How cells respond to deleterious effects imposed by disrupted nuclear pore complex (NPC) assembly is not well defined. Lord and Wente demonstrate that nuclear envelope–vacuole interactions expand in response to perturbed NPC assembly to promote viability, nuclear envelope remodeling, and proper NPC biogenesis.
Abstract
The intricacy of nuclear pore complex (NPC) biogenesis imposes risks of failure that can cause defects in nuclear transport and nuclear envelope (NE) morphology; however, cellular mechanisms used to alleviate NPC assembly stress are not well defined. In the budding yeast Saccharomyces cerevisiae, we demonstrate that NVJ1- and MDM1-enriched NE–vacuole contacts increase when NPC assembly is compromised in several nup mutants, including nup116ΔGLFG cells. These interorganelle nucleus–vacuole junctions (NVJs) cooperate with lipid droplets to maintain viability and enhance NPC formation in assembly mutants. Additionally, NVJs function with ATG1 to remodel the NE and promote vacuole-dependent degradation of specific nucleoporins in nup116ΔGLFG cells. Importantly, NVJs significantly improve the physiology of NPC assembly mutants, despite having only negligible effects when NPC biogenesis is unperturbed. These results therefore define how NE–vacuole interorganelle contacts coordinate responses to mitigate deleterious cellular effects caused by disrupted NPC assembly.
Introduction
The nuclear envelope (NE) forms a semipermeable barrier surrounding genomic DNA, with its two lipid bilayers fused at points where nuclear pore complexes (NPCs) are embedded. As multisubunit assemblies composed of nucleoporins (Nups), NPCs are essential for nucleocytoplasmic transport. A set of scaffolding Nups provide a core structural platform on which phenylalanine-glycine (FG) and glycine-leucine-phenylalanine-glycine (GLFG) Nups assemble and serve to facilitate the transport of different types of cargos (Beck and Hurt, 2017). FG Nups contain domains rich in intrinsically disordered FG repeats (Denning et al., 2003; Hough et al., 2015), whereas GLFG domains are a subset that contain GLFG repeats (Wente et al., 1992). Both FG and GLFG Nups transiently interact with specific nuclear transport receptors (NTRs) to allow movement of NTR–cargo complexes through NPCs (Strawn et al., 2004). Through cohesive binding and heterotypic interactions with scaffolding Nups, GLFG Nups may also promote NPC biogenesis by stabilizing the formation of assembly intermediates (Onischenko et al., 2017).
NPC assembly is an elaborate process, particularly when NPCs are inserted into the intact NE during interphase. Unlike metazoan cells, which also reform NPCs when the NE is reconstituted after cell division, Saccharomyces cerevisiae cells undergo a closed mitosis and therefore only directly assemble NPCs into the NE. Disrupting the function of several Nups (Wente and Blobel, 1993; Chadrin et al., 2010; Makio et al., 2009; Boehmer et al., 2003; Aitchison et al., 1995; Doye et al., 1994), the GTPase Ran (Ryan et al., 2003; Walther et al., 2003), and NTRs (Ryan et al., 2007; Lusk et al., 2002) inhibits NPC formation, suggesting that a functional nuclear transport system is required for further NPC production. Reticulons (Dawson et al., 2009), lipid-regulating proteins (Hodge et al., 2010; Zhang et al., 2018), and Torsins (Rampello et al., 2020; Pappas et al., 2018; VanGompel et al., 2015) are also involved, likely by regulating properties of the NE to promote NPC biogenesis. Additionally, Vps4 and Heh1/2 cooperatively function as a surveillance system that promotes the clearance of misassembled NPCs in a Chm7-dependent manner (Webster et al., 2014, 2016; Thaller et al., 2019).
The NE in S. cerevisiae cells contacts vacuoles at regions termed nucleus–vacuole junctions (NVJs; Pan et al., 2000; Kvam and Goldfarb, 2006). Although morphologically distinct, vacuoles are functionally similar to metazoan lysosomes, serving as acidic organelles involved in protein degradation and nutrient storage. Several outer-NE membrane proteins including Nvj1 (Pan et al., 2000) and Mdm1 (Henne et al., 2015) are present at NVJs and tether the NE to the vacuole through specific interactions with corresponding vacuolar species including Vac8 (Pan et al., 2000; Jeong et al., 2017) or phosphatidylinositol 3-phosphate (Henne et al., 2015). NVJs expand when cells are grown using carbon sources other than glucose (Hariri et al., 2018; Bean et al., 2018) or to promote piecemeal autophagy of the nucleus in response to nutrient starvation (Roberts et al., 2003), which involves autophagy-dependent degradation of specific nuclear proteins in the vacuole (Krick et al., 2008). Proteins that promote lipid droplet (LD) formation are also enriched at NVJs (Kohlwein et al., 2001) and serve as sites for LD synthesis. LDs are required for the viability of NPC assembly-defective brr6-1 mutant cells (Hodge et al., 2010), suggesting that LDs and NPCs are functionally linked.
There is a significant lack of information about how cells respond to sustained interphase NPC assembly defects, which often manifest as clusters of misassembled intermediates that herniate the NE (Thaller and Patrick Lusk, 2018; Laudermilch et al., 2016) and disrupt nuclear transport and growth (Wente and Blobel, 1993; Aitchison et al., 1995; Hodge et al., 2010). Not only are assembly defects caused by inherent complexities associated with NPC biogenesis, but they are also observed in replicatively aged S. cerevisiae cells (Rempel et al., 2019) and genetic models of early-onset torsion dystonia (Laudermilch et al., 2016; Pappas et al., 2018). Here we investigate S. cerevisiae mutants with altered NPC assembly as models to determine how cells respond to NPC assembly stress. We find that NPC biogenesis mutants exhibit dramatic expansion of NVJ1- and MDM1-dependent NE–vacuole contacts and increased levels of LDs. Furthermore, NVJs and LDs function in a cooperative manner to promote viability and NPC formation in NPC assembly mutants. NE–vacuole contacts also function with autophagy factors to remodel the NE when NPC assembly is disrupted, promoting degradation of specific Nups. Collectively, these data reveal novel mechanisms coordinated by NE–vacuole interactions that promote proper NPC formation when NPC biogenesis is compromised.
Results
Nup116’s GLFG domain is required for NPC assembly and modulates NE–vacuole interactions
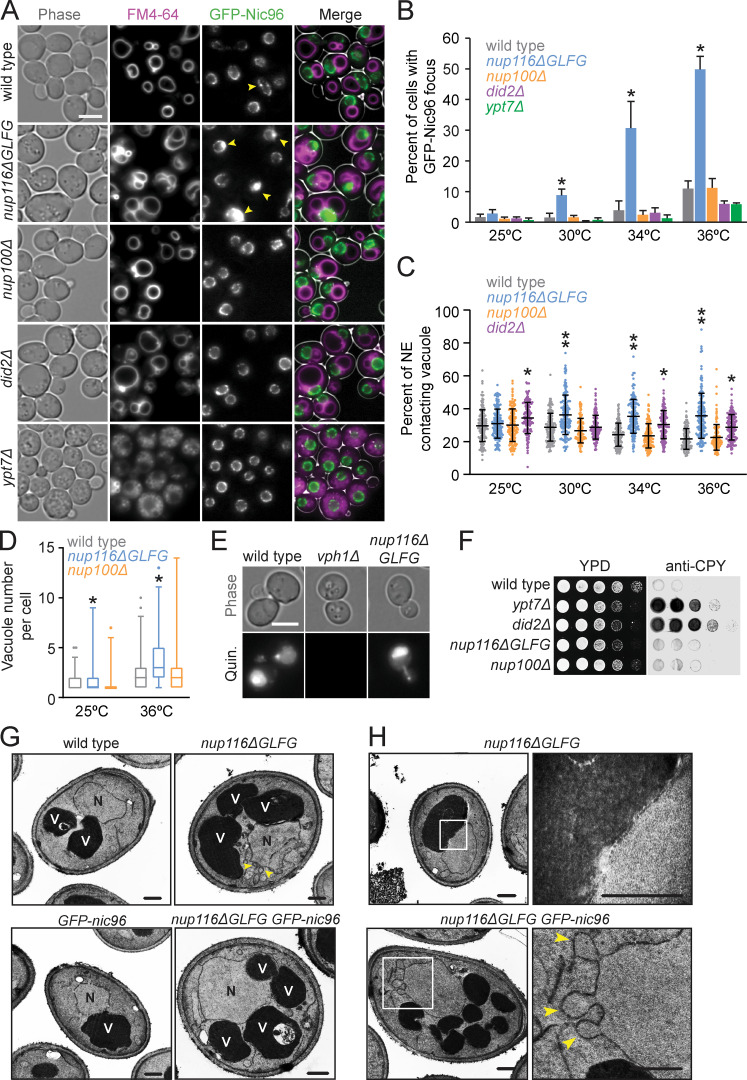
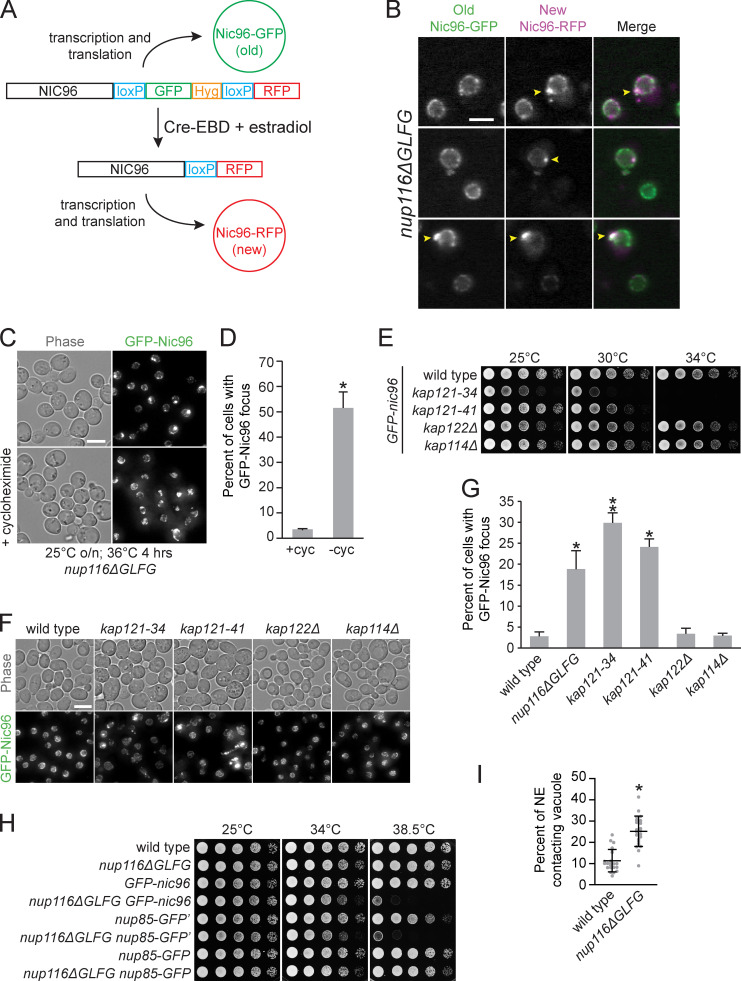
Mutant nup116ΔGLFG cells in a W303 strain background display NPC biogenesis defects in the absence of Nup188 (Onischenko et al., 2017). To further ascertain how Nup116’s GLFG domain regulates NPC formation, a GFP-nic96 nup116ΔGLFG strain produced in an S288C background was analyzed for GFP-Nic96 foci (indicative of NPC assembly defects) at temperatures ranging from 25° to 36°C. GFP-nic96 nup116ΔGLFG mutants displayed temperature-dependent increases in NPC foci formation, with ∼50% of these cells containing foci at 36°C compared with ∼10% of GFP-nic96 cells (Fig. 1, A and B). Although NPC clustering in GFP-nic96 nup116ΔGLFG cells was most pronounced at 36°C, significant levels of foci were present at 30° and 34°C. Use of the recombination-induced tag exchange (RITE) system (Verzijlbergen et al., 2010) allowed for discrimination between old (GFP) and newly formed (RFP) Nic96 (Fig. S1 A). Consistent with the hypothesis that Nic96 foci result from disrupted NPC assembly, newly formed Nic96-RFP foci were visible in regions of the NE lacking old Nic96-GFP foci after increasing the temperature from 25° to 36°C or, to a lesser degree, exclusively localized at preexisting Nic96-GFP foci (Fig. S1 B). Additionally, inhibiting protein synthesis with cycloheximide prevented GFP-Nic96 foci formation after a temperature shift (Fig. S1, C and D).
Figure 1.
Increased levels of GFP-Nic96 foci and NE–vacuole contacts in nup116ΔGLFG mutants. (A) FM4-64–stained GFP-nic96 yeast grown at 36°C. Arrows point to GFP-Nic96 foci. Scale bar, 5 µm. (B) Percentage of cells with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.05 compared with wild type at same temperature using unpaired two-tailed Student’s t test (n ≥ 100 cells from three independent trials). Additionally, P ≤ 0.01 when comparing nup116ΔGLFG at 25° and 36°C. (C) Percentage of NE-contacting vacuole membranes (detailed in Materials and methods); each dot represents value from one cell (n = 120 from three independent trials). Error bars represent SD. *, P ≤ 0.001 compared with wild type at same temperature using Dunn’s post hoc test; **, P ≤ 0.03 when also compared with did2Δ. Additionally, P ≤ 0.01 when comparing nup116ΔGLFG at 25° and 36°C. (D) Vacuole number per cell; *, P ≤ 0.01 compared with wild type at same temperature using a two-tailed Mann–Whitney U test (n = 90 from three independent trials). Box and whiskers show 1st–99th percentile. (E) Quinacrine-stained cells to assess vacuole acidity. Quinacrine fluorescence indicates vacuoles are acidic; vph1Δ cells served as a positive control for vacuoles lacking acidity (Preston et al., 1989). Scale bar, 5 µm. (F) Yeast were serially diluted onto two YPD plates and grown at 28°C; one was covered with nitrocellulose to absorb secreted proteins and blotted with anti-CPY. Secreted CPY results from blocks in Golgi-vacuole transport (Robinson et al., 1988). (G) TEM images of listed strains at 36°C. N, nuclei; V, vacuoles. Arrows point to NE herniations. Scale bars, 500 nm. (H) TEM images of nup116ΔGLFG cells at 36°C zoomed into regions of NE–vacuole contacts (top) or NE herniations (bottom). Arrowheads point to NE herniations. Scale bars, 500 nm.
Figure S1.
