Abstract
Restless legs syndrome (RLS) is a common sleep-related sensory-motor disorder. It is characterized by uncomfortable sensations in the legs during the evening or at night. The symptoms can be partially relieved by movement, so typically affected individual needs to walk during rest time; this interferes with sleep. GWAS have identified 19 RLS-associated loci. Among the first to be reported and most significant and robustly replicated reports are variants in the SKOR1 noncoding regions. SKOR1 is highly expressed in the CNS of humans and mice. Skor1 acts as a corepressor of Lbx1 transcription factor in mice and these two genes act together to regulate the cell fate of interneurons in the dorsal horn of the spinal cord. Based on this data we investigated the regulatory role of SKOR1 using a global RNA-sequencing approach in human cell lines where SKOR1 was either overexpressed or silenced. For this work we generated and validated a new poly-clonal anti-SKOR1. Pathway and gene set enrichment analyses of the differentially expressed genes showed, among others, enrichment of genes involved in neurodevelopment and iron metabolism, two RLS relevant pathways that were previously found to be enriched in the latest RLS GWAS meta-analysis. Analysis of our different datasets further supports and highlights the regulatory role of SKOR1, which when dysregulated might represent a key pathogenic element of RLS. A better understanding of SKOR1 and its activity could open new avenues of investigation for the development of a much-needed therapy.
Subject terms: Genetics of the nervous system, RNA sequencing, Genetics of the nervous system, RNA sequencing
Introduction
Despite recent advances in the genetics of restless legs syndrome (RLS), the underlying biology of this disorder is far from being fully understood. The heritability of RLS is estimated to be ~60% [1–3] and to date, 8 and 19 RLS loci were reported from linkage and genome wide association studies (GWAS), respectively [4]. Follow-up studies aiming to elucidate the pathogenic defect related to these genetic findings are in progress for some but not all the genetic risk factors. Variants positioned in the intergenic region located between MAP2K5/SKOR1 were found to be associated with RLS and the association was replicated in different populations [4]. While the association signal was intergenic, we previously reported that the variants appear to involve SKOR1 and not MAP2K5 (Catoire et al., 2018) [5–8]. The Genotype-Tissue Expression (GTEx) database [9] shows that SKOR1 expression is highest in the cerebellum and cerebellar hemispheres. In the embryonic central nervous system (CNS) of mice, Skor1 is expressed in a certain subset of postmitotic neurons posterior to the midbrain–hindbrain border. In the developing spinal cord of mice, Skor1 is selectively expressed in the dorsal horn interneurons where the homeodomain transcription factor, Lbx1, is also expressed. These two proteins together regulate the cell fate of interneurons in this region [10]. Skor1 interacts with Lbx1 and cooperatively they repress transcription, suggesting that Skor1 acts as a transcriptional corepressor with Lbx1 in regulating cell fate in the dorsal spinal cord [10]. Despite the significance of SKOR1 as a genetic risk factor associated with RLS, very little is known about its contribution to pathways that would be relevant to RLS pathogenicity. In this study, we used an RNA-sequencing (RNA-Seq) approach to investigate the transcriptional regulatory action of SKOR1 using two different cell lines in which SKOR1 was either overexpressed or silenced. A rabbit polyclonal antibody recognizing human SKOR1 was generated for the characterization of SKOR1 in these cells. The analysis of the different RNA-Seq datasets showed that SKOR1 acts as a transcriptional repressor of genes involved in neurodevelopment, in keeping with the transcriptional corepressor function previously reported in mice dorsal roots. Our data also showed a positive regulatory role for SKOR1 on genes involved in ferroptosis, a pathway related to iron metabolism. Past RLS literature supports a link between SKOR1 and MEIS1, another gene identified by GWAS that has links with iron metabolism [8]. The role that these two RLS-associated genes have in iron metabolism, in RLS, as well as their other possible interactors (that are common in the regulatory networks of these two transcription factors) can be subject to future investigations.
Methods
Human cell lines with SKOR1 overexpression or knockout
The ORF of SKOR1 gene (ENST00000341418.5, Open biosystem, Clone ID: 100068272) with high expression in Brain-Cerebellum and Brain-Cerebellar Hemisphere based on GTEx was sub-cloned in pcDNA3.1+ plasmid with the strong CMV promotor. The cDNA sequence of the plasmid was validated using Sanger sequencing. Human HAP1 and HEK293 cells were transfected with this vector using jetPRIME® (Polyplus). G418 antibiotic was used to select stable cell lines overexpressing SKOR1; 48 h post-transfection, the cells were transferred to their culture media containing 500 ug/ml G418 and the media were changed every other day for a period of 4 weeks to obtain stable cell lines overexpressing SKOR1. The case and the control conditions were done in at least triplicates. On the other hand, SKOR1-KO HAP1 cells were also used for RNA-Seq in parallel with the SKOR1-OE cells. HAP1 is a partially-haploid human cell line. These cells are derived from the male chronic myelogenous leukemia (CML) cell line KBM-7. They contain a single copy of each chromosome, except for a heterozygous 30-megabase fragment of chromosome 15 [11]. Both, HAP1 wild-type and HAP1 SKOR1-KO cells were commercially obtained from Horizon Discovery which provides additional information about the cell line and the experimental approach to generate a particular KO strain on the following website: https://www.horizondiscovery.com. HAP1 cells were grown in IMDM (Iscove’s Modifed Dulbecco’s Medium) medium (Gibco) and HEK293 cells in DMEM (Dulbecco’s Modified Eagle Medium) medium (Gibco); both were supplemented with 10% fetal bovine serum (FCS, Biochrom) and 1% penicillin/streptomycin (Pen/Strep, Sigma) at 37 °C and 5% CO2.
