Significance
Allostery is a fundamental mechanism by which proteins recognize environmental cues and elicit a response at a distal site. Allosteric signaling is essential in the regulation of myriad cellular processes, yet its molecular basis is poorly understood. Compared to traditional approaches that rely on structure to understand allostery, we chose a high-throughput function-centric approach. Using systematic mutagenesis, we found allosteric signaling to be highly adaptable where a mutation that inactivates allostery can be functionally compensated by another distal mutation. Although functionally important, allosteric hotspots, residues critical for signaling, were poorly conserved. In contrast, residues important for structural stability are significantly conserved, suggesting evolution selects fold over function. Our approach can lead to broad molecular principles of allostery.
Keywords: allostery, deep mutational scanning, functional plasticity, molecular dynamics simulation
Abstract
Allostery is a fundamental regulatory mechanism of protein function. Despite notable advances, understanding the molecular determinants of allostery remains an elusive goal. Our current knowledge of allostery is principally shaped by a structure-centric view, which makes it difficult to understand the decentralized character of allostery. We present a function-centric approach using deep mutational scanning to elucidate the molecular basis and underlying functional landscape of allostery. We show that allosteric signaling exhibits a high degree of functional plasticity and redundancy through myriad mutational pathways. Residues critical for allosteric signaling are surprisingly poorly conserved while those required for structural integrity are highly conserved, suggesting evolutionary pressure to preserve fold over function. Our results suggest multiple solutions to the thermodynamic conditions of cooperativity, in contrast to the common view of a finely tuned allosteric residue network maintained under selection.
Cellular processes are mediated by intermolecular and intramolecular interactions of proteins. Allostery is the intramolecular modulation of protein activity through perturbation at a distal site and constitutes a dominant mode of posttranslational regulation of proteins. Over the decades, we have made major strides in gaining an atomic-level understanding of how proteins fold, catalyze reactions, and interact with other biomolecules. However, understanding the molecular rules governing allostery, a fundamental property of proteins, remains an elusive goal 60 y after its discovery (1–3). The knowledge gap exists because the decentralized character of allostery makes it challenging to intuitively understand and predict how a distal residue affects an active site some 40–50 Å away (4). Mechanisms of catalysis or binding are routinely explained by mutating a limited set of residues as these processes are driven by local interactions. This classical reductionist approach of studying function with a limited set of mutations does not scale for a systemic, protein-wide property like allostery as it explores only a small fraction of available sequence space. Therefore, our current understanding of allostery is principally shaped by a structure-centric paradigm based either on conformational heterogeneity (induced fit and conformational selection) (5–8), comparison of crystallographic snapshots to infer residues linking allosteric and active sites (9), mapping residues undergoing correlated motion by NMR (10, 11), or identifying coevolving residues (12). In rare instances, when functional screens were painstakingly carried out, they revealed complex allosteric networks that cannot be gleaned by examining the structure alone (13). Therefore, while structure offers vital clues, validating the functional contribution of a residue is the clearest evidence of its role in allostery.
Here, we reframe the problem by advancing a function-centric approach guided by structure and free energy calculations to elucidate the molecular basis and the functional landscape of allostery. Allosteric switchability is defined as the ability to switch between inactive and active states in a ligand-dependent manner. To investigate the underlying functional landscape, we disrupted allosteric switchability of a bacterial transcription factor (TF) and restored function through alternative paths by systematic, protein-wide mutational scanning. This revealed remarkable functional plasticity as allosteric switchability could be reconstituted after disruption through myriad mutational combinations. While the degree of functional plasticity is site-specific, structural models indicate that recovery of function may be commonly achieved through modulation of DNA or ligand interactions. Phylogenetic analysis revealed that residues critical for allosteric signaling are surprisingly poorly conserved while those required for structural integrity are highly conserved. This suggests stronger evolutionary pressure to preserve fold over function. Molecular dynamics (MD) simulations showed conformational distributions of wildtype are distinct from those of a disrupted mutant but strikingly similar to a rescued mutant, suggesting different mutational paths lead to the same functional state. Our comprehensive function-centric framework is applicable to other allosteric proteins and can lead to a biochemical understanding of disease-associated mutations, discovery of druggable allosteric sites, and broad molecular principles of allostery.
Plasticity of Allostery
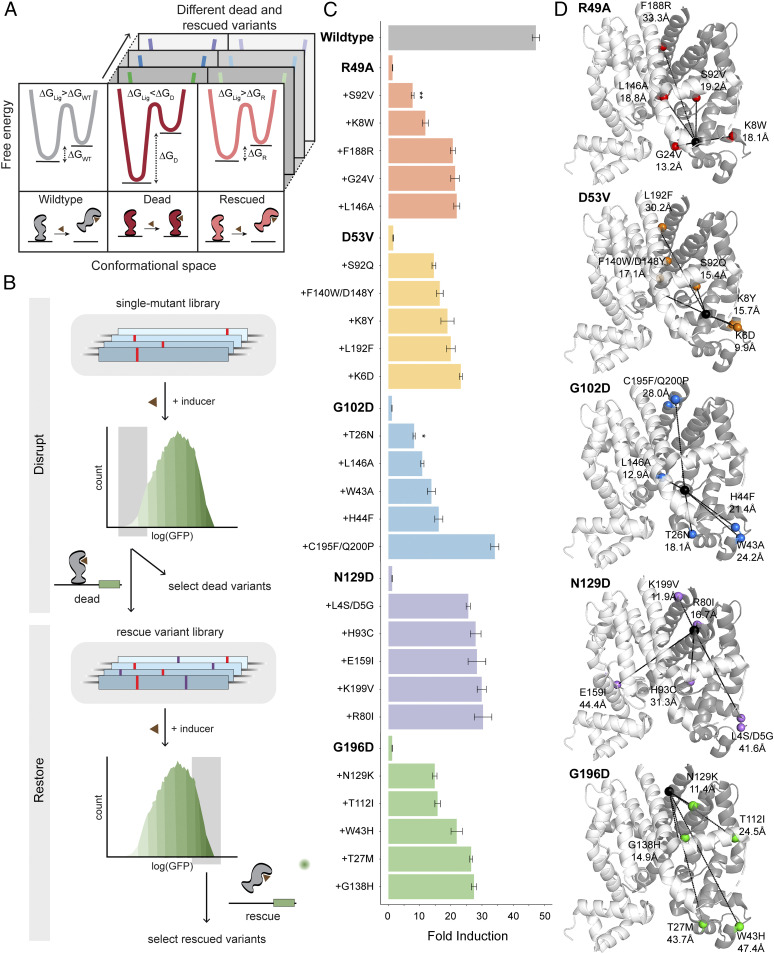
Our model system is tetracycline repressor (TetR, 207 residues), an all-helical (α1–α9), dimeric bacterial TF comprised of ligand- and DNA-binding domains (LBD and DBD) (SI Appendix, Fig. S1). As with all allosteric proteins, inactive and active states of TetR correspond to distinct free energy minima (14). TetR represses gene expression by binding to a promoter (inactive state), and ligand induction releases TetR from that promoter (active state) resulting in transcription. In simplified terms, allosteric switching occurs because free energy of ligand binding is greater than the free energy difference between inactive and active states, which provides the necessary driving force for conformational stabilization (Fig. 1A, ΔGLIG > ΔGDIFF,WT) (15). Detailed thermodynamics are described in SI Appendix, Fig. S2. A mutated TetR may no longer be ligand-inducible if the mutation, without reducing ligand affinity, increases free energy difference (ΔGDIFF) between inactive and active states by stabilizing the inactive state, destabilizing the active state, or both. We term these variants “locked” in a constitutively inactive state as “dead” (Fig. 1A, ΔGLIG < ΔGDIFF,D). A dead variant may be rescued by a compensatory mutation(s) that restores wildtype-like free energy difference (Fig. 1A, ΔGLIG > ΔGR); this we term a “rescued” variant.
