Abstract
Background:
Antimicrobial resistance (AMR) is one of the most significant health threats to society. A growing body of research demonstrates selection for AMR likely occurs at environmental concentrations of antibiotics. However, no standardized experimental approaches for determining selective concentrations of antimicrobials currently exist, preventing appropriate environmental and human health risk assessment of AMR.
Objectives:
We aimed to design a rapid, simple, and cost-effective novel experimental assay to determine selective effect concentrations of antibiotics and to generate the largest experimental data set of selective effect concentrations of antibiotics to date.
Methods:
Previously published methods and data were used to validate the assay, which determines the effect concentration based on reduction of bacterial community (wastewater) growth. Risk quotients for test antibiotics were generated to quantify risk.
Results:
The assay (SELection End points in Communities of bacTeria, or the SELECT method) was used to rapidly determine selective effect concentrations of antibiotics. These were in good agreement with quantitative polymerase chain reaction effect concentrations determined within the same experimental system. The SELECT method predicted no effect concentrations were minimally affected by changes in the assay temperature, growth media, or microbial community used as the inoculum. The predicted no effect concentrations for antibiotics tested ranged from for ciprofloxacin to for erythromycin.
Discussion:
The lack of evidence demonstrating environmental selection for AMR, and of associated human health risks, is a primary reason for the lack of action in the mitigation of release of antibiotics into the aquatic environment. We present a novel method that can reliably and rapidly fill this data gap to enable regulation and subsequent mitigation (where required) to lower the risk of selection for, and human exposure to, AMR in aquatic environments. In particular, ciprofloxacin and, to a lesser extent, azithromycin, cefotaxime, and trimethoprim all pose a significant risk for selection of AMR in the environment. https://doi.org/10.1289/EHP6635
Introduction
By 2050, it is estimated that one person will die from an antimicrobial-resistant (AMR) infection every 3 s (O’Neill 2016). The environment is a recognized reservoir of AMR bacteria and genes. However, antimicrobial compounds are also discharged into the environment from a range of sources, including (but not limited to) industrial, hospital, and domestic wastewater, as well as runoff from agricultural land (Kümmerer 2004).
The U.S. Environmental Protection Agency (U.S. EPA 2017) and the European Chemicals Agency (ECHA 2013) have outlined the stages required for human health risk assessment. The intended outcome is complete risk characterization, which is informed by exposure assessment, hazard identification, and hazard characterization (ECHA 2013; U.S. EPA 2017). In terms of human health risk assessment of AMR, there are significant knowledge gaps within this framework preventing complete risk characterization, particularly in terms of hazard identification and characterization.
For example, in terms of hazard characterization of AMR, dose–response relationships between exposure to AMR bacteria or genes and adverse health outcomes have not yet been clearly defined. However, recent research has found high exposure to coastal waters to be associated with higher human gut carriage of clinically important, AMR Escherichia coli (E. coli) responsible for serious, extra-intestinal infections (Leonard et al. 2018). Identified hazards to human health relevant for risk assessment of AMR are the presence of AMR bacteria and genes or the presence of antimicrobial agents and other selective compounds (which directly influence the prevalence of AMR genes and AMR bacteria). The number of studies that focus on the prevalence of AMR genes in bacteria in environmental compartments has increased rapidly in recent years (Ashbolt et al. 2013), but there are still very few experimental data on selective concentrations of antibiotics that can cause an increase in numbers of AMR bacteria or AMR genes.
The first experimental study to show selection for AMR at environmentally relevant antibiotic concentrations used isogenic-resistant and susceptible E. coli in competition assays to determine the minimal selective concentration of a test antibiotic (Gullberg et al. 2011). Since then, several studies have investigated the selection for AMR in complex bacterial communities to take into account competition within and between different bacterial taxa that may occur in the natural environment and potentially alter such effect concentrations (Klümper et al. 2019; Kraupner et al. 2018; Lundström et al. 2016; Murray et al. 2018; Stanton et al. 2020). However, all of these approaches require specialist equipment and personnel and are prohibitively expensive to be conducted routinely.
Currently, an environmental risk assessment of antibiotics is required when the predicted environmental concentration exceeds in Europe (EMA 2006) or in the United States (FDA 1998). The standard approach is to calculate a risk quotient (RQ) by dividing a predicted environmental concentration or measured environmental concentration (MEC) by a predicted no effect concentration (PNEC). PNECs are derived by applying an assessment factor to a no observed effect concentration (NOEC), which is determined through standardized ecotoxicological tests (EMA 2006) and represents the test concentration directly below the lowest observed effect concentration (LOEC). NOECs for antibiotics are currently determined using several different ecotoxicological tests, including the activated sludge respiration inhibition test (ASRIT) and toxicity tests on species such as Daphnia magna, fish, cyanobacteria, or green algae (Le Page et al. 2017). None of these experimental methods directly consider the selective potential of an antibiotic, and most do not focus on the target organism (bacteria). As a result, the risk of selection for AMR occurring in aquatic environments is neither understood nor addressed within current environmental risk assessments. A standardized experimental approach is therefore required to inform safe release limits of antibiotics to prevent environmental selection for AMR.
Such an experimental method would be invaluable for human health risk assessment hazard identification by characterizing the hazard posed by a specific concentration of an antibiotic or other selective agent in terms of selecting for AMR. For example, it could identify hotspots in the environment that contain levels of antibiotics likely to select for antibiotic-resistant bacteria or antibiotic resistance genes to which humans may be exposed (Ashbolt et al. 2013). Together these data could be used to lower the likelihood of environmental transmission of resistant bacteria or resistance genes to humans and animal-associated bacteria, thereby protecting the efficacy of novel antibiotics and promoting appropriate stewardship of antibiotics currently in use. Currently, no such standardized experimental method has been designed or validated that can quantify the risk that antibiotics in the environment pose in terms of selecting for AMR in environmental and human health risk assessments. Development of such a method was the aim of the present study.
The minimal selective concentration is defined as the lowest antibiotic concentration at which the growth rate of resistant and susceptible bacteria are equal (Gullberg et al. 2011). Therefore, when a reduction in bacterial growth is observed over time as a result of antibiotic exposure, this may indicate selection is occurring. Furthermore, a mathematical model was recently described that estimated the minimal selective concentration and which took into account the growth rates of resistant and susceptible bacterial strains under different antibiotic pressures (Greenfield et al. 2018). A loss in net growth, particularly of the susceptible strain, was shown to be the most sensitive parameter for minimal selective concentration prediction (Greenfield et al. 2018). Therefore, we hypothesized any significant reduction in growth of a complex bacterial community over time under a given antibiotic exposure concentration could be used as an end point to predict selection for AMR.
In the present study, we present the largest data set of experimentally derived selective effect concentrations of antibiotics currently available, using a novel growth-based approach. This SELection End points in Communities of bacTeria (or SELECT method) determines the LOECs of antibiotics as the concentration during the exponential growth phase where the net growth of the bacterial community is significantly reduced. PNECs for resistance [ (Bengtsson-Palme and Larsson 2016)] and RQs for all test antibiotics were determined, demonstrating this new approach can be used to rapidly fill crucial data gaps, enabling appropriate environmental and human health risk assessment of antibiotics in terms of their potential to select for AMR. The SELECT method was validated by comparing SELECT to genotypic that were derived using a previously published method (Murray et al. 2018).
