Abstract
The emergence of Neisseria gonorrhoeae strains with resistance (R) to extended-spectrum cephalosporins (ESCsR) represents a public health threat of untreatable gonococcal infections. This study was designed to determine the prevalence and molecular mechanisms of ESCR of Shanghai N. gonorrhoeae isolates. A total of 366 N. gonorrhoeae isolates were collected in 2017 in Shanghai. Susceptibility to ceftriaxone (CRO), cefixime (CFM), azithromycin (AZM), ciprofloxacin (CIP), spectinomycin, penicillin, and tetracycline was determined using the agar dilution method. A subset of 124 isolates was subjected to phylogenetic analysis for nine antimicrobial resistance-associated genes, i.e., penA, porB, ponA, mtrR, 23S rRNA, gyrA, parC, 16S rRNA, and rpsE. Approximately 20.0% of the isolates exhibited CFMR [minimum inhibitory concentration (MIC) >0.125 mg/L], and 5.5% were CROR (MIC > 0.125 mg/L). In total, 72.7% of ESCR isolates were clonal and associated with mosaic penA 10 and 60 alleles. Non-mosaic penA 18 allele and substitutions of PenA A501T, G542S, and PorB1b G213S/Y were observed in non-clonal ESCR. Approximately 6.8% of the isolates showed AZM MIC above the epidemiological cutoff (ECOFF, 1 mg/L), were associated with 23S rRNA A2059G mutation, and did not exhibit clonal distribution. Almost all isolates were CIPR (resistance to ciprofloxacin) and associated with GyrA-91/92 and ParC-85/86/87/88/89/91 alterations. Isolates with ParC S88P substitution were clustered into the ESCR clade. The Shanghai isolates exhibited a high level of ESCR and distinct resistant patterns.
Keywords: Neisseria gonorrhoeae, extended-spectrum cephalosporins, multidrug resistance, resistance determinants, phylogenetic analysis
Introduction
Neisseria gonorrhoeae is the causative agent of gonorrhea. The World Health Organization (WHO) estimated that N. gonorrhoeae causes more than 86.9 million new infections worldwide annually (World Health Organization [WHO], 2018). Meanwhile, gonococcal antimicrobial resistance (AMR) continues to spread worldwide and could lead to a pandemic of extensively drug-resistant gonococci (World Health Organization [WHO], 2018). Of particular concern is the fact that ESCRS [reduced susceptibility to the extended-spectrum cephalosporins (ESCs), i.e., cefixime (CFM), and ceftriaxone (CRO)], which is the first-line empirical treatment for N. gonorrhoeae infections (Unemo and Shafer, 2014; Wi et al., 2017), is becoming widely spread. According to the WHO Global Gonococcal Antimicrobial Surveillance Programme (GASP), in 2016, about one-third of the participating countries reported that ≥5% of isolates are resistant to ESCs (CRO and/or CFM), and half reported ≥5% resistance to azithromycin (AZMR). Of the 59 countries reporting ciprofloxacin resistance (CIPR), 95% reported ≥5% resistance and 17% reported >90% resistance (Wi et al., 2017; World Health Organization [WHO], 2018). In China, from 2013 to 2016, high prevalence of decreased susceptibility to CRO (CRORS) (9.7–12.2%, MIC ≥ 0.125 mg/L) and AZMR (18.6%, MIC ≥ 1.0 mg/L) has been reported (Yin et al., 2018). In Shanghai, the proportion of CRORS (MIC ≥ 0.125 mg/L) ranged from 7 to 13% during 1988–2013 (Gu et al., 2014).
Drug-resistant N. gonorrhoeae has been attributed to several molecular mechanisms. The primary mechanism for ESCR (resistance to ESC) is mutations of the penA gene (encodes penicillin (PEN)-binding protein 2, PBP2, PenA), including a recombinant mosaic allele from commensal Neisseria (Ameyama et al., 2002; Lee et al., 2010). Mutations in the Mtr repressor genes mtrR and porB have been shown to contribute to ESCR (Barry and Klausner, 2009; Unemo and Shafer, 2014). Loci involved in other AMR include mutations of 23S rRNA (Ng et al., 2002) and mtrR (Zarantonelli et al., 1999) for AZMR, mutations in gyrA and parC for CIPR (Yang et al., 2006), and mutations in 16S rRNA and rpsE for spectinomycin (SPT) (Galimand et al., 2000; Unemo et al., 2013). Currently, the identified resistance determinants do not fully account for the observed drug resistance, and thus, other factors may be involved (Unemo and Shafer, 2014).
Genetic analysis has provided insight into outbreaks and transmission networks for several pathogens with greater resolution than traditional methods (Diep, 2013). Using genetic methods, researchers found that ESCRS in Canada first emerged from a group of diverse isolates in the 1990s with non-mosaic penA alleles, followed in 2000/2001 with the mosaic penA 10 allele and then in 2007 with the mosaic penA 34 allele (Demczuk et al., 2015). ESCRS strains in the United States are mainly clonal and associated with the mosaic penA 34 allele and derivatives, whereas AZMR strains have arisen through multiple mechanisms and show limited clonal spread (Grad et al., 2014, 2016). To date, reported cases of CROR are sporadic, except for the FC428 strain, which was first identified in Japan in 2015 and has since then been observed in other countries (Lahra et al., 2018; Lee et al., 2019).
Genetic analysis has been used to study strain distribution along with multi-locus sequence typing (MLST) (Unemo and Dillon, 2011), N. gonorrhoeae multi-antigen sequence typing (NG-MAST) (Unemo and Dillon, 2011), N. gonorrhoeae sequence typing for AMR (NG-STAR) (Demczuk et al., 2017), and whole-genome sequencing (De Silva et al., 2016; Harris et al., 2018; Lee et al., 2018). We have reported NG-STAR analysis of seven loci in 124 N. gonorrhoeae isolates (Yang et al., 2020); specific NG-STAR genotypes are found to be associated with ESCR and AZMR.
The objectives of this study were to assess whether nine loci can increase the resolution of genetic analysis and to determine the association of genetic characterization and ESCR phenotypes in N. gonorrhoeae isolates in Shanghai. This is the first in-depth genomic analysis based on nine AMR-associated loci in N. gonorrhoeae in a high-level AMR setting. This study provides solid information on the molecular mechanisms and genetic characteristics of AMR in N. gonorrhoeae in Shanghai.
Materials and Methods
Neisseria gonorrhoeae Isolate Collection and Antimicrobial Susceptibility Testing
Neisseria gonorrhoeae isolates were collected from male patients with uncomplicated urogenital gonorrhea (symptoms may include pain or a burning sensation when urinating, a greater frequency or urgency of urination, and abnormal urinary discharge) at the Shanghai Skin Disease Hospital in conjunction with the China GASP. Patient consent was obtained, and ethics approval was received from the Shanghai Skin Disease Hospital. The first 30 N. gonorrhoeae isolates of each month in 2017 (except for 36 isolates collected in July to avoid recovery failure, making a total of 366 isolates) were used in this study. Basic demographic data of all patients were collected. The median age was 34 years (range: 18–69). Of the 366 subjects, 363 (99.2%) were ethnic Han. All patients were heterosexual. A majority of the patients had abnormal urinary discharge (98.9%). Approximately 16.4% of the patients had previous history of gonorrhea, and 12.8% received antibiotic treatment in the past month. One isolate was collected from one patient. Briefly, one urogenital specimen was collected using sterile Dacron swab and streaked on Thayer–Martin (T–M) medium (Oxoid; GuangZhou LOSO Science, Ltd.) supplemented with 1% IsoVitaleX (Oxoid; GuangZhou LOSO Science, Ltd.). N. gonorrhoeae was identified using criteria that included an oxidase test, Gram staining, and glucose utilization test (WHO Western Pacific Gonococccal Antimicrobial Surveillance Programme, 2008). One identified N. gonorrhoeae isolate for each patient was collected and stored at −70°C. Minimum inhibitory concentrations (MICs) for seven antimicrobials, including PEN, tetracycline (TET), ciprofloxacin (CIP), azithromycin (AZM), CFM, CRO, and SPT, were determined using the agar dilution method. Antimicrobial agents were purchased from Shanghai ANPEL Scientific Instrument, Co., Ltd. (Shanghai, China; distributors of Sigma-Aldrich, United States). Each MIC determination was performed in duplicate, and N. gonorrhoeae ATCC 49226 strain was used as a reference strain. Antimicrobial susceptibility testing results were interpreted using the EUCAST (2020) breakpoints.
