Abstract
Fbw7 is a tumor suppressor that regulates the degradation of oncogenic substrates such as c-Jun, c-Myc, Notch1 intracellular domain (ICD), and cyclin E by functioning as the substrate recognition protein in the Skp1-Cullin-F-box (SCF) ubiquitin ligase complex. Consequently, low expression or loss of FBXW7 in breast cancer has been hypothesized to result in the accumulation of oncogenic transcription factors that are master regulators of proliferation, apoptosis, and ultimately transformation. Despite this, the direct effect of Fbw7 loss on mammary gland morphology and tumorigenesis has not been examined. Here, we demonstrate that conditional deletion of Fbxw7 in murine mammary tissue initiates breast tumor development and also results in lactation and involution defects. Further, while Fbxw7 loss results in the overexpression of Notch1-ICD, c-Jun, cyclin E, and c-Myc, the downstream transcription factor pathways associated with c-Myc and cyclin E are the most dysregulated, including at the single-cell level. These pathways are dysregulated early after Fbxw7 loss, and their sustained loss results in tumorigenesis and reinforced c-Myc and cyclin E-E2F pathway disruption. We also find that loss of Fbxw7 is linked to the acquisition of Trp53 mutations, similar to the mutational spectrum observed in patients. Our results demonstrate that the loss of Fbxw7 promotes the acquisition of Trp53 mutations and that the two cooperate in breast tumor development. Targeting c-Myc, E2F, or p53 may therefore be a beneficial treatment strategy for FBXW7-altered breast cancer patients.
Keywords: Fbw7, P53, Breast cancer, Tumor cell biology, Cell cycle
Introduction
Targeted degradation through the ubiquitin–proteasome system plays a critical role in the regulation of cell homeostasis, helping to constrain the enzymatic or transcriptional activity of substrate proteins to an appropriate time and location. Many proteins that are tightly controlled by ubiquitination are master regulators of proliferation, growth, apoptosis, and cell fate decisions, making the proteins involved in their degradation tumor suppressors. One such example is the evolutionarily conserved F-box protein Fbw7 (encoded by the gene FBXW7), which partners with Skp1 and Cullin1 to form the E3 ubiquitin ligase complex SCFFbw7 [1]. Fbw7 is the substrate recognition subunit and binds proteins containing the canonical “Cdc4 phosphodegron motif” (CPD; 1). Binding enables the assembly of ubiquitin chains on the substrate, marking it for degradation. Targets of Fbw7 include the oncoproteins c-Myc, c-Jun, cyclin E, and Notch1-intracellular domain (Notch1-ICD; 1). Dysregulation of individual substrates can promote tumor development. For example, increased expression of cyclin E results in genomic instability, cell cycle dysregulation, mammary gland hyperplasia, and tumorigenesis [2], [3], [4], [5], [6]]. Dysregulation of Fbw7 targets in other cancer types has also been observed [7], [8], [9], [10]], implying that Fbw7-mediated degradation is commonly altered during transformation. Additionally, point mutations in the CPD motif of some targets can occur [8], [11], [12]], suggesting that the dysregulation of Fbw7 substrates in cancer may be widespread.
Alterations in either the expression level or mutational status of FBXW7 have also been observed in multiple cancer types. While point mutations in FBXW7 that disrupt substrate binding have been identified in some cancers, such mutations are found less often in breast cancer [13], where loss of FBXW7 expression is much more common. Deletion of chromosome 4q31, which contains FBXW7, is seen in approximately 30% of primary breast cancers [9]. Gene silencing through FBXW7 promoter methylation is also widely observed in breast cancer [14], and lower levels of FBXW7 expression have been correlated with worse disease-free survival [9], [15]]. As such, loss of FBXW7 may be a critical “first-hit” during the development of breast cancer. Despite this evidence, the direct consequences of FBXW7 expression loss on breast tumorigenesis have not been directly examined. While many studies have focused on the contribution of individual Fbw7 substrates to breast tumor development, the effects of FBXW7 loss on substrate accumulation can vary significantly by tissue type. In an effort to identify the downstream consequences of reduced Fbxw7 expression on mammary gland development and tumorigenesis, we utilized a Balb/c mammary mouse model of conditional Fbxw7 loss. We find that Fbxw7 loss triggers breast tumorigenesis that is associated with Trp53 mutation and early/sustained upregulation of the E2F and c-Myc transcription factor pathways.
Materials and methods
Mice and genotyping
Animals were housed at the Medical College of Wisconsin. Procedures used were approved by the Institutional Animal Care and Use Committees at the Medical College of Wisconsin. Generation of the Fbxw7flox/flox mouse was published by Thompson et al. (2008) [16]. Mice were purchased from The Jackson Laboratory (Stock #017563). LGB-Cre mice are described by Selbert et al. (1998) [17] and were purchased from The Jackson Laboratory (Stock #017836). Fbxw7flox/flox mice were independently backcrossed six times to WT Balb/c mice (The Jackson Laboratory Stock #000651). To obtain Fbxw7flox/flox;LGB-Cre+ females, Fbxw7flox/flox males were mated with LBG-Cre+ females. Fbxw7flox/flox mice were genotyped as described in Thompson et al. (2008) [16]. LGB-Cre positivity was genotyped with the primers F: 5′ GCC TGC ATT ACC GGT CGA TGC AAC GA 3′ and R: 5′ GTG GCA GAT GGC GCG GCA ACA CCA TT 3′. Excision of Fbxw7 exons 5 and 6 in Fbxw7flox/flox;LGB-Cre+ tissues was assayed by PCR as described in Thompson et al. (2008) [16]. Survival was calculated using GraphPad Prism v. 8.0.1.
Immunohistochemistry
Tissue sections were deparaffinized in xylene for 2 × 10 min each followed by 100, 90, and 70% ethanol solutions for 10 min each. Sections were rinsed in water for 5 min and boiled for 20 min in 10 mM citric acid, pH 6.0. After cooling (15 min), sections were placed in 3% H2O2 in methanol in the dark for 20 min. Tissues were rinsed in PBS for 5 min and then blocked in 10% normal goat serum in PBS-0.025% Triton X-100 for 1hr. Sections were washed in water for 2 min and then blocked using the Avidin/Biotin blocking kit (Vector Laboratories, SP-2001). Tissues were stained overnight in 10% NGS, PBS-0.025% Triton X-100 with the following antibodies and dilutions: Fbw7 1:200 (gift of Bruce E. Clurman and described in [18], Ki67 1:200 (Abcam, ab15580), Cleaved caspase 3 1:200 (Cell Signaling Technologies, 9661S), c-Myc 1:100 (Abcam, ab32072), cyclin E 1:200 (Sigma, HPA018169), Notch1 1:50 (Cell Signaling Technologies, 3268S), c-Jun 1:100 (Abcam, ab32137), β-casein 1:100 (Santa Cruz, sc-166684), p53 1:200 (2524S), ERα 1:100 (EMD-Millipore, 06-935), and PR 1:100 (Abcam ab16661). After 3 × 5 min washes in PBS-0.025% Triton X-100, samples were processed using the Vectastain rabbit or mouse peroxidase IgG kits (Vector Laboratories, PK-4001 and PK-4002) and visualized with DAB reagent (Vector Laboratories, SK-4100). After washing in PBS, samples were stained with hematoxylin (Sigma, MHS16-500ML) and dehydrated in 70, 90, and 100% ethanol washes for 5 min each followed by 2 × 5 min xylene washes. Sections were mounted and imaged using a Nikon Eclipse 50i scope equipped with a Nikon Digital Sight DS-L3 camera.
Isolation of mammary epithelial single cells
Following euthanasia, all mammary glands were removed and minced into a fine paste with razors. The tissue was weighed and placed into collagenase solution (0.2% trypsin, 0.2% collagenase A, 5% FBS, 5 μg/mL gentamycin in DMEM/F12). 10 mL of collagenase solution was used for each gram of tissue. Tissue was digested with shaking at 37 °C for 2 h, with pipetting every 30 min. Digested tissue was centrifuged at 1500 rpm for 5 min, and the pellet was resuspended in 10 mL serum free DMEM/F12. To reduce the number of fibroblasts, the mixture was pulse-spun at 500 g for 7 s and the supernatant was removed. This was repeated 5–8 times until the supernatant appeared clear. The pellet was resuspended in 5 mL of red cell lysis buffer (Sigma, R7757), placed in a clean 15 mL conical tube, and incubated for 5 min. After a 5 min spin at 500 g, the pellet was resuspended in serum free DMEM/F12 and centrifuged. The pellet was resuspended in 2 mL 0.25% Trypsin-EDTA with 10 μL of DNAse and transferred to a 6 cm dish. Cells were pipetted rigorously for 5 min or until a complete dissociation of organoids into single cells was observed under a microscope. 5 mL of DMEM/F12 with 10% FBS was added, and cells were filtered through a 40 μM cell strainer twice. Cells were centrifuged at 200g for 5 min, and resuspended in 3–6 mL of DMEM/F12 with 10% FBS. Epithelial enrichment was performed using the EasySep Mouse Epithelial Cell Enrichment Kit (Stemcell Technologies, 13309).
Single cell qPCR
Single cell qPCR was performed using the Fluidigm C1/BiomarkHD systems. 5–10 μm pre-amp IFCs were used to isolate single cells and for subsequent cDNA generation. Cells were loaded at a concentration of 200,000 cells/mL and were visualized with a Nikon EVOS microscope. Dead cells or chambers containing more than one cell were excluded. cDNA was prepared according to the Preamplification Using Delta Gene Assays guide (available on Fluidigm’s website). Delta gene assays for c-Myc, E2F, c-Jun, and Notch-ICD targets were purchased from Fluidigm (See Supplementary Table 1 for details) to measure mRNA levels, and RNA spike controls were included for normalization of expression (ArrayControl RNA Spikes, ThermoFisher Scientific, AM1780). qPCR was performed on a 96.96 Dynamic Array using the BiomarkHD system. Data were analyzed using the Fluidigm Real-Time PCR Analysis software. Data points for each gene (single cell values) lacking amplification or with poor melt curves were removed. Ct values were normalized to RNA spike controls and subtracted from the limit of detection (Ct of 24). Final values are expressed as Log2Expression. Differences between groups were analyzed using GraphPad Prism v. 8.0.1 and the Mann-Whitney U test using a p value cutoff of <0.05. See Supplementary Tables 2–5 for all single cell qPCR data.
RNA sequencing
For epithelial RNA isolation, cells were resuspended in 1 mL Trizol (ThermoFisher Scientific, 15596018). RNA was isolated according to the manufacturer’s recommended protocol. RNA quality was measured by NanoDrop 2000 (ThermoFisher Scientific) and on an Agilent 2200 TapeStation analyzer. RNA samples of lower quality (260/280 <1.9 or RIN value <7) were further purified using the RNeasy Plus kit (Qiagen, 74134). For mammary gland and tumor samples, tissues were homogenized in Trizol and similarly processed.