Methods
Test Antibiotics
Antibiotics used in the present study were cefotaxime (Acros Organics; 454950010), ciprofloxacin (Sigma-Aldrich; 17850), trimethoprim (Sigma-Aldrich; T7883-5G), azithromycin (Sigma-Aldrich; PHR1088), clarithromycin (Molekula; 37077446), erythromycin (Acros Organics; AC227330050), gentamicin (Acros Organics; 455310010), and chloramphenicol (Acros Organics; 227920250). Solvents were sterile water, dilute hydrochloric acid (; Fisher Chemical; 10080210), dimethyl sulfoxide (100%; Sigma-Aldrich; D8418-50ML), ethanol (100%; Fisher Bioreagents; BP2818-4), acetone (100%; Acros Organics; 444150050), ethanol (100%), sterile water, and sterile water, respectively. All antibiotics were stored in single-use aliquots at until use.
Sewage Samples
Influent and effluent samples were collected from wastewater treatment plant (WWTP) A (serving a population of approximately 43,000) in October 2016 and February 2018. Influent samples were collected from WWTP B (serving a population of approximately 77,000) in February 2018. Samples were transported in cool boxes, then mixed 1:1 with 40% glycerol (Fisher; G/0600/17) and stored at until use.
Experimental and Statistical Approach for Determination of Genotypic NOECs
Thawed sewage samples were spun down at for 10 min and resuspended in 0.85% sterile sodium chloride (NaCl; Fisher Chemical; S/3,160/60) twice to remove chemical and nutrient carryover. The resuspended pellet was used to inoculate at 10% (vol/vol) in Iso-Sensitest™ broth (Oxoid; CM0473). Microcosms spiked with antibiotic were shaken at for 24 h at 37°C. Each day, of overnight culture was transferred into fresh medium and fresh antibiotic, for a total of 7 d. Two aliquots of each culture were spun down at for 2 min, resuspended in 20% glycerol (Fisher; G/0600/17), and frozen at until DNA extraction at the beginning (Day 0) and at the end of the experiment (Day 7). The following antibiotic concentrations were used, each with five biological replicates: and 2-fold dilutions down to for trimethoprim, and 2-fold dilutions down to for gentamicin, and and 2-fold dilutions down to for chloramphenicol.
Effect concentrations were determined using the intI1 gene target given that the genes conferring resistance (i.e., dfr or aac genes) to these compounds are commonly associated with class 1 integrons, which are genetic platforms that can readily integrate a diverse range of mobile AMR genes (Partridge et al. 2009). The LOEC was determined as the test concentration at which the intI1 prevalence was significantly higher than the no-antibiotic control.
The cefotaxime (Murray et al. 2018), ciprofloxacin (Stanton et al. 2020), and macrolide (azithromycin, clarithromycin, and erythromycin) (Stanton et al. 2020) week-long experiments were conducted as part of previously published studies. The concentration range in the previous study for cefotaxime was , with 2-fold dilutions down to (Murray et al. 2018). For cefotaxime, the compound-specific gene end point () LOEC was taken directly from the previous study by Murray et al. (2018). The present study also reanalyzed the cefotaxime data from Murray et al. (2018) with additional quantitative polymerase chain reaction (qPCR) to determine the LOEC of cefotaxime using the intI1 gene target. The concentrations used in the macrolide experiments conducted previously were , , , , , , and (Stanton et al. 2020). The concentration range for the ciprofloxacin experiment conducted previously was , with 2-fold dilutions down to (Stanton et al. 2020). The LOEC data for the macrolides (determined using both the intI1 gene target and the compound-specific gene target, ermF) and the LOEC data for ciprofloxacin (intI1 gene target) were taken directly from the previously published study by Stanton et al. (2020).
DNA extraction was performed using the DNeasy Ultra-Clean Microbial kit (Qiagen; 12,224-250, previously sold by MBio as the Ultra-Clean kit) according to the manufacturer’s instructions. DNA was diluted 5-fold (Day-0 samples) to 20-fold (Day-7 samples) to dilute out qPCR inhibitors. DNA quantification was not performed because standard curves were used for absolute quantification. Standard curves were generated with custom synthetic gBlocks™ (Table S1) provided by IDTDNA, prepared according to the manufacturer’s instructions and stored in single-use aliquots at . qPCR was performed using the PrimerDesign Precision Plus SYBR™ Green Master Mix (), on the Applied Biosystems StepOne™ machine (catalog number 4,376,357, serial number 272,007,340). Reactions comprised Master Mix, template, primer ( each of forward and reverse primers, all except 16S rRNA, which was ), bovine serum albumin (, Sigma-Aldrich; A2153-10G), and water up to a final volume of . Cycling parameters used were a 2-min initial Hot Start activation at 95°C, followed by 40 cycles of data collection with 10 s at 95°C and 60 s at 60°C. All the primers used in the present study, as well as the primers used to generate the () prevalence data in the previous study on cefotaxime (Murray et al. 2018) and the primers used to generate all reported (ermF and intI1) prevalence data for ciprofloxacin and the macrolides (Stanton et al. 2020) are listed in Table S1 and were provided by IDTDNA.
Kruskal-Wallis tests were used to check that there were no significant differences between treatments at Day 0. LOECs were determined as the lowest antibiotic concentration where the prevalence at Day 7 was significantly higher than the Day-7 no-antibiotic control, according to Dunn’s test or generalized linear models (GLMs; whichever was the most protective, i.e., yielded the LOEC). GLMs were used to calculate qPCR LOECs for ciprofloxacin (Gamma; identity link), trimethoprim, chloramphenicol, and gentamicin (Gamma; log link for all three); all other data were analyzed using Dunn’s test. If the prevalence at Day 7 was greater than the prevalence at Day 0, then positive selection was considered to have occurred; if less, then significant persistence was considered to have occurred (Murray et al. 2018; Stanton et al. 2020). NOECs were assigned as the test concentration directly below the LOECs, at which no significant differences in resistance were observed according to either Dunn’s Tests or GLMs.
Experimental and Statistical Approach for Determination of SELECT NOECs
SELECT method tests were conducted in 96-well plates with washed sewage as described above and inoculated into Iso-Sensitest™ broth (Oxoid; CM0473) at 10% (vol/vol). There were six replicates per antibiotic or control treatment. There were no significant differences between the solvent controls and the no-antibiotic controls (all ; Excel Table S1). All raw data for the solvents and the corresponding no-solvent controls, means, standard deviations, and standard errors of the mean are listed in Excel Table S2. Therefore, the experimental runs contained only a no-antibiotic control and a sterile-broth control.
The rationale behind the concentration ranges was as follows. For cefotaxime, ciprofloxacin, trimethoprim, gentamicin, and chloramphenicol, the highest concentration used was the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical break point concentration for Enterobacteriaceae (EUCAST 2019). These were 2,000 (cefotaxime), (ciprofloxacin), (trimethoprim), (gentamicin), and (chloramphenicol), followed by 2-fold dilutions down to the low micrograms-per-liter range and until a NOEC could be determined. For the macrolides (azithromycin, clarithromycin, and erythromycin), EUCAST clinical break point concentrations for Enterobacteriaceae were not available (EUCAST 2019). For azithromycin, the EUCAST clinical break point concentration for Salmonella was used () (EUCAST 2019), followed by 2-fold dilutions down to the low micrograms-per-liter range and until a NOEC could be determined. Nominal starting concentrations for clarithromycin and erythromycin were selected as 10,000 and , respectively, followed by 2-fold dilutions down to the low micrograms-per-liter range and until a NOEC could be determined. The higher starting concentration for erythromycin was based on previous research that indicated erythromycin is less potent than azithromycin (Jelić and Antolović 2016).
