Abstract
Bardet–Biedl syndrome (BBS) is a pleiotropic ciliopathy caused by dysfunction of primary cilia. More than half of BBS patients carry mutations in one of eight genes encoding for subunits of a protein complex, the BBSome, which mediates trafficking of ciliary cargoes. In this study, we elucidated the mechanisms of the BBSome assembly in living cells and how this process is spatially regulated. We generated a large library of human cell lines deficient in a particular BBSome subunit and expressing another subunit tagged with a fluorescent protein. We analyzed these cell lines utilizing biochemical assays, conventional and expansion microscopy, and quantitative fluorescence microscopy techniques: fluorescence recovery after photobleaching and fluorescence correlation spectroscopy. Our data revealed that the BBSome formation is a sequential process. We show that the pre-BBSome is nucleated by BBS4 and assembled at pericentriolar satellites, followed by the translocation of the BBSome into the ciliary base mediated by BBS1. Our results provide a framework for elucidating how BBS-causative mutations interfere with the biogenesis of the BBSome.
Keywords: Bardet-Biedl Syndrome, BBSome, assembly, cilium, ciliopathy, protein sorting, protein assembly, primary cilium, microscopic imaging, genetic disease, Bardet-Biedl syndrome
Bardet–Biedl Syndrome (BBS) is a multiorgan genetic disorder caused by the dysfunction of the primary cilia, microtubule-based sensory organelles. BBS is primarily characterized by retinopathy, polydactyly, genital and renal anomalies, obesity, and cognitive impairment (1). Twenty-four causative BBS genes have been identified so far, eight of which form a stable protein complex called the BBSome (2, 3). The BBSome is an adaptor protein complex consisting of BBS1, -2, -4, -5, -7, -8, -9, and -18, possessing structural similarities to coat/adaptor proteins involved in vesicular trafficking (3, 4). The BBSome works in concert with the intraflagellar transport machinery to facilitate the trafficking of particular transmembrane cargoes into and out of the primary cilia (5–9). The role of the BBSome is pleiotropic as it mediates proper outcomes of multiple signaling pathways, including the Sonic Hedgehog signaling (10), leptin signaling (11), photoreceptor signaling (12), and neuronal signaling by G protein–coupled receptors (13–15).
Mutations in any of the BBSome subunits can cause BBS, suggesting that every subunit is essential for complete BBSome function (2). The structure of the BBSome was a long-standing enigma until recently, because the indirect approaches, such as the yeast two-hybrid system (16), co-precipitation (17), co-expression of the individual subunits followed by low-resolution cryo-EM (18), and structural analysis of individual BBSome subunits (19, 20), provided only a very limited insight into the overall BBSome structure. Moreover, some authors proposed that BBS2, -7, and -9 form the BBSome core enabling the subsequent recruitment of other subunits (21), whereas others showed that BBS2 and BBS7 are dispensable for the assembly of the remaining six subunits (18, 22).
Three independent studies resolved the BBSome structure using cryo-EM with molecular modeling using the BBSome isolated from bovine retina (23) or using high-resolution cryo-EM analysis of bovine BBSome (24) or of human BBSome (lacking BBS2 and BBS7) expressed in insect cells (22). These studies showed a high level of interconnectivity among the BBSome subunits, raising the question of how these BBSome proteins assemble in the cells.
Quantitative fluorescence microscopy techniques can reveal the formation and dynamics of functional protein complexes in living cells (25, 26). In this study, we applied multiple microscopy and biochemical techniques to a large library of genetically engineered human RPE1 cells to describe spatially resolved steps of BBSome formation in cells.
Results
Generation of a cell line library for studying BBSome formation
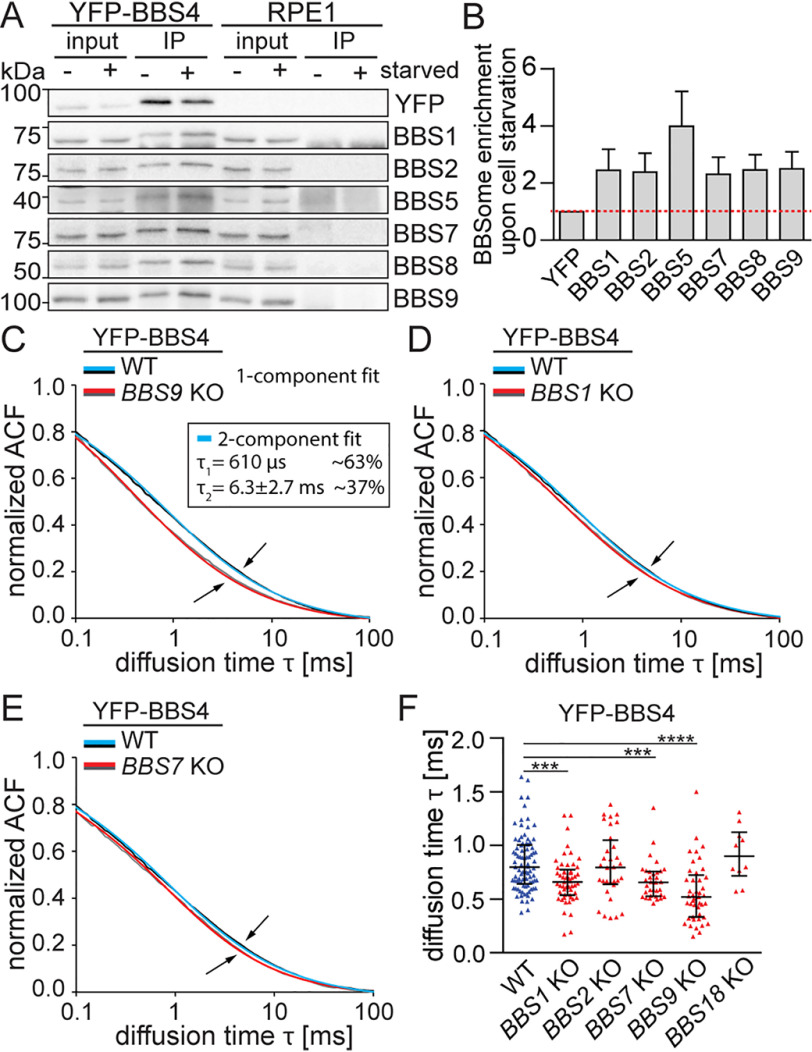
To study BBSome formation in cells, we initially established stable WT RPE1 cell lines expressing BBSome subunits fused with super yellow fluorescent protein SYFP2 (YFP) (27). BBS1, BBS4, BBS8, and BBS18 were tagged at the N termini, whereas the BBS7 and BBS9 were tagged at C termini, because the N-terminal tag interfered with their function (data not shown). All YFP-tagged BBSome subunits localized to the primary cilia of WT RPE1 cells and showed weak diffuse signals throughout the cytoplasm (Fig. 1A). The pattern of the diffused cytoplasmic localization of the BBSome subunits was not affected by the cell density. For all the following experiments, the cells were kept 80–100% confluent. YFP-BBS4 and YFP-BBS18 additionally localized to the pericentriolar satellites (PS) around the basal body as shown before (Fig. 1A) (28, 29).
Figure 1.