NPC assembly is disrupted in nup116ΔGLFG and kap121 mutants. (A) Diagram of RITE cassette used to distinguish between old (GFP) and newly produced (RFP) Nic96 in cells expressing Cre-estradiol-binding domain (EBD). In the absence of β-estradiol, Nic96-GFP is produced. When β-estradiol is introduced, the GFP tag is excised via the surrounding LoxP sites, causing Nic96-RFP to be produced. (B) Images of nup116ΔGLFG yeast expressing both new (RFP) and old (GFP) forms of Nic96 after a 3-h incubation with 5 µM β-estradiol at 25°C, followed by incubation at 36°C for 3 h to induce production of Nic96 foci (arrowheads). RFP foci formed either without any preexisting GFP foci (top two panels) or exclusively at preexisting GFP foci (bottom panel) that likely began to form at 25°C. Scale bar, 5 µm. (C) Maximum projections of GFP-nic96 nup116ΔGLFG yeast that were grown to midlog phase at 25°C, then shifted to 36°C for 4 h in the absence (top) or presence (bottom) of 10 µg/ml cycloheximide. Scale bar, 5 µm. (D) Percentage of cells ± cycloheximide with GFP-Nic96 focus; *, P ≤ 0.001 using unpaired two-tailed Student’s t test (n ≥ 100 from three independent trials). Error bars represent SD. (E) Serial dilutions of different GFP-nic96 kap mutants on YPD plates. (F) Maximum projections of GFP-nic96 yeast strains grown to at 32°C. Scale bar, 5 µm. (G) Percentage of cells grown at 32°C with GFP-Nic96 focus; *, P ≥ 0.05 using unpaired two-tailed Student’s t test compared with wild type (n ≥ 100 cells from three independent trials); **, P ≤ 0.05 when additionally compared with nup116ΔGLFG. Error bars represent SD. (H) Serial dilutions of the listed strains on YPD plates. (I) Quantification of EM images to determine physical interactions between the NE and vacuoles in cells (within 20 nm of one another) grown at 36°C. Error bars represent SD. *, P ≤ 0.001 compared with wild type using a two-tailed Student’s t test (n = 24). These values are likely lower than those derived from fluorescence microscopy, because we did not necessarily quantify the regions with maximal contacts in each cell and quantified only what was observed in the specific plane these images were captured in.
Karyopherin subunit β-3 (Kap121) is one of several NTRs whose transport is regulated by Nup116’s GLFG domain (Lord et al., 2015; Terry and Wente, 2007). If foci were forming in GFP-nic96 nup116ΔGLFG cells owing to disrupted Kap121-dependent transport, we predicted that GFP-nic96 kap121 mutants would also show foci formation. Consistent with this possibility, kap121-34 and kap121-41 mutants displayed increased GFP-Nic96 foci formation at the semipermissible temperature of 32°C, while kap122Δ and kap114Δ mutants had no effect (Fig. S1, E–G). We therefore concluded that deletion of Nup116’s GLFG domain in S288C cells causes temperature-dependent NPC assembly defects that may result from compromised Kap121-dependent nucleocytoplasmic transport.
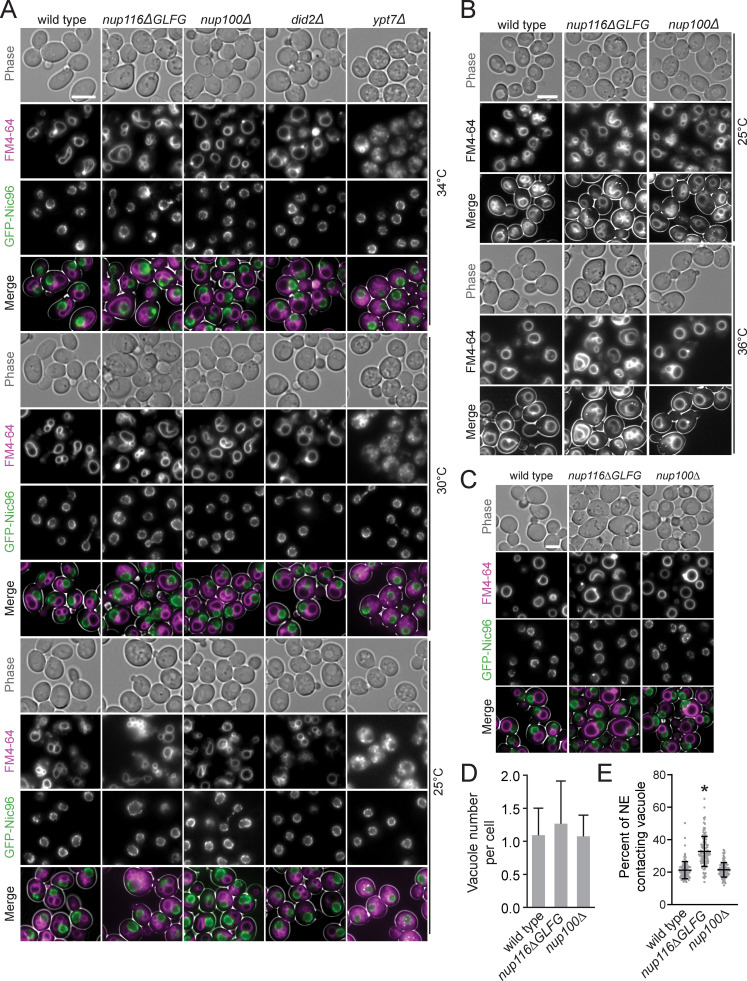
Given their relatively modest growth defects (Fig. S1 H), nup116ΔGLFG mutants were leveraged to determine how perturbed NPC assembly affects other aspects of cell function. Phase-contrast images of GFP-nic96 nup116ΔGLFG cells indicated possible changes in vacuole morphology; the endomembrane-staining dye FM4-64 confirmed that these mutants exhibited elevated contacts between the NE and vacuole membrane (Fig. 1, A and C; Fig. S1 I; and Fig. S2, A and B) and significantly more vacuoles per cell (Fig. 1 D). Increased NE–vacuole interactions were particularly evident at higher temperatures that correlated with disrupted NPC assembly. Mutant cells lacking the GLFG Nup100 (nup100Δ) displayed neither increased GFP-Nic96 foci nor altered vacuole morphology/number, indicating that these types of defects are not caused by all nup mutants. The morphological changes in nup116ΔGLFG vacuoles did not appear to significantly impact vacuolar function, as they remained acidic (Preston et al., 1989) and the peptidase CPY was properly transported to the vacuole from the Golgi (Fig. 1, E and F; Robinson et al., 1988).
Figure S2.
Images of GFP-nic96 strains at 25°, 30°, and 34°C and in synthetic medium. (A) FM4-64–stained GFP-nic96 strains grown at 25°, 30°, or 34°C that were used to quantify data in Fig. 1 (B–D). Scale bar, 5 µm. (B) FM4-64–stained wild-type, nup116ΔGLFG, and nup100Δ cells grown at 25° or 36°C; similar vacuole morphology phenotypes indicate that NE–vacuole contacts are not significantly impacted by GFP-nic96. (C) Images of GFP-nic96 strains grown at 36°C in synthetic complete medium. Scale bar, 5 µm. (D) Average vacuole number per cell at 36°C in synthetic medium. P ≥ 0.06 when nup116ΔGLFG cells are compared with wild type using a two-tailed Mann–Whitney U test. Error bars represent SD. (E) Percentage of NE-contacting vacuole membranes from cells grown in synthetic complete medium at 36°C. Each dot represents one cell (n = 120 from three independent trials). Error bars represent SD. *, P ≤ 0.001 compared with wild type using Dunn’s post hoc test.
Vacuole fragmentation and transport-deficient ypt7Δ and did2Δ mutants (Michaillat and Mayer, 2013) showed no increase in GFP-Nic96 foci formation, indicating that vacuole number does not impact NPC assembly (Fig. 1, A and B). Because of severe vacuole fragmentation in ypt7Δ cells, NE–vacuole interactions were not quantified; did2Δ mutants displayed increased NE–vacuole contacts at most temperatures tested, but unlike nup116ΔGLFG cells, the phenotype was not exacerbated in a temperature-dependent manner (Fig. 1, A and C). Average vacuole number per cell was not significantly increased in nup116ΔGLFG cells when grown in synthetic medium at 36°C, yet NE–vacuole interactions remained elevated (Fig. S2, C–E), suggesting that vacuole fragmentation is not the main reason these contacts are observed in nup116ΔGLFG mutants. Transmission EM (TEM) images of nup116ΔGLFG cells grown at 36°C confirmed extensive direct contacts between the NE and vacuole membranes (Fig. 1, G and H). NE herniations that were structurally similar to those observed in nup116Δ mutants (Wente and Blobel, 1993) were frequently detected in nup116ΔGLFG cells at 36°C. Overall, at elevated temperatures, nup116ΔGLFG cells exhibited NPC biogenesis defects that correlated with increased physical interactions between the NE and vacuole membrane.
NE–vacuole interactions expand in other NPC assembly mutants
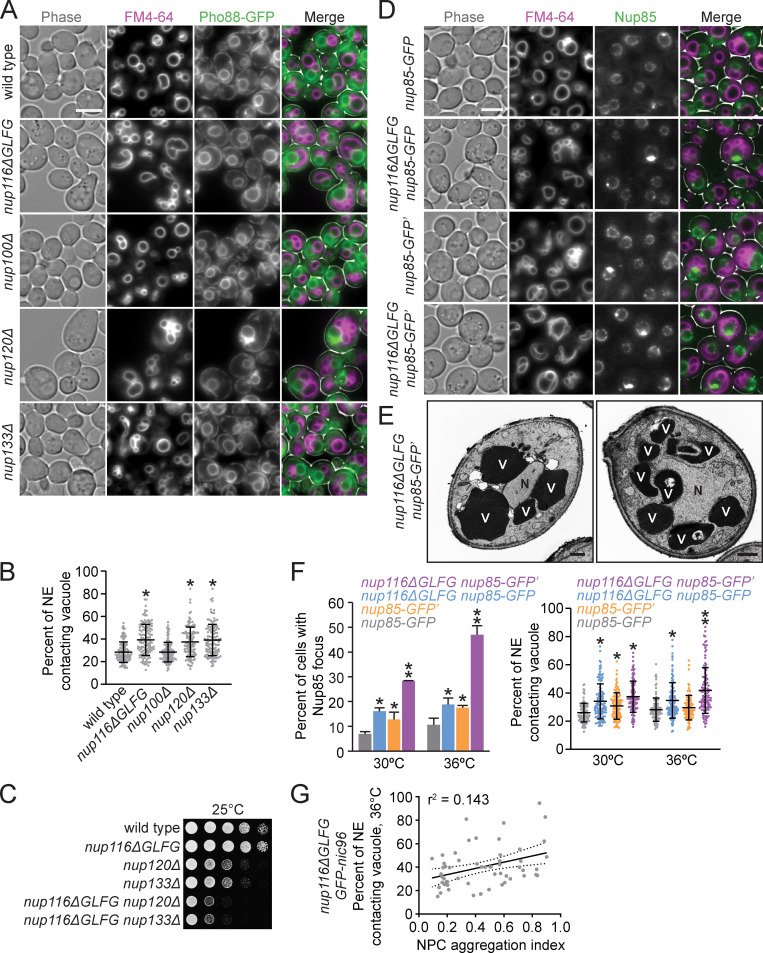
Based on the lack of NPC foci and vacuole morphology phenotypes in nup100Δ cells, we hypothesized that increased NE–vacuole contacts could result from disrupted NPC assembly or associated NE morphological changes (i.e., herniations). NE–vacuole interactions were measured in nup120Δ and nup133Δ mutants at 30°C, a temperature at which both strains are viable yet also exhibit severe NPC assembly defects (Aitchison et al., 1995; Doye et al., 1994). The predicted integral NE/ER protein Pho88-GFP (Aviram et al., 2016) was used a marker because the NPC clustering defects are so strong in nup120Δ and nup133Δ mutants that most of their nuclear rim areas are devoid of NPCs (Aitchison et al., 1995; Doye et al., 1994). FM4-64 labeling revealed extensive contacts between the NE and vacuoles in both mutants (Fig. 2, A and B). Together, these data supported a model wherein mutations that perturb NPC assembly and NE integrity also promote changes in vacuolar morphology. Consistent with their NPC assembly defects, nup116ΔGLFG mutants exhibited synthetic growth defects with nup120Δ and nup133Δ mutants (Fig. 2 C).
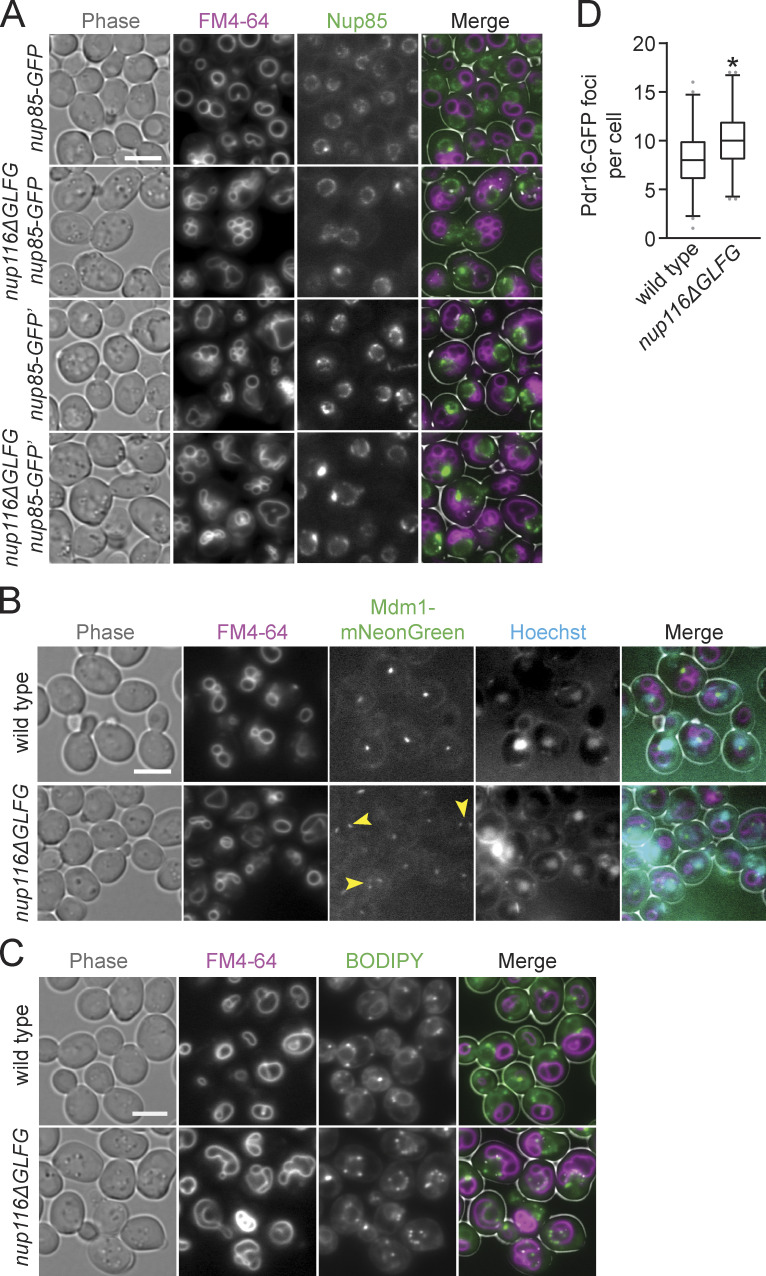
Figure 2.