Polyclonal anti-SKOR1 generation and characterization
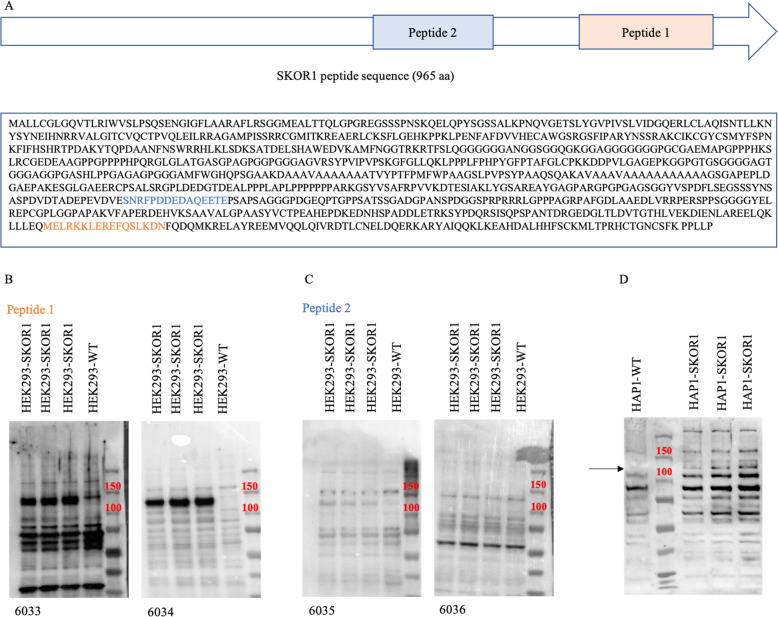
Two different peptide sequences were used to make SKOR1 polyclonal antibodies (peptide 1: MELRKKLEREFQSLKDN and peptide 2: SNRFPDDEDAQEETE, Fig. 1a). Peptide 1 was designed by our group. SKOR1_HUMAN protein sequence (UniProt identifier P84550-1) shown in Fig. 1a was used as reference sequence for epitope analysis. We implemented the Jameson-Wolfe Antigenic index profile prediction track from the software Protean (DNASTAR v5.0.5) to identify the immune-reactive regions of the SKOR1 protein. The design was based on the following criteria: lack of N-terminal glutamine or asparagine and C-terminal proline or glycine, no internal cysteine and no multiple serine, proline, or glutamine residues in the sequence. This peptide sequence is common to all SKOR1 isoforms. Peptide 2 sequence was suggested and used in a western blot analysis in a study by Arndt et al. [12]. For each of the peptide sequences, two New Zealand white rabbits were immunized at Capralogics Inc.; detailed protocol can be found at https://www.capralogics.com/rabbit-antibodies. After 73 days, final bleeds were used for antibody characterization by western blot analysis using HEK293 cells overexpressing SKOR1 ORF. The western blot conditions were optimized as follows: 1:100 final bleed dilution in PBS-T buffer with skim milk, primary incubation at room temperature for 1 h and secondary incubation using goat anti-rabbit IgG overnight at 4 °C. Endogenous levels of the SKOR1 protein cannot be detected on western blots due to the low abundance of the protein (Fig. 1b, c).
Fig. 1. Anti-SKOR1 generation and characterization.
a Schematic of the locations of peptides 1 and 2 in the SKOR1 protein and the complete peptide sequence of SKOR1 used as the reference for the epitope analysis. b Western blot analyses of the polyclonal anti-SKOR1 generated in New Zealand rabbits. Rabbits #6033 and #6034 were immunized by peptide 1. c Western blot analyses of the polyclonal anti-SKOR1 generated in New Zealand rabbits. Rabbits #6035 and #6036 were immunized by peptide 2. d Western blot analysis of HAP1 cells overexpressing SKOR1 using #6034 antibody. The black arrow indicates the SKOR1 overexpression band in the transfected cells compared with their parental wild-type cells (SKOR1 apparent MW is ~100 kDa).
