ABSTRACT
The current model for spindle positioning requires attachment of the microtubule (MT) motor cytoplasmic dynein to the cell cortex, where it generates pulling force on astral MTs to effect spindle displacement. How dynein is anchored by cortical attachment machinery to generate large spindle-pulling forces remains unclear. Here, we show that cortical clustering of Num1, the yeast dynein attachment molecule, is limited by its assembly factor Mdm36. Overexpression of Mdm36 results in an overall enhancement of Num1 clustering but reveals a population of dim Num1 clusters that mediate dynein anchoring at the cell cortex. Direct imaging shows that bud-localized, dim Num1 clusters containing around only six Num1 molecules mediate dynein-dependent spindle pulling via a lateral MT sliding mechanism. Mutations affecting Num1 clustering interfere with mitochondrial tethering but do not interfere with the dynein-based spindle-pulling function of Num1. We propose that formation of small ensembles of attachment molecules is sufficient for dynein anchorage and cortical generation of large spindle-pulling forces.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Num1, Dynein, Mitochondria, Spindle positioning
Summary: Using Mdm36 as a tool to cluster Num1 at the cell cortex, this study reveals that there are two distinct populations of Num1, each mediating its role in spindle positioning and mitochondrial tethering, respectively.
INTRODUCTION
Orienting the mitotic spindle is paramount to controlling the outcome of asymmetric cell division, which is critical for determining daughter-cell size, fate and location. In most animals and fungi, pulling forces for orienting the spindle are generated by the microtubule (MT) motor cytoplasmic dynein and are dependent on the anchoring of the dynein motor at the cell periphery (Bowman et al., 2006; Couwenbergs et al., 2007; Du and Macara, 2004; Heil-Chapdelaine et al., 2000; Kotak et al., 2012; Kotak et al., 2014; Nguyen-Ngoc et al., 2007). Current evidence suggests that cortical dynein-anchoring proteins might enhance spindle-pulling function by forming clusters on the cell membrane (Okumura et al., 2018). A favored hypothesis is that clustering of anchoring proteins contributes to the generation of large cooperative pulling forces by increasing the number of interacting motors per cortical MT contact site (Kiyomitsu, 2019; Okumura et al., 2018), analogous to how lipid microdomains on phagosomes facilitate motor clustering to achieve cooperative force generation by dynein during intracellular trafficking (Rai et al., 2016). However, despite identification of the domain required for clustering activity and punctate localization in both yeast and mammalian dynein-anchoring proteins (Harborth et al., 1999; Okumura et al., 2018; Tang et al., 2012), factors influencing cluster assembly, size and distribution along the cortex are poorly understood. Additionally, whether cluster enhancement is correlated with an increase in dynein recruitment or higher cortical dynein activity remains unknown.
The budding yeast dynein-anchoring protein Num1 is a 313 kDa protein composed of a short N-terminal CC domain (amino acids 95–303), followed by a central TR domain containing thirteen 64-residue tandem repeats (amino acids 592–1776) and a C-terminal phosphatidylinositol (4,5)-bisphosphate-binding PH domain (amino acids 2563–2683) (Greenberg et al., 2018). The CC domain is necessary and sufficient for cluster formation, in addition to being required for mediating an interaction with dynein and dynactin at the cell cortex (Tang et al., 2012). The CC domain also binds mitochondria and Mdm36, a 65 kDa protein implicated in promoting Num1 cluster formation and mitochondrial division (Hammermeister et al., 2010; Lackner et al., 2013; Ping et al., 2016). Although it is well-accepted that dynein exerts spindle-pulling force at cortical Num1 sites, the abundance and heterogeneity of Num1 patches along the cell cortex (Heil-Chapdelaine et al., 2000; Omer et al., 2018; Schmit et al., 2018) has made it impossible to follow the effects of astral MT plus end interaction with individual cortical Num1 sites, a prerequisite for understanding how clustering might impact dynein force amplification. To our knowledge, contacts between astral MT plus end and individual Num1 foci have not been observed for MT sliding, the in vivo hallmark of dynein-mediated spindle pulling (Adames and Cooper, 2000; Yeh et al., 2000), hence the size of Num1 clusters required for this classic dynein-dependent microtubule–cortex interaction remains unknown.
Additionally, recent studies have begun to reveal that organelles such as mitochondria and the endoplasmic reticulum (ER) are involved in regulating Num1 cluster formation, with a subset of clusters requiring mitochondria for their assembly in the bud (Kraft and Lackner, 2017), and another subset requiring the ER tethering proteins Scs2 and Scs22 for their formation throughout the cell cortex (Chao et al., 2014; Omer et al., 2018). The Num1 patches assembled by mitochondria can in turn bind and tether the organelle to the cell cortex (Kraft and Lackner, 2017; Ping et al., 2016). Intriguingly, like Num1, the fission yeast homologue Mcp5 also forms cortical foci for dynein anchoring and employs a similar CC domain for interaction with both dynein and mitochondria (Chacko et al., 2019; Kraft and Lackner, 2019; Saito et al., 2006; Thankachan et al., 2017; Yamashita and Yamamoto, 2006), suggesting that the mechanism regulating cortical cluster formation might be conserved. The general hypothesis emerging from these studies is that distinct populations of Num1 clusters might exist at the cell periphery, but whether different pools of Num1 could be performing different Num1 functions – namely, dynein anchoring and mitochondrial tethering – remains unclear.
Here, using Mdm36 as a tool to cluster Num1, we found that, in contrast to the prevailing notion for dynein-anchoring proteins, enhancing Num1 clustering unexpectedly reduces dynein recruitment to the cell cortex, but without affecting dynein function in spindle positioning. We report direct observation of MT sliding that occurs when an astral MT plus end encounters a cortical Num1 cluster containing only a small number of Num1 molecules. The observed sliding events do not appear to require Mdm36 and mitochondria. Furthermore, mutations that interfere with Num1 clustering disrupt mitochondrial-tethering activity but not dynein-based spindle-pulling activity of Num1, highlighting a more critical role for clustering in mitochondrial-anchoring rather than dynein-anchoring function.
RESULTS
Overexpression of Mdm36 dramatically enhances Num1 clustering
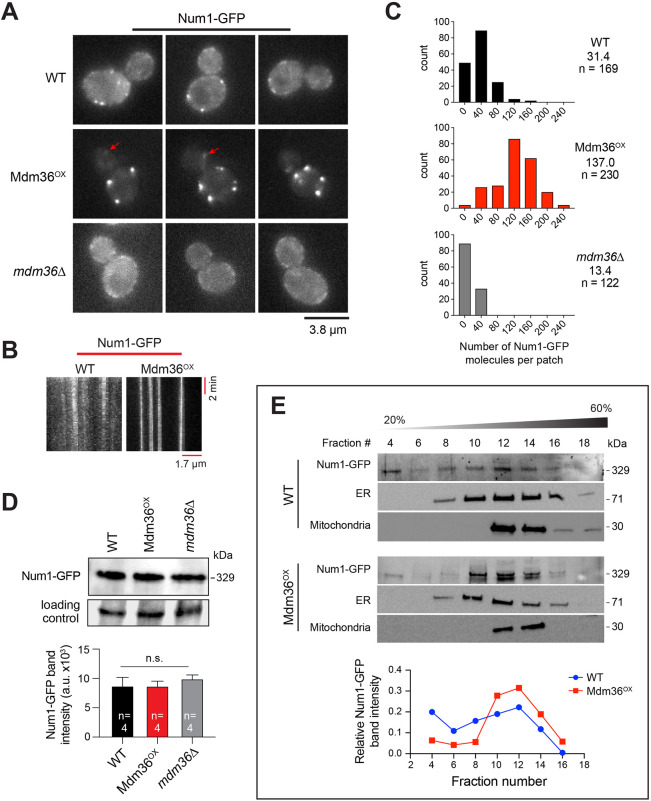
We first investigated how Num1 distribution is affected by Mdm36 at the cell cortex. As previously shown, deletion of Mdm36 resulted in smaller and dimmer Num1 patches (Lackner et al., 2013) (Fig. 1A), consistent with a clustering role for Mdm36. To determine whether Mdm36 level is limiting for Num1 clustering, we examined the effects of overexpressing Mdm36. We integrated the inducible MET3 promoter at the 5′ end of the endogenous chromosomal locus of the MDM36 gene and assayed for Num1–GFP intensity and distribution. In Mdm36-overexpressing cells, hereafter referred to as Mdm36OX cells, we observed a striking enhancement in the intensity of cortical Num1–GFP patches (Fig. 1A; Fig. S1A). The level of cytoplasmic Num1–GFP fluorescence in Mdm36OX cells was dramatically reduced compared to the levels in mdm36Δ mutant and wild-type (WT) cells (Fig. 1A), suggesting that Mdm36 overexpression enhances the recruitment of free Num1 from the cytoplasm into cortical patches. We also observed that, similar to Mdm36 overexpression, overexpression of Num1 (using the same MET3 promoter) resulted in an enhancement in the intensity of Num1 patches (Fig. S1A). However, cortical patches in Num1-overexpressing cells were markedly more dispersed compared to those in Mdm36OX cells (Fig. S1B), indicating that Mdm36 is a clustering factor for Num1 at the cell membrane.