DNA Sequencing and Analysis
As previously reported, a total of 124 N. gonorrhoeae isolates (first 10 isolates of each month, one CIP susceptible isolate, and three isolates with CRO MICs ≥ 1.0 mg/L) were subjected to genetic analysis (Yang et al., 2020). Genomic DNA from each isolate was extracted using the Genomic DNA Purification Kit (Shanghai Promega Biological Products, Ltd., Shanghai, China). Seven loci (penA, mtrR, porB, ponA, gyrA, parC, and 23S rRNA) were PCR amplified as described previously (Yang et al., 2020). 16S rRNA was amplified by PCR (Perkin Elmer 9600 Thermocycler; Perkin Elmer, Wellesley, MA United States) using primer pair 16S-F (5’-TGATCCARCCGCASSTTC-3’) and 16S-R (5’-AGAGTTTGATCYTGGYTYAG-3’), while rpsE was amplified by PCR using primer pair rpsE-F (5’-TGGCAAAACATGAAATTGAAG-3’) and rpsE-R (5’-GCCATGGTTAACTCCCAAAA-3’). All primers were purchased from Invitrogen. PCR products were purified using a PCR Purification Kit (Sangon Biotech Co., Shanghai, China). DNA sequencing was performed at Sangon Biotech Co., using 3730XL (Applied Biosystems, United States) using the Sanger sequencing method. DNA sequences were verified and edited using Geneious (11.1.4)1 and Vector NTI Advance 11.5.3 (Lu and Moriyama, 2004). Sequences were compared with the corresponding sequences of an antimicrobial-susceptible N. gonorrhoeae strain FA1090. PenA amino acid sequences were compared to a wild-type PenA (Spratt, 1988) (GenBank accession number M32091). NG-STAR and NG-MAST were also performed and reported previously (Yang et al., 2020). The DNA sequences of penA, mtrR, porB, ponA, gyrA, parC, and 23S rRNA were also as previously reported (Yang et al., 2020). The DNA sequences of 16S rRNA and rpsE were submitted to GenBank (accession numbers MK620715–MK620729 for 16S rRNA and MN823292–MN823293 for rpsE).
Phylogenetic Analysis
The sequences of all nine loci were concatenated for each strain. IQ-TREE (v1.6.12) (Nguyen et al., 2015) was used for constructing maximum-likelihood phylogenies with 1,000 bootstraps, and the best-fit model was autodetected. Phylogenies were assessed using midpoint rooting. Phylogenetic clades were determined by cluster analysis using ClusterPicker (Ragonnet-Cronin et al., 2013) with the following settings: initial and main support thresholds of 90 and genetic distance threshold of 4.5. Phylogenies and metadata (including MICs, AMR phenotypes, and molecular profiles associated with AMR) were visualized in FigTree2 and phandango (Hadfield et al., 2017).
Statistical Analysis
The χ2 test was used to identify AMR determinants associated with CFMR, CROR, and MICs above ECOFF for AZM using R (version 3.4.1). Multiple linear regression analysis was performed using R (version 3.4.1), to determine the relationship of log10 (CRO/CFM/AZM MICs) or CIP MIC intervals as the dependent variable to the presence of gene mutations.
Results
Antimicrobial Susceptibility of N. gonorrhoeae Isolates
Among the 366 N. gonorrhoeae isolates, 5.5% of the isolates were CROR (MICs > 0.125 mg/L), and 18.6% of isolates had CRO MICs ≥ 0.125 mg/L (Table 1 and Supplementary Table S1). About 19.4% of the 366 isolates were CFMR (MICs > 0.125 mg/L). About 6.8% of the isolates showed AZM MIC above the epidemiological cutoff (ECOFF, 1 mg/L), and 99.5% of the isolates were CIPR. The percentages of PENR and TETR were 82.5% and 60.9%, respectively. One isolate was SPTR. Demographic/clinical information including age, ethnicity, abnormal urinary discharge, previous history of gonorrhea, and antibiotic use in the past month was not associated with resistance to CFM, CRO, and AZM (Supplementary Table S2).
TABLE 1.
MIC distribution of seven antimicrobial agents for 366 N. gonorrhoeae isolates from Shanghai.
| MICs (mg/L) | ||||||||||||||||||
| Antimicrobials | 0.002 | 0.004 | 0.008 | 0.016 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | ≥256 |
| Ceftriaxone | ||||||||||||||||||
| na | 5 | 11 | 39 | 135 | 110 | 46 | 16 | 1 | 3 | |||||||||
| Cum%b | 1.4 | 4.4 | 15 | 51.9 | 82 | 94.5 | 98.9 | 99.2 | 100 | |||||||||
| Cefixime | ||||||||||||||||||
| n | 2 | 5 | 22 | 63 | 125 | 78 | 37 | 23 | 7 | 1 | 3 | |||||||
| Cum% | 0.5 | 1.9 | 7.9 | 25.1 | 59.3 | 80.6 | 90.7 | 97 | 98.9 | 99.2 | 100 | |||||||
| Azithromycin | ||||||||||||||||||
| n | 1 | 2 | 14 | 36 | 101 | 116 | 63 | 8 | 4 | 2 | 9 | 3 | 7 | |||||
| Cum% | 0.3 | 0.8 | 4.6 | 14.5 | 42.1 | 73.8 | 91 | 93.2 | 94.3 | 94.8 | 97.3 | 98.1 | 100 | |||||
| Ciprofloxacin | ||||||||||||||||||
| n | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | 20 | 53 | 81 | 173 | 31 | |||
| Cum% | 0 | 0.3 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 2.2 | 7.7 | 22.1 | 44.3 | 91.5 | 100 | |||
| Penicillin | ||||||||||||||||||
| n | 0 | 0 | 0 | 0 | 2 | 5 | 4 | 53 | 95 | 63 | 17 | 75 | 52 | |||||
| Cum% | 0 | 0 | 0 | 0 | 0.5 | 1.9 | 3 | 17.5 | 43.4 | 60.7 | 65.3 | 85.8 | 100 | |||||
| Tetracycline | ||||||||||||||||||
| n | 0 | 0 | 0 | 0 | 2 | 12 | 27 | 102 | 107 | 17 | 1 | 29 | 69 | |||||
| Cum% | 0 | 0 | 0 | 0 | 0.5 | 3.8 | 11.2 | 39.1 | 68.3 | 73 | 73.2 | 81.1 | 100 | |||||
| Spectinomycin | ||||||||||||||||||
| n | 0 | 18 | 65 | 170 | 106 | 6 | 1 | |||||||||||
| Cum% | 0 | 4.9 | 22.7 | 69.1 | 98.1 | 99.7 | 100 | |||||||||||
Vertical lines indicate the breakpoint concentrations for each antimicrobial. Breakpoints: MICs > 0.125 mg/L for ceftriaxone or cefixime, ECOFF value is 1 mg/L for azithromycin, MICs > 0.06 mg/L for ciprofloxacin MICs > 1 mg/L for penicillin or tetracycline, and MICs > 64 mg/L for spectinomycin (EUCAST, 2020). aNumber of isolates. bCumulative percentage.
