Abstract
The world is currently facing a novel viral pandemic (SARS-CoV-2), and large-scale testing is central to decision-making for the design of effective policies and control strategies to minimize its impact on the global population. However, testing for the presence of the virus is a major bottleneck in tracking the spreading of the disease. Given its adaptability regarding the nucleotide sequence of target regions, RT-qPCR is a strong ally to reveal the rapid geographical spreading of novel viruses. We assessed PCR variations in the SARS-CoV-2 diagnosis taking into account public genome sequences and diagnosis kits used by different countries. We analyzed 226 SARS-CoV-2 genome sequences from samples collected by March 22, 2020. Our work utilizes a phylogenetic approach that reveals the early evolution of the virus sequence as it spreads around the globe and informs the design of RT-qPCR primers and probes. The quick expansion of testing capabilities of a country during a pandemic is largely impaired by the availability of adequately trained personnel on RNA isolation and PCR analysis, as well as the availability of hardware (thermocyclers). We propose that rapid capacity development can circumvent these bottlenecks by training medical and non-medical personnel with some laboratory experience, such as biology-related graduate students. Furthermore, the use of thermocyclers available in academic and commercial labs can be promptly calibrated and certified to properly conduct testing during a pandemic. A decentralized, fast-acting training and testing certification pipeline will better prepare us to manage future pandemics.
Electronic supplementary material
The online version of this article (10.1007/s00109-020-01992-x) contains supplementary material, which is available to authorized users.
Keywords: COVID-19, Coronavirus, Infrastructure, Polymerase chain reaction, Testing
Introduction
The World Health Organization (WHO) received notification on December 31, 2019 of pneumonia cases of unknown etiology in Wuhan, China. On January 11, 2020, the Chinese authorities identified a new type of coronavirus, which was isolated on January 7, 2020, which allowed its rapid sequencing and the public data deposition on January 12 to help with the development of diagnostic kits [1]. WHO declared on March 11, 2020, a pandemic the coronavirus disease 2019 (COVID-19), which is caused by SARS-CoV-2. In March 21, after nearly 3 months of the first notification, over 291,000 thousand cases were confirmed, and 12,776 deaths were reported worldwide [2]. As of May 1, 2020, over 3.2 million cases have been confirmed globally, with a death toll surpassing 234,000 people [3].
In addition to mitigation approaches (e.g., promotion of hygiene, social distancing, isolation of infected people, and restricting traveling), comprehensive testing of the infection in the population is central to track the disease spreading as well as inform public policies. It has been suggested that the demand for health services can only be maintained at manageable levels through a prompt adoption of public health measures to suppress virus spreading [4]. Indeed, countries that have adopted broad testing strategies early have better succeeded in limiting the spread of the disease, such as South Korea, Vietnam, and New Zealand [5].
Ideally, tests should be easy to sample and analyze, quick to return results, accurate and precise, scalable, and inexpensive. Often, antibody-based point-of-care tests (POCT) fit this description. However, rapidly evolving epidemics due to novel viruses do not allow the timely development of antibody-based tests. Thus, viral load tests based on real-time, quantitative RT-PCR (referred herein as RT-qPCR) are an ideal platform for the rapid development of test kits due to the easy adaptability to the nucleotide sequence of the target.
Currently, RT-qPCR is a reliable test widely used for the detection of symptomatic and asymptomatic patients infected with SARS-CoV-2 [6, 7] with a technical limit of detection (LOD) <10 copies/reaction [8], and a detection threshold of 3.8 RNA molecules per reaction, depending on the amplified region and the primers and probes used in the analysis [9]. Indeed, although RT-qPCR requires special equipment, it allows for a relatively simple and rapid diagnosis by amplifying segments of the coronavirus genetic material with high specificity and reliability. Multiple research and clinical institutions around the world have developed molecular assays to diagnose SARS-CoV-2 and made the sets of RT-qPCR primers and probes publicly available (Table 1).
Table 1.
Summary of available protocols (adapted from WHO–House Assays, 2020)
| Country | Institute | Target genes |
|---|---|---|
| China | Center for Disease Control and Prevention (CDC) | ORF1ab, N |
| France | Institut Pasteur | RdRP (2 targets) |
| Germany | Charité - Universitätsmedizin Berlin | RdRP, E, N |
| Hong Kong SAR | The University of Hong Kong (HKU) | ORF1b-nsp14, N |
| Japan | National Institute of Infectious Diseases (NIID) | Pancorona, multiple targets, spike protein |
| Thailand | National Institute of Health (Thai NIH) | N |
| USA | Centers for Disease Control and Prevention (CDC) | N (3 target regions) |
Extensive testing of the population (and the subsequent isolation of those infected) is essential to containing a pandemic and avoiding a premature collapse of entire national health systems. For example, Li et al. [10] used infection data to model the spreading of the novel coronavirus in China before travel restrictions were imposed. They concluded that undiagnosed SARS-CoV-2 carriers accounted for 79% of the cases and the primary source of infection, which rapidly spread the disease across the globe [10]. However, as the global demand for RT-qPCR testing kits surges abruptly during a fast-spreading pandemic, commercial suppliers are unable to deliver kit components in a timely manner, which prevents the effective assessment of the disease spreading in the population in order to identify and isolate those infected and avoid further spreading.
