Abstract
Tyrosine sulfation is an important post-translational modification found in higher eukaryotes. Here we report an engineered tyrosyl-tRNA synthetase/tRNA pair that co-translationally incorporates O-sulfotyrosine in response to UAG codons in E. coli and mammalian cells. This platform enables recombinant expression of eukaryotic proteins homogeneously sulfated at chosen sites, which was demonstrated by expressing human heparin cofactor II in mammalian cells in different states of sulfation.
Sulfation of tyrosine residues is a post-translational modification (PTM) that occurs exclusively in multicellular eukaryotes.1–4 Golgi-resident tyrosylprotein sulfotransferases (TPST1 and TPST2) use 3’-phosphoadenosine-5’-phosphosulfate (PAPS) as the sulfate donor to install this PTM (Supplementary Figure 1). Consequently, only secreted and membrane-associated proteins, which are processed through the trans-Golgi network, are sulfated.1–4 Tyrosine sulfation is believed to be irreversible and it facilitates numerous protein-protein and protein-ligand interactions important in diverse physiological processes such as immunity, hormone function, blood-coagulation, and host-pathogen interaction.1–7
It is estimated that ~1% of all tyrosine residues in the eukaryotic proteome are sulfated, but the physiological roles for the most remain poorly understood.1–3 A major factor contributing to this knowledge gap is the difficulty of expressing target proteins in a homogeneously sulfated state. Synthetic approaches have been developed to access sulfated peptides and small proteins but cannot be used to generate most full-length proteins.8,9 Recombinant expression in common eukaryotic hosts often leads to incomplete sulfation of native sites.7,10 Additionally, many proteins are sulfated at multiple tyrosine residues, and the difficulty of only modifying a chosen subset makes it challenging to evaluate the roles of individual sulfations. The ability to site-specifically incorporate O-sulfotyrosine (sTyr) into proteins using genetic code expansion technology can overcome these challenges (Figure 1a).11–13 Indeed, the M. jannaschii tyrosyl-tRNA synthetase (MjTyrRS)/tRNA pair has been engineered to site-specifically incorporate sTyr into proteins expressed in E. coli, which has been useful for investigating the roles of tyrosine sulfation.7,14–16 However, this pair cannot be used for non-canonical amino acid (ncAA) mutagenesis in eukaryotes due to cross-reactivity.17 This significantly limits its utility, given that tyrosine sulfation is exclusively restricted to eukaryotic proteins (secreted and membrane-associated proteins) that are frequently incompatible with recombinant expression in E. coli. Furthermore, the ability to express eukaryotic proteins in its native environment is essential for probing how their sulfation affects the cellular pathways it participates in. Genetically encoding sTyr in eukaryotic cells would overcome these limitations.
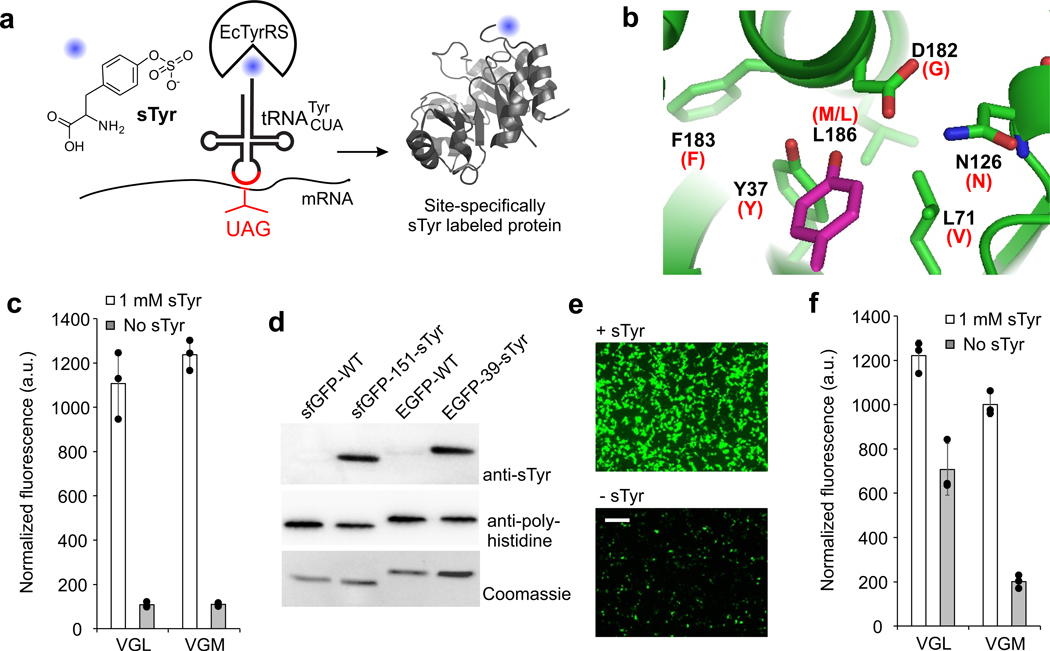
Figure 1. Genetically encoding sTyr.