Several nup mutations that disrupt NPC assembly also increase NE–vacuole contacts. (A) FM4-64–stained pho88-GFP strains grown at 30°C. Scale bar, 5 µm. (B) NE–vacuole contacts at 30°C. Each dot represents contacts from one cell (n = 120 from three independent trials). Error bars represent SD. *, P ≤ 0.001 compared with wild type using Dunn’s post hoc test. (C) Serial dilutions of cells onto YPD plates. (D) FM4-64 labeled nup85-GFP or nup85-GFP′ (refer to Methods for details regarding strain construction) cells grown at 36°C. Scale bar, 5 µm. (E) TEM images of nup116ΔGLFG nup85-GFP′ strains grown at 36°C. N, nuclei; V, vacuoles. Scale bars, 500 nm. (F) Left: Percentage of cells with GFP focus at 30° or 36°C. Error bars represent SD. *, P ≤ 0.05 compared with nup85-GFP; **, P ≤ 0.05 compared with all other values using an unpaired two-tailed Student’s t test for a particular temperature (n ≥ 100 cells from three independent trials). Right: NE–vacuole contacts from strains grown at 30° or 36°C. Each dot represents one cell (n = 120 from three independent trials). Error bars represent SD. *, P ≤ 0.01 compared with nup85-GFP; **, P ≤ 0.05 compared with all other values using Dunn’s post hoc test for a particular temperature. (G) NE–vacuole contacts versus NPC aggregation index for GFP-nic96 nup116ΔGLFG cells grown at 36°C. Each dot represents values from one cell (n = 50). Solid line represents linear regression, while dotted lines represent 95% confidence interval.
To confirm Nup116-dependent NPC clustering and NE–vacuole interactions using a different NPC marker than GFP-Nic96, nup85-GFP′ from the yeast GFP collection (Huh et al., 2003; arbitrarily and henceforth referred to as nup85-GFP′), was introduced into nup116ΔGLFG cells. Surprisingly, greater amounts of NE–vacuole contacts were detectable in nup116ΔGLFG nup85-GFP′ cells compared with nup116ΔGLFG GFP-nic96 cells at 36°C, with some nup116ΔGLFG nup85-GFP′ cells having nuclei that were nearly completely surrounded by vacuoles (Fig. 2, D–F). Even nup85-GFP′ cells exhibited somewhat increased NE–vacuole contacts, indicating that altering the function of NUP85 with this particular GFP epitope impacts vacuole morphology; nup85-GFP′ was at least partially compromised given its modest growth defect at 38.5°C and its synthetic growth defect with nup116ΔGLFG mutants (Fig. S1 H).
As a comparison, the pYM25 (Janke et al., 2004) vector was used to generate nup116ΔGLFG nup85-GFP strains with a shorter linker between the nup and GFP and with yeast codon-optimized GFP sequence. At both 30° and 36°C, nup116ΔGLFG nup85-GFP mutants exhibited less-severe NPC clustering relative to nup116ΔGLFG nup85-GFP′ mutants and fewer NE–vacuole interactions at 36°C (Fig. 2, D–F; and Fig. S3 A). We concluded that partially inhibiting the function of NUP85 can impact NPC clustering as well as NE–vacuole interactions, and increasing the severity of NPC clustering tends to cause greater changes in vacuole morphology.
Figure S3.
LDs and Mdm1 are enriched at NE–vacuole interfaces in nup116ΔGLFG cells. (A) FM4-64–stained nup85-GFP or nup85-GFP′ strains grown at 30°C; images were used to quantify data in Fig. 2 F. Scale bar, 5 µm. (B) FM4-64– and Hoechst-stained mdm1-mNeonGreen strains grown at 30°C. Arrowheads point to cells where Mdm1-mNeonGreen localizes around multiple regions of NE–vacuole contacts. Scale bar, 5 µm. (C) Images of FM4-64– and BODIPY-stained wild-type and nup116ΔGLFG cells grown at 34°C. Scale bar, 5 µm. These cells, as well as other independent experiments, were used to quantify data in Fig. 3 F. (D) Number of Pdr16-GFP foci per cell for the listed strains grown at 30°C (n = 120 cells from three independent trials). Box and whiskers represent 2.5th–97.5th percentile; *, P ≤ 0.001 compared with wild type using two-tailed Mann–Whitney U test.
We note, however, that the exact level of NPC clustering did not completely correlate with increased NE–vacuole contacts. For example, nup85-GFP′ and nup116ΔGLFG nup85-GFP strains both exhibited roughly similar proportions of the cell population with GFP foci, but NE–vacuole contacts were more abundant in nup116ΔGLFG nup85-GFP cells at 36°C (Fig. 2 F). Additionally, when comparing the severity of NPC clustering defects using an aggregation index (Casey et al., 2015; Niepel et al., 2013; higher index indicates more severe clustering) with the extent of NE–vacuole interactions in nup116ΔGLFG GFP-nic96 cells at the single-cell level (Fig. 2 G), there was no obvious correlation between these two variables. Thus, increased NE–vacuole contacts were observed in several nup mutations that coincide with NPC assembly defects and NE herniations, although at the single-cell level, the presence and severity of Nup clustering did not necessarily correlate with the extent of NE–vacuole interactions.
NVJ factors and LDs are enriched at NE–vacuole contacts in NPC assembly mutants
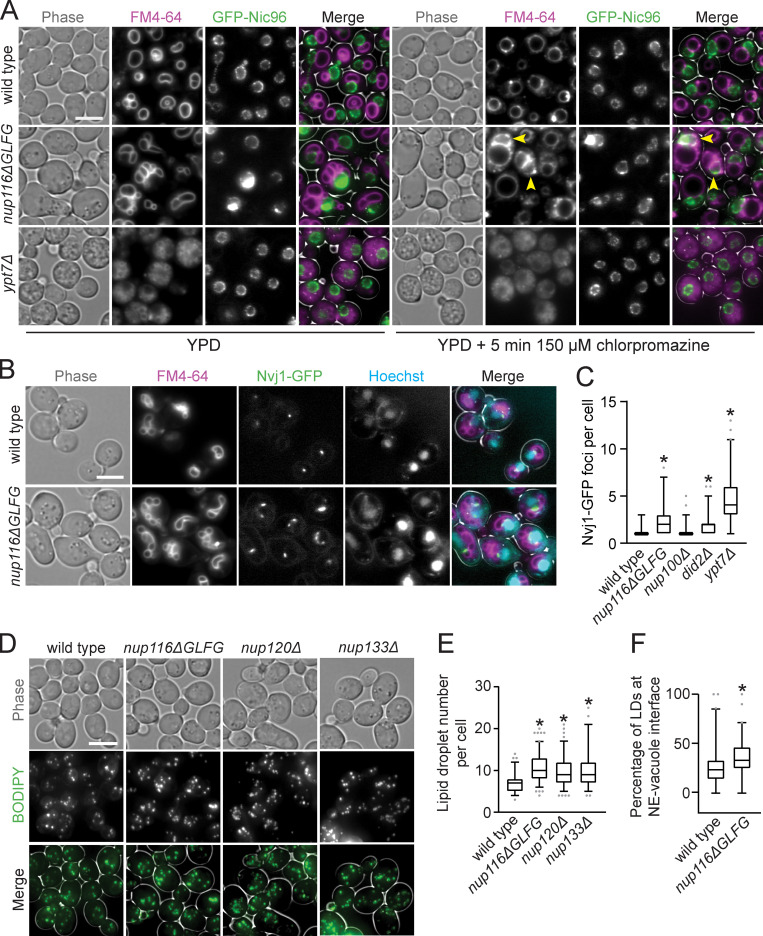
We hypothesized the extensive NE–vacuole contacts in nup116ΔGLFG mutants necessitated changes in vacuole membrane composition that could be reversed using chlorpromazine, which increases membrane fluidity and tends to enhance vacuole fusion in vitro (Fratti et al., 2007). After a 5-min chlorpromazine treatment, GFP-nic96 and GFP-nic96 nup116ΔGLFG vacuoles tended to form one large sphere that minimally interacted with the NE (Fig. 3 A), unlike chlorpromazine-insensitive GFP-nic96 ypt7Δ mutants. Despite these physical changes to the vacuole, we observed some GFP-nic96 nup116ΔGLFG cells that retained FM4-64-positive membranes around the NE (Fig. 3 A, arrows), suggesting that NE-associated factors can interact with vacuole lipids after treatment.
Figure 3.
Compositional changes to NE–vacuole interfaces and increased LD levels in NPC assembly mutants. (A) FM4-64–stained GFP-nic96 yeast grown at 36°C were imaged before (left) and after (right) incubation with 150 µm chlorpromazine for 5 min. Arrowheads point to cells that still have significant FM4-64 signal around NE following treatment. Scale bar, 5 µm. (B) FM4-64- and Hoechst-stained nvj1-GFP yeast grown at 36°C. Scale bar, 5 µm. (C) Nvj1-GFP foci per cell in the listed strains. Box and whiskers represent 2.5th–97.5th percentile. *, P ≤ 0.05 compared with wild type using Dunn’s post hoc test (n = 120 from three independent trials). (D) Max project images of BODIPY-stained yeast grown at 30°C. Scale bar, 5 µm. (E) Total LD number per cell in the listed strains grown at 30°C. Box and whiskers represent 2.5th–97.5th percentile. *, P ≤ 0.001 compared with wild type using Dunn’s post hoc test (n = 90 from three independent trials). (F) Percentage of LDs that are at NE–vacuole interface in the listed strains at 34°C. Box and whiskers represent 2.5th–97.5th percentile. *, P ≤ 0.001 compared with wild type using a two-tailed Mann–Whitney U test (n = 90 from three independent trials).
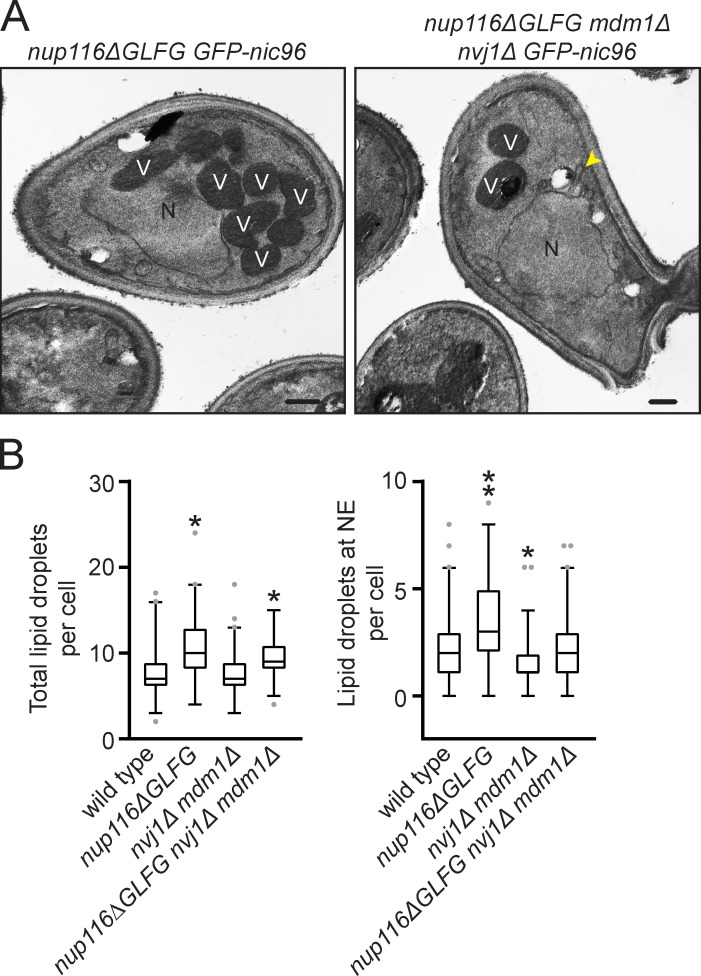
The NVJ proteins Nvj1-GFP and Mdm1-mNeonGreen were visualized to determine whether NVJs expand in nup116ΔGLFG mutants. Unlike wild-type cells, which generally displayed a single focus of Nvj1-GFP or Mdm1-mNeonGreen at NE–vacuole interfaces, nup116ΔGLFG mutants often exhibited extensive regions of both proteins at NE–vacuole contacts (Fig. 3 B and Fig. S3 B). Nvj1-GFP foci number generally correlated with the extent of NE–vacuole interactions in different mutant strains (Fig. 3 C), with nup116ΔGLFG cells displaying significantly more than wild-type and nup100Δ cells. Because NVJs can serve as sites of LD biogenesis (Hariri et al., 2018), and increased LD levels have been previously observed in NPC assembly mutants (Hodge et al., 2010), LDs per cell were measured using BODIPY dye in nup116ΔGLFG, nup120Δ, and nup133Δ mutants. LD number was significantly increased in these mutant strains (Fig. 3, D and E), and LDs were enriched at NE–vacuole contact sites (Fig. 3 F and Fig. S3 C). Levels of Pdr16-positive LDs, which tend to cluster near NVJs (Eisenberg-Bord et al., 2018), were also increased in nup116ΔGLFG mutants (Fig. S3 D). These results indicated that NVJs expand and LD number increases when NPC assembly is compromised, producing significant compositional changes to the NE that enable vacuole interactions.