The present work aimed to contribute to the study of SARS-CoV-2 diagnosis through the analysis of the virus genome sequences that were deposited at the NCBI GenBank as it spread throughout the world as well as to study the various testing approaches recommended by official government authorities from several countries in order to assess methodological testing differences and to indicate prospects of effective diagnosis via RT-PCR. Our approach underscores the importance of analyzing the evolution of genome sequences of novel viruses as it spreads around the globe, allows for the revision of adopted testing reagents, and contributes to avoiding the development of imprecise molecular tests based on mutated regions, as already observed [11, 12].
Methods
Phylogenetic analysis
The keyword “SARS-CoV-2” was used to search for complete genomes of the novel coronavirus deposited to the National Center for Biotechnology (NCBI) nucleotide database (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/). We analyzed 226 SARS-CoV-2 genome sequences deposited at the NCBI GenBank from samples collected by March 22, 2020 (Online Resource 1). Following, we selected the complete genome sequences and sorted them according to the collection dates (Table 2). We only considered the first complete genome deposited from each country for further analyses, since this study did not intend to propose new molecular assays for the diagnosis of SARS-CoV-2 but rather demonstrate the importance of genomic analysis of the virus, especially regarding disease dissemination and validation of assays already developed via RT-PCR.
Table 2.
Complete genome sequences of the novel coronavirus (SARS-CoV-2) deposited at the NCBI GenBank by March 22, 2020
| GenBank ID | Collection date | Location | Genome size (bp) |
|---|---|---|---|
| MN908947 | 2019-12 | China: Wuhan | 29,903 |
| LC529905 | 2020-01 | Japan | 29,903 |
| MT039890 | 2020-01 | South Korea | 29,903 |
| MT072688 | 2020-01-13 | Nepal | 29,811 |
| MN985325 | 2020-01-19 | USA: WA | 29,882 |
| MT192772 | 2020-01-22 | Vietnam | 29,891 |
| MT007544 | 2020-01-25 | Australia: Victoria | 29,893 |
| MT192759 | 2020-01-25 | Taiwan | 29,862 |
| MT066156 | 2020-01-30 | Italy | 29,867 |
| MT050493 | 2020-01-31 | India: Kerala State | 29,851 |
| MT093571 | 2020-02-07 | Sweden | 29,886 |
| MT233519 | 2020-02-27 | Spain: Valencia | 29,782 |
| MT126808 | 2020-02-28 | Brazil | 29,876 |
| MT240479 | 2020-03-04 | Pakistan: Gilgit | 29,836 |
Nucleotide sequences were utilized to calculate the phylogenetic distances through the tool ClustalW [13] using default parameters. The tree was generated through the MEGA 7.0 software [14], with branches inferred using the Maximum Likelihood method based on the Hasegawa-Kishino-Yano model [15].
Virus mutations and molecular assays developed in-house
For the analysis of amplified regions used in diagnostics, we used the tool ClustalW [13] to align all regions of the 14 SARS-CoV-2 complete genome sequences against the primer and probe sequences recommended for diagnosis by various research and clinical institutions around the world (Table 1). The images displaying nucleotide sequences alignments were generated by GeneDoc (http://www.nrbsc.org/gfx/genedoc/; Online Resource 2). Additionally, we searched the Web of Science database for studies on PCR variations used to diagnose viruses and compared the methods.
Results
Phylogenetics
Through the analysis of the 14 complete genomes available (Fig. 1) available during the early stages of the pandemic, we could trace the geographical spreading of SARS-CoV-2 since the beginning of the pandemic [1]. Accordingly, the phylogenetic classification of complete genome sequences can be used to trace sources of infection and inform clinical and outcomes of novel viruses and should be considered during the design of treatments and, eventually, vaccines [12]. While several therapeutic approaches to treat COVID-19 are being developed in the course of the pandemic [16], tracking the evolution of the virus may also be helpful to determine the impact on the host, including high-risk populations and genetic variations associated with different responses, as well as to inform vaccine development and interventions (cf. [17]).
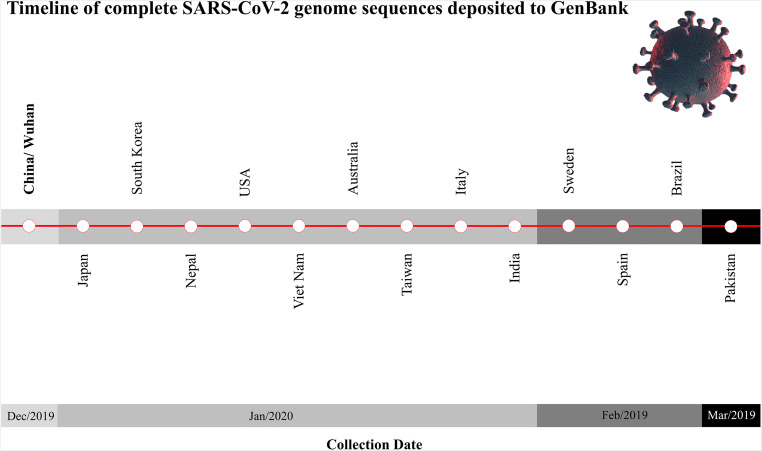
Fig. 1.