Fig. 1.
Mdm36 enhances Num1 clustering. (A) Representative unadjusted images of Num1–GFP in WT, Mdm36OX and mdm36Δ cells acquired under the same conditions using the same microscope settings. Arrows indicate dim cortical foci visible in the bud of Mdm36OX cells. (B) Kymograph of Num1–GFP foci in WT and Mdm36OX cells. (C) Histogram of protein copy number per cortical patch for Num1–GFP. Median copy number is shown for each strain. (D) Western blot showing Num1–GFP protein level in whole-cell lysates of the indicated strains. The non-specific band was used as a loading control. Plot depicts mean intensity of the Num1–GFP band for four loading replicates. Error bars represent s.e.m. n.s., not statistically significant (one-way ANOVA). (E) Sucrose gradient sedimentation analysis of Num1–GFP in WT and Mdm36OX strains. Whole-cell lysate was loaded onto a 20–60% sucrose gradient, sedimented and analyzed by western blotting using anti-GFP (for Num1–GFP), anti-Sac1 (for ER) and anti-Por1 (for mitochondria) antibodies. The intensity of the Num1–GFP band is plotted against fraction number. Blots are representative of two experiments.
Kymograph analysis showed that Num1–GFP patches in Mdm36OX cells were stationary, similar to Num1 patches in WT cells (Fig. 1B), and were remarkably stable, with some observed to persist for more than two cell division cycles (Movie 1). We next used a ratiometric comparison approach to estimate the number of Num1–GFP molecules per cortical patch. Cells expressing Cse4–GFP, a fluorescence standard for protein counting, were imaged under identical conditions with WT, Mdm36OX and mdm36Δ cells expressing Num1–GFP. The fluorescence intensities of Num1–GFP patches were quantified and compared to those of anaphase Cse4–GFP clusters (which contain ∼96 molecules/cluster) (Verdaasdonk et al., 2014) to determine the Num1 protein counts. In WT cells, we found 31 copies of Num1–GFP per cortical patch (Fig. 1C), a level higher than we had previously reported (Tang et al., 2009), given the revised protein copy number for the Cse4–GFP standard (Coffman et al., 2011; Lawrimore et al., 2011; Verdaasdonk et al., 2014). The intensities of Num1–GFP patches in Mdm36OX cells indicated 137 copies of Num1–GFP per cortical patch (Fig. 1C), a 4.4-fold increase from the number calculated for WT cells. In contrast, in mdm36Δ cells, we found 13 copies of Num1–GFP per cortical patch (Fig. 1C), a 2.3-fold decrease from that calculated for WT cells.
To verify the copy numbers determined by using the Cse4–GFP standard, we repeated our measurements using a different fluorescence standard, Mif2–GFP (Verdaasdonk et al., 2014). We found copy numbers that were consistent with those estimated using the Cse4–GFP standard (38, 139 and 15 for WT, Mdm36OX and mdm36Δ cells, respectively; Fig. S2A). Additionally, we carried out stepwise photobleaching experiments to measure the average step size of intensity loss during bleaching using the STEPFINDER software (Fig. S2B; see Materials and Methods), as described previously (Thankachan et al., 2017). Determining the step size corresponding to the photobleaching of one Num1–GFP molecule enabled the calculation of the total number of Num1–GFP molecules per patch based on the initial intensity of the patch (Coffman and Wu, 2012; Verdaasdonk et al., 2014). Using this approach, we found 36 molecules of Num1–GFP per cortical patch in WT cells (Fig. S2C), further corroborating the value obtained by ratiometric comparison to Cse4–GFP.
We next asked whether Num1 stability is affected in Mdm36OX and mdm36Δ cells. Immunoblot analysis of total cell lysates showed that Num1 levels in Mdm36OX and mdm36Δ were similar to that in WT (Fig. 1D), indicating that the observed difference in the copy number could not be attributed to changes in the expression levels or the stability of Num1 protein. Thus, our data indicate that the copy number of Num1 per cortical patch is limited by the levels of Mdm36 in the cell.
Similar to Num1 patches in WT cells (Omer et al., 2018), Num1–GFP patches in Mdm36OX cells redistributed to the polarized ends of the cell (the distal bud tip and the mother cell apex) upon deletion of the ER tethering proteins Scs2 and Scs22 (Fig. S2D). To further assay for association with ER, we analyzed sedimentation profiles of Num1 in sucrose density gradients. The ER-localized phosphoinositide phosphatase Sac1 was used as a marker for ER. We found that Num1 in Mdm36OX lysate co-fractionated with ER, more so than Num1 in WT lysate (Fig. 1E; compare Num1–GFP levels in fractions 10, 12 and 14 to fractions 4, 6 and 8). Additionally, mitochondria appeared to co-sediment with Num1 and ER in both lysates. These data indicate that the association of Num1 with the ER is enhanced by Mdm36 overexpression.
Dynein function is intact in Mdm36OX cells despite asymmetric distribution of Num1
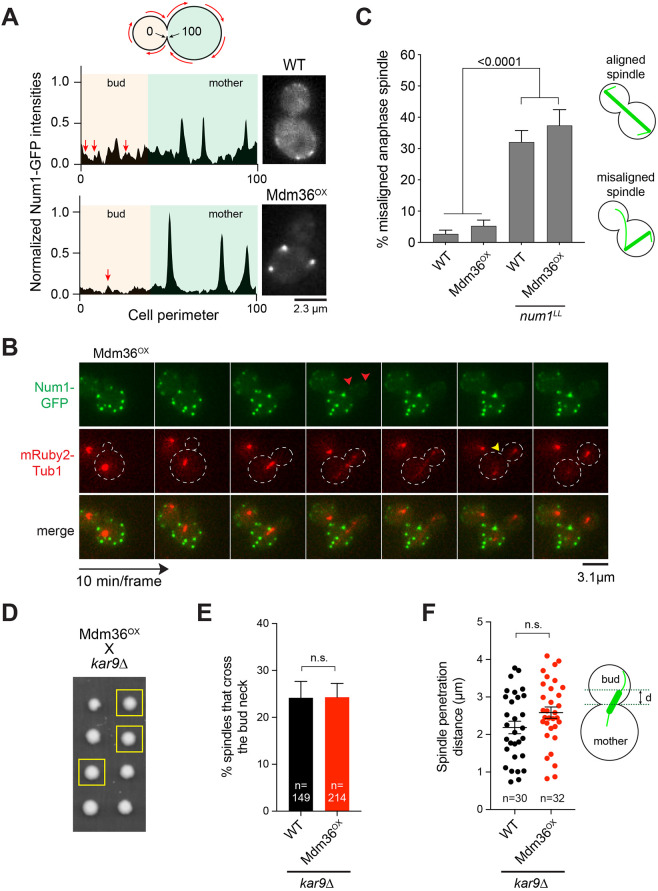
We next wondered whether the observed change in Num1 clustering in Mdm36OX cells would affect dynein pathway function. The prevailing model for dynein function proposes that dynein is recruited from the dynamic plus ends of astral MTs to cortical patches of Num1 in the bud; once anchored, dynein uses its minus-end-directed motor activity to pull the attached spindle into the bud neck (Lee et al., 2005; Lee et al., 2003; Sheeman et al., 2003). In Mdm36OX cells, however, we noticed that the enhanced Num1 patches were observed mostly in the mother cell compartment (Fig. 1A). The bud often exhibited only a few dim patches (Fig. 2A,B). Although the reason for the apparent asymmetry in Num1 distribution is unclear at this point, this phenomenon raises the possibility that dynein pathway function might be defective in the Mdm36OX background.
Fig. 2.
Dynein pathway function in Mdm36OX cells. (A) Normalized intensity profile along the cell cortex (left) and single focal plane image (right) of a representative WT and Mdm36OX cell expressing Num1–GFP. Arrows indicate peaks corresponding to dim foci in the bud of both WT and Mdm36OX profiles. (B) Asymmetry of Num1 localization between mother and daughter cell during the budding cycle in the Mdm36OX background. Red arrowheads, dim Num1–GFP patches in the bud; yellow arrowhead, end of mitosis as indicated by spindle disassembly. Dashed lines indicate cell outlines. (C) Percentage of anaphase spindles with a misoriented phenotype for the indicated strains (91≤n≤514 spindles per strain). Error bars represent the standard error of proportion (s.e.p.). P-value calculated using a one-way ANOVA test comparing each strain with every other strain. Alignment phenotypes are presented in the diagram on the right. (D) Representative tetrad progeny of a cross between Mdm36OX (MET3p:MDM36) and kar9Δ. Yellow boxes indicate MET3p:MDM36 kar9Δ progeny as determined by marker analysis. (E) Percentage of HU-arrested spindles that crossed the bud neck during a 10-min movie in WT and Mdm36OX cells in the kar9Δ background. (F) Spindle penetration distance in HU-arrested cells of the indicated strains. The distance of spindle penetration (d) is defined as the farthest distance traveled by a spindle pole moving across the bud neck during a 10-min movie. n.s., not statistically significant.