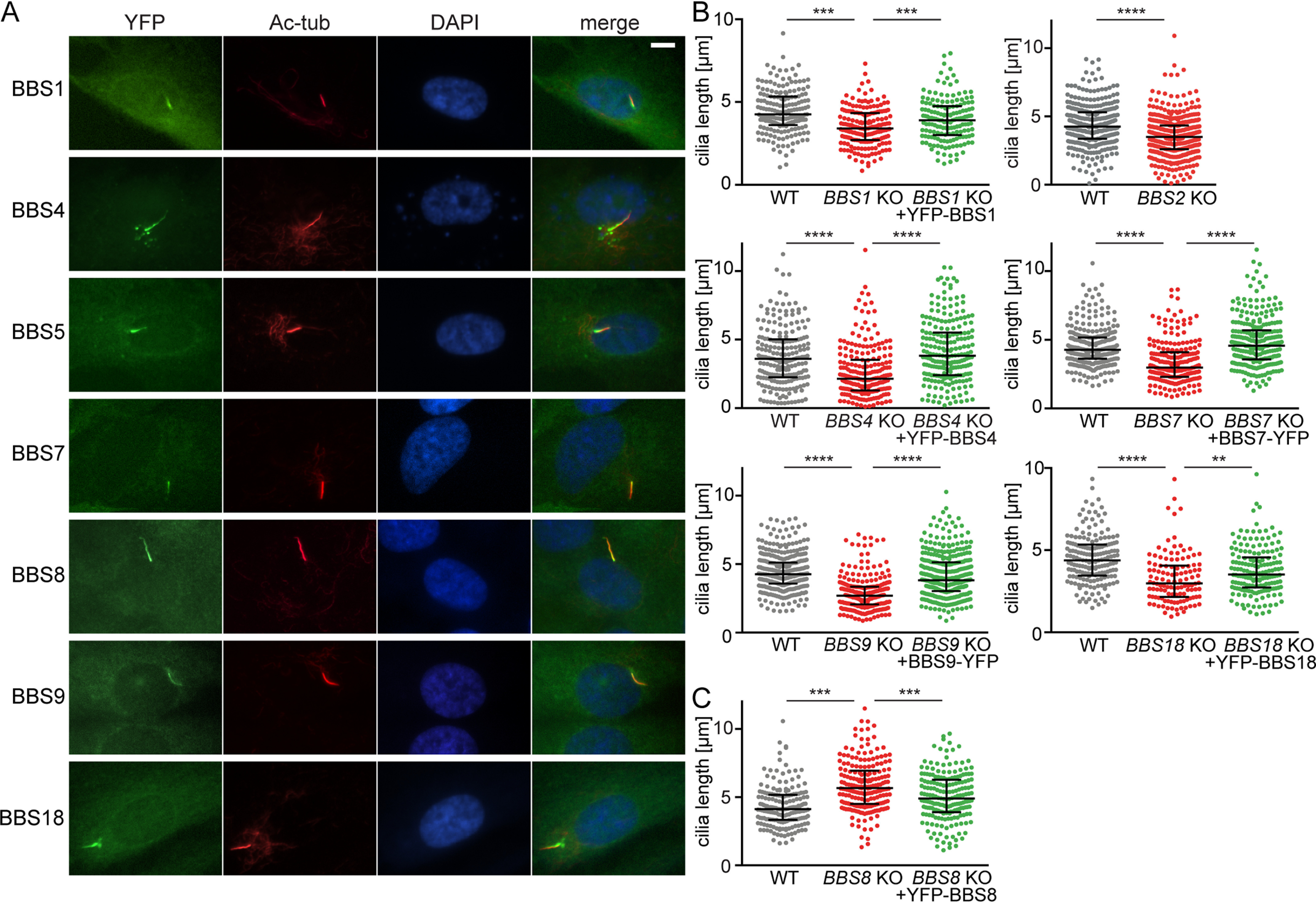
YFP-tagged BBSome subunits localize to the primary cilia and rescue the BBSome deficient cell lines. A, YFP-tagged BBSome subunits localize to the ciliary base and primary cilia upon serum starvation in WT RPE1 cells. Antibody against Ac-tub was used to stain the primary cilia. The nucleus was stained with DAPI. Scale bar, 5 μm. B, plots show the comparison of ciliary length between WT, BBS KO, and reconstituted BBS KO RPE1 cell lines. Ciliary length was rescued upon expression of the respective YFP-tagged BBSome subunit. Cilia length measurements were carried out using the Fiji ImageJ software. Medians with interquartile range from three independent experiments of n > 100 are shown. Statistical significance was calculated using two-tailed Mann–Whitney test. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C, plot shows the comparison of ciliary length between WT, BBS8 KO, and reconstituted BBS8 KO RPE1 cell lines. Ciliary length was restored to WT length upon expression of YFP-BBS8 in BBS8 KO cell line. Cilia length measurements were carried out using the Fiji ImageJ software. Medians with interquartile range (error bars) from three independent experiments of n > 100 are shown. Statistical significance was calculated using two-tailed Mann–Whitney test. ***, p < 0.001.
In the next step, we generated RPE1 cell lines deficient in BBS1, BBS2, BBS4, BBS7, TTC8/BBS8, BBS9, or BBIP1/BBS18 using the CRISPR/Cas9 technology (Table S1). Each of the seven BBS-deficient cell lines was subsequently transduced with retroviral vectors to express YFP-tagged BBS1, -4, -5, -7, -8, -9, or -18, giving rise to a library of 63 stable cell lines in total (Table S2). Lack of any of the BBSome subunits prevented the ciliary localization of YFP-tagged BBS1, BBS4, BBS5, BBS7, BBS8, BBS9, and BBS18 (Fig. S1). Likewise, the endogenous BBS9 was rather diffused throughout the cytoplasm in the KO cells, whereas it localized to primary cilia in WT cells and KO cells reconstituted with the missing YFP-tagged subunits (e.g. YFP-BBS1 in BBS1 KO) (Fig. S2A). These data suggested that only the intact BBSome, and not BBSome intermediates or subunits alone, could enter cilia.
Despite substantial overexpression of the YFP-tagged subunits over their endogenous counterparts (Fig. S2B), the fluorescence signal was still rather weak and mostly comparable in the respective WT and KO cell lines (Figs. S1 and S2C).
Cells deficient in BBS1, BBS4, BBS5, BBS7, BBS9, or BBS18 formed significantly shorter cilia compared with WT cells (Fig. 1B), corresponding to a previous observation in BBS4-deficient cells (30). This phenotype was rescued by expressing YFP-tagged variants of the missing genes (Fig. 1B), documenting both that the cilia shortening was indeed caused by the gene deficiencies and that the YFP-tag does not interfere with the function of the BBSome subunits. Intriguingly, deletion of BBS8 resulted in prolonged cilia, which was reverted by the expression of YFP-BBS8 (Fig. 1C). The unexpected phenotype of the BBS8 KO cell line could be caused by a unique role of BBS8 or could be an artifact of the particular mutation.
BBSome subunits interact in the cytoplasm
Deficiency of any BBSome subunit reduced the endogenous levels of the other subunits in RPE1 KO cell lines (Fig. S3 (A and B) and Table S1), indicating that the BBSome and/or BBSome intermediates are more stable than free individual subunits. In particular, deficiency in any of the subunits forming the proposed core of the BBSome (i.e. BBS2, BBS7, or BBS9) (21) substantially reduced the cellular levels of other subunits, whereas the absence of BBS1, BBS4, BBS8, or BBS18 had less dramatic effects on the stability of other subunits. The interdependence of the individual BBSome subunits suggests that they are present predominantly in the form of the BBSome or BBSome intermediates in WT cells.
We addressed whether ciliogenesis (stimulated by serum starvation) is coupled with de novo formation of the BBSome or whether the BBSome is preformed in nonciliated cells. BBS1, -2, -5, -7, -8, and -9 co-immunoprecipitated with YFP-BBS4 in the nonstarved cells (Fig. 2A). However, the serum starvation increased the amount of co-precipitated BBSome subunits ∼2–4-fold, indicating that the BBSome formation is slightly augmented upon induction of ciliogenesis in RPE1 cells (Fig. 2 (A and B) and Fig. S4A). Under these conditions, only 5% of cells formed cilia, which suggested that the BBSome assembles also in the nonciliated cells (Fig. S4A).
Figure 2.
BBSome subcomplexes are present in the cytoplasm. A, co-immunoprecipitation (IP) of the BBSome subunits from YFP-BBS4 WT RPE1 cell lines using anti-GFP antibodies before and after 24-h serum starvation. A representative immunoblot out of three experiments is shown. B, quantification of the co-precipitated BBSome subunits was performed using the Fiji ImageJ software. The amount of the BBSome subunits was normalized to the YFP-BBS4 amount detected on the respective membrane. The mean with S.D. (error bars) of three experiments is shown. C–E, FCS measures the in vivo mobility of the BBSome subcomplexes in the cytoplasm. Plots show the autocorrelation functions (ACFs) obtained from FCS measurements in the cytoplasm of YFP-BBS4 in WT and in BBS9 KO (C), BBS1 KO (D), and BBS7 KO (E). The ACFs were fitted with a one-component anomalous 3D diffusion model (Table S3). The means of n > 20 measurements are shown. Note the elevated mobility of YFP-BBS4 in the BBS9 KO compared with BBS1 KO and BBS7 KO cells (arrows). YFP-BBS4 WT ACFs were additionally fitted by a two-component fit (inset). Diffusion time τ1 of the fast component was fixed to the mean diffusion time τ obtained from BBS9 KO cells, yielding the diffusion time τ2 of YFP-BBS4 engaged in a putative BBS4-BBS9 subcomplex. F, plot shows the diffusion times, τ, obtained by fitting the ACFs acquired from FCS measurements in YFP-BBS4 WT and BBS KO cell lines. Medians with interquartile range of n > 10 are shown. Statistical significance was calculated using a two-tailed Mann–Whitney test. ***, p < 0.001; ****, p < 0.0001.
The presence of the BBSome in nonciliated cells suggests that the BBSome or BBSome intermediates are present in the cytoplasm. Using fluorescence correlation spectroscopy (FCS), we estimated the diffusion speed of YFP-tagged subunits in WT and KO cell lines (Table S3). Because the large proteins and complexes diffuse slower than small complexes, these measurements reveal the information about the relative size of the respective complexes. We observed that the diffusion speed of YFP-BBS4 was significantly faster in BBS9 KO cells than in WT cells, providing evidence that a fraction of BBS4 resides in a BBS9-dependent complex in the cytoplasm (Fig. 2, C and F). We performed a two-component fitting of the WT data and found that the diffusion speed of this complex is about 10 times slower than the free YFP-BBS4 (Fig. 2C). Significant but less pronounced effects were observed in BBS1 KO and BBS7 KO (Fig. 2, D–F). These results show that cytoplasmic BBS4 exists in a form of a complex with BBS9 and likely other BBSome subunits.
YFP-tagged BBSome subunits, other than BBS4, showed significant differences in their mobility between WT and KO cells only in some rare cases (Fig. S4, B–E). It is possible that their overexpression masked the effect of BBSome disruption by favoring the monomeric forms (Fig. S3, B and C). In the case of YFP-BBS1, we could observe a mild loss in the slower fraction in BBS7 or BBS9 KO cells (Fig. S4C). Overall, these data imply the existence of heterogenic complexes of BBSome subunits in the cytoplasm.
