Summary
Background
Molecular markers for antimalarial drug resistance can be used to rapidly monitor the emergence and spatial distribution of resistance to artemisinin-based combination therapies (ACTs). Little has been done to analyse molecular surveillance efforts or to assess surveillance coverage. This study aimed to develop an evidence map to characterise the spatial-temporal distribution and sampling methodologies of drug resistance surveillance in sub-Saharan Africa, specifically focusing on markers associated with ACT partner drugs.
Methods
By use of a systematic search, we identified studies that reported data on the following mutations associated with ACT partner drug resistance: pfmdr1 Asn86Tyr, Tyr184Phe, Asp1246Tyr, and copy number variation and pfcrt Lys76Thr, with sample collection occurring in sub-Saharan Africa between Jan 1, 2004, and Dec 31, 2018, corresponding to the uptake of ACTs. For each identified study, we extracted information on its sampling and laboratory methods, author and publication affiliations, years of sampling and of publication, geographic coordinates of the study sites, and prevalence of the partner drug resistance-associated markers. We used linear models to test whether urbanicity, population density, and endemicity were predictors of drug resistance survey sites and linear regressions to identify associations between the number of resistance surveys within a given country and the at-risk malaria population in 2010, the per-capita GDP in 2010, and the mean amount of funding directed to malaria and to determine trends in marker prevalence over time. For country case studies with three or more datapoints, we assessed global spatial autocorrelation using Moran’s I.
Findings
Our search yielded 254 studies encompassing 492 year-specific and location-specific surveys from 35 malaria-endemic countries, the most complete set of molecular partner drug surveillance data to date. We observed a median time lag of 3·1 years (95% CI 1·0–7·7) from final sample acquisition to publication. 22 (49%) of the 44 countries in the study region conducted, on average, one or fewer studies every 3 years. The locations of surveillance sites were positively associated with urbanicity (p<0·0001), and the abundance of country-level data was associated with reported donor funding in 2004–18 (p=0·0011) and local government funding in 2004–09 (p=0·014). Nearly all molecular markers displayed significant regional trends over time and global spatial autocorrelation in space. For selected countries with more widespread coverage of surveillance data, some markers also displayed spatial heterogeneity.
Interpretation
In most sub-Saharan countries, molecular data on antimalarial resistance might not be representative of the temporal and geographic heterogeneity of partner drug resistance, and likely do not represent the true spatially dependent distribution of partner drug resistance. Our results highlight several inefficiencies that can be improved upon to develop more accurate data landscapes, including the expansion of sentinel surveillance systems, syndemic usage of research samples, and increased participation in reporting published and unpublished data to centralised platforms.
Introduction
Antimalarial drug resistance is an enduring challenge in the global effort to control malaria. Defined as the ability of a Plasmodium parasite strain to survive or multiply despite the absorption of a drug at tolerable doses, antimalarial drug resistance has occurred with every antimalarial deployed to date.1 The emergence of parasites resistant to chloroquine and sulfadoxine-pyrimethamine occurred along similar pathways, spreading westward from southeast Asia through Africa.1 Malaria treatment relies primarily on the use of artemisinin-based combination therapies (ACTs), which combine the rapid potent activity of an artemisinin derivative with a partner drug with a longer half-life. But resistance to artemisinin and its partner drugs have again spread through southeast Asia, and a few instances of artemisinin-resistant parasites have been reported in other regions.2–6 Because ACTs are now the only remaining universally effective treatment option available in Africa, well designed surveillance systems are needed to protect both artemisinin and the partner drug throughout the continent.7
Methods for tracking the emergence and spread of antimalarial drug resistance include treatment efficacy studies, in-vitro or ex-vivo drug studies, and assessment of molecular marker prevalence from infected human participants.8 Although only treatment efficacy studies provide direct information on clinical drug failure, molecular markers are increasingly used for real-time surveillance of resistance.2,8 Molecular markers are mutations or copy number variations in the parasite genome that provide information on its resistance status. Markers in the Plasmodium falciparum multidrug resistance 1 (pfmdr1) and P falciparum chloroquine resistance transporter (pfcrt) genes have been implicated in resistance to the ACT partner drugs, amodiaquine, lumefantrine, and mefloquine.9 Systematic reviews9–11 have found that infection with P falciparum with the pfmdr1 Asn86Tyr mutation is associated with 5·4 times the odds of amodiaquine treatment failure, compared with wild-type pfmdr1, and infection with parasites without the mutation is associated with 4·7 times the odds of artemether-lumefantrine treatment failure compared with those with the mutation.
