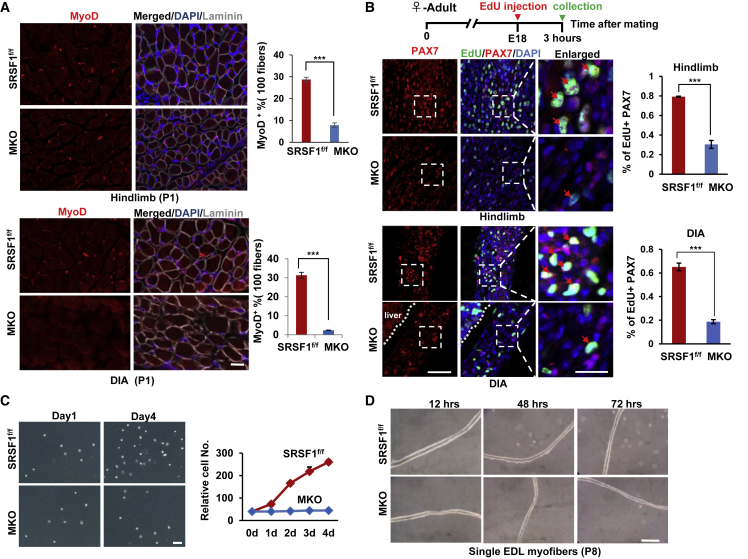
During postnatal skeletal muscle development, satellite cells provide myonuclei to developing myofibers to support their transcriptional demands. Next, we wanted to examine whether satellite cell activity was impaired in the MKO mice. First of all, we performed immunostaining with antibodies against MyoD and laminin using skeletal muscle sections isolated from the P1 mice, and then the MyoD+ cells quantified. The number of MyoD+ satellite cells was decreased by approximately 70% in the hindlimb muscles and by nearly 90% in DIA muscles of MKO mice compared with control animals (Figures 3A and S5). Then we studied the effect of SRSF1 deletion on satellite cell proliferation in vivo. We treated embryos at the age of embryonic day 18 (E18) with EdU solution by injecting the adult pregnant mice. Fully in line with the above results, we observed that the total number of EdU/PAX7+ cells was significantly reduced in both hindlimb and DIA muscles from MKO mice when compared with controls (Figure 3B). These findings demonstrated that absence of SRSF1 caused a significant decline in satellite cell numbers.
Figure 3.
SRSF1 Is Indispensable for Postnatal Satellite Cell Proliferation In Vivo and In Vitro
(A) Hindlimb and DIA muscles from P1 control and MKO mice was prepared and stained for MyoD (red), and merged images with DAPI (blue) and laminin (gray) are shown on the right. Quantification of MyoD+ nuclei between control and MKO animals is shown on the right histogram. Error bars depicted mean ± SEM (n = 3). Scale bar, 50 μm.
(B) Brief diagram of EdU in vivo labeling and detection on the top. EdU detection and calculation in labeled hindlimb muscles and DIA muscles on the bottom. EdU was detected by labeling with Alexa 488 (green) and nuclei were stained with DAPI (blue), proliferating satellite cells were labeled with PAX7 (red). Error bars depicted mean ± SEM (n = 3). Scale bars, 50 μm (or 20 μm enlarged).
(C) Satellite cells were purified from P6 control and MKO mice using FACS, and cultured in growth medium in vitro. Representative microscopic images of control and MKO satellite cells at culture day 0 and day 4 were shown. Growth curve is shown on the right (n = 3).
(D) Single EDL myofibers were isolated from P8 control and MKO mice. Myofibers were plated on Matrigel-coated dishes and cultured in proliferation medium. Representative bright-field pictures are shown at the indicated times (n = 3). Scale bar, 50 μm.
To further assess the proliferation potential of satellite cells lacking SRSF1, we FACS-purified satellite cells from hindlimbs of P6 MKO or control mice. In culture, control satellite cells could proliferate and expand, resulting in a rapid increase in cell numbers; however, the number of MKO satellite cells remained unchanged during the subsequent 4 days (Figure 3C). Next, we isolated single myofibers from EDL muscles of MKO or control mice in three independent experiments. As shown in Figure 3D, satellite cells associated with control fibers could be activated to proliferate and produce myoblasts. In contrast, MKO satellite cells on the single fiber could not be activated and proliferate efficiently as control cells, as only a few myoblasts were visible after 72 h of culture. These findings revealed an indispensable role for SRSF1 in regulation of satellite cell proliferation in vitro and in vivo.
