Abstract
Phosphate-solubilizing fungi (PSF) play an important role in increasing the bioavailability of phosphorus in soils for plants. Thirteen fungal strains, one collected from air and 12 from soil, were screened and described here in detail. These fungal strains were tested for their ability to solubilize tricalcium phosphate (TCP) on both solid and liquid Pikovskaya (PVK) media in vitro. The airborne fungal strain KUMCC 18-0196 (Aspergillus hydei sp. nov.) showed the most significant phosphate solubilizing activity on a solid PVK medium with the solubilization index (SI) (2.58 ± 0.04 cm) and the highest solubilized phosphates (1523.33 ± 47.87 μg/mL) on a liquid PVK medium. To the best of our knowledge, A. hydei sp. nov. is the first phosphate-solubilizing fungus reported from air. We also provide the identification especially for Aspergillus, Penicillium and Talaromyces, generally reported as PSF. It is important to not only screen for PSF but also identify species properly so that researchers have a clearer taxonomic picture for identifying potential taxa for future plant growth-promoting applications. Herein, A. hydei (section Nigri), Gongronella hydei, Penicillium soli (section Lanata-Divaricata) and Talaromyces yunnanensis (section Talaromyces) are fully described and introduced as new to science. These four new species are identified based on both morphological characteristics and multigene phylogenetic analyses, including the genealogical concordance phylogenetic species recognition method where necessary. Penicillium austrosinense is considered to be a synonym of P. guaibinense.
Keywords: Aspergillaceae, Cunninghamellaceae, phylogeny, Quercus spp., taxonomy
Introduction
Fungi are widely distributed across terrestrial, marine, and freshwater environments (Jones et al., 2019; Phookamsak et al., 2019; Dayarathne et al., 2020; Dong et al., 2020). Fungi play important roles in both economics and ecology as sources of food and medicine that also provide decomposition services (De Silva et al., 2012a,b; Lange, 2014; Hyde et al., 2019b). Moreover, fungal research can lead to breakthroughs in microbial biotechnology and other industries (Hyde et al., 2018; Solomon et al., 2019). However, fungi can also cause diseases in humans, animals, and plants, posing numerous health challenges in need of addressing (Giacomazzi et al., 2016; Hyde et al., 2018). Given the overlooked position of studies on phosphate-solubilizing fungi (PSF), there are likely many novel fungal species still awaiting discovery.
Phosphate-solubilizing fungi (PSF) are able to enhance the solubilization of insoluble phosphate (P) compounds. They also have the capacity to mobilize and increase nutrient uptake, produce organic acids and increase the efficiency of phosphate fertilizers, such as superphosphate and rock phosphate (Jyoti et al., 2013; Li et al., 2015). Some PSF promote plant growth by secreting indole-3-acetic acid (IAA) and siderophore (Rahi et al., 2009; Zhang et al., 2018). They have enormous potential in enhancing the release of phosphorous from fertilizer (Zhang et al., 2018). PSF in soils, particularly filamentous fungi such as Aspergillus and Penicillium, have been widely investigated (Elias et al., 2016; Li Z. et al., 2016), but airborne fungi remain to be explored. In addition, previous identifications of PSF (e.g., Aspergillus, Penicillium, and Talaromyces) have been based on morphology, nucleotide similarities in MegaBLAST searches of NCBI’s GenBank nucleotide database and/or phylogenetic analysis of ITS sequence data (e.g., Vyas et al., 2007; Nelofer et al., 2015; Qiao et al., 2019).
In this paper, 13 fungal strains from air and soil samples were screened for their ability to solubilize phosphate. Known species are identified on the basis of phylogenetic analyses. New species are introduced based on morphological characteristics and phylogenetic analyses. This study provides new scientific insights into novel PSF isolated from the air. In addition, new fungal taxa isolated from the soil of cultivated oak seedlings (Quercus rubra) in Honghe County, Yunnan Province, China are introduced.
Materials and Methods
Collection and Isolation of Fungi From Air Using the Settle Plate Method
Five Petri plates (9 cm diam.) with opened lids containing PVK agar with 5% tricalcium phosphate [TCP, Ca3(PO4)2] were placed under a Quercus variabilis tree in Kunming City, Yunnan Province, China and left for 20 min. Quercus variabilis is a selected host in this study as it is a native oak in China. Lids were then closed over the plates and carefully transferred to the laboratory where they were incubated for 2 days at room temperature. The fungal colony with the widest clear zone around the colony on PVK agar was selected and transferred to fresh potato dextrose agar (PDA) to obtain a phosphate-solubilizing fungus.
Collection and Isolation of Fungi From Soil Using Soil Dilutions
Twenty grams of rhizospheric soil samples (10 cm depth) were collected from Quercus rubra in Honghe County, Yunnan Province, China. Quercus rubra is also a selected host herein as it is the most valuable timber species in red oak group. Soil samples were separated from roots, stored in ziplock bags and carefully transferred to the laboratory. One gram of each soil sample was dissolved into 9 mL of sterilized distilled water in 15 mL test tubes and mixed well. The serial soil dilutions were made for 10–4, 10–5, 10–6. Soil dilutions were used to isolate fungi from soil samples. Half a milliliter (0.5 mL) of each dilution was drawn using a micropipette and placed onto a PDA plate supplemented with 100 mg/mL streptomycin and spread using a sterile L-shaped spreader. The Petri plates were then inoculated for 2–5 days at room temperature. Fungal colonies were purified by transferring a single hyphal tip to PDA. Pure cultures of each fungal isolate were preserved on PDA agar slants in 2 mL vial tubes at 4°C for further screening.
Determination of Solubilization Index on PVK Agar
Thirteen fungal isolates obtained from pure cultures were preliminarily screened for their potential to solubilize TCP as insoluble inorganic phosphate sources on PVK agar. One liter (1 L) of PVK agar contained the following (g/L): 0.5 g (NH4)2SO4, 0.1 g MgSO4⋅7H2O, 0.02 g NaCl, 0.02 g KCl, 0.003 g FeSO4⋅7H2O, 0.003 g MnSO4⋅H2O, 5 g Ca3 (PO4)2, 10 g glucose, 0.5 g yeast extract, 15 g agar, and 1000 mL distilled water (Pikovskaya, 1948). The medium was autoclaved at 121°C for 15 min. Sterilized PVK agar was poured into sterilized Petri plates. Fungal mycelium plugs of each isolate grown on PDA at 28°C for 7 days were cut from the edges of each actively growing colony using a sterile cork borer (5 mm3). Fungal mycelium plugs were then placed on Petri plates containing PVK agar supplemented with 0.5% TCP for 7 days at 28°C. Plugs of sterile PDA were used as controls. Three replicates were tested for each fungal isolate. The diameter of clear zones around the colony of each isolate was measured after the 1st, 3rd, 5th, and 7th day of incubation. The phosphate solubilization index was calculated according to the formula below (Premono et al., 1996).
Phosphate Solubilization Efficiency of Fungal Isolates to Tricalcium Phosphate in PVK Broth
The phosphate solubilization activity test was carried out in a 150 mL conical flask with 100 mL PVK broth supplemented with 0.5% TCP. The initial pH of the medium was adjusted to 7.0 before sterilization. The preparation of spore suspensions from each fungal isolate was done according to the method described in Elias et al. (2016). Ten milliliter (10 mL) spore suspensions of each fungal culture (107 spores/mL) were inoculated in the sterilized PVK broth of each conical flask. Ten milliliters (10 mL) of sterile distilled water in the sterilized PVK broth was treated as the control. Three replicates were maintained for each fungal test. The cultures were incubated on a rotary shaker (Sanco, India) at 28°C at 130 rpm for 7 days. Aliquots of 1.5 mL culture supernatant were aseptically collected from each conical flask on the 1st, 3rd, 5th, and 7th day. The samples were centrifuged (Sigma, Germany) at 12,000 rpm for 2 min to discard any suspended solids and mycelial fragments. Supernatants (0.1 mL) of each culture were then taken out to estimate the amount of phosphorus released from TCP of fungal strains. The available soluble phosphate in culture supernatants was estimated using the molybdenum blue method under 700 nm (Ryan et al., 2001). The pH of the culture supernatant in PVK broth was also measured with a digital pH meter FE 28 equipped with a glass electrode (Shanghai, Co., Ltd.).
Statistical Analysis
Data from different treatments were calculated and statistically analyzed with a one-way analysis of variance (ANOVA) using the Statistical Package for the Social Sciences (SPSS) for Windows version 22 (SPSS, Inc., Chicago, IL, United States). Duncan’s multiple range tests were used to determine significant differences at p < 0.05 between the mean values.
Morphological Studies
The fungi were identified using morphological characteristics and phylogenetic analyses. New taxa are introduced with full descriptions. Fungal strains grown on PDA and MEA were mounted in lactic acid for microscopic examination. Morphological observations and photomicrographs were made with a Nikon ECLIPSE 80i compound microscope fitted with a Canon 600D digital camera. Morphological characteristic measurements were made with the Tarosoft (R) Image Frame Work. Images used for figures were processed with Adobe Photoshop CS6 (Adobe Systems, United States).
Type herbarium materials were deposited in the herbarium of Mae Fah Luang University (MFLU) Chiang Rai, Thailand. Ex-type living cultures were deposited in the Kunming Institute of Botany Culture Collection (KMUCC) Yunnan, China.
DNA Extraction, PCR Amplification, and Sequencing
The DNA was extracted from the mycelium grown on PDA at 25°C for 7 days using a Biospin Fungus Genomic DNA Extraction Kit–BSC14S1 (BioFlux®, China) following the manufacturer’s protocol. The 5.8S nrRNA gene with the two flanking internal transcribed spacers (ITS) regions were first amplified and sequenced using the primers ITS1/ITS4 (White et al., 1990) for all fungal strains. They were preliminarily identified to genera level as Aspergillus, Penicillium, Talaromyces, Gongronella, and Fusarium based on the BLAST searches of ITS in the nucleotide database of GenBank1. Partial calmodulin gene (CaM) using primers CMD5/CMD6 (Hong et al., 2005), partial beta-tubulin gene (BenA) using primers Bt2a/Bt2b (Glass and Donaldson, 1995) and partial RNA polymerase II second largest subunit (RPB2) using primers 5F/7CR (Liu et al., 1999) were then amplified and sequenced for strains of Aspergillus, Penicillium, and Talaromyces. The LSU gene region was amplified and sequenced using the primers LR0R and LR5 (Vilgalys and Hester, 1990; Rehner and Samuels, 1994) for the strains of Gongronella. Translation elongation factor 1-alpha (TEF1-α) was amplified and sequenced using primers TEF1/TEF2 (O’Donnell et al., 1998) for the strain of Fusarium but was unsuccessful even after several efforts.
The PCR amplifications were conducted in 25 μL final volumes which consisted of 12.5 μL of 2 × Power Taq PCR MasterMix (a premix and ready to use solution, including 0.1 Units/μL Taq DNA Polymerase, 500 μM dNTP Mixture each (dATP, dCTP, dGTP, dTTP), 20 mM Tris-HCl pH 8.3, 100 mM KCl, 3 mM MgCl2, stabilizer and enhancer), 1 μL of each primer (10 μM), 1 μL genomic DNA extract and 9.5 μL distilled-deionized water. Thermocycling was completed on a 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, United States) under the following conditions: initial denaturation of 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 2 min at 52°C, 90 s at 72°C and a final extension period of 10 min at 72°C for ITS and RPB2. The annealing temperature was changed to 55°C for CaM, LSU, TEF, and BenA. The amplified PCR products were purified and sequenced by BGI Tech Solutions, Co. Ltd. (BGITech), China.
Phylogenetic Analyses
Consensus sequences were generated using Geneious® R7 (Biomatters, Ltd., New Zealand) and BioEdit (Hall, 1999). Sequences were BLAST searched in the nucleotide database of GenBank1 to examine their closely related taxa. The reference sequences and outgroups were selected from recent relevant literature and GenBank (Supplementary Table S1). The datasets of genomic regions were aligned individually using the MAFFT online version 7.450 server2. Phylogenetic trees were inferred with maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI), according to the details described in Wanasinghe et al. (2018).
Genealogical Concordance Phylogenetic Species Recognition (GR)
The new Aspergillus taxon and closely related species were analyzed separately using the GCPSR model. The pairwise homoplasy index (PHI) was determined using SplitsTree v. 4.15.1 (Huson and Bryant, 20063) as described by Taylor et al. (2000) and Quaedvlieg et al. (2014) to determine the recombination level within phylogenetically closely related species. A PHI value below 0.05 (Φw < 0.05) indicates the presence of significant recombination in the dataset. Split graphs were constructed to ease visualization of the relationship between closely related species. The relationships between this new taxon and closely related species were visualized by constructing splits graphs using both the LogDet transformation and splits decomposition options.
