Abstract
We investigated the biochemical and biophysical properties of one of the four alternative exon-encoded regions within the Drosophila myosin catalytic domain. This region is encoded by alternative exons 3a and 3b and includes part of the N-terminal β-barrel. Chimeric myosin constructs (IFI-3a and EMB-3b) were generated by exchanging the exon 3–encoded areas between native slow embryonic body wall (EMB) and fast indirect flight muscle myosin isoforms (IFI). We found that this exchange alters the kinetic properties of the myosin S1 head. The ADP release rate (k-D) in the absence of actin is completely reversed for each chimera compared with the native isoforms. Steady-state data also suggest a reciprocal shift, with basal and actin-activated ATPase activity of IFI-3a showing reduced values compared with wild-type (WT) IFI, whereas for EMB-3b these values are increased compared with wild-type (WT) EMB. In the presence of actin, ADP affinity (KAD) is unchanged for IFI-3a, compared with IFI, but ADP affinity for EMB-3b is increased, compared with EMB, and shifted toward IFI values. ATP-induced dissociation of acto-S1 (K1k+2) is reduced for both exon 3 chimeras. Homology modeling, combined with a recently reported crystal structure for Drosophila EMB, indicates that the exon 3–encoded region in the myosin head is part of the communication pathway between the nucleotide binding pocket (purine binding loop) and the essential light chain, emphasizing an important role for this variable N-terminal domain in regulating actomyosin crossbridge kinetics, in particular with respect to the force-sensing properties of myosin isoforms.
Keywords: muscle, myosin, kinetics, actin, fluorescence, homology modeling, force-sensing, protein structure-function, sequence alignment
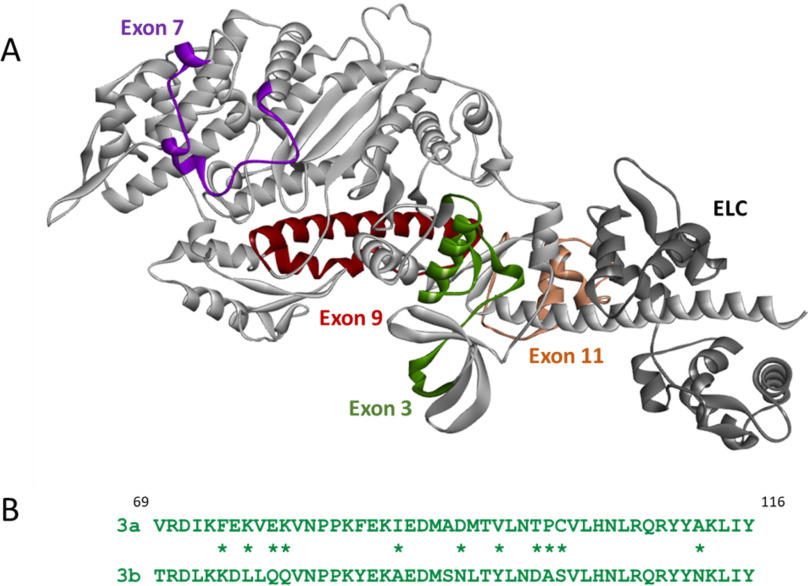
Muscle myosin isoforms display a large variety in kinetic properties and force production, despite their sequences being highly conserved. The various isoforms of Drosophila mel-an-o-gas-ter muscle myosin heavy chain (MHC) are encoded by a single gene (Mhc) and expressed using alternative splicing (1). The first four of the six alternative exon sets (exons 3, 7, 9, and 11) encode regions located in the myosin head domain (2), and the use of alternative domains in the myosin head allows for fine-tuning of myosin properties (see Fig. 1A). Two native myosin isoforms that differ in all four alternative regions in the head domain display very different kinetic and mechanical properties. The EMB (embryonic) myosin isoform is found in the embryonic body wall muscle, which is used for slow locomotion of the larvae, whereas the IFI (indirect flight muscle isoform) myosin is present in the muscle that can generate very high wing beat frequencies and enables flight. Transgenic expression of EMB in the indirect flight muscle resulted in loss of flight ability (3), and subsequent studies using isolated muscle fibers and/or myosin proteins confirmed the striking differences in kinetic and mechanical properties between IFI and EMB myosin isoforms (4–6). Exchange of the variable regions between IFI and EMB has been used as a strategy to estimate the effect of each alternative domain on muscle myosin kinetics and mechanics. Here we focus on the variable region near the N terminus of MHC, encoded by exon 3. Two alternative regions, encoded by exons 3a and 3b, are expressed in Drosophila myosin and their sequences are shown in Fig. 1B. EMB contains the region encoded by exon 3a, whereas the exon 3b encoded sequence is normally expressed within IFI. Previous work found that exchange of the exon 3 regions between IFI and EMB significantly changes the steady-state kinetic properties of both Drosophila muscle myosin isoforms. Inserting the exon 3a area into IFI resulted in significantly reduced ATPase rates and Vmax for IFI-3a (compared with IFI), whereas inserting the exon 3b region into EMB had surprisingly little effect on ATPase and Vmax of EMB-3b (compared with EMB) (7). The same study showed that in vitro actin sliding velocity was increased for EMB-3b, although not restored to IFI levels, and unaltered for IFI-3a, compared with wild type (WT).
Figure 1.
A, structure of the EMB isoform in the rigor-like conformation, with the location of the four variable regions in the myosin head indicated: exon 3 region (green), exon 7 region (purple), exon 9 region (dark red), and exon 11 region (light brown). The ELC is shown in dark gray (the coordinates of the EMB crystal structure (PDB ID: 5W1A) were used to generate this image). B, alternative sequences encoded by the exon 3a and 3b regions, with nonconservative differences denoted by asterisks: 3a is found in the slow EMB isoform and 3b is present in the fast IFI isoform.
Exchange of the exon 3 area also affects the mechanical properties of the indirect flight muscle. Flight ability of IFI-3a Drosophila is slightly decreased, compared with WT, whereas introduction of the exon 3b region in an embryonic background does not restore flight ability for EMB-3b Drosophila (7). A follow-up study found that IFI-3a Drosophila showed a reduction in both maximum power generation (Pmax) and optimal frequency for power production (fmax), whereas for EMB-3b Drosophila an increase in both Pmax and fmax was found (8). Based on these results it was suggested that the exon 3 region can influence at least two steps of the crossbridge cycle independently, thereby fine-tuning myosin muscle kinetics for optimal force generation. To understand the biochemical kinetics of various steps in the crossbridge cycle that are influenced by the exon 3 domain, we have now performed steady-state and transient kinetics measurements using the exon 3 chimeric myosin S1 isoforms (IFI-3a and EMB-3b). Our data show that exchange of this variable N-terminal area significantly alters the kinetic properties of the myosin S1 head. The catalytic activity kcat of the myosin S1 samples generated from all full-length molecules (IFI, EMB, EMB-3b, and IFI-3a) followed the trend of activities so far observed for the full-length molecules. Interestingly, the ADP release rate (k-D) in the absence of actin is completely reversed for both chimeras, compared with their WT backbones. In the presence of actin, the ADP affinity (KAD) is unchanged for IFI-3a, whereas ADP affinity for EMB-3b is increased and shifted toward IFI values. ATP-induced dissociation of acto-S1 (K1k+2) is reduced for both exon 3 chimeras. Detailed analysis of a recently published crystal structure for EMB (9), combined with homology modeling, indicates that the exon 3 encoded area is part of the communication pathway between the nucleotide binding pocket (purine binding loop) and the essential light chain, emphasizing an important role for exon 3 in regulating actomyosin crossbridge kinetics, in particular with respect to the force-sensing properties of the myosin isoforms.
Results
ATP-induced dissociation of acto-S1 (K1k+2) is reduced for both exon 3 mutants
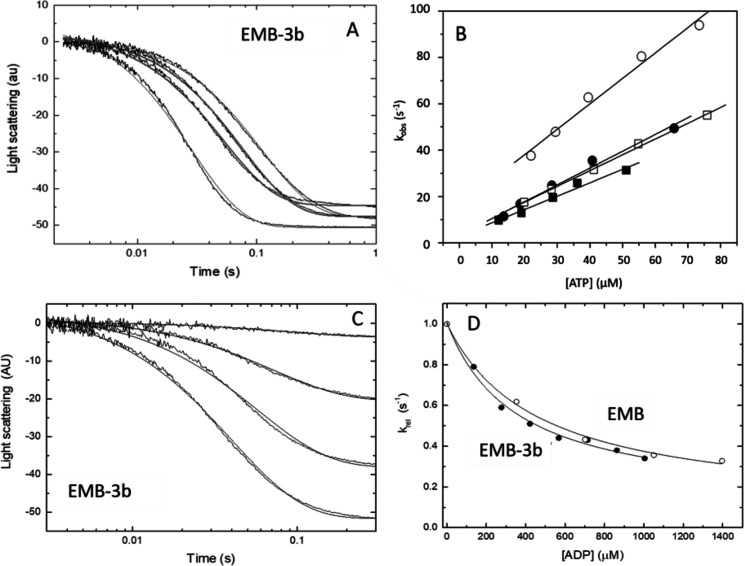
The ATP-induced dissociation of the acto-S1 complex was measured as described previously (10) using the flash photolysis method. Plotting the observed rate constant kobs versus ATP concentration allows the apparent second-order rate constant K1k+2 to be determined from the slope of the graph. Example traces of the measured light scattering data are shown in Fig. 2A for EMB-3b after ATP release. The analyzed data are depicted in Fig. 2B for the two exon 3 chimeras, together with IFI and EMB WT (see also Table 1). The measured K1k+2 value of EMB-3b (0.71 ± 0.12 μm−1s−1) is significantly lower compared with EMB (p < 0.007) (Table 1) but not significantly different from the value previously recorded for IFI (0.75 ± 0.08 μm−1s−1). For IFI-3a, a significantly lower K1k+2 value (0.66 ± 0.08 μm−1s−1) compared with IFI was recorded using an unpaired t test (p < 0.04) (Table 1), although this difference did not reach statistical significance using a more stringent one-way Welch's ANOVA that compared all four constructs. Thus, introduction of the IFI version of the exon 3 region into EMB shifted the ATP-induced dissociation of EMB-3b toward IFI values, whereas the presence of the EMB version of the exon 3 region did not shift IFI-3a toward WT EMB levels.
Figure 2.
