Introduction
The SARS-CoV-2 virus that causes COVID-19 enters cells via the angiotensin converting enzyme 2 (ACE2) receptor. The spike protein of the virus binds to ACE2 [32; 33] and can be primed by a number of proteases (TMPRSS2 [13] and Furin [8]) that are sometimes co-expressed by ACE2 positive cells [37]. A set of genes that are involved in the response to SARS-CoV-2, SARS-CoV-2 and coronavirus-associated factors and receptors (SCARFs) [27], have emerged allowing for assessment of virus effects on tissues through analysis of RNA sequencing or other high-throughput datasets.
Neurological symptoms are common in COVID-19 patients. Loss of smell (anosmia) and taste is an early symptom which is explained by non-neuronal ACE2 expression within the olfactory epithelium[6]. This is sometimes accompanied by loss of chemical sensation, chemesthesis [21]. This chemesthesis includes loss of capsaicin and menthol sensitivity [21], sensations that are mediated by nociceptive sensory neurons [4]. Other neurological effects of SARS-CoV-2 associated with nociceptors have been described, including headache and nerve pain [16; 18; 36] and may be involved in neuro-immune interactions in the lung [25]. Finally, as the pandemic has progressed it is now clear that many patients do not have a complete recovery from COVID-19 and experience continuing symptoms like dyspnea, joint pain, chest pain and cough [7], all of which are mediated, at least in part, by nociceptors. Additionally, severe CNS effects like encephalopathies and delirium have also been observed in COVID-19 patients [10; 22]. Molecular mechanisms underlying these effects are not currently known, although studies with human brain organoids demonstrate the ACE2-dependent neuro-invasive potential of the SARS-CoV-2 virus [28].
Bulk RNA sequencing datasets support the conclusion that ACE2 is expressed in human DRG [20; 23], but cellular resolution is lacking. Single cell sequencing experiments from mouse DRG show weak Ace2 expression in a subset of neurons that also express the Mrgprd and Nppb genes [30]. MrgprD is selectively expressed in a nociceptor population that forms free nerve endings in the skin [38], cornea and luminal organs such as the colon[12] as well as the meninges [31]. We tested the hypothesis that ACE2 mRNA is expressed in a subset of human DRG neurons using tissues obtained from organ donors or vertebrectomy surgery based on RNAscope in situ hybridization. Our findings support the conclusion that approximately a quarter of human DRG neurons express ACE2 mRNA and that ACE2 protein is also found in the human DRG. Most of these ACE2 mRNA expressing neurons are nociceptors that likely form free nerve endings in skin or other organs creating a potential entry point for the virus into the peripheral nervous system.
Materials and Methods
Tissue preparation
All human tissue procurement procedures were approved by the Institutional Review Boards at the University of Texas at Dallas and University of Texas MD Anderson Cancer Center. Human dorsal root ganglion (at levels T4, T5, and L5) were collected, frozen on dry ice (L5) or liquid nitrogen (T4 and T5) and stored in a −80°C freezer. Donor and/or patient information is provided in Table 1. The human DRGs were gradually embedded with OCT in a cryomold by adding small volumes of OCT over dry ice to avoid thawing. All tissues were cryostat sectioned at 20 μm onto SuperFrost Plus charged slides. Sections were only briefly thawed in order to adhere to the slide but were immediately returned to the −20°C cryostat chamber until completion of sectioning. The slides were then immediately utilized for histology.
Table 1. Human DRG tissue Information.
Donor or patient information is given for all the samples that were used for RNAscope in situ hybridization shown in Fig 1.
| Donor # | DRG | Sex | Age | Race | Surgery/Cause of Death | Collection Site |
|---|---|---|---|---|---|---|
| 1 | T4 | Male | 77 | White, non-Hispanic | Vertebrectomy | MD Anderson |
| 2 | L5 | Female | 33 | White, non-Hispanic | Opioid overdose | Southwest Transplant Alliance |
| 3 | T5 | Male | 56 | White, non-Hispanic | Vertebrectomy | MD Anderson |
RNAscope in situ hybridization
RNAscope in situ hybridization multiplex version 1 was performed as instructed by Advanced Cell Diagnostics (ACD). Slides were removed from the cryostat and immediately transferred to cold (4°C) 10% formalin for 15 minutes. The tissues were then dehydrated in 50% ethanol (5 min), 70% ethanol (5 min) and 100% ethanol (10 min) at room temperature. The slides were air dried briefly and then boundaries were drawn around each section using a hydrophobic pen (ImmEdge PAP pen; Vector Labs). When hydrophobic boundaries had dried, protease IV reagent was added to each section until fully covered and incubated for 5 minutes at room temperature. The protease IV incubation period was optimized as recommended by ACD for the specific lot of Protease IV reagent. Slides were washed briefly in 1X phosphate buffered saline (PBS, pH 7.4) at room temperature. Each slide was then placed in a prewarmed humidity control tray (ACD) containing dampened filter paper and a mixture of Channel 1, Channel 2, and Channel 3 probes (50:1:1 dilution, as directed by ACD due to stock concentrations) was pipetted onto each section until fully submerged. This was performed one slide at a time to avoid liquid evaporation and section drying. The humidity control tray was placed in a HybEZ oven (ACD) for 2 hours at 40°C. A table of all probes used is shown in Table 2. Following probe incubation, the slides were washed two times in 1X RNAscope wash buffer and returned to the oven for 30 minutes after submersion in AMP-1 reagent. Washes and amplification were repeated using AMP-2, AMP-3 and AMP-4 reagents with a 15-min, 30-min, and 15-min incubation period, respectively. AMP-4 ALT C (Channel 1 = Atto 550, Channel 2 = Atto 647, Channel 3 = Alexa 488) was used for all experiments. Slides were then washed two times in 0.1M phosphate buffer (PB, pH7.4). Human slides were incubated in DAPI (ACD) for 1 min before being washed, air dried, and cover-slipped with Prolong Gold Antifade mounting medium.