Concentrations used were the same as in the week-long genotypic experiments but with extra 2-fold dilutions to lower concentrations where necessary to determine a LOEC. Where a lowest effect concentration could not be observed in a single run (i.e., testing 14 different concentrations), a second plate was run. These additional plates started at the lowest one or two test concentrations from the previous plate and down until a LOEC could be observed.
Plates were incubated at 37°C in a Varisokan Flash (catalog number N06354, serial number 3001–1778) or BioTek Synergy (serial number 254,462) plate reader at medium shaking or , respectively. Optical density (OD) at readings were performed every hour for 12 h up to 60 h (depending on culturing conditions) to allow the bacterial community to reach the stationary growth phase.
Variations of the SELECT assay were performed—using WWTP A (2016), WWTP A (2018), and WWTP B (2018) influent, as well as WWTP A (2018) effluent as the inoculum. For these inoculum variations, experiments were conducted as described above for azithromycin, cefotaxime, ciprofloxacin, and trimethoprim. To determine the potential effects culture conditions had on effect concentration, the medium was replaced with artificial sewage and experiments were run at room temperature (), as per the ASRIT (OECD 2009), for the four antibiotics azithromycin, cefotaxime, ciprofloxacin, and trimethoprim using the WWTP A 2018 inoculum. The artificial sewage recipe is described in the ASRIT (OECD 2009) and was prepared by mixing peptone (Oxoid; LP0085), meat extract (Fluka Analytical; 70164-500G), urea (Fisher; U/0500/53), NaCl (Fisher; S/3,120/60), calcium choride dihydrate (Sigma; C7902-500g), magnesium sulfate heptahydrate (Sigma-Aldrich; 230391-500g), and dipotassium phosphate (Acros Organics; 205925000) in deionized water. This artificial sewage concentrate was autoclaved and stored in the dark at 4°C. Hundredfold dilutions were prepared from the stock concentrate by mixing concentrate with autoclaved water. Cefotaxime, ciprofloxacin, and trimethoprim were chosen for these additional tests because they resulted in the lowest determined using the SELECT method standard culturing conditions (see the “Results” section). Azithromycin was also included as the macrolide with the lowest SELECT (see the “Results” section) of the macrolides currently on the Water Framework Directive (WFD) Watch List.
To determine the LOEC, first, the time point that exhibited the strongest dose–response relationship between overall community growth () and antibiotic concentration was determined using Pearson’s correlation or Spearman’s rank correlation (depending on whether the data were distributed normally or not, respectively). All LOECs were determined using Spearman’s rank, except for trimethoprim using WWTP B influent. Dunn’s test was then performed on data at this time point to identify the concentration of antibiotic that significantly reduced the growth of the community compared with the no-antibiotic control ().
Statistical Comparison of the Two Experimental Approaches
The Bland-Altman analysis is a method commonly used in clinical studies to assess the level of agreement between measurements generated through two different methods (Bland and Altman 1986). This is a preferred analysis method to correlation, which considers only positive or negative relationships between data measurements (Giavarina 2015). To account for the large range in NOECs (i.e., the large range in measurements included in the analysis), the Bland-Altman analysis was performed using logged as previously recommended (Giavarina 2015). Significant agreement between the two experimental approaches is indicated by all differences lying within the upper and lower limits of agreement (i.e., within 95% confidence intervals around the mean for all measurements).
Calculation of and RQs
For both SELECT and qPCR methods, were determined by applying an assessment factor of 10 to the NOEC (EMA 2006). Median MEC () and maximum MEC () values for all antibiotics included in the present study (except gentamicin) were extracted from the Umweltbundesamt (UBA) database (UmweltBundesamt 2019). that excluded nondetects are presented in the main text; that included the nondetects are presented in Figure S1. Nondetect MECs were included in the median calculation as from studies where quantification was attempted but could not be performed due to concentrations being below the limit of detection or quantification. MECs included all human sewage and WWTP types of sample (i.e., livestock and industrial samples were excluded, as were unknowns). Only MECs where the emission source was listed as urban or hospital wastewater were included, as were MECs where the original measurement unit was per liter (i.e., only aquatic samples). All MECs were rounded to two decimal places before RQ calculations. For erythromycin MECs, erythromycin, erythromycin hydrate, and erthryomycin A dihydrate were included. All MEC values are presented in Excel Table S3; maximum and median values are presented in Excel Table S4.
For gentamicin, MEC data from only a single study were available, so predicted environmental concentration data were used. European consumption data (retail, prescription, and hospital data, in kilograms) for 2015 were obtained from IMS Health for gentamicin use in 22 European countries. Country-specific total substance predicted environmental concentrations were estimated assuming the following: even use across the whole population for each country (calculated as mass per person per day), 100% patient use and no wastage, no patient metabolism or sewage treatment removal, and each person generating of wastewater per day [as defined for a Phase-1 predicted environmental concentration determination (EMA 2006), but without a final dilution factor of 10 so as to represent effluent concentrations as opposed to surface-water concentrations].
Median RQs () and maximum RQs () were derived by dividing and values, respectively, by the lowest SELECT determined in the present study. Risk of selection for AMR was classified as low, medium, or high based on RQs of , and , respectively.
Results
Determined through qPCR in the Present Study and Previously Published Studies
In the present study, qPCR data were generated to calculate cefotaxime intI1 prevalence, trimethoprim intI1 prevalence, gentamicin intI1 prevalence, and chloramphenicol intI1 prevalence (Figure 1; raw data are presented in the Excel file alongside the averages, standard deviations, and standard errors of the mean). qPCR data for clarithromycin, azithromycin, and erythromycin (to calculate prevalence of both ermf and intI1) were reported previously (Stanton et al. 2020). qPCR data for cefotaxime [to calculate () prevalence] were also reported previously (Murray et al. 2018). A summary of all the qPCR effect concentrations determined in the present study and previous studies (Murray et al. 2018; Stanton et al. 2020) using the genotypic (qPCR) end points are presented in Table 1.
Figure 1.