Results
Formation of Clear Zones and Phosphate Solubilization Index (SI) on PVK Agar
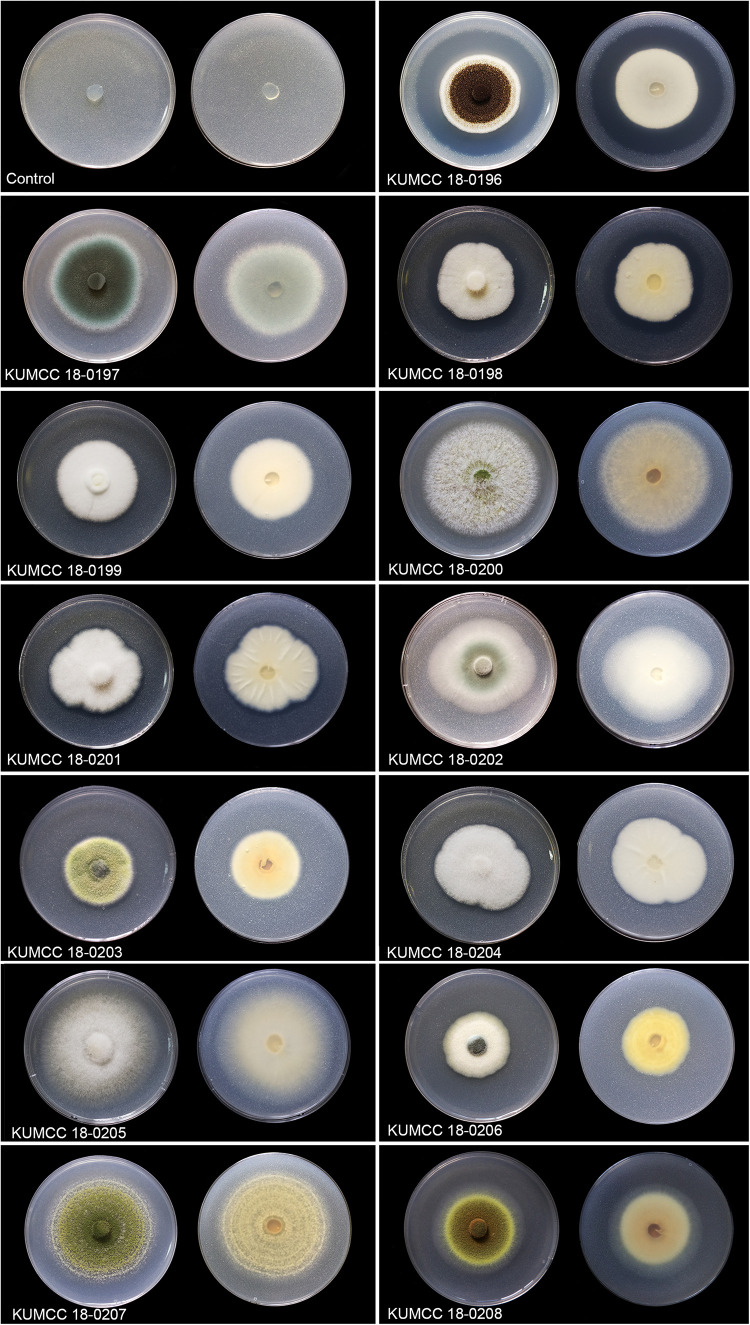
The clear zone became visible in the isolates KUMCC 18-0196 and KUMCC 18-0206 on the 1st day. The clear zone of the other fungal isolates became visible on the 3rd day and did not show the clear zone around colonies after the 7th day but solubilized TCP by removing white spots on PVK agar (Figure 1). The isolate KUMCC 18-0196 showed the widest clear zone around the fungal colony (Figure 1).
FIGURE 1.
Colony features and clear zones formation for TCP solubilization on PVK agar after 7 days Except isolate KUMCC 18-0196 after 5 days (left = above view, right = below).
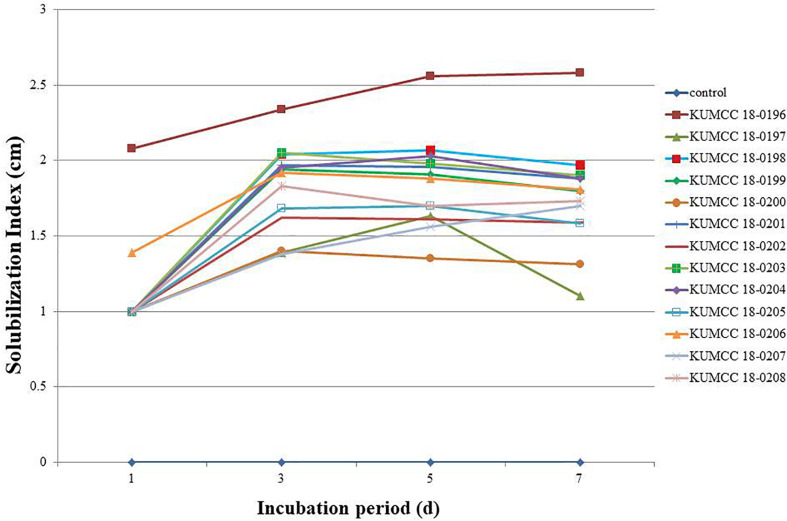
The phosphate solubilization index began increasing in isolates KUMCC 18-0196 and KUMCC 18-0206 on the 1st day, while in other fungal isolates it began increasing on the 3rd day (Figure 2). The highest phosphate solubilization index was seen in KUMCC 18-0196 (SI = 2.58 ± 0.04 cm) compared with other fungal strains and the control on the 7th day, followed by KUMCC 18-0198 with SI = 1.97 ± 0.15 cm and KUMCC 18-0203 with SI = 1.90 ± 0.01 cm (Supplementary Table S2). It was observed that isolate KUMCC 18-0197 had the smallest SI = 1.10 ± 0.17 cm. The phosphate solubilization of isolate KUMCC 18-0196 was significantly different (p < 0.05) from other fungal strains (Supplementary Table S2).
FIGURE 2.
Solubilization index (SI) of 13 fungal strains and control on PVK agar containing TCP after the 1st, 3rd, 5th, and 7th day of incubation (each value represents the mean of the three replicates).
Phosphate Solubilization and pH in PVK Broth
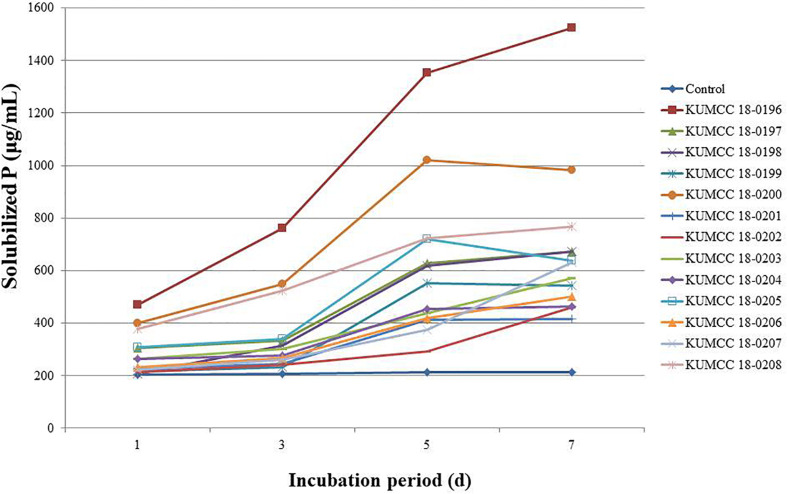
All fungal strains released phosphate on the 1st day and gradually increased when compared with the control. Isolates KUMCC 18-0199, KUMCC 18-0200 and KUMCC 18-0205 later decreased on the 7th day of incubation, while isolates KUMCC 18-0196, KUMCC 18-0197, KUMCC 18-0198, KUMCC 18-0201, KUMCC 18-0202, KUMCC 18-0203, KUMCC 18-0204, KUMCC 18-0206, KUMCC 18-0207, and KUMCC 18-0208 increased (Figure 3). Isolate KUMCC 18-0196 released solubilized phosphate (1523.33 ± 47.87 μg/mL) with significant difference (p < 0.05) compared to other fungal strains (Supplementary Table S2). The smallest amount of solubilized phosphate was detected in isolate KUMCC 18-0201 (417.33 ± 22.00 μg/mL) (Supplementary Table S2).
FIGURE 3.
Solubilized P concentrations of 13 fungal strains and control in PVK broth containing TCP after the 1st, 3rd, 5th, and 7th day of incubation (each value represents the mean of the S3). pH values of PVK broth containing TCP inoculated with each 13 fungal isolate and control after the 1st, 3rd, 5th, and 7th day of incubation (each value represents the mean of the three replicates).
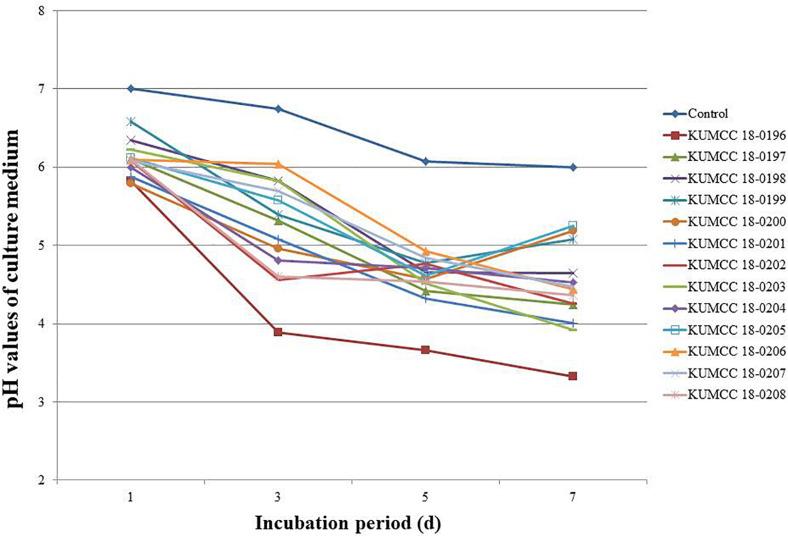
The pH values of all 13 fungal isolates decreased from 7.01 after the 1st day of incubation. The pH values continued to decrease after the 3rd day and increased or decreased to variable levels in the PVK broth containing TCP during the 5th and 7th days of incubation, depending on the fungal isolates (Figure 4). The isolate KUMCC 18-0196 showed the highest decrease in pH from an initial pH of 7.01 to 3.33 ± 0.01 on the 7th day of incubation, which was significantly different compared to the control and other fungal strains (Supplementary Table S2).
FIGURE 4.
pH values of PVK broth containing TCP inoculated with each 13 fungal isolate and control after the 1st, 3rd, 5th, and 7th day of incubation (each value represents the mean of the three replicates).
Taxonomic and Phylogenic Studies
In total, 13 fungal strains screened for TCP solubilization on both solid and liquid PVK media are listed in Supplementary Table S3. Five strains are introduced as new species viz. Aspergillus hydei, Gongronella hydei, Penicillium soli and Talaromyces yunnanensis, based on morphological characteristics and phylogenetic analyses. Penicillium guaibinense collected from soil in China is also illustrated and described. Their morphological characteristics and phylogenetic analyses are provided below.
Aspergillus Section Nigri W. Gams, M. Chr., Onions, Pitt, and Samson
Type: Aspergillus niger Tiegh.
Notes: Aspergillus section Nigri is the black aspergilli. This section includes species with smooth conidiophores and hyaline or pigmented below the vesicle, globose, subglobose to pyriform vesicles, typically radiate conidial heads, or divergent columns in some species, and black conidia (Gams et al., 1986). They have been isolated from various habitats, such as plants, contaminated materials, indoor air environments and soil samples (Sørensen et al., 2011; Visagie et al., 2014a; Fungaro et al., 2017; Serra et al., 2017; this study). Many strains of Aspergillus niger are used in biotechnology for the production of various metabolites, such as in antibiotics, organic acids, bioenergy or as microbial fermentation in food (Hasegawa et al., 2007; Show et al., 2015; Chuppa-Tostain et al., 2018). They have also been reported as the pathogen responsible for invasive pulmonary aspergillosis (Walsh et al., 2007) and other plant pathogens (Gautam et al., 2011; Sharma, 2012; Guo et al., 2017). Black aspergilli are hard to differentiate solely by morphological characteristics due to high species variation. We introduce the new species A. hydei (KUMCC 18-0196) from air based on morphological characteristics and multi-loci phylogenetic analyses, including the GCPSR method.