a, A sulfotyrosine residue can be co-translationally incorporated into proteins in response to the UAG nonsense codon using an engineered EcTyrRS/tRNA pair in both ATMY E. coli and mammalian cells. b, The active site of EcTyrRS showing the bound substrate in magenta, and highlighting residues that were randomized. Mutations found in the sTyr selective variants are noted in parenthesis in red. c, Two EcTyrRS mutants facilitate sfGFP-151-TAG reporter expression in ATMY4 E. coli in the presence of sTyr (fluorescence in resuspended cells). d, Purified wild-type and sTyr-incorporated sfGFP (ATMY4-expressed) and EGFP (HEK293T-expressed) reporter proteins analyzed by anti-sTyr and anti-polyhistidine tag Western blot, as well as Coomassie staining following SDS-PAGE (full gels in Supplementary Figure 11). e, Expression of EGFP-39-TAG reporter in HEK293T cells using VGM-EcTyrRS/tRNA in the presence and absence of sTyr (fluorescence microscopy image, scale bar: 200 µm). f, EGFP-39-TAG expression in HEK293T cell using VGL- and VGM-EcTyrRS (fluorescence in clarified cell-free extract). Data in b and f shown as mean ± s.d. (n = 3 independent experiments). Experiments in c-f were replicated at least 3 times with similar results.
The E. coli derived tyrosyl-tRNA synthetase (EcTyrRS)/tRNA pair represents a promising platform to this end.17,18 However, the repertoire of ncAAs genetically encoded using this platform has been significantly limited relative to its M. jannaschii derived counterpart (Supplementary Figure 2).17,19 While the substrate specificity of MjTyrRS can be engineered using a facile E. coli based directed evolution system, the engineering of EcTyrRS relies on a cumbersome yeast-based system, which has experienced limited success.12,17,19,20 Recently, we have established a novel approach to facilitate the directed evolution of E. coli derived aminoacyl-tRNA synthetase (aaRS)/tRNA pairs in E. coli (Supplementary Figure 3).12,17,20 First, an endogenous aaRS/tRNA pair of E. coli is functionally substituted by an orthogonal counterpart from archaea/eukaryote. Next, the liberated endogenous pair is reintroduced in the resulting ‘altered translational machinery’ (ATM) E. coli strain as an orthogonal nonsense suppressor and engineered. We have used this strategy to create ATMY strains of E. coli, where the endogenous EcTyrRS/tRNA pair is substituted by an archaeal TyrRS/tRNA pair (Supplementary Figure 3b),17 and demonstrated the feasibility of engineering the EcTyrRS/tRNA pair in ATMY E. coli strains to genetically encode new ncAAs. This platform provides an exciting opportunity to genetically encode sTyr in eukaryotes.
We started by constructing a library of EcTyrRS mutants encoded in the pBK vector (pBK-EcYRS1) by randomizing six active site residues (Y37 to FLIMVSTAYHCG, L71 to NBT, N126 to NSPTACGDH, D182 to NST, F183 to NNK, L186 to NNK) surrounding the phenolic hydroxyl of the bound tyrosine substrate (Figure 1b). The pBK-EcYRS1 library was subjected to a double-sieve selection system in ATMY3 E. coli.17 Just after a single round of positive and negative selection each, the library demonstrated sTyr-dependent survival in the presence of chloramphenicol, indicating the enrichment of sTyr-selective EcTyrRS mutants (Supplementary Figure 4a). Several individual clones from this selected pool of mutants also had the same phenotype (Supplementary Figure 4b). DNA sequencing of eight such clones revealed the presence of two distinct but highly convergent clones, where Y37, N126, and F183 are conserved, L71 and D182 are mutated to V and G, respectively, and L186 is either conserved (VGL) or is mutated to M (VGM)(Figure 1b). The enlarged active site of these mutants were consistent with the need to accommodate the additional sulfate group of sTyr.
To evaluate the sulfotyrosine incorporation efficiency of these two EcTyrRS mutants (VGL and VGM), we individually co-transformed them with a pEvolT5-sfGFP-151-TAG reporter plasmid in ATMY4 E. coli strain (encodes two genomic copies of tRNAEcTyrCUA).17 These cells expressed sfGFP only in the presence of sTyr upon induction with IPTG (Figure 1c). Purification of this reporter protein using a C-terminal polyhistidine tag (8–10 mg/L) followed by ESI-MS analysis showed a mass consistent with the incorporation of sTyr (Supplementary Figure 6a). Western-blot analysis using an anti-sTyr antibody further corroborated the presence of sTyr in this protein (Figure 1d, Supplementary Figure 11). These observations confirm that we have generated an engineered EcTyrRS/tRNA pair that selectively incorporates sTyr in response to UAG.