SRSF1 Regulates AS of Genes Functionally Involved in Cell Proliferation and NMJ Formation
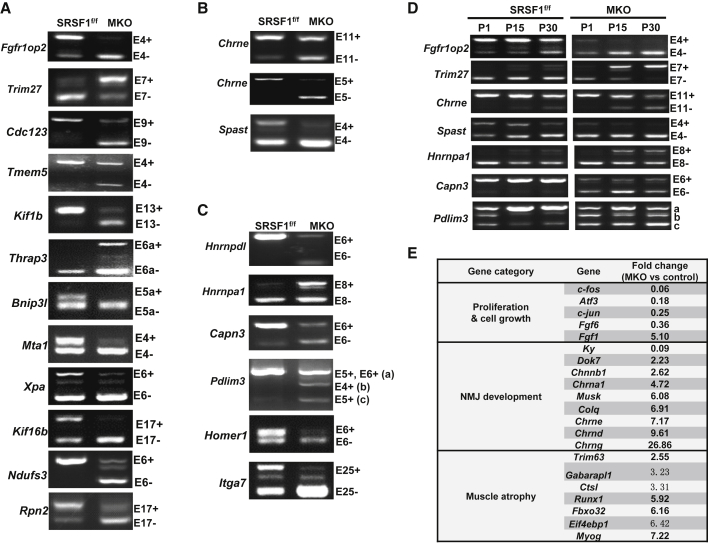
Given that SRSF1 acts as an important regulator of AS, we decided to take advantage of RNA sequencing (RNA-seq) analysis to identify SRSF1-regulated AS events. RNA-seq analysis revealed that SRSF1 ablation caused more than 100 AS events significantly altered in MKO vastus muscles compared with controls, and the majority of them were cassette exons.
We selected 100 targets for experimental validation, based mainly on their p values. Among them, 50 splicing events were successfully confirmed and examples were roughly classified by function into 3 groups. About half of the validated genes have been reported to be related with cell proliferation, and SRSF1 deficiency caused significant exon skipping or inclusion compared with their control patterns (Figure 4A). Among other validated splicing targets, one target attracted our attention (Figure 4B). Chrne gene encodes the adult ε-subunit of AChR. Mutations in CHRNE gene have been implicated in human CMS (Ohno, et al., 2003). SRSF1 deletion caused skipping of both E5 and E11 of Chrne pre-mRNA, producing non-functional Chrne isoforms. The third group included several well-known mutation-prone genes, which have been observed in different human skeletal muscle diseases (Figure 4C). For example, SRSF1 deletion resulted in an increased inclusion of Hnrnpa1 E8, and the human E8-containing isoform was related with amyotrophic lateral sclerosis (Deshaies, et al., 2018).
Figure 4.
Representative AS Events and Genes Altered Following Loss of SRSF1 in Skeletal Muscle
(A–C) Representative AS events regulated by SRSF1 in the P30 skeletal muscle. Validated RT-PCR results are shown with alternative exons included or skipped in response to loss of SRSF1 (n = 3).
(D) Seven AS events shown in (A–C) were further examined in muscles isolated from P1, P15, or P30 control and MKO mice (n = 3).
(E) Three groups of genes with significant changes in MKO mice. Fold changes of RNA-seq reads mapping to related genes were calculated between MKO and control samples.
We next wanted to examine how these AS events were modified during postnatal development. We selected seven targets for examinations, and found that all had the similar isoform switching at P15 and P30 of MKO skeletal muscles compared with controls, and several targets, such as Fgfr1op2 and Capn3, showed AS changes even at P1 (Figure 4D). In addition to AS analysis, we also analyzed differentially expressed genes in MKO mice compared with control mice. Among the most obvious genes shown in Figure 4E, proliferation-related genes, such as c-fos and Atf3, were significantly downregulated in MKO mice. Genes involved in NMJ formation and maintenance were greatly upregulated, such as Musk, ColQ (encoding the collagen-tail subunit of acetylcholinesterase), Dok7, and AChR subunit genes. Notably, all known markers of denervation were increased between ∼2- and 7-fold over the controls (Moresi, et al., 2010). These data probably suggested that altered AS events and concomitant changes in gene expression together contributed to pathological features observed in SRSF1-deficient mice.
SRSF1 Mediated Its Effects in Satellite Cell Proliferation by Regulating AS Events
We next wanted to test whether SRSF1 mediates its effects in satellite cell proliferation by regulating AS events. We decided to focus on the Fgfr1op2 gene for testing. Fgfr1op2 E4 was regulated by SRSF1 to generate two isoforms: full-length (L) and short (S) (Figure S6A). We designed two independent small interfering RNAs (siRNAs) (siL-1 or siL-2) targeted against E4 for isoform-specific knockdown. After transfection of primary myoblasts with siRNAs, we observed that treatment with these siRNAs significantly diminished levels of the L isoform, and the effect is comparable with the treatment with siRNA against SRSF1 (siSRSF1) (Figure S6B). Then we analyzed cell proliferation by EdU incorporation assay. As shown in Figure S6C, knockdown of the L isoform and SRSF1 considerably suppressed myoblast proliferation. Taken together, SRSF1 could mediate its effects in satellite cell proliferation by regulating AS events.
