Abstract
Background
We previously reported on the interplay between miR156/SPL13 and WD40–1/DFR to improve response to drought stress in alfalfa (Medicago sativa L.). Here we aimed to investigate whether the role of miR156/SPL13 module in drought response is tissue-specific, and to identify SPL13-interacting proteins. We analyzed the global transcript profiles of leaf, stem, and root tissues of one-month old RNAi-silenced SPL13 (SPL13RNAi) alfalfa plants exposed to drought stress and conducted protein-protein interaction analysis to identify SPL13 interacting partners.
Result
Transcript analysis combined with weighted gene co-expression network analysis showed tissue and genotype-specific gene expression patterns. Moreover, pathway analysis of stem-derived differentially expressed genes (DEG) revealed upregulation of genes associated with stress mitigating primary and specialized metabolites, whereas genes associated with photosynthesis light reactions were silenced in SPL13RNAi plants. Leaf-derived DEG were attributed to enhanced light reactions, largely photosystem I, II, and electron transport chains, while roots of SPL13RNAi plants upregulated transcripts associated with metal ion transport, carbohydrate, and primary metabolism. Using immunoprecipitation combined with mass spectrometry (IPMS) we showed that SPL13 interacts with proteins involved in photosynthesis, specialized metabolite biosynthesis, and stress tolerance.
Conclusions
We conclude that the miR156/SPL13 module mitigates drought stress in alfalfa by regulating molecular and physiological processes in a tissue-dependent manner.
Keywords: Alfalfa, Drought, IPMS, Medicago sativa, miR156, SPL13, Transcriptome
Background
The frequent and extreme weather events in present-day are correlated with climate change, which aggravates crop losses [38, 42]. To cope with these weather events, plants respond by developing different resilience strategies at the phenotypic, physiological, and molecular levels [23, 51]. Among the molecular strategies, the role of microRNAs in regulating various plant processes to enhance stress tolerance has been reported in a variety of crops [21]. Of the hundreds of plant microRNAs, microRNA156 (miR156) is highly conserved across species and regulates plant development as well as tolerance to biotic and abiotic stresses [2, 4, 5, 15, 17, 21, 23, 35].
Recent findings showed moderate levels of miR156 overexpression enhances drought tolerance in alfalfa by silencing SPL13 [2] and enhancing the downstream dihydroflavonol-4-reductase (DFR) which mediates flavonoid biosynthesis [15]. Low to moderate levels of miR156 and WD40–1 overexpression [15], and silencing of SPL13 [2] and WD40–2 [3] resulted in drought tolerance with enhanced anthocyanin levels, and improved photochemistry and physiological adjustments. A recent plant microRNA profiling study revealed that the previously known 21 highly conserved microRNA families [49] were present in three alfalfa genotypes, in addition to 79 present in both alfalfa and Medicago truncatula [39]. Furthermore, two microRNAs (miR160 and miR408) were expressed in a tissue-specific manner, whereas six (miR156, miR159, miR166, miR319, miR396, and miR398) were abundantly expressed in all tissues [39].
Despite the conserved presence of miR156 in different tissues, previous findings showed that increased miR156 expression affected alfalfa plant parts in different ways, including an increased number of lateral branches, improved root development, delayed flowering time and reduced plant height [4, 5, 17]. Hence, we hypothesize that the miR156/SPL13 module is regulated in a tissue-specific manner to orchestrate the necessary adjustments needed to cope with drought stress in alfalfa. We sought to investigate this hypothesis by using high throughput techniques of mRNA-based global transcriptomic analysis in leaf, stem, and root tissues of SPL13RNAi compared to empty vector alfalfa plants. Moreover, immunoprecipitation-based mass spectrometry (IPMS) combined with filter-assisted proteomics (FASP) was performed using 35S::SPL13-GFP alfalfa plants to identify SPL13-interacting proteins that may contribute to drought stress.
Results
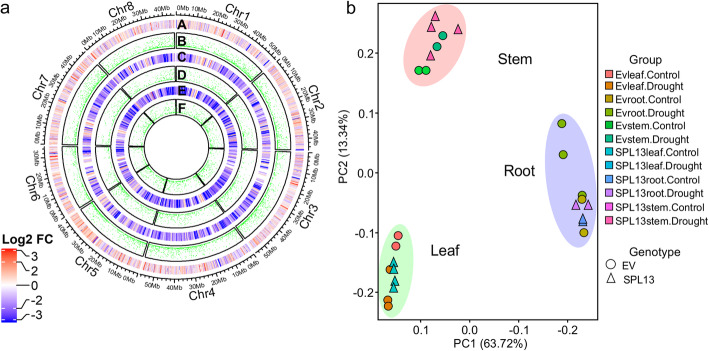
To identify miR156/SPL13-regulated genes contributing to drought tolerance, high throughput transcriptomic analysis was conducted on alfalfa plants with reduced expression of SPL13 (SPL13RNAi-65), compared to empty vector (EV) plants. On average, 7 million exon-region library sizes were generated from each replicate (Fig. S1a,b). Uniformly distributed DEG (corrected p value of p < 0.05 and an at least 2-fold changes) were observed across the eight chromosomes of the reference Medicago truncatula genome for each tissue type (Fig. 1a). Of the differentially expressed genes (DEG) derived from leaf, stem, and root tissues of drought-stressed SPL13RNAi and EV, more coverage was observed in leaf tissue followed by stem and root, respectively (Fig. 1a). The fold-changes from leaf tissues were greater towards increasing while stem and root tissues showed more decreased values in SPL13RNAi plants (Fig. 1a). Tissue-specific gene expression patterns were determined using total mRNA obtained from leaf, stem, and root tissues of these alfalfa genotypes (Fig. 1b). The major difference for the changes in transcript abundance is contributed by tissue type as explained by 63.7% of the variance in the Principal Component Analysis (PCA) (Fig. 1b).
Fig. 1.
Principal Component Analysis (PCA) depicted tissue-dependent expression patterns. Circos plot (a) visualization of transcript fold changes and their significance levels in different tissues of alfalfa plants in response to drought stress. (A) Heat map of leaf-derived log2 transcript fold changes, (B) p_values of the DEG in leaf tissues, (C) Heat map of stem-derived log2 transcript fold changes, (D) p_values of the DEG in stem tissues, (E) Heat map of root-derived log2 transcript fold changes, (F) p_values of the DEG in root tissues. b PCA plots are constructed using total exon read counts from ‘.bam’ extension RNA sequenced samples irrespective of genotype and tissue. Transcript level comparisons in ‘a’ were between leaf, stem, and root tissues of drought-stressed SPL13RNAi and EV alfalfa plants. Red and blue colours from the heat map in ‘a’ represent an increased and decreased transcript log2 fold-changes, respectively. P_values are represented with green colours. Circlize (V 0.4.11), an R-software package was used for data visualization [19]. n = 3 biological replicates
Genotype–specific transcript response of alfalfa to drought stress
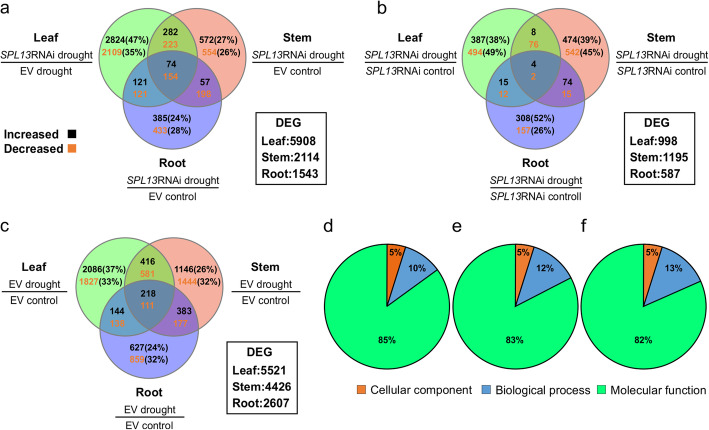
Previous reports showed that the miR156/SPL13 module regulates physiological, metabolic and phenotypic adjustments in alfalfa under drought [2, 15]. To understand SPL13-driven drought stress tolerance strategy, the global transcriptomic profile of leaf, stem, and root tissues of RNAi-silenced SPL13 and EV plants were investigated under drought stress. A total of 5908, 2114, and 1543 DEG were found in leaf, stem, and root tissues, respectively (Fig. 2a). Of the DEG, 74 were commonly increased in SPL13RNAi plants regardless of tissue source, while 154 transcripts were commonly decreased (Fig. 2a). A list of the DEG is available in the supplementary file Table S1. Among the commonly increased 74 genes, the highest fold-change corresponds to vacuolar ion transporter-like protein (Medtr2g008110) followed by gibberellin-regulated family protein (Medtr6g007897), fasciclin-like arabinogalactan protein (FLAP) (Medtr5g098420), proline dehydrogenase (PDH) (Medtr7g020820), Pmr5/Cas1p GDSL/SGNH-like acyl-esterase family protein (Medtr4g079700), LRR receptor-like kinase (Medtr5g090100), and abscisic acid receptor (Medtr7g070050) (Table S1). Contrariwise, ABC transporter family protein (Medtr2g095440), plasma membrane H+ ATPase (Medtr3g108800), and PLAT-plant-stress protein (Medtr3g087490) were among the commonly reduced 154 genes in SPL13RANi plants under drought, irrespective of tissue source (Table S1).
Fig. 2.
Tissue and genotype-specific expression patterns in genotypes of SPL13RNAi and EV alfalfa plants in response to drought. a Differentially expressed genes between drought stressed SPL13RNAi and EV plants; b SPL13RNAi-specific gene expression plasticity in response to drought stress; c EV-specific gene expression plasticity in response to drought stress; Gene Ontology (GO-term) –based percent representation of DEGs in cellular components, biological process, and molecular functions between SPL13RNAi and EV in d leaf, e Stem, and f Root tissues. The increased and decreased DEG percent provided in ‘a’, ‘b’, and ‘c’ vendiagramm were calculated from the total DEG of the specific tissue comparison. Upper and bottom values in ‘a’, ‘b’, and ‘c’ vendiagramm represent the number of significantly increased and decreased genes, respectively
To understand the drought-responsive transcript plasticity reflected by genotype, transcripts of drought-stressed and non-stressed tissues of SPL13RNAi and EV plants were analyzed. It was determined that SPL13RNAi had 998, 1195, and 587 DEG, whereas a considerably higher number (an average 4.5 fold) of DEG were observed in EV plants with 5521, 4426, and 2607 DEG in leaf, stem and root tissues, respectively (Fig. 2b,c; Table S2, S3).