Next, we explored if these mutant EcTyrRS/tRNA pairs can be used in mammalian cells for co-translational sTyr incorporation. We created a mammalian expression plasmid pB1U-Sulfo-16xtYR-TAG that expresses the VGL or the VGM EcTyrRS as well as 16 copies of the tRNAEcTyrCUA. Co-transfection of these plasmids with pAcBac1-EGFP-39-TAG led to robust expression of EGFP in the presence of sTyr, while significantly reduced reporter expression was observed in its absence (Figure 1e-1f, Supplementary Figure 5). The VGM mutant exhibited lower levels of UAG suppression in the absence of sTyr (Figure 1e-1f, Supplementary Figure 5). The reporter protein expressed in the presence of sTyr was isolated from HEK293T cells (100–120 μg from ~107 cells) using a C-terminal polyhistidine tag and analyzed by ESI-MS, which showed a mass consistent with sTyr incorporation (Supplementary Figure 6b). We also found that in the absence of sTyr, tyrosine is incorporated at the UAG codon (Supplementary Figure 6c). Western-blot using an anti-sTyr antibody further confirmed the presence of sTyr in EGFP-39-sTyr, but not in wild-type EGFP (Figure 1d) that was expressed and purified in a similar manner.
Next, we used human heparin cofactor II (HCII) as a model system to demonstrate that our platform enables facile expression of eukaryotic proteins homogeneously sulfated at native sites. HCII, a large secreted glycoprotein, is a serine protease inhibitor (serpin) that irreversibly inhibits thrombin, a key player in executing blood coagulation.21,22 This anticoagulant activity of HCII is triggered by glycosaminoglycans (GAGs) such as heparin.21,22 In the absence of GAGs, the acidic N-terminal domain (AND) of HCII binds its glycosoaminoglycan binding domain (GBD), resulting in an auto-inhibited state (Figure 2a). GAGs activate HCII by binding its GBD and displacing the AND, which then recruits thrombin by binding its exosite 1 (Figure 2a). The AND of HCII, which can bind both thrombin exosite I and GBD, is sulfated at two distinct sites (Tyr60 and Tyr73) whose roles in HCII activity is poorly understood.23 The absence of ER-Golgi processing precludes bacterial expression of HCII in its native glycosylated state, while overexpression in eukaryotic hosts can result in incomplete sulfation. We introduced UAG codons at 60 and 73 positions of full-length human HCII and overexpressed it in HEK293T cells in the presence of our sTyr incorporation system. Full-length HCII was successfully isolated from the culture medium using a C-terminal polyhistidine tag (Supplementary Figure 7). Whole-protein ESI-MS of this large protein was challenging, but we confirmed the presence of sTyr at both sites through protease digestion followed by LC-MS analysis (Supplementary Figure 8, 9). Glycosylase (PNGase) treatment significantly reduced the molecular weight of the protein (Supplementary Figure 10), suggesting the presence of robust N-linked glycosylation. These results confirm that our platform can be used to express endogenous eukaryotic proteins precisely sulfated at multiple sites.
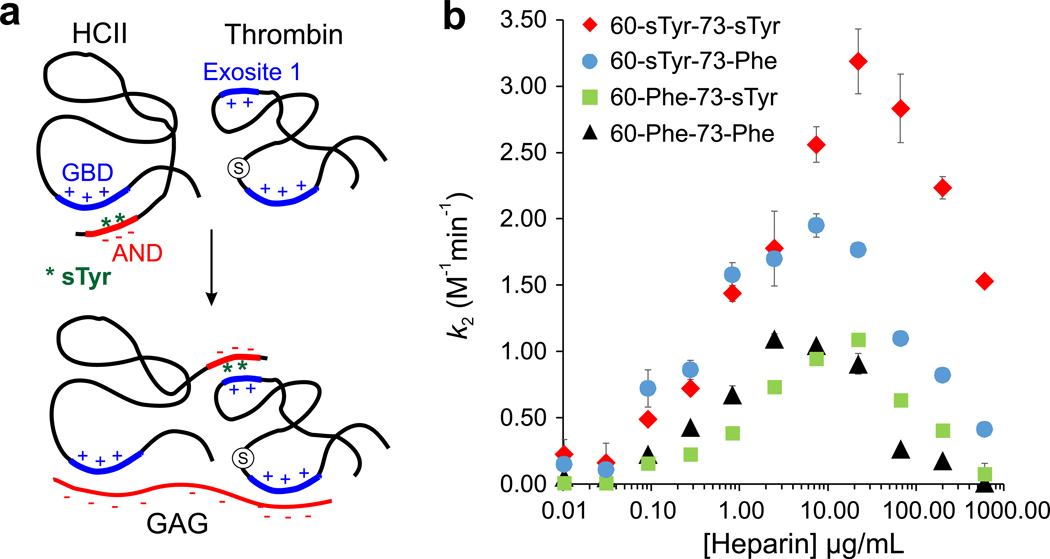
Figure 2. Expression and biochemical analysis of precisely sulfated HCII.
a, The model for GAG-activated thrombin inhibition of HCII, which is sulfated at Tyr60 and Tyr73 (shown as green stars). b, Second-order rate constant of thrombin inhibition by different HCII mutants at various heparin concentrations. Data points in b are shown as mean ± s.d. (n = 3 biologically independent samples).
