Abstract
Motivation
Hydrogen–deuterium mass spectrometry (HDX-MS) is a rapidly developing technique for monitoring dynamics and interactions of proteins. The development of new devices has to be followed with new software suites addressing emerging standards in data analysis.
Results
We propose HaDeX, a novel tool for processing, analysis and visualization of HDX-MS experiments. HaDeX supports a reproducible analytical process, including data exploration, quality control and generation of publication-quality figures.
Availability and implementation
HaDeX is available primarily as a web-server (http://mslab-ibb.pl/shiny/HaDeX/), but its all functionalities are also accessible as the R package (https://CRAN.R-project.org/package=HaDeX) and standalone software (https://sourceforge.net/projects/HaDeX/).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The understanding of interactions between proteins and other molecules is crucial for studying complex biological systems. Among the methods for characterization of conformational dynamics of proteins and their complexes, hydrogen–deuterium mass spectrometry (HDX-MS) has proven to be both rapid and sensitive (Konermann et al., 2011). This technique is especially important in the case of proteins that are difficult to study with other methods such as membrane proteins, oligomerizing proteins or intrinsically disordered proteins (Goswami et al., 2013).
Hydrogen–deuterium exchange monitors an exchange of amide hydrogens in peptide bonds. Protein incubation in D2O leads to the exchange of hydrogen to deuterium atoms in amides. The speed of such exchange depends mostly on the stability of hydrogen bonds formed by the hydrogen amides and also by the accessibility to the solvent. The importance of the influence of these factors is discussed, but it is considered that the dominant component is the stability of hydrogen bonds and not the availability of solvent. Thus, this process may be slowed due to structural factors: stability, flexibility and accessibility. Therefore the method, by measuring the level of protection of amides at different times of incubation in D2O, recognizes the stability of hydrogen bonding networks and regions of protein with limited solvent accessibility. HDX probes the dynamic nature of proteins systems, as opposed to static structures, offered by X-ray crystallography. It also allows mapping the regions affected by the interaction between the proteins concerned.
The main scheme of local (continuous-labeling, bottom-up) HDX-MS experiments consist of: the incubation of a protein in a D2O solution, buffered to native or native-like conditions, followed by exchange quench, proteolytic digestion and mass spectrometry measurement of resulting peptide’s masses.
Results generated by HDX-MS are complex and demanding in terms of analysis, interpretation and visual presentation. While there are many open-source and free to use software packages addressing the challenges of HDX-MS data analysis (Hourdel et al., 2016; Kavan and Man, 2011; Lau et al., 2019; Lumpkin and Komives, 2019), HaDeX aims to cover post-processing workflow, where results of the experiment are analyzed and presented in a publication-friendly format (for comparison of mentioned tools see Supplementary Information SI1). It forces HDX-MS users to rely on several pieces of software, thus making the already laborious process even more time-consuming. Yet, another challenge of the HDX-MS data analysis is a proper visualization of results on 3D protein structure which is supported by HDX-Viewer (Bouyssié et al., 2019). As the field HDX-MS is still growing, researchers introduce new standards, including data analysis and reporting (Masson et al., 2019). The available software does not allow automatic generation of reports according to the new guidelines, which adds work for experimentalists.
To alleviate these issues, we propose HaDeX, a comprehensive software suite for analysis of HDX-MS data. The aim of HaDeX is not only to provide a comprehensive way to study results of HDX-MS experiments but also to report their results in a reproducible way by including all parameters relevant to data analysis as the size of confidence intervals.
2 Materials and methods
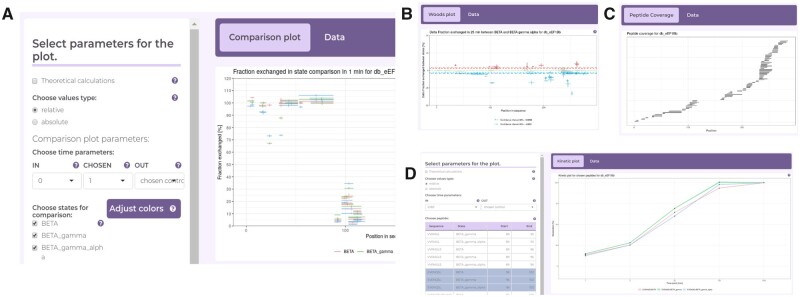
HaDeX dissects work into three steps: (i) general properties of a sequence reconstructed from measured peptides, (ii) uncertainty of measurements and their significance and (iii) visualization of results (Fig. 1).
Fig. 1.
The core functionalities of the HaDeX GUI. (A) Multistate discriminative analysis of peptides; (B) Woods plots; (C) coverage of the protein sequence by peptides measured with mass spectrometry and (D) kinetics of hydrogen–deuterium exchange
The only input necessary to start work with HaDeX is an exported data as a.csv datafile (in the Cluster format) produced by the DynamX™ 2.0 or 3.0 (Waters Corp.) Our software does not require any external preprocessing, which not only streamlines the whole workflow but also increases its reproducibility.