Supplementary Table S3 shows that 24.0% (88/366) of the sequenced isolates exhibited multidrug-resistant (MDR) phenotypes (resistance to ESC or AZM plus resistance to at least two other antimicrobials) (Martin et al., 2019). Among these phenotypes, ESC-associated phenotypes accounted for 17.7%, and AZM-associated phenotype accounted for 6.3%. Extensively drug-resistant phenotypes (resistance to ESC and AZM plus resistance to at least two other antimicrobials) (Martin et al., 2019) were noted in two isolates, namely, CFMR (MIC = 0.25 mg/L)–AZMR (MIC = 2 mg/L)–CIPR (MIC ≥ 16 mg/L)–PENR (MIC = 4 mg/L)–TETR (MIC = 4 mg/L) and CROR (MIC = 0.25 mg/L)–CFMR (MIC = 0.25 mg/L)–AZMR (MIC ≥ 8 mg/L)–CIPR (MIC ≥ 16 mg/L)-PENR (MIC ≥ 16 mg/L)–TETR (MIC = 2 mg/L).
Genotyping of N. gonorrhoeae ESCR Isolates
Mosaic penA and Substitutions in PenA and Association With NG-STAR Types
Approximately 76.2% of the CFMR isolates (16/21) had mosaic penA alleles, and only 2.9% of the CFMS isolates (3/103) possessed mosaic penA alleles (Table 2). Mosaic penA 10 and 60 alleles and substitutions in the mosaic penA coding region such as D101E and A549T were significantly associated with CFMR. Specifically, 10 out of 11 (90.9%) penA-10.001 isolates were CFMR, and four out of four (100%) penA-60.001 isolates were CFMR. Substitutions of F374V, H541N, P552V, KI555QV, I566V, and A574V were also statistically associated with CFMR.
TABLE 2.
Molecular profiles associated with resistance to cefixime, ceftriaxone, and azithromycin in N. gonorrhoeae.
| All isolates | Non-mosaic penA isolates | All isolates | |||||||||||||
| Molecular markers | CFMS n (%) | CFMR n (%) | p | CROS n (%) | CROR n (%) | p | CFMS n (%) | CFMR n (%) | p | CROS n (%) | CROR n (%) | p | <AZM ECOFF n (%) | >AZM ECOFF n (%) | p |
| NG-STAR genotypea (penA type) | |||||||||||||||
| ST-202 (penA 2) | 4 (3.9) | 0 (0) | 1 | 4 (3.4) | 0 (0) | 1 | 0 (0) | 4 (50.0) | <0.0001 | ||||||
| ST-233 (penA 60) | 0 (0) | 3 (14.3) | 0.004 | 0 (0) | 3 (37.5) | <0.001 | 3 (2.6) | 0 (0) | 1 | ||||||
| ST-348 (penA 10) | 1 (1.0) | 4 (19.0) | 0.003 | 5 (4.3) | 0 (0) | 1 | 5 (4.3) | 0 (0) | 1 | ||||||
| ST-428 (penA 18) | 3 (2.9) | 2 (9.5) | 0.199 | 3 (2.6) | 2 (25.0) | 0.033 | 3 (3.0) | 2 (40.0) | 0.017 | 3 (3.0) | 2 (40.0) | 0.017 | 5 (4.3) | 0 (0) | 1 |
| penA type | |||||||||||||||
| Mosaic | 3 (2.9) | 16 (76.2) | <0.0001 | 16 (13.8) | 3 (37.5) | 0.196 | |||||||||
| penA 10 | 1 (1.0) | 10 (47.6) | <0.0001 | 11 (9.5) | 0 (0) | 1 | |||||||||
| penA 34 | 2 (1.9) | 1 (4.8) | 0.430 | 3 (2.6) | 0 (0) | 1 | |||||||||
| penA 60 | 0 (0) | 4 (19.0) | <0.001 | 1 (0.9) | 3 (37.5) | <0.001 | |||||||||
| penA 71 | 0 (0) | 1 (4.8) | 0.169 | 1 (0.9) | 0 (0) | 1 | |||||||||
| Non-mosaic | 100 (97.1) | 5 (23.8) | <0.0001 | 100 (86.2) | 5 (62.5) | 0.196 | |||||||||
| penA 18 | 15 (14.6) | 3 (14.3) | 0.759 | 14 (12.1) | 4 (50.0) | 0.015 | 15 (15.0) | 3 (60.0) | 0.034 | 14 (14.0) | 4 (80.0) | 0.003 | |||
| Substitutions in mosaic penA | |||||||||||||||
| D101E plusb | 3 (2.9) | 11 (52.4) | <0.0001 | 14 (12.1) | 0 (0) | 0.599 | |||||||||
| YGED201HAGE, Q214E | 4 (3.9) | 12 (57.1) | <0.0001 | 16 (13.8) | 0 (0) | 0.562 | |||||||||
| A311V, T483S, T485I | 0 (0) | 4 (19.0) | <0.001 | 1 (0.9) | 3 (37.5) | <0.001 | |||||||||
| I312M, V316T, N512Y plusc | 3 (2.9) | 16 (76.2) | <0.0001 | 16 (13.8) | 3 (37.5) | 0.196 | |||||||||
| P341S | 3 (2.9) | 12 (57.1) | <0.0001 | 15 (12.9) | 0 (0) | 0.594 | |||||||||
| GA375TP | 3 (2.9) | 13 (61.9) | <0.0001 | 16 (13.8) | 0 (0) | 0.562 | |||||||||
| A549T | 1 (1.0) | 15 (71.4) | <0.0001 | 13 (11.2) | 3 (37.5) | 0.061 | |||||||||
| PenA Substitutions | |||||||||||||||
| F374V | 0 (0) | 4 (19.0) | <0.001 | 1 (0.9) | 3 (37.5) | <0.001 | |||||||||
| A501Td | 15 (14.6) | 3 (14.3) | 0.759 | 14 (12.1) | 4 (50.0) | 0.015 | 15 (15.0) | 3 (60.0) | 0.034 | 14 (14.0) | 4 (80.0) | 0.003 | |||
| A501V | 58 (56.3) | 1 (4.8) | <0.0001 | 58 (50.0) | 1 (12.5) | 0.091 | |||||||||
| A516G | 100 (97.1) | 5 (23.8) | <0.0001 | 100 (86.2) | 5 (62.5) | 0.119 | |||||||||
| H541N | 23 (22.3) | 16 (76.2) | <0.0001 | 36 (31.0) | 3 (37.5) | 0.990 | |||||||||
| G542S | 27 (26.2) | 3 (14.3) | 0.245 | 26 (22.4) | 4 (50.0) | 0.182 | 27e (27.0) | 3f (60.0) | 0.277 | 26g (26.0) | 4h (80.0) | 0.036 | |||
| P552V, KI555QV | 17 (16.5) | 15 (71.4) | <0.0001 | 29 (25.0) | 3 (37.5) | 0.716 | |||||||||
| I566V, A574NV | 47 (45.6) | 18 (85.7) | <0.001 | 58 (50) | 7 (87.5) | 0.091 | |||||||||
| porB genotype | |||||||||||||||
| porB1a | 7 (6.8) | 1 (4.8) | 1 | 8 (6.9) | 0 (0) | 1 | |||||||||
| porB1b | 96 (93.2) | 20 (95.2) | 1 | 108 (93.1) | 8 (100) | 1 | |||||||||
| PorB1b Substitutions | |||||||||||||||
| T87A | 8 (8.3) | 2 (10.0) | 0.844 | 7 (6.5) | 3 (37.5) | 0.021 | |||||||||
| T89S | 10 (10.4) | 3 (15.0) | 0.816 | 10 (9.3) | 3 (37.5) | 0.045 | |||||||||
| GA120KD | 52 (54.2) | 15 (75.0) | 0.086 | 60 (55.6) | 7 (87.5) | 0.163 | |||||||||
| G213S/Y | 11 (11.5) | 4 (20.0) | 0.503 | 10 (9.3) | 5 (62.5) | <0.001 | 11 (11.8) | 2 (40.0) | 0.1294 | 10 (10.8) | 3 (60.0) | 0.016 | |||
| Q214L | 7 (7.3) | 2 (10.0) | 0.962 | 6 (5.6) | 3 (37.5) | 0.015 | |||||||||
| G259A | 61 (63.5) | 17 (85.0) | 0.063 | 70 (64.8) | 8 (100) | 0.041 | |||||||||
| PonA Substitutions | |||||||||||||||
| T375A | 100 (97.1) | 21 (100) | 1 | 113 (97.4) | 8 (100) | 1 | |||||||||
| L421P | 99 (96.1) | 21 (100) | 1 | 112 (96.6) | 8 (100) | 1 | |||||||||
| MtrR Substitutions | |||||||||||||||
