Significance
The emergence of noxious weeds poses a serious threat to agricultural production. Understanding their origin and evolution is therefore of major importance. Here we analyzed the intriguing case of teosinte, a wild relative of maize originating from Mexico that recently emerged as an invasive weed in maize fields in Europe. Patterns of genetic variation revealed extensive genetic introgression from maize adapted to temperate latitudes into European teosintes. Introgressed genomic regions harbored a key flowering time gene and an herbicide resistance gene. Our results exemplify how adaptive introgression can drive the evolution of a crop’s wild relative into a weed. Hybridization is an evolutionary force that should not be underestimated when forecasting invasiveness risks.
Keywords: plant invasion, rapid adaptation, genetic introgression, flowering time, herbicide resistance
Abstract
Global trade has considerably accelerated biological invasions. The annual tropical teosintes, the closest wild relatives of maize, were recently reported as new agricultural weeds in two European countries, Spain and France. Their prompt settlement under climatic conditions differing drastically from that of their native range indicates rapid genetic evolution. We performed a phenotypic comparison of French and Mexican teosintes under European conditions and showed that only the former could complete their life cycle during maize cropping season. To test the hypothesis that crop-to-wild introgression triggered such rapid adaptation, we used single nucleotide polymorphisms to characterize patterns of genetic variation in French, Spanish, and Mexican teosintes as well as in maize germplasm. We showed that both Spanish and French teosintes originated from Zea mays ssp. mexicana race “Chalco,” a weedy teosinte from the Mexican highlands. However, introduced teosintes differed markedly from their Mexican source by elevated levels of genetic introgression from the high latitude Dent maize grown in Europe. We identified a clear signature of divergent selection in a region of chromosome 8 introgressed from maize and encompassing ZCN8, a major flowering time gene associated with adaptation to high latitudes. Moreover, herbicide assays and sequencing revealed that French teosintes have acquired herbicide resistance via the introgression of a mutant herbicide-target gene (ACC1) present in herbicide-resistant maize cultivars. Altogether, our results demonstrate that adaptive crop-to-wild introgression has triggered both rapid adaptation to a new climatic niche and acquisition of herbicide resistance, thereby fostering the establishment of an emerging noxious weed.
Globalization of trade and transports has considerably accelerated the rate of dispersal of species outside their native range (1). In Europe, the rate of introduction of alien plants has increased exponentially during the last century (2). This rate is expected to further increase in all temperate regions of the Northern Hemisphere due to climate changes (3). Alien plants are a serious threat to native wildlife and its associated ecosystem services and can have direct detrimental consequences on agriculture production or human health (4). Understanding the origin and establishment of invasive plants is therefore of major importance. This includes deciphering the dynamic of the genetic composition of populations associated with founding events and geographical expansion and identifying the adaptive genetic changes sustaining their habitat shifts (5, 6). Such inferences are however challenging when introductions are ancient, histories of invasion complex, and when admixture between multiple source populations has taken place (e.g., ref. 7). It is therefore particularly valuable to access the very early colonization steps in recently introduced species (sensu ref. 8).
Among invasive species, those that colonize agricultural areas are interesting in several respects: they have immediate consequences on crop production sustainability; they may spread rapidly via human-mediated dispersal and farming activities (9); and they display a suite of specific adaptive characteristics also described as “the agricultural weed syndrome” (10). This syndrome includes traits such as seed dormancy, short life cycle, and high fecundity. Two broad categories of agricultural weeds can be distinguished: those that evolved from crop relatives and those that evolved from wild species unrelated to any crop (11). Crop-related weeds display particular mechanisms of adaptation including adaptive genetic introgression from the crop leading to the acquisition of crop-mimicry traits (12, 13). Many crop-related wild species are among the most problematic weeds worldwide. Well-known examples are weedy rice, wild sorghum species, and wild sunflower species (12, 13).
Here we focused on the extremely recent invasion of Europe by emerging noxious weeds related to maize (Zea mays ssp. mays), i.e., the annual teosintes. The European Food Authority has officially reported the presence of teosinte as weeds in maize production areas in Spain and France in 2016 (14). In Spain, teosintes have invaded an area in the provinces of Aragon and Catalonia where they cause important yield loss in maize fields (15). In France, teosintes are present in the north of the Nouvelle Aquitaine region. According to a technical report, French teosintes were first observed in the early 1990s (16). In their native range, teosintes most closely related to maize (i.e., from the Z. mays species) encompass three annual subspecies: ssp. huehuetenangensis with a narrow distribution in western Guatemala (17), ssp. parviglumis (hereafter: parviglumis), and ssp. mexicana (hereafter: mexicana) both encountered in Mexico, the cradle of maize domestication. Parviglumis is considered the ancestor of maize (18, 19) and grows in the west coast lowlands of Mexico under warm and humid tropical conditions. Mexicana grows in the central highlands of Mexico, at elevations up to 2,800 m, under cooler and drier conditions (17). The geographical distributions of these two subspecies slightly overlap and hybridization occurs (20). Interestingly, gene flow from mexicana to maize has contributed to highland adaptation of maize landraces (21). Field observations in Mexico describe parviglumis as forming large populations in natural and seminatural habitats, whereas mexicana is mainly observed as a weed within maize fields, where it can cause severe yield loss (22–24).
While genetic assessment of French weedy teosintes is currently lacking, two previous studies have attempted to establish the origin of Spanish teosintes. Their genetic characterization through single nucleotide polymorphism (SNP) genotyping combined with existing SNP datasets for maize and Mexican teosinte populations has however failed to clearly group Spanish teosintes with either parviglumis or mexicana. Instead, Spanish teosintes were found intermediate between maize and mexicana (25). Microsatellite markers further confirmed the importance of maize contribution to the genetic makeup of Spanish teosintes (26). Here, we collected French teosinte populations and describe their genetic diversity. This dataset was combined with previously published ones in order to 1) elucidate the taxonomic origin of French teosintes and identify the source populations, 2) assess their genetic similarity with Spanish teosintes, 3) describe the extent of genetic admixture between European teosintes and cultivated maize, and 4) identify genomic regions that have contributed to the successful adaptation of teosintes as weeds in European maize fields.
Results
Spanish and French Teosintes both Originate from Z. mays ssp. mexicana.
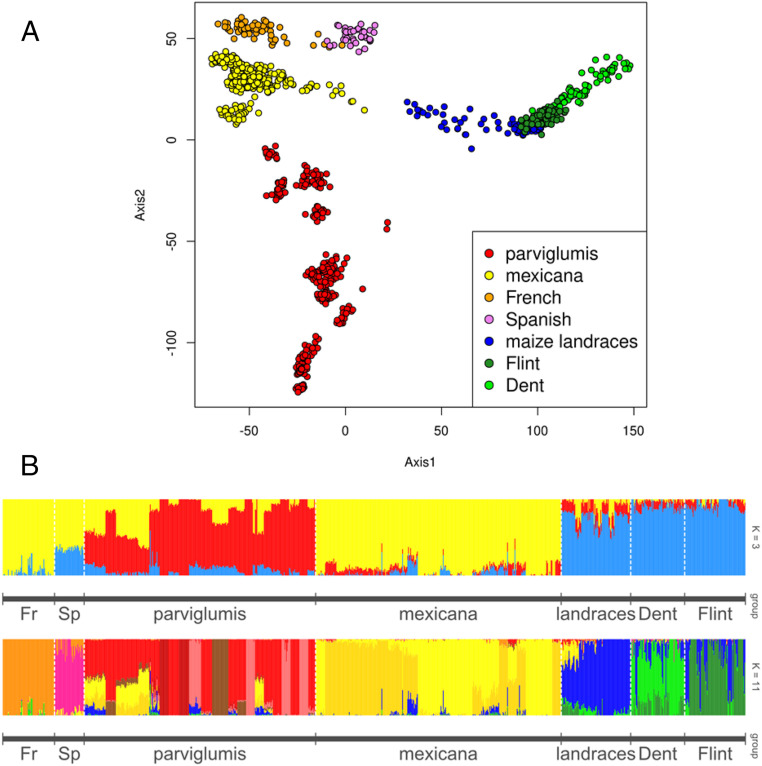
Phenotypic data from a common garden experiment conducted in Dijon demonstrated that French teosintes displayed two morphological characteristics, sheath pubescence (52% of plants) and red-colored sheaths (75% of plants), that were observed also in some mexicana plants but not in parviglumis, as previously described (20) (SI Appendix, Fig. S1). Genetic variation at 24,544 SNP data were first investigated using principal component analysis (PCA). A clear genetic structuring between teosintes and maize appeared along the first axis that explained 8% of the variation (Fig. 1A). The second axis representing 4.6% of the variation, separated parviglumis, mexicana, and the European teosintes into three nonoverlapping groups (Fig. 1A). This second axis revealed a much closer proximity of European teosintes to mexicana than to parviglumis.
