Significance
Deregulated global mRNA translation is a feature of various cancers and considered important in oncogenic transformation. In colorectal cancer (CRC), the role of the most common driver mutations in APC, KRAS, SMAD4, and TP53 on the global translational capacity are incompletely understood. Here, using mouse and human intestinal organoids, we found that each mutation governs the global translational capacity of the epithelial cell. Global translation is linked to known oncogenic hallmarks, including cell proliferation and growth upon accumulation of these mutations, posing the translational apparatus as a potential therapeutic target in CRC.
Keywords: global translation, protein synthesis, colorectal cancer, driver mutations
Abstract
Deregulated global mRNA translation is an emerging feature of cancer cells. Oncogenic transformation in colorectal cancer (CRC) is driven by mutations in APC, KRAS, SMAD4, and TP53, known as the adenoma-carcinoma sequence (ACS). Here we introduce each of these driver mutations into intestinal organoids to show that they are modulators of global translational capacity in intestinal epithelial cells. Increased global translation resulting from loss of Apc expression was potentiated by the presence of oncogenic KrasG12D. Knockdown of Smad4 further enhanced global translation efficiency and was associated with a lower 4E-BP1-to-eIF4E ratio. Quadruple mutant cells with additional P53 loss displayed the highest global translational capacity, paralleled by high proliferation and growth rates, indicating that the proteome is heavily geared toward cell division. Transcriptional reprogramming facilitating global translation included elevated ribogenesis and activation of mTORC1 signaling. Accordingly, interfering with the mTORC1/4E-BP/eIF4E axis inhibited the growth potential endowed by accumulation of multiple drivers. In conclusion, the ACS is characterized by a strongly altered global translational landscape in epithelial cells, exposing a therapeutic potential for direct targeting of the translational apparatus.
The adenoma-carcinoma sequence (ACS) is defined by a set of recurrent driver mutations in APC, KRAS, SMAD4, and TP53 genes, that accumulate during adenoma formation and progression to sporadic colorectal cancer (CRC), often correlating with specific stages of the developmental process (1). Since its early description by Vogelstein and colleagues, a body of research has further supported and elaborated on this well-established paradigm (2), and the putative oncogenic potential of each mutation has been demonstrated in various genetic rodent models (3). It is well known that driver mutations deregulate specific cell signaling pathways, but how this ultimately leads to oncogenic transformation of a healthy intestinal cell remains to be understood. Over the past decade, mounting evidence has indicated that quantitative and qualitative alterations in mRNA translation are a prominent feature of various cancers (4). Global mRNA translation can be defined as the capacity of the cell’s translation apparatus to produce nascent polypeptides in order to maintain the cellular proteome. The translational apparatus consists of the mRNA transcripts, ribosomes, translation factors, and aminoacyl-tRNAs. Cancer cells exploit the translational apparatus by deregulating these components, which is supported by multiple lines of evidence, including altered expression translation factors (4), ribosomal proteins and ribosomal RNA (rRNA) (5), and the presence of oncogenes such as PIK3CA and c-MYC, that modulate translational control (6). Thus, through aberrant signaling of pathways that converge on the translational apparatus, specific oncogenes may utilize entire proteomic programs in order to drive oncogenic transformation of a normal cell (7).
In the majority of sporadic CRCs, accumulation of a few driver mutations is considered to be sufficient in coordinating many of the cell-intrinsic aspects of oncogenic transformation, which epithelial cancer cells must adopt to clonally expand, invade, and metastasize (8). Although the transcriptomic layers underpinning this complex process have been studied extensively, effects on specific translational programs, as well as the global translational capacity itself, are largely unknown. Recently, it has been shown that Apc-deficient mouse intestinal cells harbor increased global translation rates via mTORC1-mediated modulation of translation elongation (9). This study clearly demonstrated that interfering with global translation through the mTORC1/S6K/eEF2K axis strongly impaired intestinal epithelial hyperproliferation and tumorigenesis caused by deficiency of Apc. Moreover, we have recently shown that heterozygous expression of chaperone GRP78, which leads to repression of p-eIF2α-mediated global translation, reduced adenoma formation in Apc-deficient mice (10). Interestingly, these studies have shown that genetically interfering with global translation predominantly affected Apc-deficient epithelium, with no obvious discernible phenotype of the epithelium during homeostasis (9, 10). This suggests that epithelial cells that exhibit functional loss of APC rely on ample global translational output during adenoma formation, displaying the importance of translational efficiency in transformed cells. Furthermore, efficient mRNA translation, which mainly determines the mammalian cellular proteome (11), may be particularly important for maintaining the oncogenic proteome in a cancer cell that must overcome repressed global translation when exposed to cellular stress (12).
It remains to be investigated whether the main driver mutations associated with CRC development affect the global translational capacity of the intestinal epithelial cell. To address this, we studied the impact of oncogenic mutations in APC, KRAS, SMAD, and TP53 on the global translational capacity of primary intestinal epithelial cells. Our data reveal a highly altered global translational landscape throughout the ACS, in which multiple driver mutations contribute to further enhance the global translational capacity.
Results
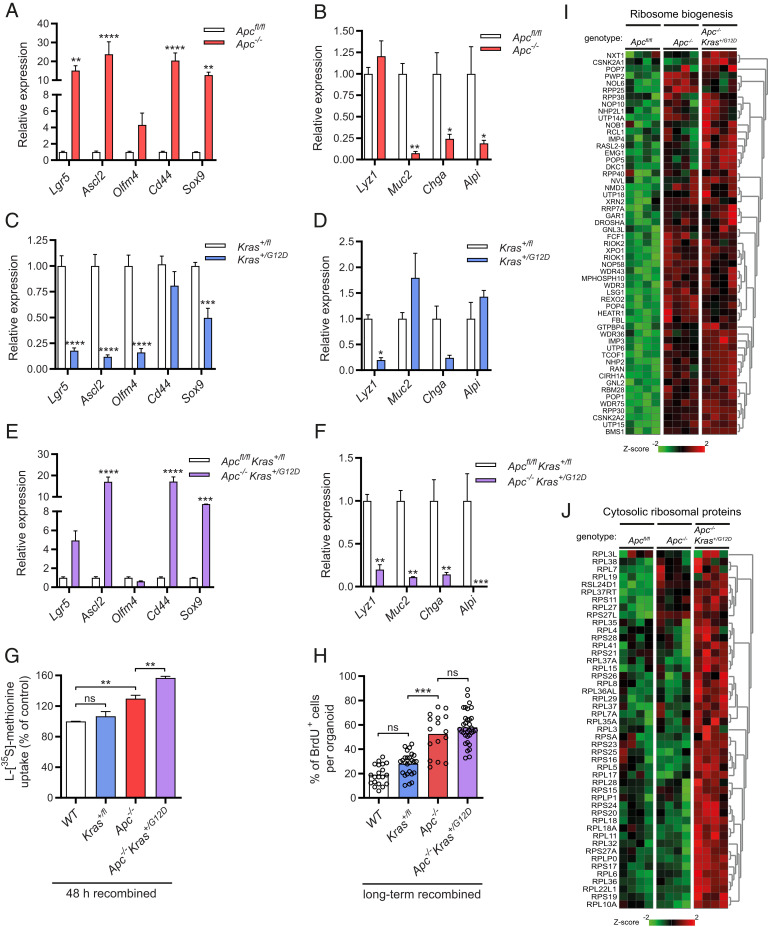
Mutations in Apc and Kras Drive Epithelial Stemness and Enhance the Rate of Global Translation, Proliferation, and Ribogenesis.