Fig. 1.
”Disrupt-and-restore” strategy to characterize plasticity of TetR allostery. (A) A simplified thermodynamic model of allosteric signaling (detailed model SI Appendix, Fig. S2). (Left) Ligand binding provides the driving force to switch from inactive to active state in wild type. (Center) A disrupting mutation (dead variant) makes TetR constitutively inactive due to larger energy difference. (Right) Allosteric function is restored (rescued variant) by energetic rebalancing via distal compensatory mutations. (B) Two-stage protein-wide mutational scanning to first disrupt and subsequently restore allosteric signaling using a GFP-based transcriptional reporter system. (C) Fold induction of individual variants (mean ± SEM) in the presence in 1 µM aTC of three biological replicates. All rescue variants are significantly different from the corresponding dead (P < 0.001 unless noted; *P < 0.05, **P < 0.01). (D) Five dead variants (black spheres), their corresponding rescued variants (colored spheres), and distances between them (dashed lines).
To characterize the plasticity of allosteric networks, we devised a “disrupt-and-restore” strategy. This two-stage, high-throughput, GFP-based mutational screen (16) of TetR involves first disrupting and subsequently restoring allosteric signaling (Fig. 1B). We used commercially available chip oligonucleotides (Twist Biosciences) to encode a library of point mutants by single-site saturation mutagenesis of each TetR residue (207 residues × 19 mutants/residue ∼ 3,900 variants). Compared to random mutagenesis, which gives rise to multiple mutations per variant, chip oligonucleotides enable systematic and comprehensive examination by prespecifying all single mutations at each residue. Due to limitations in the length of oligonucleotides, the full library of single-site variants was built by cloning nonoverlapping tiled libraries across the length of TetR. First, we sought to disrupt signaling by screening for dead variants. We sorted cells expressing low levels of GFP when incubated with ligand (anhydrotetracycline; aTC) such that their fluorescence level was comparable to uninduced wildtype TetR. We then clonally confirmed that the variants are not ligand-inducible. Their ability to repress transcription confirmed that the dead variants were well-folded proteins bound to DNA. After excluding mutations at ligand-contacting residues, we found allosterically inactivating dead variants were distributed all across TetR, including one (G102D) previously identified (17). Since rescuing each dead variant required separate mutational screens, we chose to focus only on five different dead variants. To avoid regional biases, the five dead variants were chosen from different regions of TetR for the rescue screen, although, in principle, any dead variant could have been chosen. The five dead variants were R49A (α4 near DBD), D53V (α4 near LBD), G102D (α6 at LBD-DBD interface), N129D (surface exposed on α8), and G196D (α9 at dimerization interface) (SI Appendix, Fig. S1). Next, on each dead variant’s background, we constructed another protein-wide single-site saturation mutant library and devised a sorting scheme to expressly enrich allosterically activatable rescued variants (SI Appendix, Fig. S3A). Cells containing the rescue library were incubated with ligand to enrich for potential rescued variants by sorting cells expressing high levels of GFP (Fig. 1B).
To quantify allosteric coupling, we calculated fold induction, the ratio of GFP expression with and without inducer of individual clones after sorting. The fold induction of wildtype TetR was 47 and that of all dead variants (R49A, D53V, G102D, N129D, and G196D) was ∼1.0. Clonal testing confirmed that allosteric switchability was indeed restored by compensatory mutations in all five dead backgrounds (Fig. 1C and SI Appendix, Fig. S4). Although induced reporter expression of some variants was comparable to wild type (SI Appendix, Fig. S4), fold induction of all rescued variants was below wild type, showing compensatory mutations only partially rescued function (Fig. 1C). It is possible that multiple compensatory mutations may be required to regain function to the level of wild type. In fact, the gratuitous triple mutant (G102D/C195F/Q200P), likely present due to synthesis or cloning errors, gave the highest fold induction of 34. Dead variants N129D and, to a lesser extent, D53V gave largely uniform fold induction for all rescuing mutations, suggesting that there could be an upper limit to reconstitution of function for individual dead variants (with double mutant screening). Sites for compensatory mutations were within 10–20 Å and others as far as 40–50 Å away from the site of mutation in the dead variant (Fig. 1D), suggesting that allosterically coupled residues in TetR are distributed across the protein. Since the five dead variants have no unique attribute except that they are in different regions of the protein, we concluded that other dead variants might also be rescued by distal compensatory mutations. Such a distributed network of allosterically coupled pairs of residues with no apparent spatial relationship suggests that satisfying thermodynamic conditions of cooperativity may be sufficient to maintain allostery in TetR.
Several important insights emerged from these results. First, TetR exhibits a high degree of allosteric plasticity evidenced by the ease of disrupting and restoring function through several mutational paths. This suggests the functional landscape of allostery is dense with fitness peaks, unlike binding or catalysis where fitness peaks are sparse. Second, allosterically coupled residues may not lie along the shortest path linking allosteric and active sites but can occur over long distances because thermodynamic balancing does not require spatial connectivity (18, 19). Third, allosteric signaling occurs through redundant and robust networks instead of a finely tuned unique pathway.