The pre-BBSome assembles at the pericentriolar satellites
BBS4 interacts with PCM-1 (pericentriolar material-1) (28), which leads to its enrichment at the PS in both ciliated and nonciliated cells (Fig. S5A). BBS9 localizes to PS as well, albeit to a much lesser extent than BBS4 (Fig. 3, A and B), indicating that a fraction of BBS9 and potentially other BBSome subunits reside at the PS in starved WT cells. Surprisingly, in BBS1 KO cells, both the BBS4 and BBS9 were highly enriched at the PS (Fig. 3, A and B). In addition, we observed that all other BBSome subunits (i.e. YFP-tagged BBS5, BBS7, BBS8, BBS9, and BBS18 and endogenous BBS2 and BBS9) localize to PS in BBS1 KO cells (Fig. 3C and Fig. S5B). The enrichment of the BBSome subunits at the PS depended on BBS4, because it was not observed in BBS4-deficient cells and BBS1/BBS4 DKO cells (Fig. 3D). To analyze the localization of BBS9 with higher resolution, we employed expansion microscopy (31). In line with the previous observations, BBS9 localized to the cilium in WT cells, localized to PS in BBS1 KO cells and was absent from the ciliary/centrosomal proximity in BBS4 KO cells (Fig. 3E).
Figure 3.
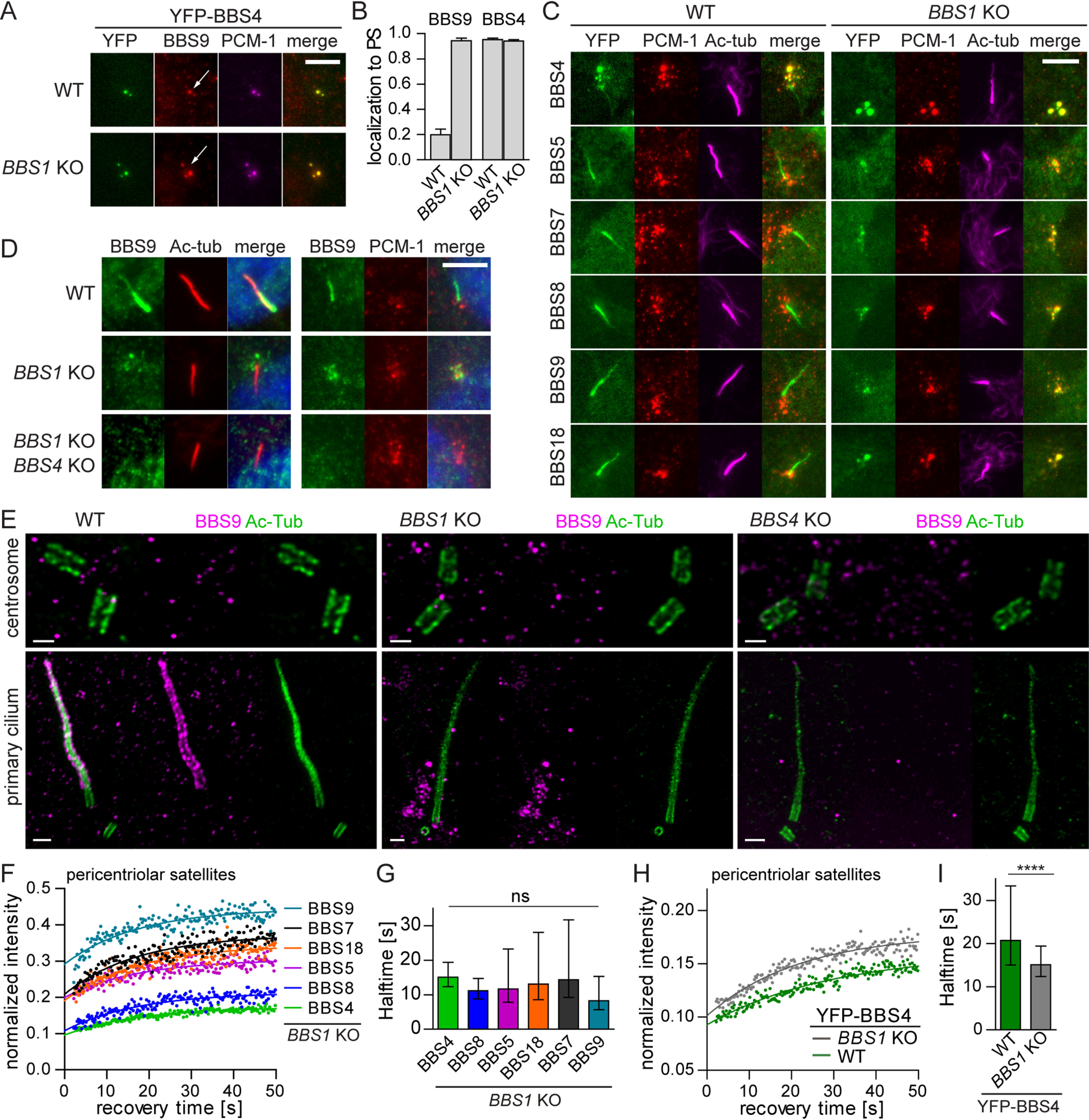
Loss of BBS1 stalls the pre-BBSome at the pericentriolar satellites. A, YFP-BBS4 along with endogenous BBS9 localize to the PS both in WT and BBS1 KO RPE1 cells (white arrows). BBS9 localization at the PS is pronounced in the BBS1 KO RPE1 cells compared with WT RPE1 cells. Antibody against PCM-1 was used to mark the PS. Scale bar, 5 μm. B, quantification of localization of YFP-BBS4 and endogenous BBS9 to the PS in WT and BBS1 KO RPE1 cells. Means and S.D. of two and three independent experiments, respectively, are shown. C, YFP-tagged BBSome subunits localize to the primary cilia (Ac-tub) in WT RPE1 cells but are accumulated at the PS (PCM-1) in BBS1 KO RPE1 cell lines. Scale bar, 5 μm. D, endogenous BBS9 localizes to the primary cilia (Ac-tub) and the PS (PCM-1) in WT and BBS1 KO RPE1 cells, respectively. Deletion of BBS4 on top of BBS1 deficiency results in BBS9 dispersal form PS to the cytoplasm. Scale bar, 5 μm. E, micrographs of BBS9 localization using expansion microscopy. BBS9 localizes to the cilia (Ac-tub) in WT RPE1 cells, around the ciliary base, and at the PS in BBS1 KO and is diffused in BBS4 KO. Scale bar, 2 μm. F, FRAP analysis of the dynamic turnover of the YFP-tagged BBSome subunits at the PS in BBS1 KO RPE1. Recovery curves were fitted at once using one-phase association fit. The mean of 20–30 measurements from three independent experiments is shown. Estimated half-times t½ and immobile fractions are shown in Table S4. G, bar graph depicts recovery half-times (s) of YFP-tagged BBSome subunits in BBS1 KO cell lines obtained from FRAP analysis in F. Means of 20–30 measurements from three independent experiments are shown. Error bars, 95% confidence interval. ns, not significant; p > 0.05. H, FRAP analysis of the dynamic turnover of the YFP-BBS4 at PS in WT and BBS1 KO RPE1 cells. Recovery curves were fitted using one-phase association fit. The mean of 20–30 measurements from three independent experiments is shown. Estimated half-times t½ and immobile fractions are shown in Table S4. I, bar graph depicts recovery half-times (s) of YFP-BBS4 in WT and BBS1 KO cell lines obtained from FRAP analysis in H. Means of 20–30 measurements from three independent experiments are shown. Error bars, 95% confidence interval. The statistically significant difference was calculated by Mann–Whitney test. ****, p < 0.0001.
Fluorescence recovery after photobleaching (FRAP) analysis revealed comparable recovery half-times for all YFP-tagged BBSome subunits in BBS1 KO cells at the PS (Fig. 3 (F and G) and Table S4), possibly because they are predominantly engaged in a single complex. YFP-BBS4 showed the highest immobile fraction, which is consistent with its direct binding to the PS via PCM-1 (Fig. 3F). Most likely, BBS4 recruits other subunits to the PS, leading to the formation of a pre-BBSome complex. The accumulation of the BBSome intermediates at the PS in BBS1 KO cells results in the mobilization of BBS4 (Fig. 3 (H and I) and Table S4), indicating that the pre-BBSome binding to PS is less stable than binding of BBS4 alone. BBS1 is crucial for the completion of the full BBSome, which prevents the BBSome subunits, with the exception of BBS4, from the arrest at the PS.
BBS1 facilitates BBSome translocation from the basal body to the cilium
As all BBSome subunits, BBS1 localizes to the cilia in WT cells (Fig. 1A). However, we observed also quite frequent BBS1 abundance at the centrosome in nonciliated cells and at the centrosome/basal body in some cells with short nascent cilia (Fig. 4A). We calculated that BBS1 localizes to the centrosome in ∼80% nonciliated cells, whereas the other BBSome subunits localize only in 10–30% nonciliated cells (Fig. S5C). Moreover, BBS1 localizes to the centrosome/basal body in BBS4 KO cells, whereas other BBSome subunits show diffuse localization in the cytoplasm of these cells (Fig. 4B). Deficiency of BBS4 increased the turnover of BBS1 at the centrosome/basal body (Fig. 4, C and D), suggesting that the interaction of BBS1 with the pre-BBSome stabilizes BBS1 at the centrosome/basal body, possibly directing the whole complex toward the cilium.
Figure 4.