RNA extraction from human cell lines
Total RNA from human cell lines were extracted using the RNeasy® mini kit (Qiagen). The concentration of RNA was measured using the Synergy H4 Hybrid Multi-Mode Microplate reader from BioTek and ~250 ng RNA was provided to Macrogen Inc. RNA Integrity Number (RIN) was assessed using the 2100 Bioanalyzer instrument, together with the 2100 Expert Software and Bioanalyzer assays (RIN > 9 for all the RNA samples). Eukaryotic long noncoding and coding transcripts (rRNA-depleted libraries) were targeted for sequencing using the Illumina Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat).
High throughput transcriptome sequencing and bioinformatic analyses
Illumina HiSeq 4000 platform at 150 bp paired-end reads with a total of 100 million reads per sample (at Macrogen Inc) were implemented for RNA-sequencing [13]. STAR v2.5.1b was used to align the FASTQ files of the paired-end reads to the human genome reference (GRCh37/hg19 assembly) [14]. Picard v1.123 was used to mark duplicates and calculate exonic/intronic/intergenic rates (https://broadinstitute.github.io/picard/). HTSeq-count was used for gene-level quantification and all the genes with <1 read per sample were removed (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). The library size for each sample was estimated using the number of mapped reads in the BAM file (ttp://samtools.sourceforge.net/). Rnaseq.py v3.0.0 pipeline from GenPipes was used for all the steps mentioned above [15]. EdgeR v3.5.2 was used to determine DEGs [16]. Quasi-likelihood F-tests was implemented with batches as covariates in an additive model. BiomaRt v2.14.0 R package (http://www.bioconductor.org/packages/release/bioc/html/biomaRt.html) was incorporated for gene annotations. The list of significantly DEGs was defined at FDR < 0.05 and were used in the pathway analysis [17, 18].
Pathway analysis
Enrichment analysis for predicted target genes was conducted using EnrichR database (http://amp.pharm.mssm.edu/Enrichr/), which is a comprehensive gene set enrichment analysis web server [19, 20]. KEGG pathway analysis and three categories of GO (http://geneontology.org) functional annotation, including biological process (BP), cellular component (CC), and molecular function (MF), were included in this study. The significantly enriched pathways with FDR < 0.05 were selected and their interactions were visualized using ClueGO and CluePedia (Cytoscape plug-in) [21].
Study approval
This study was approved by Comité d’éthique de la recherche du Centre hospitalier de l′Université de Montréal and McGill University ethics, all methods were performed in accordance with the relevant guidelines and regulations of McGill University (REB NEU-14-051).
Results
Generation and characterization of a polyclonal anti-SKOR1
The generation of polyclonal anti-SKOR1 was carried out using two different peptide sequences specific for human SKOR1 to immunize duplicates of New Zealand white rabbits. Peptide 1: MELRKKLEREFQSLKDN (rabbits #6033 and #6034) has 100% identity in human/mouse/rat, while peptide 2: SNRFPDDEDAQEETE (rabbits #6035 and #6036) is specific to human (the sequence of peptide 2 was adapted from a study by Arndt et al. [12]). The locations of the two peptides in the SKOR1 protein, and the complete peptide sequence of SKOR1 are shown in Fig. 1a. Thirteen weeks post-immunization of the rabbits, their bleeds were used in a western blot analysis to test the affinity and specificity of the antibodies within the different antisera. This characterization was performed using HEK293 cells overexpressing SKOR1 open reading frame (ORF); this ORF was sub-cloned in pcDNA3.1+ vector under CMV promoter. The final bleeds obtained from both replicates of the peptide sequences were tested and the western blot results are shown in Fig. 1b, c. The antiserum derived from Peptide 1 (rabbit #6034) showed a strong overexpression of SKOR1 with a low background noise produced from nonspecific bindings and was selected for the follow-up experiments (SKOR1 molecular weight (MW) is ~100 kDa). Endogenous levels of SKOR1 protein could not be detected on western blots because the HEK293 cells express a low level of the protein.
Generation of human cell lines with SKOR1 dysregulation
In order to identify genes that are regulated by SKOR1, two different human cell lines were used: HAP1 and HEK293. The HEK293 cells from the previous section (tested by anti-SKOR1 #6034) were used for the generation of cell lines overexpressing SKOR1 in a stable manner. Moreover, the ORF of SKOR1 was also overexpressed in HAP1 cells using the same pcDNA3.1+ vector with CMV promoter and they were characterized by western blot analysis (Fig. 1d).