In endemic countries in sub-Saharan Africa, a single or dual first-line therapy for uncomplicated malaria is generally adopted, usually artemether-lumefantrine or artesunate-amodiaquine. When the prevalence of clinical resistance exceeds 10%, WHO advises a change in first-line therapy. The prevalence of molecular markers can be a good indicator of clinical resistance or tolerance to drug regimens in a population and can be used to monitor changes in drug resistance landscapes.2,8 An increasing prevalence of alleles of markers can provide an early warning of developing resistance, and a decreasing prevalence might be an indication of returning sensitivity after a drug has been withdrawn.10,12 The prevalence of mutations associated with sulfadoxine-pyrimethamine resistance has been used as support for modification of national policies for intermittent preventive treatment in infants, although the treatment has not been widely adopted.13 Several validated molecular markers associated with partner drug resistance can also be used as cost-effective tools to rapidly inform national treatment and prevention policies.14,15
The Worldwide Antimalarial Resistance Network (WWARN), launched in 2010, is an international collaboration in the scientific community to provide reliable and timely information on drug treatment and resistance. Although WWARN has served as a repository for drug resistance data, the wider malaria community has yet to develop guidelines as to their implementation in surveillance (appendix 1 p 5).15,16 To optimise surveillance and resource distribution, the practices that are in place first need to be determined. We aimed to assess the spatial, temporal and genetic coverage by examining the available literature on ACT partner drug-related molecular monitoring in sub-Saharan Africa. Our goal was to provide an overview of survey coverage and elucidate the potential implications of the gaps in spatial and temporal surveillance.
Methods
Search strategy and selection criteria
We did an evidence mapping exercise to identify available literature assessing molecular partner drug resistance in sub-Saharan Africa. Evidence maps are a type of evidence synthesis tool that can systematically aggregate and assess data to identify research gaps and facilitate evidence-informed decision making.17 The search protocol is outlined in appendix 1 (p 2). In brief, we identified published literature in PubMed and Embase using a search strategy that included two elements: a reference to any African context and a reference to either pfcrt or pfmdr1 specifically or to resistance to antimalarial drugs. Each element was operationalised with multiple keywords to account for alternate words and orthographic variations.
Studies were included if they collected samples from at least one country in sub-Saharan Africa and Sudan (hereafter, sub-Saharan Africa) between Jan 1, 2004, and Dec 31, 2018, corresponding to the period of introduction and widespread uptake of ACTs. Included studies assessed at least one clinical isolate or specimen for pfmdr1 (gene ID 813045) Asn86Tyr, Tyr184Phe, or Asp1246Tyr, pfmdr1 copy number variation, pfcrt (gene ID 2655199) Lys76Thr, or pfcrt 72–76 haplotype analysis. These markers are commonly assessed in partner antimalarial resistance studies, and although pfmdr1 Asn86Tyr, pfmdr1 copy number variation, and pfcrt Lys76Thr and the CVIET haplotype are strongly associated with susceptibility to multiple antimalarials, less conclusive data exist regarding pfmdr1 Tyr184Phe and Asp1246Tyr.9–11 Studies were also required to report original data on baseline or pretreatment infection in English, French, or Spanish. Studies were excluded if they were conference abstracts or case reports, they could not be linked to publicly available information, or they did not specify years of sampling.
The ACT Partner Drug Molecular Surveyor (PDMS), an online application provided by WWARN, also summarises data from published and unpublished studies on molecular markers associated with resistance to four of the most common ACT partner drugs: amodiaquine, lumefantrine, mefloquine, and piperaquine. The literature search process used to identify data for the PDMS is largely similar to this study.14 We assessed the completeness of the PDMS database by comparing our included literature to that available in the PDMS during the same time period. Our most recent searches of PubMed, Embase, and the PDMS were done on Dec 10, 2019.
We reviewed each study that met our selection criteria and extracted information regarding its sampling and laboratory methods, geographic and temporal characteristics, and author and publication affiliations (appendix 1 p 2). For temporal information, we extracted the start and end year of sampling and year of publication; in calculating time lags, we did not exclude surveys whose samples had been previously collected for other purposes. For spatial information, we extracted the geographic coordinates of the study site. Surveys were excluded from spatial analyses if samples were taken from a geographic area larger 100 m2 and could not be separated by site (appendix 1 p 2). Traveller studies were included only in the study method summaries and prevalence time series owing to limited spatial granularity. We also determined the number of unique surveys per study, designated as the assessment of markers at a specific location and year.
We extracted the prevalence of partner drug resistance-associated markers for each survey by calculating the proportion of samples that had the pfmdr1 Asn86Tyr, Tyr184Phe, Asp1246Tyr, copy number variation, or pfcrt Lys76Thr mutations or that were mixed (wild-type and mutant) by any assay. Each survey was associated with a specific year or midpoint year of sampling and its geographic location, if applicable.