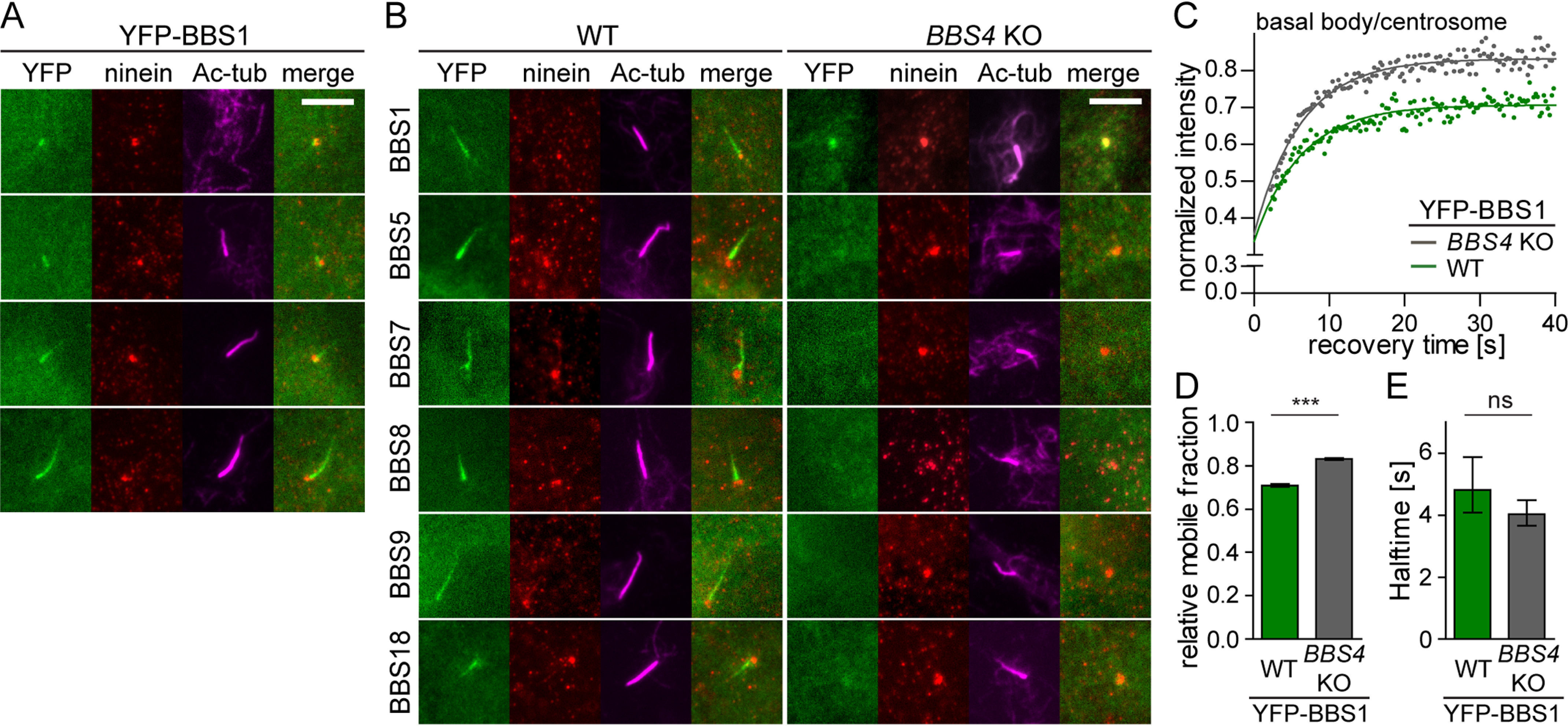
BBS1 acts as a gatekeeper at the basal body to facilitate BBSome completion and its entry to cilia. A, YFP-tagged BBS1 transits through the centrosome into the ciliary base and the cilia in WT RPE1 cells. Ninein and acetylated tubulin staining visualizes the centrosome and cilia, respectively. Scale bar, 5 μm. B, YFP-tagged BBSome subunits localize to the primary cilia (Ac-tub) in WT RPE1 cell lines. YFP-BBS1 localizes to the centrosome (Ninein) in the BBS4 KO cells, whereas the other YFP-tagged BBSome subunits are diffused in the cytoplasm. Scale bar, 5 μm. C, FRAP analysis of the dynamic turnover of the YFP-BBS1 at the centrosome or basal body in WT and BBS4 KO RPE1 cells. Recovery curves were fitted using one-phase association fit, and the means of 20–30 FRAP measurements from three independent experiments are shown. Estimated half-times t½ and immobile fractions are shown in Table S4. D, bar graph depicts the relative mobile fraction of YFP-BBS1 in WT and BBS4 KO cells extracted from FRAP analysis in C. YFP-BBS1 has a greater mobile fraction in BBS4 KO than in WT RPE1 cells. Means of 20–30 measurements from three independent experiments are shown. Error bars, 95% confidence interval. ***, p < 0.001. E, bar graph depicts the recovery half-time of YFP-BBS1 in WT and BBS4 KO cells obtained from FRAP analysis (C). Means of 20–30 measurements from three independent experiments are shown. Error bars, 95% confidence interval. ns, not significant; p > 0.5.
We performed an analysis of the dynamic behavior of the BBSome subunits at the ciliary tip and the basal body and the transition zone using FRAP (Fig. 5 (A–C) and Table S5). If the subunits are incorporated in the whole BBSome quantitatively, their recovery rate should be comparable. However, BBS1 behaved differently from the other subunits, because it was very mobile at the base of the cilium (Fig. 5C and Fig. S5E). Indeed, BBS1 was the only subunit that was more dynamic in the ciliary base than in the ciliary tip (Fig. 5D and Fig. S5 (D and E)). These data indicate that BBS1 exists in two forms at the ciliary base, as a monomeric protein with fast turnover between the ciliary base and the cytoplasm and as a part of the BBSome, whereas other BBSome subunits are recruited to the ciliary base only in the form of the complete BBSome complex. Interestingly, the absence of BBS1 not only stalls BBSome at the PS, but alters the structure of the ciliary base, resulting in a slightly prolonged basal body and transition zone (Fig. 5, E and F), suggesting a role of the BBS1 in the organization of the ciliary base.
Figure 5.
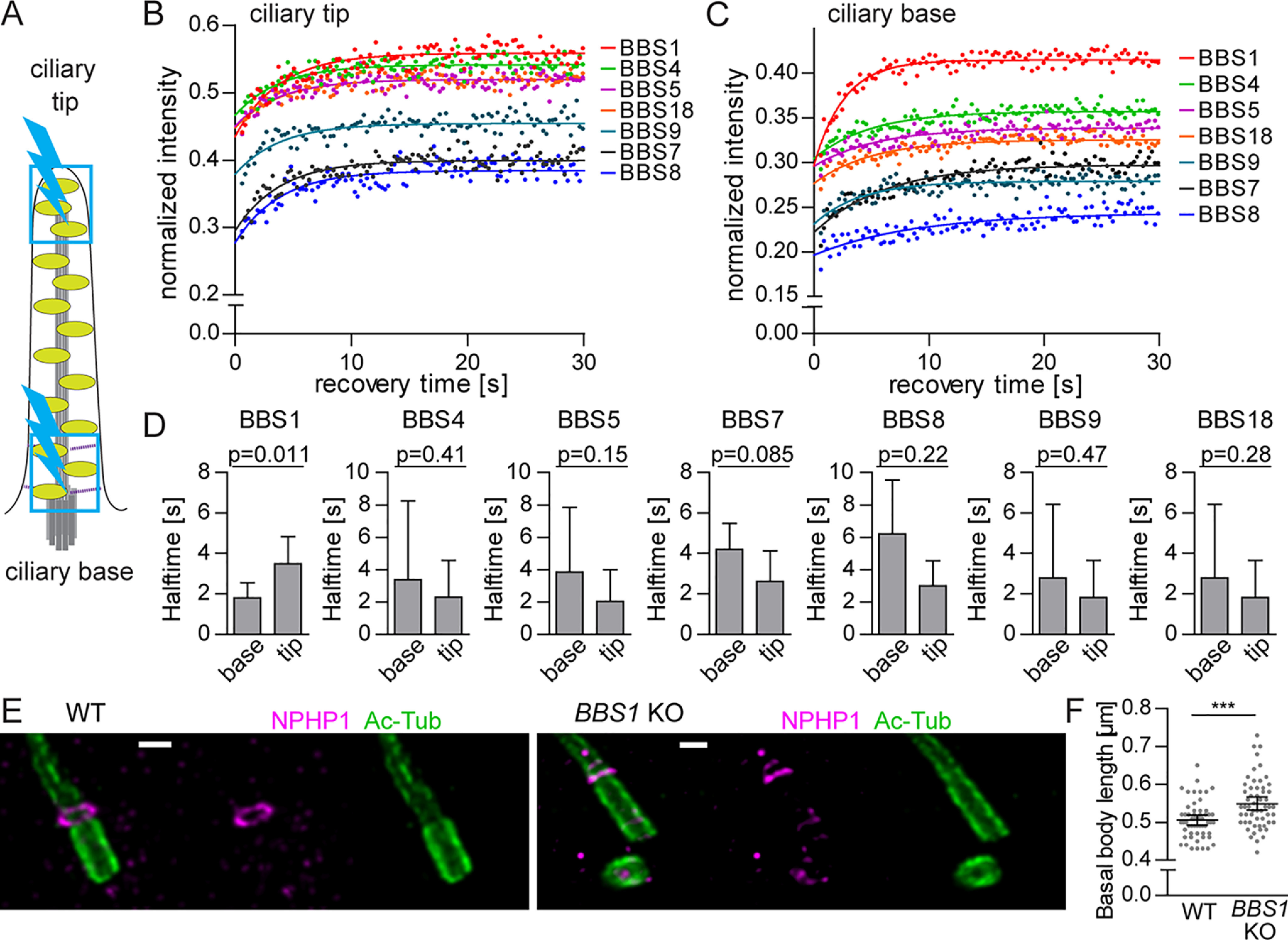
BBS1 has a distinct dynamic behavior at ciliary base. A, schematic representation of the FRAP experiment in the primary cilia. Ciliary base and tip (blue boxes) were bleached for 0.3 s using a 20-milliwatt 488-nm solid-state laser at 100% intensity (blue lightning bolt). B and C, FRAP analysis of the dynamic turnover of the YFP-tagged BBSome subunits at the ciliary base (B) and tip (C). FRAP curves were fitted with one-phase association fit. Means of 20–30 measurements from three independent experiments are shown. Estimated half-times t½ and immobile fractions are shown in Table S5. D, bar graphs depict the recovery half-times of BBSome subunits at the ciliary tip and base. Means of 20–30 measurements from three independent experiments are shown. Error bars, 90% confidence interval. E, micrographs of basal body (Ac-tub) and transition zone (NPHP1) staining in WT and BBS1 KO RPE1 cells using expansion microscopy. Scale bar, 2 μm. F, plot of the basal body lengths quantified based on the expansion microscopy performed in WT and BBS1 KO cell lines in E. BBS1 KO cells have significantly longer basal bodies compared with WT. Statistical significance was calculated using two-tailed Mann–Whitney test. Means with S.D. (error bars) of n = 56 are shown. ***, p < 0.001.