| A39T | 10 (9.7) | 3 (14.3) | 0.816 | 13 (11.2) | 0 (0) | 1 | 13 (11.20) | 0 (0) | 1 | ||||||
| A40D | 4 (3.9) | 8 (38.1) | <0.0001 | 12 (10.3) | 0 (0) | 1 | 12 (10.3) | 0 (0) | 1 | ||||||
| G45D | 33 (32) | 1 (4.8) | 0.011 | 32 (27.6) | 2 (25) | 1 | 28 (24.1) | 6 (75.0) | 0.007 | ||||||
| T86A | 10 (9.7) | 8 (38.1) | 0.002 | 18 (15.5) | 0 (0) | 0.493 | 18 (15.5) | 0 (0) | 0.493 | ||||||
| H105Y | 49 (47.6) | 8 (38.1) | 0.427 | 51 (44) | 6 (75) | 0.181 | 56 (48.3) | 1 (12.5) | 0.110 | ||||||
| promoter -35A | 88 (85.4) | 10 (47.6) | <0.001 | 91 (78.4) | 7 (87.5) | 0.873 | 90 (77.6) | 8 (100) | 0.290 | ||||||
| 23S rRNA Mutations | |||||||||||||||
| A2059G | 0 (0) | 7 (87.5) | <0.0001 | ||||||||||||
χ2 test was used to identify molecular profiles associated with resistance to cefixime (CFMR), ceftriaxone (CROR), and azithromycin (>AZM ECOFF). aNG-STAR genotypes were reported previously (Yang et al., 2020). bAlso including substitutions V160A, N173S, A280V, D285E, R288K, and R291Q. cAlso including substitutions A323S, T326V, L328A/P, NERL329TDTF, Q335L, SP342AT, RD345Q, S352T, R373M, E377K, E385D, I388V, N406S, RP411QK, A437V, V443E, L447V, IF461VI, E464A, Q457K, RE468KK, N472E, P480A, and G545S. d,f,hAll isolates were penA 18 allele. eIncluding 10 isolates with penA 5, 1 isolate with penA 17, 15 isolates with penA 18, and 1 isolate with penA 41. gIncluding 10 isolates with penA 5, 1 isolate with penA 17, 14 isolates with penA 18, and 1 isolate with penA 41. Bold values indicate p < 0.05.
Mosaic penA alleles were detected in 37.5% of CROR isolates and in 13.8% of CROS isolates. Only the mosaic penA 60 allele was significantly associated with CROR (Table 2). PenA substitutions F374V and A501T showed significantly higher frequencies in CROR isolates than in CROS isolates.
NG-STAR ST-233 (penA 60) was associated with CFMR and CROR (Table 2), while ST-348 (penA 10, exhibited by NG-MAST ST5308, ST7554, and ST12784) was associated with CFMR and ST-428 (penA 18) was associated with CROR.
The metadata of four mosaic penA 60 isolates are listed in Table 3. Demographic and clinical information revealed that four patients were young (age range: 18–44), all of them had abnormal urinary discharge, and none reported previous history of gonorrhea or any antibiotic use in the past month. Three of four mosaic penA 60 isolates were NG-STAR genotype 233, whereas one was NG-STAR genotype 1143. Four penA 60 isolates had the same pattern of PenA substitutions, which contained A311V and T483S alterations, the key CROR substitution. All penA 60 isolates have identical ponA, mtrR, 23S rRNA, gyrA, parC, 16S rRNA, and rpsE patterns and different PorB substitutions. Additional MICs and the molecular profiles of four isolates are summarized in Table 3.
TABLE 3.
Metadata of four mosaic penA 60.001 isolates in Shanghai.
| Demographic/clinical information | MICs (mg/L) | |||||||||||||||
| Isolate id | Age | Gender | Ethnicity | Transmission | Infection site | Abnormal urinary discharge | Previous history of gonorrhea | Antibiotic use in the past month | Date of clinic visit | CRO | CFM | AZM | CIP | PEN | TET | SPT |
| 17–256 | 44 | Male | Han | Hetero | Urethra | Yes | No | No | 02/12/2017 | 0.125 | 0.5 | 0.06 | 16 | ≥32 | 2 | 8 |
| SH-40 | 18 | Male | Han | Hetero | Urethra | Yes | No | No | 25/11/2017 | 1 | ≥4 | 0.125 | ≥32 | 4 | 2 | 16 |
| SH-41 | 35 | Male | Han | Hetero | Urethra | Yes | No | No | 27/11/2017 | 1 | ≥4 | 0.125 | ≥32 | 2 | 1 | 8 |
| SH-48 | 28 | Male | Han | Hetero | Urethra | Yes | No | No | 06/12/2017 | 1 | ≥4 | 0.125 | ≥32 | 4 | 2 | 8 |
| Molecular profiles | ||||||||||||||||
| Isolate ID | NG-STAR genotype | PPNG | penA | PorB1b | PonA | MtrR | 23S rRNA | GyrA | ParC | 16S rRNA | rpsE | |||||
| 17–256 | 1143 | PPNG | 60.001 | G120K, A121G, Q143K, T215A, I218M, M257R, S258R, G259A | T375A, L421P | Promoter −35A, H105Y | WT | S91F, D95A | S87R | T1458C | WT | |||||
| SH-40 | 233 | Non-PPNG | 60.001 | T87A, T89S, G120K, A121D, V151A, I209M, YD211GY, G213Y, Q214L, T215, S217N, V242A, A256T, M257S, G259A, A272V | T375A, L421P | Promoter −35A, H105Y | WT | S91F, D95A | S87R | T1458C | WT | |||||
| SH-41 | 233 | Non-PPNG | 60.001 | G120K, A121D, Q143K, T215V, M257R, S258R, G259A | T375A, L421P | Promoter −35A, H105Y | WT | S91F, D95A | S87R | T1458C | WT | |||||
| SH-48 | 233 | Non-PPNG | 60.001 | T87A, T89S, G120K, A121D, V151A, I209M, YD211GY, G213Y, Q214L, T215, S217N, V242A, A256T, M257S, G259A, A272V | T375A, L421P | Promoter −35A, H105Y | WT | S91F, D95A | S87R | T1458C | WT | |||||
Among the non-mosaic penA allele isolates, the penA 18 allele and substitutions of PenA A501T and G542S were associated with ESCR (Table 2). The proportion of non-mosaic penA ESCR isolates harboring the PenA A501T substitution (for CFM, 60%, 3/5; for CRO, 80%, 4/5) was significantly higher than the proportion of non-mosaic penA ESCS isolates harboring that substitution (for CFM, 15%, 15/100; for CRO, 14%, 14/100). Eighty percent (4/5) of non-mosaic penA CROR isolates had the G542S substitution, which was significantly higher than non-mosaic penA CROS isolates (26%, 26/100). Interestingly, all ESCR with the PenA double substitutions of A501T and G542S (three CFMR and four CROR) exhibited a penA 18 allele.
porB1a and porB1b Genes
Among the 124 isolates, genotypes porB1a and porB1b accounted for 6.5% and 93.5%, respectively. CROR isolates had a higher percentage of PorB1b substitutions T87A, T89S, G213S/Y, Q214L, and G259A than CROS isolates (Table 2).
