Abstract
The genomes of mammalian neurons are enriched for unique forms of DNA methylation, including exceptionally high levels of non-CG methylation. Here, we review recent studies defining how non-CG methylation accumulates in neurons and is read out by the critical regulator of neuronal transcription, MeCP2. We discuss the role of gene expression and genome architecture in establishing non-CG methylation and highlight emerging mechanistic insights into how non-CG methylation and MeCP2 control transcription. Further, we describe the cell type-specific functions of this methylation and explore growing evidence that disruption of this regulatory pathway contributes to neurodevelopmental disorders. These findings uncover how the distinctive epigenome in neurons facilitates the development and function of the complex mammalian brain.
DNA Methylation Guides Genomic Regulation
Eukaryotic gene expression is guided by covalent chromatin modifications that facilitate temporal and spatial control of transcription in diverse cell types during development and across dynamic processes [1]. The addition of a methyl group to the 5′ position of cytosine nucleotides (mC) is a major epigenetic modification that contributes to gene regulation across phylogeny [1]. mC can block the binding of gene regulatory proteins such as transcription factors, or recruit ‘reader’ proteins that affect chromatin structure and alter transcription. There is evidence that DNA methylation participates in both activation and repression of gene expression; however, in mammals it is predominantly associated with repeat and transposable element silencing, as well as gene repression (reviewed in [2]). While mC at CG dinucleotides contributes to gene regulation in all cell types, in the last decade it has become clear that DNA methylation has unique and essential roles in the nervous system. Here, we review the discovery and characterization of prevalent mC at non-CG sequences in neurons and discuss the identification of Methyl CpG-binding Protein 2 (MeCP2), the protein disrupted in the neurodevelopmental disorder Rett syndrome (see Glossary), as an essential reader of this mark. We explore new results shedding light on mechanisms of neurodevelopmental disorders caused by disruption of DNA methylation and gene regulation mediated by MeCP2.
The Unique Neuronal Methylome
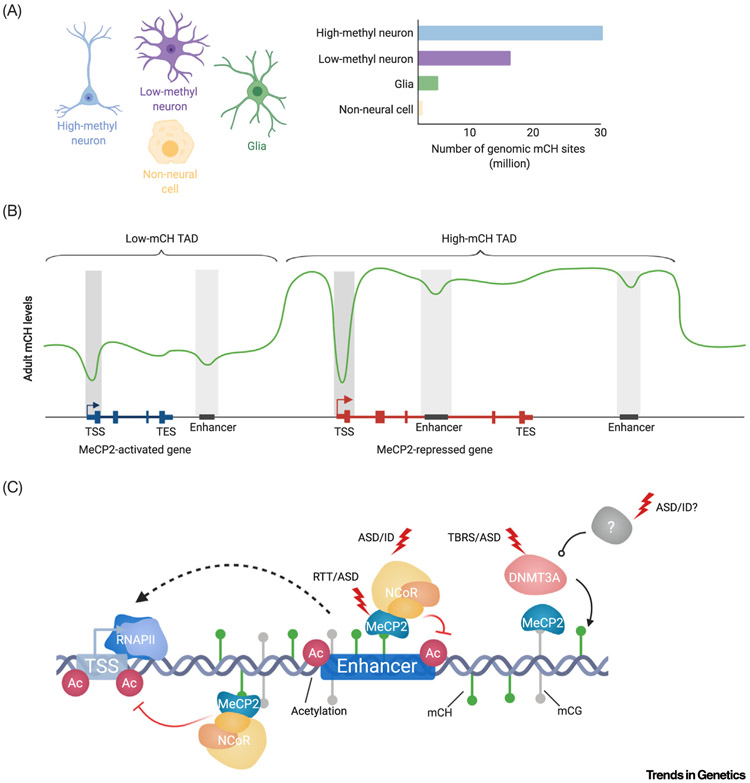
Classically, DNA methylation in mammals was described almost exclusively at cytosines followed by guanines (mCG), with symmetric methylation occurring at cytosines on both strands. mCG is the predominant form of DNA methylation in most tissue types (reviewed in [2]). However, in the brain, alternative forms of methylation are abundant. The neuronal methylome was first recognized as unique when high levels of oxidation of mCG sites to hydroxymethylation (hmCG) were discovered in Purkinje cells of the cerebellum and in brain tissue (Box 1) [3]. More recently, prevalent non-CG DNA methylation (mCH; where H = A, C, or T) has been identified, with this methylation primarily occurring at cytosines followed by adenine (mCA). mCH is highly enriched in neurons compared with other cell types in mouse and humans, and while the methylation rate of CH is lower than that of CG, in some classes of neurons the number of modified CH sites (~1.5–3% for 1.1 billion CH sites = 16–30 million modified CH sites) is equivalent to, or higher than, total modified CG sites (~80% for 21 million CG sites = ~17 million modified CG sites) [Figures 1A (Key Figure) and 2A; discussed in Box 1] [4-6].
Box 1. Neuron-Enriched DNA Modifications and Their Enzymes.
DNA methylation in mammals is deposited at unmethylated CG and non-CG sites (CH) by the de novo methyltransferases DNMT3A and DNMT3B. Additionally, DNMT1 methylates DNA at existing hemi-methylated CG sites to maintain symmetric CG methylation after new strand synthesis, or to create a fully methylated CG site after de novo methylation of one strand [87,88]. The ten-eleven translocation enzymes (TET 1,2,3) oxidize DNA, creating 5-hydroxymethylcytosine (hmC), 5-forymylcytosine (fcC), or 5-carboxylcytosine (caC). These oxidized forms of DNA can drive active demethylation through the base excision repair pathway. However, hmC primarily leads to passive demethylation (dilution) in mitotic cells upon DNA replication by blocking DNMT1 from maintaining hemi-methylated CG dinucleotides [89].
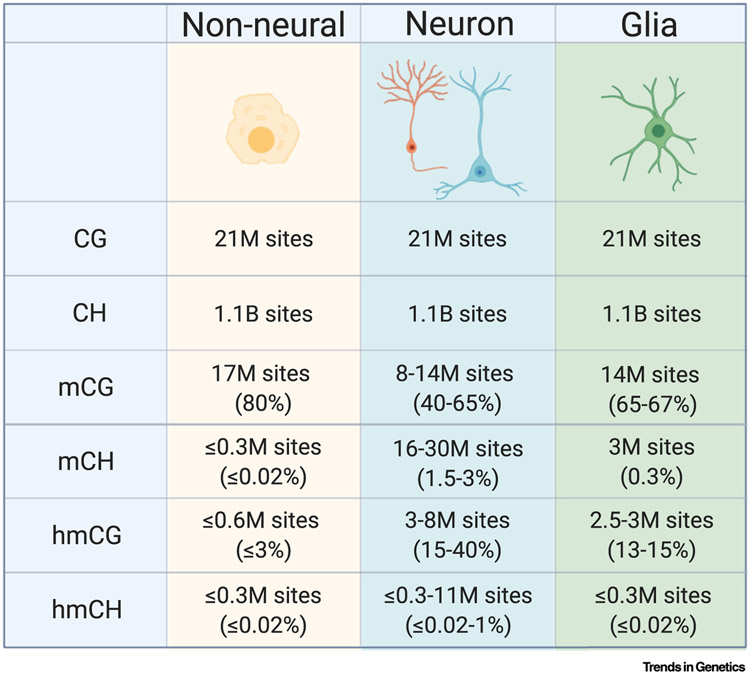
In all cells, CG sites are highly methylated due to early activity of de novo methyltransferases and subsequent active maintenance of mCG by DNMT1 (Figure I) [87]. However, both mCH and hmC are very low in most dividing cells. This is likely due to both low expression of the DNMT3A/B [90] and TET [89] enzymes and the lack of an efficient mechanism to maintain mCH and hmC after DNA replication. In contrast, it appears that increased expression of TET [91] and DNMT3A enzymes [4,6], in conjunction with a lack of DNA replication, leads to accumulation of hmC and mCH in neurons. Substantial evidence now suggests that hmC is a stable epigenetic mark in neurons, reaching high levels compared with non-neural cell types (Figure I) [3,4,89,92]. Therefore, while levels of modified CG sites are maintained in neurons, TET enzymes drive large-scale conversion of mCG into hmCG (e.g., total modified CG = mCG + hmCG, Figure I). Additionally, there appears to be limited turnover of mCH in neurons due to low hmC conversion or active demethylation at these sites.
