Abstract
Nuclear processes such as DNA replication, transcription, and RNA processing each depend on the concerted action of many different protein and RNA molecules. How biomolecules with shared functions find their way to specific locations has been assumed to occur largely by diffusion-mediated collisions. Recent studies have shown that many nuclear processes occur within condensates that compartmentalize and concentrate the protein and RNA molecules required for each process, typically at specific genomic loci. These condensates have common features and emergent properties that provide the cell with regulatory capabilities beyond canonical molecular regulatory mechanisms. We describe here the shared features of nuclear condensates, the components that produce locus-specific condensates, elements of specificity, and the emerging understanding of mechanisms regulating these compartments.
Nuclear Condensates
Diverse nuclear processes function in dynamic compartments where the tens to hundreds of different protein and RNA molecules involved are concentrated, often at specific DNA loci. Early cytologists observed the largest and most stable of these compartments over a century ago, the nucleolus and Cajal bodies [1–3]. The list of nuclear compartments has expanded substantially since and have been described as biomolecular condensates (see Glossary), membraneless organelles, nuclear bodies, non-membrane-bound bodies, factories, hubs, and clusters. We refer to them here as biomolecular condensates. The term condensate makes no assumption regarding either the physical mechanism through which assembly is achieved nor the material state of the resulting assembly. Rather, it allows us to discuss both bodies that form through phase separation, as well as bodies where the physical origins of assembly are unknown. This term also provides a link to condensed matter physics, which has become instructive for investigating the formation and regulation of these previously enigmatic compartments [4]. Many excellent reviews have been published covering the general principles of condensate biology and its links to concepts and theory adopted from physics and chemistry [4–11]. In this review, we focus on the shared and distinguishing features of nuclear condensates, and their regulation.
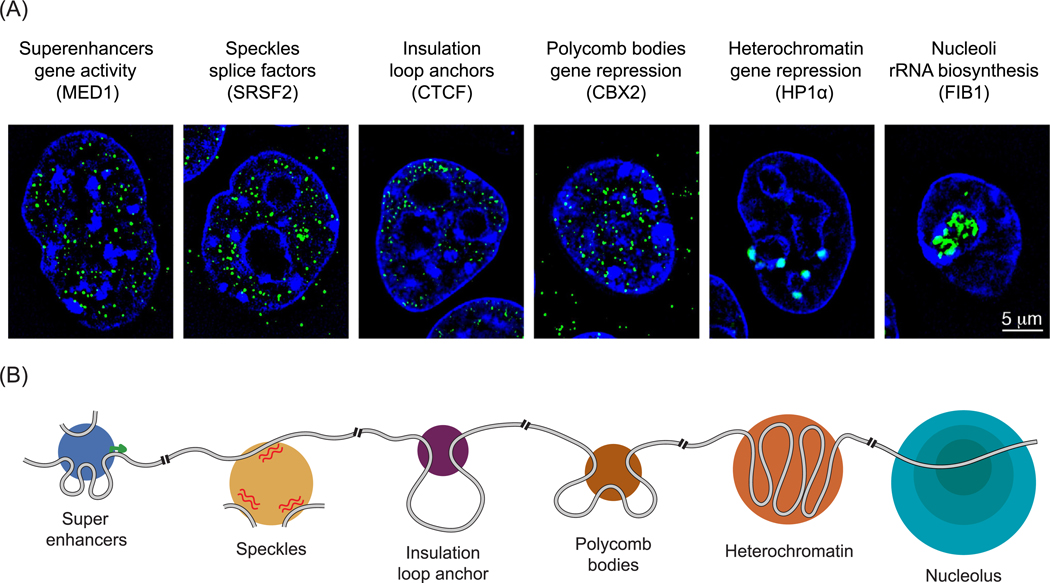
The nuclear condensates that have been described as of this writing are listed in Table 1 and images for various nuclear condensates found in embryonic stem cells are shown in Figure 1. The remarkable feature of these condensates is that they encompass most of the nuclear regulatory processes that have been the subject of detailed genetic, biochemical, and structural studies, and thus processes where there is considerable knowledge of the regulatory apparatus and the molecular mechanisms by which regulation of the process occurs. These nuclear processes include, but are not limited to, regulation of chromosome structure and maintenance [12–16], DNA replication [17], DNA repair [18,19], transcription [20–24], RNA processing [25,26], and preribosome assembly [27–30]. The reviews that have described these processes have historically focused on regulatory mechanisms by which individual proteins and multisubunit protein complexes perform their functions, but lack the additional perspectives that come from considering these processes in the context of condensates. It is our goal to provide that context by describing features shared by nuclear condensates and the additional layers of regulation these can provide to nuclear processes.
Table 1.
Nuclear Condensates
| Name | Function | Refs | Condensate features | Regulation |
|---|---|---|---|---|
| Nucleolus | rRNA transcription, processing, and preribosome assembly | [27–30,40] | Multilayered organization with functional division of labor: rRNA transcription, rRNA processing, and preribosome assembly. nucleated by local RNA synthesis | -NPM1 has two conserved acidic tracts necessary for R-motif engagement -NPM1 has a nucleic acid binding domain necessary for binding rDNA |
| Cajal body | snRNA transcription and processing | [3,58,114–117] | Concentrates RNA processing enzymes responsible for spliceosomal RNA maturation. | Nucleated by local RNA synthesis |
| Superenhancers | Transcription of cell-identity mRNA | [20,21,23] | Concentrates relevant cofactors, phosphorylation-regulated partitioning of RNA Pol II, seeded by specific enhancer features, DNA-dependent assembly, and reduces search space for signaling factors | -Serine residues in MED1 necessary for phase separation -Acidic residues in OCT4 help interactions with MED1 droplets -Length of RNA Pol II CTD is important -DNA sequence is important -Aromatic residues in b-cat are required to partition |
| Histone locus body | Histone mRNA transcription and processing | [118–121] | Coordinated expression of arrayed repeats of replication-dependent histone genes during early S phase. Both the RNA Pol II transcriptional machinery and histone mRNA processing enzymes are concentrated | -CDK phosphorylation of NPAT -Size dependent on underlying number of histone genes |
| Estrogen receptor | Estrogen-responsive gene regulation | [122] | Concentrates the RNA Pol II machinery in an estrogen-dependent manner | Chronic estrogen stimulation leads to changes in material properties and irreversibility |
| YAP | Response to hyperosmotic stress | [123] | Selective partitioning of TEAD and TAZ for target gene activation upon hyperosmotic shock | |
| Speckles | Cotranscriptional mRNA processing and processing | [25,26,48,124] | Concentrates the mRNA processing machinery. Phosphorylation-regulated partitioning of RNA Pol II, seeded by RNA machinery storage | Phosphorylation of RNA Pol II CTD leads to association synthesis |
| Paraspeckle | Storage of RNA and protein | [38,69,70] | NEAT1 RNA and various RNA-binding proteins are essential components. Core shell architecture. | Synthesis of NEAT1 RNA isoforms regulates formation |
| CTCF clusters | Genome structure | [125,126] | Reduced search space for CTCF, nucleated by RNA binding | RNA binding domain required for CTCF clustering |
| DNA damage | Double strand break repair | [18,19] | Seeded by PARP, 53BP1 phase separation selectively partitions p53 | Seeded by DNA damage induced PARylation |
| DNA replication origins | Specification of DNA replication origin | [17] | Coordinates replication initiation of clusters sites. ORC, Cdc6, and Cdt1 are proposed scaffold. DNA-dependent formation. Selective partitioning of Mcm2–7 promotes loading and replication origin choice. | Phosphorylation regulated |
| Chromatin | Physiological form of genetic material | [16] | Regulates access to underlying regulatory and coding sequence | -Acetyl-lysine-driven dissolution -Nucleosome-spacing-driven compaction |
| Constitutive heterochromatin | Repression of repetitive elements | [12,14–16,88] | HP1α proposed as scaffold, creates highly dynamic concentrations of chromatin. | -Phosphorylation of HP1 promotes phase separation -Dynamic PTM regulation: H3K9me3 promotes HP1/swi6 binding and condensate formation, Lysine acetylation promotes chromatin decompaction |
| Facultative heterochromatin (PcG bodies) | Repression of cell-type specific and developmental mRNA | [127] | Concentrates CBX2. Amino acids that drive phase separation also drive chromatin compaction and are necessary for proper segmentation during development | Basic patch in CBX2 is required |
| PML bodies | Various functions proposed | [128] | PML protein is essential component. Partitions several chromatin-associated factors. | SUMOylation regulates partitioning by binding to SUMO-interaction motifs found on PML |
| SPOP/DAXX | Protein ubiquitination | [129] | Concentrates cullin3–RING ubiquitin ligases and substrates | Substrate mediated co-condensation |
| Lge1/Bre1 | Gene body histone ubiquitination | [77] | Concentrates ubiquitination enzymes and nucleosomal substrate | |
| Barr body | Mammalian dosage compensation | [71,72] | lncRNA | Local synthesis of lncRNA initiates X inactivation |
Abbreviations: CBX2, chromobox protein homolog 2; CTCF, CCCTC-binding factor; NPAT, nuclear protein, coactivator of histone transcription; NPM1, nucleophosmin 1; OCT4, octamer-binding transcription factor 4; PARP, poly (ADP-ribose) polymerase; PML, promyelocytic leukemia; SPOP, speckle-type POZ protein; SUMO, small ubiquitin-like modifier; YAP, Yes-associated protein
Figure 1. Biomolecular Condensates in the Nucleus.