NVJs and LDs cooperatively promote viability and NPC formation in assembly mutants
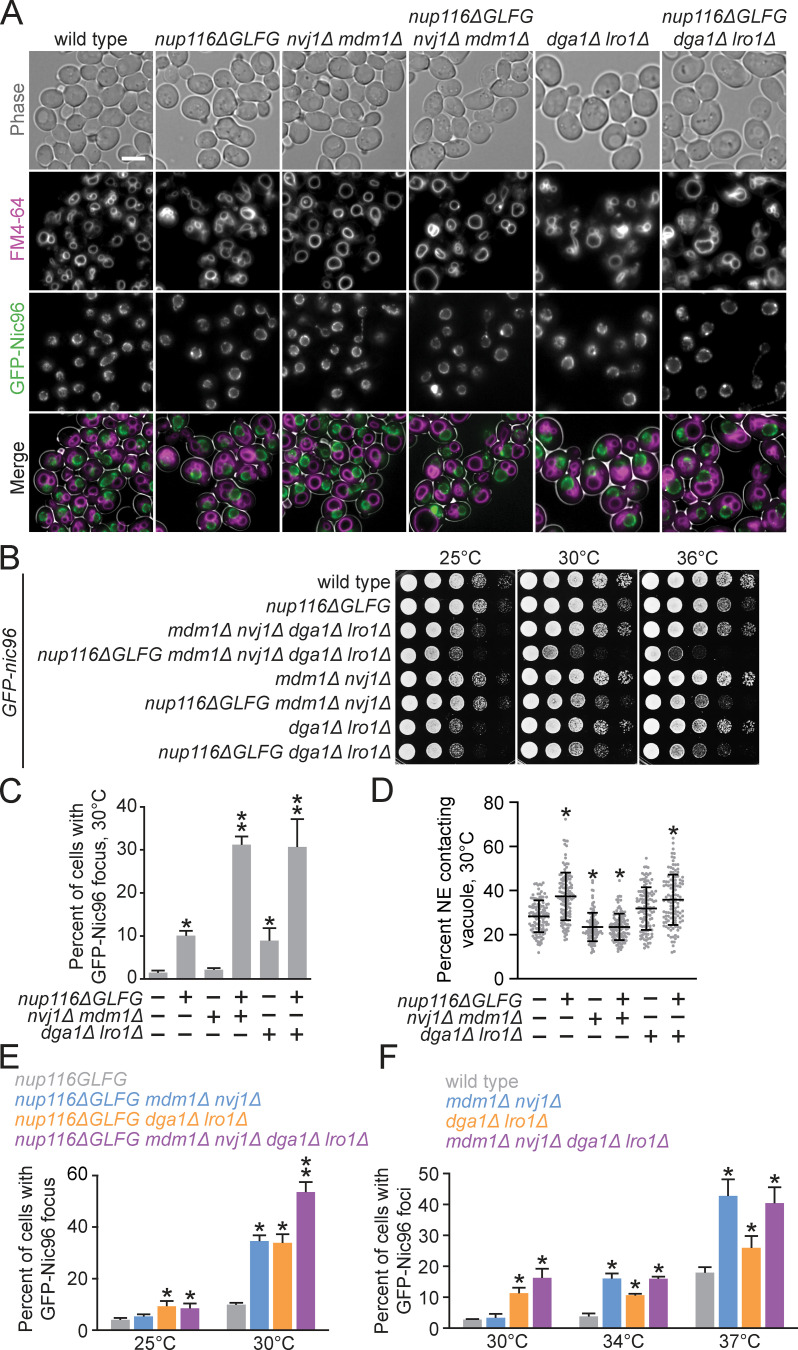
Increased NE–vacuole contacts and LDs in nup mutants could be either a deleterious/neutral effect caused by altered NPC function or a secondary protective response that mitigates complications that arise from disrupted NPC assembly. To investigate these competing hypotheses, NVJ1 and MDM1 were deleted in nup116ΔGLFG GFP-nic96 cells, as were LRO1 and DGA1, two redundant diacylglycerol acetyltransferase genes required for LD formation (Oelkers et al., 2002; Petschnigg et al., 2009). Synthetic growth defects in mutants would be consistent with a model wherein LDs or NVJs ameliorate NPC assembly stress, whereas other growth phenotypes would suggest that NVJs or LDs are neutral/harmful effects. Deletion of NVJ1 and MDM1 abrogated NE–vacuole contacts in GFP-nic96 and GFP-nic96 nup116ΔGLFG cells (Fig. 4 A), and DGA1- and LRO1-deficient strains exhibited reduced LD numbers (Fig. S4 A).
Figure 4.
NVJs and LDs promote NPC formation and proper growth in NPC assembly mutants. (A) FM4-64–stained GFP-nic96 strains grown at 30°C. Scale bar, 5 µm. (B) Serial dilutions of yeast cells on YPD plates. (C) Percentage of cells grown at 30°C with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.005 compared with wild type using an unpaired two-tailed Student’s t test (n ≥ 100 cells from three independent trials); **, P ≤ 0.03 when also compared with nup116ΔGLFG. (D) NE–vacuole interactions from GFP-nic96 strains grown at 30°C. Each dot represents value from one cell (n = 120 from three independent trials). Error bars represent SD. *, P ≤ 0.01 compared with wild type using Dunn’s post hoc test. (E) Percentage of cells grown at 25° or 30°C with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.05 compared with nup116ΔGLFG at the same temperature using an unpaired two-tailed Student’s t test (n ≥ 100 cells from three independent trials); **, P ≤ 0.05 when also compared with all other strains. (F) Percentage of cells grown at 30°, 34°, or 37°C with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.05 compared with wild type at the same temperature using a two-tailed Student’s t test (n ≥ 100 cells from three independent trials); mdm1Δ nvj1Δ dga1Δ lro1Δ mutants do not exhibit P ≤ 0.05 when compared with other strains.
Figure S4.
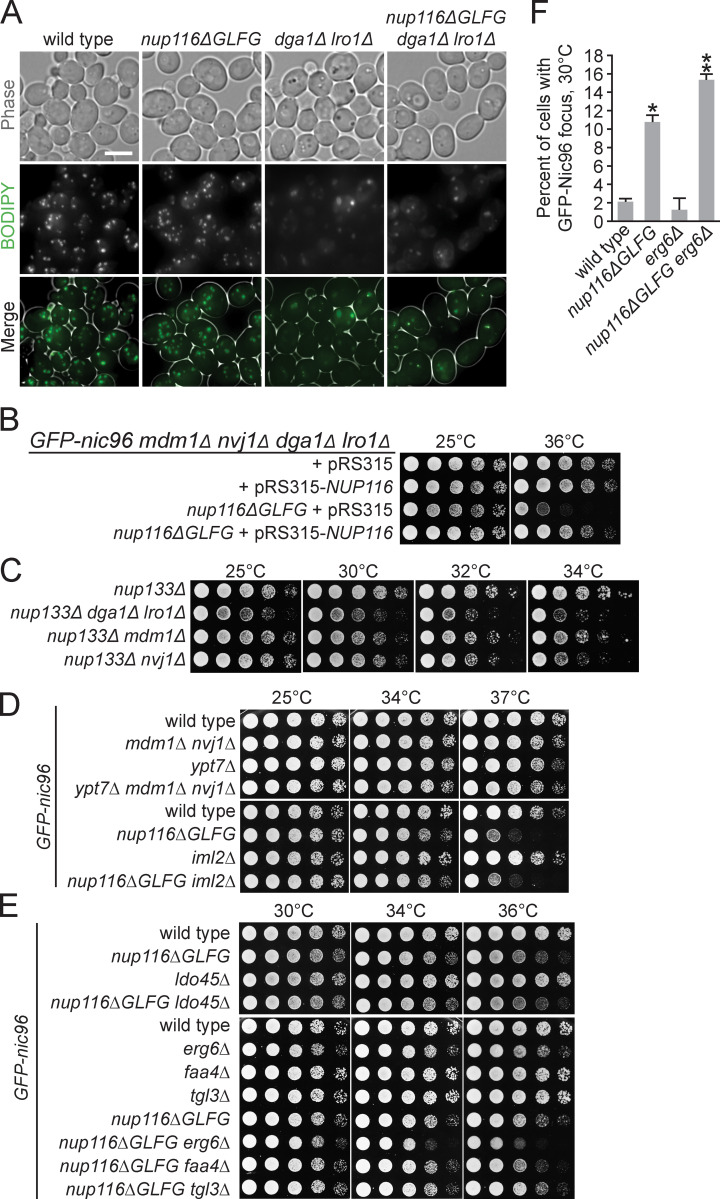
NVJs impact the viability of nup133Δ cells, and Erg6 impacts NPC assembly mutant viability. (A) BODIPY-stained strains grown at 30°C. Scale bar, 5 µm. (B) Serial dilutions of GFP-nic96 mdm1Δ nvj1Δ dga1Δ lro1Δ or nup116ΔGLFG GFP-nic96 mdm1Δ nvj1Δ dga1Δ lro1Δ cells transformed with pRS315-NUP116 or pRS315, grown in synthetic complete leu and then plated onto YPD plates. (C–E) Serial dilutions of cells on YPD plates. LDO45 is required for loading of Pdr16 into LDs (Eisenberg-Bord et al., 2018; Teixeira et al., 2018). (F) Percentage of cells with GFP-Nic96 focus grown at 30°C. Error bars represent SD. *, P ≤ 0.01 compared with wild type using an unpaired two-tailed Student’s t test (n ≥ 100 cells from three independent trials); **, P ≤ 0.01 when also compared with nup116ΔGLFG.
Both GFP-nic96 nup116ΔGLFG nvj1Δ mdm1Δ and GFP-nic96 nup116ΔGLFG dga1Δ lro1Δ mutants displayed synthetic growth defects relative to GFP-nic96 nup116ΔGLFG cells at 30° and 36°C (Fig. 4 B). Interestingly, LD- and NVJ-deficient GFP-nic96 cells had only minor growth phenotypes at higher temperatures but produced additive growth defects in GFP-nic96 nup116ΔGLFG mutants in a NUP116-dependent manner (Fig. S4 B), suggesting that NVJs and LDs regulate viability at least partially independently of one another. Similar synthetic growth defects were observed in nup133Δ mdm1Δ, nup133Δ nvj1Δ, and nup133Δ dga1Δ lro1Δ mutants (Fig. S4 C). Thus, NVJs and LDs were required for maximal viability in several nup mutants with disrupted NPC assembly, consistent with a model wherein LDs and NE–vacuole contacts cooperate to ameliorate NPC assembly stress. NVJ1 and MDM1 were not required for viability of GFP-nic96 ypt7Δ cells (Fig. S4 D), suggesting that NVJs do not promote growth in nup116ΔGLFG mutants by ameliorating stress associated with disrupted vacuole function/fusion. We concluded that LDs regulate NPC assembly stress separately from their role in clearing misfolded proteins, as the misfolded protein clearance factor IML2 (Moldavski et al., 2015) had no impact on the growth of GFP-nic96 nup116ΔGLFG cells (Fig. S4 D). While the LD-associated Tgl3, Faa4, and Pdr16 (Grillitsch et al., 2011) did not modulate viability in GFP-nic96 nup116ΔGLFG cells, erg6Δ GFP-nic96 nup116ΔGLFG mutants displayed growth defects (Fig. S4, E and F), indicating that LD-associated Erg6 (Grillitsch et al., 2011) or ergosterol is required for full viability of NPC assembly mutants.
GFP-Nic96 foci formation was measured in NVJ- or LD-deficient GFP-nic96 nup116ΔGLFG mutants at 30°C, a temperature at which subtle changes in NPC assembly could be detected. While deletion of MDM1 and NVJ1 did not impact NPC formation in GFP-nic96 cells despite decreasing NE–vacuole interactions, 31% of GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ mutants displayed GFP-Nic96 foci, a threefold increase compared with GFP-nic96 nup116ΔGLFG cells (Fig. 4, A–D). 9% of GFP-nic96 dga1Δ lro1Δ mutants exhibited NPC assembly defects, and potentially because of this or to compensate for their decreased LD content, may also have increased NE–vacuole contacts (Fig. 4 D; P < 0.05 by Mann–Whitney U test compared with wild type); 30% of GFP-nic96 nup116ΔGLFG dga1Δ lro1Δ mutants had GFP-Nic96 foci. Additionally, NVJ-deficient nup116ΔGLFG mutants tended to exhibit more NE herniations (Fig. S5 A). Overall, NE–vacuole contacts promoted NPC assembly and proper NE morphology in nup116ΔGLFG cells despite having no discernable impact in wild-type cells at 30°C. Inhibiting LD formation did impair NPC formation in otherwise wild-type cells but produced synthetic assembly defects in nup116ΔGLFG mutants. Disrupting NVJs did not significantly impact total numbers of LDs, although fewer were present at the nuclear rim (Fig. S5 B).
Figure S5.
NVJs impact NE morphology in nup116ΔGLFG strains. (A) TEM images of cells grown at 30°C. N, nuclei; V, heavily stained vacuoles. Arrowhead points to NE herniation. Scale bars, 500 nm. (B) Quantification of total LD number (left) and LDs at the NE (right) per cell in the listed strains grown at 34°C. Box and whiskers represent 2.5th–97.5th percentile. *, P ≤ 0.01 compared with wild type using Dunn’s post hoc test; **, P ≤ 0.02 when also compared with nup116ΔGLFG mdm1Δ nvj1Δ.
Foci formation was then assessed in strains lacking both NVJ and LDs at several temperatures to determine how NVJs and LDs might regulate NPC assembly together. While NVJ- and LD-deficient nup116ΔGLFG mutants did not have additive NPC clustering defects at 25°C, nearly 55% of the mutant cells displayed GFP-Nic96 foci at 30°C compared with ∼30% of nup116ΔGLFG cells deficient for LDs or NVJs (Fig. 4 E), consistent with their additive growth defects (Fig. 4 B). Thus, LDs and NVJs separately inhibited GFP-Nic96 foci formation in the context of perturbed NPC assembly. Interestingly, NVJs and LDs did not appear to function in a partially redundant manner in wild-type cells, as GFP-nic96 mdm1Δ nvj1Δ dga1Δ lro1Δ mutants did not exhibit clustering defects that were statistically more severe than GFP-nic96 mdm1Δ nvj1Δ or GFP-nic96 dga1Δ lro1Δ mutants at all temperatures tested (Fig. 4 F). We speculated that the NPC assembly defects at 34° and 37°C in GFP-nic96 mdm1Δ nvj1Δ cells result from mild assembly stress caused by elevated temperature and/or GFP-tagging NIC96, though NVJs could simply be more critical to regulating NPC assembly in wild-type cells at higher temperatures.