To study the effect of specific driver mutations on the global translational capacity, we made use of small intestinal organoids to model the ACS. First, we assessed loss of APC function, a strongly predisposing event that occurs in early transformation of intestinal epithelium in almost 80% of CRC cases (13). Organoids that were derived from inducible Villin-CreERT2-Apc580S/580S mice were recombined in vitro with 4OH-tamoxifen. As expected, these organoids adopted a cystic morphology and grew in the absence of WNT agonist R-spondin (SI Appendix, Fig. S1 A, Left). Stem cells were enriched at the cost of differentiated cell types, as indicated by transcriptional up-regulation of crypt base columnar stem cell markers Lgr5, Ascl2, Olfm4, Cd44, and Sox9, together with down-regulation of the lineage-specific differentiation markers Lyz1, Muc2, Chga, and Alpi (Fig. 1 A and B). Expansion of the proliferative stem cell compartment following Apc loss has been well described and results from constitutive autonomous Wnt/β-catenin signaling that promotes stemness and prevents normal differentiation (14). Using l-[35S]-methionine incorporation, we compared the rate of global translation between Apc−/− cells in organoids recombined for several weeks to their nonrecombined Apcfl/fl counterparts, and observed a striking increase, consistent with recently published findings (SI Appendix, Fig. S1B) (9). To validate the methodology, in which quantification of global translation was normalized to the proteomic biomass, we used flow cytometry-based l-azidohomoalanine (AHA) incorporation in order to measure global translation rates per individual cell. Similarly, AHA incorporation rates in Apc−/− organoids were strongly increased (SI Appendix, Fig. S1C). To assess whether altered translation is a direct consequence of the introduced mutation, or whether it follows secondary events emanating from the transformation by deletion of Apc, we measured global translation directly after recombination. Already at 48 h, when Cre-mediated gene-to-protein turnover has just been established (15), there was a 26% increase in the global translation rate (SI Appendix, Fig. S1D). This is in line with recent findings in which Apc deletion was achieved by recombination in vivo, and a similar impact on global translation was observed in organoid cultures assessed directly ex vivo (9). We also found the translation rate to increase over time after deletion of Apc, comparing 48-h recombination to several weeks (SI Appendix, Fig. S1 B and D). To assess if reduced epithelial WNT signaling affected global translation in wild-type cells, we withdrew canonical WNT agonist R-spondin for 24 h from the culture medium of wild-type organoids. As expected, when WNT signaling could not be sustained, the rate of global translation declined (SI Appendix, Fig. S1E).
Fig. 1.
Loss of Aps and additional transgenic expression of oncogenic KrasG12D organoids leads to stem cell enrichment, increased rates of global translation, proliferation, and ribogenesis. (A) Quantitative RT-PCR analysis of crypt base columnar stem cell markers in long-term (>2 wk) recombined Apc−/− organoids (n = 3). (B) Analysis of differentiation markers in long-term recombined (>2 wk) Apc−/− organoids (n = 3). (C) Quantitative RT-PCR analysis of crypt base columnar stem cell markers in long-term (>2 wk) recombined Kras+/G12D organoids (n = 3). (D) Quantitative RT-PCR analysis of differentiation markers in long-term (>2 wk) recombined Kras+/G12D organoids (n = 3). (E) Quantitative RT-PCR analysis of crypt base columnar stem cell markers in long-term (>2 wk) recombined Apc−/−Kras+/G12D organoids (n = 3). (F) Quantitative RT-PCR analysis of differentiation markers in long-term (>2 wk) recombined Apc−/−Kras+/G12D organoids (n = 3). (G) l-[35S]-methionine incorporation assay in Kras+/G12D, Apc−/−, and Apc−/−Kras+/G12D organoids, 48 h after recombination. (H) Quantification of BrdU immunostaining, showing the percentage of BrdU-positive cells per organoid in long-term (>2 wk) recombined Kras+/G12D, Apc−/−, and Apc−/−Kras+/G12D organoids. (I) Heatmap of microarray mRNA expression data containing genes related to rRNA production, processing, and ribosome assembly in Apc−/− and Apc−/−Kras+/G12D organoids (n = 4). (J) Heatmap of microarray mRNA expression data containing genes encoding for cytosolic ribosomal protein in wild-type, Apc−/−, and Apc−/−Kras+/G12D organoids (n = 4). Data are represented as means ± SEM. Significance (one-way ANOVA) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Clinically, human loss-of-function mutations in the APC gene usually precede or coincide with gain-of-function mutations in the oncogene KRAS, as illustrated by KRAS point mutations present in 40 to 50% of CRCs at early stages (16). The most frequent mutation is a base substitution KRASG12D, a conversion of glycine (G) to aspartic acid (D), that renders RAS in an active GTP‐bound state causing continuous activation of RAS signaling (17). To model this event, we studied whether KrasG12D modulated global translation in wild-type and Apc-deleted epithelium. Using inducible heterozygous organoids from Villin-CreERT2-Kras+/G12D transgenic mice, we generated organoids that grew independent of culture factor EGF after in vitro recombination (SI Appendix, Fig. S1 A, Middle). Kras+/G12D organoids exhibited an enlarged central lumen with maintained pericentral budding, a phenotype that differed from both Apc−/− and wild-type organoids (SI Appendix, Fig. S1 A, Middle). Interestingly, further characterization revealed significant reduction of WNT-related stemness marker genes (Fig. 1C). Differentiation appeared skewed toward a goblet cell phenotype with loss of other secretory cell types, as indicated by higher Muc2 expression in parallel with very low expression of Lyz1 and Chga (Fig. 1D). This was confirmed by immunohistochemistry (IHC) which showed increased numbers of periodic acid–Schiff (PAS)- and MUC2-positive cells in Kras+/G12D organoids (SI Appendix, Fig. S1F). Consistent with altered stem cell fate, NOTCH target gene Hes1, a fate regulator that suppresses precocious differentiation to the secretory lineage, was decreased (SI Appendix, Fig. S1G) (18). Importantly, these findings are consistent with increased goblet cell formation seen in transgenic KrasG12D mice, underscoring the similarities with ex vivo organoid models when studying epithelial mutations (19). Subsequently, we analyzed organoids where Apc was deleted in the presence of KrasG12D, using Apcfl/flKras+/fl organoids that were recombined in vitro (Apc−/−Kras+/G12D) (SI Appendix, Fig. S1 A, Right). Although stemness was markedly reduced in the Kras+/G12D organoids, Apc−/−Kras+/G12D organoids harbored clear stem cell enrichment with barely detectable differentiation markers (Fig. 1 E and F). Next, we studied how KrasG12D affected the global translational capacity in the presence and absence of deleted Apc. Interestingly, KrasG12D only enhanced the global translation rate in combination with loss of Apc (Fig. 1G). To show that epithelial RAS signaling controls global translation, we withdrew culture factor EGF, a direct activator of RAS, which led to a reduction of the global translational capacity in wild-type cells (SI Appendix, Fig. S1H). Thus, KrasG12D-mediated activation of RAS signaling further enhances the global translational capacity of Apc-deleted intestinal epithelial cells.
mRNA translation is a cell biological process that is intimately linked to proliferation under both homeostatic conditions and in cancer (20), although these processes can be uncoupled (21). To correlate global translation to proliferation rates, we assessed incorporation of 5-bromo-2-deoxyuridine (BrdU) normalized to the number of DAPI˗positive cells per organoid, in order to quantify the percentage of proliferating cells of Kras+/G12D, Apc−/−, and Apc−/−Kras+/G12D organoids. Changes in proliferation paralleled the rate of global translation, implying that these properties are linked, even under these hyperproliferative conditions (Fig. 1H). Continuous, elevated levels of protein synthesis are needed to fuel proliferation, requiring coordinated production of components of the translational apparatus, including cytoplasmic ribosomal proteins and ribosomal RNAs (22). Using microarray analysis, we searched for transcriptional reprogramming that could facilitate the increased global translation observed in Apc−/− and Apc−/−Kras+/G12D cells. Gene ontology analysis revealed that genes associated with rRNA production, processing and ribosome assembly were strongly up-regulated, particularly at the step of Apc loss (Fig. 1I). Furthermore, additional KrasG12D led to up-regulation of a majority of all genes encoding for cytoplasmic large and small ribosomal proteins (RPLs and RPSs), the protein constituents of the ribosome (Fig. 1J). KrasG12D also induced genes encoding for small nucleolar RNAs (snoRNAs), which are known for their essential role in nucleolytic processing and rRNA maturation (SI Appendix, Fig. S1I) (23). To measure rRNA synthesis we used real-time qPCR to measure 47S pre-rRNA external transcribed spacers (ETSs), a proxy for the pre-rRNA transcriptional activity of RNA polymerase I (24). In accordance with the transcriptional profile, we observed accelerated rRNA production upon loss of Apc and the additional presence of KrasG12D (SI Appendix, Fig. S1J). The oncoprotein c-MYC is a well-known regulator of ribogenesis and highly expressed in Apc-deficient cells, where c-MYC expression can be further stabilized in a RAS-dependent manner (25, 26). Indeed, both c-MYC mRNA and protein levels in Apc-deleted cells were further increased upon additional KrasG12D (SI Appendix, Fig. S1 K and L). We also assessed how interfering with ribogenesis affected organoid growth, by treatment with actinomycin D (Act D) which blocks the activity of RNA polymerases at a low dose (27). Given that c-MYC directly controls activity of the RNA polymerases I and III (Pol I and III), resulting in induced rRNA synthesis essential for protein synthesis, mutant cells could be more sensitive to Pol inhibition (28). However, contrary to what we expected, we noticed that the growth of wild-type organoids, and to a lesser extend Apc−/− organoids, was strongly sensitive to nanomolar concentrations of actinomycin D, whereas growth of Apc−/−Kras+/G12D organoids was strongly resistant to this (SI Appendix, Fig. S1 M and N). Thus, although mutations in Apc and Kras increase ribogenesis rates, the existence of a therapeutic window to specifically target this increase in ribogenesis remains to be determined.