(A) Structured illumination microscopy images of immunofluorescence for the protein indicated in parentheses in murine embryonic stem cells. Immunofluorescence for indicated protein is colored green, and signal from Hoechst, a DNA stain, is colored dark blue (unpublished results AD and RAY). Condensates are denoted by their name (e.g., superenhancers), their function (e.g., gene activity), and the protein that provides the immunofluorescent signal (e.g., MED1). (B) Cartoon depiction of how various nuclear condensates organize and are organized by different chromatin substrates. The grey line represents the chromatin fiber, green arrow designates active transcription start site, and red squiggled lines represent RNA. For a more complete list of nuclear condensates see Table 1.
Abbreviations: CBX2, chromobox protein homolog 2; CTCF, CCCTC-binding factor; HP1α, heterochromatin protein 1α.
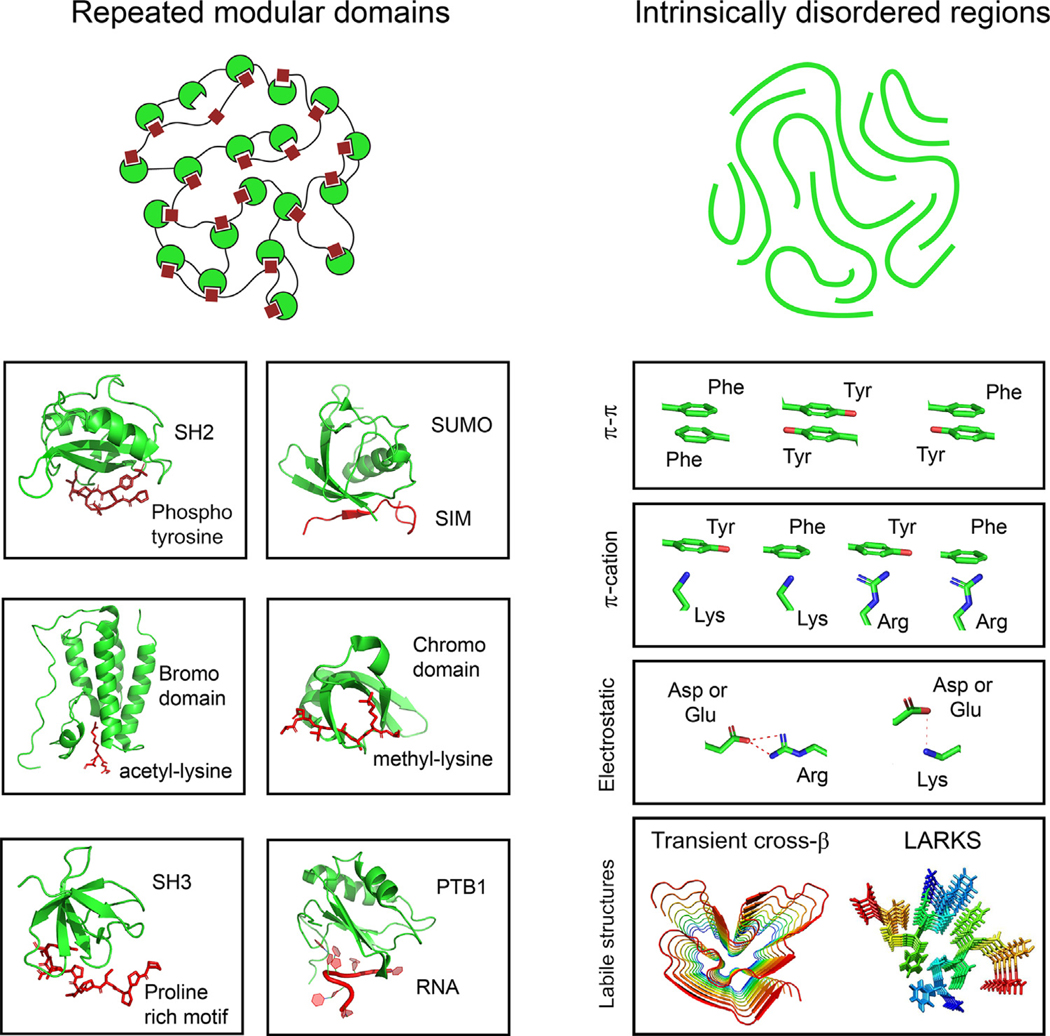
Biomolecular condensates in the nucleus and cytoplasm of cells can compartmentalize and concentrate functionally related components. They are thought to accomplish this through weak, multivalent, and dynamic interactions among proteins and other biopolymers in the absence of a bounding membrane (Figure 2) [4,5]. These weak multivalent interactions can involve intrinsically disordered regions (IDRs) [31], structured modular domains [32], oligomerization domains [33], and other features enabling one protein to engage with multiple proteins simultaneously [8]. Weak cooperative interactions are thought to facilitate evolutionary change leading to their positive selection in diverse cellular processes [34]. Most nuclear condensates interact with specific DNA loci, and this imposes some degree of organization for the biomolecular condensates within the nucleus (Figure 1B).
Figure 2. Types of Multivalent Interactions Thought to Contribute to Formation of Biomolecular Condensates.
These types and associated references are SH2-Yph [105] (PDB: 1SPS), SIM-SUMO [83] (PDB: 2ASQ), bromodomain-acetyl-lysine [16] (PDB: 3JVK), chromodomain-methyllysine [15,88] (PDB: 3FDT), SH3-PRM [83,106] (PDB: 5QU2), PTB1-RNA [83,106] (PDB: 2AD9), pi-pi interactions [78,81,82,107], pi-cation interactions [78,108], electrostatic nteractions [79,80,109,110], and labile structures [111–113] (PDB: 5W3N and 6BZM). Abbreviations:
We first consider the shared functional benefits of three features common to condensates: compartmentalization, selective partitioning, and concentration. We then discuss the different ways that condensates are formed at specific genomic loci, with a focus on bifunctional proteins with both stable structured domains and condensate-promoting domains that contribute to loci-specific condensates. The ability of biomolecules and drugs to partition selectively into specific condensates has been described, and we explore what is known about the determinants of specificity. Condensates typically contain RNA molecules and we note the nuclear RNA components in these bodies. Finally, we discuss the various ways that nuclear condensates are regulated and the various disease contexts where nuclear condensates are dysregulated.