Table 2.
Summary table of Advanced Cell Diagnostics (ACD) RNAscope probes.
| mRNA | Gene name | ACD Probe Cat No. |
|---|---|---|
| CALCA | Calcitonin gene-related peptide | 605551-C2 |
| P2RX3 | Purinergic Receptor P2X3 | 406301-C3 |
| SCN10A | Sodium Voltage-Gated Channel Alpha Subunit 10; Nav1.8 | 406291-C2 |
| MRGPRD | MAS Related GPR Family Member D | 524871-C3 |
| NPPB | Natriuretic Peptide B | 448511-C2 |
| ACE2 | Angiotensin I Converting Enzyme 2 | 848151-C1 |
Tissue Quality Check
All tissues were checked for RNA quality by using a positive control probe cocktail (ACD) which contains probes for high, medium and low-expressing mRNAs that are present in all cells (ubiquitin C > Peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). All tissues showed signal for all 3 positive control probes (Supplementary Fig 1). A negative control probe against the bacterial DapB gene (ACD) was used to check for non-specific/background label.
Image Analysis
DRG sections were imaged on an Olympus FV3000 confocal microscope at 20X or 40X magnification. For the CALCA/P2RX3/ACE2 and SCN10A/ACE2 experiments, 3–4 20X images were acquired of each human DRG section, and 3–4 sections were imaged per human donor. For the MRGPRD/NPPB/ACE2 experiments, 8 40x images were acquired of each human DRG section, and 3–4 sections were imaged per donor. The sections imaged were chosen at random, but preference was given to sections that did not have any sectioning artifact, or sections that encompassed the entire DRG bulb. The acquisition parameters were set based on guidelines for the FV3000 provided by Olympus. In particular, the gain was kept at the default setting 1, HV ≤ 600, offset = 4 (based on HI-LO settings; doesn’t change between experiments), and laser power ≤ 10% (but generally the laser power was ≤ 5% for our experiments). The raw image files were brightened and contrasted in Olympus CellSens software (v1.18), and then analyzed manually one cell at a time for expression of each gene target. Cell diameters were measured using the polyline tool. Total neuron counts for human samples were acquired by counting all of the probe-labeled neurons and all neurons that were clearly outlined by DAPI (satellite cell) signal and contained lipofuscin in the overlay image.
Large globular structures and/or signal that auto-fluoresced in all 3 channels (488, 550, and 647; appears white in the overlay images) was considered to be background lipofuscin and was not analyzed. Aside from adjusting brightness/contrast, we performed no digital image processing to subtract background. We attempted to optimize automated imaging analysis tools for our purposes, but these tools were designed to work with fresh, low background rodent tissues, not human samples taken from older organ donors. As such, we chose to implement a manual approach in our imaging analysis in which we used our own judgement of the negative/positive controls and target images to assess mRNA label. Images were not analyzed in a blinded fashion.
Immunohistochemistry
DRGs were sectioned, fixed, and dehydrated as described above. Hydrophobic boundaries were drawn around each section, and then the slides were submerged in blocking buffer for 1 hour at room temperature. For sections stained using goat ACE2 primary antibody (details below), the blocking buffer was 10% Normal Donkey Serum (Sigma Aldrich; Cat # S30-M), 0.3% Triton X-100 (Sigma Aldrich; Cat # X100) in 0.1M PB and for sections stained using rabbit ACE2 primary antibody (details below), the blocking buffer was 10% Normal Goat Serum (Atlanta Biologicals; Cat # S13150H), 0.3% Triton X-100 in 0.1M PB. Sections were then incubated overnight at 4°C with one of the following primary antibody cocktails: 1) goat-anti-ACE2 (5 μg/mL; R&D Systems; AF933; RRID:AB_355722) and rabbit-anti-peripherin (1:1000; Sigma Aldrich; SAB4502419; RRID:AB_10746677) diluted in donkey serum blocking buffer or, 2) rabbit-anti-ACE2 (1:1000; Abcam; ab15348; RRID:AB_301861) and chicken-anti-peripherin (1:1000; EnCor Biotechnology; CPCA-Peri; RRID:AB_2284443) diluted in goat serum blocking buffer. The next day, sections were washed in 0.1M PB and then incubated in their respective secondary antibody cocktails for 1 hour at room temperature. The secondary antibody cocktails used were 1) donkey-anti-goat H&L 555 (1:2000; Thermo Fisher Scientific; Cat # A-21432; RRID: AB_2535853), donkey-anti-rabbit H&L 488 (1:2000; Thermo Fisher Scientific; Cat# A32790; RRID:AB_2762833) and DAPI (1:5000; Cayman Chemical; Cat # 14285) in donkey blocking buffer, and 2) goat-anti-rabbit H&L 555 (1:2000; Thermo Fisher; A21428; RRID:AB_2535849), goat-anti-chicken H&L 488 (1:2000; Thermo Fisher; A-11039; RRID:AB_2534096) and DAPI (1:5000) in goat blocking buffer. Sections were washed in 0.1M PB, air dried and coverslipped with Prolong Gold Antifade reagent. DRG sections were imaged on an Olympus FV3000 confocal microscope at 10X or 40X magnification.