In addition to 60-sTyr-73-sTyr, we also expressed and purified HCII mutants 60-Phe-73-Phe (to prevent sulfation), 60-sTyr-73-Phe, and 60-Phe-73-sTyr (Supplementary Figure 7) and evaluated their thrombin inhibition activities using an established biochemical assay.24,25 For each HCII mutant, second-order rate constants (k2) of thrombin inhibition were measured at different heparin concentrations to find the optimal [heparin], at which maximal inhibition rate is observed (Figure 2b). 60-sTyr-73-sTyr exhibited a maximal rate constant of 3×108 M−1min−1 at ~20 µg/mL heparin, which is in close agreement with previous reports.25 Interestingly, the absence of sTyr at site 73 (60-sTyr-73-Phe) led to a slightly lower maximal k2 but a substantially reduced (~3 fold) optimal [heparin], whereas the 60-Phe-73-sTyr mutant (no sTyr at site 60) had an unchanged optimal [heparin] but a significantly lower maximal k2 (Figure 2b). The 60-Phe-73-Phe mutant showed both a low maximal k2, and a reduced optimal [heparin]. The preliminary biochemical evaluation of precisely sulfated HCII mutants suggests important – yet distinct – roles the two sulfation PTMs play in fine-tuning its GAG-triggered thrombin inhibition activity: while the 73-sTyr appears to contribute more to AND-GBD association, the 60-sTyr might be more important for thrombin recruitment.
In summary, we have developed a platform for site-specific incorporation of sTyr into proteins expressed in eukaryotic cells with high fidelity and efficiency, which would be a valuable tool for investigating the consequences of tyrosine sulfations found in the eukaryotic proteome. This platform can also be used to express therapeutically relevant proteins homogeneously modified with functionally important sulfations.7,10 Additionally, the ability to incorporate sTyr into virtually any site of any protein in eukaryotic cells offers intriguing opportunities for novel synthetic biology applications.
Online methods
General Biological Reagents, Strains, and Protocols
E. coli strain DH10B (Life Technologies) was used for plasmid propagation and cloning. E. coli strains were cultured on LB-agar plates with appropriate antibiotic concentrations as follows: 95 μg/mL spectinomycin, 50 μg/mL chloramphenicol, 30 μg/mL kanamycin. Phusion high fidelity DNA polymerase (Thermo-Fisher) was used for PCR amplifications and restriction enzymes were obtained from New England Biolabs. DNA oligonucleotides were purchased from Integrated DNA Technologies (see Supplementary Table 1 for the list of oligonucleotides), while Sanger sequencing was performed by Eton Bio. Engineered E. coli strain ATMY3 (contains one genomic copy of tRNAEcTyrCUA; no genomic EcTyrRS) was used as the selection host for the directed evolution of EcTyrRS.17 Engineered E. coli strain ATMY4 (contains two genomic copies of tRNAEcTyrCUA; no genomic EcTyrRS) was used as the expression host for expressing recombinant proteins incorporating sTyr.
HEK293T cell line was purchased from ATCC (ATCC CRL-3216) and maintained in DMEM (high glucose) supplemented with 10% FBS and Penicillin/Streptomycin. Cells were grown in a 37 °C 100% humidity, 5% CO2.
Statistical methods.
For assessing sfGFP or EGFP reporter expression in ATMY4 E. coli or HEK293T cells, respectively, the mean of three independent experiments was reported, and error bars represent s.d. For the kinetic analysis of HCII inhibition of thrombin activity, average of three independent measurements were reported (error represent s.d.).
EcTyrRS Library Construction
In the EcYRS1 library (pBK-EcYRS1), six residues were randomized as follows: Y37-FLIMVSTAYHCG, L71-NBT, N126-NSPTACGDH, D182-NST, F183-NNK, L186-NNK. We used a previously reported library (pBK-EcYRS1a), which contains the desired Y37, D182, F183, and L186 randomizations, as the template to generate pBK-EcYRS1 by sequential overlap of extension PCR. Piece A was amplified with primers pBK seqT-F and EcYRS-L71-oR. Piece B was amplified with EcYRS-L71-NBT-F and EcYRS-N126-oR, and subsequently overlapped with piece A using terminal primers pBK seqT-F and EcYRS-N126oR to create piece AB. Lastly, piece C was amplified with EcYRS-N126x-F (x corresponds to nine different codons) and pBK MCS JIsqR for all desired N126 variants. Piece C variants were combined in equal distribution and were subsequently overlapped with piece AB to form the full-length aaRS PCR product.
After amplification, the aaRS PCR product was digested with NdeI/NcoI (NEB) and ligated by T4 DNA Ligase (NEB) into the pBK vector digested with the same restriction enzymes. The ligation mixture was ethanol precipitated with yeast-tRNA (Ambion) and transformed into electrocompetent DH10B cells. Greater than 108 transformants were obtained to ensure library coverage.
Directed evolution of EcTyrRS-SulfoY variant in ATMY3
Positive selection 1:
The pBK-EcYRS1 library was transformed into ATMY3 containing the positive selection plasmid pRepTrip2.3P-EcQtR-2x. The pRep plasmid expresses a chloramphenicol acetyl transferase (CAT) reporter containing a Q98TAG, an ampicillin resistance gene containing a 3TAG, an arabinose inducible T7 RNA polyermase containing two TAG codons (site 8 and 14), a T7 promoted GFPuv, and two copies of the E. coli tRNAGln expressed from its endogenous promoter. Approximately 9×108 colony forming units were plated on LB + 0.5x spectinomycin, tetracyclin, and kanamycin + 0.02% arabinose + 30 μg/mL ampicillin + 30 or 50 μg/mL chloramphenicol in the presence of 1 mM sTyr for 18 h at 37 °C. After 18 h, colonies from plates were harvested with 15 mL LB, centrifuged and selected pBK plasmid pool (pBK-EcYRS1a-P1) was purified via miniprep followed by gel purification.