Shared Features of Nuclear Condensates
Compartmentalization
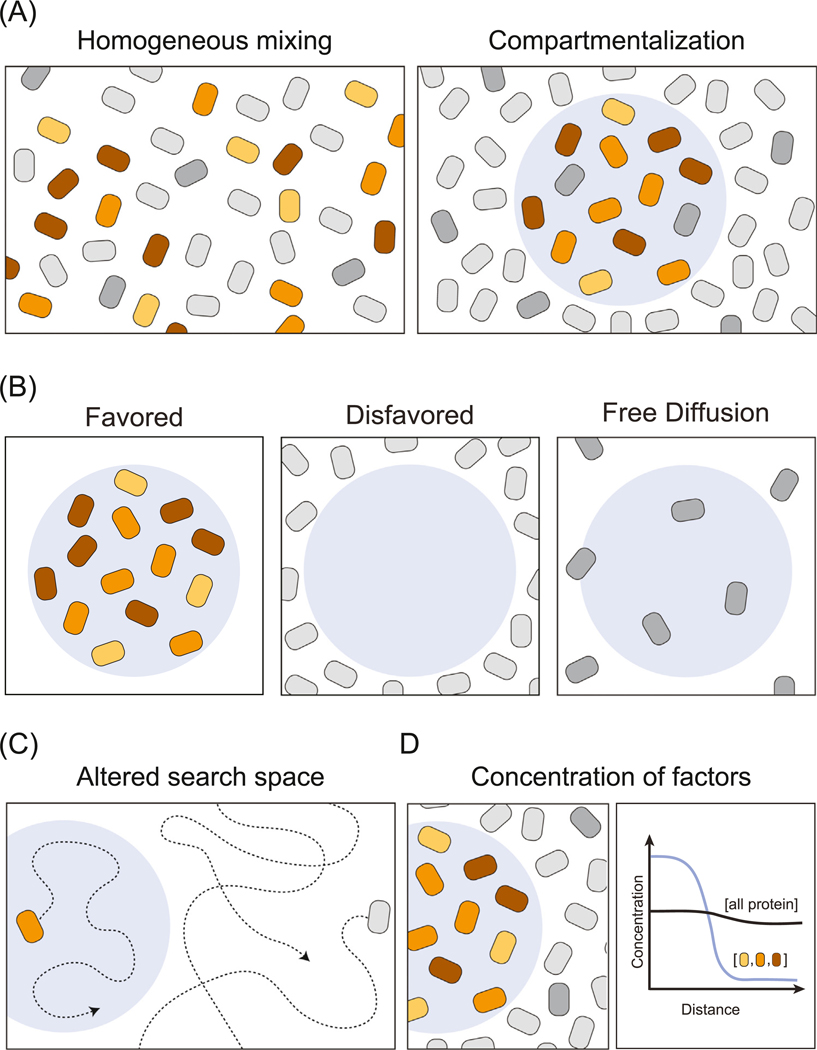
Nuclear regulatory processes typically engage large numbers of protein molecules. For example, the process of transcription initiation at a protein-coding gene involves over a hundred protein molecules, including transcription factors (TFs), cofactors, and the transcription apparatus itself [35,36]. Compartmentalization of proteins involved in specific nuclear regulatory processes provides a means to separate them from the thousands involved in other activities (Figure 3A). Compartmentalization has additional benefits when considering how 5–10 billion protein molecules in a cell find others that must cooperate in a functional process. For example, the effective search space for proteins to find their binding partners or substrates is smaller when they partition selectively into specific condensates, and are excluded from others (Figure 3B,C).
Figure 3. Features Common to Condensates: Compartmentalization, Selective Partitioning, and Concentration.
(A) Condensates compartmentalize functionally related factors. A cartoon depiction of three functionally related factors (colored) depicted homogenously mixed (left) or compartmentalized within a condensate (right). (B) Functionally related factors can be compartmentalized by selective partitioning, where the condensate physicochemical environment may favor or disfavor interactions with such factors. (C) The search space for a molecule can be reduced in two ways. First, for factors which partition into the condensate the search space is reduced to the condensate. The factor can diffuse in and out of the condensate, but will spend more time within. Second, for factors disfavored to partition into condensates that collectively take up a large volume of the nucleoplasm the search space is reduced to the remaining volume of the nucleus. (D) The concentration of the compartmentalized factors is higher inside the condensate than outside, but the absolute concentration of total cellular protein may not be higher inside the condensate than outside.
Condensates are not homogeneous entities and can consist of substructures that permit spatiotemporal regulation [37,38]. This feature, which can create an ‘assembly line’, is best characterized in the most prominent nuclear condensate, the nucleolus [28,39,40]. rRNA transcription, processing, and assembly into preribosomes all occur within three distinct domains of the nucleolus, organized to ensure proper division and order of labor [30,41]. RNA polymerase (Pol) I and its associated factors are compartmentalized in one domain of the nucleolar condensate (fibrillar center) with tandem arrays of 45S rRNA genes. Transcribed 45S rRNA is cleaved and processed into rRNA subunits in a second layer (dense fibrillar center) and then assembled into preribosomes in a third outer layer (granular component). Disruption of this spatiotemporal condensate organization leads to cell death [42,43], demonstrating the functional importance of compartmentalizing this nuclear activity.
Compartmentalization also enables the nonstoichiometric accumulation of functionally related components and can thereby enhance the efficiency of a process. For example, the presence of hundreds of molecules of RNA Pol II within a superenhancer condensate [23] allows multiple molecules of the enzyme to load back to back on promoter DNA and may facilitate the transcriptional bursting phenomenon – the production of multiple transcripts within a short timeframe – that is observed at many genes [44–47]. In the absence of a condensate mechanism, the process of accumulating multiple molecules of the transcription apparatus by one-to-one binding of the diverse proteins necessary for transcription might render the process far less efficient.
The compartmentalization of functionally related proteins in biomolecular condensates provides the cell with an additional useful feature: it allows the formation of a reservoir for components that can be efficiently ‘borrowed’ to form a separate condensate when needed temporarily at a separate site. As an example, the diverse components of the mRNA splicing apparatus occur in nuclear condensates called speckles (Figure 1) [48]. Separately, at sites of active transcription, there is evidence that sufficient amounts of splice apparatus can be recruited to form cotranscriptional splicing condensates on the nascent RNAs [23]. The cotranscriptional splicing condensates appear to borrow material from the speckles because inhibition of transcription causes the speckles to grow larger and rounder, presumably due to the collection of additional material that is no longer engaged in the splicing condensates that occur at active genes [25,48].
Selective Partitioning
The ability of condensates to selectively partition biomolecules is essential for functional compartmentalization (Figure 3B). The nucleolus again provides a striking example of this feature, where products of the first step in the process become disfavored components of an inner condensate and favored for an outer condensate, creating a vector of transport [39,40]. In a similar way, RNA Pol II is dynamically partitioned between enhancer–promoter condensates and elongation-splicing condensates, in a process regulated by phosphorylation of its disordered C-terminal domain (CTD) [25]. RNA Pol II is selectively partitioned into mediator condensates and TATA-box binding protein associated factor 15 hydrogels by its disordered CTD, but excluded upon CTD phosphorylation [25,49], whereupon it becomes a client for splicing condensates [25]. This is one illustration of how post-translational modifications, long understood to alter the preference for specific binding partners, can alter preference for the community of molecules that constitute a specific condensate or a condensate substructure.
Concentration
Selective partitioning of specific molecules into a compartment produces a higher concentration of the biomolecules involved in each process. The density of proteins within the condensate is not expected to be higher than the density of proteins outside the condensate. Rather the concentration of the functionally related proteins that selectively partition into the condensate can be higher within versus outside the condensate (Figure 3D). For example, the concentration of G3BP is estimated to be ~10-fold higher within stress granules than outside [50]. In droplet assays with one or a few components, the relative concentrations within and outside the condensate can be higher; for example, the concentration of nucleosomes in a droplet can be ~10 000-fold higher than outside the droplet [16].