ATP-induced dissociation (K1k+2) and ADP affinity (KAD) of the two myosin S1 exon 3 chimeras. A, example of light-scattering traces for acto-S1 dissociation with EMB-3b S1. B, the second-order rate constant (K1k+2) for the ATP-induced dissociation of S1 from actin is determined from a linear fit to the plot of the kobs versus [ATP] (see “Experimental Procedures”). The linear fits yielded values of 0.71 ± 0.12 μm−1 s−1 for EMB-3b (filled circles) as compared with 0.91 ± 0.13 μm−1 s−1 for EMB (open circles). For IFI-3a the linear fits yielded mean values of 0.66 ± 0.08 μm−1 s−1 (filled squares) as compared with 0.75 ± 0.08 μm−1 s−1 for IFI S1 (open squares). C, light-scattering traces for acto-S1 dissociation with EMB-3b S1 at various ADP concentrations. D, comparison of the affinity of ADP for acto-S1 (KAD) for EMB and EMB-3b, with the relative kobs (krel) values shown. Hyperbolic fits resulted in KAD values of 587 ± 48 μm for EMB (open circles) and 496 ± 79 μm for EMB-3b S1 (filled circles).
Table 1.
Kinetic parameters measured for myosin S1 isoforms and the exon 3 chimeras using flash photolysis
Values are mean ± S.D. based on a minimum of three preparations except kcat (n = 2). K1k+2 is the second-order rate constant for ATP-induced dissociation of acto-S1. KD and KAD are dissociation equilibrium constants determined by division of the dissociation rate constant by the association rate constant (e.g. KD = k-D/k+D). k-D and k-AD are the ADP dissociation rate constants in the absence and presence of actin, respectively. KAD/KD is the thermodynamic coupling constant describing the relationship between actin and ADP affinities. kcat is the catalytic activity.
| IFI† | EMB† | IFI-3a | EMB-3b | |
|---|---|---|---|---|
| K1k+2 (μm−1s−1) | 0.75 ± 0.08 | 0.91 ± 0.13 | 0.66 ± 0.08a | 0.71 ± 0.12b |
| KAD (μm) | 409 ± 26 | 587 ± 48 | 409 ± 18 | 496 ± 79c |
| k-AD (s−1)d | 4090 | 5870 | 4090 | 4960 |
| KD (μm)e | 7.5 | 1.8 | 2.1 | 7.0 |
| k-D (s−1) | 7.5 ± 1.3 | 1.8 ± 0.3 | 2.1 ± 0.41f | 7.0 ± 1.3g |
| KAD/KD | 54.5 | 326 | 194 | 70.9 |
| kcat (s−1)h | 0.170 ± 0.006i | 0.028 ± 0.006i | 0.057 ± 0.003 | 0.021 ± 0.009 |
†Data are from Ref. 5, except kcat.
aStatistically different compared with IFI, p < 0.04 by t test, but did not reach statistical significance using Welch's one-way ANOVA.
bStatistically different compared with EMB, p < 0.007.
cStatistically different compared with EMB, p < 0.04 by t test, but did not reach statistical significance using Welch's one-way ANOVA.
dData are estimated from KAD assuming an association rate constant of 107 M−1 s−1.
eData are estimated from k-D assuming an association rate constant of 106 m−1 s−1.
fStatistically different compared with IFI, p < 0.0001.
gStatistically different compared with EMB, p < 0.0001.
hActin is not at saturating conditions.
iData from Ref. 10.
ADP-affinity (KAD) is unchanged for IFI-3a but increased for EMB-3b compared with WT
The ADP affinity for S1 in the presence of actin, described by the equilibrium dissociation constant KAD, was determined according to established methods (5). ATP-induced dissociation of acto-S1 was measured in the presence of increasing amounts of ADP. Fig. 2C shows example traces of the light scattering data for EMB-3b, recorded with increasing ADP concentration. The amplitude of the signal drops as the ADP concentration increases. From the light scattering signals, kobs can be obtained by fitting to a single exponential and plotting kobs versus ADP concentration, which allows KAD to be estimated. The data show that the measured value for the embryonic chimera EMB-3b (KAD = 496 ± 79 μm) is significantly lower, as assessed by t test, compared with the WT EMB value (KAD = 587 ± 48 μm), suggesting a tighter ADP-binding affinity for EMB-3b (Fig. 2C), although this did not reach statistical significance using Welch's one-way ANOVA. The measured value of KAD for IFI-3a (KAD = 409 ± 18 μm) is not significantly different from the WT IFI value (see Table 1).
The ADP-release rate of S1 (k-D) is reversed for both IFI-3a and EMB-3b chimeras
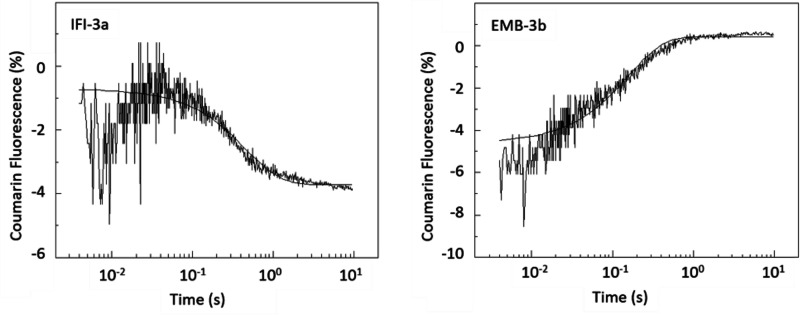
To estimate the rate constant of ADP dissociation from S1 in the absence of actin, the change in fluorescence of a coumarin-labeled ADP analog (eda-deac ADP) (11) was measured upon displacement of eda-deac ADP (cmADP) by ATP-binding to S1 (12). A single laser flash released 15–20 μm ATP from cATP (100 μm) and the fluorescence change resulting from cmADP release was recorded (Fig. 3). The fluorescence signal could be fitted to a single exponential, which gives the ADP-release rate (k-D). The small increase in fluorescence at the start of the transient is an artifact resulting from the laser flash which is difficult to eliminate entirely when working with low fluorescence signals as used here. Note the direction of the change is the same in both transients.
Figure 3.
Rate of ADP release (k-D) from Drosophila S1 isoforms. The rate constant for cmADP dissociation (k-D) from S1 in the absence of actin was determined using flash photolysis. After release of ATP (15 μm) from caged-ATP (100 μm), a fluorescent ADP analog (eda-deac ADP) bound to S1 was displaced by ATP. The change in fluorescence upon release of eda-deac ADP from S1 was used to determine k-D. Exchange of either the exon 3a or 3b domain resulted in a complete reversal of the ADP release rate (see also Table 1). The dissociation rate for EMB-3b (k-D, 7.0 s−1) is not significantly different to IFI (k-D, 7.5 s−1) whereas that of IFI-3a (k-D, 2.1 s−1) is similar to EMB (k-D, 1.8 s−1).
The two exon 3 chimeras, IFI-3a and EMB-3b, show an almost complete reversal of their k-D values compared with their WT counterparts (Table 1). Insertion of the embryonic exon 3a area into IFI results in an ADP release rate typically found in WT EMB (k-D = 2.1 ± 0.41 s−1), whereas the introduction of the fast IFI exon 3b region into EMB increases the ADP release rate of EMB-3b to WT IFI levels (k-D = 7.0 ± 1.3 s−1). Reversal of k-D rates after exon exchange was previously found for one of the other variable domains, the relay loop encoded by exon 9, as introduction of the exon 9a domain into IFI reduced the ADP release rate to embryonic levels, whereas the substitution of the exon 9b region into EMB increased the ADP release rate to IFI-levels (10).
The directionality of the observed fluorescence traces is variable and can even be reversed as seen here for EMB-3b. The fluorescence trace of IFI-3a shows a signal decrease after release of cmADP, similar to what was seen previously for IFI and EMB (5). However, EMB-3b fluorescence increases upon cmADP release and this reversal of the fluorescence signal has been observed in other studies. We previously reported a similar reversal of fluorescence change for a series of probes and myosin isoforms (12) and also for other chimeric Drosophila myosin isoforms after exchange of exon 7 (13) and exchange of exon 9 (10). We currently have no explanation of why this occurs, except that this must reflect a change in the environment of the fluorescent probe that will require analysis of a high-resolution structure to resolve.
Catalytic activity is reduced for IFI-3a and increased for EMB-3b
Basal and actin-activated ATPase activity of WT IFI S1 and the two exon 3 mutants were measured (see Fig. 4 and Table 2). IFI-3a S1 showed a significant reduction in basal Ca-ATPase (4.92 ± 1.05 s−1) compared with WT IFI S1 (9.79 ± 1.39 s−1), whereas Ca-ATPase activity of EMB-3b S1 (5.16 ± 2.01 s−1) is significantly increased compared with values for WT EMB (1.83 ± 0.08 s−1) (Fig. 4A and Table 2). The same trends were seen for basal Mg-ATPase activity, as well as actin-activated ATPase values (Vmax), although differences did not reach statistical significance (Table 2 and Fig. 4, B and C). Although previous Ca-ATPase, Mg-ATPase and Vmax measurements for full-length myosin (7) showed significantly reduced values for IFI-3a compared with IFI WT, the full-length protein showed an increase only in Ca-ATPase levels for EMB-3b compared with EMB.
Figure 4.
