Abstract
The study investigated the regulation of Smad2 by miR-18a and its role in preeclampsia (PE). Bioinformatics analysis showed that both Smad2 and Smad3 were the predicted targets for miR-18a. Mass spectrum analysis showed that two mature Smad2 isoforms existed in human placenta: full length, Smad2(FL), and that lacking exon3, Smad2(Δexon3). The protein level of Smad2(FL), but not Smad2(Δexon3) or Smad3, was significantly increased in severe PE (sPE) placenta, which was inversely correlated with the level of miR-18a. Elevated Smad2(FL) phosphorylation level appeared in sPE placenta, and Smad2 was colocalized with miR-18a in various subtypes of trophoblasts in human placenta. Smad2(FL) was validated as the direct target of miR-18a in HTR8/SVneo cells. miR-18a enhanced trophoblast cell invasion, which was blocked by the overexpression of Smad2(FL). Furthermore, overexpression of miR-18a repressed Smad2 activation and the inhibition of trophoblast cell invasion by transforming growth factor-β (TGF-β). In conclusion, our results suggest that miR-18a inhibits the expression of Smad2(FL), but not Smad2(Δexon3) or Smad3, which can reduce TGF-β signaling, leading to the enhancement of trophoblast cell invasion. A lack of miR-18a, which results in the upregulation of Smad2(FL), contributes to the development of PE.
Keywords: miR-18a, Smad2, placental trophoblast, cell invasion, preeclampsia
Graphical Abstract
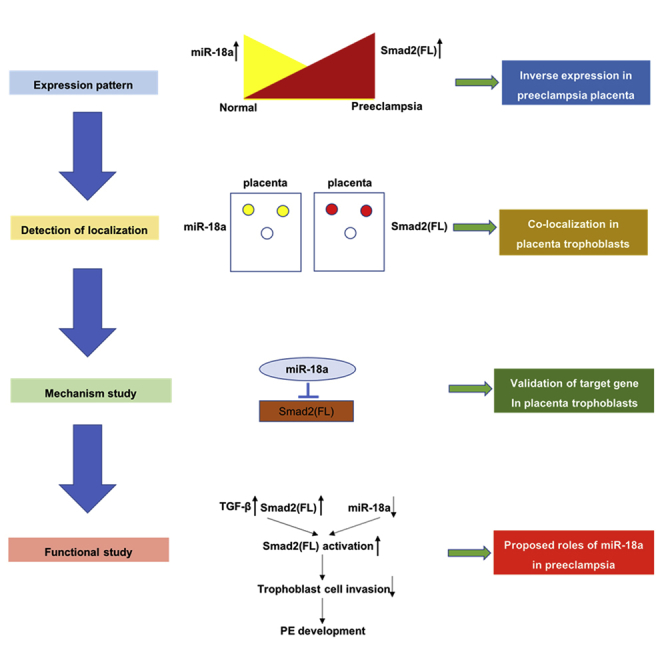
A lack of miR-18a in human placenta may contribute to the development of preeclampsia. Its underlying mechanism is that miR-18a inhibits the expression of Smad2(FL), but not Smad2(Δexon3) or Smad3, which can reduce TGF-β signaling, leading to the enhancement of trophoblast cell invasion.
Introduction
Preeclampsia (PE) is a multisystem disorder that affects about 2%–7% pregnancies worldwide and is considered one of the leading causes of maternal and neonatal mortality as well as morbidity.1,2 Although the etiology and the pathogenesis of PE are not well understood, it is generally accepted that placental defects, especially the dysregulation of the trophoblast behaviors, are the underlying mechanisms that lead to the disease. For instance, reduced trophoblast proliferation, excessive trophoblast cell apoptosis, as well as the insufficient trophoblast invasion have all been linked to the occurrence and the development of PE.3,4
MicroRNAs (miRNAs) are a subset of small, noncoding RNAs that are involved in numerous important biological events.5 A major mechanism by which miRNAs regulate gene expression is the partial base pairing to the 3′ untranslated region (3′ UTR) of the target mRNAs to repress their translation and/or to induce their degradation.6 Our7 and others’8,9 previous work have demonstrated that a great number of miRNAs are differentially expressed in PE placentas, and some of these differentially expressed miRNAs may participate in the regulation of various trophoblast cell functions.7,10,11 miR-18a is predominantly downregulated in placental tissues and maternal plasma of patients with PE and is mainly localized in the various subtypes of trophoblast cells in the placenta.7 A series of in vitro and in vivo studies have demonstrated that miR-18a plays an important role in different cell functions via downregulating FGF1, Smad4, HIF-1α, and other target genes.12, 13, 14 Therefore, elucidation of the underlying mechanisms by which miR-18a regulates the functions of trophoblast cells may provide a better understanding of the pathophysiology of this disorder and uncover new targets for therapeutic intervention.
With the use of bioinformatics tool Targetscan,15,16 we found that Smad2 and Smad3 were both among the most predicted targets of miR-18a. Smad2 and Smad3 are both central cytoplasmic mediators that can be activated by transforming growth factor-β (TGF-β) and its specific serine/threonine kinase receptors.17,18 TGF-β is a pleiotropic factor that plays essential roles in regulating numerous physiological and pathological processes.19 Furthermore, it is highly expressed in PE placentas,20,21 and the dysregulation of its signaling pathway is responsible for the dysfunction of trophoblast cells and the pathophysiology of PE.21, 22, 23, 24, 25, 26, 27 We therefore speculated that miR-18a interferes with the TGF-β signaling in PE placentas via targeting Smad2 and/or Smad3.
Smad2 is composed of 12 exons, and exon3 is spliced out in about 10% of Smad2 in several human tissues, including heart and placenta.28 There exists two mature Smad2 isoforms in human placenta: full-length Smad2, Smad2(FL), and Smad2 lacking exon 3, Smad2(Δexon3). Perhaps due to the specificity of the antibody and/or the protein extraction method,29 our previous work just detected the expression patterns of the Smad2(FL) in human placenta.7 Despite that more than 90% sequences are similar between Smad2(FL) and Smad2(Δexon3), these two proteins have different expression patterns and distinguishing function features in several pathologies.29,30 However, the expression pattern and function of these two Smad2 isoforms in severe PE (sPE) placenta remain unknown.
In the present study, based on the evidence noted above, we have proposed that miR-18a regulated trophoblast cell function by targeting at one or more of the Smad proteins, which impaired the TGF-β signaling and led to the disease. To test our hypothesis, we used human trophoblast cell line HTR8/SVneo to investigate whether miR-18a attenuated the TGF-β signaling and inhibited the expression of Smad protein(s), the disturbance of which favors the disease development.
Results
miR-18a Was Significantly Downregulated in the Chorionic Plates, but Not the Basal Plates of the sPE Placentas
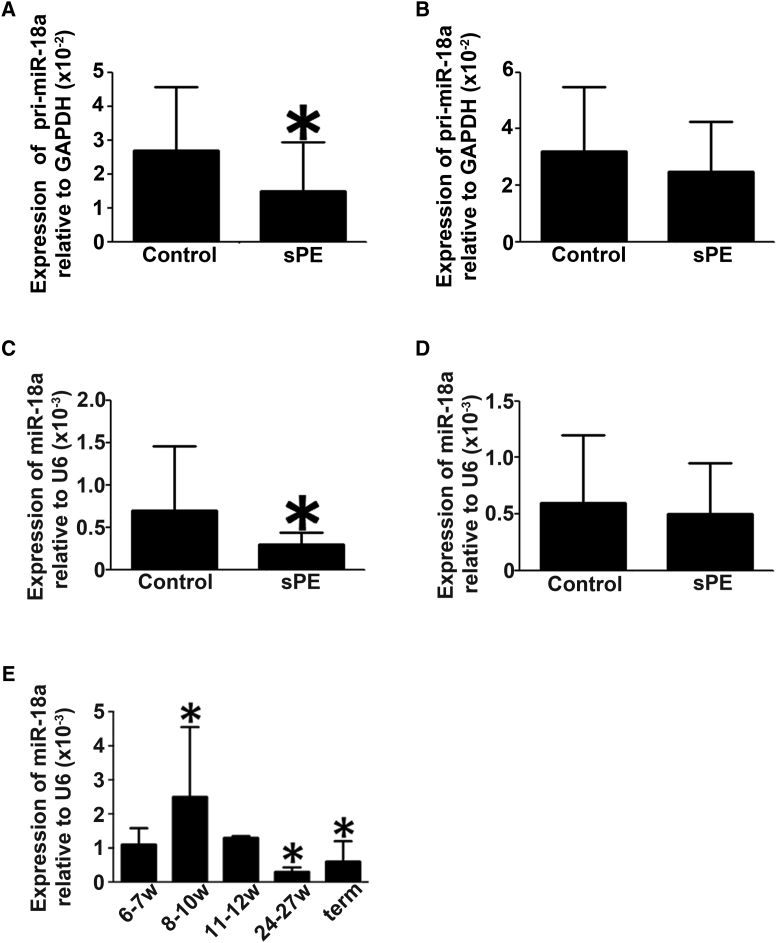
With the use of quantitative real-time PCR technology, we examined pri-miR-18a and miR-18a expression levels in the control and sPE placentas. The levels of pri-miR-18a and miR-18a were significantly downregulated in the chorionic plates (Figures 1A and 1C) but not in the basal plates (Figures 1B and 1D) of the sPE placentas.
Figure 1.