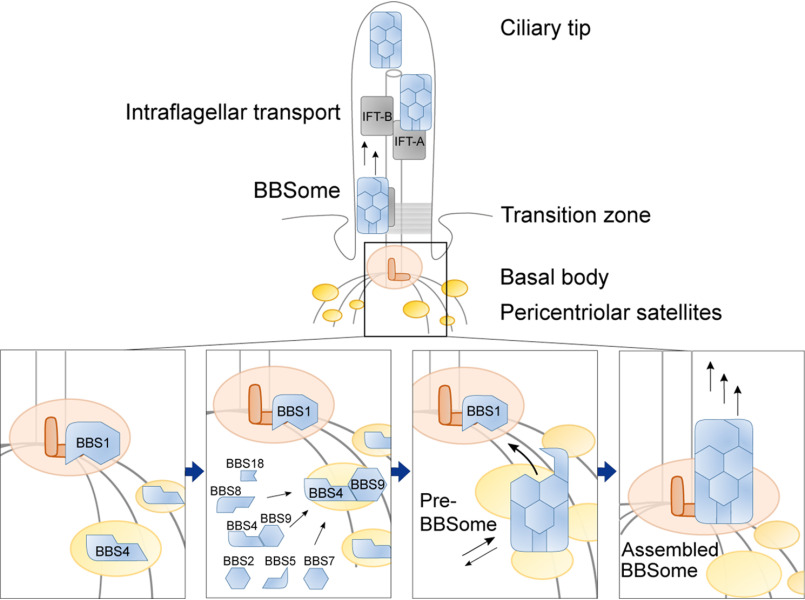
Based on our data, we propose a model of the BBSome formation in cells with the main roles for BBS4 and BBS1 in the spatial regulation of the full complex assembly (Fig. 6). BBS4 resides on PS, and via its association with BBS9, it recruits other BBSome subunits to form the pre-BBSome complex. BBS1 localizes to the basal body and guides the pre-BBSome to the ciliary base, where it facilitates the BBSome translocation to the cilium.
Figure 6.
Schematic model of the spatially resolved formation of the BBSome in human cells. BBS4 is a resident protein at PS, whereas BBS1 resides at the basal body. BBS4 associates with BBS9 in the cytoplasm, which directs the BBSome subunits to the PS to assemble the pre-BBSome. Pre-BBSome is stabilized by BBS1 at the basal body, resulting in the full BBSome competent to enter the cilia. In cilia, full BBSome moves toward the ciliary tip and back to the ciliary base to exert its cargo adaptor function.
Discussion
Because the BBSome consists of eight subunits, it has been proposed that it assembles in a stepwise manner (21). However, in vitro experiments provided contradictory data concerning the sequence of the individual steps (18, 21, 22). Although the resolution of the structure of the BBSome complex represented a breakthrough in the field (22–24), these studies could not reveal how the BBSome assembles in living cells, including the spatial regulation of individual steps. We generated a library of 64 RPE1-derived cell lines using a combination of CRISPR/Cas9 knockouts of individual BBSome subunits and retroviral expression of YFP-tagged subunits. The only misses in the library were BBS5 KO and BBS2-YFP, because we were unsuccessful in their preparation. We used these lines to address the localization of individual BBSome subunits in WT cells and in cells lacking another subunit. The signal from YFP-tagged proteins was complemented with the detection of endogenous BBS9 and BBS2 using immunofluorescence. Moreover, we used two microscopy techniques, FCS and FRAP, to elucidate the dynamics of individual subunits in our cell lines. Altogether, our data enabled us to propose a model of BBSome formation in living cells.
It has been shown that some BBSome subunits stabilize each other in cells (21, 32). Our systematic analysis demonstrated that actually all BBSome subunits are interdependent. Deficiency in any of the subunits (with the exception of BBS5, which was not tested) leads to the substantial decrease of the cellular levels of other subunits, indicating that only a minor fraction of the BBSome subunit molecules are monomeric. Deficiency of BBS2, BBS7, or BBS9 had the most pronounced effects on the abundance of other subunits, suggesting that these three structurally related proteins form the core of the BBSome in vivo (21). We showed that BBSome is readily assembled in nonciliated cells, and ciliogenesis only modestly augments the BBSome formation. Our FCS approach detected some direct or indirect cytoplasmic interactions of BBSome subunits, most notably BBS4-BBS9. Altogether, our data show that BBSome and/or its intermediates are present in the cytoplasm and that the cilia is not required for the BBSome formation.
The PS organize proteins involved in the centrosome maintenance and ciliogenesis (33, 34). It has been shown previously that BBS4 localizes to the PS, where it interacts with resident proteins PCM-1, Cep290, and AZI1 (28, 29, 35). Our data indicated that BBS4 recruits other BBSome subunits to the PS, where a pre-BBSome complex is formed. Indeed, BBS4 together with BBS2, BBS5, BBS7, and BBS9, but not BBS1, were detected in the proteome of the PS (33). Absence of PCM-1 disrupts the PS and reduces ciliogenesis (3) but does not prevent the ciliary localization of BBS4 (35). However, other PS proteins, Cep72 and Cep290, mediate ciliary localization of BBS4 and BBS8, suggesting that they orchestrate the pre-BBSome formation (35). Our data show that the pre-BBSome is released from the PS and targeted to the basal body via BBS1, the only subunit that shows an intrinsic affinity for the centrosome/basal body.
Depletion of AZI1 leads to enhanced localization of BBS4 into the cilium and restores the ciliary localization of BBS9 in the absence of BBS5 and partially in the absence of BBS2 and BBS8, but not in the absence of BBS1 (29). Although these experiments used RNAi, which usually does not lead to the complete loss of the target protein, the data are in line with the fact that BBS1 is essential for the ciliary BBSome targeting. AZI1 might negatively regulate the release of the pre-BBSome from the PS. Interestingly, deficiency of AZI1 in zebrafish leads to the typical BBS symptoms (29), suggesting that the role of AZI1 is important for the BBSome function. LZTFL1 is a BBSome-interacting partner showing a similar loss-of-function phenotype to AZI1, including the ciliary localization of the incomplete BBSome (36) and the development of BBS symptoms, in this case in humans and mice (37, 38). It is tempting to speculate that AZI1 and/or LZTFL1 mediate a quality control mechanism blocking the release of incomplete pre-BBSome from the PS. As BBS5 is a relatively peripheral subunit of the BBSome (22–24) that is substoichiometrically incorporated into the complex in vitro (22), a control mechanism checking the incorporation of BBS5 into the pre-BBSome in cells seems to be plausible.
Earlier studies suggested that the sequestration of the BBS4 at the PS might deplete the pool of BBS4 accessible for the BBSome formation (29, 35). However, the experimental data presented in these studies are consistent with our model of the pre-BBSome assembly at the PS and its subsequent release. Moreover, the putative quality control function of LZTFL1 and AZI1 might explain why the depletion of these proteins causes a seemingly paradoxical coincidence of enhanced ciliary localization of BBSome subunits as well as the induction of BBS-like phenotypes. Last, but not least, it would be difficult to reconcile the model of BBS4 sequestration at the PS with our observation that all BBSome subunits are strongly enriched at the PS in the absence of BBS1.
Our model proposes that BBS1 is incorporated into the BBSome as the last subunit, because of its affinity to the centrosome/basal body. However, we cannot exclude the possibility that the last step of the BBSome formation is not the incorporation of BBS1 itself but rather another BBS1-dependent event. It could be a conformational change in the BBSome, caused by a BBS1-interacting GTPase, BBS3/ARL6 (9, 19, 24). In any case, BBS3 seems to be involved in the translocation of the (pre-)BBSome from the PS to the ciliary base, as the BBSome-BBS3-GTP does not co-purify with PCM-1 (3, 9), and BBS3 was proposed to mediate the interaction of the BBSome with ciliary membranes (9, 24). BBS1 has been shown to interact with RABIN8, a guanine-exchange factor for RAB8, which is involved in vesicular trafficking and might regulate BBSome targeting to the ciliary base (3). Overall, it seems likely that BBS1 facilitates an energy-consuming conformational change of the complex that eventually confines the BBSome in the cilia.
The slight prolongation of the basal body in the BBS1-deficient cells suggests a role of BBS1 or the BBSome in the structural organization of the basal body. One potential explanation is that the BBSome regulates the activity of CP110, a protein controlling the length of the basal body (39), via their common interactor CEP290 (40, 41). However, the corresponding mechanism should be addressed in further studies.