Among non-mosaic penA allele isolates, 60% (3/5) of CROR non-mosaic penA isolates harbored the PorB1b G213S/Y substitution, which was significantly higher than the CROS non-mosaic penA isolates (10.8%, 10/93) (Table 2).
mtrR Gene and Promoter
MtrR G45D and mtrR promoter -35A were significantly lower in CFMR isolates than in CFMS isolates. MtrR A40D and T86A were significantly associated with CFMR. No mtrR mutations was found to be associated with CROR.
Characteristic Genotypes of N. gonorrhoeae Isolates With MICs Above AZM ECOFF
In isolates with MICs above the AZM ECOFF value (1 mg/L), 87.5% (7/8, AZM MICs ≥ 256 mg/L) harbored the A2059G mutation in 23S rRNA, which is significantly higher than in isolates with MICs below AZM ECOFF (0/116, AZM MIC range: ≤0.03–1 mg/L). MtrR G45D and NG-STAR ST-202 (NG-MAST ST1866) were significantly higher in isolates above AZM ECOFF than in isolates below AZM ECOFF (p = 0.007 for MtrR G45D, p < 0.0001 for NG-STAR ST-202).
Characteristic Genotypes in N. gonorrhoeae SPTR Isolates
There was only one SPTR isolate identified. This SPTR isolate was the only strain that harbored 16S rRNA C1192U mutation in our dataset (Figure 1), which has earlier been reported to be associated with SPTR. The K26E substitution in RpsE was not detected.
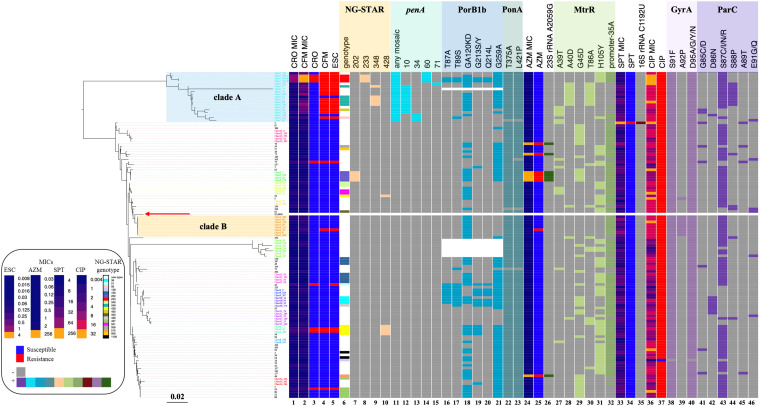
FIGURE 1.
Phylogenetic reconstruction of nine genes, patterns of antimicrobial resistance, and genetic polymorphisms in 124 N. gonorrhoeae isolates. Left: Phylogenetic reconstruction of 124 isolates based on maximum likelihood. The red arrow indicates the reference strain FA1090. Heatmap. Columns 1 and 2: MIC values of CRO and CFM. Columns 3–5: Susceptible/resistant categories according to the EUCAST MIC breakpoints of CRO, CFM, and ESCs. Column 6: NG-STAR genotype (white band indicates NG-STAR genotypes new to the NG-STAR database). Columns 7–10: A specific NG-STAR genotype. Column 11: Any mosaic penA allele. Columns 12–15: A specific mosaic penA allele. Columns 16–21: Non-synonymous amino acid changes from wild type in PorB1b. Columns 22 and 23: Non-synonymous amino acid changes from wild type in PonA. Column 24: MIC values of AZM. Column 25: Susceptible/resistant categories according to the EUCAST MIC breakpoints of AZM. Column 26: A2059G mutation in 23S rRNA. Columns 27–31: Non-synonymous amino acid changes from wild type in MtrR. Column 32: The –35A deletion in the mtrR promoter. Column 33: MIC values of SPT. Column 34: Susceptible/resistant categories according to the EUCAST MIC breakpoints of SPT. Column 35: C1192U mutation in 16S rRNA. Column 36: MIC values of CIP. Column 37: Susceptible/resistant categories according to the EUCAST MIC breakpoint of CIP. Columns 38–40: Non-synonymous amino acid changes from wild type in GyrA. Columns 41–46: Non-synonymous amino acid changes from wild type in ParC. The purple and orange rectangles indicate clades A and B described in the text, respectively. Sequence names are colored by cluster.
Clonal Distribution of N. gonorrhoeae ESCR Isolates by Phylogenetic Analysis
Phylogenetic analysis of nine genes was performed. Compared to a seven-gene phylogeny (Supplementary Figure 1), the inclusion of two SPTR genes (16S rRNA and rpsE) did not change the structure or resolution of the phylogeny. Cluster analysis results showed that a nine-gene phylogeny had 14 clusters, while a seven-gene phylogeny had 16 clusters. Most ESCR strains were classified as one clade that had the mosaic penA alleles (Figure 1, clade A). NG-STAR ST-233, ST-348, and ST-90 belonged to clade A. Approximately 76% (16 of 21) of CFMR were included in clade A (Figure 1). CROR appeared sporadically across the phylogeny.
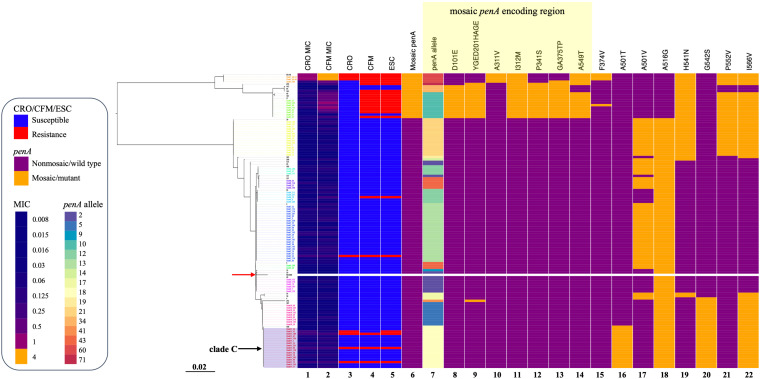
Four of the six ESCR that did not possess a mosaic penA allele harbored a penA 18 allele and was classified into clade C (Figure 2). The penA 18 allele consisted of the A501T and G542S double substitutions.
FIGURE 2.
penA gene phylogeny, patterns of antimicrobial resistance, and genetic polymorphisms of 124 N. gonorrhoeae isolates. Left: Phylogenetic reconstruction of 124 isolates based on the penA gene and using the maximum likelihood. The red arrow indicates reference strain FA1090. Heatmap. Columns 1 and 2: MIC values of CRO and CFM. Columns 3–5: Susceptible/resistant categories according to the EUCAST MIC breakpoints of CRO, CFM, and ESCs. Column 6: Any mosaic penA allele. Column 7: penA alleles. Columns 8–22: Non-synonymous amino acid changes from wild type in PenA. The purple rectangle indicates clade C as described in the text. The yellow rectangle indicates substitutions in the mosaic penA coding region. Sequence names are colored by cluster.
Multiple linear regression analysis revealed that the mosaic penA, PenA A501T/V, and PorB1b G213S/Y substitutions were strongly associated with increased MICs of CRO or CFM (Supplementary Table S4).
Phylogenetic Analysis of N. gonorrhoeae Isolates With MICs Above AZM ECOFF
Isolates with MICs above AZM ECOFF appeared sporadically across the phylogenetic tree (Figure 1) and were highly associated with the 23S rRNA A2059G mutation (Table 2 and Figure 1). All NG-STAR ST-202 isolates harbored the 23S rRNA A2059G mutation. Multiple linear regression analysis indicated that the 23S rRNA A2059G mutation was strongly associated with increased AZM MICs (Supplementary Table S5).