More efficient methylation at CG sites compared with CH sites leads to a substantially higher percent of mCG compared with mCH (Figure I). However, the depletion of CG dinucleotides from the genome, resulting from mutation of methylated cytosine to thymine over evolution [93], results in nearly equivalent numbers of mC events occurring at CG and non-CG sites in neuronal genomes (Figure I) [4]. Together, the high, stable levels of hmC and mCH create a unique environment for epigenetic regulation in neurons that can affect the binding of regulatory factors to DNA and impact gene expression.
Figure 1. Key Figure. Non-CG Methylation and MeCP2 in Neuronal Gene Regulation.
(A) Non-CG DNA methylation is enriched in neurons compared with glia and other cell types and can show substantial variations in global levels across neuronal subtypes [4,10,13]. (B) Summary of mCH profiles detected at genes most impacted by mCH and MeCP2 mediated gene regulation. Two examples ‘meta genes’ and TADs depict local depletion of mCH within gene bodies and enhancer sequences. MeCP2-repressed genes (red) are enriched for mCH within their gene bodies, at associated enhancers, and throughout the TAD they are located within, while MeCP2-activated genes (blue) and enhancers associated with them are in regions of mCH depletion [8,9,44]. (C) Neuronal gene regulation by the mC-MeCP2-NCoR axis is impacted by disease-associated mutations at multiple levels: mCH deposition, MeCP2 expression, and NCoR complex components. Dysregulation of enhancers and transcriptional activity resulting from these mutations may contribute to disease pathology. The susceptibility of this pathway to disruption suggests that it may be affected in additional neurodevelopmental disorders caused by mutations of epigenetic regulatory genes. Abbreviations: ASD, Autism spectrum disorder; ID, intellectual disability; MeCP2, methyl CpG-binding protein 2; NCoR, nuclear co-repressor complex; RTT, Rett syndrome; TAD, topologically associating domain; TBRS, Tatton-Brown Rahman syndrome; TES, transcription end site; TSS, transcription start site.
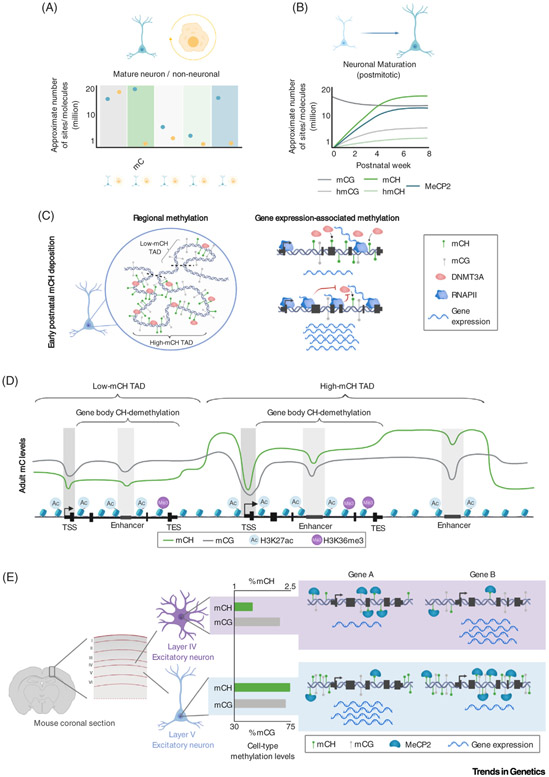
Figure 2. The Unique Neuronal Epigenome.
(A) Summary of reported levels of DNA modification sites and MeCP2 molecules between neurons and non-neural cell types, illustrating that mCH, hmCG, and MeCP2 are uniquely enriched within neurons. (B) During postnatal neuronal maturation, non-CG methylation, hydroxymethylation, and MeCP2 build up, reaching high levels in mature neurons at the young adult stage. (C) Left, a model depicting the early postnatal deposition of non-CG methylation by DNMT3A, establishing megabase-scale domains of high and low methylation associated with TADs. Right, a model illustrating how high levels of gene expression block DNMT3A activity, as highly expressed genes exhibit less DNMT3A binding and mCH accumulation compared with lowly expressed genes. (D) Illustration of typical methylation profiles in adult brain tissue. Both mCG and mCH are depleted at promoters and enhancers, but only mCH exhibits robust gene body demethylation and megabase-scale variations. H3K27ac and H3K36me3 histone modifications illustrate the relationship between DNA methylation and regulatory elements and transcriptional activity. (E) Illustration of differential global levels of mCH detected in subtypes of neurons. A layer V excitatory neuron shows substantially more mCH than a layer IV excitatory neuron [13]. These differences in methylation, combined with differential methylation of genes, set the stage for cell type-specific repressive effects by MeCP2 (e.g., gene A is more strongly expressed in layer V neurons than in layer IV neurons and gene B more so in layer IV than in layer V). Abbreviations: DNMT3A, DNA methyltransferase 3A; MeCP2, methyl CpG-binding protein 2; RNAPII, RNA polymerase II; TAD, topologically associating domain; TES, transcription end site; TSS, transcription start site.
mCH is deposited by the de novo DNA methyltransferase 3A (DNMT3A) (Box 1), which is upregulated in neurons at birth and reaches peak expression at ~2 weeks in mice, before declining to lower expression levels in adulthood [4,6,7]. In frontal cortex, this expression leads to postnatal accumulation of mCH, which plateaus by 4–6 weeks in mice. In humans, mCH builds up primarily during the first 2 years, but requires 16 years to fully accumulate (Figure 2B) [4]. Like mCG, mCH occurs broadly across the genome. Postnatal mCH accumulation across the neuronal genome is influenced by pre-existing gene expression and chromatin structure [4,8,9]. Little to no mCH is deposited at completely silent genes and inaccessible regions of constitutive heterochromatin (e.g., olfactory receptor genes clusters) [4]. Within euchromatic regions, DNMT3A is readily recruited and deposits mCH at repeated sequences, extragenic regions, lowly transcribed genes, and inactive regulatory elements. In contrast, DNMT3A binding and mCH accumulation are depleted from the transcribed region of highly expressed genes and active regulatory elements (Figures 1B and 2C,D) [4,8,10-13]. Experimental manipulation of gene expression in mouse cerebral cortex during the postnatal period indicates that high transcriptional activity blocks DNMT3A binding in genes and results in low mCH accumulation, which persists throughout adulthood [8]. Readout and repression of genes through mCH in adult neurons likely reinforces low expression of highly methylated genes, while lowly methylated genes escape repression and are highly transcribed [8] (discussed later). Genes expressed at moderate levels postnatally build up intermediate levels of mCH, resulting in balanced activation by transcription-promoting machinery and repression by mCH in adult neurons that may tune gene expression levels [8].
Non-CG methylation depletion at genes and regulatory elements largely parallels patterns of mCG at a local scale (kilobase), but mCH shows unique variations on a large scale (megabase) that have been linked to the folding of chromosomes within the nucleus [9]. Megabase-sized genomic regions show enrichment and depletion of mCH that correlate with topologically associating domains (TADs) of chromatin folding [9]. TADs appear to be regions of consistent DNMT3A binding and accumulation of mCH, such that sequences found within individual TADs share similar mCH levels, or an ‘mCH set-point’, while sequences in neighboring TADs can have very different mCH levels [9]. This consistency of TAD mCH impacts genes and enhancer elements within TADs. For example, genes and enhancers in a high-mCH TAD tend to have higher methylation than genes and enhancers in a low-mCH TAD (Figure 2D) [9]. Importantly, these differences in TAD methylation influence the regulation of genes in TADs by mCH (discussed later).