Components That Promote Condensate Formation at Specific Genomic Loci
TFs, Enhancers, and Promoters
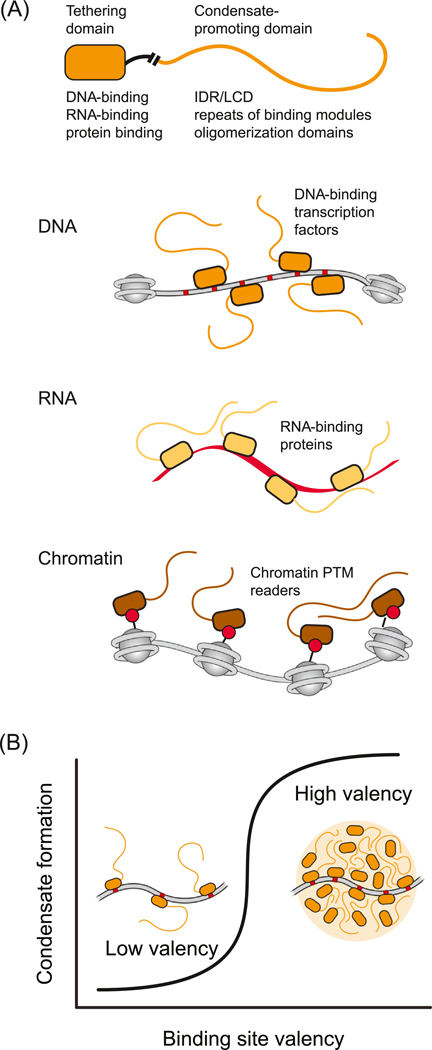
TFs bind to enhancers and promoters and stimulate transcription from specific genes. TFs typically consist of a stable structured domain involved in selective DNA binding, and an activation domain consisting of an IDR [51] that weakly interacts with IDRs in cofactors to form dynamic assemblies [21]. This type of bifunctional protein – one that has a structured domain capable of relatively high-affinity binding to a specific DNA, RNA, or protein sequence, coupled to an IDR or other condensate-promoting domain, is typical of proteins that tether nuclear condensates to specific regions of the genome (Figure 4A).
Figure 4. Bifunctional Proteins Promote Condensate Formation at Specific Genomic Loci.
A) representation of a protein that contains one domain that binds specifically and with high affinity to a DNA, RNA, or protein partner (tethering domain) and another domain that engages in multivalent interactions (condensate-promoting domain). These bifunctional proteins can promote condensate formation when crowded by binding a sufficient number and density of sites in DNA (e.g., regulatory element), RNA (e.g., nascent RNA or long noncoding RNA), or protein (e.g., modified nucleosomal histones). Condensate-promoting domains are depicted here as IDRs, but can be any domain capable of weak multivalent interactions (Box 1). (B) With increasing numbers of binding sites on a polymer substrate, the bifunctional protein will become more locally crowded and can cros a threshold where the multivalent interaction domains promote condensate formation. Abbreviations: IDR, intrinsically disordered region; LCD,; PTM, post-translational modifications.
Enhancers and promoters are DNA elements that contain large numbers of binding sites for TFs, and recent studies suggest that these elements have evolved to crowd TFs to a threshold for condensate formation with coactivators [24]. These studies indicate that such elements create the threshold density and number of TF binding sites necessary to locally concentrate coactivators, which tend to be highly disordered, thus forming transcriptional condensates (Figure 4B). They also suggest that activation of transcription depends on reaching this threshold [24]. This type of localized phase separation is similar to the diffusive-capture model used to describe condensates formed around engineered multivalent seeds [52,53] and the localized-induction model for regulation of condensate formation and size [54].
RNA and RNA-Binding Proteins
Transcription produces an RNA species that is tethered to the elongating RNA Pol, during which various processing steps are accomplished. This nascent RNA, and the phosphorylated CTD of RNA Pol II, can be bound directly by structured domains of RNA-processing enzymes, which can also self-associate via IDRs rich in arginine and serine (RS domains) [55]. This is another example of a bifunctional protein that has a structured domain capable of relatively high-affinity binding to a specific molecule, coupled to a condensate-promoting domain that participates in weak multivalent interactions of the type that occur in condensates (Figure 4). It is also an example of a nucleic acid polymer providing multiple binding sites that locally concentrate proteins with condensate-promoting domains, thus providing threshold concentrations of molecules that form networks of weak multivalent and dynamic interactions.
RNA molecules are among the components of nearly all well-studied biomolecular condensates. Some of these RNA species may mediate nucleated condensate formation by crowding RNA binding proteins and components of the RNA processing machinery that are rich in condensate-promoting domains [56–58]. RNA species also promote condensate formation through electrostatic interactions with proteins [59,60] and by forming secondary structures that facilitate RNA–RNA [61] or RNA–protein interactions [62].
Nascent RNA associated with the active transcription apparatus is thought to nucleate the formation of various nuclear condensates, including the nucleolus, histone locus bodies, and Cajal bodies [58,63]. This view is supported by the observation that artificially tethering specific RNAs to the genome leads to the formation of specific condensates at a locus [64]. Based on this knowledge, it is likely that the RNA species transcribed from active enhancers [65], sites of DNA damage [66], and repetitive regions of the genome [67,68] also contribute to the formation of nuclear condensates.
Thousands of noncoding RNAs are transcribed in cells, the vast majority of which do not have defined functions, but some are essential components of nuclear condensates. For example, nuclear paraspeckle assembly transcript (NEAT)1 is a highly expressed nuclear long noncoding RNA (lncRNA) and an essential component of paraspeckles [69]. NEAT1 promotes condensate formation by recruitment and local crowding of proteins capable of self-interaction to repeats of binding sites [38,70]. Xist is a lncRNA required for X chromosome inactivation in mammalian dosage compensation, and Xist-driven X inactivation yields a compacted X chromosome called a Barr body, which has been suggested to form by locally crowding RNA-binding proteins with condensate-promoting domains, similar to the mechanism described for NEAT1 [71,72]. The lncRNA DIGIT, required for definitive endoderm differentiation, forms condensates with bromodomain-containing protein (BRD)3 at key endoderm-specifying genes during differentiation [73]. It seems likely that many additional lncRNAs will be found to contribute to condensate formation and regulation, and that the dysregulation of lncRNAs commonly observed in diverse tumor cells [74] may contribute to the oncogenic state of these cells by altering the condensate environment.
Nucleosomal Histones
Histones are another example of bifunctional proteins that form well-characterized structures and engage in condensate formation. The core histone proteins each have a structured domain that together with DNA forms a stoichiometric complex, the nucleosome. Each histone also contains N- and C-terminal IDRs (often referred to as histone tails). These histone tails participate in weak multivalent interactions regulated by an array of post-translational modifications (PTMs), which contribute to different chromatin states associated with different gene activities [75,76]. For example, the chromatin fiber can condense via multivalent nucleosome–nucleosome interactions mediated by the unstructured N-terminal tails of the histone proteins and form a silent transcriptional state [16]. These interactions are altered by acetylation of lysine residues in the N-terminal tail, which reverses chromatin condensation, and this form of chromatin is associated with an active transcriptional state [16]. Such acetylation is carried out by transcriptional coactivators that are recruited to specific regulatory elements by TFs. Nucleosomes within the gene body are modified by a different set of histone marks, including monoubiquitinated on histone H2B. Condensate-forming proteins associated with gene bodies serve to concentrate specific H2B ubiquitin ligases, thereby enhancing H2B ubiquitination on these portions of active genes [77].