In conclusion, loss of Apc and oncogenic KrasG12D, associated with early adenomagenesis, enhances the global translational capacity of epithelial cells. This is accompanied by accelerated proliferation rates and expansion of the stem cell pool. Furthermore, both mutations synergistically induce a transcriptional program that results in increased c-MYC levels and activation of ribogenesis.
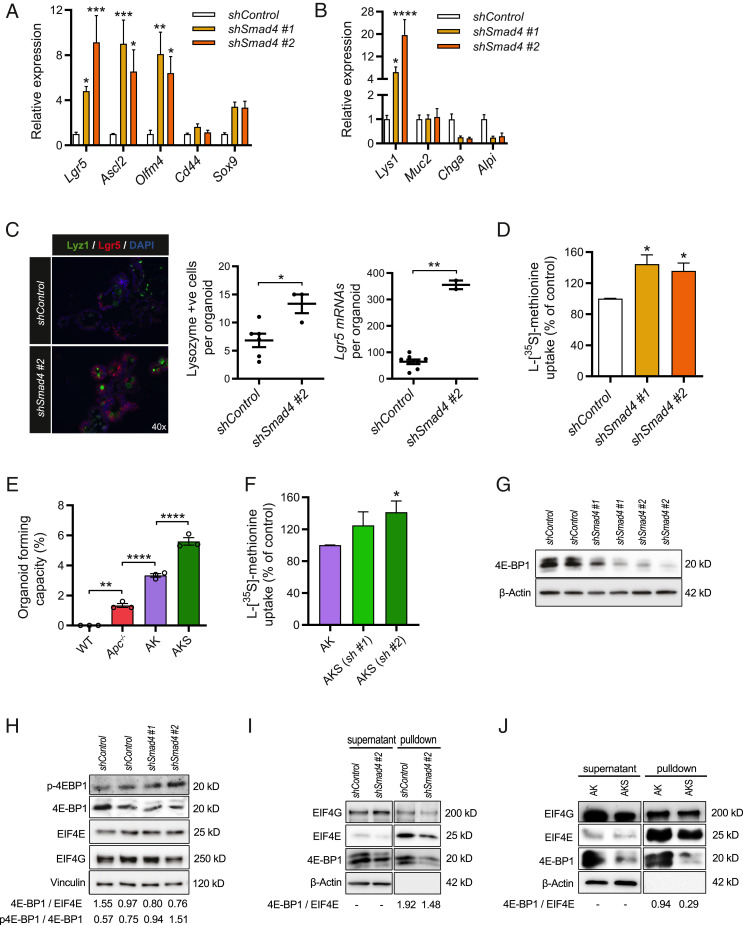
Reduced SMAD4 Expression Increases the Global Translational Capacity and Is Accompanied by Lower Levels of the CAP-Dependent Translational Repressor 4E-BP1.
To extend these findings beyond genetic events associated with early adenomagenesis, we investigated the role of SMAD4. Inactivating mutations in TGFβ signaling components, including complete or partial loss of tumor suppressor SMAD4, correlate strongly with late-stage adenomas and the carcinoma transition phase (29). To include this in our model, we generated small intestinal organoids with constitutive knockdown using stable expression of short hairpin RNAs against Smad4 (shSmad4 organoids). This resulted in ∼50% reduction of Smad4 mRNA expression (SI Appendix, Fig. S2A). shSmad4 organoids were resistant to BMP-mediated growth repression, as indicated by long-term culturing (>15 d) independent of BMP antagonist Noggin, showing that SMAD4 was functionally depleted (SI Appendix, Fig. S2B). When observing the growth of shSmad4 organoids we noticed increased bud formation, suggestive of accelerated crypt formation (Movies S1 and S2). Accordingly, mRNA levels of Lgr5, Ascl2, Olfm4, as well as Lyz1, were elevated in shSmad4 organoids, indicating enrichment of crypt-resident stem cells and Paneth cells (Fig. 2 A and B). We confirmed this by combining in situ hybridization for Lgr5 and IHC for lysozyme (Fig. 2C). Expansion of stem- and Paneth cells in the crypt indeed strongly resembles the recently reported small intestinal phenotype of Smad4 knockout mice (30). Interestingly, when comparing the global translational capacity of shSmad4 organoids to organoids with the wild-type organoids transduced with a scrambled control shRNA (shControl), we observed a 30 to 40% increase (Fig. 2D). Withdrawal of Noggin from the culture medium of wild-type organoids resulted in a small but significantly declined global translation rate, indicating involvement of BMP signaling herein (SI Appendix, Fig. S2C).
Fig. 2.
Knockdown of Smad4 results in enrichment of the stem and Paneth cell compartments, which is accompanied by increased global translation and reduced expression of translational repressor 4E-BP1. (A) Quantitative RT-PCR analysis of crypt base columnar stem cell markers in shSmad4 organoids (n = 3). (B) Analysis of small intestinal differentiation markers in shSmad4 organoids (n = 3). (C) Combined staining of in situ hybridization of stem cell marker lgr5 and immunostaining for Paneth cell marker lysozyme in shSmad4 organoids, including quantification of the mRNA particles and lysozyme-positive cells per organoid. *P < 0.05, Student’s t test. (D) l-[35S]-methionine incorporation assay of shSmad4 organoids assessed at day 4 after passaging (n = 3). (E) Clonogenic capacity of single cells that grow out to fully developed organoids, presented as a quantification of the organoid number per 20,000 seeded cells (n = 3). (F) l-[35S]-methionine incorporation assay in Apc−/− Kras+/G12D shSmad4#2 (AKS) and Apc−/− Kras+/G12D shControl (AK) organoids assessed at day 3 after passaging (n = 4). (G) Representative immunoblotting analysis of 4E-BP1 expression in shSmad4 organoids. β-Actin served as a loading control (n = 3). (H) Representative immunoblotting analysis of phospho-4E-BP1 (Thr70), eIF4E, and eIF4G in shSmad4 organoids. Optical density ratios of phospho-4E-BP1/4E-BP1 and 4E-BP1/eIF4E were also calculated (n = 2). (I) m7GTP-agarose pulldown assay in shSmad4 organoids to assess the levels of cap-bound eIF4E, eIF4G, and 4E-BP1 (n = 3). Unbound levels of β-actin 4E-BP1 and eIF4G were detected in supernatants. (J) m7GTP-agarose pulldown assay comparing AKS (shSmad4 #2) to AK organoids (shControl) (n = 3). Data are represented as means ± SEM. Significance (one-way ANOVA) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine whether the higher global translational capacity following knockdown of Smad4 was exerted by mechanisms additional to hyperactivation of WNT and RAS signaling, we introduced Smad4 knockdown in cells with preexisting Apc and Kras mutations. Recombined Apc−/−Kras+/G12D organoids were either transduced with a scrambled shRNA, from here on referred to as AK organoids, or shRNAs against Smad4, from here on referred to as AKS organoids. This organoid model is relevant, since mutations in the human SMAD4 gene often mark late-stage events in carcinoma formation that follow preexisting mutations in APC and KRAS (31). As expected, AKS organoids grew completely independent of all medium growth factors (EGF, R-spondin, and Noggin) (32). Additional selection for reduced SMAD4 function was ensured using high-dose and long-term incubation with mouse recombinant TGFβ (33), which caused disintegration of AK organoids, whereas AKS organoids were completely resistant and showed clear regenerative potential after repassaging (SI Appendix, Fig. S2D). This effect has been described and involves the lack of SMAD4 which provides resistance to TGFβ/SMAD4-induced proapoptotic effects in the Apc-deficient stem cell (34). Interestingly, the clonogenic capacity, a functional stem cell property, was further increased in single cells derived from AKS organoids (Fig. 2E). Furthermore, Smad4 knockdown in cells with mutations in Apc and Kras resulted in a higher global translational capacity (Fig. 2F).