While transcription of genes and TAD structure are clearly associated with mCH deposition, molecular mechanisms controlling DNMT3A activity to create these patterns are not yet defined. Analysis of diverse histone modifications in mouse cortex suggests that chromatin structure during early postnatal development impacts mCH deposition [8], and studies of mCG deposition by DNMT3A outside the nervous system may provide clues to mCH deposition mechanisms within neurons. DNMT3A can bind to unmethylated lysine 4 on histone H3 (H3K4me0) through its ATRX-DNMT3-DNMT3L (ADD) domain, which releases an autoinhibitory conformation to allow cytosine methylation [14,15]. This mechanism could restrict mCH deposition from active regulatory elements, which are marked by H3K4 methylation. The Pro-Trp-Trp-Pro (PWWP) domain of DNMT3A can bind methylated lysine 36 on histone H3 (H3K36me) [15,16]. Based on studies of the close paralog, DNMT3B, this domain has been thought to bind H3K36me3 [2], but recent studies indicate similar or more robust binding to H3K36me2 compared with H3K36me3. Notably, H3K36me2 has been shown to accumulate in broad euchromatic regions and facilitate CG methylation of these domains in dividing mouse and human cells [17-19], while H3K36me3 is associated with the gene bodies of highly expressed genes. If similar patterns of H3K36me2 exist in neurons, DNMT3A may bind to broadly distributed H3K36me2 to guide TAD-scale methylation, while conversion of H3K36me2 to H3K36me3 in the gene body could result in lower levels of DNMT3A recruitment and less mCH deposition in highly expressed genes. Future studies examining the effects of disruption of these histone marks on DNMT3A localization and activity in neurons will help to define the precise mechanisms that govern the deposition of mCH in the brain.
mCH as a Hallmark of Cell Types
An intriguing feature of non-CG methylation is its high degree of cell type specificity, both in global levels of mCH and in local patterns of demethylation at genes and regulatory elements. In both mouse and human, levels of mCH can vary by up to twofold between brain regions [4,6,7,20,21] and 1.5-fold among neuron subtypes in the same brain region [10,12,13]. For example, somatostatin- (SST+) and parvalbumin-positive (PV+) inhibitory interneurons in the cerebral cortex are enriched approximately 30% for mCH compared with vasoactive intestinal polypeptide-positive (VIP+) neurons [10,13]. mCH is also enriched 30–50% in deep layer cortical excitatory neurons compared with their upper layer counterparts [13]. These large variations contrast with smaller global differences in mCG across brain regions and cell classes (Figure 2E) [10,13]. Studies of single neuron methylomes in hippocampus and cortex suggest that both the subtype of a neuron and its location influence mCH levels. For example, inhibitory neuron classes (e.g., PV+) from both cortex and hippocampus share similar mCH levels, but total amounts can be different for cells within the same class in different layers within a region [13,22]. How these global variations in mCH originate has not been determined, but differential expression or activation of DNMT3A in cell types during postnatal development is a potential mechanism that future studies can explore. In all, the varying global mCH levels across cell types suggest that mCH may play a larger regulatory role in some brain regions and neuronal classes compared with others.
Differential CG methylation at genes and regulatory sequences has historically been known to contribute to differentiation and maintenance of distinct cell types (reviewed in [2]). In addition to global variations, local mCH profiles at genomic loci show even more robust cell type-specific patterning than canonical mCG [10,13]. Gene body mCH patterns across cell types are tightly associated with gene expression [8,10,13], such that mCH within genes varies to a greater degree and is more highly correlated with cell type-specific expression than mCG or open chromatin signatures, which show a less dynamic range in signal at genes across cell types [10,13]. Emerging compendiums of single-cell methylomes across multiple mouse brain regions are further defining cell type-specific patterns of mCH [22,23]. These data show that mCH profiles of individual neurons can be used to predict the precise location of a given neuron within one of five different brain regions and the laminar position within that region. The data also reveal patterns of gene expression and enhancer activity across increasingly refined neuronal subtypes. Cell type-specific mCH patterns appear to be dictated by existing gene expression patterns in the early postnatal period through the mechanisms described earlier (Figure 2C). Once established, these mCH patterns function with canonical mCG to maintain cell type-specific gene programs in the adult brain (Figure 2D) [8] (discussed later).
Readout of mCH by MeCP2
Insights into the functional importance of mCH in the brain have emerged through studies establishing MeCP2 as a major reader of this methyl mark. MeCP2 accumulates dramatically in neurons during postnatal development in parallel with the build-up of mCH (Figure 2B) [24-26]. In mature neurons, MeCP2 protein reaches expression levels nearly equivalent to that of histone H4 (Figure 2A) [25], and the expression of Mecp2 in neurons has been shown to be essential for nervous system function [27,28]. While MeCP2 was originally identified as a reader of mCG sites, the discovery of high levels of mCH in the brain prompted close examination of its affinity for this methyl-mark. Indeed, several independent studies identified high-affinity binding of MeCP2 to mCH sites, specifically mCA [6,7,29]. MeCP2 binds to methylated DNA through its methyl-binding domain (MBD), a motif common amongst other methyl-binding proteins [30,31], and shows the strongest binding to mCA compared with the other members of this protein family [31,32]. Notably, mCAC is the most common site for non-CG methylation in the neuronal genome [4,5] and is the highest affinity trinucleotide non-CG site for MeCP2 binding [33]. The preference of MeCP2 for the most prevalent mCH site suggests a functional evolution of non-CG binding for MeCP2 [33]. Further support for read-out of mCA as a critical player in MeCP2 function comes from an emerging study of mice carrying an engineered MeCP2 protein that can bind mCG, but not mCA. These mice recapitulate many neurologic phenotypes and gene expression changes seen in MeCP2 knockouts [34], indicating that mCG binding is not sufficient for normal nervous system function. Together, these studies emphasize read-out of mCA by MeCP2 as essential in the brain.
Interestingly, the high levels of hydroxymethylation in neurons (Box 1) may increase the functional importance for mCH as a site of MeCP2 binding. Biochemical and structural studies indicate that MeCP2 has a lower affinity for hmCG than mCG, while conversion of mCH to hmCH appears to have little effect on MeCP2 binding [7,20,32,33,35-37]. The large-scale conversion of mCG to hmCG (Box 1) [11] has been proposed to inactivate (‘functionally demethylate’) high-affinity CG binding sites of MeCP2. Given that the number of MeCP2 molecules in neurons appears to be substantially lower than total numbers of mCG and mCH binding sites (Figure 2A; also see Figure I in Box 1) [4,25], hmCG accumulation could shift MeCP2 binding in favor of mCH or hmCH sites [7,20,32,33,35-37].
Figure I. Neuron-Enriched DNA Modifications and Their Enzymes.
Approximate levels of CG and CH dinucleotides and their levels of modifications across cell types. Numbers of mC sites are estimated based on measurements made in [2-4,10,11,13,20,97,98]. mC versus hmC levels are inferred by combining results quantifying all modified mC (using bisulfite-sequencing) with studies using hmC-sensitive detection methods (TAB-seq, OxBS). Values are based on the mouse genome, but similar numbers occur in the human genome [99].
Chromatin immunoprecipitation-sequencing (ChIP-seq) studies of MeCP2 in mouse and human brain and isolated neuronal cell populations have detected extremely broad occupancy of the protein across the genome, with relative enrichment of binding at methylated DNA [7,8,20,25,29,33,38-43]. At approximately 16 million molecules of MeCP2 per neuronal nucleus [25], there are sufficiently high numbers of MeCP2 molecules to engage a substantial percentage of the ~24–44 million total mCG and mCH binding sites for the protein in typical neurons (Box 1). Indeed, ChIP signals from multiple studies reflect near-ubiquitous binding with high levels of enrichment (~10–100-fold) compared with Mecp2 knockout controls at all sites in the genome assessed [40,44]. Within the context of broad binding, MeCP2 ChIP signal is enriched at regions with high levels of mCG and mCH (e.g., extragenic regions) and is depleted at regions with low mCH and mCG and high levels of hmCG (e.g., promoters, enhancers, gene bodies for highly expressed genes). However, the magnitude of this depletion is minimal (~1–2-fold), even at regions that are essentially devoid of mC sites [7,8,20,25,29,33,38-43]. It is unclear if this limited dynamic range is a technical limitation of the ChIP method or if it indicates that MeCP2 binds substantially to unmethylated DNA in vivo. In vitro studies have shown that MeCP2 is capable of binding unmethylated DNA, with a preference for GTG residues, albeit with lower binding affinity than methylated DNA [45,46]. However, recent analyses of MeCP2 binding in cells indicate that ChIP-seq and footprint signals for MeCP2 are not enriched at unmethylated GT-rich DNA sequences [47]. A new study suggests that MeCP2 may undergo phase separation with DNA [48,49] (discussed later), raising the possibility that condensates of MeCP2 may drive multivalent contacts with regions of the genome. These associations may contribute to the observed ubiquitous binding pattern of MeCP2, even at sites of low methylation. Together, the largely ubiquitous and low dynamic range patterns of MeCP2 ChIP-seq signal have not allowed researchers to definitively classify specific ‘target genes’ of MeCP2 based on binding profiles alone. Rather, some genes and regulatory elements display modest enrichment of MeCP2 binding compared with those with modest depletion [7,9,41,43,44]. This suggests MeCP2 may play a regulatory role at virtually every region of the genome (Box 2).