Aspergillus hydei Doilom, sp. nov., Figure 5
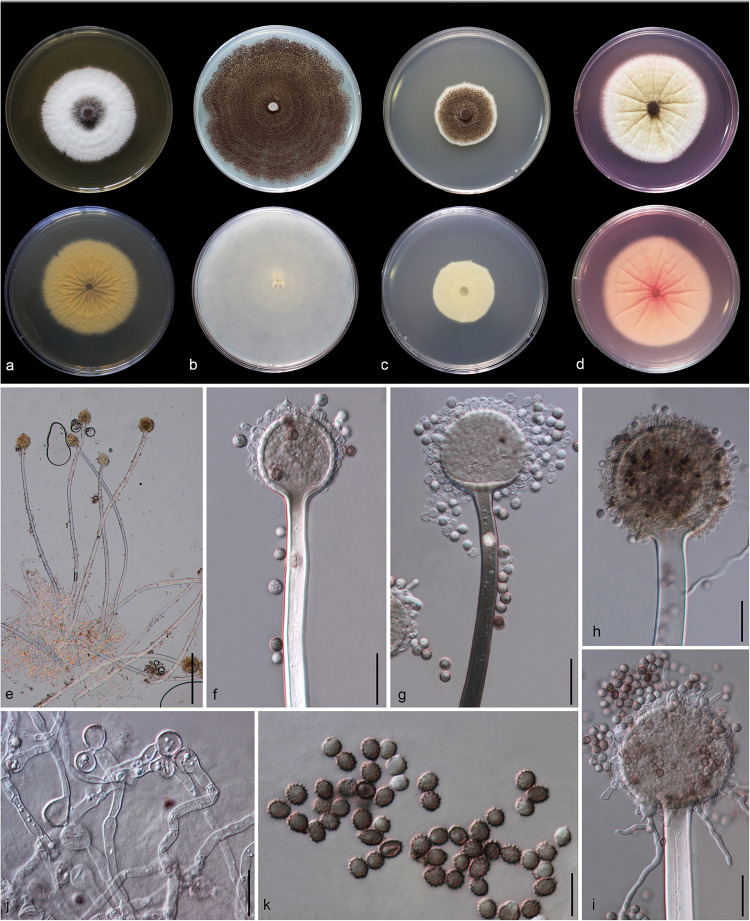
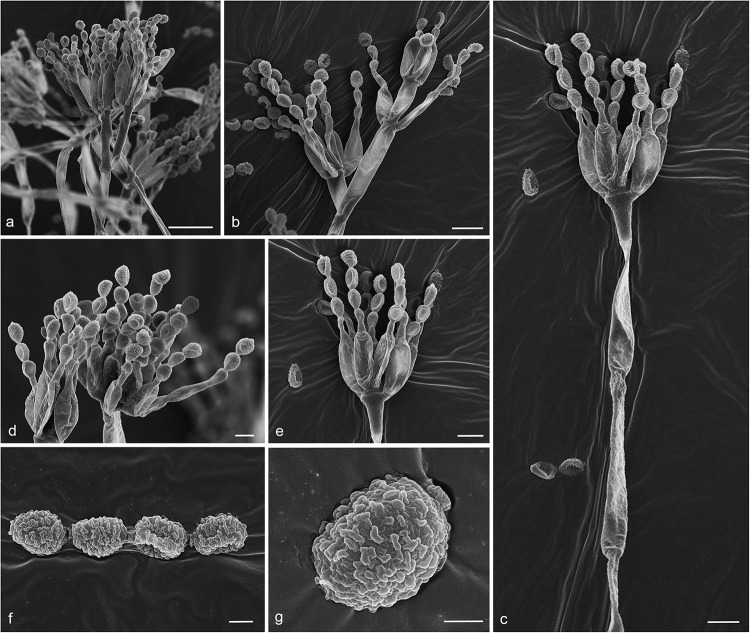
FIGURE 5.
Aspergillus hydei (MFLU 20-0430, holotype). (a–d) Colonies on MEA, CYA, PDA and Rosebengal at 25°C, respectively. (e) Conidiophores. (f–i) Close-up of conidiophores, vesicle, phialides, and conidia (f,g: exhibiting colors of polarized light under DIC illumination). (j) Chlamydospores. (k) Conidia. Scale bars: (e) = 200 μm, (f–j) = 20 μm, (k) = 10 μm.
Index Fungorum number: IF 557860; Facesoffungi number: FoF 08837
Etymology: Named in honor of Kevin D. Hyde for his excellent contributions to mycology.
Holotype: MFLU 20-0430
Macromorphology: Colonies on MEA reaching 50–55 mm diam. after 7 days in the dark at 25°C, circular to filamentous, undulate to filamentous, flat or effuse to raise, dense, compact white basal felt covered by a dense layer of dark brown to black conidial heads in the center, white at the edge from above; yellow or creamy, wrinkled and cracked from below. Colonies on Czapeck Yeast Agar (CYA) reaching 75–80 mm diam. after 7 days in the dark at 25°C, irregular in shape, curled to undulate, flat or effuse, compact white basal felt covered by a dense layer of black conidial heads, with concentric rings from above; white from below. Colonies on PDA reaching 35–40 mm diam. after 7 days in the dark at 25°C, filamentous shape, undulate to filamentous, flat or effuse to raise, dense, compact white basal felt covered by a dense layer of dark brown to black conidial heads, white at the edge from above; white or creamy from below. Colonies on WA reaching 50–55 mm diam. after 7 days in the dark at 25°C, slightly circular, edge entire to undulate, flat, loose, white with black conidial heads at the center (mycelium plug) from above; white from below. Colonies on Rosebengal reaching 60–65 mm diam. after 7 days in the dark at 25°C, slightly circular, edge entire to undulate, flat, loose, compact white basal felt covered by a lose layer of cream conidial heads with gray conidial heads at the center (mycelium plug), with wrinkled from the center to the edge, white at the edge from above; white to cream from below.
Micromorphology: Conidial heads globose, dark brown, becoming black, radiate, split into several loose columns with age, uniseriate. Conidiophores up to 1200 μm long, 14–18 μm wide, smooth-walled, hyaline or pigmented, aseptate, unbranched, straight to curved. Vesicles 32–64 μm wide ( = 56.3 × 52.2 μm, n = 15), globose to subglobose, uniseriate, fertile over entire surface, without metula; phialides 4–8 × 3.5–9 μm ( = 5.3 × 4.9 μm, n = 20), hyaline, flask-shaped, ampulliform. Conidia 4.3–6 × 4–6.2 μm ( = 4.7 × 4.8 μm, n = 20), globose to subglobose, sometime ellipsoidal, initially hyaline, becoming reddish brown to black, with coarsely roughened to echinulate surface, rough-walled.
Material examined: CHINA, Yunnan Province, Kunming City, from air under Quercus variabilis, 20 March, 2018, M. Doilom (MFLU 20-0430, holotype); ex-type living culture (KUMCC 18-0196).
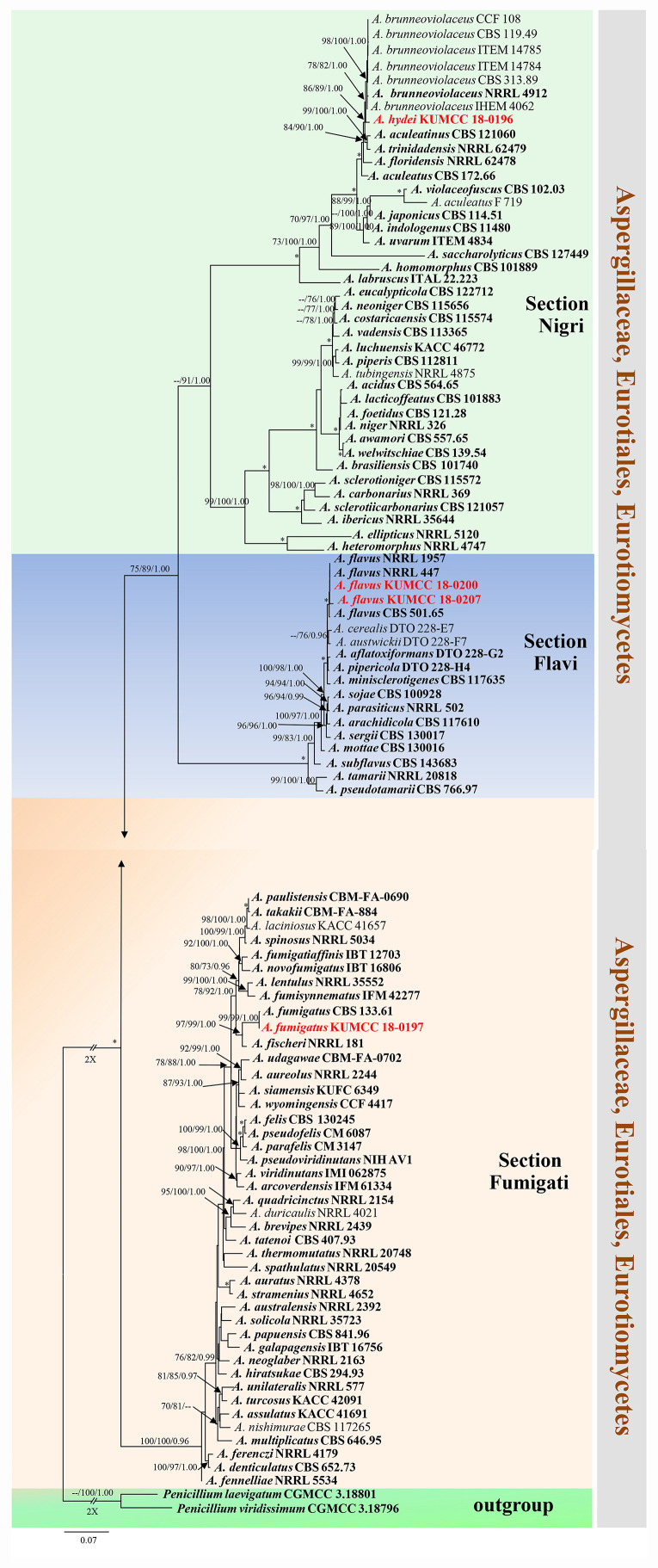
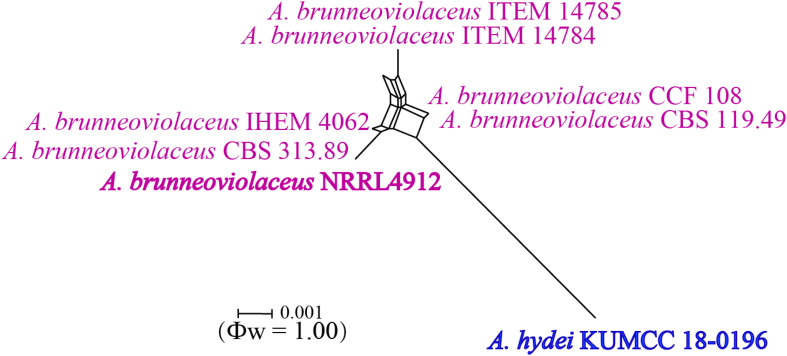
Notes: Colony diameter after 7 days of incubation at 25°C on CYA of A. hydei is obviously smaller than A. brunneoviolaceus (75–80 vs. > 85 mm) (Jurjević et al., 2012). A. hydei (MFLUCC 18-0196) had a close phylogenetic affinity with A. brunneoviolaceus based on individual ITS and CaM sequence data (Supplementary Figures S1, S2). However, A. hydei formed an independent lineage to the ex-type (NRRL 4912) and other strains of A. brunneoviolaceus based on combined CaM, BenA, ITS and RPB2 sequence data with significant support (78% MP, 82% ML and 1.00 PP) (Figure 6). A. hydei also had close phylogenetic affinity with A. aculeatinus, A. aculeatus, A. floridensis, and A. trinidadensis (Supplementary Figures S3, S4) but formed a distinct lineage from these four species (Figure 6). The GCPSR test was applied to a combined dataset including A. hydei and its closely related A. brunneoviolaceus, containing eight isolates. The result revealed that there was no significant recombination between A. brunneoviolaceus and A. hydei (Φw = 1.00) (Figure 7). Thus, A. hydei can be recognized as a new species.
FIGURE 6.
Phylogram generated from maximum likelihood analysis based on combined CaM, BenA, ITS and RPB2 sequence data. One hundred and three sequences are included in the combined analysis, which consisted of 2805 characters including alignment gaps. The tree is rooted to Penicillium viridissimum (CGMCC 3.18796) and P. laevigatum (CGMCC 3.18801). The MP analysis for the combined dataset had 1199 parsimony informative, 1432 constant, 174 parsimony uninformative and yielded 356 most parsimonious trees. The best-fit model HKY + I + G was selected for CaM, BenA, GTR + I + G for ITS and RPB2 in BI analysis. Maximum parsimony and maximum likelihood bootstrap values ≥ 70% and Bayesian posterior probabilities ≥ 0.90 (MPBS/MLBS/BYPP) are indicated at the nodes. Branches with 100% MPBS, 100% MLBS and 1.00 BYPP values are indicated as an asterisk (∗). The ex-type strains are bolded black, and the new isolates are in red.
FIGURE 7.
Split graph showing the results of the pairwise homoplasy index (PHI) test of the combined CaM, BenA, ITS and RPB2 sequence data between Aspergillus brunneoviolaceus and A. hydei using LogDet transformation and splits decomposition. PHI test result (Φw) < 0.05 indicates significant recombination within the dataset. Ex-type strains are indicated in bold.