Site-Specific Rescuability of Allosteric Dysfunction
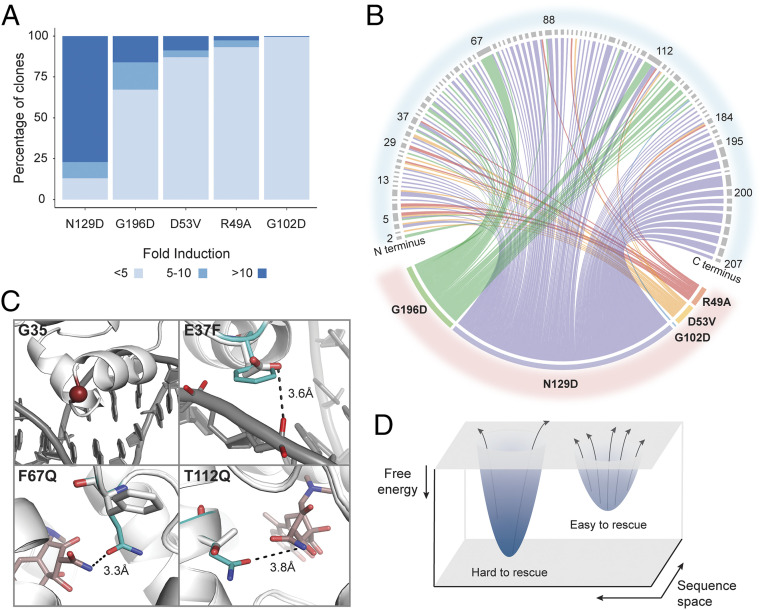
Two key questions emerged from the screen. Are some dead variants easier to rescue than others and why? Are there common structural mechanisms of rescue? To answer these questions, we sought to exhaustively map and quantify rescuing mutations for each dead variant and examine possible common rescue modes. We define “rescuability” as the ease of rescuing function and quantify rescuability using the strength of allosteric response measured by fold induction and the number of unique rescuing mutations. We sorted each rescue variant library for low GFP cells without inducer to enrich DNA-bound, repressed variants (SI Appendix, Fig. S3B). Whereas the previous screening scheme expressly enriched allosterically active variants by sorting GFP-positive cells after ligand induction, here we only enrich DNA-bound variants. This allows us to estimate rescuability of each variant by clonally testing the DNA-bound population and counting the number of functional variants. For each library, we then clonally induced 192 variants and determined the number of unique ligand-responsive clones and their induction level. We classified rescued clones into three groups based on their fold induction: inactive (<5-fold), moderate (5- to 10-fold), or strong rescues (>10-fold) (Fig. 2A).
Fig. 2.
Site-specific differences and structural mechanisms of rescue of function. (A) Percentage of rescued clones whose allosteric activity, based on fold induction, is strong (>10), moderate (5–10), or inactive (<5). (B) Distribution of rescuing mutations (Upper) for each dead variant (Lower) shown in different colors. Each line connects dead and rescuing variants, and its thickness represents number of rescuing mutations at that position. (C) Rosetta structural models of LBD and DBD rescue mutations. Potential common mechanisms of rescue include weaker interaction between DBD and DNA (G35 and E37F) or stronger interactions between LBD and ligand (F67Q and T112Q). (D) Free energy landscape hypothesizing relationship between rescuability and energy gap between inactive and active states. Larger gap (deeper well) is harder to rescue than smaller gap (shallower well).
A clear gradient in rescuability existed from easy to hard as follows based on the number of unique rescuing variants: N129D > G196D > D53V > R49A > G102D (Fig. 2A and 2B). The same ranking emerged when the five variants were ordered based on average fold induction of all 192 screened clones (SI Appendix, Fig. S5). Since the number of clones screened is only a fraction of all possible rescuing variants for each dead variant, the clonal screens only indicate trends in rescuability. Comprehensive double mutant cycling screen by sort-sequencing would fully reveal differences in rescuability between dead variants. Leveraging the wealth of dead-rescue allosteric coupling data, we wanted to infer potential structural mechanisms leading to reconstitution of function. Instead of individually sequencing hundreds of colonies, we deep sequenced all active variants (≥5-fold induction) as a barcoded pool to identify all rescues but cannot pair genotype and phenotype.
We exhaustively mapped each dead variant and their corresponding rescuing mutations to look for focal regions of rescue (Fig. 2B). We observed two broad types of rescuing patterns: those specific to an individual dead variant and those common to multiple dead variants akin to allele-specific and global suppressors observed before (20, 21). The first group were unique rescuing mutations for each dead variant converging at a few key regions indicating that variant-specific regional bias may exist (Fig. 2B and SI Appendix, Table S1). However, no obvious structural mechanism of rescue could be gleaned from the regions where these rescuing mutations were found. An exception was N129D, for which a high concentration of rescuing mutations fell along α9, suggesting restoration of allosteric signaling through the dimerization interface. The second, more striking group, were rescuing mutations for multiple dead variants converging on specific residues in LBD or DBD. This tantalizingly suggested a common structural mechanism of rescue irrespective of the dead variant. Using Rosetta software, we generated structural models of rescuing mutations at these sites for closer structural inspection. Rescuing mutations at the DBD appeared to generally weaken protein–DNA interactions. In the DBD, mutation of capping glycine G35 will likely reduce helix-turn-helix stability and E37F result in loss of key amino acid–nucleobase interaction (Fig. 2C). In the LBD, mutants such as F67Q and T112Q appeared to strengthen interactions with ligand by additional hydrogen bonds (Fig. 2C). Common rescuing variants (global suppressors) appear to counteract greater stability of the repressed state of the dead variant by weakening affinity for DNA or strengthening affinity for ligand. Suppressor mutations have been shown to restore function, binding, or stability of an inactivating mutation in various proteins (20–23). Our results are consistent with a limited screen of suppressor mutations of induction-deficient TetR variants, which were found largely in the dimerization interface and at ligand- or DNA-binding positions (20).
Taken together, these results suggest that many structural mechanisms may lead to rescued function, and dominant modes of rescue appear to be mediated through modulation of ligand, DNA, and dimerization interface interactions. Furthermore, we posit that the ease of rescuing a dead variant may correlate with the degree of stabilization of the inactive state (Fig. 2D) i.e., a more stable dead variant could be harder to rescue than a less stable dead variant. This argument is consistent with N129D and G196D being relatively exposed residues (smaller energy perturbation) and, hence, easier to rescue while the remaining three are more buried and consequently harder to rescue (Fig. 2A).
Structural Hotspots More Conserved than Allosteric Hotspots
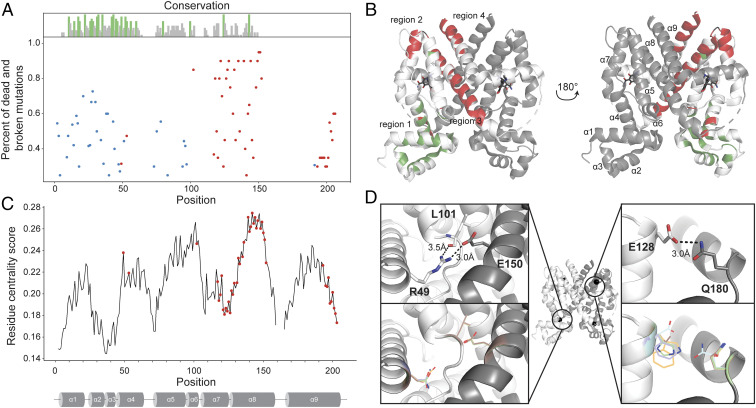
Next, we examined evolutionary conservation and structural context of residues critical for allosteric signaling (hotspots). To comprehensively map allosteric hotspots, we deep sequenced single-site TetR mutants that repressed GFP but were not ligand-inducible (SI Appendix, Fig. S3C, Table S2, and Dataset S1). We found hundreds of inactivating mutations nearly seven times greater than what was previously known (SI Appendix, Fig. S6) (13). We classified 57 residues as allosteric hotspots (47 after excluding ligand-contacting residues) if 25% or more mutations at that position inactivated function. The hotspots clustered into four regions. Region 1 is at the interface of the DBD and LBD on α4 and α6, region 2 is a short motif connecting α7 and α8, region 3 is at the dimer interface on α8, and region 4 is at the C-terminal end on α9 (Fig. 3B). Earlier studies identified hotspots in region 1, although not comprehensively, but hotspots in regions 2, 3, and 4 were previously unknown.