Box 2. Global, Graded Gene Regulation by mC and MeCP2.
Unlike transcription factors, where sparse genomic binding sites can be linked to ‘target’ genes, MeCP2 binds to millions of mCH and mCG sites, which are present in varying amounts at every gene and regulatory element. This genome-wide binding suggests that MeCP2 influences transcription of all genes to some extent. Indeed, genome-wide analysis of RNA changes in Mecp2 knockout and missense mutants, MECP2 overexpressing mice, and brain-specific Dnmt3a conditional knockout mice have detected trends in which the degree of gene dysregulation is proportional to the number of mCH and mCG sites found in the gene body [7-9,33,38,42,44,54]. In addition, genome-wide upregulation of expression associated with gene body length has been detected upon loss of MeCP2 [7,33,38,69,70], providing the first clues that intragenic binding by MeCP2 is an important aspect of its regulatory mechanism [7,8]. Care must be taken to ensure that apparent genome-wide effects do not result from technical noise [94], but studies have now verified these trends in large-replicate datasets, using multiple RNA quantification methods [9,39,42,44]. Notably, the investigation of these trends has led to the generation of new molecular models for gene regulation by mCH, mCG, and MeCP2 [8,9,39,42,44].
While these global trends provide mechanistic clues, limitations of gene expression analysis also present challenges for interpretation of transcriptomic data. For instance, normalization procedures used to quantify relative RNA levels between samples assume that no global changes in the distribution of gene expression values occur across conditions [95]. As a result, genome-wide fold-changes will effectively be recentered around zero during data processing and this can possibly switch the sign of perceived changes in gene expression. Similar effects can occur in RT-qPCR experiments, where total RNA and house-keeping gene normalization are employed. Thus, in one plausible model, loss of repression in the Mecp2 knockout leads to upregulation of nearly all genes in the genome, with the most highly methylated regions being most derepressed. However, upon normalization, genes that are lowly methylated and the least derepressed are recentered below zero and quantified as being downregulated. In such a paradigm, a substantial portion of MeCP2-activated genes that appear to decrease in expression when MeCP2 is lost may in fact represent genes that normally escape repression by MeCP2 rather than being genes that are directly activated by the protein.
In addition to these issues, standard transcriptomic approaches do not detect changes in global RNA levels per cell and several reports indicate that Mecp2-null neurons, which are reduced in size, contain less total RNA than normal cells [33,96]. In all, relative quantification methods can combine with secondary effects on RNA levels and gene expression, as well as disruption of potential direct gene-activating functions for MeCP2 [50], to result in the overall changes in gene expression observed in MeCP2 mutants. These complexities highlight the need to integrate transcriptomic findings with biochemical insights into the direct function of MeCP2 to build accurate models of transcriptional regulation by DNA methylation and MeCP2.
Despite these challenges, in vitro and in vivo binding studies clearly indicate that MeCP2 binds with high affinity to mC and does exhibit enriched binding patterns by ChIP-seq [6,7,33,43]. Building on this knowledge, researchers have employed genomic analysis of high-affinity MeCP2 binding sites, mCA and mCG, together with transcriptomic studies to establish a functional role of MeCP2 in controlling transcription of methylation-rich genes [7,8,20,25,29,33,38-43].
Gene Regulation by MeCP2 and Neuronal DNA Methylation
Since its initial identification as a reader of mCG, a myriad of putative protein binding partners for MeCP2 have been identified. These include proteins involved in transcriptional repression and activation, splicing regulation, and microRNA processing, suggesting diverse molecular functions of MeCP2 (reviewed in [27,50]). In addition, MeCP2 is heavily phosphorylated in response to neuronal stimulation, which modulates its activity [51]. A large body of evidence supports gene repression as a major function of MeCP2 [27] and this direct, repressive function of MeCP2 is the focus of discussion here. The best characterized interactor for MeCP2 is the nuclear co-repressor complex (NCoR). NCoR binding is critical for the repressive function of MeCP2 in in vitro assays, and mutations of MECP2 that specifically disrupt this interaction have been shown to drive Rett syndrome [27,52]. Identifying the mechanisms by which the MeCP2-NCoR complex affects gene expression is a major outstanding challenge for the field.
The function of MeCP2 in gene regulation has been intensely studied, but remains difficult to decode [27,53]. Hundreds to thousands of genes can be detected as significantly dysregulated in transcriptomic studies of brain tissue and isolated cells from Mecp2 knockout and missense mutants or MECP2 overexpressing mice, as well as humans with Rett syndrome. However, the magnitude of dysregulation for these genes is subtle (less than twofold), and the near-ubiquitous binding patterns of MeCP2 do not provide sufficient evidence to suggest that these genes are the exclusive targets of regulation by the protein (Box 2).
Recent integrated analysis of whole-genome bisulfite-sequencing DNA methylation maps and transcriptomic changes in Mecp2 knockout and missense mutants or MECP2 transgenic mice has detected methylation signatures on genes most highly affected by MeCP2 and has provided clues to the mechanism of MeCP2-mediated gene regulation [7-9,29,33,38,42-44,54] (Figure 1B). These studies identify a reproducible enrichment of mCH compared with the genome average within the gene body and flanking sequences of MeCP2-repressed genes, those that show significant increases in expression when MeCP2 is disrupted and decreases when MeCP2 is overexpressed. The high levels of mCH in and around MeCP2-repressed genes arise from high mCH in the TADs of which these genes are found, indicating that genome topology plays a role in establishing high methylation at these genes [9]. MeCP2-activated genes, those that show significant decreases in expression when MeCP2 is lost and increases when it is overexpressed, are often found within lower mCH TADs, although they can show moderate gene body mCH enrichment in some studies (Figure 1B) [9,29,44]. Though it is low in dynamic range, the MeCP2 ChIP-signal in and around MeCP2-repressed genes is enriched [29,33,42,44], further supporting a direct role of MeCP2 in repression of these genes.
The function of mCH in MeCP2-mediated gene repression is supported by studies in which mCH accumulation was blocked through perinatal conditional deletion of Dnmt3a in the brain or specifically in neurons of mice [7-9,38,44,54] (Figure 3A). These analyses detected alterations in gene expression in the absence of mCH that partially recapitulate the effects observed in Mecp2 knockout and missense mutants and demonstrated a loss in MeCP2 ChIP-signal at sequences that lose the most mCH upon deletion of Dnmt3a [9,38]. Importantly, because mCG is pre-established early in development by DNMT3A/B and is largely maintained in neurons by DNMT1 [55], the effects observed after Dnmt3a conditional deletion can be attributed to the absence of mCH [7-9,38,44,54]. This provides substantial in vivo evidence supporting mCH as a key site through which MeCP2 affects transcription.