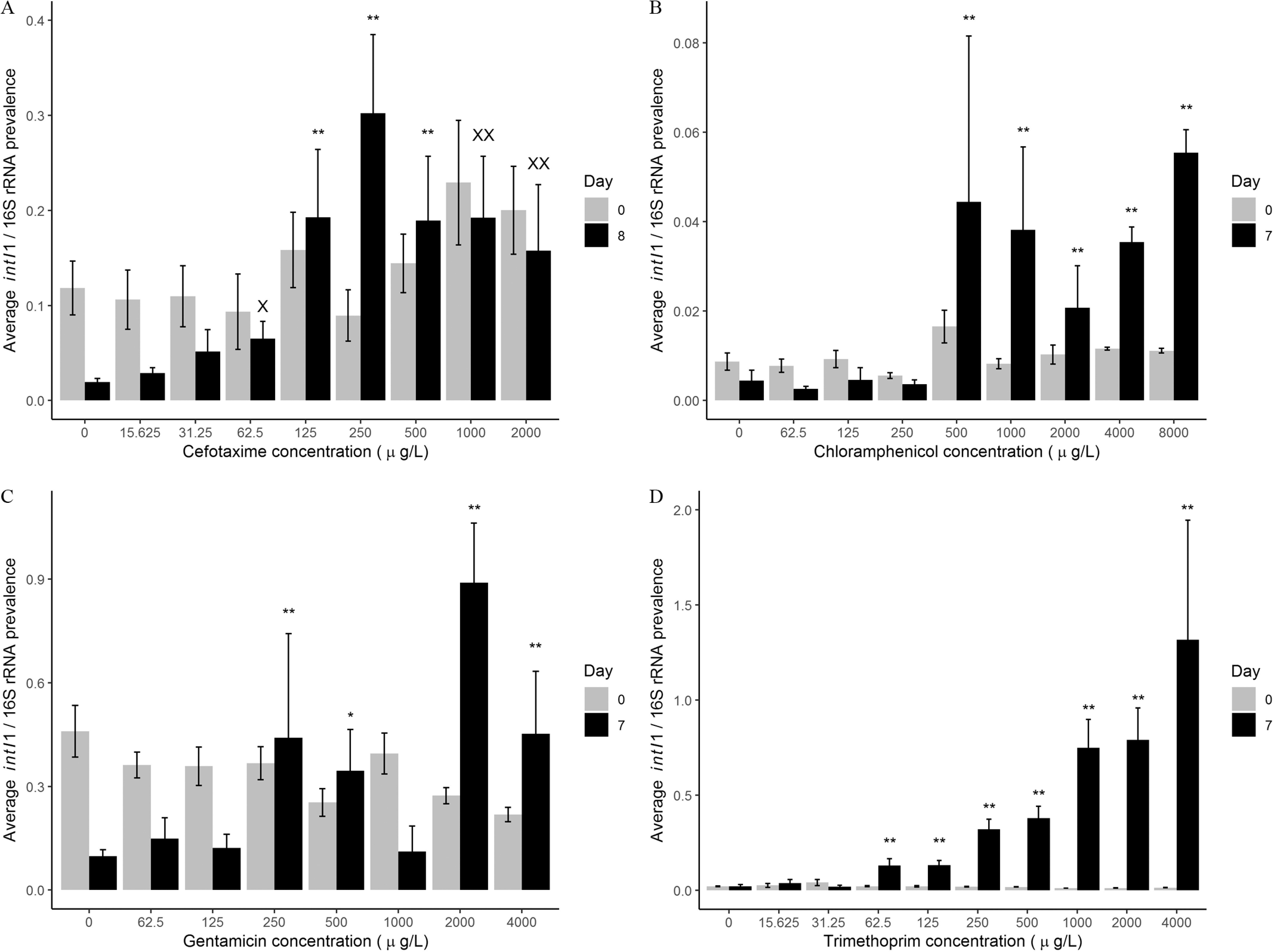
Average intI1 prevalence (intI1 copy number/16S rRNA copy number) at the beginning (Day 0, gray bar) and end (Day 7 or Day 8, black bar) of the evolution experiments for (A) cefotaxime, (B) chloramphenicol, (C) gentamicin, and (D) trimethoprim. for each antibiotic treatment for each day, except gentamicin Day 7 () and trimethoprim Day 7 (). For cefotaxime (A): **, (significantly different) to Day 8 no-antibiotic control, Dunn’s test—positive selection; , (significantly different); , (significantly different) to Day 8 no-antibiotic control, Dunn’s test—significant increased persistence. -Values were 0.0888 (), 0.0050 (), 0.0003 (), 0.0070 (), 0.0033 (), and 0.0374 (). For chloramphenicol (B): **, (significantly different) to Day 7 no-antibiotic control, gamma general linearized model, log link—positive selection. -Values were 0.0012 (), 0.0021 (), 0.02,348 (), 0.0029 (), and 0.0004 (). For gentamicin (C): **, (significantly different) to Day 7 no-antibiotic control, *, (significantly different), gamma general linearized model with log link—positive selection. -Values were 0.0213 (), 0.0508 (), 0.0012 (), and 0.0266 (). For trimethoprim (D): **, (significantly different) to Day 7 no-antibiotic control, gamma general linearized model, log link—positive selection. -Values were 0.0002 (), 0.0002 (), (), (), (), (), and ().
Table 1.
All lowest observed effect concentrations (LOECs), no observed effect concentrations (NOECs) and corresponding predicted no effect concentrations for resistance () reported in the present study ().
| Antibiotic | qPCR LOEC | qPCR NOEC | qPCR | SELECT LOEC | SELECT NOEC | SELECT | SELECT T LOECa | SELECT T NOECa | SELECT T a | SELECT T/AS LOECb | SELECT T/AS NOECb | SELECT T/AS b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZ | 750c | 500c | 50c | 1,000 | 500 | 50 | 1,000 | 500 | 50 | 1,000 | 500 | 50 |
| TAX | 125d | 62.5d | 6.25d | 31.25 | 15.63 | 1.56 | 15.63 | 7.81 | 0.78 | 15.63 | 7.82 | 0.78 |
| CHL | 500 | 250 | 25 | 250 | 125 | 12.5 | ND | ND | ND | ND | ND | ND |
| CIP | 15.63c | 7.81c | 0.78c | 1.95 | 0.98 | 0.1 | 15.63 | 7.82 | 0.78 | 0.976 | 0.49 | 0.05 |
| CLA | 750c | 500c | 50c | 5,000 | 2,500 | 250 | ND | ND | ND | ND | ND | ND |
| ERY | 750c | 500c | 50c | 25,000 | 12,500 | 1,250 | ND | ND | ND | ND | ND | ND |
| GEN | 250 | 125 | 12.5 | 250 | 125 | 12.5 | ND | ND | ND | ND | ND | ND |
| TRMP | 62.5 | 31.25 | 3.13 | 31.25 | 15.63 | 1.56 | 125 | 62.5 | 6.25 | 250 | 125 | 6.25 |
Note: All data were generated in the present study unless otherwise indicated (c,d). The LOEC is the lowest concentration where a significant difference was observed compared with the control. The NOEC is the test concentration directly below the LOEC. All were determined by dividing the NOEC by an assessment factor of 10. Corresponding graphs of average intI1 prevalence for cefotaxime, chloramphenicol, gentamicin, and trimethoprim are shown in Figure 1A–D, respectively. SELECT assays where the temperature was reduced from 37 deg to 21 deg. SELECT T/AS refers to SELECT assays where the temperature was reduced from 37 deg to 21 deg AND artificial sewage (AS) was used instead of Iso-Sensitest Broth. AZ, azithromycin; CHL, chloramphenicol; CIP, ciprofloxacin; CLA, clarithromycin; ERY, erythromycin; GEN, gentamicin; LOEC, lowest observed effect concentration; ND, not determined; NOEC, no observed effect concentration; , predicted no effect concentrations for resistance; qPCR, quantitative polymerase chain reaction; SELECT, SELection End points in Communities of bacTeria assay; TAX, cefotaxime; TRMP, trimethoprim.
SELECT T LOECs, NOECs, and refer to SELECT assays conducted in Iso-Sensitest™ broth at 21°C.
SELECT T/AS LOECs, NOECs, and refer to SELECT assays conducted in artificial sewage at 21°C.
qPCR LOECs, NOECs, and for AZ, CLA, ERY, and CIP were from Stanton et al. (2020). All macrolide values reported are based on ermF (compound-specific gene target); CIP values reported are based on the intI1 target.