Negative selection:
The isolated plasmid was subsequently transformed into ATMY3 containing pNeg2–2xQtR (contains arabinose dependent barnase with 3TAG, 45TAG, and two copies of the E. coli tRNAGln). Approximately 108 cells were plated on LB-agar plates containing 0.5x spectinomycin, ampicillin, and kanamycin + 0.02% arabinose in the absence of sTyr for 12 h at 37 °C. After 12 h, colonies from plates were harvested with 15 mL LB, centrifuged and the pBK library subjected to one positive selection and one negative selection (pBK-EcYRS1a-P1N1) was purified via miniprep.
Positive selection 2:
pBK-EcYRS1a-P2N1 was subjected to a second round of positive selection (106 cfu plated). 96 single colonies from the second round of positive selection plates containing 1 mM sTyr were picked into 500 μL LB supplemented with spectinomycin, tetracyclin, kanamycin in a 96 deep-well plate and grown to confluence overnight. These overnight cultures were diluted 100 fold and 3 μL were individually spot plated on LB-Agar plates containing spectinomycin, tetracyclin, kanamycin + 0.02% arabinose and 30 or 50 μg/mL chloramphenicol in the presence or absence of 1 mM sTyr. Eight pBK variants showing the most sTyr-dependent survival were picked for further characterization.
Characterization of tRNA/aaRS activity in E. coli using sfGFP reporter
ATMY4 containing pEvolT5-sfGFP151TAG was transformed with pBK-EcTyrRS variants. Overnight starter cultures were diluted 100 fold in 10 mL LB containing required antibiotics and grown at 37 °C while shaking at 250 rpm in 50 mL flasks. Upon reaching 0.55 OD600, 1 mM final IPTG was added to induce protein expression. 1 mL aliquots of induced cultures were placed in 15 mL culture tubes with or without 1 mM sulfotyrosine and grown for 18–20 h at 30 °C. Afterwards, cells were pelleted, resuspended in PBS, and diluted 10 fold. Dilutions were transferred to a 96-well clear bottom plate. Expression of full-length sfGFP was measured using the associated characteristic fluorescence by a SpectraMAX M5 (Molecular Devices) multimode plate reader (Ex. 488 nm; Em. 534 nm; 515 cutoff) and normalized with respect to OD600.
Purification of sfGFP-TAG from bacterial expression
Protein expression was performed in 10 mL culture as described above (sfGFP151-TAG reporter assay). Afterwards, the cells were pelleted at 5000 x g, resuspended in lysis buffer [B-PER Bacterial Protein Extraction Reagent (Thermo Scientific), 1x Halt Protease Inhibitor Cocktail (Thermo Scientific), 0.01% Pierce Universal Nuclease (Thermo Scientific), and incubated for 10 min on ice. After incubation, the crude lysate was clarified at 22,000 x g. The full-length sfGFP containing a C-terminal 6x HisTag was purified using HisPur Ni-NTA resin (Thermo Scientific) according to the manufacturers protocol. SDS-PAGE and Bradford analysis were used to assess protein purity, while the molecular weight was confirmed by ESI-MS (Agilent Technologies 1260 Infinity ESI-TOF).
Site-specific incorporation of sTyr into protein expressed in mammalian cells
The pB1U-Sulfo-16xtYR-TAG plasmids were constructed from previously reported pAcBac1 plasmids (maps provided in the supporting information section).26 The VGL and VGM EcTyrRS mutants were amplified using primers EcYRS-NheI-F and EcYRS-NheI-R (Supplementary Table 1) and cloned into NheI/XhoI sites.
HEK293T cells were maintained as described above. pB1U-SulfoA1–16xtYR-TAG (VGL) or pB1U-SulfoB7–16xtYR-TAG (VGM) contain 16 copies of alternating U6/H1 promoted E. coli tRNATyrCUA and UbiC promoted EcTyrRS mutants. pAcBac1-EGFP-39TAG was used as a reporter plasmid. 0.7×106 cells per well were seeded one day prior to transfection in a 12 well plate. At 70% confluence, the transfection mixture (500 ng each of suppressor and reporter plasmid, 18 μL DMEM, 3.5 μL Sigma PEI (1 mg/mL), 10 min incubation prior to addition) was added to each well and gently mixed. A final concentration of 2 mM sTyr was added to the wells at the time of transfection. After 48 h, cells were harvested by centrifugation at 5000 x g and residual media was removed. 50 μL lysis buffer (10 mL CellLytic M, 1x Halt Protease inhibitor, 0.01% Pierce universal nuclease) was added per well and incubated for 10 min. After incubation, cells were clarified by centrifugation and lysate was analyzed for fluorescence in the SpectraMAX M5 (Molecular Devices) under the same conditions as sfGFP.
