Abstract
Human stanniocalcin-1 (STC1) is a paracrine factor associated with inflammation and carcinogenesis. The role of STC1 in the pro- and anti-inflammatory functions of differentiating macrophage, however, is not clear. In this study, our data showed that phorbol 12-myristate 13-acetate (PMA) treatment induced human leukemia monocytic cells (ThP-1) differentiation to M0 macrophages. The differentiation was accompanied by a significant increase in the mRNA expression levels of STC1, the pro-inflammatory cytokine TNFα, and anti-inflammatory markers, CD163 & CD206. An intermitted removal of PMA treatment reduced the mRNA levels of STC1 and TNFα but had no noticeable effects on the anti-inflammatory markers. The correlation in the expression of STC1 and pro-inflammatory markers in differentiating macrophages was investigated, using siRNASTC1-transfected PMA-induced cells. Consistently, the transcripts levels of TNFα and IL-6 were significantly reduced. Moreover, LPS/IFNγ-induced M1-polarization showed remarkably higher expression levels of STC1 than IL-4/IL-13-induced M2-macrophages and PMA-induced M0-macrophages. Transcriptomic analysis of siRNASTC1-transfected M1-polarized cells revealed an upregulation of TBC1 domain family member 3 (TBC1D3G). The gene regulates the payload of macrophage-released extracellular vesicles to mediate inflammation. The conditioned media from siRNASTC1-transfected M1-polarized cells were found to reduce Hep3B cell motility. The data suggest that the expression of STC1 were associated with macrophage differentiation, but preferentially to M1 polarization.
Introduction
Inflammation is one of the cancer hallmarks and is a crucial factor in the regulation of tumor progression. In tumor microenvironment (TME), cancer cells maintain intricate interactions with surrounding stroma [1]. The infiltration of immune cells and secretion of various growth factors/cytokines modulate the process of tissue remodeling in cancers. One crucial component of tumor stroma is macrophages that multiple aspects of coordination in immune responses. Besides, macrophages can reversibly alter their endotypes to augment antitumor reactions or antagonize cytotoxic actions [2]. Macrophages can be classified based on their activation states. These include the resting (M0), and the polarizing states [the classically activated macrophages (CAM/M1) and alternatively activated macrophages (AAM/M2)]. In general, M1 macrophages are considered to be pro-inflammatory and exhibit antitumor effects. On the contrary, M2 macrophages assist a pro-tumoral role by suppressing the local immune response, to promote angiogenesis, invasion, and metastasis [[3], [4], [5]]. Nonetheless, clinical and experimental data indicated that cancer tissues with high infiltration of tumor-associated macrophages were associated with poor prognosis and exhibited resistance to therapeutics [6]. An understanding of the factors responsible for macrophage homing and differentiation is recognized to be an essential immunotherapeutic strategy.
Human stanniocalcin-1 (STC1), an autocrine/paracrine factor, is suggested to play roles in inflammation and carcinogenesis. Early clinicopathological studies showed its differential expressions in paired normal and tumor tissues [7]. Experimental characterization on the role of STC1 in some cancer hallmarks, like proliferation, apoptosis, and angiogenesis in different cancer cell models, was reported [8]. Later studies revealed stimulation of STC1 expression under oxidative stress in the tumor microenvironment [9,10]. The involvement of STC1 in wound healing [11], and cardiovascular inflammation were demonstrated [12]. The anti-inflammatory action of STC1 was shown via the suppression of superoxide production at glomerulonephritis [13]. The effect of STC1 on inhibition of ROS production via mitochondrial uncoupling protein 2 in renal ischemia/reperfusion injury in mice was reported [14]. Moreover, the counteracting effects of STC1 on LPS-induced lung injury [15] or ischemic cardiac injury in mice [16], via inhibition of inflammatory cascades or suppression on monocytes/macrophages recruitment were revealed respectively. The anti-inflammatory effects of STC1 on other models, like osteoarthritis [17], and retinopathy [18] were reported.
Our previous studies showed the inhibitory effects of STC1 on the growth of STC1-overexpressing human hepatocellular carcinoma (HCC) cells in a mouse xenograft model [19]. The inhibitory effects of STC1 on p70S6K/p-rpS6 signaling in the HCC cells to reduce tumor growth was demonstrated [20]. In considering the intricate interactions at the TME, the role of STC1 in macrophage differentiation has not been addressed. With hindsight, STC1 and macrophages are known to be involved in inflammation and carcinogenesis. In this study, we hypothesized that STC1 might play a role in differentiation and inflammatory functions of macrophages and thus modulate tumorigenicity through macrophage-cancer cell interactions. Using human leukemia monocytic cell line ThP-1, we aimed to characterize the role of STC1 in macrophage differentiation and functions.
Materials and methods
Cell culture and macrophage differentiation
Human leukemia monocytic cell line (ATCC), ThP-1 was cultured in RPMI1640, supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (25 U/ml penicillin and 25 μg/ml streptomycin) (Gibco, Life Technologies). The cells were maintained in a CO2 (5%) incubator at 37 °C.
The cells were treated with phorbol 12-myristate 13-acetate (PMA) (Abcam) for 24–48 h to stimulate macrophage differentiation (M0). An increasing concentration of PMA (2.5–100 nM) was applied to the cells to determine the optimal dose of the stimulation. Once the optimal PMA concentration was identified, the differentiated cells (M0) could then be further treated with selected stimulators to obtain M1 and M2 polarized macrophages. For M1 polarization, ThP-1 cells were treated with 5 nM PMA for 6 h, followed by 20 ng/ml IFNγ (Gibco, Life Technologies), and 100 ng/ml LPS (Invitrogen; Thermo Fisher Scientific) stimulation for another 18 h. For M2 polarization, ThP-1 cells were treated with 5 nM PMA for 6 h, followed by 20 ng/ml IL-4, and 20 ng/ml IL-13 (Gibco, Life Technologies) stimulation for another 18 h. The identities of M0, M1, and M2 polarized cells were characterized using both pro- and anti-inflammatory markers.
Total RNA extraction and real-time PCR
Cellular RNA was extracted by TRIzol Reagent (Gibco/BRL) according to the manufacturer's instruction. The A260/A280 of total RNA was >1.8, which was utilized to synthesize cDNA using SuperScript VILO Master Mix (Invitrogen, Life Technologies). Real-time PCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems). The primer sequences were listed in Table 1. The data were normalized to the mRNA levels of human GAPDH. Control amplifications were conducted without either RNA or reverse transcriptase.
Table 1.