Fig. 1.
Genetic structure based on SNP data for teosinte samples collected in France combined with available SNP data for Spanish teosintes, wild teosintes populations from Mexico, and cultivated maize accessions (24,544 SNPs and 1,005 individuals). (A) Principal Component Analysis with axes 1 and 2 (12.6% of the variation explained). (B) Population structure and admixture patterns revealed by fastStructure. Each color represents a genetic group and individuals (vertical lines) are partitioned into segments whose length represents the admixture proportions from K genetic groups. Ancestry proportions are shown for K = 3 genetic groups (Top) and K = 11 genetic groups (Bottom). At K = 11, the following nine reference genetic groups were identified in Mexican teosintes and maize: PARV1 (red), PARV2 (dark red), PARV3 (light pink), PARV4 (brown), MEX1 (yellow), MEX2 (gold), TROP (blue), DENT (light green), and FLINT (dark green).
Results from fastStructure (27) at the subspecies level (K = 3, ssp. parviglumis, ssp. mexicana, ssp. mays) further supported this observation, with European teosintes being of predominant mexicana ancestry (Fig. 1B). Increasing the number of genetic groups to K = 11 (Fig. 1B and Dataset S1) confirmed previous reports defining, in addition to the Spanish and French teosintes, nine reference genetic groups (28–32): 1) parviglumis accessions clustered into four geographical genetic groups (hereafter, PARV1, PARV2, PARV3, and PARV4) (Fig. 1B and SI Appendix, Fig. S2); 2) some parviglumis populations were highly admixed with mexicana (29); 3) mexicana accessions grouped into two genetic clusters (hereafter MEX1 and MEX2) corresponding to geographical races “Chalco” and “Central Plateau,” respectively (17, 29, Fig. 1B, and SI Appendix, Fig. S2); and 4) in maize, the three observed genetic clusters corresponded to three major germplasm pools (Fig. 1B), the tropical landraces, the Dent inbred lines, and the Flint inbred lines (hereafter TROP, DENT, and FLINT), with admixture among them (31, 32). In agreement with the PCA results, the results of fastStructure at K = 11 separated French and Spanish teosinte populations in two distinct genetic clusters, different from the nine reference genetic groups found across parviglumis, mexicana, and maize clusters.
Consistently with results at K = 3, the average pairwise genetic differentiation FST between the French/Spanish teosintes with mexicana accessions (0.138/0.195) was on average smaller than with the parviglumis populations (0.195/0.213) (SI Appendix, Table S1). Pairwise FST between French and Spanish teosinte populations was 0.237, a value greater than that observed between mexicana and parviglumis populations (0.105). Genetic diversity within groups as measured by Nei’s heterozygosity was similar for Spanish (0.251) and French teosintes (0.221). These values stand within the range of genetic diversity estimates both within mexicana (0.273 for MEX1 and 0.258 for MEX2) and within parviglumis clusters (ranging from 0.131 to 0.305; SI Appendix, Table S1).
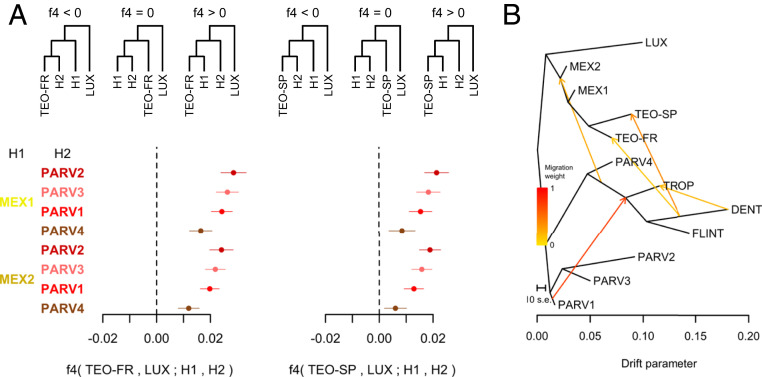
We further employed the f-statistics framework (33) to test histories of divergence among parviglumis, mexicana, and the European teosintes, using Zea luxurians as an outgroup. Observed values of f4(French or Spanish teosinte, Z. luxurians; mexicana, parviglumis) were consistently significantly positive, again arguing in favor of a tree topology where both the French and the Spanish teosinte populations are more closely related to mexicana than to parviglumis (Fig. 2A). The f4 values observed for the two mexicana genetic clusters, MEX1 and MEX2, however, were similar, so that the origin of European teosinte could not be more precisely refined using this statistic.
Fig. 2.
Origin of European teosintes and admixture with maize. (A) Genetic relationships between European teosintes and their two putative ancestors, mexicana (H1) and parviglumis (H2), inferred using the four-populations (f4) test. Each of the reference populations inferred by fastStructure for mexicana (two groups: MEX1 and MEX2) and parviglumis (four groups: PARV1, PARV2, PARV3, and PARV4) were tested for ancestry relationship. Z. luxurians (LUX) is used as the outgroup. Theoretical tree topologies and the corresponding sign of the f4-statistics are shown at the Top. Points indicate observed f4 values for each pair of mexicana and parviglumis reference populations, with horizontal bars showing 3.3 SEs. Inference was made separately for French teosinte (TEO-FR, on the Left) and Spanish teosinte (TEO-SP, on the Right). (B) TreeMix analysis: maximum-likelihood tree showing the relationships among French teosintes (TEO-FR), Spanish teosintes (TEO-SP), and the nine reference groups identified from the fastStructure analysis for parviglumis accessions (PARV1, PARV2, PARV3, and PARV4), mexicana accessions (MEX1 and MEX2), and cultivated maize: tropical landraces (TROP), Dent inbred lines (DENT), and Flint inbred lines (FLINT). Five migration events were inferred and are shown on the tree as arrows connecting genetic groups. Yellow to red color indicates the intensity (weight) of each migration event.
Footprints of Admixture from Maize to the European Teosintes.
The fastStructure analysis detected footprints of maize admixture within French and Spanish teosintes (Fig. 1B). In order to examine admixture patterns in more detail, we used TreeMix (34) to reconstruct phylogenetic relationships among the nine reference genetic groups defined by fastStructure (ancestry > 0.8, Dataset S1) and the European teosintes. Without migration, the topology inferred was in agreement with the known relationships among subspecies (SI Appendix, Fig. S3A). European teosintes were most closely related to mexicana. This topology explained 98.4% of the observed covariance among populations. Adding five migration events increased the proportion of variation explained (99.7%), with the likelihood reaching an asymptote (SI Appendix, Fig. S3 C and D). In the maximum-likelihood tree with five migration events (Fig. 2B), both French and Spanish teosintes were closest to the mexicana reference group MEX1, the Chalco mexicana group.