Analysis of N. gonorrhoeae CIPR Isolates
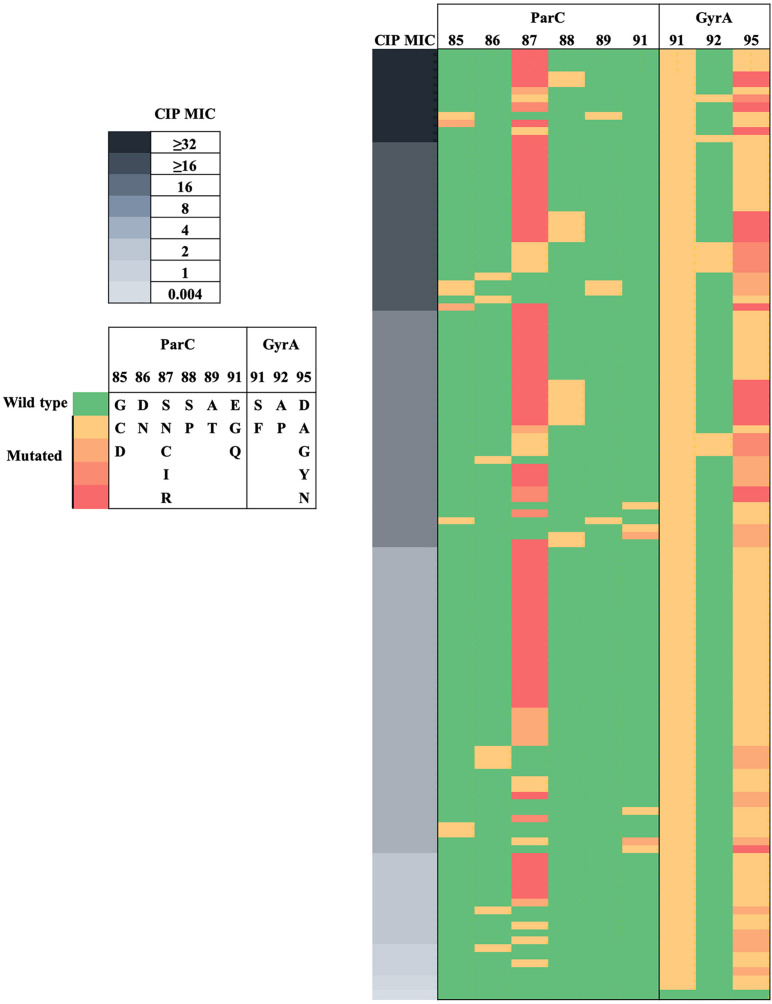
The 124 isolates included 123 CIPR and 1 CIPS. Substitutions at GyrA-91 and GyrA-95 were highly predictive of the resistant phenotype (Figure 3). CIPS did not harbor GyrA or ParC substitutions. CIPR with CIP MICs > 0.06 mg/L had both GyrA-91 (S91F) and GyrA-95 (D95A/G/Y/N) substitutions. N. gonorrhoeae isolates with triple GyrA substitutions at positions 91, 92, and 95 exhibited a high level of quinolone resistance (CIP MICs ≥ 16 mg/L). Phylogenetic analysis (Figure 1) revealed that GyrA A92P and ParC S88P substitutions could be clustered into two clades, whereas other GyrA and ParC substitutions were distributed across the phylogeny. Specifically, eight out of nine (88.9%) isolates with GyrA A92P substitution were clustered into clade B, whereas 9 out of 14 (64.3%) isolates with ParC S88P substitution were clustered into clade A. Multiple linear regression analysis indicated that ParC-85/86/87/88/89/91 and GyrA-91/92 substitutions heavily contributed to CIP MIC increments (Supplementary Table S6).
FIGURE 3.
Heat map visualization of CIP MICs with ParC and GyrA substitutions in N. gonorrhoeae. MICs of CIP are indicated on the left panel. ParC and GyrA substitutions are indicated on the middle and right panels, respectively.
Discussion
A large proportion of N. gonorrhoeae isolates in Shanghai in 2017 exhibited resistance to ESCs. Approximately 19.4% of 366 N. gonorrhoeae isolates were CFMR (MICs > 0.125 mg/L), and 40.7% of the isolates had CFM MICs ≥ 0.125 mg/L. About 5.5% of the isolates were CROR (MICs > 0.125 mg/L), and 18.0% of isolates had CRO MICs ≥ 0.125 mg/L. About 6.8% of the isolates had MICs above AZM ECOFF. One isolate was SPTR. N. gonorrhoeae CFMR isolates exhibited clonal distribution of one cluster containing mosaic penA alleles, whereas CROR isolates appeared sporadically across the phylogeny.
The resistant percentages of N. gonorrhoeae isolates to CRO, CFM, or AZM in Shanghai exceeded the WHO cutoff of 5%, indicating a need to review recommended treatments (World Health Organization [WHO], 2012). Over 18.0% of N. gonorrhoeae isolates had CRO MICs ≥ 0.125 mg/L, higher than that reported in previous years in Shanghai and other places in China (Gu et al., 2014; Yin et al., 2018) as well as several other countries such as 0.1% in the United States in 2013 and 2014 (Kirkcaldy et al., 2016), 1.8% in Canada in 2016 (Martin et al., 2019), and 10.7% in Japan in 2012–2013 (Hamasuna et al., 2015). The proportion of N. gonorrhoeae CFMR isolates (MICs > 0.125 mg/L, 19.4%) in Shanghai in 2017 was much higher than that in the United States (0.4–0.8% in 2013–2014) (Kirkcaldy et al., 2016), Europe (1.7–2.0% in 2014–2015) (Cole et al., 2017), and Canada (0.3% in 2016) (Martin et al., 2019). These findings indicate that unlike those in the United States, European countries, Japan, and Canada, CRO and CFM may need to be reviewed as a treatment for gonorrhea in Shanghai. N. gonorrhoeae isolates in Shanghai remain susceptible to SPT (Yang et al., 2006), suggesting that SPT may have potential as a first-line therapy for the treatment of uncomplicated urogenital gonorrhea in Shanghai. SPT is available in China. However, SPT is not suitable for the treatment of pharyngeal gonorrhea, as its efficacy rate is approximately 80% (Moran and Levine, 1995). Furthermore, SPTR isolates have been reported in several countries such as the Netherlands, the Philippines, South Korea, and the United Kingdom (Unemo and Shafer, 2014). There is concern that drug resistance would be rapidly selected when SPT is introduced as a first-line monotherapy. Therefore, SPT should be considered as a first-line treatment in combination with CRO or AZM in Shanghai.
Neisseria gonorrhoeae ESCR Isolates Tend to Be Clonal
Previous studies have shown that N. gonorrhoeae CFMR isolates in Japan (Yahara et al., 2018) and ESCRS isolates in the United States (Grad et al., 2014, 2016), Europe (Chisholm et al., 2013), and Canada (Demczuk et al., 2015) are predominantly clonal and associated with the mosaic penA allele. We also found that N. gonorrhoeae ESCR isolates were predominantly clonal in this study. In addition, we observed that ESCR isolates without the penA mosaic alleles were distributed sporadically across the phylogenetic tree, which is also concordant with a previous report (Grad et al., 2016). In our study, of the six ESCR isolates that did not possess the mosaic penA allele, four contained the penA 18 allele that included the PenA A501T and G542S double substitutions. However, ESCRS lineages in Canada were associated with non-mosaic penA 12 and 13 alleles (Demczuk et al., 2015), while ESCRS isolates with non-mosaic penA reported in the United States have sporadically emerged even in the penA gene phylogeny (Grad et al., 2016). The sample size of N. gonorrhoeae isolates in this study could be expanded, and whole-genome sequencing should be examined to further confirm this difference.