List of PCR primers.
| Gene | Sequences |
|---|---|
| GAPDH | 5′-GGACCTGACCTGCCGTCTAG-3′, |
| 5′-TAGCCCAGGATGCCCTTGAG-3′ | |
| Actin | 5′-GACTACCTCATGAAGATCCTCACC-3′, |
| 5′-TCTCCTTAATGTCACGCACGATT-3′ | |
| STC1 | 5′-TGAGGCGGAGCAGAATGACT-3′, |
| 5′-CAGGTGGAGTTTTCCAGGCAT-3′ | |
| CD163 | 5′-CCAACAAGATGCTGGAGTGAC -3′, |
| 5′-TGACAGCACTTCCACATTCAAG −3′ | |
| CD206 | 5′-GCGGAACCACTACTGACTA - 3′, |
| 5′-GTTGTTGGCAGCTTTTCCTC-3′ | |
| TNFα | 5′-GGGCCTGTACCTCATCTACT - 3′, |
| 5′-TAGATGGGCTCATACCAGGG-3′ | |
| IL-6 | 5′-AGCCCACCGGGAACGAAAGA - 3′, |
| 5′-TGTGTGGGGCGGCTACATCT-3′ | |
| TBC1D3G | 5′-AACCCCGGAAGATACCAGA-3′, |
| 5′-TTCCTTAATGTCCCGCTTATG-3′ |
STC1 Knock-down by siRNA
On-Target human STC1 siRNA (Dharmacon) was used to knock down the STC1 transcript. Non-targeting siRNA: UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUAC AUGUUUUCUGA, UGGUUUACAUGUUUUCCUA, and human STC1 siRNA: AAACGCACAUCCCAUGAGA, GGGAAAAGCAUUCGUCAAA, GUACAGCG CUGCUAAAUUU, CAACAGAUACUAUAACAGA were used in this study. Briefly, 1 × 105 ThP-1 cells were seeded in 24-well plates, then transfected with 50 nM siRNA using 0.5 μl Lipofectamine 2000 transfection reagent (Invitrogen; Life Technologies) in antibiotic-free complete medium. After 6 h incubation, the siRNA transfected cells were treated with 5 nM PMA to induce M0 macrophage differentiation. In some experiments, the siRNA transfected M0 macrophages were further stimulated to obtain M1 or M2 polarized cells. The decrease in the expression levels of STC1 mRNA and protein were validated by real-time PCR and western blotting. Western blot was conducted using a rabbit antibody to STC1 (Sigma), followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody. Specific bands were visualized with chemiluminescent reagent (Western-lightening Plus, PerkinElmer Life Sciences). Blots were then washed in PBS and re-probed with rabbit anti-actin serum (Sigma).
Boyden chamber-based cell migration assay
ThP-1 cells were stimulated to differentiate into M0, M1, and M2 polarization in 24-well companion plates in quadruplicate. After 24 h, the cells were washed with a complete RPMI medium. The human hepatoma cell line, HepB3 cells (ATCC) were seeded onto 24-well cell-culture inserts (Falcon) with a membrane pore size of 8 μm in serum-free DMEM medium on top of the polarized ThP-1. In siRNA transfection experiments, ThP-1 cells were treated with either siRNACTRL or siRNASTC1 before the stimulation of M1 or M2 polarization in quadruplicate. The conditioned media of M1 and M2 macrophages were collected, filtered, and used in the cell migration assay. The conditioned media were transferred to 24-well companion plates. HepB3 cells were seeded onto 24-well cell-culture inserts (Falcon) with a membrane pore size of 8 μm in serum-free DMEM medium. After 24 h incubation at 37 °C, cells on the top side of the membrane were removed by cotton swab, whereas cells migrated to the bottom of the membrane were fixed with ice-cold methanol, followed by 0.5% crystal violet (Farco Chemical Supplies) at room temperature for 15 min. The membranes were then mounted. Migrated cells were captured by a microscope and counted by Image J.
Transcriptomic analysis
Total RNA of siRNACTRL or siRNASTC1-transfected M1 macrophages was extracted. The RNA quality was measured by Agilent 2100 Bioanalyzer system. Four replicates per treatment with RNA Integrity Number (RIN) > 8 were used for library construction. DNA sequencing was conducted at the Beijing Genomics Institute (Wuhan, China) using the BGISEQ-500RS sequencer. Single-end reads of 50 bp read-length were sequenced and trimmed according to BWA's–q algorithm. Quality-trimmed sequence reads were mapped to human genome reference (GRCh38/hg38). Read-count data were then subjected to differential expression analysis using the edgeR package [21]. Genes with posterior probability of equal expression (PPEE) < 0.05 were considered as differentially expressed genes (DEGs).
Statistical analysis
Statistical analysis was conducted using SigmaPlot version 12.0. Data were evaluated by the Student's t-test or one-way analysis of variance (ANOVA) followed by Duncan's multiple range test. All data are presented as a statistical mean ± SD. A p-value < 0.05 was used as the cutoff for statistical significance.
Results
Dose- and time-dependent effects of PMA on ThP-1 differentiation and STC1 expression
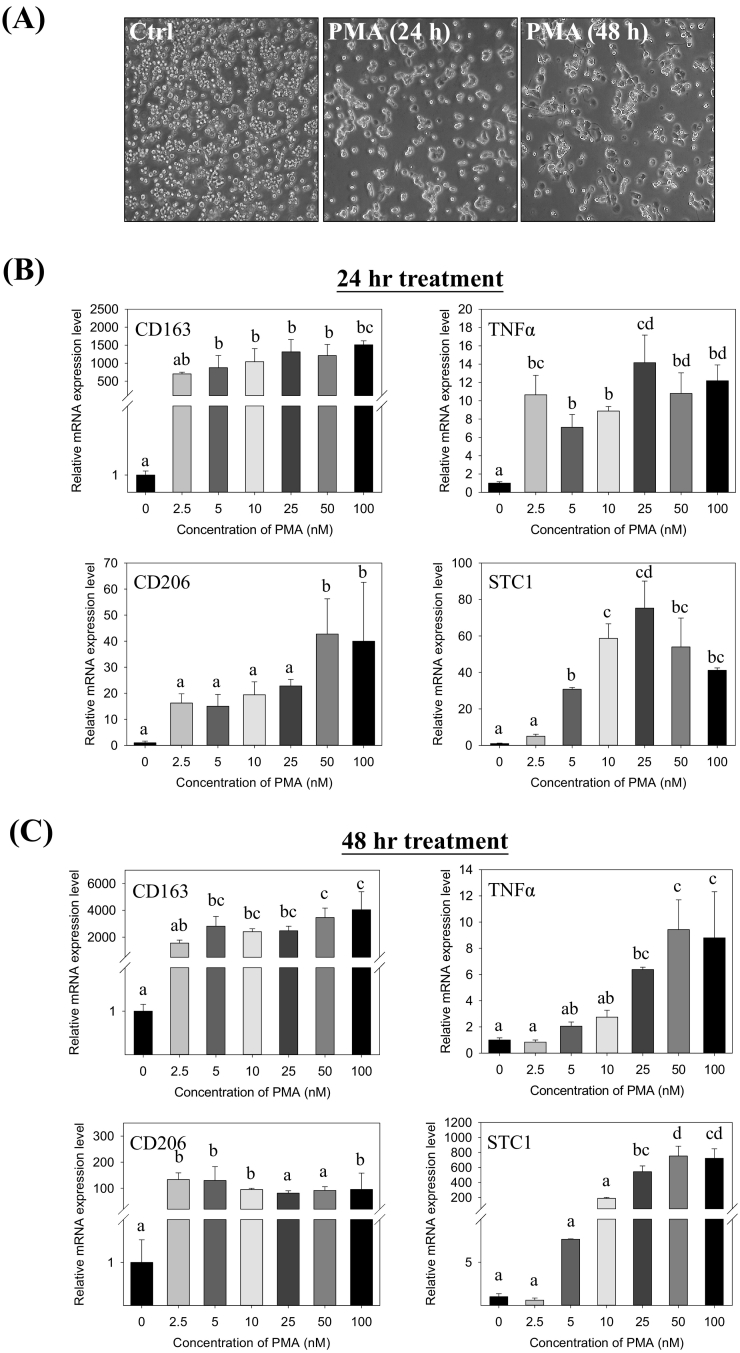
ThP-1 monocytes were stimulated to differentiate into M0 macrophages by the treatment with an increasing dose (2.5–100 nM) of phorbol 12-myristate 13-acetate (PMA) for 24–48 h. The cells were transformed from a suspension to an adherent form, demonstrating a differentiated phenotype (Fig. 1A). To characterize the molecular phenotypes of the differentiation, the mRNA expression levels of pro- and anti-inflammatory markers, TNFα, CD163 and CD206 were measured. Dose- and time-dependent inductions in the expression levels of the markers were noted at 24 h (Fig. 1B) and 48 h post-treatment (Fig. 1C). Significant increases of STC1 mRNA (Fig. 1B–C) was observed.