Both HEK293 and HAP1 stable cell lines showed overexpression (OE) of SKOR1. The overexpression was stronger in HEK293 derived cells, probably because these cells were more efficiently transfected than HAP1 cells. The decision to use two different human cell lines was made to minimize the likelihood of identifying expression changes that would be specific to a single cell type. Hence, changes that would be observed in the datasets of the two cell types would be more likely to be linked to SKOR1 regulatory functions. We considered that genes which are up- or downregulated in these OE cells are possibly activated or repressed by SKOR1 transcriptional activity, respectively. In addition to the RNA-Seq datasets derived from the SKOR1-OE cells, datasets derived from SKOR1 knockout (SKOR1-KO) HAP1 cells and their parental wild-type (WT) cells (commercially obtained from Horizon) were also generated. The direction of SKOR1 regulation on genes that were up- or downregulated in SKOR1-OE cells should be observed in the opposing directions in SKOR1-KO.
Differential gene expression analysis of human cell lines with SKOR1 dysregulation
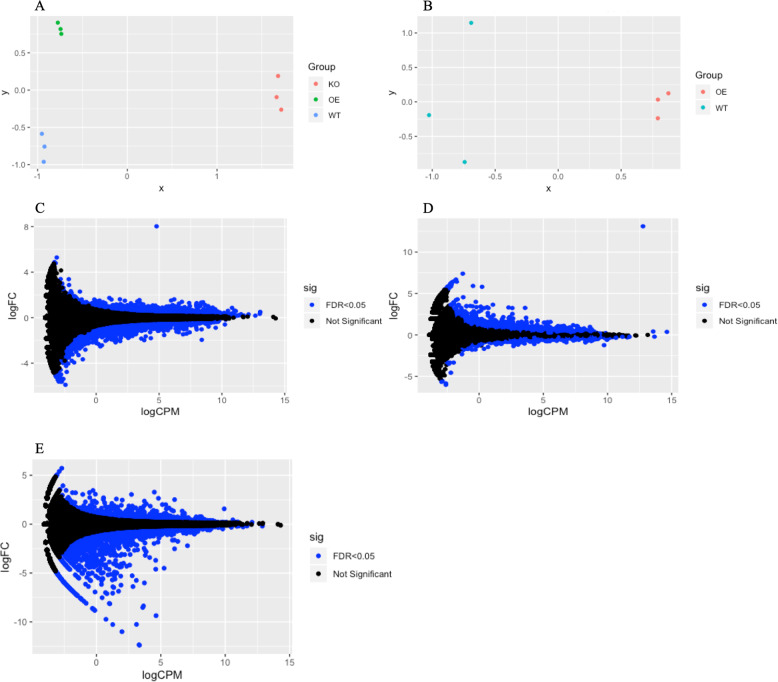
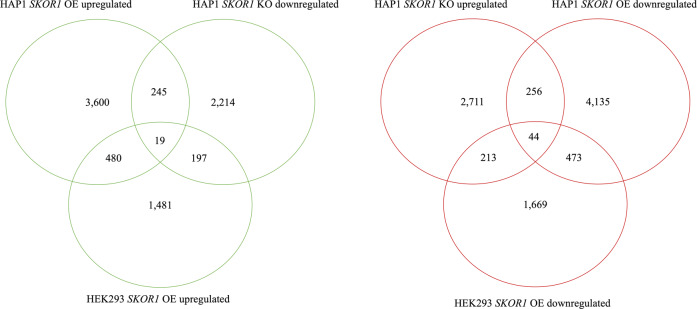
RNA-Seq was performed using total RNA samples extracted from the human cell lines in which SKOR1 expression was dysregulated (OE and KO). Differential gene expression analysis was performed using edgeR algorithm [16] with the following contrast designs: HAP1-OE vs. HAP1-WT; HAP1-KO vs. HAP1-WT; and HEK293-OE vs. HEK293-WT. The MDS (multidimensional scale) plots of the RNA-Seq analyses in the two cell lines, separately show clusters of each condition’s replicates (Fig. 2a, b). MD (mean-difference) plots also show the distribution of the differentially expressed genes (DEGs), and both HAP1 and HEK293 cells overexpressing SKOR1 show the data point corresponding to SKOR1 as the most differentially expressed gene (Fig. 2c–e). The DEGs that are upregulated in OE cells and downregulated in KO cells were considered to be possibly activated by SKOR1 (19 genes). Conversely, the DEGs that are downregulated in OE cells and upregulated in the KO cells were deemed to be possibly repressed by SKOR1 (44 genes, Fig. 3). The DEGs in each cell line, their corresponding p value and false discovery rate (FDR) can be found in the supplemental Table).
Fig. 2. RNA-Seq data analysis of human cell lines with SKOR1 gene overexpressed or silenced.
a MDS plot of the RNA-Seq data analyzed in HAP1 with clusters of HAP1 SKOR1-KO (in red), HAP1 SKOR1-OE (in green) and HAP1 parental WT cells (in blue). b MDS plot of the RNA-Seq data analyzed in HEK293 cells with clusters of HEK293 SKOR1-OE (in red) and HEK293 WT cells (in blue). c MD plot of the DEGs comparing HAP1 cells overexpressing SKOR1 with the parental wild-type cells; the single data point with a high LogFC indicates SKOR1. d MD plot of the DEGs comparing HEK293 cells overexpressing SKOR1 with the wild-type cells; the single data point with a high LogFC indicates SKOR1. e MD plot of the DEGs comparing HAP1 SKOR1-KO cells with their parental WT cells (blue data points in c, d, and e correspond to DEGs with FDR < 0.05).