TreeMix analyses pinpointed migration between Dent and Tropical maize lines, likely reflecting the admixed origin of Corn Belt Dents between Northern Flint ancestors and tropical material (31). There was also evidence for admixture between maize (ancestral node or edge) and both parviglumis and mexicana. Those events are well documented (18, 20, 21). More importantly, both French and Spanish teosintes displayed admixture from the Dent maize reference group (Fig. 2B). Note that the migration event between Dent maize and Spanish teosintes was the most supported, with an estimated weight of 0.39, while the migration edge between Dent maize and French teosintes was added in third (SI Appendix, Fig. S4, estimated weight = 0.14).
Admixture between European teosintes and each of the three maize reference group was further tested using a four-population test where each mexicana reference group was used as a sibling population: f4(European teosinte group; mexicana reference group; maize reference group, parviglumis). We first verified that none of the reference mexicana group was itself admixed with either maize or parviglumis by estimating f4(MEX1, MEX2; maize, parviglumis), which was consistently not significantly different from zero (SI Appendix, Table S2). The four-population tests for admixture in French or Spanish teosinte instead were all significant (SI Appendix, Table S2). In agreement with estimated weights for migration edges, Z-scores were greater for Spanish teosintes in comparison to French teosintes, and for the Dent maize group in comparison to Tropical and Flint. The proportion of Dent maize ancestry estimated using the f4 ratio estimator was 0.122 (95% confidence interval 0.114 to 0.131) for French teosintes and 0.422 (95% confidence interval 0.411 to 0.434) for Spanish teosintes.
Introgression from Maize Has Contributed to the Adaptation of Teosintes in Europe.
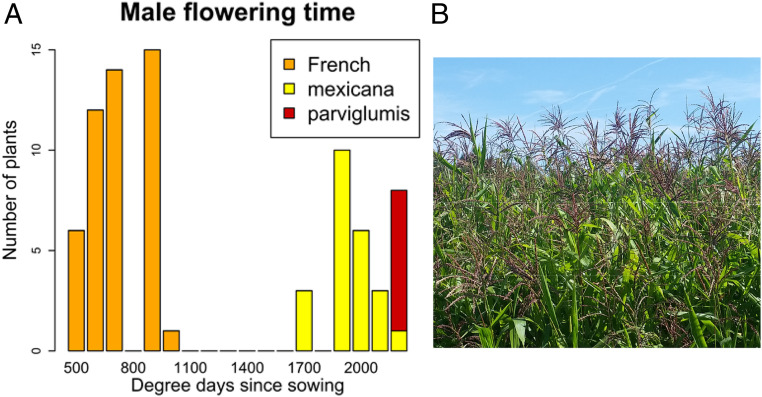
As tropical, short-day plants, native teosintes flower very late or not at all at higher latitudes (35, 36). A shift toward long-day flowering was therefore necessary for European teosintes to adapt to temperate latitudes. To verify these predictions, we grew plants from all French and from 12 populations of native teosintes in Dijon, France. French teosintes initiated their male flowering from the end of June to the end of July (534 to 1,059 growing degree days after sowing). In contrast, we observed a much-delayed transition to flowering in native teosintes. A single plant out of 24 Mexican mexicana and a majority of Mexican parviglumis plants (17 out of 24) remained vegetative until the end of the experiment (at the beginning of November). When occurring, flowering started at the beginning of September (1,703 degree days) in mexicana and at the end of October (2,221 degree days) in parviglumis (Fig. 3A). The flowering of most French teosintes overlapped with the flowering of the three European maize varieties used as controls (which flowered respectively at 782, 904, and 987 degree days). Synchronous flowering with maize was also observed in infested fields (Fig. 3B). Altogether, these results suggested that flowering-time genes played a prominent role in teosinte adaptation to European day length.
Fig. 3.
Flowering phenology in European teosintes. (A) Time to male flowering assessed from a common garden experiment in Dijon, France. The histogram shows the number of degree days from sowing to tassel initiation in 48 plants of French teosintes, 24 Mexican teosintes of the subspecies mexicana, and 24 Mexican teosintes of the subspecies parviglumis. One mexicana plant and 17 parviglumis are not represented as they were still in a vegetative stage at the end of the experiment. (B) A teosinte population within a maize field in France. Flowering is synchronous between teosintes and maize.
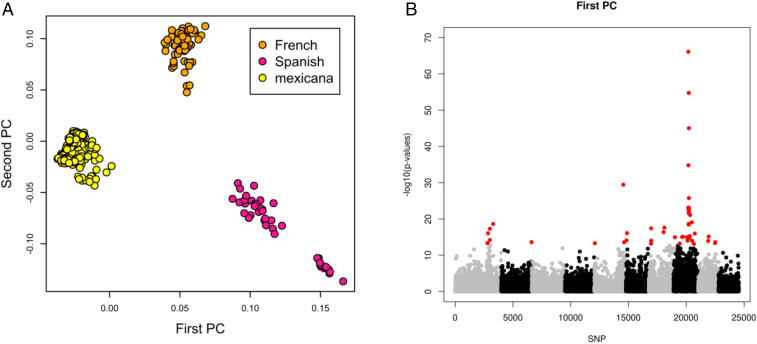
We sought signatures of selection using our SNP data, paying specific attention to flowering time genes. We first conducted a PCA using mexicana and European accessions. Second, we performed a genome scan using pcadapt (37) based on squared loadings of the two first principal components (SI Appendix, Fig. S5). The two first principal components together explained 15% of the variation. The first component captured the differentiation between native mexicana and European teosintes (Fig. 4A), and we detected 45 outlier SNPs significantly associated with it at a false discovery rate (FDR) of 0.1% (Fig. 4B). The second component mainly differentiated the French teosintes from the Mexicana and Spanish teosintes (Fig. 4A) and revealed only 2 significant outlier SNPs (SI Appendix, Fig. S6). Twenty-five out of the 45 outliers detected with component 1 (>55%) were found on chromosome 8 (Fig. 4B). Interestingly, we observed elevated levels of genetic differentiation (FST > 0.60) of both French teosintes and Spanish teosintes with native mexicana at a subset of 12 outlier SNPs all located on the same region of chromosome 8 (SI Appendix, Fig. S7).
Fig. 4.
Detection of outlier SNPs along the 10 chromosomes of the genome based on a Principal Component Analysis of mexicana accessions, French teosintes and Spanish teosintes using pcadapt. (A) Projection of accessions onto axes 1 and 2 of the Principal Component Analysis. (B) Manhattan plot of the P values of SNPs with the first principal component of the PCA. The 45 top SNPS having q-values less than 0.1% are displayed in red.