Western blot
Frozen sections of lumbar DRG and spinal cord (donor 2) were submerged in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, pH 8.0, and 1% Triton X-100) containing protease and phosphatase inhibitors (Sigma-Aldrich; Cat #s P8340, P0044, P5726), and then sonicated briefly. Samples were centrifuged at 14,000 rpm for 15 min at 4°C and the supernatants were transferred to new tubes. Protein concentration was assessed using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; Cat # 23225). 10 μg of protein was loaded into each lane of a 10% SDS-PAGE gel and then separated using electrophoresis. Proteins were transferred to a 0.45 μm PVDF membrane (Millipore) at 30 V overnight at 4°C. The next day, the membranes were blocked for 1 hour at room temperature. The blocking buffer for the goat-anti-ACE2 antibody was 5% Normal Donkey Serum diluted in 1X Tris buffer solution containing Tween 20 (TTBS) and the blocking buffer used for the rabbit-anti-ACE2 antibody was 5% Bovine Serum Albumin (BSA; Biopharm; Cat # 71–040) in TTBS. Membranes were then incubated in their respective primary antibodies overnight at 4°C, either goat-anti-ACE2 at 5 μg/mL in donkey blocking buffer or rabbit-anti-ACE2 at 1:1000 in BSA blocking buffer. The following day, membranes were washed 3 times in TTBS for 5 min each, then incubated with their corresponding secondary antibody, either donkey-anti-goat HRP (1:5000; Jackson ImmunoResearch; Cat # 705-035-003) or goat-anti-rabbit HRP (1:10000; Jackson ImmunoResearch; Cat # 111-035-003). Membranes were then washed with TTBS 5 times for 5 min each. Signals were detected using Immobilon Western Chemiluminescent HRP substrate (Millipore). Bands were visualized using a Bio-Rad ChemiDoc Touch.
RNA Sequencing Sources
We used previously published RNA sequencing datasets to generate Table 2 and Supplementary file 1. Human DRG sequencing data was described previously[20; 23] and mouse single cell sequencing data from DRG was described previously[30; 34].
Data Analysis
Graphs were generated using GraphPad Prism version 7.01 (GraphPad Software, Inc. San Diego, CA USA). Given that the percentage of ACE2-expressing neurons was assessed in each experiment, we averaged these numbers for each donor to generate the final data values. A total of 2,224 neurons were analyzed between all three donors and all three experiments. A relative frequency distribution histogram with a fitted Gaussian distribution curve was generated using the diameters of all ACE2-positive neurons detected in all experiments.
Results
We conducted RNAscope for ACE2 mRNA on human DRG to assess cellular expression of the SARS-CoV-2 receptor. We found that ACE2 mRNA was localized to some neurons, many of which expressed calcitonin gene-related peptide gene, CALCA (Fig 1A–B), and/or the P2X purinergic ion channel type 3 receptor gene, P2RX3. Both CALCA and P2RX3 have been widely utilized to delineate nociceptor subpopulations in rodent DRG [4]. Expression of ACE2 was consistent in lumbar DRG from a female organ donor (Table 2, demographic data for all DRGs analyzed) and in two thoracic DRGs taken from male patients undergoing vertebrectomy surgery (Fig 1B). Across samples, ACE2 mRNA was present in 19.8 – 25.4% of all sensory neurons (Fig 1C), however, mRNA puncta for ACE2 was sparse suggesting a low level of expression. Consistent with mouse single cell RNA sequencing data, ACE2 neurons co-expressed MRGPRD and NPPB mRNA (Fig 1D). They also co-expressed Nav1.8 (SCN10A gene) mRNA (Fig 1E). SCN10A is a nociceptor-specific marker in rodent DRG at the protein and mRNA levels [1; 9; 26]; therefore, these findings are consistent with the notion that ACE2 mRNA is mostly expressed by nociceptors in the human DRG. Summary statistics for the ACE2 population and ACE2 mRNA-positive neuron cell sizes are shown in Fig 1F and G.