Specificity
The canonical model of high-affinity interactions between structured portions of proteins is based on well-documented features of shape, charge, and hydrophobicity, and these features provide a solid foundation for understanding the determinants of specificity between interacting biomolecules. By contrast, our understanding of the features of condensates that produce selective partitioning is less well understood. In the nucleus, where specific DNA sequences and RNA molecules can contribute to locus-specific interactions, the bifunctional proteins discussed above can provide a scaffold for both condensate location and formation. What are the determinants of specificity that cause preferential partitioning of a protein into one condensate (e.g., a transcriptional condensate) versus another (e.g., a heterochromatin condensate)? We have clues from studies of condensate-forming proteins and small-molecule drugs that selectively partition into specific condensates.
Detailed study of several condensate-promoting proteins has revealed that interactions among charged and aromatic residues (Figure 2), the valence of those interactions, and the patterning of those interactions are key molecular determinants of phase separation in vitro and condensate formation in cells [78–81]. For example, the condensate-forming protein FUS has been subjected to extensive mutagenesis to identify amino acid side chains that contribute to condensate formation. The results highlight contributions by collections of side chains contributing to electrostatic, pi–pi and pi–cation interactions [78]. pi–pi interactions have been proposed to be a general feature for condensate formation [82]. Other condensates are dominated by charge–charge interactions, occurring either between negative and positive patches of the same protein [80] or between two differently charged proteins [79]. Associative polymer models can now predict the specific phase separation capacity of proteins based on protein sequence alone by considering type, valence, and pattern of interacting amino acids [8,81]. As much as partitioning is determined by binding sites in the underlying multivalent network [83], these studies provide clues as to how molecules are selectively partitioned.
Additional clues to the determinants of specificity of partitioning have come from the study of anticancer drugs [84]. Some of these compounds preferentially concentrate in transcriptional coactivator condensates and then act on their targets at concentrations far higher (600-fold) than anticipated in conventional assays. In the case of cisplatin, the coactivator amino acids that contribute to this high preferential partitioning suggest that pi–pi and pi–cation interactions are important. Interestingly, coactivator condensate formation does not depend on these amino acids, but rather on a separate set of amino acids that compose a large serine patch.
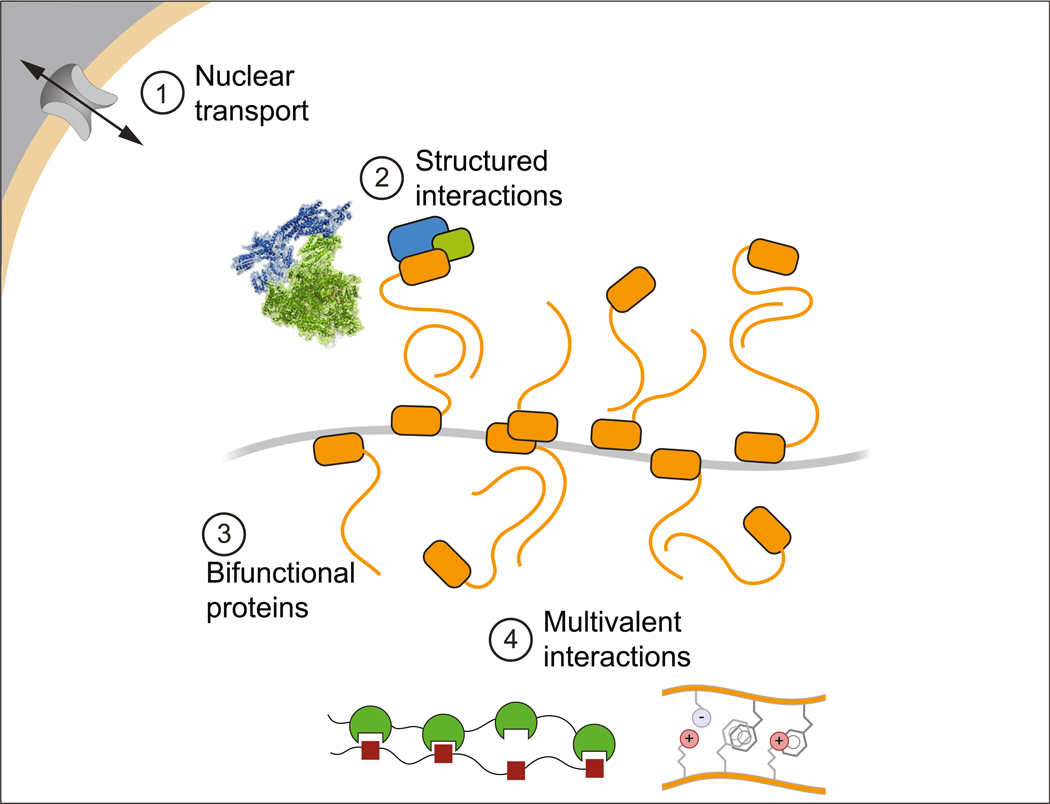
Although much remains to be discovered about the features of condensates that produce selective partitioning, we infer that there are at least four types of contributions: canonical partitioning among membrane-bound organelles, conventional interactions among biomolecules, bifunctional biomolecules that interact with specific genomic loci through sequence specificity, and the milieu formed by amino acid side chains intermingling dynamically with conventional electrostatic, pi–pi and hydrophobic interactions (Figure 5).
Figure 5. Sources of Compositional Specificity in Nuclear Condensates.
This model depicts four types of contributions to compositional specificity: (1) nuclear trafficking; (2) conventional high-affinity structured interactions among proteins; (3) the ability of bifunctional proteins with a condensate-promoting domain to be crowded by binding to multiple sites on a DNA, RNA, or protein substrate; and (4) the various weak multivalent interactions.
Regulation of Nuclear Condensates
The multivalent interaction networks that form nuclear condensates are regulated by reversible covalent modifications of specific regions of chromatin, local synthesis of RNA, and by kinases whose activities can dissolve condensates (Figure 6). Each of these regulatory mechanisms operates by enhancing or reducing multivalent interactions among components.
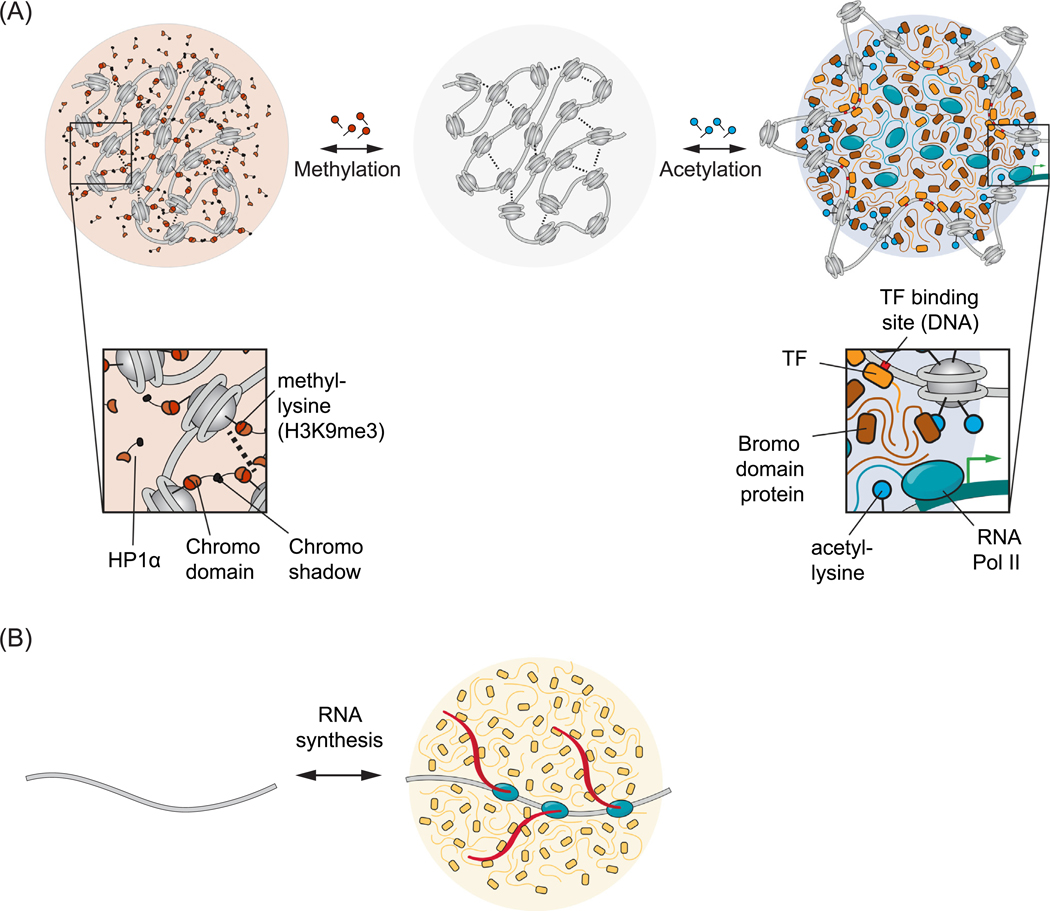
Figure 6. Some Examples of Nuclear Condensate Regulation.