Leaf-specific transcript profile of alfalfa plants under drought
To understand the gene co-expression pattern upon drought stress across tissues and genotypes, exon read-counts from both genotypes and all tissues were subjected to weighted gene co-expression network analysis (WGCNA) (Fig. S2c) followed by principal component analysis (PCA) (Fig. 1). Transcript profiles from leaf, stem, and root tissues of SPL13RNAi and EV responded to drought in a tissue-specific manner; presenting different clusters based on tissue type (Fig. 1). Of the 5908 DEG present between drought-stressed SPL13RNAi and EV leaf tissues, 47% of the genes were significantly increased and were leaf-specific in SPL13RNAi plants (Fig. 2a). On the other hand, considering tissue plasticity between well-watered and drought-stressed leaf tissues, 38% (of the 998) of DEG were increased in SPL13RNAi leaves (Fig. 2b) while 37% (of 5521) of DEG were increased in EV leaves (Fig. 2c). Moreover, 49% (494 of 998) of leaf-specific DEG were decreased in SPL13RNAi leaf tissues while 33% (1827 of 5521) were decreased uniquely in EV leaves (Fig. 2b,c). GO-terms of the DEG were analyzed and categorized into molecular function, biological process, and cellular components to understand the role of DEG between leaf tissues of drought-stressed SPL13RNAi and EV plants. GO-term analysis from leaf tissues showed 85% corresponding to molecular function followed by 10 and 5% to biological process and cellular components, respectively (Fig. 2d). Graphical representation of the components of GO-term analysis is provided in supplementary file Fig. S3. The top three highly represented leaf-derived GO-terms for the molecular function category correspond to transcription activity (phosphorelay response regulator activity, sequence-specific DNA binding transcription factor activity, and transcription cofactor activity) (Table S4.1). Likewise, the most highly represented three biological processes correspond to telomere maintenance, translation and alcohol metabolic process in addition to glutamine catabolic process and porphyrin-containing compound biosynthetic process (includes chlorophyll biosynthesis) (Table S4.1).
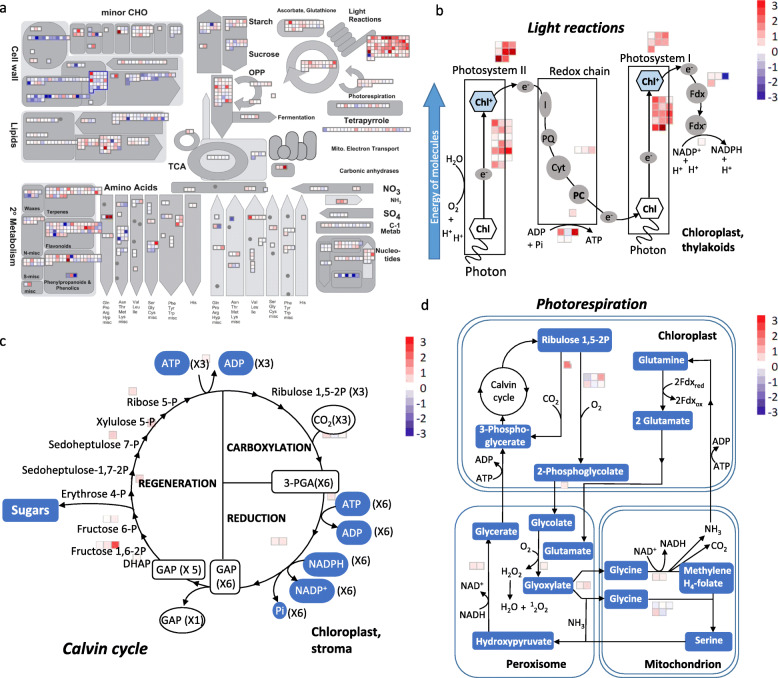
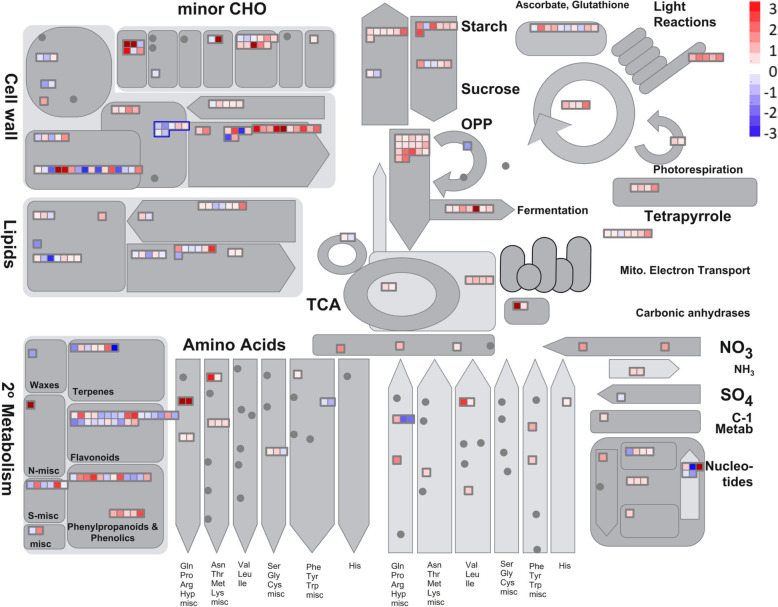
The differentially regulated genes were mapped to the M. truncatula genome using MapMan pathway analysis tool to understand their functional associations. Accordingly, leaf-specific DEG between drought-stressed SPL13RNAi and EV showed that various metabolic pathways were significantly affected, including carbohydrate metabolism, photosynthesis (mainly light reaction), and primary metabolism-related genes (Fig. 3a). Most importantly, photosynthesis-associated transcripts were highly increased in SPL13RNAi plants (Fig. 3a). Further investigation of transcripts associated with photosynthesis revealed that light-dependent reaction centers, namely of photosystem I and photosystem II, were expressed higher in SPL13RNAi (Fig. 3b). Unlike the light-dependent reaction centers, the Calvin cycle (Fig. 3c) and photorespiration-associated transcripts (Fig. 3d) were either slightly increased or not altered. Specifically, the carboxylation- and photorespiration-associated transcript of Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase) was slightly increased in SPL13RNAi plants under drought stress (Fig. 3c,d).
Fig. 3.
Leaf-specific DEG attributed to photosynthesis are enhanced in SPL13RNAi plants. a Summary of affected metabolites and pathways between drought-stressed EV and SPL13RNAi leaf tissues, b transcripts coding for light-dependent photosynthetic reaction in the chloroplast thylakoids, c carbon dioxide fixation in Calvin cycle in chloroplast stroma region, d photorespiration-associated transcripts involving chloroplast, mitochondria and peroxisome differentially regulated between drought-stressed SPL13RNAi and EV plants. Transcript fold changes are provided in log 2 with red and blue colours representing increased and decreased transcript levels relative to EV. Minor CHO corresponds to minor charbohydrate; TCA, Tricarboxylic acid cycle; OPP, oxidative pentose phosphate pathway. N = 3 biological replicates for each genotype and treatment condition
Stem-specific transcript changes in alfalfa plants under drought
Expression levels of 27% of the 2114 DEG between drought-stressed stem tissues of SPL13RNAi and EV plants were increased exclusively in stems of SPL13RNAi plants (Fig. 2a). On the other hand, genotype- and stem-specific drought stress-responsive transcript plasticity revealed 39% of the 1195 DEGs in SPL13RNAi plants were increased while EV plants showed a 26% increase out of 4426 DEGs (Fig. 2b,c). Comparable to DEG in leaf tissues, the majority (83%) of the DEG in stem are associated with molecular functions followed by biological processes (12%) and cellular components (5%), despite DEG profile composition differences between leaves and stems (Fig. 2e; Fig. S4). The most represented three molecular functions which are differentially affected between SPL13RNAi and EV stem tissues are acyl-CoA dehydrogenase activity, ubiquinol-cytochrome-c reductase activity, and hydroxymethylglutaryl-CoA reductase (NADPH) activity (Table S4.2). On the other hand, the highly enriched categories of the affected biological processes include ATP catabolic process, response to stress, defense response, intercellular signal transduction and response to desiccation (Table S4.2).