Figure 1. Distribution of ACE2 mRNA in human dorsal root ganglia.
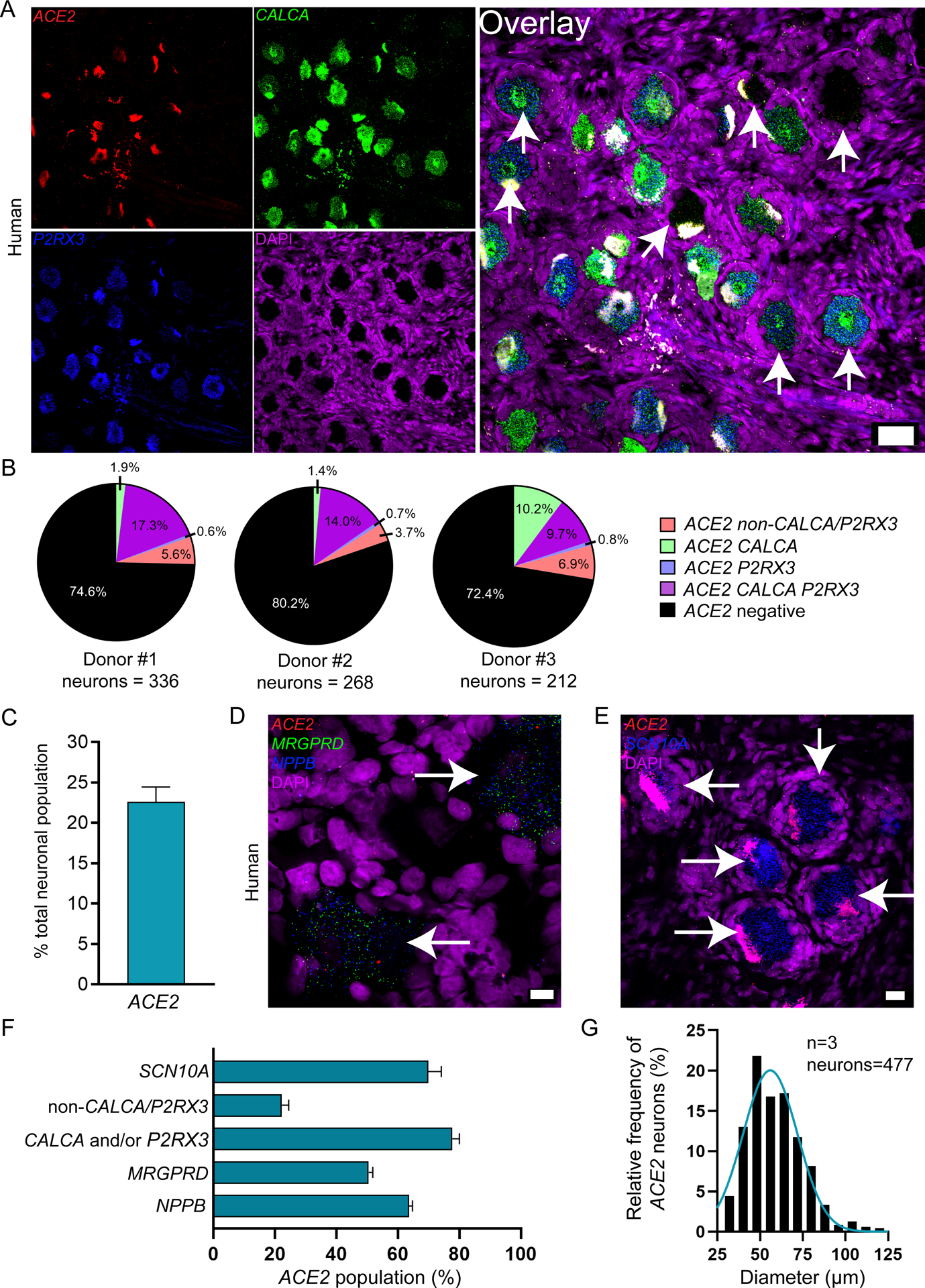
A) Representative 20x images of human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and ACE2 (red) mRNA and co-stained with DAPI (purple). Lipofuscin (globular structures) that autofluoresced in all 3 channels and appear white in the overlay image were not analyzed as this is background signal that is present in all human nervous tissue. B) Pie charts showing distribution of ACE2 neuronal subpopulations for each human donor DRG. ACE2 was found in neurons expressing solely CALCA (green), solely P2RX3 (blue) or both (purple), and a smaller population of neurons negative for both CALCA and P2RX3 (red). Some mRNA was detected in non-neuronal cells that are likely vascular or satellite glia but not immune cells (Supplementary file 1). C) ACE2 was expressed in 22.6% of all sensory neurons in human DRG. D) Representative 100x overlay image showing MRGPRD (green), NPPB (blue), ACE2 (red) and DAPI (purple) signal in human DRG. E) Representative 40x overlay image showing SCN10A (blue), ACE2 (red) and DAPI (purple) signal in human DRG. F) 69.9% of ACE2 neurons expressed SCN10A, 77.7% expressed CALCA and/or P2RX3, 22.2% were negative for CALCA and/or P2RX3, 50.4% expressed MRGPRD, and 63.6% expressed NPPB. Scale bars: 20x = 50μm, 40x = 20μm, 100x = 10μm. White arrows point toward ACE2-positive neurons.
We next investigated whether ACE2 protein was present in human DRG. We conducted immunohistochemistry and western blot analysis of human lumbar and thoracic DRGs using two commercially available ACE2 antibodies (Abcam ab15348 and R&D Systems AF933) which are both extensively cited (see product datasheets for publication lists) and recently utilized in several SARS-CoV-2-related research papers [13; 15; 19; 28; 35]. It is important to note that these antibodies are both polyclonals and have not been knock-out validated, therefore, their reliability as ACE2 specific markers is unverified using mouse gene knockout. To our knowledge, no commercial ACE2 antibody has been validated for specificity in human tissue. We observed similar protein staining for both antibodies in both lumbar and thoracic human DRGs which was found in both neurons and non-neuronal cells including some satellite glial cells (Fig 2A–C and Supplementary Fig 2A–C). Both antibodies gave several non-specific bands on a western blot, however, a band at the predicted molecular weight for ACE2 was observed for both antibodies (Fig 2D and Supplementary Fig 2D). The western blot signal for ab15348 was cleaner and matched the expected differential expression of ACE2 between human spinal cord and DRG in which the mRNA is virtually undetectable in spinal cord [17; 24], but present in DRG (Figure 1). These findings support the notion that ACE2 protein is found in human DRG where it is likely secreted, binding to membranes of surrounding cells, as observed in human brain organoids [2; 28].
Figure 2. ACE2 protein is detected in human dorsal root ganglia using the abcam ab15348 antibody.