Chaperonins BBS6, BBS10, and BBS12 assist the formation of the BBSome (21, 42), and the loss of any of them leads to the BBS (43–45). Because the deficiency in BBS1 or BBS3 generally presents with a less severe BBS disease than the deficiency in BBS2, BBS7, or BBS9 core proteins or in the BBS chaperonins (2), it is probable that the chaperonin complex assists any of the initial steps of the BBSome formation as proposed previously (21) rather than the BBS1-dependent translocation into the ciliary base. However, this should be experimentally addressed in further studies.
Overall, our study reveals that the BBSome formation is a sequential process with spatially compartmentalized steps. The formation of the pre-BBSome is nucleated by BBS4 at the PS, whereas BBS1 drives the translocation of the BBSome to the ciliary base. It shall be addressed in follow-up studies whether some of the multiple reported causative mutations (2) interfere with the process of the BBSome assembly.
Experimental procedures
Antibodies
Mouse anti-acetylated tubulin was kindly provided by Dr. Vladimir Varga (Institute of Molecular Genetics of the Czech Academy of Sciences), and rabbit anti-BBS4 was kindly provided by Prof. Maxence Nachury (University of California, San Francisco, CA, USA). Rabbit antibodies against BBS1 (ab222890) and acetylated tubulin (ab179484) were purchased from Abcam. Mouse antibodies against BBS2 (sc-365355), BBS5 (sc-515331), BBS8 (sc-271009), PCM-1 (sc-398365), and Ninein (sc-376420) were purchased from Santa Cruz Biotechnology, Inc. Rabbit antibody against BBS5 (14569-1-AP) was purchased from Proteintech. Rabbit antibodies against BBS7 (HPA044592), BBS9 (HPA021289), and GFP (SAB4301138) and mouse antibody against actin (A1978) were purchased from Sigma–Aldrich. Rabbit antibody against actin (catalog no. 4967) was purchased from Cell Signaling Technology. Rabbit antibody against NPHP1 (E-AB-61290) was purchased from Elabscience.
Secondary antibodies were as follows: anti-mouse Alexa Fluor 488 (Invitrogen, A11001; 1:1000), anti-rabbit Alexa Fluor 488 (Invitrogen, A11008; 1:1000), anti-mouse Alexa Fluor 555 (Invitrogen, A21422; 1:1000), anti-rabbit Alexa Fluor 555 (Invitrogen, A21428; 1:1000), anti-mouse Alexa Fluor 647 (Invitrogen, A21235; 1:1000), and anti-rabbit Alexa Fluor 647 (Invitrogen, A21245; 1:1000).
Cell cultures
Immortalized human retinal pigment epithelium cells, hTERT-RPE-1 (ATCC, CRL-4000), henceforth RPE1, were kindly provided by Dr. Vladimir Varga (Institute of Molecular Genetics of the Czech Academy of Sciences), and Phoenix Ampho cells were kindly provided by Dr. Tomas Brdicka (Institute of Molecular Genetics of the Czech Academy of Sciences). Cells were cultured in complete Dulbecco's modified Eagle's medium (Sigma, D6429-500ML) supplemented with 10% FBS (Gibco, 10270-106), 100 units/ml penicillin (BB Pharma), 100 μg/ml streptomycin (Sigma–Aldrich), and 40 μg/ml gentamicin (Sandoz) at 37 °C and 5% CO2. Cells were seeded to reach 80–100% confluence for all experiments.
Cloning, gene transfections and deletions
BBS ORFs were amplified from cDNA obtained from RPE1 cells, fused with the SYFP2 coding region using recombinant PCR, and cloned into pMSCV-Thy-IRES 1.1 vector (Clontech) using EcoRI/XhoI and ClaI restriction sites.
30 µg of plasmid DNA was transfected into Phoenix Ampho cells using the polyethyleneimine to generate YFP-BBS retroviruses. RPE1 cells were spinfected using 2 ml of virus aliquot in the presence of 2.4 µg/ml Polybrene using standard protocols. RPE1 cells stably expressing YFP-tagged BBSome subunits were sorted using the 488-nm laser on FACSAria IIu (BD Biosciences).
BBS knockout cell lines were generated using the CRISPR/Cas9 approach. Single guided RNAs (sgRNAs) targeting BBS genes were designed using the web tool CHOPCHOP (46). sgRNA was cloned into pSpCas9(BB)-2A-GFP (PX458) vector kindly provided by Feng Zhang (Addgene plasmid 48138) (47). sgRNA sequence with PAM motif (3′ end) for respective BBS genes are as follows: human BBS1, GCAATGAGGCCAATTCGAAGTGG; human BBS2, GTGGCCATAGGGCGCTACGACGG; human BBS4, GGCTGAGGAGAGAGTCGCGACGG; human BBS7, CAGTGTTCAAGACTTTACCCGGG; human BBS8, GGCCGGTACCTGGTCATAAGGGG; human BBS9, GATTGGTGGTCTACTATTCTGGG; human BBS18, CAGAAGTGAAGTCAATGTTCCGG.
RPE1 cells were transfected with PX458 vector with specific BBS sgRNA using polyethyleneimine (Polysciences, Inc., 23966-2). After 48 h, cells expressing GFP were sorted as single cells in 96-well plates using the 488-nm laser on FACSAria IIu (BD Biosciences). Clones were then analyzed for the expression of target BBS proteins by immunoblotting and sequencing of DNA surrounding the sgRNA target site.
Co-immunoprecipitation and Western blotting
RPE1 cells were grown in 15-cm cell culture dishes and lysed in lysis buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 2 mm EDTA, pH 8, 0.5% Triton X-100) supplemented with protease inhibitor mixture (complete, Roche Applied Science, catalog no. 05056489001). Lysates were cleared by centrifugation at 15,000 × g for 15 min at 4 °C. Protein concentration was adjusted using the Pierce™ BCA Protein Assay Kit (Thermo Scientific). For the co-immunoprecipitation assay, protein lysates were immunoprecipitated with anti-GFP antibody conjugated to protein A–coupled polyacrylamide beads (catalog no. 53142, Thermo Scientific) for 2 h at 4 °C. Beads were washed three times in the lysis buffer, and co-precipitated proteins were denatured in 1× Laemmli buffer. For protein expression analysis, protein lysates were immediately denatured in reducing 4× Laemmli buffer. Denatured protein samples were analyzed by Western blotting following standard protocols. Membranes were probed with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature and developed using the chemiluminescence immunoblot imaging system Azure c300 (Azure Biosystems, Inc.).
Flow cytometry
RPE1 cells expressing the YFP-tagged subunits were cultivated in 24-well dishes, trypsinized, and centrifuged at 1000 × g for 1 min. The pellets were washed in PBS and resuspended in FACS buffer (2 mm EDTA, 2% FBS, and 0.1% sodium azide). Measurements were taken on Aurora™ (Cytek Biosciences). Geometric means of fluorescence intensity of YFP-positive cells were obtained and used to examine the protein expression. Flow cytometry data were analyzed using FlowJo (BD Biosciences).
Immunofluorescence
RPE1 cells were cultured on 12-mm coverslips and serum-starved for 24 h. Cells were fixed (4% formaldehyde) and permeabilized (0.2% Triton X-100) for 10 min. Blocking was done using 5% goat serum (Sigma, G6767-100ML) in PBS for 15 min and incubated with primary antibody (1% goat serum/PBS) and secondary antibody (PBS) for 1 h and 45 min, respectively, in a wet chamber. The cells were washed after each step in PBS three times. At last, the cells were washed in distilled H2O, air-dried, and mounted using ProLong™ Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific).
Fluorescence microscopy
Image acquisition for cilia length rescue assays was performed on the Delta Vision Core microscope using the oil immersion objective (Plan-Apochromat ×60, NA 1.42) and filters for DAPI (435/48), FITC (523/36), and TRITC (576/89). Z-stacks were acquired in 1024 × 1024–pixel format and Z-steps of 0.2 μm. The cilia length was measured using the Fiji ImageJ.
Imaging of the YFP-tagged cell lines was performed using a Zeiss Axioplan2 microscope equipped with an oil immersion objective (Plan-Apochromat Zeiss ×100, NA 1.4) and filters for DAPI (435/48), YFP (559/38), TRITC (576/89), and Cy5 (632/22).
Expansion microscopy and basal body length quantification
The protocol is based on Ref. 31. RPE1 cells were cultured on 12-mm coverslips and serum-starved for 24 h. Coverslips with cells were fixed with 4% formaldehyde, 4% acrylamide in PBS overnight and then washed twice with PBS. The gelation was performed by incubating coverslips face-down with 45 μl of monomer solution (19% (w/w) sodium acrylate, 10% (w/w) acrylamide, 0.1% (w/w) N,N´-methylenbisacrylamide in PBS supplemented with 0.5% TEMED and 0.5% APS, prepared as described (31) in a precooled humid chamber. After 1 min on ice, the chamber was incubated at 37 °C in the dark for 30 min. Samples in the gel were denatured in denaturation buffer (200 mm SDS, 200 mm NaCl, 50 mm Tris in ddH2O) at 95 °C for 4 h. Gels were expanded in ddH2O for 1 h until they reached full expansion factor (4.2×) and then cut into 1 × 1–cm pieces. Pieces of gel were incubated with primary antibodies diluted in 2% BSA in PBS overnight at room temperature. After staining, shrunk pieces of gel were incubated in ddH2O for 1 h, during which time they re-expanded. After reaching their original expanded factor, pieces of gel were incubated with secondary antibodies diluted in 2% BSA in PBS for 3 h at room temperature. The last expansion in ddH2O with exchange every 20 min was for 1 h until pieces of gel reached full size. Samples were imaged in 35-mm glass-bottom dishes (CellVis) precoated with poly-l-lysine. During imaging, gels were covered with ddH2O to prevent shrinking.