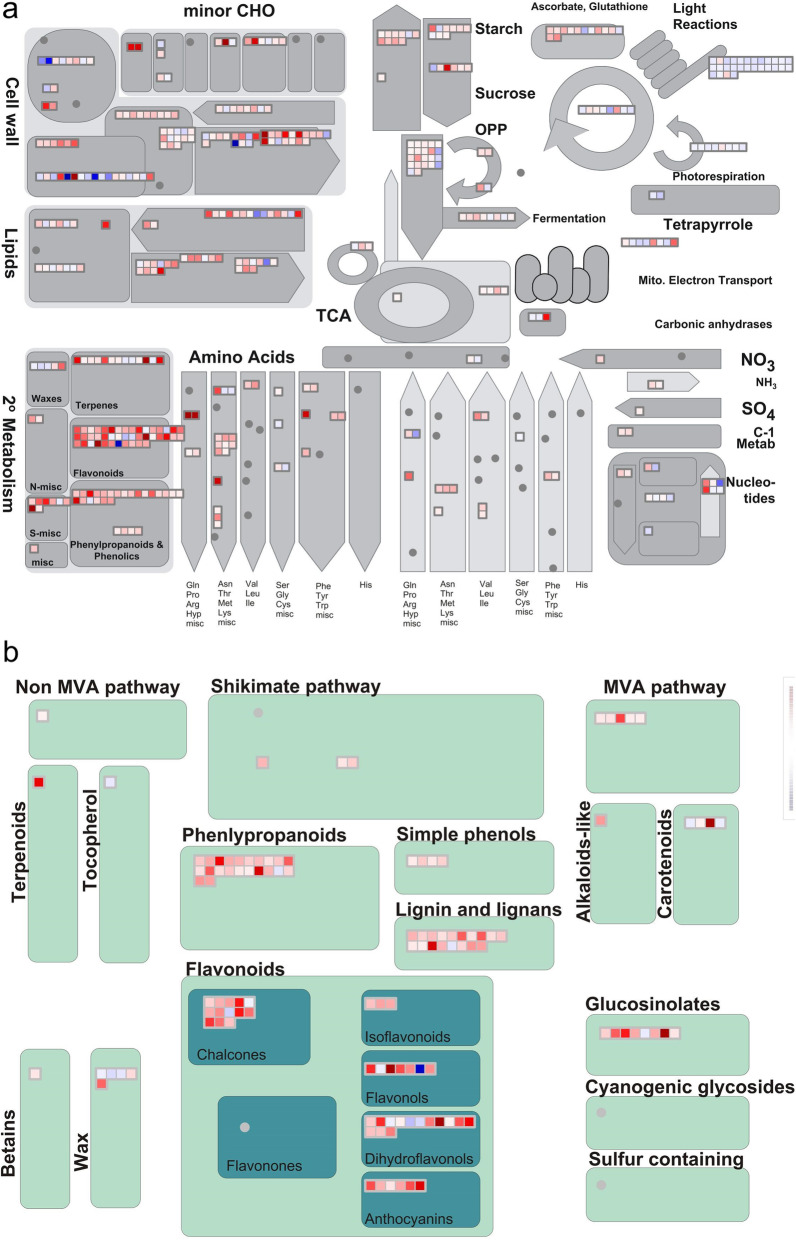
Furthermore, to understand the association of DEG of stem with metabolic pathways, the DEG were subjected to MapMan-based pathway analysis (Fig. 4a). DEG of stem tissues corresponded mainly with increased flavonoid biosynthesis, carbohydrate metabolism and response to desiccation in SPL13RNAi plants (Fig. 4a, Table S1). On the other hand, DEG associated with photosynthesis were decreased significantly in SPL13RNAi plants (Fig. 4a). Transcriptomic analysis of DEG obtained from stem tissues combined with MapMan-based pathway analysis revealed an activation of the phenylpropanoid pathway in SPL13RNAi plants under drought stress (Fig. 4b).
Fig. 4.
Stem-specific DEGs are associated with secondary metabolism. a Summary of affected metabolites and pathways between drought stressed EV and SPL13RNAi stem tissues, b distribution of the differentially expressed genes associated with secondary metabolism. Pathway analysis was performed using MapMan. Transcript fold changes are provided in log 2 with red and blue colours representing an increased and decreased transcript levels relative to EV. Minor CHO corresponds to minor charbohydrate; TCA, Tricarboxylic acid cycle; OPP, oxidative pentose phosphate pathway; MVA, mevalonate pathway. N = 3 biological replicates for each genotype and treatment condition
Root-specific transcript profile of alfalfa plants under drought
A total of 1543 DEG were detected between roots of drought-stressed SPL13RNAi and EV plants, with 24 and 28% of them increased and decreased, respectively, exclusively in the roots (Fig. 2a). Further analysis of the plasticity between well watered and drought-stressed root samples showed 52% of 587 DEG in SPL13RNAi roots were upregulated while 24% of 2607 DEG were increased only in EV roots (Fig. 2b,c). Moreover, GO-term analysis of the root-specific DEG showed a similar proportion of components to that of stem and leaf tissues where the majority (82%) of transcripts belong to molecular function followed by biological process (13%) and cellular components (5%), but with a varied profile composition (Fig. 2f; Fig. S4). The top highly represented biological processes encompass ATP catabolic process, response to stress, defense response, intercellular signal transduction, phosphorelay signal transduction system, metabolic process, metal ion transport and transmembrane transport (Table S4.3). On the other hand, the major representation from molecular functions are attributed to phosphorelay response regulator activity, sequence-specific DNA binding transcription factor activity, catalytic activity, GTPase activity, secondary active sulfate transmembrane transporter activity (Table S4.3). Moreover, to further understand the DEG association, the DEG were subjected to MapMan-based pathway analysis. We found that metal ion transport, carbohydrate and primary metabolism were significantly and differentially affected between SPL13RNAi and EV plants in response to drought (Fig. 5). Moreover, cell wall and lipid biosynthesis were increased in roots of SPL13RNAi plants as compared to EV.
Fig. 5.
Distribution of root-specific differentially expressed genes between EV and SPL13RNAi plants. Summary of differentially affected metabolites and pathways between drought stressed EV and SPL13RNAi root tissues. Transcript fold changes are provided in log 2 with red and blue colours representing increased and decreased transcript levels relative to EV. Minor CHO corresponds to minor charbohydrate; TCA, Tricarboxylic acid cycle; OPP, oxidative pentose phosphate pathway. N = 3 biological replicates for each genotype and treatment conditions
Identification of SPL13 interacting proteins in alfalfa
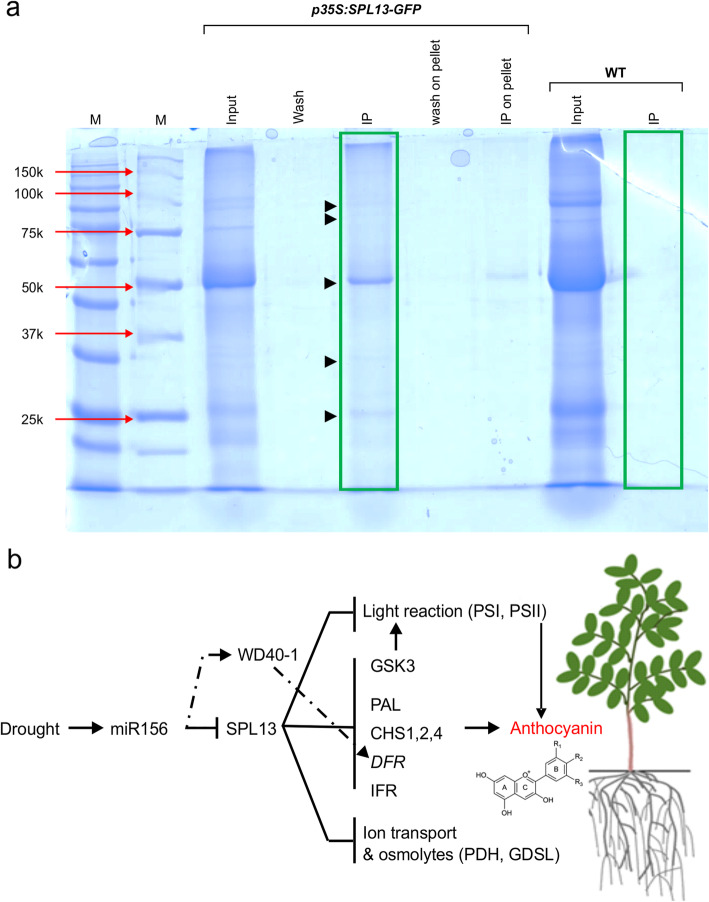
IPMS was used to identify candidate proteins that interact with SPL13 in alfalfa under drought stress. In this experiment, alfalfa plants overexpressing SPL13 tagged with a green fluorescent protein, 35S::SPL13-GFP [17] and wild-type plants grown under control and drought stress conditions were used. Isolation of SPL13-interacting proteins followed by Coomasie staining showed the presence of unique protein bands in 35S::SPL13-GFP plants that were lacking in wild-type (Fig. 6a). There were some prominent and many faint bands. Therefore, to identify all the proteins present, a FASP proteomics method was employed on the total IP eluent. Subsequently, the precipitated proteins were digested with trypsin and identified by mass spectrometry. To account for the lack of a complete M. sativa proteome database, a combined use of contig database and partial Uniprot database identified unique candidate proteins that would have been missed by the M. truncatula database alone. We were able to identify candidate SPL13-interacting proteins involved in the photosynthesis process, specialized metabolite biosynthesis, ROS scavenging, and abiotic stress tolerance in addition to normal cellular activity-involved peptides (Table 1; Table S6).
Fig. 6.
SPL13 interacts with proteins involved in photosynthesis process, stress alleviation, and specialized metabolite biosynthesis. a Coomasie stained SDS-PAGE gel, b a proposed tissue-specific drought tolerance model modulated by miR156/SPL13. The diagrammatic representation shows a tissue-specific miR156/SPL13 regulation module in response to drought tolerance. miR156 is induced by drought stress, which in turn silences SPL13 [2]. Reduced expression of SPL13 driven by miR156 and enhanced level of WD40–1 enhances DFR [15], together with less inactivation of GSK3 signalling with PAL, CHSs, and DFR result in accumulation of anthocyanins mainly in stem tissues. In moderate miR156OE plants, primary metabolites such as GABA, proline and sugars also accumulate for carbon-to-nitrogen balance and osmotic homeostasis. Induction of miR156 during drought stress also enhances phenotypic plasticity, such as longer roots and higher biomass to access more water from the rhizosphere. With reduced SPL13 expression and miR156OE, higher photosynthesis efficiency is also achieved during drought stress. The most prominent differential protein bands present in immuno-precipitated 35S::SPL13-GFP alfalfa plants are indicated with black arrow in ‘a’. Abbreviations in ‘b’ represent: CHS, Chalcone synthase; DFR, dihydroflavonol-4-reductase; GDSL, Gly-Asp- Ser-Leu –lipase; GSK, Glycogen synthase kinase-3; IFR, Isoflavone reductase; PAL, Phenylalanine Ammonia-Lyase; PDH, Proline Dehydrogenase
Table 1.