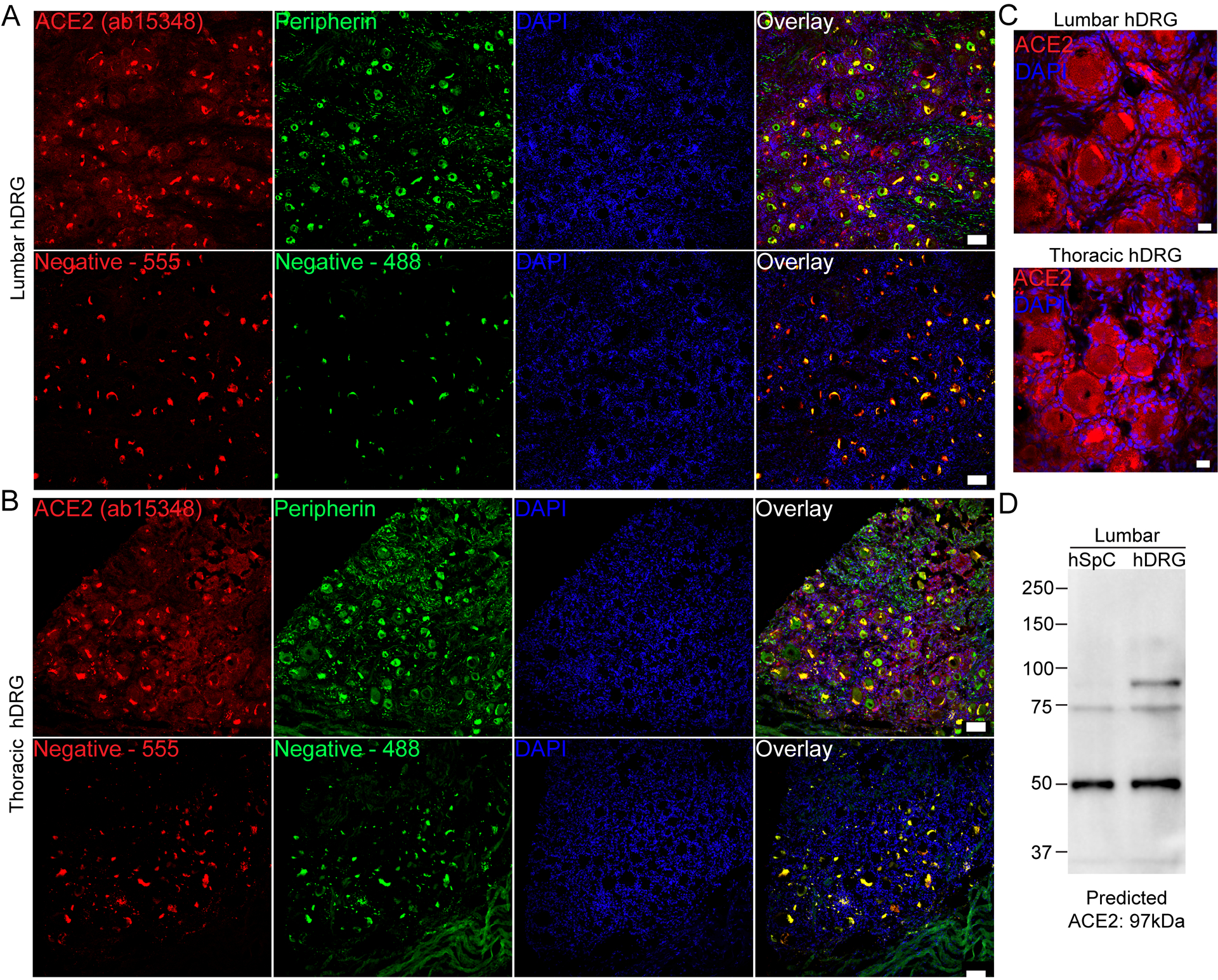
A) Representative 10x images of protein staining for ACE2 (red; Abcam ab15348), peripherin (green; a sensory neuron marker) and DAPI (blue) in lumbar and B) thoracic human dorsal root ganglia. C) 40x images showing ACE2 signal in both neurons and non-neuronal cells including satellite glial cells. D) Western blot of ACE2 using ab15348 on lumbar spinal cord and DRG revealed a strong band at the predicted molecular weight of ACE2 protein in human DRG but not spinal cord, and other non-specific bands. Scale bars: 10x = 100μm, 40x = 20μm.
Having established that the major receptor for SARS-CoV-2 is likely expressed by a specific subset of human nociceptors, we sought to use existing sequencing datasets to assess expression of other SCARFs [27] in human DRG. We grouped these into receptors, proteases, replication factors, trafficking factors (shown only in supplementary file 1 because they were all ubiquitous) and restriction factors (Table 3 and supplementary file 1) as described in other surveys of RNA sequencing databases, none of which included DRG. While ACE2 expression is low in these bulk RNA sequencing datasets, it was reliably detectable above 1 transcript per million (TPM) in certain thoracic DRG samples (Supplementary file 1). Some other potential receptors for SARS and MERS viruses were also found in human DRG and expressed in mouse DRG neurons, suggesting they may also be neuronally expressed in human DRG. Among proteases, the two main proteases for the SARS-CoV-2 spike protein priming, TMPRSS2 and FURIN, were robustly expressed in human DRG and they were also expressed by mouse DRG neurons (Table 3). Most other SCARFs were also expressed in human DRG and, again, their expression in mouse DRG neurons suggests that most of them are likely to also be found in human DRG neurons. Many of these genes showed high variation between samples, an effect that may explain variable effects of SARS-CoV-2 on the nervous system in patients.
Table 3. Expression of SCARF genes in human DRG from bulk RNA sequencing experiments.
Genes are shown with mean +/− standard deviation (SD) expression in TPMs in 3 lumbar DRG samples and 5 thoracic DRG samples. Mouse DRG neuronal expression denotes detection in the neurofilament (NF), peptidergic (PEP), nonpeptidergic (NP) and tyrosine hydroxylase (TH) subsets of DRG neurons. Human DRG data is from [20; 23] and mouse single cell data is from [30]. ND: Not Detectable.