For RNA sequencing, ERCC Spike-In Mix (ThermoFisher Scientific, 4456840) was added. Libraries were prepped in accordance with the NEBNext Ultra II RNA library prep kit (New England Biolabs, E7770) in combination with the NEBNext Poly(A) mRNA magnetic isolation module (New England Biolabs, E7490), the NEBNext multiplex oligos set 1 (New England Biolabs, E7335), and Ampure XP magnetic beads (Beckman Coulter, A6388). Libraries were quantified using the NEBNext library quant kit (New England Biolabs, E7630) and combined with PhiX control v3 (Illumina, 15017666). Pooled libraries were run on an Illumina NextSeq 500 system using paired end sequencing (38 × 2).
Raw RNA sequence reads were mapped to mouse reference genome mm9 using Salmon (v1.2.0) and default parameters, and were normalized using ERCC spike-ins. Differential Expression analysis was done using the DESeq2 (v1.24.0) package in R. Differentially expressed genes were identified as significant at a Benjamini-Hochberg adjusted p-value <0.05 and an absolute fold change of 2 (|Log2FC|>1). Heatmaps were done in R using pheatmap (v1.0.12) with the Euclidean method for distance and k-means clustering. The MMTV-PyMT 8 week tumor time point RNA sequencing data published by Cai et al. (2017) [19] were downloaded from GEO Omnibus (Accession number GSE76772). We analyzed GSM2037468, GSM2037476, and GSM2037484, which represent 3 replicates of the 8 week time point. DESeq analysis comparing our expression data with those of Cai et al. (2017) [19] was performed as described above.
For GSEA, gene expression data for each condition alongside a mouse to human gene symbol conversion file were loaded into GSEA 4.0.3 (Broad Institute). The human derived cancer (h.all.v7.0) dataset was queried using 1000 as the set number of permutations, and permutation type was set to gene_set. Only those genesets that met the false discovery rate of 0.25 and NOM p < 0.01 were considered.
Analysis of Trp53 mutations
Genomic DNA was isolated from tissue samples in 75 μL of 25 mM NaOH/0.2 mM EDTA for 1 h at 98 °C. After cooling to room temperature, 75 μL of 40 mM Tris HCl (pH 5.5) was added to neutralize the samples. After centrifugation, 2 μL of resulting DNA was used in PCR amplification of exons 5–6 or 7–8 using LongAmp Taq mastermix (New England Biolabs, M0287S) and the following primers:
Exons 5–6: 5′-CGTTACTCGGCTTGTCCCCGACCT-3′and 5′-CAACTGTCTCTAAGACGCACAAC-3′
Exons 7–8: 5′-GAGGTAGGGAGCGACTTCACCTGG-3′and 5′-TGAAGCTCAACAGG CTCCTCCGCCTCC-3′
Amplicons were cloned using the TA Cloning Kit (Invitrogen/ThermoFisher, K204001). To determine mutations in Fbxw7−/− cells or tumors, comparisons were made with WT tail samples taken from the same mouse as the cells or tumor.
Results
Fbxw7 loss initiates mammary tumor development
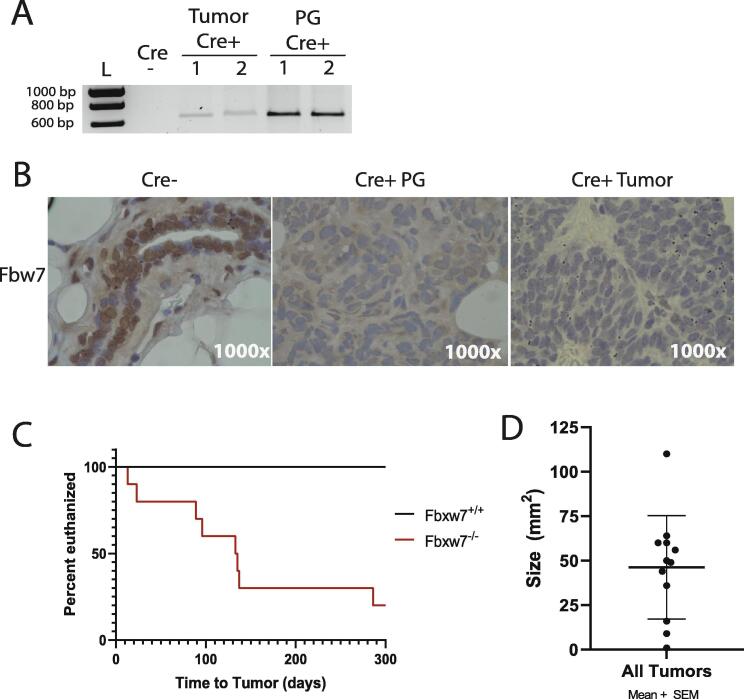
To determine if loss of Fbxw7 would initiate the development of breast tumors, we modified a published mouse model of inducible Fbxw7 gene excision previously used to study hematopoiesis [16]. In this model, exons 5 and 6 in both alleles of Fbxw7 are flanked by LoxP sites (Fbxw7fl/fl). Thompson et al. [16] previously showed that Cre-mediated excision of exons 5 and 6 results in a complete loss of Fbw7 protein expression. To direct Fbxw7 excision specifically within the mammary gland, we mated Fbxw7fl/fl mice with beta-lactoglobulin (LGB)-Cre transgenic mice [17]. The LGB promoter drives Cre expression in the mammary gland epithelial cells during pregnancy and subsequent lactation, and thus Fbxw7 excision in the breast epithelium is inducible by pregnancy. We examined the effect of Fbxw7 loss in 10 Fbxw7fl/fl;LGB Cre+ and 8 control Fbxw7fl/fl;LGB Cre- animals. Excision of Fbxw7 in Cre+ animals was verified in any tumors that developed and in nearby healthy mammary glands using PCR primers specific to the excised allele (representative results shown in Fig. 1A; [16]). While excision was observed following the first pregnancy (Fig. 1A, PG Cre+ lanes), the majority of animals underwent two pregnancies. Loss of Fbw7 protein expression in the mammary glands of pregnant Cre+ mice and any resulting tumors was verified by IHC (representative tissues shown in Fig. 1B). These data indicate that Fbxw7 expression is lost upon pregnancy in the mammary glands of Fbxw7fl/fl;LGB Cre+ mice.
Fig. 1.
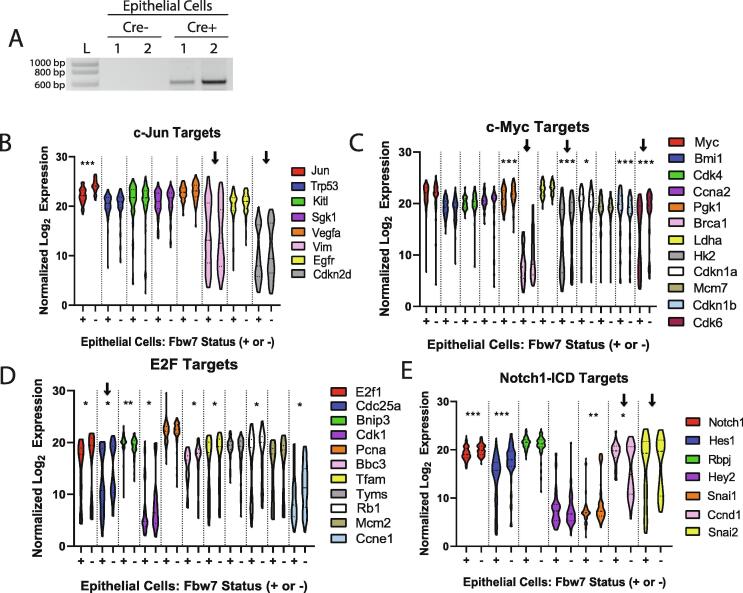
Fbxw7 loss triggers mammary tumorigenesis. A. PCR analysis for excision of exons 5–6 in murine Fbxw7 was performed on DNA extracted from control (Cre-) mammary tissue, tumor samples from Cre+ mice, and mammary glands from Cre+ mice in their third week of pregnancy. Proper excision results in a band of ∼650 bp. Representative results shown from 1 control (Cre-), 2 tumors (Tumor Cre+ 1 and 2), and 2 pregnant Cre+ mice (PG Cre+ 1 and 2). L = molecular weight ladder. B. IHC for Fbw7 showing expression in control Cre- tissue and loss of expression in one example Cre+ PG animal and one example Cre+ tumor. Magnification = 1000×. C. Mice were sacrificed at the time of tumor discovery, and the time to tumor for each animal is shown. D. The size (area) of each tumor was measured at the time of sacrifice and the mean size + SEM was plotted. Note that 8/10 mice developed tumors, with 4 developing more than 1 tumor, thus N = 12.
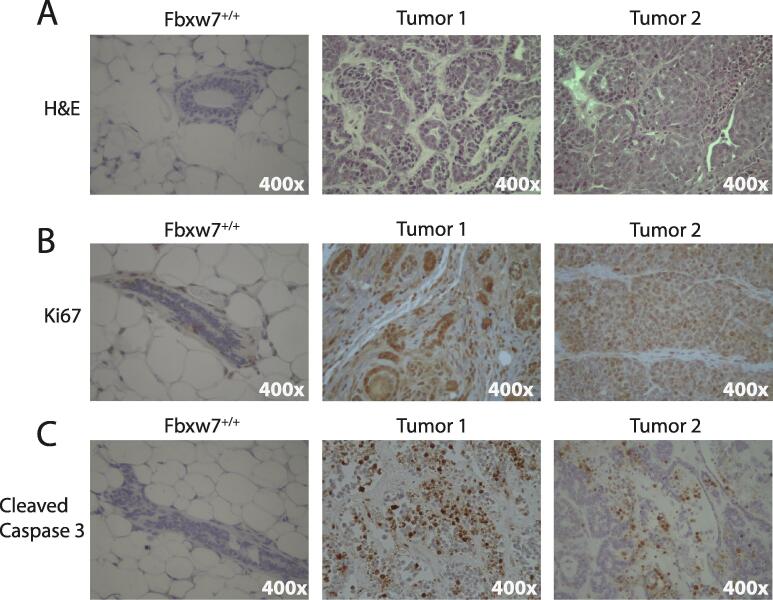
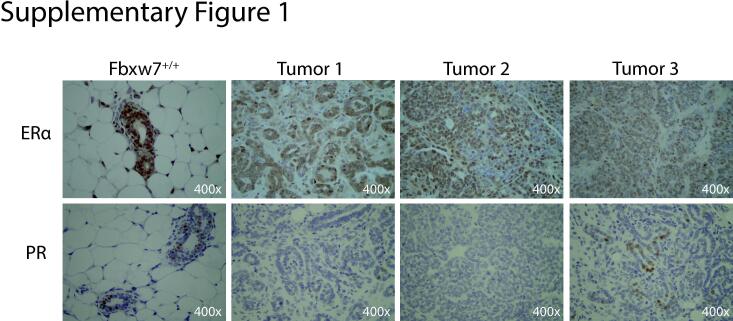
We next determined whether Fbxw7 loss promotes breast tumor development. Beginning at the time of first pregnancy, all animals were palpated weekly to identify tumors. As our goal was to establish if Fbxw7 loss alone would promote tumor development, and multiple genetic hits or the use of immunocompromised mice are typically required to observe metastasis in breast cancer models [20], [21], [22]], identification of palpable tumors was defined as the experimental endpoint. No tumors were noted in any of the Fbxw7+/+ control mice. In contrast, 8/10 (80%) of Fbxw7−/− animals developed palpable tumors, with the time to tumor detection from the date of the first pregnancy ranging from 13 to 286 days (Fig. 1C). 3/10 mice developed tumors after a single pregnancy, with an additional 5/10 undergoing a second pregnancy prior to tumor development. Four mice had tumors identified simultaneously in distinct locations. Mice were sacrificed upon tumor palpation and the area of each tumor was measured following excision to obtain a more accurate size measurement, as localized swelling or other factors may contribute to the palpable mass size. Tumor size ranged from 1 mm2 for the smallest to 110 mm2 for the largest, with an average tumor size of ∼46.3 mm2 (Fig. 1D). Tumor location was variable, with tumors found in thoracic, abdominal, and inguinal mammary glands. No additional non-palpable tumors were observed following dissection, with no gross evidence of metastatic disease in the lungs or livers (data not shown). As the longest time to tumor from first pregnancy was 286 days (∼40 weeks), the two remaining Fbxw7−/− animals and all control animals were aged to 40 weeks post primary pregnancy before sacrifice. No mammary tumors were observed in these animals upon dissection. 1/8 control animals developed a large intestine mass which was not positive for Fbxw7 excision and deemed an unrelated tumor. By H&E, large areas of epithelial cell hyper-proliferation were noted in the tumors, with excessive filling of the fat pad (Fig. 2A). Tumors were moderately- to well-differentiated, as evidenced by the presence of retained normal breast architecture or collapsed/filled-in tubules. No local invasion of tumors into the surrounding muscle tissue was noted in any of the specimens, consistent with their relatively well-differentiated appearance by histology. Consistent with hyper-proliferation, prevalent staining of the proliferation marker Ki67 was observed in the tumors (Fig. 2B). Areas of apoptosis were also observable, as indicated by cleaved caspase 3 staining (Fig. 2C). We also examined estrogen receptor (ERα) and progesterone receptor (PR) status by IHC (Supplementary Fig. 1). While some intra-tumor heterogeneity was observed, the majority of tumors were ERα-positive and PR negative (Supplementary Fig. 1). The phenotype of ERα+, PR-, and high Ki67 staining is most consistent with a luminal B-like subtype. We conclude that loss of Fbxw7 expression promotes mammary tumor development and results in increased proliferation and apoptosis.