We first assessed dynein pathway function using a simple spindle orientation assay, observing anaphase spindle position at a single timepoint in a population of asynchronous cells. Remarkably, the Mdm36OX strain exhibited only 5.2% of cells with a misoriented anaphase spindle phenotype, quantitatively similar to that observed for the WT strain (2.7%; Fig. 2C), indicating that dynein pathway function is not defective. In contrast, Mdm36OX cells expressing Num1LL, which harbors two point mutations that abolish the Num1–dynein interaction (Tang et al., 2012), exhibited a high level of misoriented anaphase spindle phenotype (37.4%; Fig. 2C), confirming that Num1–dynein interaction is required for proper spindle orientation in the Mdm36OX background.
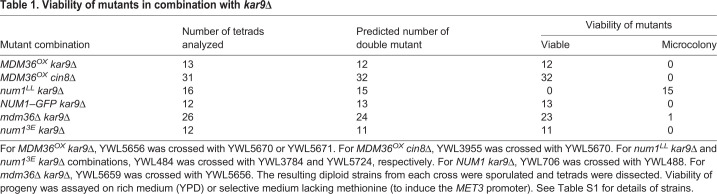
We further evaluated dynein function by assaying for synthetic growth defects with kar9Δ and cin8Δ. Budding yeast harboring KAR9 or CIN8 deletions require the dynein pathway for normal growth (Geiser et al., 1997; Gerson-Gurwitz et al., 2009; Miller and Rose, 1998). Tetrad dissection analysis showed that MDM36OX kar9Δ and MDM36OX cin8Δ progeny formed viable haploid colonies (Table 1), exhibiting no growth defects when compared with growth of kar9Δ and cin8Δ single mutants (Fig. 2D), consistent with the dynein pathway being functional in the Mdm36OX background.
Table 1.
Viability of mutants in combination with kar9Δ
We next quantitated dynein function using a spindle crossing assay, scoring for preanaphase spindle movements through the bud neck in a kar9Δ background. We found that Mdm36OX kar9Δ cells exhibited the same frequency of spindle traversing the bud neck when compared with kar9Δ cells (24.3% versus 24.2%; Fig. 2E). Moreover, in cells where the spindle was able to penetrate the bud neck, it moved for a similar distance compared with that in kar9Δ cells (Fig. 2F), consistent with an intact dynein pathway function. These results are surprising given the observed asymmetry in Num1 distribution in Mdm36OX cells.
Dynein and dynactin localization are altered in Mdm36OX cells
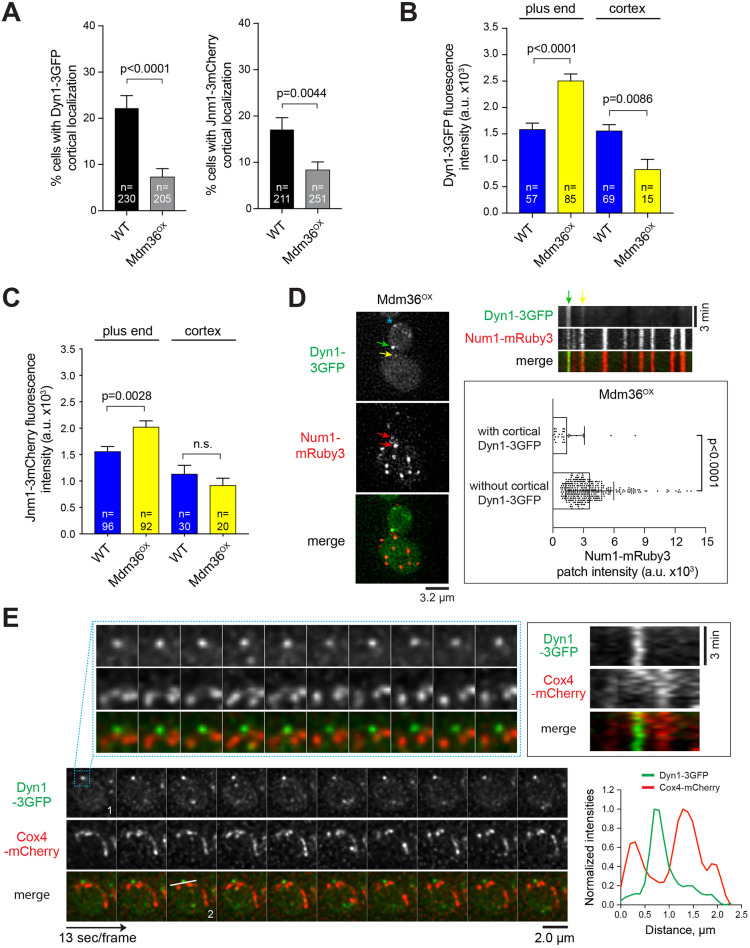
We asked whether the observed change in Num1 localization in Mdm36OX cells affects dynein and dynactin targeting to the astral MT plus ends and the cell cortex. Strikingly, compared to the frequency observed in WT cells, the frequency of observing cortical Dyn1–3GFP (dynein heavy chain tagged with three copies of GFP) and Jnm1–3mCherry (dynactin p50dynamitin subunit tagged with three copies of mCherry) foci was significantly reduced in the Mdm36OX background (Fig. 3A). The mean fluorescence intensity of Dyn1–3GFP and Jnm1–3mCherry foci at the MT plus ends was significantly enhanced in Mdm36OX cells relative to that in WT (Fig. 3B,C). These data indicate that the delivery of dynein and dynactin from the MT plus ends to the cell cortex via the offloading mechanism is reduced, even though Num1 clustering is enhanced in Mdm36OX cells. One possible explanation for this seemingly paradoxical result is that the sites for dynein offloading might be constituted by a distinct population of Num1 patches, the availability of which is affected when MDM36 is overexpressed. In agreement with the observed reduction in dynein offloading, the mean intensity of the cortical Dyn1–3GFP foci in Mdm36OX cells was significantly decreased (Fig. 3B). The mean intensity of the cortical Jnm1–3mCherry foci was also decreased; however, because the cytoplasmic background fluorescence for Jnm1–3mCherry was high, the observed change was not significantly different from the intensity of WT cells (Fig. 3C; P=0.368). As a control, Mdm36OX cells expressing Num1LL did not exhibit any cortical Dyn1–3GFP foci (0 out of 288 cells; Movie 2), confirming that Num1–dynein interaction is required for proper dynein anchorage in the Mdm36OX background. These results uncover compromised dynein targeting in Mdm36OX cells, albeit without causing a spindle misorientation phenotype.
Fig. 3.
Defects in dynein and dynactin localization in Mdm36OX cells. (A) Frequency of observing cortical Dyn1–3GFP (left) and Jnm1–3mCherry (right) patches in the indicated strains. Error bars represent the standard error of proportion (s.e.p.). P-value calculated using an unpaired t-test. (B,C) Dyn1–3GFP and Jnm1–3mCherry mean fluorescence intensity at the MT plus end and cell cortex. Error bars represent s.e.m. P-value calculated using an unpaired t-test. n.s., not statistically significant. (D) Left, deconvolved wide-field images of a representative Mdm36OX cell expressing Dyn1–3GFP and Num1–mRuby3. Green and yellow arrows mark stationary cortical Dyn1–3GFP foci (see kymograph, top right). Asterisk marks a motile plus-end Dyn1–3GFP focus (based on time-lapse sequence). Red arrows mark Num1–mRuby3 clusters possessing Dyn1–3GFP signal. Top right, kymograph of Dyn1–3GFP and Num1–mRuby3 foci from the same cell shown on the left. Bottom right, mean intensity of Num1–mRuby3 foci that possess or lack Dyn1–3GFP signal in Mdm36OX cells (n=338). Error bars represent s.d. P-value calculated using an unpaired t-test. (E) Deconvolved time-lapse images of Dyn1–3GFP and Cox4–mCherry in an Mdm36OX cell. A region of the cortex (indicated by the box in frame 1) is enlarged to illustrate the lack of colocalization between dynein and mitochondria. Top right, kymograph showing the behavior of dynein and mitochondria along the cortex of the enlarged region. Stationary dynein (green) and motile mitochondria (red) did not appear to be anchored to the same cortical spot. Bottom right, intensity profile plot (along the line drawn in frame 2) showing non-overlapping peaks of Dyn1–3GFP and Cox4–mCherry signals.
We next asked whether cortical dynein colocalizes with Num1 patches in the Mdm36OX background, as would be expected if Num1 anchors dynein to the cell cortex. We scored for cortical Dyn1 by analyzing time-lapse two-color images of Dyn1–3GFP and Num1–mRuby3. To be considered cortical, the Dyn1–3GFP foci had to remain stationary at the cell cortex for 3 min or longer. We observed that all cortical Dyn1–3GFP foci colocalized with a Num1–mRuby3 patch; however, to our surprise, the intensity of the Num1–mRuby3 patches that possessed Dyn1–3GFP was strikingly lower than those lacking Dyn1–3GFP (by ∼2.6-fold; Fig. 3D; n=338 patches). Colocalization of Dyn1–3GFP with a bright Num1–mRuby3 cluster was rarely observed, suggesting that Mdm36OX-mediated Num1 clustering negatively regulates cortical dynein targeting.