| Human gene | Lumbar DRG (mean +/− SD) TPM | Thoracic DRG (mean +/− SD) TPM | Neuronal expression in mouse DRG? | SCARF Classification |
|---|---|---|---|---|
| ACE2 | 0.09 +/− 0.02 | 0.94 +/− 0.88 | NF | Receptors |
| BSG | 304.82 +/− 82.3 | 239.42 +/− 79.23 | PEP, NP, NF, TH | Receptors |
| ANPEP | 14.56 +/− 7.27 | 13.73 +/− 4.26 | PEP, TH | Receptors |
| CD209 | 0.57 +/− 0.43 | 1.60 +/− 1.28 | Multiple mouse orthologs | Receptors |
| CLEC4G | 0.79 +/− 0.34 | 0.22 +/− 0.45 | ND | Receptors |
| CLEC4M | 0.02 +/− 0.03 | 0.19 +/− 0.17 | No ortholog | Receptors |
| DPP4 | 0.34 +/− 0.23 | 3.75 +/− 4.05 | ND | Receptors |
| TMPRSS2 | 3.05 +/− 1.04 | 6.69 +/− 2.65 | NF, TH | Proteases |
| TMPRSS11A | ND | 0.06 +/− 0.13 | NF, TH | Proteases |
| TMPRSS11B | ND | 0.03 +/− 0.04 | No ortholog | Proteases |
| FURIN | 20.86 +/− 5.69 | 34.11 +/− 10.44 | PEP, NP, NF, TH | Proteases |
| TMPRSS4 | 0.77 +/− 0.30 | 0.08 +/− 0.09 | NF | Proteases |
| CTSL | 154.53 +/− 21.71 | 105.19 +/− 13.36 | PEP, NP, NF, TH | Proteases |
| CTSB | 1116.17 +/− 132.45 | 697.50 +/− 96.96 | PEP, NP, NF, TH | Proteases |
| TOP3B | 6.46 +/− 0.88 | 18.93 +/− 5.45 | PEP, NP, NF, TH | Replication Factors |
| MADP1 | 35.13 +/− 8.08 | 32.74 +/− 7.44 | PEP, NP, NF, TH | Replication Factors |
| LY6E | 446.65 +/− 234.34 | 218.17 +/− 161.15 | PEP, NP, NF, TH | Restriction Factors |
| IFITM1 | 828.53 +/− 425.04 | 322.86 +/− 104.16 | PEP, NP, NF, TH | Restriction Factors |
| IFITM2 | 880.05 +/− 340.01 | 2241.85 +/− 91.24 | PEP, NP, NF, TH | Restriction Factors |
| IFITM3 | 2339.37 +/− 943.43 | 744.00 +/− 224.85 | PEP, NP, NF, TH | Restriction Factors |
| CALCA | 1634.46 +/− 665.44 | 524.86 +/− 111.60 | PEP, NP, NF, TH | Neuronal Markers |
| P2RX3 | 44.65 +/− 42.69 | 144.54 +/− 47.60 | PEP, NP, NF, TH | Neuronal Markers |
| NPPB | 41.20 +/− 16.37 | 21.29 +/− 8.99 | NP, PEP | Neuronal Markers |
| MRGPRD | 3.01 +/− 4.62 | 10.47 +/− 7.64 | PEP, NP, NF, TH | Neuronal Markers |
Discussion
Our work highlights neuronal expression of ACE2 in a select subset of nociceptors that express CALCA, P2RX3, MRGPRD, NPPB and SCN10A. While we cannot state with certainty the anatomical projections of these neurons because tracing studies cannot be done in humans, the neurochemical signature of these neurons is consistent with nociceptors that form free nerve endings in the skin [38], luminal organs [12] and meninges [31]. Therefore, one potential consequence of this ACE2 expression could be infection of nociceptors through the nasal passages, cornea, or upper or lower airway. To this end, we noted higher expression of ACE2 in thoracic DRGs, and these DRGs contain nociceptors that innervate the lungs [14; 29], a major site for proliferation of the SARS-CoV-2 virus [36]. It is now clear that many COVID-19 patients have persistent symptoms lasting for months after initial infection. These symptoms include joint and chest pain, cough, headache and dyspnea [7], symptoms that involve activation of nociceptors. Sensory neuronal infection by SARS-CoV-2 may be a causative factor in some of these persistent symptoms.
An interesting early symptom of SARS-CoV-2 is chemesthesis [21]. This symptom is less frequent than loss of smell and cannot be explained via the same mechanisms as anosmia [5] because the sensing of noxious chemicals in the oral cavity is mediated by nociceptors. Our data suggests that a possible explanation of chemesthesis in COVID-19 is silencing of nerve endings by the initial viral infection. If pulmonary afferents are also infected by SARS-CoV-2, as our data would predict, this could have important consequences for disease severity. Previous studies have shown that ablation of airway nociceptors increases the severity of respiratory disease due to a loss of the trophic action of CGRP within the airway [3]. Widespread, early silencing of nociceptors innervating the oral and nasal cavity as well as the upper and lower airway may have important impacts on COVID-19 disease severity.
There are many currently unexplained neurological features in COVID-19 patients [16; 18], including persistent symptoms like joint pain and headache linked to nociceptor function [7]. It is mysterious how the virus enters the nervous system as most neurons do not express ACE2 [11; 27]. Our work presents a plausible entry point for the peripheral nervous system – through a select subset of ACE2 expressing nociceptors. While our findings can be taken as putative evidence for this idea, ACE2 mRNA expression is very low in these neurons, and we have not tested whether ACE2 protein can be detected in free nerve endings in appropriate organs, or whether such expression can be harnessed by the virus to gain entry. We did observe ACE2 protein expression within the human DRG. Studies in brain organoids demonstrate that SARS-CoV-2 can infect neurons, and the virus does so in an ACE2-dependent fashion [28]. Interestingly, ACE2 mRNA expression is low in human brain organoids but this low mRNA expression leads to widespread ACE2 protein staining within the organoids [28]. We obtained similar results using 2 independent antibodies, one of which was used in the brain organoid study [28]. Therefore, our results in human DRG are consistent with the idea that low-level mRNA expression can lead to broader protein expression, likely because ACE2 is a secreted protein that is bound to the extracellular part of the cellular plasma membrane [2]. This finding, coupled with the strong expression of many SCARFs in human DRG, suggests that sensory neurons can be targeted by SARS-CoV-2.
More work will be needed to better understand the impact of SARS-CoV-2 on nociceptors. Our results lay a foundation for beginning to understand the symptoms, pathology and long-term outcomes of COVID-19 as they relate to the sensory nervous system. As this pandemic continues to spread it will be important to study these neurobiological findings in the context of long-term patient outcomes, in particular as they relate to pain and other forms of chemo-sensation that rely on nociceptors.
Supplementary Material
Supplemental Figure 1 Caption. mRNA quality check for each human donor DRG. All tissues were checked for mRNA quality by using a positive control probe cocktail (ACD) which contains probes for high, medium and low-expressing mRNAs that are present in all cells (ubiquitin C > Peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). All DRGs showed signal for all 3 positive control probes indicating that the mRNA quality is excellent and that low-expressing transcripts such as ACE2 should be able to be detected. 20x scale bar = 50 μm.