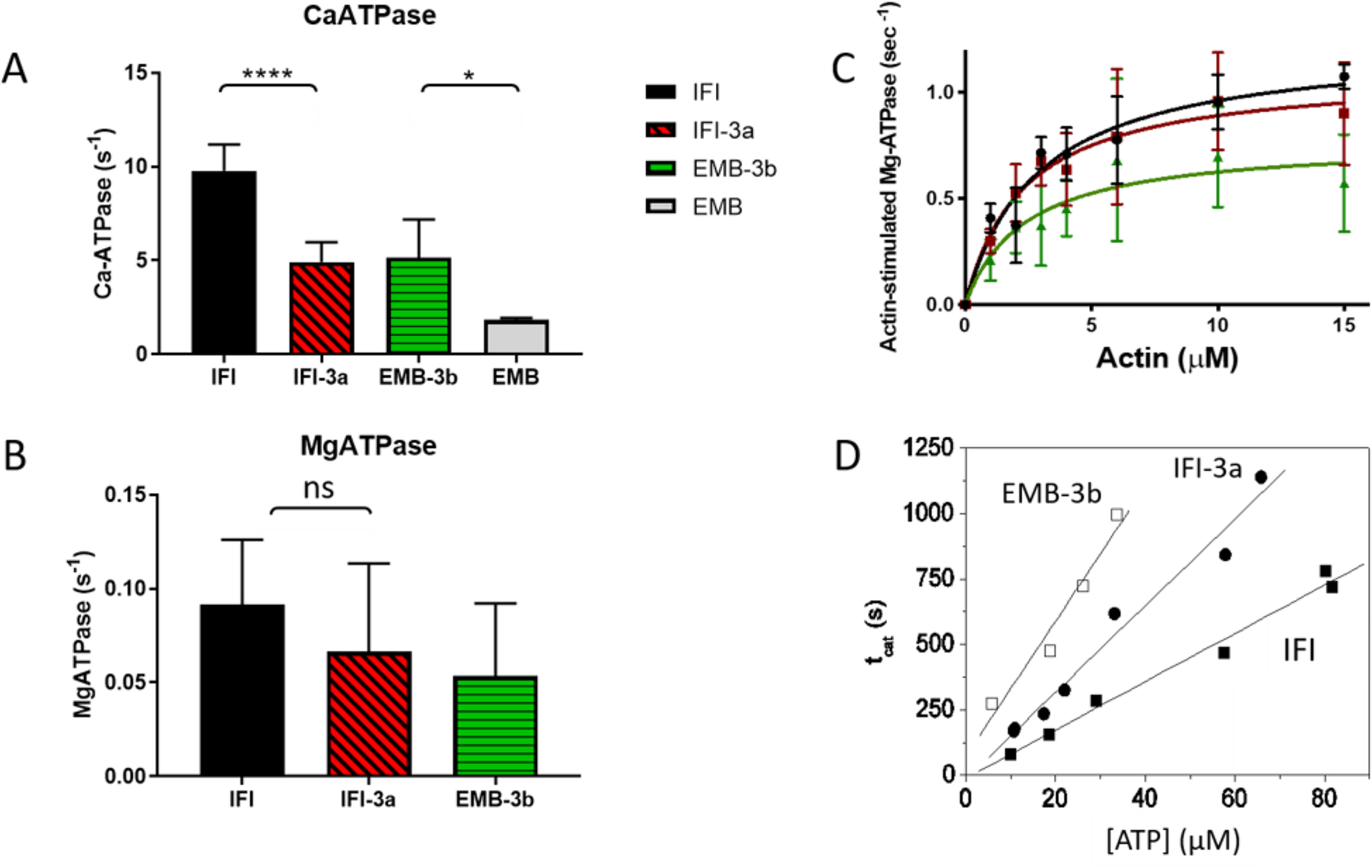
Steady-state ATPase activity of IFI, EMB, and exon 3 chimeric S1 isoforms. A–D, basal Ca-ATPase activity (A), basal Mg-ATPase (B), actin-activated Mg-ATPase activity (Vmax) (C), and the turnover number (kcat) for acto-S1 (D) were determined as described under “Experimental Procedures.” Notations above histograms indicate the level of statistically significant differences (*, p < 0.05; ****, p < 0.0001; ns, not statistically significant). Significant differences were assumed for p < 0.05.
Table 2.
Steady-state kinetic parameters measured for IFI, IFI-3a, EMB, and EMB-3b Drosophila myosin S1
Values are mean ± S.D. based on a minimum of four preparations (except for EMB Ca-ATPase, which is three).
| Myosin S1 | Basal Ca-ATPase (s−1) | Basal Mg-ATPase (s−1) | Vmax (s−1) | Km (μm) |
|---|---|---|---|---|
| IFI | 9.79 ± 1.39 | 0.092 ± 0.035 | 1.25 ± 0.29 | 3.26 ± 2.36 |
| IFI-3a | 4.92 ± 1.05a | 0.066 ± 0.047 | 1.07 ± 0.26 | 2.38 ± 0.91 |
| EMB-3b | 5.16 ± 2.01b | 0.053 ± 0.039 | 0.83 ± 0.37 | 2.95 ± 2.95 |
| EMB | 1.83 ± 0.08 | 0.016 ± 0.002ǂ | 0.67 ± 0.06ǂ | 2.54 ± 0.46ǂ |
ǂData from Ref. 10.
aStatistically different compared with IFI, p < 0.0001.
bStatistically different compared with EMB, p < 0.05.
Using flash photolysis, the turnover number (kcat) was measured at a fixed actin concentration for S1 of both exon 3 exchange mutants and compared with IFI WT (Fig. 4D). The time taken to hydrolyze all released ATP (tcat) was estimated from the time at which the dissociation reaction was 50% complete (tdiss) to the time for 50% recovery of light scattering (tass). This time period (tcat) is linearly dependent upon the amount of released ATP (see Fig. 4D). The steady-state rate of ATP hydrolysis is the inverse of the slope (steady-state rate = [ATP]/tcat), allowing the catalytic activity (kcat) to be determined. For IFI a steady-state rate of 0.104 (μM s−1) was found, resulting in a kcat of 0.18 s−1. This value is essentially the same as the kcat value reported previously for IFI S1 using the same experimental setup (kcat = 0.17 ± 0.006 s−1) (10). The catalytic activity of EMB-3b (kcat = 0.021 ± 0.009s−1) is nearly identical to the value measured previously for EMB (0.028 s−1), whereas the kcat value for IFI-3a (0.057 ± 0.003 s−1) shows a decrease compared with IFI (10) (see Table 1). Previously reported Vmax values for full-length myosin constructs showed similar behavior (7), with unchanged Vmax values for EMB-3b compared with EMB, and a 2.36-fold decrease in Vmax for IFI-3a compared with IFI.
Homology models and general description of the exon 3–encoded area
Sequence alignment of various myosin isoforms show that the exon 3 area is highly variable, with only five residues that are fully conserved: Asp90, Asn105, Arg109, Ile115, and Tyr116 (Fig. S1). The recently published crystal structure of embryonic Drosophila myosin in the rigor-like conformation (PDB ID: 5W1A) shows that the exon 3 region (residues 69–116) starts with two β-strands (β4: 69–73 and β5: 77–79), which form part of the SH3-like domain found in many other myosins (Fig. 5A). The β4 and β5 strands are followed by two short helices (HA: 83–85 and HB: 91–93) and a longer helix C (residues 98–111). The C-terminal residues of the exon 3 area are at the start of the next β-strand (S1B) (Fig. 5B) which forms the first β-strand of the central seven-stranded β-sheet in the myosin head.
Figure 5.
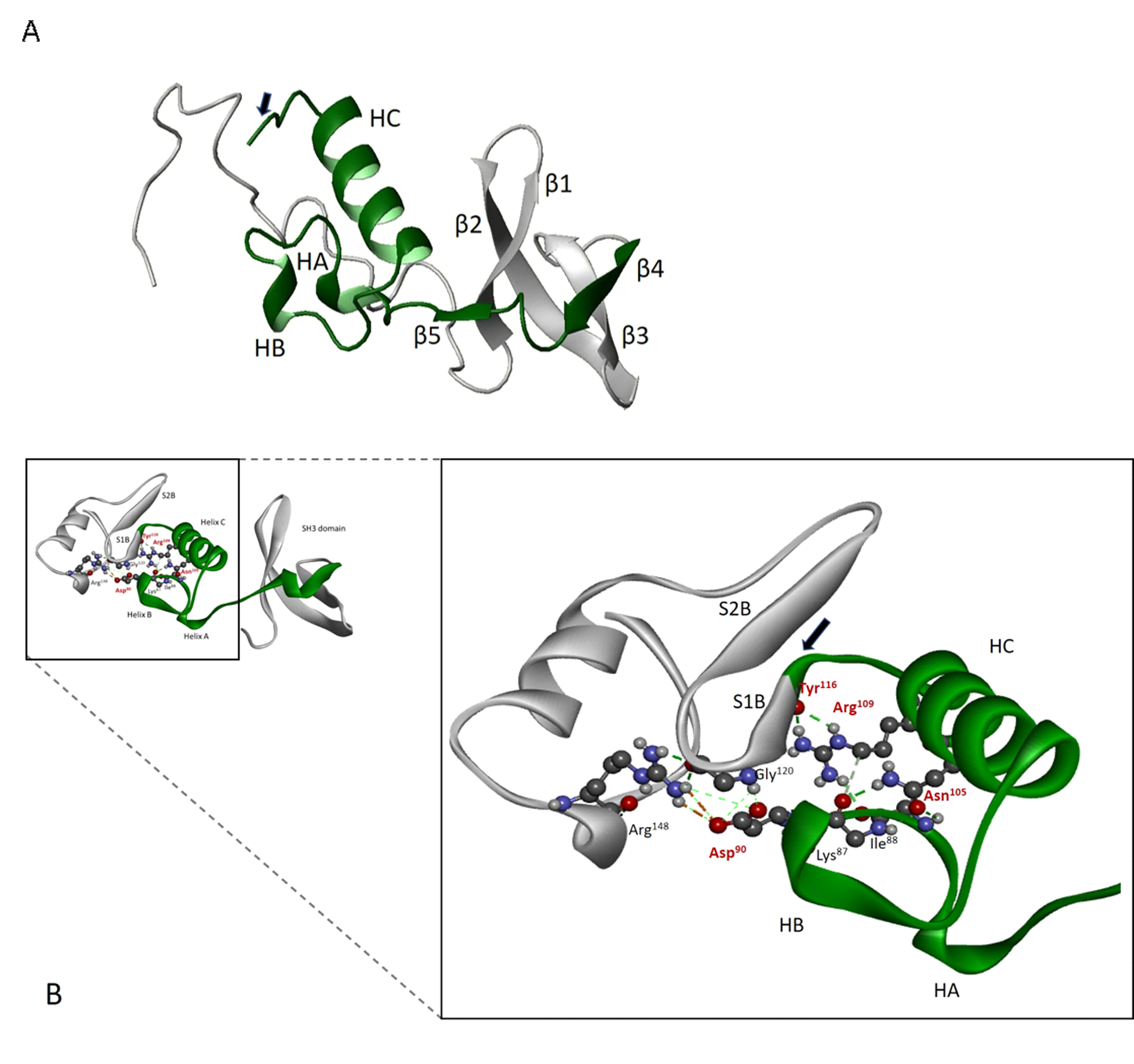
Location and structure of exon 3 domain (green) within the myosin heavy chain N terminus. A, secondary structure elements encoded by exon 3 (residues 69–116) include β4 (69–73), β5 (77–79), helix A (HA, 83–85), helix B (HB, 91–93), helix C (HC, 98–111), and the first β-strand of the seven-stranded β-sheet S1β (starts at 115, indicated with an arrow). The full SH3 element is also depicted (β1, Lys36–Glu43; β2, Glu45–Lys56; β3, Ile59–Gln65). B, conserved interactions within exon 3 regions involve residues Asp90, Asn105, Arg109, and Tyr116. Asn105 forms hydrogen bonds with backbone oxygens of three residues close to Asp90 (Lys87, Ile/Ala88, and Met91) whereas Arg109 forms H bonds with Tyr116.