Expanded cells were imaged by confocal microscopy on Leica TCS SP8 using a ×63, 1.4 NA oil objective with closed pinhole to 0.4 Airy Units. Cilia and detail of basal bodies were acquired in Z-stacks at 0.1-μm stack size with pixel size 30–50 nm according to the cilia length. Images were computationally deconvolved using Huygens Professional software.
For basal body length, only those perfectly or nearly perfectly oriented longitudinally to the image focal plane were selected for measurements. The line scan and plot profile tools in Fiji ImageJ were used to determine the width at the proximal end of the basal body marked with acetylated tubulin (Ac-tub) antibody. Length of the basal body was measured as a distance between proximal end (Ac-tub signal) and distal end stained by NPHP1 antibody. At least four independent experiments were measured in each condition (WT, n = 56; BBS1 KO, n = 56 in total). For quantification of basal body length, in every experiment, the average value of widths was compared with ideal basal body width in full expansion (4.2×) to define the difference between ideal and real expansion factor (note that the range of expansions was between 3.35× and 4.1×). Then, as the length is subject to variability, all lengths were standardized by recounting the difference from the full 4.2 expansion factor in particular experiment, and second, the average length was counted.
Fluorescence recovery after photobleaching, data processing, and analysis
Cells were seeded in glass-bottom 8-well chambers (CellVis) and serum-starved for 24–48 h to induce ciliogenesis. Prior to FRAP experiments, cells were washed and supplemented with Hanks' balanced salt solution medium (+Ca and +Mg) with 20 mm HEPES. FRAP measurements were performed using a Leica TCS SP8 confocal microscope equipped with an oil immersion objective HC Plan-Apochromat 63×, NA 1.4 oil, CS2 at room temperature. Data acquisition was performed in 512 × 512–pixel format with pinhole 2.62 Airy, at a speed of 1000 Hz in bidirectional mode and 8-bit resolution. Photobleaching (0.3 s) was performed with a circular spot 1 μm in diameter (centrosome, PS) or with a rectangle spot 1 μm in length (ciliary base and tip) at 100% intensity using a 20-milliwatt 488-nm solid-state laser. Fluorescence recovery was monitored at low laser intensity (3–10%) at 0.26-s intervals until reaching the plateau of recovery, in total for 60–80 s after photobleaching. A total of 25–30 separate FRAP measurements were performed for each sample. All FRAP curves were normalized to fluorescence loss during acquisition following the subtraction of background fluorescence. Curve fitting was performed in GraphPad Prism software using the one-phase association fit. All individual curves were fitted at once to obtain the mean and 90 or 95% confidence intervals of the desired parameters, half-time t½, and the mobile fraction Fm (see Tables S4 and S5). Because the initial recovery after bleaching at the centrosome/basal body and PS was contaminated by the diffusion toward these compartments, for simplicity and to estimate only the half-time of the protein fraction bound at these compartments, we restricted fitting of our data to times t > 2 s as we did in our previous study (26).
Fluorescence correlation spectroscopy, data processing, and analysis
Cells were seeded in glass-bottom 8-well chambers (CellVis) and serum-starved for 24 h to induce ciliogenesis. Prior to FCS experiments, cells were washed and supplemented with Hanks' balanced salt solution medium (+Ca and +Mg) with 20 mm HEPES. FCS measurements were performed using a Leica TCS SP8 SMD confocal microscope equipped with three SMD HyD detectors, FCS module HydraHarp400 (PicoQuant, Germany), water objective HC Plan-Apochromat ×63, NA 1.2, and WLL 470–670 nm at 37 °C. Samples were spot-illuminated with the 488-nm laser line with 10-microwatt laser power at the sample plane, using pinhole size 1 Airy unit and detector range 505–600 nm. Data acquisition was performed using simultaneously the LAS-X software for operating the microscope and Symphotime64 software (PicoQuant) to online visualize FCS curves and save TTTR data. Illumination spots were selected based on the reference images of the cells. Spots were selected in the cytoplasm, depending on the cell line analyzed. Spots were positioned close to the PS in YFP-BBS4–expressing cells or BBS1 KO cell lines. In the WT cells, spots were selected in the cytoplasm in close proximity to the primary cilia. At least 10–30 FCS measurements with an acquisition time of 60 s were taken per cell line.
The instrument was aligned, and the detection volume was calibrated using 20 nm Abberior STAR 488 carboxylic acid (Abberior GmbH, Göttingen, Germany) calibration dye. TTTR offline data processing and fitting was performed in custom-made TTTR Data Analysis software. The analysis pipeline starts with splitting each recording into 10 equivalent parts, calculation of an autocorrelation curve for each part, removal of outlier curves that contain signal from aggregates, estimation of S.D. for each time point of the correlation curve, and weighted nonlinear least-square fitting by a standard one- or two-component anomalous 3D diffusion in a Gaussian shape detection volume model (48) with triplet state fraction (49). The equation used corresponds to Equation 10 in Chapter 5 of the online FCS manual (50). Initially, the correlation curves were all fitted with one-component anomalous diffusion fit to obtain the average diffusion time τ (see Table S3). The fits enabled us to normalize the amplitude of correlation curves to 1 for a lag time of 10 μs, which allowed us to visually examine the differences between the measurements obtained from WT and KO cell lines and thus evaluate the presence of any potential second component. Only in the case of YFP-BBS4 in WT cells did we observe such dynamic behavior, and therefore for these data we performed fitting also with the two-component anomalous diffusion model (see Fig. 2C, inset).
Statistical analysis
Statistical analysis was performed using built-in calculations of GraphPad Prism version 5.04. All statistical tests were two-tailed. The statistical tests are indicated in the respective figure legends.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Dr. Vladimir Varga for sharing the reagents and the Zeiss microscope. We thank Dr. Ales Benda for help with the FCS data analysis. We acknowledge the Light Microscopy Core Facility, IMG CAS, Prague, Czech Republic, supported by MEYS (LM2015062, CZ.02.1.01/0.0/0.0/16_013/0001775), OPPK (CZ.2.16/3.1.00/21547), and MEYS (LO1419), for support with obtaining the widefield and FRAP data presented herein. In addition, we acknowledge the Imaging Methods Core Facility at BIOCEV, Prague, Czech Republic, supported by the MEYS CR (Large RI Project LM2018129 Czech-BioImaging) and ERDF (project CZ.02.1.01/0.0/0.0/16_013/0001775) for support with obtaining and analyzing FCS data presented in this paper.
This article contains supporting information.
Author contributions—A. P., M. S. C., K. R., M. A., O. S., and M. H. formal analysis; A. P., M. S. C., K. R., V. N., M. A., and M. H. investigation; A. P. and M. H. visualization; A. P., M. S. C., V. N., P. D., and M. H. methodology; A. P., O. S., and M. H. writing-original draft; A. P., M. S. C., K. R., V. N., M. A., P. D., O. S., and M. H. writing-review and editing; O. S. and M. H. conceptualization; O. S. and M. H. supervision; O. S. and M. H. funding acquisition; O. S. and M. H. project administration.
Funding and additional information—This study was supported by Czech Science Foundation Grant 17-20613Y (to M. H.). The Group of Adaptive Immunity is supported by EMBO Installation Grant 3259 (to O. S.) and Institute of Molecular Genetics of the Czech Academy of Sciences core funding (RVO 68378050). O. S. is supported by the Purkyne Fellowship provided by the Czech Academy of Sciences. A. P. and V. N. are students partially supported by the Faculty of Science, Charles University, Prague.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- BBS
- Bardet–Biedl syndrome
- YFP
- super yellow fluorescent protein SYFP2
- PS
- pericentriolar satellites
- sgRNA
- single guided RNAs
- DAPI
- 4′,6-diamidino-2-phenylindole
- NA
- numerical aperture
- TRITC
- tetramethylrhodamine isothiocyanate
- ddH2O
- double-distilled H2O
- FRAP
- fluorescence recovery after photobleaching
- Ac-tub
- acetylated tubulin
- ACF
- autocorrelation function
- TEMED
- N,N,N',N'-tetramethylethylenediamine.