To identify signaling transduction pathways related to regulation of global mRNA translation that were affected by Smad4 knockdown, we compared the transcriptional profiles of AK and AKS organoids. Gene set enrichment analysis (GSEA) comparing gene sets from the Oncogenic Signatures database (Broad Institute), revealed that Smad4 knockdown was associated with a transcriptional profile that matched a set (M2847) of up-regulated transcripts associated with cells with active eukaryotic translation initiation factor G1 (eIF4G1) (SI Appendix, Fig. S2E) (35). eIF4G is an essential translation initiation factor of the eIF4F complex, comprising the cap-binding protein eIF4E, the scaffold protein eIF4G, and the helicase eIF4A. This complex facilitates the interaction between the 43S-preinitiation complex and the capped mRNA (36). The entire complex is a critical regulatory nexus of dep-dependent translation initiation through upstream signaling pathways, including mTORC1 via eIF4E-binding protein 1 (4E-BP1) and RAS signaling via MNK1/2 (37). Upon hypophosphorylation, 4E-BP1 binds to eIF4E and prevents it from interacting with eIF4G to promote ribosome recruitment (38). Interestingly, recent reports have demonstrated that SMAD4 binds the promoter region of mouse and human 4EBP1, stimulating translation initiation due to disinhibition of eIF4E (30, 39). In line with this observation, the transcriptome of AKS organoids was enriched for genes related to the gene set (M1919) of Eif4ebp1/Eif4ebp2 double knockout mouse embryonic fibroblasts, cells that exhibit elevated protein synthesis rates (SI Appendix, Fig. S2F) (40). We thus hypothesized that knockdown of Smad4 could reduce available 4E˗BP1 levels that compete with eIF4G to bind and enhance the activity of eIF4E (41), which in turn would enhance cap-dependent translation activity (42). In support of this hypothesis, we found mRNA levels of both mouse homologs 4ebp1 and 4ebp2 to be reduced in shSmad4 organoids (SI Appendix, Fig. S2 G and H). Moreover, 4E-BP1 protein expression was also at a reduced level in shSmad4 organoids, while expression of the cap-binding proteins EIF4E and EIF4G were unchanged (Fig. 2 G and H). The ratio of 4E-BP1 to EIF4E was lower in shSmad4 organoids, while any 4E-BP1 was largely phosphorylated (Fig. 2H). To show that less 4E-BP1 was bound to the cap-binding complex upon Smad4 knockdown, we performed m7GTP pulldown assays in shSmad4 organoids and quantified protein levels of bound 4E-BP1 relative to eIF4E. We consistently found relatively less 4E-BP1 bound to the mRNA cap, normalized to eIF4E, upon knockdown of Smad4 in epithelial cells of wild-type and Apc−/−Kras+/G12D backgrounds (Fig. 2 I and J). These data suggest that reduced SMAD4 levels could facilitate global translation via loss of 4E-BP1-mediated repression of functional eIF4E, increasing the cap-dependent translation efficiency.
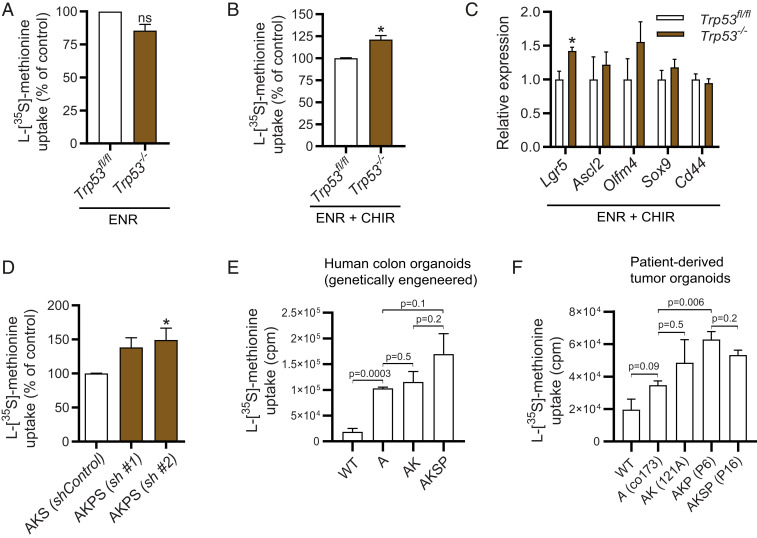
Mouse and Human Epithelial Cells with Multiple Driver Mutations in APC, KRAS, SMAD4, and TP53 Are Characterized by the Highest Capacity for Global Translation.
To study a combination of all driver mutations related to the ACS, we continued investigating loss of p53, which hallmarks a majority of the metastasizing carcinomas (43). We first used small intestinal organoids isolated from Trp53fl/fl mice, which we subsequently transduced with the pRetro-MSCV-CreERT2 vector in order to obtain inducible Trp53−/− organoids. After in vitro recombination, mRNA levels of Trp53 were undetectable and organoids proved completely insensitive to cell death induced by the MDM2-antagonist Nultin-3, confirming p53 loss of function (SI Appendix, Fig. S3 A and B). Trp53−/− organoids were undiscernible from nonrecombined (Trp53fl/fl) controls, and global translation rates were unaffected (Fig. 3A). However, upon pretreating Trp53−/− organoids with the GSK3β inhibitor CHIR-99021, which creates a condition of high WNT activity at the level of the β-catenin destruction complex that resembles deletion of Apc, global translation rates were significantly higher in Trp53−/− organoids (Fig. 3B). Lgr5 was up-regulated, although expression of other WNT-related stem cell markers were unchanged in Trp53−/− cells (Fig. 3C). To further investigate the role of p53 in cells with the additional ACS mutations, we transduced shRNAs against Trp53 in AKS organoids, from now on referred to as AKSP organoids (SI Appendix, Fig. S3C). AKSP organoids were resistant to simultaneously administered high-dose and long-term incubation with recombinant TGFβ and Nutlin-3 (SI Appendix, Fig. S3D). As expected, transcriptional profiling of AKSP cells was characterized by decreased expression of a large set of proapoptotic genes (SI Appendix, Fig. S3E). Strikingly, we found that the global translational capacity in AKSP organoids was even further elevated (Fig. 3D). To assess if these findings are relevant in the setting of human colon epithelium, we used previously published genetically engineered colon organoids with mutations in the APC, KRAS, and TP53 genes (32). Here we observed a pattern similar to murine small intestinal cells, where the global translational capacity was highest in colonic cells with quadruple driver mutations (Fig. 3E). To further validate this in human CRC cells that carry naturally occurring oncogenic mutations, we analyzed the global translation rate of patient-derived tumor organoids that were previously characterized (44, 45). Again, tumor cells with multiple driver mutations in APC, KRAS, SMAD4, and TP53 harbored the highest global translational capacity, indicating that each of these mutations controls global translation in CRC cells (Fig. 3F).
Fig. 3.
A combination of mutations in APC, KRAS, SMAD4, and TP53 results in the highest global translational capacity in mouse small intestinal organoids, human colon organoids, and patient-derived tumor organoids. (A) l-[35S]-methionine incorporation assay of MSCV-Cre Trp53−/− organoids assessed at day 3 after passaging in ENR (n = 3). (B) l-[35S]-methionine incorporation assay of MSCV-Cre Trp53−/− organoids assessed at day 3 after passaging pretreated for 96 h in ENR with 5 μM CHIR-99021 (n = 3). (C) Quantitative RT-PCR analysis of crypt base columnar stem cell markers in MSCV-Cre Trp53−/− organoids (n = 3). One-way ANOVA. (D) l-[35S]-methionine incorporation assay of AKSP compared to AKS organoids assessed at day 3 after passaging (n = 4). (E) l-[35S]-methionine incorporation assay in genetically engineered human colon organoids with mutations in APC, KRAS, SMAD4, and TP53 as previously described (32). (n = 3) (F) l-[35S]-methionine incorporation assay in FAP patient-derived organoids (co173, 121A) and sporadic CRC tumor organoids (P6, P16) that carry mutations in APC, KRAS, SMAD4, and TP53 as previously described (n = 3) (37, 38). FAP, familial adenomatous polyposis; counts per minute (cpm), cpm relative to protein biomass. Data are represented as means ± SEM. Significance (Student’s t test/one-way ANOVA) *P < 0.05. ns, not significant.