Detected candidate proteins interacting with SPL13 by IPMS analysis
| Proteins | Reported roles and description | Sequence coverage (%) | # Peptides | # Spectra | Molecular weight (KDa) |
|---|---|---|---|---|---|
| 50S ribosomal protein L22 | Early stages of 50S ribosomal protein assembly | 18 | 4 | 7 | 21.88 |
| 60s ribosomal protein L11 | Biogenesis of 60S ribosomal subunits | 49.72 | 6 | 11 | 20.67 |
| Arginine biosynthesis bifunctional protein | Involved in the cyclic version of arginine biosynthesis | 26.72 | 7 | 21 | 48.6 |
| Beta-xylosidase/alpha-L-arabinofuranosidase | Releases xylose and arabinose from cell walls | 25.32 | 15 | 30 | 83.67 |
| Cafeic acid 3-O-methlyltransferase (COMT1) | Catalyzes the conversion of caffeic acid to ferulic acid and of 5-hydroxyferulic acid to sinapic acid. | 17.81 | 6 | 31 | 39.92 |
| Cell division control protein 2 homolog 1 | control of the eukaryotic cell cycle | 17.87 | 4 | 11 | 33.46 |
| Chalcone synthase 1 | naringenin-chalcone or chalcone biosynthesis | 15.17 | 5 | 14 | 42.74 |
| Chalcone synthase 2 | 31.62 | 9 | 38 | 42.68 | |
| Chalcone synthase 4 | 28.53 | 8 | 34 | 42.63 | |
| Cyclic beta-(1,2)-glucan synthase | biosynthesis of cyclic beta-(1,2)-glucan | 15.11 | 29 | 86 | 315.55 |
| Dihydroflavonol 4-reductase | anthocyanin biosynthesis | 33.18 | 6 | 12 | 24.37 |
| Eukaryotic translation initiation factor 5A-1 | mRNA-binding protein involved in translation elongation | 32.3 | 3 | 32 | 17.65 |
| Glutamate synthasee [NADH] amyloplastic | assimilation of symbiotically fixed nitrogen into amino acids | 19.51 | 30 | 99 | 240.22 |
| Glutamine synthetase cytosolic isozyme | homeostatic control of glutamine synthesis | 16.85 | 4 | 9 | 39.08 |
| Glutamine synthetase leaf isozyme, chloroplastic | Re-assimilation of the ammonia generated by photorespiration | 14.95 | 6 | 15 | 47.09 |
| Glycogen synthase kinase-3 homolog − 1 | Brassinosteroids signaling and abiotic stress response | 21.41 | 6 | 11 | 46.74 |
| Glycogen synthase kinase-3 homolog − 2 | 25.3 | 7 | 94 | 46.03 | |
| Glycogen synthase kinase-3 homolog − 3 | 37.23 | 11 | 27 | 46.24 | |
| Isoflavone reductase, IFR | Reduces achiral isoflavones to chiral isoflavanones | 13.52 | 4 | 5 | 35.43 |
| Long-distance movement protein | facilitate the cell-to-cell movement | 17.93 | 3 | 3 | 26.67 |
| Malate dehydrogenase, chloroplastic | Catalyzes a reversible NAD-dependent dehydrogenase reaction involved in central metabolism | 16.87 | 3 | 3 | 35.52 |
| Methylthioribose-1-phosphate isomerase | Catalyzes methylthioribose-1-phosphate (MTR-1-P) into methylthioribulose-1-phosphate (MTRu-1-P) interconversion | 38.14 | 11 | 15 | 42.24 |
| Mitogen-activated protein kinase homolog | mitogenic induction of symbiotic root nodules | 21.19 | 6 | 32 | 44.37 |
| Nodulation protein D1 | Regulates other nodulation proteins | 15.2 | 3 | 3 | 34.86 |
| Nodulation protein E | synthesize NOD factor fatty acyl chain | 26.68 | 7 | 9 | 41.64 |
| Nodulin-25 | development and function of nodules | 19.51 | 3 | 6 | 27.51 |
| Phenylalanine ammonia-lyase | catalyzes the deamination of phenylalanine to cinnamate and ammonia | 21.38 | 12 | 21 | 78.82 |
| Phosphoenolpyruvate carboxylase | catalyzes the addition of bicarbonate to phosphoenolpyruvate | 17.08 | 11 | 16 | 110.82 |
| Polygalacturonase | depolymerization of pectates | 15.44 | 6 | 29 | 43.93 |
| Replication protein 1A | stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. | 33.13 | 25 | 170 | 111.95 |
| Ribulose bisphosphate carboxylase large chain | Catalyzes the carboxylation of D-ribulose 1,5-bisphosphate and oxidative fragmentation of the pentose substrate | 24.05 | 12 | 21 | 52.59 |
| Ribulose bisphosphate carboxylase small chain | Catalyzes the carboxylation of D-ribulose 1,5-bisphosphate and oxidative fragmentation of the pentose substrate | 46.67 | 7 | 34 | 20.24 |
| RNA-directed RNA polymerase | catalyses synthesis of the RNA strand complementary to a given RNA template | 15.14 | 9 | 34 | 93.6 |
| RNA-directed RNA polymerase 2a | required for cytoplasmic and chromatin RNA silencing pathways | 16.91 | 9 | 13 | 93.46 |
| Serine/threonin-protein phosphatase PP1 | essential for cell division and glycogen metabolism by dephosphorylating substrates | 29.6 | 8 | 97 | 36.23 |
| Serine/threonin-protein phosphatase PP2A | Dephosphorylates and activates the actin-depolymerizing factor ADF1 | 18.85 | 3 | 5 | 35.69 |
| Sucrose synthase | Sucrose-cleaving enzyme that provides UDP-glucose and fructose | 15.65 | 8 | 31 | 92.27 |
| Tyrosine recombinase XerC | Catalyze the cutting and rejoining of the recombining DNA molecules | 15.72 | 3 | 3 | 33.75 |
| Vacuolar membrane protease | vacuolar sorting and osmoregulation | 21.96 | 14 | 45 | 112.35 |
| Vacuolar protein sorting/targeting protein 10 | sorting receptor in the Golgi compartment required for the intracellular sorting and delivery of soluble vacuolar proteins | 18.45 | 22 | 67 | 161.33 |
Candidate proteins were considered when at least three peptides with six spectra were present from the protein while undergoing the database search as specified in the method part of the article. Thermo Q-Exactive Orbitrap mass spectrometer coupled to an Easy-nLC 1000 nano LC system were used to analyze samples
SPL13 interacts with photosynthesis- and phenylpropanoid pathway -related proteins
The identification of SPL13-interacting proteins indicated proteins involved in photosynthesis are among the top candidates. For example, there were multiple occurrences of both small and large chains of the ribulose bisphosphate carboxylase (Rubisco), photosynthesis I p700 chlorophyll a apoprotein A2, and phytochrome A (Table 1; Table S6).
A previous study reported that reduced binding of SPL13 to DFR promoter enhances anthocyanin biosynthesis [15]. Using the current IPMS analysis, we were able to identify DFR and other candidate proteins interacting with SPL13, such as phenylalanine ammonia-lyase1(PAL1) which catalyses the first step in the phenylpropanoid pathway by converting phenylalanine to cinnamate [37]. Other candidate peptides interacting with SPL13 from this pathway are chalcone synthase1, 2 and 4 (CHS1, CHS2, CHS4), which catalyzes the specialization from the general phenylpropanoid pathway into flavonoids converting 4-coumaroyl-CoA and malonyl-CoA [10]. Other specialized metabolite biosynthesis mediating proteins such as cafeic acid-3-o-methyltransferase (COMT1) and isoflavone reductase (IFR) were also found to interact with SPL13 (Table 1).
SPL13 interacts with proteins involved in stress signal transduction and resilience
Perception of environmental changes followed by signal transduction are a prerequisite for orchestrating the necessary measures to alleviate the stress effects through various kinases. In our study, Glycogen synthase kinase-3 and Mitogen-activated protein kinase homologs were detected in 35S::SPL13-GFP plants under drought stress (Table 1). Moreover, N+/H+ antiporter, which is involved in ion balance to mediate stress tolerance and cellular activity [58], was also detected by IPMS analysis as an interacting partner of SPL13 under drought stress (Table S6).
Discussion
Genotype-specific gene expression patterns in response to drought stress
Silencing of SPL13, a target of miR156, in alfalfa affected a plethora of genes leading to improved drought tolerance [2]. Among the commonly and significantly increased transcripts across the different tissues of SPL13RNAi alfalfa, FLAP was reported to have primarily a cell adhesion function and to play a role in abiotic stress tolerance in Arabidopsis [27, 60]. Moreover, proline dehydrogenase (PDH) was increased in SPL13RNAi. PDH is known for its role in drought tolerance by either scavenging ROS, balancing the carbon to nitrogen ratio through GABA shunt [18], or serving as an osmolyte [22]. Proline and its catabolic products, mediated through PDH, are more important to drought tolerance than the high accumulation of proline itself [7]. The other commonly increased transcripts with higher fold-changes were transcripts of ABA receptors known for their role in abiotic stress tolerance [32, 34]. Accordingly, the increased transcript levels of FLAP, PDH, and ABA receptors along with other transcripts across the three tissue types (leaf, stem and root) in SPL13RNAi alfalfa relative to EV suggests that these proteins play a major role in the response of SPL13RNAi plants to drought stress.
Photosynthesis-related DEG are upregulated in leaves of SPL13RNAi plants during drought
Physiological investigation of SPL13RNAi plants under drought stress showed maintenance of vital physiological processes, such as photosynthesis and leaf water holding capacity [15]. This was accompanied by an upregulation of photosynthesis-related genes (PSI and PSII) in SPL13RNAi plants [15]. In the current study, comparison of transcript profiles of leaves of both genotypes under control and drought conditions revealed that various metabolic pathways were affected, with photosynthesis being predominantly higher in SPL13RNAi plants. The observed upregulation of photosynthesis-associated DEG was mainly in the light-dependent reaction, consistent with the physiological data report of the previous study [15] which showed that SPL13RNAi plants maintained the photosynthesis process during drought stress, while photosynthesis was reduced in EV plants. Other studies have shown that the light-dependent reaction centers were significantly affected by drought in maize [61, 63]. The maintained or slightly increased photorespiration-associated transcripts in SPL13RNAi plants may serve as an energy sink to prevent over-reduction of NADPH and photo inhibition in SPL13RNAi plants. Moreover, an increased level of photo-inhibition; unreduced NADPH with lower photosynthetic assimilation rate, was reported previously in drought-stressed barley plants [56]. A higher level of photosynthesis assimilation rate reported in the previous study [2, 15] along with the current transcript-based evidence in SPL13RNAi plants suggest a mechanism to potentially lower the reductive power of NADPH that could affect the photosynthesis otherwise.