Timeline of complete SARS-CoV-2 genome sequences deposited to GenBank, according to their collection dates. The viral structure image was designed using resources at Freepik.com
Our phylogenetic analysis (Fig. 2) shows the formation of three distinct clades. In line with our findings, a recent study that analyzed 160 complete SARS-CoV-2 genomes also found three central variants named A, B, and C. The A variant was the ancestral type and closest to a coronavirus from bats and pangolins. The B variant had a higher incidence in East Asia, while the C variant was found first in patients from France, Italy, and Sweden and is the prevalent version in Europe [12]. This analysis also revealed the main origins of the virus in other countries, such as Brazil, which reported its first sequenced genome from a patient who had returned from Italy. Indeed, in our phylogenetic reconstruction, Brazil and Italy fell in the same clade (variant C), alongside Australia, Sweden, and South Korea, which in line with the report by Forster et al. [12]. In a recent study, 54.8% of the SARV-CoV-2 cases in Brazil by March 5, 2020, were estimated to be originated from contaminated travelers arriving from Italy [22].
Fig. 2.
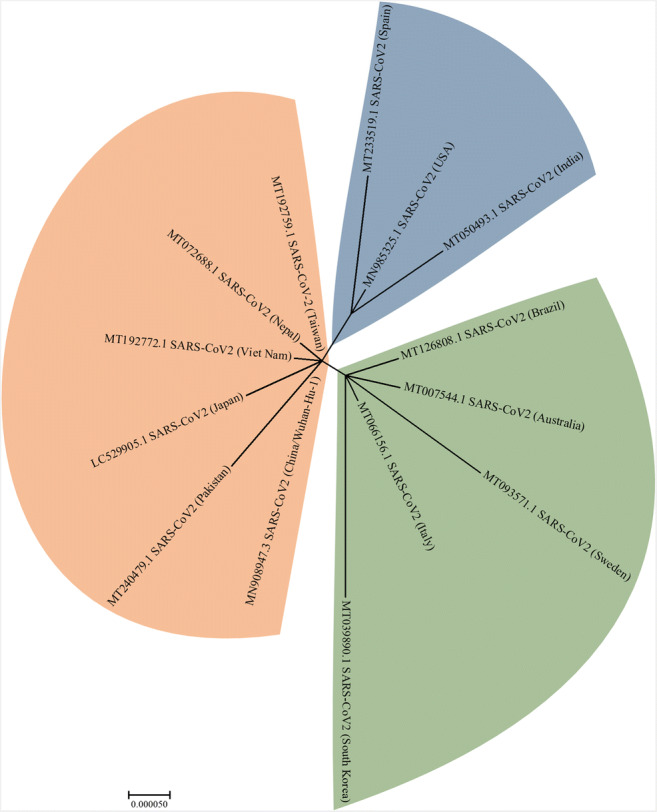
Phylogenetic analysis of 14 complete SARS-CoV-2 genome sequences published by several countries. The molecular phylogenetic analysis was conducted in MEGA7 and inferred by the maximum likelihood method on the Hasegawa-Kishino-Yano model. For more complete studies on the evolution of SARS-CoV-2 during the pandemic and follow-up discussions on proper methodologies of phylogenetic analyses, please refer to [18–21]
Phylogenetic analyses assessing the origin of the novel coronavirus have already been published. These studies revealed that bats might be the primary reservoir of SARS-CoV-2 [23–25]. Therefore, phylogeny is an essential tool to monitor the evolution of pandemic pathogens and can help, considering its limitations, in tracing the spreading of viruses. However, such studies must utilize representative datasets and the correct analysis methods to produce results with the necessary robustness for monitoring lethal outbreaks, such as for the novel coronavirus [18–21].
Mutations and molecular assays developed in-house
The study of genome mutations of pathogens as epidemics spread can help us better understand emerging outbreaks [26]. This type of data is essential, as it can indicate the frequency and extent of the genetic variation of novel viruses. The alignment of complete SARS-CoV-2 genomes revealed that the new coronavirus had undergone several mutations (Online Resource 2). Korber et al. [27] developed an analysis pipeline to facilitate real-time mutation tracking of SARS-CoV-2. This analysis initially focused on the spike (S) protein because it mediates cell infection, and it is the focus of most vaccine strategies and antibody-based therapeutics. Using this method, the authors identified fourteen mutations that are accumulating in S [27]. In the host, a major focus of study is the gene ACE, which is directly involved with SARS-CoV-2 infection and which genetic variations may be involved with the severity of the disease and may help explain why the virus is hitting southern Europe so hard [28]. In our study, however, we focused our analysis on the regions used for diagnosis via PCR.