Fig. 3.
Allosteric hotspots are not highly conserved. (A) Comparison of allosteric hotspots (red dot, ligand-contacting residues excluded) with highly conserved residues (green lines). Residues critical for structural integrity and activity (DNA-binding) by experimental validation are shown as blue dot. Each dot represents the number of unique point mutants at a position that make TetR constitutively inactive (red) or active (blue). (B) Allosteric hotspots (red, ligand-contacting residues excluded) and conserved residues (green) separate into distinct groups on TetR structure with only two overlapping residues (black). (C) Location of allosteric hotspots (red) overlaid on residue centrality score of every residue (black line). (D) Loss of allosteric signaling at hotspots R49 (Upper Left) and E128 (Upper Right) is likely due to loss of salt bridge or hydrogen bond interactions with inactive mutations (Lower Left and Lower Right).
Catalytic or binding sites are under strong evolutionary selection as reflected in their high sequence conservation even among distant homologs. We aligned TetR-family homologs to determine if allosteric hotspots too are under evolutionary selection. To our surprise, allosteric hotspots showed low sequence conservation in the TetR family. In fact, highly conserved residues and allosteric hotspots neatly separated into distinct, nonoverlapping groups (Fig. 3A, red points and green lines). To assess the sensitivity of this result to different thresholds for classifying a residue as a hotspot, we determined the number and location of hotspots at different thresholds. At 10%, 25%, and 50% thresholds, there are 67, 57, and 34 hotspots, respectively. However, changing the definition of a hotspot to be more or less strict affects the overall number of hotspots in the dataset, but the regions of functional importance are consistent at all thresholds (SI Appendix, Fig. S7). Based on their location in the DBD (Fig. 3B, green regions), we hypothesized that conserved residues may be required for structural integrity and DNA binding. Since TetR is a repressor, any mutation that destabilizes, reduces DNA-binding, or interferes with dimerization will constitutively express GFP. To evaluate the functional role of conserved residues, we used deep sequencing to determine single-site mutants constitutively expressing reporter without ligand (SI Appendix, Fig. S6 and Dataset S1). We term these “broken” hotspots as they impair TetR’s ability to repress gene expression. Indeed, the location of broken hotspots overlapped with conserved sites, confirming the role of conserved residues in maintaining structural integrity and DNA binding (Fig. 3A, blue points). Compared to allosteric hotspots, the broken hotspots were significantly more conserved on average (SI Appendix, Fig. S8A). An earlier study on known mutations that allosterically inactivate TetR suggested that these residues have higher conservation scores than other residues in the protein (24). A plausible reason for this difference in conclusion could be our high throughput screen revealed three times more allosteric hotspots than was previously known. A computational study comparing known catalytic and allosteric site residues in 56 enzymes similarly concluded that allosteric sites are less conserved than functionally essential catalytic sites (25).
Protein topology plays an important role in determining structure and function. A simplified network representation of protein structures as nodes and edges has been effective understanding residue contributions to protein folding (26), protein–protein interactions (27), and functionally important residues in enzymes (28). We investigated if allosteric hotspots could be recognized based on their local structural context. Computational approaches predict allosteric hotspots based on the connectivity of a residue to all other residues (residue centrality score), which is rooted in the premise that residues with dense interactions act as preferred transmission nodes for signal propagation (24, 28, 29). Structure-based analysis of several protein families showed that central residues were crucial for efficiency of signaling (29). We compared residue centrality score at every position and location of allosteric hotspots and found that although hotspots had a higher centrality score on average than all other positions in the protein (SI Appendix, Fig. S8B), residues with lower centrality score were also allosteric hotspots (Fig. 3C). To understand the relationship between allosteric hotspots and centrality score, we classified all residues of TetR based on their centrality score as “high,” “medium,” or “low” and counted the number of allosteric hotspots within each group (SI Appendix, Fig. S9). One out of 30 low centrality score residues (3.3%), 32 of 134 medium centrality score residues (23.4%), and 21 of 33 high centrality score residues (63.6%) are allosteric hotspots. Residues with low centrality score include floppy loops and peripheral ends of the DBD and do not appear to play a critical role in allosteric signaling. Residues with medium centrality score include many surface-exposed sites on helices, which likely represent sites that are sources or sinks of allosteric signaling. Residues with high centrality scores are buried deep in the dimerization interface and between and LBD and DBD. These likely play a key role in signal transmission. To understand at an atomic level what makes a residue an allosteric hotspot, we modeled using Rosetta two hotspots with high (R49) and low (E128) residue centrality scores (30). R49 of one monomer makes a critical salt bridge with D150 in the second monomer. Allosteric signaling may be lost because mutations at this position break the salt bridge (Fig. 3D). Surface-exposed residue E128 forms a hydrogen bond with Q180 in α9 of the second monomer. Mutations at E128 lose the hydrogen bond, potentially disrupting signaling between dimers (Fig. 3D).
Low conservation of allosteric hotspots, although surprising at first, is consistent with high functional plasticity. Evolution appears to favor preserving structural integrity and activity (DNA binding) because their disruption by mutational drift may be harder to restore than allostery. Clustering of hotspots in distinct regions suggests that signaling occurs through coordinated, decentralized action instead of the shortest path between LBD and DBD. Separation of allosteric hotspots from structural and DBD residues indicates each property could evolve independently, leading to orthologs adapted to new niches.