Fig. 3. Venn diagram of DEGs common between HAP1 and HEK293 cells.
The left panel shows the overlaps between DEGs upregulated in HAP1 SKOR1-OE cells, HEK293 SKOR1-OE cells and downregulated in HAP1 SKOR1-KO; 19 genes common to all the three datasets were considered as possibly regulated by SKOR1. The right panel shows the overlaps between DEGs upregulated in HAP1 SKOR1-KO cells and downregulated in HAP1 SKOR1-OE and HEK293 SKOR1-OE cells; 44 genes common to all the three datasets were considered as possibly repressed by SKOR1.
Pathway and gene set enrichment analysis shows pathways related to known RLS mechanisms
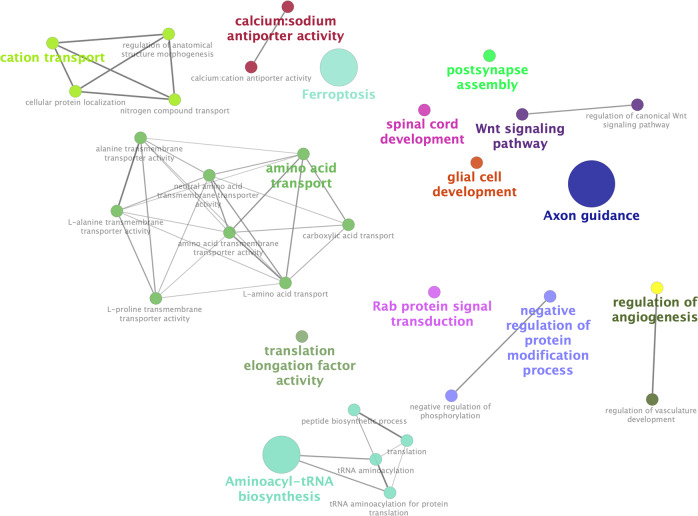
Gene Ontology (GO) annotation and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment information was compiled for the DEGs using Enrichr and their interactions were visualized by Cytoscape (Fig. 4) [19, 20]. The 44 DEGs which appear to be repressed by SKOR1 showed a significant enrichment in pathways involved in neurodevelopmental processes (Table 1). These pathways include, axon guidance, spinal cord development, glial cell development, and post-synapse assembly. This result is consistent with previous observations made by Mizuhara et al. who profiled the expression of Skor1 in mice [10]. Based on their work, Skor1 acts as a transcriptional corepressor for Lbx1 in regulating cell fate determination in the dorsal horn spinal cord [10].
Fig. 4. Interactions between the pathways significantly enriched in the DEGs possibly repressed or activated by SKOR1.
The interactions are visualized by ClueGO and CluePedia (Cytoscape software plug-in). The size of the circles corresponds to their p value. The pathways are colored based on the groups.
Table 1.
Significantly enriched pathways in the DEGs possibly repressed by SKOR1.
| Pathway | Description | p value | FDR |
|---|---|---|---|
| KEGG:04360 | Axon guidance | 2.33E-06 | 1.28E-04 |
| GO:0021782 | Glial cell development | 1.20E-05 | 7.80E-03 |
| GO:0016055 | Wnt signaling pathway | 9.06E-05 | 0.02 |
| GO:0060828 | Regulation of canonical Wnt signaling pathway | 9.25E-05 | 0.02 |
| GO:0099068 | Post-synapse assembly | 2.93E-04 | 0.04 |
| GO:0021510 | Spinal cord development | 2.93E-04 | 0.04 |
In addition to the neurodevelopmental pathways, other pathways were found, including the ferroptosis pathway that was significantly enriched in the 19 DEGs possibly activated by SKOR1 (Table 2). Ferroptosis is an iron-related form of non-apoptotic programmed cell death, where the accumulation of reactive oxygen species (ROS) leads to oxidative stress and death [22]. This process is only dependent on intracellular iron and not the other metals. Inhibiting ferroptosis can protect the organism from neurodegenerative disorders [23] and it has been implicated in several neurological diseases including Parkinson’s and Alzheimer’s disease [24–26]. Based on the current literature, brain iron abnormalities are the most robust observation in RLS [27]. Furthermore, we also observed HMOX1 (heme oxygenase 1) to be present in this ferroptosis pathway. HMOX1 is a heme degrading protein that produces free iron in addition to carbon monoxide and biliverdin [28]. This gene has been suggested to play roles in brain aging and neurodegeneration [29]. In a recent publication our group also reported HMOX1 to be part of the MEIS1 regulatory network (MEIS1 has a positive regulatory role on HMOX1 expression) [30]. In addition to ferroptosis, other pathways were also found to be significantly enriched in the genes possibly activated by SKOR1 (Table 2). Most of these pathways are involved in protein translation or transport of ions or amino acids, among others. Current knowledge of RLS does not allow us to link these pathways to RLS. Given the fact that SKOR1 functions are poorly studied and much remains to be known about its roles, these other pathways should be taken into consideration in the future SKOR1 studies. The new antibody reported in this article will facilitate additional studies to unravel the roles of SKOR1, either related to RLS or in a more general aspect.