Conserved residue Asp90 is located between exon 3 helix A and helix B, residues Asn105 and Arg109 are located on the same side of helix C, and Ile115 and Tyr116 are part of the first β-strand of the central 7-stranded β-sheet, a conserved feature of all myosins. The EMB crystal structure shows that Asp90 forms a strong salt-bridge with Arg148 and that the latter also forms a hydrogen bond with Gly120, located between the first two β-strands of the central 7-stranded β-sheet (Fig. 5B). Conserved residue Asn105 forms hydrogen bonds with the backbone oxygen of three residues close to Asp90 (Lys87, Ile/Ala88, and Met91), whereas conserved residue Arg109 contacts Tyr116.
The available crystal structure of embryonic Drosophila myosin in the rigor-like conformation (PDB ID: 5W1A) was used as a template to generate a homology model for IFI myosin. Overlay of the IFI model and the EMB crystal structure showed that the backbone topology for both is very similar (root mean square deviation of 1.59 Å). Inspecting the homology models using scallop myosin templates that represent conformations at different steps of the mechanochemical cycle shows that some of these conserved interactions are maintained throughout the cycle, whereas others are lost (see “Discussion” for details).
Exon 3 interacts with the nucleotide binding site
The kinetic data show that exchange of the exon 3 area alters the nucleotide binding properties of Drosophila myosins. A large body of research has established that nucleotide binding and processing in the active site of myosin involves four highly conserved loops: P-loop (GESGAGKT), switch 1 (AKTXXN(N D)NSSR), switch 2 (DIXGFE), and the purine binding loop (NPXXXXXXY) (14). The exon 3 region (residues 69–116) could potentially alter nucleotide binding properties by interacting with any of these highly conserved loops. Inspection of the EMB crystal structure (Fig. 6A) shows that the purine binding loop (residues 127–135) interacts with the exon 3 region; for example, exon 3 residue Tyr110 forms a hydrogen bond with purine binding loop residue Asn127 (Fig. 6B). Asn127 also has a hydrogen bond with the backbone carbonyl of P-loop residue Gly182. These contacts suggest a role for the exon 3 region of the myosin head in regulating nucleotide binding and release via interaction with the purine binding loop.
Figure 6.
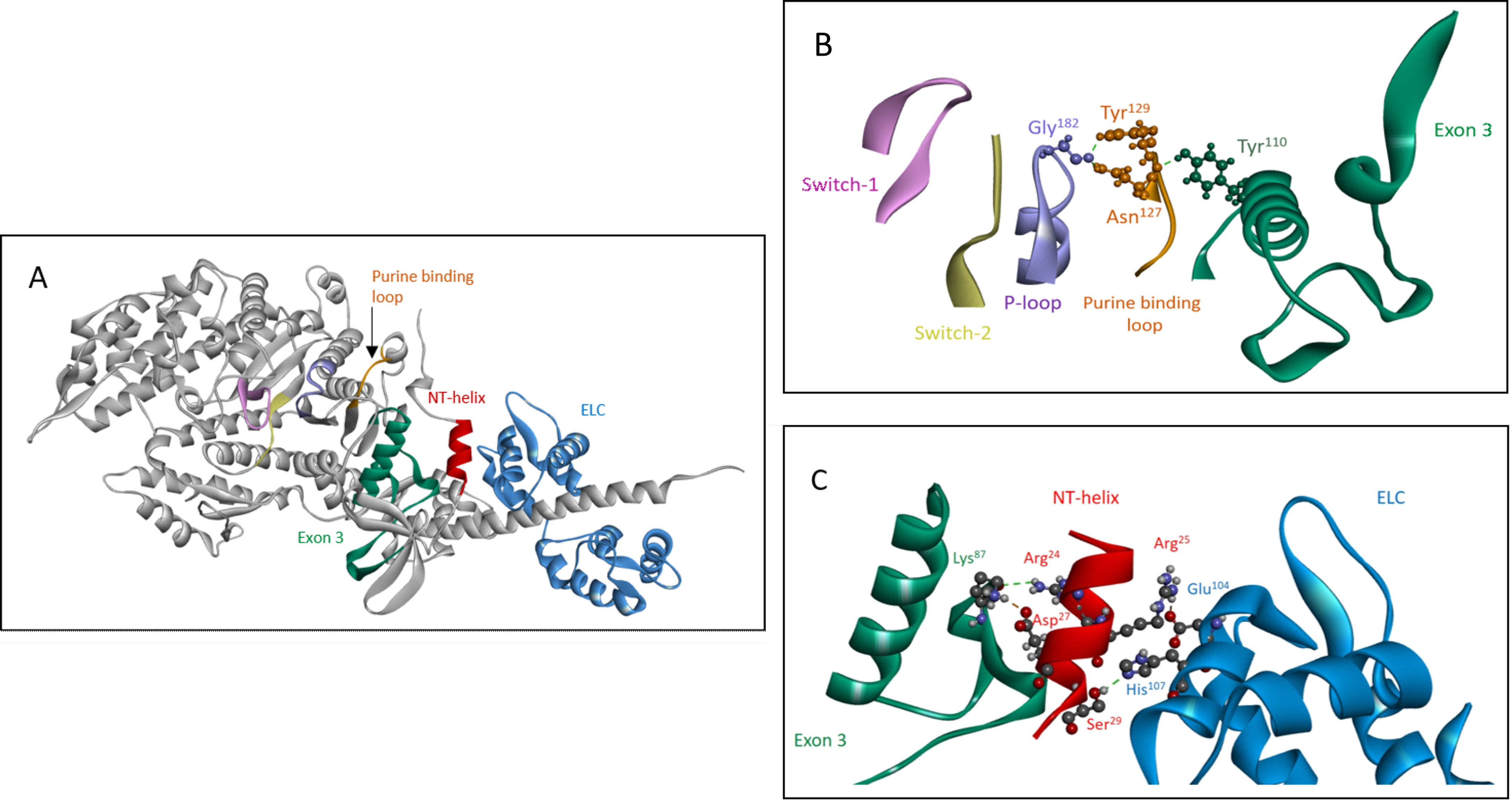
The exon 3 area is part of the communication pathway between the essential light chain and the purine binding loop. A, overview of elements in the communication pathway between the ELC (blue) and purine binding loop (127–135, orange), shown for rigor-like EMB myosin S1 (PDB ID: 5W1A). In addition to the exon 3 area (69–116, green), the small N-terminal helix (21–30, red) is also involved in signal transfer from the ELC toward the nucleotide binding pocket. B, detailed view of purine binding loop interactions with the exon 3 area and the P-loop (PDB ID: 5W1A). Fig. 6B is reused and extended in the supplementary section to demonstrate that interaction of the exon 3 region with the purine binding loop depends on conformational state of the myosin head (Figs. S2 and S3). C, close-up of N-terminal helix interactions with the ELC (Glu104 and His107) and the exon 3 region (Lys87) (PDB ID: 5W1A).
Homology models derived from scallop myosin structures at various states of the crossbridge cycle were generated for both IFI and EMB and used for further analysis, because the EMB crystal structure is only available for a rigor-like configuration that has no nucleotide bound. The homology models confirm the interaction between the purine binding loop and the exon 3 area and suggest this interaction depends on the myosin state involved (see Fig. S2 for a detailed description of these interactions for different myosin conformations). However, the interactions between the purine binding loop and the exon 3 area are very similar for IFI and EMB (see Figs. S2 and S3 for details).
The EMB crystal structure shows that the exon 3 region has no direct contacts with any of the other variable domains in the myosin head. However, the SH1-SH2 helix is wedged between the exon 3 and exon 9 (relay loop) regions and makes contacts with both variable domains. Because Drosophila EMB and IFI share the same SH1-SH2 sequence, the two variable regions could potentially interact differently with this element, thereby altering the myosin properties. However, detailed analysis showed that contacts between exon 3 residues and the highly conserved SH1-SH2 region in the myosin head were also found to be very similar for both chimeras (see Fig. S4 for details). In summary, the conserved interaction of the exon 3 region with both the nucleotide binding loop and the SH1-SH2 area suggest that alternative versions of exon 3 differentially affect myosin properties via another mechanism.
Exon 3 interaction with the myosin essential light chain
Part of the exon 3 area is embedded in the SH3 element, a β-barrel found at the N terminus of the MHC. In vertebrates this SH3 element is thought to interact with the extended N terminus of the myosin essential light chain (ELC) in the presence of actin, thereby modulating myosin ATPase kinetics (15). The EMB crystal structure does not show any direct contacts between the exon 3 region and the ELC, as the ELC of Drosophila lacks this N-terminal extension (16). However, two ELC residues (Glu104 and His107) form salt-bridge/H-bond contacts with two MHC residues located outside the SH3 domain (Arg25 and Ser29). The latter two are part of a small helix near the N terminus (helix NT), which is wedged in between the ELC and the SH3-exon 3 area (Fig. 6, A and C). Another residue located on this small helix (Asp27) forms a strong salt-bridge with exon 3 residue Lys87 (Fig. 6C). Lys87 is part of a network of highly conserved interactions in the exon 3 area, as it makes contacts with conserved residues Arg109 and Asn105 (see Fig. 5B). Lys87 (backbone oxygen) also interacts with the side chain of helix NT residue Arg24 (Fig. 6C). Therefore, the exon 3 region is likely to be part of a communication pathway between the essential light chain and the purine binding loop, which also involves helix NT.