Free Energy Landscapes of Allosteric Mutants
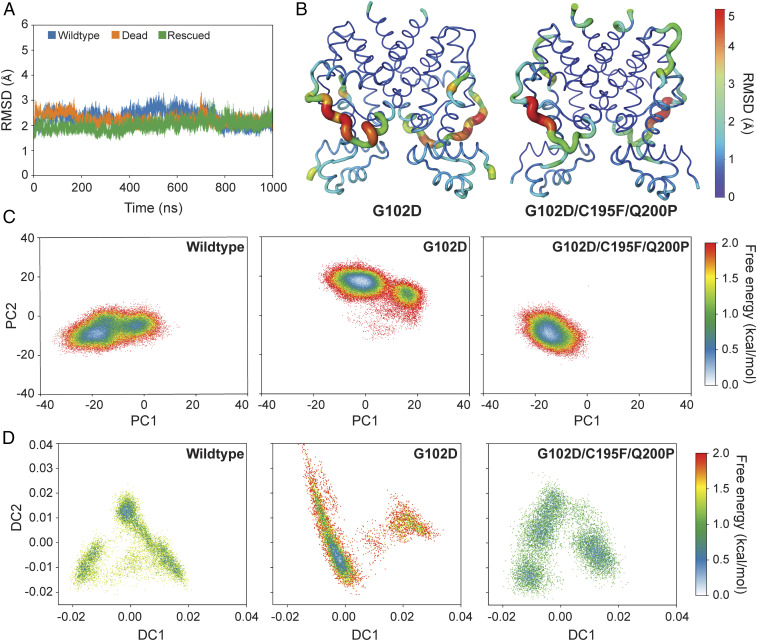
To further investigate if functional similarities between wildtype and rescued variant, and functional differences between wildtype and dead variant, are reflected in global structural properties and conformational distributions, we conducted explicit solvent MD simulations at the microsecond time scale. The simulations were carried out for ligand-bound TetR (without DNA) to understand how remote mutations impact the distribution of active and inactive conformations in the ligand-bound state for all three TetR systems (wildtype, dead, and rescued). We chose wildtype, G102D (dead), and G102D/C195F/Q200P (rescued) for MD simulations. We expected to see pronounced differences in conformational ensembles for this dead-rescue pair because G102D was hardest to rescue, but G102D/C195F/Q200P gave highest fold induction among all rescued variants (Figs. 1C and 2A). All three TetR systems are structurally stable with a similar backbone rmsd of 2–2.9 Å relative to wildtype crystal structure and similar magnitudes of thermal fluctuations (Fig. 4A). To further calibrate the conformational ensembles sampled in our MD simulations, we conducted additional apo state simulations for the wildtype TetR and computed the SAXS profiles for both the apo and ligand-bound states (SI Appendix, Fig. S10). The computed profiles showed excellent agreement with experimental data (31), providing further support to the conformations sampled in our microsecond-scale simulations. Compared to G102D, the triplet mutant features larger displacements relative to wild type near the additional mutation sites (C195F/Q200P), including a large fraction of the α4/α5 loop (Fig. 4B). The region in the DNA-binding domain that connects α3/α4 also undergoes a larger average displacement relative to wild type in the triplet mutant compared to G102D (Fig. 4B). Evidently, the additional remote mutations (C195F/Q200P) lead to subtle but persistent structural differences in the DNA-binding site more than 35 Å away. We have also examined a range of properties relevant to the motional coupling between protein residues (SI Appendix, Figs. S11 and S12), such as positional covariance matrix (SI Appendix, Fig. S13), conformational entropy (SI Appendix, Fig. S14), distribution of community and hub residues (SI Appendix, Fig. S15), and suboptimal pathways that connect ligand- and DNA-binding sites (SI Appendix, Fig. S16). These properties do not exhibit any robust or distinct features among the systems simulated here at the microsecond time scale, consistent with our experimental finding that allostery in TetR does not involve a unique, finely tuned pathway.
Fig. 4.
Free energy landscapes of wildtype, dead, and rescued mutants consistent with function. (A) Backbone RMSDs of wildtype TetR, dead (G102D), and rescued (G102D/C195F/Q200P) variants over time shows stable structures. (B) Average structures of dead and rescued variants over the last 800 ns of simulation. Thickness of ribbon and color scale reflect RMSD per residue relative to average wildtype TetR structure over this time window. (C) Two-dimensional free energy landscapes projected onto first and second principal components show strong similar conformational distributions of wildtype and rescued variant, but distinct from dead variant. (D) Two-dimensional free energy landscapes from the locally scaled diffusion map analysis of wildtype, dead, and rescued variants also observe similar conformational distributions for the wildtype and rescued variant, with the dead variant being distinct.
However, the underlying free energy landscapes projected onto principal components and locally scaled diffusion maps (LSDMaps) (32) revealed remarkable differences among the three TetR systems (Fig. 4 C and D). The principal components describe large amplitude motions while the LSDMaps aim to capture slow motions, thus the two types of analyses complement each other (33). The free energy landscape projected onto the first two principal components (Fig. 4C and see SI Appendix, Fig. S17 for projections along individual principal components) showed a higher degree of similarity between wild type and the triple mutant, with G102D being clearly different. Similarly, the LSDMap landscapes of the wild type and the rescued mutant resemble each other in shape and locations of free energy basins, while the landscape of the dead mutant is significantly different (Fig. 4D). Projections along higher principal components or diffusion coordinates, by contrast, are similar for all three systems. The first two principal component eigenvectors (SI Appendix, Figs. S18 and S19), especially the second one, involve pendulum type of motions of DNA-binding domains that were proposed to affect DNA binding affinity (Movies S1 and S2) (34, 35). Therefore, the different conformational distributions along the primary principal components in different TetR variants are likely functionally relevant as observed in NMR studies of a TetR homolog, QacR (36). However, we caution that it remains challenging to assign quantitative DNA-binding affinities to the different conformations sampled in these simulations. Even with 1 μs of unbiased simulation, it is unlikely that we have completed the sampling of a protein system like TetR. Thus, one should not equate Fig. 4 C and D with the schematic free energy landscapes shown in Fig. 1A or SI Appendix, Fig. S2. Therefore, within the framework of these simulations, we focused on a qualitative comparison of free energy landscapes of wildtype, dead, and rescued variants. For more quantitative comparisons, such as relative populations of key conformational basins outlined in SI Appendix, Fig. S2, enhanced sampling techniques are needed. Nevertheless, these results clearly highlight that mutations remote from the ligand- and DNA-binding sites may lead to sequence-specific changes in the conformational distribution that ultimately get manifested as significant perturbation in function. It is encouraging that these changes are observed in unbiased MD simulations at the microsecond time scale, and we anticipate that additional insights can be gleaned with enhanced sampling simulations in the future. Combined with biophysical characterization, these simulations will help better define the nature of change in conformational ensembles of TetR upon inducer binding (37) or mutations.
