Abstract
With the recent spread of severe acute respiratory syndrome coronavirus (SARS-CoV-2)_ infecting >16 million people worldwide as of 28 July 2020, causing >650 000 deaths, there is a desperate need for therapeutic agents and vaccines. Building on knowledge of previous outbreaks of SARS-CoV-1 and Middle East respiratory syndrome (MERS), the development of therapeutic antibodies and vaccines against coronavirus disease 2019 (COVID-19) is taking place at an unprecedented speed. Current efforts towards the development of neutralizing antibodies against COVID-19 are summarized. We also highlight the importance of a fruitful antibody development pipeline to combat the potential escape plans of SARS-CoV-2, including somatic mutations and antibody-dependent enhancement (ADE).
Keywords: SARS-CoV-2, COVID-19, neutralizing antibodies, antibody-dependent enhancement, viral escape, drug resistance
Structural and Functional Overview of SARS-CoV-2
Since the first reported hospitalization attributed to COVID-19 on 12 December 2019 in Wuhan, China, >16 million patients have been afflicted worldwide, causing >650 000 deaths as of 28 July 2020 [1., 2., 3., 4.]. The official declaration of COVID-19 as a pandemic by the WHO, the worldwide societal lockdown in an attempt to thwart the progression of the disease, and the vast number of lives affected all underline the dire need for a treatment or vaccine. SARS-CoV-2 (the virus implicated in COVID-19), as well as SARS-CoV-1 and MERS-CoV (see Glossary) coronavirus (MERS-CoV), belong to the Betacoronavirus genus in the Coronaviridae family [5,6]. These viruses have a positive-sense RNA genome that in SARS-CoV-2 encodes four structural proteins and 16 nonstructural proteins (NSPs). The four structural proteins encoded are spike (S), envelope (E), membrane (M), and nucleocapsid (N) [7]. The structural proteins are largely responsible for receptor recognition on the host cell, membrane fusion, and subsequent viral entry. The NSPs are essential for replicative functions such as RNA polymerization by the RNA-dependent RNA polymerase (RdRp, NSP12) [8]. The structural S protein forms homotrimers on the viral membrane in which each monomer is composed of two subunits – the N-terminal S1, that is largely responsible for receptor recognition, and the C-terminal S2, that is implicated in membrane fusion and viral entry (Figure 1 ). The S1 subunit contains the receptor-binding domain (RBD), the region of the protein that makes direct contact with the host cell receptor, angiotensin-converting enzyme 2 (ACE2) [6]. Two conformations of RBD have been observed – 'down/closed' and 'up/open' – wherein the latter conformation reveals the full extent of the RBD that allows ACE2 binding [9,10]. The S2 subunit contains the fusion peptide (FP), the heptad repeat 1 (HR1), and heptad repeat 2 (HR2).
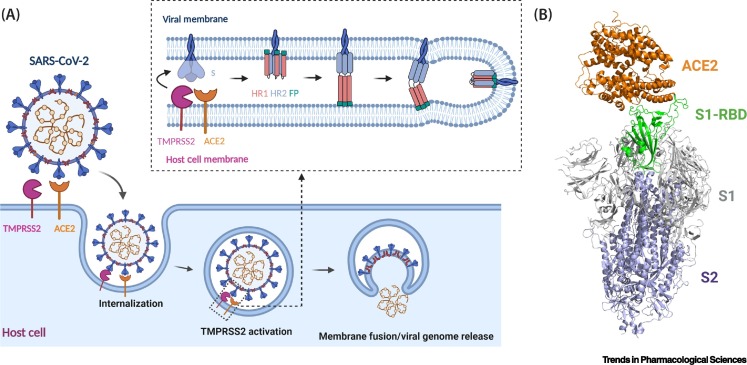
Figure 1.
SARS-CoV-2 Infection Depends on the Host Cell Receptor ACE2.
(A) Cartoon representation of spike protein binding to ACE2 of the host cells. SARS-CoV-2 spike protein S binds to ACE2 through the receptor-binding domain (RBD) and is proteolytically activated by the human protease TMPRSS2, which loosens the structural constraints on the fusion peptide (FP) and initiates a cascade of refolding events (e.g., formation of the three-stranded coiled-coil) and facilitates membrane fusion and release of the viral genome. S protein, FP, HR1, HR2, ACE2, and TMPRSS2 are not drawn to scale. (B) Superimposition of ACE2–RBD structural complex (PDB 6M17) onto the spike protein trimer of SARS-CoV-2 (PDB 6VSB) with the RBD in the 'up' conformation. The spike protein is shown in ribbons with the RBD in green, the S1 domain in grey, and the S2 domain in blue. Abbreviations: HR1, heptad repeat 1; HR2, heptad repeat 2. Figure generated in Biorender (https://biorender.com/).
Upon binding of S protein to ACE2, transmembrane protease serine 2 (TMPRSS2), a host cell serine protease, cleaves the S1 subunit from the S protein, revealing the S2 subunit FP (Figure 1A) [11., 12., 13.]. The FP then initiates membrane fusion by inserting into the host cell membrane, allowing HR1 and HR2 to refold and form a post-fusion conformation that drives membrane fusion of the virus and target cell [14]. Similar to SARS-CoV-1, the SARS-CoV-2 S protein is extensively glycosylated, a quality that may facilitate immune escape of the virus [15]. The interaction between the RBD and ACE2 is the first event in cellular entry of SARS-CoV-2, and is thus an attractive prospect for the development of therapeutics against COVID-19, both for treating infected patients and for preventing infection [16,17]. Targeting the RBD of the SARS-CoV-2 S protein to inhibit its binding to ACE2 can potentially be achieved with limited side effects on surrounding host cells [18].
Current Landscape of Preventative and Therapeutic Strategies for COVID-19
Cambridge-based Moderna Inc. was the first to launch a vaccine clinical trial, initiating Phase I of their mRNA-based vaccine trial only 67 days (mRNA-1273, Clinical Trial Numberi NCT04283461, 16 March 2020) after inception of the program. Promising data from their Phase I trial were recently published in which vaccination induced strong virus-neutralizing activity [19]. The Chinese company CanSino also announced their Phase I study of an adenovirus 5 (Ad5) vector carrying the SARS-CoV-2 S protein gene (Ad5-nCoV, NCT04341389) on 16 March 2020, and has since reported significant humoral and specific T cell responses against SARS-CoV-2 in their Phase I [20] and II studies [21]. A host of other agents have since emerged, ranging from additional viral vector- and nucleic acid-based designs to protein-based S protein subunit vaccines, as well as the traditional inactivated/attenuated viruses [22., 23., 24.]ii.
