Abstract
The long flexible tail tube of bacteriophage lambda connects its capsid to the tail tip. On infection, a DNA ejection signal is passed from the tip, along the tube to the capsid that triggers passage of the DNA down the tube and into the host bacterium. The tail tube is built from repeating units of the major tail protein, gpV, which has two distinctive domains. Its N-terminal domain has the same fold as proteins that form the rigid inner tubes of contractile tail phages, such as T4, and its C-terminal domain adopt an Ig-like fold of unknown function. We determined structures of the lambda tail tube in free tails and in virions before and after DNA ejection using cryoelectron microscopy. Modeling of the density maps reveals how electrostatic interactions and a mobile loop participate in assembly and also impart flexibility to the tube while maintaining its integrity. We also demonstrate how a common protein fold produces rigid tubes in some phages but flexible tubes in others.
Keywords: Bacteriophage, Cryoelectron microscopy, Helical, Siphoviridae, Tail
Introduction
Double-stranded DNA (dsDNA)–tailed bacteriophages are among the most abundant organisms on earth and show enormous diversity [1]. Morphologically they are composed of either an icosahedrally symmetric or prolate icosahedral capsid that contains their genetic information, a tail that shows variable shapes, and a tail tip or baseplate that recognizes specific host bacteria and triggers DNA ejection. Despite variations in virion size and shape, the major structural proteins of the capsid and tail each have a common fold, and the virion assembly pathway also follows a general paradigm.
Tailed phages are divided into three families according to the shape of their tail: Myoviridae have a rigid tail surrounded by a contractile sheath; Siphoviridae have a flexible noncontractile tail; and Podoviridae have a short, noncontractile “spike”. For Siphoviridae and Myoviridae, the tail tube connects the capsid to the tail tip and transmits the signal for DNA release to the capsid when the tip binds to the host; subsequently, it becomes the channel through which the viral DNA is transferred from the capsid to the host. The tails of phages T4 (Myoviridae) and lambda (Siphoviridae) have both been the subject of numerous studies focusing on their assembly [2–8]. The rigidity and highly symmetrical organization of the T4 tail tube coupled with advances in cryo-EM (cryoelectron microscopy) have yielded a detailed view of the tail’s complex organization [9]. However, the flexibility of the lambda tail poses a unique challenge for structural analysis. Nevertheless, recent work on the similarly flexible phage T5 tail tube demonstrated that detailed structural information of such a particle may be determined successfully by cryo-EM [10].
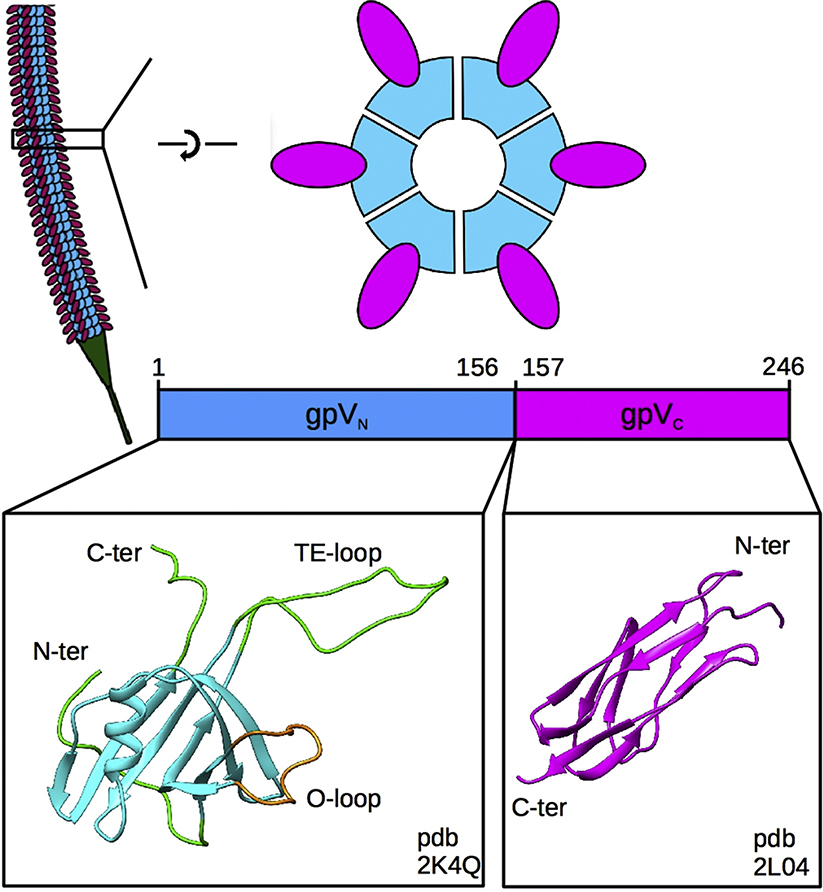
The tubular part of the lambda tail is 135 nm long and composed of 32 stacked hexameric rings of the major tail protein (gpV) that are wrapped around a polymer of tape measure protein (gpH) that regulates the tail length during assembly [11]. gpV is a 246-amino acid protein that possesses two structurally and functionally distinct domains whose respective monomeric structures have been solved by NMR (Fig. 1) [12,13]. The N-terminal domain (residues 1 to 156) of gpV (gpVN) comprises the core of the tail tube and folds as a beta sandwich with an embedded alpha-helix and an extended-loop (tail extended-loop or TE-loop). This fold is duplicated in the major tail protein of the Siphoviridae phage T5 pseudohexameric tail tube [10] and has also been found in tube proteins of the Myoviridae phage T4 tail tube protein gp199; 14, the Pseudomonas aeruginosa type VI secretion system (T6SS) protein Hcp1 [15], the P. aeruginosa R-type pyocin tube [16], and the Photorhabdus asymbiotica extracellular contractile injection system [17], suggesting that they all come from a common ancestor [12]. Interestingly, the NMR solution structure of lambda gpVN revealed that the N-terminal-most region (N-ter, residues 1 to 14), the TE-loop (residues 50 to 78) and the C-terminal-most region (C-ter, residues 149 to 153) were disordered. This NMR structure also shows an extra loop on the outside of the fold (Outer-loop or O-loop, residues 24 to 38) that is not present on known homologs. The C-terminal domain (residues 157 to 246) of gpV (gpVC) shows an Ig-like fold, commonly found on the surfaces of many phage structural proteins [18]. While most phage Ig-like domains do not have clearly defined functions, lambda mutants with truncations of all or parts of the gpV Ig-like domain have measurable biological defects [13,19] suggesting that it has a role in tail assembly.