The LOEC for ciprofloxacin was determined as by Stanton et al. (2020) using the intI1 gene target. For all other antibiotics with two gene target effect concentrations (i.e., the macrolides), the compound-specific gene target data were determined as the most protective by Stanton et al. (2020). The exception was cefotaxime, where the () LOEC determined previously by Murray et al. (2018) was identical to the intI1 LOEC generated in the present study (Figure 1). ranged from [ciprofloxacin for gene target intI1 (Stanton et al. 2020)] to [azithromycin, clarithromycin, and erythromycin for gene target ermF (Stanton et al. 2020)] (Table 1).
SELECT and Comparison to qPCR
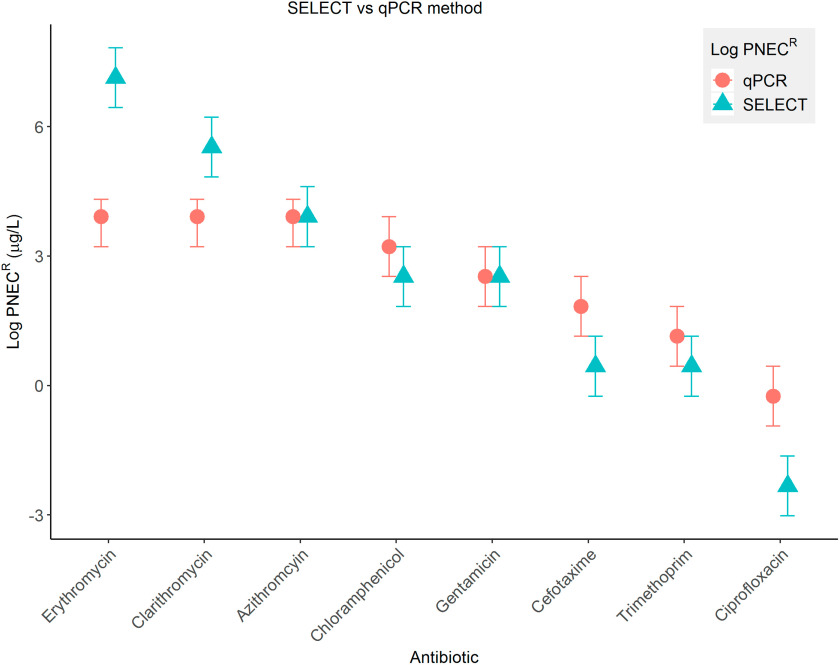
A summary of all the SELECT data determined are shown in Table 1. ranged from for ciprofloxacin to for erythromycin. The most protective qPCR and SELECT determined to date are shown in Figure 2. The error bars represent the span of the test concentrations above and below the NOEC. Overall, there was very good agreement between the two methods, with the span for all overlapping except for two macrolides (erythromycin and clarithromycin) and ciprofloxacin. For the macrolides, this was using the compound-specific qPCR target (i.e., ermF) (Stanton et al. 2020). However, when using the intI1 gene targets, the qPCR (Stanton et al. 2020) and SELECT for erythromycin and clarithromycin were more comparable (Figure S2). derived using the SELECT method were identical or more protective than the qPCR method in all cases, except for erythromycin and clarithromycin, when using the more protective compound-specific gene target ermF reported previously (Stanton et al. 2020) (Figure 2).
Figure 2.
Predicted no effect concentrations for resistance (, logged) determined using the SELECT (blue triangle) and qPCR methods (pink circle). Error bars represent the test concentrations directly above and directly below the NOECs used to calculate the . For qPCR , the most protective gene target is presented. This was ermF for the macrolides and intI1 for the remaining antibiotics. All data used to generate this figure are shown in Table 1. qPCR for azithromycin, clarithromycin, erythromycin, and ciprofloxacin were taken from Stanton et al. (2020). Note: , predicted no effect concentrations for resistance; qPCR, quantitative polymerase chain reaction; SELECT, SELection End points in Communities of bacTeria assay.
Bland-Altman statistical analysis was also performed to determine the level of agreement between derived using the two experimental methods (Figure S3). All data points lay within the upper and lower limits of agreement, with the exception of erythromycin. However, the erythromycin was still within the absolute limits of agreement, demonstrating that the two methods provided statistically very similar measurements.
Spatiotemporal Effects on SELECT
Several variations in bacterial community inoculum were tested to determine whether this affected the derived using the SELECT method. These variations included comparing influent samples from the same WWTP (i.e., A) at two times points (2016 and 2018), influent and effluent from the same WWTP (i.e., A), and influent from two different WWTPs (A and B; the latter of which served a larger population in a different part of the UK). In many cases, SELECT were the same (Figure S4). There were some differences in SELECT , depending on sampling year and sampling site, but all SELECT were within a factor of 2, with the exception of ciprofloxacin—where the SELECT for the 2016 WWTP A influent sample was one-fourth that of the of the WWTP B influent 2018 sample—and for trimethoprim—where the for the 2016 WWTP A influent sample was one-fourth that of the of all other SELECT with WWTP variations for this antibiotic.
Assessing Effects of Culturing Conditions on SELECT
Low-temperature, artificial sewage SELECT (Table 1; Figure S5) were, in some cases, identical to the SELECT determined at a higher temperature and in Iso-Sensitest™ broth. were within a factor of two (with the exception of trimethoprim, where the artificial sewage, 21°C was one-fourth that of the Iso-Sensitest™ broth, 37°C and ciprofloxacin, where all were within the same order of magnitude). No one condition was more protective across all antibiotics; that is, for cefotaxime and ciprofloxacin, the artificial sewage and 21°C condition was more protective, but for azithromycin and trimethoprim, the Iso-Sensitest™ broth and 37°C condition was more protective.
The Risk of Selection for AMR Posed by Different Antibiotics
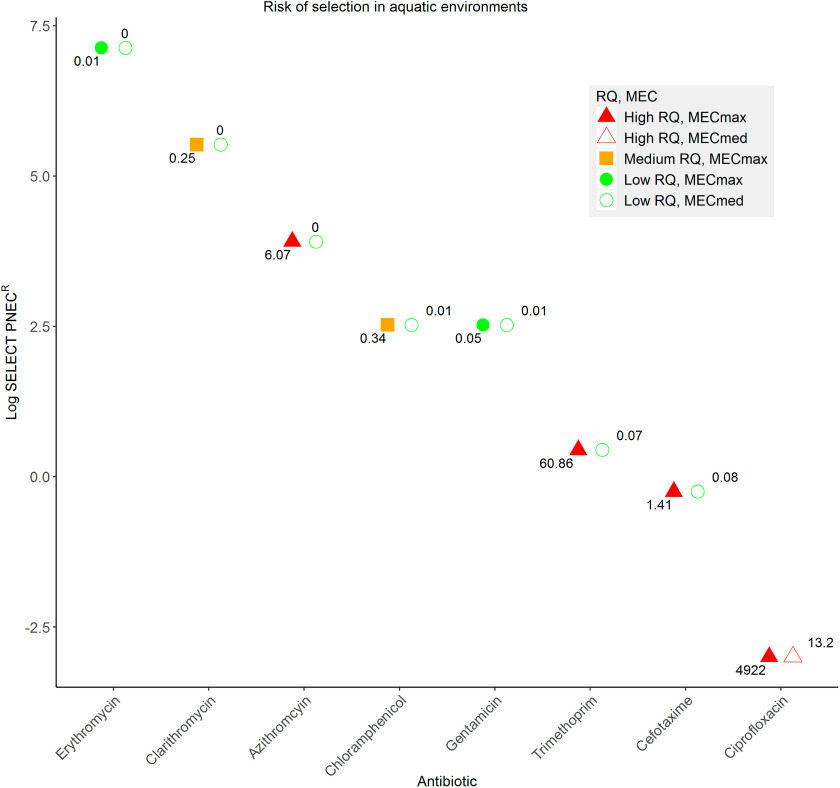
For the present study, were determined by dividing (medium/maximum predicted environmental concentration, see the “Methods” section; Figure 3, and Figure S1) by the lowest SELECT determined in the present study. Overall, for most antibiotics were within the acceptable range (i.e., RQ ). However, several antibiotics posed unacceptable risks. These were azithromycin (), cefotaxime (), trimethoprim (), and ciprofloxacin (both and ). Interestingly, even including the nondetects for calculation of the , ciprofloxacin still posed an unacceptable risk (; Figure S1).