Conclusions
The conclusions of this study compel us to reexamine the evolutionary, structural, and biophysical nature of allostery. On the one hand, structural biology studies suggest a physically contiguous pathway transmitting allosteric signals across the protein. However, our functional screen reveals that allosteric coupling exists between distal, physically disconnected pairs of residues, which are close to neither the ligand- or the DNA-binding domains. Furthermore, the surprising preponderance of mutational combinations that preserve allosteric signaling, the majority of which do not fall along a contiguous pathway, raises the question of why does such a high degree of functional plasticity exist? One possibility is these rescued double mutants are conditionally neutral mutations that permit TetR to drift in mutational space without appreciable loss of fitness under existing selection pressure (38, 39). However, if the selection pressure changes, a population of genetic variants accumulated by neutral drift could readily adapt under the new conditions. The conditionally neutral mutations may allow TetR to sample new conformational states that may confer new ligand specificities without abrogating native function (40). This may partly explain the extraordinary diversity of ligands binding to TetR-family proteins (41). These results also have important implications for design of allosteric proteins. It may be beneficial to use all of the conditionally neutral mutants as starting scaffolds for structure-guided design to diversify sampling in structural and functional space. A critical assumption in statistical coupling analysis and other coevolution-based approaches is that nature imposes evolutionary pressure at specific sites to preserve allosteric coupling. Our case study of TetR challenges this assumption because we find that allosteric sites are not necessarily conserved, although allostery by itself may be conserved (42). Conservation of specific sites may be a feature of catalysis or binding, which strongly depend on key local molecular interactions. In contrast, since allostery is systemic, balancing of thermodynamic forces can be satisfied by decentralized functional constraints in many possible ways without the need for conserving specific sites. This idea is qualitatively consistent with the mechanistic models (18, 37) that highlight the ensemble nature of allostery, although further studies are required to better define, at the molecular level, the changing conformational ensembles upon inducer binding or mutation. Future mutational studies on other TetR-family members may reveal if common lynchpin sites and couplings exist within the family that can be recognized from sequences alone. Together with machine learning, large datasets like ours linking structural effects of mutations to function can help develop heuristic molecular rules, such as gain/loss of salt bridges or hydrogen bonds (R49 and E128, Fig. 3D), for allosteric communication (43). Furthermore, simulating molecular trajectories of tens or hundreds of mutants will help us interpret how conformational heterogeneity of protein ensembles is related to protein function. This is an exciting direction for the field of MD simulations where MD is used not just as a validation and explanatory tool but also for function prediction.
Materials and Methods
Plasmid Construction.
We constructed a sensor plasmid with TetR(B) (Uniprot P04483) cloned into a low-copy backbone (SC101 origin of replication) carrying spectinomycin resistance. The tetRb gene was driven by a variant of promoter apFAB61 and Bba_J61132 RBS (44). On a second plasmid, superfolder GFP (45) was cloned into a high-copy backbone (ColE1 origin of replication) carrying kanamycin resistance under the control of the ptetO promoter. To control for plasmid copy number, RFP was constitutively expressed with the BBa_J23106 promoter and Plotkin RBS (44) in a divergent orientation to sfGFP.
Library Synthesis.
A comprehensive single-mutant TetR library was generated by replacing wildtype residues at positions 2–207 of TetR to all other 19 canonical amino acids (3,914 total mutant sequences). Oligonucleotides encoding each single-point mutation were synthesized as single-stranded Oligo Pools from Twist Bioscience and organized into subpools spanning the tetRb gene: residues 2–39, 40–77, 78–115, 116–153, 154–191, 192–207. Oligo pools were encoded as a concatemer of the forward priming sequence, a BasI restriction site (5′-GGTCTC), six-base upstream constant region, tetR mutant sequence, six-base downstream constant region, a BsaI site (5′-GAGACC), and the reverse priming sequence. Subpools were resuspended to 25 ng/μL and amplified using primers specific to each oligonucleotide subpool with KAPA SYBR FAST qPCR (KAPA Biosystems; 1-ng template). A second PCR amplification was performed with KAPA HiFi (KAPA Biosystems; 1-μL qPCR template, 15 cycles maximum). We amplified corresponding regions of pSC101_tetR_specR with primers that linearized the backbone, added a BsaI restriction site, and removed the replaced wildtype sequence. Vector backbones were further digested with DpnI, BsaI, and Antarctic phosphatase before library assembly.
We assembled mutant libraries by combining the linearized sensor backbone with each oligo subpool at a molar ratio of 1:5 using Golden Gate Assembly Kit (New England Biolabs; 37 °C for 5 min and 60 °C for 5 min, repeated 30 times). Reactions were dialyzed with water on silica membranes (0.025-μm pores) for 1 h before transformed into DH10B cells (New England Biolabs). Library sizes of at least 100,000 colony-forming units (cfu) were considered successful. DH5α cells (New England Biolabs) containing the reporter pColE1_sfGFP_RFP_kanR were transformed with extracted plasmids to obtain libraries of at least 100,000 cfu.
Rescue variant libraries were synthesized as described above using sensor plasmid of each dead variant as the linearized backbone. To avoid mutations close in sequence space, the oligo subpool containing the mutation was not cloned into the library—for example, the G102D library did not contain mutations spanning residues 78–115 in TetR. All double-mutant libraries contained 3,192 possible sequences, except for the G196D rescue library, which contained 3,610 sequences.
Fluorescence Activated Cell Sorting.
All library cultures and clonal variants were grown for 16 h at 37 °C in lysogeny broth (LB) containing 50 μg/mL kanamycin (kan) and 50 μg/mL spectinomycin (spec). Libraries were seeded from a 50-µL aliquot of glycerol stocks and grown to an OD600 ∼0.2 before induction with 1 µM aTC. Saturated library cultures were diluted 1:50 in 1× phosphate saline buffer (PBS), and fluorescence intensity was measured on a SH800S Cell Sorter (Sony). Remaining uninduced cultures were spun down and plasmids were extracted for next-generation sequencing to represent the presorted library distribution. We first gated cells to remove debris and doublets and selected for variants constitutively expressing RFP. The induction profile of wildtype TetR was used as reference in drawing gates on GFP fluorescence to identify DNA-bound and rescued variants in the double-mutant rescue libraries (SI Appendix, Figs. S3 A and B). Libraries were sorted in the absence of inducer between ∼10–1,000 RFU (fluorescence distribution of repressed wildtype TetR, DNA-bound) to identify DNA-bound variants or in the presence of 1 µM aTC ∼30,000–200,000 RFU (fluorescence distribution of inducted wildtype TetR, active) to identify rescues. A total of 500,000 events were sorted for each gated population, and cells were recovered in 5 mL of LB for 1 h before cultures were plated. Rescue variants identified in Fig. 1 were identified by clonally screening ∼200–400 colonies from the sorted population for each double-mutant rescue library.
To identify dead variants in the TetR single-mutant library, DNA-bound variants were sorted in the presence and absence of 1 µM aTC using the distribution of repressed wildtype TetR to define the sorting gate (Fig. 3C). Cells were sorted as above before antibiotics were added and cultures grown for an additional 6 h until an OD600 ∼0.2 was reached for plasmid extraction and sequencing. Each library was grown, sorted, and sequenced in duplicate. Dead variants analyzed in Fig. 1 were identified by sorting and plating DNA-bound variants in the presence of 1 µM aTC and clonally screening ∼300 colonies from the sorted population.
Clonal Screening and Flow Cytometry.