HaDeX uses a well-established method to compute confidence intervals for measured peptides (Houde et al., 2011) (see Supplementary Information SI2.5). Additionally, we enhanced this functionality by providing uncertainties derived by error propagation (Joint Committee for Guides in Metrology, 2008) (see Supplementary Information SI2.2).
Known in the literature as Woods charts (Woods and Hamuro, 2001), these types of plots are used to visually inspect results of HDX-MS studies (Kupniewska-Kozak et al., 2010). A user can further enhance these charts by indicating specified confidence levels (Fig. 1B). All figures are exportable in vector formats.
HaDeX provides a highly customizable report generation module, which increases the reproducibility of its analytic workflow. The report not only contains partial results of the analysis, but also the additional input provided by the user (e.g. altered significance levels).
3 Conclusion and availability
HaDeX supports a large part of the analytic workflow of HDX-MS data (Claesen and Burzykowski, 2017), from the quality control of the data to publication-ready visualizations. However, our tool does not provide any high-resolution output (as 3D visualizations or deuteration heatmaps based on single residues) thus we refer the user to methods addressing this problem (Gessner et al., 2017). Thanks to the input of HDX-MS users, our software is not only a convenient re-implementation of already existing methods but also provides unique functionalities unavailable elsewhere: novel, ISO-based uncertainty computations, multistate analysis and downloadable reports produced according to the novel guidelines (Masson et al., 2019). As our tool is targeted at experimentalists, it is available as a web server and a standalone GUI (Windows only). However, bioinformaticians can access HaDeX functions programatically, as it is also available as an R package. We hope that thanks to its comprehensiveness and reproducibility-oriented features HaDeX can satisfy requirements of users from both academia and industry.
Supplementary Material
Acknowledgements
The authors would like to thank Julien Marcoux (IPBS-Toulouse) and members of the Mass Spectrometry Core Facility (Institute of Biochemistry and Biophysics Polish Academy of Sciences) for suggestions with software features.
Funding
Foundation of Polish Science TEAM TECH CORE FACILITY/2016-2/2 Mass Spectrometry of Biopharmaceuticals—improved methodologies for qualitative, quantitative and structural characterization of drugs, proteinaceous drug targets and diagnostic molecules.
Conflict of Interest: none declared.
Contributor Information
Weronika Puchała, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw 02-106, Poland.
Michał Burdukiewicz, Faculty of Mathematics and Information Science, Warsaw University of Technology, Warsaw 00-662, Poland.
Michał Kistowski, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw 02-106, Poland.
Katarzyna A Dąbrowska, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw 02-106, Poland.
Aleksandra E Badaczewska-Dawid, Faculty of Chemistry, Biological and Chemical Research Center, University of Warsaw, Warsaw 02-093, Poland.
Dominik Cysewski, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw 02-106, Poland.
Michał Dadlez, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw 02-106, Poland.
References
- Bouyssié D. et al. (2019) HDX-Viewer: interactive 3D visualization of hydrogen–deuterium exchange data. Bioinformatics, 35, 5331–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J., Burzykowski T. (2017) Computational methods and challenges in hydrogen/deuterium exchange mass spectrometry. Mass Spectr. Rev., 36, 649–667. [DOI] [PubMed] [Google Scholar]
- Gessner C. et al. (2017) Computational method allowing hydrogen–deuterium exchange mass spectrometry at single amide resolution. Sci. Rep., 7, 3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D. et al. (2013) Time window expansion for HDX analysis of an intrinsically disordered protein. J. Am. Soc. Mass Spectr., 24, 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde D. et al. (2011) The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J. Pharm. Sci., 100, 2071–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourdel V. et al. (2016) MEMHDX: an interactive tool to expedite the statistical validation and visualization of large HDX-MS datasets. Bioinformatics, 32, 3413–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint Committee for Guides in Metrology. (2008) JCGM 100: evaluation of measurement data – guide to the expression of uncertainty in measurement. Technical report, JCGM.
- Kavan D., Man P. (2011) MSTools—web based application for visualization and presentation of HXMS data. Int. J. Mass Spectr., 302, 53–58. [Google Scholar]
- Konermann L. et al. (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev., 40, 1224–1234. [DOI] [PubMed] [Google Scholar]
- Kupniewska-Kozak A. et al. (2010) Intertwined structured and unstructured regions of exrage identified by monitoring hydrogen–deuterium exchange. J. Mol. Biol., 403, 52–65. [DOI] [PubMed] [Google Scholar]
- Lau A.M.C. et al. (2019) Deuteros: software for rapid analysis and visualization of data from differential hydrogen deuterium exchange-mass spectrometry. Bioinformatics, 35, 3171–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin R., Komives E.A. (2019) DECA, a comprehensive, automatic post-processing program for HDX-MS data. Mol. Cell. Proteomics., 18, 2516–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson G.R. et al. (2019) Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods, 16, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods V.L., Hamuro Y. (2001) High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell. Biochem., 37, 89–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.