Figure 3.
Risk quotients measured environmental concentration () or median measured environmental concentration ()/lowest determined SELECT-predicted no effect concentration for resistance (). Here, values do not include the nondetects (for these data, see Figure S1). MECs were extracted for all antibiotics except gentamicin from the Umweltbundesamt (UBA) Pharmaceuticals in the Environment database. For gentamicin, predicted environmental concentration data were used. Risk was broadly classified in a traffic light color system. Red, bold (), using ; red, empty (), using ; orange, bold ( and ), using ; green, bold (), using ; green, empty (), using . Note: , predicted no effect concentration for resistance; SELECT, SELection End points in Communities of bacTeria assay.
Comparison of SELECT to Previous and Ecotoxicological PNECs
Environmental PNECs (; derived using standard ecotoxicological tests on cyanobacteria) (Tell et al. 2019) and modeled using minimum inhibitory concentration data from the EUCAST database [ (Bengtsson-Palme and Larsson 2016)] were collated and compared with the SELECT determined in the present study (Table 2). were overall more protective, although some were lower. Ciprofloxacin was the only antibiotic where the SELECT (WWTP A influent, artificial sewage at 21°C) was most protective and very similar to the . Otherwise, qPCR and SELECT were the least protective for all antibiotics, with the exception of trimethoprim, where the was the least protective.
Table 2.
Summary of environmental predicted no effect concentrations () derived using ecotoxicological tests, modeled predicted no effect concentrations for resistance based on MIC data () and experimental predicted no effect concentrations for resistance determined in the present study (qPCR and SELECT ) and previously published studies [(some qPCR c,d)] for all test antibiotics in the present study.
| Antibiotic | () (Tell et al. 2019) | () (Bengtsson-Palme and Larsson 2016) | Lowest qPCR ()a | Lowest SELECT ()b |
|---|---|---|---|---|
| Azithromycin | 0.02 | 0.25 | 50c | 50 |
| Cefotaxime | 0.1 | 0.13 | 6.25d | 0.78 |
| Chloramphenicol | NA | 8 | 25 | 12.5 |
| Ciprofloxacin | 0.57 | 0.06 | 0.78c | 0.05 |
| Clarithromycin | 0.08 | 0.25 | 50c | 250 |
| Erythromycin | 0.5 | 1 | 50c | 1,250 |
| Gentamicin | 0.15 | 1 | 12.5 | 12.5 |
| Trimethoprim | 100 | 0.5 | 3.125 | 1.56 |
Note: Original data citations are in the column headers or denoted by footnotes (c,d), as appropriate. LOEC, lowest observed effect concentration; NA, not available; NOEC, no observed effect concentration; , environmental predicted no effect concentration; , predicted no effect concentration for resistance modeled using minimum inhibitory concentration; , predicted no effect concentrations for resistance; qPCR, quantitative polymerase chain reaction; SELECT, SELection End points in Communities of bacTeria assay; WWTP, wastewater treatment plant.
Lowest qPCR reported for the gene target that gave the lowest, lowest observed effect concentration. were calculated by dividing the no observed effect concentration by an assessment factor of 10.
Lowest SELECT reported for the experimental conditions for any influent sample (i.e., from WWTP A or B) that gave the lowest, lowest observed effect concentration. were calculated by dividing the no observed effect concentration by an assessment factor of 10.
Data from Stanton et al. (2020).
Data from Murray et al. (2018). This is identical to the cefotaxime for the intI1 gene targeted, which was calculated as part of the present study.
Discussion
The present study used a new growth-based assay (the SELECT method) to determine for a total of eight antibiotics, spanning 6 of 9 single antibiotic classes [3rd subgroup, according to the Anatomical Therapeutic Chemical classification (WHO 2019)]. In addition, qPCR generated in the present study and others (Murray et al. 2018; Stanton et al. 2020) with the same experimental system were collated and compared with SELECT . There was very good agreement, indicating the SELECT method to be a reliable, rapid, and cost-effective method to rapidly generate selective end point data. This has been repeatedly identified as a significant knowledge gap in environmental and human health risk assessment (Ashbolt et al. 2013) given that currently it is unclear whether ecotoxicological end points are protective of selection for AMR (Le Page et al. 2017). We showed that the SELECT method outlined in the present study can be used to rapidly determine experimental for antibiotics that can a) inform release limits of antibiotics that aim to reduce the evolution of AMR in situ; b) provide data that can be used to study the evolution of different AMR mechanisms in greater detail; and c) identify likely environmental hotspots of AMR to which humans may be exposed.
Both the intI1 gene and compound-specific resistance genes were used as qPCR end points to determine for the macrolides, ciprofloxacin and cefotaxime. For the remaining antibiotics (chloramphenicol, gentamicin, and trimethoprim), only intI1 was used because the qPCR target because genes conferring resistance to these antibiotics are frequently integron associated (Partridge et al. 2009). A metagenomic approach could be used to identify additional compound-specific targets for qPCR quantification (Murray et al. 2018); however, it has also been shown that exposure of a sewage-derived microbial community to even relatively high levels of ciprofloxacin and trimethoprim resulted in more co-selection than selection for compound-specific resistance genes (Murray et al. 2019). Macrolide-specific gene targets generated lower than intI1, which may be because macrolide-resistance genes are not commonly integron associated (Stanton et al. 2020); however, interestingly, cefotaxime using compound-specific () (Murray et al. 2018) and intI1 gene targets (the present study) were identical. This suggests that compound-specific gene targets to determine qPCR will not always be more protective than using intI1. Class 1 integrons are considered a good indicator of environmental pollution owing to their ability to integrate different resistance gene cassettes (Gillings et al. 2015), making them an appropriate gene target to quantify the potential for selection as well as co-selection. A single gene target such as intI1 that summarizes or estimates total selection may also be an attractive regulatory end point. This end point also has utility in human health risk assessment, given that class 1 integrons are often clinically associated with AMR (Ghaly et al. 2017).
For all antibiotics, with the exception of erythromycin and clarithromycin, the SELECT was either identical or more protective than the qPCR . This may be because, in the previous study that determined the macrolide reported here, metagenome analyses showed that the predominant members of the community were Gram-negative opportunistic pathogens belonging to Enterobacteriaceae, such as E. coli, Proteus mirabilis, and Klebsiella pneumoniae (Stanton et al. 2020), and macrolides are generally used to treat Gram-positive infections. Therefore, it may be that there is minimal reduction in growth because the majority of bacteria within the sample were already (intrinsically) resistant.