On the therapeutics side, small molecules are receiving significant attention, and >300 candidates are in development, primarily based around the strategy of drug repurposing. Repurposing of remdesivir [25] (initially trialed against Ebola virus, MERS coronavirus, and other RNA viruses [26,27]) has recently shown some promising data in a Phase III trial [28] in patients with moderate COVID-19. In another clinical trial (NCT04381936), researchers from the University of Oxford have also reported success with another repurposed drug, dexamethasone (primarily used as a general anti-inflammatory medicine) in reducing the mortality rate of COVID-19 patients [28]. Another treatment option that has shown some promise is convalescent plasma transfer [29., 30., 31., 32.]. Despite US FDA approval and widespread use as a therapeutic, a recent report [33] conflicts with earlier studies on the efficacy of convalescent plasma therapy, suggesting that this strategy may not confer significant clinical improvement in COVID-19 patients.
Although the above highlights a multitude of different paths towards COVID-19 treatments, one class of therapeutics that has not yet shown success are neutralizing antibodies and their functional fragments [18,34]. We discuss these neutralizing antibodies in COVID-19 and highlight how the robust antibody pipeline can help to combat the escape strategies presented by SARS-CoV-2.
Crossreactivity between SARS-CoV-1 and SARS-CoV-2 Antibodies
Genomic identity between the S protein coding sequences of SARS-CoV-1 and SARS-CoV-2 is ~76% [5,35]. In addition, structural comparisons of SARS-CoV-1 and SARS-CoV-2 RBD binding to ACE2 illustrate a high degree of similarity in overall conformation [9]. Thus, antibodies against SARS-CoV-1 serve as an initial pool of candidates with potentially neutralizing potency against SARS-CoV-2 [36,37]. In Wrapp et al., the landmark paper highlighting the first solved structure of the prefusion spike of SARS-CoV-2, initial testing of previously established antibodies S230, m396, and 80R against SARS-CoV-1 RBD was conducted, but failed to show any level of binding to SARS-CoV-2 at concentrations as high as 1 μM [16]. In another study, T62, a preparation of polyclonal rabbit anti-SARS-CoV-1 S1 antibodies, showed potent neutralization activity against SARS-CoV-1, but only limited neutralization activity against SARS-CoV-2 pseudovirions [38].
Other antibodies such as VHH-72 and S309 (both originally developed for SARS-CoV-1) have shown significant neutralization activity against both SARS-CoV-1 and SARS-CoV-2 pseudovirions [39,40]. The difference in the cross-neutralizing activities of different antibodies could be explained by whether their epitopes span regions that are conserved between SARS-CoV-1 and SARS-CoV-2. The RBD of SARS-CoV-2 S (residues 387–516) consists of a core domain and a receptor-binding subdomain, also referred to as a receptor-binding motif (RBM, residues 438–505), that loops out of the core domain structure to directly engage the host receptor [36]. Although the protein sequence identity of the RBD core domain between SARS-CoV-1 and SARS-CoV-2 is 86.3%, it is substantially lower for the RBM at 46.7% [41]. In addition, the RBM is looped in nature and is thus subject to the conformational variation of looped sequences, a quality that may further decrease structural homology. Antibodies such as CR3022, VHH-72, and S309 (discussed in detail in the next section), that target regions of S protein that are conserved throughout the Sarbecovirus subgenus (Figure 2 ), show cross-neutralizing potential with SARS-CoV-2, whereas S230 [42], m396 [43], and 80R [44] hold epitopes directly on the lesser conserved SARS-CoV-1 RBM. Likewise, SARS-CoV-2 antibodies B38 and H4 both have epitope residues that span the RBM, and show no potential cross-binding to SARS-CoV-1 RBD [45].
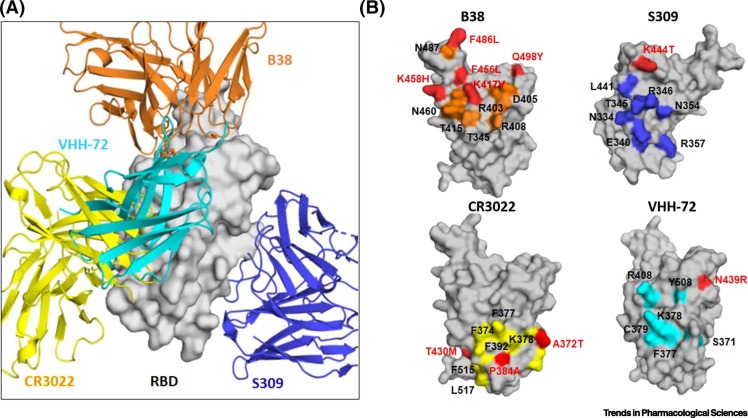
Figure 2.
Crossreactivity of Antibodies S309, CR3022, and VHH-72 against the Receptor-Binding Domain (RBD) of SARS-CoV-1 and SARS-CoV-2 and Their Epitopes.
(A) Superimposition of the RBD with four antibodies. The structures used for superimposition of S309 and CR3022 are with RBD of SARS-CoV-2 (PDB 6WPT [39] and PDB 6W41 [68]), whereas that of VHH-72 is with RBD of SARS-CoV-1 (PDB 6WAQ [70]). B38 is a SARS-CoV-2 specific antibody (PDB 7BZ5 [45]). RBD is shown in surface representation and antibodies are shown in ribbons. The molecular operating environment (MOE) program (Chemical Computing Group ULC, Montreal, QC, Canada; www.chemcomp.com/Products.htm) was used for structural superimposition. (B) Mapping of the epitopes of the four superimposed antibodies on the SARS-CoV-2 RBD surface. Epitope residues conserved between SARS-CoV-1 and -2 RBDs are shown in color corresponding to the respective antibody, and residue differences are shown in red. Note that the residue differences are depicted as residue numbers in the SARS-CoV-2 RBD. For example, K in position 444 in SARS-CoV-2 RBD is T in SARS-CoV-1 RBD, and is shown as K444T in the binding epitope of S309.
Further analysis of the currently available structures of RBD–antibody complexes reveals several interesting antibody-binding sites surrounding the RBD protein surface in the 'up/open' conformation (Figure 2A). Mapping of the epitope residues for antibodies CR3022, VHH-72, and S309 across SARS-CoV-1 and SARS-CoV-2 indicates that, together with many of the highly conserved residues, non-conserved residues are also involved in antibody binding. For example, the non-conserved SARS-CoV-2 RBD epitope residue K444 (T in SARS-CoV-1 RBD) is involved in binding to S309. Likewise, the non-conserved SARS-CoV-2 RBD epitope residues T430 (M in SARS-CoV-1 RBD), P384 (A in SARS-CoV-1 RBD), and A372 (T in SARS-CoV-1 RBD) are involved in binding to CR3022, and the non-conserved SARS-CoV-2 RBD epitope residue N439 (R in SARS-CoV-1 RBD) is involved in binding to VHH-72 (Figure 2B). Such interactions may explain the weakened binding affinities and crossreactivity with SARS-CoV-2 S protein [16]. It is important to note here that neutralizing antibody induction is a key feature of many effective vaccines and hence is a crucial component to be considered in the design of a successful COVID-19 vaccine. Further understanding of the conserved neutralizing epitopes and associated conformational changes in SARS-CoV-2 S protein is urgently needed [46] because such information will promote effective vaccine design for SARS-CoV-2.