We next asked whether cortical Dyn1–3GFP colocalizes with mitochondria in Mdm36OX cells, as would be expected if dynein is anchored at sites of mitochondria-assembled Num1 clusters (Kraft and Lackner, 2017; Schmit et al., 2018). We acquired time-lapse images of Mdm36OX cells expressing Dyn1–3GFP and mitochondria-targeted Cox4–mCherry. As shown in Fig. 3E, we observed that stationary cortical Dyn1–3GFP foci rarely colocalized with mitochondria. Kymograph analysis showed that mitochondria exhibited lateral movement and/or diffusion at the cell cortex, but they did not appear to be tethered to the same cortical site containing Dyn1–3GFP. These observations suggest that cortical dynein and mitochondria might not occupy the same Num1 clusters in Mdm36OX cells.
Direct observation of MT sliding initiated by dim Num1 clusters
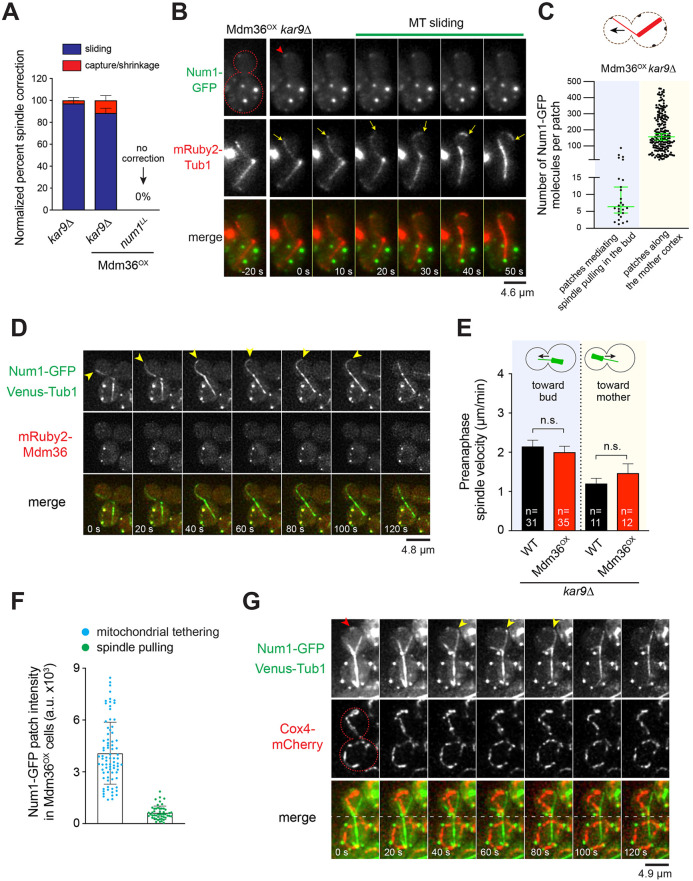
We next sought to determine how dynein mediates spindle positioning in Mdm36OX cells. In WT cells, cortical dynein pulls the spindle into the bud cell compartment via a lateral MT sliding mechanism (Adames and Cooper, 2000). More recently, however, several studies have shown that cortical dynein can also pull on the astral MTs via an end-on MT capture–shrinkage mechanism (Laan et al., 2012; Omer et al., 2018). To assess the mechanism in Mdm36OX cells, we monitored dynein-dependent astral MT interaction with the bud cortex using a spindle correction assay, scoring for MT behavior during anaphase spindle re-alignment from a misoriented position. In control kar9Δ cells, spindle correction was primarily mediated by MT sliding along the bud cortex (97.2%, n=36 events; Fig. 4A), as previously reported (Omer et al., 2018; Yeh et al., 2000). Remarkably, in kar9Δ Mdm36OX cells, the mechanism of spindle correction was unaffected, despite the apparent asymmetry in Num1 distribution between the mother and bud compartments (Fig. 4A; 46 out of 52 misaligned spindles were corrected by the MT sliding mechanism). Disrupting dynein anchoring in Mdm36OX cells using the num1LL allele abolished lateral MT sliding and prevented spindle correction (0 out of 50 spindles were corrected in num1LL Mdm36OX cells; Fig. 4A). These results indicate that dim Num1 patches in the bud of Mdm36OX cells are sufficient for dynein attachment and generation of spindle-pulling forces. As a control, we found that spindle correction was also mediated by the MT sliding mechanism in kar9Δ mdm36Δ cells (18 out of 22 events, 82%), suggesting that the observed phenotype in kar9Δ Mdm36OX cells was not due to an artificial effect of Mdm36 overexpression.
Fig. 4.
Dim Num1 patches mediate spindle pulling via MT sliding. (A) Quantification of spindle correction mechanisms for the indicated strains (22≤n≤52 events per strain). Error bars represent the standard error of proportion (s.e.p.). (B) Time-lapse sequence showing spindle correction via MT sliding in an Mdm36OX kar9Δ cell expressing Num1–GFP and mRuby2–Tub1. Red arrowhead marks initiation of MT sliding at a dim cortical Num1 cluster. Yellow arrows mark the position of the astral MT plus end. Red dashed line indicates the cell outline. (C) Copy number of Num1–GFP for individual foci found along the mother cell cortex versus those found at the bud cortex mediating spindle pulling during a spindle correction assay in Mdm36OX kar9Δ cells (n=201 and 26, respectively). Median±95% confidence interval is indicated. Diagram shows bud and mother Num1–GFP patches that were quantified in this assay. (D) Deconvolved two-color image sequence showing spindle correction via MT sliding in a NUM1–GFP VENUS–TUB1 kar9Δ cell overexpressing mRuby2-tagged Mdm36. Arrowheads mark the position of the astral MT plus end. (E) Mean preanaphase spindle velocity toward the bud or mother cell observed during a 7-min movie. Error bars represent s.e.m. n.s., not statistically significant by unpaired t-test. (F) Mean fluorescence intensity of Num1–GFP patches seen mediating spindle pulling or mitochondrial tethering in Mdm36OX cells. Patches mediating spindle pulling were observed in the kar9Δ background. Error bars represent s.d. (G) Deconvolved two-color image sequence showing a spindle pulling event in an Mdm36OX kar9Δ cell expressing Num1–GFP, Venus–Tub1 and Cox4–mCherry. Yellow arrowheads indicate spindle pulling by MT sliding (frames 40 s to 80 s) in a cortical region lacking mitochondria. Red arrowhead marks Num1 foci involved in mitochondrial tethering. Red dashed line indicates cell outline and the white dashed line indicates position of the bud neck.
To confirm this idea, we captured two-color time-lapse movies of kar9Δ Mdm36OX cells that also expressed Num1–GFP. In these movies, we observed that MT sliding occurred upon the plus end encountering a dim Num1–GFP focus at the bud cortex (Fig. 4B; Movie 3). The astral MT slid over the Num1–GFP focus, pulling the minus-end-attached spindle into the bud, causing spindle correction. We also sometimes observed MT sliding occurring over a cortical region without any apparently visible Num1–GFP focus (Movie 4), suggesting that the signals from the dynein-anchoring Num1 foci were at or below our detection limit. Co-imaging with Cse4–GFP cells followed by ratiometric comparison of fluorescence intensity revealed that the Num1 foci in the bud that mediated spindle correction contained a significantly smaller copy number of Num1 molecules compared to the Num1 patches found along the mother cell cortex. The median copy number for the Num1 foci mediating spindle correction was 6 (n=26), whereas the median copy number for the Num1 patches in the mother cell was 158 (n=201; Fig. 4C). Thus, Num1 does not need to assemble into a large focal patch or structure to enable dynein to generate cortical spindle-pulling forces. Additionally, further localization analysis revealed that, during spindle-pulling events in kar9Δ Mdm36OX cells, Mdm36 appeared to be absent at the site where MT sliding was initiated in the bud (Fig. 4D; 17 out of 24 events occurring without Mdm36). These results implicate the existence of a morphologically and functionally distinct population of Num1 foci that mediate dynein-based spindle-pulling activity along the bud cell cortex.
Num1 clustering enhances mitochondrial tethering but not dynein-mediated spindle-pulling function
To further examine whether patch enhancement has any effect on cortical dynein activity, we quantified spindle movements during spindle oscillation across the bud neck in hydroxyurea (HU)-arrested kar9Δ Mdm36OX cells. We considered the possibility that the bright Num1 patches in the mother compartment of kar9Δ Mdm36OX cells might enable a stronger spindle-pulling force by dynein, potentially retarding the spindle traversing the neck from the mother to the daughter or, conversely, enhancing its movement from the daughter to the mother. However, we found that spindle oscillation across the bud neck was unaffected in kar9Δ Mdm36OX cells compared with that in kar9Δ cells (Fig. 4E). Notably, the preanaphase spindle velocity for retrograde movements from the bud to the mother in kar9Δ Mdm36OX cells was indistinguishable from that in kar9Δ cells (Fig. 4E), indicating that the bright Num1 patches observed in the mother cortex of kar9Δ Mdm36OX cells did not correlate with an enhancement of cortical dynein activity.