Statistical analysis
To assess the hypothesis that molecular resistance survey sites were concentrated in urban regions, we compared covariates of drug resistance survey sites with malaria prevalence surveys extracted from the Malaria Atlas Project, done in the same period.18 Although an imperfect control, we believed Malaria Atlas Project sites were superior to the use of randomly selected points in malaria endemic regions; Malaria Atlas Project survey locations represent feasible sites for P falciparum sample acquisition and thus locations where surveillance of molecular markers of resistance could be theoretically done. We fit a series of binomial generalised linear models with the binary response variable of survey classification and the following singular explanatory variables extracted at survey sites: log-transformed population density in 2010, P falciparum parasite rate in children aged 2–10 years (PfParasite Rate2–10) in the respective survey year, and rural or urban site classification.18–20
We used linear regressions to identify associations between the number of resistance surveys within a given country and the at-risk malaria population in 2010, the per-capita GDP in 2010, and the mean amount of funding directed to malaria by donors (as reported by donors) and by the National Malaria Program (as reported by countries) in 2004–09 and 2010–17; the split time segments accounted for substantial increases in malaria investments around the middle of the study period.3,21 We also fit linear regressions to determine significant trends in marker prevalence over time, weighting the surveys by the standard error of the prevalence estimates. We also explored nonlinear trends in the data (appendix 1 pp 2–3). For the study region and for country case studies with three or more datapoints, we assessed global spatial autocorrelation using Moran’s I.22 The thresholds for significance for all analyses were p values of less than 0·05. All statistical tests were done in R version 3.5.1.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
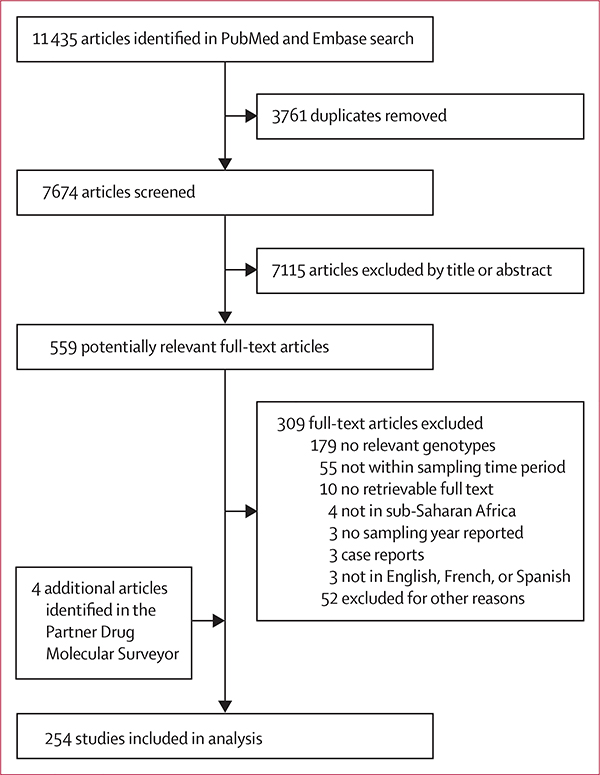
Our search yielded 254 studies reporting molecular data on selected pfmdr1 or pfcrt markers between Jan 1, 2004, and Dec 31, 2018, representing 492 location-specific and year-specific surveys from 35 malaria endemic countries (figure 1). We captured an additional 50 studies not included in the WWARN PDMS database that are described in more detail in appendix 1 (p 5) and appendix 2. The PDMS included four publications not identified in our search that were added to our analysis and five data summaries that were excluded because they could not be linked to publicly available information. Summaries of study design and publication affiliations can be found in appendix 1 (pp 4).
Figure 1:
Study profile
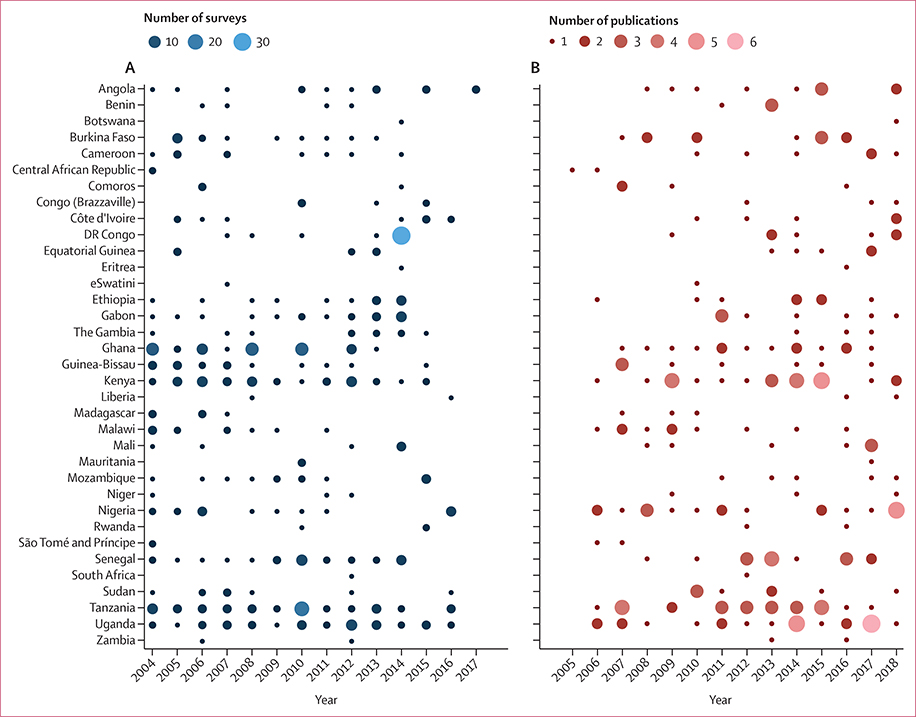
We observed a median time lag of 3·1 years (95% CI 1·0–7·7) between the last year of sample collection and the year of online publication, also visualised by the time lag between survey points (figure 2A) and publications (figure 2B). 35 countries conducted at least one partner drug-associated molecular survey in 2004–18 but published less frequently (figures 2, 3). We were unable to identify any published data that met our search criteria from nine malaria-endemic countries: Burundi, Cape Verde, Chad, Guinea, Namibia, Sierra Leone, South Sudan (or the equivalent geographic region of Sudan before 2011), Togo, and Zimbabwe. In contrast, six countries conducted surveys and published, on average, at least once annually: Ghana, Kenya, Nigeria, Senegal, Tanzania, and Uganda (figure 2). Eight additional countries conducted a survey, on average, at least once every other year: Angola, Burkina Faso, Cameroon, Ethiopia, Gabon, The Gambia, Guinea, and Mozambique (figure 2A). When dividing the study period into 3-year increments, of the countries reporting any data, 15 (43%) of 35 conducted at least one survey within all of the 3-year segments. Benin, Central African Republic, eSwatini, Madagascar, São Tomé and Príncipe, and South Africa did not publish any survey data in the most recent 3-year segment (2016–18).