For purification and further charectrization, EGFP-39-sTyr was expressed in 10 cm dishes (8.5×106 seeded 24 h prior to transfection). 5 μg suppressor plasmid and 5 μg reporter plasmid were incubated with 180 μL DMEM (no FBS) and 40 μL PEImax (Polysciences; 1 mg/mL). 2 mM sTyr and 2 mM Sodium Butyrate was added at the time of transfection. After 48 hr, cells were harvested at 5000 x g. 600 μL lysis buffer (CellLytic M, 1x Halt protease inhibitor, 0.01% Pierce universal nuclease) was used to lyse the cells. After 10 min incubation, the lysate was clarified by centrifugation and the protein was purified using HisPur Ni-NTA resin (Thermo-Scientific). Purity and the molecular weight of the expressed protein was analyzed by SDS-PAGE and ESI-MS (Agilent Technologies 1260 Inifinity ESI-TOF).
Anti-His and anti-Sulfotyrosine western blot of GFP reporters
Western blot was used to confirm the presence of a polyhistidine tag (via anti-HisTag blot) and the presence of sulfotyrosine (via anti-sTyr blot) in reporter proteins expressed above. 500 ng each of purified wild-type or sTyr-incorporated mutant of sfGFP or EGFP reporter proteins were resolved by SDS-PAGE, and transferred to a PVDF membrane (Life Technologies) using a Trans-Blot Turbo Transfer Sytem 15 (BioRad) in Towbin Transfer Buffer (at 12V for 30min, twice). After complete transfer, membrane was blocked in 10 mL 5% milk in TBST (HisBlot) or 10 mL Pierce Superblocker (Fisher Scientific) at 4 °C overnight with constant agitation. Membranes were subsequently incubated in 1:3000 anti-HisTag mouse mAb (Invitrogen, MA1–21315, in 5% milk TBST) or 1:6000 anti-Sulfotyrosine mouse mAb (Millipore Sigma, Clone: Sulfo-1C-A2, in Pierce Superblocker) overnight. Next, the membrane was washed six times, 10 min per wash, using TBST at room temperature. Afterwards, 1:6000 dilution of chicken anti-mouse secondary antibody (Invitrogen, SA1–72021, in 5% milk TBST) was incubated for 2 h at room temperature. The membrane was washed and activated using SuperSignal West Dura Kit (Fisher Scientific). The activated blot was imaged on the ChemiDoc MP imaging system (BioRad).
Expression and purification of Heparin Cofactor II (HCII)
The pB3-SulfoRS-16xYtR-TAG-HCII plasmid was constructed from previously reported pAcBac3 plasmids (maps provided in the Supporting Information section).26 The VGL and VGM EcTyrRS mutants were amplified using primers EcYRS-NheI-F and EcYRS-NheI-R (Supplementary Table 1) and cloned into NheI/XhoI sites. The SfiI sites were used to clone HCII after the CAG promoter. HCII-SfiI-F and 10xHis-TGA-SfiI-R were used to amplify HCII from pCMV-SerpinD1 (Origene, SC120039). Mutations were introduced via overlap extension PCR (see Supplementary Table 1 for primers used).
HEK293T cells were maintained as described above. pB3-SulfoRS-16xYtR-TAG-HCII contains the following three components: 16 copies of alternatingly H1/U6 promoted E. coli tyrosine tRNACUA, a UbiC promoted EcTyrRS mutant, and HCII mutants under a CAG promoter. 10 cm dishes were seeded with 8.5 × 106 cells 24 h prior to transfection. Afterwards, DMEM +FBS media was aspirated and replaced with DMEM without FBS. A transfection mixture (10 μg pB3 plasmid, 180 μL DMEM, 50 μL PEI Max) was incubated for 10 min prior to addition. 2 mM sTyr and 2 mM sodium butyrate were added at the time of transfection. Since HCII is a secreted protein, the media was harvested on days 2 and 3 post transfection, stored at 4 °C for up to 2 days, and adherent HCII expressing cells were re-supplemented with DMEM (no FBS) + 2 mM sTyr + 2 mM sodium butyrate. Collected media containing overexpressed HCII (20 mL total per 10 cm plate) were pooled and subjected to purification.
HCII containing media was centrifuged at 5,000 x g at 4 °C for 30 min to remove any residual debris. The supernatant was concentrated with Amicon 30 kDa MWCO centrifugal filters to approximately 2 mL. For concentrated media harvested from five 10 cm dishes, 1 mL Ni-NTA (Thermo-Scientific) resin was used for protein purification. Bound protein was washed with 50 mL of wash buffer containing PBS + 45 mM imidazole. HCII was eluted with 10 mL elution buffer, concentrated down to 1 mL using a 30 kDa MWCO filter, and buffer exchanged into HNPN –PEG buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% NaN3). Protein yields and purity were analyzed by Bradford, SDS-PAGE, anti-His tag dot blot, and tryptic/elastase mass spectrometry.