Table 2.
Significantly enriched pathways in the DEGs possibly activated by SKOR1.
| Pathway | Description | p value | FDR |
|---|---|---|---|
| GO:0015175 | Neutral amino acid transmembrane transporter activity | 2.20E-04 | 0.01 |
| GO:0015807 | L-amino acid transport | 2.40E-04 | 0.03 |
| GO:1901342 | Regulation of vasculature development | 3.10E-04 | 0.03 |
| GO:0043043 | Peptide biosynthetic process | 4.90E-04 | 0.03 |
| GO:0045765 | Regulation of angiogenesis | 5.10E-04 | 0.03 |
| KEGG:04216 | Ferroptosis | 5.80E-04 | 0.02 |
| GO:0043039 | tRNA aminoacylation | 6.10E-04 | 0.03 |
| GO:0004812 | Aminoacyl-tRNA ligase activity | 6.40E-04 | 0.01 |
| GO:0006418 | tRNA aminoacylation for protein translation | 8.10E-04 | 0.03 |
| GO:0006865 | Amino acid transport | 8.80E-04 | 0.03 |
| GO:0003924 | GTPase activity | 9.90E-04 | 0.01 |
| GO:0046942 | Carboxylic acid transport | 9.80E-04 | 0.03 |
| GO:0006412 | Translation | 1.10E-03 | 0.03 |
| GO:0015171 | Amino acid transmembrane transporter activity | 1.20E-03 | 0.01 |
| GO:0031400 | Negative regulation of protein modification process | 1.50E-03 | 0.04 |
| KEGG:00970 | Aminoacyl-tRNA biosynthesis | 1.60E-03 | 0.03 |
| GO:0032482 | Rab protein signal transduction | 1.80E-03 | 0.04 |
| GO:0071705 | Nitrogen compound transport | 2.00E-03 | 0.04 |
| GO:0042326 | Negative regulation of phosphorylation | 2.10E-03 | 0.04 |
| GO:0006812 | Cation transport | 2.30E-03 | 0.04 |
| GO:0022603 | Regulation of anatomical structure morphogenesis | 2.50E-03 | 0.04 |
| GO:0042803 | Protein homodimerization activity | 2.50E-03 | 0.02 |
| GO:0034613 | Cellular protein localization | 2.80E-03 | 0.05 |
| GO:0017111 | Nucleoside-triphosphatase activity | 6.10E-03 | 0.03 |
| GO:0005432 | Calcium:sodium antiporter activity | 6.20E-03 | 0.03 |
| GO:0015193 | L-proline transmembrane transporter activity | 7.10E-03 | 0.03 |
| GO:0015180 | L-alanine transmembrane transporter activity | 7.10E-03 | 0.03 |
| GO:0022858 | Alanine transmembrane transporter activity | 7.10E-03 | 0.03 |
| GO:0015368 | Calcium:cation antiporter activity | 0.01 | 0.05 |
| GO:0003746 | Translation elongation factor activity | 0.01 | 0.05 |
| GO:0008483 | Transaminase activity | 0.01 | 0.05 |
Discussion
Restless legs syndrome is a sleep-related movement disorder. Despite being a highly prevalent condition in the elderly populations of western countries, the pathways underlying its onset and progression are poorly defined. RLS is a complex disease and as such genetic and environmental conditions (e.g., brain iron dysregulation, kidney disease, and pregnancy) must regulate its onset and progression [31, 32]. This study aims to shed light on the contribution of SKOR1 to the mechanisms that underlie RLS. We have previously reported links between SKOR1 and MEIS1 (another RLS-associated gene). In that study we showed that reduced expression of MEIS1 in lymphoblastoid cell lines and thalamus of RLS patients was associated with a reduced SKOR1 expression [8]. Based on the corepressor transcriptional activity of SKOR1 in the nervous system, we sought to assess its regulatory roles in a transcriptome-wide context in RLS [8]. For this aim, we manipulated SKOR1 expression in HAP1 and HEK293 cells and used an RNA-Seq approach to investigate the transcriptional regulatory function of SKOR1. The transcriptome of HEK293 SKOR1-OE was compared with the transcriptome of wild-type HEK293 cells. For the partially-haploid HAP1 cells (Horizon) [11], their single allele of SKOR1 has been inactivated using CRISPR-Cas9. The transcriptome of SKOR1-KO HAP1 cells, as well as SKOR1-OE HAP1 cells were compared with the one of their parental wild-type cells which retained an expressed allele of SKOR1. It is a reasonable assumption that the DEGs that are shared between HEK293 and HAP1 datasets are downstream targets of SKOR1. The decision to use two unrelated human cell lines was made to reduce the cell specific transcriptomic changes when interpreting the DEGs.