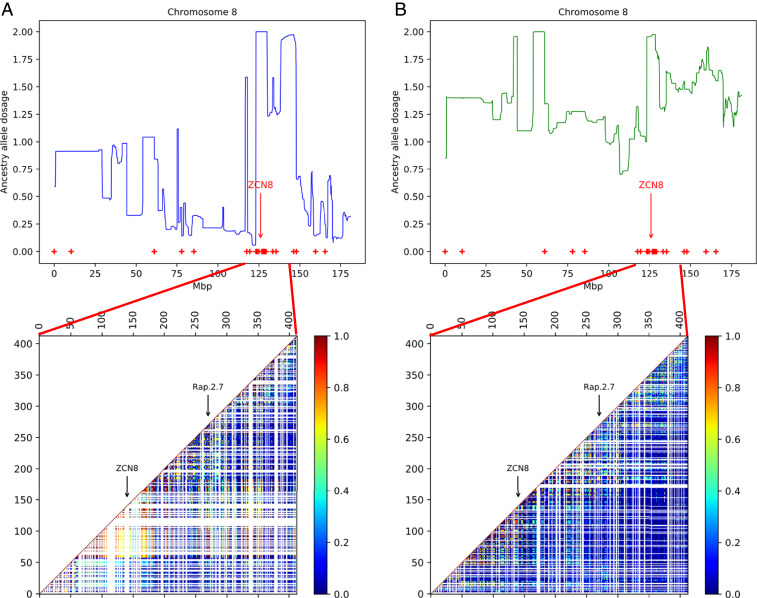
We hypothesized that this pattern resulted from a recent adaptive introgression of a maize chromosomal fragment into the European teosintes. Searching for maize introgression using the ELAI software indeed revealed a peak of introgression from maize to the European teosintes on chromosome 8 (Fig. 5). A cluster of nine adjacent outlier SNPs distant by less than 10 kb was located under this peak. Remarkably, this cluster included the major maize flowering time gene ZCN8 (38). Note that a second major flowering time gene, RAP2.7, and its regulator, VGT1 (39), were located in the introgressed region but not in close vicinity to any of the outlier SNPs. Patterns of linkage disequilibrium (LD) in the region surrounding these two candidate genes uncovered a signal of elevated LD around ZCN8 in French teosintes and to a lesser extent in Spanish teosintes (Fig. 5). We recovered no such pattern around RAP2.7. A detailed examination of genotypes along chromosome 8 further confirmed the presence of a high-frequency extended haplotype at ZCN8 in French teosintes. This same haplotype seems present in the Spanish teosinte but is still segregating, suggesting weaker or partial selection at this position if any (SI Appendix, Fig. S8).
Fig. 5.
Patterns of local ancestry as inferred by ELAI along chromosome 8 and the matrix of LD in a region that includes the candidate genes ZCN8 and RAP2.7 for (A) French teosintes and (B) Spanish teosintes. The ELAI plots show ancestry allele dosages (y axis) for Dent cultivated maize along chromosome 8 (x axis). Positions of the outlier SNPs identified with pcadapt are marked with red dots. LD was estimated using the r2 statistics for all pairs of SNPs in a region of about 30 Mb starting 10 Mb upstream of ZCN8 and ending 10 Mb downstream of RAP2.7 (positions 116,880,531 to 146,012,084 on reference genome B73 v4). SNP positions at which LD could not be calculated (absence of polymorphism or missing data) are marked in white.
Acquisition by French Teosintes of an Herbicide Resistance Gene Introgressed from Maize.
In France, maize cultivars resistant to the herbicide cycloxydim carry an allele of the gene encoding acetyl-CoA carboxylase 1 (ACC1) with a mutation that confers resistance to herbicides inhibiting this enzyme (Duo System, ref. 40). Because French teosintes grow in the vicinity of such cultivars, we assessed their sensitivity to cycloxydim. Out of 200 French teosinte seedlings assayed, 86 (43%) were rated resistant to cycloxydim and 114 (57%) were rated sensitive. As controls, we used a sensitive (RGT TetraXX) and a resistant (RGT CarmiDuo) maize cultivar. As expected, all plants from the former cultivar were rated sensitive while all plants from the latter were rated resistant. To explore the possibility that resistant alleles were transferred from maize to teosintes, we genotyped the ACC1 gene in teosintes and maize at all known codons involved in herbicide resistance. We detected one mutation at codon 1781 (Ile to Leu) in the resistant maize variety (RGT CarmiDuo) and in some of the French teosintes. All CarmiDuo maize plants were homozygous mutant. All plants from the sensitive maize cultivar (TetraXX) were homozygous wild type. Among the French teosinte plants assayed for herbicide sensitivity, all 86 herbicide-resistant plants were homozygous mutants at codon 1781. Among the 114 herbicide-sensitive teosinte plants, 78 were heterozygous and 36 were homozygous wild type.
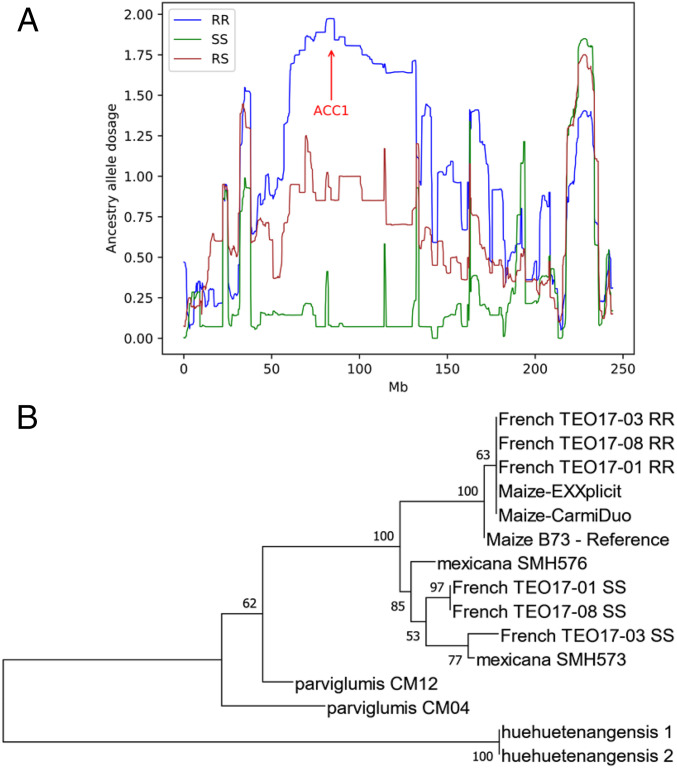
ACC1 is located on chromosome 2, in a region where a peak of squared loadings was observed with the second principal component of the PCA differentiating the French from the Spanish teosintes (SI Appendix, Fig. S6). Although it did not pass the FDR at 0.1%, this region was associated with a large divergence between French and Spanish teosintes (Fig. 4A). When examining the pattern of introgression over chromosome 2 using ELAI, it was clear that a large genomic region encompassing ACC1 and spanning more than 500 SNPs from our genotyping chip had been introgressed from maize into homozygous ACC1-mutant teosinte plants, whereas wild-type plants showed no introgression and heterozygotes showed the expected halved ancestry dosage (Fig. 6A). Sequencing the full genomic DNA corresponding to the coding sequence of ACC1 revealed that all mutant teosintes carried one same ACC1 allele that was exactly identical to the allele carried by herbicide-resistant maize cultivars, whereas wild-type ACC1 alleles carried by nonmutant French teosintes clustered with the alleles carried by the mexicana accessions (Fig. 6B). Taken together, these results demonstrate that the mutant ACC1 allele present in French teosinte was introgressed from cultivated, herbicide-resistant Duo System maize.
Fig. 6.
Introgression of a mutant ACC1 gene from maize into French teosintes. (A) Pattern of local ancestry in French teosintes along chromosome 2 as inferred by ELAI. The plot shows ancestry allele dosages (y axis) for Dent maize along the chromosome (x axis). Individuals homozygous for the mutant allele (RR) at ACC1 are shown in blue, heterozygous (RS) in brown, and homozygous for the nonmutant (SS) allele are shown in green. The red arrow points to the SNP that is closest to the ACC1 gene. (B) Neighbor-joining tree showing the relationships among Acetyl-CoA carboxylase gene sequences in two herbicide-tolerant maize varieties (EXXplicit and CarmiDuo), in the reference B73 maize inbred line, in French teosinte accessions homozygous for the wild-type ACCase allele (SS) or homozygous for the resistant mutant allele (RR) and in Mexican teosinte accessions (two mexicana, two parviglumis, and two Z. mays ssp. huehuetenangensis).