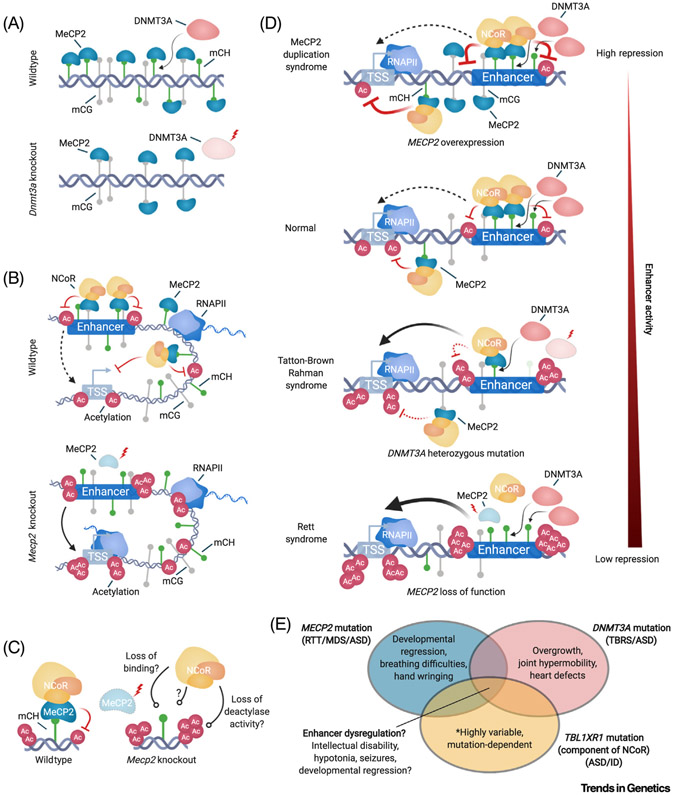
Figure 3. Molecular Mechanisms of the mC-MeCP2-NCoR Axis and Its Disruption in Neurodevelopmental Disorders.
(A) Top, a model depicting that MeCP2 binds to mCH deposited by DNMT3A. Bottom, upon complete or conditional knockout of Dnmt3a, mCH binding sites for MeCP2 are lost, but binding of MeCP2 to mCG persists. (B) Top, model of MeCP2 binding to mC at enhancers and gene bodies to reduce the acetylation of enhancers, genes, and promoters, resulting in a reduction of transcription initiation. Bottom, upon Mecp2 knockout, restriction on acetylation and transcription are reduced. (C) Left, the MeCP2-NCoR complex bound to mC represses enhancer acetylation. This may occur directly through the HDAC component of the complex or indirectly through other undefined activities of NCoR. Right, loss of MeCP2 can lead to loss of NCoR recruitment to the genome, loss of co-repressor activities that result in reduced acetylation, or potentially a mechanism yet to be defined. (D) Schematic depicting a spectrum of disruption for MeCP2 repressive effects at enhancers across neurodevelopmental disorders; ranging from hyper repression in MeCP2 duplication, to intermediate disruption of repression occurring when heterozygous loss of DNMT3A leads to global reduction in mCH, to complete loss of repression when MeCP2 is knocked out. (E) Venn diagram illustrating mechanistic and phenotypic overlaps of molecular pathologies of neurological disorders. Disruption of enhancer repression by the mC-MeCP2-NCoR axis may be shared across these disorders and contribute to pathology. Notably, each genetic lesion results in unique phenotypes that are likely to drive loss of molecular functions outside of the overlapping mC-MeCP2-NCoR axis. Abbreviations: ASD, Autism spectrum disorder; DNMT3A, DNA methyltransferase 3A; ID, intellectual disability; MDS, MeCP2 duplication syndrome; MeCP2, methyl CpG-binding protein 2; NCoR, nuclear co-repressor complex; RNAPII, RNA polymerase II; RTT, Rett syndrome; TBRS, Tatton-Brown Rahman syndrome; TSS, transcription start site.
Notably, while signatures of methylation can be detected at lists of genes significantly dysregulated in Mecp2 and Dnmt3a mutants, neuronal DNA methylation and MeCP2 are present at every gene in the genome to varying degrees and therefore may impact all genes. Indeed, gradients of gene dysregulation associated with genic methylation levels have been detected across all genes genome-wide in Dnmt3a and Mecp2 knockout, conditional knockout, and missense mutants (Box 2) [7-9,33,38,39,42,44,54]. This suggests that disruption of mCH or MeCP2 has subtle yet global effects on neuronal transcriptomes and therefore can have far-reaching impacts on circuit function.
Mechanistic Insights into Gene Regulation by Neuronal Methylation and MeCP2
The identification of DNA methylation signatures associated with MeCP2-mediated regulation has provided a starting point for studies dissecting the mechanism of this regulation. The observation that MeCP2-repressed genes are enriched for methylation within the gene body and flanking regions, rather than sequences at the transcription start site, has suggested that MeCP2 regulates transcription through binding to mC outside of the core promoter region (Figure 1B) [7,29,38]. This finding, combined with the fact that MeCP2-repressed genes also tend to be expressed through extremely long pre-mRNAs, led to the initial hypothesis that MeCP2 binds mC and acts as a ‘speed-bump’, inhibiting processivity of RNA polymerase II (RNAPII) [33,38,39]. However, recent studies in mice analyzing intronic RNA-seq and GRO-seq data, as well as ChIP-seq data for RNAPII and histone modifications associated with transcription (H3K4me3, H3K27ac, H3K36me3) [9,44] did not support this prediction. Rather than finding altered rates of RNAPII processivity, these studies detected altered promoter activity and transcription initiation in genes dysregulated upon mutation of Mecp2 [9,44].
How can binding of MeCP2 to methylation outside of the promoter control transcription initiation? One possibility is that MeCP2 bound to mCH and mCG within and outside of genes can loop to contact promoters, recruiting the NCoR complex and repressing transcription initiation (Figure 3B) [44]. In support of this possibility, recent high-throughput sequencing (Hi-C) analysis in mice indicates that contacts between these regions and promoters do occur in the brain [44]. Substantial additional evidence suggests that MeCP2-NCoR binding to mC at distal sites can broadly block histone acetylation [9,25,44,56] and genomic looping could bring this function to promoters.
In a parallel mechanism, MeCP2 bound to mC at enhancers locally represses the capacity of these elements to activate their cognate genes [9]. Loss of Mecp2 in mice leads to an increase in histone acetylation at sequences with highly methylated TADs, particularly within MeCP2-repressed genes [9,44]. These effects are most robust at enhancer elements, where derepression of acetylation in the absence of MeCP2 is correlated with the number of mCG and mCH sites. Notably, intragenic enhancers are more highly repressed by MeCP2 than extragenic enhancers, providing one explanation for the original identification of enriched intragenic methylation in MeCP2-repressed genes. A role for both mCG and mCH in mediating these effects at enhancers has been further supported through analysis of Dnmt3a conditional knockout mice lacking mCH [9]. In addition, a recent study in mouse cortex found that DNA methylation accumulates postnatally in embryonic-specific enhancers and MeCP2 represses these elements [57]. Thus, deacetylation at enhancers and potentially other genomic sequences is an important consequence of mC binding by MeCP2 that contributes to gene regulation in neurons.
The biological role of NCoR in the context of MeCP2 binding to mC is also coming into focus. The NCoR complex is known to possess deacetylase activity mediated by its HDAC3 subunit [58]. It is therefore intuitive to theorize that this activity could mediate repression by MeCP2. Specific disruption of the MeCP2-NCoR interaction through an MeCP2 point mutation in mice leads to similar effects on histone acetylation and gene expression as the Mecp2 knockout, underscoring the role of NCoR in these effects [7,44]. In addition, loss of MeCP2 and loss of HDAC3 result in shared social and motor impairments as well as an overlap of dysregulated genes associated with neuronal function in mice, supporting a role for HDAC3 in MeCP2-NCoR regulation [59]. However, a recent study tested the importance of NCoR-associated HDAC activity by assessing the severity of phenotypes when Mecp2 is overexpressed in vivo. This study found that introducing an R306C missense mutation associated with Rett syndrome into overexpressed MeCP2 disrupts the NCoR-MeCP2 interaction and blocks the toxicity of MeCP2 overexpression. In contrast, introducing mutations into NCoR components that inhibit activation of HDAC3 did not rescue lethality [60], suggesting the deacetylase activity of the NCoR complex is not required for the key functions of MeCP2-NCoR. This study relied primarily on gross organismal phenotypes for its interpretations. Future studies directly testing changes in histone acetylation in similar mutants can confirm that deacetylation by NCoR is not the direct activity needed for MeCP2’s repressive function (Figure 3C).
