Abstract
Simple Summary
Honey bees are vital to the agriculture of the world, but like all managed organisms, disease control has become challenging due to the overuse and misuse of antibiotics. Alternate solutions with potential to control disease include natural compounds and probiotic supplements. Probiotic supplements in honey bees have been praised by industry, but studies applying probiotics to honey bee larval disease are lacking and technically challenging. In this study we tested the effectiveness of a demonstrated probiotic (Parasacharribacter apium strain C6) to mitigate a damaging larval disease called European Foul Brood (EFB). Based on a controlled laboratory study and two separate trials, the probiotic had no effect on EFB disease. The control groups performed as expected, validating the very sensitive lab procedure used to artificially rear honey bee larvae. Surprisingly, the probiotic provided no survival benefit to larvae in the absence of disease, contradicting past results. We discuss the difficult technique of larval rearing in the laboratory with reference to an improved experimental design introducing disease agents and potential remedies. In summary, our findings indicate that the representation of honey bee health and disease in the laboratory setting requires repeatable validation with reference to rigorous control and natural colony context.
Abstract
European honey bees (Apis mellifera Linnaeus) are beneficial insects that provide essential pollination services for agriculture and ecosystems worldwide. Modern commercial beekeeping is plagued by a variety of pathogenic and environmental stressors often confounding attempts to understand colony loss. European foulbrood (EFB) is considered a larval-specific disease whose causative agent, Melissococcus plutonius, has received limited attention due to methodological challenges in the field and laboratory. Here, we improve the experimental and informational context of larval disease with the end goal of developing an EFB management strategy. We sequenced the bacterial microbiota associated with larval disease transmission, isolated a variety of M. plutonius strains, determined their virulence against larvae in vitro, and explored the potential for probiotic treatment of EFB disease. The larval microbiota was a low diversity environment similar to honey, while worker mouthparts and stored pollen contained significantly greater bacterial diversity. Virulence of M. plutonius against larvae varied markedly by strain and inoculant concentration. Our chosen probiotic, Parasaccharibacter apium strain C6, did not improve larval survival when introduced alone, or in combination with a virulent EFB strain. We discuss the importance of positive and negative controls for in vitro studies of the larval microbiome and disease.
Keywords: larval microbiome, honey bee microbiome, EFB disease, larvae, Melissococcus plutonius, in vitro larval rearing, Parasaccharibacter apium strain C6, probiotic
1. Introduction
Honey bees are valuable pollinators of agriculture and ecosystems worldwide [1,2]. Recent and sustained colony loss has necessitated a review of pollination ecology and its relation to agriculture [3]. Recent colony loss has been described as multifactorial, involving combinations of environmental stress and disease agents [4,5]. Often associated with colony decline, bacterial diseases of honey bee larvae have become more prevalent worldwide [6]. To confront this growing threat, the patterns and processes that cause or encourage honey bee larval disease are being explored in greater detail [7,8].
The interplay between disease states and the native microbiome is a growing field of study [9,10,11,12,13,14,15]. Protection from pathogens is considered the primary function of the eukaryotic microbiome, and changes to the native honey bee gut microbiota can range from mildly anti-commensal to pathogenic [9,10,15]. Many non-communicable and chronic disease states are associated with microbiome variation, highlighting the importance of microbiome integrity or taxonomic membership in disease susceptibility [10,11,12,13]. In an applied context, bacteria native to the honey bee are being developed as probiotics to provide resistance to pathogens [16]. Species of Lactobacillus and Bifidobacterium showed an inhibitory effect against larval pathogens [17,18]. Similarly, in Apis cerana (Eastern honey bee) Bacillus subtilis inhibits a larval pathogen in vitro and when fed to larvae [19]. Another native bacteria, Parasaccharibacter apium (strain C6), was also reported to benefit survival when introduced to larvae in vitro [20]. Collectively, these works suggest that the native microbiome can provide solutions to mitigate disease, but the demonstration of biological relevance and colony-level application remains experimentally challenging [16,21].
In the honey bee, disease states and transmission routes have evolved to exploit social context and distinct developmental and reproductive phenotypes [10,22,23]. Honey bee colonies contain a single reproductive queen, developing larvae, functionally sterile workers and stored food; honey and pollen. Queens produce the eggs, but parental care of developing larvae is performed by non-reproductive adult workers called nurse bees; young adults that consume stored pollen, convert it to lipid and protein rich jelly in their hypopharyngeal glands, and secrete this jelly via mouthparts to nourish the queen and developing larvae. Similar to mammalian parental care, these glandular secretions have co-evolved to control microbial growth and support commensal or putatively beneficial bacteria [20,24]. Time from egg to worker pupation is only 7 days, and the larval microbiota is specialized to capitalize on this narrow time window [24]. As larvae develop, they are exposed to a wide variety of commensal and pathogenic microbes vectored from the pollination environment, food stores, nurse worker glands or mouthparts [9,20,25]. Larvae defecate in their wax cells prior to pupation, concurrent with a marked decrease in bacterial load [26]. These same wax cells are cleaned by newly emerged adults that quickly transition into nurse workers and begin to feed developing larvae [27,28].
The two main bacterial diseases of larvae include American Foulbrood (AFB), caused by Paenibacillus larvae [4,5], and European Foulbrood (EFB), attributed to Melissococcus plutonius [29]. While AFB disease is overt, highly virulent, and caused by a singular bacterial species, the causative factor of EFB and EFB-like symptomology is less well known. With the recent increase and severity of EFB disease worldwide, the last few years has seen great progress elucidating the mechanisms of virulence [8,30,31,32]. Historically, EFB was considered an opportunistic disease, affecting stressed hives. EFB is anecdotally associated with particular crops or environmental conditions and, while economically important, often clears up on its own [33]. Although considered the primary cause of EFB disease, M. plutonius often goes undetected when EFB-like symptoms are present. Many other bacteria are detected in association with EFB symptoms, suggesting a disease state associated with opportunistic microbes typically found in the hive environment [8,33]. As a treatment for larval disease, antibiotics damage the native microbiome, select for antibiotic resistance [34], and are now tightly regulated or banned in some countries. Thus, an understanding of the native microbiome may facilitate the development of a long-term EFB management strategy.
A healthy larval microbiota can be dominated by Lactobacillus spp. (including L. kunkeei) and an Acetobacteraceae referred to as Alpha 2.2, Parasaccharibacter apium, more recently renamed Bombella apis [20,24,35,36]. Although Bombella apis is the scientifically valid nomenclature [36], we use P. apium in this manuscript to maintain continuity with past work [20]. Lactobacillus kunkeei and P. apium reflect a deep ecological and evolutionary history with bee species and the pollination environment in general [24,37] in that particular species and strains are intimately associated with honey bees [38]. Consistently, the growth of flower-derived Lactobacillus kunkeei is inhibited by royal jelly, but strains of L. kunkeei isolated from honey bee larvae are unaffected [24]. Parasaccharibacter apium isolated from honey bee larvae flourished when cultured with royal jelly, but its sister species Saccharibacter floricola, also isolated from flowers, is inhibited [24]. It is hypothesized that both of these species have co-evolved with the production of royal jelly, a substance produced only by honey bees [10]. This claim is supported by the prevalence and abundance of these bacterial species in royal jelly, nurse head (hypopharyngeal) glands that produce royal jelly, nurse worker foreguts (a social stomach) and queen guts; a reproductive phenotype fed exclusively royal jelly [10,20,39] (Table 1).