Mosaic penA Alleles Are Associated With N. gonorrhoeae ESCR and NG-STAR Clusters
Mosaic penA alleles have been associated with N. gonorrhoeae ESCRS (Ameyama et al., 2002; Lee et al., 2010). We observed that ESCR is highly associated with mosaic penA 10, whereas reports in the United States and Canada indicated that N. gonorrhoeae ESCRS is highly associated with mosaic penA 34 (Grad et al., 2014, 2016; Demczuk et al., 2015). All of the N. gonorrhoeae isolates with an NG-STAR ST-348 genotype (n = 5) contained the mosaic penA 10 allele. Several mosaic penA alleles (penA 60, penA 71, and penA 34) are associated with ESCR in this study.
penA 60 is significantly associated with both CROR and CFMR and occurs in a single cluster; thus, it is of great concern when it spreads. None of the carriers of the penA 60 isolates reported a previous history of gonorrhea or any antibiotic use in the past month, which indicates that they were recently infected with penA 60 ESCR strains. It is important to monitor the clonal expansion of penA 60 ESCR strains to contain its spread. The reported CRO-resistant cluster FC428 has a mosaic penA 60 genotype with a NG-STAR sequence type ST-233 (Lee et al., 2019). Three of the four penA 60 N. gonorrhoeae isolates also have an NG-STAR ST-233. Links between the penA 60 isolates in this study and FC428 strains remain to be elucidated.
Novel PorB1b Substitutions Associated With N. gonorrhoeae ESCR
In this study, we found that in contrast to CFMR, CROR is apparently associated with PorB1b substitutions other than mosaic penA or PenA substitutions. This is concordant with the results of a previous study that CRO is more severely affected by PorB1b than CFM (Unemo and Shafer, 2014), suggesting that either CFM does not readily diffuse into the periplasm through PorB1b or such diffusion is not altered by the porB determinant (Unemo and Shafer, 2014). To our knowledge, our study is the first to report that PorB1b substitutions T87A, T89S, S213S/Y, Q214L, and G259A are associated with CROR. Similar to a previous report, although certain mutations in porB can contribute to N. gonorrhoeae resistance, most mutations in porB do not (Goire et al., 2014). In vitro selection by introducing porB mutations into CROS isolates should be considered in the future to confirm the role of these mutations in the formation of CROR (Johnson et al., 2014). Previous studies have reported substitutions at amino acid positions 120 and 121 in putative loop 3 of PorB1b, which reduce the permeability of ESCs (Olesky et al., 2002). Interestingly, none of the substitutions detected in the present study are situated in any loops of PorB1b, and whether these substitutions could perturb protein structure remains unknown. Electrophysiological and biochemical studies of PorB1b proteins to reveal the mechanism of CROR conferred by these substitutions are warranted.
ParC S88P Substitution Clustered in ESCR Clade
We noticed that among the 14 isolates with ParC S88P substitution, nine were clustered in the ESCR clade (clade A), suggesting that this substitution may be associated with ESCR. Intriguingly, it was reported that various MDR bacteria such as methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae, and ESBL-producing Escherichia coli were demonstrated to have been selected by favorable fitness balance associated with high-level resistance to fluoroquinolones, principally attained by the mutations of some serine residues in gyrA and parC/grlA (Fuzi et al., 2017, 2020). The association of the ParC S88P substitution and that of other QRDR serine replacements with fitness gain, the promotion of particular clades, and the acquisition of the MDR phenotype warrant further investigation.
Limitations
This study only investigated a small percentage of N. gonorrhoeae isolates in Shanghai, with a total of 5,711 reported cases in this city in 2017. However, it is representative of the institution where the isolates were collected. A study with a larger sample size is required to extrapolate a broader strain distribution and to provide convincing evidence for the clonal distribution of ESCR with mosaic penA alleles and the sporadic distribution of ESCR with non-mosaic penA alleles. Specimens were obtained only from male patients, which may cause a higher proportion of CRO-resistant isolates, as reported by a Chinese national surveillance (Yin et al., 2018). Transmission of gonorrhea via different behaviors may result in infection of other mucosal sites, and isolates from other sites may exhibit different AMR phenotypes and genotypes. In the future, whole-genome sequencing should be considered to examine the population structure in Shanghai N. gonorrhoeae isolates, which would provide a significantly higher resolution for phylogenetic reconstruction.
Conclusion
This study observed a high percentage of N. gonorrhoeae isolates with reduced susceptibility to ESCs in Shanghai in 2017. Phylogenetic analysis of resistance determinants revealed that CFMR isolates tend to be clonal. Mosaic penA alleles and certain substitutions in PenA and PorB1b are associated with N. gonorrhoeae ESCR. CRO and CFM may need to be reviewed as treatment for gonorrhea in Shanghai. Monitoring clonal expansion and development of novel antimicrobials for gonorrhea treatment are urgently needed.
Data Availability Statement
The sequence data generated for this study has been submitted to GenBank and accession numbers can be found in the article.
Ethics Statement
The studies involving human participants were reviewed and approved by Shanghai Skin Disease Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YD analyzed the data and wrote the manuscript. YY performed the experiments and collected the data. YW, IM, and WD revised the manuscript. WG designed experiments, performed the experiments, collected the data, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Mingmin Liao (University of Saskatchewan, Canada) for assistance in data analysis and in the preparation of this manuscript.
Funding. The Wu Jieping Medical Foundation of China (grant number 320.6750.17339), the National Natural Science Foundation of China (grant number 81602905), the Outstanding Youth Training Program of Shanghai Municipal Health Commission (grant number 2018YQ57), and Foundation of Shanghai Municipal Health Commission (grant number 20164Y0245) supported this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.580399/full#supplementary-material
References
- Ameyama S., Onodera S., Takahata M., Minami S., Maki N., Endo K., et al. (2002). Mosaic-like structure of penicillin-binding protein 2 Gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46 3744–3749. 10.1128/aac.46.12.3744-3749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry P. M., Klausner J. D. (2009). The use of cephalosporins for gonorrhea: the impending problem of resistance. Expert. Opin. Pharmacother. 10 555–577. 10.1517/14656560902731993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S. A., Unemo M., Quaye N., Johansson E., Cole M. J., Ison C. A., et al. (2013). Molecular epidemiological typing within the European gonococcal antimicrobial resistance surveillance programme reveals predominance of a multidrug-resistant clone. Euro. Surveill. 18:20358. [PubMed] [Google Scholar]
- Cole M. J., Spiteri G., Jacobsson S., Woodford N., Tripodo F., Amato-Gauci A. J., et al. (2017). Overall low extended-spectrum cephalosporin resistance but high azithromycin resistance in Neisseria gonorrhoeae in 24 European countries, 2015. BMC Infect. Dis. 17:617. 10.1186/s12879-017-2707-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D., Peters J., Cole K., Cole M. J., Cresswell F., Dean G., et al. (2016). Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect. Dis. 16 1295–1303. 10.1016/S1473-3099(16)30157-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk W., Lynch T., Martin I., Van Domselaar G., Graham M., Bharat A., et al. (2015). Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J. Clin. Microbiol. 53 191–200. 10.1128/JCM.02589-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk W., Sidhu S., Unemo M., Whiley D. M., Allen V. G., Dillon J. R., et al. (2017). Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae Strains. J. Clin. Microbiol. 55 1454–1468. 10.1128/JCM.00100-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep B. A. (2013). Use of whole-genome sequencing for outbreak investigations. Lancet Infect. Dis. 13 99–101. 10.1016/S1473-3099(12)70276-1 [DOI] [PubMed] [Google Scholar]
- EUCAST (2020). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0 [Online]. Available online at: https://eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf (accessed August 14, 2020). [Google Scholar]