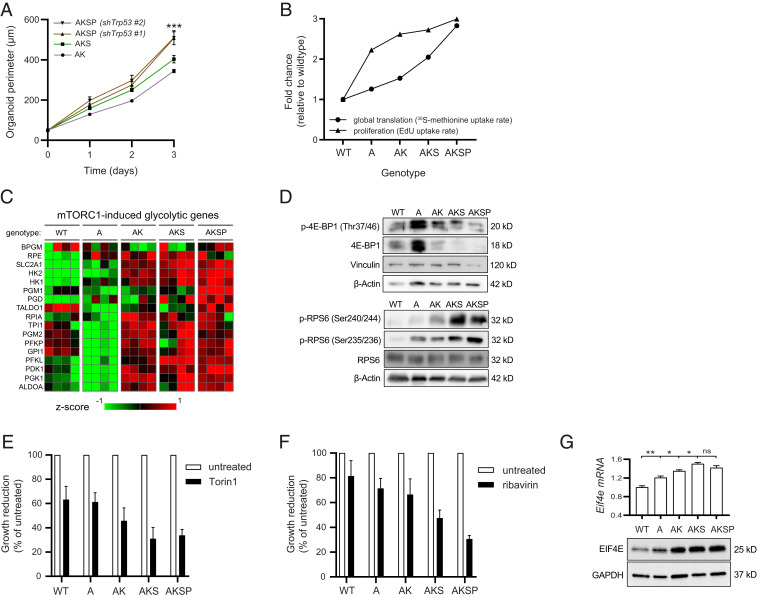
Increased Proliferation and Growth Can Be Targeted by Interfering with Global mRNA Translation Via mTOR Signaling or the Function of Downstream eIF4E.
Sustained global translation provides the biosynthetic needs for cell growth, which explains why these biological processes are often linked, at least in immortalized mammalian cell lines (42). To assess whether the parameters of global translation, growth, and proliferation are correlated in intestinal epithelial cells, we seeded single cells from AK, AKS, and AKSP organoids and quantified growth over time. AKSP organoids grew exceptionally fast, reaching their optimal size in as fast as 3 d, exceeding the rate of AKS and AK organoids (Fig. 4A). Proliferation rates, as measured by incorporation of 5-ethynyl-2′-deoxyuridine (EdU) in the S phase, were also highest in AKSP organoids, demonstrating that global translation, cell division, and growth remain associated during oncogenic transformation (Fig. 4B). mRNA translation and proliferation are likely to be codependent processes, in which translation rates are adjusted to meeting the demands of proliferation (46–48). However, whether enhanced rates of global translation due to the concerted action of multiple driver mutations enables greater proliferation, and whether there exists a window to inhibit this process by selectively targeting the translational apparatus, remains to be determined. To address this, we directly blocked global translation at the level of initiation or elongation and measured the effect on cellular proliferation, using ribavirin and cycloheximide. After short-term exposure to both inhibitors, proliferation was already strongly reduced, indicating that global translation is required for these cells to divide (SI Appendix, Fig. S3 F and G). Next, we aimed to identify which signaling transduction pathways involved in control of global translation were activated during the ACS. To this end we compared transcriptional profiles of each organoid genotype, and looked for genes that follow a stepwise increase, similar to the relative increase in global translation rates, using R2 software. Unbiased pathway analysis on the resulting list of genes revealed that mTOR signaling components were overrepresented (P = 6.1 × 10−6). mTOR is a pivotal cellular transduction pathway that coordinates eukaryotic cell growth and metabolism with environmental inputs, by regulating global translation via several downstream effector kinases (SI Appendix, Fig. S3H) (49). Moreover, when looking at a set of previously described genes related to glucose metabolism that are transcriptionally regulated by mTORC1 (50), a pattern of increased expression was observed, suggesting mTORC1 hyperactivation throughout the ACS (Fig. 4C). We examined the posttranslational phosphorylation status of the target effector proteins RPS6 and 4EBP1, which can be phosphorylated by mTORC1 (49). In line with the transcriptional data, levels of phosphorylated RPS6 (Ser240/244 and Ser235/236) were increased upon accumulation of ACS mutations (Fig. 4D). On the other hand, expression levels of phosphorylated 4E-BP1, which are initially up-regulated in the context of Apc loss, are subsequently extinguished by additional mutations in Kras, Smad4, and P53. This was paralleled by loss of total 4EBP1 levels, which are undetectable in AKS and AKSP organoids (Fig. 4D). Interestingly, similar observations of lost 4EBP1 expression were seen in tissue of progressed human pancreatic cancers, tumors hallmarked by mutations in KRAS, SMAD4, and TP53 (51). Thus mTORC1 signaling may drive global translation during oncogenic transformation throughout the ACS, potentially steering growth and proliferation of intestinal epithelial cells as a consequence.
Fig. 4.
High global translational capacity enables growth in cells with multiple ACS mutations in a mTOR/eIF4E-dependent manner. (A) Representative time curve of organoid growth starting from 10,000 single-seeded cells and grown for 3 d. Quantified as mean organoid perimeter per well, four wells per condition (n = 2). (B) Plotted rates of global translation (from l-[35S]-methionine incorporation assays), and proliferation (from EdU incorporation assays), in organoids of all ACS genotypes. Data are presented as fold change relative to the proliferation and translation rate of wild-type organoids. (C) Heatmap of microarray mRNA expression data in organoid of all ACS genotypes, containing a subset of transcriptional target genes associated with mTOR signaling described previously (50). (n = 4). (D) Western blotting analysis of phosphorylated and total 4E-BP1 and RPS6, effectors downstream of mTORC1 signaling. (E) Quantification of growth in organoids treated for 48 h with 50 nM Torin 1 and presented as the growth reduction after treatment in each genotype relative to untreated controls (n = 4). (F) Quantification of growth in organoids treated for 48 h with 10 µM ribavirin and presented as the growth reduction after treatment in each genotype relative to untreated controls (n = 3). (G) Western blotting analysis of EIF4E levels in organoid of all ACS genotypes as well as mRNA expression levels derived from the microarray analysis. Data are represented as means ± SEM. Significance (one-way ANOVA) *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
To assess the vulnerability to mTOR inhibition and the effects on growth, we used multiple mTOR inhibitors that vary in specificity and potency (52). First we tested rapamycin, a well-studied compound that inhibits mTORC1/S6K signaling, and has been proven effective in CRC mouse models with loss of APC, although tumor cells with high RAS signaling due to additional mutations in KRAS or PIK3CA have been proven rapamycin insensitive (9, 53, 54). We indeed found that rapamycin affected epithelial cells with loss of Apc, consistent with previous literature, but had little to no effect on growth of AK and AKS organoids, respectively (SI Appendix, Fig. S3I) (9). We then used Torin1, a more potent inhibitor of mTORC1, that possesses dual effectiveness in blocking both the mTORC1/S6K and mTORC1/4E-BP1 effector branches of the pathway (49, 55). Interestingly, contrary to rapamycin, Torin1 was able to also inhibit growth of cells with multiple ACS mutations, indicating that these cells remain dependent on activation of mTORC1 signaling (Fig. 4E and SI Appendix, Fig. S3J). Moreover, sensitivity to Torin1 corresponded with a stepwise reduction in mRNA expression of stemness marker Lgr5 (SI Appendix, Fig. S3K). Because we observed reduced 4E-BP1 interaction to the cap-binding complex in our cancer models with knockdown of Smad4, we examined the effect of inhibiting functional eIF4E using ribavirin, a pharmacological 4E-BP1 mimetic (56). Strikingly, after dose titration, we identified a therapeutic window at which the growth of wild-type and Apc-deficient organoids was relatively unaffected, whereas AK, AKS, and AKSP organoids were increasingly more susceptible (Fig. 4F and SI Appendix, Fig. S3L). Most notable was the effect of ribavirin on growth of AKSP organoids that carry quadruple mutations (Fig. 4F). It was previously reported that growth of cancer cells with high eIF4E expression, including breast cancer and leukemia cell lines, were increasingly ribavirin sensitive (56, 57). In line with these studies, we observed elevated baseline mRNA and protein expression levels of eIF4E in our ACS organoid models (Fig. 4G). Collectively, our data thus demonstrate increased global translation enables proliferation and growth of cells with triple and quadruple ACS mutations, via a deliberate and targetable increase in downstream mTORC1 target eIF4E.