Table 1.
Relative abundance of Parasaccharibacter apium by niche and study.
| Study | Site | Tissue or Niche | Developmental Stage or Caste | Total Sequences | % P. apium |
|---|---|---|---|---|---|
| Relative abundance of P. apium from whole worker gut samples | |||||
| Martinson et al. 2012 [40] | AZ | whole guts | 9-day-old nurses (N = 3) | 78,595 | 0 |
| Martinson et al. 2012 [40] | AZ | whole guts | 30-day-old forager (N = 1) | 17,910 | 0 |
| Sabree et al. 2012 [41] | MA | whole guts | 12 days old | 106,344 | 1 |
| Moran et al. 2012 [42] | AZ, MD | whole guts | in hive, outer frames | 329,550 | 1 |
| Corby-Harris et al. 2014a [43] | AZ | whole guts | pollen foragers | 354,505 | 6 |
| Anderson et al. 2016 [28] | AZ | whole guts | 3-day-old bees A | 90,893 | 0 |
| Anderson et al. 2016 [28] | AZ | whole guts | 7-day-old bees A | 166,113 | 0 |
| Relative abundance of P. apium from honey-rich food storage environments | |||||
| Anderson et al. 2014 [44] | AZ | corbicular pollen | forager corbiculae | 85,043 | 6 |
| Anderson et al. 2014 [44] | AZ | stored pollen | hive environment | 116,593 | 3 |
| present study | AZ | ripening honey | hive environment | 784,635 | 4 |
| present study | AZ | stored pollen | hive environment | 314,705 | 6 |
| Relative abundance of P. apium from niches with >20% P. apium abundance | |||||
| Corby-Harris et al. 2014a [43] | AZ | crop (foregut) | pollen forager | 195,264 | 43 |
| Corby-Harris et al. 2014a [43] | AZ | crop (foregut) | nurse | 113,405 | 32 |
| Corby-Harris et al. 2014b [20] | AZ | nurse headglands | nurse | 127,157 | 34 |
| Corby-Harris et al. 2014b [20] | AZ | royal jelly | worker head glands | 105,820 | 40 |
| Kapheim et al. 2015 [45] | IL | dissected guts | queen (N = 4) | 78,355 | 38 |
| Tarpy et al. 2015 [46] | NC | dissected guts | queen B | 5000 | 24 |
| Anderson et al. 2018 [39] | AZ, CA | mouthparts | queen C | 1,305,332 | 89 |
| Anderson et al. 2018 [39] | AZ, CA | midgut | queen C | 1,572,752 | 86 |
| Anderson et al. 2018 [39] | AZ, CA | ileum | queen C | 1,419,416 | 38 |
| present study | AZ | mouthparts | nurse worker | 502,473 | 22 |
| present study | AZ | whole body | worker larvae 2nd instar | 330,556 | 25 |
A Only bees fed a natural diet are included. B Libraries were subsampled to 5000. C Corrected for rRNA copy number and community size.
Previous in vitro work suggests that distinct strains of P. apium may have varying effects on larval survival, and that P. apium strain C6 significantly increased larval survival [20]. In this manuscript, we characterize the niche and ecology of bacteria associated with disease transmission routes using 16S rRNA gene amplicon sequencing of larvae, nurse worker mouthparts, and stored food consumed by nurse workers. Next, we test the putative benefit of P. apium strain C6 on developing larvae in vitro by introducing it in a quantitative disease context. We infected honey bee larvae with a pathogenic strain of European Foul Brood (EFB) and determined an in vitro dose response (LD50) for larval survival. We then inoculated EFB infected larvae with the putatively beneficial bacterium; P. apium strain C6. We hypothesized that P. apium would rescue the larvae from EFB disease; that the survival rate of larvae exposed to both P. apium and EFB would increase compared to larvae exposed to only the EFB disease agent.
2. Materials and Methods
2.1. Characterization of Disease Transmission Routes
To characterize the microbiome associated with larval disease transmission, we examined sources of inoculum associated with the task of feeding larvae. To produce nutritional and antibiotic secretions, adult nurse workers consume stored pollen and honey, then feed larvae via their mouth parts. We examined 16S rRNA gene sequences from honey, stored pollen, nurse mouthparts and larvae (N = 5 libraries per sample type). We extracted DNA from the following substrates from five hives housed at the Carl Hayden Bee Research Center (CHBRC) in Tucson, AZ, USA: Stored pollen, honey, 2nd instar larvae (50 larvae pooled within hive), and nurse bee mouthparts (dissected from 3 nurses pooled within hive) using established techniques [9,20]. Honey samples were obtained by washing ripening, uncapped honey with 100 µL of dH20 in 50 cells per hive. These samples were combined into a 5 mL water/honey sample that was centrifuged at maximum speed for 10 min. The supernatant was removed and the resulting pellet was subjected to DNA extraction [9]. Prior to DNA extraction, all samples were placed into 2 mL bead-beating tubes containing 0.25 g of 0.1-mm silica beads and 1.8 mL of lysis buffer (20 mM Tris-HCl, 2 mM EDTA, 1.2% Triton X-100, pH 8.0). Samples were bead-beaten for 2 min in 30 s intervals, cooled on wet ice for 30 s between each cycle. Samples were then vortexed for 5 s, and the supernatant was removed into 1.5 mL Eppendorf tubes. We then extracted total genomic DNA using a Fermentas Gene Jet Genomic DNA Purification Kit following the protocol for gram-positive bacteria.
2.1.1. PCR and MiSeq
The V1–V2 region of the 16S rRNA gene was amplified using PCR primers 27F (AGRGTTTGATCMTGGCTCAG) and 338R (AGTGCTGCCTCCCGTAGGAGT). Amplification was performed using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 1 min, with a final elongation step at 72 °C for 5 min. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Samples were purified using calibrated Ampure XP beads. Purified PCR product was used to prepare DNA library following Illumina TruSeq DNA library preparation protocol. Sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on a MiSeq following the manufacturer’s guidelines.
2.1.2. MiSeq Sequence Analysis
Sequences were processed using MOTHUR v.1.35.1 [47]. Forward and reverse reads were joined using the make.contigs command. After the reads were joined the first and last five bases pairs were removed using the SED command in unix. Sequences were then screened, using the screen.seqs command, to remove any sequences containing ambiguous bases. Unique sequences were generated using the unique.seqs command. A count file containing group information was generated using the count.seqs command. Sequences were aligned to Silva SSUREF database v102 [48] using the align.seqs command. Sequences not overlapping in the same region and columns not containing data were removed using the filter.seqs command. Sequences were pre-clustered using the pre.culster command. Chimeras were removed using UCHIME [49] and any sequences that were not of known bacterial origin were removed using the remove.seqs command. All remaining sequences were classified using the classify.seqs command. All sequences with only one or two (single/doubletons) associated reads were removed using the AWK command in unix. A distance matrix was constructed for the aligned sequences using the dist.seqs command. Sequences were classified with the RDP Naive Bayesian Classifier [50] using a manually constructed training set containing sequences sourced from the greengenes 16S rRNA database (version gg_13_5_99 accessed May 2013 [51], the RDP version 9 training set, and all full length honeybee-associated gut microbiota listed in NCBI. Operational taxonomic units (OTUs) were generated using the cluster command. Representative sequences for each OTU were generated using the get.oturep command. To further confirm taxonomy, resulting representative sequences were subject to a BLAST query using the NCBI nucleotide database.