A challenge for researchers going forward is to integrate findings on protein–protein interactions and chromatin modifying activity of MeCP2-NCoR with potential structural or biophysical roles of MeCP2 in chromatin that are re-emerging. Historic findings have shown that MeCP2 localizes to regions of heterochromatin and drives nucleosome aggregation in vitro [61-63]. Recent studies have shed new light on this activity by showing that MeCP2 can undergo liquid–liquid phase separation, forming condensates with nucleosomal DNA in vitro [48,49]. Rett syndrome causing mutations of MECP2 reduced condensate forming activity, suggesting that MeCP2 might affect compartmentalization of heterochromatic droplets in cells and that disruption of this activity may be indicative of MECP2 inactivation in disease. High resolution nuclear imaging in Mecp2 knockout mice has also observed altered heterochromatin volume in neurons lacking MeCP2, suggesting altered chromatin condensation [64]. However, these findings have not yet been linked to the epigenomic and transcriptomic consequences of mCH-MeCP2 disruption. Recent analyses of chromatin looping by 3C and Hi-C detected no dramatic changes in chromosome topology in Mecp2 knockout mouse brain tissue [9,44]. The lack of changes in chromosome topology does not support changes in nuclear compartmentalization occurring upon loss of MeCP2, as might be predicted by the phase-separation experiments. However, a study of human embryonic stem cell-derived interneurons carrying the MECP2 R133C Rett syndrome mutation detected altered global topology [65]. Notably, these condensation and human topology studies analyzed MECP2 truncation and missense mutants, while the mouse studies focused on complete loss of the protein. Different MECP2 mutations result in differential clinical severity [27,66] and these differences may account for contrasting effects across studies. Additional analysis will be needed to determine how condensation characteristics of MeCP2 may impact genome topology and gene expression to manifest cellular dysfunction when disrupted.
Future studies can build on these recent findings to further decode the mechanism of gene regulation by the mC-MeCP2-NCoR pathway. For example, if epigenomic profiling of MeCP2 detects alterations in enhancer activation that occur upstream of histone acetylation at highly methylated enhancers upon loss of MeCP2 (e.g., transcription factor binding, nucleosome removal, H3K4me1 deposition), it would suggest that MeCP2-NCoR indeed acts outside of histone acetylation in controlling chromatin structure. A study of cultured human neurons suggests that MeCP2 represses binding of BRD4 [65], a coregulatory protein that forms condensates with acetylated enhancers to mediate gene activation [67]. Future studies can examine if alterations in BRD4 occur downstream of the mC-MeCP2-NCoR pathway in adult neurons. In addition, assessing if the phase-separation characteristics of MeCP2 can influence mechanisms of molecular crowding and condensation of BRD4 [68] at enhancers to impact gene regulation could link biophysical properties of MeCP2 to these epigenomic effects.
Biological Function of mCH-MeCP2-Mediated Gene Regulation
In addition to these mechanistic insights, recent studies have shed light on the functional impact of mCH-MeCP2-mediated gene regulation. The highly cell type-specific nature of mCH profiles suggests an important role for MeCP2 is to mediate neuron subtype-specific gene expression. In support of this, an initial study of hand-sorted cell populations from Mecp2 knockout mouse brains detected larger changes in mRNA levels in purified cell types compared with whole tissue and identified distinct sets of dysregulated genes in different cell types [69]. Recent integrated analyses of mCH profiles and RNA changes in isolated cell populations [8,54,70] and single cells [8,42] from Mecp2 and Dnmt3a knockout, conditional knockout, and missense mouse mutants and humans with Rett syndrome have identified cell type-specific derepression of genes enriched for mCH. Notably, the genes most derepressed in cell-specific and tissue-based studies of mCH and MeCP2 disruption tend to be long genes that encode protein with important roles in the establishment and maintenance of synaptic connectivity (e.g., cell-adhesion molecules, ion channels, and synaptic receptors) [7,9,33,42,44,69]. Thus, mCH and MeCP2 appear to regulate genes with essential roles in establishing connectivity in a cell type-specific fashion.
These recent findings, coupled with the knowledge that MeCP2 reads postnatal mCH patterns to repress transcription in the adult, suggest a functional model: during development, when MeCP2 expression is rising and mCH is being deposited in each neuronal subtype, genes and enhancers that are robustly expressed escape accumulation of mCH and subsequent repression by MeCP2. Conversely, genes that are lowly expressed accumulate mCH and MeCP2, which in turn maintains them in a repressed state later in life [8]. The absence of mCH-MeCP2 repression in the early postnatal period may allow for flexible expression of critical protein components of synaptic connectivity, as cells respond to extrinsic cues and integrate into circuits. Build-up of mCH and MeCP2 occurs primarily during the closure of postnatal hyperplastic periods, which could then maintain these gene expression patterns to allow for consolidation and refinement of cellular functions in the circuit. Thus, mCH-MeCP2 repression might effectively close an ‘epigenomic critical period’ of plastic gene expression to stabilize functional circuits, in much the same way that build-up of extracellular matrix closes critical periods of plastic connectivity in the brain during this same period [71]. Once global patterns of mCH are established, stimulus-dependent inactivation of MeCP2-NCoR-mediated repression that results from MeCP2 phosphorylation [51] and activity-dependent alterations in DNA methylation [72] could facilitate more limited, but important, dynamic gene expression during adult plasticity. In support of this role for mCH and MeCP2 in brain function, loss of MeCP2 disrupts critical period timing and synaptic plasticity [50,71,73,74].
Expanding Roles for mCH and MeCP2 in Disease
For over a decade, disruption of MeCP2 due to loss-of-function mutations or overexpression has been recognized as the cause of Rett syndrome and MeCP2 duplication syndrome, respectively (reviewed in [53]). The fact that either too much or too little MeCP2 manifests in severe neurologic dysfunction suggests that circuits require precise, dose-sensitive tuning of gene regulation by the mC-MeCP2-NCoR pathway [27,53] and raises the possibility that this pathway may be susceptible to additional insults in disease (Figure 1C). Indeed, exome sequencing studies have recently uncovered mutations in individuals with intellectual disability, autism, and related disorders that disrupt this pathway up- and downstream of MeCP2. Heterozygous mutations in DNMT3A cause Tatton-Brown-Rahman syndrome and autism [75-77]. Strikingly, Dnmt3a heterozygous knockout mice that model this disorder exhibit ~50% global reductions in mCH across multiple brain regions [21,78]. This reduction in mCH drives alterations in enhancer histone acetylation and gene expression in the cerebral cortex that partially recapitulate MeCP2 loss of function [21]. These findings indicate that mCH deposition and its role in neuronal regulation are highly sensitive to reduction in DNMT3A protein (Figure 3D).
Mutations of a component in the NCoR complex, TBL1XR1, have also emerged as causal for neurodevelopmental disease [77,79,80]. Notably, some missense mutations identified in TBL1XR1 have been shown to specifically disrupt the NCoR-MeCP2 interaction and a patient with a missense mutation in this interacting domain has been diagnosed with Rett syndrome, based on clinical criteria [81,82]. In further support of overlapping pathology arising from an absence of NCoR, loss-of-function mutations of the deacetylase activating domain of Ncor1 within GABAergic neurons resulted in 492 cognitive-, social-, and anxiety-related phenotypes in mice that have some similarities with Mecp2 493 knockout models [83].
It is important to note that mutations in DNMT3A and NCOR also manifest multiple distinct clinical features from those of Rett syndrome (e.g., overgrowth, heart defects, joint hypermobility). Undoubtedly, these factors have gene regulatory roles early in development and outside of the mC-MeCP2-NCoR axis that contribute to the nonoverlapping aspects of these disorders [82,84] (Figure 3E). The precise degree of overlapping molecular etiology between these disorders may have implications for the development of treatments. A striking feature of Mecp2 mutations is that reintroduction of exogenous MeCP2 in adults can dramatically reverse symptoms in mice [85], likely because restored MeCP2 reads out mCH and mCG patterns that were appropriately laid down during postnatal development. This finding has fueled development of gene therapies for Rett syndrome [86]. If DNMT3A mutations cause deficits in DNA methylation during critical temporal windows of embryonic and postnatal development, these effects may not be as reversible as the absence of MeCP2. Likewise, changes in cellular compositions of the brain or structural changes that occur due to loss of early roles for NCoR or DNMT3A may be difficult to reverse later in life. Nonetheless, disruption of epigenomic regulation through the mC-MeCP2-NCoR axis in adults is likely a shared deficit that contributes to neurologic dysfunction in these disorders and may be a viable candidate to explore therapeutic approaches.