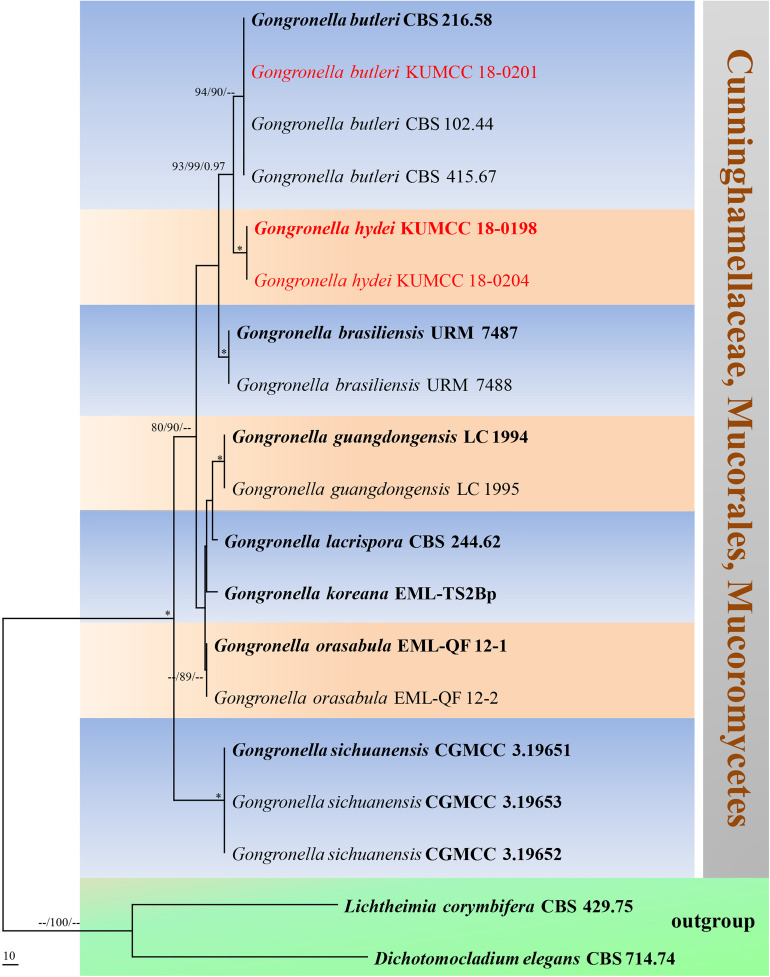
Gongronella Ribaldi
Type: Gongronella urceolifera Ribaldi
Notes: Species of Gongronella have mainly been isolated from soil (Akone et al., 2014; Adamčík et al., 2015; Ariyawansa et al., 2015). Morphological characteristics of Gongronella species are diverse in shape, making it difficult to clearly delineate the species belonging to this genus. In addition, Gongronella species have been described from different media (Hesseltine and Ellis, 1961, 1964; Adamčík et al., 2015; Ariyawansa et al., 2015; Zhang et al., 2019). However, Gongronella species can be separated by phylogenetic analysis based on ITS sequences as shown in Li G. J. et al. (2016) and Tibpromma et al. (2017) and this study. Multi-locus gene regions would well-resolve to determine the species. The genus comprises nine epithets in Index Fungorum (2020). G. hydei is introduced as a new species herein.
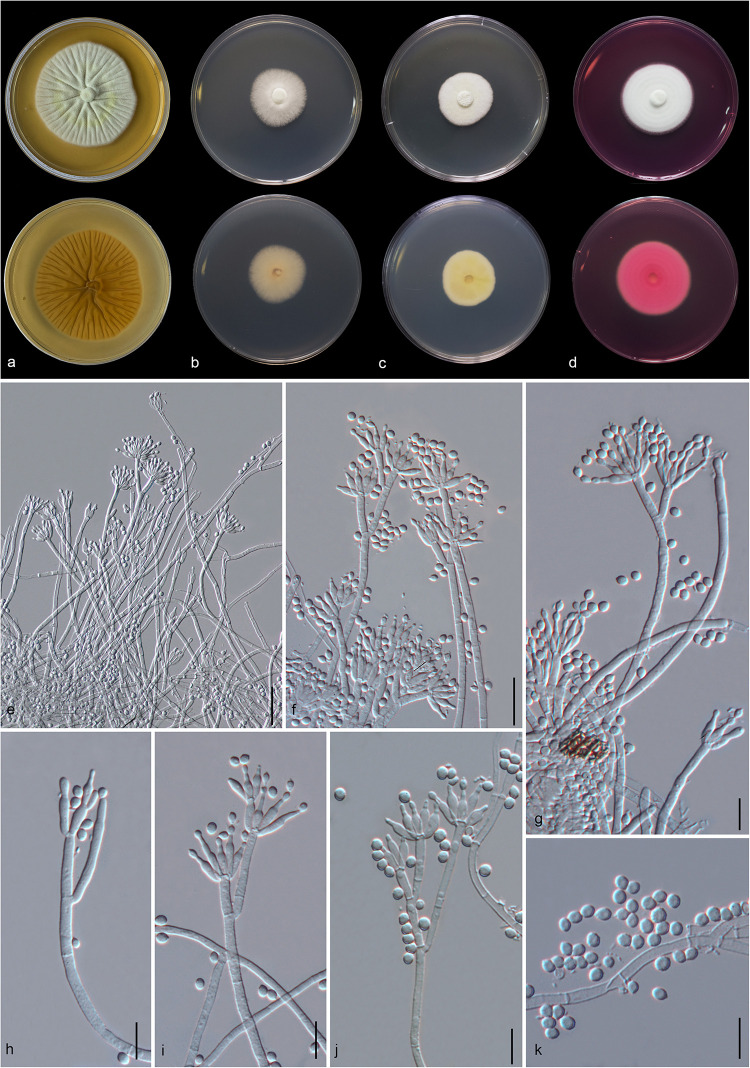
Gongronella hydei Doilom, sp. nov., Figure 8
FIGURE 8.
Gongronella hydei (MFLU 20-0431, holotype). (a–d) Colonies on PDA, CYA, rosebengal and WA at 25°C, respectively. (e) Branched sporangiophore with sporangia. (f–h) Sporangiophores with sporangia and apophyses. (i,j) Aggregated sporangiospores in sporangia with variously shaped apophysis (black arrows). (k,m,n) Septate sporangiophores with apophyses, columella (blue arrows) and collarette (green arrow). (l) Developed sporangiophore from hypha. (o) Sporangiospores. Scale bars: (e,h) = 20 μm, (f,g,i–n) = 10 μm, (o) = 5 μm.
Index Fungorum number: IF 557861; Facesoffungi number: FoF 08838
Etymology: Named in honor of Kevin D. Hyde for his excellent contributions to mycology.
Holotype: MFLU 20-0431
Macromorphology: Colonies on PDA reaching 60–65 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat or effuse, to raise, dense, white from above and below. Colonies on CYA reaching 55–65 mm diam. after 7 days in the dark at 25°C, irregular in shape, curled to undulate, flat or effuse, medium dense, white from above and below. Colonies on Rosebengal reaching 45–47 mm diam. after 7 days in the dark at 25°C, slightly circular, edge entire to undulate, filamentous, flat, dense, cottony, concentric ring, white from above and below. Colonies on WA reaching 30–35 mm diam. after 7 days in the dark at 25°C, irregular in shape, curled, flat, loose, with obviously white filamentous around the center (mycelium plug) from above; white from below.
Micromorphology: Rhizoids absent. Sporangiophores on PDA up to 120 μm long, 1.6–3.2 μm wide, erect, arising from hyphae, straight or flexuous, smooth- to minutely rough-walled, hyaline, unbranched, occasionally branched, 1–2-septate, mostly 1-septate below the apophysis. Apophyses variable in shape, cuboid-shaped with truncate at the base: 2.5–3.9 × 3.5–5.1 μm; cup-shaped with rounded at the base: 2.7–6.2 × 3.8–7.8 μm; cup-shaped with truncate at the base: 3.7–7.3 × 3.8–7.3 μm, hyaline, smooth-walled. Sporangia 10.5–18.8 × 10–17.5 μm, hyaline, globose to subglobose, thin-walled, occasionally thick-walled, smooth-walled, multi-spored, with an apophyses. Columellae 1.7–4.7 × 2.2–6.3 μm, hemispherical with a collarette, smooth-walled, sometime tiny, almost inconspicuous, mostly with an evident collar. Sporangiospores variable in shape, reniform: 2.4–3.8 × 1.5–2.3 μm, ellipsoidal to fusiform: 2.6–3.4 × 1.8–3.4 μm, hyaline, smooth-walled, aseptate, guttulate. Chlamydospores present in the aerial mycelium, globose. Giant cells up to 25 μm diam., hyaline, globose, guttulate. Zygosporangia not observed.
Material examined: CHINA, Yunnan Province, Honghe County, from rhizosphere soil of Quercus rubra, 16 April 2018, M. Doilom (MFLU 20-0431, holotype); ex-type living culture (KUMCC 18-0198), ibid., living culture KUMCC 18-0204.
Notes: The novel G. hydei lacks rhizoids, which are present in G. butleri (Hesseltine and Ellis, 1964). However, G. hydei was observed on PDA, while G. butleri observed on SMA (synthetic mucor agar). Phylogenetic analysis of ITS sequences confirmed that our isolates (KUMCC 18-0198 and KUMCC 18-0204) grouped clearly distinct from other known species (Figure 9). An isolate KUMCC 18-0198 grouped with KUMCC 18-0204 (100% MP, 100% ML and 1.00 PP) but separated from other G. butleri strains with 93% MP, 99% ML and 0.97 PP (Figure 9). Thus, we introduce G. hydei as a new species.
FIGURE 9.
Phylogram generated from maximum parsimony analysis based on ITS sequence data. Nineteen sequences are included in the ITS analysis, which consisted of 772 characters including alignment gaps. The tree is rooted to Lichtheimia corymbifera (CBS 429.75) and Dichotomocladium elegans (CBS 714.74). The MP analysis for the ITS dataset had 187 parsimony informative, 315 constant, 270 parsimony uninformative characters and yielded two most parsimonious trees. The best-fit model GTR + G was selected for ITS. Maximum parsimony and maximum likelihood bootstrap values ≥ 70% and Bayesian posterior probabilities ≥ 0.90 (MPBS/MLBS/BYPP) are given at the nodes. The ex-type strains are bolded, and the new isolates are in red. Branches with 100% MPBS, 100% MLBS and 1.00 BYPP values are indicated as an asterisk (∗).
Penicillium Section Lanata-Divaricata Raper and Thom ex Pitt
Type: Penicillium janthinellum Biourge
Notes: Species of Penicillium in the section Lanata-Divaricata are characterized by divaricate conidiophores and broadly scattering colonies (Houbraken and Samson, 2011). Members of Penicillium are widespread in food products, soil, plant-decaying materials, animals and indoor environments (Visagie et al., 2016; Diao et al., 2018; Heo et al., 2019). Many recent studies reported Penicillium strains belonging to the section Lanata-Divaricata (Visagie et al., 2016; Lorenzini et al., 2019; Hyde et al., 2019a; Guevara-Suarez et al., 2020). Delimiting species in this section based only on morphological characteristics is difficult; accordingly, ITS, BenA, CaM and RPB2 are used to delimit species boundaries (Visagie et al., 2016). P. soli is introduced as a new species herein.