Biomolecular condensates can be regulated by modifying the underlying multivalent interactions. (A) Reversible covalent modifications of chromatin. Nucleosomal histones can be reversibly modified, leading to changes in chromatin state. A model is depicted where chromatin alone can form a condensate (center, gray) mediated by internucleosomal contacts (dotted lines). Methylation of histones at histone H3K9 can recruit HP1α via chromodomain binding and produce a condensate rich in HP1α and other heterochromatin factors (left). In contrast, acetylation of histones at multiple lysine residues can reduce internucleosomal interactions, exposing TF-binding sites on DNA and recruit bromodomain-containing factors, leading to a condensate rich in components of the transcriptional machinery (right). B) Local RNA synthesis. A nascent transcript tethered to the elongating polymerase can be bound by many RNA processing enzymes, leading to a condensate rich in RNA processing machinery. Abbreviations: HP1α, heterochromatin protein 1α; TF, transcription factor.
Reversible Covalent Modifications of Chromatin
Protein, DNA, and RNA are subjected to diverse reversible covalent modifications that can enhance or reduce interactions among macromolecules. Studies of histones provide the richest picture of the types of modifications that occur in proteins. Histones can be modified by acetylation, methylation, phosphorylation, ubiquitylation, GlcNAcylation, citrullination, crotonylation, and more [85]. These modifications are added or removed from specific amino acid residues by specific sets of enzymes, often called writers and erasers. The modification of histones regulates the binding of a class of proteins called readers, which bind to or are ejected by specifically modified residues. These reader proteins that bind with weak affinity to histones in a modification-dependent fashion can, in turn, recruit enzymes that contribute to local gene activity or repression [86,87]. The weak multivalent interactions among readers and histone PTMs are associated with condensate formation and dissolution [15,16,88]. Thus, we now understand that genomic loci with specific chromatin marks, patterns of histone modifications across a locus, might represent different dynamic condensate states [15,16,88] (Figure 6A).
As examples, we now know that large numbers of BRD4 molecules, readers of acetylated nucleosomes, occur in active transcriptional condensates [20]. Similarly, large numbers of heterochromatin protein (HP)1α bind nucleosomes marked by H3K9me3 in heterochromatin condensates [88] (Figure 6A). In addition to these examples of euchromatic and heterochromatic condensates, chromatin and chromatin-associated proteins present at sites of DNA damage are modified by diverse PTMs, including poly-ADP ribosylation, which promotes concentration of components in condensates at sites of DNA damage [18,78]. Poly-ADP ribosylation has also been implicated in promoting compartmentalization of components necessary for transcriptional activity [89,90]. Cells have thus evolved a large diversity of enzymatically regulated covalent modifications to produce dynamic control of multivalent interaction networks that regulate condensate assembly and disassembly.
Reversible covalent modifications of both RNA and DNA have also been implicated in condensate regulation. The m6A modification of RNA creates binding sites for DF proteins, which undergo phase separation at threshold numbers of m6A-induced DF binding sites [91]. Methylation of CpG dinucleotides in DNA creates binding sites for MeCP2 and other methyl-CpG binding proteins, which can promote phase separation of protein-DNA complexes (Li et al., unpublished). MeCP2 is another example of a bifunctional protein (Figure 4) with a DNA-binding domain and a large C-terminal IDR. This IDR is often mutated or truncated in patients with the neurological disorder, Rett syndrome, and is crowded at domains of CpG methylation, thus modulating condensation of chromatin (Li et al., unpublished).
Local RNA Synthesis
Proteins with RNA-binding domains and multivalent-interaction domains become locally concentrated when multiple protein subunits bind a single RNA molecule and the multivalent-interaction regions self-associate [4,5]. Many such cases have been documented for cytoplasmic RNA granules [62,92–94]. In the nucleus, nascent RNAs tethered to RNA Pol provide templates for local condensate formation at specific loci (Figure 6B) [58,63]. The regulatory role of RNA in nuclear condensates is multifaceted and context dependent. While we have focused here on the role of RNAs in condensate formation, RNA has also been proposed to buffer the solubility of RNA-binding proteins in the nucleus [95]. The amount of RNA seems to be important to its regulation of condensate formation. More specifically, the ratio of the negatively charged RNA polymer to a positively charged protein will impact complex coacervation, a specific class of phase separation involving oppositely charged polymers, with equal ratios promoting and unequal ratios disfavoring condensates in a process known as re-entrant phase transition [96,97]. It will be of interest to explore the extent to which these polymer properties have a regulatory role in cellular condensates.
Multicondensate Dissolution by Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK3)
Many nuclear condensates dissolve during mitosis and reform in the daughter cells, suggesting that general regulatory mechanisms exist to rapidly dissolve multiple types of nuclear condensates. The kinase DYRK3 has been proposed to exhibit this general dissolvase activity [98]. Chemical inhibition of DYRK3 leads to the persistence of several nuclear condensates through nuclear envelope breakdown, leading to the aberrant mixing of several cytoplasmic and nuclear condensates. A sharp threshold of DYRK3 to substrate concentrations was identified over which condensates dissolved and below which they persisted, suggesting a tightly regulated switch for controlling multiple condensates simultaneously [98]. Some condensates are unaffected by DYRK3 inhibition, including nucleoli and Cajal bodies, suggesting that other mechanisms exist to dissolve these nuclear condensates during mitosis. The reduction in concentration experienced by nuclear condensate components after nuclear envelope breakdown might contribute to condensate dissolution [99]. The evidence that PTMs can have powerful effects on condensates suggests that additional protein- and RNA-modifying enzymes may contribute to multicondensate regulation.
Dysregulation of Nuclear Condensates
The mechanisms that contribute to formation and composition of condensates at specific loci are dysregulated by mutations in various cancers and neurodegenerative diseases, which reinforces the idea that condensates play important regulatory roles in cell biology and suggest new approaches to disease therapy [10]. The types of mutations that contribute to nuclear condensate dysregulation include DNA rearrangements that produce oncogenic fusion proteins [22,100], small base insertions or deletions that enhance condensate formation [101], DNA repeat expansions that produce aberrant condensate-forming protein [42,43,102] or RNA species [61], and mutations that cause the loss of function of one of the two domains in the bifunctional proteins shown in Figure 4 (Li et al., unpublished). Specific examples of such dysregulation are described later.
The driver of Ewing sarcoma, the oncogenic protein EWS-FLI, is the fusion product of the IDR of EWS (an RNA binding protein of the FET family) and the DNA binding domain of FLI1 (an ETS-family transcription factor). In tumor cells, EWS-FLI forms condensates at new sites in the genome, redistributing transcriptional activity and activating a proliferative gene program [22,100], likely due to compartmentalization of RNA Pol II at these sites [49].