Fig. 1.
Dose- and time-dependent effects of PMA on ThP-1 differentiation to M0 macrophage and STC1 expression. (A) Cell morphology of ThP-1 cells changed from suspension to adherent phenotype after 5 nM PMA treatment for 24 and 48 h. Gene expressions of CD163, TNFα, CD206, and STC1 in ThP-1 were up-regulated after (B) 24-h and (C) 48-h PMA treatments. Data were presented as the mean ± S.D. Bars with the same letter are not significantly different according to the results of one-way ANOVA followed by Duncan's multiple ranges tests (p < 0.05).
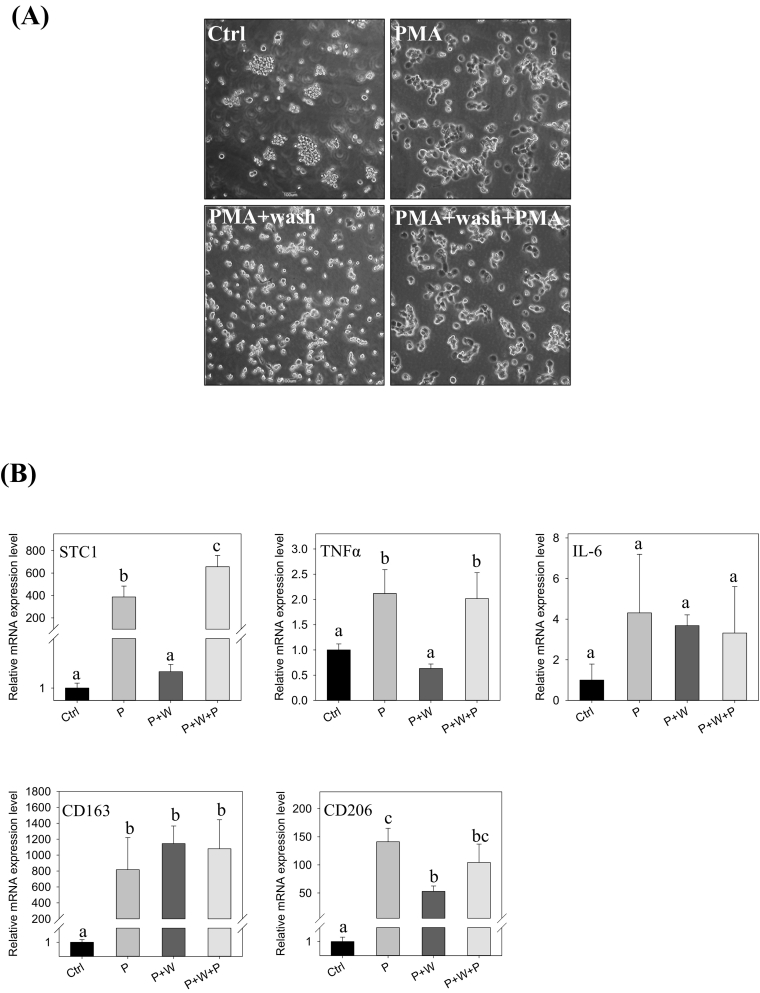
An experiment with an intermitted removal of PMA in the treatment was conducted. In the cells treated with 5 nM PMA for 24 h, followed by another 24 h of incubation in a PMA-free medium. The cells maintained a lesser differentiated phenotype, as compared with the cells maintained in the PMA-medium for 48 h, with or without washing step (Fig. 2A, the right panels). The observation revealed that the intermitted removal of PMA led to a reduction of cell spreading, suggesting further differentiation was halted. Moreover, the expression levels of the pro- and anti-inflammatory markers were determined. The cells with the intermitted removal of PMA exhibited significantly lower expression levels of the pro-inflammatory cytokine TNFα and STC1 transcripts (Fig. 2B). The expression levels of the anti-inflammatory markers CD163 and CD206, however, were not noticeably affected.
Fig. 2.
Intermitted PMA removal abrogated STC1 gene expression. (A) Cell morphologies of the (i) control, (ii) PMA, (iii) PMA withdrawal after 24-h (PMA + wash), and (iv) PMA withdrawal after 24 h + replenishment of fresh PMA (PMA + wash + PMA). (B) The expressions of STC1, cytokine markers (TNFα, IL6, CD163, CD206) in differentiating cells after the treatments for 48 h (P: PMA; W: wash). Data were presented as the mean ± S.D. Bars with the same letter are not significantly different according to the results of one-way ANOVA followed by Duncan's multiple ranges tests (p < 0.05).