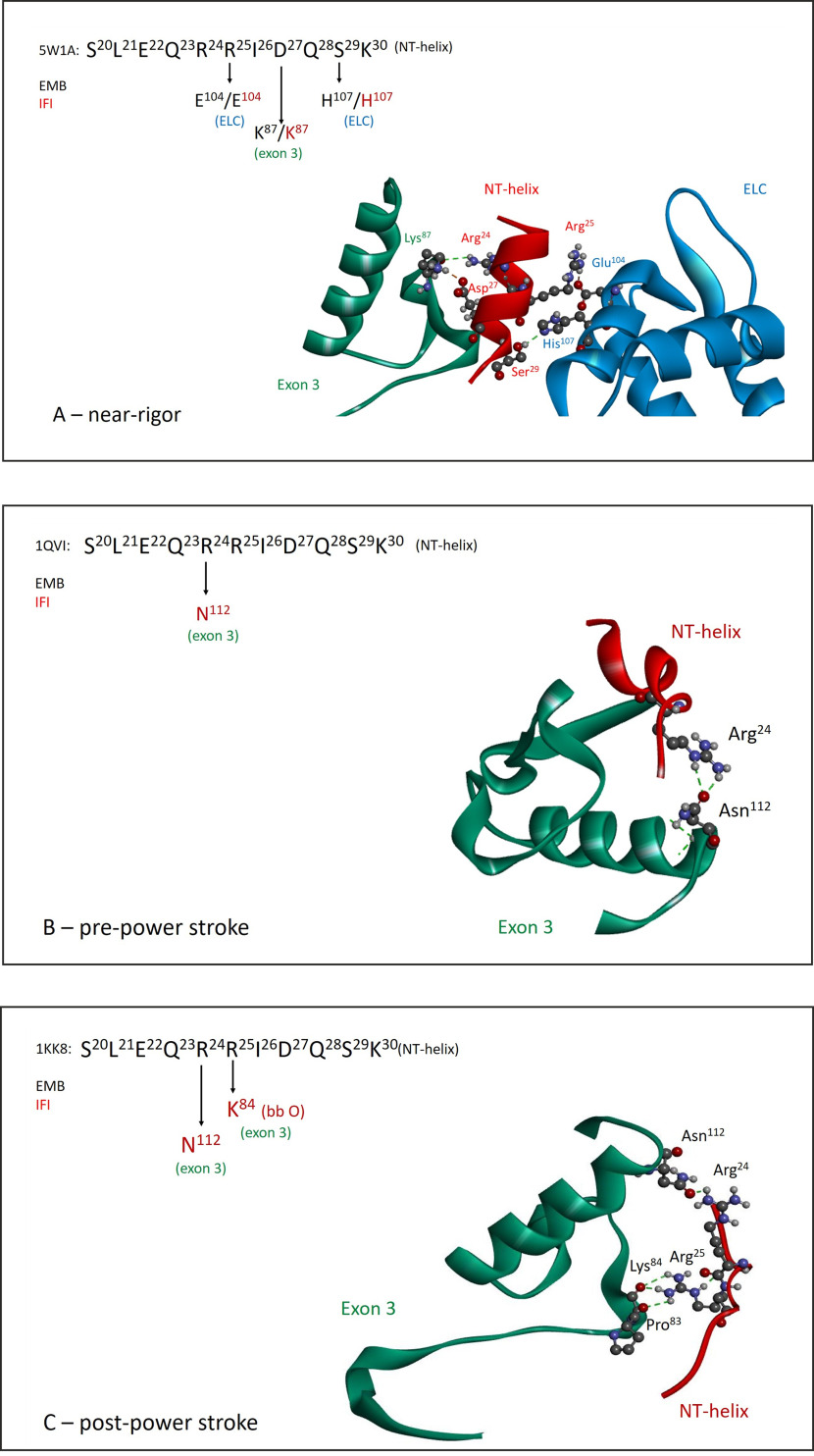
Homology models for IFI show a similar picture when using the EMB crystal structure PDB ID 5W1A as a template, with no direct ELC–exon 3 region contacts, but indirect contacts via helix NT (Fig. 7A). Homology models representing other states in the crossbridge cycle indicate that for IFI, helix NT maintains contacts with the exon 3 region throughout the cycle, whereas for EMB these contacts are lost. For instance, the pre-power stroke state of IFI (using PDB ID 1QVI as template) shows interactions between exon 3 residue Asn112 and helix NT residue Arg24 (Fig. 7B). Exon 3 residue 112 is a variable residue, Ala112 for EMB, and therefore the pre-power stroke state of EMB lacks this exon 3 contact with helix NT. The post-power stroke state of IFI (using PDB ID 1KK8 as template) maintains the Asn112–Arg24 contact and has additional interactions between the side chain of Arg25 (helix NT) and the backbone oxygen of exon 3 residues Pro83 and Lys84 (Fig. 7C). For EMB the post-power stroke state again does not show any contacts between exon 3 and helix NT, indicating the interaction between the exon 3 region and helix NT is fundamentally different between the IFI and EMB isoforms.
Figure 7.
Exon 3 interaction with helix NT is distinct between IFI and EMB and depends on conformational state. Comparison of helix NT interaction with exon 3 residues and the ELC throughout the crossbridge cycle for IFI and EMB myosin isoforms, with exon 3 area (residues 69–116) in green, the NT–helix (residues 21–30) in red and the ELC in blue. A, near rigor state: EMB crystal structure (PDB ID: 5W1A) and corresponding IFI homology model show similar interaction of helix NT with exon 3 region and ELC. B, pre-power stroke state: interaction of helix NT (Arg24) with the exon 3 area is present for IFI (Asn112), whereas for EMB (Ala112) this interaction is absent (PDB ID: 1QVI). C, post-power stroke state: IFI maintains contacts between the sidechains of variable exon 3 residue Asn112 and helix NT residue Arg24. Additional contacts are found for exon 3 residues Pro83 and Lys84 (backbone oxygen) with helix NT residue Arg25 (side chain). No exon 3–helix NT contacts were found for EMB (PDB ID: 1KK8).
Discussion
The use of alternative domains in the myosin head is thought to play a role in fine-tuning the kinetic and mechanical properties of myosin isoforms (2), but the exact mechanisms whereby alternative structures affect these properties are not well understood. Herein we investigate the structure-function relationship of the area encoded by exon 3, a variable domain that forms part of the SH3-fold found near the N terminus of the myosin head. Our results show that exchange of the exon 3 region between the indirect flight isoform (IFI) and the embryonic isoform (EMB) alters various kinetic properties of the generated myosin S1 chimeras (IFI-3a and EMB-3b). The most profound effect is on the ADP release rates in the absence of actin, as the k-D values for the chimeras are completely reversed, compared with their WT counterparts. In the presence of actin, the ADP affinity for IFI-3a is not affected compared with WT IFI. However, the ADP affinity of EMB-3b is significantly tighter, compared with WT EMB and shifts toward IFI values. ATP-induced dissociation is also affected after exchange of the exon 3 area between IFI and EMB, although the effect is less dramatic compared with ADP release. Introduction of exon 3b into EMB shifted the ATP-induced dissociation toward IFI values, however, the insertion of the exon 3a region into an IFI background did not shift the ATP-induced dissociation toward WT EMB levels. The steady-state kinetic parameters measured for the IFI, EMB, and exon 3 chimeric S1 isoforms show a similar pattern for kcat, with values of IFI-3a shifting toward the EMB donor isoform, but values for EMB-3b being very similar to EMB. Although this pattern holds for IFI versus IFI-3a values for Vmax and basal Ca- and Mg-ATPase, increases in these parameters were seen for EMB-3b relative to EMB.
Correlation between K1k+2 or KAD and motility
It is generally accepted that ADP release is rate-limiting for slow sarcomere myosin isoforms (17). In contrast, for faster myosins, ATP-induced dissociation can be rate-limiting, depending on the particular conditions used (18, 19). Plotting the measured ADP affinity (KAD) for EMB, EMB-3b, IFI, and IFI-3a against previously reported in vitro motility values, with and without the actin-binding protein tropomyosin (Tpm) (7), shows a decrease in velocity with increasing KAD values (lower ADP affinity) (see Fig. S5 and Table 3). This implies that ADP release is not rate-limiting for these Drosophila myosin isoforms, as the actin sliding velocity is higher for isoforms with higher ADP affinity. This agrees with previous biochemical studies that reported ADP affinity measured in solution did not correlate with actin sliding velocity of other Drosophila myosin isoforms (5, 10, 13). Mechanical studies on isolated Drosophila muscle fibers also found no correlation between ADP affinity and frequency of maximum work production (4, 8, 20, 21). Plotting in vitro motility versus ATP-induced dissociation (K1k+2) for IFI, EMB, and the two exon 3 chimeras shows reduced motility with increasing K1k+2 values, although the correlation is not particularly strong (R2 = 0.86) (Fig. S5).
Table 3.
Summary of measured and calculated kinetic and mechanical data for Drosophila myosin isoforms
| Isoform | Velocity (−Tpm) (μm/s)a | Velocity* (+Tpm) (μm/s)a | KAD (μm) | K1k+2 (μm−1s−1) | K1k+2 (μm−1s−1)c | k-AD (s−1)d | Kmin,5 (s−1) | Kmin,5* (s−1) | Kdissn,2 (s−1)e | Kdissn,1 (s−1)f |
|---|---|---|---|---|---|---|---|---|---|---|
| IFI-3a | 6.5 | 6.2 | 409 | 0.66 | 1.32 | 4090 | 1300 | 1240 | 2640 | 1320 |
| IFI | 6.4 | 5.7 | 409b | 0.75b | 1.50 | 4090 | 1280 | 1140 | 3000 | 1500 |
| EMB-3b | 3.8 | 5.2 | 496 | 0.71 | 1.42 | 4960 | 760 | 1040 | 2840 | 1420 |
| EMB | 0.7 | 4.0 | 587b | 0.91b | 1.82 | 5870 | 140 | 800 | 3640 | 1820 |
aFrom Ref. 7; *indicates parameters determined in the presence of tropomyosin.
bFrom Ref. 5.
cValue of K1k+2 corrected for inhibition of the ATP reaction by cATP (correction factor 2) (18).
dAssumes k+AD = 107 m−1s−1 and k-AD = KAD × k+AD.
eAssumes kdiss = K1k+2 × [ATP] and [ATP] is 2 mm (ATP concentration used in motility assays).
fAssumes kdiss = K1k+2 × [ATP] and [ATP] is 1 mm.
ADP release (k-AD) is faster than ATP-induced dissociation of AM (kdiss)
To estimate if either ADP release or ATP-induced dissociation could be rate-limiting for these Drosophila myosins, the theoretical rate-limiting step kmin was calculated using the equation kmin = V/d, assuming a working stroke (d) of 5 nm (18) and using the published motility data for velocity in the presence (V*) and absence (V) of tropomyosin (Tpm) (7). The calculated values for the rate-limiting step kmin are listed in Table 3, together with the motility data and estimated values for ADP release (k-AD) and ATP-induced dissociation (kdiss). The values associated with ATP-induced dissociation (kdiss,2) are within 2600–3600 s−1, whereas values estimated for ADP release (k-AD) are within 4000–5900 s−1. Thus, both ATP-induced dissociation and ADP release are significantly faster than the rate-limiting step kmin (800–1200 s−1). Therefore, it seems unlikely that either ADP-release or ATP-induced dissociation is rate-limiting under these conditions.