Fig. 2.
Fbxw7−/− tumors are highly proliferative and apoptotic. A. Representative results of H&E staining showing normal breast architecture in a control Fbxw7+/+ animal and excessive filling of the fat pad and epithelial proliferation in two independent Fbxw7−/− tumors. B. Representative IHC results showing Ki67 staining (marking proliferation), which is low in control tissues and high in tumors. C. Representative IHC results for cleaved caspase 3 marking increased apoptosis in tumors. Little apoptosis was observed in control animals. All images 400×.
Fbxw7−/− mice exhibit impaired lactation and involution
All mice were able to carry their litters to term, and no differences in litter size were observed. Importantly, the Fbxw7−/− mice were unable to nurse their pups long-term. All pups born from Fbxw7−/− mothers died between 1 and 7 days of age. However, pups were successfully nursed by WT foster mothers, indicating Fbxw7−/− mothers have a lactation defect. This phenotype is similar to that reported by Hu et al. in mice overexpressing the Fbw7 target Notch1 [23]. Defects in lactation have also been observed upon c-Myc overexpression [24].
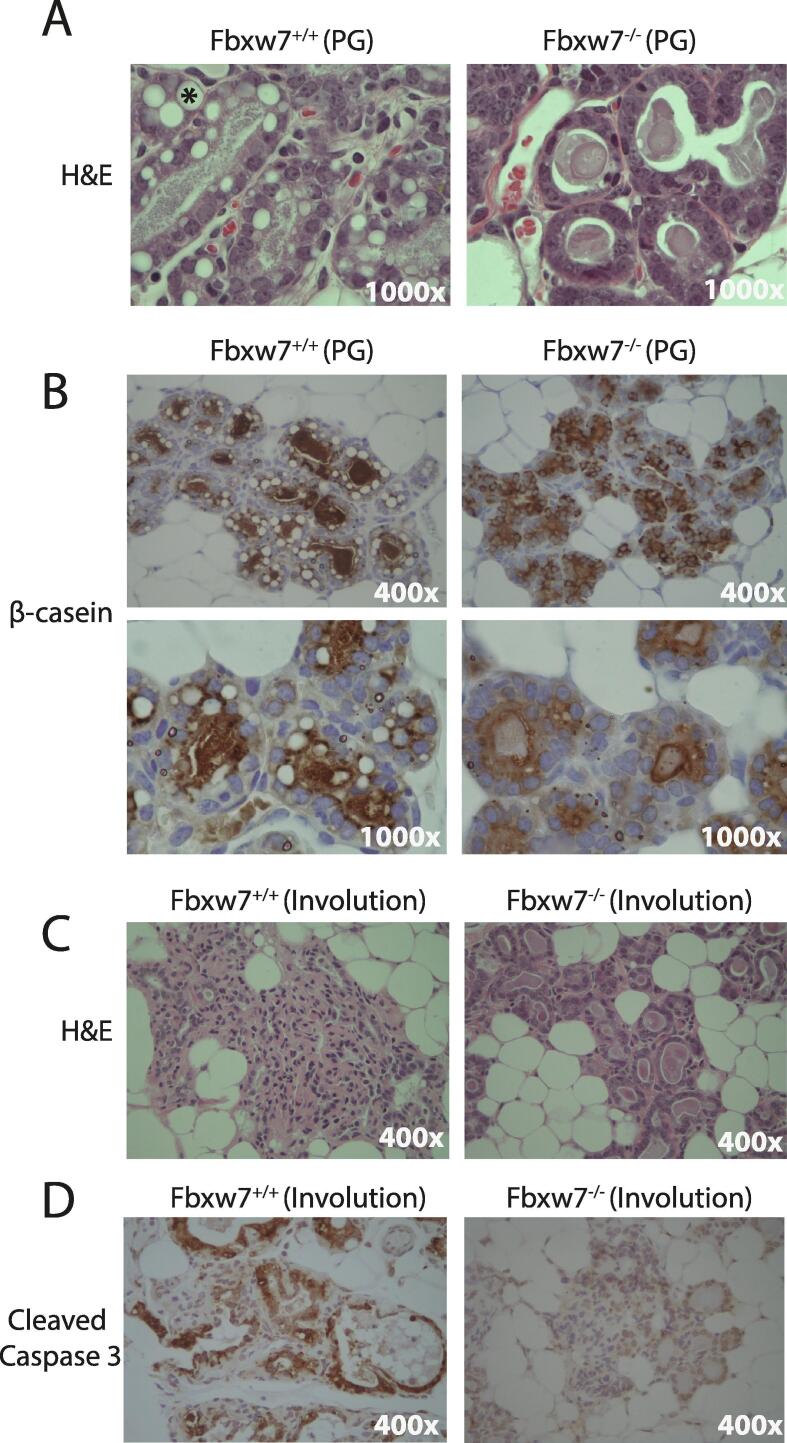
To better delineate the observed lactation defect in Fbxw7−/− mothers, we examined mammary gland development in virgin mice, during late pregnancy, and 5 days post involution. No differences were observed in the mammary glands in virgin mice (data not shown), which was expected due to intact Fbxw7 alleles in both Cre- and Cre+ mice. In contrast, changes were readily observed in pregnant Fbxw7+/+ vs. Fbxw7−/− mice. Although no differences in alveolar or lobular size was noted, glands in Fbxw7−/− pregnant mice contained fewer lipid droplets (Fig. 3A). Milk secretions were observable in the lumina in both genotypes, however the staining was more granular in the Fbxw7+/+ animals, indicating differences in milk composition between the two genotypes (Fig. 3A). We next performed IHC for the milk protein β-casein and observed abnormal staining in the Fbxw7−/− alveoli. Although β -casein was readily detectable, higher magnification revealed little β -casein accumulation in the lumina of Fbxw7−/− mice compared to Fbxw7+/+ mice (Fig. 3B). These data suggest that Fbxw7−/− mice are unable to nurse their litters due to defects in milk composition and secretion.
Fig. 3.
Fbxw7 loss results in lactation deficiency and disrupted involution. A. H&E staining from representative control Fbxw7+/+ as well as Fbxw7−/− mammary tissues. Tissue sections were taken from mice during the third week of pregnancy. Fbxw7−/− mice show a lack of lipid droplets (marked with * in Fbxw7+/+ panel) and a less granular staining in the lumina than Fbxw7+/+ mice. Images taken at 1000×. B. IHC for β-casein shows a reduction in Fbxw7−/− versus Fbxw7+/+ lumina. Upper panel = 400×, lower panel = 1000×. C. H&E staining of mammary tissue from representative Fbxw7+/+ and Fbxw7−/− mice 5 days into involution. Images taken at 400×. D. IHC staining for cleaved caspase 3 showing weaker staining in the Fbxw7−/− glands at 5 days involution vs. control glands. Images 400×.
In addition to lactation defects, dysregulation of the Fbw7 targets Notch-ICD, c-Myc, and cyclin E have been associated with aberrant involution. As defects were reported as early as 5 days post-weaning in these studies [23], [24], [25]], we examined the mammary glands of Fbxw7+/+ and Fbxw7−/− mice at this time point. Weaning occurred at 3 weeks post-partum for Fbxw7+/+ mice and was defined as 3 weeks post-partum for Fbxw7−/− mice due to their inability to nurse. Five days following this time point, acinar collapse was observable in the Fbxw7+/+ animals, indicating normal involution (Fig. 3C). In contrast, the 5 day post-involution glands from Fbxw7−/− mice were indistinguishable from pregnant glands, with no observable alveolar collapse (Fig. 3C). As involution requires apoptosis, we performed IHC for cleaved caspase 3, and found weaker staining in the Fbxw7−/− animals compared to the Fbxw7+/+ controls (Fig. 3D). Collectively, these data indicate that Fbxw7 loss results in both lactation and involution defects, similar to what has been reported with dysregulation of the Fbw7 substrates Notch1-ICD, cyclin E, and c-Myc.