Differential Expression Patterns of pri-miR-18a and Mature miR-18a in sPE Placentas
(A–D) Quantitative real-time PCR experiments were performed to measure the expression levels of pri-miR-18a (A and B) and mature miR-18a (C and D) in the chorionic plates (A and C) and basal plates (B and D) of the placentas derived from sPE patients (n = 13) and normal pregnant women (n = 32). Data are presented as mean ± SD. ∗p < 0.05. (E) miR-18a expression levels change across gestation. At least 3 samples were collected and analyzed in each gestational week. Data are presented as mean ± SD. ∗Compared to the miR-18a level at gestational weeks 6–7 (6–7w), p < 0.05.
The Expression Pattern of Placental miR-18a at Different Gestational Stages during Normal Gestation
The expression pattern of miR-18a at different gestational stages of normal pregnancy was examined by quantitative real-time PCR. At least 3 samples were collected and analyzed in each gestational week. As shown in Figure 1E, the level of miR-18a during gestational weeks 8–10 was significantly higher than that at gestational weeks 6–7, and the level of miR-18a began to decrease after gestational week 10. The level of miR-18a at mid-term and term was significantly decreased compared to that at gestational weeks 6–7.
Expressions of Smad2(FL) and Phosphorylated (p)-Smad2(FL) Proteins Are in Inverse Correlation with miR-18a in sPE Placentas
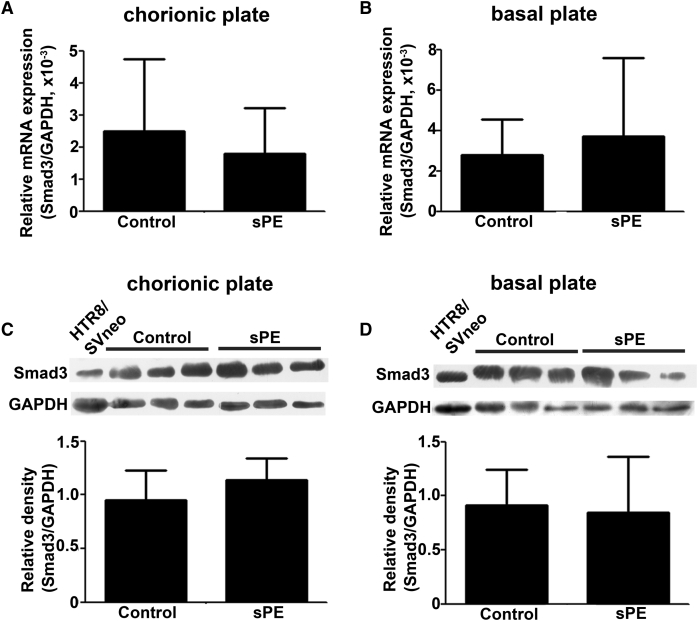
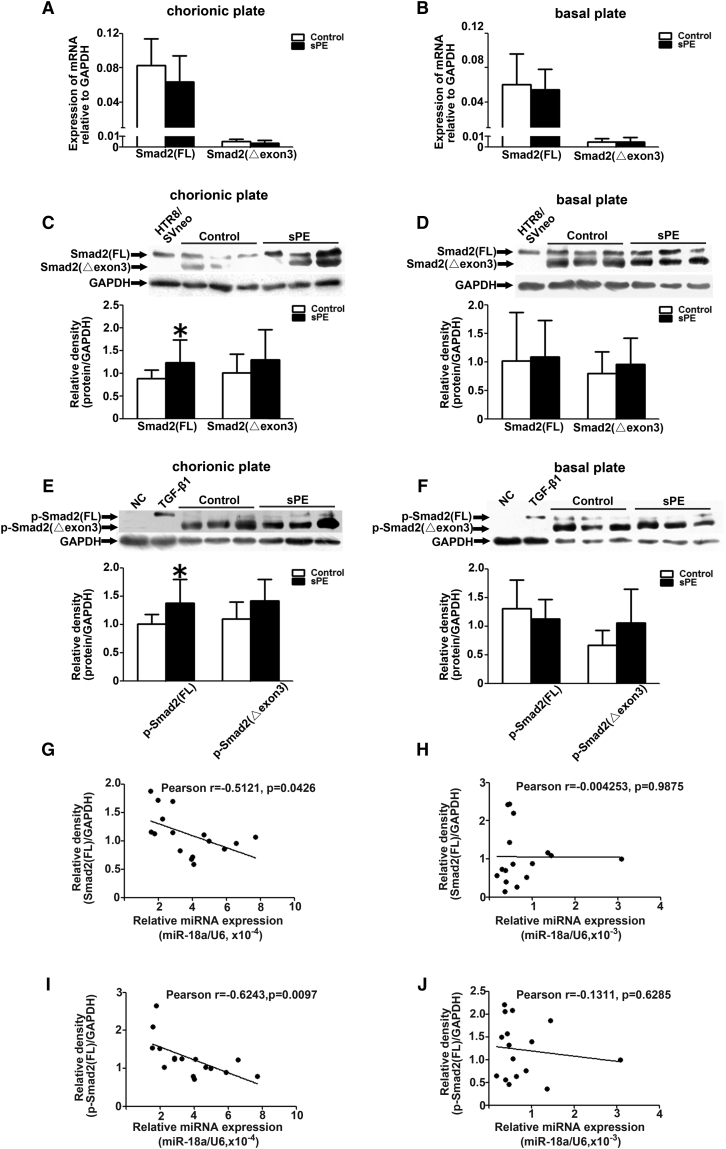
We compared the expression levels of Smad2(FL), Smad2(Δexon3), and Smad3 in placentas from 13 sPE patients and 32 normal pregnant women, both at mRNA and protein levels. The mRNA expression of Smad3, Smad2(FL), and Smad2(Δexon3) had no obvious difference between sPE and control placentas in neither chorionic nor basal plates (Figures 2A, 2B, 3A, and 3B). The protein levels of Smad3 and Smad2(Δexon3) also exhibited no significant difference in sPE and normal placentas in either chorionic or basal plates (Figures 2C, 2D, 3C, and 3D). However, the Smad2(FL) protein level was increased in the chorionic plate, but not basal plates, of sPE placentas (Figures 3C and 3D). Interestingly, the p-Smad2(FL) protein level in the sPE placentas was also significantly increased in the chorionic plate but not the basal plates. There were no significant differences of the p-Smad2(Δexon3) between normal and sPE placentas either in the chorionic plate or in the basal plate (Figures 3E and 3F). p-Smad2(Δexon3) and Smad2(Δexon3) could not be detected in the Htr8/SVneo cells (Figure 3C–3F).
Figure 2.
Differential Expression Patterns of Smad3 in Placentas Derived from sPE Patients and Normal Pregnant Women
(A–D) Quantitative real-time PCR (A and B) and western blotting (C and D) were performed to measure the expression of Smad3 in chorionic (A and C) and basal plates (B and D) of placentas derived from sPE patients (n = 13) and normal pregnant women (n = 32). Data are presented as mean ± SD.
Figure 3.
Differential Expression Patterns of Smad2 Isoforms and p-Smad2 Isoforms in Placentas Derived from sPE Patients and Normal Pregnant Women
(A and B) Quantitative real-time PCR were performed to measure the expression of the Smad2(FL) and Smad2(Δexon3) in chorionic (A) and basal plates (B) of placentas derived from sPE patients (n = 13) and normal pregnant women (n = 32). (C–F) Western blotting was performed to measure the expression of Smad2(FL), Smad2(Δexon3), p-Smad2(FL), and p-Smad2(Δexon3) in chorionic (C and E) and basal plates (D and F) of placentas derived from sPE patients (n = 8) and normal pregnant women (n = 8). (G–J) The inverse correlation between the miR-18a and Smad2(FL) (or p-Smad2(FL)) expression in the placental chorionic (G and I) and basal (H and J) plates of the studied individuals was shown. Data are presented as mean ± SD. ∗p < 0.05.
A correlation analysis of miR-18a concentration and Smad2(FL) (or p-Smad2(FL)) level was also performed accordingly. The data revealed an inverse correlation between miR-18a and Smad2(FL) (or p-Smad2(FL)) expression in the placental chorionic but not in the basal plates of the studied individuals (Figures 3G–3J).
The antibody against Smad2 detected two bands on the western blots (Figure 3), which raises a question of whether it is due to an antibody-specificity issue or whether there are two variants of the protein. In addition, apart from Smad2(Δexon3) (composed of 437 amino acids), three other Smad2 variants (composed of 425 amino acids, 418 amino acids, and 414 amino acids, respectively) could also be retrieved in the NCBI database (https://www.ncbi.nlm.nih.gov). To verify that the lower band is Smad2(Δexon3), the corresponding protein spot was cut from the middle of the lanes and used for mass-spectrum analysis. As shown in Figure S1, five conserved peptides and one Smad2(Δexon3)-specific peptide were found, respectively. The five conserved peptide motifs were SLDGR, NATVEMTRR, RHIGR, MSFVK, and MSFVKGWGAEYR, respectively. The Smad2(Δexon3)-specific peptide motif was LDELEK. These results confirmed that there are two variants of Smad2, wild-type (WT) and mutant (MUT), and the MUT Smad2 isoform is Smad2(Δexon3).