With the exception of erythromycin, clarithromycin, and ciprofloxacin, all qPCR and SELECT were within one test concentration of each other. For comparison, when performing the ASRIT, large variability is expected and results are often reported within orders of magnitude [e.g., to (OECD 2009)], whereas most of the qPCR and SELECT values did not differ by a factor . The SELECT method is also more rapid and cost effective than the ASRIT test, which is currently recommended for environmental risk assessment of antibiotics (EMA 2018) despite evidence showing that it is insensitive to antibiotics (Le Page et al. 2017).
The lower SELECT observed for ciprofloxacin was likely due to the fact that resistance to ciprofloxacin is conferred through multiple mechanisms, all of which are captured in the SELECT method, as opposed to a single target with qPCR. Presumably, one or more of these mechanisms has a lower fitness cost than integron carriage. Previously, gyrA1 mutants have been selected at concentrations of as low as , which is the lowest minimal selective concentration (MSC) determined to date (Gullberg et al. 2011). In more elaborate biofilm experimental systems, the LOEC of ciprofloxacin was , based on higher qnrD relative abundance and significant effects on community structure (Kraupner et al. 2018). Selection for this mobile resistance gene in the community occurred at a lower concentration than selection for gyrA mutations in E. coli isolated from the same community (Kraupner et al. 2018). Therefore, mobile quinolone resistance genes may have a lower fitness cost compared with chromosomal mutations in housekeeping genes. The lowest SELECT for ciprofloxacin was , which is more comparable to these values than the previously determined qPCR of (Stanton et al. 2020). This lends further support to reliability of the SELECT method and indicates selective effect concentrations may be very similar, even across different experimental systems (at least in the case of ciprofloxacin, and possibly other fluoroquinolones).
This is further exemplified through our investigation of spatiotemporal factors and culturing conditions. Spatiotemporal factors affecting the risk of selection were investigated by comparing SELECT determined using wastewater samples from different time points, geographical locations, or type of sample (i.e., influent or effluent). were very similar overall, which suggests that a single for each antibiotic could be applicable to many different environments. However, the samples tested were all from the UK, serving rural or semi-rural populations, and urban sewage serving much greater populations may yield different results. No significant differences in determined using influent or effluent indicates that influent samples may be sufficient for future testing. This would enable a to be determined in less than 15 h (including setup, the experimental run, and data analyses), which is significantly quicker than many current ecotoxicological tests, including those currently recommended for assessment of antibiotics.
Potential limitations of the experimental conditions used for the determination of qPCR and SELECT are that the temperature (37°C) and nutrient content (Iso-Sensitest™ broth) are high (compared with environmental conditions). To address this potential issue and to improve the environmental realism of the SELECT assay, for four antibiotics were determined with the SELECT assay conducted at room temperature () and with artificial sewage used as the growth medium. This medium and temperature range are used in the widely used Organisation for Economic Co-operation and Development (OECD)-approved ASRIT (OECD 2010). SELECT were almost all identical or very similar (i.e., within one test concentration), independent of temperature or culture medium used. This suggests that the SELECT method is also representative of more environmentally relevant conditions (i.e., lower temperature and nutrient levels). The largest discrepancy was observed for trimethoprim, where the artificial sewage, lower-temperature PNECR was four times that of the Iso-Sensitest™, higher-temperature treatment. No single experimental condition consistently gave the lowest for all four tested antibiotics. This may be a reflection of changes in relative fitness of bacteria harboring resistance or of preferential enrichment of different species, which may be differentially favored under different conditions.
The determined in the present study were used to calculate RQs for aquatic environments (hospital effluent, wastewater influent and effluent) based on MEC data extracted from the UBA database (UmweltBundesamt 2019) (except for gentamicin, which used predicted environmental concentration data). Ciprofloxacin posed the highest risk of selection for AMR, given that both the and were (derived using the low-temperature, artificial sewage SELECT ), and, in the case of the , this value approached 5,000. Trimethoprim, azithromycin, and cefotaxime posed the next highest risks (), indicating selection is likely to occur in particularly impacted environments, which has implications for human exposure and human health risk assessment. For all antibiotics, with the exception of ciprofloxacin, the was , which translates to a lower risk of selection. Our data support the recommendation to include ciprofloxacin and retain azithromycin on the second version of the European WFD Watch List (Loos et al. 2018) but also suggest that trimethoprim should be added on the basis of risk of selection for AMR. The other two macrolides (clarithromycin and erythromycin) are also included on the WFD Watch List, but only clarithromycin exerts a medium risk of selection occurring in the most impacted environments ().
A comparison with previously published PNECs was also performed. were most protective in most cases, except for chloramphenicol, ciprofloxacin, and trimethoprim, where a was more protective. In these three cases, SELECT were highly comparable to the , indicating a good predictive value of the method. However, because our data are experimental as opposed to modeled, this also indicates that the method may be overestimating selective effects in some cases. This comparison also strengthens the case for adding trimethoprim to the WFD Watch List on the basis of risk for selection, given that the was two orders of magnitude greater than all (, qPCR , or SELECT ), as well as resulting in an . This also highlights the need for continued generation of data on selective end points, especially in cases where the is not determined (as with chloramphenicol). This is particularly pertinent because AMR is a potential human health hazard not currently considered by standard ecotoxicological approaches. The SELECT method will be useful for rapid data generation for antibiotics such as chloramphenicol, where ecotoxicological PNEC data do not exist.
Our data also support a recent meta-analysis that compared different ecotoxicological PNECs and (Le Page et al. 2017). PNECs in cyanobacteria were the most protective of the ecotoxicological end points included; however, they were not always more protective than . The reasons for this are unclear but may indicate higher sensitivity to some antibiotics in some cyanobacterial species, as suggested previously (Le Page et al. 2017), compared with the sewage communities used in the SELECT method. Therefore, as in the previous study, we recommend cyanobacterial PNECs be used in conjunction with experimental (such as those generated using the SELECT method) to ensure maximum protection of both the environment and human health.
The current environmental risk assessment is based on single compounds, and an issue so far unaddressed is the fact that bacterial communities in sewage, or in the environment, are exposed to a complex mixture of antibiotics and other co-selective compounds such as biocides or heavy metals. Therefore, additive or synergistic effects of compounds with selective effects are likely to result in lower and higher RQs. Additional research into the effects of mixtures of selective compounds is required to understand the true risk of selection/co-selection for AMR occurring in aquatic environments. In addition, emerging data on MECs in low- and middle-income countries (J. Wilkinson and A. Boxall, personal communication) will result in significantly higher , , and RQ values. Therefore, the global risk of selection will be greater than reported here.
In conclusion, the present study reports the largest set of empirical selective endpoint data determined within the same experimental system. For all antibiotics tested except clarithromycin and erythromycin, the novel SELECT method generated more protective end points than those derived using qPCR in previously published experimental studies (Murray et al. 2018; Stanton et al. 2020). The SELECT method is recommended for rapid generation of further selective endpoint data for antibiotics and other antimicrobials, including the effects of complex mixtures. These data can be used to inform environmental and human health risk assessment of AMR. The data presented support the inclusion of azithromycin and ciprofloxacin as priority substances and suggest that trimethoprim and possibly cefotaxime should also be considered for the next WFD Watch List on the basis of risk of selection for AMR. By reducing selection for AMR in the natural environment, the risk of human exposure to AMR is also likely to be reduced.