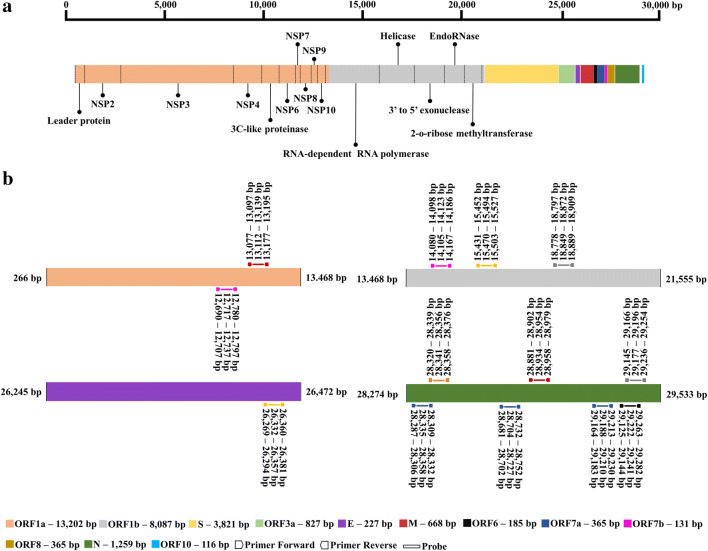
We examined the regions used for the design of SARS-CoV-2 primer and probe sets by leading official institutions around the world (Fig. 3). We noticed that the most common genomic region of the virus used for diagnosis comprises the gene “N”, followed by ORF1b, ORF1a, and the gene “E” (Fig. 3). Five institutions from four countries (Thailand, China (The University of Hong Kong (HKU) and Center for Disease Control and Prevention (CDC), the United States, and Japan) recommended the “N” gene region for diagnosis, with at least one set of primers and probe. The US Centers for Disease Control and Prevention (CDC) developed a SARS-CoV-2 Detection Panel composed of three sets of primers and probes for that region, which comprise a universal set for SARS beta-coronaviruses, and two other sets specific for SARS-CoV-2 [29]. Chu et al. [8] developed two quantitative assays via RT-qPCR to detect two different regions of the viral genome (ORF1b and gene ‘N’—HKU-Hong Kong/China) and demonstrated that could detect SARS-CoV-2 < 10 copies/reaction [8]. On the other hand, Corman et al. [9] developed an assay targeting the genes N, E and ORF1b (RNA-dependent RNA polymerase/RdRP) (Charité/Germany) and obtained the best sensitivity with the respective limits of detection (LOD): 8.3, 5.2 and 3.8 copies/reaction, at the 95% detection probability [9].
Fig. 3.
a Representation of the SARS-CoV-2 genome sequence depicting annotated genes. This scheme uses the nucleotide coordinates of a virus sequenced in China (NC_045512). b Positions of the primer/probe targets used for detection by institutions in six countries. Colors of primers and probes correspond to countries: China, CDC (red); USA, CDC (blue); Germany (yellow); China/Hong Kong (gray); France (pink); Japan (black); and Thailand (orange). The numbers above each primer and probe indicate nucleotide position (coordinate) in the genome. Important details: France developed two assays for the RdRP gene (identified in the figure as ORF1a and ORF1b, according to the reference sequence [NC_045512] used). The assay developed by Germany for gene E was also tested in France. Germany developed two probes for the same RdRP gene region indicated in the image, one being specific for SARS-CoV-2 and another common for SARS-CoV-2, SARS-CoV, and bat SARS-related coronavirus, but only one assay is indicated in the image. Japan also indicated assays for nested RT-PCR, but the image above only represents RT-qPCR assays. An important aspect of RT-qPCR assay development for viral detection is confirming that primers and probes are specific to the virus of interest and do not detect viruses from the same family. This is particularly important for coronaviruses, since members of this group already circulate in the human population
Our SARS-CoV-2 genome alignment also revealed that the mutations (Online Resource 2) were outside of the annealing sequences designed for primers and probes (Fig. 3), that is except for the virus circulating in South Korea, which holds a mutation in the region of a probe recommended by two European organizations, the Charité - Universitätsmedizin Berlin and the Pasteur Institute of France. Therefore, the tests developed by these institutions may be ineffective to accurately detect that virus version. Indeed, mutations lying within primer annealing sites have already been reported for SARS-CoV-2 [30].
Variations of PCR techniques and diagnosis methods
Since the genetic code of coronaviruses consists of RNA, a critical step of RT-qPCR protocols is RNA purification from the testing samples. Specific RNA isolation kits for SARS-CoV-2 detection were officially recommended by institutions in six countries (Table 3). All the protocols endorsed included RNA extraction by affinity columns that allow automation to optimize time and minimize errors.
Table 3.
Kits for viral RNA extraction and detection used by institutions from six countries listed by WHO
| Kits utilized (company) | Country | |
|---|---|---|
| Virus RNA extraction | MagNA Pure 96 System (Roche) | Germany |
| QIAamp Viral RNA Mini Kit (Qiagen) | Hong Kong, Japan, USA | |
| NucleoSpin RNA Virus - Macherey (Nagel) | France, Thailand | |
| QIAamp DSP Viral RNA Mini Kit (Qiagen) | USA* | |
| Virus detection: RT reaction and qPCR platform | SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (ThermoFisher) | Germany |
| TaqMan Fast Virus 1-Step Master Mix (ThermoFisher) | Hong Kong | |
| SuperScript III Platinum One-Step qRT-PCR Kit (ThermoFisher) | France, Thailand | |
| QuantiTect Probe RT-PCR Kit (Qiagen) | Japan** | |
| TaqPath 1-Step RT-qPCR Master Mix, CG (ThermoFisher) | USA | |
*The US CDC indicates the possibility of using several RNA extraction kits. This table only lists the first kit indicated by each country
**The Japan NIID also indicates the reagents for nested RT-PCR
All official institutions (Table 3) recommended using one-step RT-qPCR (the reverse transcriptase reaction automatically precedes the PCR phase in the same tube), which is regarded as the gold standard for viral detection and diagnosis (Fig. 4). Most of the endorsed assays were designed to detect two regions of the SARS-CoV-2 genome. However, the diagnosis of SARS-CoV-2 can also be performed by conventional PCR reaction after reverse transcription (RT-PCR), nested RT-PCR reaction (i.e., two successive PCRs with the second reaction using primers that anneal internally to the first amplicon) or via two-step RT-qPCR reaction (two successive reactions with the cDNA synthesis via reverse transcription preceding the PCR).