- Fuzi M., Rodriguez Baño J., Toth A. (2020). Global evolution of pathogenic bacteria with extensive use of fluoroquinolone agents. Front. Microbiol. 11:271. 10.3389/fmicb.2020.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzi M., Szabo D., Csercsik R. (2017). Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 8:2261. 10.3389/fmicb.2017.02261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Gerbaud G., Courvalin P. (2000). Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob. Agents Chemother. 44 1365–1366. 10.1128/aac.44.5.1365-1366.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goire N., Lahra M. M., Chen M., Donovan B., Fairley C. K., Guy R., et al. (2014). Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat. Rev. Microbiol. 12 223–229. 10.1038/nrmicro3217 [DOI] [PubMed] [Google Scholar]
- Grad Y. H., Harris S. R., Kirkcaldy R. D., Green A. G., Marks D. S., Bentley S. D., et al. (2016). Genomic epidemiology of gonococcal resistance to extended-spectrum Cephalosporins, Macrolides, and Fluoroquinolones in the United States, 2000-2013. J. Infect. Dis. 214 1579–1587. 10.1093/infdis/jiw420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y. H., Kirkcaldy R. D., Trees D., Dordel J., Harris S. R., Goldstein E., et al. (2014). Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect. Dis. 14 220–226. 10.1016/S1473-3099(13)70693-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. M., Chen Y., Yang Y., Wu L., Hu W. Z., Jin Y. L. (2014). Twenty-five-year changing pattern of gonococcal antimicrobial susceptibility in Shanghai: surveillance and its impact on treatment guidelines. BMC Infect. Dis. 14:731. 10.1186/s12879-014-0731-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Croucher N. J., Goater R. J., Abudahab K., Aanensen D. M., Harris S. R. (2017). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34 292–293. 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasuna R., Yasuda M., Ishikawa K., Uehara S., Hayami H., Takahashi S., et al. (2015). The second nationwide surveillance of the antimicrobial susceptibility of Neisseria gonorrhoeae from male urethritis in Japan, 2012-2013. J. Infect. Chemother. 21 340–345. 10.1016/j.jiac.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Harris S. R., Cole M. J., Spiteri G., Sanchez-Buso L., Golparian D., Jacobsson S., et al. (2018). Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect. Dis. 18 758–768. 10.1016/S1473-3099(18)30225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. R., Grad Y., Ganakammal S. R., Burroughs M., Frace M., Lipsitch M., et al. (2014). In Vitro selection of Neisseria gonorrhoeae mutants with elevated MIC values and increased resistance to cephalosporins. Antimicrob. Agents Chemother. 58 6986–6989. 10.1128/AAC.03082-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkcaldy R. D., Harvey A., Papp J. R., Del Rio C., Soge O. O., Holmes K. K., et al. (2016). Neisseria gonorrhoeae antimicrobial susceptibility surveillance - the gonococcal isolate surveillance project, 27 Sites, United States, 2014. MMWR Surveill. Summ. 65 1–19. 10.15585/mmwr.ss6507a1 [DOI] [PubMed] [Google Scholar]
- Lahra M. M., Martin I., Demczuk W., Jennison A. V., Lee K. I., Nakayama S. I., et al. (2018). Cooperative recognition of internationally disseminated ceftriaxone-resistant Neisseria gonorrhoeae Strain. Emerg. Infect. Dis. 24 735–740. 10.3201/eid2404.171873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Nakayama S. I., Osawa K., Yoshida H., Arakawa S., Furubayashi K. I., et al. (2019). Clonal expansion and spread of the ceftriaxone-resistant Neisseria gonorrhoeae strain FC428, identified in Japan in 2015, and closely related isolates. J. Antimicrob. Chemother. 74 1812–1819. 10.1093/jac/dkz129 [DOI] [PubMed] [Google Scholar]
- Lee R. S., Seemann T., Heffernan H., Kwong J. C., Goncalves Da Silva A., Carter G. P., et al. (2018). Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J. Antimicrob. Chemother. 73 353–364. 10.1093/jac/dkx405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. G., Lee H., Jeong S. H., Yong D., Chung G. T., Lee Y. S., et al. (2010). Various penA mutations together with mtrR, porB and ponA mutations in Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime or ceftriaxone. J. Antimicrob. Chemother. 65 669–675. 10.1093/jac/dkp505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Moriyama E. N. (2004). Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform. 5 378–388. 10.1093/bib/5.4.378 [DOI] [PubMed] [Google Scholar]
- Martin I., Sawatzky P., Allen V., Lefebvre B., Hoang L., Naidu P., et al. (2019). Multidrug-resistant and extensively drug-resistant Neisseria gonorrhoeae in Canada, 2012-2016. Can. Commun. Dis. Rep. 45 45–53. 10.14745/ccdr.v45i23a01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J. S., Levine W. C. (1995). Drugs of choice for the treatment of uncomplicated gonococcal infections. Clin. Infect. Dis. 20(Suppl. 1), S47–S65. 10.1093/clinids/20.supplement_1.s47 [DOI] [PubMed] [Google Scholar]
- Ng L. K., Martin I., Liu G., Bryden L. (2002). Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46 3020–3025. 10.1128/aac.46.9.3020-3025.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesky M., Hobbs M., Nicholas R. A. (2002). Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46 2811–2820. 10.1128/aac.46.9.2811-2820.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragonnet-Cronin M., Hodcroft E., Hue S., Fearnhill E., Delpech V., Brown A. J., et al. (2013). Automated analysis of phylogenetic clusters. BMC Bioinform. 14:317. 10.1186/1471-2105-14-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. (1988). Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature 332 173–176. 10.1038/332173a0 [DOI] [PubMed] [Google Scholar]
- Unemo M., Dillon J. A. (2011). Review and international recommendation of methods for typing neisseria gonorrhoeae isolates and their implications for improved knowledge of gonococcal epidemiology, treatment, and biology. Clin. Microbiol. Rev. 24 447–458. 10.1128/CMR.00040-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Golparian D., Skogen V., Olsen A. O., Moi H., Syversen G., et al. (2013). Neisseria gonorrhoeae strain with high-level resistance to spectinomycin due to a novel resistance mechanism (mutated ribosomal protein S5) verified in Norway. Antimicrob. Agents Chemother. 57 1057–1061. 10.1128/AAC.01775-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Shafer W. M. (2014). Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbiol. Rev. 27 587–613. 10.1128/CMR.00010-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Western Pacific Gonococccal Antimicrobial Surveillance Programme (2008). Surveillance of antibiotic resistance in Neisseria gonorrhoeae in the WHO Western Pacific Region, 2006. Commun. Dis. Intell. Q. Rep. 32 48–51. [DOI] [PubMed] [Google Scholar]
- Wi T., Lahra M. M., Ndowa F., Bala M., Dillon J. R., Ramon-Pardo P., et al. (2017). Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med. 14:e1002344. 10.1371/journal.pmed.1002344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2012). Global Action Plan to Control the Spread and Impact of Antimicrobial Resistance in Neisseria gonorrhoeae. Geneva: World Health Organization. [Google Scholar]
- World Health Organization [WHO] (2018). Report on Global Sexually Transmitted Infection Surveillance, 2018. Geneva: World Health Organization. [Google Scholar]
- Yahara K., Nakayama S. I., Shimuta K., Lee K. I., Morita M., Kawahata T., et al. (2018). Genomic surveillance of Neisseria gonorrhoeae to investigate the distribution and evolution of antimicrobial-resistance determinants and lineages. Microb. Genom. 4:205. 10.1099/mgen.0.000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liao M., Gu W. M., Bell K., Wu L., Eng N. F., et al. (2006). Antimicrobial susceptibility and molecular determinants of quinolone resistance in Neisseria gonorrhoeae isolates from Shanghai. J. Antimicrob. Chemother. 58 868–872. 10.1093/jac/dkl301 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yang Y., Martin I., Dong Y., Diao N., Wang Y., et al. (2020). NG-STAR genotypes are associated with MDR in Neisseria gonorrhoeae isolates collected in 2017 in Shanghai. J. Antimicrob. Chemother. 75 566–570. 10.1093/jac/dkz471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. P., Han Y., Dai X. Q., Zheng H. P., Chen S. C., Zhu B. Y., et al. (2018). Susceptibility of Neisseria gonorrhoeae to azithromycin and ceftriaxone in China: a retrospective study of national surveillance data from 2013 to 2016. PLoS Med. 15:e1002499. 10.1371/journal.pmed.1002499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarantonelli L., Borthagaray G., Lee E. H., Shafer W. M. (1999). Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 43 2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data generated for this study has been submitted to GenBank and accession numbers can be found in the article.