Fig. 1. Organization of the lambda tail.
Top left, representation of the tail tube showing identical disks of the gpV tail tube protein stacked along the length of the tail and the tail tip in green. Top right, top, and side views of a single hexameric disk of the tail tube protein showing the organization of the N-terminal gpVN (blue) and C-terminal gpVC (magenta) domains. Bottom, the monomeric NMR structures of the gpVN domain (PDB ID: 2K4Q), including the rigid (blue), flexible (green) and O-loop (tan) regions, and the Ig-like gpVC domain (PDB ID: 2L04).
Because the structures of gpV domains in solution are known, we are particularly interested in studying the structure of the tail to identify the regions of gpV that contribute to its assembly. In this work, we have determined cryo-EM structures of the tubular part of the lambda tail in three different contexts: in the virion before and after ejection and in a tail purified from plasmid expression (free tail). Despite the challenges of tail flexibility and, in the case of the virion sample, ice thickness, we have been able to solve these structures at resolutions sufficient for identifying the gpV fold, building a model of the in vivo tail structure and detailing the structural reorganization of gpV that occurs during tail polymerization (6.8, 6.4, and 5.4 Å for the tail tube of the virion, the ejected particles and the free tails, respectively). The three structures also give clues about how the DNA is transported through the tube and the structural role that the Ig-like domain of gpV may play.
Results and Discussion
Lambda tail tube structures
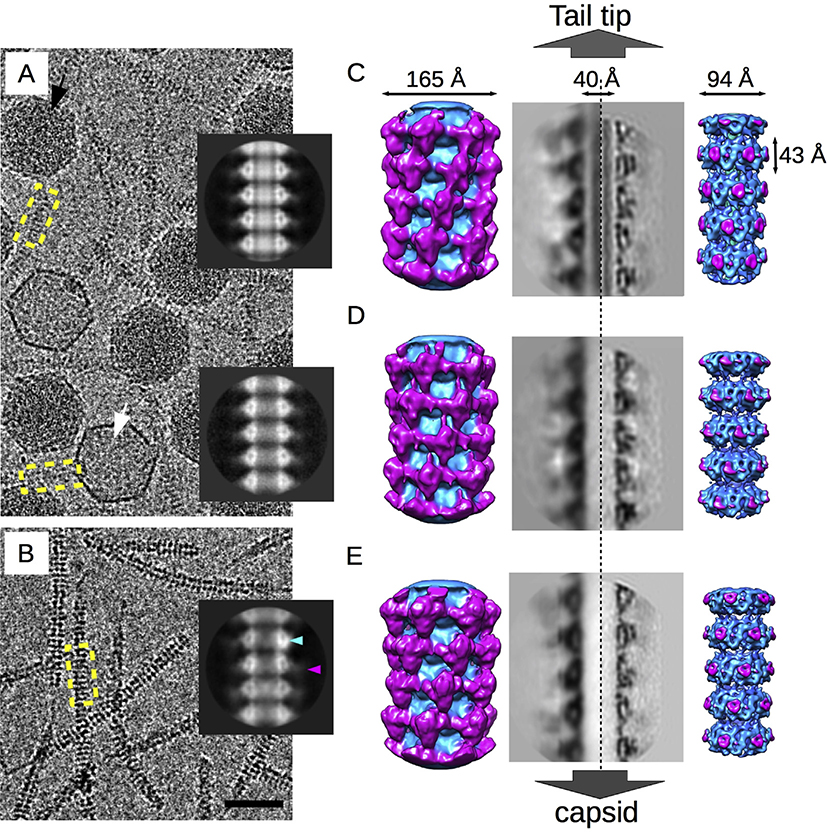
Purified virions and free tails were vitrified separately for imaging by cryo-EM. In addition to normal DNA-containing phages, the virion sample also included empty particles (Fig. 2A and B) that we interpreted as viruses that have ejected their DNA. Images of the full virions and empty particles were separated into independent data sets. While the capsids were readily visible in this sample, contrast for the tails was low because of the relatively thick ice embedding the ~70 nm diameter capsid. In comparison, the free tail sample was embedded in thinner ice and showed much higher contrast as demonstrated by the highly visible striations because of the tail tube disks (Fig. 2C). The three types of tail tube were analyzed by 2D averaging (Fig. 2A and B, inset) and 3D reconstruction (Fig. 2C–E). As expected, the 2D average of the virion tail tube showed strong inner-density filling the lumen (Fig. 2A, top inset) that we attribute to the tape measure protein (gpH). The luminal density was absent in the ejected tail (Fig. 2A, bottom inset) consistent with gpH being released during DNA ejection. Surprisingly, the free tail also lacked the inner luminal density (Fig. 2B, inset) suggesting that gpH was lost during preparation, perhaps because of the absence of one or more proteins on the plasmid that may act to retain gpH in the tail or from purification steps that strip gpH from the tail.