Effects of STC1-knockdown on the expression of pro- and anti-inflammatory markers in PMA-induced differentiation (M0)
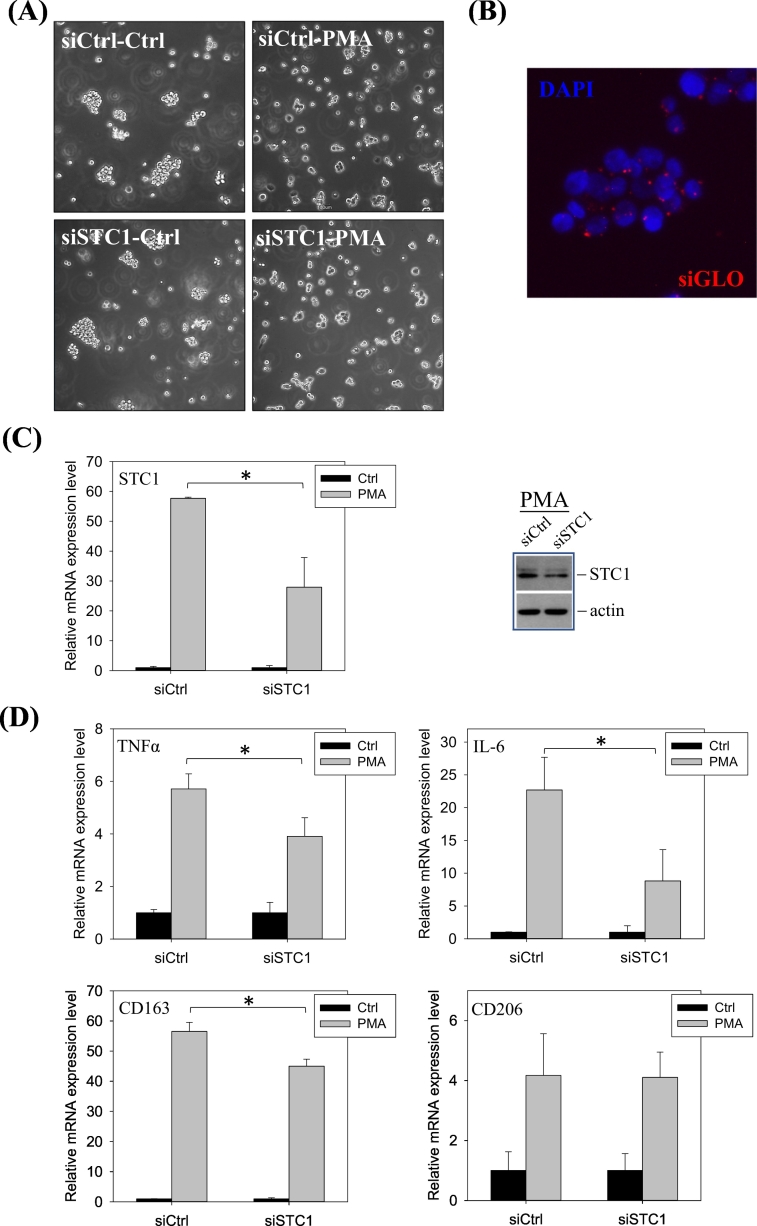
ThP-1 cells were transfected with either siRNACTRL or siRNASTC1 before the PMA treatment. After transfection, there were no noticeable differences in the cell phenotypes, when comparing the siRNACTRL and siRNASTC1 in the control (Fig. 3A, the left panels) or in the PMA-treatment (Fig. 3A, the right panels). The transfection efficiency was illustrated in the siGLO co-transfection (Fig. 3B). The transfection of siRNASTC1 in PMA-induced ThP-1 cells (M0) showed a significant reduction in the expression levels of STC1 (Fig. 3C) and the pro-inflammatory (TNFα, IL-6) and anti-inflammatory (CD163) markers (Fig. 3D).
Fig. 3.
Effects of STC1-knockdown on the expression of STC1, pro- and anti-inflammatory markers in PMA-induced M0 macrophages. (A) Cell morphology of THP-1 transfected with siRNACTRL or siRNASTC1, in the presence or absence of PMA treatment. (B) Transfection efficiency of siRNA was shown by siGLO (blue: DAPI; red: siGLO). (C) The downregulation of STC1 mRNA and protein expression after siRNASTC1 treatment. (D) Gene expressions of the pro- and anti-inflammatory markers after siRNA treatments. Significant reductions in the expression of STC1, IL-6, TNFα, and CD163 in siRNASTC1 transfected PMA-treated cells were observed. Data were presented as the mean ± S.D. Asterisks denote statistical significance relative to the respective control (*p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
STC1 expression in M1- and M2-polarized cells
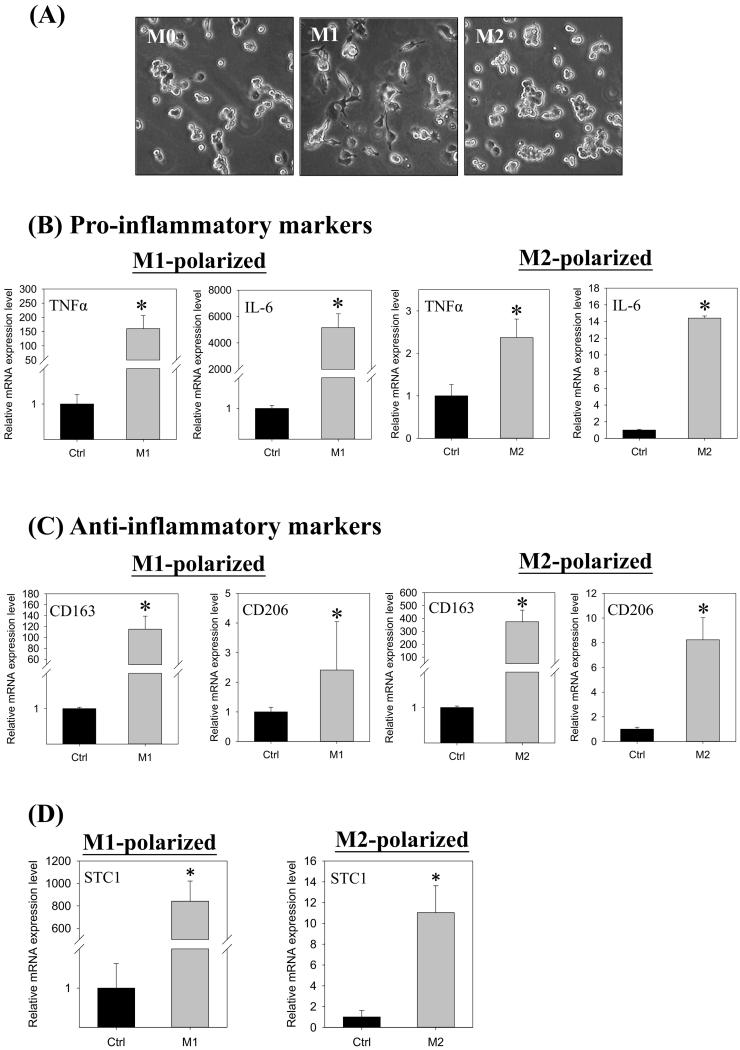
To elucidate further the association of STC1 expression in macrophage differentiation, PMA treated ThP-1 cells (M0) were further stimulated to differentiate into M1- or M2-polarized cells. Fig. 4A shows the cell morphology of the M0 (PMA), M1 (LPS/IFNγ), and M2 (IL-4/IL-13) polarization. M1-polarized cells exhibited epithelial-like phenotype, whereas M2-polarized cells showed adherent round-shaped morphology. The M1- and M2-polarization were characterized by measuring the expression levels of the pro- and anti-inflammatory markers. The expressions of pro-inflammatory markers (TNFα & IL-6) were remarkably induced in M1-polarized cells (TNFα: ~150-fold; IL-6: ~5000-fold) as compared with the levels measured in M2-polarized cells (TNFα: ~2-fold; IL-6: ~14-fold) (Fig. 4B). On the other hand, the expression levels of the anti-inflammatory markers (CD163: ~400 folds; CD206: ~8 fold) in M2-polarized cells were significantly higher, as compared with the M1-polarized cells (CD163: ~100-fold; CD206: ~2-fold) (Fig. 4C). In M1-polarization, STC1 gene expression was remarkably increased by 800-fold, whereas in M2-polarization, the induction was about 10-fold (Fig. 4D). Significant stimulation of STC1 mRNA expression was measured in the differentiation, in which ~800- and ~ 10-fold inductions were measured in M1- and M2-polarized cells, respectively. When comparing the gene expression between M0 and M2 cells, similar expression levels of the anti-inflammatory markers and STC1 were observed (Supplementary Fig. 1). M0 cells exhibited significantly higher expression levels of TNFα and lower levels of IL6, as compared with M2 cells.