To screen for dead and rescuing variants, individual colonies were picked and grown to saturation in a 96-well plate for 8 h. Nonfluorescent colonies under blue light were selected in these screens to select for variants with DNA-binding capability in the absence of inducer. Saturated cultures were diluted 1:50 in LB-kan/spec and grown in the presence and absence of 1 µM aTC for 16 h before OD600 and GFP fluorescence (Gain: 40; Excitation: 488/20; Emission: 525/20) were read on a multiplate reader (Synergy HTX, BioTek). Fluorescence was normalized to OD600, and the fold inductions for each variant were computed as the ratio of induced and noninduced fluorescences. Variants with at least 10-fold induction were plated, confirmed in triplicate, and Sanger sequenced (Functional Bioscience). Selected variants were diluted 1:50 into 1× PBS before being measured on the Attune NxT Flow Cytometer (Thermo Fisher Scientific). A one-way ANOVA followed by a Tukey-HSD (honest significant difference) post hoc test was performed for each group of dead and rescue variants to confirm rescues were functionally more active than the dead.
Quantifying Rescuability.
Rescue variant libraries were grown in the absence of inducer, and nonfluorescent cells were sorted to select for DNA-bound variants. For each rescue variant library, we randomly selected and screened 192 nonfluorescent colonies after sorting to determine the percentage of cells that recovered some degree of function. Fold induction for each individual variant was measured on the plate reader, and variants with at least fivefold induction were pooled and prepped for deep sequencing. To identify the number of unique variants in the screen, an initial group of all 192 variants was prepped for sequencing. The average fold induction of all 192 screened clones for each dead variant was calculated and a one-way ANOVA with a Tukey-HSD post hoc test performed to compare rescuability.
Next-Generation Sequencing and Analysis.
Libraries were prepared from plasmids extracted from the presorted, sorted, and pooled libraries to identify dead and rescued variants. Libraries were sequenced with a 2 × 250 sequencing run in which the TetR gene was broken into two fragments to cover the entire encoding region. Libraries were amplified with two primer sets, one specific to the encoding region of interest (residues 2–115 or 116–207) adding the next-generation sequencing priming region, and a second outer pair adding the unique barcode and library adapter. Libraries were amplified in a two-step PCR. First, inner primers were added at a final concentration of 125 nM each and reaction run for 11 cycles before 7× the concentration of the outer stem primers was added (0.9 μM) and another 8 cycles were run. Inactive variant libraries were sequenced with a 2 × 250 MiSeq run through the University of Wisconsin–Madison Biotechnology Center and Genewiz (Amplicon-EZ).
Paired-end Illumina sequencing reads were merged with FLASH (Fast Length Adjustment of SHort reads) using the default software parameters (46). Phred quality scores (Q) were used to compute the total number of expected errors (E) for each merged read (47). Reads exceeding the maximum expected error threshold (Emax) of 1 were removed. To identify inactive variants, nonfluorescent cells in the single-mutant library were sorted in duplicate in both the presence and absence of 1 µM aTC and prepped for sequencing along with the initial, unsorted library. Sequencing reads within each barcoded sample were normalized to 200,000 total reads and then by the number of variants within each sample before a cutoff of 10 reads was applied to reduce noise. Raw and normalized sequencing reads show good correlation between replicates for all samples (SI Appendix, Fig. S20).
Variants with at least 25 reads in both the presence and absence of ligand in both replicates were identified as “dead.” Altering the threshold used to define a mutation as dead to 10, 25, or 50 read counts changes the total number of dead mutations present in the library (1,057, 728, and 452, respectively) but does not change the regions of functional importance within the protein (SI Appendix, Fig. S21). Positions with 25% or more dead variants were termed allosteric, or dead, hotspots. Variants present in the presorted libraries but not in either nonfluorescent (±1 µM aTC) were assumed to be largely fluorescent. We termed these variants “broken” as they are predicted to destabilize the protein and/or affect DNA binding. Positions in which 25% or more mutations broke the protein were termed broken hotspots. Sequencing of variants from the rescuability screen were prepared as above and a cutoff of 10 reads was applied to identify the presence of a variant. Rescued variants with two or more compensatory mutations were found in the screen, but only single-mutant compensatory mutations were used for further analysis.
Sequence Conservation.
TetR homologs were generated using HMM search (https://www.ebi.ac.uk/Tools/hmmer/) (48) against UniProtKB database with TetR(B) sequence as query. Sequences with alignment coverage less than 95% of full-length TetR(B) were removed from consideration. The remaining sequences were aligned using Clustal Omega (49). After applying a redundancy cutoff of 90%, we were left with ∼1,900 sequences, which was used to evaluate sequence conservation score within Jalview (50). Conservation score in Jalview is computed by AMAS tool (51), and positions with a score of seven or more were termed highly conserved. A two-sample t test was used to compare the average conservation score of residues classified as dead or broken (P = 0.0002).
ddG Calculations and Structural Models of Mutations.
The crystal structure of TetR(B) with bound [Minocycline:Mg]+ and DNA-bound TetR(D) dimer structures were obtained (PDB ID codes: 4ac0 and 1qpi) and water molecules removed before calculations run; the bound ligand was also removed from TetR(B). All modeling calculations were performed using the Rosetta molecular modeling suite v3.9. Single-point mutants were generated using the standard ddg_monomer application (30), which enables local conformational to minimize energy. For TetR(B), calculations were run at every position in protein for all 20 amino acids, generating 50 possible mutant and wildtype structural models for each protein variant. Structures with the lowest total energy from the 50 mutant and wildtype models were used to calculate ddG and served as models for structural analysis. Calculations for DNA-bound TetR(D) variants were prepared and analyzed in similarly, but only select rescuing mutations at G35 and E37 were run.
MD Simulations.
The missing residues 160–164 of the TetR(B) crystal structure (PDB ID code: 4ac0) were modeled in CHARMM-GUI. To ensure a stable hydrogen bond interaction between residue 64 and aTC, the protonation state of residue 64 was set to be HSE, as was done in a previous computational analysis of the TetR system (52). For mutants, corresponding residues were mutated.
For each TetR system, the initial structure was solvated in a rectangular box solvated with ∼27,300 TIP3P water molecules with a 15.0 Å of edge distance. There were 87–89 Na+ and 77 Cl- counter ions added to ensure neutrality at an ionic strength of 0.15 M, resulting in a box of around 88 × 88 × 88 Å3 using periodic boundary conditions. All simulations (∼88,700 total atoms) were performed in OpenMM using the CHARMM36m force field. CHARMM-GUI was used to generate input files. Particle mesh Ewald with the Ewald error tolerance of 0.0005 was used to calculate electrostatic interactions; the tolerance stands for the average fractional error in forces that is acceptable. The van der Waals interaction was treated with a switching scheme for distances between 10 and 12 Å. Energy minimization was carried out with 10,000 steps of the l-BFGS algorithm. The system was then equilibrated in the NVT ensemble at 303.15 K for 250 ps. Langevin dynamics was used with a collision frequency of 1 ps−1. During minimization and equilibration, small harmonic restraints were applied to both protein backbone and sidechain atoms, with force constants of 400 kJ/(mol·nm−2) and 40 kJ/(mol·nm−2), respectively. After equilibration, all atoms were released and no restraint was applied. Production simulations were carried out in the NPT ensemble at 303.15 K using Langevin dynamics with a friction coefficient of 1 ps−1. MonteCarloBarostat was used with the pressure of 1 bar and the pressure coupling frequency of 100 steps. In equilibration and production runs, all water molecules were rigid and all bonds involving hydrogen atoms were constrained using HBonds constraints in OpenMM, allowing a time step of 2 fs.