Fig. 2. Cryo-EM structures of lambda tails.
A. Representative micrograph of the virion sample showing DNA-filled virions (black arrow) and empty particles (white arrow). B. Representative micrograph of the free tails. The bar is 50 nm. Inset from top to bottom, 2D averages of the tail tube from the virions, the empty particles and the free tails. The blue and magenta arrowheads point to the inner tube density and the blurry external density, respectively. C–E. Three-dimensional reconstructions of the virions, empty particles, and the free tails, respectively. From left to right, surface views of the low resolution outer-density reconstructions, showing the external Ig-like gpVC domain organization (magenta); cross-section through the outer-density reconstructions; cross-section through the high-resolution inner-density reconstructions; and surface views of the inner-density reconstruction showing the organization of the gpVN domain (blue).
The 2D averages of the three tail data sets confirmed that the overall morphology of the tubes and individual disks are similar. The tail tube density of the 2D averages was divided into two parts corresponding to the two domains of gpV: an intense triangular-shaped density surrounding the lumen that we ascribed to the N-terminal tube domain, gpVN, and a weaker external density with a poorly defined shape that we attributed to the C-terminal Ig-like gpVC domain (Fig. 2A and B, inset). The poor definition of the external density may be due to the flexibility of the Ig-like domain relative to the gpVN domain. To account for this flexibility, we performed two 3D reconstructions for each tail sample using cylindrical masks of different radial limits. For the external tube structure composed of the C-terminal Ig-like domain, only density between diameters of 98–160 Å was included during the refinement (Fig. 2C–E, left). For the internal tube structure comprised the N-terminal tube domain, we included density between diameters of 40–98 Å (Fig. 2C–E, right). The same helical rise (42.8 Å) and rotation (17.5°) as well as the imposition of six-fold symmetry were applied to each reconstruction. Maps that focused on the external structure did not yield detailed structural information about the fold as it is probably too flexible to reach high resolution. However, clues about the quaternary structure of the domain emerged (Fig. 2C–E, left). The external density that we attributed to the Ig-like domain (in pink) forms a quaternary filamentous structure through interdisk contacts and wraps around the tail along its axis. Furthermore, while the Ig-like domains of both virion and free tail tubes appear to be similarly organized, that of the ejected tail tube shows a disrupted organization, suggesting that DNA ejection triggers conformational changes that affect the outer surface of the tube. Because gpH is present in the virion and absent in the free tail, it also suggests that this protein does not significantly influence organization of the Ig-like domain.
Density maps focused on the inner tube structure were solved to resolutions of 6.8, 6.4, and 5.4 Å (gold standard FSC = 0.143—see Table 1) for the tail tube of the virion, the ejected particles and the free tails, respectively (Fig. 2C–E, cross-sections and surfaces on the right; Table 1; Fig. S1). The cross-sections show that the three maps show a similar level of detail and that the gpVN domain is slightly different in each reconstruction (movie S1, Fig S2). The most obvious differences are between the virion tail tube before and after ejection, including a slight rotation of the inner density toward the interior of the tube and a clear reorganization of the outer density—O-loop—that may be linked to reorganization of the Ig-like domain. Although our density maps have insufficient resolution for modeling subtle atomic conformational changes our data show that the conformation of gpV is not rigid but adjusts to environmental modifications including the presence or absence or of gpH, and passage of DNA through its lumen.
Table 1.
Details of reconstructions.
| Particle counts |
|||
|---|---|---|---|
| Source of tail tubes | total | in final map | Resolutiona (Å) |
| Virions | 9761 | 4668 | 6.4 |
| Ejected empty particles | 1504 | 1138 | 6.8 |
| Plasmid expressed | 7934 | 5543 | 5.4 |
Gold standard Fourier Shell Correlation curves are shown in Fig. S1.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.jmb.2019.10.031.
In contrast, the free tail tube structure is almost identical to that of the virion with the latter being subtly contracted closer to the gpH density. Because the free tails lacked gpH, the comparison suggests that ejection of gpH is unlikely to cause the conformational changes to gpV seen in the tail tubes of the emptied particles. We conclude that the interactions between gpV and gpH in the virion tail are very limited, facilitating efficient ejection of gpH. The interaction between gpV and gpH during assembly is apparently mediated by tail chaperone protein gpGT and may obviate the need for gpH to interact specifically with gpV to regulate tail length [8]. While recent work on the phage T5 tail tube structure suggested that the tape measure protein is the signal transmitter [10], a different study on phage SPP1 by negative-stain EM suggested instead that the signal is transmitted through the major tail protein [20]. In the present work comparing the tails of virions before and after DNA ejection, we detected only subtle conformational differences in gpV that are insufficient for identifying a protein or a mechanism responsible for the transmission of the DNA ejection signal.