To better define the niche of P. apium, we compared the average abundance of P. apium in honey, stored pollen, larvae, and nurse mouthparts to the abundances of P. apium observed previously for other related honey bee niches including royal jelly, the glands that produce royal jelly (hypopharygeal glands), gut compartments of queens and worker castes; cell cleaners, nurses and foragers [20,28,40,41,42,43,44,45,46].
2.1.3. Sequence Data Deposition
All sequence data were deposited in GenBank under Sequence Read Archive (SRA) accession PRJNA640829.
2.2. General Bacterial Culturing
To identify a virulent strain of EFB and acquire isolates associated with EFB disease, we cultured bacteria from larvae associated with EFB disease acquired from the Beltsville Bee Research Lab collection, and an outbreak of EFB diagnosed near Crestone, Colorado during the summer of 2018. Larvae were collected into sterile 2 mL tubes and shipped on ice to the Carl Hayden Bee Research Center in Tucson, AZ, USA. Saline (500 µL) was added to the tube and the larvae were homogenized with a cotton swab. This aqueous mixture was streaked onto plates containing KSBHI or Medium 1 from Arai et al. [52]. Plates were incubated for 7 days at 34 °C under anaerobic conditions using anaerobic jars, gaspacks, and anaerobic indicator strips. Resulting isolates were picked and banked, PCR amplified and Sanger sequenced at the 16S rRNA gene as described previously [9].
From the Carl Hayden Bee Research Center microbe collection, we selected Parasaccharibacter apium strain C6 as a putative rescue bacterium. P. apium strain C6 is a honey bee-specific strain of Acetobacteraceae considered beneficial to larval survival [20], and contains genes associated with bacterial competition and survival in the insect gut [20,53]. We cultured strain C6 according to Corby-Harris et al. [20].
2.3. Bacterial Inhibition Assay
Royal and worker jelly are highly antimicrobial, but bacteria co-evolved with the honey bee can flourish in royal jelly and the in vitro larval diet [20,24]. We tested the ability of our two experimental strains to survive in commercially available royal jelly. We plated both M. plutonius and P. apium with varying concentrations of the same royal jelly used in the rescue experiment, from full strength to highly dilute (diluted with sterile deionized water). Each bacterium was plated in their respective media type. We cultured M. plutonius strain AF5 in KSBHI, and P. apium strain C6 in SDA [20]. Filter paper discs were saturated with royal jelly of different dilutions (100%, 50% and 10%) and placed onto the inoculated plates. A disc was placed in each quarter of the plate. The assay was performed in triplicate resulting in 12 reps for each bacterium and dilution.
While P. apium strain C6 grows rapidly in both royal jelly and the larval diet [20], the effects of the larval diet on M. plutonius growth and survival are unknown. We verified the survival of M. plutonius (strain AF5) in the larval diet by culturing a time series on KSBHI media. We first cultured the physiological saline solution containing the M. plutonius in triplicate to verify initial inoculant concentrations (time 0). We then cultured the inoculated larval diet at 6, 12, 24, 48, and 72 h to detail bacterial growth and survival.
2.4. Rearing of Host (A. mellifera) Larvae In Vitro
We used in vitro larval rearing to identify a virulent strain of EFB bacteria that reliably killed honey bee larvae. We then determined the lethal dose required to kill approximately 50% of the larvae (LD50). Using specially designed grafting tools, newly hatched larvae were removed from the wax frames of 10 healthy robust colonies, and floated spiracle side up into 2 mL wells of sterile 24-well plates typically used for in vitro larval rearing. The larval diet consisted of royal jelly, yeast exact, fructose, glucose and water according to [54]. Larvae were incubated at 34 °C and 90% humidity for 6 days. On day 6, prior to pupation, larvae were moved to a petri dish with filter paper to defecate, then returned to the incubator at 34 °C and 80% humidity. After 24 h the larvae were moved back into a clean 24-well plate to pupate and emerge in the incubator.
2.5. LD50 Determination M. plutonius
Melissococcus plutonius virulence varies by strain [30,31,32], and is often detected in asymptomatic hives. Culturing M. plutonius as defined by cell morphology and 16S sequence does not guarantee strain virulence [29,52,55]. We therefore tested the virulence of multiple M. plutonius isolates by inoculating larvae in vitro. We determined an LD50 for the chosen M. plutonius strain (AF5), by introducing an inoculant dilution series to larvae. A scrape of 7-day-old growth on KSBHI agar was mixed into 1000 µL of saline. An arbitrary 10% dilution was equivalent to 64 CFU/µL in the diet. From this initial concentration, an order of magnitude serial dilution was prepared ranging from 64 to 0.0064 CFUs/µL. To define an LD50, each inoculant concentration was fed in 225 µL of diet (see below) to 24 larvae in vitro. After 7 days growth we determined CFU (colony forming unit) counts for each concentration as the average of three plates. We used a spectrophotometer and optical density (OD) at 600 nm to construct a standard curve for CFU counts, and then relied on OD600 readings to determine M. plutonius concentrations.
2.6. In Vitro Rescue Experiment
To test whether P. apium strain-C6 [20] was able to rescue larvae infected with M. plutonius, we fed both M. plutonius and P. apium to larvae in vitro. We used the quantified M. plutonius (strain AF5) as the disease agent in the P. apium rescue experiment. We introduced as independent variables; (1) the lowest M. plutonius concentration that killed 100% of the larvae (6.4 CFU/µL, positive control), (2) the M. plutonius concentration that killed 50% of the larvae (0.64 CFU/µL a treatment concentration most likely to reveal rescue potential), and (3) no M. plutonius bacteria (a negative control). To each of these 3 conditions we added the same concentration of P. apium strain C6 used in past experiments demonstrating a benefit (3 CFUs/µL; [20]). A total of 288 larvae were assayed, 48 for each of the six experimental conditions. We ran two independent trials with 24 larvae per condition per trial. Larvae were fed all diet and inoculant a single time point, as first instars. We distributed each of the six treatment conditions equally among the plates to remove the potential for a “plate effect”.
The larval diet was comprised of 34 mL sterile distilled water, 6 g of D-glucose (6%), 6 g of D-fructose, 1 g yeast extract and 53 g fresh commercially available RJ from GloryBee [54]. The negative control contained no bacteria and was comprised only of the above ingredients and saline solution as a control for bacterial introduction. We used the OD600 standard curve to estimate inoculant concentration, resulting in a final concentration of 6.4 CFU/µL and 0.64 CFU/µL in the diet. For the three treatments containing P. apium (C6), glycerol stock was grown up on SDA agar plates for 48 h at 34C. Bacteria was then scraped into saline and plated in ten-fold dilutions onto SDA agar plates. CFUs were counted to determine concentration of the bacteria in saline. A 60 CFUs/µL dilution was added to the diet resulting in a final concentration of 3 CFUs/µL of P. apium, the same concentration used for previous in vitro larval rearing trials [20].
Statistical Analysis—Mortality
We assessed total larval mortality at the end of the seven-day period using a chi-square contingency table to test for treatment differences. P-values were adjusted based on a multiple comparison false discovery rate adjustment (FDR).