In Wilms tumor, small gain-of-function insertion mutations introduce three amino acids into the transcription elongation factor ENL, enhancing self-association and condensate formation, which coincides with enhanced occupancy and transcription of key proliferative genes [101]. While the enhanced self-association requires the IDR of ENL, the three amino acids that cause this enhancement are introduced into a structured region of the ENL protein [101], which high-lights the fact that amino acid residues that modify the condensate forming behaviors of proteins do not have to occur within IDRs.
Nucleotide repeat expansions are causative factors in amyotrophic lateral sclerosis, muscular dystrophy, and Huntington disease. Aggregation of proteins is thought to be the cause of many neurodegenerative disorders, but aggregation of RNA is now thought to be a culprit in patients with nucleotide repeat expansions. In most repeat expansion disorders, the repeat-containing RNA forms punctate aggregates, called RNA foci, that are thought to be neurotoxic. The multivalent base-pairing interactions in the repeat-containing nucleic acids cause aggregation of these RNA species, producing the neurotoxic RNA foci [61].
Mutations in MeCP2 cause Rett syndrome, a postnatal progressive neurodevelopmental disorder. MeCP2 is a dynamic component of heterochromatin condensates and when altered by Rett syndrome-causing mutations is disrupted in its ability to form condensates. The protein contains a DNA-binding domain and a C-terminal IDR, and both domains contribute to condensate formation and are found mutated in patients (Li et al., unpublished). Thus, MeCP2 condensate disruption may be a common consequence of patient mutations that cause Rett syndrome.
Concluding Remarks
We now understand that most nuclear regulatory processes are compartmentalized in condensates. In this manner, the many different biomolecules that are necessary to carry out a process such as transcription are efficiently localized and concentrated. Bifunctional proteins with both structured and condensate-promoting domains localize condensates to specific genomic loci, and components with shared functions can partition selectively into specific condensates. Diverse RNA species and RNA-binding proteins promote formation of particular condensates and thereby contribute to specificity. Further study is needed to advance our understanding of the mechanisms by which condensates contribute to biological regulatory phenomena and their dysregulation (see Outstanding questions). In some diseases, it is now evident that the mechanisms that contribute to the formation and composition of condensates at specific loci are dysregulated. Insights into condensate properties and condensate dysregulation have suggested new approaches to disease therapy. For example, it may be possible to develop drugs that specifically suppress formation of disease-related aggregates due to mutant proteins or RNAs. Furthermore, evidence that condensates can selectively partition and concentrate small molecule cancer therapeutics and thereby alter their pharmacodynamic properties, could lead to advances in disease therapy for a broad spectrum of diseases [84].
Highlights
Most nuclear regulatory processes are compartmentalized in condensates.
Components with shared functions partition selectively into specific condensates.
Bifunctional proteins with both structured and condensate-promoting domains localize condensates to specific genomic loci.
Diverse RNA species and RNA-binding proteins promote formation of specific condensates.
Further understanding of condensates may provide new therapeutic opportunities for diseases.
Outstanding Questions
What are all the components of each condensate?
What are the features of molecules that contribute to selective partitioning into particular condensates?
How does the physicochemical environment within condensates differ from that outside?
How do nonequilibrium events drive the dynamic formation and dissolution of condensates?
Acknowledgments
We thank Salman Banani, Jon Henninger, and Adam Klosin for comments on the manuscript. This work was supported by NIH grants GM123511, MH104610, CA213333 and CA155258 (R.A.Y.), NSF grant PHY1743900 (R.A.Y.), St. Jude Children’s Research Hospital (R.A.Y.), Novo Nordisk (R.A.Y.), Damon Runyon Cancer Research Foundation Fellowship 2309–17 (B.R.S.), and CPRIT grant RR190090 (B.R.S.). R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics.
Glossary
- Biomolecular condensate
a membraneless cellular compartment where specific biomolecules (e.g., protein, RNA, and DNA) are concentrated. Condensates are composed of higher-order assemblies of biomolecules which engage in dynamic weak multivalent interactions and the biomolecules that are recruited to these assemblies
- Condensate-promoting domain
protein domains that enable dynamic weak multivalent interactions with other proteins leading to condensate formation. Examples of these domains include intrinsically disordered regions, repeated motifs, and oligomerization domains. This is to distinguish these domains from protein domains which engage in high-affinity and low valence interactions often associated with complex formation
- Intrinsically disordered region
a region of a protein computationally predicted or experimentally verified to lack a fixed 3D structure. IDRs typically exhibit high conformational flexibility, allowing them to engage dynamically in weak multivalent interactions. While IDRs have been implicated as condensate-promoting domains, they can also provide additional functions to proteins [103,104]
- pi–pi and pi–cation interactions
noncovalent molecular interactions involving an electron-rich pi system found in aromatic amino acid residues. pi–pi interactions occur between two aromatic residues and pi–cation interactions occur between an aromatic residue and cation present on positively charged amino acid residues. These two molecular interactions among specific amino acid residues (Figure 2), together with a specific number and spacing of these residues within IDRs, have been implicated as molecular determinants of protein phase separation
- Selective partition
Once a condensate is formed, other molecules either prefer to be inside the condensate, disfavor being inside the condensate, or remain unaffected. The degree to which molecules are preferred or disfavored within the condensate environment, the degree to which they partition, defines the community of molecules compartmentalized and concentrated. Condensate selectively partition molecules by a range of chemical, physical, and material properties of the underlying multivalent network
- Weak multivalent interactions
valence in this context refers to the number of interactions a biomolecule can engage in simultaneously. Multivalent interactions are defined as a single factor being able to interact with at least three other factors simultaneously, thereby enabling networks of interactions. Weak refers to the low affinity of the interaction or relatively high dissociation constant relative to interactions often found in stable complexes. Weak interactions with high valence promote the formation of condensates
References
- 1.Wagner R. (1835) Einige bemerkungen und fragen über das keimbläschen (vesicular germinativa). Müllers Archiv. Anat. Physiol. Wissenschaft. Med 373–377 [Google Scholar]
- 2.Montgomery TH (1898) Comparative cytological studies, with especial regard to the morphology of the nucleolus. J. Morphol 15, 265–582 [Google Scholar]
- 3.Cajal SR (1903) Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. Madrid 2, 129–221 [Google Scholar]
- 4.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 [DOI] [PubMed] [Google Scholar]
- 6.Hyman AA et al. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30, 39–58 [DOI] [PubMed] [Google Scholar]
- 7.Holehouse AS and Pappu RV (2018) Functional implications of intracellular phase transitions. Biochemistry 57, 2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J-M et al. (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys 49, 107–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley EP et al. (2019) Physical chemistry of cellular liquid-phase separation. Chem. Eur. J 25, 5600–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberti S and Dormann D. (2019) Liquid-liquid phase separation in disease. Annu. Rev. Genet 53, 171–194 [DOI] [PubMed] [Google Scholar]
- 11.Dignon GL et al. (2020) Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu. Rev. Phys. Chem 71, 53–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson AG et al. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson AG and Narlikar GJ (2018) The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strom AR et al. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanulli S et al. (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson BA et al. (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker MW et al. (2019) A new class of disordered elements controls DNA replication through initiator self-assembly. Elife 8, e48562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altmeyer M et al. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun 6, 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilic S et al. (2019) Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabari BR et al. (2018) Coactivator condensation at superenhancers links phase separation and gene control. Science 361, eaar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boija A et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong S et al. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho W-K et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrinivas K et al. (2019) Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo YE et al. (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y and Belmont AS (2019) Genome organization around nuclear speckles. Curr. Opin. Genet. Dev 55, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brangwynne CP et al. (2011) Active liquid-like behavior of nucleoli determines their size and shape in xenopus laevis oocytes. Proc. Natl. Acad. Sci 108, 4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feric M et al. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165,1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitrea DM et al. (2016) Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, e13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pederson T. (2011) The nucleolus. Cold Spring Harb. Perspect. Biol 3, a000638-a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato M et al. (2012) Cell-free Formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders DW et al. (2020) Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao A et al. (2018) Evolution of weak cooperative interactions for biological specificity. Proc. Natl. Acad. Sci 115, E11053–E11060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeder RG (2019) 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol 26, 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cramer P. (2019) Organization and regulation of gene transcription. Nature 573, 45–54 [DOI] [PubMed] [Google Scholar]