Supplementary Material
Acknowledgments
This research was supported by a Biotechnology and Biological Research Sciences Council and AstraZeneca Collaborative Awards in Science and Engineering studentship (grant BB/L502509/1) and a Natural Environment Research Council (NERC) Industrial Innovation Fellowship (NE/R01373X/1). A.K.M supported by BBSRC and NERC fellowship awarded to A.K.M. I.S. was supported by a Biotechnology and Biological Research Sciences Council (BBSRC) and AstraZeneca Collaborative Awards in Science and Engineering studentship (grant BB/N504026/1), L.Z. was supported by NERC NE/M011259/1 and NE/N019717/1, and J.W. was supported on placement by the Nuffield Foundation in 2017.
References
- Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, et al. . 2013. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ Health Perspect 121(9):993–1001, PMID: 23838256, 10.1289/ehp.1206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ. 2016. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ Int 86:140–149, PMID: 26590482, 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1(8476):307–310, PMID: 2868172, 10.1016/S0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- ECHA (European Chemicals Agency). 2013. Guidance on biocides legislation. Helsinki, Finland: ECHA; https://echa.europa.eu/guidance-documents/guidance-on-biocides-legislation [accessed 2 October 2020]. [Google Scholar]
- EMA (European Medicines Agency). 2006. Guideline on the Environmental Risk Assessment of Medicinal Products for Human Use. First version. London, UK: Committee for Medicinal Products for Human Use; https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-environmental-risk-assessment-medicinal-products-human-use-first-version_en.pdf [accessed 2 October 2020]. [Google Scholar]
- EMA. 2018. Guideline on the environmental risk assessment of medicinal products for human use. https://www.ema.europa.eu/en/documents/scientific-guideline/draft-guideline-environmental-risk-assessment-medicinal-products-human-use-revision-1_en.pdf [accessed 2 October 2020].
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf [accessed 2 October 2020].
- FDA (U.S. Food and Drug Administration). 1998. Guidance for Industry: Environmental Assessment of Human Drug and Biologics Applications. Washington, DC: U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research, Center for Biologics Evaluation and Research; https://www.fda.gov/media/70809/download [accessed 2 October 2020]. [Google Scholar]
- Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. 2017. Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS One 12(6):e0179169, PMID: 28586403, 10.1371/journal.pone.0179169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavarina D. 2015. Understanding Bland Altman analysis. Biochem Med (Zagreb) 25(2):141–151, PMID: 26110027, 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu Y-G. 2015. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J 9(6):1269–1279, PMID: 25500508, 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield BK, Shaked S, Marrs CF, Nelson P, Raxter I, Xi C, et al. . 2018. Modeling the emergence of antibiotic resistance in the environment: an analytical solution for the minimum selection concentration. Antimicrob Agents Chemother 62(3):e01686-17, PMID: 29263062, 10.1128/AAC.01686-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al. . 2011. Selection of resistant bacteria at very low antibiotic concentrations. PloS Pathog 7(7):e1002158, PMID: 21811410, 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelić D, Antolović R. 2016. From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics (Basel) 5(3):29, PMID: 27598215, 10.3390/antibiotics5030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klümper U, Recker M, Zhang L, Yin X, Zhang T, Buckling A, et al. . 2019. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J 13(12):2927–2937, PMID: 31384011, 10.1038/s41396-019-0483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraupner N, Ebmeyer S, Bengtsson-Palme J, Fick J, Kristiansson E, Flach C-F, et al. . 2018. Selective concentration for ciprofloxacin resistance in Escherichia coli grown in complex aquatic bacterial biofilms. Environ Int 116:255–268, PMID: 29704804, 10.1016/j.envint.2018.04.029. [DOI] [PubMed] [Google Scholar]
- Kümmerer K. 2004. Resistance in the environment. J Antimicrob Chemother 54(2):311–320, PMID: 15215223, 10.1093/jac/dkh325. [DOI] [PubMed] [Google Scholar]
- Le Page G, Gunnarsson L, Snape J, Tyler CR. 2017. Integrating human and environmental health in antibiotic risk assessment: a critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environ Int 109:155–169, PMID: 28964562, 10.1016/j.envint.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Leonard AFC, Zhang L, Balfour AJ, Garside R, Hawkey PM, Murray AK, et al. . 2018. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ Int 114:326–333, PMID: 29343413, 10.1016/j.envint.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Loos R, Marinov D, Sanseverino I, Napierska D, Lettieri T. 2018. Review of the 1st Watch List under the Water Framework Directive and recommendations for the 2nd Watch List. EUR 29173 EN. Luxembourg: Publications Office of the European Union; http://publications.jrc.ec.europa.eu/repository/bitstream/JRC111198/wl_report_jrc_2018_04_26_final_online.pdf [accessed 2 October 2020]. [Google Scholar]
- Lundström SV, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, et al. . 2016. Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci Total Environ 553:587–595, PMID: 26938321, 10.1016/j.scitotenv.2016.02.103. [DOI] [PubMed] [Google Scholar]
- Murray AK, Zhang L, Snape J, Gaze WH. 2019. Comparing the selective and co-selective effects of different antimicrobials in bacterial communities. Int J Antimicrob Agents 53(6):767–773, PMID: 30885807, 10.1016/j.ijantimicag.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AK, Zhang L, Yin X, Zhang T, Buckling A, Snape J, et al. . 2018. Novel insights into selection for antibiotic resistance in complex microbial communities. mBio 9(4):e00969-18, PMID: 30042197, 10.1128/mBio.00969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. 2016. Tackling drug-resistant infections globally: Final report and recommendations, https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf.
- OECD (Organisation for Economic Co-operation and Development). 2009. OECD Guidelines for the Testing of Chemicals. Proposal for a revised Guideline 209—Inhibition of Respiration of Activated Sludge (Carbon and/or Ammonium Oxidation). http://www.oecd.org/chemicalsafety/testing/43735667.pdf [accessed 2 October 2020].
- OECD. 2010. OECD Guidelines for the Testing of Chemicals. Activated Sludge, Respiration Inhibition Test (Carbon and Ammonium Oxidation). https://www.oecd-ilibrary.org/docserver/9789264070080-en.pdf?expires=1601672303&id=id&accname=guest&checksum=DE420B1160802275C987254FBB1C2632 [accessed 2 October 2020].
- Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33(4):757–784, PMID: 19416365, 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- Stanton IC, Murray AK, Zhang L, Snape J, Gaze WH. 2020. Evolution of antibiotic resistance at low antibiotic concentrations: selection below the minimal selective concentration. Commun Biol 3(1):467, PMID: 32884065, 10.1038/s42003-020-01176-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell J, Caldwell DJ, Häner A, Hellstern J, Hoeger B, Journel R, et al. . 2019. Science-based targets for antibiotics in receiving waters from pharmaceutical manufacturing operations. Integr Environ Assess Manag 15(3):312–319, PMID: 30884149, 10.1002/ieam.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2017. Conducting a Human Health Risk Assessment. https://www.epa.gov/risk/conducting-human-health-risk-assessment#tab-2 [accessed 12 June 2019].
- UmweltBundesamt. 2019. Pharmaceuticals in the Environment. [Database.]. https://www.umweltbundesamt.de/en/database-pharmaceuticals-in-the-environment-0 [accessed 2 October 2020].
- WHO (World Health Organisation) Collaborating Centre for Drug Statistics Methodology. 2019. https://www.whocc.no/atc_ddd_index/?code=J01 [accessed 16 December 2019].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.