Colocalization of miR-18a and Smad2 in Human Placenta
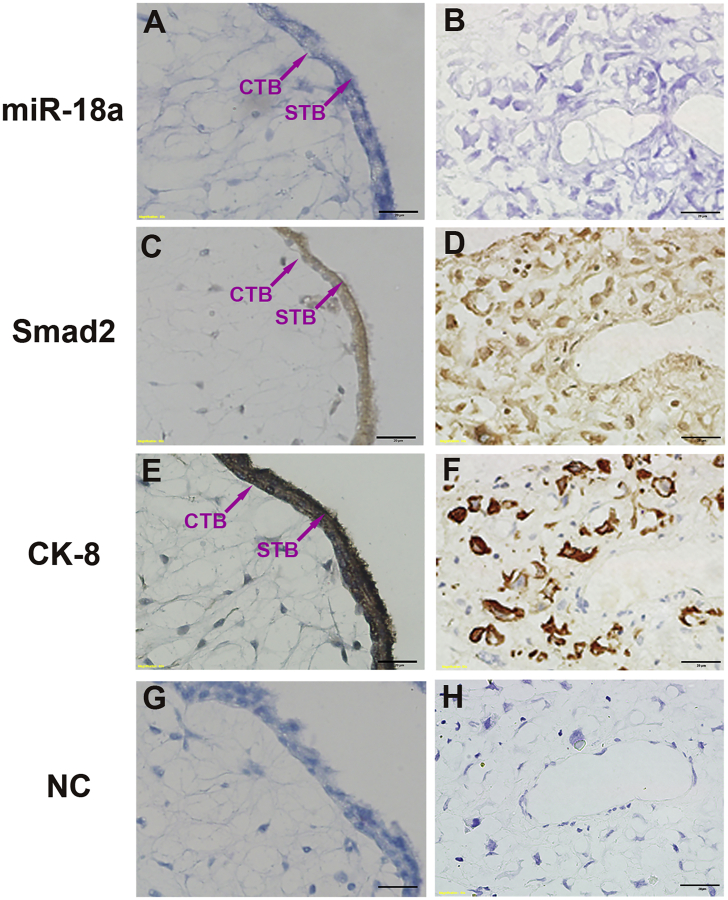
The localization of miR-18a and Smad2 in normal human placentas was examined by in situ hybridization and immunohistochemistry. Immunostaining for cytokeratin (CK)-8 was used as a marker for trophoblast cells (Figures 4E and 4F). miR-18a and Smad2 were both localized in the in placental tissues, derived from gestational weeks 7–8, in similar patterns. They were predominantly localized in various subtypes of trophoblasts, including villous cytotrophoblast cells, syncytiotrophoblasts (Figures 4A and 4C), and interstitial trophoblasts cells invading the decidual stroma (Figures 4B and 4D). They were also weekly expressed in some villous mesenchymal cells (Figures 4A and 4C). No hybridization signals were detected in decidual cells (Figures 4B and 4D). In the negative control (NC) hybridization and immunochemistry without probe or primary antibody, no positive signals were observed (Figures 4G and 4H), showing the specificity of the experiments.
Figure 4.
Colocalization of miR-18a and Smad2 in Human Placenta
(A–D) In situ hybridization and immunochemistry experiments showing the colocalization of miR-18a (A and B) and Smad2 (C and D) in human normal placentas at gestational weeks 7–8. (E and F) Immunostaining for CK-8 was used to mark the interstitial trophoblasts in placentas using a section adjacent to that used in (A)–(D). STB, syncytiotrophoblasts; CTB, cytotrophoblast cells; iEVT, interstitial extravillous trophoblast cells. Scale bars, 20 μm.
Verification of Primary Syncytiotrophoblasts and Cytotrophoblasts
We performed western blot analysis to detect syncytin-2 and human chorionic gonadotropin (hCG)-β of the primary syncytiotrophoblasts and cytotrophoblasts, which are markers for the cell type, which confirmed that syncytin-2 and hCG-β were present only in the primary cytotrophoblasts and syncytiotrophoblasts, respectively (Figure S2).
HTR8/SVneo Cell Line Was Selected as the Suitable Trophoblast Cell Line in This Study
We compared the expression level of miR-18a in three trophoblast cell lines (HTR8/SVneo, JEG-3, and B6-tert), as well as primary cultured cytotrophoblasts and syncytiotrophoblasts. We found that the HTR8/SVneo cells and the primary trophoblasts had comparable expression levels of miR-18a, whereas other two cell lines (JEG-3 and B6-tert) had a higher expression level than the primary cells (Figure S3), suggesting that HTR8/SVneo, rather than other two cell lines, had a miR-18a homeostasis similar to that in trophoblasts. Thus, the HTR8/SVneo cell line was selected as the experimental model to study the function of miR-18a in trophoblasts in vitro.
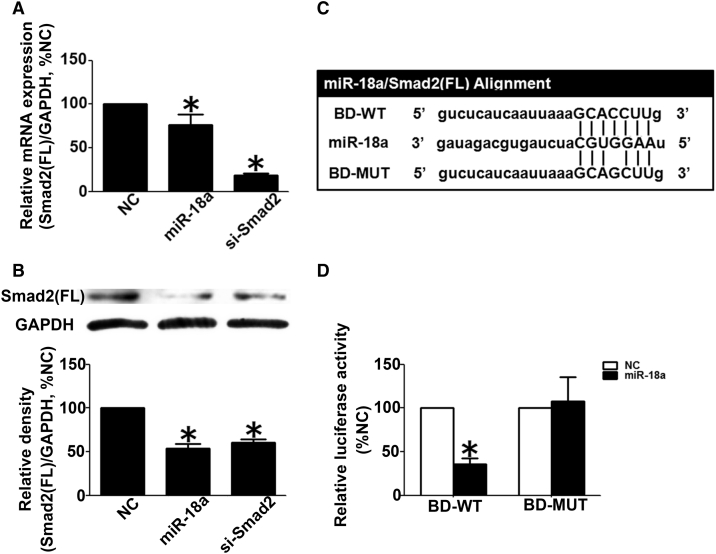
Smad2(FL) Is the Direct Target of miR-18a in Human Trophoblast Cells
As shown in Figures 5A and 5B, the transfection of miR-18a downregulated the expression of Smad2(FL) at both mRNA and protein levels, up to 76% and 54%, respectively. According to bioinformatics analysis, the seed sequence of miR-18a was complementary to 136 to 142 nt of 3ʹ UTR in Smad2(FL) mRNA. To further confirm that Smad2(FL) was the target of miR18a, the luciferase reporter construct carrying a 300-bp DNA fragment, including binding site (BD) of the 3ʹ UTR in human Smad2 mRNA (the vector was named BD-WT) or the point-mutated report construct (the vector was named BD-MUT) was transfected into HTR8/SVneo cells together with miR-18a mimics and thymidine kinase promoter-Renilla luciferase reporter plasmid (pRL-TK). 48 h after transfection, miR-18a mimics could evidently reduce the relative luciferase activity of the BD-WT construct by ≈35%, compared with scramble control, but could not affect the relative luciferase activity of the BD-MUT construct (Figures 5C and 5D).
Figure 5.
Validation of Smad2(FL) as the Direct Target of miR-18a in Human Trophoblast Cells
(A) Quantitative real-time PCR to reveal change of the Smad2(FL) mRNA level in HTR8/ SVneo cells transfected with negative control (NC), miR-18a mimics, or Smad2 siRNA. (B) Western blot analysis to show change of Smad2(FL) protein level in HTR8/SVneo cells transfected with NC, miR-18a, or si-Smad2. (C) Schematic representation of luciferase constructs used for reporter assays. The construct containing the region complementary to the seed sequence for miR-18a in the 3′ untranslated region (UTR) segment of the human Smad2(FL) gene is shown as BD-WT, and mutant construct is shown as BD-MUT (mutation sites). (D) Luciferase activity measured in HTR8/SVneo cells transfected with BD-WT or BD-MUT luciferase constructs together with miR-18a or NC. All experiments were performed with triplicate in at least 3 independent experiments. Data above are presented as mean ± SD. ∗p < 0.05.
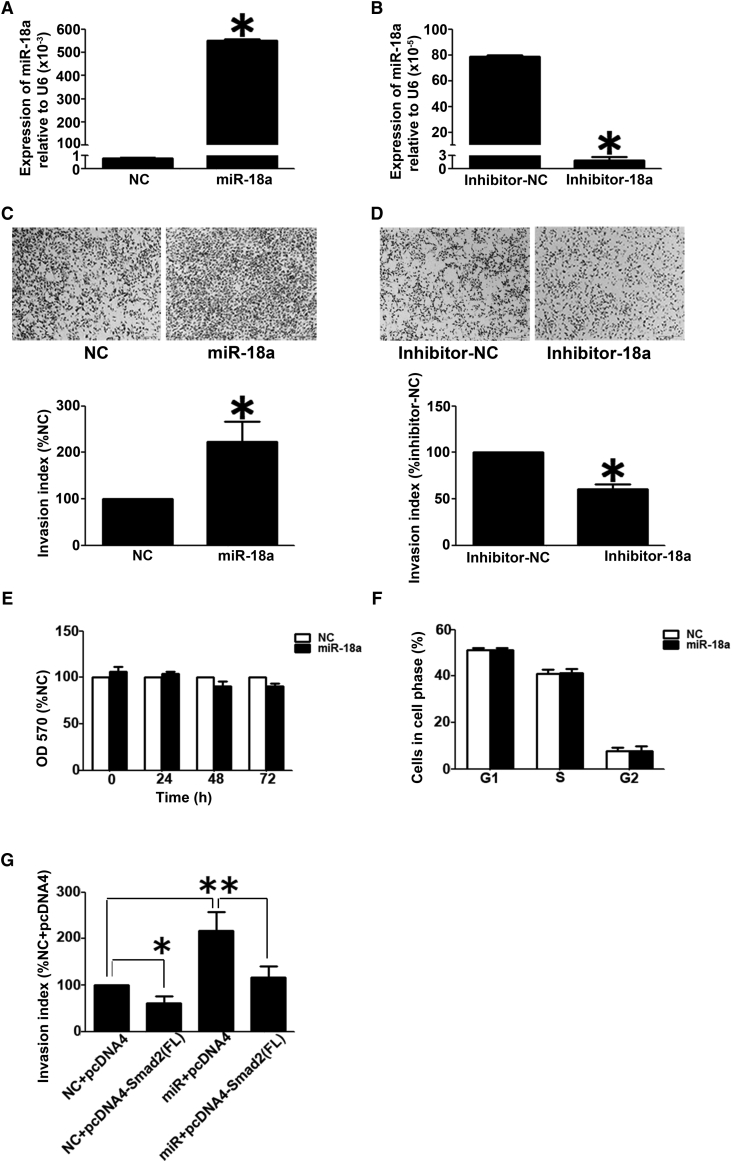
miR-18a Promoted Trophoblast Cell Invasion but Had No Effect on Cell Growth or Cell Cycle
The effects of miR-18a on trophoblast cell invasion and proliferation were examined. Data from quantitative real-time PCR experiments showed that the level of miR-18a increased to ∼690-fold of that of the scramble control cells after miR-18a mimics transfection (Figure 6A). miR-18a inhibitor decreased the level of miR18a by ∼98% (Figure 6B). As shown in Figures 6C and 6D, overexpression of miR-18a in Htr8/SVneo cells significantly promoted cell invasiveness (Figure 6C). Conversely, inhibition of miR-18a could inhibit the cell-invasive potential (Figure 6D). In addition, miR-18a did not have an effect on cell growth (Figure 6E) or cell cycle (Figure 6F).