Deglycosylation assay of HCII
PNGase F was purchased from Promega (V4831). 18 μL (10 μg, in 0.5 mM Tris-HCl, pH 7.8) purified recombinant HCII was incubated at 37 °C with or without 2 μL PNGase for 18 hrs. After incubation, mixtures were resolved by SDS-PAGE and imaged via ChemiDoc imaging.
Tryptic & Elastase mass spectrometry characterization of HCII
An in gel digestion was performed to prepare peptides for MS analysis. 1000–2000 ng HCII was resolved by SDS-PAGE. Gel was stained for 1 hr, and destained overnight. After destain, HCII bands were sliced and cut into approximately 1 mm2 pieces. Pieces were placed in microcentrifuge tubes containing 500 μL 100 mM ammonium bicarbonate. Gel bands were frozen at −80°C overnight in the 500 μL ammonium bicarbonate. Gel bands were thawed, supernatant was removed, and gel bands were washed 1–2x for 15 min with 500 μL 100 mM ammonium bicarbonate. After washes, supernatant was removed and 200 μL 10 mM TCEP was added to completely cover gel bands. Samples were placed in a 60 °C water bath for 30 min. Samples were quickly spun and TCEP was aspirated. 200 μL 55 mM iodoacetamide was added to cover the gel bands. Tubes were placed in the dark for 30 min at RT. Supernatant was removed and gel bands were washed 3x for 15 min in 500 μL 50:50 acetonitrile:100 mM ammonium bicarbonate. After washes, supernatant was removed and 50 μL acetonitrile was added to completely dehydrate the gel bands (turned opaque). Acetonitrile was removed and residual solvent was removed using a SpeedVac for 5 min.
Sequencing grade trypsin (V5111) and neutrophil elastase (V1891) was purchased from Promega. Sample was resuspended in either trypsin (for 60 site) or trypsin+elastase (for 73 site). For trypsin, 200 ng trypsin (20 μL, resuspended in 25 mM ammonium bicarbonate) was added to dehydrated gel slices. For trypsin+elastase, 300 ng (30 μL, 25 mM ammonium bicarbonate) trypsin was added, immediately followed by 35 μL elastase (30 ng, resuspended in double distilled water according to manufacturer protocol) + 50 μL 50 mM Tris-HCl. In both cases, enzymes were incubated with gel sample for 10 min before 200 μL 50 mM ammonium bicarbonate was added and placed at 37 °C incubator overnight. Next, the supernatant was transferred to a clean tube and 100 μL formic acid was added to the gel bands followed by a 15 min incubation at RT. The supernatant was aspirated and combined with the supernatant from the last step. Formic acid washes of the gel slices were repeated two more times. Next, 150 μL acetonitrile was added to cover the gel slices, incubated at RT for 15 min, and combined with all previous washes. Acetonitrile washes were repeated two more times until bands became opaque. Lastly, the peptide sample (~500 μL consisting of the overnight incubation supernatant, formic acid washes, and acetonitrile washes) was evaporated down to 10 μL using SpeedVac and stored at −80 °C until subjected to HPLC-MS analysis.
Digested peptides were analyzed by LC-MS using an LTQ Orbitrap XL mass spectrometer (Thermo Fisher) coupled to an EASY-nLC 1000 nanoLC (Thermo Fisher). 18 μL of sample was loaded onto 100 μm fused silica column with a 5 μm tip packed with 10 cm of Aqua C18 reverse-phase resin (Phenomenex) using the EASY-nLC 1000 autosampler. Peptides were eluted with a gradient 0–55% buffer B in buffer A (buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; buffer B; 20% water, 80% acetonitrile, 0.1% formic acid). The flow rate through the column was set to 400 nl/min and the spray voltage was set to 3.5 kV. One full MS scan (400–1800 MW) was followed by seven data dependent scans. For the data dependent scans, a mass list was used to target the predicted peptides with sTyr at residues 60 and 73. In the absence of a targeted peptide, data dependent scans were performed on the nth most intense ions in the MS1. MS1 spectra and total ion chromatograms were manually analyzed for peptide identification and presence of sulfation at each residue.
HCII-Thrombin activity assay
To calculate the second order rate constant of thrombin inhibition by HCII, thrombin was incubated with excess HCII under pseudo-first order conditions in the presence of different heparin concentrations (details below). The reaction was quenched after 1 minute and the residual thrombin activity (kinhibited) was measured using a chromogenic substrate. The pseudo-first order rate constant (k1) was calculated from this using the equation k1 = –ln(kinhib/kuninhib)/t, where kuninhib is the activity of thrombin in the absence of HCII inhibition under identical treatment. The second order inhibition rate constant (k2) was calculated from k1using the equation k2 = k1/[HCII] with units of M−1min−1. The second order rate constant at each heparin concentration was plotted against the corresponding heparin concentration.