Discussion
Deregulated translational control is considered to be a crucial component in various cancers, although not much is known about CRC in this respect. We set out to investigate the role of key oncogenic driver mutations on the global translational capacity of intestinal epithelial cells. Using various murine and human intestinal organoid models, we show that global translation can be strongly enhanced upon the acquisition of mutations in the genes for APC, KRAS, SMAD4, and TP53. Activated WNT signaling by deletion of Apc appears to be a prerequisite for KrasG12D to further increase global translation and proliferation. Indeed, synergic mechanisms operating between WNT and RAS signaling have been reported (58, 59). In mice, transgenic KrasG12D alone is insufficient to initiate tumor formation, but it significantly promotes CRC formation and progression in an Apc-deleted background (19, 53, 58, 59). Whether enforced global translation is actually required to exert the oncogenic potential of KrasG12D in cells is an issue that warrants further investigation. With regards to (partial) loss of SMAD4 and p53, we observed that both events can act as enhancers of the global translational capacity. Interestingly, synergy in terms of their oncogenic potential has been reported between these driver mutations as well (60, 61). It is plausible that deregulated global translation, through the cross-talk of multiple oncogenic pathways, is the main determinant of sustaining the cancer cell proteome. APC and KRAS may facilitate this process by activation of ribogenesis, through stabilizing the activity of c-MYC (25). Additional loss of SMAD4, via reduced 4E-BP expression, could unleash maximal efficiency of eIF4E-mediated cap-dependent translation of tumorigenic mRNAs that promotes oncogenic transformation (62). Loss of p53, which we have not further assessed here, could be crucial for cancer cells to cope with prolonged proteotoxic stress that arises from processing nascent polypeptides by preventing apoptosis (63), allowing high expression of oncogenes such as c-MYC (64).
It remains challenging to interfere therapeutically with global translation in cancer cells, without affecting healthy cell function concurrently. We first tried to indirectly modulate global translation by inhibition of ribogenesis using a Pol1 inhibitor. However, this was not possible without significantly retarding growth of wild-type cells too, probably because the therapeutic window was too narrow for partial suppression of ribogenesis. Secondly, because we found hyperactivated mTORC1 signaling, we tried to modulate global translation by blocking mTORC1 function and assess organoid growth. However, additional mutations in Kras and Smad4 conferred rapamycin resistance in Apc-deficient cells, possibly due to lower 4E-BP1/eIF4E ratios, which has been described to be a determinant for responsiveness in other cancers (65, 66). Furthermore, rapamycin has a differential effect on 4E-BP1 versus S6K, and cap-dependent translation can be maintained through 4EBP1 phosphorylation (67). Torin1 on the other hand, a more potent inhibitor that also suppresses rapamycin-resistant functions of mTORC1 that are necessary for cap-dependent translation (68), could effectively inhibit growth of cells with ACS mutations, although we found wild-type cells to be sensitive too. Since we found lower ratios of 4E-BP1/eIF4E in triple mutant cells, we reasoned that directly interfering with the eIF4G-eIF4E interaction using the 4E-BP1 mimetic ribavirin could be effective. Indeed, we identified a therapeutic window of opportunity where selectively impeding with cap-dependent translation initiation predominantly affected quadruple mutant cells, while minimal effect on wild-type cells was seen. Apc-deficient cells were also relatively insensitive to ribavirin in these concentrations, potentially because these cells rely primarily on increased translation elongation rather than initiation (9). However, our results point toward a central role for translation initiation at later stages during the ACS, which suggests that adenocarcinoma cells might not be able to fully circumvent cap-dependent translation by alternative modes of initiation (69).
A few points of discussion must be mentioned with regards to this study. First of all, since the role of SMAD4 and p53 was partly assessed using noninducible siRNA strategies, the nature of these models did not allow us to examine any direct effects of complete loss of function, although these models are still relevant (70). Gene dosage and temporal aspects associated with knockdown strategies may have a significant effect, as they gradually change the distribution of cell types in the multicellular organoids, and thus the global translational capacity that is measured. Second of all, relation between global translation and proliferation in organoids was nonlinear. This may be explained by different cell ratios in a multicellular system, particularly after introducing a deletion in Apc, which is known to enrich the number of activated stem cells at the cost of differentiated cells. Better techniques for accurate single cell measurement of global translation in organoids must delineate how different intestinal cell types harbor different translational capacities. Third, an important caveat of using in vitro epithelial organoid culture systems to study global translation is that the effects of nonepithelial cells remain unknown. The cellular context normally provided by stroma and immune cells, as well as nutrient deficiency due to poorly vascularized tumor regions, will likely have a major impact on cancer cell behavior, including translation and proliferation. Additional research will unravel the degree of dependency on global translation of CRC cells in vivo, in order to explore the feasibility of developing treatment strategies that specifically target the translational apparatus in CRC. Because mRNA translation is such an energetically expensive process, future research should also address how cancer cells meet the high metabolic demands of global translation (71).
Materials and Methods
Generation and Culture of Organoids.
All experiments were approved by the relevant local ethical committees. When experimental tissue from human subjects were used informed consent was obtained and the study was approved by the ethical committee. Mouse small intestinal organoids were generated according to methods previously described (72). Organoids were cultured in media containing N2, B27 supplements (Invitrogen), 1.25 mM n-acetylcysteine, 50 ng/mL mouse EGF (Invitrogen), Noggin-Fc-conditioned medium (20%, equivalent to 200 ng/mL), and Rspo1-Fc-conditioned medium, unless specifically stated otherwise. Noggin-Fc-conditioned medium was obtained through collection of supernatant from HEK293T cells transfected with a Noggin-Fc expression vector as previously described (73). Human genetically engineered colon organoids (32), familial adenomatous polyposis (FAP) patient-derived organoids (44), and tumor organoids from CRC patients (45), were cultured in mouse small intestinal organoid medium with addition of 2-mercaptoethanol (Sigma-Aldrich), gentamicin (BioWhittaker), 50 mM TGF-βRI inhibitor A38-01 (Tocris Bioscience), 30 mM p38 inhibitor SB202190 (Sigma-Aldrich), and 500 µM (Leu15)-gastrin (Sigma-Aldrich).
Experiments with inducible organoids using the Villin-CreER system were performed on a C57BL/6J background. In vitro Cre-mediated recombination was achieved using 0.75 µg/mL 4-hydroxy tamoxifen for 24 h. Organoids with mutations in APC, KRAS, and TP53 were derived from previously published Apc580S/580S conditional mice, Kras+/G12D conditional mice, and Trp53F2-10/F2-10 conditional mice, respectively (74–76). AKS and AKSP organoids were generated by stable lentiviral cotransduction of two short hairpin RNAs against Smad4 and Trp53 in Apc−/−Kras+/G12D organoids, according to previously described methodology (77, 78). Subsequently, selection for mutated cells was performed using growth factor depletion from the culture medium (EGF depletion to select for Kras+/G12D cells, R-spondin depletion to select for Apc−/− cells, and Noggin depletion to select for cells with knockdown of Smad4). A 5-d incubation with (±)-Nutlin-3 (10 µM, Cayman Chemical) was used to select for cells with knockout and knockdown of Trp53. Knockdown of Smad4 in Apc−/−Kras+/G12D organoids was achieved by at least a 7-d incubation with recombinant human TGFβ (10 ng/mL, Bioscience).
Quantification of organoid growth was done by microscopic assessment of individual organoids that were cultured in a 48-well plate, by measuring the perimeter or area with ImageJ software. In quantifications presented in the figures, each biological replicate consists of the mean quantified growth of three wells.