Figure 6.
Effects of miR-18a on Cell Invasion and Proliferation in HTR8/SVneo Cells
(A and B) Quantitative real-time PCR results to reveal the miR-18a expression in HTR8/SVneo cells transfected with miR-18a mimics and scramble control (NC) (A) and miR-18a inhibitor or scramble inhibitor (inhibitor-NC) (B). The level of miR-18a was adjusted by the value of U6, and the relative value is presented as the mean ± SD, based on 3 independent experiments. ∗p < 0.05. (C and D) Transwell insert assay to examine cell invasiveness in HTR8/SVneo cells transfected with NC and miR-18a (C) and inhibitor NC or miR-18a inhibitor (D). The invasion index is presented as mean ± SD, based on 3 independent experiments. ∗p < 0.05. (E) The effect of miR-18a on the cell growth of HTR8/SVneo cells. The data are presented as the mean ± SD, according to the results of three independently repeated experiments. (F) The effect of miR-18a on the cell cycle of HTR8/SVneo cells. The data are presented as the mean ± SD, according to the results of three independently repeated experiments. (G) Smad2(FL) rescued the effect of miR-18a on cell invasion in HTR8/SVneo cells. The cells were transfected with miR-18a and Smad2 alone or in combination with scramble siRNA (NC) or pcDNA4 vector (pcDNA4) as corresponding NC. The Transwell insert assay was performed to monitor cell invasiveness. The bar chart represents statistical analysis based on 3 independently repeated experiments. Data are presented as mean ± SD. ∗p < 0.05.
Overexpression of Smad2(FL) Rescued the Invasion-Promoting Effect of miR-18a in HTR8/SVneo Cells
We performed a rescue experiment by transfecting HTR8/SVneo cells with miR-18a together with pcDNA4-Smad2(FL). Interestingly, the invasion-promoting effect of miR-18a was inhibited by the overexpressed Smad2(FL) (Figures 6G, S4A, and S4B).
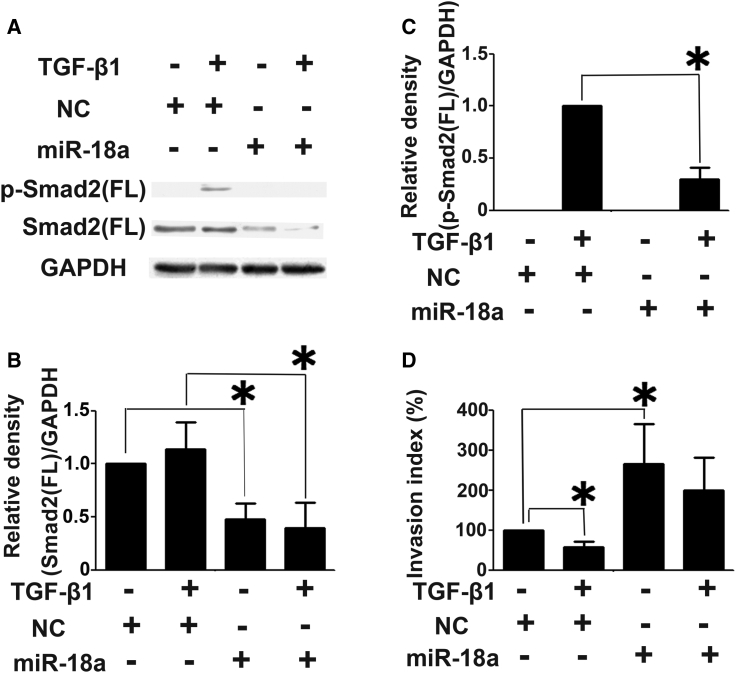
miR-18a Inhibits the Invasion Suppression by TGF-β Induced in Htr8/SVneo Cells
With the consideration of the antagonistic potential of TGF-β signaling on trophoblast cell invasion, we investigated whether miR-18a could counteract the invasion suppression activity of TGF-β. The Htr8/SVneo cells were transfected with miR-18a mimics and then treated with recombinant TGF-β protein. We observed that TGF-β stimulated Smad2(FL) activation and inhibited the invasion in HTR8/SVneo cells (Figures 7A–7D). More importantly, transfection of miR-18a mimics inhibited the upregulation of p-Smad2(FL) and suppression of cell invasion by TGF-β (Figures 7A–7D).
Figure 7.
miR-18a Blocks TGF-β Signaling in HTR8/SVneo Cells
(A–C) miR-18a inhibited Smad2(FL) expression and activation induced by TGF-β1. (D) miR-18a reversed the inhibitory effect of TGF-β1 on cell invasion. All experiments were carried out with triplicate in at least three independent experiments. The data above were presented as mean ± SD. ∗p < 0.05.
Discussion
Since the first major study conducted by Pineles et al.,8 more and more differentially expressed miRNAs have been identified in the placentas from PE patients. However, despite the wealth of studies that have suggested an association of certain miRNAs with PE, consensus has not yet been reached on which appears to be the key contributor to the pathogenesis of the condition and what the underlying mechanisms are. miR-18a is among the top 20 members of a list of differentially expressed miRNAs in PE placentas.31 With the consideration of the primary expression of miR-18a in various subtypes of placental trophoblasts,7 it is likely that miR-18a may have functional roles in the regulation of trophoblast cell behaviors.
Despite previous studies have demonstrated that this small RNA has biological functions in different cells,7,32, 33, 34, 35, 36 the existing results are conflicting. We proposed that such discrepancies might be due to the differences in microenvironments and regulatory networks in different kinds of cells. For instance, human placenta trophoblast cells share properties, such as active proliferation and invasion with tumor cells. However, these tumor-like behaviors in trophoblasts are tightly organized in unique temporal and spatial manners during pregnancy. In this study, we examined the expression profile of miR-18a in placentas at different gestational weeks and found that miR-18a was significantly upregulated during gestational weeks 8–10, followed by a decrease until term (Figure 1C), which is consistent with the previous results.37 Since trophoblast cells are highly invasive at gestational weeks 8–10 during the whole gestation,38,39 miR-18a may regulate trophoblast cell invasion. However, unlike the report in tumor cells, miR-18a substantially improved cell invasion (Figures 6C and 6D), but had no influence on cell growth and cell cycle in trophoblast cells (Figures 6E and 6F). These observations indicate that this small molecule has unique functions in placental trophoblast cells.
Our results have shown that miR-18a promoted trophoblast invasion via targeting Smad2(FL): (1) The expression of Smad2(FL) was markedly upregulated in PE placentas, whereas miR-18a was decreased (Figures 1A, 1B, 3C, and 3D).7 (2) miR-18a inhibited the expression of Smad2(FL) (Figures 5A and 5B).7 (3) The luciferase reporter assay demonstrated the direct binding of miR-18a to the 3′ UTR of Smad2(FL) gene (Figures 5C and 5D).7 (4) miR-18a was colocalized with Smad2 in human trophoblast cells (Figures 4A–4D). (5) The invasion-promoting effect of miR-18a could be partly attenuated by the overexpression of Smad2(FL) (Figure 6G).7
Notably, there exists a great difference in the miR-18a expression levels of control placentas (Figure 1C), and this difference may be due to the different genetic backgrounds of these control placentas. The mRNA levels of Smad2(Δexon3) in sPE and control placentas were much lower than those of Smad2(FL) (Figures 3A and 3B). In contrast, the protein levels of these two molecules were comparable in both sPE and control placentas (Figures 3C and 3D). The inconsistency may be due to the differences of translation efficiency, mRNA, and protein stability of these two molecules in the sPE and control placenta tissues. In addition, the protein level of p-Smad2(Δexon3) in control and sPE placentas was much higher than those of p-Smad2(FL) (Figures 3E and 3F). This discrepancy may be caused by the activities and specificities of phosphokinases and/or phosphatases, as well as the stabilities of these two phosphor proteins in the sPE and control placenta tissues.