References
- 1. Forsythe E., and Beales P. L. (2013) Bardet-Biedl syndrome. Eur. J. Hum. Genet. 21, 8–13 10.1038/ejhg.2012.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niederlova V., Modrak M., Tsyklauri O., Huranova M., and Stepanek O. (2019) Meta-analysis of genotype-phenotype associations in Bardet-Biedl syndrome uncovers differences among causative genes. Hum. Mutat. 40, 2068–2087 10.1002/humu.23862 [DOI] [PubMed] [Google Scholar]
- 3. Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., and Jackson P. K. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- 4. Loktev A. V., Zhang Q., Beck J. S., Searby C. C., Scheetz T. E., Bazan J. F., Slusarski D. C., Sheffield V. C., Jackson P. K., and Nachury M. V. (2008) A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell 15, 854–865 10.1016/j.devcel.2008.11.001 [DOI] [PubMed] [Google Scholar]
- 5. Wingfield J. L., Lechtreck K. F., and Lorentzen E. (2018) Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 62, 753–763 10.1042/EBC20180030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dilan T. L., Singh R. K., Saravanan T., Moye A., Goldberg A. F. X., Stoilov P., and Ramamurthy V. (2018) Bardet-Biedl syndrome-8 (BBS8) protein is crucial for the development of outer segments in photoreceptor neurons. Hum. Mol. Genet. 27, 283–294 10.1093/hmg/ddx399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nachury M. V. (2018) The molecular machines that traffic signaling receptors into and out of cilia. Curr. Opin. Cell Biol. 51, 124–131 10.1016/j.ceb.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu P. W., and Lechtreck K. F. (2018) The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. U. S. A. 115, E934–E943 10.1073/pnas.1713226115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., and Nachury M. V. (2010) The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q., Seo S., Bugge K., Stone E. M., and Sheffield V. C. (2012) BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum. Mol. Genet. 21, 1945–1953 10.1093/hmg/dds004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo D. F., Cui H., Zhang Q., Morgan D. A., Thedens D. R., Nishimura D., Grobe J. L., Sheffield V. C., and Rahmouni K. (2016) The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet. 12, e1005890 10.1371/journal.pgen.1005890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimura D. Y., Fath M., Mullins R. F., Searby C., Andrews M., Davis R., Andorf J. L., Mykytyn K., Swiderski R. E., Yang B., Carmi R., Stone E. M., and Sheffield V. C. (2004) Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 101, 16588–16593 10.1073/pnas.0405496101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McIntyre J. C., Hege M. M., and Berbari N. F. (2016) Trafficking of ciliary G protein-coupled receptors. Methods Cell Biol. 132, 35–54 10.1016/bs.mcb.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 14. Loktev A. V., and Jackson P. K. (2013) Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5, 1316–1329 10.1016/j.celrep.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 15. Berbari N. F., Lewis J. S., Bishop G. A., Askwith C. C., and Mykytyn K. (2008) Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. U. S. A. 105, 4242–4246 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodsmith J., Apelt L., Casado-Medrano V., Özkan Z., Timmermann B., and Stelzl U. (2017) Protein interaction perturbation profiling at amino-acid resolution. Nat. Methods 14, 1213–1221 10.1038/nmeth.4464 [DOI] [PubMed] [Google Scholar]
- 17. Katoh Y., Nozaki S., Hartanto D., Miyano R., and Nakayama K. (2015) Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362 10.1242/jcs.168740 [DOI] [PubMed] [Google Scholar]
- 18. Klink B. U., Zent E., Juneja P., Kuhlee A., Raunser S., and Wittinghofer A. (2017) A recombinant BBSome core complex and how it interacts with ciliary cargo. Elife 6, e27434 10.7554/eLife.27434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mourão A., Nager A. R., Nachury M. V., and Lorentzen E. (2014) Structural basis for membrane targeting of the BBSome by ARL6. Nat. Struct. Mol. Biol. 21, 1035–1041 10.1038/nsmb.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knockenhauer K. E., and Schwartz T. U. (2015) Structural characterization of Bardet-Biedl syndrome 9 protein (BBS9). J. Biol. Chem. 290, 19569–19583 10.1074/jbc.M115.649202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Q., Yu D., Seo S., Stone E. M., and Sheffield V. C. (2012) Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J. Biol. Chem. 287, 20625–20635 10.1074/jbc.M112.341487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klink B. U., Gatsogiannis C., Hofnagel O., Wittinghofer A., and Raunser S. (2020) Structure of the human BBSome core complex. Elife 9, e53910 10.7554/eLife.53910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chou H. T., Apelt L., Farrell D. P., White S. R., Woodsmith J., Svetlov V., Goldstein J. S., Nager A. R., Li Z., Muller J., Dollfus H., Nudler E., Stelzl U., DiMaio F., Nachury M. V., et al. (2019) The molecular architecture of native BBSome obtained by an integrated structural approach. Structure 27, 1384–1394.e4 10.1016/j.str.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh S. K., Gui M., Koh F., Yip M. C., and Brown A. (2020) Structure and activation mechanism of the BBSome membrane protein trafficking complex. Elife 9, e53322 10.7554/eLife.53322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huranová M., Ivani I., Benda A., Poser I., Brody Y., Hof M., Shav-Tal Y., Neugebauer K. M., and Stanek D. (2010) The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J. Cell Biol. 191, 75–86 10.1083/jcb.201004030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huranova M., Muruganandam G., Weiss M., and Spang A. (2016) Dynamic assembly of the exomer secretory vesicle cargo adaptor subunits. EMBO Rep. 17, 202–219 10.15252/embr.201540795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kremers G. J., Goedhart J., van Munster E. B., and Gadella T. W. Jr. (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45, 6570–6580 10.1021/bi0516273 [DOI] [PubMed] [Google Scholar]
- 28. Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J., Leroux M. R., Katsanis N., and Beales P. L. (2004) The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36, 462–470 10.1038/ng1352 [DOI] [PubMed] [Google Scholar]
- 29. Chamling X., Seo S., Searby C. C., Kim G., Slusarski D. C., and Sheffield V. C. (2014) The centriolar satellite protein AZI1 interacts with BBS4 and regulates ciliary trafficking of the BBSome. PLoS Genet. 10, e1004083 10.1371/journal.pgen.1004083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hernandez-Hernandez V., Pravincumar P., Diaz-Font A., May-Simera H., Jenkins D., Knight M., and Beales P. L. (2013) Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Hum. Mol. Genet. 22, 3858–3868 10.1093/hmg/ddt241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gambarotto D., Zwettler F. U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J. G., Reuss M., Unser M., Boyden E. S., Sauer M., Hamel V., and Guichard P. (2019) Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Q., Nishimura D., Vogel T., Shao J., Swiderski R., Yin T., Searby C., Carter C. S., Kim G., Bugge K., Stone E. M., and Sheffield V. C. (2013) BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci. 126, 2372–2380 10.1242/jcs.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quarantotti V., Chen J. X., Tischer J., Gonzalez Tejedo C., Papachristou E. K., D'Santos C. S., Kilmartin J. V., Miller M. L., and Gergely F. (2019) Centriolar satellites are acentriolar assemblies of centrosomal proteins. EMBO J. 38, e101082 10.15252/embj.2018101082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dammermann A., and Merdes A. (2002) Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159, 255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stowe T. R., Wilkinson C. J., Iqbal A., and Stearns T. (2012) The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell 23, 3322–3335 10.1091/mbc.E12-02-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seo S., Zhang Q., Bugge K., Breslow D. K., Searby C. C., Nachury M. V., and Sheffield V. C. (2011) A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 7, e1002358 10.1371/journal.pgen.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marion V., Stutzmann F., Gérard M., De Melo C., Schaefer E., Claussmann A., Hellé S., Delague V., Souied E., Barrey C., Verloes A., Stoetzel C., and Dollfus H. (2012) Exome sequencing identifies mutations in LZTFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet–Biedl syndrome with situs inversus and insertional polydactyly. J. Med. Genet. 49, 317–321 10.1136/jmedgenet-2012-100737 [DOI] [PubMed] [Google Scholar]
- 38. Jiang J., Promchan K., Jiang H., Awasthi P., Marshall H., Harned A., and Natarajan V. (2016) Depletion of BBS protein LZTFL1 affects growth and causes retinal degeneration in mice. J. Genet. Genomics 43, 381–391 10.1016/j.jgg.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y. D., and Nigg E. A. (2009) Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y., Seo S., Bhattarai S., Bugge K., Searby C. C., Zhang Q., Drack A. V., Stone E. M., and Sheffield V. C. (2014) BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum. Mol. Genet. 23, 40–51 10.1093/hmg/ddt394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsang W. Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., and Dynlacht B. D. (2008) CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell 15, 187–197 10.1016/j.devcel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seo S., Baye L. M., Schulz N. P., Beck J. S., Zhang Q., Slusarski D. C., and Sheffield V. C. (2010) BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. U. S. A. 107, 1488–1493 10.1073/pnas.0910268107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stoetzel C., Muller J., Laurier V., Davis E. E., Zaghloul N. A., Vicaire S., Jacquelin C., Plewniak F., Leitch C. C., Sarda P., Hamel C., de Ravel T. J., Lewis R. A., Friederich E., Thibault C., et al. (2007) Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am. J. Hum. Genet. 80, 1–11 10.1086/510256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stoetzel C., Laurier V., Davis E. E., Muller J., Rix S., Badano J. L., Leitch C. C., Salem N., Chouery E., Corbani S., Jalk N., Vicaire S., Sarda P., Hamel C., Lacombe D., et al. (2006) BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 38, 521–524 10.1038/ng1771 [DOI] [PubMed] [Google Scholar]
- 45. Katsanis N., Beales P. L., Woods M. O., Lewis R. A., Green J. S., Parfrey P. S., Ansley S. J., Davidson W. S., and Lupski J. R. (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat. Genet. 26, 67–70 10.1038/79201 [DOI] [PubMed] [Google Scholar]
- 46. Labun K., Montague T. G., Krause M., Torres Cleuren Y. N., Tjeldnes H., and Valen E. (2019) CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwille P., Haupts U., Maiti S., and Webb W. W. (1999) Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys. J. 77, 2251–2265 10.1016/S0006-3495(99)77065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Widengren J., Mets U., and Rigler R. (1995) Fluorescence correlation spectroscopy of triplet states in solution: a theoretical and experimental study. J. Phys. Chem. 99, 13368–13379 10.1021/j100036a009 [DOI] [Google Scholar]
- 50. Machán R., and Hof M. (2016) Fluorescence Correlation Spectroscopy (FCS). in Practical Manual for Fluorescence Microscopy Techniques (Ahmed S., Thankiah S., Machan R., Hof M., Clayton A. H. A., Wright G., Sibarita J.-B., Korte T., and Herrmann A., eds) p. 5–9, PicoQuant GmbH, Berlin [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.