3. Results
3.1. Niches Associated with Disease Transmission
We sequenced the 16S rRNA gene from various niches implicated in disease transmission. Next-generation sequencing returned 1,777,744 quality trimmed reads (300 bp) from 25 amplicon libraries (summarized in Supplementary Material, Table S1). Beebread was represented by 314,705 reads averaging 31,470 per library, honey by 630,010 reads averaging 126,002 per library, mouthparts by 502,473 reads averaging 100,495 per library and larvae by 330,556 reads averaging 66,111per library.
Across all libraries, a total of 2285 OTUs were resolved at 97% similarity. Average library coverage was high across all samples. Good coverage was 0.99 (±0.004) for the beebread samples, 1.00 (±0.0003) for mouthpart samples, 1.00 (±0.0003) for honey samples and 1.00 (±0.0001) for the larval samples. We used the inverse Simpson’s diversity index to portray niche diversity (larger vales indicate greater diversity) resulting in a 7.98 (±7.17) for beebread, 6.04 (±5.51) for mouthparts, 2.27 (±1.32) for honey and 2.15 (±0.47) for larvae (Supplementary Material, Table S2).
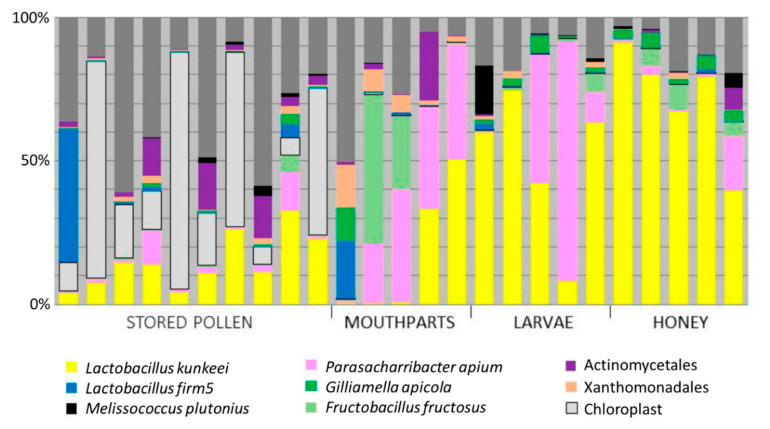
Sequences corresponding to the EFB disease agent, Melissococcus plutonius, were detected in all 25 libraries, but had the greatest relative abundance in honey and larvae, and the least relative abundance on nurse worker mouthparts (Figure 1). The number of sequence reads designated as M. plutonius is significantly associated with total sampling effort across all libraries (Pearson’s R = 0.45, n = 25, p = 0.02).
Figure 1.
Bacterial communities associated with potential hive refugia (food storage) and transmission from adult mouthparts to larvae. We sequenced the 16S rRNA gene from four niches associated with larval feeding (see methods). Colors represent various bacterial species, genera or broad groups. The chosen primer set (V1–V3 region) also amplifies chloroplast genes from pollen (light grey). Dark grey represents the relative proportion of bacterial groups not listed in the figure legend (see Table S1).
Also detected in all libraries, P. apium comprised 12.6% of the total sequences obtained from nurse mouthparts, honey, and second instar larvae (Table 1). The larval libraries were 25% P. apium on average, the honey libraries 4%, and the mouthparts 22%. Results from the current survey and previous publications indicate that P. apium is abundant (>20% relative abundance) in larvae, nurse worker mouthparts, worker foreguts (social stomach), nurse worker hypopharyngeal glands, royal jelly, and queen guts, and less abundant in adult worker guts and honey rich food stores (Table 1).
3.2. Bacterial Culturing
Based on BLASTn (Supplementary Material, Table S3), the bacteria recovered by our culturing effort were most similar to Melissococcus plutonius DAT 561 (n = 49), a variety of Enterococcus species (n = 22), primarily Enterococcus faecalis (n = 15), Lactobacillus kunkeei (n = 16), four strains of Paenibacillus alvei (n = 12), primarily Paenibacillus alvei strain Oa17A (n = 9). The remaining sequences were Bifidobacterium spp. (n = 1), and Parasaccharibacter apium strain G7_7_3c (n = 1). The EFB strain chosen to challenge larvae in vitro was named Mp5, and originated from an outbreak in Colorado in 2018.
3.3. Bacterial Inhibition Assay
To verify that M. plutonius strain AF5 survives in the larval environment, we cultured it in the presence of royal jelly and the in vitro diet. Both M. plutonius and P. apium grew in their preferred culture media in the presence of royal jelly. We observed no inhibition zone around royal jelly-containing discs, and no growth inhibition for either bacterium at royal jelly concentrations from dilute (10%) to concentrate (100%). P. apium strain C6 is known to proliferate (increase by a factor of 5) in the larval diet [20]. When M. plutonius was inoculated into the larval diet, it decreased quickly but survived at very low concentration for 72 h (Table 2).
Table 2.
M. plutonius strain AF5 decreases in the in vitro larval rearing diet.
| Time | * CFU 1 | CFU 2 | CFU 3 | Avg | CFU/µL in Diet |
|---|---|---|---|---|---|
| 0 h | 285 | 261 | 253 | 266 | 33.29 |
| 6 h | 114 | 142 | 152 | 136 | 13.6 |
| 24 h | 91 | 74 | 57 | 74 | 1.48 |
| 48 h | 59 | 85 | 53 | 66 | 1.31 |
| 72 h | 5 | 9 | 7 | 7 | 0.14 |
* CFU colony forming units, plated in triplicate.
3.4. LD50 Determination M. plutonius
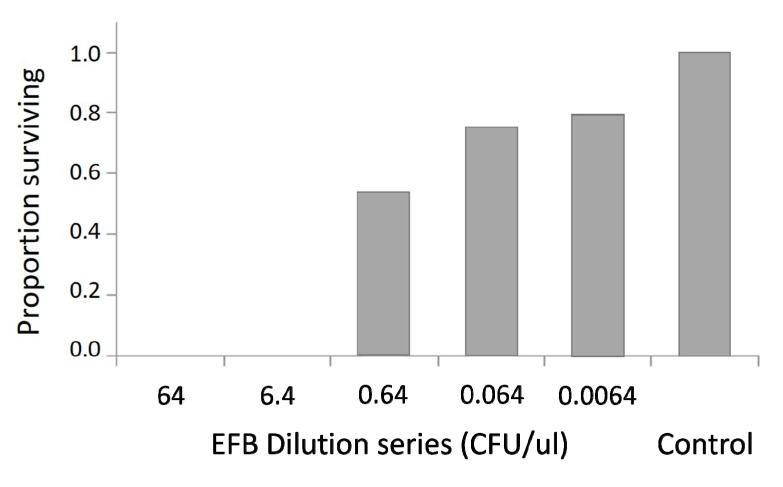
We tested four strains of M. plutonius for virulence against larvae in vitro, and found that three of the four resulted in larval mortality. We chose one of these virulent strains (AF5) to determine an LD50 value and perform further experiments. Consistent with expectations, in vitro larval survival decreased with increasing concentration of M. plutonius strain AF5. (Figure 2). Following infection, M. plutonius dilutions were plated in triplicate to confirm CFUs introduced to larvae in the diet.
Figure 2.