NVJs and autophagy factors mediate PEP4-dependent NE remodeling in nup116ΔGLFG mutants
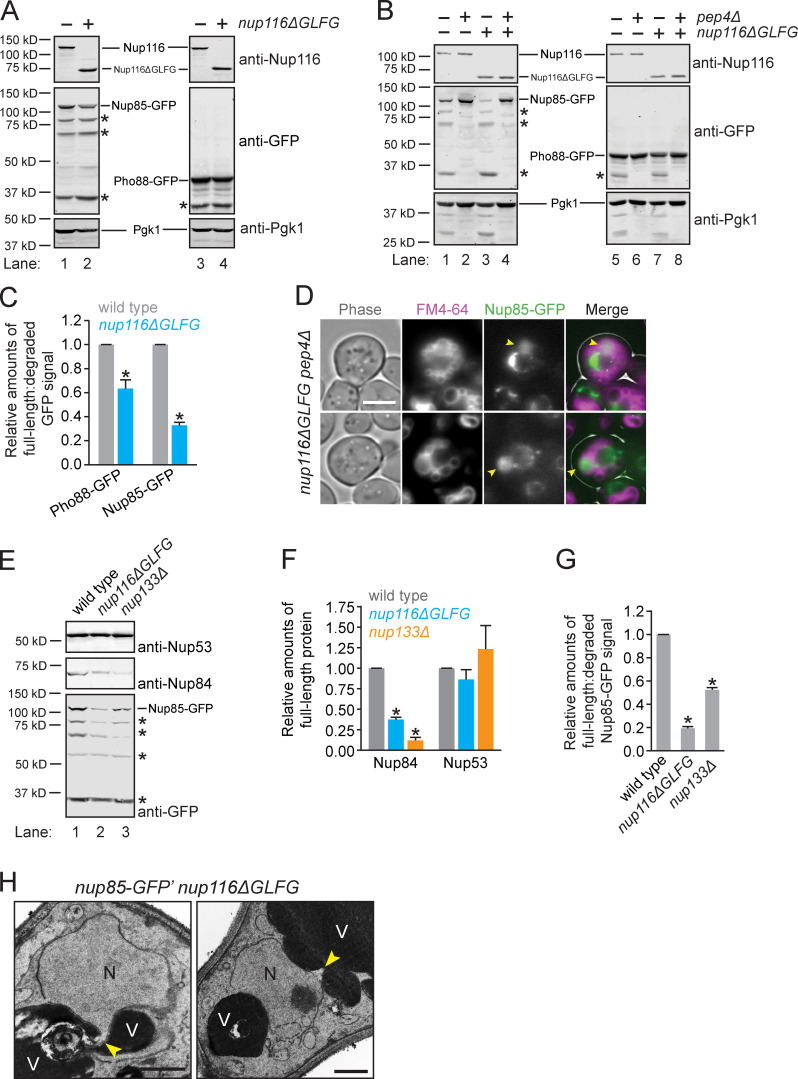
Given recent work demonstrating that Nups are degraded upon nitrogen starvation or rapamycin treatment by autophagy factors (Lee et al., 2020; Tomioka et al., 2020), we hypothesized that regions of the NE could be undergoing vacuole-mediated remodeling in NPC assembly mutants that result in degradation of NPCs or specific Nups. We found that Nup85-GFP was significantly more degraded in nup116ΔGLFG mutants in a PEP4-dependent manner, suggesting that processing occurs in vacuoles (Fig. 5, A–C; compare intensities of degraded versus full-length Nup85-GFP). Consistent with this possibility, we observed significant Nup85-GFP signal in the lumens of vacuoles adjacent to the NE in ∼10% of nup116ΔGLFG pep4Δ cells (Fig. 5 D); GFP was likely detected in these cells because their vacuoles remained fragmented, which prevented diffusion of GFP throughout the lumens of other vacuoles. That Pho88-GFP was also more degraded in nup116ΔGLFG mutants suggested that regions of the NE containing Nup85 were undergoing a remodeling event (Fig. 5, A–C), as opposed to Nup85 that had not been incorporated into the NE.
Figure 5.
Specific Nups are degraded in NPC assembly mutants by Pep4. (A and B) Western blots of lysates from nup85-GFP or pho88-GFP cells grown at 36°C. Full-length proteins are labeled; asterisks correspond to major PEP4-dependent degradation products. (C) Relative amounts of full-length:degraded Nup85-GFP and Pho88-GFP. Error bars represent SD. *, P ≤ 0.01 compared with wild type using unpaired two-tailed Student’s t test (n = 3 independent trials). (D) Images of FM4-64-labeled nup116ΔGLFG nup85-GFP pep4Δ cells grown at 36°C. Arrowheads point to vacuoles with significant GFP signal. Scale bar, 5 µm. (E) Western blots of lysates from cells grown at 32°C. Full-length Nup85-GFP is labeled; asterisks correspond to major degradation products. Degradation products for endogenous Nups could not be detected. (F) Relative amounts of full-length Nup53 and Nup84 in the listed nup85-GFP strains grown at 32°C. Error bars represent SD. *, P ≤ 0.01 compared with wild type using unpaired two-tailed Student’s t test (n = 4 independent trials). (G) Relative amounts of full-length:degraded Nup85-GFP in strains grown at 32°C. Error bars represent SD. *, P ≤ 0.01 compared with wild type using unpaired two-tailed Student’s t test (n = 4 independent trials). (H) TEM images of nup85-GFP′ nup116ΔGLFG cells grown at 36°C. N, nuclei; V, vacuoles. Potential sites of NE remodeling are labeled with a yellow arrowhead. Scale bars, 500 nm.
Nup85-GFP, Nup84, and Nup53 were assessed in nup133Δ and nup116ΔGLFG cells to determine whether levels of endogenous Nups were also decreased, and whether other NPC assembly mutants cause similar degradation phenotypes. Interestingly, while full-length Nup84 levels were also decreased in NPC assembly mutants, Nup53 levels were unaffected (Fig. 5, E–G), suggesting that specific Nups or Nup subcomplexes were degraded. We were unable to detect large blebs of nuclei in vacuoles, although smaller regions of the NE were infrequently observed that potentially were directly remodeled near vacuoles (Fig. 5 H). We concluded that NPC assembly mutants undergo vacuole-dependent NE remodeling that involves the degradation of specific Nups or nuclear pore subcomplexes that include Nup85 and Nup84.
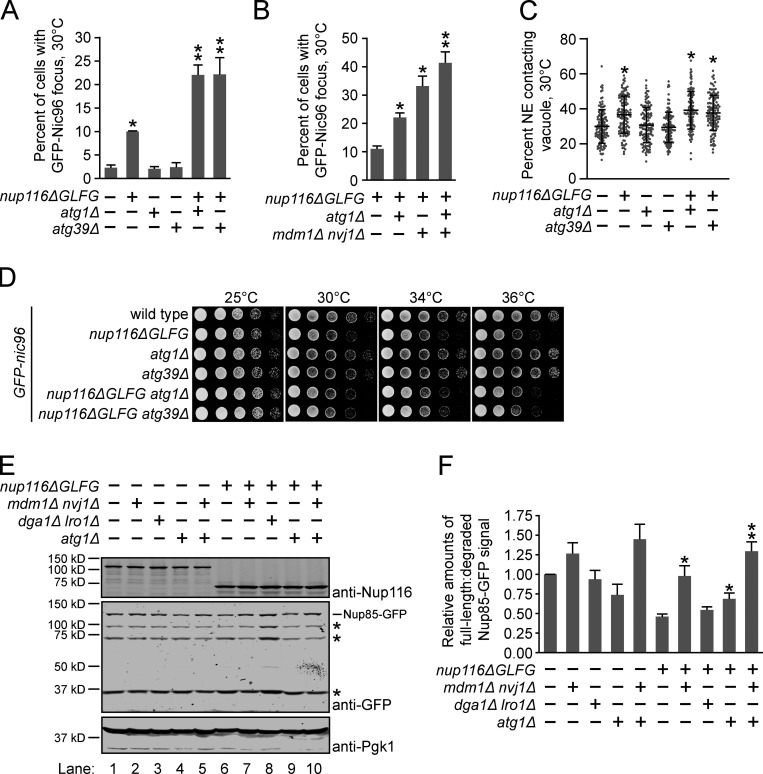
ATG1 and ATG39 are among several genes required for rapamycin-induced Nup degradation (Lee et al., 2020; Tomioka et al., 2020). We tested whether these autophagy proteins could be mediating NE remodeling to prevent NPC clustering in nup116ΔGLFG mutants. We predicted that, like NVJs and LDs, significantly more GFP-Nic96 foci would be observed in combinatorial mutants if ATG1 and/or ATG39 was necessary to prevent formation of NPC clusters. While neither ATG1 nor ATG39 significantly impacted NPC assembly, deletion of either gene along with Nup116’s GLFG domain doubled the levels of GFP-Nic96 foci at 30°C (Fig. 6 A). Interestingly, these autophagy factors likely function independently of NVJs, because GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ atg1Δ mutants exhibited additive NPC clustering defects compared with GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ cells (Fig. 6 B). Additionally, NE–vacuole interactions in nup116ΔGLFG mutants were unaffected by deletion of ATG1 or ATG39 (Fig. 6 C). Unlike NVJ and LD genes, GFP-nic96 nup116ΔGLFG atg1Δ and GFP-nic96 nup116ΔGLFG atg39Δ mutants did not display additive growth defects (Fig. 6 D), indicating that the extent of NPC clustering does not necessarily correlate with growth rate. Thus, ATG1 and ATG39 were necessary to limit NPC clustering in nup116ΔGLFG mutants along with NVJs, and neither gene impacted the viability of NPC assembly mutants under normal growth conditions.
Figure 6.
NE–vacuole interactions and autophagy factors promote NE remodeling in nup116ΔGLFG mutants. (A) Percentage of cells grown at 30°C with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.01 compared with wild type using an unpaired two-tailed Student’s t test (n ≥ 100 cells from three independent trials); **, P ≤ 0.05 when also compared with single mutants. (B) Percent of cells with GFP-Nic96 focus. Error bars represent SD. *, P ≤ 0.001 when compared with nup116ΔGLFG using an unpaired two-tailed Student’s t test (n ≥ 100 cells from five independent trials); **, P ≤ 0.01 compared with all other strains. (C) Quantification of NE–vacuole interactions in the listed GFP-nic96 strains at 30°C. Each dot represents value from one cell (n = 120 cells from three independent trials). Error bars represent SD. *, P ≤ 0.01 when compared with wild type using Dunn’s post hoc test. (D) Serial dilutions of cells on YPD plates. (E) Western blots of nup85-GFP lysates from cells grown at 36°C; this data and four other independent experiments were used to generate data in F. Asterisks label major degradation products. (F) Quantification of relative degradation levels of Nup85-GFP in the listed strains at 36°C. Error bars represent standard error of the mean. *, P ≤ 0.05 when compared with nup116ΔGLFG (n = 5 independent trials) using an unpaired two-tailed Student’s t test; **, P ≤ 0.05 when also compared with nup116ΔGLFG atg1Δ.
To directly assess whether autophagy, NE–vacuole contacts, and LDs were required for NE remodeling in nup116ΔGLFG cells, Nup85-GFP degradation was compared in strains lacking MDM1 and NVJ1, DGA1 and LRO1, or ATG1 at 36°C (Fig. 6, E and F). Significantly less Nup85-GFP degradation occurred in nup116ΔGLFG mdm1Δ nvj1Δ and nup116ΔGLFG atg1Δ lysates compared with nup116ΔGLFG lysates, although degradation was unchanged in nup116ΔGLFG dga1Δ lro1Δ lysates (compare lanes 6–109, especially intensity of degradation products). Consistent with the fact GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ atg1Δ mutants exhibit additive NPC assembly defects, Nup85-GFP degradation was significantly decreased in nup116ΔGLFG mdm1Δ nvj1Δ atg1Δ lysates compared with nup116ΔGLFG atg1Δ lysates (Fig. 6 F); there was no statistically significant difference between nup116ΔGLFG mdm1Δ nvj1Δ atg1Δ lysates and nup116ΔGLFG mdm1Δ nvj1Δ lysates, although we suspect this is likely because immunoblotting is inherently more variable than foci quantification. Notably, none of these factors played a significant role in regulating Nup85-GFP degradation in otherwise wild-type cells (lanes 1–5). These results indicated that NVJ and autophagy factors, but not LDs, are required for degradation of Nup85 in nup116ΔGLFG vacuoles.
We speculated that NVJ-mediated remodeling could be occurring in nup116ΔGLFG cells at a relatively constant level that prevents formation and/or expansion of NPC clusters, or in a manner that promotes direct removal of clustered NPCs. To distinguish between these possibilities, GFP-nic96 nup116ΔGLFG cells were monitored for several hours using a microfluidic chamber to determine if GFP-Nic96 foci were ever observed being cleared from the NE (Video 1). Since we did not observe the removal of any obvious NPC clusters over 4.5 h, we concluded that NVJ1- and MDM1-mediated NE–vacuole contacts continuously remodel the NE under conditions of NPC assembly stress to prevent NPC cluster formation/expansion.
Video 1.
GFP-Nic96 foci are not removed from the NE in nup116ΔGLFG cells. GFP-nic96 nup116ΔGLFG cells grown overnight at 32°C, then imaged using a microfluidic chamber at 36°C for 4.5 h. Frames are imaged 20 min apart for a total of 18 time points, while maximum projections are shown. Scale bar, 5 µm.
CHM7 differentially affects nup116ΔGLFG and vps4Δ mutants
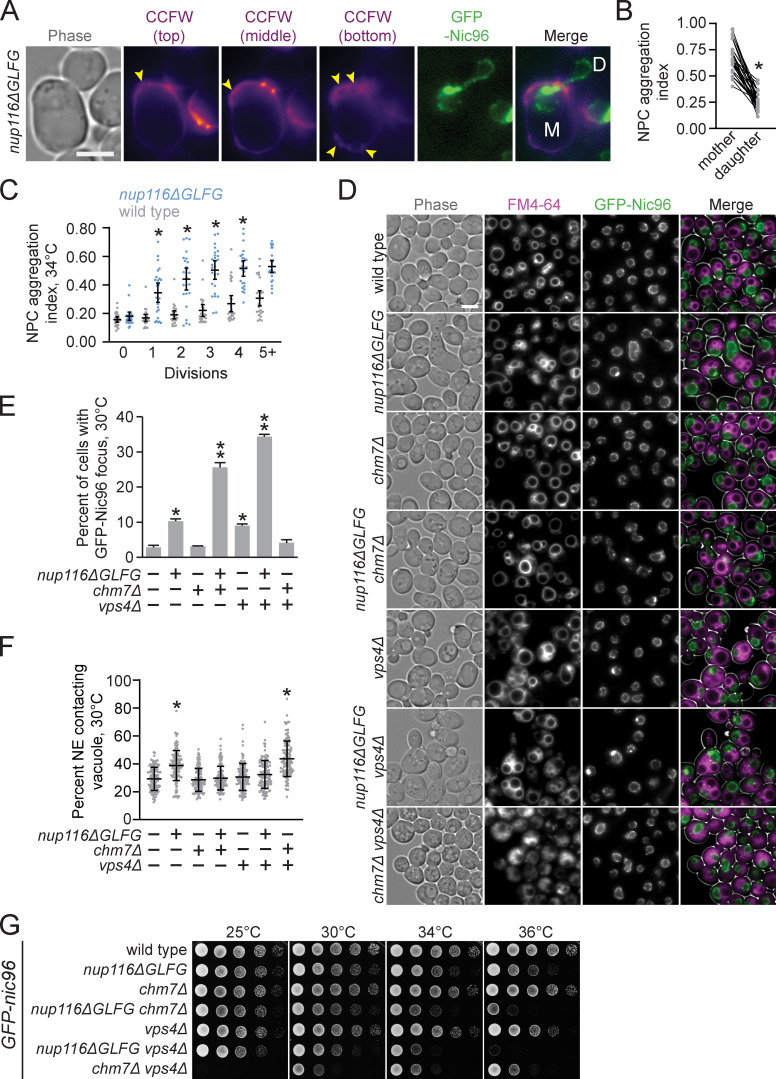
The endosomal sorting complexes required for transport subunit Vps4 functions in a surveillance pathway that removes misassembled NPC intermediates that can form in the NE when NPC biogenesis is disrupted (Webster et al., 2014). When this surveillance pathway is inhibited in vps4Δ cells, foci of misassembled NPC intermediates can form along the NE in a structure termed storage of improperly assembled NPCs compartment (SINC); these SINC structures are retained in mother yeast cells during the replicative aging process (Webster et al., 2014). Because Nup foci that form in nup116ΔGLFG mutants also result from disrupted NPC assembly, we tested whether nup116ΔGLFG foci are retained in mother cells. Comparison of NPC aggregation in mother versus daughter GFP-nic96 nup116ΔGLFG nuclei in cells that had not yet fully divided and contained at least one focus at 34°C revealed that NPC aggregation indices were greater in mother cells (Fig. 7, A and B). Consistent with these data, NPC aggregation increased as nup116ΔGLFG mutants replicatively aged at 34°C relative to wild-type cells (Fig. 7 C). Importantly, these experiments indicated that NPC aggregates in nup116ΔGLFG cells tend to be retained in mother cells.