Cross-neutralization has also been reported with respect to convalescent studies. Ou et al. conducted cross-neutralization tests using SARS-CoV-1 and SARS-CoV-2 patient sera, which could neutralize their own pseudovirions but had limited ability to cross-neutralize [40]. By contrast, other convalescent studies showed successful cross-neutralization by serum antibodies. One study utilizing serum from SARS-CoV-1 convalescent patients, as well as rabbit sera raised against SARS-CoV-1 S protein, showed complete inhibition of SARS-CoV-1 S pseudotyped virus and slightly less efficient inhibition of SARS-CoV-2 S pseudotype infection of Vero E6 cells (kidney epithelial cell line from African green monkey) [17]. In another study, murine polyclonal antibodies produced against SARS-CoV-1 S protein reduced transduction by murine leukemia virus expressing SARS-CoV-2 S protein (S-MLV) to ~10%, thus exhibiting potent cross-neutralizing activity [47].
It is interesting to note that, in a recent study performing a thorough epitope T cell analysis, SARS-CoV-2-reactive CD4+ T cells were detected in 40–60% of unexposed individuals, suggesting that immune memory induced by earlier infections with seasonal coronaviruses can crossreact with SARS-CoV-2 [48,49].
Current Pipeline for Antibodies against COVID-19
Both pharmaceutical and academic laboratories are on an expedited schedule to push the development of therapeutic antibodies against COVID-19 by exploring almost every available platform [40,45,50., 51., 52., 53.]. The efforts can be broadly divided into exploring antibodies that target the virus itself (neutralizing antibodies, Table 1 ) or the hyperinflammatory effects associated with disease progression. The hyperinflammatory response in COVID-19 is characterized by excessive production of proinflammatory cytokines such as tumor necrosis factor-α (TNF-α), IL-1β, IL-6, IL-8, and interferon-γ (IFN-γ) that cause a ‘cytokine storm’ in severe cases [1,54., 55., 56.]. Limiting the effects of these abundant proinflammatory cytokines thus offers a viable therapeutic option. Recent reviews have discussed the viability of targeting various aspects of SARS-CoV-2-induced hyperinflammation as a therapeutic avenue [57., 58., 59., 60.], whereas other studies have evaluated established anti-IL-6 tocilizumab in the treatment of COVID-19 to varying degrees of success [61., 62., 63.]. We focus below on neutralizing antibodies that directly target the virus.
Table 1.
Summary of Current Efforts towards Developing Neutralizing Antibodies against SARS-CoV-2 (as of 28 July 2020)a
| Lead | Target | Platform/technology/source | Format | Investigator(s) for COVID-19 | Status (clinical trial number) | Refs |
|---|---|---|---|---|---|---|
| LY-CoV555 | Spike protein | DARPA pandemic prevention platform | Human IgG1 | Abcellera Biologics/Eli Lilly/VRC-NIAID | Phase I/II (NCT04411628 and NCT04427501) | iv |
| JS016 | RBD | Convalescent patients | Human antibody | Institute of Microbiology CAS/Junshi Biosciences/Eli Lilly | Phase I (NCT04441918) | iv |
| REGN-COV2 | Spike protein | VelociMab/convalescent patients | Dual human antibodies | Regeneron | Phase I/II/III (NCT04425629, NCT04426695, and NCT04452318) | [88,89] |
| TY027 | Spike protein | Convergent analytics | NA | Tychan | Phase I (NCT04429529) | iv |
| SCTA01 | SARS-CoV-2 | NA | NA | Sinocelltech | Phase I (NCT04483375) | iv |
| BRII-196/198 | SARS-CoV-2 | Convalescent patients | Human antibody | Tsinghua University/Brii Biosciences | Phase I (NCT04479631 and NCT04479644) | iv |
| CT-P59 | SARS-CoV-2 | NA | Antibody/cocktail | Celltrion | Phase I (NA) | iv |
| COVI-GUARD | SARS-CoV-2 | Convalescent patients/human libraries | Human antibody | Mount Sinai Health System/Sorrento | Phase I expected | iv |
| AZD7442 | SARS-CoV-2 | Patients/humanized mice/display | Dual human antibodies | AstraZeneca/Vanderbilt U | Phase I expected | iv |
| COVI-SHIELD | SARS-CoV-2 | Convalescent patients/human libraries | Three human antibodies | Mount Sinai Health System/Sorrento | Phase I expected | iv |
| NA | SARS-CoV-2 | Individual B cell isolation | Human antibody | AbCellera/Eli Lilly | Phase I expected | iv |
| VIR-7831 and VIR-7832 | SARS-CoV-2 | Convalescent patients | Human antibody | GSK/Vir Biotechnology | Phase I expected | iv |
| NA | SARS-CoV-2 | RTMTM technology platform | Human antibody | Neurimmune/Ethris | Phase I expected | iv |
| NA | SARS-CoV-2 | Fully human antibody library/patients | Human antibody | YUMAB and its CORAT partners | Phase I expected | iv |
| NA | SARS-CoV-2 | Vanderbilt custom antibody libraries | NA | Vanderbilt U/Ology Bioservices | Phase I expected | iv |
| 47D11 | SARS-CoV-2 | Harbour's H2L2 Harbour mice | Human antibody | AbbVie/Harbour Biomed/Utrecht U/Erasmus Med Center | Phase I expected | [41] |
| SAB-185 | SARS-CoV-2 | Convalescent patients | Polyclonal | Sab Biotherapeutics/DOD/BARDA | Phase I expected | iv |
| 4A8 | NTD | Convalescent patients | Human antibody | Academy of Military Medical Sciences | Preclinical | [73] |
| NA | ACE2/spike | AI/high-speed mutagenesis | Single-domain | Bioduro LLC | Preclinical | iv |
| NA | NA | Adaptive's Immune Medicine | NA | Amgen Inc./Adaptive Biotechnologies Inc. | Preclinical | iv |
| CR3022 | RBD | Convalescent patients/phage display | Human IgG1 | Scripps Research Institute | Preclinical | [68] |
| S309 | RBD | Convalescent patients | Human IgG1 | Vir Biotechnology | Preclinical | [70] |
| BD-386-2 | RBD | Individual B cell isolation | Human antibody | Peking U/Sino Biological/WuXi Biologics | Preclinical | [50] |
| CA1 and CB6-LALA | RBD | Convalescent patients | Human antibody | CAS/NCRCIF/SMS-UCAS | Preclinical | [71] |
| P2C-1F11/P2B-2F6/P2A-1A3 | RBD | Convalescent patients | Human antibody | Shenzhen TPH/SUST/Tsinghua U | Preclinical | [72] |
| H11-D4/H11-H4 | RBD | Phage display | Single-domain | U of Oxford | Preclinical | [115] |
| 311mab-31B5311/32D4 | RBD | Convalescent patients | Human antibody | Peking Union Medical College | Preclinical | [64] |