At lower ATP concentrations, one predicts that kdiss could be limiting the velocity for these myosins, in particular for IFI, which has an unusual low affinity for ATP, compared with EMB (20). At 1 mm ATP concentration, kdiss,1 is around 1300-1800 s−1, close to the values calculated for kmin,5*, indicating that at such low ATP concentrations kdiss can indeed be rate-limiting for these myosins. At saturating ATP levels, another step in the crossbridge cycle could become rate-limiting, for instance, phosphate release (IFI) or an isomerization step before ADP release (EMB) as proposed by Swank et al. (20). However, EPR studies of Drosophila myosins investigating the isomerization step between the open to closed conformation of the nucleotide binding pocket found that the pocket is predominantly closed for both IFI and EMB with ADP bound. Therefore, this isomerization step is unlikely to control velocity for these fast myosin isoforms (22). Overall, our calculations suggest that ADP release is too fast to be rate-limiting, but ATP-induced dissociation could be rate-limiting at lower [ATP].
Biological implications
Homology modeling, combined with the crystal structure for Drosophila EMB, indicates that the exon 3 encoded region in the myosin head is part of a communication pathway between the nucleotide binding pocket (purine binding loop) and the essential light chain via helix NT, emphasizing an important role for this variable N-terminal domain in regulating actomyosin crossbridge kinetics. IFI maintains the interaction between helix NT and exon 3 during the crossbridge cycle for different conformational states, whereas for EMB these interactions are lost. The presence of helix NT-exon 3 contacts throughout the crossbridge cycle allows the IFI isoform to rapidly communicate any conformational change from the nucleotide binding site toward the ELC and vice versa, whereas for the EMB isoform this process is likely to be slower, because of the loss of exon 3–helix NT contacts.
The presence of additional stabilizing interactions between the exon 3 region and helix NT for IFI throughout the crossbridge cycle might be expected to stabilize the structure and make transitions between states more difficult, thus reducing the reaction rates of its chemomechanical cycle compared with EMB, which is opposite to experimental data. In contrast to these expectations, the increased stability may restrict the number of conformational states the myosin head can adopt and actually improve its function. In this regard, a recent paper by Schmid and Hugel (23) explores this concept and introduces the idea of “conformational confinement of proteins,” in which the authors argue that restricting conformational states enhances the function of a protein “by limiting the nonproductive degrees of freedom.” For myosin IFI, the presence of helix NT–exon 3 contacts throughout the crossbridge cycle may therefore ensure that the conformational states of myosin are confined to those that enhance its function.
If the exon 3 region plays an important role in the communication pathway between the essential light chain and the purine binding loop via helix NT, any mutations along this route are expected to have a serious effect on the biological function of myosin. An extensive literature search (24) revealed that various cardiomyopathy mutations have been reported along this pathway, involving the exon 3 region, helix NT, and the essential light chain. Mutations have been reported for exon 3 residues Pro81, Ala100, and Tyr115 (the equivalent exon 3–encoded residues in Drosophila are Pro82, Ser101/Cys101, and Tyr116). Tyr115 can interact with the purine binding loop (Tyr116 in Fig. S2, B and C) and therefore a mutation of Tyr115 is expected to alter this interaction and affect the nucleotide binding properties of myosin. The helix NT equivalent in human β-myosin heavy chain also contains a cardiomyopathy site, Ala26 (Glu27 in both Drosophila EMB and IFI myosin) which could disrupt the signal transfer between the essential light chain and the purine binding loop. The essential light chain residue Arg154 (Gln106 equivalent in Drosophila) can result in serious cardiomyopathy when mutated into His (25). Gln106 is close to ELC residues Glu104 and His107, which both interact with the NT helix (Fig. 6C). Based on kinetic measurements, and molecular modeling, we propose that these mutations alter myosin function by disrupting the relay pathway between the nucleotide binding pocket and the essential light chain. A recent structure of β-cardiac myosin (PDB ID: 6FSA) shows a strong contact (salt-bridge) between exon 3 residue Asp85 and helix NT residue Arg29, in addition to contacts between exon 3 residue Tyr115 and purine binding loop residue Tyr134 (26), strongly supporting our hypothesis that the exon 3 area is part of the relay pathway toward the purine binding loop via helix NT.
Disruption of the above-mentioned pathway not only affects the crossbridge kinetics but also is expected to alter the mechanical properties of myosin. The N-terminal region (NTR) of myosin has been identified as an important element in tuning the mechanical properties of myosin-I (27). Exchange of the NTR from Myo-1b, a highly tension-sensitive motor, onto Myo-1c, which is less tension-sensitive, converts Myo-1c into a highly tension-sensitive motor, resulting in sensitivity to forces <2 pN. Overlay of the crystal structures of Myo-1b (PDB ID: 4L79) (28) and the EMB crystal structure (PDB ID: 5W1A) shows a remarkable similarity between the secondary structure elements of the N-terminal region of EMB and Myo-1b, except for the SH3 domain of EMB (Fig. 8). The NTR of Myo-1b and a small helix immediately following the NTR are located near helix A and helix B of EMB, whereas the longer helix C of EMB nearly overlaps with an equivalent longer helix of Myo-1b (see Fig. 8, A and B). Overlay of EMB with Myo-1c shows a similar picture (Fig. 8C) with the NTR and the first two helices of Myo-1c following the backbone topology of the exon 3 area secondary structure elements (helices A, B, and C). The similarity between the N termini of EMB and the Myo-1b/Myo-1c structures suggests a similar functional role, implicating the exon 3 area as a mechano-sensing element.
Figure 8.
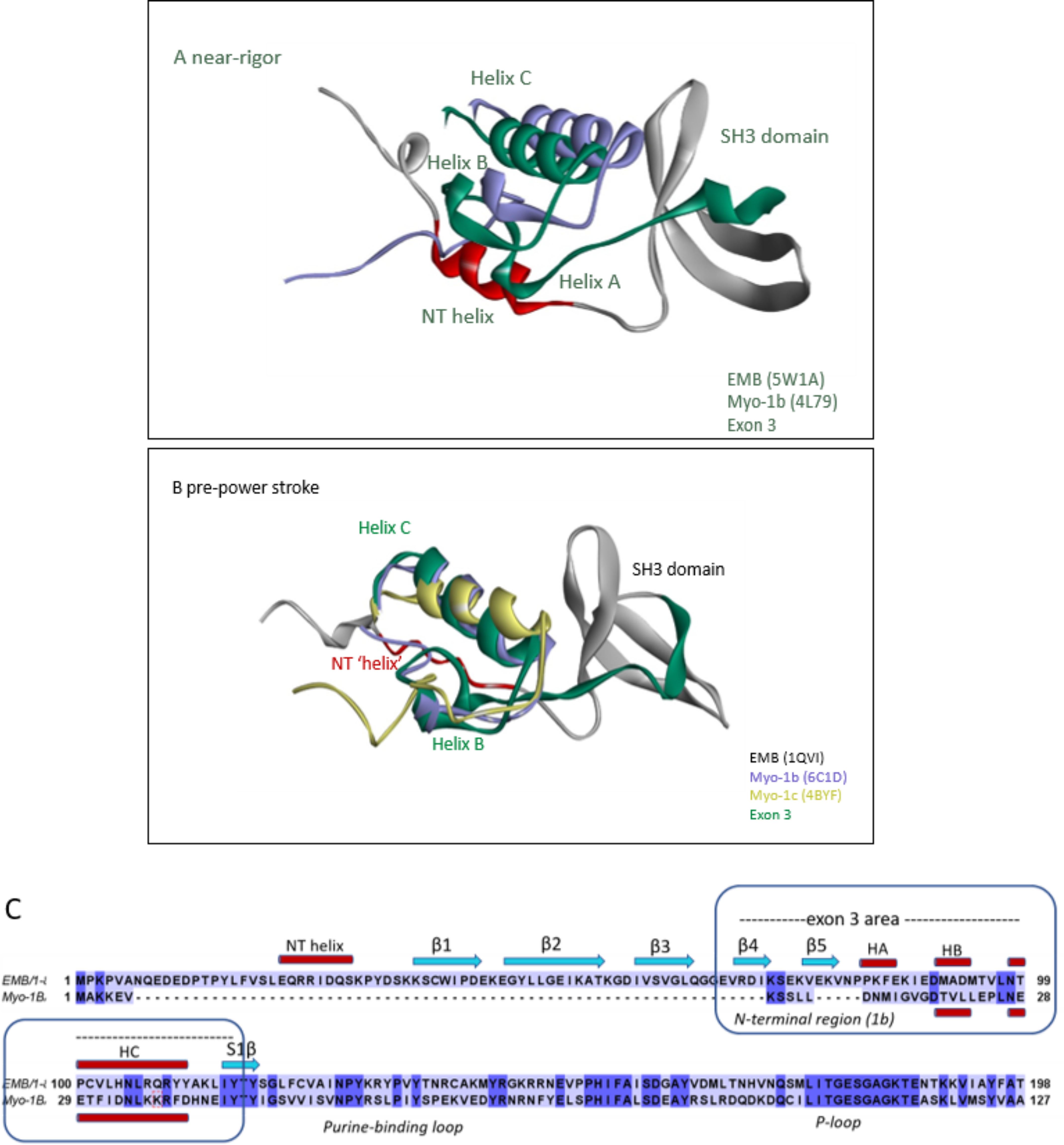
Overlay of N-terminal regions of EMB and Myo-1b or Myo-1c. A and B, EMB (gray) with the exon 3 area (green) and the N-terminal helix (NT) shown in red, Myo-1b (purple), and Myo-1c (yellow). Note the similar orientation of exon 3 secondary structure elements (helix B and C) of EMB with respect to the NTR of Myo-1b and/or Myo-1c in both near-rigor and pre-power stroke state. A, overlay of crystal structures of Myo-1b (PDB ID: 4L79) with EMB (rigor-like, PDB ID: 5W1A). B, overlay of Myo-1b (PDB ID: 6C1D) and Myo-1c (PDB ID: 4BYF) with EMB homology model (pre-power stroke, PDB ID: 1QVI template). C, sequence alignment of N-terminal regions of EMB and Myo-1b showing conserved secondary structure elements (HB and HC) for the exon-3 encoded region (EMB) and the NTR (Myo-1b).