Assembly results in a reorganization of the tail tube fold
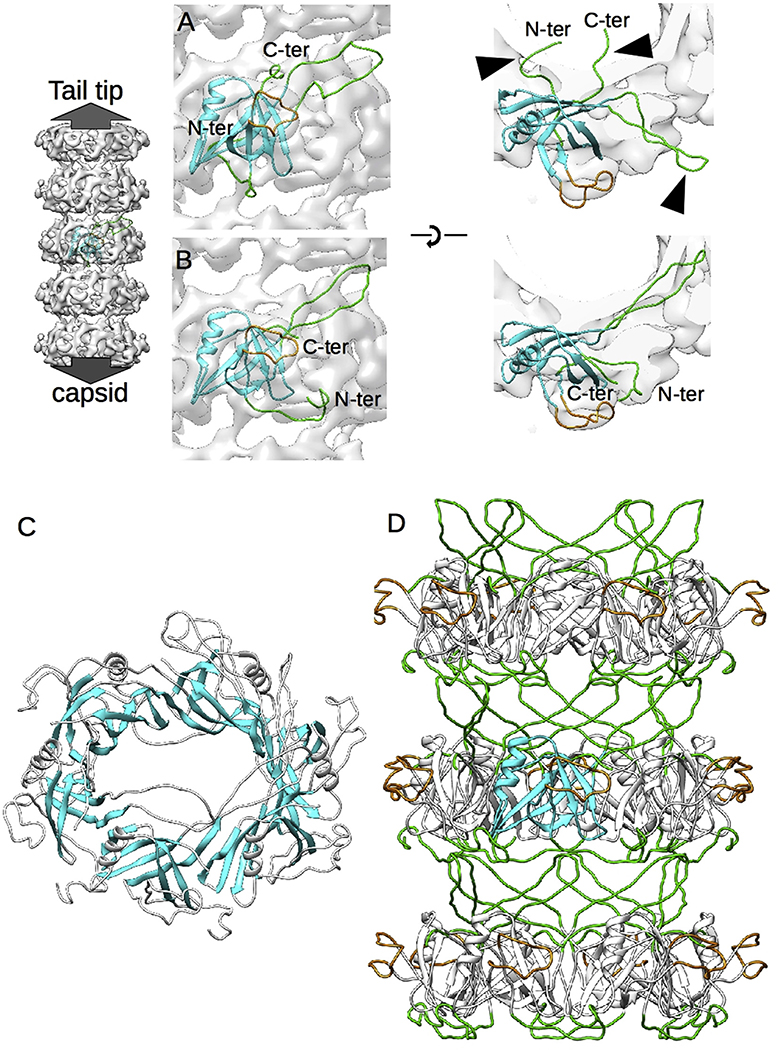
The fold of gpVN is readily apparent in the density maps of each of the three tail samples, and the NMR model of the monomer (PDB Id: 2K4Q 12) can be fit into all three maps. We used our highest quality map from free tails to build a complete model of gpVN in the context of the assembled lambda tail tube. The initial rigid body fit accommodated most of the gpVN model into the tail tube density map (Fig. 3A), but not the three highly flexible regions identified in the NMR model: the N-terminal region, the TE-loop and the C-terminal region (indicated by arrowheads in Fig. 3A). Nevertheless, density corresponding to those regions is present in the tail tube map and may be assigned, through flexible fitting, to reveal the assembled state of gpVN (Fig. 3B). Indeed, the strength of the density corresponding to those regions indicates a very well ordered conformation in comparison to the flexibility observed in the NMR model, and shows that a significant transition in gpVN conformation occurs during tail tube assembly. This transition includes reorganization of the fold within individual disks (Fig. 3C), where the beta strands of all six subunits form a continuous intradisk beta barrel-like motif. We conclude that this intradisk beta barrel likely stabilizes fully assembled tail tube disks. Noting that an increase of beta content was found after the SPP1 tail tube protein assembles [21], we propose that the formation of intradisk beta barrels may be a common strategy for structural reinforcement of tail tubes.
Fig. 3. Modeling of the N-terminal gpVN domain.
A. Rigid body fitting of the gpVN domain model (PDB Id: 2K4Q) into the free tail density map viewed from the side of the tube (left) and the top (right). One representative NMR model of the 20 available has been chosen. B. Same views as (A) after refinement of the gpVN structure by flexible fitting. C. Formation of an intradisk beta barrel-like structure mediated by the rigid beta strands of the gpVN domain. D. Final model of the inner tail tube structure, showing that the interdisk contacts are mediated by the flexible loops (green) identified in the NMR model of gpVN.
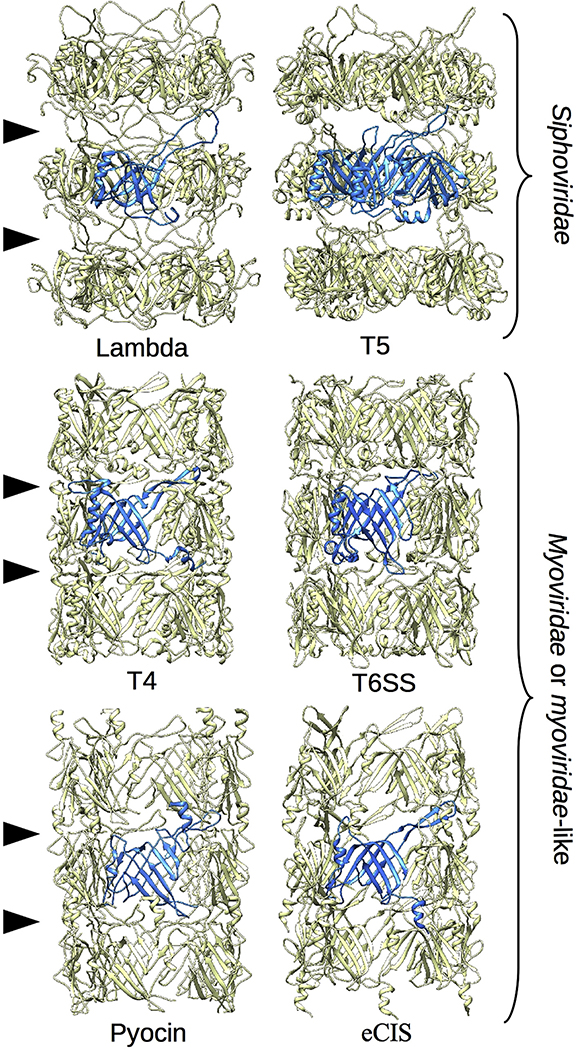
Another set of conformational transitions immobilize formerly flexible regions, including 1) trapping of the N-terminal region by interactions with the next subunit in the capsid-end direction; 2) capture of the TE-loop on the tail-tip proximal surface of the adjacent subunit so that it lies along the tail tube shell where it can interact with other subunits; and 3) forced location of the gpVN C-terminal residues and connected gpVC domain to the exterior of the tail tube. Interestingly, a model of the tube structure made with three disks of gpVN hexamers shows that the interdisk interface is composed solely of the refolded N-terminal and TE-loop regions (Fig. 3D). The lack of other structures at the disk interfaces could explain the intrinsic flexibility of the tail tube. To verify this hypothesis, we compared our lambda tail tube structure with the similarly flexible T5 tail tube and to the more rigid inner tubes of T4, T6SS, pyocin, and extracellular contractile injection system (eCIS) (Fig. 4) [10,14,16,17,22]. All these structures show the same N-terminal-TE-loop interaction at the disk interface. However, in the case of the rigid tubes additional loops are involved, the interaction networks are more intricate and the interdisk grooves are completely filled. Each of these features very likely contributes to the observed rigidity of these structures.