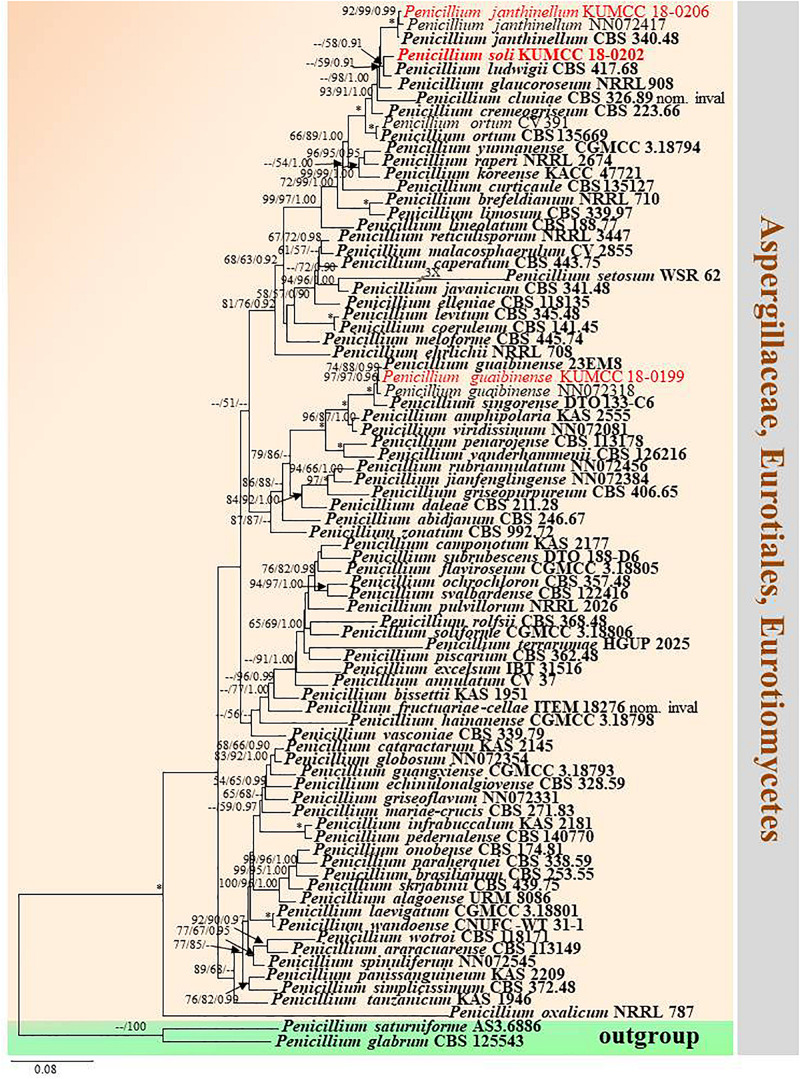
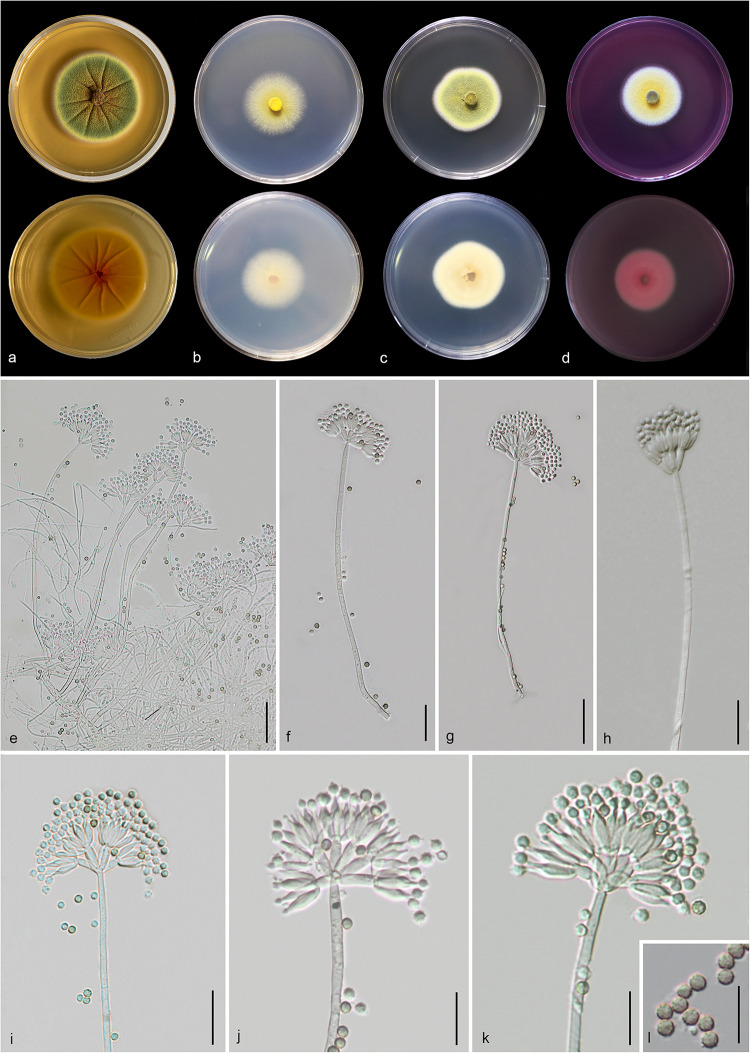
Penicillium guaibinense J. P. Andrade, C. N. Figueiredo, R. P. Nascimento, P. A. S. Marbach, and J. T. De Souza, in Crous et al., Persoonia 41: 389 (2018), Figures 10, 11
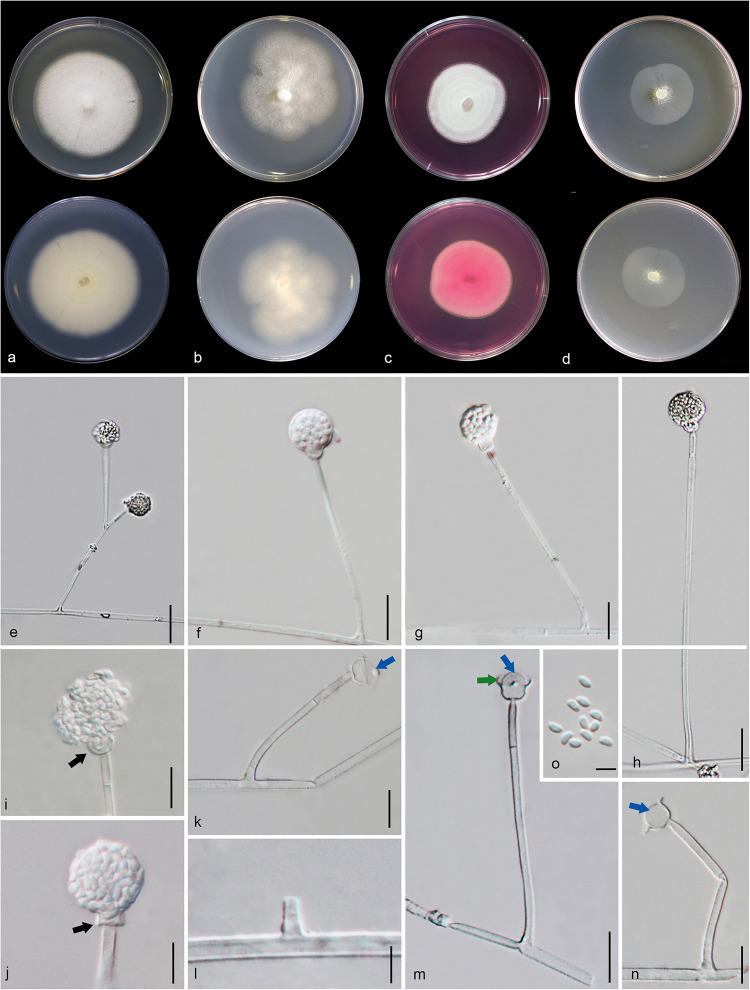
FIGURE 10.
Penicillium guaibinense (MFLU 20-0433). (a–d) Colonies on MEA, CYA, PDA and rosebengal at 25°C, respectively. (e–j) Conidiophores and conidia. (k) Conidia. Scale bars: (e,f) = 20 μm, (g–k) = 10 μm.
FIGURE 11.
Penicillium guaibinense (MFLU 20-0433). (a–c) Conidiophores and conidia. (d,e) Catenate conidia forming on phialides. (f) Conidia. (g) Close-up of conidia showing conidial wall. Scale bars: (a) = 10 μm, (b,c) = 5 μm, (d) = 2 μm, (e) = 3 μm, (f,g) = 1 μm.
= Penicillium austrosinense L. Cai, Houbraken and X. Z. Jiang, in Diao, Chen, Jiang, Houbraken, Barbosa, Cai and Wu, Cladistics: 10.1111/cla.12365, 12 (2018)
Facesoffungi number: FoF 08839
Macromorphology: Colonies on MEA reaching 55–60 mm diam. after 7 days in the dark at 25°C, circular to slightly undulate, with entire margin, flat or effuse, dense, with wrinkled from center to edge, white with pale yellow circle from above, pale yellow from below. Colonies on CYA reaching 30–35 mm diam. after 7 days in the dark at 25°C, circular, filamentous, low convex in the center, flat at the edge, medium dense, cottony, white from above, orange-cream from below. Colonies on PDA reaching 25–32 mm diam. after 7 days in the dark at 25°C, circular, entire, flat or effuse, dense, cottony, white from above, yellow to cream from below. Colonies on Rosebengal reaching 40–45 mm diam. after 7 days in the dark at 25°C, circular, edge entire to filamentous, flat, dense, cottony, white from above and below.
Micromorphology: Conidiophores on MEA up to 200 μm long, 2.7–3.5 μm wide, divaricate, biverticillate to terverticillate, sometime quaterverticillate, stipes smooth- to finely verruculose-walled, septate, with branched 15–38 μm long × 2.6–3 μm wide. Metulae 11–20.3 × 2.2–4.1 μm ( = 13.7 × 3 μm, n = 20), 1–3 per branch, divergent, each bearing 3–10 phialides. Phialides 6.8–14.6 × 2.5–3.4 μm ( = 9.8 × 2.8 μm, n = 20), ampulliform to cylindrical. Conidia 2.8–4 × 2.5–3.9 μm ( = 3.6 × 3.2 μm, n = 30), hyaline, globose, subglobose to ellipsoidal, catenate, verruculose, finely rough-walled.
Material examined: CHINA, Yunnan Province, Honghe County, from rhizosphere soil of Quercus rubra, 16 April 2018, M. Doilom (MFLU 20-0433); living culture (KUMCC 18-0199).
Notes: Conidia of our specimen MFLU 20-0433 are slightly wider and longer than the holotype of Penicillium guaibinense [2.8–4 × 2.5–3.9 μm (av. 3.6 × 3.2) vs. 2–3 × 2–3 μm (av. 2.3 × 2.2)]. However, it is identified as P. guaibinense based on phylogenetic analysis of combined ITS, BenA, CaM, and RPB2 sequences (Figure 12) and nucleotide comparison of ITS, BenA and CaM (Supplementary Table S4).
FIGURE 12.
Phylogram generated from maximum likelihood analysis based on combined ITS, BenA, CaM, and RPB2 sequence data. Eighty-one sequences are included in the combined analysis, which consisted of 2590 characters including alignment gaps. The tree is rooted to Penicillium glabrum (CBS 125543) and P. saturniforme (AS3.6886). The MP analysis for the combined dataset had 815 parsimony informative, 1358 constant, 417 parsimony uninformative characters and yielded 39 most parsimonious trees. The best-fit model GTR + I + G was selected for ITS, BenA and RPB2, and SYM + I + G for CaM in BI analysis. Maximum parsimony and maximum likelihood bootstrap values ≥ 50% and Bayesian posterior probabilities ≥ 0.90 (MPBS/MLBS/BYPP) are indicated at the nodes. Branches with 100% MPBS, 100% MLBS and 1.00 BYPP values are indicated as an asterisk (∗). The ex-type strains are bolded black, and the new isolates are in red.
Crous et al. (2018) established Penicillium guaibinense from soil in the Guaibim sandbank in Bahia, Brazil. Diao et al. (2018) introduced P. austrosinense from acidic soil in Hainan, China. The conidia of P. guaibinense (CCDCA 11512 = 23EM8: ex-type) are finely rough and broadly subglobose, whereas they are smooth-walled and globose to ellipsoidal in P. austrosinense (CGMCC 3.18797: ex-type); despite this, conidial sizes between P. guaibinense and P. austrosinense are slightly similar [2–3 × 2–3 μm (av. 2.3 × 2.2) vs. 2–4 × 2–3 μm (av. 2.87 × 2.59)]. Penicillium austrosinense is considered as a synonym of P. guaibinense on the basis of phylogenetic analyses and nucleotide comparison of ITS, BenA, CaM, and RPB2 (Supplementary Table S4). Penicillium guaibinense was prior published, and we treat P. austrosinense as a synonym of P. guaibinense.
Penicillium soli Doilom, C. F. Liao and D. Pem, sp. nov., Figure 13
FIGURE 13.
Penicillium soli (MFLU 20-0432, holotype). (a–d) Colonies on MEA, CYA, PDA and rosebengal at 25°C, respectively. (e–g) Conidiophores and conidia. (h–k) Close-up of conidiophores and conidia. (l) Conidia. Scale bars: (e–g) = 20 μm, (h–l) = 10 μm.
Index Fungorum number: IF 557862; Facesoffungi number: FoF 08840
Etymology: The specific epithet “soli” From Latin, refers to soil, where the holotype was collected.
Holotype: MFLU 20-0432
Macromorphology: Colonies on MEA reaching 55–60 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat or effuse, dense, with wrinkled from center to edge, green with pale green to white at the edge from above, yellow or creamy with white at the edge from below. Colonies on CYA reaching 35–40 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat, medium dense, gray in the center, white at the edge from above and white from below. Colonies on PDA reaching 53–55 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat or effuse, dense, grayish in the center gradually green and white at the edge from above and white from below. Colonies on Rosebengal reaching 40–45 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat, dense, cottony, with slightly wrinkled from center to edge, white from above and below.
Micromorphology: Sclerotia absent. Conidiophores on MEA up to 205 μm long, 2.7–4.2 μm wide, divaricate, biverticillate, sometime terverticillate to quaterverticillate, stipes smooth-walled, septate, with branches 17–51 × 3–4.7 μm. Metulae 11–23 × 2.3–4.5 μm ( = 15 × 3.1 μm, n = 30), divergent, 1–4 per branch, each bearing 3–9 phialides, with 2–3 whorls per branch. Phialides 6.5–12.3 × 2.2–3.5 μm ( = 8.7 × 2.8 μm, n = 30), ampulliform to cylindrical. Conidia 2.5–4.5 × 2.3–3.4 μm ( = 3.4 × 2.8 μm, n = 30), hyaline, globose, subglobose to broadly ellipsoidal, chain, smooth-walled.
Material examined: CHINA, Yunnan Province, Honghe County, from rhizosphere soil of Quercus rubra, 16 April 2018, M. Doilom (MFLU 20-0432, holotype); ex-type living culture (KUMCC 18-0202).
Notes: In the phylogram generated from maximum likelihood analysis based on ITS sequence data, P. soli was closely related to Penicillium spp., e.g., P. cremeogriseum (CBS 223.66) (Supplementary Figure S5). The new taxon P. soli clustered with P. ludwigii (CBS 417.68) in the analysis based on BenA sequences (Supplementary Figure S6). Upon analysis of the CAM and RPB2 sequence data, P. soli clustered as a sister to P. cluniae (CBS 326.89) (Supplementary Figures S7, S8), and showed 0.96% (4/415) and 1.46% (11/755) nucleotide differences, respectively. However, phylogenetic analyses across a combined ITS, BenA, CAM and RPB2 sequence data showed that P. soli (KUMCC 18-0202) formed a distinct lineage and was sister to P. ludwigii (CBS 417.68), with bootstrap support in ML analysis (58% ML, 0.91 PP) (Figure 12). Comparison of CaM and RPB2 sequence data between P. soli (KUMCC 18-0202) and P. ludwigii (CBS 417.68) indicated 5.3% (22/415) and 2.38% (18/755) nucleotide difference, respectively. Accordingly, following the guidelines of Jeewon and Hyde (2016), we introduce P. soli as a new species.