Supplemental Figure 2 Caption. ACE2 protein is detected in human dorsal root ganglia using the R&D Systems AF933 antibody. A) Representative 10x images of protein staining for ACE2 (red; R&D Systems AF933), peripherin (green; a sensory neuron marker) and DAPI (blue) in lumbar and B) thoracic human dorsal root ganglia. C) 40x images showing ACE2 signal in both neurons and non-neuronal cells including satellite glial cells. D) Western blot of ACE2 using AF933 on lumbar spinal cord and DRG revealed a band at the predicted molecular weight of ACE2 protein in both human DRG and spinal cord, but also several other non-specific bands. Scale bars: 10x = 100μm, 40x = 20μm.
Acknowledgements
The authors declare no conflicts of interest for this work. The work was funded by NIH grants NS111929 to PMD and TJP and NS 065926 to TJP. The UTD authors dedicate this work to the memory of our friend and colleague Zhiyue “Mark” Wang. We are grateful to the patients and the families of organ donors who provided the DRGs for this study.
References Cited
- [1].Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 1996;379(6562):257–262. [DOI] [PubMed] [Google Scholar]
- [2].Antalis TM, Conway GD, Peroutka RJ, Buzza MS. Membrane-anchored proteases in endothelial cell biology. Curr Opin Hematol 2016;23(3):243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Baral P, Umans BD, Li L, Wallrapp A, Bist M, Kirschbaum T, Wei Y, Zhou Y, Kuchroo VK, Burkett PR, Yipp BG, Liberles SD, Chiu IM. Nociceptor sensory neurons suppress neutrophil and gammadelta T cell responses in bacterial lung infections and lethal pneumonia. Nat Med 2018;24(4):417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009;139(2):267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brann DH, Tsukahara T, Weinreb C, Lipovsek M, Van den Berge K, Gong B, Chance R, Macaulay IC, Chou H-j, Fletcher R, Das D, Street K, de Bezieux HR, Choi Y-G, Risso D, Dudoit S, Purdom E, Mill JS, Hachem RA, Matsunami H, Logan DW, Goldstein BJ, Grubb MS, Ngai J, Datta SR. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. bioRxiv 2020:2020.2003.2025.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Butowt R, Bilinska K. SARS-CoV-2: Olfaction, Brain Infection, and the Urgent Need for Clinical Samples Allowing Earlier Virus Detection. ACS Chem Neurosci 2020;11(9):1200–1203. [DOI] [PubMed] [Google Scholar]
- [7].Carfi A, Bernabei R, Landi F, Gemelli Against C-P-ACSG. Persistent Symptoms in Patients After Acute COVID-19. JAMA 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dib-Hajj SD, Tyrrell L, Cummins TR, Black JA, Wood PM, Waxman SG. Two tetrodotoxin-resistant sodium channels in human dorsal root ganglion neurons. FEBS Lett 1999;462(1–2):117–120. [DOI] [PubMed] [Google Scholar]
- [10].Hernandez-Fernandez F, Valencia HS, Barbella-Aponte RA, Collado-Jimenez R, Ayo-Martin O, Barrena C, Molina-Nuevo JD, Garcia-Garcia J, Lozano-Setien E, Alcahut-Rodriguez C, Martinez-Martin A, Sanchez-Lopez A, Segura T. Cerebrovascular disease in patients with COVID-19: neuroimaging, histological and clinical description. Brain 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. bioRxiv 2020:2020.2003.2031.016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hockley JRF, Taylor TS, Callejo G, Wilbrey AL, Gutteridge A, Bach K, Winchester WJ, Bulmer DC, McMurray G, Smith ESJ. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181(2):271–280 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kummer W, Fischer A, Kurkowski R, Heym C. The sensory and sympathetic innervation of guinea-pig lung and trachea as studied by retrograde neuronal tracing and double-labelling immunohistochemistry. Neuroscience 1992;49(3):715–737. [DOI] [PubMed] [Google Scholar]
- [15].Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, Dorscheid DR, Sin DD. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J 2020;55(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segre AV, Djebali S, Niarchou A, Consortium GT, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigo R. Human genomics. The human transcriptome across tissues and individuals. Science 2015;348(6235):660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Montalvan V, Lee J, Bueso T, De Toledo J, Rivas K. Neurological manifestations of COVID-19 and other coronavirus infections: A systematic review. Clin Neurol Neurosurg 2020;194:105921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nicin L, Abplanalp WT, Mellentin H, Kattih B, Tombor L, John D, Schmitto JD, Heineke J, Emrich F, Arsalan M, Holubec T, Walther T, Zeiher AM, Dimmeler S. Cell type-specific expression of the putative SARS-CoV-2 receptor ACE2 in human hearts. Eur Heart J 2020;41(19):1804–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, Dussor G, Kim TH, Price TJ, Dougherty PM. Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 2019;142(5):1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Parma V, Ohla K, Veldhuizen MG, Niv MY, Kelly CE, Bakke AJ, Cooper KW, Bouysset C, Pirastu N, Dibattista M, Kaur R, Liuzza MT, Pepino MY, Schöpf V, Pereda-Loth V, Olsson SB, Gerkin RC, Domínguez PR, Albayay J, Farruggia MC, Bhutani S, Fjaeldstad AW, Kumar R, Menini A, Bensafi M, Sandell M, Konstantinidis I, Di Pizio A, Genovese F, Öztürk L, Thomas-Danguin T, Frasnelli J, Boesveldt S, Saatci Ö, Saraiva LR, Lin C, Golebiowski J, Hwang L-D, Ozdener MH, Guàrdia MD, Laudamiel C, Ritchie M, Havlícek J, Pierron D, Roura E, Navarro M, Nolden AA, Lim J, Whitcroft K, Colquitt LR, Ferdenzi C, Brindha EV, Altundag A, Macchi A, Nunez-Parra A, Patel ZM, Fiorucci S, Philpott CM, Smith BC, Lundström JN, Mucignat C, Parker JK, van den Brink M, Schmuker M, Fischmeister FPS, Heinbockel T, Shields VDC, Faraji F, Enrique Santamaría E, Fredborg WEA, Morini G, Olofsson JK, Jalessi M, Karni N, D’Errico A, Alizadeh R, Pellegrino R, Meyer P, Huart C, Chen B, Soler GM, Alwashahi MK, Abdulrahman O, Welge-Lüssen A, Dalton P, Freiherr J, Yan CH, de Groot JHB, Voznessenskaya VV, Klein H, Chen J, Okamoto M, Sell EA, Singh PB, Walsh-Messinger J, Archer NS, Koyama S, Deary V, Roberts SC, Yanik H, Albayrak S, Novákov LM, Croijmans I, Mazal PP, Moein ST, Margulis E, Mignot C, Mariño S, Georgiev D, Kaushik PK, Malnic B, Wang H, Seyed-Allaei S, Yoluk N, Razzaghi S, Justice JM, Restrepo D, Hsieh JW, Reed DR, Hummel T, Munger SD, Hayes JE. More than smell. COVID-19 is associated with severe impairment of smell, taste, and chemesthesis. medRxiv 2020:2020.2005.2004.20090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Paterson RW, Brown RL, Benjamin L, Nortley R, Wiethoff S, Bharucha T, Jayaseelan DL, Kumar G, Raftopoulos RE, Zambreanu L, Vivekanandam V, Khoo A, Geraldes R, Chinthapalli K, Boyd E, Tuzlali H, Price G, Christofi G, Morrow J, McNamara P, McLoughlin B, Lim ST, Mehta PR, Levee V, Keddie S, Yong W, Trip SA, Foulkes AJM, Hotton G, Miller TD, Everitt AD, Carswell C, Davies NWS, Yoong M, Attwell D, Sreedharan J, Silber E, Schott JM, Chandratheva A, Perry RJ, Simister R, Checkley A, Longley N, Farmer SF, Carletti F, Houlihan C, Thom M, Lunn MP, Spillane J, Howard R, Vincent A, Werring DJ, Hoskote C, Jager HR, Manji H, Zandi MS, Neurology UCLQSNHf, Neurosurgery C-SG. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018;159(7):1325–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ray PR, Khan J, Wangzhou A, Tavares-Ferreira D, Akopian AN, Dussor G, Price TJ. Transcriptome Analysis of the Human Tibial Nerve Identifies Sexually Dimorphic Expression of Genes Involved in Pain, Inflammation, and Neuro-Immunity. Front Mol Neurosci 2019;12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ray PR, Wangzhou A, Ghneim N, Yousuf MS, Paige C, Tavares-Ferreira D, Mwirigi JM, Shiers S, Sankaranarayanan I, McFarland AJ, Neerukonda SV, Davidson S, Dussor G, Burton MD, Price TJ. A pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain Behav Immun 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sangameswaran L, Delgado SG, Fish LM, Koch BD, Jakeman LB, Stewart GR, Sze P, Hunter JC, Eglen RM, Herman RC. Structure and function of a novel voltage-gated, tetrodotoxin-resistant sodium channel specific to sensory neurons. J Biol Chem 1996;271(11):5953–5956. [DOI] [PubMed] [Google Scholar]
- [27].Singh M, Bansal V, Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. bioRxiv 2020:2020.2005.2008.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Song E, Zhang C, Israelow B, Lu P, Weizman O-E, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Wilen CB, Horvath TL, Louvi A, Farhadian SF, Bilguvar K, Iwasaki A. Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model. bioRxiv 2020:2020.2006.2025.169946. [Google Scholar]
- [29].Springall DR, Cadieux A, Oliveira H, Su H, Royston D, Polak JM. Retrograde tracing shows that CGRP-immunoreactive nerves of rat trachea and lung originate from vagal and dorsal root ganglia. J Auton Nerv Syst 1987;20(2):155–166. [DOI] [PubMed] [Google Scholar]
- [30].Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18(1):145–153. [DOI] [PubMed] [Google Scholar]
- [31].von Buchholtz LJ, Lam RM, Emrick JJ, Chesler AT, Ryba NJP. Assigning transcriptomic class in the trigeminal ganglion using multiplex in situ hybridization and machine learning. Pain 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of virology 2020;94(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020;367(6485):1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S. Molecular Architecture of the Mouse Nervous System. Cell 2018;174(4):999–1014.e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao X, Chen D, Szabla R, Zheng M, Li G, Du P, Zheng S, Li X, Song C, Li R, Guo J-T, Junop M, Zeng H, Lin H. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. bioRxiv 2020:2020.2004.2019.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395(10229):1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J, lung-network@humancellatlas.org HCALBNEa, Network HCALB. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 2005;45(1):17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Caption. mRNA quality check for each human donor DRG. All tissues were checked for mRNA quality by using a positive control probe cocktail (ACD) which contains probes for high, medium and low-expressing mRNAs that are present in all cells (ubiquitin C > Peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). All DRGs showed signal for all 3 positive control probes indicating that the mRNA quality is excellent and that low-expressing transcripts such as ACE2 should be able to be detected. 20x scale bar = 50 μm.
Supplemental Figure 2 Caption. ACE2 protein is detected in human dorsal root ganglia using the R&D Systems AF933 antibody. A) Representative 10x images of protein staining for ACE2 (red; R&D Systems AF933), peripherin (green; a sensory neuron marker) and DAPI (blue) in lumbar and B) thoracic human dorsal root ganglia. C) 40x images showing ACE2 signal in both neurons and non-neuronal cells including satellite glial cells. D) Western blot of ACE2 using AF933 on lumbar spinal cord and DRG revealed a band at the predicted molecular weight of ACE2 protein in both human DRG and spinal cord, but also several other non-specific bands. Scale bars: 10x = 100μm, 40x = 20μm.