| COVA 2-15 | RBD | Convalescent patients | Human antibody | U of Amsterdam/Cornell U | Preclinical | [53] |
| 414-1 | RBD | Convalescent patients | Human antibody | Fudan U/Active Motif China | Preclinical | [52] |
| H014 | RBD | Hybridoma | Humanized antibody | U of CAS/CAS/Academy of Military Medical Sciences | Preclinical | [37] |
| NA | RBD | Hybridoma | Single-domain | VIB/Ghent U | Preclinical | iv |
| NA | SARS-CoV-2 | Convalescent patients | Human antibody | Tekara/Pennsylvania-based CSL Behring | Preclinical | iv |
| B38 and H4 | SARS-CoV-2 | Convalescent patients | Human antibody | Institute of Microbiology CAS/Junshi Biosciences/Lilly | Preclinical | [45] |
| rCIG | SARS-CoV-2 | Convalescent patients | Polyclonal | Gigagen Inc. | Preclinical | iv |
| XAV-19 | SARS-CoV-2 | Humanized animal | Antibody cocktail | LFB SA/Xenothera SAS | Preclinical | iv |
| NA | SARS-CoV-2 | Omniab(transgenic animal)/AI | PolyTope mAb | Immunoprecise Antibodies | Preclinical | iv |
| NA | SARS-CoV-2 | Convalescent patients | Human antibody | Fairjourney Biologics SA/Iontas | Preclinical | iv |
| NA | SARS-CoV-2 | Convalescent patients | Human antibody | Just-Evotec Biologics/Ology Bioservices | Preclinical | iv |
| NA | SARS-CoV-2 | NA | IgM/IgA | Atreca/Beigene/IGM Biosciences | Preclinical | iv |
| VHH-72 | Spike protein | Llama immunization | Nanobody-Fc | Ghent U/U of Texas at Austin | Preclinical | [39] |
| n3088/3130 | Spike protein | Phage display | Humanized nanobody | Fudan U | Preclinical | [111] |
| 80R | Spike protein | Phage display | Human IgG1 | Dana-Farber Cancer Institute | Preclinical | [41] |
| ADI-55689/56046 | Spike protein | Convalescent patients | Human antibody | Adimab LLC | Preclinical | [113] |
| NA | Spike protein | VNAR phage display | Single-domain | Ossianix | Preclinical | iv |
| NA | Spike protein | Beacon platform | Human antibody | Ablexis/AlivaMab Discovery Services/Berkeley Lights | Preclinical | iv |
aAbbreviations: AI, artificial intelligence; BARDA, Biomedical Advanced Research and Development Authority; CAS, Chinese Academy of Sciences; DARPA, Defense Advanced Research Projects Agency; DOD, Department of Defense; NA, not available; NTD, N-terminal domain (SARS-CoV-2 spike protein); NCRCIF, National Clinical Research Center for Infectious Diseases; RBD, receptor-binding domain (SARS-CoV-2 spike protein); RTMTM, Translational MedicineTM; Shenzhen TPH, Shenzhen Third People’s Hospital; SMS–UCAS, Savaid Medical School, University of the Chinese Academy of Sciences; SUST, Southern University of Science and Technology; U, university; VIB, Flanders Institute for Biotechnology; VNAR, shark variable domain; VRC-NIAID, Vaccine Research Center, National Institute of Allergy and Infectious Diseases.
Notably, eight SARS-CoV-2-neutralizing antibodies (LY-CoV555, JS016, REGN-COV2, TY027, BRII-196, BRII-198, CT-P59, and SCTA01) have recently entered clinical studies (Table 1). We discuss below early efforts to develop neutralizing antibodies directly targeting different parts of SARS-CoV-2 S protein, highlighting methodologies of screening, efficacy, and epitopes.
Early work from Wang et al. [41] utilized SARS-CoV-1 hybridomas for the development of a SARS-CoV-2 cross-neutralizing antibody. Their lead antibody, 47D11, showed potent binding affinity to S protein ectodomain (Secto, the portion of the spike that extends into the extracellular space) in ELISA and biolayer interferometry (BLI) studies as well as neutralizing efficacy against both SARS-CoV-1 and SARS-CoV-2 in a pseudotyped vesicular stomatitis virus (VSV) assays (IC50 = ~0.19 and ~0.57 μg/ml respectively). The antibody does not necessarily inhibit the interaction between S protein and ACE2, and might mediate virus neutralization through alternative mechanisms. Although the epitope is not fully characterized, 47D11 is believed to bind to part of the conserved core of the RBD [41].
Chen et al. [64] developed a set of antibody leads by isolating SARS-CoV-2 RBD-specific memory B cells from recovered COVID-19 patient sera. The sorting processes yielded three unique IgG1 monoclonal antibodies (mAbs), 311mab-31B5, 311mab-32D4, and 311mab-31B9. Further analysis showed two leads, 311mab-31B5 and 311mab-32D4, that had neutralizing activity against SARS-CoV-2 S pseudotyped virus infection by blocking the interaction between SARS-CoV-2 RBD and ACE2 receptor (IC50 = ~0.034 and ~0.070 μg/ml, respectively).
Tian et al. [65] made the first efforts to characterize the cross-binding capabilities of the SARS-CoV-1 antibody, CR3022, that was first isolated in 2005 from a phage display library constructed from the plasma of a SARS-CoV-1 patient [66]. Other SARS-CoV-1 antibodies including CR3014 [67], m396, and m336 [43] were also tested but failed to show strong binding to SARS-CoV-2 RBD, whereas CR3022 showed significantly higher binding (Kd = 6.28 × 10−9). Yuan et al. [68] reported that CR3022 has little neutralizing capabilities (at concentrations as high as 0.4 mg/ml). However, in another recent report by Huo et al., CR3022 was shown to potently neutralize live SARS-CoV-2 virus in a plaque-reduction neutralization test (PRNT), with 50% neutralization at ~0.114 μg/ml (~1 nM) [69]. The discrepancy between the neutralizing activities in different tests can potentially be explained by differing methodologies employed in the in vitro assays, more specifically in whether the antibody–virus complex was removed after adsorption to cells in the neutralization tests in each study. In addition, Huo et al. validated their neutralization assays with multiple rounds of PRNT against three different batches of CR3022 (with and without washing after adsorption). CR3022 has been shown to bind to a conserved epitope distinct from the RBM [68,69] (Figure 2A). Thus, CR3022 can be used in combination with antibodies directly targeting the SARS-CoV-2 RBM to provide synergy in potency as well as to limit the likelihood of escape mutants of the virus [66].