Figure 7.
NPC clustering in nup116ΔGLFG cells is CHM7-independent. (A) Calcofluor white (CCFW)-stained GFP-nic96 nup116ΔGLFG cell grown at 34°C that underwent 6 divisions. Mother cell is labeled with an “M”, while undivided daughter is labeled with a “D”; arrowheads point to bud scars. Scale bar, 5 µm. (B) NPC aggregation indices from undivided mother cells with mother and daughter nuclei. Each left dot represents one mother nucleus, connected with its corresponding daughter nucleus on the right. *, P ≤ 0.001 using a paired two-tailed Wilcoxon test. (C) NPC aggregation index versus division number for GFP-nic96 strains at 34°C. Each dot represents values from one cell (n ≥ 20); error bars represent SD. *, P ≤ 0.05 compared with wild type using Dunn’s post hoc test for the given division number. (D) FM4-64–stained GFP-nic96 cells grown at 30°C. Scale bar, 5 µm. (E) Percentage of cells with at least one GFP-Nic96 focus from cells grown at 30°C. Error bars represent SD. *, P ≤ 0.01 compared with wild type using an unpaired two-tailed Student’s t test (n ≥ 100 from three independent trials); **, P ≤ 0.01 when also compared with nup116ΔGLFG or vps4Δ. (F) NE–vacuole contacts in the listed GFP-nic96 strains grown at 30°C. Error bars represent SD. Each dot corresponds to the value from an individual cell (n = 120 from three independent trials). *, P ≤ 0.01 compared with wild type using Dunn’s post hoc test. (G) Serial dilutions of GFP-nic96 strains on YPD plates.
Formation of SINC foci in vps4Δ mutants is abolished in the absence of the endosomal sorting complexes required for transport subunit Chm7 (Webster et al., 2016), likely because clustering is promoted by improper compartmentalization of Chm7 (Thaller et al., 2019). GFP-Nic96 foci and NE–vacuole contacts were measured in GFP-nic96 nup116ΔGLFG chm7Δ cells at 30°C to test whether foci formation in nup116ΔGLFG mutants also required CHM7. Surprisingly, instead of abrogating GFP-Nic96 foci formation as it does in vps4Δ cells (Fig. 7, D and E), deletion of CHM7 increased GFP-Nic96 foci formation ∼150% in nup116ΔGLFG cells without impacting NPC assembly in otherwise wild-type cells. This correlated with a significant loss of NE–vacuole contacts in GFP-nic96 nup116ΔGLFG chm7Δ mutants (Fig. 7 F); similar effects were observed when VPS4 was deleted in GFP-nic96 nup116ΔGLFG mutants. The loss of NE–vacuole interactions by deletion of CHM7 or VPS4 suggested that Chm7 and Vps4 are somehow required to alter NE/vacuole composition so that these interorganelle interactions can occur in nup116ΔGLFG mutants. Consistent with a loss of NE–vacuole interactions, GFP-nic96 nup116ΔGLFG chm7Δ and GFP-nic96 nup116ΔGLFG vps4Δ cells exhibited significant synthetic growth defects (Fig. 7 G). In sum, NPC clusters in nup116ΔGLFG mutants formed in a CHM7-independent manner distinct from those observed in vps4Δ mutants, suggesting that NVJ-mediated regulation of NPC assembly functions in a separate pathway than VPS4-mediated assembly surveillance.
Discussion
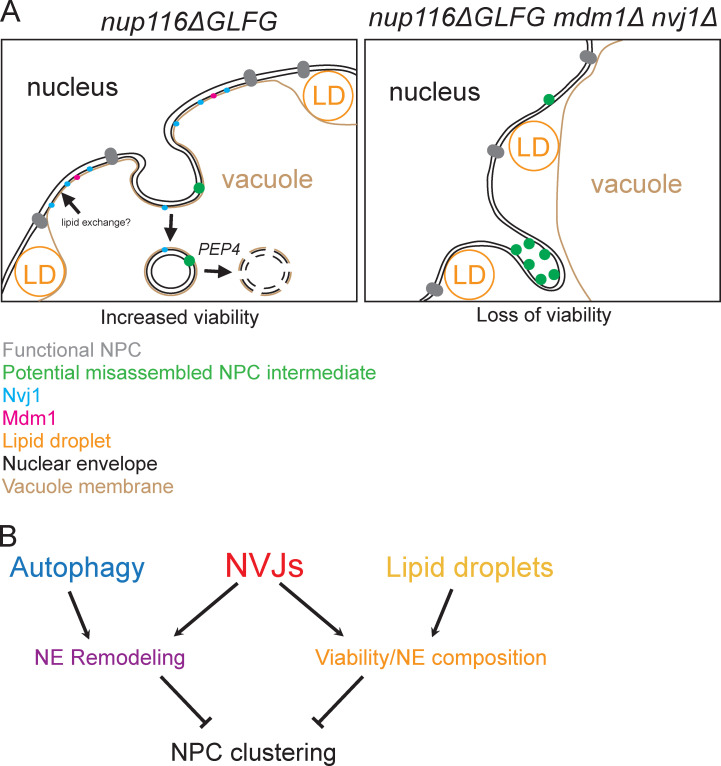
Results presented herein suggest a model wherein NPC assembly is compromised in nup116ΔGLFG mutants and other specific nup mutants, particularly at elevated temperatures, leading to expansion of NVJs that cooperate with LDs to maintain cell viability as well as degrade regions of the NE that contain specific Nups. This occurs in a PEP4-dependent manner with the autophagy factors Atg1 and Atg39 (Fig. 8). Based on their close apposition and potential intermediates observed using TEM, we propose that NVJs can serve as sites to directly promote removal of NE domains that contain Nup84 and Nup85, potentially as misassembled NPC intermediates (Fig. 8); NPC clusters are then less likely to form by removal of these domains in a fairly continuous manner (Video 1). Our results also suggest that both NVJ-dependent events are necessary to promote proper NPC assembly when this process is already compromised in some way.
Figure 8.
NE–vacuole interactions mediate dual responses to NPC assembly stress. (A and B) Cartoons depicting proposed functions of NVJs in nup116ΔGLFG cells. NVJs containing Nvj1 and Mdm1 expand around the NE in response to misassembled NPCs, and LDs also are enriched at these contacts. NVJs and LDs promote viability together, potentially through lipid exchange that stabilizes the NE. Additionally, NVJs promote continuous vacuole-mediated remodeling of the NE, potentially degrading misassembled NPC intermediates that form in the inner NE. Separately, the autophagy factors ATG1 and ATG39 also mediate remodeling of the NE. Together, these dual functions of NVJs enhance proper NPC formation in mutants where NPC assembly is perturbed. In the absence of NVJs, more misassembled NPC intermediates accumulate in NE herniations that appear as clusters using fluorescence microscopy.
The observation that NE–vacuole contacts do not expand in nup100Δ mutants, which display increased NPC permeability (Lord et al., 2015; Timney et al., 2016; Popken et al., 2015) as well as tRNA export defects (Lord et al., 2017), suggests that NVJ expansion in nup116ΔGLFG, nup120Δ, nup133Δ, and nup85-GFP′ mutants is related to perturbation of NPC assembly and not a general loss of NPC function. We speculate that NVJs are not used in wild-type cells under normal growth conditions, because Vps4-mediated surveillance (Webster et al., 2016, 2014; Thaller et al., 2019) is likely sufficient to deal with defects that infrequently arise during normal NPC biogenesis. When mutations or other stress conditions are imposed that further disrupt NPC assembly, NVJs become important factors to mitigate stress on the NE. Alternatively, the biogenesis defects in the nup mutants we analyzed may impart some kind of additional stress on the NE not typically observed when NPC assembly is compromised in wild-type cells.
We propose that expansion of NE–vacuole contacts and increased LD content constitutes a general cellular response to mitigate several types of sustained NPC assembly stress. For example, TEM images of other NPC assembly mutants clearly show significant amounts of LDs relative to wild-type cells, and less obviously, show accompanying changes in vacuole morphology (Makio et al., 2009; Ryan et al., 2003). Additionally, when the temperature-sensitive brr6-1 allele is inactivated for several hours, NPC assembly is compromised and LD number increases; genetic experiments indicate that these LDs are necessary for full viability of the mutant (Hodge et al., 2010). Short-term inactivation of Brr6, however, inhibits NPC assembly without altering lipid composition (Zhang et al., 2018), suggesting that LDs and potentially NVJs serve as a long-term cellular mechanism to compensate for perturbations in NPC formation.
We speculate that NVJs and LDs promote viability in nup116ΔGLFG and nup133Δ strains by serving as interfaces to promote lipid exchange that beneficially alters NE composition in response to perturbations in NPC assembly. Although NVJs have not been specifically shown to mediate lipid transfer between the vacuole and NE, interorganelle contact sites often serve as sites of lipid exchange (Kornmann et al., 2009; Elbaz-Alon et al., 2014) owing to their close apposition and enrichment ]of lipid-exchanging factors. The phosphatidylinositol-4-phosphate– and ergosterol-binding protein Osh1 (Manik et al., 2017), which is found at NVJs and interacts with Nvj1 (Jeong et al., 2017; Kvam and Goldfarb, 2004; Levine and Munro, 2001), belongs to the oxysterol-binding homology protein family that can aid in lipid transfer between membranes (de Saint-Jean et al., 2011). Additionally, LDs are thought to promote lipid transfer between organelles, potentially through “lipid bridges” (Schuldiner and Bohnert, 2017). Because deletion of NVJs or LDs significantly increases NPC clustering in nup116ΔGLFG cells (Fig. 4), we hypothesize that preventing changes in lipid composition by abrogating NVJs or LDs further inhibits NPC formation, resulting in more misassembled clustered NPCs and NE herniations.
We hypothesize that, in addition to maintaining viability with LDs, NVJs function separately from the autophagy factors Atg1 and Atg39 to remodel and degrade portions of the NE that contain specific Nups or NPC subcomplexes. From our tests to date, at least Nup84 and Nup85 are specifically degraded, but not Nup53 or Nup116. Interestingly, the mammalian orthologues of Nup84 and 85 regulate initial stages of interphase NPC biogenesis (Otsuka et al., 2016). It is intriguing to speculate that some Nup84 subcomplexes (that also contain Nup85) might be forming misassembled intermediates in nup116ΔGLFG cells that lead to their clustering and degradation. However, we have no direct evidence that such misassembled intermediates form. Nitrogen starvation or rapamycin treatment were recently found to trigger autophagy pathways that degrade NPCs in vacuoles (Lee et al., 2020; Tomioka et al., 2020); starvation exposes a Nup159 motif that interacts with Atg8 to degrade NPCs (Lee et al., 2020) or non–NPC-associated Nup159 (Tomioka et al., 2020), likely depending on the strain background. Deletion of NUP120 or NUP133 increases Nup degradation not only during starvation, but also in rich medium (Lee et al., 2020), consistent with our results.
While changing Nup159’s Atg8-interacting motif impairs starvation-induced Nup degradation in nup120Δ cells, it does not decrease degradation in rich medium (Lee et al., 2020), suggesting that different factors may recognize and degrade NPCs/Nups in these different conditions. Additionally, deletion of NVJ1 has no apparent impact on starvation-induced degradation (Lee et al., 2020; Tomioka et al., 2020), unlike what we observed in our experiments. ATG39 is partially required for degradation of several Nups during rapamycin treatment (Tomioka et al., 2020), and our genetic evidence showing that deletion of ATG39 increases NPC assembly defects in nup116ΔGLFG cells suggests that Atg39 could function as a receptor to recognize misassembled NPC intermediates. Phosphorylation (Laurell et al., 2011) and ubiquitylation (Niño et al., 2016) are potential posttranslational modifications that can be added to specific Nups, which may allow autophagy machinery and NVJs to distinguish them from assembled and transport-competent NPCs. Fully defining the cellular machinery required to deform the NE along with NVJs to promote remodeling will be an important future endeavor.
NPC assembly and nuclear transport play essential roles in modulating yeast replicative life span (Lord et al., 2015, 2017; Rempel et al., 2019; King et al., 2019), and disrupted NPC function is associated with aging and neurological disorders in metazoans (D’Angelo et al., 2009; Toyama et al., 2013; Da Cruz and Cleveland, 2016). Given their impact on promoting proper NPC formation and viability in mutants with disrupted NPC assembly, it will be important to define how NE–vacuole interactions regulate the replicative aging process and potentially other stress conditions. Although metazoans have no obvious orthologous system to NVJs, their cells can produce nuclear LDs (Ohsaki et al., 2016), and regions of their nuclei can be degraded via autophagy (Dou et al., 2015), both of which could potentially be used to respond to perturbations in interphase NPC assembly. Importantly, our experiments establish novel and critical roles for NVJs in ameliorating NPC assembly defects and provide mechanistic insights into how interorganelle contacts can be used to respond to stress/disease states.