Recent crystal structures of cardiac myosin with the myosin activator omecamtiv mecarbil (OM) bound also hint at the possibility that the exon 3 area is involved in fine-tuning the mechano-sensing properties of myosin (29, 30). OM is a selective, small-molecule cardiac myosin activator that binds to the myosin head domain and can increase the power output of the cardiac muscle (31). OM is currently in clinical trials for the treatment of heart failure. Two cardiac myosin–binding sites have been reported for OM (29, 30), one of which shows the OM-binding site in a narrow cleft between the N-terminal domain and lower 50 K domain (29). Three out of six N-terminal domain residues that interact with OM are located in the exon 3 area (Ala91, Met92, Leu96), whereas the other three residues are just outside the exon 3 area on the loop between the first (S1β) and second β-strand (S2β) of the seven-stranded β-sheet (Ser118, Gly119, and Phe121). Because OM can alter the power output of myosin, the involvement of exon 3–encoded residues in this process is another indication of this region's potential in fine-tuning mechanical properties of myosin isoforms.
In conclusion, we find that the alternatively expressed domain encoded by exon 3 in muscle myosin is critical for optimal myosin performance. Various steps in the crossbridge cycle are affected by exchange of the exon 3 regions, in particular ADP release and ATP binding. Based on the recently reported EMB crystal structure, combined with the homology modeling presented here, we propose that this exon 3 area is part of the communication pathway between the nucleotide binding pocket (purine binding loop) and the essential light chain and plays an important role in modulating actomyosin crossbridge kinetics and load-dependent mechanics.
Experimental Procedures
Generation of subfragment-1 (S1) from isolated full-length myosin
Myosin was isolated from the indirect flight muscles of 140 WT (PwMhc2 transgenics), 200 EMB, 140 IFI-3a, or 300 EMB-3b transgenic flies (those expressing the IFI, EMB, EMB-3b, or IFI-3a myosin isoforms in the indirect flight muscles, respectively) as described previously (6). The production of S1 by α-chymotrypsin digestion was carried out using a method based on Silva et al. (32) with the following modifications: The final myosin pellet was dissolved in digestion buffer (120 mm NaCl, 20 mm Na2PO4, pH 7.0, 1 mm EDTA, and 4 mm DTT). The myosin was incubated at 20°C for 5 min to equilibrate, and then incubated with 0.2 mg/ml α-chymotrypsin for 6 min. To quench the reaction, phenylmethylsulfonyl fluoride (PMSF) was added to a final concentration of 1.5 mm. The reaction was subsequently centrifuged at 60,000 rpm (TLA 100.3 rotor) for 20 min in a Beckman ultracentrifuge to pellet the undigested myosin and myosin rods. The supernatant containing the S1 was removed and diluted to 1 ml with low-salt buffer (30 mm KCl, 15 mm MgCl2, 20 mm MOPS, pH 7.0, and 4 mm DTT). To concentrate the S1, the samples were centrifuged at 15,000 rpm in a Sorvall MC 12V microcentrifuge at 4°C using a Millipore Ultrafree 0.5 μm centrifugal filter with a 5000 kDa cut-off. The final volume of the supernatant was 30–40 µl, containing an S1 concentration of roughly 1.5–2 µg/µl. S1 concentration was determined from the absorbance at 280 nm (E1% = 0.73 cm−1) and the molecular mass of 115 kDa.
Flash photolysis
Because of the small amounts of protein available, flash photolysis was used to measure the kinetics of the mutant Drosophila myosin S1 (5, 33). The ATP-induced dissociation of the acto-S1 complex was followed by measuring changes in the light-scattering signal. The dissociation of nucleotide from S1 alone was detected by changes in fluorescence using fluorescently labeled analogues. In brief, the 20 µl sample was held in a quartz cuvette and ATP was released by a single 5-ns flash at 353 nm from a neodymium-yttrium-aluminum-garnet laser (Surelite I-10, 70 mJ maximum power) along the vertical axis of the cell at a rate of 90 s−1 to start the reaction. Both absorbance (to determine the ATP concentration) and light scattering (to monitor the acto-S1 complex) or fluorescence (to determine cmADP release) were measured from the cuvette simultaneously following the laser flash. White light >389 nm was introduced to the sample from a 100-watt halogen lamp and the change in the amount of light scattered 90° was monitored after each flash. The absorbance at 405 nm was measured with a monochromator to determine the amount of ATP liberated from caged ATP during each laser flash. Coumarin fluorescence changes were detected by monitoring the emission through a 455-nm cut-off filter after excitation at 434 nm (75-watt xenon/mercury lamp).
All light-scattering experiments were conducted in a low-salt buffer (pH 7.0, 30 mm KCl, 5 mm MgCl2, 20 mm MOPS, and 4 mm DTT) with 1 μm actin, 1–3 μm S1, 500 μm cATP, 10 mm DTT, and either apyrase (2 units/ml, ATP-induced dissociation of acto-S1) or ADP (various concentrations) and a glucose-hexokinase system (0.03 units/ml hexokinase, 1 mm glucose, and 100 μm Ap5A, KAD determination). Each sample was subjected to multiple laser flashes. During KAD determination, ADP and cATP were added after each flash. The cmADP dissociation experiments were also performed in this low-salt buffer and contained 4 μm S1, 10 μm cmATP (source of cmADP) and 100 μm cATP. For the determination of kcat, the acto-S1 sample, incubated without apyrase, was irradiated by a series of laser pulses of different intensities, which released a range of ATP concentrations. The time taken to hydrolyze all of the ATP (tcat) was estimated from the time at which the dissociation reaction was 50% complete (tdiss) to the time for 50% recovery of the light scattering (tass) (33).
Analysis of the kinetic data
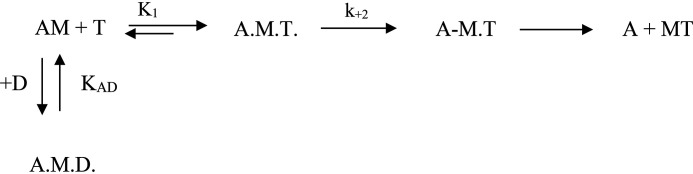
Equation 1 was derived from the interaction of actin and S1 with ATP and ADP shown in Scheme 1 and was used to determine KAD.
| (Eq. 1) |
where kobs is the observed rate constant for the ATP-induced dissociation of acto-S1; K1k+2 is the second-order rate constant for ATP binding to acto-S1; KAD is the equilibrium dissociation constant for the binding of ADP to acto-S1. The equation krel = kobs/ko was used to determine the relative rate constant (krel) shown in Fig. 2D, where ko is the value when [ADP] = 0. Plotting tcat versus ATP concentration allows one to estimate the steady-state rate of ATP hydrolysis according to Equation 2:
| (Eq. 2) |
Scheme 1.
The interaction of S1 with actin, ATP, and ADP. M, A, T, and D symbolize myosin S1, actin, ATP, and ADP, respectively.
From which one can determine the kcat (Equation 3):
| (Eq. 3) |
Values reported for flash photolysis studies are mean ± S.D. based on a minimum of three preparations, except kcat, which is based on two preparations, and therefore kcat is not included in the statistical analysis that was carried out by unpaired t tests and Welch's correction, with significant differences assumed at p < 0.05. Significance was also assessed using one-way Welch's ANOVA.
Basal and actin-stimulated Mg-ATPase assays
ATPase activities for myosin S1 were determined using 2 µg S1 samples and [γ-32P]ATP. Two technical samples were run for each biological replicate. Ca-ATPase activity was determined as described previously (6). Basal and actin-activated Mg-ATPase activities were determined using chicken skeletal muscle actin as described previously (7), with modifications (10). Notably, no salt (KCl) was added to the ATPase assay solution. G-actin was isolated from acetone powder of chicken skeletal muscle (34). After one cycle of polymerization-depolymerization, soluble G-actin obtained after dialysis against 2 mm Tris-HCl, pH 8.0, 0.2 mm ATP, 2 mm CaCl2, and 1 mm DTT was quantified spectrophotometrically using an extinction coefficient of 0.62 cm−1 (A310 nm–A290 nm) for 1 mg ml−1. F-actin was prepared by adding 1 volume of 10× polymerization buffer (50 mm Tris-HCl, pH 8.0, 0.5 M KCl, 20 mm MgCl2, and 10 mm ATP) to 9 volumes of G-actin. The working F-actin solution had a concentration of ∼300 μm. For the Mg-ATPase activity assays, myosin was added to Mg-ATPase solution (10 mm imidazole, pH 6.0, 0.1 mm CaCl2, 1 mm MgCl2, 1 mm [γ-32P]ATP) with increasing concentrations of F-actin (0–15 μm). The reaction was quenched using 1.8 N HClO4 after 25 min at room temperature prior to extraction and scintillation counting. Vmax and Km values were obtained by fitting data, after subtraction of basal Mg-ATPase values, with the Michaelis-Menten equation using Prism (GraphPad) software. ATPase values are presented as mean ± S.D. Statistical significance was assessed using unpaired t tests and Welch's correction, with significant differences assumed at p < 0.05.
Homology modelling
Three-dimensional homology models were generated for the Drosophila IFI myosin motor domain and exon 3 chimeras using the SWISS-MODEL automatic comparative protein modeling server (35–37). The embryonic myosin crystal structure (PDB ID: 5W1A) was used as a template to build a homology model of the IFI isoform, after pairwise alignment of the primary sequences of the Drosophila IFI and EMB S1 domains (96% conserved). To represent various states from the crossbridge cycle, scallop myosin structures were also used as templates: PDB IDs 1KK8 (actin-detached state), 1QVI (pre-power stroke state), and 1S5G (near-rigor ADP-bound state) using the CLUSTALW alignment protocol. The alignments were submitted to the alignment interface of SWISS-MODEL. The chosen templates allowed us to generate 3D homology models of the IFI and EMB myosin heads and of the chimeras obtained after swapping the exon 3 region (IFI-3a and EMB-3a).
Data availability
All data described in the manuscript are either contained within the manuscript or will be shared upon request to corresponding author.