Fig. 4.
The gene expression levels of STC1 and cytokine markers in LPS/IFNγ-induced M1 or IL-4/IL-13-induced M2 macrophages. (A) The differentiating morphologies of M0, M1, and M2 polarized cells. (B) Messenger RNA expression levels of pro-inflammatory markers in M1- (the left panel) and M2- (the right panel) polarized cells. The induction of the pro-inflammatory markers in M1-polarized cells was remarkably higher than in the M2 cells. Noted the different scales in the y-axes. (C) Messenger RNA expression levels of anti-inflammatory markers in (i) M1- (the left panel) and (ii) M2- (the right panel) polarized cells. The induction of anti-inflammatory marker expression in M2-polarized cells was remarkably higher than in the M1 cells. Noted the different scales in the y-axes. (D) The induction of STC1 expression in M1-polarized cells was remarkably higher than in the M2 cells. Noted the different scales in the y-axes. Data were presented as the mean ± S.D. Asterisks denote statistical significance relative to the respective control (*p < 0.05).
Effects of STC1 knockdown on global gene expression in M1-polarized cells
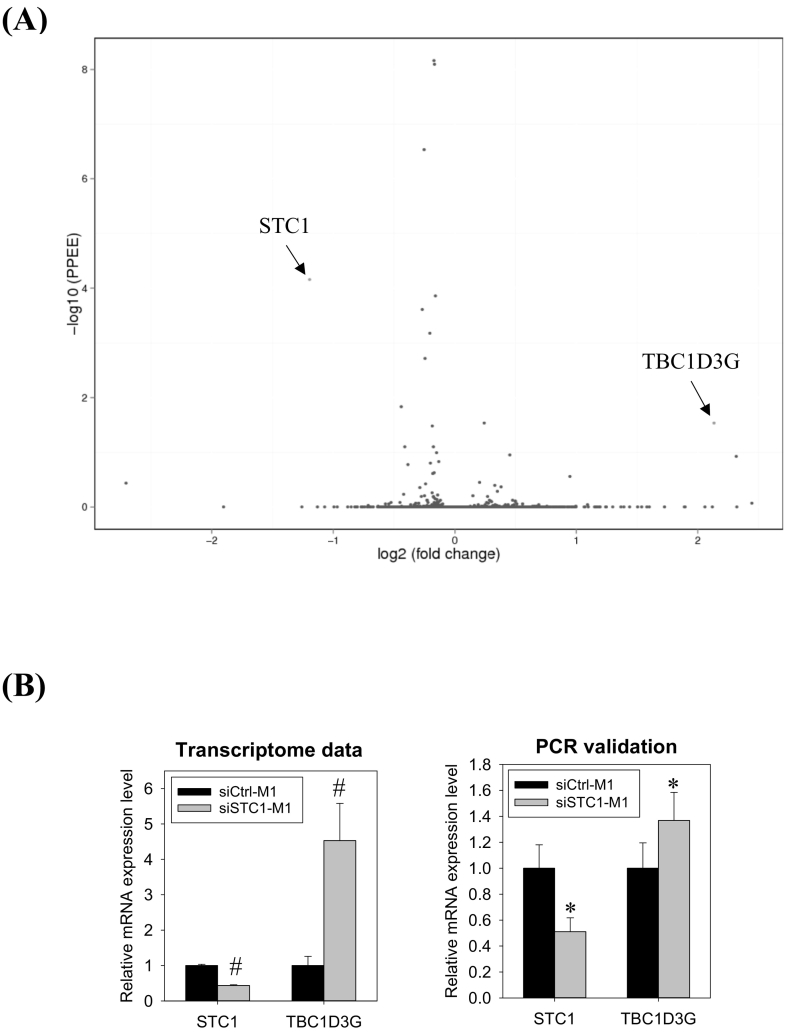
In the transcriptomic analysis of STC1-knockdown, quality-trimmed Illumina reads of about 26.3 M were acquired from each sample of siRNACTRL and siRNASTC1-transfected M1-polarized cells. Supplementary file 1 shows the list of differentially expressed genes (DEGs). A volcano plot showed that only two genes were significantly dysregulated (Fig. 5A). The STC1 transcript was downregulated, while the TBC1D3G mRNA was up-regulated (Fig. 5B, the left panel). Real-time PCR analysis validated the changes of STC1 and TBC1D3G mRNA expression (Fig. 5B, the right panel).
Fig. 5.
Transcriptomic analysis of STC1 knockdown M1 cells. (A) Volcano plot of differentially expressed genes (DEGs). The x-axis represents the differential gene expression (log2). Y-axis represents the p-value (−log10). The red dots represent the significantly up-regulated DEG - TBC1D3G and the down-regulated DEG - STC1. (B) Messenger RNA levels of STC1 and TBC1D3G, as revealed by transcriptome (the left panel) and validated by real-PCR (the right panel). Data were presented as the mean ± S.D. Pounds (#) indicate statistical significance relative to the respective control (posterior probability of equal expression, PPEE, Supplementary file 1). Asterisks denote statistical significance relative to the respective control (*p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Transcriptomic analysis of STC1 knockdown M1 cells. (A) Volcano plot of differentially expressed genes (DEGs). The x-axis represents the differential gene expression (log2). Y-axis represents the p-value (−log10). The red dots represent the significantly up-regulated DEG - TBC1D3G and the down-regulated DEG - STC1. (B) Messenger RNA levels of STC1 and TBC1D3G, as revealed by transcriptome (the left panel) and validated by real-PCR (the right panel). Data were presented as the mean ± S.D. Pounds (#) indicate statistical significance relative to the respective control (posterior probability of equal expression, PPEE, Supplementary file 1). Asterisks denote statistical significance relative to the respective control (*p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Effects of conditioned media from siRNASTC1 transfected M1-polarized cells on Hep3B migration
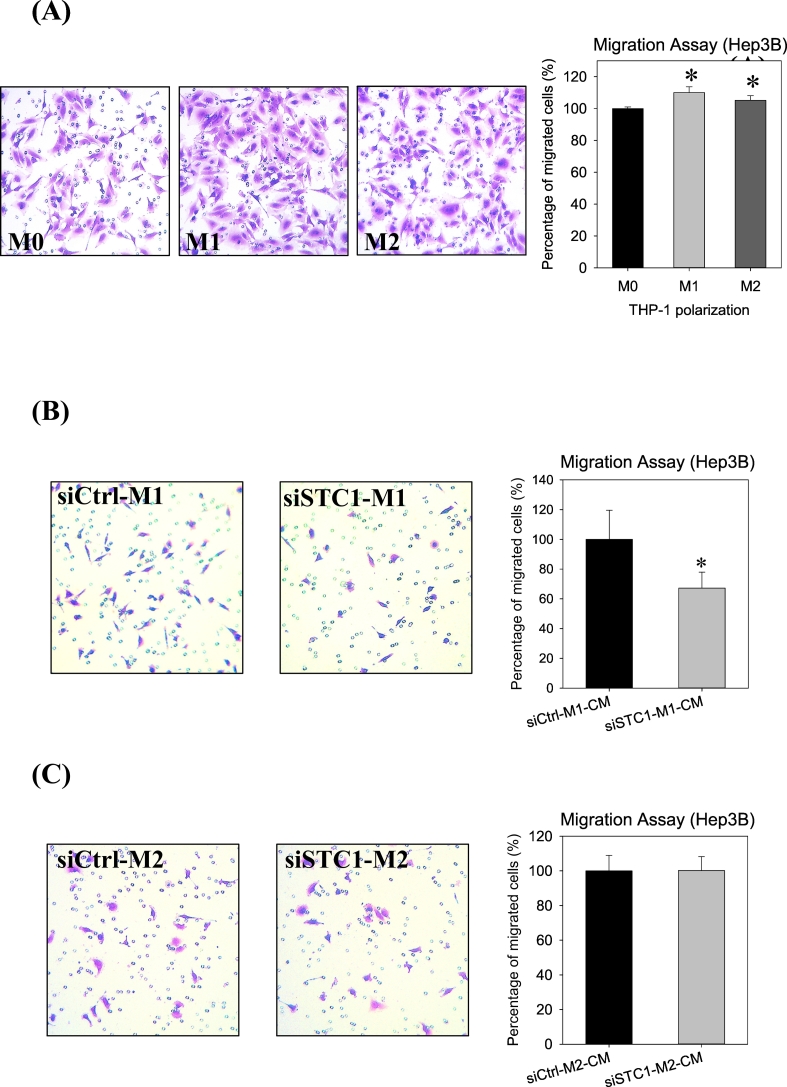
M1 or M2 cells showed significant inhibition on Hep3B motility as compared with M0 cells (Fig. 6A). Conditioned media from the siRNACTRL- and siRNASTC1-transfected M1-polarized cells were collected. The efficiency of siRNA delivery was checked by siGLO transfection, and the reduction of STC1 expression was measured using real-time PCR (data not shown). The conditioned media of the M1-polarized cells (siRNASTC1-transfected) showed inhibitory effects on the migration of Hep3B cells (Fig. 6B) as compared with the condition media of the siRNACTRL-transfected M1 cells. On the other hand, conditioned media from STC1 knockdown M2-polarized cells showed no noticeable effects on the migration, as compared with the respective control (Fig. 6C).
Fig. 6.
Cell Migration assay. (A) Effects of M0, M1 and M2 polarization on the motility of Hep3B cells. M1 and M2 macrophages significantly stimulated the migration of Hep3B cells. Effects of conditioned media from siRNASTC1 transfected (B) M1-polarized cells and (C) M2-polarized cells on Hep3B migration. A significant reduction in the motility of STC1-knockdown M1 cells was noted as compared with the respective siCtrl M1 cells. Data were presented as the mean ± S.D. Asterisks denote statistical significance relative to the respective control (*p < 0.05).