- 37.Boisvert F-M et al. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 38.Fox AH et al. (2018) Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci 43,124–135 [DOI] [PubMed] [Google Scholar]
- 39.Mitrea DM et al. (2018) Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun 9, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riback JA et al. (2020) Composition-dependent thermodynamics of intracellular phase separation. Nature 357, eaaf4382–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strom AR and Brangwynne CP (2019) The liquid nucleome - phase transitions in the nucleus at a glance. J. Cell Sci 132, jcs235093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White MR et al. (2019) C9orf72 Poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol. Cell 74, 713–728.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon I et al. (2014) Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345,1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tantale K et al. (2016) A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun 7, 12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukaya T et al. (2016) Enhancer control of transcriptional bursting. Cell 166, 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsson AJM et al. (2019) Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho W-K et al. (2016) RNA Polymerase II cluster dynamics predict mRNA output in living cells. Elife 5, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spector DL and Lamond AI (2011) Nuclear speckles. Cold Spring Harb. Perspect. Biol 3, a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon I et al. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillén-Boixet J et al. (2020) RNA-induced conformationa switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sigler PB (1988) Acid blobs and negative noodles. Nature 333, 210–212 [DOI] [PubMed] [Google Scholar]
- 52.Bracha D et al. (2018) Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin Y et al. (2018) Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Söding J et al. (2019) Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol. 30, 4–14 [DOI] [PubMed] [Google Scholar]
- 55.Greig JA et al. (2020) Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol. Cell 77, 1237–1250.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boeynaems S et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burke KA et al. (2015) Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawyer IA et al. (2017) Specific genomic cues regulate Cajal body assembly. RNA Biol. 14, 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aumiller WM Jr. et al. (2016) RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32,10042–10053 [DOI] [PubMed] [Google Scholar]
- 60.Drobot B et al. (2018) Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun 9, 15947–15949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain A and Vale RD (2017) RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langdon EM et al. (2018) MRNA structure determines specificity of a polyQ-driven phase separation. Science 157, eaar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berry J et al. (2015) RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci 112, E5237–E5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shevtsov SP and Dundr M. (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol 13, 167–173 [DOI] [PubMed] [Google Scholar]
- 65.Arner E et al. (2015) Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pessina F et al. (2019) Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol 21, 1286–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frank L and Rippe K. (2020) Repetitive RNAs as regulators of chromatin-associated subcompartment formation by phase separation. J. Mol. Biol Published online April 19, 2020. 10.1016/j.jmb.2020.04.015 [DOI] [PubMed] [Google Scholar]
- 68.Novo CL et al. (2020) Satellite repeat transcripts modulate heterochromatin condensates and safeguard chromosome stability in mouse embryonic stem cells. bioRxv. Published online June 8, 2020. 10.1101/2020.06.08.139642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clemson CM et al. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamazaki T et al. (2018) Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 [DOI] [PubMed] [Google Scholar]
- 71.Pandya-Jones A et al. (2020) An Xist-dependent protein assembly mediates Xist localization and gene silencing. bioRxiv 2 2020.03.09.979369 [Google Scholar]
- 72.Cerase A et al. (2019) Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol 26, 331–334 [DOI] [PubMed] [Google Scholar]
- 73.Daneshvar K et al. (2020) lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmitt AM and Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jenuwein T and Allis CD (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 76.Allis CD and Jenuwein T. (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet 17, 487–500 [DOI] [PubMed] [Google Scholar]
- 77.Gallego LD et al. (2020) Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579, 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J et al. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pak CW et al. (2016) Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nott TJ et al. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin EW et al. (2020) Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vernon RM et al. (2018) Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Banani SF et al. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein IA et al. (2020) Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang H et al. (2014) SnapShot: histone modifications. Cell 159, 458–458.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Musselman CA et al. (2012) Perceiving the epigenetic land-scape through histone readers. Nat. Struct. Mol. Biol 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruthenburg AJ et al. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L et al. (2019) Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76, 646–659.e6 [DOI] [PubMed] [Google Scholar]
- 89.Benabdallah NS et al. (2019) Decreased enhancer-promoter proximity accompanying enhancer activation. Mol. Cell 76, 473–484.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zobeck KL et al. (2010) Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol. Cell 40, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ries RJ et al. (2019) m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H et al. (2015) RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith J et al. (2016) Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bounedjah O et al. (2014) Free mRNA in excess upon polysome dissociation is a scaffold for protein multi-merization to form stress granules. Nucleic Acids Res. 42, 8678–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maharana S et al. (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Banerjee PR et al. (2017) Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. Engl 18, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milin AN and Deniz AA (2018) Reentrant phase transitions and non-equilibrium dynamics in membraneless organelles. Biochemistry 57, 2470–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rai AK et al. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 [DOI] [PubMed] [Google Scholar]
- 99.Weber SC and Brangwynne CP (2015) Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol 25, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boulay G et al. (2017) Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wan L et al. (2020) Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Basu S et al. (2020) Unblending of transcriptional condensates in human repeat expansion disease. Cell 181, 1062–1079.e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wright PE and Dyson HJ (2014) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol 16, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van der Lee R et al. (2014) Classification of intrinsically disordered regions and proteins. Chem. Rev 114, 6589–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su X et al. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li P et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frey S et al. (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 [DOI] [PubMed] [Google Scholar]
- 108.Qamar S et al. (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elbaum-Garfinkle S et al. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U. S. A. 112, 7189–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wei M-T et al. (2017) Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem 9, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murray DT et al. (2017) Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hughes MP et al. (2018) Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kato M and McKnight SL (2018) A solid-state conceptualization of information transfer from gene to message to protein. Annu. Rev. Biochem 87, 351–390 [DOI] [PubMed] [Google Scholar]
- 114.Kaiser TE et al. (2008) De novo formation of a subnuclear body. Science 322, 1713–1717 [DOI] [PubMed] [Google Scholar]
- 115.Wang Q et al. (2016) Cajal bodies are linked to genome conformation. Nat. Commun 7,10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shevtsov SP and Dundr M. (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol 13, 167–173 [DOI] [PubMed] [Google Scholar]
- 117.Gall JG (2000) Cajal Bodies: The First 100 Years. Annu. Rev. Cell Dev. Biol 16, 273–300 [DOI] [PubMed] [Google Scholar]
- 118.Hur W et al. (2019) CDK-regulated phase separation seeded by histone genes ensures precise growth and function of Histone Locus Bodies. bioRxiv 34, 789933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koreski KP et al. (2020) Drosophila Histone Locus Body assembly and function involves multiple interactions. bioRxiv. 47 2020.03.16.994483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Duronio RJ and Marzluff WF (2017) Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 14, 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao J et al. (2000) NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14, 2283–2297 [PMC free article] [PubMed] [Google Scholar]
- 122.Nair SJ et al. (2019) Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol 26, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai D et al. (2019) Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol 21, 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim J et al. (2019) Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci 132, jcs226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hansen AS et al. (2020) Guided nuclear exploration increases CTCF target search efficiency. Nat. Chem. Biol 16, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hansen AS et al. (2019) Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell 76, 395–411.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Plys AJ et al. (2019) Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lallemand-Breitenbach V and de The H. (2010) PML nuclear bodies. Cold Spring Harb. Perspect Biol. 2, a000661–a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bouchard JJ et al. (2018) Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]