Specialized metabolism-related DEG are upregulated in stems of SPL13RNAi plants during drought
Specialized metabolites are important in plant growth and development, and their association with reduced ROS levels was reported in in vivo [1] and in vitro assays [41]. Accordingly, increasing the abundance of specialized metabolites and associated primary metabolites, such as ascorbates and proline, are considered as a marker for enhanced biotic and abiotic stress tolerance [24]. In our earlier study, we observed enhanced levels of primary and specialized metabolites related to stress tolerance, such as anthocyanin, in miR156 overexpressing alfalfa genotypes [15]. This suggests anthocyanin and possibly other ROS scavenging metabolites are regulated by SPL13 in a miR156-dependent manner involving DFR. To further investigate the involvement of SPL13 in regulating levels of specialized metabolites, especially phenylpropanoids, the global transcript levels in leaf, stem and root tissues of SPL13RNAi plants was investigated relative to tissues from EV. Importantly, stem-derived samples showed an enhanced abundance of transcripts associated mainly with the phenylpropanoid pathway, unlike leaf and root tissues that showed an increase in mainly photosynthesis- and ion transport- associated transcripts, respectively. The enhanced abundance of transcripts associated with the phenylpropanoid pathway is consistent with the observed colour development in stems of miR156 overexpressing plants [15]. An earlier study showed that the accumulation of anthocyanin was positively correlated with the level of DFR expression in potato in a non-tissue specific manner [54]. The ChIP-qPCR analysis revealed that while SPL13 indeed binds to DFR promoter presumably to regulate its expression [15], enhanced anthocyanin accumulation was observed only in the stem. RNAseq analysis also indicated upregulation of transcripts belonging to the biosynthesis of anthocyanin and other polyphenols from the phenylpropanoid pathway mainly in stem tissues during drought stress. Whether SPL13 also regulates the phenylpropanoid pathway in tissues other than the stems remains to be investigated further.
The upregulated root-specific DEG are mainly attributed to ion transport in SPL13RNAi plants during drought stress
Roots are the first to encounter low moisture in the soil, but maintenance of plant water potential is not completely dependent on roots but rather on a continuum that involves soil, root, leaf, and the atmosphere via the transpiration stream [13, 36]. To maintain water potential, drought tolerant plants use different strategies to affect the osmotic balance and/or hydrostatic force governed by the transpiration stream. Osmolytes, such as sugars and proline, have been reported to adjust osmotic balance in different plants [24, 45]. Previously, we showed drought tolerant genotypes of miR156OE had higher levels of the osmolytes proline and sugars [2, 15]. To understand the involvement of SPL13 in maintaining water potential under drought, transcripts from SPL13RNAi and EV root tissues were profiled under control and drought conditions. The DEG showed increased levels of transcripts associated with GABA shunt and membrane integrity, such as GDSL, in SPL13RNAi plants. Primary metabolites, such as ascorbate and glutathione, and phenylpropanoid specialized metabolites known to scavenge ROS were also significantly increased. To validate the transcript-based metabolite pathway analysis and identify non-enzymatic metabolite conversions [28], primary and specialized metabolite analysis in SPL13RNAi plants is important. The eventual release of the alfalfa genome sequence should allow for pathway analysis to potentially identify novel metabolite pathways unmapped to the M. truncatula genome in the current study, but which may contribute to drought stress response in alfalfa.
SPL13 interacts with proteins involved in photosynthesis, specialized metabolite biosynthesis, stress signalling and ROS scavenging processes
Previously, we showed that silencing of SPL13 in alfalfa led to the maintenance of photosynthesis under drought stress by affecting genes involved in this process [15]. In the current study, an enhanced level of transcripts associated with photosynthesis, mainly light reaction, was observed in leaf tissues. Consistent with this finding, IPMS analysis showed that SPL13 interacts with photosynthesis- related proteins such as RUBISCO (small and large chain) and PSI. Rubisco is composed of eight small sub units encoded by the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RBCS) and eight large subunits encoded by RBCL [50]. Posttranslational modification of both the small and large subunits of RUBISCO is required for functional activity [25] and functional-holoenzyme assembly [12]. Accordingly, modifications to either subunits of RUBSICO result in reduced photosynthetic activity and retarded growth [46], and any interactions these subunits have may influence the enzyme activity. In this regard, the increased binding of the small and large chains of RUBISCO to SPL13 under drought stress in SPL13 overexpressing genotypes may interfere with the subunits assembly. Furthermore, alfalfa plants with reduced level of SPL13 (SPL13RNAi plants) were previously reported to have a higher level of maximum rate of rubisco carboxylase activity (Vcmax) under drought stress [15]. Investigating SPL13 binding sites in both small and large chain subunits of RUBISCO would provide knowledge on how the miR156/SPL13 module maintains photosynthesis efficiency under drought stress.
PAL, DFR and CHSs were among the protein candidates identified that interact with SPL13 as detected by IPMS in the current study. These enzymes mediate the biosynthesis of specialized metabolites in the phenylpropanoid pathway, and alteration of their expression levels affects abundance of these metabolites in plants. For examples, Arabidopsis plants with reduced expression of PAL (PAL1 to PAL4) had reduced content of proanthocyandins in the seed coats, and developed ultraviolet light sensitivity [26]. Likewise, enhanced levels of CHS, which mediates the step-wise condensation of malonyl-CoA with coumaroyl-CoA to form naringenin chalcone (flavonoids), was associated with increased accumulation of anthocyanin abundance and stress tolerance in plants [11, 48]. The regulation of PAL, DFR, and CHSs, may explain the enhanced biosynthesis of phenolic compounds needed to scavenge ROS in miR156OE and SPL13RNAi alfalfa plants [15].
Plants use various stress signal transduction mechanisms, which among others involve mitogen-activated protein kinase and mitogen-activated protein kinase kinase [16, 20, 43]. In the current study, we detected the Glycogen synthase kinase-3 homologs (GSK3_1, 2,and 3) along with one of its phosphorylated target Serine/threonin-protein phosphatase (PP1 and PP2A) as an interacting partner of SPL13. Brassinosteroid-involving GSK3s are involved in cell signalling [9, 47, 59] and regulate mitogen-activated protein kinase during abiotic stress response for stomatal regulation [29, 30]. The IPMS data showed that the above stress signal transduction pathway and its regulators interact with SPL13, suggesting a strategy used by alfalfa plants to respond to drought.
Conclusions
Based on previous reports on the role of miR156/SPL13 module in regulating drought response in alfalfa [15], we investigated whether the role of miR156/SPL13 in drought response is tissue-specific. In this study, the global transcriptomic profiles of SPL13RNAi plants showed tissue-specific regulation of transcripts and associated pathways. In leaf tissues, there was an increase in the transcript levels of mainly photosynthesis- and photorespiration-related genes in SPL13RNAi plants. On the other hand, the stem tissues of SPL13RNAi plants exposed to drought showed an increase in transcripts related to the phenylpropanoid pathway. Furthermore, roots of SPL13RNAi plants under drought had an increase in transcripts associated with ion transporters, as well as primary and specialized metabolism, presumably to transport osmolytes and scavenge ROS while maintaining membrane integrity through GDSL. Moreover, SPL13 was found to interact with proteins involved in photosynthesis process, stress tolerance, and specialized metabolite biosynthesis.
We propose a model for a drought tolerance mechanism in which moderate levels of miR156 overexpression regulate SPL13 in a tissue-dependent manner (Fig. 6b). miR156 is induced by drought stress, which in turn silences SPL13 [2]. Reduced expression of SPL13 driven by miR156 and enhanced level of WD40–1 enhances DFR [15], resulting in the accumulation of anthocyanins mainly in stem tissues. Similarly, silencing of SPL13 would allow for continued activity of GSK3 signalling along with PAL, CHS, and DFR due to reduced interaction of these proteins with SPL13, further fueling the biosynthesis of drought stress-alleviating specialized metabolites, and facilitating ROS scavenging in SPL13RNAi and miR156OE alfalfa plants under drought stress. Future structural and enzymatic investigation of the candidate SPL13-interacting proteins could elucidate the active sites of the proteins, and their modifications needed to regulate drought stress in alfalfa. It is also important to validate the protein-protein interactions detected in this study using other techniques, such as yeast-two-hybrid assay.
Methods
Plant material
SPL13 overexpressing and SPL13RNAi alfalfa plants
Alfalfa (Medicago sativa L.) genotypes with reduced expression levels of SPL13, SPL13RNAi-6 genotypes [2], empty vector control (EV) [5], 35S::SPL13-GFP [17], and wild-type (WT) were used in this study. The transgenic alfalfa plants were generated previously in Dr. Hannoufa’s laboratory using the WT clone N4.4.2 [6] that was obtained from Dr. Daniel Brown (Agriculture and Agri-Food Canada). The transgenic alfalfa plants are not deposited in a public herbarium, so requests for any plant material should be directed to the corresponding author. The plants were grown in a fully automated greenhouse with 16-h light (380–450 W/m2), relative humidity (RH) of 70% and temperature of 25 ± 2 °C at the Agriculture and Agri-Food Canada London Research and Development Center, London, Ontario, Canada. Given that alfalfa is an obligatory outcross, we used vegetative cuttings for propagation according to Aung et al. [5] to maintain genotypes throughout the study.
Imposing drought stress
Drought stress was imposed on alfalfa plants devoid of water for 2 weeks at 30 days post vegetative propagation (juvenile vegetative stage) during which time plants were kept in a completely randomized design. Equal soil moisture levels were maintained before starting the experiment using a SM 100 soil moisture sensor (Spectrum Technologies Inc., Jakarta, Indonesia). Three biological replicates were used per genotype per treatment for transcript analysis (each replicate being an individual plant). Leaves (newly developed upper leaves), stems (lower 5 cm internode close to soil) and roots (7.5 cm of main and auxiliary root tips) were harvested from SPL13RNAi and EV plants for total mRNA extraction. Samples were flash frozen with liquid nitrogen and kept at − 80 °C for later transcriptomic analyses.
RNA extraction
Lower basal stem internode, young top leaves and root tip samples were collected and flash frozen in liquid nitrogen and kept in a -80 °C freezer until used for qRT-PCR analysis and RNA sequencing. Approximately 50 mg fresh weight was used for total RNA extraction using a PowerPlant® RNA isolation kit (Cat # 13500) for leaf samples, a QIAGEN RNeasy® Plant mini kit for stem and root tissues (Cat # 74904), and a PowerLyzer®24 bench top bead-based homogenizer (Cat # 13155) following manufacturers protocols. Total RNA quality was verified using BioRad Bioanalyzer for integrity and Nanodrop concentration before RNAseq analysis.