Fig. 4.
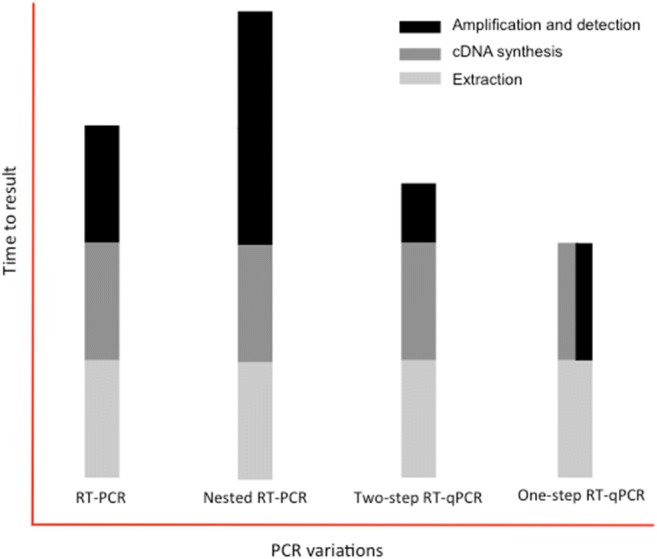
Variations of the polymerase chain reaction (PCR) used for virus diagnoses. In conventional PCR after reverse transcription (RT-PCR), the target DNA is detected at the end of PCR amplification, requiring a post-PCR process for visualization (e.g., DNA electrophoresis). The nested RT-PCR significantly enhances the sensitivity of conventional PCR, but it nearly doubles the amplification time. All qPCR protocols detect and measure target DNA after each polymerization (extension) cycle during the exponential amplification through fluorescence and do not require post-PCR processing. While two-step RT-qPCR involves two distinct stages of reverse transcription followed by qPCR, one-step RT-involves a single reaction in which cDNA synthesis and amplification occur successively
Thus, given the scarcity of detection kits officially recommended by each institution, analyzing the effectiveness of technique variations could help diagnosis, since the isolation of infected patients and epidemiological models depend on the availability of such data.
Much of the PCR work published on the development of detecting other viruses can be adapted for the diagnosis of SARS-CoV2. These studies include the use or comparison between PCR variations for the detection of viruses [31, 32], the use of PCR and the ELISA serological method [33, 34], commercial kits for virus detection [35, 36], and the chemistry (e.g., the general double-strand DNA intercalating dye SYBR Green, or post-amplification exonuclease-based probes, such as the TaqMan system) used for detecting and quantifying DNA amplification [37, 38]. Moreover, these studies aimed to determine simple or multiplex reaction assays for virus detection and serotyping [39–42] and the development of tests for simultaneous detection of viruses [43–46].
Currently, several integrated, random-access, point-of-care molecular devices are under development for the diagnosis of SARS-CoV-2 infections. These assays are expected to be simple, fast, accurate, safe, and amenable to be used in local hospital and clinic settings bearing the burden of testing and treating patients [47]. Recent studies evaluated different tests available and explored the possibility of improving SARS-CoV-2 diagnosis [47–49]. Tests based on biomarkers (e.g., serum porphobilinogen and aminolaevulinic acid) can be sensitive, specific, and low cost, and could even be used to monitor the response to treatments [50]. The international, non-profit organization Foundation for Innovative New Diagnostics (FIND) has identified almost 800 testing pipelines proposed to detect SARV-CoV-2 (for updates, cf.: https://www.finddx.org/covid-19/pipeline). As of September 1, 2020, these included 403 immunoassays, 362 molecular assays, 17 sample collections/inactivation, and 7 digital solutions.
Discussion
Interpretation
Most of the internal diagnostic assays developed by several groups around the globe were designed to detect two regions of the SARS-CoV-2 genome. Here we present a workflow to validate the internal tests indicated for the novel coronavirus through a phylogenetic approach. It becomes evident that monitoring the evolution of the virus is crucial for the validation of internal tests and even for the development of new protocols.
Furthermore, we suggest there is viability in using PCR variations (RT-PCR, nested RT-PCR, and two-step RT-qPCR) for the diagnosing of SARS-CoV-2 as a way to face one of the major bottlenecks that emerged with the new pandemic, namely the availability of inputs to carry out one-step RT-qPCR, which is the gold standard of virus diagnosis. Moreover, the investment to utilize the infrastructure already existent in many university campuses around the world, and especially in developing countries, of laboratories fully equipped to carry out RT-PCR or nested RT-PCR analyses to diagnose SARS-CoV-2 is much lower and amenable than creating at speed new labs with qPCR equipment while many academic labs are at a standstill situation during the adoption of quarantine restrictions. Express laboratory certifications could be issued by regulatory agencies by using resources in local offices, and personnel teams could be trained for virus diagnosis to enable comprehensive testing of the population.
All in all, the validation of other PCR methods, mainly regarding sensitivity and specificity for the diagnosis of SAR-CoV-2, could help countries handle higher volumes of diagnosis by using alternative supplies and amplify the laboratory network for the diagnosis, thus also reducing the test turnaround time.