Survival of larvae exposed to a dilution series of M. plutonius strain AF5. Larvae were inoculated in vitro (N = 24 for each concentration) and survival was recorded at Day 6. The control received diet and saline with no bacteria. We designated the 6.4 CFU/µL dilution as LD100, and the 0.64 CFU/µL dilution as LD50 for use as positive controls in the in vitro rescue assay.
3.5. In Vitro Rescue Experiment
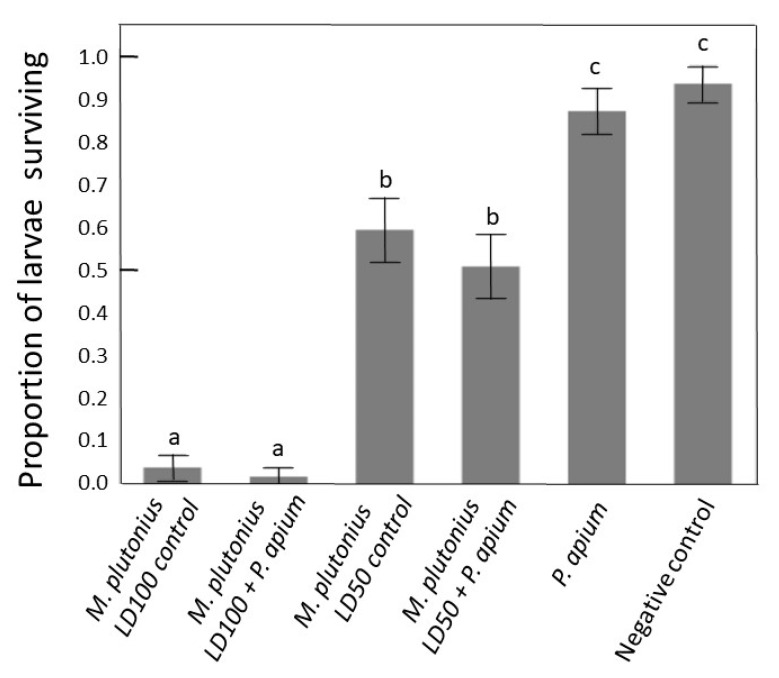
The negative control produced >90% survival in both rescue trials validating the in vitro rearing procedure (Figure 3). The two positive controls also produced larval survival at expected proportions; low survival in the high M. plutonius concentration and survival not significantly different from 50% for the LD50 M. plutonius concentration. We introduced P. apium strain C6 as a rescue bacterium for EFB infection. Treatment groups inoculated with both P. apium strain C6 and a high concentration of M. plutonius strain AF5 did not differ, indicating no effect of P. apium on larval survival. The same was true for strain C6 and the lower concentration of strain AF5. The treatment group inoculated with only strain C6 did not differ significantly from the negative control group in either trial (Figure 3).
Figure 3.
Proportion of larvae surviving M. plutonius challenge and P. apium rescue. Error bars represent two independent trials (see methods). From Figure 1, LD100 represents (6.4 CFU/µL) of M. plutonius AF5, and LD50 is (0.64 CFU/µL) killing approximately 50% of the larvae by 6 days of age. P. apium was introduced at 3 CFU/µL, the concentration used in Corby Harris et al. [20]. Columns with the same letter do not differ for larval survival.
4. Discussion
Honey bee foul brood diseases have serious economic consequences for beekeeping and pollination services globally [56,57]. Although various assays can detect larval pathogens in hives [56,58] there is much to be learned about the ecology and epidemiology of M. plutonius, the causative agent of European Foul Brood (EFB) disease. Based on broad sampling throughout the United Kingdom, M. plutonius is comprised of at least three genetically distinct groups [7] and virulence differs markedly by strain [30,31,32]. Melissococcus plutonius is often found in the guts of workers so may be easily transmitted among hives when they are placed in proximity [13]. Recent increases in antibiotic resistance and incidence of EFB and EFB-like brood disease have spurred efforts to understand virulence including genetic sequencing, comparative genomic analysis, the contribution of secondary bacteria and targeted molecular biology [8,30,31,32,52,55,59,60,61,62]. Understanding the microbial ecology associated with M. plutonius will provide a functional context of disease progression and suggest potential solutions to mitigate disease. Here, we expand on past work to describe and discuss the general context of EFB disease transmission in hives, identify virulent strains and biologically relevant inoculant concentrations of M. plutonius, and test whether a putatively beneficial bacterium (P. apium strain C6) can rescue or diminish EFB related larval mortality.
4.1. Context of Disease Transmission
Results from our next-generation 16S rRNA amplicon survey suggests that the prevalence and abundance of M. plutonius is diminished in niches with higher bacterial diversity (beebread and mouthparts; Figure 1). Melissococcus plutonius is relatively abundant when abiotic factors are extreme (e.g., low pH, high acidity, low water availability), as is the case with 2nd instar larvae and the tops of ripening honey cells (Figure 1). That M. plutonius might best survive in extreme environments with low competition is consistent with recently described gene sets [60]. It is hypothesized that 2nd instar larvae are most vulnerable to EFB infection, concurrent with the presumed diet transition from royal to worker jelly. Our results agree with culture-based assays [24] that 1st and 2nd instar larvae can be dominated by P. apium or L. kunkeei, both fast-growing bacteria often considered at least commensal if not beneficial [20]. Both of these species groups are comprised of multiple strains that possess functional differences and occupy a variety of honey bee niches [10]. Given the genetic variation within each species group, it is likely that function varies by niche and strain. Consistent with our findings and the literature, P. apium is clearly co-evolved with royal and worker jelly, and considering the totality of available data, P. apium proliferates in a wide variety of in-hive niches seemingly defined by the social network of food sharing and the potential to encounter royal jelly (Table 1).
Based on spatial occurrence patterns, M. plutonius may be somewhat ubiquitous in disease free hives. It was represented in every niche and library with relative abundance correlated significantly with sequencing effort (Figure 1, Table S1). The relatively low representation of M. plutonius on the nurse bee mouthparts may reflect an oxygenated niche and/or highly competitive environment. Also considered ubiquitous in past investigations [29,57] the putative secondary invaders Paenibacillus alvei and Enterococcus faecalis were detected at very low prevalence and abundance throughout our survey, despite the deep sequencing effort (Table 2). We detected multiple strains of P. alvei, but based on 16S gene sequences, Enterococcus spp. were poorly defined taxonomically, with variability likely inflated by the chosen 16S gene region. Perhaps as suggested, P. alvei is a saprophyte on dead larvae [8] and may only be detected in abundance during a disease outbreak.
4.2. Bacterial Competition In Vitro
When cultured in their preferred growth media, neither P. apium strain C6 nor M. plutonius strain AF5 were inhibited by royal jelly. When P. apium strain C6 was added to the in vitro rearing diet, it increased by a factor of five [20], but results from the present experiment show that M. plutonius strain AF5 was diminished by the larval diet, but it was still detected after 72 h (Table 2). These contrasting results may highlight virulence associated factors that vary among M. plutonius strains and/or larval microbial ecology that may influence disease progression. Strain AF5 likely belongs to clade CC12, because it is highly virulent [30], exhibits similar growth requirements [59,60,61,62], and shares 99.5% 16S rRNA gene sequence similarity with M. plutonius strain DAT561, a member of the CC12 clade [7] (Supplemental information Table S3). Although M. plutonius strain AF5 may not survive for long in the environment of the larval diet, even small numbers can successfully find their preferred niche in the larval gut, multiply and cause disease (Figure 2). Consistent with this hypothesis, the introduction in larval diet of relatively small doses of M. plutonius strain AF5 still produced significant larval mortality relative to the negative control (Figure 3).