To test whether patch enhancement correlates with mitochondrial-tethering activity, we imaged Mdm36OX cells expressing Num1–GFP and mitochondria-targeted Cox4–mCherry. We observed that Mdm36OX cells displayed a branched and tubular mitochondrial network that localized close to the cell periphery, as observed in WT cells (Movie 5; Fig. S3A); however, the mitochondrial network in Mdm36OX cells exhibited an enhanced tethering phenotype: 88.9% of Mdm36OX cells versus 55.0% of WT cells showed more than three persistent mitochondrial tether points over the course of a 3-min video (P<0.0003, n≥40 cells; Movie 6). Additionally, full 3D stacks showed that every bright Num1 patch at the cell cortex of Mdm36OX cells was associated with mitochondria (Movie 7). Using the same camera settings and imaging conditions to capture mitochondrial tethering events in MDM36OX NUM1–GFP COX4–mCherry cells and spindle correction events in MDM36OX NUM1–GFP mRuby2–TUB1 kar9Δ cells, we found a striking relationship between the intensity of the Num1 patch and the function of the patch in the cell. When the intensity of 130 Num1 patches was plotted against the activity that they performed in the cell, we observed a strong association between the bright Num1 patches and mitochondrial-tethering function, and between the dim Num1 patches and spindle-pulling function (Fig. 4F). On average, the Num1 patches mediating mitochondrial tethering were 6.4-fold brighter than the Num1 patches mediating spindle correction function. Furthermore, during spindle-pulling events in kar9Δ Mdm36OX cells, we observed that MT sliding was initiated at a cortical site devoid of mitochondria (Fig. 4G). These results, combined with the observation that patch enhancement in the mother compartment has no effect on the spindle velocity in either anterograde or retrograde directions (Fig. 4E), reveal a correlation between the size of the Num1 patch and mitochondrial-tethering activity but not dynein-based spindle-pulling activity.
3E mutations disrupt Num1 clustering but not cortical dynein activity
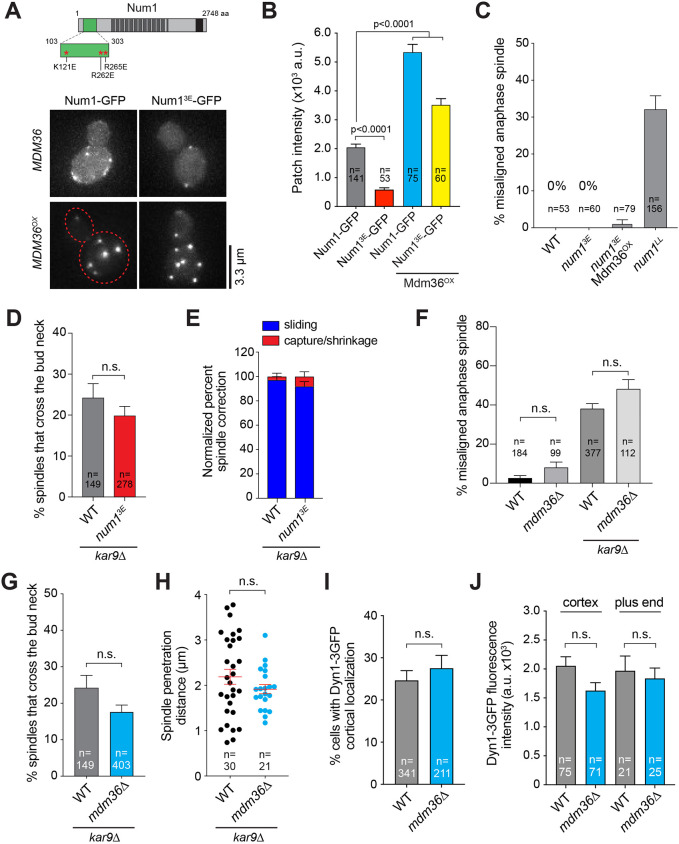
Notably, recent studies (Kraft and Lackner, 2017; Ping et al., 2016; Schmit et al., 2018) proposed that the assembly of functional Num1 patches in the bud is dependent on an interaction between Num1 and mitochondria. In light of our data suggesting differential functions of Num1 patches, we wondered whether the interaction with mitochondria is required for the formation of the two morphologically and functionally distinct types of patches observed in Mdm36OX cells. If mitochondria are indeed required for the formation of both types of patches, Mdm36OX cells harboring disrupted Num1–mitochondria interaction should be defective in mitochondrial-tethering and spindle-pulling functions. To test this, we used a well-characterized mitochondria-binding mutant, Num1K121E+R262E+R265E (hereafter referred to as Num13E; Fig. 5A), which harbors three point mutations that disrupt the Num1–mitochondria interaction but do not interfere with the Num1–Mdm36 interaction (Ping et al., 2016). The 3E mutations were originally identified in a mutagenesis screen for mutations in the CC domain of Num1 that diminish its affinity for liposomes mimicking the phospholipid composition of the mitochondrial outer membrane (Ping et al., 2016); however, how they affect Num1 cluster formation and dynein pathway function have not been characterized.
Fig. 5.
Num13E forms cortical patches that are functional in the dynein pathway. (A) Schematic diagram indicating the location of 3E mutations in Num1. Bottom, representative images of Num1–GFP and Num13E–GFP in WT (MDM36) and Mdm36OX backgrounds. Each image is a maximum intensity projection of five optical sections spaced 0.5 µm apart. Red dashed line indicates cell outline. (B) Mean intensity of Num1–GFP patches for the strains indicated in A. Error bars represent s.e.m. P-values calculated using unpaired t-tests. (C) Percentage of anaphase spindles with a misoriented phenotype for the indicated strains. Error bars represent the standard error of proportion (s.e.p.). (D) Percentage of HU-arrested spindles that crossed the bud neck during a 10-min movie. Error bars represent s.e.p. (E) Quantification of spindle correction mechanisms in the indicated strains (36≤n≤50 events per strain). Error bars represent s.e.p. (F) Percentage of misoriented anaphase spindles for the indicated strains. Error bars represent s.e.p. n.s., not statistically significant by one-way ANOVA test. (G) Frequency of observing HU-arrested spindles crossing the bud neck during a 10-min movie. Error bars represent s.e.p. (H) Distance of spindle penetration. Horizontal lines indicate the mean. Error bars represent the s.e.m. (I) Frequency of observing cortical Dyn1–3GFP patches. Error bars represent s.e.p. (J) Mean fluorescence intensity of Dyn1–3GFP at the MT plus end and cell cortex. Error bars represent s.e.m. For D and G–J: n.s., not statistically significant by unpaired t-test.
We introduced the 3E mutations into full-length GFP-tagged Num1 and expressed the fusion protein from its endogenous locus in the WT and Mdm36OX backgrounds. WT cells expressing Num13E–GFP displayed fewer (Fig. S3B) and dimmer cortical patches (Fig. 5A,B) compared with those expressing Num1–GFP. However, similar to WT Num1 patches, the intensity of Num13E–GFP patches was dramatically enhanced upon Mdm36 overexpression (Fig. 5A,B), consistent with the notion that the 3E mutations do not disrupt the interaction of Num1 with Mdm36 (Ping et al., 2016). Surprisingly, spindle orientation analysis revealed that dynein pathway function was unaffected in Num13E–GFP cells (0% versus 0% and 32.1% of misaligned anaphase spindle phenotype for num13E versus WT and the control strain num1LL, respectively; Fig. 5C) regardless of Mdm36 overexpression (compare num13E with num13E Mdm36OX; Fig. 5C), supporting the idea that small and dim Num13E foci are sufficient for dynein-based spindle-pulling activity. Moreover, tetrad dissection analysis showed that num13E kar9Δ progeny formed viable haploid colonies (Table 1), whereas num1LL kar9Δ progeny were inviable, indicating that small and dim Num13E foci are sufficient for rescuing the synthetic lethality with kar9Δ. Thus, our genetic and cell biological data argue that although Num1–mitochondria interaction is required for Num1 clustering (namely the formation of bright Num1 patches), the assembled patches are dispensable for dynein pathway function.
To interrogate dynein function in num13E cells more closely, we monitored preanaphase spindle movements through the bud neck using a spindle crossing assay (Fig. 5D) and assessed dynein-dependent astral MT interaction with the bud cortex using a spindle correction assay (Fig. 5E). Consistent with intact dynein pathway function, the frequency of observing the preanaphase spindle traversing the bud neck in num13E kar9Δ cells in the spindle crossing assay was indistinguishable from that in the control kar9Δ cells (19.8% versus 24.2% for num13E kar9Δ and kar9Δ, respectively, P=0.287; Fig. 5D). Additionally, as observed in kar9Δ cells, the re-alignment of anaphase spindles from a misoriented position in num13E kar9Δ cells was predominantly mediated by MT sliding along the bud cortex (92.0% versus 97.2% for num13E kar9Δ and kar9Δ, respectively, P=0.3080; Fig. 5E), further supporting the notion that Num1 does not need to assemble into a large focal patch to enable cortical force generation by dynein. Consistent with this idea, two-color time-lapse movies of spindle correction showed that MT sliding occurred upon interaction of the plus end with a dim Num13E–GFP focus at the bud cortex (Movie 8). Furthermore, co-imaging with Cse4–GFP cells revealed that the median protein copy number for Num13E–GFP foci mediating spindle correction in Mdm36OX kar9Δ cells was 5.6 (Fig. S3C), a number strikingly similar to that observed for WT Num1–GFP foci (Fig. 4C). These results show that a single mitochondria-independent Num1 patch containing a few Num1 molecules is sufficient for dynein-based spindle-pulling activity at the bud cortex.