Substrate proteins and their downstream transcriptional targets are dysregulated following loss of Fbxw7
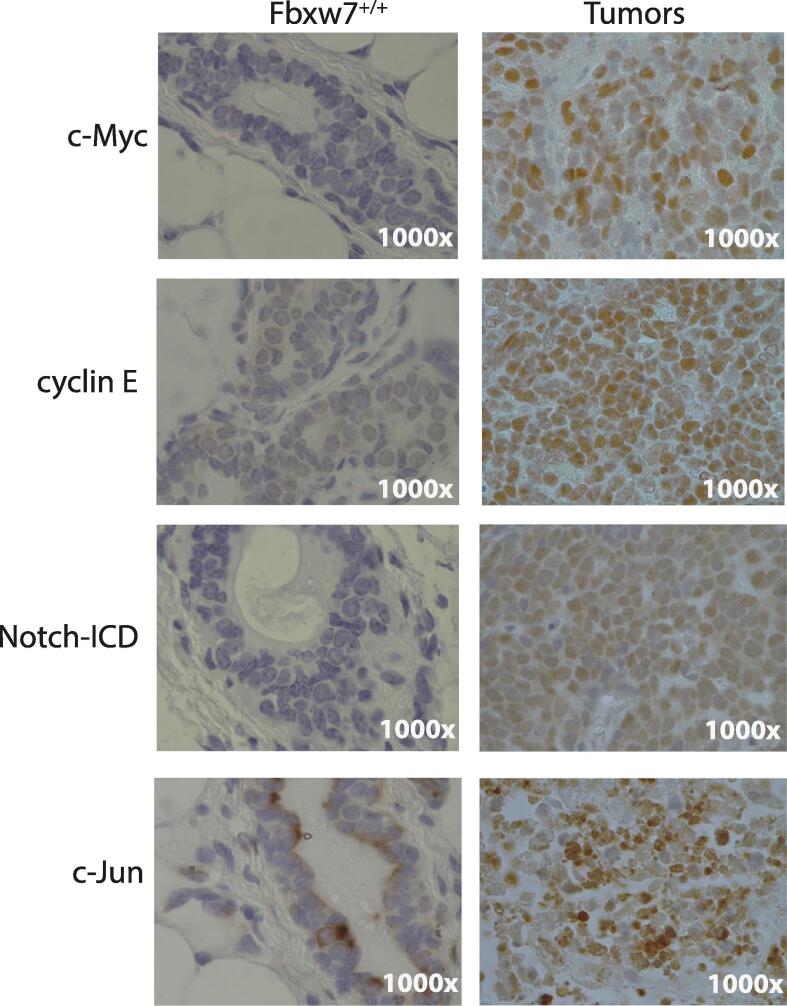
Fbw7 loss can result in differential accumulation of substrate proteins depending on tissue type. We hypothesized that transformation following loss of Fbxw7 may be due to the accumulation of substrates with critical roles in mammary gland morphogenesis and tumor biology such as c-Myc, cyclin E, Notch1-ICD, and c-Jun. First, we performed IHC on tumor and control mammary sections for c-Myc, cyclin E, c-Jun and Notch1-ICD. Staining for all four substrates was strongly observed in tumor sections, with minimal staining in corresponding control mammary gland sections (Fig. 4). These data suggest that multiple substrate proteins accumulate in the mammary tumors that develop upon Fbxw7 loss, consistent with decreased Fbw7-mediated degradation. The overexpression of cyclin E, c-Myc, and Notch1-ICD is also consistent with the observed effects of Fbxw7 loss on lactation and involution [23], [24], [25]] and suggests that loss of Fbxw7 broadly disrupts normal mammary development and function and promotes tumorigenesis through the sustained dysregulation of key substrates.
Fig. 4.
Oncogenic substrates of Fbw7 are overexpressed in Fbxw7/− mammary glands. IHC was performed on mammary tissue sections from control Fbxw7+/+ animals vs. Fbxw7−/− tumors. Representative results are shown. Increased staining for c-Myc, cyclin E, Notch1-ICD, and c-Jun was observed in tumor tissues vs. control. All images 1000×.
c-Myc, Notch1-ICD, and c-Jun are transcription factors that control the expression of many genes involved in tumor development. Additionally, active cyclin E-Cdk2 and c-Myc can induce the activation or expression of the E2F transcription factor family, particularly E2F1 and E2F3A, which are activated in late G1 and S phases and overexpressed in breast cancer [26], [27], [28]]. As we observed a strong accumulation of c-Myc, Notch1-ICD, c-Jun, and cyclin E in our tumor tissues, we hypothesized that dysregulation of these substrates and associated transcription factor pathways may be a critical driver of tumor development. Further, we hypothesized that such dysregulation would occur early after Fbxw7 loss. To address this hypothesis, we performed single cell qPCR analysis on non-cancerous, purified mammary epithelial cells isolated from two control Fbxw7fl/fl;LGB Cre- and two Fbxw7fl/fl;LGB Cre+ mice during late pregnancy. Fbxw7 gene excision was verified in the isolated Cre+ cells (Fig. 5A). For qPCR, we assayed published downstream targets for c-Myc (12 genes), E2F (11 genes), Notch1-ICD (7 genes), and c-Jun (8 genes; References provided in Supplementary Table 1) using the Fluidigm C1 and Biomark HD platforms. The values obtained for each cell and gene shown in Fig. 5 are provided in Supplementary Tables 2–5. With the exception of c-Myc, mRNA levels of the transcription factors themselves were increased early after Fbxw7 loss (Fig. 5B-E). Surprisingly, of the c-Jun targets we assayed, only Jun itself was significantly affected by Fbxw7 loss (Fig. 5B).
Fig. 5.
Early loss of Fbxw7 results in dysregulated c-Myc, E2F, and Notch-ICD target expression. A. Mammary epithelial cells were isolated from two control (Cre-) and two Fbxw7−/− (Cre+) mice during the third week of pregnancy. PCR analysis for the excision of exons 5–6 of Fbxw7 was performed as in Fig. 1A. L = molecular weight ladder. B-E. Single cells were isolated from the populations shown in A using the Fluidigm C1 system and qPCR assays were performed using a Biomark HD (Fluidigm). Target amplification was performed with Delta Gene assays (primer sets) from Fluidigm for (B) c-Jun, (C) c-Myc, (D) E2F, and (E) Notch1-ICD target genes of interest. Lanes marked with + indicate Fbxw7+/+ control cells and lanes marked with - indicate Fbxw7−/− cells. Statistical analysis was performed using the Fluidigm Real-Time PCR Analysis software. Data are expressed as Log2Expression normalized to RNA spike-in controls. Significance was measured using the Mann-Whitney test with a p value cutoff of p < 0.05. *p < 0.05, **p < 0.01, ***p < 0.001. Arrows indicate genes exhibiting a distinct bimodal distribution. See Supplementary Tables 2–5 for data included in these analyses.
We observed statistically significant changes in the expression of 5/12c-Myc targets in Fbxw7 null cells (Fig. 5C). These genes were upregulated with the exception of Cdkn1b (encoding the cyclin/Cdk inhibitor p27), which is known to be repressed by c-Myc [26]. These data are consistent with c-Myc transcription factor activity following Fbxw7 loss. The majority of the E2F targets we examined (8/11) were also affected by Fbxw7 loss (Fig. 5D). These 8 genes are known to be upregulated by E2F (see Supplementary Table 1), and most were observed to be upregulated in our Fbxw7 knockout cells. With regard to Notch1-ICD, we observed dysregulation of 4/7 targets in the Fbxw7 null cells (Fig. 5E). Along with Notch1 itself, we found that Hes1 and Snai1 were, as expected, upregulated following Fbxw7 loss. However, Ccnd1 is reported to be upregulated by Notch1-ICD, and we instead saw a downregulation of this gene in the Fbxw7 null cells. This is particularly surprising as Ccnd1 is also a target of c-Myc and E2F [26], [27]]. Thus, not all target genes in a particular pathway are activated upon Fbxw7 loss. This may be due to differences in the dosage level of each substrate following Fbxw7 deletion, or alternative pathways may prevent changes in downstream target genes.
We did observe some cell-to-cell heterogeneity in substrate accumulation in our tumor staining (Fig. 4). Consistent with this, some genes displayed a prominent bimodal distribution in mRNA levels (Vim, Cdkn2d, Brca1, Hk2, Cdk6, Cdc25A, Ccnd1, Snai2; Marked by arrows in Fig. 5B-E), suggesting that cell-to-cell variation occurs in nontumor cells as well. However, Fbxw7 loss resulted in mostly uniform, whole-population shifts in gene expression. Minimal differences from Fig. 5 were observed if individual cells were stratified into “high” and “low” expressing cells based on median gene expression, confirming that the observed changes were relatively uniform (data not shown).
Collectively, these data suggest that gene dysregulation in response to Fbxw7 loss occurs prior to tumor development, and that genes in the c-Myc and E2F pathways are predominantly affected. Several of the Notch1-ICD targets we assayed were also influenced by Fbxw7 loss, while deregulation of the c-Jun pathway as a whole was not strongly impacted. These data indicate that altered regulation of the E2F and c-Myc pathways may be critical first steps to mammary tumor development following Fbxw7 loss.
E2F and c-Myc pathway dysregulation dominate in Fbxw7−/− mammary tumors
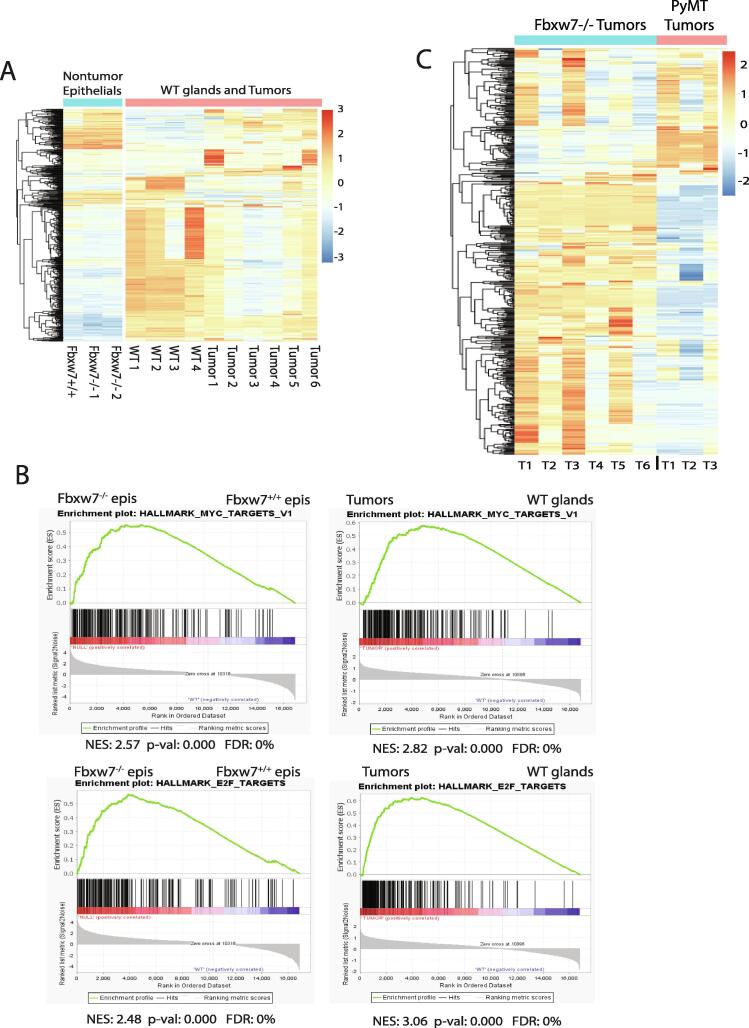
We next hypothesized that dysregulation of Fbw7 substrates would induce broad transcriptome changes consistent with increased proliferation. To address this, we performed unbiased bulk RNA sequencing on non-cancerous, purified epithelial cell populations, WT mammary glands, and tumor samples. Compared to the changes in gene expression observed between WT gland and tumor samples, fewer changes in the gene expression profiles were observed between the non-cancerous Fbxw7+/+ and Fbxw7−/− epithelial populations (Fig. 6A, nontumor epithelials), although 559 genes were significantly differentially expressed between the two groups (at least 2-fold change, p-val adj <0.05). Differential expression (DE) analysis revealed the upregulation of a multitude of cell cycle related genes, many of which are well-described E2F or c-Myc targets, including Cdk6, Cdk1, Cdca3, Cdc20, Ccna2, Ccnb1, Cdk2, and Cdc6. These genes were all upregulated approximately 2.5–4 fold upon Fbxw7 deletion. We also observed a 3–4 fold suppression of the cell cycle inhibitor genes Cdkn2c and Cdkn1c in the Fbxw7 null cells. This reinforces our single cell qPCR data indicating that, even prior to cancer development, loss of Fbxw7 promotes high level expression of cell cycle genes, driving proliferation in epithelial cells.