Figure 2: Molecular surveys and publications reporting any data.
(A) Number of molecular surveys per year by country. (B) Number of publications per year by country. Using our search method, we were unable to identify any applicable published data during the study period from nine malaria-endemic countries in sub-Saharan Africa: Burundi, Cabo Verde, Chad, Guinea, Namibia, Sierra Leone, South Sudan (or the equivalent geographic region of Sudan before 2011), Togo, and Zimbabwe.
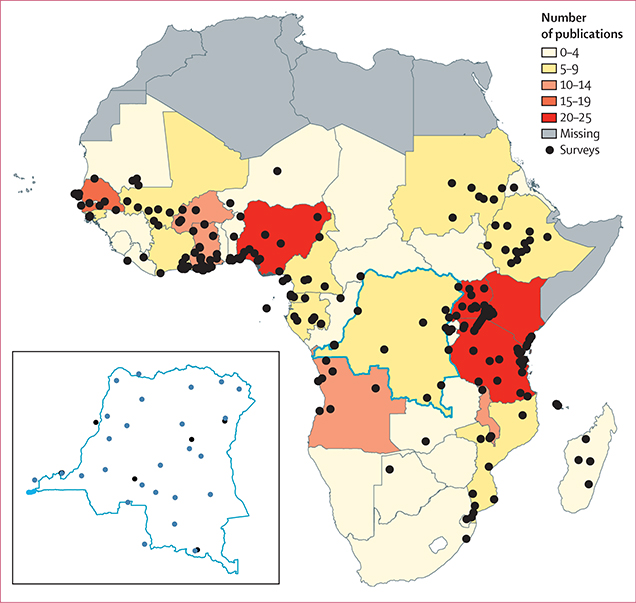
Figure 3: Location of molecular marker surveys with sampling from 2004–18.
Survey locations are represented by black points. The colour scale corresponds to the number of publications for the respective country. One study20 in the Democratic Republic of the Congo included samples from 26 Demographic and Health Surveys; these are shown only in the inset map of the country in blue.
The annual number of publications slightly increased over the study period, with an additional 1·4 total publications per year in 2005–18 (R2=0·67, p=0·0004), with no significant change in the annual number of surveys done. On average, 2·6 surveys were done per publication. One study, in the Democratic Republic of the Congo, used samples collected from 26 Demographic and Health Surveys (DHS) in 2013–14 (figure 3).23
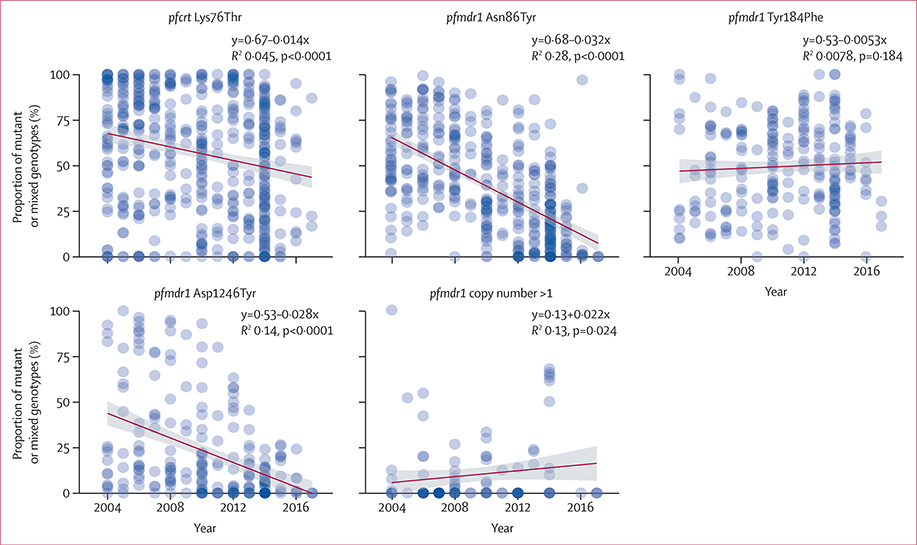
Aggregated across sub-Saharan Africa, the prevalence of pfmdr1 Asn86Tyr, Asp1246Tyr, and pfcrt Lys76Thr alleles all decreased significantly by a rate of 0·032, 0·028, and 0·014 per year (p<0·0001), suggesting an increase in the proportion of parasites with reduced lumefantrine sensitivity and a re-emergence of parasites with increased chloroquine sensitivity (figure 4). The prevalence of pfmdr1 copy numbers of more than one increased by a rate of 0·022 per year (p=0·024). In subdividing the region, the same significant trends were almost always observed in all four subregions, except for pfcrt Lys76Thr in southern Africa and pfmdr1 Asp1246Tyr in southern and west Africa. pfmdr1 Tyr184Phe increased in east Africa and decreased in central Africa, and pfmdr1 copy number variation increased in east Africa only (appendix 1 p 6).