Discussion
Current evidence suggests the involvement of STC1 in inflammation, based on its inhibitory effects on the production of pro-inflammatory cytokines and chemokines, and suppressive effects on macrophage responses to chemoattractant [8,22]. In this study, the connection of STC1 to macrophage differentiation, through studying its association with M0-, M1- and M2-polarization was revealed. The effect of STC1-knockdown in macrophages on transwell migration of the hepatocellular carcinoma cell-line Hep3B was studied.
PMA treatment on ThP-1 cells stimulated the expression of STC1 and inflammatory markers. PMA is a protein kinase C (PKC) activator, a standard chemical used to induce differentiation of monocytic ThP-1 into macrophages via PKC activation [23]. Multiple downstream signaling pathways were reported to be responsible for PMA-induced leukemic differentiation. Those include phosphatidylinositol 3-kinase/Ca2+, Raf-1, MAPK, and NF-κB signaling pathways and superoxide production [24,25]. The Ca2+-signaling and stress-induced pathways were also demonstrated to stimulate STC1 expression in various studies [10,13,[26], [27], [28]]. With hindsight, STC1 expression is known to be induced in cell differentiation. These include neural cells [28,29], hematopoietic cells [30,31], osteoblast cells [32], ovarian cells [33] and adipocyte [34]. Therefore, the observation of increased STC1 expression in macrophage differentiation is consistent. Macrophage activation comprises the resting (M0), and polarizing M1 and M2 states. The classical M1 and alternative M2 macrophages that confer pro- or anti-inflammatory activities. Our following question is on the connection of STC1 to these heterogeneous populations of macrophages.
Our data show that the differentiation of ThP-1 to M0, M1, and M2 macrophages induced STC1 expression, which was considerably higher in M1 polarized cells. In examining the effects of intermitted PMA-withdrawal, macrophage differentiation (M0) was arrested. STC1 expression was significantly lower as compared with the continuous PMA-treated cells. Our observation was consistent with the report by Spano and co-workers, which showed that PMA-withdrawal caused cell de-differentiation [35]. In the measurement of cytokine markers in the intermitted PMA treated cells, a significant reduction in the expression of the pro-inflammatory marker, TNFα were noted. In the STC1 knockdown experiment, the transcript levels of pro-inflammatory markers (TNFα, IL-6) were downregulated. The data suggested a possible correlation in the expression levels of STC1 and pro-inflammatory markers. To further elucidate this correlation, we adopt the standard protocols to stimulate M1- and M2-polarization. The identity of the differentiated macrophages was characterized using the standard markers [36]. Our data show that a remarkably higher expression of STC1 was measured in M1 polarization, as compared with M2. M1 macrophages are known to be pro-inflammatory to produce cytokines to exert tumoricidal activities [1]. The biological function of STC1, however, was suggested to be anti-inflammatory [22]. The pro-inflammatory role of M1-polarized cells and the reported function of STC1 seemed to be inconsistent. However, in considering a possible negative feedback circuity in controlling pro-inflammatory signals, the expression of endogenous anti-inflammatory mediators (i.e., STC1) may help to minimize the persistent activation of immune cells in inflammatory responses. In fact, the negative feedback regulation of inflammation has been reported for over 20 years [37]. For instance, the negative feedback regulation was reported in various inflammatory pathways, like NKκB, Smad, and JAK/STAT by the suppressors of cytokine signaling or cytokine-inducible SH2 protein [38,39]. In our previous study in characterizing the effects of STC1 on tumorigenicity of hepatocellular carcinoma Hep3B cells, IL-6 and IL-8 treatment induced STC1 expression. However, STC1 exhibited inhibitory action on the pro-migratory functions of IL-6 and IL8 [19]. Other examples of reciprocal regulation in inflammation responses include glucocorticoid-cytokine [40] and inducible NO synthase-connexin 43 [41]. Retrospectively, this suggests that the high expression level of STC1 in M1-polarized cells may serve as a negative mediator in inflammation.