Limitations
Studies on the use of RT-qPCR have sought to optimize the assays, aiming mainly at increasing the efficiency of viral detection [51]. However, due to the short time in which the virus appeared and spread, few studies aimed to analyze the assay variables, such as the PCR variations for viral load detection, and the comparison between the molecular genetic methods and serological tests. Some studies aimed at evaluating and comparing already established RT-qPCR tests [52] or molecular point-of-care tests (e.g., RT-LAMP—reverse transcriptase loop-mediated isothermal amplification) with RT-qPCR [53]. Notwithstanding, the broad spectrum of PCR inputs and methodologies potentially available to efficiently detect SARS-CoV2 still warranted further analysis. Consequently, studies developed for other viruses show some avenues that can be adapted to diagnosing SARS-CoV-2.
Therefore, it is clear that we can still improve the RT-qPCR technique, although it is currently considered the gold standard for diagnosing patients with an ongoing viral infection. Another PCR variant called digital droplet PCR (ddPCR) has great potential in this area, especially to detect low viral loads in samples, which is a major limitation of the RT-qPCR [54]. Recently, Yu et al. [55] reported a study in which they analyzed viral loads of SARS-CoV-2 in infected patients and concluded that although the RT-qPCR is sensitive and reliable, ddPCR was better to detect samples with low viral loads [55]. Falzone et al. [56] also evaluated the ddPCR sensitivity and specificity for detecting SARS-CoV-2 comparatively to RT-qPCR and reported that ddPCR was superior in both parameters. They noticed that ddPCR could also be used to detect SARS-CoV-2 in blood and saliva samples, which have not been optimally established for RT-qPCR. ddPCR could indeed improve the diagnostic procedures since rhino-pharyngeal swabs are often not practical or even possible in some patients [56]. Suo et al. [57] pointed to the superiority of ddPCR for clinical diagnosis of SARS-CoV-2 to reduce false-negative results [57]. However, we argue that the limited availability of ddPCR instrumentation and expertise worldwide, as well as the higher analysis costs, still makes RT-PCR as the gold standard for diagnosing and monitoring viral pandemics, at least for the time being.
Another technique based on nucleic acid detection that has been successfully used to detect SARS-CoV-2 [58–63] but does not depend on sophisticated equipment is the reverse transcription-loop-mediated isothermal amplification (RT-LAMP) [64, 65]. Indeed, quantitative RT-LAMP has been utilized for diagnosing and the surveillance of viruses because it is a highly sensitive and specific method allied to being simple (one-step, single tube), fast, and low-cost [66]. Commercial RT-LAMP kits have already been developed for SARS-CoV-2 during the course of the pandemic (cf. https://www.finddx.org/covid-19/pipeline). We speculate that RT-LAMP may soon become standard in comprehensive point-of-care testing and surveillance of ongoing pandemics.
Conclusion
An efficient strategy to enable and increase the efficiency of molecular testing via PCR for viral detection is the examination of the protocols endorsed by WHO collaborating institutions and the technique variations. Since many tests have been developed or are under development, the range of options will eventually increase and can alleviate the scarcity of diagnostic kits, thus quickly relieving the demand for testing kits. The present study underscores the importance of using phylogenetic approaches not only to understand the evolution of the virus but also to designing and reviewing primers and probes used for SARS-CoV-2 diagnosis via PCR. We also show the versatility of RT-qPCR for the viral diagnosis during a pandemic and identified testing alternatives that can be used in the SARS-CoV-2 diagnosis.
Electronic supplementary material
(PDF 266 kb)
(PDF 64313 kb)
Acknowledgments
The authors thank their respective institutions for time and scientific resources available to conduct this work.
Authors’ contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Horllys Gomes Barreto, Matheus Martins Daúde, and Vagner Augusto Benedito. Flávio Augusto de Pádua Milagres and Gessi Carvalho de Araújo Santos contributed with critical methodological concepts in the development of the study. The first draft of the manuscript was written by Horllys Gomes Barreto and all authors contributed with editing. All authors read and approved the final manuscript.
Data availability
All data used in this study is available to the scientific community upon request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
This manuscript does not contain clinical studies or patient data.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (2020) Coronavirus disease (COVID-2019) situation reports - 1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4. Accessed 22 Mar 2020
- 2.World Health Organization Coronavirus disease (COVID-2019) situation reports - 62. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200322-sitrep-62-covid-19.pdf?sfvrsn=f7764c46_2. Accessed 22 Mar 2020
- 3.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker P, Whittaker C, Watson O, et al (2020) Report 12: the global impact of COVID-19 and strategies for mitigation and suppression. https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-12-global-impact-covid-19/. Accessed 22 Mar 2020
- 5.McKinsey Company COVID-19. https://www.mckinsey.com/~/media/mckinsey/business functions/risk/our insights/covid 19 implications for business/covid 19 may 6/covid-19-facts-and-insights-may-6.ashx. Accessed 7 May 2020