We next investigated whether Num1's mitochondrial-tethering function is affected by the 3E mutations in Mdm36OX cells. The majority of num13E cells displayed a collapsed mitochondrial network (Fig. S3A) (Ping et al., 2016), in contrast to the reticulated and cortically tethered network observed in WT cells. Remarkably, the majority of num13E Mdm36OX cells (containing enhanced Num13E patches; see Fig. 5A) displayed a WT mitochondrial network (Fig. S3A). The percentage of num13E Mdm36OX cells showing a collapsed mitochondrial phenotype was significantly less than that exhibited by num13E cells (5.3% versus 78.0% for num13E Mdm36OX and num13E, respectively), indicating that mitochondrial-tethering function was restored by enhancing Num13E–GFP patches. This result indicates that Mdm36-mediated Num1 clustering is critical for the function of Num1 in mitochondrial tethering.
Dynein pathway function is intact in mdm36Δ cells
Given that Num1 patches with a small ensemble of Num1 molecules are sufficient for dynein-based spindle-pulling activity, we next wondered whether dynein pathway is functional in mdm36 deletion mutant cells, because these cells exhibited 13 copies of Num1 molecules per cortical patch (Fig. 1C). To test this, we performed dynein functional assays and examined dynein localization in cells lacking Mdm36.
We found that deletion of MDM36 did not result in a significant increase in the percentage of cells with misoriented spindles (Fig. 5F; 8.0% versus 2.7% for mdm36Δ and WT, respectively, P=0.695, 99≤n≤184). In the absence of Kar9, loss of Mdm36 did not further increase the number of misoriented spindles (Fig. 5F; 48.2% versus 38.2% for mdm36Δ kar9Δ and kar9Δ, respectively, P=0.2303, 112≤n≤377). We also found that, in a spindle crossing assay, the frequency of observing preanaphase spindles traversing the bud neck for an mdm36Δ kar9Δ strain was statistically indistinguishable from that of a kar9Δ strain (Fig. 5G; 18.0% versus 24.0% for mdm36Δ kar9Δ and kar9Δ, respectively, P=0.0838, 149≤n≤403). For spindles that were able to penetrate the bud neck, the distance traveled was also not significantly different between mdm36Δ kar9Δ and kar9Δ (Fig. 5H). Additionally, tetrad dissection analysis showed that mdm36Δ kar9Δ progeny formed viable haploid colonies (Table 1), exhibiting no significant growth defects when compared with kar9Δ single mutant. These results indicate that dynein pathway function is not defective in mdm36Δ cells.
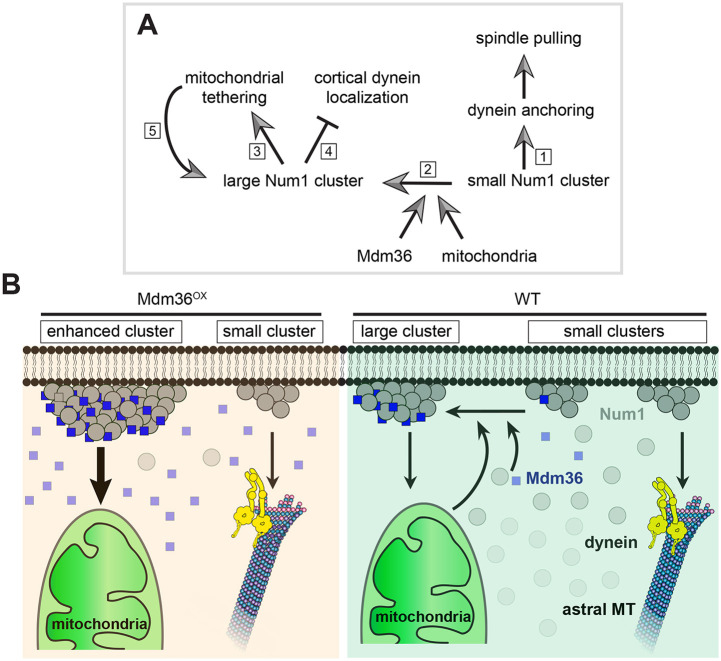
We also found that dynein localization was not affected by MDM36 deletion. The percentage of cells exhibiting cortical Dyn1–3GFP foci in the mdm36 deletion mutant was not significantly different from that in WT (Fig. 5I; 27.5% versus 24.6% for mdm36Δ and WT, respectively, P=0.453, 211≤n≤341). The mean fluorescence intensity of Dyn1–3GFP foci at the cell cortex and the MT plus ends in mdm36Δ cells was quantitatively similar to that in WT cells (Fig. 5J; P=0.149 for cortical foci, 71≤n≤75; P=0.982 for plus end foci, 21≤n≤25). These results show that Mdm36 is dispensable for dynein targeting, lending further support to our model that small and dim patches with a small ensemble of Num1 molecules are sufficient for cortical dynein anchorage and spindle positioning function (Fig. 6A).
Fig. 6.
Proposed function and regulation of Num1 clustering. (A) Differential functions of small and large Num1 clusters. Small Num1 clusters mediate dynein anchoring and cortical spindle-pulling function (1). Mdm36 and mitochondria cooperatively mediate formation of large Num1 clusters (2) for mitochondrial-tethering function (3). Enhancing the number of Num1 molecules in the patch reduces cortical dynein targeting (4), probably because Mdm36 and mitochondria both bind to the same CC domain of Num1 as dynein. As proposed previously (Kraft and Lackner, 2017), mitochondria in turn promotes large cluster formation and stabilization (5). (B) Model for Mdm36- and mitochondria-dependent enhancement of Num1 clustering. Diagram illustrates scenarios in WT and Mdm36OX cells. In both cases, two morphologically and functionally distinct populations of Num1 clusters (i.e. small and large clusters) mediate spindle pulling and mitochondrial-tethering functions at the cell cortex.
DISCUSSION
Here, we provide evidence demonstrating that Mdm36 levels determine the size of Num1 clusters at the cell cortex. We speculate that proper clustering of Num1 is mediated through cooperative actions of Mdm36 and mitochondria (Fig. 6A,B), because both mdm36Δ deletion (Fig. 1A; Fig. S3D) and 3E mutations (Fig. 5A; Fig. S3D) resulted in a Num1 clustering phenotype, and the phenotype of 3E mutations was rescued by Mdm36 overexpression (Fig. 5A,B; Fig. S3A,D). Importantly, the current view on Num1 assembly (Kraft and Lackner, 2017; Ping et al., 2016; Schmit et al., 2018) postulates that cortical clustering of Num1 serves to simultaneously anchor mitochondria and dynein to the plasma membrane. Our results, however, show that enhancing Num1 clustering only intensifies mitochondrial-tethering function of Num1, not dynein-based spindle-pulling function, which is inconsistent with a simultaneous interaction with dynein and mitochondria. This inconsistency is underscored by (1) the lack of strong colocalization between cortical Dyn1 and enhanced Num1 patches in Mdm36OX cells (Fig. 3D), (2) the fact that astral MT sliding was rarely observed to occur on a brightly enhanced Num1 patch, (3) the lack of spindle misorientation defects in num13E cells where Num1 clustering and mitochondria tethering function were disrupted (Fig. 5B,C; Fig. S3A) and (4) the lack of dynein targeting defects in mdm36Δ cells (Fig. 5I,J). Our results appear to be more consistent with a recent study by Chacko et al. (2019) in fission yeast, where they showed that dynein and mitochondria do not associate with the same Mcp5 foci at the meiotic cortex of S. pombe cells undergoing horsetail nuclear oscillation. Why and how dynein and mitochondria bind to different Num1 or Mcp5 foci will require further investigation. Interestingly, Chacko et al. (2019) showed that the presence of dynein on an Mcp5 cluster precluded the attachment of mitochondria to the same cluster, suggesting that steric hindrance might prevent dynein and mitochondria from simultaneously interacting with the same Num1 or Mcp5 spot.
One possible explanation for our results is that dynein anchorage and cortical force generation might be performed by a distinct population of dim Num1 clusters, an idea consistent with the lack of correlation between patch enhancement and hyperactivation of cortical dynein activity (Fig. 4E). The existence of this population of Num1 is supported by direct observation of individual encounters between a dynamic astral MT plus end and a dim Num1 cluster at the bud cortex (Fig. 4B) and with the ensuing MT sliding occurring without Mdm36 being present at the cluster, despite the fact that Mdm36 was overexpressed in the cell. We do not think that these observations are caused by artificial overexpression of Mdm36, because we also observed MT sliding occurring in mdm36Δ cells, where Num1 foci are similarly small in size. We speculate that the redistribution of Num1 molecules observed in Mdm36OX cells, as compared to the distribution in WT cells (Fig. 2A), makes it possible to follow astral MT interaction with individual cortical Num1 sites. Such encounters would have been otherwise impossible to observe in WT cells due to the abundance and heterogeneity of Num1 clusters along the cell cortex (see Omer et al., 2018; Heil-Chapdelaine et al., 2000; Kraft and Lackner, 2017; Schmit et al., 2018).