Cross-validation of SKOR1 transcriptomic data identified 44 and 19 genes that were repressed or activated, respectively. Functional annotations of these two datasets were made and the results were investigated based on their links with known RLS related pathways. Among the 44 genes possibly repressed by SKOR1, there was a significant enrichment of genes involved in neurodevelopmental processes like axon guidance, glial cell development, spinal cord development, and postsynaptic assembly. A 2017 meta-analysis of RLS GWAS, which included 15,126 cases and 95,725 controls of European ancestry, conducted a similar functional annotation analysis [7] and it also observed an enrichment of neurodevelopmental genes also linked to axon guidance, synapse formation, and neuronal specification [7]. The similarity between the enrichment analyses made from our cell models and the one made from the large-scale meta-analysis supports the contribution of SKOR1 to the pathogenic events leading to RLS. Moreover, there is evidence in the literature supporting the fact that RLS includes aspects of a neurodevelopmental disorder. For instance, a study by Spieler et al. showed that RLS-associated risk factors in mice can reduce their enhancer activity. This reduction takes place in lateral/medial ganglionic eminences, which give rise to basal ganglia [33]. Dysfunction of basal ganglia have been suggested to play roles in primary RLS [34]. The enrichment of RLS-associated genes in neurodevelopment [7], their involvement in the development of basal ganglia [33] and the enrichment of SKOR1 regulated genes in neurodevelopmental processes (found in this work) are all in support of neurodevelopment as a new focus for RLS research.
In regard to the enrichment analysis made from the 19 genes activated by SKOR1 in our cell models, the ferroptosis pathway was found to be significantly enriched. Ferroptosis is an iron-related form of programmed cell death, which results from the accumulation of ROS and oxidative stress [23]. Brain iron dysregulation is known to be present in RLS patients, hence the identification of this iron-related pathway in the SKOR1 regulated genes is meaningful. The protein encoded by HMOX1 (heme oxygenase 1) is part of the ferroptosis pathway where it degrades heme to biliverdin, carbon monoxide, and ferrous ion [28]. It has been proposed that HMOX1 is involved in brain aging and neurodegeneration [29], an observation which might be relevant to RLS considering that a small association has been reported to exist between this gene and RLS in a Spanish cohort [35]. HMOX1 has also been reported to be downstream of MEIS1 transcription factor [30]. It is noteworthy that we also observed a positive regulatory role of MEIS1 over SKOR1 in our earlier publication [8]. These results altogether suggest the presence of a regulatory network involving these three genes, a network which we believe warrants further investigation.
Use of human cell lines in this study provided a standard in vitro model to investigate the effects and implications of dysregulated SKOR1 in RLS. Overall, our observations support the contribution of SKOR1 transcriptional regulatory action in the onset and/or progression of RLS. This novel finding opens new venues for future investigations. However, in vitro studies have some limitations. For instance, genes and pathways that are exclusively expressed in the neuronal cells may have been missed in this study. Future follow-up studies can take advantage of neurological cell lines, like SK-N-SH cells, or actual brain samples of RLS individuals. These samples can also be extremely useful to validate the findings of this article and relate them to RLS. These in vitro and future in vivo investigations will help researchers to more precisely understand the pathophysiology of RLS and eventually move forward to develop more effective and long-lasting treatments.
Supplementary information
Acknowledgements
GAR holds the Canada’s Research Chair in Neurogenetics and the Wilder Penfield Chair in Neuroscience. This study was funded by Canadian Institute of Health Research (RN254517-332736). FA was funded by the Fonds de Recherche du Québec–Santé.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-0670-4) contains supplementary material, which is available to authorized users.