- 6.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R, Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, Behrens P, Böddinghaus B, Götsch U, Naujoks F, Neumann P, Schork J, Tiarks-Jungk P, Walczok A, Eickmann M, Vehreschild MJGT, Kann G, Wolf T, Gottschalk R, Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science (80- ) 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park WB, Kwon N-J, Choi S-J et al (2020) Virus isolation from the first patient with SARS-CoV-2 in Korea. J Korean Med Sci 35. 10.3346/jkms.2020.35.e84 [DOI] [PMC free article] [PubMed]
- 12.Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 16.Hossein-khannazer N, Shokoohian B, Shpichka A, Aghdaei HA, Timashev P, Vosough M. Novel therapeutic approaches for treatment of COVID-19. J Mol Med (Berl) 2020;1:789–803. doi: 10.1007/s00109-020-01927-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olwenyi OA, Dyavar SR, Acharya A, Podany AT, Fletcher CV, Ng CL, Reid SP, Byrareddy SN. Immuno-epidemiology and pathophysiology of coronavirus disease 2019 (COVID-19) J Mol Med. 2020;98:1–15. doi: 10.1007/s00109-020-01961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chookajorn T. Evolving COVID-19 conundrum and its impact. Proc Natl Acad Sci U S A. 2020;117:12520–12521. doi: 10.1073/pnas.2007076117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Pacheco SJ, Kong S, Pulido-Santacruz P, Murphy RW, Kubatko L. Median-joining network analysis of SARS-CoV-2 genomes is neither phylogenetic nor evolutionary. Proc Natl Acad Sci. 2020;117:12518–12519. doi: 10.1073/pnas.2007062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavian C, Pond SK, Marini S, Magalis BR, Vandamme AM, Dellicour S, Scarpino SV, Houldcroft C, Villabona-Arenas J, Paisie TK, Trovão NS, Boucher C, Zhang Y, Scheuermann RH, Gascuel O, Lam TTY, Suchard MA, Abecasis A, Wilkinson E, de Oliveira T, Bento AI, Schmidt HA, Martin D, Hadfield J, Faria N, Grubaugh ND, Neher RA, Baele G, Lemey P, Stadler T, Albert J, Crandall KA, Leitner T, Stamatakis A, Prosperi M, Salemi M. Sampling bias and incorrect rooting make phylogenetic network tracing of SARS-COV-2 infections unreliable. Proc Natl Acad Sci. 2020;117:12522–12523. doi: 10.1073/pnas.2007295117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forster P, Forster L, Renfrew C, Forster M. Reply to Sánchez-Pacheco et al., Chookajorn, and Mavian et al.: explaining phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;117:12524–12525. doi: 10.1073/pnas.2007433117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candido DDS, Watts A, Abade L, et al. Routes for COVID-19 importation in Brazil. J Travel Med. 2020;27:taaa042. doi: 10.1093/jtm/taaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Zai J, Zhao Q, Nie Q, Li Y, Foley BT, Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P, Yang X-L, Wang X-G et al (2020) Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 10.1101/2020.01.22.914952
- 25.Boni MF, Lemey P, Jiang X et al (2020) Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. bioRxiv. 10.1038/s41564-020-0771-4 [DOI] [PubMed]
- 26.Grubaugh ND, Petrone ME, Holmes EC. We shouldn’t worry when a virus mutates during disease outbreaks. Nat Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber B, Fischer W, Gnanakaran SG et al (2020) Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 10.1101/2020.04.29.069054
- 28.Bellone M, Calvisi SL (2020) ACE polymorphisms and COVID-19-related mortality in Europe. J Mol Med:1–5. 10.1007/s00109-020-01981-0 [DOI] [PMC free article] [PubMed]
- 29.(CDC) C for DC and P (2020) Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primer and probe information. In: Centers Dis. Control Website. Accessed March. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed 22 Mar 2020
- 30.Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, Meex C, Melin P, Hayette MP, Bours V, Durkin K (2020) A recurrent mutation at position 26,340 of SARS-CoV-2 is associated with failure of the E-gene qRT-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol 58. 10.1128/JCM.01598-20 [DOI] [PMC free article] [PubMed]
- 31.Tsai J-J, Liu L-T, Lin P-C, Tsai CY, Chou PH, Tsai YL, Chang HFG, Lee PYA. Validation of the Pockit dengue virus reagent set for rapid detection of dengue virus in human serum on a field-deployable PCR system. J Clin Microbiol. 2018;56:e01865–e01817. doi: 10.1128/JCM.01865-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abachin E, Convers S, Falque S, Esson R, Mallet L, Nougarede N. Comparison of reverse-transcriptase qPCR and droplet digital PCR for the quantification of dengue virus nucleic acid. Biologicals. 2018;52:49–54. doi: 10.1016/j.biologicals.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Ambrose JH, Sekaran SD, Azizan A. Dengue virus NS1 protein as a diagnostic marker: commercially available ELISA and comparison to qRT-PCR and serological diagnostic assays currently used by the state of Florida. J Trop Med. 2017;2017:1–6. doi: 10.1155/2017/8072491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla MK, Singh N, Sharma RK, Barde PV. Utility of dengue NS1 antigen rapid diagnostic test for use in difficult to reach areas and its comparison with dengue NS1 ELISA and qRT-PCR. J Med Virol. 2017;89:1146–1150. doi: 10.1002/jmv.24764. [DOI] [PubMed] [Google Scholar]