Talaromyces Section Talaromyces C. R. Benj.
= Penicillium subgenus Biverticillium section Simplicium series Mini- olutea Pitt, The genus Penicillium: 419. 1980.
= Penicillium subgenus Biverticillium section Coremigenum series
Duclauxii Raper and Thom ex Pitt, The genus Penicillium: 404. 1980.
= Talaromyces section Talaromyces series Flavi Pitt, The genus Penicillium: 471. 1980.
Type: Talaromyces flavus (Klöcker) Stolk and Samson
Notes: Talaromyces section Talaromyces was introduced by Stolk and Samson (1972) and characterized by yellow ascomata, as well as occasionally white, creamish, pinkish or reddish and yellow ascospores. Conidiophores are usually of the biverticillate-symmetrical type, and some species are reduced to conidiophores with solitary phialides. Phialides are usually acerose, with a minor proportion of species having wider bases (Stolk and Samson, 1972). Phylogenetic analyses of ITS, BenA, CaM, and RPB2 sequence data were used to determine species delimitation (Barbosa et al., 2018; Guevara-Suarez et al., 2020). Section Talaromyces species are usually isolated from soil, indoor air environments and humans (Chen et al., 2016; Yilmaz et al., 2016; Lau et al., 2017; Pangging et al., 2019; Sun et al., 2020). We introduce the new species T. yunnanensis into the section.
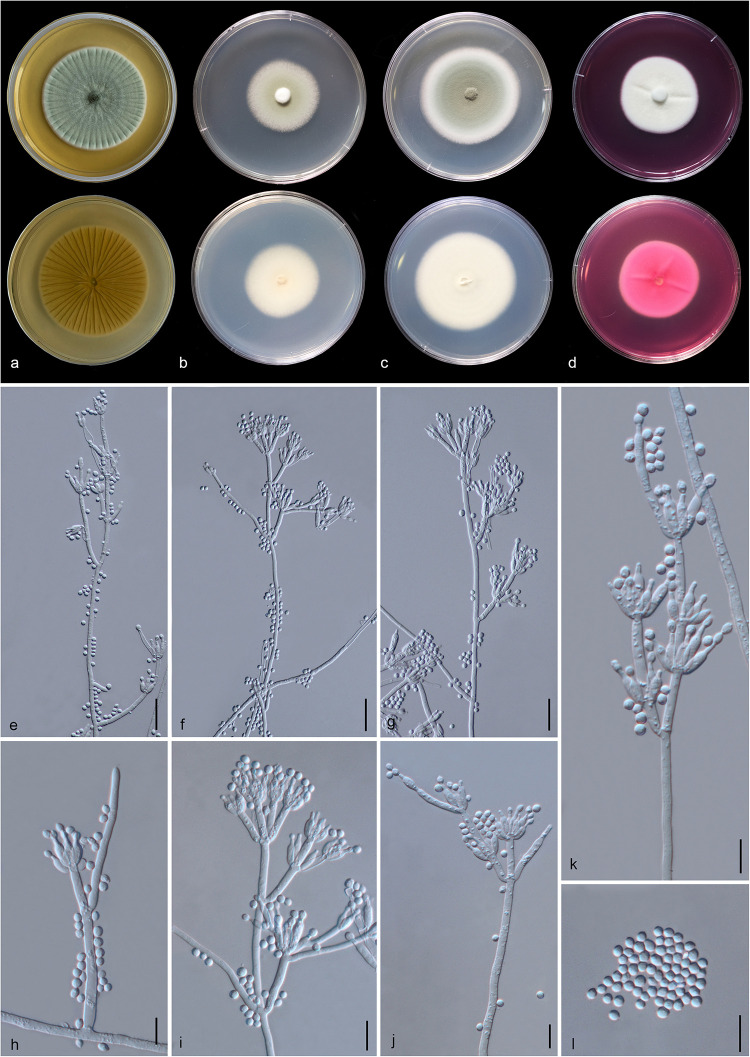
Talaromyces yunnanensis Doilom and C. F. Liao, sp. nov., Figure 14
FIGURE 14.
Talaromyces yunnanensis (MFLU 20-0434, holotype). (a–d) Colonies on MEA, CYA, PDA and rosebengal at 25°C, respectively. (e–h) Conidiophores and conidia. (i–k) Close-up of conidiophores and conidia. (l) Conidia. Scale bars: (e,g) = 30 μm, (f,h,i) = 20 μm, (j–l) = 5 μm.
Index Fungorum number: IF 557863; Facesoffungi number: FoF 08841
Etymology: Referring to the province, where the holotype was collected.
Holotype: MFLU 20-0434
Macromorphology: Colonies on MEA reaching 50–53 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat or effuse, dense, wrinkled from center to edge, green with orange mass (exudate) in the center, gradually yellow and white at the edge from above, yellow from below. Colonies on CYA reaching 35–37 mm diam. after 7 days in the dark at 25°C, circular, edge entire to filamentous, flat, medium dense, yellow at the center and white at the edge from above, pale yellow at the center and white at the edge from below. Colonies on PDA reaching 30–40 mm diam. after 7 days in the dark at 25°C, irregular in shape, curled, flat, dense, yellowish green in the center, yellow and white from above, yellowish cream from below. Colonies on Rosebengal reaching 35–38 mm diam. after 7 days in the dark at 25°C, circular, edge entire, flat, dense, yellow in the center, white at the edge from above, white from below.
Micromorphology: Sclerotia absent. Conidiophores on MEA up to 275 μm long, 2.2–3.7 μm wide, biverticillate, stipes finely verruculose-walled, septate, mostly unbranched, occasionally with branched 9.5–10 μm long × 2.5–3.1 μm wide. Metulae 7.3–11.5 × 2.3–4.2 μm ( = 9.3 × 3.2 μm, n = 20), 4–8 per stipe, divergent, each bearing 3–10 phialides. Phialides 7.3–11.5 × 2.2–3.8 μm ( = 9.4 × 2.9 μm, n = 30), ampulliform to cylindrical. Conidia 2.3–3.4 × 2.1–3.2 μm ( = 2.8 × 2.6 μm, n = 30), hyaline when immature, pale brown to dark brown when mature, globose to subglobose, chain, finely verruculose-walled. Ascomata not observed.
Material examined: CHINA, Yunnan Province, Honghe County, from rhizosphere soil of Quercus rubra, 16 April 2018, M. Doilom (MFLU 20-0434, holotype); ex-type living culture (KUMCC 18-0208).
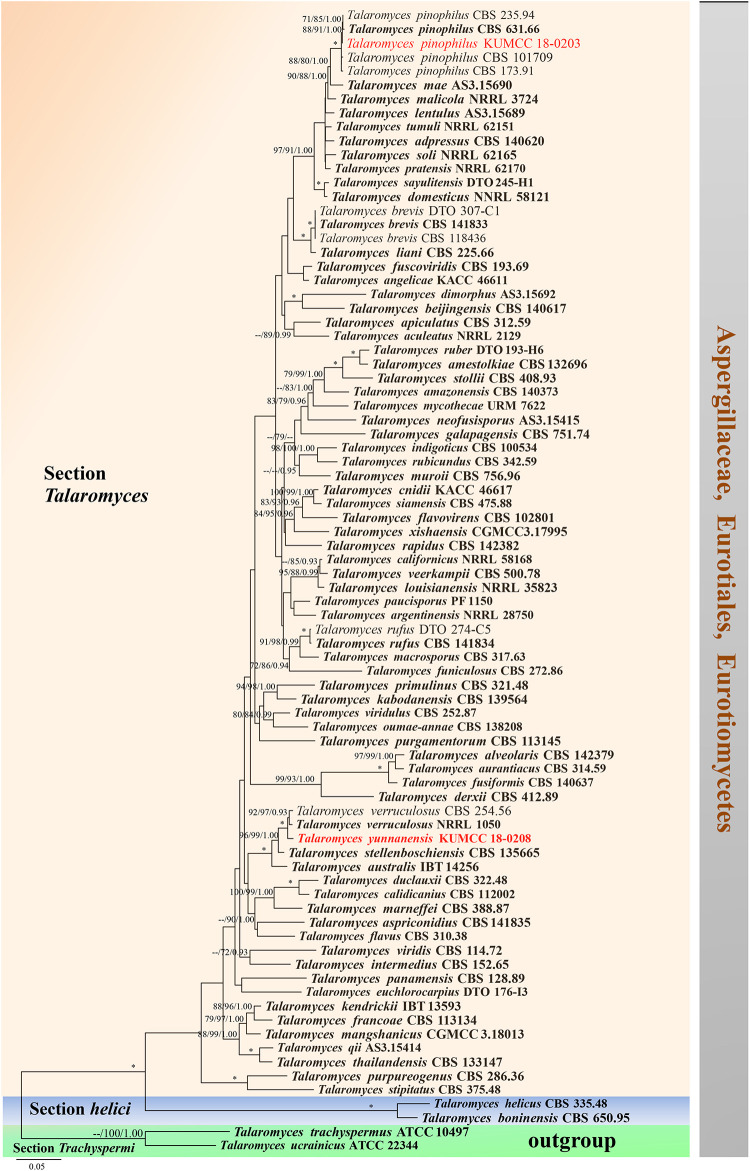
Notes: Talaromyces yunnanensis, T. verruculosus, and T. stellenboschiensis have similar morphological characteristics featuring biverticillate conidiophores, ampulliform phialides tapering into thin necks and globose conidia; however, they can be differentiated by colony characteristics. T. yunnanensis grows faster than T. stellenboschiensis (50–53 vs. 40–42 mm) and T. verruculosus (50–53 vs. 35–36 mm) after 7 days of incubation on MEA at 25°C (Visagie et al., 2015). T. yunnanensis (KUMCC 18-0208) was phylogenetically related to T. verruculosus (NRRL 1050: ex-type) and (CBS 254.56) but had its own distinct linear with 100% MP, 100% ML, 1.00 PP (Figure 15). Phylogenetic analysis based on individual ITS sequence data was unsuccessful in separating T. yunnanensis and T. verruculosus (Supplementary Figure S9). However, BenA and CaM were able to separate these two species (Supplementary Figures S10, S11). The RBP2 gene was amplified and sequenced but also unsuccessful. Thus, comparison was unable to be performed. Comparison of BenA and CaM sequence data between T. yunnanensis (KUMCC 18-0208) and T. verruculosus (NRLL 1050: ex-type) revealed 1.94% (9/465) and 2.08% (10/480) nucleotide difference, respectively. This justifies these two isolates as different taxa according to the guidelines proposed by Jeewon and Hyde (2016).
FIGURE 15.
Phylogram generated from maximum likelihood analysis based on combined ITS, BenA, CaM, and RPB2 sequence data. Eighty-two sequences are included in the combined analysis, which consisted of 2858 characters including alignment gaps. The tree is rooted to Talaromyces trachyspermus (ATCC 10497) and T. ucrainicus (ATCC 22344). The MP analysis for the combined dataset had 857 parsimony informative, 1552 constant, 176 parsimony uninformative characters and yielded 46 most parsimonious trees. The best-fit model SYM + I + G was selected for CaM, GTR + I + G for ITS and HKY + I + G for RPB2 and BenA in BI analysis. Maximum parsimony and maximum likelihood bootstrap values ≥ 70% and Bayesian posterior probabilities ≥ 0.90 (MPBS/MLBS/BYPP) are indicated at the nodes. Branches with 100% MPBS, 100% MLBS and 1.00 BYPP values are indicated as an asterisk (∗). The ex-type strains are bolded black, and the new isolates are in red.
Discussion
We carried out phosphate solubilization of 13 fungal strains in both solid and liquid PVK media in vitro. This study indicates that the airborne isolate KUMCC 18-0196 (A. hydei sp. nov.) was a highly efficient strain in solubilizing phosphate. Isolate KUMCC 18-0196 also showed the greatest drop in pH in the PVK broth containing TCP, suggesting that it might produce organic acids. The A. hydei is the most efficient P-solubilizing than other strains tested in this study. Our results are similar to the study of Pradhan and Sukla (2006), which concluded that Aspergillus spp. show a much higher drop in pH in the PVK broth containing TCP and high P solubilization when compared to Penicillium spp. Many previous studies reported PSF mostly from soils, including rhizospheric and non-rhizospheric soil samples from cultivated plants and asymptomatic plant roots (Nelofer et al., 2015; Elias et al., 2016; Adhikari and Pandey, 2019; Qiao et al., 2019, Supplementary Table S5). To date, PSF from air have not yet been reported. Thus, A. hydei sp. nov. described herein is the first report of PSF from air. However, the determination of phosphate solubilization has been tested on solid and liquid media under laboratory conditions. The ability to solubilize phosphate under field conditions should be closely examined to confirm P dissolving activity. PSF did not lose P solubilizing ability under laboratory conditions, although sub-culturing was repeated (Kucey, 1983; Sharma et al., 2013).