Deletion of SRSF1 Caused Severe NMJ Defects in Mice
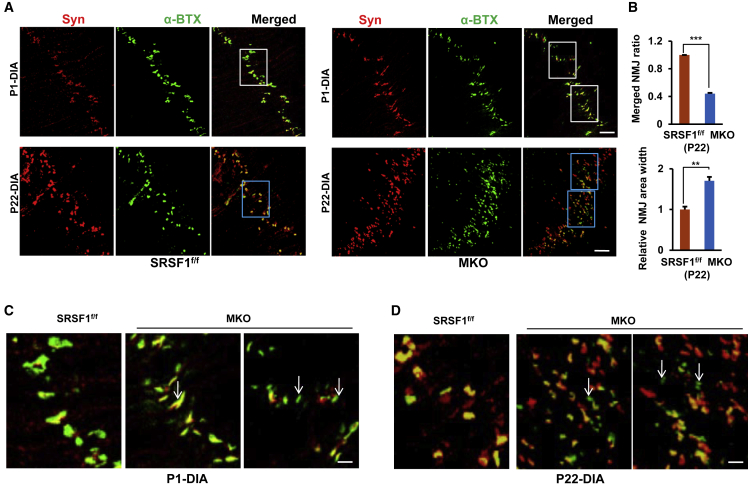
Given that SRSF1 regulated AS of Chrne pre-mRNA and that MKO mice displayed abnormal phenotypes, such as droopy eyelids and muscle weakness, we decided to test whether NMJs were impaired in MKO mice. To address this issue, we isolated DIA muscles from P1 or P22 mice and performed whole-mount staining with synaptophysin antibodies to label the presynaptic nerve terminals or ATTO 488-conjugated α-bungarotoxin (α-BTX) to label the postsynaptic AChRs. As shown in Figure 5A, the number of NMJs and coverage of presynaptic nerve terminals over AChR clusters were comparable in mice at P1 irrespective of SRSF1. In contrast, NMJs were much more broadly distributed in MKO mice at P22. More strikingly, a significant number (40%–50%) of nerve terminals were not co-localized with AChRs in the DIA muscles (Figure 5B). Moreover, AChR clusters were quite small and displayed various shapes in MKO mice compared with those observed in control mice (Figures 5C and 5D, arrows), and the majority of clusters lost their normal architecture and shrunk into small dot-like areas in P22 MKO mice (Figure 5D). All the findings strongly indicated that loss of SRSF1 caused severe NMJ defects in DIA muscles.
Figure 5.
SRSF1 Deficiency Dramatically Impaired NMJ Maturation in the DIA Muscles
(A) Whole-mount DIA staining at P1 or P22. DIA prepared from SRSF1f/f (left) or MKO (right) at P1 or P22 were stained with synaptophysin (Syn, red) and α-BTX (green). Merged pictures are shown on the right. Scale bars, 50 μm for the P1 group or 100 μm for the P22 group.
(B) The number of merged NMJs and NMJ area width were measured between the P22 group described in (A) (n = 3). The mean value of the merged NMJ in SRSF1f/f mice was arbitrarily defined as 1.
(C) Enlarged view of white boxes in the merged panel of P1 DIA in (A). Scale bar, 15 μm.
(D) Enlarged view of blue boxes in the merged panel of P22 DIA in (A) (n = 3). Scale bar, 30 μm.
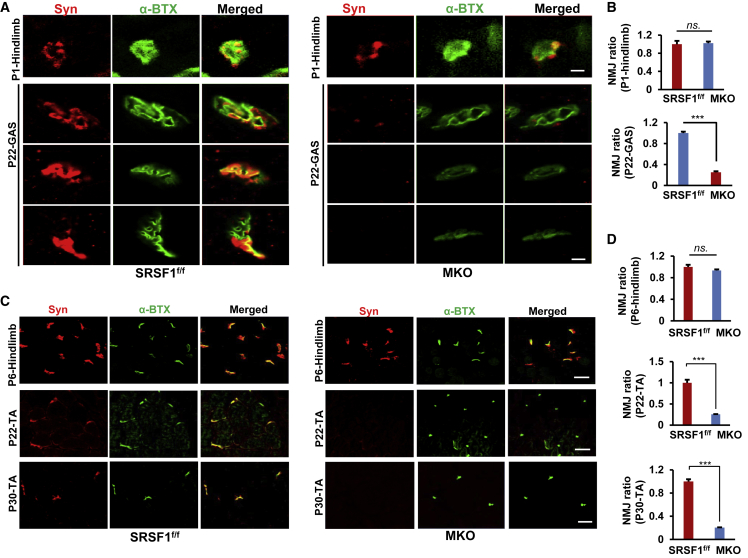
To further address the difference of NMJ morphology between control and MKO mice during early postnatal development, we then performed synaptophysin and BTX double staining on the longitudinal section of hindlimb and GAS muscles isolated from control and MKO mice at P1 or P22. Under this period (P1–P22), NMJs usually undergo the transition from plaque-like shape to a complex, pretzel-like structure. Such transition was observed in control mice, but not in MKO mice (Figure 6A). Instead NMJs in MKO mice presented a variety of abnormal structures at P22, and NMJs were almost fully denervated at P22 in MKO mice, in contrast to those fully innervated in age-matched control mice (Figures 6A and 6B).