Materials and methods
Yeast strains and growth
A complete list of strains used for this study are listed in Table S1. BY4741 and 4742 S288C strains (Brachmann et al., 1998) were used for all experiments. Unless otherwise noted, yeast strains were grown in 1% yeast extract/2% peptone/2% dextrose (YPD) medium overnight at listed temperatures to midlog phase. An S288C nup116ΔGLFG strain was generated by cloning the nup116ΔGLFG genome sequence from SWY2791 into SnaBI- and XhoI-digested pAG306-GPD-empty chr I (41895; Addgene) using Gibson Assembly master mix (E2611L; NEB). The resulting vector was digested with SpeI (R0133S; NEB) and transformed into BY4742 yeast. URA3-positive transformants were then plated onto 5-fluoroorotic acid to excise full-length NUP116, which was confirmed using PCR and Western blots. Other gene deletions were generated using PCR products derived from pFA6a-kanMX6 (39296; Addgene), pFA6a-hphNT1 (P30347; Euroscarf), or the yeast deletion library (Giaever et al., 2002). nup85-GFP′ strains were derived from the GFP collection (Huh et al., 2003), itself derived from pFA6a-GFP(S65T)-His3MX6 (41598; Addgene), as were nvj1-GFP strains. pYM25 (P30237; Euroscarf) was used to create nup85-GFP and mdm1-GFP strains. DNA sequencing revealed no differences in the NUP85 coding region between nup85-GFP and nup85-GFP′. pFA6a-link-ymNeongreen-SpHis5 (125704; Addgene) was used to generate Mdm1-mNeonGreen cells. pSS146 (64770; Addgene) and pKV015 (64766; Addgene) were used to create strains with nic96-GFP/RFP RITE tags. GFP-nic96 strains were constructed by transforming strains with AflII-digested pSW950 (Bucci and Wente, 1998). Most combinatorial mutants were generated using mating and tetrad dissections.
Plasmids
Other than the plasmids used to generate yeast strains described above, pRS315 (77144; ATCC) and pRS315-NUP116 (Wente et al., 1992) were used for complementation.
FM4-64 staining
1 ml of cells grown to midlog phase in YPD were transferred to an amber 1.5-ml tube along with 1 µl of 8 mM FM4-64/DMSO for a final cell staining concentration of 8 µM. Cells were then stained at the temperature at which they grew overnight, along with shaking for 20–30 min. After staining, cells were pelleted and washed two times with 1 ml fresh medium (prewarmed if necessary) and allowed to grow for an additional 2 h before imaging, again at overnight growth temperature, unless otherwise noted for temperature-shift experiments.
Fluorescence microscopy
With the exception of Video 1, all images were acquired using a BX53 Olympus microscope equipped with a 100×/1.35-NA or 40×/1.30-NA Olympus UPlan oil lens and Hamamatsu Photonics Orca-R2 camera under immersion in Olympus Immoil-F30CC oil using Nikon Elements software at RT (cells were moved from temperatures listed in figure legends within 5 min of imaging). Unless otherwise stated in the figure legend, single planes of images are shown, but for quantification, 15 successive 400-nm slices were imaged to observe foci and NE–vacuole interactions near the edges of cells. Images in Video 1 were acquired using a GE Healthcare Personal DeltaVision equipped with an Olympus IX71 microscope, a 60× 1.42-NA Plan Apochromat objective, and a Photometrics CoolSnap HQ2 camera at 36°C under immersion in Olympus Immoil-F30CC oil using softWoRx software. Time-lapse imaging was performed using an EMD Millipore CellASIC Onix Perfusion system with Y04C plates; successive Z stacks were imaged every 20 min for a total of 18 time points. Fiji was used to generate images after data acquisition.
Quantification of NE–vacuole interactions
FM4-64–stained cells that were imaged using 15 successive 400-nm Z stacks were examined to find the region of the cell, generally near the center, where the GFP-tagged nuclear rim was largest. Using this slice, the total length of the nuclear rim was calculated using the freehand line tool in Fiji. Then the length of the NE that directly contacted the NE was measured, again using the freehand line tool. The contact length was divided by the total length to quantify the amount of NE–vacuole interactions for a single cell. This same method was also used for TEM images to quantify NE–vacuole interactions in Fig. S1 I; here, a 20-nm distance between the NE and vacuoles was considered a physical interaction between the organelles.
EM
The glutaraldehyde/potassium permanganate protocol was performed largely according to published protocols (Wright, 2000). 20 ml yeast was grown overnight to midlog phase, then mixed with 20 ml 2× fixation buffer (200 mM Pipes, pH 6.8, 200 mM sorbitol, 2 mM MgCl2, 2 mM CaCl2, and 4% glutaraldehyde) with rocking for 5 min in a 50-ml conical tube. The cells were then centrifuged for 3 min at 3,000 rpm and resuspended in 1× fixation buffer, where the cells were left overnight at 4°C while rocking. The following day, cells were washed three times for 10 min in 25 ml double-distilled H2O. Cells were transferred to 1.5-ml tubes and washed with 1 ml of freshly made 2% potassium permanganate for 5 min, then incubated with 1 ml 2% potassium permanganate for 1 h while rocking. Cells were washed five times with 1 ml of double-distilled H2O for 1 min, transferred to new 1.5-ml tubes, then incubated with Uranyless (22409; EMS) for 1 h while rocking. After cells were washed three times with 1 ml water for 1 min, the cells were dehydrated with increasing amounts of ethanol (25%, 50%, 75%, and 95%) for 5 min each. The cells were then washed three times with 100% ethanol for 5 min and incubated overnight with 50% acetone/50% EMbed 812 (14120; EMS) while rocking at RT. This was replaced with 100% EMbed 812 for 2 h, which was repeated before finally resuspending cells in 500 µl EMbed 812. The mixture was solidified by baking overnight at 60°C. After sectioning, cells were stained with uranyl acetate and lead citrate before analyzing using a Philips/FEI T-12 transmission electron microscope equipped with a 2,000 × 2,000 AMT charge-coupled device camera system. Fiji was used to produce final images after data acquisition.
Cell lysates and Western blotting
3–5 ml of cells was grown to midlog phase at temperatures listed in the figure legends and then centrifuged at 3,000 rpm in a tabletop centrifuge to pellet cells. Cells were transferred to 1.5-ml tubes and washed once with 1 ml YPD. After all the remaining YPD was carefully removed, cell pellets were resuspended evenly in 40 µl 1× SDS sample buffer (60 mM Tris, pH 6.8, 1% SDS, 10% glycerol, 1.67% β-mercaptoethanol, and 0.003% bromophenol blue) and immediately boiled for 5 min. After boiling, acid-washed glass beads were added so that they encompassed the entire volume of liquid/cell suspension. Tubes were then vortexed heavily for 2 min. Finally, 60 µl 1× SDS sample buffer was added, and the samples were quickly boiled for 30 s and loaded onto polyacrylamide gels. SDS-PAGE and transfers were completed using established protocols and nitrocellulose membranes. After transfer, membranes were stained with Ponceau S (0.1% Ponceau S and 5% acetic acid) to ensure that protein levels were similar among samples. Ponceau S stain was removed by briefly washing membranes in 0.01 N NaOH, and then washed three times with 50 ml water to remove residual NaOH. Membranes were blocked for 1 h in TBS plus Tween (TBST)/7% milk, then incubated overnight at 4°C with primary antibodies used at dilutions listed below. The next day, membranes were washed three times with 25 ml TBST for 5 min at RT, then incubated with TBST/7% milk for 1 h at RT with 1:10,000 dilutions of secondary antibodies. Membranes were finally washed three times with 25 ml TBST for 5 min at RT and imaged using a Li-Cor Odyssey system. Antibodies included 1:1,000 affinity-purified polyclonal rabbit anti-Nup116 C-term (Iovine et al., 1995), 1:500 monoclonal mouse anti-GFP (11814460001; Millipore Sigma) reconstituted at 0.4 mg/ml in water/0.05% sodium azide, 4 µg/ml goat polyclonal anti-Nup84 (ab122883; AbCam), 1:1,000 polyclonal rabbit anti-Nup53 (01-675-390; Thermo Fisher Scientific), 1:1,000 monoclonal mouse anti-Pgk1 (459250; Thermo Fisher Scientific), 1:10,000 IRDye 680LT donkey anti-mouse IgG (926-68022; Li-Cor), 1:10,000 IRDye 800CW goat anti-Mouse IgG (926-32210; Li-Cor), 1:10,000 IRDye 680RD donkey anti-goat IgG (926-68074; Li-Cor), and 1:10,000 IRDye 800CW donkey anti-rabbit IgG (926-32213; Li-Cor).
NPC aggregation indices
Aggregation indices were quantified similarly to previous studies (Casey et al., 2015; Niepel et al., 2013). The average GFP-Nic96 fluorescence intensity at the nuclear rim was calculated by using the freehand line tool in Fiji to make a curved line that covered the entire rim in one plane of an image, which was then converted to quantifiable data using the plot profile tool; these numbers were averaged after background GFP intensity was subtracted. Average GFP-Nic96 fluorescence intensity was then subtracted from each plot profile measurement, and the SD of these absolute values was divided by the average GFP-Nic96 fluorescence intensity to produce an aggregation index.
Quantification of protein degradation
Li-Cor Image Studio was used to quantify the amounts of full-length Nup85-GFP or Pho88-GFP (top labeled bands in anti-GFP blots in Fig. 5, A, B, and E; and Fig. 6 E), as well as the three major GFP degradation products below full-length Nup85-GFP or the one major degradation product below Pho88-GFP (marked with asterisks in blots). The amount of full-length GFP protein was divided by the sum of the degradation products, and this value was then compared with the value from wild-type lysates to produce relative degradation amounts in Fig. 5 C (n = 3 independent trials), Fig. 5 G (n = 4 independent trials), and Fig. 6 F (n = 5 independent trials). Li-Cor Image Studio was also used to quantify bands for Fig. 5 F, except in this case the values of full-length Nup84 and Nup53 were compared with wild type, as degradation products were not detectable (n = 4 independent trials).
Serial dilutions
1 OD unit of yeast cells grown overnight at 25°C (unless temperature is otherwise noted in legends) to midlog phase was pelleted via centrifugation, resuspended in 500 µl sterile water, and serially diluted fivefold in water four times. 4–5 µl of the mixture was spotted onto plates, which were grown at temperatures listed in the figure legends. Images were taken once wild-type cells started to show growth at the greatest dilution.
RITE experiment
Mutant nup116ΔGLFG CYC1term::pTDH3-CRE-EBD78-HIS3 nic96-LoxP-GFP-HYG-LoxP-RFP cells were grown overnight at 4°C to midlog phase in YPD, wherein the majority solely expressed Nic96-GFP. To induce production of Nic96-RFP while simultaneously ending Nic96-GFP formation, cells were incubated for 3 h at 25°C in the presence of 5 µM β-estradiol. Finally, cells were incubated at 36°C for 3 h to induce formation of GFP/RFP foci and were analyzed using fluorescence microscopy.
CPY secretion assay
Cells were serially diluted as described onto two YPD plates. One plate was covered with nitrocellulose to absorb secreted CPY, and both were grown overnight at 28°C. The nitrocellulose was removed and washed several times in water, whereas the plate without nitrocellulose was imaged. The nitrocellulose was then blocked for 1 h in TBST/5% milk and incubated overnight at 4°C in 2 ml TBST/5% milk with a 1:1,000 dilution of anti-CPY monoclonal antibody 10A5B5 (A-6428; Invitrogen). The nitrocellulose was then washed three times in 20 ml TBST at RT for 10 min and incubated with 5 ml TBST/5% milk with a 1:10,000 dilution of IRDye 680LT donkey anti-mouse IgG (926-68022; Li-Cor) for 1 h at RT in a light-sensitive box. After washing three times with 20 ml TBST for 10 min at RT, the nitrocellulose was imaged on a Li-Cor Odyssey system.
BODIPY 493/503 staining
1 ml of cells grown to mid-log phase in YPD was transferred to an amber 1.5-ml tube, along with 1 µl of 2 mM BODIPY 493/503/DMSO, for a final concentration of 2 µM. Cells were incubated for 5 min at the temperature at which they were initially grown, washed two times with 1 ml YPD, then imaged.
Quantification of NE-associated LDs
The total amounts of LDs were measured in FM4-64– and BODIPY-labeled yeast strains by counting in maximum projections. In addition to staining LDs, BODIPY also weakly stains the NE and ER membranes, which is particularly evident when examining individual Z stacks. LDs at the NE were quantified by counting the amount at the NE that colabeled with FM4-64 in the same Z stack; this number was then divided by the total number of LDs.
Quinacrine staining
1 ml of cells grown to midlog phase in YPD was transferred to an amber 1.5-ml tube. The cells were centrifuged; resuspended in YPD/50 mM sodium phosphate, pH 7.6/100 µM quinacrine; and shaken at 36°C for 5 min. Cells were centrifuged and washed two times with prewarmed YPD/50 mM sodium phosphate, pH 7.6, before analyzing using fluorescence microscopy.
Hoechst 33342 staining
Cells were incubated with 10 µg/ml Hoechst 33342 for 20 min in YPD amber tubes at specified temperatures, quickly washed twice with 1 ml YPD, and imaged.
Calcofluor white staining
Cells were incubated for 5 min in 1 ml of 0.5 µg/ml calcofluor white/YPD in amber tubes. Cells were then washed two times with 1 ml YPD and imaged.
Statistical analysis
Data are depicted as mean with SD error bars, except for bar-and-whisker plots, which are shown with mean and percentiles listed in figure legends, and Fig. 6 F, in which mean with SEM are shown. Unpaired two-tailed Student’s t tests were used to compare GFP-Nic96, Nup85-GFP, and Nup85-GFP′ foci formation data, as well as Western blot quantifications. For NE–vacuole interaction data, as well as data in Figs. 3 E, 7 C, and S4 F, Dunn’s post hoc test was used after Kruskal–Wallis ANOVA. Mann–Whitney U tests were performed for data in Figs. 1 D, 3 F, and S3 D. Microsoft Excel and Prism 8 were used to perform statistical tests. Data were considered significant if P values were < 0.05 for a particular statistical test. Data distribution was assumed to be normal for Student’s t tests, but this was not formally tested.
Online supplemental material
Fig. S1 contains nup116ΔGLFG RITE cassette experiments, GFP-nic96 nup116ΔGLFG cycloheximide treatment data, GFP-nic96 kap mutant NPC clustering data; nup116ΔGLFG GFP-nic96/nup85-GFP/nup85-GFP′ serial dilutions, and quantification of NE–vacuole interactions in nup116ΔGLFG cells using TEM. Fig. S2 contains images of FM4-64–stained GFP-nic96 nup116ΔGLFG, nup100Δ, did2Δ, and ypt7Δ cells at 25°, 30°, and 34°C; images of FM4-64–stained nup116ΔGLFG and nup100Δ cells; and images and quantification of FM4-64–stained GFP-nic96 nup116ΔGLFG and nup100Δ cells grown in synthetic medium. Fig. S3 contains images of FM4-64–stained nup116ΔGLFG nup85-GFP/nup85-GFP′ cells grown at 30°C, images of FM4-64– and Hoechst-stained mdm1-mNeonGreen nup116ΔGLFG cells, FM4-64– and BODIPY-stained nup116ΔGLFG cells, and Pdr16-GFP foci quantification. Fig. S4 contains images of BODIPY-stained GFP-nic96 dga1Δ lro1Δ cells, serial dilutions of several strains, and NPC assembly defects in erg6Δ GFP-nic96 nup116ΔGLFG cells. Fig. S5 contains TEM images of GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ cells and LD quantification for GFP-nic96 nup116ΔGLFG mdm1Δ nvj1Δ cells. Video 1 shows GFP-nic96 nup116ΔGLFG cells over 4.5 h in 20-min increments after shifting from 32° to 36°C. Table S1 lists all the yeast strains used in this study, along with genotypes.