The early growth patterns of M. plutonius and competing bacteria that typically colonize newly hatched larvae may be important factors in the progression and treatment of EFB disease. Results presented here and in the literature indicate that M. plutonius has evolved in strong niche proximity with P. apium (Figure 1). Because P. apium (strain C6) has been reported to increase larval survival [20], we hypothesized that it may provide benefits in a disease context, protecting larvae from the establishment of M. plutonius. However, our results show that P. apium strain C6 does not rescue larvae from the effects of EFB disease associated with M. plutonius strain AF5 (Figure 3). Larvae inoculated with both M. plutonius and P. apium strain C6 did not survive significantly better than larvae inoculated with only M. plutonius. Moreover, the in vitro procedure was validated by the consistent performance of both positive and negative control groups (Figure 3). We speculate that M. plutonius avoids competition with P. apium strain C6 in the larval gut via niche partitioning or competitively inhibition. Future studies would benefit by using MLST genotyping to characterize M. plutinious strains [60], varying the introduction times and concentrations of probiotic and pathogen, and quantifying the proliferation and establishment of M. plutonius and potential probiotics. To place our sequencing results (Figure 1) in a broader context, our microbiome survey revealed >30 OTUs classified as P. apium, many abundant in larval guts (Supplementary Materials, Table S1). A single P. apium OTU accounted for 85% of all P. apium in larvae, but the OTU matching P. apium strain C6 accounted for less than 1% of all P. apium in larvae, suggesting that P. apium strain C6 is not a dominant larval strain. As detailed earlier (Table 1), various P. apium strains occupy many niches throughout the hive and colony. Testing more strains and more combinations of strains will better reveal the potential of the native larval microbiome to mitigate disease.
4.3. Larval Assay In Vitro
Our in vitro results do not agree with our past study [20]. In the present study, we found that negative control survival (no introduced bacteria) did not differ from survival of larvae inoculated with P. apium strain C6 (Figure 3). Thus, in contrast to our 2014 study, we found no effect of P. apium strain C6 on larval survival in the absence of disease. Our continued lab experience suggests that differences between the studies were a consequence of the in vitro rearing technique. In our 2014 study, the low and variable nature of control and treatment survival between trials within the study suggested a lack of reproducibility as discussed by the authors [20]. More specifically, negative control survival was exceedingly low, and differed significantly between the two trials at 40 and 70 percent survival. However, the statistical model in [20] did not consider trial variation, thus, it was difficult to conclude whether treatment effects or trial variation was the greatest contributor to the statistical differences reported in [20].
In the present study, we used the same in vitro larval diet containing the same bacterial strain (C6) and inoculum concentration used in the 2014 study. However, in 2014, the diet and inoculate was introduced multiple times during larval development; uneaten diet and inoculate were removed with a pipette at each daily feeding interval, and replaced with fresh diet and inoculate [20]. While daily feeding more closely replicates the continuous nature of feeding in the colony, our continued lab experience suggests that multiple disturbances over time can influence larval survival in a plate-specific manner, increasing assay error. Specifically, in the 2014 study, (1) we segregated treatments and controls by plate, allowing for a “plate-specific effect” on survival, and (2) we disturbed plates multiple times for feedings and inoculations (repeated exposure), increasing the probability of larval contamination, damage or drowning. In contrast, the methods of the present study limit these random factors by, (1) mixing treatments across plates to minimize plate-specific effects and, (2) minimally disturbing larvae (single exposure) to reduce the opportunity for contamination, damage or drowning.
In the honey bee system, larval rearing in vitro has been continuously refined to approximate what occurs in colonies under natural conditions; high and repeatable larval survival [54,63,64,65]. Accordingly, an a priori threshold for high and repeatable survival of the negative control (Figure 2) drove the present experimental design. In-hive larval survival under natural conditions is generally greater than 90% [64,65], and represents the established standard for in vitro rearing. If negative control survival in vitro falls below this natural threshold, it indicates a failure of the in vitro rearing protocol or larvae sourced from an unhealthy colony. Given a priori survival expectations in the absence of a positive control, a survival benefit attributed to an introduced treatment could only attain statistical relevance if survival of the negative control falls significantly below that of the putatively beneficial treatment. In turn, the beneficial treatment would be difficult, if not impossible to distinguish from repeatable in vitro rearing technique. In other words, when recording survival as the only dependent variable, the demonstration of a benefit would rely on poor survival of the negative control, an inherent design flaw. Given the established expectation of high and repeatable survival in the negative control, introducing a factor that is hypothesized to benefit larval survival requires a positive control for adequate design.
5. Conclusions
The results and methodology presented here contribute to the development of a microbiome-centric model of health and disease in honey bees. Assessing the microbial ecology of larvae including Melissococcus plutonius and associated larval microbiomes will promote a better understanding of the infection process and suggest potential management strategies beyond broad-spectrum antibiotics. Within the context of an improved experimental design, P. apium strain C6 was commensal, producing no effect on larval survival. Our results highlight the importance of both positive and negative controls when designing ecological studies involving disease agents and potential remedies. Because we performed our experiments under controlled lab conditions, we exposed larvae to less variety and abundance of naturally occurring hive microbiota and fresh royal jelly components. For future in vitro experiments, the ability to classify native vs. non-native microbiota and represent the associated microbiome and social dynamics during larval development will be critical for modeling larval health and disease in the natural hive setting.
Acknowledgments
We thank Mark Carroll, Vincent Ricigliano, and William Meikle for manuscript review, particularly discussions concerning experimental design and statistical analysis. We thank Jay Evans, Samuel Abban, and Jose Villa for providing diseased larval samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/9/555/s1, Table S1. Microbiome raw read count (relative abundance) by niche and taxonomy; Table S2. Alpha diversity of microbiome samples by niche; Table S3. Results culturing diseased larvae from various locales experiencing EFB outbreaks.