Figure 6.
Loss of SRSF1 Led to Disassembly of AChRs and Denervation in GAS and TA Muscles
(A) Double staining of NMJs in the longitudinal section of hindlimb (P1) or GAS (P22) prepared from SRSF1f/f (left) or MKO mice (right). Merged pictures were shown on the right. Scale bar, 5 μm for both P1 and P22 groups.
(B) NMJ ratio was compared between control and MKO mice described in (A) (n = 3).
(C) Double staining of NMJs on the cross-section of TA prepared from SRSF1f/f (left) or MKO mice (right) at P6, P22, or P30. Scale bar, 30 μm for the P6 group and 40 μm for both P22 and P30 groups.
(D) NMJ ratio was compared between control and MKO mice described in (C) (n = 3).
In the end, we wanted to examine whether the progression of NMJ defects occur during the development. We performed double staining on the cross-sections of TA muscles isolated from P6, P22, or P30 mice. While there was no obvious difference observed between control and MKO mice at P6, nearly 80%–90% degeneration of nerve terminals and small dot-like AChR clusters were progressively observed in MKO TA muscles at P22 and P30 (Figures 6C and 6D). Taken together, these data strongly demonstrated that loss of SRSF1 severely impaired functional NMJ formation during early postnatal development.
Abnormal Levels of NMJ Markers Were Observed in the MKO Mice
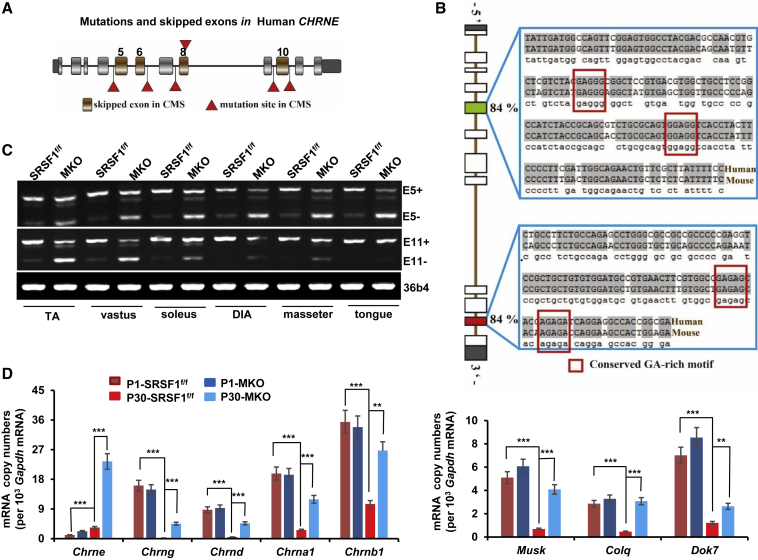
Next we wanted to analyze molecular changes underlying NMJ defects in MKO mice. Mutations observed in CHRNE account for most cases of CMS, and some mutations can lead to exon skipping, including E5 (Figure 7A) (Ohno, et al., 2005). Sequence alignment analysis revealed that human and mice shared about 84% sequence identity in both E5 and E11 of Chrne. Moreover, predicted SRSF1 binding motifs were also conserved between human and mice (Figure 7B). Now we wanted to test whether SRSF1 regulated inclusion of E5 and/or E11 in different skeletal muscles isolated from MKO mice at P30. SRSF1 deletion significantly increased E5 skipping from 40% to 80% in the different muscles isolated from MKO mice in relative to controls (Figure 7C). By comparison, loss of SRSF1 caused more skipping of E11 in TA, vastus, and soleus muscles, but much less in DIA, masseter, and tongue muscles, indicating that SRSF1 was mainly responsible for E5 inclusion in skeletal muscles.
Figure 7.
Abnormal Levels of NMJ Markers after Loss of SRSF1
(A) Diagram of human CHRNE gene structure. Brown boxes indicated skipping exons reported in CMS; red triangles indicated point mutations that trigger CMS-related exon skipping. UTRs were marked as black boxes; constitutive exons were marked as gray boxes.
(B) Alignment of human CHRNE E5, E11 and mouse Chrne E5, E11. Note that predicted binding motifs for SRSF1 were conserved between human and mice and are indicated by red rectangles.
(C) Total RNA was isolated from TA, vastus, soleus, DIA, masseter, or tongue muscles from SRSF1f/f and MKO mice at P30. RT-PCR was performed to detect the splicing pattern of Chrne E5 and E11. 36b4 was used as loading control (n = 3).
(D) qPCR was performed using total RNA isolated from hindlimb muscle of P1 mice or from vastus muscles of P30 mice. Comparison of mRNA levels of NMJ markers were performed using mRNA copy numbers (per 103Gapdh mRNA). Error bars depicted mean ± SEM (n = 3).