Wrapp et al. [39] immunized a llama against MERS-CoV and SARS-CoV-1 S proteins in successive rounds to develop a lead cross-neutralizing camelid nanobody VHH-72, with efficacy against both SARS-CoV-2 and SARS-CoV-1. Although VHH-72 showed relatively high binding affinity to SARS-CoV-2 RBD, as determined by surface plasmon resonance (SPR), neutralization of SARS-CoV-2 S VSV pseudotypes by VHH-72 had a higher IC50 than for SARS-CoV-1 pseudotypes. The team reasoned this is because of the high dissociation rate constant between VHH-72 and RBD. To circumvent this issue, a bivalent format of VHH-72, VHH-72-Fc (fragment crystallizable region), was constructed [39]. The new format showed improved binding affinity to SARS-CoV-2 RBD in ELISA and succeeded in neutralizing SARS-CoV-2 S VSV pseudoviruses, exhibiting an IC50 of ~0.2 μg/ml. Structural analysis showed that VHH-72 binds to a region of RBD that partially overlaps with the CR3022 binding site (Figure 2A). Unlike CR3022, VHH-72 prevents ACE2 binding to SARS-CoV-2 [39].
An antibody reported by Pinto et al. [70], S309, was developed from memory B cell screening of a SARS-CoV-1 recovered patient. Of eight potential candidates, S309 was shown to potently neutralize both SARS-CoV-2 and SARS-CoV-1 pseudoviruses as well as authentic SARS-CoV-2 (2019n-CoV/USA_WA1/2020, IC50 = ~79 ng/ml) through a mechanism that did not rely on host receptor (ACE2) blocking. The cryo-electron microscopy (cryo-EM) structure revealed that S309 recognizes a highly conserved N343-glycan-containing epitope that is not located on the ACE2–RBD interface, and is also distinct from the CR3022 epitope binding site (Figure 2). S309 was tested in combination with a panel of anti-SARS-CoV-2 antibodies against different epitopes, resulting in enhanced neutralization potency (compared to any mAb alone) against both pseudovirions and authentic SARS-CoV-2 [70]. The observed synergy highlights the potential of using antibody cocktails to treat COVID-19.
Wu et al. [45] also recently identified neutralizing antibodies from COVID-19 patients (IC50 values ranging from 0.177 to 1.375 μg/ml). B38, as well as H4, showed blocking of ACE2 receptor recognition by RBD (Figure 2) of SARS-CoV-2 in a BLI assay. In an epitope competition assay, both B38 and H4 bound to RBD, with partial inhibition against each other, suggesting that they target different epitopes on the RBD that only partially overlap. In vivo studies using human (h)ACE2 transgenic mice confirmed reduction of viral titer in the lungs and improved histopathology scores of mice administered with B38 compared to control and H4-treated groups [45]. Subsequent structural validation through cryo-EM confirms B38 binding to RBD, with an epitope that spans 36 residues on the RBD, of which 15 lie within the RBD/ACE2 interface (Figure 2).
Shi et al. [71] isolated one lead antibody, CB6, from convalescent COVID-19 patients. CB6 was found to recognize an epitope overlapping with the ACE2-binding site on the RBD of SARS-CoV-2. CB6 exhibits strong neutralizing activity against live SARS-CoV-2 infection of Vero E6 cells, with an observed IC50 of ~0.036 μg/ml. Further study in a rhesus macaque SARS-CoV-2 infection model demonstrated that CB6 (50 mg/kg) inhibits viral titer (decreased from ~32 million to <1000 RNA copies per ml) with reduction of lung damage in both prophylactic and therapeutic settings [71]. Using a similar strategy, 206 anti-RBD antibodies were isolated and characterized by Ju et al. [72]. Because these antibodies target different epitopes on RBD, combinations may be explored to enhance antiviral effects.
Antibody 4A8 against SARS-CoV-2 has a unique proposed mechanism of action. The 4A8 antibody exhibited neutralization activity against pseudotyped HIV-vectored SARS-CoV-2 and could also protect Vero E6 cells against live SARS-CoV-2 (IC50 = ~0.61 μg/ml). Characterization of binding through BLI showed no interference of ACE2 binding to S protein of SARS-CoV-2 by 4A8 [73]. Structural characterization confirmed that the epitope of 4A8 is on the N-terminal domain (NTD), and established that epitopes outside the RBD can be viable targets for neutralizing antibodies, hinting at possible mechanisms in addition to traditional receptor blocking. It was proposed that 4A8 blocks conformational changes from the pre- to post-fusion state of SARS-CoV-2 S protein [73], thus inhibiting the initial step of fusion and preventing virus entry.
Challenges in the Successful Development of Neutralizing Antibodies
Although the current pipeline of COVID-19 antibodies has identified a large number of neutralizing antibodies, there are also challenges. We discuss two that affect the development of these antibodies: (i) somatic mutations in SARS-CoV-2 that may dampen the efficacy of antibodies, and further lead to new viral strains that may gradually develop resistance to existing antibodies, and (ii) unwanted enhancement of immune response via a phenomenon known as ADE.
SARS-CoV-2 Mutations
Although retroviruses such as HIV use reverse transcriptase (estimated error rate at 10−3 misincorporations per nucleotide position per genome replication) during their replication [74], SARS-CoV-2 uses a higher-fidelity RdRp (with an estimated error rate of 10−4 to 10−5 misincorporations per nucleotide position per genome replication) [75]. Hence, SARS-CoV-2 may be less prone to incorporate mutations [76]. However, a broad mutation spectrum of SARS-CoV-2 has been observed, which could lead to vaccine and drug resistance [77., 78., 79.]. In one analysis of 7710 assemblies of SARS-CoV-2 genomes available on the Global Initiative on Sharing All Influenza Data (GISAID)iii database, 198 sites of mutations were identified [80]. Furthermore, Starr et al. showed that most of the mutations are deleterious for RBD expression and ACE2 binding [81]. However, mutation V367F was found to substantially increase RBD expression. Another analysis of the GISAID dataset revealed a number of enriched mutations in SARS-CoV-2 S protein, a major target for neutralizing antibody development: G476S in the RBD; V367F and V483A in RBD 'up/down' conformations; L5F and L8V/W in the signal peptide; H49Y, Y145H/del, and Q239K in the S1 NTD domain; D614G and V615I/F in the S1 subunit; A831V in the potential FP in S2 subunit; D839Y/N/E in the S2 subunit; P1263L in the cytoplasmic tail; and a mutational cluster 937–943 in the fusion core of HR1 (Figure 3A) [69].
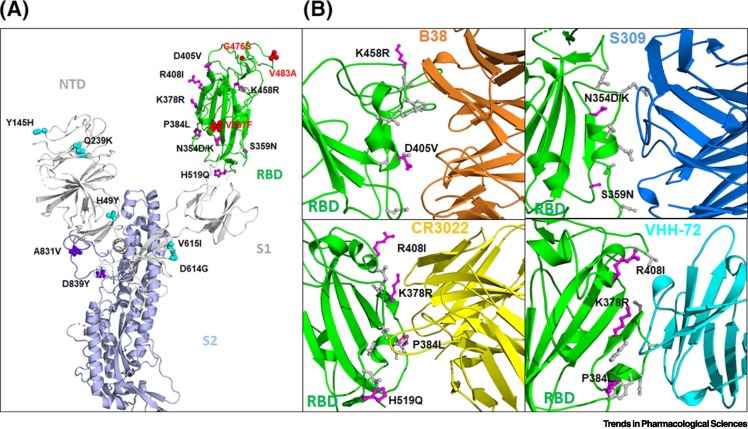
Figure 3.