The budding yeast model is uniquely suited for in vivo assessment of spindle pulling under load. The movement of the spindle into the narrow aperture of the bud neck introduces a load burden that antagonizes dynein pulling at the bud cortex. How much force is needed to pull the spindle across the bud neck remains a total mystery. Our molecule-counting data (Fig. 4C; Fig. S3C) suggest that a small ensemble of approximately six Num1 molecules is sufficient for dynein anchorage and cortical force generation under load. Assuming Num1 binds dynein with a 1:1 stoichiometry, an ensemble of six Num1 molecules, which probably exist as three Num1 dimers (Ping et al., 2016; Tang et al., 2012), can potentially anchor up to three dynein dimers. Because each dynein dimer produces 4–7 pN of force (Belyy et al., 2016; Cho et al., 2008; Gennerich et al., 2007), an ensemble of Num1 anchoring three dynein dimers can generate a force of 12–21 pN, which we speculate likely corresponds to the minimum force needed to pull the spindle across the bud neck. By comparison, in Dictyostelium cells, 8–16 pN of dynein-generated force is needed for rapid transport of maturing phagosomes to lysosomes (Rai et al., 2016). In these cells, the phagosomes acquire increasingly greater minus-end-directed movements (as they mature) through clustering of multiple dynein motors (up to 16) into a microdomain on the phagosomal surface (Rai et al., 2016). Thus, unlike the evidence for Dictyostelium dynein in phagosomal transport, our data suggest that clustering of a large number of motors into a single site does not appear to be a key regulatory step for the function of yeast dynein in spindle positioning. In cultured human cells, although cortical clustering behavior has recently been demonstrated for the dynein-anchoring homologue NuMA (Okumura et al., 2018), the size of the NuMA clusters and the force required for spindle displacement remain to be determined. In future studies, it would be interesting to test whether small NuMA clusters can generate large spindle-pulling forces, as observed in yeast.
MATERIALS AND METHODS
Media and strain construction
Yeast media were obtained from Sunrise Science Products (San Diego, CA). All strains (Table S1) were generated in the YEF473A background (Bi and Pringle, 1996) by standard genetic crosses or PCR-product-mediated homologous recombination at the chromosomal locus (Longtine et al., 1998), unless otherwise noted. Yeast was transformed using the lithium acetate method (Knop et al., 1999). To overexpress Mdm36, we replaced 50 nucleotides of the MDM36 promoter immediately upstream of the start codon with a hygromycin resistance marker (HPH) and 562 bp of the MET3 promoter, using the plasmid pHPH::MET3p (Zhu et al., 2017) as PCR template. To generate Num13E (K121E, R262E and R265E) expressed from the endogenous chromosomal locus, we used the site-specific genomic mutagenesis approach (Gray et al., 2004). The CC domain (amino acids 25–290) was first replaced in a Num1–GFP strain with a URA3 marker from pRS306 (Sikorski and Hieter, 1989). Next, we generated a PCR fragment containing the mutated residues using an overlap extension PCR method. We included flanking sequences for homologous recombination targeting the corresponding chromosomal region that has been replaced by the URA3 marker. Transformants were plated on medium containing 5-fluoroorotic acid (5-FOA) to select for substitution of URA3 with the PCR fragment. We then verified all point mutations by DNA sequencing of the genomic locus. To overexpress mRuby2–Mdm36 (tagged at the N-terminus of Mdm36) using the MET3 promoter, a 1347-bp 5′ fragment of the MDM36 open reading frame was subcloned into a LEU2 plasmid (pBJ78; Sikorski and Hieter, 1989) containing MET3p:mRuby2. The resulting plasmid was linearized using BamHI (within the MDM36 open reading frame) and transformed into a haploid strain to integrate at the endogenous MDM36 locus. To label MTs, strains were transformed with Tub1-tagging plasmids for integration into the TUB1 locus, as previously described (Markus et al., 2015).
Image acquisition and analysis
Widefield fluorescence images were acquired at room temperature using a 1.49 NA 100× objective on a Nikon TiE inverted microscope equipped with a Nikon LUN4 laser unit (405 nm, 488 nm, 561 nm and 640 nm), an EMCCD camera (iXon 888; Andor) and a sCMOS camera (Zyla 4.2; Andor). We used a multi-pass quad filter cube set (C-TIRF; Chroma) for imaging GFP, Venus, mRuby2, mRuby3 and mCherry fluorescence. The microscope system was controlled using NIS-Elements imaging software (Nikon). Image stacks were deconvolved where indicated using 3D Deconvolution in NIS-Elements with the Automatic method. We performed standard photobleaching correction where indicated using Bleach Correction in Fiji (https://imagej.net/Fiji) with the Simple Ratio method. Cells were grown to mid-log phase in synthetic defined (SD) medium at 30°C and mounted on a 1.7% agarose pad containing SD or SD lacking methionine for imaging. To induce the MET3 promoter, overnight cultures grown at 30°C in SD medium were harvested, washed once with water, diluted into fresh SD medium lacking methionine, and grown for 5–6 h at 30°C before imaging. Confocal images were acquired using a 1.4 NA 60× oil immersion objective on an Andor W1 spinning-disk confocal microscope housed in the Life Sciences Center Imaging Facility at Dartmouth College.
To determine the copy number of Num1–GFP at individual cortical patches, we used the ratiometric comparison approach as described in Verdaasdonk et al. (2014). Strains expressing Num1–GFP were mixed with the Cse4–GFP strain and imaged together on the same slide. For experiments using the Mif2–GFP strain, images of the standard and experiments were taken consecutively under the same conditions on the same microscope system on the same day. We used Fiji to draw a square encompassing 4×4 or 3×3 pixels to quantify the integrated intensity of individual foci of Cse4–GFP, Mif2–GFP and Num1–GFP clusters. To subtract the background intensity from each measurement, we moved the square from the cluster to a nearby cytoplasmic area within the same cell. The mean integrated intensity of Cse4–GFP and Mif2–GFP clusters was assigned a value of 96 and 58 molecules (Lawrimore et al., 2011; Verdaasdonk et al., 2014), respectively, and was used to calculate the copy number of Num1–GFP.
To verify the protein copy number obtained using the ratiometric comparison approach, we performed bleaching step analysis to estimate the step size correlating to bleaching a single GFP fluorophore using the STEPFINDER software (Kerssemakers et al., 2006). Briefly, WT cells expressing Num1–GFP were imaged continuously in a single plane with a low laser power (10% of a 15 mW 488 nm laser) and with a 250 ms exposure for each frame. The intensities of individual Num1–GFP patches were quantified over the course of the fluorescence decay (using a circle encompassing 10×10 pixels in Fiji), subjected to a five-frame rolling average to reduce background noise and then analyzed by STEPFINDER to detect bleaching steps and to determine associated step sizes, as described previously (Kerssemakers et al., 2006; Thankachan et al., 2017). The starting intensity of the patch was divided by the average step size to obtain the number of molecules.
To measure the number of bright cortical Num1 patches per cell, we performed particle analysis using the Analyze Particles tool in Fiji with the minimum particle size set at 9 pixel units and the threshold set at the background intensity of the cytoplasm. To measure the total fluorescence intensity of Dyn1–3GFP and Jnm1–3mCherry at the plus end and cell cortex, we used the circle selection tool in Fiji to encompass the 3GFP or 3mCherry spot and measured the intensity from the maximum intensity projection images. Background intensity from a nearby cytoplasmic area was subtract from each measurement.
To measure spindle velocity, we used the mTrack plugin in Fiji to track the position of the spindle pole body (SPB) (determined from the mRuby2–Tub1 image) during a 7-min movie in cells arrested with hydroxyurea (HU). We grew the cells to mid-log phase and arrested them by treatment with 200 mM HU for 1–1.5 h before imaging. For the spindle penetration assay, we measured the farthest distance traveled by a spindle pole moving across the bud neck during a 10-min movie.
To plot the intensity profile of Num1–GFP along the cell cortex, we used the segmented line tool in Fiji to trace the bud and mother cell perimeter. Next, we used the Plot Profile tool to graph the intensity along the segmented line.
For spindle correction assay, we scored for misaligned anaphase spindles that moved into the bud neck and became aligned along the mother-bud axis during a 10-min movie in kar9Δ background. Spindle correction was scored as mediated by ‘sliding mechanism’ or ‘capture–shrinkage mechanism’ based on the interaction displayed by the astral MT with the bud cortex while the spindle moved into the bud neck during its re-alignment, as previously described (Omer et al., 2018).