Discussion
We report here the genetic description of the very recent settlement in France of teosintes, which are recognized in their native tropical range as a major threat to agricultural production (41). We addressed three main questions: Where do they originate? How did they adapt to Europe? To which extent has introgression from maize facilitated their establishment?
Our results clearly assigned both French and Spanish teosintes (25) to Z. mays ssp. mexicana, and suggested a single geographical origin for all invasive populations of teosintes reported to date in Europe. We identified the source genetic group as the mexicana race Chalco (MEX1). This finding is interesting in at least two respects: First, Chalco teosintes are located at elevations ∼2,300 m and higher (29). They are therefore adapted to moderate rainfall and low temperatures (17), environmental conditions that are closer to the European climate than the Mexican tropical lowlands. Second, Chalco teosintes are the ones that hybridize the most frequently with maize (42, 43). Plants carrying hybrid-like cobs have been reported frequently in Mexico for Chalco teosintes growing within or near maize fields (23, 24). In fact, Chalco teosintes have consistently been described as weeds infesting cultivated maize fields (22, 24, 41). This parallels the sites colonized in France and Spain, which are chiefly maize fields (15).
However, the native and introduced ranges differ strongly in their latitude and hence their photoperiod, short days in the native range versus long days in the introduced one. In Mexico, mexicana occurs at latitudes comprised roughly between 18° North and 20° North. In Europe, invasive mexicana are observed at latitudes comprised between 42° North (in Spain) and 46° North (in France), which would correspond to an area north of Chicago in the United States. Flowering is accelerated by short days in native teosintes (35). We indeed confirmed that native mexicana populations flowered very late in France and were unable to produce seeds before maize harvest time. On the contrary, the flowering period of French teosintes was much earlier and overlapped that of maize. The establishment of mexicana in Europe therefore most likely involved a substantial genetic shift in the control of flowering time.
Given the narrow time window for adaptation to occur from de novo mutations (two to three decades), we combined outlier detection in European teosintes and the examination of introgression patterns from maize to test whether preadapted local maize varieties had contributed to teosinte adaptation. In line with this hypothesis, we detected introgression in both Spanish and French teosintes. Interestingly, we observed a marked pattern of introgression in a genomic region that contains ZCN8, a gene that underlies one of the largest maize flowering time quantitative trait loci (QTL). Consistently, this region was enriched for outlier SNPs displaying high differentiation between native and European mexicana populations. ZCN8 is a key floral activator of the maize flowering time pathway also known to be involved in photoperiod sensitivity (44). Guo et al. (38) have shown that two polymorphisms with an additive effect in the promoter of ZCN8 are associated with early flowering time under long days. These polymorphisms exist as standing variation in mexicana. They have been under strong selection during early maize domestication and contributed to latitudinal adaptation in this crop (31, 38). While introduction to Europe of a preadapted, early-flowering mexicana population is a possibility, here we propose that introgression from a maize early flowering variant at ZCN8 opened up a new niche for weedy teosintes in Europe. Indeed, a clear signal of selective sweep was observed around ZCN8 in French teosintes, consistent with a single event of adaptive introgression, i.e., hard sweep signature. Interestingly, a similar haplotype was observed in Spanish teosintes, albeit with a greater level of heterozygosity, which suggests an ongoing, incomplete selective sweep (SI Appendix, Fig. S8). Genetic introgression was not limited to this genomic region but was pervasive in all European teosintes (SI Appendix, Fig. S9). Given the complexity and number of genes involved in the regulation of maize flowering time, we suspect several genes other than ZCN8 to contribute to the substantial shift in flowering time in European teosintes (see SI Appendix, Fig. S9 and Dataset S2 for a proposed list of candidate genes). Our experimental design, however, recovered no specific selection signal at any of the a priori flowering time candidate genes.
We pinpointed the Dent genetic group as the most likely donor of the introgressed segments. Because modern maize varieties deriving from the Dent germplasm are widely cultivated in Europe but not in Mexico, where tropical germplasm is dominant (32, 45), this suggests that hybridization has occurred after the introduction of mexicana to Europe. In other words, our results are consistent with a scenario where European maize varieties adapted to temperate latitudes have contributed to the establishment of teosintes in Europe. Note that hybridization is seemingly still occurring, as plants carrying hybrid-like cobs are regularly observed in infested maize fields in Spain (25, 26) as well as in France (SI Appendix, Fig. S10). One surprising outcome of our study is the much lower global introgression rate in the French as compared to the Spanish teosintes (SI Appendix, Fig. S9), despite the seemingly earlier French introduction. Introgression segments can convey alleles that are deleterious for the maintenance of weediness traits and eliminated over time by purifying selection (13), perhaps explaining subtler introgression patterns in French teosintes. Along this line, we detected very little introgression from maize to European teosintes on an extended portion of the short arm of chromosome 4 (SI Appendix, Fig. S9). This region encompasses a large domestication QTL hotspot containing several loci involved in the variation of domestication traits between maize and teosinte, as well as the incompatibility locus TCB1 (46, 47, see SI Appendix, Fig. S9 and Dataset S2). Our results, together with those of a previous study (43) strongly suggest that introgression from maize to mexicana is counterselected in this genomic region. In European teosintes, such selection most likely contributes to preserve wild alleles necessary for the maintenance of the weedy phenotype.
Last, but not least, we detected a second adaptive introgression specific to the French teosintes and involving a large region of chromosome 2. This region encompasses an allele of the herbicide-target gene ACC1 carried by herbicide-resistant maize cultivars. Such cultivars have been authorized in France since 2001 and are cultivated in the area where teosintes occur (16). Teosinte plants homozygous for this mutant allele are herbicide resistant, which is clearly an adaptive trait in agricultural fields. Since the history of cultivation of resistant maize cultivars in France is quite recent, it follows that the introgression of the ACC1 region in French teosintes is also very recent, consistent with the fact that a large region of chromosome 2 encompassing ACC1 was introgressed.
In conclusion, while introgression has been proposed as a key source of adaptive genetic variation (48, 49), establishing it has been particularly challenging with only a handful of reported cases. Notorious examples often illustrate the contribution of wild relatives to domesticated gene pools (50), more rarely the reverse (but see 51–53). This is because crop-to-wild gene flow may not always be beneficial since many characteristics making crops suitable to cultivated environments are detrimental for wild or weedy forms (e.g., nonshattering seeds, lack of dormancy, bolting time) (10). In some instances, however, a crop allele can provide a clear advantage to a wild or weedy form, e.g., by conferring a given resistance or by allowing a niche shift. Here, we present two instances of clear evidence of adaptive introgression from locally adapted crop varieties to a wild relative. One introgression facilitated reproduction under temperate latitudes; the other enabled plants to thrive in herbicide-treated fields. Together, those introgressions contributed to the establishment of a new weed. Previous studies have reported a low rate of spontaneous hybridization between mexicana and cultivated maize, less than 1% per generation (54). However, first-generation hybrids display a great vigor, and are highly male fertile (55). We propose that the rare first-generation hybrids served as a bridge for the transfer of maize genes into mexicana populations, fostering their local adaptation. This result nicely parallels the contribution of mexicana alleles to highland maize landraces adaptation (42). In sum, we demonstrate that crop-wild introgression can be a two-way street, allowing the transfer of beneficial variants to both partners. Our work highlights the importance of introgression in allowing large evolutionary shifts or even opening up new niches. In the case of maize and teosinte, the common consensus was that given their ecology and biology, the risk of seeing teosinte emerge as a problematic weed under a temperate climate was remote (17, 20). Here we not only show that such risk exists, but more generally that crop-wild introgression should not be underestimated when forecasting invasiveness risks.