Clonogenicity Assay.
We assessed the clonogenic potential of single cells from organoids. Briefly, organoids were grown up to 4 d before repassaging. First Matrigel was fractionated using a narrowed Pasteur pipette and consequently removed through centrifuging (800 rpm). Crypts were dissociated to single cells after 10 min of 37 °C incubation with TrypLE (Gibco). The absolute number of cells per condition was equalized using a Beckman Coulter particle counter (Z-series). For each condition, 20,000 cells in 20 µL Matrigel were seeded per well (three wells) and the number of outgrowing organoids was quantified after 3 to 5 d.
Assessment of Global Translation.
To measure all newly synthesized proteins, we quantified the incorporation of 35S-labeled methionine and cysteine in organoids. Organoids were grown for 3 to 4 d after passaging and seeded into 48-well plates. In the experiments using human organoids, growth factors (EGF, Noggin, and R-spondin) and TGFβ and p38 inhibitors were depleted for 48 h prior to the assay.
After a 15-min starvation period to deplete endogenous methionine, cells were exposed to a pulse phase of 45 min to label all newly synthesized proteins using 1 µL (1.25 µCi/mL) of EasyTag l-[35S]-methionine (PerkerElmer) per well at 37 °C, 95% humidity, and 5% CO2 conditions. Organoids were then washed two times in cold PBS, harvested, and spun down in cold PBS to remove supernatant and any Matrigel fragments. Cell pellets were lysed in cell lysis buffer (Cell Signaling Technology) and spun down to remove any unresolvable phospholipids. A 15-µL aliquot of radioactive lysate was blotted on labeled 24-mm diameter glass microfiber filters (GF/C Whatman) that were presoaked in 20% trichloroacetic acid (TCA). Filters were replaced in a vacuum manifold and incubated in 10% ice cold TCA for another 15 min, followed by 10% TCA at 90 °C for 8 min to break any aminoacyl-tRNA bonds. Filters were washed twice in 2% TCA and twice in 96% ethanol. Air dried filters (>20 min) were incubated in liquid scintillation mixture (Ultima Gold, PerkerElmer) for 2 h and radioactive disintegration per minute (dpm) was quantified using a scintillation counter (Tri-Carb 2900TR, PerkerElmer). Counts were presented as percentage relative to control samples, after normalization to total protein, using bicinchoninic acid assay according to manufacturer’s instructions (BCA Protein Assay Kit, Pierce).
Alternatively, incorporation of AHA-labeled methionine was used to label newly synthesized protein according to manufacturer’s instructions (AHA Click-it kit for Alexa Fluor 488, Thermo Fisher). Fluorescence was quantified on a flow cytometer (LSR Fortessa) and analyzed with FlowJo 8.0 software.
Assessment of Proliferation.
To assess proliferation in organoids, we used BrdU labeling for DNA synthesis and S phase analysis. Organoids were incubated with 10 µg/mL BrdU 2 h before harvesting for paraffin embedding and immunohistochemistry using a mouse anti-BrdU antibody (11170376001, Roche). Alternatively, EdU staining was performed to analyze cell proliferation by flow cytometry using the Click-iT EdU Alexa Fluor 647 kit (Invitrogen). In some experiments, 2 h of 10 µg/mL cycloheximide (Sigma-Aldrich) or 4 h of 1 mM ribavirin (Sigma-Aldrich) was added prior to and during the labeling step. Organoids were incubated with 10 μM EdU for 4 h followed by TrypLE-mediated cell dissociation. Cell were fixated for 20 min with 3.7% formaldehyde, washed, and incubated with Click-iT reaction mixture according to the manufacturer’s protocol. Fluorescence was quantified on a flow cytometer (LSR Fortessa) and the percentage of EdU-positive cells was calculated with FlowJo 8.0 software.
RNA Extraction and RT-qPCR.
All organoids were harvested at the indicated times, and RNA was extracted using the ISOLATE II RNA Mini Kit (BIO-52073, Bioline). cDNA was synthesized from 1 µg of purified RNA with Oligo-dT primer and Random Hexamers Primer using Revertaid reverse transcriptase according to protocol (Fermentas). Quantitative RT-PCR was performed using sensifast SYBR No-ROX Kit (GC-biotech Bio-98020) according to the manufacturer’s protocol on a BioRad iCycler.
In Situ Hybridization.
RNAscope experiments were performed using RNAscope, an RNA in situ hybridization technique. RNAscope was performed according to the “Formalin-Fixed Paraffin-Embedded (FFPE) Sample Preparation and Pretreatment for RNAscope 2.5 assay” and “RNAscope 2.5 HD Detection Reagent—RED” protocols as provided by the manufacturer.
Microarray Analysis.
Organoids were grown to day 3 and RNA was harvested as described above. A total of 400 ng of purified RNA was amplified and labeled using the 3′ IVT Pico Kit (Affymetrix) and RNA Amplification Kit (Nugene) according to manufacturers’ protocols. Microarray analysis of mouse organoids was performed using Affymetrix Clariom S mouse array by the Dutch Genomics Service and Support Provider (MAD, Science Park, University of Amsterdam, The Netherlands). Washing and staining were performed by the GeneChip Fluidics Station 450 and the scanning was performed using the GeneChip Scanner 3000 7G (both Thermo Fisher Scientific). Data normalization, statistical testing, and extraction of differentially expressed genes was performed using the R2 platform. A complete description of the bioinformatics tool R2 may be found at https://hgserver1.amc.nl/cgi-bin/r2/main.cgi.
Immunoblotting.
Cells were lysed in cell lysis buffer (Cell Signaling Technology) and heated to 95 °C for 5 min in sample buffer containing 0.25 M Tris⋅HCl (pH 6.8), 8% sodium dodecyl sulfate (SDS), 30% glycerol, 0.02% bromophenol blue, and 1% β-mercaptoethanol. Separation was done on 6 to 12% SDS/polyacrylamide gel electrophoresis (PAGE), and proteins were transferred to a polyvinylideenfluoride membrane. Specific detection was done by incubating the blot overnight in Tris-buffered saline (TBS) with 0.1% Tween-20 and 1% bovine serum albumin (BSA). Antibody binding was visualized using the Lumi-Light Western blotting substrate (Roche). Bands were quantified using ImageJ software. Antibodies are as follows: β-Actin (Sigma, Ab1978), Vinculin (Cell Signaling, 18799S), GAPDH (Cell Signaling, 2118S), 4E-BP1 (Cell Signaling, 9644), p4E-BP1 Thr37/46 (Cell Signaling, 2855), EIF4E (Cell Signaling, 9644), EIF4G (Cell Signaling, 8701), pS6 (Cell Signaling, 4857S), and c-MYC (Santa Cruz, sc-764).
Immunohistochemistry.
Organoids were fixed overnight in 4% formaldehyde, embedded in paraffin, and sectioned. For staining, sections were deparaffinized with xylene and gradually rehydrated in ethanol. After blocking the endogenous peroxidase (0.01% H2O2 in methanol), slides were boiled for 20 min at 100 °C on a heat block in 0.01 M sodium citrate buffer (pH 6) for antigen retrieval. Slides were incubated overnight with primary antibody diluted in phosphate buffered saline (PBS) with 1% BSA and 0.1% Triton X‐100. Powervision secondary antibody (Immunologic) was added for 30 min followed by incubation with detection antibody. Antibody binding was visualized by adding chromagen substrate diaminobenzedine (Sigma‐Aldrich) according to the manufacturer’s protocol. Antibody used was Muc2 (Santa Cruz, sc-15334).
m7GTP Pulldown Assay.
Organoids from 24 wells of a 48-well plate were pooled and pellets were lysed in buffer containing 10 mM Tris⋅HCl (pH 7.6), 140 mM KCL, 4 mM MgCl2, 1 mM dithiothreitol, 1 mM ethylenediaminetetraacetic acid, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, Complete Protease Inhibitor Mixture (Roche), 0.2 mM sodium orthovanadate. 30 to 50 µL of 7-methyl-GTP-agarose beads (Immobilized γ-Aminophenyl-m7GTP-C10-spacer, Jena Bioscience) was added and samples were incubated overnight. After washing two times in lysis buffer and two times in PBS, beads were resuspended in 1× SDS loading buffer. Samples were loaded on 6% gradient SDS/PAGE gel and immunoblotted for EIF4E and 4E-BP1. Quantification of the 4E-BP1/EIF4E ratio was calculated using ImageJ software.