Fig. 4. Tube flexibility depends on interdisk organization.
The structures of two flexible tubes and four rigid inner tubes are shown as marked: the tail tubes of phage lambda (this work) and phage T5 (PDB 5NGJ, EMDB- 3692), and the inner tubes of phage T4 (PDB 5W5F), the Type 6 secretion system (PDB 50JQ), a pyocin (PDB 3J9Q) and the extracellular contractile injection system (PDB 6J0B).
The TE-loop also appears to play a dominant role in facilitating tail tube polymerization and intersubunit connectivity. In the tail tube, the TE-loop is oriented toward the tail tip and interacts with the N-terminal regions of two gpVN subunits located in the adjacent tip-proximal disk. In addition to these interdisk interactions, the TE-loop interacts with adjacent subunits in the same disk, and together these multiple interactions make the TE-loop a central part of the tail tube’s overall structure. The significant structural reorganization of the gpVN tube domain that occurs during assembly could explain why purified gpV is monomeric in solution, but requires a tail tip/tape measure protein-chaperone complex to initiate polymerization into a lambda tail tube [23].
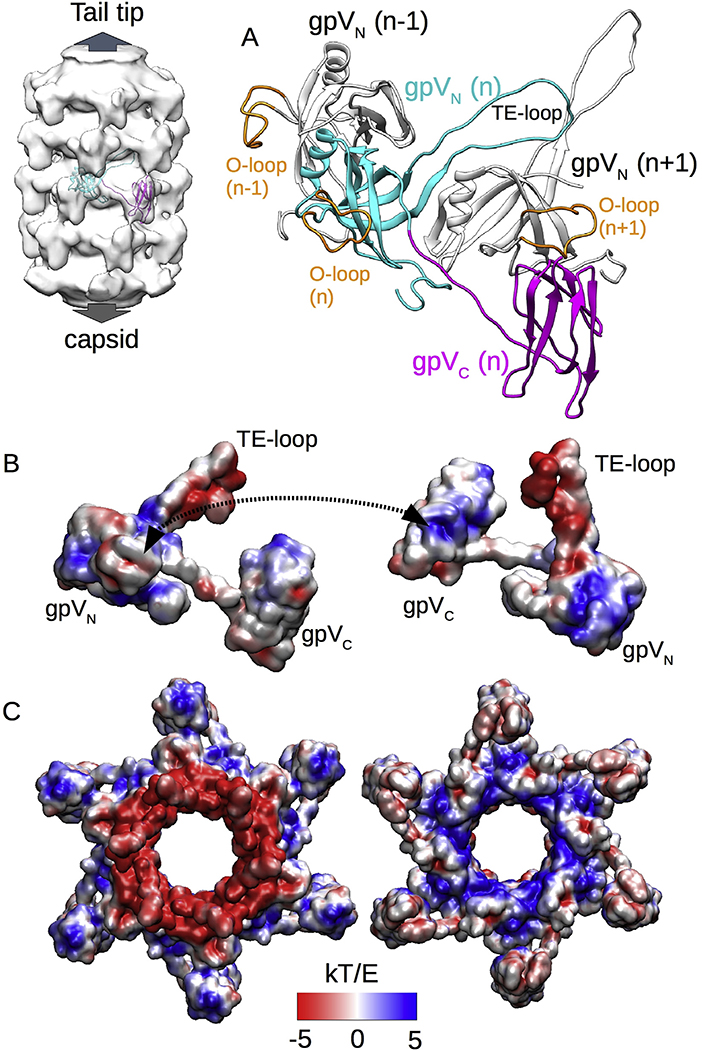
Roles for electrostatics in tail tube assembly and stability
From the high-resolution density map of the tail tube inner structure, we could locate the linker between the N-terminal and C-terminal domains of gpV (Fig S3) and thus determine the connectivity of the Ig-like gpVC domain and its orientation in the virion tail. As shown in Fig. 5, the linker between gpVN and gpVC is extended so that the gpVC domain nestles against the gpVN O-loop of an adjacent subunit on the same disk. According to our fitting, albeit in a relatively low-resolution map, the long axis of the Ig-like domain is oriented along the tail tube axis so that each Ig-like domain contacts two other Ig-like domains in the disks immediately above and below. The resulting coaxial network of Ig-like domain connectivity may contribute to tail tube stability and could also function during assembly. Interestingly, the electrostatic pattern of gpV shows striking polarities (Fig. 5B): the TE-loop (tip proximal) is very electronegative, while the N-terminal domain (capsid proximal) is mostly electropositive. In contrast, the Ig-like domain shows inverted polarity, i.e., the tip proximal side is mostly electropositive, while the capsid proximal side is mostly electronegative. Electrostatic interactions seem likely to be involved in the interface between the Ig-like and N-terminal tube domains because the mostly electropositive part of the Ig-like domain is in contact with the electronegative O-loop. Similar charge complementarities are even more evident in our electrostatic analysis of an entire disk (Fig. 5C). The disk is clearly divided into two regions with inverted charge distributions: the opposite surfaces of the central core are strongly opposed in charge, while the Ig-like domains are similarly opposed but inverted in charge, suggesting an electrostatic mechanism favoring stabilization of disk stacking. Little is known about the events that occur during lambda tail tube polymerization, but according to our model the TE-loop likely plays an important role in transitioning from a flexible to a rigid conformation, making both interdisk and intradisk interactions, and carrying most of the negative charges that are exposed on the top of the disk—7 aspartates and 3 glutamates. Thus, conformational reorganization of the TE-loop could be the trigger for tail tube polymerization.