Author Contributions
Conceptualization, A.S.F., B.M.M. and K.E.A. methodology, A.S.F., B.M.M., P.M., D.C.C., Q.S.M. and K.E.A software, A.S.F. and P.M.; validation, A.S.F., B.M.M., P.M., D.C.C., Q.S.M. and K.E.A.; formal analysis, A.S.F., B.M.M., P.M., D.C.C., Q.S.M. and K.E.A; investigation, A.S.F., B.M.M. and K.E.A.; resources, K.E.A.; data curation, P.M. and K.E.A.; writing—original draft preparation, A.S.F. and K.E.A.; writing—review and editing, B.M.M. and K.E.A. visualization, A.S.F., P.M. and K.E.A.; supervision, K.E.A.; project administration, K.E.A.; funding acquisition, K.E.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USDA parent project 2022-21000-021-00D.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chopra S.S., Bhavik R.B., Vikas K. Economic Dependence of U.S. Industrial Sectors on Animal-Mediated Pollination Service. Environ. Sci. Technol. 2015;49:14441–14451. doi: 10.1021/acs.est.5b03788. [DOI] [PubMed] [Google Scholar]
- 2.Klein A.M., Vaissiere B.E., Cane J.H., Steffan-Dewenter I., Cunningham S.A., Kremen C., Tscharntke T. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricigliano V.A., Mott B.M., Maes P.W., Floyd A.S., Fitz W., Copeland D.C., Meikle W.G., Anderson K.E. Honey bee colony performance and health are enhanced by apiary proximity to US Conservation Reserve Program (CRP) lands. Sci. Rep. 2019;9:4894. doi: 10.1038/s41598-019-41281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genersch E. American foulbrood in honey bees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010;103:S10–S19. doi: 10.1016/j.jip.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Genersch E. Honeybee pathology: Current threats to honeybees and beekeeping. Appl. Microbiol. Biotechnol. 2010;87:87–97. doi: 10.1007/s00253-010-2573-8. [DOI] [PubMed] [Google Scholar]
- 6.Engelsdorp D.V., Lengerich E., Spleen A., Dainat B., Cresswell J., Baylis K., Nguyen B.K., Soroker V., Human H., Le Conte Y., et al. Standard epidemiological methods to understand and improve Apis mellifera health. J. Apicult. Res. 2015;8839:1–16. doi: 10.3896/IBRA.1.52.1.08. [DOI] [Google Scholar]
- 7.Budge G.E., Shirley M.D.F., Jones B., Quill E., Tomkies V., Feil E.J., Brown M.A., Haynes E.G. Molecular epidemiology and population structure of the honey bee brood pathogen Melissococcus plutonius. ISME J. 2014;8:1588–1597. doi: 10.1038/ismej.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewkowski O., Erler S. Virulence of Melissococcus plutonius and secondary invaders associated with European foulbrood disease of the honey bee. Microbiol. Open. 2018;8:e00649. doi: 10.1002/mbo3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maes P.W., Rodrigues P.A.P., Oliver R., Mott B.M., Anderson K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera) Mol. Ecol. 2016;25:5439–5450. doi: 10.1111/mec.13862. [DOI] [PubMed] [Google Scholar]
- 10.Anderson K.E., Ricigliano V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017;22:125–132. doi: 10.1016/j.cois.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Erban T., Ledvinka O., Kamler M., Nesvorna M., Hortova B., Tyl J., Titera D., Markovic M., Hubert J. Honeybee (Apis mellifera)-associated bacterial community affected by American foulbrood: Detection of Paenibacillus larvae via microbiome analysis. Sci. Rep. 2017;7:5084. doi: 10.1038/s41598-017-05076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubert J., Bicianova M., Ledvinka O., Kamler M., Lester P.J., Nesvorna M., Kopecky J., Erban T. Changes in the Bacteriome of Honey Bees Associated with the Parasite Varroa destructor, and Pathogens Nosema and Lotmaria passim. Microb. Ecol. 2017;73:685–698. doi: 10.1007/s00248-016-0869-7. [DOI] [PubMed] [Google Scholar]
- 13.Sopko B., Zitek J., Nesvorna M., Markovic M., Kamler M., Titera D., Erban T., Hubert J. Detection and quantification of Melissococcus plutonius in honey bee workers exposed to European foulbrood in Czechia through conventional PCR, qPCR, and barcode sequencing. J. Apic. Res. 2020;59:503–514. doi: 10.1080/00218839.2019.1685148. [DOI] [Google Scholar]
- 14.Raymann K., Coon K.L., Shaffer Z., Salisbury S., Moran N.A. Pathogenicity of Serratia marcescens Strains in Honey Bees. MBio. 2018;9:e01649-18. doi: 10.1128/mBio.01649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buford T.W. (Dis)Trust your gut: The gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5:80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberoni D., Gaggìa F., Baffoni L., Gioia D.D. Beneficial microorganisms for honey bees: Problems and progresses. Appl. Microbiol. Biotechnol. 2016;100:9469–9482. doi: 10.1007/s00253-016-7870-4. [DOI] [PubMed] [Google Scholar]
- 17.Killer J., Dubná S., Sedláček I., Švec P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol. 2014;64:152–157. doi: 10.1099/ijs.0.053033-0. [DOI] [PubMed] [Google Scholar]
- 18.Forsgren E., Olofsson T.C., Vasquez A., Fries I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie. 2010;41:99–108. doi: 10.1051/apido/2009065. [DOI] [Google Scholar]
- 19.Wu M., Sugimura Y., Iwata K., Takaya N., Takamatsu D., Kobayashi M., Taylor D., Kimura K., Yoshiyama M. Inhibitory effect of gut bacteria from the Japanese honey bee, Apis cerana japonica, against Melissococcus plutonius, the causal agent of European foulbrood disease. J. Insect. Sci. 2014;14:129. doi: 10.1093/jis/14.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corby-Harris V., Snyder L.A., Schwan M.R., Maes P., McFrederick Q.S., Anderson K.E. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol. 2014;80:7460–7472. doi: 10.1128/AEM.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephan J.G., Lamei S., Pettis J.S., Riesbeck K., de Miranda J.R., Forsgren E. Honeybee-Specific Lactic Acid Bacterium Supplements Have No Effect on American Foulbrood-Infected Honeybee Colonies. Appl. Environ. Microbiol. 2019;85:e00606-19. doi: 10.1128/AEM.00606-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch S., Ashiralieva A., Hedtke K., Genersch E. Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the etiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 2009;75:3344–3347. doi: 10.1128/AEM.02839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojvodic S., Jensen A.B., James R.R., Boomsma J.J., Eilenberg J. Temperature dependent virulence of obligate and facultative fungal pathogens of honeybee brood. Vet. Microbiol. 2011;149:200–205. doi: 10.1016/j.vetmic.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Vojvodic S., Rehan S.M., Anderson K.E. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS ONE. 2013;8:e72106. doi: 10.1371/journal.pone.0072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson K.E., Sheehan T.H., Mott B.M., Maes P., Snyder L., Schwan M.R., Walton A., Jones B.M., Corby-Harris V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PLoS ONE. 2013;8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hroncova Z., Havlik J., Killer J., Doskocil I., Tyl J., Kamler M., Titera D., Hakl J., Mrazek J., Bunesova V., et al. Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS ONE. 2015;10:e118707. doi: 10.1371/journal.pone.0118707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seeley T.D. Adaptive significance of the age polyeithism schedule in honeybee colonies. Behav. Ecol. Sociobiol. 1982;11:287–293. doi: 10.1007/BF00299306. [DOI] [Google Scholar]
- 28.Anderson K.E., Rodrigues P.A., Mott B.M., Maes P., Corby-Harris V. Ecological succession in the honey bee gut: Shift in Lactobacillus strain dominance during early adult development. Microb. Ecol. 2016;71:1008–1019. doi: 10.1007/s00248-015-0716-2. [DOI] [PubMed] [Google Scholar]