Supplementary Material
Acknowledgments
We thank Martin Webb (National Institute for Medical Research, Mill Hill, London, UK) for the coumarin ATP/ADP used in this work; Aileen Knowles (San Diego State University) for help with Ca-ATPase data; Corey Dambacher, Jennifer Suggs, and Floyd Sarsoza (San Diego State University) for dissecting indirect flight muscles and preparing S1 samples; and Sam Lynn (University of Kent) for excellent technical support.
This article contains supporting information.
Author contributions—M. J. B., K. H. H., and S. I. B. data curation; M. J. B., K. H. H., and S. I. B. formal analysis; M. J. B. and K. H. H. investigation; M. J. B. and K. H. H. methodology; M. J. B. writing-original draft; K. H. H., M. A. G., and S. I. B. writing-review and editing; M. A. G. and S. I. B. conceptualization; M. A. G. resources; M. A. G. and S. I. B. supervision; M. A. G. and S. I. B. funding acquisition; M. A. G. and S. I. B. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grant GM32443 (to S. I. B.) and by Wellcome Trust Grant 085309 (to M. J. B. and M. A. G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MHC
- myosin heavy chain
- EMB
- embryonic muscle myosin isoform in Drosophila
- IFI
- adult indirect flight muscle isoform of myosin
- ELC
- essential light chain
- cmADP
- coumarin-ADP
- cATP
- caged ATP
- Tpm
- tropomyosin
- NTR
- N-terminal region
- OM
- omecamtiv mecarbil
- ANOVA
- analysis of variance.
References
- 1. George E. L., Ober M. B., and Emerson C. P. (1989) Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol. Cell Biol. 9, 2957–2974 10.1128/mcb.9.7.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernstein S. I., and Milligan R. A. (1997) Fine tuning a molecular motor: The location of alternative domains in the Drosophila myosin head. J. Mol. Biol. 271, 1–6 10.1006/jmbi.1997.1160 [DOI] [PubMed] [Google Scholar]
- 3. Wells L., Edwards K. A., and Bernstein S. I. (1996) Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 15, 4454–4459 10.1002/j.1460-2075.1996.tb00822.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swank D. D. M., Knowles A. F. A., Suggs J. A. J., Sarsoza F., Lee A., Maughan D. W., and Bernstein S. I. (2002) The myosin converter domain modulates muscle performance. Nat. Cell Biol. 4, 312–316 10.1038/ncb776 [DOI] [PubMed] [Google Scholar]
- 5. Miller B. M., Nyitrai M., Bernstein S. I., and Geeves M. A. (2003) Kinetic analysis of Drosophila muscle myosin isoforms suggests a novel mode of mechanochemical coupling. J. Biol. Chem. 278, 50293–50300 10.1074/jbc.M308318200 [DOI] [PubMed] [Google Scholar]
- 6. Swank D. M., Bartoo M. L., Knowles A. F., Iliffe C., Bernstein S. I., Molloy J. E., and Sparrow J. C. (2001) Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J. Biol. Chem. 276, 15117–15124 10.1074/jbc.M008379200 [DOI] [PubMed] [Google Scholar]
- 7. Swank D. M., Knowles A. F., Kronert W. A., Suggs J. A., Morrill G. E., Nikkhoy M., Manipon G. G., and Bernstein S. I. (2003) Variable N-terminal regions of muscle myosin heavy chain modulate ATPase rate and actin sliding velocity. J. Biol. Chem. 278, 17475–17482 10.1074/jbc.M212727200 [DOI] [PubMed] [Google Scholar]
- 8. Swank D. M., Kronert W. A., Bernstein S. I., and Maughan D. W. (2004) Alternative N-terminal regions of Drosophila myosin heavy chain tune muscle kinetics for optimal power output. Biophys. J. 87, 1805–1814 10.1529/biophysj.103.032078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caldwell J. T., Mermelstein D. J., Walker R. C., Bernstein S. I., and Huxford T. (2020) X-ray crystallographic and molecular dynamic analyses of Drosophila melanogaster embryonic muscle myosin define domains responsible for isoform-specific properties. J. Mol. Biol. 432, 427–447 10.1016/j.jmb.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bloemink M. J., Dambacher C. M., Knowles A. F., Melkani G. C., Geeves M. A., and Bernstein S. I. (2009) Alternative exon 9-encoded relay domains affect more than one communication pathway in the Drosophila myosin head. J. Mol. Biol. 389, 707–721 10.1016/j.jmb.2009.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Webb M. R., and Corrie J. E. T. (2001) Fluorescent coumarin-labeled nucleotides to measure ADP release from actomyosin. Biophys. J. 81, 1562–1569 10.1016/S0006-3495(01)75810-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clark R. J., Nyitrai M., Webb M. R., and Geeves M. A. (2003) Probing nucleotide dissociation from myosin in vitro using microgram quantities of myosin. J. Muscle Res. Cell Motil. 24, 315–321 [PubMed] [Google Scholar]
- 13. Miller B. M., Bloemink M. J., Nyitrai M., Bernstein S. I., and Geeves M. A. (2007) A variable domain near the ATP-binding site in Drosophila muscle myosin is part of the communication pathway between the nucleotide and actin-binding sites. J. Mol. Biol. 368, 1051–1066 10.1016/j.jmb.2007.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vale R. D., and Milligan R. A. (2000) The way things move: Looking under the hood of molecular motor proteins. Science 288, 88–95 10.1126/science.288.5463.88 [DOI] [PubMed] [Google Scholar]
- 15. Lowey S., Saraswat L. D., Liu H., Volkmann N., and Hanein D. (2007) Evidence for an interaction between the SH3 domain and the N-terminal extension of the essential light chain in class II myosins. J. Mol. Biol. 371, 902–913 10.1016/j.jmb.2007.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Falkenthal S., Parker V. P., Mattox W. W., and Davidson N. (1984) Drosophila melanogaster has only one myosin alkali light-chain gene which encodes a protein with considerable amino acid sequence homology to chicken myosin alkali light chains. Mol. Cell Biol. 4, 956–965 10.1128/mcb.4.5.956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siemankowski R. F., Wiseman M. O., and White H. D. (1985) ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc. Natl. Acad. Sci. U. S. A. 82, 658–662 10.1073/pnas.82.3.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nyitrai M., Rossi R., Adamek N., Pellegrino M. A., Bottinelli R., and Geeves M. A. (2006) What limits the velocity of fast-skeletal muscle contraction in mammals? J. Mol. Biol. 355, 432–442 10.1016/j.jmb.2005.10.063 [DOI] [PubMed] [Google Scholar]
- 19. Walklate J., Ujfalusi Z., and Geeves M. A. (2016) Myosin isoforms and the mechanochemical cross-bridge cycle. J. Exp. Biol. 219, 168–174 10.1242/jeb.124594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swank D. M., Vishnudas V. K., and Maughan D. W. (2006) An exceptionally fast actomyosin reaction powers insect flight muscle. Proc. Natl. Acad. Sci. U. S. A. 103, 17543–17547 10.1073/pnas.0604972103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang C., Kaplan C. N., Thatcher M. L., and Swank D. M. (2010) The influence of myosin converter and relay domains on cross-bridge kinetics of Drosophila indirect flight muscle. Biophys. J. 99, 1546–1555 10.1016/j.bpj.2010.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eldred C. C., Naber N., Pate E., Cooke R., and Swank D. M. (2013) Conformational changes at the nucleotide site in the presence of bound ADP do not set the velocity of fast Drosophila myosins. J. Muscle Res. Cell Motil. 34, 35–42 10.1007/s10974-012-9331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmid S., and Hugel T. (2020) Controlling protein function by fine-tuning conformational flexibility. Elife 9, e57180 10.7554/eLife.57180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colegrave M., and Peckham M. (2014) Structural implications of β-cardiac myosin heavy chain mutations in human disease. Anat. Rec. 297, 1670–1680 10.1002/ar.22973 [DOI] [PubMed] [Google Scholar]
- 25. Poetter K., Jiang H., Hassanzadeh S., Master S. R., Chang A., Dalakas M. C., Rayment I., Sellers J. R., Fananapazir L., and Epstein N. D. (1996) Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat. Genet. 13, 63–69 10.1038/ng0596-63 [DOI] [PubMed] [Google Scholar]
- 26. Robert-Paganin J., Auguin D., and Houdusse A. (2018) Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 9, 4019 10.1038/s41467-018-06191-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greenberg M. J., Lin T., Shuman H., and Ostap E. M. (2015) Mechanochemical tuning of myosin-I by the N-terminal region. Proc. Natl. Acad. Sci. U. S. A. 112, E3337–E3344 10.1073/pnas.1506633112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shuman H., Greenberg M. J., Zwolak A., Lin T., Sindelar C. V., Dominguez R., and Ostap E. M. (2014) A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning. Proc. Natl. Acad. Sci. U. S. A. 111, 2116–2121 10.1073/pnas.1321022111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winkelmann D. A., Forgacs E., Miller M. T., and Stock A. M. (2015) Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat. Commun. 6, 7974 10.1038/ncomms8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Planelles-Herrero V. J., Hartman J. J., Robert-Paganin J., Malik F. I., and Houdusse A. (2017) Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 8, 190 10.1038/s41467-017-00176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nanasi P., Komaromi I., Gaburjakova M., and Almassy J. (2018) Omecamtiv mecarbil: A myosin motor activator agent with promising clinical performance and new in vitro results. Curr. Med. Chem. 25, 1720–1728 10.2174/0929867325666171222164320 [DOI] [PubMed] [Google Scholar]
- 32. Silva R., Sparrow J. C., and Geeves M. A. (2003) Isolation and kinetic characterisation of myosin and myosin S1 from the Drosophila indirect flight muscles. J. Muscle Res. Cell Motil. 24, 489–498 10.1023/B:JURE.0000009809.69829.74 [DOI] [PubMed] [Google Scholar]
- 33. Weiss S., Chizhov I., and Geeves M. A. (2000) A flash photolysis fluorescence/light scattering apparatus for use with sub microgram quantities of muscle proteins. J. Muscle Res. Cell Motil. 21, 423–432 10.1023/A:1005690106951 [DOI] [PubMed] [Google Scholar]
- 34. Pardee J. D., and Spudich J. A. (1982) Purification of muscle actin. in Structural and Contractile Proteins Part B: The Contractile Apparatus and the Cytoskeleton (Frederiksen D. W., and Cunningham L. W., eds.), Vol. 85, pp. 164–181, Academic Press, Cambridge, MA [Google Scholar]
- 35. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 37. Schwede T., Kopp J., Guex N., and Peitsch M. C. (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are either contained within the manuscript or will be shared upon request to corresponding author.