Fig. 6.
Fbxw7 loss results in E2F and c-Myc pathway dysregulation. A. Heatmap showing significantly differentially expressed genes (2-fold change, p-val adj <0.05) in non-tumor Fbxw7+/+ and Fbxw7−/− epithelial cells (left, blue bar) and WT mammary tissue vs. Fbxw7−/− tumors (right, pink bar). B. Left panels: GSEA analysis was performed on bulk RNA sequencing data from non-tumor epithelial (epis) cells. Genesets that were enriched in the Fbxw7−/− vs. Fbxw7+/+ cells included c-Myc (top) and E2F (bottom) targets. Only those genesets that met the cutoff of p < 0.01 and an FDR of < 25% were considered. Right panels: GSEA was performed on bulk RNA sequencing data from WT mammary tissue vs. tumors. Similar genesets were enriched in the tumor samples as in the Fbxw7−/− nontumor epithelial cells, however the normalized enrichment scores (NES) increased for c-Myc (top) and E2F (bottom) targets. C. Heatmap showing significantly differentially expressed genes (2-fold change, p-val adj <0.05) in Fbxw7−/− tumors (left, blue bar) vs. MMTV-PyMT 8 week tumors from Cai et al. (2017) (right, pink bar; 19). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
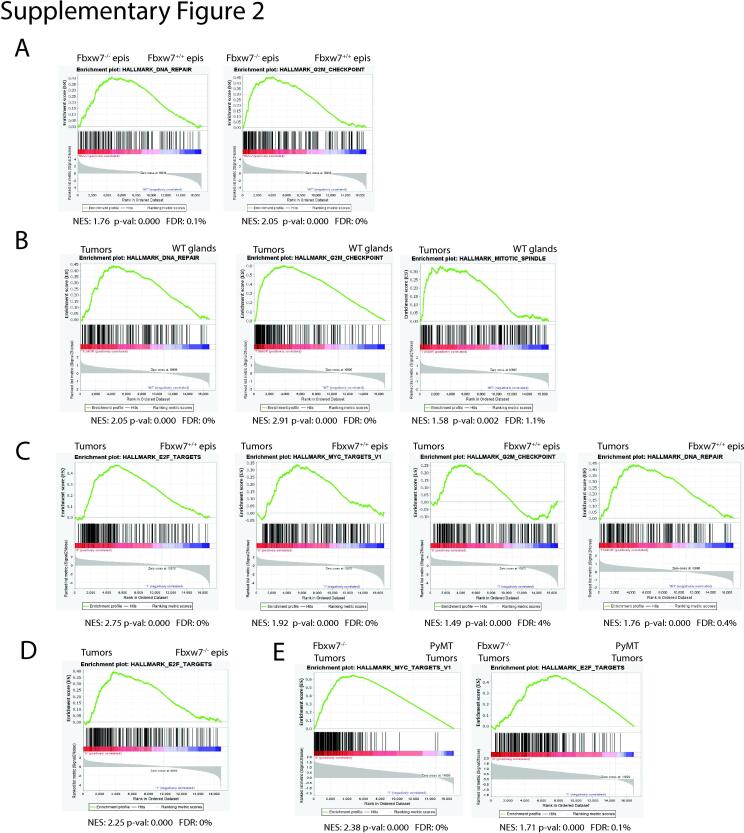
To identify additional dysregulated pathways in an unbiased fashion, we performed gene set enrichment analysis (GSEA) with the MSigDB Hallmarks genesets using a cutoff of p < 0.01 and a false discovery rate (FDR) of <25% and found an enrichment of genesets involved in proliferation. Consistent with our DE analysis, E2F and c-Myc targets were strongly enriched in Fbxw7−/− vs. Fbxw7+/+ non-cancerous epithelial cell populations, as were G2/M checkpoint and DNA repair genes (Fig. 6B left panels and Supplementary Fig. 2A). This again is consistent with deregulation of the cell cycle being an early, critical first step prior to tumor development following Fbxw7 loss. We did not observe a significant enrichment in c-Jun target genesets, consistent with our single cell data. While we were able to observe changes in a subset of Notch gene targets (Heyl, Hes1, Notch3, Fzd1, Aph1a, and Rbx1), the GSEA Notch geneset was not large enough for significance to be reached. Both our single cell and RNAseq studies are consistent with early dysregulation of the E2F and c-Myc pathways driving proliferation prior to breast tumor development.
As expected, much more variability was observed when comparing the transcriptional profiles of the WT mammary gland and tumor samples, indicating that gene expression becomes further dysregulated upon tumor formation (Fig. 6A, WT glands and tumors). We found that 3304 genes were significantly differentially expressed in tumor vs. WT mammary gland samples, implying that transformation results in additional changes that more broadly disrupt the transcriptome. However, DE seq analysis revealed the continued deregulation of several cell cycle genes seen after early Fbxw7 loss, including Cdk1, Cdca3, Cdc20, Ccna2, Ccnb1, Cdc6, and Cdkn2c as well as additional cell cycle genes such as Ccnb2, Cdkn2a, and Ccnd1. We next performed unbiased GSEA on WT glands vs. Fbxw7−/− tumors to identify pathways associated with transformation. Similar to the late-pregnancy Fbxw7−/− epithelial cells, we found an enrichment of E2F and c-Myc targets, DNA repair, and G2/M checkpoint genes in our tumor samples, with an increase in normalized enrichment score (NES) vs. that observed in the nontumor epithelial cell comparison (Fig. 6B right panels and Supplementary Fig. 2B). The increase in the NES scores implies that the early gene expression changes observed in epithelial cells following loss of Fbxw7 are reinforced as tumors develop. We also saw an enrichment of mitotic spindle genes, many of which are E2F target genes (Supplementary Fig. 2B). The mitotic spindle geneset was not significantly enriched in our non-tumor Fbxw7−/− epithelials, suggesting that long-term Fbxw7 loss impacts cell division and chromosome stability through E2F upregulation.
Because the mammary gland contains a mixture of stromal and epithelial cells, while the tumor body is predominated by epithelials, we also performed unbiased GSEA analysis comparing WT purified epithelials vs. tumor samples. We again observed a significant enrichment of the E2F, c-Myc, DNA repair, and G2M pathways in the tumor samples, with an increase in NES for E2F pathway genes (Supplementary Fig. 2C) compared to that observed for Fbxw7−/− vs. Fbxw7+/+ epithelials. We did not observe an increase in the NES over that for Fbxw7−/− vs. Fbxw7+/+ epithelials in the c-Myc, DNA repair, or G2M pathways with this comparison, however these pathways remained significantly enriched. We also performed GSEA comparing Fbxw7−/− epithelial cells and tumor samples and again observed an enrichment of the E2F pathway in the tumors (Supplementary Fig. 2D), confirming the increase in E2F pathway dysregulation from early Fbxw7 loss to tumor formation. Based upon our data, a clear progression in gene expression changes can be seen.
We next hypothesized that the Fbxw7−/−-derived tumors would show a stronger enrichment in E2F and c-Myc pathways than other well-established mammary tumor models. To address this, we turned to the MMTV-PyMT breast tumor model. This model undergoes progressive breast cancer development, starting with hyperplasia at 6 weeks of age, neoplasia at 8, and carcinoma at 12 [19]. Similar to our study, Cai et al. observed that genes in the c-Myc, E2F, mitotic spindle, and G2M checkpoint pathways were upregulated early (8 weeks) and remained upregulated at the carcinoma stage [19]. We chose to compare gene expression in our tumors with that of the 8 week (neoplasia) samples, as our tumors were likely at a similar developmental stage based on differentiation/morphology. While some similarities were observed, the two sample types exhibited overall different gene expression profiles (Fig. 6C). Unbiased GSEA analysis revealed a strong enrichment of c-Myc, E2F, and DNA repair targets in Fbxw7−/− tumors compared to the 8 week PyMT tumors (Supplemental Fig. 2E and data not shown). Thus, while enrichment of E2F and c-Myc targets were observed in the PyMT model, Fbxw7 loss results in a stronger deregulation of these targets. In contrast, GSEA revealed an enrichment of angiogenesis and K-Ras-mediated signaling in the PyMT model in comparison to our model (data not shown). Our data suggest that while some similarities exist between the two models, the Fbxw7−/− model is mechanistically distinct from the MMTV-PyMT model. Upon Fbxw7 loss, changes in cell cycle gene expression, prominently driven by E2F and c-Myc, occur early within the mammary epithelial cells. As breast tumors develop, E2F dysregulation continues, and signatures related to DNA repair, G2M checkpoint genes, and the mitotic spindle become apparent. Thus, while increased proliferation is a critical, early step in the transformation process, continued deregulation of DNA repair, G2/M checkpoint, and mitotic spindle genes likely facilitate tumorigenesis.