Figure 4: Trends in molecular markers.
Shown is the proportion of mutant or mixed genotypes, where applicable, of the total number of samples for each survey (blue points) and best-fit lines (dark red) with standard error bounds (grey). Nonlinear trends are shown in appendix 1 (pp 3, 6).
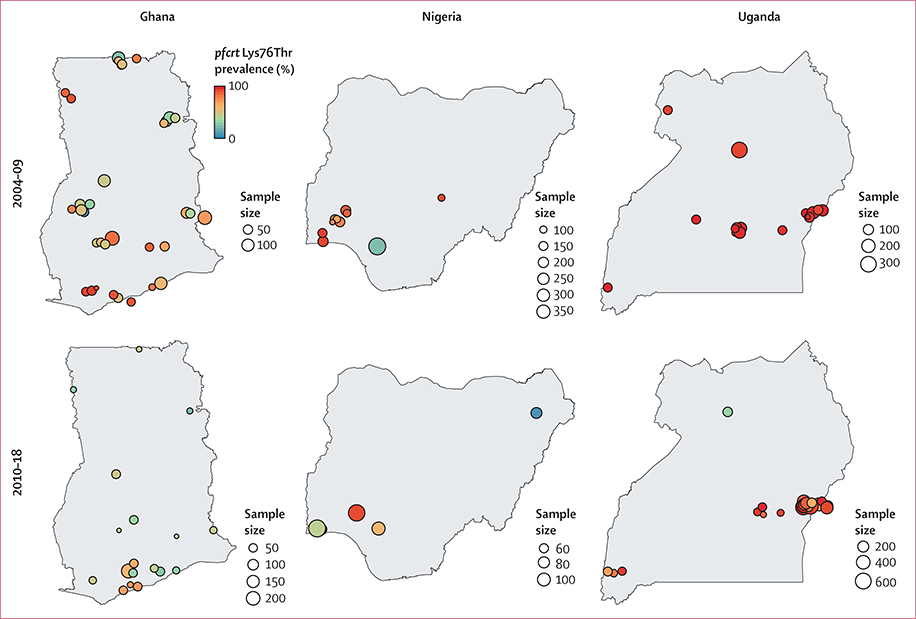
For all four markers, prevalence estimates exhibited global spatial autocorrelation in the study region (pfcrt Lys76Thr Moran’s I 0·31, pfmdr1 Asn86Tyr 0·32, Tyr184Phe 0·30, and Asp1246Tyr 0·55; p<0·0001), suggesting significant spatial trends across sub-Saharan Africa. To explore the potential within-country heterogeneity in drug resistance, we selected three countries with high frequency of surveys and high coverage for all markers: Ghana, Nigeria, and Uganda. These countries adopted new first-line treatments for falciparum malaria after 2004: artemether-lumefantrine or artesunate-amodiaquine in Ghana and Nigeria, and artemether-lumefantrine in Uganda. In Ghana, the degree of spatial heterogeneity for pfcrt Lys76Thr decreased over time, whereas it increased in Nigeria and Uganda (figure 5). In Uganda, the pfcrt Lys76Thr mutant appeared as saturated and fixed in the population from 2004–09 but decreased in frequency in some regions between 2010 and 2018. Spatial autocorrelation was significant in Ghana, with Moran’s I of 0·39 (p=0·022) for 2004–09 and 0·32 (p<0·0001) for 2010–18. Spatial autocorrelation was negligible in Nigeria and Uganda: Moran’s I was −0·025 (p=0·58) and −0·080 (p=0·60) for the two periods in Nigeria and −0·094 (p=0·79) and −0·022 (p=0·54) in Uganda.
Figure 5: Prevalence of pfcrt Lys76Thre markers.