RNAseq analysis
mRNA stranded library preparation followed by Illumina HiSeq 2500 RNA-sequencing with pair end of 126 nucleotide bases were performed as a fee-for-service at The Center for Applied Genomics, The Hospital for Sick children, Toronto, ON, Canada. RNAseq data was analyzed according to Trapnell et al. [52] on biocluster with shell scripts of Linux. The scripts used for the analysis are provided as supplementary file (Table S7). To identify expression pattern of genes and module identification, R-software (V.4.0) environment-based network analysis with weighted gene co-expression network, WGCNA, in ‘BiocManager’ package was performed according to Langfelder and Horvath [31]. Using the ‘featureCounts’ [33], the exon read counts were obtained from ‘.bam’ extension files of RNA sequenced samples followed by Principal Component Analysis in R-software.
Visualization of differentially expressed genes-based pathway analysis was carried out using MapMan free software V3.6 (https://mapman.gabipd.org/) with a M. truncatula reference genome sequence, Mt4.0 V2 (http://www.medicagogenome.org/downloads). Using an online free gene ontology analysis tool (http://revigo.irb.hr/), lists of differentially expressed genes with significance p values between genotypes and tissue types, the corresponding tree map codes for molecular function, biological process and cellular components were exported. Subsequently, the codes for generating the tree maps were visualized using R-software. Raw RNA sequencing reads can be accessed at the National Center for Biotechnology Information, NCBI, BioProject PRJNA598830.
qRT-PCR analysis to validate RNAseq results
To validate the results from RNAseq analysis, the extracted RNA was treated with Ambion®TURBO DNA-free™ DNase (Cat # AM1907) followed by iScript™ cDNA synthesis (Cat # 1708891). Medtr3g498825: basic helix-loop-helix 137 bHLH137 (F:- CAGGATAATGCAGCAGAAGG, R:- GGGCTCATCCAAACACTTTC); Medtr7g109510: basic leucine zipper bZIP (F:- AGTTCGGGTTCTGATGGTGT, R:- CTCCAACAGTTTCTGCTTCC); Medtr5g048850: flavonol synthase/flavanone 3-hydroxylase F3H (F:- TTGTTCTAGGTGTGCCTCCA, R:- GAATGACAAGGGCATTAGGG); Medtr3g091350: flavonol synthase/flavanone 3-hydroxylase F3H-2 (F:- TCTCTCCTGGTTCCTTCTGT, R:- GGTCGATAACAGGAACTTGT); Medtr5g081860: MYB transcription factor MYB51 (F:- GATGGCTTAAGGTTGCTGAG, R:- GATAGCCAGGAATCGGAACA); Medtr5g090620: UDP-glucose flavonoid 3-O-glucosyltransferase UDPFG (F:- CGTGATAACCGTCTCCGAAT, R:- TGATGACCTGGAGAGAATCG); Medtr7g071050: UDP-glucosyltransferase family protein UDPGT (F:- CTTCTGGAAAGGCAAGATGG, R:- GCTCCTTCTTTGGTTGTTGG) genes were used for validation. The gene primers were designed using publicly available Primer3 software (http://primer3.ut.ee/) based on M. truncatula genome sequence and amplified product was sequenced for an identity check. Primer efficiency was checked before proceeding using for qRT-PCR analysis. qRT-PCR was performed using the CFX96™ Real-Time PCR detection system and SsoFast™ EvaGreen® Supermixes (Bio-Rad Cat # 1725204). Specifically, 2 μL cDNA (equivalent to 200 ng cDNA), 1 μL forward and reverse gene-specific primers (10 μM each), 5 μL SsoFast Eva green Supermix, and 2 μL of nuclease-free water was used to make the final reaction volume of 10 μL. PCR amplification was performed at: cDNA denaturation at 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s, 58 °C for 30 s and 72 °C for 30 s (denaturation, annealing and extension, respectively) followed by a melting curve that runs from 65 °C to 95 °C with a gradual increment of 0.5 per 5 s. All reactions were performed with three technical replicates. Transcript levels were analysed relative to acetyl-CoA carboxylase1 (F:- GATCAGTGAACTTCGCAAAGTAC, R:- CAACGACGTGAACACTACAAC) and ACTIN (F:- TCAATGTGCCTGCCATGTATGT, R:- ACTCACACCGTCACCAGAATCC) housekeeping genes designed based on alfalfa sequence [15]. The selected differentially expressed genes from the RNAseq result were subjected to fold-change in different tissues of SPL13RNAi-6 and EV plants and compared to qRT-PCR results as described in Wang et al., [55]. The values are presented with regression analysis (Fig. S5) indicating a positive correlation (R2 = 0.74) with some differences associated with sensitivity of the two platforms.
Isolation of SPL13-interacting proteins
To determine SPL13-interacting proteins, we used alfalfa plants expressing SPL13 tagged with GFP proteins (35S::SPL13-GFP) and wild-type alfalfa plants. Plants were subjected to drought stress for 2 weeks prior to protein extraction. Subsequently, samples were immediately flash frozen using liquid nitrogen and kept in − 80 °C until use. μMACS GFP Isolation Kit (product number 130–091-125) was used to separate SPL13 interacting proteins using the manufacturer’s instructions (Miltenyi Biotec, USA). In brief, plant tissue was ground using a mortar and a pestle in liquid nitrogen and 1 g tissue was used for analysis. Two ml of pre-cooled lysis buffer was added along with 50 μL proteinase inhibitor cocktail and kept on ice for 30 min with occasional mixing. The samples were centrifuged at 10000 g for 10 min and 50 μL of anti-GFP tag microbeads was added to the supernatant to magnetically label the epitope-tagged SPL13, mixed and kept on ice for 30 min. A total of 50 μL of the extract was kept as input control while the rest of the sample was eluted on μMCAs separator. After four rounds of stringent wash to remove non-interacting proteins with 200 μL of wash buffer 1, a final 100 μL wash buffer was used to remove the remaining unbound proteins and the eluent was used as the negative control. Finally, 20 μL of elution buffer (heated at 95 °C) was added to the column, and the column was incubated for 5 min before adding the final 50 μL of heated elution buffer and recovering the eluent.
SDS-PAGE electrophoresis
Proteins were resolved using SDS-polyacrylamide gel electrophoresis (PAGE). Samples were heated at 80 °C for 5 min to better resolve proteins. The electrophoresis was set for 30 min with 110 V followed by 125 v until the loading dye reached the bottom of the gel (~ 1.5 h) using vertical Mini-PROTEAN® Tetra cell system (BioRad, Catalog # 1658000, Mississauga, Canada). Gels were washed twice with distilled water and treated with Coomasie solution (Acetone: Methanol: distilled water 10:45:45 with 0.25% Brilliant Blue G-250), de stained with de-staining solution (Acetone: Methanol: distilled water 10:45:45) until protein bands were observed. The protein gels were then visualized using Gel DocTM EZ imager system (BioRad, Catalog # 1708270).
FASP – LC-MSMS-based protein identification
Proteins pulled down by immunoprecipitation were identified by LC-MS/MS with filter aided sample preparation (FASP), according to Wiśniewski et al., [57] with modifications. Briefly, 50 μl of the proteins eluted in the IP step were loaded onto a 10 kDa size exclusion Amicon® pro filter (Millipore Sigma Catalog # ACS501024). The solvent was exchanged with 150 μl of 8 M urea (Sigma, U5128), incubated at 40 °C for 25 min followed by centrifugation (14,000 G, 15 min). The elute was reduced with 100 mM DTT in Urea and alkylated with 100 μl of 50 mM iodoacetamide in a dark for 20 min. The proteins were washed twice with 100 μl of 50 mM ammonium bicarbonate in water and digested with 2 μg trypsin overnight at 34 °C. The digested peptides were then pulled through the molecular weight cut-off filter and recovered by washing with 1 ml 0.1% formic acid in LC-MS grade water. Solid-Phase Extraction (SPE) was performed using Waters OASIS HLB cartridge (Waters, SKU: 186003365) in a vacuum chamber. Subsequently, peptides were dried on a centrivap and reconstituted in 100 μL acetonitrile:water (95:5) with 0.1% formic acid and analyzed by a Thermo Q-Exactive Orbitrap mass spectrometer coupled to an Easy-nLC 1000 nanoLC system according to Fan et al. [14].
Database development and peptide identification for IPMS
To identify peptides in alfalfa, previous reports used either the Medicago truncatula proteome database [8, 62] or combinations of green plants (viridiplantae) [40, 44], despite relatively low identity matches. In the current study, we took a different approach by constructing a new peptide database derived from alfalfa transcriptomic contigs, followed by confirmation with a partial (2201 amino acid sequences) M. sativa database and M. truncatula protein database. Due to the absence of information on where the contig sequences are located relative to the genes transcription start sites (TSS), first, we translated 112,626 alfalfa transcript contigs followed by two additional frameshifts. When the translation process reached an early stop codon, the following sequences were assigned to the same contig but with a unique extension number (e.g. contig_1, contig_1_2). Moreover, any contig, or its extension, containing less than five amino acids were removed from the database. Subsequently, we were able to generate a 5,859,496 alfalfa peptide sequence database, from now onwards referred to as ‘contig database’ (Table S5). Peptide identity search from the samples was performed using the contig database and candidates containing at least three peptides and six spectra were considered for further analysis. The top SPL13-interacting candidate peptides were functionally annotated using the M. truncatula and M. sativa protein databases, by best fit. To confirm the results, peptide identification was repeated with the candidate peptide contigs replaced by M. truncatula homolog sequences in the contig database. Additionally, as of January 8, 2020, there are 2201 M. sativa protein sequences on the Uniprot protein database (https://www.uniprot.org/uniprot/) derived from manual annotation (140) and computational analysis (2061). Hence, we used the Uniprot database to determine whether the contig database-identified peptides are detectable with this analysis, and also to search for other SPL13-interacting proteins.