Transcriptomic analysis of STC1-knockdown M1-polarized cells identified a deregulated gene, Tre-2/Bub2/Cdc16 (TBC1) domain family member 3 (TBC1D3). The gene is reported to regulate the payload (i.e., RNA-binding protein, secretory pathways regulatory proteins) of released extracellular vesicles (EVs) from macrophages to mediate inflammation and tissue repair [42]. The high expression of TBC1D3 was associated with decreased STAT3 phosphorylation in recipient cells [42]. A decreased phosphorylated level of STAT3 was found to inhibit the proliferation and migration of HCC [43]. Consistently in this study, the incubation of Hep3B cells in conditioned media of siRNASTC1-transfected M1-polarized cells showed a significant reduction in cell motility. Alternatively, the high expression level of STC1 in M1-polarized cells might have low expression of TBC1D3, resulting in higher phosphorylation of STAT3 in cancer cells, which associated with poor prognosis in HCC [19,44].
In summary, we unravel the connection of STC1 expression in macrophage differentiation, and its association with the expression of pro-inflammatory markers. STC1 may serve as a negative mediator to regulate inflammation, which might be associated with the expression of TBC1D3. The data support further investigation, using clinical HCC samples to unravel the intricate association of STC1, macrophages, and tumor cells in carcinogenesis.
The following are the supplementary data related to this article.
The list of differentially regulated genes in the transcriptiomic analysis of STC1-knockdown M1-polarized cells.
Comparisons on the gene expression levels of (A) pro-inflammatory, (B) anti-inflammatory markers and (C) STC1 in M0, M1 and M2 macrophages. (A) The induction of the pro-inflammatory markers in M1-polarized cells was remarkably higher than in the M0 and M2 cells. (B) The induction of anti-inflammatory marker expression in M0 and M2-polarized cells was sifnificantly higher than in the M1 cells. (C) The induction of STC1 expression in M1-polarized cells was remarkably higher than in the M0 and M2 cells. Data were presented as the mean ± S.D. Bars with the same letter are not significantly different according to the results of one-way ANOVA followed by Duncan's multiple ranges tests (p < 0.05).
Supplementary material
Author contribution statement
CKCW was responsible for the design and planning of the experiments. CCTL was responsible for experimental works. Both CKCW and CCTL are responsible for data analysis and manuscript writing.
Declaration of competing interest
The authors declare they have no actual or potential competing financial interests.
Acknowledgments
This work was supported by the General Research Fund [HKBU12103817], Research Grant Council, Hong Kong.
References
- 1.Poh A.R., Ernst M. Targeting macrophages in cancer: from bench to bedside. Front Oncol. 2018;8:49. doi: 10.3389/fonc.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., Locati M., Mantovani A., Martinez F.O., Mege J.L., Mosser D.M., Natoli G., Saeij J.P., Schultze J.L., Shirey K.A., Sica A., Suttles J., Udalova I., van Ginderachter J.A., Vogel S.N., Wynn T.A. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas S.K., Allavena P., Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin.Immunopathol. 2013;35:585–600. doi: 10.1007/s00281-013-0367-7. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat.Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 5.Sica A., Larghi P., Mancino A., Rubino L., Porta C., Totaro M.G., Rimoldi M., Biswas S.K., Allavena P., Mantovani A. Macrophage polarization in tumour progression. Semin.Cancer Biol. Ther. 2008;18:349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Petty A.J., Yang Y. Tumor-associated macrophages: implications in cancer immunotherapy. Immunotherapy. 2017;9:289–302. doi: 10.2217/imt-2016-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A.C., Jellinek D.A., Reddel R.R. Mammalian stanniocalcins and cancer. Endocr. Relat Cancer. 2003;10:359–373. doi: 10.1677/erc.0.0100359. [DOI] [PubMed] [Google Scholar]
- 8.Yeung B.H., Law A.Y., Wong C.K. Evolution and roles of stanniocalcin. Mol.Cell Endocrinol. 2012;349:272–280. doi: 10.1016/j.mce.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Prockop D.J., Oh J.Y. Mesenchymal stem/stromal cells (MSCs): role as guardians of inflammation. Mol.Ther. 2012;20:14–20. doi: 10.1038/mt.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung H.Y., Lai K.P., Chan H.Y., Mak N.K., Wagner G.F., Wong C.K. Hypoxia-inducible factor-1-mediated activation of stanniocalcin-1 in human cancer cells. Endocrinology. 2005;146:4951–4960. doi: 10.1210/en.2005-0365. [DOI] [PubMed] [Google Scholar]
- 11.Yeung B.H., Wong C.K. Stanniocalcin-1 regulates re-epithelialization in human keratinocytes. PLoS.One. 2011;6:e27094. doi: 10.1371/journal.pone.0027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C., Jamaluddin S., Yan S., Sheikh-Hamad D., Yao Q. Human stanniocalcin-1 blocks TNF-{alpha}-induced monolayer permeability in human coronary artery endothelial cells. Arterioscler.Thromb.Vasc.Biol. 2008;28:906–912. doi: 10.1161/ATVBAHA.108.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L., Garcia G., Lou Y., Zhou Q., Truong L.D., Dimattia G., Lan X.R., Lan H.Y., Wang Y., Sheikh-Hamad D. Anti-inflammatory and renal protective actions of stanniocalcin-1 in a model of anti-glomerular basement membrane glomerulonephritis. Am.J.Pathol. 2009;174:1368–1378. doi: 10.2353/ajpath.2009.080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L., Belousova T., Chen M., Dimattia G., Liu D., Sheikh-Hamad D. Overexpression of stanniocalcin-1 inhibits reactive oxygen species and renal ischemia/reperfusion injury in mice. Kidney Int. 2012;82:867–877. doi: 10.1038/ki.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang S.E., Wu C.P., Wu S.Y., Peng C.K., Perng W.C., Kang B.H., Chu S.J., Huang K.L. Stanniocalcin-1 ameliorates lipopolysaccharide-induced pulmonary oxidative stress, inflammation, and apoptosis in mice. Free Radic.Biol.Med. 2014;71:321–331. doi: 10.1016/j.freeradbiomed.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Mohammadipoor A., Lee R.H., Prockop D.J., Bartosh T.J. Stanniocalcin-1 attenuates ischemic cardiac injury and response of differentiating monocytes/macrophages to inflammatory stimuli. Transl.Res. 2016;177:127–142. doi: 10.1016/j.trsl.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Li Z., Jia W., Li M., Tang M. Upregulation of stanniocalcin-1 inhibits the development of osteoarthritis by inhibiting survival and inflammation of fibroblast-like synovial cells. J Cell Biochem. 2019;120:9768–9780. doi: 10.1002/jcb.28257. [DOI] [PubMed] [Google Scholar]