Materials and Methods
Plant Material and Genotyping.
Teosinte seeds were collected from eight cultivated fields in the region of Nouvelle Aquitaine, France, in autumn 2017 (SI Appendix, Table S3). Geographic distances between fields varied from 0.25 kilometers to 11 kilometers. In spring 2018, seeds were germinated in growth chambers at 25 °C and 16-h day length. Leaf samples were harvested from a total of 70 French teosintes individuals (4 to 14 individuals per field population). Leaf fragments were ground in liquid nitrogen and DNA was extracted using NucleoSpin Plant II (Macherey-Nagel). Genotyping was performed by Eurofins Genomics using the Illumina MaizeSNP50 BeadChip array (Illumina). SNPs were called using the GenomeStudio algorithm (Illumina). Out of 56,110 markers contained on the chip, 49,574 could be successfully genotyped on all plants.
Phenotypic Assays.
A common garden experiment was conducted in 2018 at INRAE in Dijon, France (47.32°N; 05.10°E) to compare phenotypic variation in French and Mexican teosintes. Seed material from six populations of the subspecies mexicana and six populations of the subspecies parviglumis (collection described in 56, 57) was used as reference material in the common garden (see below).The experiment included 48 plants from the eight populations of French teosinte (six plants per population), 24 plants from the six mexicana populations (three to five plants per population) and 24 plants from the six parviglumis populations (four plants per population). We included three maize varieties commercialized in France: ES Gallery (36 plants), RGT CarmiDuo (12 plants), and RGT TetraXX (12 plants). All seedlings were transplanted 1 wk after sowing and arranged in a semirandomized single block design with alternate rows of teosinte and maize. The experiment was set up on May 24 and ended on November 6. We measured traits related to early growth and architecture (plant height, number of leaves on the main tiller, and number of primary tillers), leaf shape (length, width, and their ratio), the presence of trichomes on leaf sheaths, sheath color, and flowering time (time to emission of the tassel and time to silking). Due to the much-delayed transition to reproduction in Mexican teosintes (see results), postreproductive traits were not considered.
Herbicide Sensitivity Bioassay.
French teosintes have almost exclusively been observed in maize-growing fields. Growers in the area where teosintes are present have tried to control it using herbicide-resistant maize cultivars (Duo System, BASF) that withstand the application of the herbicide cycloxydim. Bioassays were conducted to assess the herbicide sensitivity of French teosinte seedlings issued from seeds from the eight populations collected in maize fields. Seedlings were grown in individual pots in a glasshouse at 22/18 °C day/night with a 14-h photoperiod. At the two-leaf stage, cycloxydim was applied as the commercial herbicide Stratos Ultra (BASF, 100 g/L cycloxydim) at the recommended French field rate (200 g/ha cycloxydim) on 200 teosinte seedlings (25 per population) and on 25 seedlings from each of one classical (RGT TetraXX) and one herbicide-resistant (RGT CarmiDuo) maize cultivar that were included as herbicide-sensitive and herbicide-resistant controls, respectively. Twenty-five additional French teosinte seedlings and 25 seedlings of each maize cultivar were sprayed with water to serve as an untreated control. After 48 h, the last 0.5 cm of the first leaf of every sprayed seedling was collected for ACCase genotyping (see below). Plant phenotypes were rated 3 wk after herbicide application, when herbicide-sensitive control maize plants were clearly dead. Plants killed by the herbicide were rated sensitive (S), while surviving plants were rated resistant (R).
SNP Array Data.
Genotype data for the 70 French teosintes was combined with published and available data for the following material: 40 accessions of Spanish teosintes (25), 314 accessions of parviglumis (28, 29), 332 accessions of mexicana (28, 29), 94 maize landraces from Meso- and Central America (58), and 155 maize inbred lines from North America and Europe (32). We only kept SNPs that were shared and correctly scored among the different datasets; the final combined dataset consisted of 24,544 SNPs genotyped on 1,005 accessions (59) (https://zenodo.org/record/3959138). For analyses requiring an outgroup, we included the SNPs data available for 12 accessions of Z. luxurians (25) using the 24,544 markers above.
Acetyl-CoA Carboxylase Genotyping and Sequencing.
Herbicide-resistant, Duo System maize cultivars grown in French fields to control teosinte populations all carry an herbicide-resistant mutant allele of ACC1, one of the two maize acetyl-CoA carboxylases (40). The mutation involved has not been published, but the major acetyl-CoA carboxylase codons involved in herbicide resistance are known (codons 1781, 1999, 2027, 2041, 2078, 2088, and 2096 as standardized in ref. 60). Two herbicide-resistant maize cultivars (RGT CarmiDuo and RGT EXXplicit) and the 200 French teosinte plants used in herbicide sensitivity bioassays were genotyped at these codons. The sequences of the two maize acetyl-CoA carboxylases homeologs (GenBank accessions XM_020548014 for ACC1 and XM_008664827 for ACC2) were aligned and gene-specific primers were designed for ACC1. Primers pairs AC1ZM3/AC1ZM3R and AC1ZM2/AC1ZM2R (SI Appendix, Table S4) were used to amplify ACC1 regions carrying codon 1781 and codons 1999 through 2096, respectively. Mutations were sought in the amplicons obtained using previously described assays (60).
The ACC1 protein-coding sequence of 12,002 nucleotide with its 32 introns was fully sequenced on both strands in 14 individual plants: one plant from each of the two herbicide-resistant maize cultivars, three French teosinte individuals homozygous mutant at ACC1, and three homozygous wild type at ACC1 as determined after genotyping, one parviglumis individual in each of two Mexican populations, one mexicana individual in each of two Mexican populations, and two Z. mays ssp. huehuetenangensis individuals that were used as an outgroup. PCR primers used for sequencing are in SI Appendix, Table S4. All sequences were aligned with the maize reference ACC1 sequence (GenBank XM_020548014). A phylogenetic tree was generated using the neigbor-joining method as implemented in Mega 10.0.5 (61) with 1,000 bootstraps.
Population Genetic Structure.
A PCA was conducted using the Adegenet R package (62). The clustering program FastStructure (27) was run to evaluate ancestry proportions for K genetic groups, with K varying from 1 to 12 with five replicates for each value of K and using the “simple prior” option (flat beta-prior over allele frequencies). To evaluate the repeatability across runs, and rule out true multimodality (as opposed to cluster labels switching), we ran the program CLUMPP v.1.1.2 using the Greedy algorithm (63). Genetic diversity within each genetic group and pairwise genetic differentiation (FST) values were calculated using the last version of the EggLib package (64).
Origin of European Teosintes.
In this analysis, we aimed at inferring the Mexican origin of European teosintes. We first defined Mexican reference groups of parviglumis and mexicana. We considered the results from FastStructure at K = 11, as this was the value for which the observed genetic clustering for Mexican teosintes and maize was in best agreement with previous studies (28–32). This clustering revealed six teosinte genetic groups (four from parviglumis, two from Mexicana) as well as three maize genetic groups (tropical landraces, Dent and Flint inbred lines). We retained individuals with an ancestry higher than 0.8 in each group. This set of 628 individuals defined our nine reference groups (Dataset S1).