Mutations on SARS-CoV-2 Spike Protein and Effects on Neutralizing Antibodies.
(A) Selected mutations of SARS-CoV-2 S protein observed in the current pandemic are highlighted. The S protein in the 'up' conformation is shown in ribbons. The enriched mutations are shown as spheres and are colored in cyan (S1 domain), red (RBD), and purple (S2). The non-enriched mutations in the RBD are shown in magenta. (B) Binding epitopes of SARS-CoV-2 RBD for antibodies B38 (PDB 7BZ5), S309 (PDB 6WPT), CR3022 (PDB 6W41), and VHH-72 (based on the superimposed structure of SARS-CoV-1, PDB 6WAQ) are shown where the residues depicted in stick representation (grey and magenta) are proposed to be involved in contacts between the RBD and the antibodies. Magenta sticks represent non-enriched mutations that have been sporadically detected in publicly available databases. Abbreviations: NTD, N-terminal domain; RBD, receptor-binding domain; S1 and S2, subunits.
Further, two other analyses of SARS-CoV-2 have suggested a trend towards enhanced structural stability as well as higher affinity for ACE2, and this may indicate acquired increased infectivity during the pandemic [82., 83., 84.]. Recent structural analysis has also shown that the D614G mutation (in the S1 domain, a mutation from a negatively charged amino acid to a flexible amino acid) induced a more stable S protein that can lead to higher infectivity [85]. This may explain why the prevalence of this particular mutation increased from 0% in February 2020 to ~70% in May 2020 [85,86].
Moreover, it is interesting to note that some of the mutations that have been sporadically detected are within the epitopes for the neutralizing antibodies B38 (K458R, D405V), S309 (N354D/K and S359N), CR3022 (R408I, K378R, P384L, H519Q), and VHH-72 (R408I, K378R, and P384L) (Figure 3B) [39,45,68,70]. Although the frequencies of these mutations are low, and none has been found to be enriched in the current pandemic, these highlight that such mutations can potentially diminish or abolish the effectiveness of the currently available and in-development antibodies [66,87]. Because spontaneously arising mutated SARS-CoV-2 strains can be positively selected, and ultimately become the dominant strain to achieve viral escape under pressure from a neutralizing antibody [88., 89., 90., 91.], strategies have been developed to protect against this. One strategy is to design broadly neutralizing antibodies that target conserved epitopes in the RBD because these are less likely to mutate [92., 93., 94.]. Another method to combat potential escape mutants is to use a combination of several neutralizing antibodies. Antibodies in cocktails would target distinct epitopes, enabling an additive effect in potency. Indeed, such strategies have been employed for earlier outbreaks, as seen in Ebola (REGN-EB3) and SARS-CoV-1 (antibody cocktail of CR3014 and CR3022) [66]. These previous successes spurred Regeneron to start the development of a dual antibody cocktail for COVID-19 (REGN-COV2, a combination of two antibodies, REGN10933 + REGN10987; Table 1) [88,89]. AstraZeneca has also confirmed a pair of mAbs (AZD8895 + AZD1061) that will be taken forward as a combination therapy, known as AZD7442, into clinical development (Table 1). It is anticipated that this cocktail strategy may be crucial in increasing the potency of antibodies for use against SARS-CoV-2 and its mutating strains.
Antibody-Dependent Enhancement
Antibodies represent a double-edged sword in that they can both neutralize and enhance viral infection [95]: highly potent neutralizing antibodies can recognize specific viral epitopes and block viral entry to the host cells, whereas poorly neutralizing antibodies can promote infection through a phenomenon known as ADE. ADE is seen in responses to many infectious diseases including dengue fever, in which the natural progression of the disease has been shown to utilize antibodies from the primary response to infection as a basis for increased severity in later secondary infections [96]. Similarly, poorly neutralizing antibodies or antibodies with low affinity can promote virus entry upon secondary infection, resulting in increased viral burden and more severe disease progression via ADE (Figure 4 ) [97., 98., 99.]. Mechanisms of ADE largely rely on the uptake of virus–antibody immune complexes into host immune cells such as macrophages and monocytes, a process that is mediated through interactions between the Fc domain on the antibodies and Fc receptor (FcR) on host cells. These virus–antibody immune complexes in turn initiate different downstream proinflammatory signaling pathways, leading to increased production of proinflammatory cytokines, decreased production of anti-inflammatory cytokines, and increased viral load (Figure 4) [97., 98., 99.].
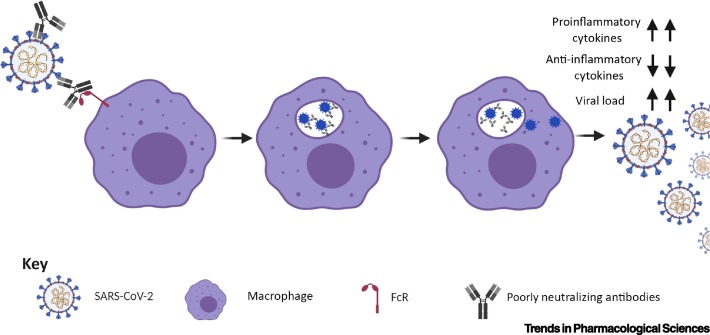
Figure 4.
Antibody-Dependent Enhancement (ADE) Observed with Poorly Neutralizing Antibodies.
ADE is induced by low-affinity, low-quantity, poorly neutralizing antibodies against SARS-CoV-2. In the process, poorly neutralizing antibodies interact with SARS-CoV-2 and also with the Fc receptors (FcRs) of macrophages/monocytes. The antibody–virus complexes are internalized into the cells and eventually lead to increased production of proinflammatory cytokines, reduction of anti-inflammatory cytokines, and increased viral load. Figure generated with Biorender (https://biorender.com/).
ADE has previously been observed for antibodies against SARS-CoV-1 [100., 101., 102.]. Upon rechallenge in SARS-CoV-1-vaccinated rhesus macaques, passive transfer of anti-spike IgG correlated with disease progression, as observed by the presence of severe diffuse alveolar damage and loss of disease tolerance [103,104]. It was observed that the same antisera against SARS-CoV-1 may confer neutralization or enhancement based solely on the concentrations of the antisera employed – where enhancement and neutralization occur at lower and higher concentrations, respectively [102]. Thus, a threshold dependent on both antibody concentration and affinity may need to be considered in aiming to confer neutralization instead of enhancement [103].