Concluding Remarks and Future Perspectives
Recent studies have discovered large quantities of mCH in neurons, defined cell type-specific profiles for this methyl mark, and linked it to the essential functions of MeCP2. These findings have demonstrated that mCH is a critical epigenetic component of the mammalian brain. However, important questions remain to be answered (see Outstanding Questions). Progress has been made in identifying how chromatin architecture and gene expression states define mCH profiles in neurons, but insights into the molecular mechanisms that recruit and activate DNMT3Aare still needed. Integrated methylomic, transcriptomic, and epigenomic studies have revealed strong candidate models for the long-enigmatic mechanism of MeCP2-mediated transcriptional control. Comprehensive assessment of epigenomic changes in MeCP2 mutants and studies exploring how the phase-separation of MeCP2 contribute to its functions in gene regulation promise to further solidify our understanding of MeCP2.
Outstanding Questions.
What are the molecular mechanisms of mCH deposition by DNMT3A? Do they differ from non-neural CG methylation mechanisms?
What are the contributions of gene expression-associated and topology-associated mechanisms to mCH profiles at genes?
Dissecting cause and consequence: Which effects on RNA expression observed in MeCP2 mutants are directly caused by MeCP2 disruption and which alterations are secondary consequences of cellular dysfunction?
How does variation in global mCH across neuronal subtypes arise and does it result in differential importance of transcriptional regulation by MeCP2 in these cells? Are subclasses of high mCH cells particularly susceptible to disruption of mCH or MeCP2 in disease?
Does MeCP2 directly regulate histone acetylation at enhancers and other genomic regions through NCoR or through upstream mechanisms?
Why are intragenic enhancers more susceptible to repression by MeCP2 than extragenic enhancers?
What is the contribution of MeCP2 condensation properties to neuronal gene regulation and neurodevelopmental disease?
Does mutation of NCoR components cause similar epigenomic effects to loss of MeCP2 or DNMT3A?
Is the mCH-MeCP2-NCoR pathway disrupted in other neurodevelopmental disorders caused by mutation of other newly identified epigenomic regulators?
A role for mCH and MeCP2 in cell type-specific gene regulation is clear, however, systematic studies across cell types are needed to understand the relative importance of mCH in each cell type and to determine the complements of genes most impacted by its regulation. For example, given the large differences in total mCH levels across the genomes of neuronal subtypes, it is possible that mCH and MeCP2 have a larger impact on gene regulation in high mCH subtypes compared with lower mCH subtypes and loss of mCH or MeCP2 in disease may disproportionately impact these high mCH subtypes, manifesting in specific circuit defects.
The identification of disease-associated mutations in genes, functioning up- and downstream of MeCP2 along the mC-MeCP2-NCoR axis, suggest an expanded role of this gene regulatory pathway across multiple distinct disorders. With a wide array of new neurodevelopmental disorder genes now identified [77,79], it will be valuable to explore if mutations of epigenetic regulators in these disorders disrupt this unique neuronal DNA methylation pathway. Through these studies, disease genetics can help uncover how neuron-specific DNA methylation uniquely contributes to the development and function of the extraordinarily complex mammalian brain.
Highlights.
Non-CG DNA methylation and its reader, MeCP2, are highly enriched in neurons over other cell types.
Genomic distribution of non-CG methylation is shaped by chromosomal structure and gene expression in early postnatal development.
Genomic profiles of non-CG methylation are highly cell type-specific and contribute to cell-specific gene programs.
Non-CG methylation and MeCP2 repress histone acetylation in neurons. Loss of this repression, including at enhancers, contributes to gene dysregulation.
Mutations disrupting MeCP2; the CH methyltransferase, DNMT3A; and the MeCP2 interacting co-repressor, NCoR, underlie neurodevelopmental disorders and may share enhancer dysregulation as a disease driver.
Acknowledgments
We thank the Gabel Laboratory, J. Goodman, and J. Yi for thoughtful comments. The figures in the manuscript were created with BioRender.com. This work was funded by the National Institutes of Health (NIH) through F31NS108574 to A.W.C and 539 grants from The Simons Foundation Autism Research Initiative 508034, the Klingenstein-Simons Fellowship Fund, and 540 NIMH R01MH117405 to H.W.G.
Glossary
- 3C and Hi-C:
chromatin conformation capture methods employing crosslinking and proximity ligation followed by PCR (3C) or Hi-C to map 3D-interactions and the architecture of genome folding within the nucleus.
- Enhancer:
regulatory element found outside of promoters that recruits transcription factors and coactivators and interacts with promoters to drive transcription.
- Gene body:
region of the gene from transcription start site (TSS) to transcription end site (TES) that is transcribed during pre-mRNA production.
- GRO-seq:
global run-on sequencing; method that utilizes cotranscriptional labeled nucleotide incorporation followed by sequencing to directly quantify gene transcription.
- Histone modifications:
denoted by histone number, amino acid, and modification (e.g., monomethylation, me1; trimethylation, me3; acetylation, ac). Associated with steps in transcriptional regulation.
- H3K4me1:
present at active enhancers.
- H3K4me3:
present at active/bivalent promoters and enhancers.
- H3K27ac:
present at active promoters and enhancers.
- H3K36me2:
precursorto H3K36me3; marks open euchromatic regions.
- H3K36me3:
marks the 3′ portion of the gene body for actively transcribed genes.
- mCA set-point:
hypothesis that levels of mCA are consistent across discrete megabase-scale domains of the genome but can vary from domain to domain. mCA set-points appear to influence the levels of mCA at kilobase-scale sequence elements such as enhancers and gene bodies.
- MeCP2-activated genes:
genes significantly downregulated when MeCP2 is inactivated and upregulated when MeCP2 is overexpressed.
- MeCP2 duplication syndrome:
neurological disorders caused by genetic duplication of MECP2. Causes developmental delay among other severe clinical features.
- MeCP2-repressed genes:
genes significantly upregulated when MeCP2 is inactivated and downregulated when MeCP2 is overexpressed.
- Methyl-binding domain (MBD):
conserved protein domain that binds specifically to methylated cytosine. Found in MeCP2 as well as MBD1/2/3/4.
- Nuclear co-repressor complex (NCoR):
a complex comprised of NCoR1 and/or its paralog SMRT, with TBL1, HDAC3, and GPS2 as its core components. It is recruited to the genome by multiple DNA binding proteins, including MeCP2.
- Rett syndrome:
X-linked recessive neurological disorder, predominantly occurring in females, that is caused by loss-of-function MECP2 mutations. Typified by phenotypically normal early development, followed by regression and decline in brain growth.
- Tatton-Brown-Rahman syndrome:
autosomal dominant neurodevelopmental disorder characterized by overgrowth, intellectual disability, and autism. Caused by heterozygous mutations of DNMT3A.
- Topologically associating domains (TADs):
linear regions of the genome that show enrichments for interactions in 3D space. Can facilitate and/or result from contacts between enhancers and promoters that drive gene expression.
- Whole-genome bisulfitesequencing:
base-resolution DNA methylation profiling method whereby methylated cytosines are protected from sodium bisulfite conversion to uracil (read as thymidine), thereby distinguishable from unmethylated cytosines during whole genome sequencing.