Shown is the prevalence in Ghana, Nigeria, and Uganda for 2004–09 and 2010–18. Circle size is proportional to sample size and the colour corresponds to the prevalence of mixed or mutant genotypes. Circles are jittered for visualisation to account for overlapping survey sites. The median sample size for Ghana was 49 (IQR 38–56), Nigeria 81 (70–104), and Uganda 104 (78–216).
304 (61·0%) of 492 surveys included in this study were done in urban regions, with 104 (21·1%) occurring within 50 km of a nation’s capital city. By contrast, 537 (11·0%) of 4900 cross-sectional malaria prevalence surveys in the Malaria Atlas Project database were located in urban regions. We compared spatial attributes of drug resistance and prevalence survey sites, designated as comparison sites in which malaria research could feasibly occur. We similarly found that population density, but not endemicity, was a positive predictor of drug resistance survey sites (log odds 0·25; p<0·0001) after controlling for country-level effects. The number of surveys per country were significantly associated with the mean amount of financial donor support provided towards malaria in 2004–09 (p=0·0041) and 2010–18 (p<0·0001) and with the amount of funding provided by local or national programme support in 2004–09 only (p=0·014). GDP per capita and the size of the population at risk for malaria were not significant predictors of survey abundance.
Discussion
The malaria research community has strongly advocated for the scale up of molecular surveillance for antimalarial resistance.2,24,25 In particular, the need to monitor partner drug resistance remains pressing, because partner drug failure might result in greater increases in morbidity than would be observed from artemisinin resistance alone.7 The purpose of this study was to characterise the spatial and temporal trends and sampling designs and laboratory assays in molecular studies of partner drug resistance prevalence done in sub-Saharan Africa in 2004–18. We used evidence mapping to identify the extent and distribution of data to identify strengths and gaps in surveillance, including summarising the number of publications and surveys per country per year and analysing factors associated with survey location and abundance. To our knowledge, this is the largest and most comprehensive compilation of literature on molecular partner drug resistance in sub-Saharan Africa, specifically regarding the resistance markers, pfmdr1 Asn86Tyr, Tyr184Phe, Asp1246Tyr, pfmdr1 copy number variation, and pfcrt Lys76Thr. 50 (20%) of the 254 studies analysed were not included in the PDMS of WWARN at the time of our search.14
Despite improved technology for the detection of molecular markers, our results suggest that molecular surveillance studies are geographically clustered in sub-Saharan Africa, with only a minority of countries conducting high coverage surveillance. Additionally, there was a time lag of more than 3 years between final sample acquisition and publishing. This time lag might not be applicable to countries that rely on surveillance data that are not publicly available, such as those with internal reporting systems. However, even in such settings, delays in publicly available data might hinder responses to the emergence and spread of drug resistance at a more regional level.
We observed clear patterns in the spatial distribution of drug resistance surveys; sampling occurred predominantly in and adjacent to urban regions, especially in countries with small numbers of surveys (appendix 1 p 5). Historically, resistance has been hypothesised to emerge in low transmission regions, which can include cities; therefore, administering early warning surveillance in such regions is necessary to detect novel resistant phenotypes.1,26,27 But surveillance efforts in these settings alone are insufficient, because our study and others28,29 have shown that drug resistance can exhibit spatial heterogeneity (eg, Ghana). Sampling in varied locations will provide comprehensive information on the population-level prevalence of resistance phenotypes to ensure that appropriate treatment regimens are used.
A minority of the studies (38 [15%] of 254) included in this analysis were done at designated sentinel sites for the explicit purpose of sentinel surveillance, rather than research. Additionally, we found that the number of surveys within a given country was positively associated with the amount of funding that country received during the study period but not with local financial contributions from the governing health department in 2010–18, and that more than half of the last authors on publications were affiliated with US or European research institutions, similar to trends in authorship for other infectious disease research done in Africa (appendix 1 p 4).30 Hence, molecular surveillance might rely on external resources, both in terms of financial support and research institutions, suggesting a lack of sustainability in longitudinal molecular and treatment efficacy surveillance efforts.
This study has limitations regarding the completeness of our data. We could not consider data from internal reporting systems that some nations use for antimalarial resistance surveillance. Additionally, we did not capture literature published in journals that are not indexed in PubMed or Embase. We excluded traveller studies and other spatially aggregated surveys from spatial analyses owing to a lack of granularity (appendix 2). We only reviewed select molecular markers associated with partner drug resistance, and thereby left out markers of sulfadoxine-pyrimethamine resistance and more recently discovered markers associated with artemisinin or partner drug piperaquine resistance, such as pfkelch13 Cys580Tyr or pfpm2–3 copy number. Notably, policy makers and researchers might be missing a substantial amount of data, some of which we were able to include here.