- 29.Forsgren E. European foulbrood in honey bees. J. Invertebr. Pathol. 2010;103:S5–S9. doi: 10.1016/j.jip.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura K., Yamazaki Y., Shiraishi A., Kobayashi S., Harada M., Yoshiyama M., Osaki M., Okura M., Takamatsu D. Virulence Differences among Melissococcus plutonius Strains with Different Genetic Backgrounds in Apis mellifera Larvae under an Improved Experimental Condition. Sci. Rep. 2016;6 doi: 10.1038/srep33329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossar D., Kilchenmann V., Forsgren E., Charrière J.-D., Gauthier L., Chapuisat M., Dietemann V. Putative determinants of virulence in Melissococcus plutonius, the bacterial agent causing European foulbrood in honey bees. Virulence. 2020;11:554–567. doi: 10.1080/21505594.2020.1768338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura K., Okumura K., Harada M., Okamota M., Okura M., Takamatsu D. Different impacts of pMP19 on the virulence of Melissococcus plutonius strains with different genetic backgrounds. Environ. Microbiol. 2020;22 doi: 10.1111/1462-2920.14999. [DOI] [PubMed] [Google Scholar]
- 33.Bailey L. The Pathogenicity for Honey-Bee Larvae of Microorganisms Associated with European Foulbrood. J. Insect Pathol. 1963;5:198–205. [Google Scholar]
- 34.Tian B., Fadhil N.H., Powell J.E., Kwong W.K., Moran N.A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. MBio. 2012;3:e00377-12. doi: 10.1128/mBio.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinson V.G., Danforth B.N., Minckley R.L., Rueppell O., Tingek S., Moran N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 36.Yun J.H., Lee J.Y., Hyun D.W., Jung M.J., Bae J.W. Bombella apis sp. nov., an acetic acid bacterium isolated from the midgut of a honey bee. Int. J. Syst. Evol. Microbiol. 2017;67:2184–2188. doi: 10.1099/ijsem.0.001921. [DOI] [PubMed] [Google Scholar]
- 37.McFrederick Q.S., Wcislo W.T., Taylor D.R., Ishak H.D., Dowd S.E., Mueller U.G. Environment or kin: Whence do bees obtain acidophilic bacteria? Mol. Ecol. 2012;21:1754–1768. doi: 10.1111/j.1365-294X.2012.05496.x. [DOI] [PubMed] [Google Scholar]
- 38.Endo A., Salminen S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 2013;36:444–448. doi: 10.1016/j.syapm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Anderson K.E., Ricigliano V.A., Mott B.M., Copeland D.C., Floyd A.S., Maes P. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome. 2018;6:108. doi: 10.1186/s40168-018-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinson V.G., Moy J., Moran N.A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012;78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabree Z., Hansen A., Moran N.A. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS ONE. 2012;7:e41250. doi: 10.1371/journal.pone.0041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran N.A., Hansen A.K., Powell J.E., Sabree Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corby-Harris V., Maes P., Anderson K.E. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE. 2014;9:e95056. doi: 10.1371/journal.pone.0095056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson K.E., Carroll M.J., Sheehan T., Mott B.M., Maes P., Corby-Harris V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014;23:5904–5917. doi: 10.1111/mec.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapheim K.M., Rao V.D., Yeoman C.J., Wilson B.A., White B.A., Goldenfeld N., Robinson G.E. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera) PLoS ONE. 2015;10:e0123911. doi: 10.1371/journal.pone.0123911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarpy D.R., Mattila H.R., Newton I.L.G. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl. Environ. Microbiol. 2015;81:3182–3191. doi: 10.1128/AEM.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W.G., Peplies J., Glockner F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinform. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arai R., Tominaga K., Wu M., Okura M., Ito K., Okamura N., Onishi H., Osaki M., Sugimura Y., Yoshiyama M., et al. Diversity of Melissococcus plutonius from honeybee larvae in Japan and experimental reproduction of European foulbrood with cultured atypical isolates. PLoS ONE. 2012;7:e33708. doi: 10.1371/journal.pone.0033708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chouaia B., Gaiarsa S., Crotti E., Comandatore F., Degli Esposti M., Ricci I., Alma A., Favia G., Bandi C., Daffonchio D. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol. Evol. 2014;6:912–920. doi: 10.1093/gbe/evu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaftanoglu O., Linksvayer T.A., Page R.E. Rearing honey bees (Apis mellifera L.) in vitro: Effects of feeding intervals on survival and development. J. Apic. Res. 2010;49:311–317. doi: 10.3896/IBRA.1.49.4.03. [DOI] [Google Scholar]
- 55.McKee B.A., David Goodman R., Alan Hornitzky M. The transmission of European foulbrood (Melissococcus plutonius) to artificially reared honey bee larvae (Apis mellifera) J. Apic. Res. 2004;43:93–100. doi: 10.1080/00218839.2004.11101117. [DOI] [Google Scholar]
- 56.Garrido-Bailón E., Higes M., Martínez-Salvador A., Antúnez K., Botías C., Meana A., Prieto L., Martín-Hernández R. The prevalence of the honeybee brood pathogens Ascosphaera apis, Paenibacillus larvae and Melissococcus plutonius in Spanish apiaries determined with a new multiplex PCR assay. Microb. biotechnol. 2013;6:731–739. doi: 10.1111/1751-7915.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fünfhaus A., Ebeling J., Genersch E. Bacterial pathogens of bees. Curr. Opin. Insect Sci. 2018;26:89–96. doi: 10.1016/j.cois.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Govan V.A., Zel V.B., Allsopp M.H., Davison A.S. A PCR Detection Method for Rapid Identification of Melissococcus pluton in Honeybee Larvae. Appl. Environ. Microbiol. 1998;64 doi: 10.1128/AEM.64.5.1983-1985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamatsu D., Okumura K., Tabata A., Okamoto M., Okura M. Transcriptional regulator SpxA1a controls the resistance of the honey bee pathogen Melissococcus plutonius to the antimicrobial activity of royal jelly. Environ. Microbiol. 2020;22:2736–2755. doi: 10.1111/1462-2920.15125. [DOI] [PubMed] [Google Scholar]
- 60.Djukic M., Erler S., Leimbach A., Grossar D., Charrière J.D., Gauthier L., Hartken D., Dietrich S., Nacke H., Daniel R., et al. Comparative genomics and description of putative virulence factors of Melissococcus plutonius, the causative agent of European foulbrood disease in honey bees. Genes. 2018;9:419. doi: 10.3390/genes9080419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erban T., Ledvinka O., Kamler M., Hortova B., Nesvorna M., Tyl J., Titera D., Markovic M., Hubert J. Bacterial community associated with worker honeybees (Apis mellifera) affected by European foulbrood. PeerJ. 2017;5 doi: 10.7717/peerj.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takamatsu D., Osawa A., Nakamura K., Yoshiyama M., Okura M. High-level resistance of Melissococcus plutonius clonal complex 3 strains to antimicrobial activity of royal jelly. Environ. Microbiol. Rep. 2017;9:562–570. doi: 10.1111/1758-2229.12590. [DOI] [PubMed] [Google Scholar]
- 63.Crailsheim K., Brodschneider R., Aupinel P., Behrens D., Genersch E., Vollmann J., Riessberger-Gallé U. Standard methods for artificial rearing of Apis mellifera larvae. J. Apic. Res. 2013;52:1–16. doi: 10.3896/IBRA.1.52.1.05. [DOI] [Google Scholar]
- 64.Schmehl D.R., Tomé H.V.V., Mortensen A.N., Martins G.F., Ellis J.D. Protocol for the in vitro rearing of honey bee (Apis mellifera L.) workers. J. Apic. Res. 2016;55:113–129. doi: 10.1080/00218839.2016.1203530. [DOI] [Google Scholar]
- 65.Mortensen A.N., Ellis J.D. A honey bee (Apis mellifera) colony’s brood survival rate predicts its in vitro-reared brood survival rate. Apidologie. 2018;49:573–580. doi: 10.1007/s13592-018-0584-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.