Fbxw7 loss triggers Trp53 mutation
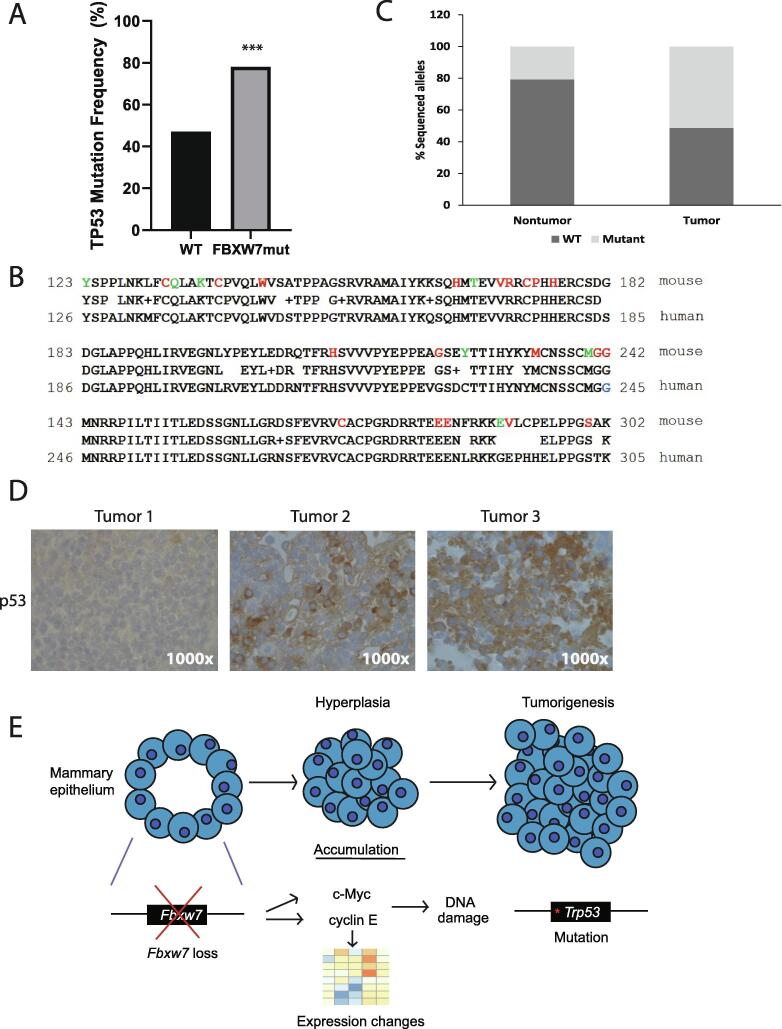
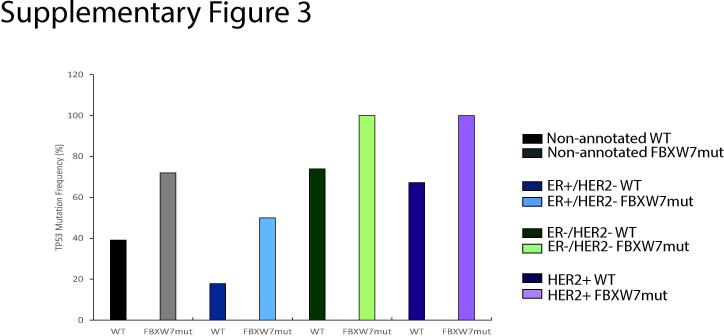
Given the changes we observed over time in gene expression and the wide range in latency from early Fbxw7 loss to tumor development, we hypothesized that perhaps a critical “second-hit” occurred which ultimately promoted tumor formation. One candidate for this second hit is TP53. Mutations in TP53 occur in approximately 23% of all breast cancers, with about 70% of these being missense mutations [29]. Often, TP53 mutation drives tumorigenesis by inactivating p53’s downstream tumor suppressive functions [29]. Such mutations have been shown to accelerate transformation in a range of tissues in combination with FBXW7 loss/mutation or cyclin E dysregulation [30], [31], [32], [33]]. In breast cancer specifically, TP53 mutation combined with overexpression of cyclin E accelerates tumorigenesis, implying genetic cooperation during transformation [6], [34]]. As such, we hypothesized there would be an enrichment for genetic alterations in both FBXW7 and TP53 in breast cancer patients. To test this hypothesis, we queried available breast cancer datasets using the cBioPortal (8874 patients from 15 studies) to determine the co-occurrence of TP53 and FBXW7 mutations. We found a statistically significant enrichment of TP53 mutations in patients harboring an FBXW7 mutation (78%) vs. those without (47%; Fig. 7A). While hormone receptor and HER2 expression status were not annotated for the majority of these samples, we were able to examine the relationship between FBXW7 and TP53 mutations in 1257 ER+/HER2- samples, 307 ER-/HER2- samples, and 198 HER2+ samples (Supplementary Fig. 3). Regardless of the category examined, the association between FBXW7 and TP53 mutations was observed, suggesting that the subtype of breast cancer does not influence the relationship between FBXW7 and TP53 mutations (Supplementary Fig. 3). No difference in overall survival was observed between patients harboring a FBXW7 single mutation and those with both FBXW7 and TP53 mutations in any category examined (data not shown). Although we were only able to use FBXW7 mutational status and not overall expression level in these analyses, these data suggest that TP53 mutations synergize with FBXW7 mutations in human breast tumorigenesis.
Fig. 7.
Fbxw7 loss results in Trp53 mutation. A. cBioPortal was used to query datasets encompassing 8874 breast cancer patients across 15 studies for FBXW7 and TP53 mutational status. Patients that harbored a FBXW7 mutation had an increased frequency of TP53 mutations compared to FBXW7 WT patients. ***p < 0.001, one-sided Fisher’s exact test. B. Alignment of the mouse and human TP53 gene sequences, with all identified mutations in the nontumor samples highlighted in green and those in the tumor samples in red. The blue reside (G245) in the human sequence indicates a “hotspot” mutation in human breast cancer. C. Graph depicting percentage of sequenced alleles that were mutated vs. nontumor (left) and tumor (right) samples. D. IHC for p53 in three separate tumor samples, with tumors 2 and 3 showing elevated expression and partial nuclear exclusion of p53. All photos 1000×. E. Model for breast tumorigenesis triggered by Fbxw7 loss. Fbxw7 depletion results in upregulation of Notch-ICD, c-Myc, and cyclin E, which impact cell cycle, DNA damage response, and G2/M checkpoint genes. Enforced proliferation in the presence of DNA damage provides a selective pressure for the acquisition of Trp53 mutations, facilitating transformation.
Given this finding of an enrichment of TP53 mutations, we hypothesized that Trp53 mutations would be found in our tumor samples. To test this hypothesis, we sequenced the most commonly mutated exons of Trp53 (exons 5–8 which include the DNA binding domain; 29) observed in breast cancer patients in our murine tumor samples. Using WT mammary tissue and matched tail samples as controls, we identified somatic missense mutations in all (6/6) tumor samples analyzed (Fig. 7B, highlighted in red). We identified 19 different missense mutations, with 10 occurring in exons 5–6 and 9 in exons 7–8. In contrast, we found 6 WT Trp53 alleles in exons 5–6, and 12 in exons 7–8, indicating that not every allele is mutated within the tumor tissue. The total percentage of mutated alleles sequenced was 51.3% vs. 48.7% WT alleles (Fig. 7C). We found 2 silent mutations in exons 5–6 and 9 in exons 7–8, however silent mutations were also uncovered in WT mammary tissues or matched Fbxw7fl/fl;LGB Cre+ tail tissues, suggesting they occur independently of Fbxw7 excision. Each tumor exhibited a different mutational spectrum, with no codon being mutated in more than one tumor sample. All mutations, with the exception of V291L, occurred at amino acids conserved between human and mouse p53 (Fig. 7B) and were observed in codons mutated in breast cancer patients ([35], [36], [37] and cBioPortal query). One mutation, G242S (G245S human), is defined as a hotspot conformational mutation in breast cancer and is found with an overall frequency of approximately 2.8% [37]. Interestingly, the exact same nucleotide change (GGC to AGC) observed here has been reported in patients, suggesting this represents a potential “hot-spot” for p53 inactivation in both species [36], [37]]. E284K has also been detected in several patient tumors [36]. While we were unable to determine if Trp53 mutations are heterozygous or homozygous, we were able to identify multiple different mutations in the majority of our tumor samples, pointing to some intratumor heterogeneity. Only a single missense mutation was found per sequenced allele, with the exception of one allele in tumor 5 that harbored 3 different mutations (G242S, E284K, V291I). Collectively, these data indicate that loss of Fbxw7 in tumors is associated with the acquisition of Trp53 missense mutations.
To determine if loss of Trp53 occurred prior to tumor development, we analyzed nontumor epithelial cells from pregnant Fbxw7−/− mice and identified mutations in a subset (3/5) of the samples tested. Importantly, the number of mutated alleles identified in normal tissue was lower than that uncovered in the tumor samples, with 3 mutated and 14 WT Trp53 alleles in exons 5–6. Similarly, we found 3 mutations and 9 WT alleles in exons 7–8, with one allele containing two separate mutations. In contrast to tumor tissues, the total percentage of mutated alleles in the nontumor samples was much lower (20.7% mutated vs. 79.3% WT, Fig. 7C). All mutations uncovered in nontumor samples were different from those in the tumor samples and are highlighted in green in Fig. 7B. These data suggest that Trp53 mutations can be acquired early following Fbxw7 loss, with additional mutations accumulating during tumorigenesis. Collectively, this implies that loss of Tp53 occurs early in the transformation process following loss of Fbxw7, but the mutational burden accelerates as tumors form.
Given that some TP53 mutations can increase p53 protein stability [38], we examined p53 protein levels by IHC, but detected overexpression in only two tumors, with most showing low-moderate expression (Fig. 7D). Although neither of the mutations in these two tumors have been previously linked to p53 mislocalization, p53 was at least partially localized to the cytoplasm in these samples, suggesting they may interfere with proper p53 function (Fig. 7D). We were unable to detect increased expression of p53 in tissues from pregnant, non-tumor bearing mice (data not shown). The detection of Trp53 mutations early after Fbxw7 loss and the accumulation of additional mutations upon tumorigenesis strongly suggests that Fbxw7 loss and Trp53 missense mutations genetically cooperate to promote breast tumorigenesis. The enrichment of TP53 mutations in FBXW7 mutant patient-derived tumors suggests the same synergy between these two pathways is critical in humans as well.
Discussion
Fbw7 has a well-known role in regulating the expression levels of a multitude of transcription factors that control cell proliferation and survival pathways and can exhibit tissue-specific effects on its substrates and their respective downstream activities. For instance, in embryonic stem cells, Fbxw7 knockdown affects only c-Myc abundance while other substrates are unaffected [39]. Additionally, ablation of Fbw7 in the brain primarily induces Notch and c-Jun accumulation, while other substrates are unchanged [40]. The effect of Fbxw7 loss on tumorigenesis has been studied in several cancer types [10], [13], [30], [32], [33]], however Fbxw7 loss is not always sufficient for tumorigenesis [33], [41]] and the contribution of Fbxw7 loss to breast tumorigenesis has not been studied. Here, we determine that loss of Fbxw7 serves as an initiating event in the induction of breast tumor development in a murine model system. A proposed mechanism is depicted in Fig. 7E. Our data suggest that the E2F and c-Myc pathways are affected early after Fbxw7 loss and remain dysregulated following tumor formation. We suggest that persistent cell cycling in the presence of DNA damage results in tumorigenesis. Additionally, the loss of Fbxw7 triggers the acquisition of Trp53 mutations in mouse mammary tumors, suggesting that Fbxw7/Trp53 alterations cooperate during transformation. Importantly, an enrichment of TP53 mutations in FBXW7-altered breast cancer samples indicates this genetic cooperativity also occurs in humans.