Our findings support multiple recommendations. We encourage the improvement of sentinel surveillance systems to increase the frequency and distribution of molecular monitoring efforts and to improve the sustainability of associated research.31 Additionally, we recommend the development and scale-up of alternative surveillance approaches, such as the analysis of blood samples from rapid diagnostic tests used in health facilities or dried blood spots from DHS and malariometric surveys, because they can provide a large amount of additional information at low cost.23,32 We also strongly urge local health departments to provide public access to their key findings on drug resistance, to improve coordination of regional monitoring and data sharing efforts generally. The availability of such data to the wider regional or international community, particularly if reported through a central platform, such as WWARN, could enhance the effect that surveillance can have at stemming and reacting to emerging trends. By lowering the barriers to reporting of relevant data or by expanding participation in alternative publishing channels, molecular monitoring studies can be more widely accessible and rapidly available for spatial or temporal mapping and for policy making.
This study attempted to identify limitations in molecular monitoring of antimalarial drug resistance in sub-Saharan Africa, especially by noting inefficiencies that can be improved upon to reduce cost and improve coverage. By acting on these limitations, specifically by implementing faster research dissemination and enhancing communication channels, the scientific malaria community can learn from the failure to contain widespread chloroquine resistance and contend with artemisinin and future antimalarial resistance. The need to improve the molecular surveillance of drug resistance across sub-Saharan Africa is crucial to maintaining the therapeutic efficacy of ACTs and will lead to advances in laboratory capacity, regional or inter-continental research networks, and early warning systems for future public health challenges.
Supplementary Material
Research in context.
Evidence before this study
To our knowledge, two previous meta-analyses have considered the distribution of evidence on molecular markers: one article reviewed the abundance of evidence available globally for partner drug resistance, and another analysed temporal trends in molecular markers associated with national drug policy changes in sub-Saharan Africa.
Added value of this study
This study provides an evidence map to systematically aggregate and assess research on molecular partner drug resistance in sub-Saharan Africa with the explicit purpose of identifying surveillance gaps. For our study region and period, we compiled an evidence base of 254 studies that was 20% larger (50 additional publications) than that of the largest publicly available database, the Worldwide Antimalarial Resistance Network. We reviewed the study designs of these articles, to encourage the development of gold standards for molecular research, and considered factors regarding the frequency, abundance, and location of drug resistance surveys. We found that a majority of studies were done in urban regions, often relying on external donor and institutional support, and with large delays in publishing times. We assessed trends over time and space across the continent and discovered micro-scale and macro-scale spatial heterogeneities in molecular markers. This study goes beyond previous work to provide a comprehensive overview of the coverage of partner drug molecular surveillance in sub-Saharan Africa.
Implications of all the available evidence
Given the urgency of surveillance efforts to preserve the efficacy of artemisinin combination therapies in sub-Saharan Africa, the consensus that molecular surveillance should be scaled up across the malaria scientific community, and the ad-hoc method with which such research is done across the region, our study aimed to identify several inefficiencies that can be addressed to develop more accurate and timely data landscapes. We believe that current surveillance efforts on partner drug resistance do not achieve sufficient coverage of the parasite landscape and remain largely unsustainable. Our results could be deployed to encourage standardisation of protocols, enhancement of sampling in rural regions, and the development of faster and more accessible reporting channels.
Acknowledgments
HYE was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases (NIAID) Ruth L Kirschstein National Research Service Award (F31AI150168) and SP was supported by NIAID grant R21AI135477. We thank Kate Nyhan (Yale School of Medicine) for her help in developing the search protocol, Ted Cohen (Yale School of Public Health) for his assistance in the conception of this study, as well as Daniel M Weinberger, Joshua L Warren, and Kayoko Shioda (Yale School of Public Health) for advising on statistical analyses.
Funding
National Institute of Allergy and Infectious Diseases.
Footnotes
Declaration of interests
We declare no competing interests.