References
- 1.Zhou J et al. (2017) Tissue-specific DNA methylation is conserved across human, mouse, and rat, and driven by primary sequence conservation. BMC Genomics 18, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg MVC and Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Ceil Biol 20, 590–607 [DOI] [PubMed] [Google Scholar]
- 3.Kriaucionis S and Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie W et al. (2012) Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo JU et al. (2014) Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci 17, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabel HW et al. (2015) Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroud H et al. (2017) Early-life gene expression in neurons modulates lasting epigenetic states. Cell 171, 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens AW et al. (2020) MeCP2 represses enhancers through chromosome topology-associated DNA methylation. Mol. Cell 77, 279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mo A et al. (2015) Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozlenkov A et al. (2018) A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci. Adv 4, eaau6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mo A et al. (2016) Epigenomic landscapes of retinal rods and cones. Elite 5, e11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo C et al. (2017) Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondelet G et al. (2016) Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol 194, 357–367 [DOI] [PubMed] [Google Scholar]
- 15.Guo X et al. (2015) Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644 [DOI] [PubMed] [Google Scholar]
- 16.Dhayalan A et al. (2010)The Dimt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dukatz M et al. (2019) H3K36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J. Mol. Biol 431, 5063–5074 [DOI] [PubMed] [Google Scholar]
- 18.Weinberg DN et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell 11, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellén M et al. (2017) 5-Hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci U. S. A 114, E7812–E7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christian DL et al. (2020) DNMT3A haploinsufficiency results in behavioral deficits and global epigenomic dysregulation shared across neurodevelopment disorders. bioRxiv Published online July 13, 2020. 10.1101/2020.07.10.195859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H et al. (2020) DNA methylation atlas of the mouse brain at single-cell resolution. bioRxiv Published online April 30, 2020. 10.1101/2020.04.30.069377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z et al. (2020) An integrated transcriptomic and epigenomic atlas of mouse primary motor cortex cell types. bioRxiv Published online March 5, 2020. 10.1101/2020.02.29.970558 [DOI] [Google Scholar]
- 24.Kishi N and Macklis JD (2004) MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell. Neurosci 27, 306–321 [DOI] [PubMed] [Google Scholar]
- 25.Skene PJ. et al. (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahbazian MD (2002) Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet 11, 115–124 [DOI] [PubMed] [Google Scholar]
- 27.Tillotson R and Bird A (2019) The molecular basis of MeCP2 function in the brain. J. Mol. Biol Published online October 17, 2019. 10.1016/j.jmb.2019.10.004 [DOI] [PubMed] [Google Scholar]
- 28.Luikenhuis S et al. (2004) Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc. Natl. Acad. Sci. U. S. A 101, 6033–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L et al. (2015) MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U. S. A 112, E2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrich B and Bird A (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Celt. Biot 18, 6538–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginder GD and Williams DC (2018) Readers of DNA methylation, the MBD family as potential therapeutic targets. Pharmacol. Ther 184, 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperlazza MJ et al. (2017) Structural basis of MeCP2 distribution on non-CpG methylated and hydroxymethylated DNA. J. Mol. Biol 429, 1581–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagger S et al. (2017) MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 13, e1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tillotson R et al. (2020) Neuronal non-CG methylation is an essential target for MeCP2 function. bioRxiv Published online July 2, 2020. 10.1101/2020.07.02.184614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valinluck V et al. (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 32, 4100–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimoto H et al. (2012) Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchmuller BC et al. (2020) Complete profiling of methyl-CpG-binding domains for combinations of cytosine modifications at CpG dinucleotides reveals differential read-out in normal and Rett-associated states. Sci. Rep 10, 4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinde B et al. (2016) DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc. Natl. Acad. Sci. U. S. A 113, 15114–15119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cholewa-Waclaw J et al. (2019) Quantitative modelling predicts the impact of DNA methylation on RNA polymerase II traffic. Proc. Natl. Acad. Sci. U. S. A 116, 14995–15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen S et al. (2011) Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker SA et al. (2013) An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 152, 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renthal W et al. (2018) Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat. Neurosci 21, 1670–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rube HT et al. (2016) Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nat. Commun 7, 11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boxer LD et al. (2020) MeCP2 represses the rate of transcriptional initiation of highly methylated long genes. Mol. Cell 77, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei M et al. (2019) Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain. Biochim. Biophys. Acta - Gene Regul. Mech 1862, 194409. [DOI] [PubMed] [Google Scholar]
- 46.Meehan RR et al. (1992) Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 20, 5085–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Connelly JC et al. (2020) Absence of MeCP2 binding to non-methylated GT-rich sequences in vivo. Nucleic Acids Res. 48, 3542–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L et al. (2020) Rett syndrome-causing mutations compromise MeCP2-mediated liquid–liquid phase separation of chromatin. Cell Res. 30, 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan C et al. (2020) Rett mutations attenuate phase separation of MeCP2. Cell Discov. 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ip JPK et al. (2018) Rett syndrome: insights into genetic, molecular and circuit mechanisms. Nat. Rev. Neurosci 19, 368–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap E-L and Greenberg ME (2018) Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron 100, 330–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong Y et al. (2020) Nuclear receptor corepressors in intellectual disability and autism. Mol. Psychiatry Published online February 7, 2020. 10.1038/s41380-020-0667-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavery LA and Zoghbi HY (2019) The distinct methylation landscape of maturing neurons and its role in Rett syndrome pathogenesis. Curr. Opin. Neurobiol 59, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavery LA et al. (2020) Losing Dnmt3a dependent methylation in inhibitory neurons impairs neural function by a mechanism impacting Rett syndrome. Elife 9, e52981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okano M et al. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 56.Shahbazian MD et al. (2002) Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35, 243–254 [DOI] [PubMed] [Google Scholar]
- 57.Stroud H et al. (2020) An activity-mediated transition in transcription in early postnatal neurons. Neuron Published online June 24, 2020. 10.1016/j.neuron.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guenther MG et al. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nott A et al. (2016) Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat. Neurosci 19, 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koerner MV et al. (2018) Toxicity of overexpressed MeCP2 is independent of HDAC3 activity. Genes Dev. 32, 1514–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nan X et al. (1996) DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol 16, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brero A et al. (2005) Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol 169, 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agarwal N et al. (2007) MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 35, 5402–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Linhoff MW et al. (2015) A high-resolution imaging approach to investigate chromatin architecture in complex tissues. Cell 163, 246–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiang Y et al. (2020) Dysregulation of BRD4 function underlies the functional abnormalities of MeCP2 mutant neurons. Mol. Cell 79, 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyst MJ et al. (2013) Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci 16, 898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bancaud A et al. (2009) Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 28, 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sugino K et al. (2014) Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci 34, 12877–12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson BS et al. (2017) Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat. Med. 23, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picard N and Fagiolini M (2019) MeCP2: an epigenetic regulator of critical periods. Curr. Opin. Neurobiol 59, 95–101 [DOI] [PubMed] [Google Scholar]
- 72.Bayraktar G and Kreutz MR (2018) Neuronal DNA methyltransferases: epigenetic mediators between synaptic activity and gene expression? Neuroscientist 24, 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krishnan K et al. (2015) MeCP2 regulates the timing of critical period plasticity that shapes functional connectivity in primary visual cortex. Proc. Natl. Acad. Sci. U. S. A 112, E4782–E4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patrizi A et al. (2019) Accelerated hyper-maturation of parvalbumin circuits in the absence of MeCP2. Cereb. Cortex 30, 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatton-Brown K et al. (2014) Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet 46, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanders SJ et al. (2015) Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satterstrom FK et al. (2020) Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sendžikaitė G et al. (2019) A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun 10, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coe BP et al. (2019) Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet 51, 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Firth HV et al. (2009) DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet 84, 524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kruusvee V et al. (2017) Structure of the MeCP2-TBLR1 complex reveals a molecular basis for Rett syndrome and related disorders. Proc. Natl. Acad. Sci. U. S. A 114, E3243–E3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaghlula M et al. (2018) Current clinical evidence does not support a link between TBL1XR1 and Rett syndrome: descripttion of one patient with Rett features and a novel mutation in TBL1XR1, and a review of TBL1XR1 phenotypes. Am. J. Med. Genet A 176, 1683–1687 [DOI] [PubMed] [Google Scholar]
- 83.Zhou W et al. (2019) Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus–CA3 projection. Nat. Neurosci 22, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tatton-Brown K et al. (2018) The Tatton-Brown-Rahman syndrome: a clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 3, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guy J et al. (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315, 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gadalla KKE et al. (2013) Improved survival and reduced phenotypic severity following AAV9/MECP2 gene transfer to neonatal and juvenile male Mecp2 knockout mice. Mol. Ther 21, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hermann A et al. (2004) The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem 279, 48350–48359 [DOI] [PubMed] [Google Scholar]
- 88.Gowher H and Jeltsch A (2018) Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans 46, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu X and Zhang Y (2017) TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet 18, 517–534 [DOI] [PubMed] [Google Scholar]
- 90.Okano M et al. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 91.Szulwach KE et al. (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci 14, 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Globisch D et al. (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Raman AT et al. (2018) Apparent bias toward long gene misregulation in MeCP2 syndromes disappears after controlling for baseline variations. Nat. Commun 9, 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI et al. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Y et al. (2013) Global transcriptional and translational represssion in human embryonic stem cells-derived Rett syndrome neurons. Cell Stem Cell 13, 446–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sardina JL et al. (2018) Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 23, 727–741 [DOI] [PubMed] [Google Scholar]
- 98.Hon GC et al. (2014) 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kent WJ et al. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]