Fig. 5. Electrostatic forces stabilize the tail tube structure.
A. Structural model of the entire gpV including both N-terminal gpVN and C-terminal gpVC domains as well as gpVN domains from two adjacent gpV subunits. B. Electrostatic pattern of the gpV subunit viewed from the outside (left) and inside (right) of the tail tube. A dashed double arrow shows the contact area of the O-loop and the Ig-like domain of an adjacent subunit. C. Electrostatic pattern of a tail tube disk viewed from the tail-tip end (left) or the capsid end (right) of the tail tube.
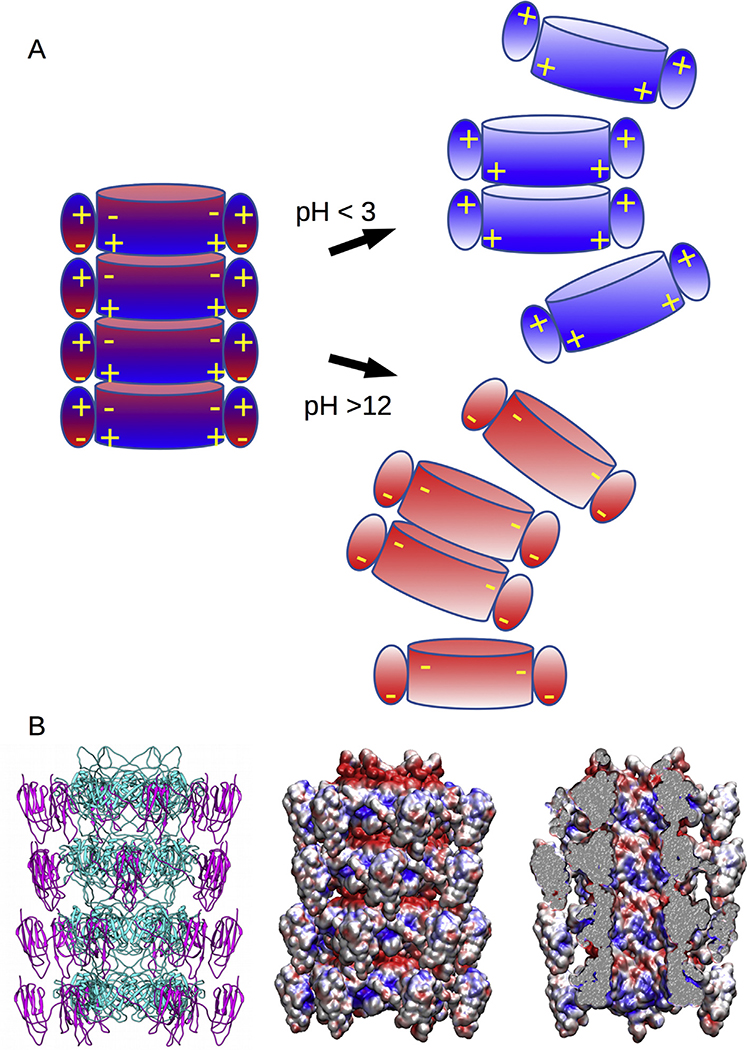
Studies of the in vitro dissociation and reassociation of lambda tails have revealed the influences of pH, ionic strength and the presence of urea on the tail [24], and the contribution of the Ig-like domain to tail stability [19]. Lambda tails are not particularly sensitive to urea (e.g., 7 M urea has no effect) or to ionic strength, but both alkaline conditions (pH > 12) and acidic conditions (pH < 3) lead to dissociation of the tail, which can subsequently reassociate into “polytails” after neutralization [24]. Interestingly, tails of gpV mutants with a truncated or missing Ig-like domain do not dissociate in acidic conditions [19]. Our structural model is compatible with those observations. At neutral pH, the complementary charge distributions of the opposite surface of the disks promote stacking (Fig. 6A, left). However, in strongly acidic conditions where the electrostatic potential of acidic residues is reduced, the inner domain positive charges on one disk may be repulsed by the Ig-like domain positive charges of the adjacent disk leading to disassembly. Similarly, in strongly alkaline conditions, the electrostatic potential of positively charged residues is diminished and now the negatively charged inner domain surface of one disk is repulsed by the negatively charged regions of the Ig-like domains, also leading to disassembly (Fig. 6A, right). In the absence of the Ig-like domain, the opposing charges are no longer present in the acidic conditions tested and the disks remain stacked through nonelectrostatic interactions. Although high salt did not induce tails to dissociate, tail tube dissociation and reassembly at high and low pH were inhibited by high concentrations of (>0.7 M) of NaCl [24]. This effect could be due to the screening of charges by high ionic strength and is consistent with our proposal that electrostatic forces drive both tube assembly and disassembly.