To get a first insight on the proximity between each European teosinte population (Spanish and French) and the reference groups of parviglumis and mexicana, we used the f-statistics first introduced by Reich (65). F-statistics provide a measure of genetic drift between populations, based on the branch length separating them on a simple phylogeny (66). The four-population f4-statistics can be used to investigate ancestry relationships and find the closest relative of a contemporary population by comparing different tree topologies (19, 33). We used f4 (European teosinte, Z. luxurians; mexicana, parviglumis), where mexicana and parviglumis are the two putative ancestors to European teosintes and Z. luxurians is an outgroup. The value of this f4 statistics is expected to be positive if the European teosinte descends from mexicana, negative if it descends from parviglumis, and null in the case of no ancestry relationship (see ref. 19 for a similar analysis). Observed f4 values were calculated using the fourpop program in TreeMix 1.13 (34). Note that we make here the implicit assumption of no gene flow between reference groups. We considered more complex scenarios in the following section.
History of Admixture among Teosintes and Maize.
We inferred the relationship between cultivated maize and teosintes using TreeMix 1.13 (34) on the nine reference genetic groups defined above. The analysis was based on SNP allele frequencies in Spanish teosintes, French teosintes, and the nine reference genetic groups. Maximum-likelihood trees were built using 200 SNP windows to account for linkage disequilibrium. We tested the addition of 0 to 10 migration events, by building 10 replicate trees for each. We considered as the most meaningful number of migration events the first value at which the mean likelihood of trees and the proportion of explained covariance among groups stabilized toward their maximum asymptotic values.
As both the comparison of f-statistics for varying tree topologies and TreeMix results assigned mexicana as the most likely ancestor of European teosintes, we performed a four-population test (65, 66) considering (test population, mexicana; maize, parviglumis). We estimated the f4 statistics for all combinations of the test population being either Spanish or French teosintes, and considering the two mexicana reference groups (MEX1, MEX2), the three maize reference groups, and all parviglumis reference groups grouped together. The expected value of this f4 statistics is zero if (test population, mexicana) and (maize, parviglumis) form two independently diverged clades. Significant deviation from zero indicates admixture. Before implementing this test, we verified that the two reference mexicana groups were not themselves admixed with either maize or parviglumis, which would confuse interpretation. We did so by estimating f4 (MEX1, MEX2; maize, parviglumis). Finally, under an admixture scenario of the test population with maize, the admixture proportion in the test population was estimated as the ratio of the two statistics f4 (parviglumis, Zea luxurians; test population, mexicana) and f4 (parviglumis, Zea luxurians; maize, mexicana). Here, in line with the reasoning in Patterson et al. (66) and Peter (33), we used Zea luxurians as the outgroup, mexicana and maize as the two potential contributors to the admixed test population, and parviglumis as a subspecies more closely related to one of the contributors, here to maize. A 95% confidence interval for the admixture proportion was obtained from a block jackknife procedure, where each block of 200 SNPs was removed in turn.
Signatures of Selection and Genomic Patterns of Introgression.
A genome-wide scan for signature of positive selection in European teosintes was performed using a principal component analysis over all European teosintes and mexicana populations as implemented in pcadapt (67). In contrast to FST-based approaches, pcadapt does not require any a priori grouping of individuals into populations. It is well suited to scenarios of population divergence and range expansion, as principal components are able to discriminate successive divergence and selection events (37). We performed the analysis for each principal component (component-wise method) and used the loadings (correlation between each PC and each SNP) as the test statistic. Outlier SNPs were identified by transforming the P values into q values with a cutoff value of 0.001, ensuring a false discovery rate lower than 0.1% using the R package qvalue (68).
We investigated genome-wide patterns of introgression from cultivated maize using the ELAI software (69). Parameters used were two upper-layer clusters and 10 lower-level clusters, 30 Expectation-Maximization steps and 10 generations of admixture between the two source populations identified in the TreeMix and f-statistics analyses (nonadmixed reference genetic groups as identified above, namely MEX1 and DENT). We thus analyzed each French (Spanish, respectively) teosinte individual as resulting from the introgression between the haplotypes of the two source populations, MEX1 and DENT. We then plotted the average ancestral allele dosage over all French (Spanish, respectively) teosinte individuals. ELAI analyses were performed separately for each chromosome.
Supplementary Material
Acknowledgments
We thank Bruno Chauvel (INRAE) for bringing to our attention the presence of teosintes in maize fields in France. We thank Séverine Michel (INRAE) for herbicide sensitivity bioassays and molecular analysis of the ACC1 gene. We thank Delphine Madur from Génétique Quantitative et Evolution (GQE)-Le Moulon for handling DNA samples used for SNP array genotyping. GQE-Le Moulon benefits from the support of Saclay Plant Sciences (ANR-17-EUR-0007). M.I.T. and Y.V. are supported by an Agence Nationale de la Recherche grant (ANR-19-CE32-0009).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006633117/-/DCSupplemental.
Data Availability.
SNP data have been deposited in Zenodo (https://zenodo.org/record/3959138) (59).
References
- 1.Hulme P. E., Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J. Appl. Ecol. 46, 10–18 (2009). [Google Scholar]
- 2.Lambdon P. W. et al., Alien flora of Europe: Species diversity, temporal trends, geographical patterns and research needs. Preslia 80, 101–149 (2008). [Google Scholar]
- 3.Seebens H. et al., Global trade will accelerate plant invasions in emerging economies under climate change. Glob. Change Biol. 21, 4128–4140 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Vilà M., Hulme P. E., Impact of Biological Invasions on Ecosystem Services, (Springer Nature, 2017), p. 359, 10.1007/978-3-319-45121-3. [DOI] [Google Scholar]
- 5.Sax D. F. et al., Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 22, 465–471 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Van Kleunen M., Bossdrof O., Dawson W., The ecology and evolution of alien plants. Annu. Rev. Ecol. Evol. Syst. 49, 25–47 (2018). [Google Scholar]
- 7.Barker B. S., Andonian K., Swope S. M., Luster D. G., Dlugosch K. M., Population genomic analyses reveal a history of range expansion and trait evolution across the native and invaded range of yellow starthistle (Centaurea solstitialis). Mol. Ecol. 26, 1131–1147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson D. M. et al., Naturalization and invasion of alien plants: Concepts and definitions. Divers. Distrib. 6, 93–107 (2000). [Google Scholar]
- 9.Petit S. et al., Weed dispersal by farming at various spatial scales. A review. Agron. Sustain. Dev. 33, 205–217 (2013). [Google Scholar]
- 10.Baker H. G., The evolution of weeds. Annu. Rev. Ecol. Syst. 5, 1–24 (1974). [Google Scholar]
- 11.Vigueira C. C., Olsen K. M., Caicedo A. L., The red queen in the corn: Agricultural weeds as models of rapid adaptive evolution. Heredity 110, 303–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L. et al., Genomic clues for crop-weed interactions and evolution. Trends Plant Sci. 23, 1102–1115 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ellstrand N. C. et al., Introgression of crop alleles into wild or weedy populations. Annu. Rev. Ecol. Evol. Syst. 44, 325–345 (2013). [Google Scholar]
- 14.European Food Safety Authority , “Relevance of new scientific evidence on the occurrence of teosinte in maize fields in Spain and France for previous environmental risk assessment conclusions and risk management recommendations on the cultivation of maize events MON810, Bt11, 1507 and GA21.” (EFSA supporting publication 2016:EN-1094, 2016) 13 pp. 10.2903/sp.efsa.2016.EN-1094. [DOI]
- 15.Martínez Y., Cirujeda A., Gómez M. I., Marí A. Y., Pardo G., Bioeconomic model for optimal control of the invasive weed Zea mays subspp (teosinte) in Spain. Agric. Syst. 165, 116–127 (2018). [Google Scholar]
- 16.Arvalis , Téosinte: Une adventice qui demande une vigilance toute particulière. 13/14 Service Communication Marketing Arvalis (Institut du vegetal, 2013), p. 4.