References
- 1.Desai AV, Cherkas LF, Spector TD, Williams AJ. Genetic influences in self-reported symptoms of obstructive sleep apnoea and restless legs: a twin study. Twin Res. 2004;7:589–95. doi: 10.1375/1369052042663841. [DOI] [PubMed] [Google Scholar]
- 2.Xiong L, Jang K, Montplaisir J, Levchenko A, Thibodeau P, Gaspar C, et al. Canadian restless legs syndrome twin study. Neurology. 2007;68:1631–3. doi: 10.1212/01.wnl.0000261016.90374.fd. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Ondo WG, Rao S, Li L, Chen Q, Wang Q. Genomewide linkage scan identifies a novel susceptibility locus for restless legs syndrome on chromosome 9p. Am J Hum Genet. 2004;74:876–85. doi: 10.1086/420772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez-Jimenez FJ, Alonso-Navarro H, Garcia-Martin E, Agundez JAG. Genetics of restless legs syndrome: an update. Sleep Med Rev. 2018;39:108–21.. doi: 10.1016/j.smrv.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 6.Yang Q, Li L, Chen Q, Foldvary-Schaefer N, Ondo WG, Wang QK. Association studies of variants in MEIS1, BTBD9, and MAP2K5/SKOR1 with restless legs syndrome in a US population. Sleep Med. 2011;12:800–4. doi: 10.1016/j.sleep.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schormair B, Zhao C, Bell S, Tilch E, Salminen AV, Putz B, et al. Identification of novel risk loci for restless legs syndrome in genome-wide association studies in individuals of European ancestry: a meta-analysis. Lancet Neurol. 2017;16:898–907. doi: 10.1016/S1474-4422(17)30327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catoire H, Sarayloo F, Mourabit Amari K, Apuzzo S, Grant A, Rochefort D, et al. A direct interaction between two Restless Legs Syndrome predisposing genes: MEIS1 and SKOR1. Sci Rep. 2018;8:12173. doi: 10.1038/s41598-018-30665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. The. J Biol Chem. 2005;280:3645–55. doi: 10.1074/jbc.M411652200. [DOI] [PubMed] [Google Scholar]
- 11.Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–65. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arndt S, Poser I, Moser M, Bosserhoff AK. Fussel-15, a novel Ski/Sno homolog protein, antagonizes BMP signaling. Mol Cell Neurosci. 2007;34:603–11. doi: 10.1016/j.mcn.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourgey M, Dali R, Eveleigh R, Chen KC, Letourneau L, Fillon J, et al. GenPipes: an open-source framework for distributed and scalable genomic analyses. Gigascience. 2019;8. [DOI] [PMC free article] [PubMed]
- 16.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids Res. 2016;44(W1):W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–79. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Do Van B, Gouel F, Jonneaux A, Timmerman K, Gele P, Petrault M, et al. Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol Dis. 2016;94:169–78. doi: 10.1016/j.nbd.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Hambright WS, Fonseca RS, Chen L, Na R, Ran Q. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol. 2017;12:8–17. doi: 10.1016/j.redox.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiland A, Wang Y, Wu W, Lan X, Han X, Li Q, et al. Ferroptosis and its role in diverse brain diseases. Mol Neurobiol. 2019;56:4880–93.. doi: 10.1007/s12035-018-1403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen RP, Earley CJ. The role of iron in restless legs syndrome. Mov Disord: Off J Mov Disord Soc. 2007;22(Suppl 18):S440–8. doi: 10.1002/mds.21607. [DOI] [PubMed] [Google Scholar]
- 28.Morse D, Choi AM. Heme oxygenase-1: the “emerging molecule” has arrived. Am J respiratory cell Mol Biol. 2002;27:8–16. doi: 10.1165/ajrcmb.27.1.4862. [DOI] [PubMed] [Google Scholar]
- 29.Schipper HM. Heme oxygenase-1: role in brain aging and neurodegeneration. Exp Gerontol. 2000;35:821–30. doi: 10.1016/S0531-5565(00)00148-0. [DOI] [PubMed] [Google Scholar]
- 30.Sarayloo F, Dionne-Laporte A, Catoire H, Rochefort D, Houle G, Ross JP, et al. Mineral absorption is an enriched pathway in a brain region of restless legs syndrome patients with reduced MEIS1 expression. PloS ONE. 2019;14:e0225186. doi: 10.1371/journal.pone.0225186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trenkwalder C, Allen R, Hogl B, Paulus W, Winkelmann J. Restless legs syndrome associated with major diseases: a systematic review and new concept. Neurology. 2016;86:1336–43.. doi: 10.1212/WNL.0000000000002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarayloo F, Dion PA, Rouleau GA. MEIS1 and restless legs syndrome: a comprehensive review. Front Neurol. 2019;10:935. doi: 10.3389/fneur.2019.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spieler D, Kaffe M, Knauf F, Bessa J, Tena JJ, Giesert F, et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyvaert L, Houdayer E, Devanne H, Bourriez JL, Derambure P, Monaca C. Cortical involvement in the sensory and motor symptoms of primary restless legs syndrome. Sleep Med. 2009;10:1090–6. doi: 10.1016/j.sleep.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Martin E, Jimenez-Jimenez FJ, Alonso-Navarro H, Martinez C, Zurdo M, Turpin-Fenoll L, et al. Heme Oxygenase-1 and 2 common genetic variants and risk for restless legs syndrome. Medicine. 2015;94:e1448. doi: 10.1097/MD.0000000000001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.