Fig. 6. Tail tube structure and disassembly model.
A. Disassembly model of the tail tube because of the masking of charges on either the inner tube at low pH, or the Ig-like domain at high pH, and the subsequent charge repulsion between disks. B. Structural model (right) of the tail tube with the electrostatic patterns shown viewed from the outside (middle) and inside (right) of the tail tube showing that its lumen surface is composed of a mix of negative and positive charges.
Finally, for both phage T4 and phage T5, the interior of the tail tube is mostly electronegative, and it was concluded that the repulsive charges of the DNA and the interior of the tube would allow the dsDNA molecule to slide through the tail during ejection [10,14]. However, analysis of the lambda tail (Fig. 6B) shows both positive and negative charges present in roughly equal proportions, suggesting that the interior of the tail tube does not need to be net electronegative to facilitate the movement of DNA. The lumen of the tail tube is about 40 Å in diameter, considerably greater than the width of dsDNA at about 20 Å and leaving a gap of 10 Å on average between the DNA and the interior surface of the tube. Furthermore, the DNA is solvated and possibly surrounded by counterions that would screen potential DNA-tube interactions. In this scenario, we consider that any potential electrostatic interactions between the DNA and the tail tube would be negligible and would not interfere with DNA ejection.
Materials and Methods
Tail and virus purification
Lambda phage particles were purified essentially as outlined previously [25]. A lysogen of λ CI857 Sam7 in Esherichia coli strain MM594 (Sup0, Smr, mal−) was grown to moderate density at 30 °C, induced at 42 °C, and grown for ~3 h at 37 °C. Cells were harvested by centrifugation, lysed with chloroform, and cell debris removed by centrifugation. Phage were purified by polyethylene glycol (PEG) precipitation (10% w/v PEG in 0.5 M NaCl), banded in cesium chloride by isopycnic centrifugation, and dialyzed into lambda dilution buffer (10 mM Tris HCl pH 7.5, 10 mM MgSO [4]).
The plasmid pT7–5-Tail4 that encodes a complete set of tail genes was used for tail production, as described previously [8] but with an additional chromatography step. After expression of the plasmid genes in BL21(DE3)plysS cells [26], the cells were harvested, lysed by sonication in 50 mM Tris HCl pH 8.0 and 5 mM EDTA buffer, and centrifuged at 14,000g to remove debris. The supernatant was precipitated by the addition of 0.7 M potassium glutamate plus 7.5% (w/v) PEG. The resulting pellet was resuspended in TKG buffer (20 mM Tris HCl, pH 7.5, 100 mM potassium glutamate) and sedimented in a 10–30% glycerol gradient for 5 h at 27k rpm in a SW28 rotor (Beckman, Fullerton, CA). The tail band was extracted then pelleted by ultracentrifugation at 100,000g and suspended in TKG buffer. Tails were further purified by adsorption to a 7.5 × 1500 mm HQ-20 anion exchange column (Applied Biosystems, Foster City, CA) in 20 mM Tris HCl buffer at pH 7.5, eluted with a gradient of sodium chloride, and concentrated by ultracentrifugation, as in the previous step.
CryoEM and image reconstruction
Three μl of purified and concentrated samples were deposited on a Quantifoil R2/1 copper grid covered by a 2 nm thick layer of continuous carbon (Quantifoil Micro Tools GmbH, Germany) that had been glow-discharged for 15 s. Grids were blotted and plunge-frozen with an FEI Vitrobot Mk III (FEI, Hillsboro OR) into a liquid nitrogen-cooled mixture of 2:1 ethane:propane[27]. Grids were mounted in an FEI Polara microscope operating at 300 kV and imaged under conditions of parallel illumination at a nominal magnification of 93,000×. Images were collected under control of the FEI “EPU” automated data collection software and recorded on an FEI Falcon 2 direct electron detecting camera with postcolumn magnifications of 1.4× yielding a calibrated pixel size at the specimen of 1.15 Å. Tail images were picked manually with the e2helixboxer.py software [28] and the contrast transfer functions estimated automatically from micrographs with CTFFIND4 [29]. Orientation determination and reconstructions were carried out with the RELION software [30,31]. Helical segments were extracted as overlapping regions 42 Å in length (roughly the helical rise). Starting from known parameters of a helical tail tube [10,14], the helical parameters were refined with RELION and determined to be 42.8 Å (helical rise) and 17.5° (helical twist). Those parameters were used for all the reconstructions in this work, and details of the final maps are shown in Table 1 with the FSC resolution estimates in Fig. EV1.
Structural modeling
We fit the NMR structures of the gpVN N-terminal (PDB ID: 2K4Q) and gpVC C-terminal (PDB ID: 2L04) domains [12,13] into our density maps as rigid bodies and used UCSF Chimera [32] and Coot [33] for manual refinement of the N-terminal domain. The gpVN model was subsequently refined using MDFF [34] followed by real space refinement in Phenix [35], the statistics of the map and the fitting are shown Fig. S4. For the electrostatic analysis, Adaptive Poisson-Boltzmann Solver (APBS) was used with a mobile ion concentration of 150 mM [36].
Accession numbers
The three maps are deposited in the EM-Data Bank with accession numbers EMD-20241, EMD-20242, and EMD-20243 for the free tail tube, the virion tail tube, and the empty particle tail tube, respectively. The structural model of gpV is deposited in the PDB with the accession number 6P3E.
Supplementary Material
Acknowledgments
The authors thank Dr. Alexander Makhov for technical support with electron microscopy. This work was supported in part by National Institutes of Health grant R01 GM047795 (RWH & RLD) and by the Office of the Director, National Institutes of Health, under Award Number S10 OD019995 (JFC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This paper is dedicated to Roger Hendrix, who inspired the current work and passed away before we achieved the results reported here.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2019.10.031.