The next step was to clarify which signal pathway was regulated by miR-18a in PE placentas. Smad2(FL) is an important downstream effector of the TGF-β superfamily.18 Aberrant expressions of several members in this superfamily have been implicated in PE pregnancies. For instance, productions of TGF-β and Nodal, as well as their receptors, were significantly enhanced in PE placentas, which may lead to over-activated signaling, excessive cell apoptosis, and less invasion.20,21,40 More importantly, in human placenta, TGF-β primarily regulates trophoblast cell invasion,22, 23, 24, 25, 26, 27 and Nodal promotes cell apoptosis.41 We found that miR-18a increased cell invasiveness (Figures 6C and 6D) but did not influence the cell growth or cell cycle (Figures 6E and 6F). Furthermore, members in the same miRNA cluster have similar functions.42 miR-18a belongs to miR-17-92 cluster,42 and our previous bioinformatic analysis demonstrated that the miR-17-92 cluster may regulate placenta development via fine tuning TGF-β signaling.7 These provide clues that miR-18a has a vital role in TGF-β signaling and PE, which is indeed supported by the experimental findings (Figures 3E, 3F, 7A, 7B, 7C, and 7D).
Our previous results have demonstrated the varying expression pattern of miRNAs in differential sampling sites of sPE placentas.7 Consistent with this, in this study, miR-18a, Smad2(FL), and p-Smad2(FL) exhibited differential expression patterns in sPE placentas, and they are differentially expressed in the chorionic plates of sPE placentas (Figures 1A, 1B, 3C, 3D, 3E, and 3F).7 miR-18a could promote trophoblast cell invasion via downregulating Smad2(FL) (Figure 6G). The improper synthesis of hCG and the dysregulated expression of HLA-C gene and PPI3 genes in the chorionic plates of human placenta are linked to the insufficient trophoblast cell invasion and the occurrence of PE.43, 44, 45 Therefore, the differential expressions of miR-18a, Smad2(FL), and p-Smad2(FL) in the chorionic plates of the sPE placentas may interfere with the synthesis of hCG, as well as the expression of the HLA-C gene and PPI3 gene, which is implicated in the control of the trophoblast cell invasion and the occurrence of sPE.
One of the most complex properties of miRNAs is the large number of targets that may exist for one miRNA. Our previous results have revealed that several target genes of miR-195 and miR-210 are involved in PE.32,46, 47, 48, 49, 50, 51 Furthermore, miR-195 can promote trophoblast cell invasion via downregulating activin receptor IIA (ActRIIA) and ActRIIB,46,48 whereas, miR-210 can inhibit trophoblast cell invasion via modulating potassium channel modulatory factor 1 and thrombospondin type I domain-containing 7A.50,51 Our results have shown that Smad2 was not activated without TGF-β treatment (Figure 7C), whereas miR-18a promoted cell invasion independent of TGF-β treatment (Figure 7D). Previous studies have also shown that miR-18a may promote the trophoblast cell invasion via targeting estrogen receptor (ER)-α.32 Therefore, we propose that miR-18a regulates the trophoblast cell invasion via modulating other target genes, and further experimental studies are warranted to elucidate the way that miR-18a and its targets are involved in causing the trophoblast cell dysfunction associated with PE.
The mechanism of miR-18a downregulation in PE placentas remains unknown. Our finding of lowered expression of pri-miR-18a in sPE placentas strongly indicates the aberrant transcription of this small RNA during the pathological process. Several transcription factors have been proven to be responsible for the transcription regulation of pri-miR-18a, such as c-myc52 and ER-α53. What transcription factors can account for the reduced pri-miR-18a and mature miR-18a expression in sPE placentas needs further investigation.
In addition, it is well known that primary cells retain most of their physical characteristics in vivo, thus mirroring the status and functionality of these cells under normal physiological conditions, making them well suited for use in studies of cell biology.42 However, for the limited number of each separation and the finite lifespan, the selection of a suitable cell line is urgently needed.42 Recently, research into the function of human trophoblasts has been largely restrained by a lack of proper cell models, despite considerable research efforts.54 In this study, by comparing the expression level of miR-18a in three trophoblast cell lines, as well as primary cultured cytotrophoblasts and syncytiotrophoblasts (Figure S2), the HTR8/SVneo cell line was selected as the suitable trophoblast cell line in this study. Previous results have showed that JEG-3 cells and the primary trophoblasts had comparable expression levels of several miRNAs.37 Such discrepancies may be due to the differences of miRNAs examined and/or the experimental settings, as well as platforms. Our results strongly suggest that the comparison of the expression levels of miRNAs among various trophoblast cell lines and the primary cells is a good approach for the selection of the suitable trophoblast cell model to study the function of miRNAs in human trophoblasts.
In conclusion, our results suggest that miR-18a inhibits Smad2(FL) expression and activation, which promote trophoblast cell invasion, which is likely via inhibition of TGF-β signaling. This, thus, plays an important role in the pathogenesis of PE (Figure 8).
Figure 8.
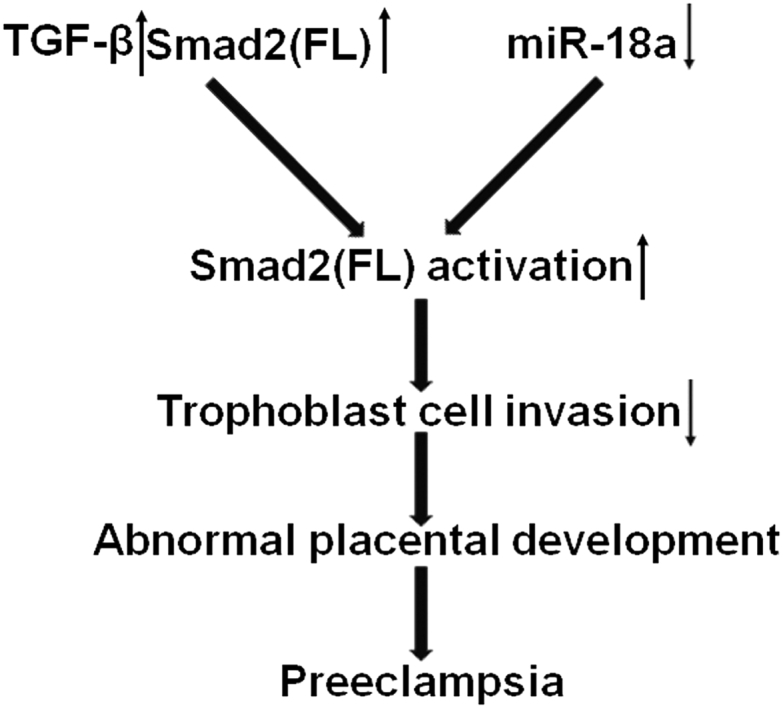
Proposed Roles of miR-18a in Placenta Development and Preeclampsia
In preeclamptic placenta, an increase in TGF-β expression and a decrease in miR-18a expression will cause overactivation of Smad2(FL), leading to insufficient invasion of trophoblast cells.
Materials and Methods
Subjects
In this study, all of the collection of human placenta tissues was performed with the permission of the Local Ethical Committee in the Institute of Zoology, Chinese Academy of Sciences, and in Peking University Third Hospital. The informed consents were obtained from all patients enrolled in this study.
Placenta samples from normal pregnancies and those from sPE women were obtained from Peking University Third Hospital. A total of 13 sPE patients and 32 normal pregnant women were enrolled in this study. sPE was diagnosed according to the definition in Williams Obstetrics (23rd edition).55 In brief, the patients had no history of pre-existing or chronic hypertension, but they showed systolic blood pressure of >160 mmHg or diastolic blood pressure of >110 mmHg on at least 2 occasions, accompanied by significant proteinuria (>2 g per 24 h or ≥3+ by dipstick in 2 random samples collected at >4-h intervals) after 20 weeks of gestation. Normal or uncomplicated pregnancy was defined as gestation in a previously normotensive woman who did not suffer from any complications during pregnancy and who delivered a healthy neonate with a weight adequate for a gestational age of more than 37 weeks of pregnancy.56 Women who developed renal disease, transient hypertension during pregnancy, multiple pregnancy, gestational diabetes, spontaneous abortion, intrauterine fetal death, fetal chromosomal or congenital abnormalities, or pregnancies conceived by fertility treatment were all excluded from this study. For each of the pregnant women in this cohort, placenta of the pregnant woman was collected within 1 h of caesarean birth, and specimens at the chorionic plate and the basal plate were separately taken from the placenta disc near the position of umbilical cord insertion. The specimens were snap frozen in liquid nitrogen until use. The clinical characteristics of the patients were summarized in Table 1.
Table 1.
Clinical Characteristics of the Pregnant Women Who Donated the Placentas for This Study
| Characteristics | Normal Pregnancy (n = 32) | Preeclampsia (n = 13) | p Value |
|---|---|---|---|
| Maternal age (years) | 31 ± 4 | 29 ± 5 | 0.211 |
| BMI (kg m−2) | 22 ± 2 | 22 ± 3 | 0.661 |
| SBP (mmHg) | 116 ± 6 | 157 ± 13a | 1.549E−07 |
| DBP (mmHg) | 77 ± 5 | 101 ± 8a | 2.094E−14 |
| 24 h urine protein (g) | N/D | 3 ± 2a | – |
| 50 g GCT (mM) | 7 ± 1 | 6 ± 2 | 0.062 |
| Gestational age at delivery (day) | 268 ± 7 | 256 ± 6a | 2.020E−06 |
Data are expressed as mean ± SD and compared by one-way ANOVA. SBP, systolic blood pressure; DBP, diastolic blood pressure; N/D, no data; GCT, glucose challenge test.
p < 0.05 versus normal pregnancy.
Tissues of human chorionic villi at gestational weeks 6–12 and placenta tissues at gestational weeks 24–27 were obtained at Beijing Haidian Hospital (Beijing, China) from patients who underwent spontaneous abortion and preterm labor, respectively. The gestational week of specimens was determined according to morphological observation and pathological examination, with the record of menstrual cycles as a reference.