A fixed modification of carbamidomethylation (57.02) and a variable Oxidation modifications (15.99) of the target compounds were considered while undertaking the database search. Moreover, other protease and fragmentation parameters: such as trypsin as a cleavage enzyme; two maximum missed cleavages; precursor m/z tolerance of 12.0 ppm; fragment m/z tolerance of 0.08 Da, precursor charge of 2–4 with one isotope level were incorporated while searching and identifying for the candidate proteins. Peak lists obtained from MS/MS spectra were identified using X!Tandem version X! Tandem Vengeance (2015.12.15.2) [PMID 14976030], Andromeda version 1.5.3.4 [PMID 21254760] and MyriMatch version 2.2.140 [PMID 17269722]. The peptide search was conducted using SearchGUI version 3.3.20 [PMID 21337703] and were inferred from the spectrum identification results using PeptideShaker version 1.16.45 [PMID 25574629] [53].
Statistical analysis
All statistical data analyses were undertaken using R-software environment 4.0. Shell scripts of Linux and R-software scripts used to analyze and visualize, respectively, the RNA sequence reads are provided as a supplementary file (Table S7).
Supplementary information
Additional file 1: Fig. S1. Visualization of total exon read counts and library sizes generated from each biological samples. (a) Exon read counts library sizes, (b) log2 transformed exon read counts for constructing PCA plots, (c) Weighted Gene Co-expression Network Analysis (WGCNA) –based transcript analysis to visualize co-expression trend between genotypes and among tissues. Fig. S2 Visualization of differentially expressed genes-associated pathways between drought-stressed leaf tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S3 Visualization of differentially expressed genes-associated pathways between drought-stressed stem tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S4 Visualization of differentially expressed genes-associated pathways between drought-stressed root tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S5 Validation of selected differentially expressed genes using quantitative real-time PCR (qRT-PCR) in different tissues of SPL13RNAi-6 and EV under drought conditions. Although an observed fold-change value differences between RNAseq data and qRT-PCR a linear regression (R2 = 0.74) indicates a positive correlation between the two platforms but reflecting a different sensitivity. Genes of Medtr3g498825: basic helix-loop-helix 137 bHLH137; Medtr7g109510: basic leucine zipper bZIP; Medtr5g048850: flavonol synthase/flavanone 3-hydroxylase F3H; Medtr3g091350: flavonol synthase/flavanone 3-hydroxylase F3H-2; Medtr5g081860: MYB transcription factor MYB51; Medtr5g090620: UDP-glucose flavonoid 3-O-glucosyltransferase UDPFG; Medtr7g071050: UDP-glucosyltransferase family protein UDPGT were used for comparisons while acetyl-CoA carboxylase1 and ACTIN were used as reference genes. (PPTX 1486 kb)
Additional file 2: Table S1. List of genes differentially expressed between drought stressed SPL13RNAi and EV plants. DEG that are (a) increased and (b) decreased in drought-stressed SPL13RNAi plants compared to drought stressed EV plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 3: Table S2. List of genes differentially expressed between control and drought stressed SPL13RNAi plants. DEG that are (a) increased and (b) decreased in drought-stressed SPL13RNAi plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 4: Table S3. List of genes differentially expressed between control and drought stressed EV plants. DEG that are (a) increased and (b) decreased in drought-stressed EV plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 5: Table S4.1 Full list of components included in molecular function, biological process and cellular component in leaf tissuesTable S4.2 Full list of components included in molecular function, biological process and cellular component in stem tissuesTable S4.3 Full list of components included in molecular function, biological process and cellular component in root tissues
Additional file 6: Table S5 Medicago sativa contig database (FASTA 237506 kb)
Additional file 7: Table S6 List of candidate peptides interacting with SPL13
Additional file 8: Table S7.. Shell scripts of Linux and R-software scripts used to analyze and visualize data.
Acknowledgments
The authors acknowledge Dr. Chen Chen from AAFC-AAC, London, Ontario, Canada for his help with contig database development. Also, we would like to acknowledge Dr. Margaret Gruber for her help in editing the manuscript and Alex Molnar for his assistance with figures.
Abbreviations
- ChIP
Chromatin Immunoprecipitation
- CHS
Chalcone synthase
- COMT
Cafeic acid-3-O-methyltransferase
- DEG
Differentially expressed genes
- DFR
dihydroflavonol-4-reductase
- EV
Empty Vector construct
- FASP
Filter-assisted proteomics
- FLAP
Fasciclin-Like Arabinogalactan Protein
- GABA
Gamma aminobutyric acid
- GDSL
Gly-Asp- Ser-Leu –lipase
- GO
Gene Ontology
- GSK
Glycogen synthase kinase-3
- IFR
Isoflavone reductase
- IPMS
ImmunoPrecipitation-based Mass Spectrometry
- miR156
microRNA156
- NCBI
National Center for Biotechnology Information
- PAGE
Polyacrylamide gel electrophoresis
- PAL
Phenylalanine Ammonia-Lyase
- PCA
Principal Component Analysis
- PDH
Proline Dehydrogenase
- PP1
Serine/threonin-protein phosphatase
- ROS
Reactive Oxygen Species
- RUBISCO
Ribulose-1,5-bisphosphate carboxylase/oxygenase
- SPE
Solid-Phase Extraction
- TSS
Transcription Start Sites
- WGCNA
Weighted Gene Co-expression Network Analysis
Authors’ contributions
BAF selected the genotypes, performed the experiments and analyzed Omics data; JR helped with FASP-IPMS proteomics; VN assisted in handling plants and physiological data collection; SEK and AH supervised the project; BAF drafted the manuscript; BAF, JR, VN, SEK and AH edited the manuscript. The author(s) read and approved the final manuscript.
Funding
The research is funded through grants from Agriculture and Agri-Food Canada and the Natural Science and Engineering Research Council of Canada to AH. The funding agencies had no role in the design of the study; collection, analysis, and interpretation of data.
Availability of data and materials
Raw RNA sequencing reads can be accessed at the National Center for Biotechnology Information, NCBI, BioProject PRJNA598830.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare there is no competing of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-07118-4.
References
- 1.Agati G, Azzarello E, Pollastri S, Tattini M. Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 2012;196:67–76. doi: 10.1016/j.plantsci.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017;258:122–136. doi: 10.1016/j.plantsci.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Arshad M, Gruber MY, Hannoufa A. Transcriptome analysis of microRNA156 overexpression alfalfa roots under drought stress. Sci Rep. 2018;8:9363. doi: 10.1038/s41598-018-27088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aung B, Gruber MY, Amyot L, Omari K, Bertrand A, Hannoufa A. Ectopic expression of LjmiR156 delays flowering, enhances shoot branching, and improves forage quality in alfalfa. Plant Biotechnology Reports. 2015;9:379–393. doi: 10.1007/s11816-015-0375-2. [DOI] [Google Scholar]
- 5.Aung B, Gruber MY, Amyot L, Omari K, Bertrand A, Hannoufa A. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol J. 2015;13:779–790. doi: 10.1111/pbi.12308. [DOI] [PubMed] [Google Scholar]
- 6.Badhan A, Jin L, Wang Y, Han S, Kowalczys K, Brown DC, Ayala CJ, Latoszek-Green M, Miki B, Tsang A, McAllister T. Expression of a fungal ferulic acid esterase in alfalfa modifies cell wall digestibility. Biotechnol Biofuels. 2014;7:39. doi: 10.1186/1754-6834-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaskara GB, Yang T-H, Verslues PE. Dynamic proline metabolism: importance and regulation in water limited environments. Front Plant Sci. 2015;6:484. doi: 10.3389/fpls.2015.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai C, Cui W, Pan J, Xie Y, Wang J, Shen W. Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J Proteome. 2017;152:109–120. doi: 10.1016/j.jprot.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, Rozhon W, Heuck A, Clausen T, Jonak C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2012;24:3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dao TTH, Linthorst HJM, Verpoorte R. Chalcone synthase and its functions in plant resistance. Phytochemistry Rev. 2011;10:397–412. doi: 10.1007/s11101-011-9211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X, Bashandy H, Ainasoja M, Kontturi J, Pietiäinen M, Laitinen RAE, Albert VA, Valkonen JPT, Elomaa P, Teeri TH. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 2014;201:1469–1483. doi: 10.1111/nph.12610. [DOI] [PubMed] [Google Scholar]
- 12.Dhingra A, Portis AR, Jr, Daniell H. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci U S A. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elfving DC, Kaufmann MR, Hall AE. Interpreting leaf water potential measurements with a model of the soil-plant-atmosphere continuum. Physiol Plant. 1972;27:161–168. doi: 10.1111/j.1399-3054.1972.tb03594.x. [DOI] [Google Scholar]
- 14.Fan W, Ge G, Liu Y, Wang W, Liu L, Jia Y. Proteomics integrated with metabolomics: analysis of the internal causes of nutrient changes in alfalfa at different growth stages. BMC Plant Biol. 2018;18:78. doi: 10.1186/s12870-018-1291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feyissa BA, Arshad M, Gruber MY, Kohalmi SE, Hannoufa A. The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa. BMC Plant Biol. 2019;19:434. doi: 10.1186/s12870-019-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiil BK, Petersen K, Petersen M, Mundy J. Gene regulation by MAP kinase cascades. Curr Opin Plant Biol. 2009;12:615–621. doi: 10.1016/j.pbi.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Gao R, Gruber MY, Amyot L, Hannoufa A. SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Mol Biol. 2018;96:119–133. doi: 10.1007/s11103-017-0683-8. [DOI] [PubMed] [Google Scholar]
- 18.Ghafoor R, Akram NA, Rashid M, Ashraf M, Iqbal M, Lixin Z. Exogenously applied proline induced changes in key anatomical features and physio-biochemical attributes in water stressed oat (Avena sativa L.) plants. Physiol Mol Biol Plants. 2019;25:1121–1135. doi: 10.1007/s12298-019-00683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 20.Hahn A, Harter K. Mitogen-activated protein kinase cascades and ethylene: signaling, biosynthesis, or both? Plant Physiol. 2009;149:1207–1210. doi: 10.1104/pp.108.132241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannoufa A, Matthews C, Feyissa BA, Gruber MY, Arshad M. Progress in Botany. Berlin, Heidelberg: Springer; 2018. Progress toward deep sequencing-based discovery of stress-related MicroRNA in plants and available bioinformatics tools. [Google Scholar]
- 22.Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997;21:79–102. doi: 10.1023/A:1005703923347. [DOI] [Google Scholar]