Sedimentation analysis, cell lysis and western blotting
Sedimentation analysis on sucrose gradients was performed as previously described (Omer et al., 2018). For analysis of Num1–GFP levels, overnight yeast cultures grown at 30°C in 15 ml SD medium lacking methionine were harvested and resuspended in ice-cold lysis buffer containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA and 1.5% Triton X-100 supplemented with a protease inhibitor tablet (Millipore Sigma). Equal amounts of cells were lysed by bead beating six times for 30 s, with 2 min on ice between each beating. Next, we centrifuged the crude lysate at 500 g for 5 min at 4°C in a microfuge (Eppendorf, model 5424R) to remove cell debris. The resulting supernatants were separated on 4–15% SDS–PAGE gels (Bio-Rad) and then electroblotted onto nitrocellulose membrane using a Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were probed with rabbit anti-GFP antibody (Cat# PABG1-20, RRID: AB_2749857; Chromotek) at 1:1000, rabbit anti-Sac1 antibody (a gift from Dr Charles Barlowe, Dartmouth College, New Hampshire, USA) at 1:2000, mouse anti-Por1 antibody (Cat# 459500, RRID: AB_2532239; Thermo Fisher Scientific) at 1:1000, HRP-conjugated goat anti-rabbit IgG antibody (Jackson ImmunoResearch) at 1:10,000 dilution, and HRP-conjugated goat anti-mouse IgG antibody (BioLegend) at 1:10,000 dilution. Chemiluminescence signals were acquired using a ChemiDoc Imaging System (Bio-Rad). Immunoblots were exposed for durations ranging from 1 s to 10 min without saturating the camera‘s pixels.
Statistical methods
All statistical significance in this study was determined by unpaired two-tailed Student's t-test or one-way ANOVA test performed using the GraphPad Prism software.
Supplementary Material
Acknowledgements
We thank Dr Jacob Kerssemakers and Dr Marileen Dogterom for the STEPFINDER software, and Samuel Greenberg and Weimin Tan for valuable help with making yeast strains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.O., W.-L.L.; Methodology: S.O., W.-L.L.; Validation: S.O., W.-L.L.; Formal analysis: S.O., W.-L.L.; Investigation: S.O., K.B., J.B., W.-L.L.; Data curation: S.O., W.-L.L.; Writing - original draft: W.-L.L.; Writing - review & editing: W.-L.L.; Supervision: W.-L.L.; Funding acquisition: W.-L.L.
Funding
This work was supported by a National Institutes of Health/National Institute of General Medical Sciences grant (GM076094) to W.L.L and in part by a Scholarly Innovation and Advancement Award to W.L. Lee at Dartmouth College. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.246363.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.246363.reviewer-comments.pdf
References
- Adames N. R., and Cooper J. A., (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149: 863-874. 10.1083/jcb.149.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyy V., Schlager M. A., Foster H., Reimer A. E., Carter A. P., and Yildiz A., (2016). The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol. 18: 1018-1024. 10.1038/ncb3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., and Pringle J. R., (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5264-5275. 10.1128/MCB.16.10.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Neumuller R. A., Novatchkova M., Du Q., and Knoblich J. A., (2006). The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10: 731-742. 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Chacko L. A., Mehta K., and Ananthanarayanan V., (2019). Cortical tethering of mitochondria by the anchor protein Mcp5 enables uniparental inheritance. J. Cell Biol. 218: 3560-3571. 10.1083/jcb.201901108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J. T., Wong A. K., Tavassoli S., Young B. P., Chruscicki A., Fang N. N., Howe L. J., Mayor T., Foster L. J., and Loewen C. J., (2014). Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158: 620-632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Cho C., Reck-Peterson S. L., and Vale R. D., (2008). Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J. Biol. Chem. 283: 25839-25845. 10.1074/jbc.M802951200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V. C., and Wu J. Q., (2012). Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37: 499-506. 10.1016/j.tibs.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V. C., Wu P., Parthun M. R., and Wu J. Q., (2011). CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195: 563-572. 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couwenbergs C., Labbé J.-C., Goulding M., Marty T., Bowerman B., and Gotta M., (2007). Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179: 15-22. 10.1083/jcb.200707085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., and Macara I. G., (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119: 503-516. 10.1016/j.cell.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Geiser J. R., Schott E. J., Kingsbury T. J., Cole N. B., Totis L. J., Bhattacharyya G., He L., and Hoyt M. A., (1997). Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell 8: 1035-1050. 10.1091/mbc.8.6.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A., Carter A. P., Reck-Peterson S. L., and Vale R. D., (2007). Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131: 952-965. 10.1016/j.cell.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Movshovich N., Avunie R., Fridman V., Moyal K., Katz B., Hoyt M. A., and Gheber L., (2009). Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell. Mol. Life Sci. 66: 301-313. 10.1007/s00018-008-8479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Kupiec M., and Honigberg S. M., (2004). Site-specific genomic (SSG) and random domain-localized (RDL) mutagenesis in yeast. BMC Biotechnol. 4: 7 10.1186/1472-6750-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S. R., Tan W., and Lee W.-L., (2018). Num1 versus NuMA: insights from two functionally homologous proteins. Biophys. Rev. 10: 1631-1636. 10.1007/s12551-018-0472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammermeister M., Schodel K., and Westermann B., (2010). Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2443-2452. 10.1091/mbc.e10-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Wang J., Gueth-Hallonet C., Weber K., and Osborn M., (1999). Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18: 1689-1700. 10.1093/emboj/18.6.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine R. A., Oberle J. R., and Cooper J. A., (2000). The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151: 1337-1344. 10.1083/jcb.151.6.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., and Dogterom M., (2006). Assembly dynamics of microtubules at molecular resolution. Nature 442: 709-712. 10.1038/nature04928 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T. (2019). The cortical force-generating machinery: how cortical spindle-pulling forces are generated. Curr. Opin. Cell Biol. 60: 1-8. 10.1016/j.ceb.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., and Schiebel E., (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963-972. [DOI] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gonczy P., (2012). Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199: 97-110. 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gönczy P., (2014). NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 33: 1815-1830. 10.15252/embj.201488147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft L. M., and Lackner L. L., (2017). Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J. Cell Biol. 216: 3061-3071. 10.1083/jcb.201702022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft L. M., and Lackner L. L., (2019). A conserved mechanism for mitochondria-dependent dynein anchoring. Mol. Biol. Cell 30: 691-702. 10.1091/mbc.E18-07-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., Lopez M. P., Vale R. D., Julicher F., Reck-Peterson S. L., and Dogterom M., (2012). Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148: 502-514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L. L., Ping H., Graef M., Murley A., and Nunnari J., (2013). Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl Acad. Sci. USA 110: E458-E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K. S., and Salmon E. D., (2011). Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195: 573-582. 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. L., Oberle J. R., and Cooper J. A., (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160: 355-364. 10.1083/jcb.200209022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.-L., Kaiser M. A., and Cooper J. A., (2005). The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168: 201-207. 10.1083/jcb.200407036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R., (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- Markus S. M., Omer S., Baranowski K., and Lee W. L., (2015). Improved Plasmids for Fluorescent Protein Tagging of Microtubules in Saccharomyces cerevisiae. Traffic 16: 773-786. 10.1111/tra.12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., and Rose M. D., (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140: 377-390. 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K., and Gönczy P., (2007). Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9: 1294-1302. 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- Okumura M., Natsume T., Kanemaki M. T., and Kiyomitsu T., (2018). Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife 7, e36745. 10.7554/eLife.36559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer S., Greenberg S. R., and Lee W. L., (2018). Cortical dynein pulling mechanism is regulated by differentially targeted attachment molecule Num1. eLife 7, e36745. 10.7554/eLife.36745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping H. A., Kraft L. M., Chen W., Nilles A. E., and Lackner L. L., (2016). Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J. Cell Biol. 213: 513-524. 10.1083/jcb.201511021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A., Pathak D., Thakur S., Singh S., Dubey A. K., and Mallik R., (2016). Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell 164: 722-734. 10.1016/j.cell.2015.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Okuzaki D., and Nojima H., (2006). Mcp5, a meiotic cell cortex protein, is required for nuclear movement mediated by dynein and microtubules in fission yeast. J. Cell Biol. 173: 27-33. 10.1083/jcb.200512129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit H. L., Kraft L. M., Lee-Smith C. F., and Lackner L. L., (2018). The role of mitochondria in anchoring dynein to the cell cortex extends beyond clustering the anchor protein. Cell Cycle. 17: 1345-1357. 10.1080/15384101.2018.1480226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M. A., and Pellman D., (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13: 364-372. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S. and Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Punch J. J., and Lee W.-L., (2009). A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 8: 3182-3190. 10.4161/cc.8.19.9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Germain B. S., and Lee W.-L., (2012). A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J. Cell Biol. 196: 743-756. 10.1083/jcb.201112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thankachan J. M., Nuthalapati S. S., Addanki Tirumala N., and Ananthanarayanan V., (2017). Fission yeast myosin I facilitates PI(4,5)P2-mediated anchoring of cytoplasmic dynein to the cortex. Proc. Natl Acad. Sci. USA 114: E2672-E2681. 10.1073/pnas.1615883114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J. S., Lawrimore J., and Bloom K., (2014). Determining absolute protein numbers by quantitative fluorescence microscopy. Methods Cell Biol. 123: 347-365. 10.1016/B978-0-12-420138-5.00019-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., and Yamamoto M., (2006). Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase. Genetics 173: 1187-1196. 10.1534/genetics.105.050062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E. D., Lew D. J., and Bloom K., (2000). Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell 11: 3949-3961. 10.1091/mbc.11.11.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., An X., Tomaszewski A., Hepler P. K., Lee W. L. (2017). Microtubule cross-linking activity of She1 ensures spindle stability for spindle positioning. J. Cell Bio. 122, 2759-2775 10.1083/jcb.201701094 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.