Statistical Analysis.
Statistical analysis was performed using Prism 8.0 (GraphPad Software). All values are represented as the mean ± SEM. Statistical tests are indicated in the figure legends. All experiments were performed as biological triplicates unless specifically stated otherwise. P value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank professor Owen Sansom of the Cancer Research UK Beatson Institute for kindly providing mouse small intestinal inducible Apc580S/580S and Kras+/G12D organoids. This work was supported in part by grants of the Amsterdam Medical Center PhD scholarship 2015, Dutch Cancer Foundation (KWF/UVA 2013-6135 and KWF/Alpe 11053/2017-1), and by a grant from the Netherlands Organization for Scientific Research (NWO-Veni 91615032).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912772117/-/DCSupplemental.
Data Availability.
All of the microarray data shown in the manuscript and SI Appendix are deposited in the Gene Expression Omnibus Database (GSE143509).
References
- 1.Fearon E. R., Vogelstein B., A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Wood L. D. et al., The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Jackstadt R., Sansom O. J., Mouse models of intestinal cancer. J. Pathol. 238, 141–151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvera D., Formenti S. C., Schneider R. J., Translational control in cancer. Nat. Rev. Cancer 10, 254–266 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Chu J., Cargnello M., Topisirovic I., Pelletier J., Translation initiation factors: Reprogramming protein synthesis in cancer. Trends Cell Biol. 26, 918–933 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Bader A. G., Vogt P. K., An essential role for protein synthesis in oncogenic cellular transformation. Oncogene 23, 3145–3150 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Truitt M. L., Ruggero D., New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomasetti C., Marchionni L., Nowak M. A., Parmigiani G., Vogelstein B., Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 112, 118–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faller W. J. et al., mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Lidth de Jeude J. F. et al., Heterozygosity of chaperone Grp78 reduces intestinal stem cell regeneration potential and protects against adenoma formation. Cancer Res. 78, 6098–6106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwanhäusser B. et al., Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Loayza-Puch F. et al., p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 14, R32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korinek V. et al., Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 275, 1784–1787 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Dow L. E. et al., Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huels D. J., Sansom O. J., Stem vs non-stem cell origin of colorectal cancer. Br. J. Cancer 113, 1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Roock W. et al., Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 11, 753–762 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Fearon E. R., Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 6, 479–507 (2011). [DOI] [PubMed] [Google Scholar]
- 18.van Es J. H. et al., Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Feng Y. et al., Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology 141, 1003–1013–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolfi S. C. et al., The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab. 1, 20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topisirovic I., Sonenberg N., mRNA translation and energy metabolism in cancer: The role of the MAPK and mTORC1 pathways. Cold Spring Harb. Symp. Quant. Biol. 76, 355–367 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tollervey D., Kiss T., Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9, 337–342 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Popov A. et al., Duration of the First Steps of the Human rRNA Processing, (Nucl., United States, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Riggelen J., Yetil A., Felsher D. W., MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Sears R., Leone G., DeGregori J., Nevins J. R., Ras enhances Myc protein stability. Mol. Cell 3, 169–179 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Girard M., Penman S., Darnell J. E., The effect OF actinomycin ON ribosome formation IN hela cells. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.51.2.205 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell K. J., White R. J., MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 4, a018408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voorneveld P. W., Jacobs R. J., Kodach L. L., Hardwick J. C. H., A meta-analysis of SMAD4 immunohistochemistry as a prognostic marker in colorectal cancer. Transl. Oncol. 8, 18–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi Z. et al., BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao X. et al., Clinicopathological characterization of SMAD4-mutated intestinal adenocarcinomas: A case-control study. PLoS One 14, e0212142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drost J. et al., Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Matano M. et al., Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Wiener Z. et al., Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-β. Proc. Natl. Acad. Sci. U.S.A. 111, E2229–E2236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramírez-Valle F., Braunstein S., Zavadil J., Formenti S. C., Schneider R. J., eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181, 293–307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong J., Lasko P., Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383–394 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Richter J. D., Sonenberg N., Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Grüner S. et al., The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell 64, 467–479 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Azar R., Alard A., Susini C., Bousquet C., Pyronnet S., 4E-BP1 is a target of Smad4 essential for TGFbeta-mediated inhibition of cell proliferation. EMBO J. 28, 3514–3522 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colina R. et al., Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Haghighat A., Sonenberg N., eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272, 21677–21680 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Jossé L., Xie J., Proud C. G., Smales C. M., mTORC1 signalling and eIF4E/4E-BP1 translation initiation factor stoichiometry influence recombinant protein productivity from GS-CHOK1 cells. Biochem. J. 473, 4651–4664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker S. J. et al., Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244, 217–221 (1989). [DOI] [PubMed] [Google Scholar]
- 44.Fessler E. et al., TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 8, 745–760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Wetering M. et al., Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crissman H. A., Steinkamp J. A., Rapid, simultaneous measurement of DNA, protein, and cell volume in single cells from large mammalian cell populations. J. Cell Biol. 59, 766–771 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haneke K. et al., CDK1 couples proliferation with protein synthesis. J. Cell Biol. 219, e201906147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polymenis M., Aramayo R., Translate to divide: Сontrol of the cell cycle by protein synthesis. Microb. Cell 2, 94–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thoreen C. C. et al., A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Düvel K. et al., Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martineau Y. et al., Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene 33, 1367–1374 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Tian T., Li X., Zhang J., mTOR signaling in cancer and mtor inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 20, E755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung K. E. et al., Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. U.S.A. 107, 1565–1570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riemer P. et al., Oncogenic β-catenin and PIK3CA instruct network states and cancer phenotypes in intestinal organoids. J. Cell Biol. 216, 1567–1577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Q. et al., Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 53, 7146–7155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kentsis A., Topisirovic I., Culjkovic B., Shao L., Borden K. L. B., Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. U.S.A. 101, 18105–18110 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersson F. et al., Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin. Cancer Res. 17, 2874–2884 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janssen K. P. et al., APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 131, 1096–1109 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Sansom O. J. et al., Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 14122–14127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takaku K. et al., Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92, 645–656 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Halberg R. B. et al., Tumorigenesis in the multiple intestinal neoplasia mouse: Redundancy of negative regulators and specificity of modifiers. Proc. Natl. Acad. Sci. U.S.A. 97, 3461–3466 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Ye Q., She Q.-B., New insights into 4E-BP1-regulated translation in cancer progression and metastasis. Cancer Cell Microenviron. 1, e331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohashi A. et al., Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun. 6, 7668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy D. J. et al., Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu H. S. et al., The 4E-BP1/eIF4E ratio is a determinant for rapamycin response in esophageal cancer cells. J. Thorac. Cardiovasc. Surg. 149, 378–385 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Grosso S. et al., Sensitivity of global translation to mTOR inhibition in REN cells depends on the equilibrium between eIF4E and 4E-BP1. PLoS One 6, e29136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choo A. Y., Yoon S. O., Kim S. G., Roux P. P., Blenis J., Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. U.S.A. 105, 17414–17419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thoreen C. C. et al., An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sriram A., Bohlen J., Teleman A. A., Translation acrobatics: How cancer cells exploit alternate modes of translational initiation. EMBO Rep. 19, e45947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alberici P. et al., Smad4 haploinsufficiency: A matter of dosage. PathoGenetics 1, 2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita M. et al., mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 14, 473–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato T. et al., Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Heijmans J. et al., ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Shibata H. et al., Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997). [DOI] [PubMed] [Google Scholar]
- 75.Jackson E. L. et al., Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jonkers J. et al., Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Koo B.-K. et al., Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 9, 81–83 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Van Lidth de Jeude J. F., Vermeulen J. L. M., Montenegro-Miranda P. S., Van den Brink G. R., Heijmans J., A protocol for lentiviral transduction and downstream analysis of intestinal organoids. J. Vis. Exp., 52531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the microarray data shown in the manuscript and SI Appendix are deposited in the Gene Expression Omnibus Database (GSE143509).