How might dysregulated levels of c-Myc and cyclin E drive tumorigenesis? The role of c-Myc in oncogenesis is well-known, as it acts as both a driver of both proliferation and apoptosis [26], [42]]. Normally c-Myc is expressed at a low, constitutive level in dividing cells, and removal of growth factor signaling results in its degradation [26], [42]]. In our study, Fbxw7 loss resulted in sustained c-Myc overexpression, which is known to promote proliferation through the up-regulation of cell cycle genes [26]. Consistent with this, we observed the upreguation of c-Myc-regulated cell cycle genes such as Ccna2, Cdk6, and Cdk4 in our single cell qPCR and/or bulk RNAseq following Fbxw7 loss (Fig. 5C and 6B). However, high levels of c-Myc can also trigger apoptosis. We were able to observe the upregulation of several pro-apoptotic factors in our bulk RNAseq data following Fbxw7 loss, including Bcl2l11, Bcl2l14, Bcl10, Bcl2l1, and Bnipl. We also observed suppression of the anti-apoptotic factor Bcl2, which is known to be repressed by c-Myc and cyclin E [43]. Consistent with both the pro-proliferative and anti-apoptotic effects of c-Myc, we observed increased proliferation and apoptosis in our tumors as evidenced by Ki67 and cleaved caspase-3 staining, respectively. Interestingly, c-Myc can induce apoptosis by inducing p53 expression and activation [44]. Thus, a loss of p53 function would shift the balance between proliferation and apoptosis. We hypothesize that the acquisition of TP53 mutations contributes to breast tumorigenesis in FBXW7-altered patients by preventing high levels of c-Myc from promoting apoptosis. Consistent with this hypothesis, B-cell lymphomas induced by c-Myc overexpression lost WT TP53 during tumor development [44]. Additional experiments to delineate the interplay between Trp53 and Fbxw7 mutation-induced c-Myc overexpression in breast cancer will be needed to directly test this hypothesis.
The relationship between cyclin E and Fbw7 in cancer has also been well-studied. Normally, cyclin E levels are strictly regulated during the cell cycle, with Fbw7 mediating cyclin E degradation during S phase [25]. During G1 phase, cyclin E-Cdk2 phosphorylates Rb protein, relieving its inhibition of the E2F1, E2F2, E2F3, and E2F4 transcription factors to promote the expression of genes regulating S phase initiation and progression [27], [45]]. We observed a prominent upregulation of cell cycle genes early after Fbxw7 loss, which remained dysregulated in resulting breast tumors. As many of these genes are known to be direct targets of E2F, this strongly suggests that loss of Fbxw7 triggers hyperplasia through E2F deregulation, resulting in tumorigenesis. Additionally, similar to c-Myc, E2F can promote the expression of pro-apoptotic genes [27], consistent with the increase in cleaved caspase 3 staining observed in our tumors. Cai et al. also observed early dysregulation of the E2F pathway in the MMTV-PyMT tumor model [19]. While E2F targets were more strongly dysregulated in our Fbxw7−/− model, the dysregulation of E2F in multiple models suggests that E2F pathway disruption is a key contributor to breast tumor development [19]. Importantly, we cannot exclude that family members other than the Rb-regulated E2Fs are also playing a role in the observed hyperplasia following loss of Fbxw7. Our RNAseq shows enriched expression of DNA repair, mitotic spindle, and G2/M checkpoint genes in the Fbxw7−/− tumors, in line with deregulation of E2F transcriptional activity. It should be emphasized that significant cross-talk exists between the c-Myc and E2F pathways [46], [47]]. Thus, these pathways likely continually reinforce one another in the absence of Fbxw7. Additional experiments will be required to precisely define the unique and overlapping molecular targets resulting from c-Myc and E2F dysregulation.
In contrast to a clear upregulation of E2F and c-Myc target genes early after Fbxw7 loss and upon tumor formation, we found little effect of Fbxw7 loss on c-Jun/AP-1 target genes in any of our analyses, despite a high expression of c-Jun itself. c-Jun is more predominantly expressed in human breast cancers vs. normal tissue, however its expression has been observed mainly at the leading edge of tumors and has been associated with increased invasiveness and angiogenesis [48], [49]]. Consistent with this finding, the tumors that formed from Fbxw7 loss in our model system were noninvasive and moderately to well-differentiated, suggesting that alterations in c-Jun may be more relevant to the transition from a more- to less-differentiated and/or aggressive tumor type. In addition, we did not uncover strong effects on Notch1-ICD signaling in our tumor samples, although early dysregulation of Notch1-ICD likely contributes to the lactation and involution defects in our Fbxw7−/− mice, as both phenotypes have been observed with sustained Notch1-ICD overexpression [23]. Hu et al. also observed that a small number of their MMTV/Notch1-ICD transgenic animals developed hyperplasia [23]. Thus, early Notch1-ICD dysregulation following Fbxw7 loss may also contribute to over-proliferation.
It is notable that Fbxw7 loss promoted Trp53 mutation in our breast cancer study. Several studies have reported a relationship between FBXW7 loss and TP53 mutation in other cancers. For instance, co-deletion or mutation of TP53 in the absence of one or more alleles of FBXW7 has been shown to drive T-ALL, intestinal adenomas, and epithelial tumors in response to ionizing radiation [9]. Fbxw7/Pten−/− uterine carcinosarcomas also accumulate Trp53 mutations [33]. The discovery of Trp53 mutations soon after Fbxw7 deletion in our model suggests that the two cooperate in breast cancer as well. Importantly, our studies mechanistically link Fbxw7 loss with the development of Trp53 mutations in breast tumorigenesis. As p53 plays a critical role in promoting apoptosis in response to DNA damage and cell stress, loss of p53 function through mutation would facilitate tumorigenesis driven by E2F and c-Myc-enforced cell cycling. Consistent with this notion, p53 activity has been shown to restrain the effects of elevated cyclin E on genomic instability in the absence of Fbxw7 [50]. p53 loss also allows for E2F1-induced S phase entry through suppression of the G1 checkpoint [51]. It is therefore likely that decreased Fbxw7 expression results in a selective pressure for the acquisition of Trp53 loss of function mutations through its effects on cyclin E and E2F.
The Fbxw7−/− tumors that developed most closely resemble the human breast cancer luminal B subtype, as they are ERα+, PR-, and exhibit high Ki67 staining. While ERα+ human breast cancers are not as frequently mutated for TP53 compared to basal-like or HER2-enriched tumors, TP53 mutations do occur in approximately 25% of luminal B cancers [52]. Consistent with our results, luminal B cancers have a higher proportion of missense mutations in the DNA binding domain of TP53 compared to other subtypes [52]. Further, Silwal-Pandit showed that TP53 mutations are predictive of reduced survival only in ERα+ disease [52]. Collectively, this suggests that the accumulation of TP53 missense mutations that are triggered by FBXW7 loss are likely to contribute to disease progression.
Conclusions
In conclusion, our results have clear implications for understanding breast cancer initiation, in which loss of FBXW7 expression is relatively common. Fbxw7 loss in the mouse mammary gland initiates tumor development, with the tumors subsequently accumulating mutations within Trp53. Significant changes to gene expression were detectable soon after Fbxw7 loss, even before tumor formation occurred. Our data suggest that targeting the c-Myc and cyclin E-E2F pathways, for example through synthetic lethal approaches or Cdk2 inhibition, may be particularly helpful in the treatment of patients with tumors characterized by diminished FBXW7 expression. Given the well-known roles of p53 in tumor suppression and the clear connection between FBXW7, TP53 status, and luminal B-type breast cancers, our study further supports the ongoing search for new therapeutic strategies to target TP53-mutant cancers [53].
Data availability
All RNAseq data are available in GEO Omnibus #GSE144690, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144690.
Funding sources
This work was supported in part by funding from Midwest Athletes against Childhood Cancer and the NIH (CA204231) to SR and an American Cancer Society Research Scholar Grant (22511-RSG-12-132-01-CCG) to ACM. Additional research support to S. Rao provided by a charitable gift from Ms. Nanette Gardetto.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to Scott Wood for technical support, Benedetta Bonacci for assistance with flow cytometry, Barbara Fleming for histology, and Bruce E. Clurman for the gift of the anti-Fbw7 antibody.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.07.001.
Contributor Information
Alison E. Meyer, Email: AMeyer@Versiti.org.
Quinlan Furumo, Email: QFurumo@Versiti.org.
Cary Stelloh, Email: CStelloh@Versiti.org.
Alex C. Minella, Email: aminella@me.com.
Sridhar Rao, Email: sridhar.rao@versiti.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Fbxw7-/- tumors are ER a -positive and PR-negative. IHC for ER a and PR was performed on mammary tissue sections from control Fbxw7+/+ animals vs. Fbxw7-/- tumors. All images 400x.
Supplementary figure 2.
Fbxw7 loss results in the dysregulation of E2F, c-Myc, DNA repair, G2/M checkpoint, and mitotic spindle genes. A. GSEA analysis was performed on bulk RNA sequencing data from non-tumor epithelial (epis) cells. Genesets that were enriched in the Fbxw7-/- cells vs. the Fbxw7+/+ cells included DNA repair genes (left) and G2M checkpoint genes (right). B. GSEA was performed on WT mammary tissue vs. Fbxw7-/- tumors. Similar to Fbxw7-/- nontumor epithelial cells, tumor samples were enriched for DNA repair genes (left) and G2M checkpoint genes (middle), however, an increase in the normalized enrichment score (NES) was observed. Enrichment of an additional geneset, mitotic spindle genes (right) was also observed in the tumor samples. C. GSEA was performed on Fbxw7+/+ epithelials vs. tumors. Genesets that were enriched in the tumor samples included E2F (left) and c-Myc targets (second from left) as well as G2M checkpoint (third from left) and DNA repair pathways (right). D. GSEA was performed on Fbxw7-/- epithelials vs. tumors. The E2F pathway was enriched in the tumor samples. E. GSEA was performed on Fbxw7-/- tumors vs. MMTV-PyMT 8 week tumors (19). The E2F and c-Myc pathways were enriched in the Fbxw7-/- tumor samples. For all GSEA analyses, only those genesets that met the cutoff of p<0.01 and an FDR of <25% were considered.
Supplementary figure 3.
TP53 mutation is associated with FBXW7 genotype regardless of hormone receptor status. cBioPortal was used to query datasets encompassing 4,413 breast cancer patients across three studies for which FBXW7 and TP53 mutational status as well as ER and HER2 status were partially annotated (Supplementary References 22-24). Of the 4,413 total samples, 2,491 were not annotated regarding ER/HER2 status. 1,257 were annotated as ER+/HER2-, 307 as ER-/HER2-, and 198 as HER2+. Regardless of category, patients that harbored a FBXW7 mutation had an increased frequency of TP53 mutations compared to FBXW7 WT patients. The low number of samples harboring both FBXW7 and TP53 mutations per category precluded statistical analysis.