Supplementary Material
Acknowledgments
We thank Dr. Elizabeth Craig and Dr. Katherine Henzler-Wildman for critical review of the manuscript. This work is funded by NIH Director’s New Innovator Award DP2GM132682 (to S.R.) a Shaw Scientist Award (to S.R.), and NIH Molecular Biophysics Training Program Grant T32 GM08293 (M.L.). The computational component of the work is supported by NIH Grant R01-GM106443.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002613117/-/DCSupplemental.
Data and Materials Availability.
All study data are included in the article and SI Appendix.
References
- 1.Wodak S. J. et al., Allostery in its many disguises: From theory to applications. Structure 27, 566–578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Changeux J. P., Edelstein S. J., Allosteric mechanisms of signal transduction. Science 308, 1424–1428 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Changeux J. P., 50 years of allosteric interactions: The twists and turns of the models. Nat. Rev. Mol. Cell Biol. 14, 819–829 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Dokholyan N. V., Controlling allosteric networks in proteins. Chem. Rev. 116, 6463–6487 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Cui Q., Karplus M., Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J., Zhou H. X., Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilser V. J., Biochemistry. An ensemble view of allostery. Science 327, 653–654 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunasekaran K., Ma B., Nussinov R., Is allostery an intrinsic property of all dynamic proteins? Proteins 57, 433–443 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Daily M. D., Gray J. J., Local motions in a benchmark of allosteric proteins. Proteins 67, 385–399 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Holliday M. J., Camilloni C., Armstrong G. S., Vendruscolo M., Eisenmesser E. Z., Networks of dynamic allostery regulate enzyme Function. Structure 25, 276–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popovych N., Sun S., Ebright R. H., Kalodimos C. G., Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 13, 831–838 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Süel G. M., Lockless S. W., Wall M. A., Ranganathan R., Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59–69 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Müller G. et al., Characterization of non-inducible Tet repressor mutants suggests conformational changes necessary for induction. Nat. Struct. Biol. 2, 693–703 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Chure G. et al., Predictive shifts in free energy couple mutations to their phenotypic consequences. Proc. Natl. Acad. Sci. U.S.A. 116, 18275–18284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai C. J., Nussinov R., A unified view of “how allostery works”. PLOS Comput. Biol. 10, e1003394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler D. M., Fields S., Deep mutational scanning: A new style of protein science. Nat. Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholz O. et al., Activity reversal of Tet repressor caused by single amino acid exchanges. Mol. Microbiol. 53, 777–789 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marzen S., Garcia H. G., Phillips R., Statistical mechanics of Monod-Wyman-Changeux (MWC) models. J. Mol. Biol. 425, 1433–1460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biburger M., Berens C., Lederer T., Krec T., Hillen W., Intragenic suppressors of induction-deficient TetR mutants: Localization and potential mechanism of action. J. Bacteriol. 180, 737–741 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuber J., Danial S. A., Connelly S. M., Naider F., Dumont M. E., Identification of destabilizing and stabilizing mutations of Ste2p, a G protein-coupled receptor in Saccharomyces cerevisiae. Biochemistry 54, 1787–1806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apanovitch D. M., Iiri T., Karasawa T., Bourne H. R., Dohlman H. G., Second site suppressor mutations of a GFPase-deficient G-protein α-subunit. J. Biol. Chem. 273, 28597–28602 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Suad O. et al., Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations. J. Mol. Biol. 385, 249–265 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Yu Z., Reichheld S. E., Savchenko A., Parkinson J., Davidson A. R., A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 400, 847–864 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Yang J.-S., Seo S. W., Jang S., Jung G. Y., Kim S., Rational engineering of enzyme allosteric regulation through sequence evolution analysis. PLOS Comput. Biol. 8, e1002612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vendruscolo M., Dokholyan N. V., Paci E., Karplus M., Small-world view of the amino acids that play a key role in protein folding. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 65, 061910 (2002). [DOI] [PubMed] [Google Scholar]
- 27.del Sol A., O’Meara P., Small-world network approach to identify key residues in protein-protein interaction. Proteins 58, 672–682 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Amitai G. et al., Network analysis of protein structures identifies functional residues. J. Mol. Biol. 344, 1135–1146 (2004). [DOI] [PubMed] [Google Scholar]
- 29.del Sol A., Fujihashi H., Amoros D., Nussinov R., Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol. 2, 2006.0019 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellogg E. H., Leaver-Fay A., Baker D., Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79, 830–838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palm G. J. et al., Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding. Biochim. Biophys. Acta. Proteins Proteomics 1868, 140404 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Rohrdanz M. A., Zheng W., Maggioni M., Clementi C., Determination of reaction coordinates via locally scaled diffusion map. J. Chem. Phys. 134, 124116 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y., Cui Q., The histone H3 N-terminal tail: A computational analysis of the free energy landscape and kinetics. Phys. Chem. Chem. Phys. 17, 13689–13698 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Aleksandrov A., Schuldt L., Hinrichs W., Simonson T., Tet repressor induction by tetracycline: A molecular dynamics, continuum electrostatics, and crystallographic study. J. Mol. Biol. 378, 898–912 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W., Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7, 215–219 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi K., Imai M., Shimada I., Conformational equilibrium defines the variable induction of the multidrug-binding transcriptional repressor QacR. Proc. Natl. Acad. Sci. U.S.A. 116, 19963–19972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichheld S. E., Yu Z., Davidson A. R., The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor. Proc. Natl. Acad. Sci. U.S.A. 106, 22263–22268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman A. S., White K. I., Ranganathan R., Origins of allostery and evolvability in proteins: A case study. Cell 166, 468–480 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Payne J. L., Wagner A., The causes of evolvability and their evolution. Nat. Rev. Genet. 20, 24–38 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Hong N. S. et al., The evolution of multiple active site configurations in a designed enzyme. Nat. Commun. 9, 3900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuthbertson L., Nodwell J. R., The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fodor A. A., Aldrich R. W., On evolutionary conservation of thermodynamic coupling in proteins. J. Biol. Chem. 279, 19046–19050 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Demerdash O. N., Daily M. D., Mitchell J. C., Structure-based predictive models for allosteric hot spots. PLOS Comput. Biol. 5, e1000531 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosuri S. et al., Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 14024–14029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Magoč T., Salzberg S. L., FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R. C., Flyvbjerg H., Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Potter S. C. et al., HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F. et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livingstone C. D., Barton G. J., Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 9, 745–756 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Aleksandrov A., Proft J., Hinrichs W., Simonson T., Protonation patterns in tetracycline:tet repressor recognition: Simulations and experiments. ChemBioChem 8, 675–685 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.