- 23.Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayat S, Hayat Q, Alyemeni MN, Wani AS, Pichtel J, Ahmad A. Role of proline under changing environments: a review. Plant Signaling Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houtz RL, Portis AR. The life of ribulose 1,5-bisphosphate carboxylase/oxygenase—posttranslational facts and mysteries. Arch Biochem Biophys. 2003;414:150–158. doi: 10.1016/S0003-9861(03)00122-X. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y-H, Yu J-Q, Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson KL, Jones BJ, Bacic A, Schultz CJ. The Fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller MA, Piedrafita G, Ralser M. The widespread role of non-enzymatic reactions in cellular metabolism. Curr Opin Biotechnol. 2015;34:153–161. doi: 10.1016/j.copbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B. Brassinosteroid-regulated GSK3/shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J Biol Chem. 2013;288:7519–7527. doi: 10.1074/jbc.M112.384453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T-W, Michniewicz M, Bergmann DC, Wang Z-Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Li G, Li Y, Kong X, Zhang L, Wang J, Li X, Yang Y. ABA receptor subfamily III enhances abscisic acid sensitivity and improves the drought tolerance of Arabidopsis. Int J Mol Sci. 2018;19:1938. doi: 10.3390/ijms19071938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Liu T, Zhou T, Lian M, Liu T, Hou J, Ijaz R, Song B. Genome-wide identification and characterization of the AREB/ABF/ABI5 subfamily members from Solanum tuberosum. Int J Mol Sci. 2019;20:311. doi: 10.3390/ijms20020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews C, Arshad M, Hannoufa A. Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant. 2019;165:830–842. doi: 10.1111/ppl.12787. [DOI] [PubMed] [Google Scholar]
- 36.Meinzer FC, Clearwater MJ, Goldstein G. Water transport in trees: current perspectives, new insights and some controversies. Environ Exp Bot. 2001;45:239–262. doi: 10.1016/S0098-8472(01)00074-0. [DOI] [PubMed] [Google Scholar]
- 37.Nuñez-Palenius HG, Ochoa-Alejo N. Effect of phenylalanine and phenylpropanoids on the accumulation of capsaicinoids and lignin in cell cultures of chili pepper (Capsicum annuum L.) In Vitro Cellular and Developmental Biology - Plant. 2005;41:801–805. doi: 10.1079/IVP2005708. [DOI] [Google Scholar]
- 38.Olesen JE, Trnka M, Kersebaum KC, Skjelvåg AO, Seguin B, Peltonen-Sainio P, Rossi F, Kozyra J, Micale F. Impacts and adaptation of European crop production systems to climate change. Eur J Agron. 2011;34:96–112. doi: 10.1016/j.eja.2010.11.003. [DOI] [Google Scholar]
- 39.Pokoo R, Ren S, Wang Q, Motes CM, Hernandez TD, Ahmadi S, Monteros MJ, Zheng Y, Sunkar R. Genotype- and tissue-specific miRNA profiles and their targets in three alfalfa (Medicago sativa L) genotypes. BMC Genomics. 2018;19:913. doi: 10.1186/s12864-018-5280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman MA, Yong-Goo K, Iftekhar A, G-s L, Hyoshin L, Joo LJ, Byung-Hyun L. Proteome analysis of alfalfa roots in response to water deficit stress. J Integr Agric. 2016;15:1275–1285. doi: 10.1016/S2095-3119(15)61255-2. [DOI] [Google Scholar]
- 41.Ramya R, Kalaiselvi M, Narmadha R, Gomathi D, Bhuvaneshwari V, Amsaveni R, Devaki K. Secondary metabolite credentials and in vitro free radical scavenging activity of Alpinia calcarata. J Acute Med. 2015;5:33–37. doi: 10.1016/j.jacme.2015.02.005. [DOI] [Google Scholar]
- 42.Ray DK, Gerber JS, MacDonald GK, West PC. Climate variation explains a third of global crop yield variability. Nat Commun. 2015;6:5989. doi: 10.1038/ncomms6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez MCS, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 44.Sergeant K, Printz B, Guerriero G, Renaut J, Lutts S, Hausman J-F, Tian L. The dynamics of the cell wall proteome of developing alfalfa stems. Biology. 2019;8:60. doi: 10.3390/biology8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slama I, Abdelly C, Bouchereau A, Flowers T, Savouré A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann Bot. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spreitzer RJ, Salvucci ME. RUBISCO: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- 47.Sun S, Wang T, Wang L, Li X, Jia Y, Liu C, Huang X, Xie W, Wang X. Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat Commun. 2018;9:2523. doi: 10.1038/s41467-018-04952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS One. 2015;10:e0119054. doi: 10.1371/journal.pone.0119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki Y, Makino A. Availability of rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol. 2012;160:533–540. doi: 10.1104/pp.112.201459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theocharis A, Clément C, Barka EA. Physiological and molecular changes in plants grown at low temperatures. Planta. 2012;235:1091–1105. doi: 10.1007/s00425-012-1641-y. [DOI] [PubMed] [Google Scholar]
- 52.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7:562. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol. 2015;33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Fan W, Li H, Yang J, Huang J, Zhang P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS One. 2013;8:e78484. doi: 10.1371/journal.pone.0078484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Zhou P, Zhu W, Wang F. De novo comparative Transcriptome analysis of genes differentially expressed in the Scion of Homografted and Heterografted tomato seedlings. Sci Rep. 2019;9:20240. doi: 10.1038/s41598-019-56563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wingler A, Quick WP, Bungard RA, Bailey KJ, Lea PJ, Leegood RC. The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant, Cell and Environment. 1999;22:361–373. doi: 10.1046/j.1365-3040.1999.00410.x. [DOI] [Google Scholar]
- 57.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 58.Xu K, Zhang H, Blumwald E, Xia T. A novel plant vacuolar Na+/H+ antiporter gene evolved by DNA shuffling confers improved salt tolerance in yeast. J Biol Chem. 2010;285:22999–23006. doi: 10.1074/jbc.M109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Youn J-H, Kim T-W. Functional iInsights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant. 2015;8:552–565. doi: 10.1016/j.molp.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Zang L, Zheng T, Chu Y, Ding C, Zhang W, Huang Q, Su X. Genome-wide analysis of the fasciclin-like arabinogalactan protein gene family reveals differential expression patterns, localization, and salt stress response in Populus. Front Plant Sci. 2015;6:1140. doi: 10.3389/fpls.2015.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zenda T, Liu S, Wang X, Liu G, Jin H, Dong A, Yang Y, Duan H. Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting inbred lines. Int J Mol Sci. 2019;20:1268. doi: 10.3390/ijms20061268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng N, Yang Z, Zhang Z, Hu L, Chen L. Comparative transcriptome combined with proteome analyses revealed key factors involved in alfalfa (Medicago sativa) response to waterlogging stress. Int J Mol Sci. 2019;20:1359. doi: 10.3390/ijms20061359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Lei L, Lai J, Zhao H, Song W. Effects of drought stress and water recovery on physiological responses and gene expression in maize seedlings. BMC Plant Biol. 2018;18:68. doi: 10.1186/s12870-018-1281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Visualization of total exon read counts and library sizes generated from each biological samples. (a) Exon read counts library sizes, (b) log2 transformed exon read counts for constructing PCA plots, (c) Weighted Gene Co-expression Network Analysis (WGCNA) –based transcript analysis to visualize co-expression trend between genotypes and among tissues. Fig. S2 Visualization of differentially expressed genes-associated pathways between drought-stressed leaf tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S3 Visualization of differentially expressed genes-associated pathways between drought-stressed stem tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S4 Visualization of differentially expressed genes-associated pathways between drought-stressed root tissues of SPL13RNA and EV plants. (a) Molecular function tree map, (b) Biological process tree map, (c) Cellular component tree map. The free online gene ontology analysis tool (http://revigo.irb.hr/) was used to generate codes to construct tree map in R-software. Fig. S5 Validation of selected differentially expressed genes using quantitative real-time PCR (qRT-PCR) in different tissues of SPL13RNAi-6 and EV under drought conditions. Although an observed fold-change value differences between RNAseq data and qRT-PCR a linear regression (R2 = 0.74) indicates a positive correlation between the two platforms but reflecting a different sensitivity. Genes of Medtr3g498825: basic helix-loop-helix 137 bHLH137; Medtr7g109510: basic leucine zipper bZIP; Medtr5g048850: flavonol synthase/flavanone 3-hydroxylase F3H; Medtr3g091350: flavonol synthase/flavanone 3-hydroxylase F3H-2; Medtr5g081860: MYB transcription factor MYB51; Medtr5g090620: UDP-glucose flavonoid 3-O-glucosyltransferase UDPFG; Medtr7g071050: UDP-glucosyltransferase family protein UDPGT were used for comparisons while acetyl-CoA carboxylase1 and ACTIN were used as reference genes. (PPTX 1486 kb)
Additional file 2: Table S1. List of genes differentially expressed between drought stressed SPL13RNAi and EV plants. DEG that are (a) increased and (b) decreased in drought-stressed SPL13RNAi plants compared to drought stressed EV plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 3: Table S2. List of genes differentially expressed between control and drought stressed SPL13RNAi plants. DEG that are (a) increased and (b) decreased in drought-stressed SPL13RNAi plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 4: Table S3. List of genes differentially expressed between control and drought stressed EV plants. DEG that are (a) increased and (b) decreased in drought-stressed EV plants. The differentially expressed genes are categorized as common to all tissues followed by tissue-specific (leaf, stem, or root tissues) expressions. N = 3 biological samples for each condition, where biological samples represent individual plants treated with similar treatment exposure.
Additional file 5: Table S4.1 Full list of components included in molecular function, biological process and cellular component in leaf tissuesTable S4.2 Full list of components included in molecular function, biological process and cellular component in stem tissuesTable S4.3 Full list of components included in molecular function, biological process and cellular component in root tissues
Additional file 6: Table S5 Medicago sativa contig database (FASTA 237506 kb)
Additional file 7: Table S6 List of candidate peptides interacting with SPL13
Additional file 8: Table S7.. Shell scripts of Linux and R-software scripts used to analyze and visualize data.
Data Availability Statement
Raw RNA sequencing reads can be accessed at the National Center for Biotechnology Information, NCBI, BioProject PRJNA598830.