List of target gene names, corresponding RefSeq numbers, and Fluidigm Design Assay IDs (Delta Gene Assay/primer ID numbers ordered from Fluidigm) used for single cell qPCR analysis shown in Figure 4B–E. References supporting target gene selection are listed in the main text as Supplementary References.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each c-Jun target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each c-Myc target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each E2F target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each Notch-ICD target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
References
- 1.Shimizu K., Nihira N.T., Inuzuka H., Wei W. Physiological functions of FBW7 in cancer and metabolism. Cell Signal. 2018;46:15–22. doi: 10.1016/j.cellsig.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spruck C.H., Won K.A., Reed S.I. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 3.Ekholm-Reed S., Spruck C.H., Sangfelt O., van Drogen F., Mueller-Holzner E., Widschwendter M., Zetterberg A., Reed S.I., Reed S.E. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.can-03-3417. [DOI] [PubMed] [Google Scholar]
- 4.Loeb K.R., Kostner H., Firpo E., Norwood T.D., Tsuchiya K., Clurman B.E., Roberts J.M. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell. 2005;8:35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Bortner D.M., Rosenberg M.P. Induction of mammary gland hyperplasia and carcinomas in transgenic mice expressing human cyclin E. Mol Cell Biol. 1997;17:453–459. doi: 10.1128/mcb.17.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith A.P.L., Henze M., Lee J.A., Osborn K.G., Keck J.M., Tedesco D., Bortner D.M., Rosenberg M.P., Reed S.I. Deregulated cyclin E promotes p53 loss of heterozygosity and tumorigenesis in the mouse mammary gland. Oncogene. 2006;25:7245–7259. doi: 10.1038/sj.onc.1209713. [DOI] [PubMed] [Google Scholar]
- 7.Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 8.Bahram F., Von Der Lehr N., Cetinkaya C., Larsson L.G. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 9.Cao J., Ge M.-H., Ling Z.-Q. Fbxw7 tumor suppressor: a vital regulator contributes to human tumorigenesis. Medicine (Baltimore) 2016;95 doi: 10.1097/MD.0000000000002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babaei-Jadidi R., Li N., Saadeddin A., Spencer-Dene B., Jandke A., Muhammad B., Ibrahim E.E., Muraleedharan R., Abuzinadah M., Davis H., Lewis A., Watson S., Behrens A., Tomlinson I., Nateri A.S. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei W., Jin J., Schlisio S., Harper J.W., Kaelin W.G. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Gregory M.A., Hann S.R. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhoondi S., Sun D., Von Der Lehr N., Apostolidou S., Klotz K., Maljukova A., Cepeda D., Fiegl H., Dofou D., Marth C., Mueller-Holzner E., Corcoran M., Dagnell M., Nejad S.Z., Nayer B.N., Zali M.R., Hansson J., Egyhazi S., Petersson F., Sangfelt P., Nordgren H., Grander D., Reed S.I., Widschwendter M., Sangfelt O., Spruck C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 14.Akhoondi S., Lindström L., Widschwendter M., Corcoran M., Bergh J., Spruck C., Grandér D., Sangfelt O. Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010;12:R105. doi: 10.1186/bcr2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibusuki M., Yamamoto Y., Shinriki S., Ando Y., Iwase H. Reduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patients. Cancer Sci. 2011;102:439–445. doi: 10.1111/j.1349-7006.2010.01801.x. [DOI] [PubMed] [Google Scholar]
- 16.Thompson B.J., Jankovic V., Gao J., Buonamici S., Vest A., Lee J.M., Zavadil J., Nimer S.D., Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbert S., Bentley D.J., Melton D.W., Rannie D., Lourenço P., Watson C.J., Clarke A.R. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 1998;7:387–396. doi: 10.1023/a:1008848304391. [DOI] [PubMed] [Google Scholar]
- 18.Grim J.E., Gustafson M.P., Hirata R.K., Hagar A.C., Swanger J., Welcker M., Hwang H.C., Ericsson J., Russell D.W., Clurman B.E. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181:913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y., Nogales-Cadenas R., Zhang Q., Lin J.R., Zhang W., O’Brien K., Montagna C., Zhang Z.D. Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. BMC Genomics. 2017;18:1–14. doi: 10.1186/s12864-017-3563-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh S., Kumar S., Srivastava R.K., Nandi A., Thacker G., Murali H., Kim S., Baldeon M., Tobias J., Blanco M.A., Saffie R., Zaidi M.R., Sinha S., Busino L., Fuchs S.Y., Chakrabarti R. Loss of ELF5–FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-γ signalling. Nat Cell Biol. 2020;22:591–602. doi: 10.1038/s41556-020-0495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balamurugan K., Mendoza-Villanueva D., Sharan S., Summers G.H., Dobrolecki L.E., Lewis M.T., Sterneck E. C/EBPδ links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene. 2019;38:3765–3780. doi: 10.1038/s41388-018-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rustighi A., Zannini A., Tiberi L., Sommaggio R., Piazza S., Sorrentino G., Nuzzo S., Tuscano A., Eterno V., Benvenuti F., Santarpia L., Aifantis I., Rosato A., Bicciato S., Zambelli A., Del Sal G. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6:99–119. doi: 10.1002/emmm.201302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C., Diévart A., Lupien M., Calvo E., Tremblay G., Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blakely C.M., Sintasath L., D’Cruz C.M., Hahn K.T., Dugan K.D., Belka G.K., Chodosh L.A. Developmental stage determines the effects of MYC in the mammary epithelium. Development. 2005;132:1147–1160. doi: 10.1242/dev.01655. [DOI] [PubMed] [Google Scholar]
- 25.Minella A.C., Loeb K.R., Knecht A., Welcker M., Varnum-Finney B.J., Bernstein I.D., Roberts J.M., Clurman B.E. Cyclin E phosphorylation regulates cell proliferation in hematopoietic and epithelial lineages in vivo. Genes Dev. 2008;22:1677–1689. doi: 10.1101/gad.1650208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bretones G., Delgado M.D., León J. Myc and cell cycle control. Biochim Biophys Acta – Gene Regul Mech. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Bracken A.P., Ciro M., Cocito A., Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Lee M.-Y., Moreno C.S., Saavedra H.I. E2F activators signal and maintain centrosome amplification in breast cancer cells. Mol Cell Biol. 2014;34:2581–2599. doi: 10.1128/MCB.01688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walerych D., Napoli M., Collavin L., Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–2017. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao J.H., Perez-Losada J., Wu D., DelRosario R., Tsunematsu R., Nakayama K.I., Brown K., Bryson S., Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 31.Grim J.E., Knoblaugh S.E., Guthrie K.A., Hagar A., Swanger J., Hespelt J., Delrow J.J., Small T., Grady W.M., Nakayama K.I., Clurman B.E. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol. 2012;32:2160–2167. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu K.T., Xu Y., Swartz K.L., Bhattacharyya M., Gurbuxani S., Hua Y., Minella A.C. Chromosome instability underlies hematopoietic stem cell dysfunction and lymphoid neoplasia associated with impaired Fbw7-mediated cyclin E regulation. Mol Cell Biol. 2014;34:3244–3258. doi: 10.1128/MCB.01528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuevas I.C., Sahoo S.S., Kumar A., Zhang H., Westcott J., Aguilar M., Cortez J.D., Sullivan S.A., Xing C., Neil Hayes D., Brekken R.A., Bae-Jump V.L., Castrillon D.H. Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2019;116:25880–25890. doi: 10.1073/pnas.1911310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl T., Landberg G., Ahlgren J., Nordgren H., Norberg T., Klaar S., Holmberg L., Bergh J. Overexpression of cyclin E protein is associated with specific mutation types in the p53 gene and poor survival in human breast cancer. Carcinogenesis. 2004;25:375–380. doi: 10.1093/carcin/bgh019. [DOI] [PubMed] [Google Scholar]
- 35.Coles C., Condie A., Chetty U., Steel C.M., Evans H.J., Prosser J. p53 mutations in breast cancer. Cancer Res. 1992;52:5291–5298. [PubMed] [Google Scholar]
- 36.Lai H., Lin L., Nadji M., Lai S., Trapido E., Meng L. Mutations in the p53 tumor suppressor gene and early onset breast cancer. Cancer Biol Ther. 2002;1:31–36. doi: 10.4161/cbt.1.1.37. [DOI] [PubMed] [Google Scholar]
- 37.Freed-Pastor W.A., Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frum R.A., Grossman S.R. Mechanisms of mutant p53 stabilization in cancer. Subcell Biochem. 2014;85:187–197. doi: 10.1007/978-94-017-9211-0_10. [DOI] [PubMed] [Google Scholar]
- 39.Buckley S.M., Aranda-Orgilles B., Strikoudis A., Apostolou E., Loizou E., Moran-Crusio K., Farnsworth C.L., Koller A.A., Dasgupta R., Silva J.C., Stadtfeld M., Hochedlinger K., Chen E.I., Aifantis I. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeck J.D., Jandke A., Blake S.M., Nye E., Spencer-Dene B., Brandner S., Behrens A. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 41.Sancho R., Jandke A., Davis H., Diefenbacher M.E., Tomlinson I., Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 42.Hanson K.D., Shichiri M., Follansbee M.R., Sedivy J.M. Effects of c-myc expression on cell cycle progression. Mol Cell Biol. 1994;14:5748–5755. doi: 10.1128/mcb.14.9.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eischen C.M., Packham G., Nip J., Fee B.E., Hiebert S.W., Zambetti G.P., Cleveland J.L. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F–1. Oncogene. 2001;20:6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- 44.Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kent L.N., Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 46.Mudryj M., Hiebert S.W., Nevins J.R. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leone G., Sears R., Huang E., Rempel R., Nuckolls F., Park C.H., Giangrande P., Wu L., Saavedra H.I., Field S.J., Thompson M.A., Yang H., Fujiwara Y., Greenberg M.E., Orkin S., Smith C., Nevins J.R. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 48.Vleugel M.M., Greijer A.E., Bos R., van der Wall E., van Diest P.J. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum Pathol. 2006;37:668–674. doi: 10.1016/j.humpath.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 49.Jiao X., Katiyar S., Willmarth N.E., Liu M., Ma X., Flomenberg N., Lisanti M.P., Pestell R.G. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J Biol Chem. 2010;285:8218–8226. doi: 10.1074/jbc.M110.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minella A.C., Grim J.E., Welcker M., Clurman B.E. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene. 2007;26:6948–6953. doi: 10.1038/sj.onc.1210518. [DOI] [PubMed] [Google Scholar]
- 51.Lomazzi M., Moroni M.C., Jensen M.R., Frittoli E., Helin K. Suppression of the p53- or pRB-mediated G1 checkpoint is required for E2F-induced S-phase entry. Nat Genet. 2002;31:190–194. doi: 10.1038/ng891. [DOI] [PubMed] [Google Scholar]
- 52.Silwal-Pandit L., Vollan H.K.M., Chin S.F., Rueda O.M., McKinney S., Osako T., Quigley D.A., Kristensen V.N., Aparicio S., Børresen-Dale A.L., Caldas C., Langerød A. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20:3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 53.Blandino G., Di Agostino S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J Exp Clin Cancer Res. 2018;37:30. doi: 10.1186/s13046-018-0705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of target gene names, corresponding RefSeq numbers, and Fluidigm Design Assay IDs (Delta Gene Assay/primer ID numbers ordered from Fluidigm) used for single cell qPCR analysis shown in Figure 4B–E. References supporting target gene selection are listed in the main text as Supplementary References.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each c-Jun target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each c-Myc target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each E2F target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Fluidigm Biomark HD qPCR values obtained from single cell analysis for each Notch-ICD target gene. The symbol before each gene (+ or -) represents values obtained from Fbxw7+/+ (+) cells or Fbxw7-/- (-) cells. Each value in a column represents data obtained from one cell. See Materials and Methods for details on data normalization and analysis.
Data Availability Statement
All RNAseq data are available in GEO Omnibus #GSE144690, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144690.