Supplementary Material
lists yeast strains and genotypes.
Acknowledgments
The authors thank members of Susan Wente’s and Yi Ren’s laboratories for useful feedback about experiments and manuscript preparation, Patrick Lusk for discussions regarding NPC clustering experiments, Sepp D. Kohlwein (University of Graz, Graz, Austria) for the dga1Δ lro1Δ strain, Kathleen Gould for use of her DeltaVision microscope and perfusion system (Vanderbilt University, Nashville, TN), Alaina Willet for assistance with the DeltaVision, and Evan Krystofiak for assistance with EM sample preparation.
EM experiments were performed in part using the Vanderbilt Cell Imaging Shared Resource (supported by National Institutes of Health grants CA68485, DK20593, DK58404, DK59637, and EY08126). S.R. Wente and this work are supported by National Institute of General Medical Sciences grant 5R37GM051219.
The authors declare no competing financial interests.
Author contributions: C.L. Lord and S.R. Wente cooperatively designed and interpreted experiments and co-wrote the manuscript. C.L. Lord performed experiments.
References
- Aitchison, J.D., Blobel G., and Rout M.P.. 1995. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131:1659–1675. 10.1083/jcb.131.6.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram, N., Ast T., Costa E.A., Arakel E.C., Chuartzman S.G., Jan C.H., Haßdenteufel S., Dudek J., Jung M., Schorr S., et al. 2016. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 540:134–138. 10.1038/nature20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, B.D.M., Dziurdzik S.K., Kolehmainen K.L., Fowler C.M.S., Kwong W.K., Grad L.I., Davey M., Schluter C., and Conibear E.. 2018. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 217:3593–3607. 10.1083/jcb.201804111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, M., and Hurt E.. 2017. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18:73–89. 10.1038/nrm.2016.147 [DOI] [PubMed] [Google Scholar]
- Boehmer, T., Enninga J., Dales S., Blobel G., and Zhong H.. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 100:981–985. 10.1073/pnas.252749899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., and Boeke J.D.. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Bucci, M., and Wente S.R.. 1998. A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol. Biol. Cell. 9:2439–2461. 10.1091/mbc.9.9.2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, A.K., Chen S., Novick P., Ferro-Novick S., and Wente S.R.. 2015. Nuclear pore complex integrity requires Lnp1, a regulator of cortical endoplasmic reticulum. Mol. Biol. Cell. 26:2833–2844. 10.1091/mbc.E15-01-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin, A., Hess B., San Roman M., Gatti X., Lombard B., Loew D., Barral Y., Palancade B., and Doye V.. 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189:795–811. 10.1083/jcb.200910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo, M.A., Raices M., Panowski S.H., and Hetzer M.W.. 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 136:284–295. 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz, S., and Cleveland D.W.. 2016. CELL BIOLOGY. Disrupted nuclear import-export in neurodegeneration. Science. 351:125–126. 10.1126/science.aad9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, T.R., Lazarus M.D., Hetzer M.W., and Wente S.R.. 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184:659–675. 10.1083/jcb.200806174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean, M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G.. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195:965–978. 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D.P., Patel S.S., Uversky V., Fink A.L., and Rexach M.. 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 100:2450–2455. 10.1073/pnas.0437902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Z., Xu C., Donahue G., Shimi T., Pan J.-A., Zhu J., Ivanov A., Capell B.C., Drake A.M., Shah P.P., et al. 2015. Autophagy mediates degradation of nuclear lamina. Nature. 527:105–109. 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye, V., Wepf R., and Hurt E.C.. 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13:6062–6075. 10.1002/j.1460-2075.1994.tb06953.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord, M., Mari M., Weill U., Rosenfeld-Gur E., Moldavski O., Castro I.G., Soni K.G., Harpaz N., Levine T.P., Futerman A.H., et al. 2018. Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217:269–282. 10.1083/jcb.201704122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz-Alon, Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Fratti, R.A., Collins K.M., Hickey C.M., and Wickner W.. 2007. Stringent 3Q.1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J. Biol. Chem. 282:14861–14867. 10.1074/jbc.M700971200 [DOI] [PubMed] [Google Scholar]
- Giaever, G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Grillitsch, K., Connerth M., Köfeler H., Arrey T.N., Rietschel B., Wagner B., Karas M., and Daum G.. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta. 1811:1165–1176. 10.1016/j.bbalip.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri, H., Rogers S., Ugrankar R., Liu Y.L., Feathers J.R., and Henne W.M.. 2018. Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep. 19:57–72. 10.15252/embr.201744815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne, W.M., Zhu L., Balogi Z., Stefan C., Pleiss J.A., and Emr S.D.. 2015. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol. 210:541–551. 10.1083/jcb.201503088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, C.A., Choudhary V., Wolyniak M.J., Scarcelli J.J., Schneiter R., and Cole C.N.. 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123:141–151. 10.1242/jcs.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, L.E., Dutta K., Sparks S., Temel D.B., Kamal A., Tetenbaum-Novatt J., Rout M.P., and Cowburn D.. 2015. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 4:e10027. 10.7554/eLife.10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Iovine, M.K., Watkins J.L., and Wente S.R.. 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131:1699–1713. 10.1083/jcb.131.6.1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jeong, H., Park J., Kim H.-I., Lee M., Ko Y.-J., Lee S., Jun Y., and Lee C.. 2017. Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure. Proc. Natl. Acad. Sci. USA. 114:E4539–E4548. 10.1073/pnas.1701030114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, G.A., Goodman J.S., Schick J.G., Chetlapalli K., Jorgens D.M., McDonald K.L., and Ünal E.. 2019. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. eLife. 8:e47156. 10.7554/eLife.47156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein, S.D., Eder S., Oh C.-S., Martin C.E., Gable K., Bacikova D., and Dunn T.. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:109–125. 10.1128/MCB.21.1.109-125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann, B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick, R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.-L., Millen J., Goldfarb D.S., and Thumm M.. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 19:4492–4505. 10.1091/mbc.e08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam, E., and Goldfarb D.S.. 2004. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 117:4959–4968. 10.1242/jcs.01372 [DOI] [PubMed] [Google Scholar]
- Kvam, E., and Goldfarb D.S.. 2006. Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site. Biochem. Soc. Trans. 34:340–342. 10.1042/BST0340340 [DOI] [PubMed] [Google Scholar]
- Laudermilch, E., Tsai P.-L., Graham M., Turner E., Zhao C., and Schlieker C.. 2016. Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol. Biol. Cell. 27:3964–3971. 10.1091/mbc.E16-07-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell, E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., and Kutay U.. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Lee, C.-W., Wilfling F., Ronchi P., Allegretti M., Mosalaganti S., Jentsch S., Beck M., and Pfander B.. 2020. Selective autophagy degrades nuclear pore complexes. Nat. Cell Biol. 22:159–166. 10.1038/s41556-019-0459-2 [DOI] [PubMed] [Google Scholar]
- Levine, T.P., and Munro S.. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 12:1633–1644. 10.1091/mbc.12.6.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.L., Ospovat O., and Wente S.R.. 2017. Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs. RNA. 23:365–377. 10.1261/rna.057612.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.L., Timney B.L., Rout M.P., and Wente S.R.. 2015. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J. Cell Biol. 208:729–744. 10.1083/jcb.201412024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, C.P., Makhnevych T., Marelli M., Aitchison J.D., and Wozniak R.W.. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267–278. 10.1083/jcb.200203079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio, T., Stanton L.H., Lin C.-C., Goldfarb D.S., Weis K., and Wozniak R.W.. 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185:459–473. 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manik, M.K., Yang H., Tong J., and Im Y.J.. 2017. Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction. Structure. 25:617–629.e3. 10.1016/j.str.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Michaillat, Lydie, and Mayer Andreas. 2013. Identification of Genes Affecting Vacuole Membrane Fragmentation in Saccharomyces cerevisiae. PLoS One. 8. 10.1371/journal.pone.0054160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldavski, O., Amen T., Levin-Zaidman S., Eisenstein M., Rogachev I., Brandis A., Kaganovich D., and Schuldiner M.. 2015. Lipid Droplets Are Essential for Efficient Clearance of Cytosolic Inclusion Bodies. Dev. Cell. 33:603–610. 10.1016/j.devcel.2015.04.015 [DOI] [PubMed] [Google Scholar]
- Niepel, M., Molloy K.R., Williams R., Farr J.C., Meinema A.C., Vecchietti N., Cristea I.M., Chait B.T., Rout M.P., and Strambio-De-Castillia C.. 2013. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell. 24:3920–3938. 10.1091/mbc.e13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño, C.A., Guet D., Gay A., Brutus S., Jourquin F., Mendiratta S., Salamero J., Géli V., and Dargemont C.. 2016. Posttranslational marks control architectural and functional plasticity of the nuclear pore complex basket. J. Cell Biol. 212:167–180. 10.1083/jcb.201506130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers, P., Cromley D., Padamsee M., Billheimer J.T., and Sturley S.L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277:8877–8881. 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- Ohsaki, Y., Kawai T., Yoshikawa Y., Cheng J., Jokitalo E., and Fujimoto T.. 2016. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212:29–38. 10.1083/jcb.201507122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko, E., Tang J.H., Andersen K.R., Knockenhauer K.E., Vallotton P., Derrer C.P., Kralt A., Mugler C.F., Chan L.Y., Schwartz T.U., and Weis K.. 2017. Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell. 171:904–917.e19. 10.1016/j.cell.2017.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., Bui K.H., Schorb M., Hossain M.J., Politi A.Z., Koch B., Eltsov M., Beck M., and Ellenberg J.. 2016. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife. 5:e19071. 10.7554/eLife.19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas, S.S., Liang C.-C., Kim S., Rivera C.O., and Dauer W.T.. 2018. TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet. 27:407–420. 10.1093/hmg/ddx405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschnigg, J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., and Kohlwein S.D.. 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284:30981–30993. 10.1074/jbc.M109.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popken, P., Ghavami A., Onck P.R., Poolman B., and Veenhoff L.M.. 2015. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell. 26:1386–1394. 10.1091/mbc.E14-07-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, R.A., Murphy R.F., and Jones E.W.. 1989. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc. Natl. Acad. Sci. USA. 86:7027–7031. 10.1073/pnas.86.18.7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampello, A.J., Laudermilch E., Vishnoi N., Prophet S.M., Shao L., Zhao C., Lusk C.P., and Schlieker C.. 2020. Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J. Cell Biol. 219:e201910185. 10.1083/jcb.201910185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel, I.L., Crane M.M., Thaller D.J., Mishra A., Jansen D.P., Janssens G., Popken P., Akşit A., Kaeberlein M., van der Giessen E., et al. 2019. Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife. 8:e48186. 10.7554/eLife.48186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., and Goldfarb D.S.. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141. 10.1091/mbc.e02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.S., Klionsky D.J., Banta L.M., and Emr S.D.. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948. 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., McCaffery J.M., and Wente S.R.. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160:1041–1053. 10.1083/jcb.200209116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., Zhou Y., and Wente S.R.. 2007. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 18:886–898. 10.1091/mbc.e06-06-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner, M., and Bohnert M.. 2017. A different kind of love - lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10 Pt B, 10 Pt B):1188–1196. 10.1016/j.bbalip.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Strawn, L.A., Shen T., Shulga N., Goldfarb D.S., and Wente S.R.. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197–206. 10.1038/ncb1097 [DOI] [PubMed] [Google Scholar]
- Teixeira, V., Johnsen L., Martínez-Montañés F., Grippa A., Buxó L., Idrissi F.-Z., Ejsing C.S., and Carvalho P.. 2018. Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217:127–138. 10.1083/jcb.201704115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, L.J., and Wente S.R.. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178:1121–1132. 10.1083/jcb.200704174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller, D.J., and Patrick Lusk C.. 2018. Fantastic nuclear envelope herniations and where to find them. Biochem. Soc. Trans. 46:877–889. 10.1042/BST20170442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller, D.J., Allegretti M., Borah S., Ronchi P., Beck M., and Lusk C.P.. 2019. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife. 8:e45284. 10.7554/eLife.45284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney, B.L., Raveh B., Mironska R., Trivedi J.M., Kim S.J., Russel D., Wente S.R., Sali A., and Rout M.P.. 2016. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215:57–76. 10.1083/jcb.201601004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka, Y., Kotani T., Kirisako H., Oikawa Y., Kimura Y., Hirano H., Ohsumi Y., and Nakatogawa H.. 2020. TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J. Cell Biol. 219:e201910063. 10.1083/jcb.201910063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama, B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R. III, and Hetzer M.W.. 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154:971–982. 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel, M.J.W., Nguyen K.C.Q., Hall D.H., Dauer W.T., and Rose L.S.. 2015. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell. 26:1752–1763. 10.1091/mbc.E14-07-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijlbergen, K.F., Menendez-Benito V., van Welsem T., van Deventer S.J., Lindstrom D.L., Ovaa H., Neefjes J., Gottschling D.E., and van Leeuwen F.. 2010. Recombination-induced tag exchange to track old and new proteins. Proc. Natl. Acad. Sci. USA. 107:64–68. 10.1073/pnas.0911164107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I.W., and Hetzer M.. 2003. RanGTP mediates nuclear pore complex assembly. Nature. 424:689–694. 10.1038/nature01898 [DOI] [PubMed] [Google Scholar]
- Webster, B.M., Colombi P., Jäger J., and Lusk C.P.. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 159:388–401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, B.M., Thaller D.J., Jäger J., Ochmann S.E., Borah S., and Lusk C.P.. 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35:2447–2467. 10.15252/embj.201694574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente, S.R., and Blobel G.. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123:275–284. 10.1083/jcb.123.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente, S.R., Rout M.P., and Blobel G.. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119:705–723. 10.1083/jcb.119.4.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Neuner A., Rüthnick D., Sachsenheimer T., Lüchtenborg C., Brügger B., and Schiebel E.. 2018. Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol. 217:877–894. 10.1083/jcb.201706024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains and genotypes.