References
- [1].Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF, Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Arisaka F, Yap ML, Kanamaru S, Rossmann MG, Molecular assembly and structure of the bacteriophage T4 tail, Biophys. Rev 8 (2016) 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Katsura I, Mechanism of length determination in bacteriophage lambda tails, Adv. Biophys 26 (1990) 1–18. [DOI] [PubMed] [Google Scholar]
- [4].Katsura I, Kuhl PW, Morphogenesis of the tail of bacteriophage lambda. III. Morphogenetic pathway, J. Mol. Biol 91 (1975) 257–273. [DOI] [PubMed] [Google Scholar]
- [5].Kikuchi Y, King J, Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway, J. Mol. Biol 99 (1975) 695–716. [DOI] [PubMed] [Google Scholar]
- [6].Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG, Morphogenesis of the T4 tail and tail fibers, Virol. J 7 (2010) 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xu J, Hendrix RW, Duda RL, A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly, J. Mol. Biol 425 (2013) 3476–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu J, Hendrix RW, Duda RL, Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length, J. Mol. Biol 426 (2014) 1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG, Structure of the T4 baseplate and its function in triggering sheath contraction, Nature 533 (2016) 346–352. [DOI] [PubMed] [Google Scholar]
- [10].Arnaud CA, Effantin G, Vives C, Engilberge S, Bacia M, Boulanger P, Girard E, Schoehn G, Breyton C, Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection, Nat. Commun 8 (2017) 1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Katsura I, Hendrix RW, Length determination in bacteriophage lambda tails, Cell 39 (1984) 691–698. [DOI] [PubMed] [Google Scholar]
- [12].Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR, The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 4160–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pell LG, Gasmi-Seabrook GM, Morais M, Neudecker P, Kanelis V, Bona D, Donaldson LW, Edwards AM, Howell PL, Davidson AR, Maxwell KL, The solution structure of the C-terminal Ig-like domain of the bacteriophage lambda tail tube protein, J. Mol. Biol 403 (2010) 468–479. [DOI] [PubMed] [Google Scholar]
- [14].Zheng W, Wang F, Taylor NMI, Guerrero-Ferreira RC, Leiman PG, Egelman EH, Refined cryo-EM structure of the T4 tail tube: exploring the lowest dose limit, Structure 25 (2017) 1436–1441 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Park YJ, Lacourse KD, Cambillau C, DiMaio F, Mougous JD, Veesler D, Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy, Nat. Commun 9 (2018) 5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH, Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states, Nat. Struct. Mol. Biol 22 (2015) 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiang F, Li N, Wang X, Cheng J, Huang Y, Yang Y, Yang J, Cai B, Wang YP, Jin Q, Gao N, Cryo-EM structure and assembly of an extracellular contractile injection system, Cell 177 (2019) 370–383 e15. [DOI] [PubMed] [Google Scholar]
- [18].Fraser JS, Yu Z, Maxwell KL, Davidson AR, Ig-like domains on bacteriophages: a tale of promiscuity and deceit, J. Mol. Biol 359 (2006) 496–507. [DOI] [PubMed] [Google Scholar]
- [19].Katsura I, Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion, J. Mol. Biol 146 (1981) 493–512. [DOI] [PubMed] [Google Scholar]
- [20].Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, Tavares P, Orlova EV, Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection, EMBO J. 26 (2007) 3720–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Langlois C, Ramboarina S, Cukkemane A, Auzat I, Chagot B, Gilquin B, Ignatiou A, Petitpas I, Kasotakis E, Paternostre M, White HE, Orlova EV, Baldus M, Tavares P, Zinn-Justin S, Bacteriophage SPP1 tail tube protein self-assembles into beta-structure-rich tubes, J. Biol. Chem 290 (2015) 3836–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang J, Brackmann M, Castano-Diez D, Kudryashev M, Goldie KN, Maier T, Stahlberg H, Basler M, Cryo-EM structure of the extended type VI secretion system sheath-tube complex, Nat. Microbiol 2 (2017) 1507–1512. [DOI] [PubMed] [Google Scholar]
- [23].Katsura I, Tsugita A, Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail, Virology 76 (1977) 129–145. [DOI] [PubMed] [Google Scholar]
- [24].Bleviss M, Easterbrook KB, Self-assembly of bacteriophage lambda tails, Can. J. Microbiol 17 (1971) 947–954. [DOI] [PubMed] [Google Scholar]
- [25].Sambrook J, Fritsch EF, Maniatis T, Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989. [Google Scholar]
- [26].Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW, Use of T7 RNA polymerase to direct expression of cloned genes, Methods Enzymol. 185 (1990) 60–89. [DOI] [PubMed] [Google Scholar]
- [27].Tivol WF, Briegel A, Jensen GJ, An improved cryogen for plunge freezing, Microsc. Microanal 14 (2008) 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ, EMAN2: an extensible image processing suite for electron microscopy, J. Struct. Biol 157 (2007) 38–46. [DOI] [PubMed] [Google Scholar]
- [29].Rohou A, Grigorieff N, CTFFIND4: fast and accurate defocus estimation from electron micrographs, J. Struct. Biol 192 (2015) 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination, J. Struct. Biol 180 (2012) 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].He S, Scheres SHW, Helical reconstruction in RELION, J. Struct. Biol 198 (2017) 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera – a visualization system for exploratory research and analysis, J. Comput. Chem 25 (2004) 1605–1612. [DOI] [PubMed] [Google Scholar]
- [33].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Trabuco LG, Villa E, Mitra K, Frank J, Schulten K, Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics, Structure 16 (2008) 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr. D Biol. Crystallogr 68 (2012) 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.