The adult ε-subunit of AChR gradually replaces the fetal γ-subunit during early postnatal development. Incorrect replacement or co-expression of fetal and adult subunits would lead to NMJ defects in mice. We next wanted to examine levels of all subunits of AChRs and other NMJ markers during early development. As shown in Figure 7D, both control and MKO animals at P1 showed pronounced but comparable levels of Musk, ColQ, Dok7, and γ/δ/α1/β1 of AChRs, but low levels of the ε-subunit. During the first postnatal month, the levels of Musk, ColQ, and Dok7 mRNA significantly declined in control mice. Meanwhile expression of the fetal γ-subunit is lost, accompanied by an increase in the adult ε-mRNA and a dramatic decline in levels of other subunits in control mice. However, such dramatic changes did not occur in the MKO mice, which resulted in a significant difference in expressed transcripts between MKO and control mice at P30, fully in line with differentially expressed genes revealed by RNA-seq analysis. Especially the fetal AChRs (γ/δ/α1/β1) were not replaced by adult receptors composed of γ/ε/α1/β1 as NMJs mature, because the fetal γ-subunit and adult ε-subunit were highly co-expressed in the MKO mice. Unexpectedly, the Chrne transcript was still 3-fold greater in the mutant vastus muscles than in controls. This probably reflected another transcriptional and compensatory regulation in the Chrne gene because of dysregulated splicing. Together dysregulated Chrne splicing combined with abnormal expression of NMJ markers likely underlined NMJ defects in MKO mice.
Discussion
In this study, we have provided strong evidence that the splicing factor SRSF1 is required for satellite cell proliferation and functional NMJ formation. SRSF1-deficient mice display reduced number of satellite cells and severe NMJ disorders. Below we discuss how SRSF1 functions as a critical regulator of cell proliferation and how regulated AS events are implicated in vivo biological processes.
Muscle fibers form during prenatal development, but they are extensively expanded after birth (Bachman, et al., 2018; White, et al., 2010). There is no debating that proliferation and fusion of satellite cells contribute to muscle growth during early stages of postnatal development. However, little is known about the mechanisms by which proliferation of satellite cells is regulated. It is well known that PAX7 could guide postnatal muscle growth and determine satellite cell number, but PAX7 seems to be required for the maintenance and survival of satellite cells, instead of for extensive proliferation of satellite cells (Oustanina, et al., 2004). Notch signaling is implicated in satellite cell activation and proliferation; for example, Numb-deficient satellite cells have proliferation defects during regeneration after muscle injury; however, depletion of Numb cells does not affect postnatal muscle growth (George, et al., 2013). In this study, our data showed that loss of SRSF1 severely impairs proliferation of satellite cells in vivo and therefore causes a significant reduction in satellite cell number during postnatal rapid growth phase. Both single-fiber cultures and primary satellite cell cultures further demonstrated that satellite cells totally lose an ability to proliferate in the absence of SRSF1. Therefore, it is reasonable to conclude that SRSF1 is the driving force for satellite cell proliferation during postnatal period.
Beyond the role of SRSF1 in satellite cell proliferation, unexpectedly we found that SRSF1 was essential for the functional NMJ formation during early postnatal development. Several genes also play indispensable roles in postnatal NMJ maintenance and maturation. Inactivation of ColQ gene affects postnatal synaptic structure and function in mice (Feng, et al., 1999). Postnatal inactivation of Musk in mice caused NMJ disassembly and formation of new NMJs (Hesser, et al., 2006). Mice lacking the Chrne gene died prematurely and displayed NMJ abnormalities (Witzemann, et al., 1996). SRSF1 deficiency caused NMJ defects in the MKO mice, which became progressively worsened with defect development, indicating direct effects of SRSF1 deletion. High levels of ColQ, Musk, and AChR subunits following the loss of SRSF1 probably reflected compensatory regulation.
Numerous cases of NMJ disorders caused by splicing defects have been reported in CMS. In this study, we observed that SRSF1 could promote exon inclusion of Chrne pre-mRNA, especially E5. E5 skipping due to mutations occurred in some CMS patients, and the human and mouse sequences were quite conservative. Therefore, NMJ defects observed in MKO mice could at least be attributed to dysregulated splicing of Chrne pre-mRNA. Besides, we also observed that SRSF1 could regulate AS of hnRNPDL, hnRNPA1, or CAPN3 pre-mRNA (Figure 4). Interestingly, mutations in these genes, and their abnormal splice variants due to SRSF1 deficiency were often observed in different types of muscular dystrophies and myopathies (Vieira, et al., 2014; Kim, et al., 2013). Thus, it would be informative to examine whether SRSF1 is involved in the pathogenesis of human CMS and other skeletal muscle disorders in the future.