Convalescent plasma therapy for infectious diseases is also known to be more effective as a prophylactic if administered shortly after symptoms arise when viral titers are thought to be lower [31,105,106], suggesting that the threshold of viral load is also an important factor to consider. Although to date (as of 28 July 2020) there have been no observations of ADE in antibodies currently in clinical trials for COVID-19 [107], in vitro and in vivo studies, as well as two early clinical studies in China, showed that patients with severe disease frequently had an increased IgG response [108., 109., 110.]. These accumulated data raise the possibility of ADE by antibodies in COVID-19, and close attention should be paid to the matter.
In addition to antibody concentration/affinity and viral load, there are two other ways in which ADE can be avoided. Both involve the Fc, the interacting partner on the virus–antibody complexes that initiates ADE. Nanobodies lacking the Fc domain would potentially avoid the pitfall of ADE [111], at the cost of a shortened in vivo half-life. Hence, in such a case, options to elongate the half-life of nanobody candidates should be taken before moving to the clinic. These may include linking a nanobody to anti-human serum albumin (HSA), a technique that has been proven to increase the serum half-life to ~6 days in cynomolgus monkeys [112]. Another strategy to circumvent ADE is the introduction of a LALA mutation (Leu234Ala together with Leu235Ala) into the Fc to minimize FcR activation and Fc-mediated toxicity of human antibody candidates against SARS-CoV-2. This has been done with JS016 (currently in clinical trial) which employs a LALA mutation in the Fc to minimize FcγR activation and Fc-mediated toxicity, and also with antibody CB6 where CB6-LALA is currently being investigated in clinical trials (Table 1) [71].
Concluding Remarks
We are currently experiencing an explosion of research into antibodies to combat COVID-19 that include neutralizing antibodies against SARS-CoV-2 (Table 1) and therapeutic antibodies against COVID-19-associated hyperinflammation. To date, 8 neutralizing antibodies against SARS-CoV-2 have entered clinical evaluation – LY-CoV555, JS016, REGN-COV2, TY027, BRII-196, BRII-198, CT-P59, and SCTA01. It is understood that SARS-CoV-2 may develop resistance to a single neutralizing antibody by accumulating spontaneous mutations [88,89]. To overcome this problem, antibody cocktails such as REGN-COV2, AZD7442, and COVI-SHIELD have entered clinical trials (Table 1). Further, multiple research programs have also started to work on antibodies against conserved epitopes that would avoid the SARS-CoV-2 mutations that are currently being observed in the pandemic. This will enable us to get ahead of the potential issue of viral resistance [53,81,88,89,113,114]. Efforts will also be necessary to discover conserved functional neutralizing epitopes that, once mutated, can cause SARS-CoV-2 to lose its infectivity. To circumvent the potential problem of ADE, several strategies are being explored, including the use of nanobodies lacking the Fc domain and the introduction of the LALA mutation into the Fc domain.
Given the development of fruitful concurrent pipelines of antibodies against COVID-19 driven by close collaboration between academia and industry, we hold optimism that broadly neutralizing candidates will emerge that induce less or no ADE, and thus play a major role in therapy and protection against COVID-19. Over the next few years we are likely to see an expansion of preclinical research and gain extensive experience in the development of antibodies against COVID-19 in the clinic. An in-depth understanding of the mechanisms of SARS-CoV-2 will better prepare us for the next pandemic (see Outstanding Questions).
Outstanding Questions.
What is the best strategy to develop broadly neutralizing antibodies against conserved epitopes of SARS-CoV-2?
What are the ways to combat antibody-dependent enhancement (ADE) from neutralizing antibodies in COVID-19?
Alt-text: Outstanding Questions
Acknowledgments
We thank Dr Shaolei Teng (Howard University) for sharing the analysis of mutations on S protein of SARS-CoV-2. We thank all colleagues from the National Center for Advancing Translational Sciences (NCATS) COVID-19 team for helpful comments and advice in the preparation of this manuscript. This research was supported in by the NCATS Intramural Research Program of the NIH.
Glossary
- Angiotensin-converting enzyme 2 (ACE2)
the target receptor of SARS-CoV-1/2 which is mainly expressed in human vascular endothelial cells and the renal tubular epithelium.
- Biolayer interferometry (BLI)
a commonly used biophysical method to characterize binding between two proteins by measuring interference wavelength changes caused by protein binding.
- Convalescent plasma transfer
a therapeutic option involving transfusion of blood plasma from recovered patients to symptomatic patients
- Ebola virus
an enveloped, negative-stranded RNA virus that causes severe bleeding, organ failure, and can lead to death.
- ELISA
a fast plate-based assay technique that can be used for detecting and quantifying virus.
- Epitope
the specific segment of an antigen to which an antibody binds.
- Fragment crystallizable region (Fc)
the tail region of an antibody that interacts with FcRs.
- Fc receptor (FcR)
Fc receptors recognize and bind to the Fc region of specific immunoglobulin classes.
- Hybridoma
cells formed by fusion between a short-lived antibody-producing B cell and an immortal myeloma cell to generate monoclonal antibody-expressing stable cell lines.
- LALA mutation
two mutations, Leu234Ala and Leu235Ala, within the antibody Fc region that reduce FcγR binding affinity.
- Middle East respiratory syndrome (MERS)
a contagious, sometimes fatal respiratory illness caused by MERS coronavirus.
- Nanobody
an antibody fragment consisting of a single monomeric variable antibody domain. Nanobodies, with a size of ~15 kDa, are smaller in size than common antibodies.
- Phage display
a screening technology in which a peptide or protein such as an antibody is displayed on the surface of a bacteriophage (primarily on pIII).
- Plaque-reduction neutralization test (PRNT)
a method to determine the titer of neutralizing antibodies for a virus by counting plaque units formed as a result of virus-infected cells.
- Pseudotyped vesicular stomatitis virus (VSV)
prototypic enveloped animal virus that is a proven useful study tool for identifying cellular receptors for viruses, as well as for screening for entry inhibitors including antibodies.
- Pseudovirions
these are closely related to viruses in structure and behavior but, unlike the parent viruses, are not contagious, and thus provide an excellent research tool.
- Surface plasmon resonance(SPR)
a method that makes use of the non-radiative electromagnetic surface wave that propagates in a direction parallel to the negative permittivity/dielectric material interface to measure the affinity between two molecules.