- 35.Najioullah F, Viron F, Césaire R. Evaluation of four commercial real-time RT-PCR kits for the detection of dengue viruses in clinical samples. Virol J. 2014;11:164. doi: 10.1186/1743-422X-11-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saengsawang J, Nathalang O, Kamonsil M, Watanaveeradej V. Comparison of two commercial real-time PCR assays for detection of dengue virus in patient serum samples. J Clin Microbiol. 2014;52:3781–3783. doi: 10.1128/JCM.01870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes-Ruiz AC, Nascimento RT, de Paula SO, da Fonseca BAL. SYBR green and TaqMan real-time PCR assays are equivalent for the diagnosis of dengue virus type 3 infections. J Med Virol. 2006;78:760–763. doi: 10.1002/jmv.20620. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Parimelalagan M, Lai YL, Lee KS, Koay ESC, Hapuarachchi HC, Ng LC, Ho PS, Chu JJH. Development and evaluation of a SYBR green–based real-time multiplex RT-PCR assay for simultaneous detection and serotyping of dengue and chikungunya viruses. J Mol Diagn. 2015;17:722–728. doi: 10.1016/j.jmoldx.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, Medina F, Colón C, Margolis H, Muñoz-Jordán JL. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis. 2013;7:e2311. doi: 10.1371/journal.pntd.0002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai J-J, Liu W-L, Lin P-C, Huang BY, Tsai CY, Chou PH, Lee FC, Ping CF, Lee PYA, Liu LT, Chen CH. An RT-PCR panel for rapid serotyping of dengue virus serotypes 1 to 4 in human serum and mosquito on a field-deployable PCR system. PLoS One. 2019;14:e0214328. doi: 10.1371/journal.pone.0214328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mun M-J, Bae J-Y, Kim JH, Kim SB, Lee I, Kim JI, Park MS, Park MS, Nam YS. One-step multiplex real-time RT-PCR for detection and typing of dengue virus. Mol Cell Probes. 2019;43:86–91. doi: 10.1016/j.mcp.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Pierro AM, Gaibani P, Guo FP, Sambri V, Balmaseda A, Karunaratne K, Harris E, Pinsky BA. Single-reaction, multiplex, real-time RT-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Negl Trop Dis. 2013;7:e2116. doi: 10.1371/journal.pntd.0002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJV, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis. 2016;22:1295–1297. doi: 10.3201/eid2207.160326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabbaraju K, Wong S, Gill K, Fonseca K, Tipples GA, Tellier R. Simultaneous detection of Zika, chikungunya and dengue viruses by a multiplex real-time RT-PCR assay. J Clin Virol. 2016;83:66–71. doi: 10.1016/j.jcv.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Santiago GA, Vázquez J, Courtney S, Matías KY, Andersen LE, Colón C, Butler AE, Roulo R, Bowzard J, Villanueva JM, Muñoz-Jordan JL. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat Commun. 2018;9:1391. doi: 10.1038/s41467-018-03772-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lura T, Su T, Brown MQ. Preliminary evaluation of Thermo Fisher TaqMan ® Triplex q-PCR kit for simultaneous detection of chikungunya, dengue, and Zika viruses in mosquitoes. J Vector Ecol. 2019;44:205–209. doi: 10.1111/jvec.12347. [DOI] [PubMed] [Google Scholar]
- 47.Loeffelholz MJ, Tang Y-W. Laboratory diagnosis of emerging human coronavirus infections – the state of the art. Emerg Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Touma M. COVID-19: molecular diagnostics overview. J Mol Med. 2020;98:1–8. doi: 10.1007/s00109-020-01931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan Y, Chang L, Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol. 2020;30:e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Comentale G, Manzo R, Pilato E. Sars-Cov-2 interference in HEME production: is it the time for an early predictive biomarker? J Mol Med. 2020;98:1053–1054. doi: 10.1007/s00109-020-01945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan JF-W, Yip CC-Y, To KK-W et al (2020) Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 58. 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed]
- 52.Uhteg K, Jarrett J, Richards M, Howard C, Morehead E, Geahr M, Gluck L, Hanlon A, Ellis B, Kaur H, Simner P, Carroll KC, Mostafa HH. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osterdahl M, Lee K, Ni Lochlainn M, Wilson S, Douthwaite S, Horsfall R, Sheedy A, Goldenberg S, Stanley C, Spector T, Steves C (2020) Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to PCR. SSRN Electron J. 10.2139/ssrn.3564906 [DOI] [PMC free article] [PubMed]
- 54.Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol. 2017;55:1621–1628. doi: 10.1128/JCM.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, Gao G, Wang S, Ma C, Xie R, Wang F, Tan C, Zhu L, Guo Y, Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo C, Scalia G, Libra M, Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suo T, Liu X, Feng J et al (2020) ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect:1–30. 10.1080/22221751.2020.1772678 [DOI] [PMC free article] [PubMed]
- 58.Huang WE, Lim B, Hsu C, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thi VLD, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12:eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buck MD, Poirier EZ, Cardoso A et al (2020) Standard operating procedures for SARS-CoV-2 detection by a clinical diagnostic RT-LAMP assay. medRxiv. 10.1101/2020.06.29.20142430
- 61.Yan C, Cui J, Huang L, du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, Yao H, Li N, Zhao H, Feng Y, Liu S, Zhang Q, Liu D, Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park G-S, Ku K, Baek S-H, Kim SJ, Kim SI, Kim BT, Maeng JS. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J Mol Diagnostics. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kitagawa Y, Orihara Y, Kawamura R et al (2020) Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol:104446. 10.1016/j.jcv.2020.104446 [DOI] [PMC free article] [PubMed]
- 64.Kim J, Easley CJ. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis. 2011;3:227–239. doi: 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- 65.Euler M, Wang Y, Heidenreich D, Patel P, Strohmeier O, Hakenberg S, Niedrig M, Hufert FT, Weidmann M. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J Clin Microbiol. 2013;51:1110–1117. doi: 10.1128/JCM.02704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thai HTC, Le MQ, Vuong CD, et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 266 kb)
(PDF 64313 kb)
Data Availability Statement
All data used in this study is available to the scientific community upon request.