Postnatal muscle development depends on satellite cell proliferation and NMJ integrity. Loss of SRSF1 caused both satellite cell defects and NMJ defects in the mice. It is possible that decreased number of satellite cells might further exaggerate NMJ defects. Previous findings have shown the connection between number of satellite cells and NMJ repair after injury, as satellite cell depletion exacerbated muscle atrophy related with NMJ disruption (Liu, et al., 2017; Liu, et al., 2015b). As the optimal driver for satellite cell-specific deletion of SRSF1 is tamoxifen-regulated PAX7 Cre, we aimed to investigate the role of SRSF1 in satellite cells and satellite cell-mediated contribution of NMJ using PAX7 Cre-crossed SRSF1 mice in the future. In addition, we observed that the MKO mice showed feeding problems and malnutrition even with meticulous care. Malnutrition was an unavoidable consequence of muscle weakness due to NMJ defects, and could further aggravate muscle atrophy. Therefore, it is reasonable to conclude that dysfunctional satellite cells, NMJ defects, and concomitant malnutrition would together contribute to phenotypes seen in the MKO mice.
In summary, our data demonstrated that SRSF1 was required for functional NMJ formation and satellite cell proliferation during early postnatal development.
Experimental Procedures
Generation of Skeletal Muscle-Specific SRSF1 Knockout Mice
SRSF1f/f mice were described previously (Xu, et al., 2005). The SRSF1f/f mice were crossed with transgenic C57BL/6J mice expressing Cre recombinase driven by the MyoD promoter. Next, resulting progeny (SRSF1f/+/MyoD-Cre) were mated with SRSF1f/f mice to generate skeletal muscle-specific SRSF1 MKO mice. Littermates lacking the Cre gene (SRSF1f/f) were used as controls. All experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences.
Histology and Immunofluorescence Analysis
Skeletal muscle tissue was dissected and frozen in dry ice, then cryo-sectioned (7 μm) and fixed in ice-cold acetone for immunofluorescence. In brief, frozen sections were permeabilized by 0.5% Triton X-100 and blocked by 5% horse serum in 0.5% Triton X-100 for 1 h. Samples were then stained with primary antibodies followed with secondary antibodies. Images were acquired with a LSM880NLO Zeiss microscope and the ZEN software. For H&E staining, dissected muscle were fixed in 4% paraformaldehyde (PFA), embedded in paraffin, sectioned and stained by standard protocol.
Single Myofiber Isolation and In Vitro Culture
Single myofiber was isolated by tendon to tendon method according to published protocols (Pasut, et al., 2013; Shefer and Yablonka-Reuveni, 2005). In brief, EDL muscle was dissected and digested in DMEM containing 0.2% collagenase type I at 37°C. Fibers were then released to pre-warmed wash medium by carefully pipetting. Single fibers were then plated into Matrigel-coated dishes with culture medium. For immunofluorescence, fibers were fixed in ice-cold 2% PFA for 10 min, quenched by 1% glycine in PBS, permeabilized with PBST and incubated in blocking solution (5% horse serum in PBST) for 40 min. Samples were then incubated with primary antibodies overnight at 4°C and secondary antibodies for 40 min at room temperature. Nuclei were labeled with DAPI. Images were visualized under a confocal laser scanning microscope (LSM880).
Satellite Cell Isolation by FACS
Satellite cell isolation by FACS was performed according to published protocols (Gromova, et al., 2015; Liu, et al., 2015a). In brief, hindlimb muscles were isolated, mechanically dissected and dissociated in digestion media I and II to get mononuclear cells. Mononuclear cells were then stained with CD45, CD31, CD11b, Sca1, and a7-integrin antibodies (Table S1). Stained cells were then analyzed and isolated through a FACSAria II cell sorter (BD Biosciences). Purified satellite cells were placed on Matrigel-coated dishes, and cultured in proliferation medium.
Cell Culture, Transfection, and EdU Incorporation
Transient siRNA transfection was performed using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer's instructions. EdU incorporation was performed by adding EdU (Invitrogen) at a final concentration of 20 μM to primary satellite cells. EdU incorporation was detected using the Click-iT EdU Kit (Invitrogen) according to the manufacturer's instructions. Nuclei were labeled with Hoechst 33342 at a final concentration of 5 μg/mL.
EdU In Vivo Labeling and Detection
For in vivo proliferation assays, pregnant mice at 18 days after mating were injected with EdU (0.1 μg/g body weight) 3 h before delivery by cesarean. DIA muscles and hindlimb muscles were then isolated and sectioned on a cryostat. EdU incorporation was detected using the Click-iT EdU Kit (Invitrogen) according to the manufacturer's instructions. PAX7 was co-stained with EdU to label satellite cells. Images were visualized under a confocal laser scanning microscope (LSM880).
NMJ Immunostaining
For whole-mount tissues staining, DIA muscles from P1 or P22 control and MKO mice were fixed in 1% PFA in PBS for 2 h at room temperature and then quenched with 1% glycine in PBS. After dissection of the connective tissue on the surface, the muscles were then permeabilized with 1% Triton X-100 in PBS overnight at 4°C and then incubated with rabbit antibody against synaptophysin (Thermo Scientific) to label presynaptic nerve terminals followed by incubation with α-BTX (Allomone) to label postsynaptic AChR clusters. For GAS NMJ detection, samples were fixed in 4% PFA for 2 h at room temperature and sectioned on a cryostat into 100-μm-thick sections. Sections were then permeabilized and incubated with antibodies and α-BTX. Images were visualized under a confocal laser scanning microscope (LSM880).