Quantitative Real-Time PCR
Total RNA was extracted from cells and placenta tissues using TRIzol reagent (Life Technologies, CA, USA) following the manufacturer’s protocol. Two micrograms of total RNA was reverse transcribed into cDNA using oligo(dT) primer (Tiangen, Beijing, China) and Moloney murine leukemia virus reverse transcriptase (Promega, WI, USA). Reverse transcription of miRNA was performed with the use of the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China).
Quantitative real-time PCR was performed using Applied Biosystems 7500 (Life Technologies, CA, USA). For the detection of cDNA, the experiment was carried out using the SYBR Premix Ex Taq Kit (Takara, Dalian, China) following the manufacturer’s instructions. The reaction for each sample was carried out in duplicate at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. Quantitation of miRNA cDNA was achieved using the miRcute miRNA qPCR Detection Kit (Tiangen, Beijing, China), according to the manufacturer’s instructions. The reaction for each sample was carried out in duplicate at 94°C for 2 min, followed by 40 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 30 s. Human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 RNA was amplified in parallel as an internal control. The fold changes in mRNA and miRNA expression were calculated using the 2−ΔΔCq method, where ΔCq indicated the subtraction of the Cq of GAPDH or U6 from the mRNA or miRNA of interest, and ΔΔCq was calculated by subtracting the ΔCq. The sequences of primers of U6, miR-18a, GAPDH, pri-miR-18a, Smad2(FL), Smad2(Δexon3), and Smad3 were listed in Table 2.
Table 2.
Sequences of the Primers Used for Plasmid Construction or Real-Time PCR
| Product | Primer Sequence (5′ to 3′) |
|---|---|
| GAPDH real-time PCR | F: AAGGTCATCCCTGAGCTGAAC |
| GAPDH real-time PCR | R: ACGCCTGCTTCACCACCTTCT |
| pri-miR-18a real-time PCR | F: CAGTAAAGGTAAGGAGAGCTCA ATCTG |
| pri-miR-18a real-time PCR | R: CATACAACCACTAAGCTAAAG AATAATCTGA |
| miR-18a real-time PCR | F: TAAGGTGCATCTAGTGCAGATAG |
| U6 real-time PCR | F: CGCAAGGATGACACGCAAATTC |
| Smad2(FL) real-time PCR | F: TGGGCAGGAAGAAAAGTGGT |
| Smad2(FL) real-time PCR | R: GGCCTGTTGTATCCCACTGA |
| Smad2(Δexon3) real-time PCR | F: GAGCAGAATGGGCAGGAAGA |
| Smad2(Δexon3) real-time PCR | R: ACGACCATCAAGAGACCTTGG |
| BD-WT | F: GGACTAGTTCATAGCATTGTG TGTGGTCC |
| BD-WT | R: CCCAAGCTTTAAACAGCACAA TACTGTGA |
| BD-MUT | F: GGTCTCATCAATTAAAGCAGC TTGTGGAATCTGTTCCT |
| BD-MUT | R: AGGAAACAGATTCCACAAGC TGCTTTAATTGATGAGACC |
| Smad2(FL)-expressing plasmid | F: GGAATTCAAGAGGCTGTTTTC CTAGCGT |
| Smad2(FL)-expressing plasmid | R: CCGCTCGAGCATTAAGTCTTT TCATGGGAC |
F, forward; R, reverse.
Western Blotting
The proteins were extracted by lysing the cells or the homogenized tissues with Radio-Immunoprecipitation Assay (RIPA) buffer containing 150 mM NaCl, 10 mM Tris (pH 7.6), 1% Nonidet P-40 (NP-40), 0.5% Na deoxylcholate, 0.1% SDS, 1 mM NaF, Na3VO4 with 100× protease inhibitor cocktail (Sigma-Aldrich, MO, USA) and harvesting the supernatants after centrifuging. Protein concentration was measured by a bicinchoninic acid (BCA) protein assay (Boster Biological Technology, Wuhan, China), and 40 μg protein from each cell lysate was subjected to 10% SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare, CT, USA). The membrane was blocked for 1 h with 5% nonfat milk in PBST (PBS and 0.1% Tween 20) and incubated at 4°C overnight with Smad2 (R&D Systems, MN, USA), p-Smad2 (Cell Signaling Technology, MA, USA), Smad3 (Cell Signaling Technology, MA, USA), syncytin-2 (Abcam, Cambridge, UK), hCG-β (Abcam, Cambridge, UK), and GAPDH (Ambion, TX, USA) primary antibodies, respectively. The membrane was washed with PBST 3 times, incubated further with horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, PA, USA) at room temperature for 2 h, and washed 3 times with PBST. Final visualization was achieved by an enhanced chemiluminescence (ECL) kit (Thermo Scientific, MA, USA), and the signals were captured on X-ray films (Kodak, NY, USA) and analyzed by ImageJ software (NIH, MD, USA). The relative densities of Smad2, p-Smad2, and Smad3 protein were determined by normalization with a density value of GAPDH.
SDS-PAGE and Mass Spectrum Assays
40 μg of proteins for each sample was separated on a 10% SDS-PAGE gel. Protein bands were visualized by Coomassie blue R-250 staining. To identify the proteins, the corresponding protein spots were cut from the middle of the lanes, washed with water, and stored at −20°C. The samples were sent to Shanghai Applied Protein Technology (Shanghai, China) for mass spectrum analysis. Experiments were carried out on a Q Exactive mass spectrometer (Thermo Scientific, MA, USA), which was coupled with Easy-nLC 1000 (Thermo Scientific, MA, USA). The data of mass spectrum were analyzed by MASCOT software.
In Situ Hybridization for miRNAs
Paraffin sections of 5 μm in thickness were subjected to routine deparaffinization and rehydration and treated with protease K, followed by hybridization with the miRCURY LNA (locked nucleic acid) miRNA detection probes (Exiqon, Copenhagen, Denmark), specifically against miR-18a and NC, at 55°C overnight. The sections were serially washed in 5×, 2×, and 0.2× saline sodium citrate (SSC) buffer and then were incubated with alkaline phosphatase (AP)-conjugated anti-digoxygenin (DIG) antibody (Roche, IN, USA) at 37°C for 2 h. The stained signals were visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitro-blue tetrazolium chloride (BCIP/NBT; Promega, WI, USA) as a substrate. The scramble miRNA (NC) probe was used as a NC. The probe sequences (5′ to 3′) were the following: miR-18a, CTATCTGCACTAGATGCACCTTA, and NC, GTGTAACACGTCTATACGCCCA.
Immunohistochemistry
Paraffin sections of 5 μm in thickness were subjected to routine deparaffinization, rehydration, and antigen retrieval before being incubated with antibodies against human Smad2 (Proteintech, IL, USA) and CK-7 (Cell Signaling Technologies, MA, USA). The sections were further incubated with secondary antibody conjugated with horseradish peroxidase (Zhongshan Goldenbridge, Beijing, China) and were visualized with diaminobenzidine (Zhongshan Goldenbridge, Beijing, China) as the substrate.
Cell Culture and Transient Transfection
The human choriocarcinoma cell line, JEG-3, was purchased from American Type Culture Collection and cultured as previously described.11 The human trophoblast cell line HTR8/SVneo was kindly provided by Dr. C.H. Graham at Queen’s University, Canada, and cultured as previously described.7 Immortalized human cytotrophoblast cell line B6Tert-1 was cultured as previously described.54
The chorionic villi tissues were spliced into small pieces of approximately 3 cm × 2 cm in size and then were agitated violently in the PBS. The primary syncytiotrophoblasts could be isolated from the tissues. After centrifugation (800 rpm for 5 min), the primary syncytiotrophoblasts could be collected for further use (such as RNA extraction). Then, the primary cytotrophoblast cells could be isolated from the remaining tissues as previously described.57 The primary cytotrophoblast cells were cultured as previously described.57 To verify that the syncytiotrophoblasts were really isolated and separated from other cell types, the expression levels of syncytin-2 and hCG-β were examined in the proteins extracted from the primary cytotrophoblast cells and the primary syncytiotrophoblasts.
After HTR8/SVneo cells were grown to 60%–70% confluence, transient transfection of small interfering (si)RNA or miRNA duplexes was carried out using Lipofectamine 2000 (Life Technologies, CA, USA) following the manufacturer’s protocol.
Sequences and Constructs
The siRNA duplex against human Smad2, mature miRNA mimics for miR-18a, miR-18a inhibitor, and the scramble control was designed and purchased from Shanghai GenePharma, China. The sequences of siRNA and miRNA are shown in Table 3.
Table 3.
The Sequences of siRNA and miRNA
| Mimics | Sense (5′ to 3′) | Antisense (5′ to 3′) |
|---|---|---|
| Scramble control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| si-Smad2 | GGUUUACUCUCCAAUGUUATT | UAACAUUGGAGAGUAAACCTA |
| hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUAG | AUCUGCACUAGAUGCACCUUAUU |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA | – |
| Inhibitor-18a | CUAUCUGCACUAGAUGCACCUUA | – |
To generate pMIR-REPORT Luciferase plasmids for Smad2(FL), 3′ UTR segments of human Smad2 mRNA (50∼349 nt) containing putative miR-18a-binding sequences (136–142 nt) were amplified and cloned into pMIR-REPORT luciferase plasmid (Ambion, TX, USA) at the SpeI and HindIII sites. A point mutation was incorporated into the 139 nt of the 3′ UTR in the BD-WT plasmid to generate a BD-MUT construct. For the construction of the Smad2(FL)-expressing plasmid (pcDNA4-Smad2(FL)), the coding sequence of Smad2 was amplified and inserted into the pcDNA4.0 vector at the EcoRI and XhoI restriction sites. The sequences of primers for the construction of vectors are also listed in Table 2. All of the constructs were confirmed by DNA sequencing.