See Online for appendix 1
See Online for appendix 2
References
- 1.Blasco B, Leroy D, Fidock DA. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 2017; 23: 917–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talisuna AO, Karema C, Ogutu B, et al. Mitigating the threat of artemisinin resistance in Africa: improvement of drug-resistance surveillance and response systems. Lancet Infect Dis 2012; 12: 888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. World malaria report 2018. Geneva: World Health Organization, 2018. https://www.who.int/malaria/publications/world-malaria-report-2018/en/ (accessed Dec 12, 2019). [Google Scholar]
- 4.Rosenthal PJ. Artemisinin resistance outside of southeast Asia. Am J Trop Med Hyg 2018; 99:1357–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu F, Culleton R, Zhang M, et al. Emergence of Indigenous artemisinin-resistant Plasmodium falciparum in Africa. N Engl J Med. 2017; 376: 991–993 [DOI] [PubMed] [Google Scholar]
- 6.Mathieu LC, Cox H, Early AM et al. (2020). Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife 2020 9: e51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slater HC, Griffin JT, Ghani AC, Okell LC. Assessing the potential impact of artemisinin and partner drug resistance in sub-Saharan Africa. Malar J 2016; 15: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nsanzabana C, Ariey F, Beck H-P, et al. Molecular assays for antimalarial drug resistance surveillance: a target product profile. PLoS One 2018; 13: e0204347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picot S, Olliaro P, de Monbrison F, Bienvenu A-L, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J 2009; 8: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okell LC, Reiter LM, Ebbe LS, et al. Emerging implications of policies on malaria treatment: genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether-lumefantrine and artesunate-amodiaquine in Africa. BMJ Glob Health 2018; 3: e000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan M, Gadalla NB, Stepniewska K, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 2014; 91: 833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003; 187: 1870–75. [DOI] [PubMed] [Google Scholar]
- 13.WHO. WHO policy recommendation on intermittent preventive treatment during infancy with sulphadoxine-pyrimethamine (SP-IPTi) for Plasmodium falciparum malaria control in Africa. Geneva: World Health Organization, 2010. https://www.who.int/malaria/publications/atoz/policy_recommendation_IPTi_032010/en/ (accessed Dec 12, 2019). [Google Scholar]
- 14.Otienoburu SD, Suay I, Garcia S, et al. An online mapping database of molecular markers of drug resistance in Plasmodium falciparum: the ACT Partner Drug Molecular Surveyor. Malar J 2019; 18: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. Malaria surveillance, monitoring & evaluation: a reference manual. Geneva: World Health Organization, 2018. https://www.who.int/malaria/publications/atoz/9789241565578/en/ (accessed Dec 12, 2019). [Google Scholar]
- 16.Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol 2013; 29: 505–15. [DOI] [PubMed] [Google Scholar]
- 17.Parkhill AF, Clavisi O, Pattuwage L, et al. Searches for evidence mapping: effective, shorter, cheaper. J Med Libr Assoc 2011; 99: 157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeffer DA, Lucas TCD, May D, et al. malariaAtlas: an R interface to global malariometric data hosted by the Malaria Atlas Project. Malar J 2018; 17: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Center for International Earth Science Information Network. Gridded Population of the World, version 4 (GPWv4): population density adjusted to match 2015 revision UN WPP country totals, revision 11. Palisades, NY: NASA Socioeconomic Data and Applications Center, 2018. https://sedac.ciesin.columbia.edu/data/set/gpw-v4-population-density-adjusted-to-2015-unwpp-countrytotals-rev11 (accessed Dec 12, 2019). [Google Scholar]
- 20.Center for International Earth Science Information Network, International Food Policy Research Institute, The World Bank, Centro Internacional de Agricultura Tropical. Global Rural-Urban Mapping Project, version 1 (GRUMPv1): urban extent polygons, revision 01. Palisades, NY: NASA Socioeconomic Data and Applications Center, 2017. https://sedac.ciesin.columbia.edu/data/set/grump-v1-urban-ext-polygons-rev01 (accessed Dec 12, 2019). [Google Scholar]
- 21.The World Bank. GDP per capita (constant 2010 US$). World Bank national accounts data and OECD National Accounts data files. World Bank, 2019. https://data.worldbank.org/indicator/NY.GDP.PCAP.KD (accessed Dec 12, 2019). [Google Scholar]
- 22.Moran PA. Notes on continuous stochastic phenomena. Biometrika 1950; 37: 17–23. [PubMed] [Google Scholar]
- 23.Aydemir O, Janko M, Hathaway NJ, et al. Drug-resistance and population structure of Plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J Infect Dis 2018; 218: 946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prosser C, Meyer W, Ellis J, Lee R. Evolutionary ARMS race: antimalarial resistance molecular surveillance. Trends Parasitol 2018; 34: 322–34. [DOI] [PubMed] [Google Scholar]
- 25.Tessema SK, Raman J, Duffy CW, Ishengoma DS, Amambua-Ngwa A, Greenhouse B. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar J 2019; 18: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabaria CW, Gilbert M, Noor AM, Snow RW, Linard C. The impact of urbanization and population density on childhood Plasmodium falciparum parasite prevalence rates in Africa. Malar J 2017; 16: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushman M, Antia R, Udhayakumar V, de Roode JC. Within-host competition can delay evolution of drug resistance in malaria. PLoS Biol 2018; 16: e2005712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrhardt S, Eggelte TA, Kaiser S, et al. Large-scale surveillance of Plasmodium falciparum crt(K76T) in northern Ghana. Antimicrob Agents Chemother 2007; 51: 3407–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andriantsoanirina V, Ratsimbasoa A, Bouchier C, et al. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob Agents Chemother 2009; 53: 4588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbaye R, Gebeyehu R, Hossmann S, et al. Who is telling the story? A systematic review of authorship for infectious disease research conducted in Africa, 1980–2016. BMJ Glob Health 2019; 4: e001855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra N, Singh JP, Srivastava B, et al. Monitoring antimalarial drug resistance in India via sentinel sites: outcomes and risk factors for treatment failure, 2009–2010. Bull World Health Organ 2012; 90: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papa Mze N, Ndiaye YD, Diedhiou CK, et al. RDTs as a source of DNA to study Plasmodium falciparum drug resistance in isolates from Senegal and the Comoros Islands. Malar J 2015; 14: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.