Resources
ihttps://clinicaltrials.gov/iiwww.bioworld.com/COVID19productsiiiwww.gisaid.org/ivwww.antibodysociety.org/covid-19-biologics-tracker/References
- 1.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E., et al. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sia S.F., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16 doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belouzard S., et al. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaimes J.A., et al. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience. 2020 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White J.M., et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y., et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho M. Perspectives on the development of neutralizing antibodies against SARS-CoV-2. Antibody Ther. 2020;3:109–114. doi: 10.1093/abt/tbaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson L.A., et al. An mRNA vaccine against SARS-CoV-2 – preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. Published online July 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F-C., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet (Lond. Engl.) 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu F-C., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31605-6. Published online July 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Q., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callaway E. The race for coronavirus vaccines. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 24.Cohen J. With record-setting speed, vaccinemakers take their first shots at the new coronavirus. Science. 2020 doi: 10.1126/science.abb9996. Published online May 31, 2020. [DOI] [Google Scholar]
- 25.Richard T.E., et al. Remdesivir: a review of its discovery and development leading to human clinical trials for treatment of COVID-19. ACS Cent. Sci. 2020;6:627–683. doi: 10.1021/acscentsci.0c00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan R., et al. Broad-spectrum investigational agent GS-5734 for the treatment of Ebola, MERS coronavirus and other pathogenic viral infections with high outbreak potential. Open Forum Infect. Dis. 2017;4 [Google Scholar]
- 27.Green N., et al. Cell-based assays to identify inhibitors of viral disease. Expert Opin. Drug Discovery. 2008;3:671–676. doi: 10.1517/17460441.3.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beigel J.H., et al. Remdesivir for the treatment of COVID-19 – preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. Published online May 22, 2020. [DOI] [PubMed] [Google Scholar]
- 29.Rajendran D.K., et al. Convalescent plasma transfusion for the treatment of COVID-19: systematic review. J. Med. Virol. 2020 doi: 10.1002/jmv.25961. Published online May 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L., et al. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen C., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L., et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020 doi: 10.1001/jama.2020.10044. Published online June 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S., et al. Neutralizing antibodies against SARS-CoV-2 and pther human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun C., et al. SARS-CoV-2 and SARS-CoV spike-RBD structure and receptor binding comparison and potential implications on neutralizing antibody and vaccine development. bioRxiv. 2020 doi: 10.1101/2020.02.16.951723. Published online February 20, 2020. [DOI] [Google Scholar]
- 36.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 37.Lv Z., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;181:1004–1015. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrapp D., et al. Structural nasis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto D., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 41.Wang C., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls A.C., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prabakaran P., et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang W.C., et al. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lv H., et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31:107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grifoni A., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dijkstra J., Hashimoto K. Expected immune recognition of COVID-19 virus by memory from earlier infections with common coronaviruses in a large part of the world population. F1000Res. 2020;9:285. doi: 10.12688/f1000research.23458.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao Y., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnes C.O., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.025. Published online June 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan J., et al. Human IgG neutralizing monoclonal antibodies block SARS-CoV-2 infection. Cell Rep. 2020;32:107918. doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brouwer P.J.M., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020 doi: 10.1126/science.abc5902. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin C., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen G., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schett G., et al. COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol. 2020;20:271–272. doi: 10.1038/s41577-020-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alijotas-Reig J., et al. Immunomodulatory therapy for the management of severe COVID-19. Beyond the anti-viral therapy: a comprehensive review. Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X., et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. 2020;117:10970. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guaraldi G., et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020 doi: 10.1016/S2665-9913(20)30173-9. Published online June 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Somers E.C., et al. Tocilizumab for treatment of mechanically ventilated patients with COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa954. Published online July 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian X., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ter Meulen J., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Brink E.N., et al. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan M., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huo J., et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.06.010. Published online June 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pinto D., et al. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv. 2020 doi: 10.1101/2020.04.07.023903. Published online April 9 . [DOI] [Google Scholar]
- 71.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. Published online May 26, 2020. [DOI] [PubMed] [Google Scholar]
- 72.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. Published online May 26, 2020. [DOI] [PubMed] [Google Scholar]
- 73.Chi X., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020 doi: 10.1126/10.1126/science.abc6952. Published online June 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Preston B.D., et al. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 75.Vignuzzi M., et al. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 76.Hillen H.S., et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. Published online May 21, 2020. [DOI] [PubMed] [Google Scholar]
- 77.Ou J., et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.15.991844. Published online April 20, 2020 . [DOI] [Google Scholar]
- 78.Teng S., et al. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor binding affinity. bioRxiv. 2020 doi: 10.1101/2020.05.21.109835. Published online May 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhan S.H., et al. SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence? bioRxiv. 2020 doi: 10.1101/2020.05.01.073262. Published online May 2, 2020. [DOI] [Google Scholar]
- 80.van Dorp L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Starr T.N., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020 doi: 10.1016/j.cell.2020.08.012. Published online August 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan Y., et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korber B., et al. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054. Published online May 5, 2020. [DOI] [Google Scholar]
- 84.Sanyal D., et al. An exploration of the SARS-CoV-2 spike receptor binding domain (RBD) – a complex palette of evolutionary and structural features. bioRxiv. 2020 doi: 10.1101/2020.05.31.126615. Published online June 4, 2020. [DOI] [Google Scholar]
- 85.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. Published online July 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang L., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. Published online June 12, 2020. [DOI] [Google Scholar]
- 87.Rockx B., et al. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2010;201:946–955. doi: 10.1086/651022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baum A., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020 doi: 10.1126/10.1126/science.abd0831. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen J., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020 doi: 10.1126/10.1126/science.abd0827. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doud M.B., et al. Complete mapping of viral escape from neutralizing antibodies. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimizu Y.K., et al. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 1994;68:1494. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Z-Y.J., et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 93.Ekiert D.C., et al. A highly conserved neutralizing epitope on group 2 influenza a viruses. Science. 2011;333:843. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ekiert D.C., et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klasse P.J., Burton D.R. Antibodies to West Nile virus: a double-edged sword. Cell Host Microbe. 2007;1:87–89. doi: 10.1016/j.chom.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 96.Dejnirattisai W., et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Halstead S.B. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 98.Diamond M.S., Pierson T.C. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell. 2015;162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin Y-S., et al. Molecular mimicry between virus and host and its implications for dengue disease pathogenesis. Exp. Biol. Med. 2011;236:515–523. doi: 10.1258/ebm.2011.010339. [DOI] [PubMed] [Google Scholar]
- 100.Wang Q., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yip M.S., et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang S-F., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu L., et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Russell L., Cecil W.D.S. The treatment of lobar pneumonia with concentrated antipneumococcus serum. JAMA. 1928;91:2035–2042. [Google Scholar]
- 106.Cecil R.L. Effects of very early serum treatment in Pneumococcus type I pneumonia. JAMA. 1937;108:689–692. [PMC free article] [PubMed] [Google Scholar]
- 107.Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao J., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. Published online March 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang B., et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. Front. Mol. Biosci. 2020;7:158. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Y., et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van Roy M., et al. The preclinical pharmacology of the high affinity anti-IL-6R Nanobody® ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res. Ther. 2015;17 doi: 10.1186/s13075-015-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wec A.Z., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020 doi: 10.1126/10.1126/science.abc7424. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rogers T.F., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020 doi: 10.1126/10.1126/science.abc7520. Published online June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huo J., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020 doi: 10.1038/s41594-020-0469-6. Published online July 13, 2019. [DOI] [PubMed] [Google Scholar]