In addition, there exists a poorly conserved site in the 7,608–7,614 nt of the 3′ UTR of Smad2(FL) mRNA. The site type of this poorly conserved site is 7-mer-A1 but not 7-mer-m8, and several score values (such as context++ score, context++ score percentile) are too low. Therefore, we did not perform a luciferase assay to confirm this BD.
Luciferase Assay
For the luciferase assay, Htr8/SVneo cells (40,000 per well) were seeded into 24-well plates, 12 h before transfection. The cells were cotransfected with 80 ng of Smad2(FL) 3′ UTR luciferase reporter plasmid, 8 ng of Renilla luciferase reporter vectors (Promega, WI, USA), and 40 nM chemically synthesized, mature hsa-miR-18a or NC miRNAs with Lipofectamine 2000, according to the manufacturer’s protocol. Luciferase activity was assayed 48 h after transfection with a dual-lucifererase reporter assay system (Promega, WI, USA) and was measured with a luminometer (Promega, WI, USA). All transfections were carried out with triplicate replicates in at least three independent experiments.
Transwell Invasion Assay
A Transwell insert invasion assay was conducted in 24-well fitted inserts (8 μm pore size; Millipore, MA, USA). Briefly, 48 h after transfection, HTR8/SVneo cells were treated with 10 μg/mL mitomycin C for another 2 h and were trypsinized and seeded into Transwell inserts, precoated with 20 μg Matrigel (BD Biosciences, NJ, USA) with 200 μL serum-free culture medium. Lower chambers were loaded with the same medium with 10% fetal bovine medium. After incubating for 28 h, cells on the upper surface of membranes were completely removed, and the migrated cells were fixed with 0.25% glutaraldehyde and stained with hematoxylin. The number of invaded cells was counted in the whole field of the membranes under light microscope. A cell-invasion index was presented as the percentage of invaded cell number compared with that in the corresponding controls.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium (MTT) Assay
HTR8/SVneo cells transfected with miR-18a or NC miRNAs for 48 h were trypsinized and plated on 24-well culture plates at a density of 3 × 104 cells/well. Then after 24, 48, and 72 h, cells were incubated in 5 mg/mL MTT solution (Sigma Aldrich, CA, USA) for 4 h. After removal of MTT solution, dimethyl sulfoxide (DMSO) was added, and the absorbance at a wavelength of 490 nm (reference wavelength 690 nm) was recorded with a microplate reader (BioTek, VT, USA).
Cell-Cycle Analysis
As described by Xia et al.,58 HTR8/SVneo cells transfected with miR-18a or NC miRNAs for 72 h were harvested with trypsinization (0.25% trypsin) and fixed with 70% ethanol overnight at 4°C. The cells were further incubated with 120 μg/mL RNase A (Sigma-Aldrich, CA, USA) and 120 μg/mL propidium iodide (Sigma-Aldrich, CA, USA) at room temperature for 30 min before cell-cycle analysis. Then the distribution of cells in the different phases of the cell cycle was determined with flow cytometry (BD Biosciences, NJ, USA) and analyzed by CellQuest software (BD Biosciences, NJ, USA).
TGF-β1 Treatment
To investigate whether miR-18a could interfere with the TGF-β1 signaling in human trophoblast cells, we measured p-Smad2 in the cells to monitor the activation of TGF-β1 signaling. After the transfection with miRNA duplexes for 24 h and then cell starvation for 24 h, cells were treated with 5 ng/mL TGF-β1 (R&D Systems, MN, USA) for 15 min before protein extraction.
We also investigated whether there existed an additive effect of miR-18a and TGF-β1 on cell invasion. Htr8/SVneo cells transfected with miRNA duplexes for 48 h were used for Transwell invasion assay, supplemented with or without 2.5 ng/mL TGF-β1.
Statistical Analysis
All statistical analyses were performed with Statistical Package for the Social Sciences software (version 17.0; SPSS, IL, USA). The comparison among groups was estimated by one-way ANOVA, and p <0.05 was considered statistically significant.
Author Contributions
P.X., Y.W., C.P., Y.Z., and Y.-L.W. conceived and designed the experiments. P.X., Z.L., X.Y., X.S., and Y.-x.L. performed the experiments. Y.W. and Y.Z. contributed reagents/materials. P.X. and Y.-L.W. wrote the paper.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank Dr. C.H. Graham at Queen’s University, Canada, for the kind gift of HTR8/SVneo cells. We thank Dr. Shao Chin Lee of Jiangsu Normal University for his comment on the manuscript. This study was supported by grants from the National Key Research and Development Program of China (2016YFC1000401, 2017YFC1001404, and 2018YFC10041002) and the National Natural Science Foundation of China (81730040).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.019.
Contributor Information
Yangyu Zhao, Email: yangaogi@163.com.
Yan-Ling Wang, Email: wangyl@ioz.ac.cn.
Supplemental Information
References
- 1.Steegers E.A., von Dadelszen P., Duvekot J.J., Pijnenborg R. Pre-eclampsia. Lancet. 2010;376:631–644. doi: 10.1016/S0140-6736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- 2.Mol B.W.J., Roberts C.T., Thangaratinam S., Magee L.A., de Groot C.J.M., Hofmeyr G.J. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 3.Lyall F., Robson S.C., Bulmer J.N. Spiral artery remodeling and trophoblast invasion in preeclampsia and fetal growth restriction: relationship to clinical outcome. Hypertension. 2013;62:1046–1054. doi: 10.1161/HYPERTENSIONAHA.113.01892. [DOI] [PubMed] [Google Scholar]
- 4.Chen J.Z., Sheehan P.M., Brennecke S.P., Keogh R.J. Vessel remodelling, pregnancy hormones and extravillous trophoblast function. Mol. Cell. Endocrinol. 2012;349:138–144. doi: 10.1016/j.mce.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Chekulaeva M., Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Xu P., Zhao Y., Liu M., Wang Y., Wang H., Li Y.X., Zhu X., Yao Y., Wang H., Qiao J. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63:1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 8.Pineles B.L., Romero R., Montenegro D., Tarca A.L., Han Y.M., Kim Y.M., Draghici S., Espinoza J., Kusanovic J.P., Mittal P. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 2007;196:261.e1–261.e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhu X.M., Han T., Sargent I.L., Yin G.W., Yao Y.Q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 2009;200:661.e1–661.e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 10.Pan Q., Niu H., Cheng L., Li X., Zhang Q., Ning Y. Invasion of trophoblast cell lines is inhibited by miR-93 via MMP-2. Placenta. 2017;53:48–53. doi: 10.1016/j.placenta.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Gao W.L., Liu M., Yang Y., Yang H., Liao Q., Bai Y., Li Y.X., Li D., Peng C., Wang Y.L. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1) RNA Biol. 2012;9:1002–1010. doi: 10.4161/rna.20807. [DOI] [PubMed] [Google Scholar]
- 12.Liu C., Chen M., Wang M., Pi W., Li N., Meng Q. MiR-18a regulates myoblasts proliferation by targeting Fgf1. PLoS ONE. 2018;13:e0201551. doi: 10.1371/journal.pone.0201551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montoya M.M., Maul J., Singh P.B., Pua H.H., Dahlström F., Wu N., Huang X., Ansel K.M., Baumjohann D. A Distinct Inhibitory Function for miR-18a in Th17 Cell Differentiation. J. Immunol. 2017;199:559–569. doi: 10.4049/jimmunol.1700170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Wu L., Li D., Xu Y., Zhang L., Niu K., Kong R., Gu J., Xu Z., Chen Z., Sun J. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1α. Cancer Med. 2018;7:3834–3847. doi: 10.1002/cam4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Heldin C.H., Miyazono K., ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 18.Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goumans M.J., Liu Z., ten Dijke P. TGF-beta signaling in vascular biology and dysfunction. Cell Res. 2009;19:116–127. doi: 10.1038/cr.2008.326. [DOI] [PubMed] [Google Scholar]
- 20.Benian A., Madazli R., Aksu F., Uzun H., Aydin S. Plasma and placental levels of interleukin-10, transforming growth factor-beta1, and epithelial-cadherin in preeclampsia. Obstet. Gynecol. 2002;100:327–331. doi: 10.1016/s0029-7844(02)02077-x. [DOI] [PubMed] [Google Scholar]
- 21.Caniggia I., Grisaru-Gravnosky S., Kuliszewsky M., Post M., Lye S.J. Inhibition of TGF-beta 3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J. Clin. Invest. 1999;103:1641–1650. doi: 10.1172/JCI6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karmakar S., Das C. Regulation of trophoblast invasion by IL-1beta and TGF-beta1. Am. J. Reprod. Immunol. 2002;48:210–219. doi: 10.1034/j.1600-0897.2002.01151.x. [DOI] [PubMed] [Google Scholar]
- 23.Cheng J.C., Chang H.M., Leung P.C.K. TGF-β1 Inhibits Human Trophoblast Cell Invasion by Upregulating Connective Tissue Growth Factor Expression. Endocrinology. 2017;158:3620–3628. doi: 10.1210/en.2017-00536. [DOI] [PubMed] [Google Scholar]