RNA-Seq Analysis
Total RNAs were isolated from vastus muscles of P30 control or MKO mice and subjected to paired-end RNA-seq using an Illumina HiSeq 2000 system according to the manufacturer's instructions. Raw reads were subjected to quality control and then trimmed of library adapters using a custom Python script. Trimmed reads were then aligned with TopHat to the mouse genome mm10. Analysis of AS events was carried out as described previously (Wu, et al., 2017; Zhou, et al., 2014b).
Data and Code Availability
The raw sequence data of RNA-seq have been submitted to the Gene Expression Omnibus database under accession number GSE111671.
Statistical Analyses
All experiments were repeated at least three times. All data presented as histograms refer to mean values ± SEM of the total number of independent experiments. Statistical analysis was performed by Student's t test, p values of ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 were considered to be statistically significant.
Author Contributions
Y.Liu and Y.F. designed the research and analyzed the data. Y.Luo, L.S., R.G., Z.Z., N.Y., R.S., W.Q., Z.W., and Z.X. performed the research. W.W. helped with RNA-seq analysis. Y.L. and Y.F. wrote the paper. All authors reviewed and edited the manuscript.
Acknowledgments
We thank X.D. Fu for the SRSF1f/f mice. This work was supported by grants from the Shanghai Scientific Research Project (19JC1416000), the National Natural Science Foundation of China (31870819, 31570818), and also from the "Personalized Medicine-Molecular Signature-based Drug Discovery and Development" Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12010100).
Published: September 3, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.004.
Contributor Information
Wenwu Wu, Email: wwwu@zafu.edu.cn.
Ying Feng, Email: fengying@sibs.ac.cn.
Supplemental Information
References
- Bachman J.F., Klose A., Liu W., Paris N.D., Blanc R.S., Schmalz M., Knapp E., Chakkalakal J.V. Prepubertal skeletal muscle growth requires PAX7-expressing satellite cell-derived myonuclear contribution. Development. 2018;145:dev167197. doi: 10.1242/dev.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Chen J.C., Mortimer J., Marley J., Goldhamer D.J. MyoD-Cre transgenic mice: a model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis. 2005;41:116–121. doi: 10.1002/gene.20104. [DOI] [PubMed] [Google Scholar]
- Chen L., Luo C., Shen L., Liu Y., Wang Q., Zhang C., Guo R., Zhang Y., Xie Z., Wei N. SRSF1 prevents DNA damage and promotes tumorigenesis through regulation of DBF4B Pre-mRNA splicing. Cell Rep. 2017;21:3406–3413. doi: 10.1016/j.celrep.2017.11.091. [DOI] [PubMed] [Google Scholar]
- Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Luo C., Wu W., Xie Z., Fu X., Feng Y. Liver-specific deletion of SRSF2 caused acute liver failure and early death in mice. Mol. Cell. Biol. 2016;36:1628–1638. doi: 10.1128/MCB.01071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danoviz M.E., Yablonka-Reuveni Z. Skeletal muscle satellite cells: background and methods for isolation and analysis in a primary culture system. Methods Mol. Biol. 2012;798:21–52. doi: 10.1007/978-1-61779-343-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Deshaies J.E., Shkreta L., Moszczynski A.J., Sidibe H., Semmler S., Fouillen A., Bennett E.R., Bekenstein U., Destroismaisons L., Toutant J. TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain. 2018;141:1320–1333. doi: 10.1093/brain/awy062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan J., Rando T.A. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Engel A.G., Shen X.M., Selcen D., Sine S.M. Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. Lancet Neurol. 2015;14:461. doi: 10.1016/S1474-4422(15)00010-1. [DOI] [PubMed] [Google Scholar]
- Feng G., Krejci E., Molgo J., Cunningham J.M., Massoulie J., Sanes J.R. Genetic analysis of collagen Q: roles in acetylcholinesterase and butyrylcholinesterase assembly and in synaptic structure and function. J. Cell Biol. 1999;144:1349–1360. doi: 10.1083/jcb.144.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R.M., Biressi S., Beres B.J., Rogers E., Mulia A.K., Allen R.E., Rawls A., Rando T.A., Wilson-Rawls J. Numb-deficient satellite cells have regeneration and proliferation defects. Proc. Natl. Acad. Sci. U S A. 2013;110:18549–18554. doi: 10.1073/pnas.1311628110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D.J., Bowen D.C., Stitt T.N., Radziejewski C., Bruno J., Ryan T.E., Gies D.R., Shah S., Mattsson K., Burden S.J. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/s0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- Gromova A., Tierney M.T., Sacco A. FACS-based satellite cell isolation from mouse hind limb muscles. Bio Protoc. 2015;5:e1558. doi: 10.21769/bioprotoc.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesser B.A., Henschel O., Witzemann V. Synapse disassembly and formation of new synapses in postnatal muscle upon conditional inactivation of MuSK. Mol. Cell Neurosci. 2006;31:470–480. doi: 10.1016/j.mcn.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xiong W.C., Mei L. Neuromuscular junction formation, aging, and disorders. Annu. Rev. Physiol. 2018;80:159–188. doi: 10.1146/annurev-physiol-022516-034255. [DOI] [PubMed] [Google Scholar]
- Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Chakkalakal J.V. The composition, development, and regeneration of neuromuscular junctions. Curr. Top. Dev. Biol. 2018;126:99–124. doi: 10.1016/bs.ctdb.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Liu W., Klose A., Forman S., Paris N.D., Wei-LaPierre L., Cortes-Lopez M., Tan A., Flaherty M., Miura P., Dirksen R.T. Loss of adult skeletal muscle stem cells drives age-related neuromuscular junction degeneration. Elife. 2017;6:e26464. doi: 10.7554/eLife.26464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wei-LaPierre L., Klose A., Dirksen R.T., Chakkalakal J.V. Inducible depletion of adult skeletal muscle stem cells impairs the regeneration of neuromuscular junctions. Elife. 2015;4:e09221. doi: 10.7554/eLife.09221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Cheng Y., Liu Y., Chen L., Liu L., Wei N., Xie Z., Wu W., Feng Y. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 2017;77:1168–1178. doi: 10.1158/0008-5472.CAN-16-1919. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Moresi V., Williams A.H., Meadows E., Flynn J.M., Potthoff M.J., McAnally J., Shelton J.M., Backs J., Klein W.H., Richardson J.A. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Milone M., Shen X.M., Engel A.G. A frameshifting mutation in CHRNE unmasks skipping of the preceding exon. Hum. Mol. Genet. 2003;12:3055–3066. doi: 10.1093/hmg/ddg334. [DOI] [PubMed] [Google Scholar]
- Ohno K., Rahman M.A., Nazim M., Nasrin F., Lin Y., Takeda J.I., Masuda A. Splicing regulation and dysregulation of cholinergic genes expressed at the neuromuscular junction. J. Neurochem. 2017;142(Suppl 2):64–72. doi: 10.1111/jnc.13954. [DOI] [PubMed] [Google Scholar]
- Ohno K., Tsujino A., Shen X.M., Milone M., Engel A.G. Spectrum of splicing errors caused by CHRNE mutations affecting introns and intron/exon boundaries. J. Med. Genet. 2005;42:e53. doi: 10.1136/jmg.2004.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Inoue A., Okada M., Murata Y., Kakuta S., Jigami T., Kubo S., Shiraishi H., Eguchi K., Motomura M. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- Oustanina S., Hause G., Braun T. PAX7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasut A., Jones A.E., Rudnicki M.A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 2013:e50074. doi: 10.3791/50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. PAX7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Sen S., Jumaa H., Webster N.J. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat. Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G., Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol. Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustanina S., Carvajal J., Rigby P., Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Vieira N.M., Naslavsky M.S., Licinio L., Kok F., Schlesinger D., Vainzof M., Sanchez N., Kitajima J.P., Gal L., Cavacana N. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G) Hum. Mol. Genet. 2014;23:4103–4110. doi: 10.1093/hmg/ddu127. [DOI] [PubMed] [Google Scholar]
- Weatherbee S.D., Anderson K.V., Niswander L.A. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- White R.B., Bierinx A.S., Gnocchi V.F., Zammit P.S. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzemann V., Schwarz H., Koenen M., Berberich C., Villarroel A., Wernig A., Brenner H.R., Sakmann B. Acetylcholine receptor epsilon-subunit deletion causes muscle weakness and atrophy in juvenile and adult mice. Proc. Natl. Acad. Sci. U S A. 1996;93:13286–13291. doi: 10.1073/pnas.93.23.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Zong J., Wei N., Cheng J., Zhou X., Cheng Y., Chen D., Guo Q., Zhang B., Feng Y. CASH: a constructing comprehensive splice site method for detecting alternative splicing events. Brief. Bioinform. 2017;19:905–917. doi: 10.1093/bib/bbx034. [DOI] [PubMed] [Google Scholar]
- Xie N., Chen M., Dai R., Zhang Y., Zhao H., Song Z., Zhang L., Li Z., Feng Y., Gao H. SRSF1 promotes vascular smooth muscle cell proliferation through a Delta133p53/EGR1/KLF5 pathway. Nat. Commun. 2017;8:16016. doi: 10.1038/ncomms16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr., Ye Z., Liu F. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Zhou X., Li X., Cheng Y., Wu W., Xie Z., Xi Q., Han J., Wu G., Fang J., Feng Y. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat. Commun. 2014;5:4581. doi: 10.1038/ncomms5581. [DOI] [PubMed] [Google Scholar]
- Zhou X., Wu W., Li H., Cheng Y., Wei N., Zong J., Feng X., Xie Z., Chen D., Manley J.L. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. 2014;42:4019–4030. doi: 10.1093/nar/gkt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data of RNA-seq have been submitted to the Gene Expression Omnibus database under accession number GSE111671.