Several phosphate-solubilizing microorganisms viz. actinomyces, bacteria, fungi, and yeasts have been screened for phosphate solubilization. They were shown to enhance the solubilization of insoluble P compounds (Gizaw et al., 2017; Nandimath et al., 2017; Mohamad et al., 2018; Wang et al., 2018; Chen and Liu, 2019; Liu et al., 2019). For fungi, Rhizopus stolonifer var. stolonifer, Aspergillus niger, and Alternaria alternata have been found in the literature to be the most effective strains with respect to the amount of TCP solubilization by Ceci et al. (2018). However, solubilization by PSF depends on the insoluble inorganic phosphate source, type of carbon, nitrogen and metal ions in soils and culture conditions (Richardson and Simpson, 2011; Jyoti et al., 2013). Vyas et al. (2007) tested the quantity of phosphate solubilized by Eupenicillium parvum (current name Penicillium parvum) from the different inorganic phosphates [TCP, aluminum phosphate (AP) and North Carolina phosphate (NCRP)]. Their results showed that the quantity of phosphate solubilized was by far the greatest for TCP among the inorganic phosphates, whereas the solubilization of iron phosphate was low. Phosphate-solubilizing microorganisms convert insoluble phosphates into a soluble form through the processes of acidification, chelation, exchange reaction, and production of organic acid (Behera et al., 2017).
Some studies of PSF identified fungal species names based on morphology, highest nucleotide similarity in MegaBLAST searches of NCBI’s GenBank nucleotide database and/or phylogenetic analysis of ITS sequence data (Nelofer et al., 2015; Wang et al., 2018; Zhang et al., 2018, Supplementary Table S5). However, these are insufficient to determine the species names, especially within species complexes (Samson et al., 2014; Doilom et al., 2017; Hyde et al., 2020a,b). A comparison of sequence data from fungal strains with the ex-type cultures of named species must be considered to confirm the species name (Dayarathne et al., 2016). Several taxonomic schemes and new molecular approaches have been proposed for fungal identification (Visagie et al., 2014b; Jeewon and Hyde, 2016; Zhao et al., 2018). We provide the fungal identification of PSF based on morphological characteristics and multi-loci phylogenetic analyses, including the GCPSR method where necessary. GenBank accession numbers of species in Aspergillus, Gongronella, Penicillium, and Talaromyces with our newly generated sequences are also provided in Supplementary Table S1 to further facilitate species identification. Fungal taxonomy is discussed in each taxonomic note.
Applications of beneficial microbes, especially phosphate-solubilizing fungi, are useful toward plant growth-promoting strategies. The isolate KUMCC 18-0196 (A. hydei sp. nov.) was a highly efficient strain in solubilizing phosphate; accordingly, we are optimistic that it can play potential roles in supporting plant growth in practical applications. This research provides important information relevant to the application of beneficial fungi to promote plant growth. This will have critical implications for the development of agriculture, industry, restoration ecology and forestry going forward.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MD, PM, J-CX, and J-WG designed the study. MD planned and conducted the experiments and wrote the manuscript. MD, KY, WD, C-FL, RP, KY, and DP conducted the experiments, analyzed the data, and revised the manuscript. PM, IP, NS, and SL contributed to research funds. All authors revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank Austin Smith [World Agroforestry (ICRAF), Kunming Institute of Botany, China] for English editing and Dhanushka N. Wanasinghe (Kunming Institute of Botany, China) for suggestions on the research. MD thanks Shaun Pennycook (Landcare Research, New Zealand) for assistance in new epithets. Gu Zhi-Jia (Kunming Institute of Botany, China) is thanked for the assistance in taking the scanning electron microscopy photographs.
Funding. MD sincerely thanks the 5th batch of Post-doctoral Orientation Training Personnel in Yunnan Province (grant no.: Y934283261) and the 64th batch of China Post-doctoral Science Foundation (grant no.: Y913082271) for financial research support. J-WG would like to thank the National Natural Science Foundation of China (grant no.: 31860026). J-CX thanks the Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (grant no.: QYZDY-SSW-SMC014). KY thanks the National Natural Science Foundation of China (grant no.: 41807524) and Yunnan Province Natural Science Foundation (grant no.: 2020FD925). PM thanks the National Science Foundation of China and the Chinese Academy of Sciences for financial support under the following grants: 41761144055, 41771063, and Y4ZK111B01. RP thanks CAS President’s International Fellowship Initiative (PIFI) for Young Staff 2019–2021 (grant no.: 2019FY0003) and the Research Fund from China Post-doctoral Science Foundation (grant no.: Y71B283261). SK would like to thank the CAS President’s International Fellowship Initiative (PIFI) young staff under the grant number: 2020FYC0002 and the National Natural Science Foundation of China (NSFC, project code 31851110759). This research work was partially supported by Chiang Mai University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.585215/full#supplementary-material
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on RPB2 sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on RPB2 sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Talaromyces strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Talaromyces strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Talaromyces strains.
Taxa and their GenBank accession numbers used in this study.
Phosphate solubilization of tricalcium phosphate (TCP) on solid and liquid Pikovskaya (PVK) media with pH values by 13 fungal strains and control.
Details of 13 phosphate-solubilizing fungal strains used in this study.
Nucleotide comparison of ITS, BenA, CaM, and RPB2 between strains of Penicillium guaibinense and P. austrosinense.
List of phosphate-solubilizing fungi.
References
- Adamčík S., Cai L., Chakraborty D., Chen X. H., Cotter H. V. T., Dai D. Q., et al. (2015). Fungal biodiversity profiles 1–10. Cryptogam. Mycol. 36 121–166. 10.7872/crym/v36.iss2.2015.121 [DOI] [Google Scholar]
- Adhikari P., Pandey A. (2019). Phosphate solubilization potential of endophytic fungi isolated from Taxus wallichiana Zucc. roots. Rhizosphere 9 2–9. 10.1016/j.rhisph.2018.11.002 [DOI] [Google Scholar]
- Akone S. H., Rahn S., Henrich B., Daletos G., Vardamides J. C., Nkengfack A. E., et al. (2014). 2-Pentenedioic acid derivatives from a soil-derived fungus Gongronella butleri. Phytochem. Lett. 10 184–188. 10.1016/j.phytol.2014.09.001 [DOI] [Google Scholar]
- Ariyawansa H. A., Hyde K. D., Jayasiri S. C., Buyck B., Chethana K. W. T., Dai D. Q., et al. (2015). Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75 27–274. 10.1007/s13225-015-0346-5 [DOI] [Google Scholar]
- Barbosa R. N., Bezerra J. D. P., Souza-Motta C. M., Frisvad J. C., Samson R. A., Oliveira N. T., et al. (2018). New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek 111 1883–1912. 10.1007/s10482-018-1081-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera B. C., Yadav H., Singh S. K., Mishra R. R., Sethi B. K., Dutta S. K., et al. (2017). Phosphate solubilization and acid phosphatase activity of Serratia sp. isolated from mangrove soil of Mahanadi river delta, Odisha, India. J. Genet. Eng. Biotechnol. 15 169–178. 10.1016/j.jgeb.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci A., Pinzari F., Russo F., Maggi O., Persiani A. M. (2018). Saprotrophic soil fungi to improve phosphorus solubilisation and release: in vitro abilities of several species. Ambio 47 30–40. 10.1007/s13280-017-0972-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. J., Sun B. D., Houbraken J., Frisvad J. C., Yilmaz N., Zhou Y. G., et al. (2016). New Talaromyces species from indoor environments in China. Stud. Mycol. 84 119–144. 10.1016/j.simyco.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Liu S. (2019). Identification and characterization of the phosphate-solubilizing bacterium Pantoea sp. S32 in reclamation soil in Shanxi, China. Front. Microbiol. 10:2171. 10.3389/fmicb.2019.02171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuppa-Tostain G., Hoarau J., Watson M., Adelard L., Shum Cheong Sing A., Caro Y., et al. (2018). Production of Aspergillus niger biomass on sugarcane distillery wastewater: physiological aspects and potential for biodiesel production. Fungal Fungal. Biol. Biotechnol. 5:1. 10.1186/s40694-018-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Luangsa-ard J. J., Wingfield M. J., Carnegie A. J., Hernández-Restrepo M., Lombard L., et al. (2018). Fungal Planet description sheets: 785–867. Persoonia 41 238–417. 10.3767/persoonia.2018.41.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarathne M., Boonmee S., Braun U., Crous P., Daranagama D., Dissanayake A., et al. (2016). Taxonomic utility of old names in current fungal classification and nomenclature: conflicts, confusion & clarifications. Mycosphere 7 1622–1648. 10.5943/mycosphere/7/11/2 [DOI] [Google Scholar]
- Dayarathne M., Jones E., Maharachchikumbura S., Devadatha B., Sarma V., Khongphinitbunjong K., et al. (2020). Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- De Silva D. D., Rapior S., Fons F., Bahkali A. H., Hyde K. D. (2012a). Medicinal mushrooms in supportive cancer therapies: an approach to anti-cancer effects and putative mechanisms of action. Fungal Divers. 55 1–35. 10.1007/s13225-012-0151-3 [DOI] [Google Scholar]
- De Silva D. D., Rapior S., Hyde K. D., Bahkali A. H. (2012b). Medicinal mushrooms in prevention and control of diabetes mellitus. Fungal Divers. 56 1–29. 10.1007/s13225-012-0187-4 [DOI] [Google Scholar]
- Diao Y. Z., Chen Q., Jiang X. Z., Houbraken J., Barbosa R. N., Cai L., et al. (2018). Penicillium section Lanata-divaricata from acidic soil. Cladistics 35 1–36. 10.1111/cla.12365 [DOI] [PubMed] [Google Scholar]
- Doilom M., Manawasinghe I., Jeewon R., Jayawardena R., Tibpromma S., Hongsanan S., et al. (2017). Can ITS sequence data identify fungal endophytes from cultures A case study from Rhizophora apiculata. Mycosphere 8 1869–1892. 10.5943/mycosphere/8/10/11 [DOI] [Google Scholar]
- Dong W., Hyde K. D., Doilom M., Yu X. D., Bhat D. J., Jeewon R., et al. (2020). Pseudobactrodesmium (Dactylosporaceae, Eurotiomycetes, Fungi) a novel lignicolous genus. Front. Microbiol. 11:456. 10.3389/fmicb.2020.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias F., Woyessa D., Muleta D. (2016). Phosphate solubilization potential of rhizosphere fungi isolated from plants in Jimma Zone, Southwest Ethiopia. Int. J. Microbiol. 2016:5472601. 10.1155/2016/5472601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fungaro M. H. P., Ferranti L. S., Massi F. P., da Silva J. J., Sartori D., Taniwaki M. H., et al. (2017). Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Sci. Rep. 7:6203. 10.1038/s41598-017-06589-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W. C. M., Onions A. H., Pitt J., I, Samson R. A. (1986). “Infrageneric taxa of Aspergillus,” in Advances in Penicillium and Aspergillus Systematics, eds Samson R. A., Pitt J. I. (New York, NY: Plenum Press; ), 55–62. [Google Scholar]
- Gautam A. K., Sharma S., Avasthi S., Bhadauria R. (2011). Diversity, pathogenicity and toxicology of A. niger: an important spoilage fungi. Res. J. Microbiol. 6 270–280. 10.3923/jm.2011.270.280 [DOI] [Google Scholar]
- Giacomazzi J., Baethgen L., Carneiro L. C., Millington M. A., Denning D. W., Colombo A. L., et al. (2016). The burden of serious human fungal infections in Brazil. Mycoses 59 145–150. 10.1111/myc.12427 [DOI] [PubMed] [Google Scholar]
- Gizaw B., Tsegay Z., Tefera G., Aynalem E., Wassie M., Abatneh E. (2017). Phosphate solubilizing fungi isolated and characterized from Teff rhizosphere soil collected from North Showa and Gojam, Ethiopia. J. Fertil. Pestic. 8:180 10.4172/2471-2728.1000180 [DOI] [Google Scholar]
- Glass N. L., Donaldson G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Suarez M., Garcia D., Cano-Lira J. F., Guarro J., Gené J. (2020). Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Syst. Evol. 5 39–75. 10.3114/fuse.2020.05.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. W., Gao Y., Li C. Y., Yang L. F., Tian X. J., Hong L., et al. (2017). First report of leaf spot disease caused by Aspergillus tubingensis on Jatropha curcas in Yunnan, China. Plant Dis. 101:505 10.1094/pdis-08-16-1183-pdn [DOI] [Google Scholar]
- Hall T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hasegawa Y., Fukuda T., Hagimori K., Tomoda H., Ōmura S. (2007). Tensyuic acids, new antibiotics produced by Aspergillus niger FKI-2342. Chem. Pharm. Bull. 55 1338–1341. 10.1248/cpb.55.1338 [DOI] [PubMed] [Google Scholar]
- Heo I., Hong K., Yang H., Lee H. B., Choi Y. J., Hong S. B. (2019). Diversity of Aspergillus, Penicillium, and Talaromyces species isolated from freshwater environments in Korea. Mycobiology 47 12–19. 10.1080/12298093.2019.1572262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseltine C. W., Ellis J. (1961). Notes on mucorales, especially Absidia. Mycologia 53 406–426. 10.2307/3756584 [DOI] [Google Scholar]