- 24.Zuo Y., Fu Z., Hu Y., Li Y., Xu Q., Sun D., Tan Y. Effects of transforming growth factor-β1 on the proliferation and invasion of the HTR-8/SVneo cell line. Oncol. Lett. 2014;8:2187–2192. doi: 10.3892/ol.2014.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J., Sivasubramaniyam T., Yinon Y., Tagliaferro A., Ray J., Nevo O., Post M., Caniggia I. Aberrant TGFβ Signaling Contributes to Altered Trophoblast Differentiation in Preeclampsia. Endocrinology. 2016;157:883–899. doi: 10.1210/en.2015-1696. [DOI] [PubMed] [Google Scholar]
- 26.Cheng J.C., Chang H.M., Leung P.C. Transforming growth factor-β1 inhibits trophoblast cell invasion by inducing Snail-mediated down-regulation of vascular endothelial-cadherin protein. J. Biol. Chem. 2013;288:33181–33192. doi: 10.1074/jbc.M113.488866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao M.R., Qiu W., Li Y.X., Zhang Z.B., Li D., Wang Y.L. Dual effect of transforming growth factor beta1 on cell adhesion and invasion in human placenta trophoblast cells. Reproduction. 2006;132:333–341. doi: 10.1530/rep.1.01112. [DOI] [PubMed] [Google Scholar]
- 28.Takenoshita S., Mogi A., Nagashima M., Yang K., Yagi K., Hanyu A., Nagamachi Y., Miyazono K., Hagiwara K. Characterization of the MADH2/Smad2 gene, a human Mad homolog responsible for the transforming growth factor-beta and activin signal transduction pathway. Genomics. 1998;48:1–11. doi: 10.1006/geno.1997.5149. [DOI] [PubMed] [Google Scholar]
- 29.Ueberham U., Lange P., Ueberham E., Brückner M.K., Hartlage-Rübsamen M., Pannicke T., Rohn S., Cross M., Arendt T. Smad2 isoforms are differentially expressed during mouse brain development and aging. Int. J. Dev. Neurosci. 2009;27:501–510. doi: 10.1016/j.ijdevneu.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Faure S., Lee M.A., Keller T., ten Dijke P., Whitman M. Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development. 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- 31.Sheikh A.M., Small H.Y., Currie G., Delles C. Systematic Review of Micro-RNA Expression in Pre-Eclampsia Identifies a Number of Common Pathways Associated with the Disease. PLoS ONE. 2016;11:e0160808. doi: 10.1371/journal.pone.0160808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X., Yang Y., Han T., Yin G., Gao P., Ni Y., Su X., Liu Y., Yao Y. Suppression of microRNA-18a expression inhibits invasion and promotes apoptosis of human trophoblast cells by targeting the estrogen receptor α gene. Mol. Med. Rep. 2015;12:2701–2706. doi: 10.3892/mmr.2015.3724. [DOI] [PubMed] [Google Scholar]
- 33.Krutilina R., Sun W., Sethuraman A., Brown M., Seagroves T.N., Pfeffer L.M., Ignatova T., Fan M. MicroRNA-18a inhibits hypoxia-inducible factor 1α activity and lung metastasis in basal breast cancers. Breast Cancer Res. 2014;16:R78. doi: 10.1186/bcr3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Wang J., Cheng L., Lu M.P. miR-18a downregulates DICER1 and promotes proliferation and metastasis of nasopharyngeal carcinoma. Int. J. Clin. Exp. Med. 2014;7:847–855. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y.J., Wu H., Zhu J.M., Li X.D., Luo S.W., Dong L., Liu T.T., Shen X.Z. MicroRNA-18a modulates P53 expression by targeting IRF2 in gastric cancer patients. J. Gastroenterol. Hepatol. 2016;31:155–163. doi: 10.1111/jgh.13041. [DOI] [PubMed] [Google Scholar]
- 36.Song Y., Wang P., Zhao W., Yao Y., Liu X., Ma J., Xue Y., Liu Y. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 2014;324:54–64. doi: 10.1016/j.yexcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Morales-Prieto D.M., Chaiwangyen W., Ospina-Prieto S., Schneider U., Herrmann J., Gruhn B., Markert U.R. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33:725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Lash G.E., Otun H.A., Innes B.A., Bulmer J.N., Searle R.F., Robson S.C. Low oxygen concentrations inhibit trophoblast cell invasion from early gestation placental explants via alterations in levels of the urokinase plasminogen activator system. Biol. Reprod. 2006;74:403–409. doi: 10.1095/biolreprod.105.047332. [DOI] [PubMed] [Google Scholar]
- 39.Genbacev O., Joslin R., Damsky C.H., Polliotti B.M., Fisher S.J. Hypoxia alters early gestation human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J. Clin. Invest. 1996;97:540–550. doi: 10.1172/JCI118447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadeem L., Munir S., Fu G., Dunk C., Baczyk D., Caniggia I., Lye S., Peng C. Nodal signals through activin receptor-like kinase 7 to inhibit trophoblast migration and invasion: implication in the pathogenesis of preeclampsia. Am. J. Pathol. 2011;178:1177–1189. doi: 10.1016/j.ajpath.2010.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munir S., Xu G., Wu Y., Yang B., Lala P.K., Peng C. Nodal and ALK7 inhibit proliferation and induce apoptosis in human trophoblast cells. J. Biol. Chem. 2004;279:31277–31286. doi: 10.1074/jbc.M400641200. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Vecchiarelli-Federico L.M., Li Y.J., Egan S.E., Spaner D., Hough M.R., Ben-David Y. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119:4486–4498. doi: 10.1182/blood-2011-09-378687. [DOI] [PubMed] [Google Scholar]
- 43.Cole L.A. hCG and hyperglycosylated hCG in the establishment and evolution of hemochorial placentation. J. Reprod. Immunol. 2009;82:112–118. doi: 10.1016/j.jri.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Xiong S., Sharkey A.M., Kennedy P.R., Gardner L., Farrell L.E., Chazara O., Bauer J., Hiby S.E., Colucci F., Moffett A. Maternal uterine NK cell-activating receptor KIR2DS1 enhances placentation. J. Clin. Invest. 2013;123:4264–4272. doi: 10.1172/JCI68991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huppertz B., Meiri H., Gizurarson S., Osol G., Sammar M. Placental protein 13 (PP13): a new biological target shifting individualized risk assessment to personalized drug design combating pre-eclampsia. Hum. Reprod. Update. 2013;19:391–405. doi: 10.1093/humupd/dmt003. [DOI] [PubMed] [Google Scholar]
- 46.Bai Y., Yang W., Yang H.X., Liao Q., Ye G., Fu G., Ji L., Xu P., Wang H., Li Y.X. Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS ONE. 2012;7:e38875. doi: 10.1371/journal.pone.0038875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Zhang L., Guo X., Bai Y., Li Y.X., Sha J., Peng C., Wang Y.L., Liu M. MiR-195 modulates oxidative stress-induced apoptosis and mitochondrial energy production in human trophoblasts via flavin adenine dinucleotide-dependent oxidoreductase domain-containing protein 1 and pyruvate dehydrogenase phosphatase regulatory subunit. J. Hypertens. 2018;36:306–318. doi: 10.1097/HJH.0000000000001529. [DOI] [PubMed] [Google Scholar]
- 48.Wu H., Wang H., Liu M., Bai Y., Li Y.X., Ji L., Peng C., Yu Y., Wang Y.L. MiR-195 participates in the placental disorder of preeclampsia via targeting activin receptor type-2B in trophoblastic cells. J. Hypertens. 2016;34:1371–1379. doi: 10.1097/HJH.0000000000000948. [DOI] [PubMed] [Google Scholar]
- 49.Wang H., Zhao Y., Luo R., Bian X., Wang Y., Shao X., Li Y.-X., Liu M., Wang Y.-L. A positive feedback self-regulatory loop between miR-210 and HIF-1alpha mediated by CPEB2 is involved in trophoblast syncytiolization: implication of trophoblast malfunction in preeclampsia. Biol. Reprod. 2020;102:560–570. doi: 10.1093/biolre/ioz196. [DOI] [PubMed] [Google Scholar]
- 50.Luo R., Shao X., Xu P., Liu Y., Wang Y., Zhao Y., Liu M., Ji L., Li Y.X., Chang C. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1. Hypertension. 2014;64:839–845. doi: 10.1161/HYPERTENSIONAHA.114.03530. [DOI] [PubMed] [Google Scholar]
- 51.Luo R., Wang Y., Xu P., Cao G., Zhao Y., Shao X., Li Y.X., Chang C., Peng C., Wang Y.L. Hypoxia-inducible miR-210 contributes to preeclampsia via targeting thrombospondin type I domain containing 7A. Sci. Rep. 2016;6:19588. doi: 10.1038/srep19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 53.Castellano L., Giamas G., Jacob J., Coombes R.C., Lucchesi W., Thiruchelvam P., Barton G., Jiao L.R., Wait R., Waxman J. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y.L., Qiu W., Feng H.C., Li Y.X., Zhuang L.Z., Wang Z., Liu Y., Zhou J.Q., Zhang D.H., Tsao G.S. Immortalization of normal human cytotrophoblast cells by reconstitution of telomeric reverse transcriptase activity. Mol. Hum. Reprod. 2006;12:451–460. doi: 10.1093/molehr/gal054. [DOI] [PubMed] [Google Scholar]
- 55.Cunningham F.G., Leveno K.J., Bloom S.L., Hauth J.C., Rouse D.J., Spong C.Y. Williams Obstetrics. Twenty-third Edition. The McGraw-Hill Companies; 2009. Pregnancy hypertension; pp. 706–756. [Google Scholar]
- 56.2000). Report of the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy. Am. J. Obstet. Gynecol. 183, S1–S22. [PubMed]
- 57.Li R.H., Luo S., Zhuang L.Z. Establishment and characterization of a cytotrophoblast cell line from normal placenta of human origin. Hum. Reprod. 1996;11:1328–1333. doi: 10.1093/oxfordjournals.humrep.a019381. [DOI] [PubMed] [Google Scholar]
- 58.Xia H., Zhang W., Li Y., Guo N., Yu C. EZH2 silencing with RNA interference induces G2/M arrest in human lung cancer cells in vitro. BioMed Res. Int. 2014;2014:348728. doi: 10.1155/2014/348728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.