- 18.Dalvin L.A., Hartnett M.E., Bretz C.A., Hann C.R., Cui R.Z., Marmorstein A.D., Sheikh-Hamad D., Fautsch M.P., Roddy G.W. Stanniocalcin-1 is a modifier of oxygen-induced retinopathy severity. Curr.Eye Res. 2020;45:46–51. doi: 10.1080/02713683.2019.1645184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung B.H., Shek F.H., Lee N.P., Wong C.K. Stanniocalcin-1 reduces tumor size in human hepatocellular carcinoma. PLoS.One. 2015;10:e0139977. doi: 10.1371/journal.pone.0139977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung C.C., Wong C.K. Effects of STC1 overexpression on tumorigenicity and metabolism of hepatocellular carcinoma. Oncotarget. 2018;9:6852–6861. doi: 10.18632/oncotarget.23566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh-Hamad D. Mammalian stanniocalcin-1 activates mitochondrial antioxidant pathways: new paradigms for regulation of macrophages and endothelium. Am. J. Physiol Renal Physiol. 2010;298:F248–F254. doi: 10.1152/ajprenal.00260.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund M.E., To J, O'Brien B.A., Donnelly S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J Immunol.Methods. 2016;430:64–70. doi: 10.1016/j.jim.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Kang C.D., Lee B.K., Kim K.W., Kim C.M., Kim S.H., Chung B.S. Signaling mechanism of PMA-induced differentiation of K562 cells. Biochem.Biophys.Res.Commun. 1996;221:95–100. doi: 10.1006/bbrc.1996.0551. [DOI] [PubMed] [Google Scholar]
- 25.Park S.J., Kang S.Y., Kim N.S., Kim H.M. Phosphatidylinositol 3-kinase regulates PMA-induced differentiation and superoxide production in HL-60 cells. Immunopharmacol.Immunotoxicol. 2002;24:211–226. doi: 10.1081/iph-120003751. [DOI] [PubMed] [Google Scholar]
- 26.Law A.Y., Lai K.P., Lui W.C., Wan H.T., Wong C.K. Histone deacetylase inhibitor-induced cellular apoptosis involves stanniocalcin-1 activation. Exp.Cell Res. 2008;314:2975–2984. doi: 10.1016/j.yexcr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen A., Chang A.C., Reddel R.R. Stanniocalcin-1 acts in a negative feedback loop in the prosurvival ERK1/2 signaling pathway during oxidative stress. Oncogene. 2009;28:1982–1992. doi: 10.1038/onc.2009.65. [DOI] [PubMed] [Google Scholar]
- 28.Yeung H.Y., Chan D.K., Mak N.K., Wagner G.F., Wong C.K. Identification of signal transduction pathways that modulate dibutyryl cyclic adenosine monophosphate activation of stanniocalcin gene expression in neuroblastoma cells. Endocrinology. 2003;144:4446–4452. doi: 10.1210/en.2003-0504. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K.Z., Westberg J.A., Paetau A., von Boguslawsky K., Lindsberg P., Erlander M., Guo H., Su J., Olsen H.S., Andersson L.C. High expression of stanniocalcin in differentiated brain neurons. Am.J.Pathol. 1998;153:439–445. doi: 10.1016/S0002-9440(10)65587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serlachius M., Alitalo R., Olsen H.S., Andersson L.C. Expression of stanniocalcin-1 in megakaryocytes and platelets. Br.J.Haematol. 2002;119:359–363. doi: 10.1046/j.1365-2141.2002.03916.x. [DOI] [PubMed] [Google Scholar]
- 31.Serlachius M., Zhang K.Z., Andersson L.C. Stanniocalcin in terminally differentiated mammalian cells. Peptides. 2004;25:1657–1662. doi: 10.1016/j.peptides.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 32.Yoshiko Y., Maeda N., Aubin J.E. Stanniocalcin 1 stimulates osteoblast differentiation in rat calvaria cell cultures. Endocrinology. 2003;144:4134–4143. doi: 10.1210/en.2003-0130. [DOI] [PubMed] [Google Scholar]
- 33.Luo C.W., Kawamura K., Klein C., Hsueh A.J. Paracrine regulation of ovarian granulosa cell differentiation by stanniocalcin (STC) 1: mediation through specific STC1 receptors. Mol.Endocrinol. 2004;18:2085–2096. doi: 10.1210/me.2004-0066. [DOI] [PubMed] [Google Scholar]
- 34.Serlachius M., Andersson L.C. Up-regulated expression of stanniocalcin-1 during adipogenesis. Exp.Cell Res. 2004;296:256–264. doi: 10.1016/j.yexcr.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 35.Spano A., Barni S., Sciola L. PMA withdrawal in PMA-treated monocytic THP-1 cells and subsequent retinoic acid stimulation, modulate induction of apoptosis and appearance of dendritic cells. Cell Prolif. 2013;46:328–347. doi: 10.1111/cpr.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orecchioni M., Ghosheh Y., Pramod A.B., Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front Immunol. 2019;10:1084. doi: 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanada T., Yoshimura A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002;13:413–421. doi: 10.1016/s1359-6101(02)00026-6. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura A., Mori H., Ohishi M., Aki D., Hanada T. Negative regulation of cytokine signaling influences inflammation. Curr.Opin.Immunol. 2003;15:704–708. doi: 10.1016/j.coi.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimura A., Nishinakamura H., Matsumura Y., Hanada T. Negative regulation of cytokine signaling and immune responses by SOCS proteins. Arthritis Res.Ther. 2005;7:100–110. doi: 10.1186/ar1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton R., Shah S., Altonsy M.O., Gerber A.N. Glucocorticoid and cytokine crosstalk: Feedback, feedforward, and co-regulatory interactions determine repression or resistance. J Biol Chem. 2017;292:7163–7172. doi: 10.1074/jbc.R117.777318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li K., Yao J., Shi L., Sawada N., Chi Y., Yan Q., Matsue H., Kitamura M., Takeda M. Reciprocal regulation between pro-inflammatory cytokine-induced inducible NO synthase (iNOS) and connexin43 in bladder smooth muscle cells. J Biol Chem. 2011;286:41552–41562. doi: 10.1074/jbc.M111.274449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin S., Dorschner R.A., Masini I., Lavoie-Gagne O., Stahl P.D., Costantini T.W., Baird A., Eliceiri B.P. TBC1D3 regulates the payload and biological activity of extracellular vesicles that mediate tissue repair. FASEB J. 2019;33:6129–6139. doi: 10.1096/fj.201802388R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y., Li J., Zhang C. STAT3 promotes the proliferation and migration of hepatocellular carcinoma cells by regulating AKT2. Oncol.Lett. 2018;15:3333–3338. doi: 10.3892/ol.2017.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan K.K., Leung C.O., Wong C.C., Ho D.W., Chok K.S., Lai C.L., Ng I.O., Lo R.C. Secretory Stanniocalcin 1 promotes metastasis of hepatocellular carcinoma through activation of JNK signaling pathway. Cancer Lett. 2017;403:330–338. doi: 10.1016/j.canlet.2017.06.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The list of differentially regulated genes in the transcriptiomic analysis of STC1-knockdown M1-polarized cells.
Comparisons on the gene expression levels of (A) pro-inflammatory, (B) anti-inflammatory markers and (C) STC1 in M0, M1 and M2 macrophages. (A) The induction of the pro-inflammatory markers in M1-polarized cells was remarkably higher than in the M0 and M2 cells. (B) The induction of anti-inflammatory marker expression in M0 and M2-polarized cells was sifnificantly higher than in the M1 cells. (C) The induction of STC1 expression in M1-polarized cells was remarkably higher than in the M0 and M2 cells. Data were presented as the mean ± S.D. Bars with the same letter are not significantly different according to the results of one-way ANOVA followed by Duncan's multiple ranges tests (p < 0.05).
Supplementary material