- 17.Sánchez González J. J. et al., Ecogeography of teosinte. PLoS One 13, e0192676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuoka Y. et al., A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos-Madrigal J. et al., Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr. Biol. 26, 3195–3201 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Hufford M. B., Martínez-Meyer E., Gaut B. S., Eguiarte L. E., Tenaillon M. I., Inferences from the historical distribution of wild and domesticated maize provide ecological and evolutionary insight. PLoS One 7, e47659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Segovia E. et al., Characterization of introgression from the teosinte Zea mays ssp. mexicana to Mexican highland maize. PeerJ 7, e6815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins G. N., Teosinte in Mexico. J. Hered. 12, 339–350 (1921). [Google Scholar]
- 23.Wilkes H. G., Hybridization of maize and teosinte, in Mexico and Guatemala and the improvement of maize. Econ. Bot. 31, 254–293 (1977). [Google Scholar]
- 24.Vibrans H., Estrada Flores J. G., Annual teosinte is a common weed in the valley of Toluca, Mexico. Maydica 43, 45–48 (1998). [Google Scholar]
- 25.Trtikova M. et al., Teosinte in Europe–searching for the origin of a novel weed. Sci. Rep. 7, 1560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díaz A., Taberner A., Vilaplana L., The emergence of a new weed in maize plantations: Characterization and genetic structure using microsatellite markers. Genet. Resour. Crop Evol. 67, 225–239 (2020). [Google Scholar]
- 27.Raj A., Stephens M., Pritchard J. K., fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197, 573–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguirre-Liguori J. A. et al., Connecting genomic patterns of local adaptation and niche suitability in teosintes. Mol. Ecol. 26, 4226–4240 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Pyhäjärvi T., Hufford M. B., Mezmouk S., Ross-Ibarra J., Complex patterns of local adaptation in teosinte. Genome Biol. Evol. 5, 1594–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukunaga K. et al., Genetic diversity and population structure of teosinte. Genetics 169, 2241–2254 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandenburg J.-T. et al., Independent introductions and admixtures have contributed to adaptation of European maize and its American counterparts. PLoS Genet. 13, e1006666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unterseer S. et al., A comprehensive study of the genomic differentiation between temperate Dent and Flint maize. Genome Biol. 17, 137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter B. M., Admixture, population structure, and F-statistics. Genetics 202, 1485–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emerson R. A., Control of flowering in teosinte: Short-day treatment brings early flowers. J. Hered. 15, 41–48 (1924). [Google Scholar]
- 36.Minow M. A. A. et al., Distinct gene networks modulate floral induction of autonomous maize and photoperiod-dependent teosinte. J. Exp. Bot. 69, 2937–2952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duforet-Frebourg N., Luu K., Laval G., Bazin E., Blum M. G. B., Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol. Biol. Evol. 33, 1082–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L. et al., Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 28, 3005–3015.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvi S. et al., Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. U.S.A. 104, 11376–11381 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gengenbach B. G. et al., Genetic relationships of alleles for tolerance to sethoxydim herbicide in maize. Crop Sci. 39, 812–818 (1999). [Google Scholar]
- 41.Balbuena-Melgarejo A. et al., Competencia entre maíz y teocintle: Efecto en el rendimiento y sus components. Field corn and teocintle competition: Effect on grain yield and grain yield components. Cent. Agríc. 38, 5–12 (2011). [Google Scholar]
- 42.Doebley J., Stec A., Wendel J., Edwards M., Genetic and morphological analysis of a maize-teosinte F2 population: Implications for the origin of maize. Proc. Natl. Acad. Sci. U.S.A. 87, 9888–9892 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hufford M. B. et al., The genomic signature of crop-wild introgression in maize. PLoS Genet. 9, e1003477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng X., Muszynski M. G., Danilevskaya O. N., The FT-Like ZCN8 Gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas-Barrera I. C. et al., Contemporary evolution of maize landraces and their wild relatives influenced by gene flow with modern maize varieties. Proc. Natl. Acad. Sci. U.S.A. 116, 21302–21311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q. et al., TeoNAM: A nested association mapping population for domestication and agronomic trait analysis in maize. Genetics 213, 1065–1078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y., Hokin S. A., Kermicle J. L., Hartwig T., Evans M. M. S., A pistil-expressed pectin methylesterase confers cross-incompatibility between strains of Zea mays. Nat. Commun. 10, 2304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson E., Introgressive hybridization. Biol. Rev. Camb. Philos. Soc. 28, 280–307 (1953). [Google Scholar]
- 49.Stebbins G. L., Relationship between adaptive radiation, speciation and major evolutionary trends. Taxon 20, 3–16 (1971). [Google Scholar]
- 50.Burgarella C. et al., Adaptive introgression: An untapped evolutionary mechanism for crop adaptation. Front. Plant Sci. 10, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snow A. A. et al., Long-term persistence of crop alleles in weedy populations of wild radish (Raphanus raphanistrum). New Phytol. 186, 537–548 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Merotto A. Jr. et al., Evolutionary and social consequences of introgression of nontransgenic herbicide resistance from rice to weedy rice in Brazil. Evol. Appl. 9, 837–846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corbi J., Baack E. J., Dechaine J. M., Seiler G., Burke J. M., Genome-wide analysis of allele frequency change in sunflower crop-wild hybrid populations evolving under natural conditions. Mol. Ecol. 27, 233–247 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Ellstrand N. C., Garnir L. C., Hedge S., Guadagnuolo R., Blancas L., Spontaneous hybridization between maize and teosinte. J. Hered. 98, 183–187 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Guadagnuolo R., Clegg J., Ellstrand N. C., Relative fitness of transgenic vs. non-transgenic maize x teosinte hybrids: A field evaluation. Ecol. Appl. 16, 1967–1974 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Díez C. M. et al., Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytol. 199, 264–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fustier M. A. et al., Signatures of local adaptation in lowland and highland teosintes from whole-genome sequencing of pooled samples. Mol. Ecol. 26, 2738–2756 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Takuno S. et al., Independent molecular basis of convergent highland adaptation in maize. Genetics 200, 1297–1312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Corre V., Siol M., Vigouroux Y., Tenaillon M. I., Delye C., Adaptive introgression from maize has facilitated the establishment of teosinte as a noxious weed in Europe [Dataset]. Zenodo. https://zenodo.org/record/3959138. Deposited 24 July 2020. [DOI] [PMC free article] [PubMed]
- 60.Délye C., Pernin F., Michel S., “Universal” PCR assays detecting mutations in acetyl-coenzyme A carboxylase or acetolactate-synthase that endow herbicide resistance in grass weeds. Weed Res. 51, 353–362 (2011). [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jombart T., adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Jakobsson M., Rosenberg N. A., CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007). [DOI] [PubMed] [Google Scholar]
- 64.De Mita S., Siol M., EggLib: Processing, analysis and simulation tools for population genetics and genomics. BMC Genet. 13, 27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patterson N. et al., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luu K., Bazin E., Blum M. G. B., pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 17, 67–77 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Storey J. D., Bass A. J., Dabney A., Robinson D., qvalue: Q-value Estimation for False Discovery rate Control. R Package Version 2.18.0, https://github.com/jdstorey/qvalue (2019).
- 69.Guan Y., Detecting structure of haplotypes and local ancestry. Genetics 196, 625–642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SNP data have been deposited in Zenodo (https://zenodo.org/record/3959138) (59).