Concentrations of different HCII protein were measured by Bradford and normalized by anti-His dot-blot assay (blot intensities quantified via ChemiDoc imaging). Clear plastic 96 well plates were coated with 2 mg/mL ovalbumin (Fisher) for 1 hr at 37 °C. Ovalbumin was removed by tapping the plate on a paper towel. A master mix of 2 mg/mL ovalbumin, 0–2 mg/mL heparin (Fisher), 0.6 nM α-thrombin (Fisher) in HNPN (20 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% NaN3) were incubated in the treated 96 wells for 1 min with 10 nM HCII. After 1 min, 10 μL of a solution of 1 mg/mL polybrene was directly added to all wells using a multichannel pipet to quench the heparin-dependent inhibition of thrombin by HCII. The plates were spun down in a bucket centrifuge for 10 min at 3,500 rpm to precipitate the heparin/polybrene complex. 100 μL supernatant was removed and 50 μL 450 μM ChromozymeTH (Sigma) substrate was added to measure the amount of residual thrombin activity by monitoring the absorbance on the SpectroMax plate reader at 405 nm for 1 hr in triplicate.
Supplementary Material
Acknowledgements
We thank Prof. D. M. Monroe III (UNC) for helpful discussions on the HCII kinetic assay. This work was supported by National Institutes of Health (R01GM124319 and R01GM126220 to A.C. and 1R01GM118431 and 1R01GM117004 to E.W.).
Footnotes
Data availability.
Data associated with this work are available upon request.
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper.
References:
- 1.Moore KL Protein tyrosine sulfation: a critical posttranslation modification in plants and animals. Proceedings of the National Academy of Sciences of the United States of America 106, 14741–14742, doi: 10.1073/pnas.0908376106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seibert C. & Sakmar TP Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 90, 459–477, doi: 10.1002/bip.20821 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Stone MJ, Chuang S, Hou X, Shoham M. & Zhu JZ Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. New biotechnology 25, 299–317 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Yang YS et al. Tyrosine sulfation as a protein post-translational modification. Molecules (Basel, Switzerland) 20, 2138–2164, doi: 10.3390/molecules20022138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farzan M. et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Huang C. c. et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proceedings of the National Academy of Sciences of the United States of America 101, 2706–2711 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Hitomi J. & Liu CC Characterization of a Sulfated Anti-HIV Antibody Using an Expanded Genetic Code. Biochemistry (2018). [DOI] [PubMed] [Google Scholar]
- 8.Stone MJ & Payne RJ Homogeneous sulfopeptides and sulfoproteins: synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Accounts of chemical research 48, 2251–2261 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Thompson RE et al. Tyrosine sulfation modulates activity of tick-derived thrombin inhibitors. Nature chemistry 9, 909 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen J, Thomsen J. & Ezban M. Heterogeneity in the tyrosine sulfation Chinese hamster ovary cell produced recombinant FVIII. Biochemistry 30, 1533–1537 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Chin JW Expanding and reprogramming the genetic code. Nature 550, 53 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Italia JS et al. Expanding the genetic code of mammalian cells. Biochemical Society transactions 45, 555–562, doi: 10.1042/bst20160336 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Young DD & Schultz PG Playing with the molecules of life. ACS chemical biology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CC, Brustad E, Liu W. & Schultz PG Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. Journal of the American Chemical Society 129, 10648–10649 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu CC & Schultz PG Recombinant expression of selectively sulfated proteins in Escherichia coli. Nature biotechnology 24, 1436–1440, doi: 10.1038/nbt1254 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Watson EE et al. Mosquito-Derived Anophelin Sulfoproteins Are Potent Antithrombotics. ACS central science 4, 468–476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Italia JS, Latour C, Wrobel CJ & Chatterjee A. Resurrecting the bacterial tyrosyl-tRNA synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes. Cell chemical biology 25, 1304–1312. e1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin JW et al. An expanded eukaryotic genetic code. Science (New York, N.Y.) 301, 964–967, doi: 10.1126/science.1084772 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Dumas A, Lercher L, Spicer CD & Davis BG Designing logical codon reassignment–Expanding the chemistry in biology. Chemical Science 6, 50–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Italia JS et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nature chemical biology 13, 446–450, doi: 10.1038/nchembio.2312 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Tollefsen DM Heparin cofactor II. Advances in experimental medicine and biology 425, 35–44 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Tollefsen DM Heparin cofactor II modulates the response to vascular injury. Arteriosclerosis, thrombosis, and vascular biology 27, 454–460 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Hortin G, Tollefsen D. & Strauss AW Identification of two sites of sulfation of human heparin cofactor II. Journal of Biological Chemistry 261, 15827–15830 (1986). [PubMed] [Google Scholar]
- 24.Ciaccia AV, Monroe DM & Church FC Arginine 200 of Heparin Cofactor II Promotes Intramolecular Interactions of the Acidic Domain IMPLICATION FOR THROMBIN INHIBITION. Journal of Biological Chemistry 272, 14074–14079 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Mitchell JW & Church FC Aspartic acid residues 72 and 75 and tyrosine-sulfate 73 of heparin cofactor II promote intramolecular interactions during glycosaminoglycan binding and thrombin inhibition. Journal of Biological Chemistry 277, 19823–19830 (2002). [DOI] [PubMed] [Google Scholar]
Reference:
- 26.Zheng Y, Lewis TL Jr, Igo P, Polleux F. & Chatterjee A. Virus-enabled optimization and delivery of the genetic machinery for efficient unnatural amino acid mutagenesis in mammalian cells and tissues. ACS synthetic biology 6, 13–18 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.