- Hesseltine C. W., Ellis J. (1964). The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 56 568–601. 10.2307/3756362 [DOI] [Google Scholar]
- Hong S. B., Go S. J., Shin H. D., Frisvad J. C., Samson R. A. (2005). Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97 1316–1329. 10.3852/mycologia.97.6.1316 [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R. A. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 70 1–51. 10.3114/sim.2011.70.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Hyde K. D., Al-Hatmi A. M. S., Andersen B., Boekhout T., Buzina W., Dawson T. L., et al. (2018). The world’s ten most feared fungi. Fungal Divers. 93 161–194. 10.1007/s13225-018-0413-9 [DOI] [Google Scholar]
- Hyde K. D., Jeewon R., Chen Y. J., Bhunjun C. S., Calabon M. S., Jiang H. B., et al. (2020a). The numbers of fungi: is the descriptive curve flattening?. Fungal Divers. 10.1007/s13225-020-00458-2 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Maharachchikumbura S. S. N., Bhat D. J., Jones E. B. G., Bundhun D., et al. (2020b). Refined families of Sordariomycetes. Mycosphere 11 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Hyde K. D., Tennakoon D. S., Jeewon R., Bhat D. J., Maharachchikumbura S. S. N., Rossi W., et al. (2019a). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde K. D., Xu J., Rapior S., Jeewon R., Lumyong S., Niego A. G. T., et al. (2019b). The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 97 1–136. 10.1007/s13225-019-00430-9 [DOI] [Google Scholar]
- Jeewon R., Hyde K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Jones E. B. G., Pang K. L., Abdel-Wahab M. A., Scholz B., Hyde K. D., Boekhout T., et al. (2019). An online resource for marine fungi. Fungal Divers. 96 347–433. 10.1007/s13225-019-00426-5 [DOI] [Google Scholar]
- Jurjević Ž, Peterson S. W., Stea G., Solfrizzo M., Varga J., Hubka V., et al. (2012). Two novel species of Aspergillus section Nigri from indoor air. IMA Fungus 3 159–173. 10.5598/imafungus.2012.03.02.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyoti S., Paramita B., Vanaja J., Shalini C. (2013). Phosphate solubilization by a few fungal strains belonging to the genera Aspergillus and Penicillium. Afr. J. Microbiol. Res. 7 4862–4869. 10.5897/ajmr2013.5991 [DOI] [Google Scholar]
- Kucey R. (1983). Phosphate-solubilizing bacteria and fungi in various cultivated and virgin Alberta soils. Can. J. Soil Sci. 63 671–678. 10.4141/cjss83-068 [DOI] [Google Scholar]
- Lange L. (2014). The importance of fungi and mycology for addressing major global challenges. IMA Fungus 5 463–471. 10.5598/imafungus.2014.05.02.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Tsang C. C., Woo P. (2017). Talaromyces marneffei Genomic, Transcriptomic, proteomic and metabolomic studies reveal mechanisms for environmental adaptations and virulence. Toxins 9:192. 10.3390/toxins9060192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. J., Hyde K. D., Zhao R. L., Hongsanan S., Abdel-Aziz F. A., Abdel-Wahab M. A., et al. (2016). Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78 1–237. 10.1007/s13225-016-0366-9 [DOI] [Google Scholar]
- Li X., Luo L., Yang J., Li B., Yuan H. (2015). Mechanisms for solubilization of various insoluble phosphates and activation of immobilized phosphates in different soils by an efficient and salinity-tolerant Aspergillus niger strain An2. Appl. Biochem. Biotechnol. 175 2755–2768. 10.1007/s12010-014-1465-2 [DOI] [PubMed] [Google Scholar]
- Li Z., Bai T., Dai L., Wang F., Tao J., Meng S., et al. (2016). A study of organic acid production in contrasts between two phosphate solubilizing fungi: Penicillium oxalicum and Aspergillus niger. Sci. Rep. 6:25313. 10.1038/srep25313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. H., Wei Y. Y., Mohamad O. A. A., Salam N., Zhang Y.-G., Guo J. W., et al. (2019). Diversity, community distribution and growth promotion activities of endophytes associated with halophyte Lycium ruthenicum Murr. 3 Biotech 9:144. 10.1007/s13205-019-1678-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lorenzini M., Cappello M. S., Perrone G., Logrieco A., Zapparoli G. (2019). New records of Penicillium and Aspergillus from withered grapes in Italy, and description of Penicillium fructuariae-cellae sp. nov. Phytopathol. Mediterr. 58 325–341. 10.14601/Phytopathol_Mediter-10619 [DOI] [Google Scholar]
- Mohamad O. A. A., Li L., Ma J. B., Hatab S., Xu L., Guo J. W., et al. (2018). Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahliae. Front. Microbiol. 9:924. 10.3389/fmicb.2018.00924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandimath A. P., Karad D. D., Gupta S. G., Kharat A. S. (2017). Consortium inoculum of five thermo-tolerant phosphate solubilizing Actinomycetes for multipurpose biofertilizer preparation. Iran. J. Microbiol. 9 295–304. [PMC free article] [PubMed] [Google Scholar]
- Nelofer R., Syed Q., Nadeem M., Bashir F., Mazhar S., Hassan A. (2015). Isolation of phosphorus-solubilizing fungus from soil to supplement biofertilizer. Arabian J. Sci. Eng. 41 2131–2138. 10.1007/s13369-015-1916-2 [DOI] [Google Scholar]
- O’Donnell K., Cigelnik E., Nirenberg H. I. (1998). Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90 465–493. 10.2307/3761407 [DOI] [Google Scholar]
- Pangging M., Nguyen T. T. T., Lee H. B. (2019). New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology 47 154–164. 10.1080/12298093.2018.1554777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R., Hyde K. D., Jeewon R., Bhat D. J., Jones E. B. G., Maharachchikumbura S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Pikovskaya R. (1948). Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 17 362–370. [Google Scholar]
- Pradhan N., Sukla L. (2006). Solubilization of inorganic phosphates by fungi isolated from agriculture soil. Afr. J. Biotechnol. 5 850–854. [Google Scholar]
- Premono M. E., Moawad A., Vlek P. (1996). Effect of phosphate-solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indonesian J. Crop Sci. 11 13–23. [Google Scholar]
- Qiao H., Sun X. R., Wu X. Q., Li G. E., Wang Z., Li D. W. (2019). The phosphate-solubilizing ability of Penicillium guanacastense and its effects on the growth of Pinus massoniana in phosphate-limiting conditions. Biol. Open 8:bio046797. 10.1242/bio.046797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg W., Binder M., Groenewald J. Z., Summerell B. A., Carnegie A. J., Burgess T. I., et al. (2014). Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 33 1–40. 10.3767/003158514x681981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi P., Vyas P., Sharma S., Gulati A., Gulati A. (2009). Plant growth promoting potential of the fungus Discosia sp. FIHB 571 from tea rhizosphere tested on chickpea, maize and pea. Indian J. Microbiol. 49 128–133. 10.1007/s12088-009-0026-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner S. A., Samuels G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98 625–634. [Google Scholar]
- Richardson A. E., Simpson R. J. (2011). Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 156 989–996. 10.1104/pp.111.175448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J., Estefan G., Rashid A. (2001). Soil and Plant Analysis Laboratory Manual, second Edn Aleppo: ICARDA. [Google Scholar]
- Samson R. A., Visagie C. M., Houbraken J., Hong S. B., Hubka V., Klaassen C. H. W., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78 141–173. 10.1016/j.simyco.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra R., Cabañes F. J., Perrone G., Castellá G., Venâncio A., Mulć G., et al. (2017). Aspergillus ibericus: a new species of section Nigri isolated from grapes. Mycologia 98 295–306. 10.1080/15572536.2006.11832702 [DOI] [PubMed] [Google Scholar]
- Sharma R. (2012). Pathogenecity of Aspergillus niger in plants. Cibtech J. Microbiol. 1 47–51. [Google Scholar]
- Sharma S. B., Sayyed R. Z., Trivedi M. H., Gobi T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587. 10.1186/2193-1801-2-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Show P. L., Oladele K. O., Siew Q. Y., Aziz Zakry F. A., Lan J. C. W., Ling T. C. (2015). Overview of citric acid production from Aspergillus niger. Front. Life Sci. 8 271–283. 10.1080/21553769.2015.1033653 [DOI] [Google Scholar]
- Solomon L., Tomii V. P., Dick A. A. (2019). Importance of fungi in the petroleum, agro-allied, agriculture and pharmaceutical industries. N. Y. Sci. J. 12 8–15. 10.7537/marsnys120519.02 29154796 [DOI] [Google Scholar]
- Sørensen A., Lubeck P. S., Lubeck M., Nielsen K. F., Ahring B. K., Teller P. J., et al. (2011). Aspergillus saccharolyticus sp. nov., a black Aspergillus species isolated in Denmark. Int. J. Syst. Evol. Microbiol. 61 3077–3083. 10.1099/ijs.0.029884-0 [DOI] [PubMed] [Google Scholar]
- Stolk A. C., Samson R. A. (1972). The genus Talaromyces. Studies on Talaromyces and related genera II. Stud. Mycol. 2 1–65. 10.3114/sim.2011.70.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. D., Chen A. J., Houbraken J., Frisvad J. C., Wu W. P., Wei H. L., et al. (2020). New section and species in Talaromyces. MycoKeys 68 75–113. 10.3897/mycokeys.68.52092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Jacobson D. J., Kroken S., Kasuga T., Geiser D. M., Hibbett D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Tibpromma S., Hyde K. D., Jeewon R., Maharachchikumbura S. S. N., Liu J. K., Bhat D. J., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 83 1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C. M., Hirooka Y., Tanney J. B., Whitfield E., Mwange K., Meijer M., et al. (2014a). Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 78 63–139. 10.1016/j.simyco.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C. M., Houbraken J., Frisvad J. C., Hong S. B., Klaassen C. H. W., Perrone G., et al. (2014b). Identification and nomenclature of the genus Penicillium. Stud. Mycol. 78 343–371. 10.1016/j.simyco.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C. M., Renaud J. B., Burgess K. M. N., Malloch D. W., Clark D., Ketch L., et al. (2016). Fifteen new species of Penicillium. Persoonia 36 247–280. 10.3767/003158516x691627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C. M., Yilmaz N., Frisvad J. C., Houbraken J., Seifert K. A., Samson R. A., et al. (2015). Five new Talaromyces species with ampulliform-like phialides and globose rough walled conidia resembling T. verruculosus. Mycoscience 56 486–502. 10.1016/j.myc.2015.02.005 [DOI] [Google Scholar]
- Vyas P., Rahi P., Chauhan A., Gulati A. (2007). Phosphate solubilization potential and stress tolerance of Eupenicillium parvum from tea soil. Mycol. Res. 111 931–938. 10.1016/j.mycres.2007.06.003 [DOI] [PubMed] [Google Scholar]
- Walsh T. J., Raad I., Patterson T. F., Chandrasekar P., Donowitz G. R., Graybill R., et al. (2007). Treatment of invasive aspergillosis with posaconazole in patients who are refractory to or intolerant of conventional therapy: an externally controlled trial. Clin. Infect. Dis. 44 2–12. 10.1086/508774 [DOI] [PubMed] [Google Scholar]
- Wanasinghe D. N., Phukhamsakda C., Hyde K. D., Jeewon R., Lee H. B., Gareth Jones E. B. G., et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- Wang X., Wang C., Sui J., Liu Z., Li Q., Ji C., et al. (2018). Isolation and characterization of phosphofungi, and screening of their plant growth-promoting activities. AMB Express 8:63. 10.1186/s13568-018-0593-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic; ), 315–322. [Google Scholar]
- Yilmaz N., López-Quintero C. A., Vasco-Palacios A. M., Frisvad J. C., Theelen B., Boekhout T., et al. (2016). Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycol. Progress 15 1041–1056. 10.1007/s11557-016-1227-3 [DOI] [Google Scholar]
- Zhang Y., Chen F. S., Wu X. Q., Luan F. G., Zhang L. P., Fang X. M., et al. (2018). Isolation and characterization of two phosphate-solubilizing fungi from rhizosphere soil of moso bamboo and their functional capacities when exposed to different phosphorus sources and pH environments. PLoS One 13:e0199625. 10.1371/journal.pone.0199625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Y., Han Y. F., Chen W. H., Liang Z. Q. (2019). Gongronella sichuanensis (Cunninghamellaceae, Mucorales), a new species isolated from soil in China. Phytotaxa 416 167–174. 10.11646/phytotaxa.416.2.4 24943631 [DOI] [Google Scholar]
- Zhao X., Tian K., Yau S. S. T. (2018). A new efficient method for analyzing fungi species using correlations between nucleotides. BMC Evol. Biol. 18:200. 10.1186/s12862-018-1330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on RPB2 sequence data of Aspergillus strains.
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on RPB2 sequence data of Penicillium strains.
Phylogram generated from maximum likelihood analysis based on ITS sequence data of Talaromyces strains.
Phylogram generated from maximum likelihood analysis based on BenA sequence data of Talaromyces strains.
Phylogram generated from maximum likelihood analysis based on CaM sequence data of Talaromyces strains.
Taxa and their GenBank accession numbers used in this study.
Phosphate solubilization of tricalcium phosphate (TCP) on solid and liquid Pikovskaya (PVK) media with pH values by 13 fungal strains and control.
Details of 13 phosphate-solubilizing fungal strains used in this study.
Nucleotide comparison of ITS, BenA, CaM, and RPB2 between strains of Penicillium guaibinense and P. austrosinense.
List of phosphate-solubilizing fungi.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.