Genetic tools are needed to understand and manipulate the biology of archaea, which serve critical roles in the biosphere. Methanogenic archaea (methanogens) are essential for the biological production of methane, an intermediate in the global carbon cycle, an important greenhouse gas, and a biofuel. The CRISPRi-dCas9 system in the model methanogen Methanosarcina acetivorans is, to our knowledge, the first Cas9-based CRISPR interference system in archaea. Results demonstrate that the system is remarkably efficient in targeted gene repression and provide new insight into nitrogen fixation by methanogens, the only archaea with nitrogenase. Overall, the CRISPRi-dCas9 system provides a simple, yet powerful, genetic tool to control the expression of target genes and operons in methanogens.
KEYWORDS: archaea, CRISPR interference, Methanosarcina, dCas9, gene regulation, methanogens
ABSTRACT
CRISPR-based systems are emerging as the premier method to manipulate many cellular processes. In this study, a simple and efficient CRISPR interference (CRISPRi) system for targeted gene repression in archaea was developed. The Methanosarcina acetivorans CRISPR-Cas9 system was repurposed by replacing Cas9 with the catalytically dead Cas9 (dCas9) to generate a CRISPRi-dCas9 system for targeted gene repression. To test the utility of the system, genes involved in nitrogen (N2) fixation were targeted for dCas9-mediated repression. First, the nif operon (nifHI1I2DKEN) that encodes molybdenum nitrogenase was targeted by separate guide RNAs (gRNAs), one targeting the promoter and the other targeting nifD. Remarkably, growth of M. acetivorans with N2 was abolished by dCas9-mediated repression of the nif operon with each gRNA. The abundance of nif transcripts was >90% reduced in both strains expressing the gRNAs, and NifD was not detected in cell lysate. Next, we targeted NifB, which is required for nitrogenase cofactor biogenesis. Expression of a gRNA targeting the coding sequence of NifB decreased nifB transcript abundance >85% and impaired but did not abolish growth of M. acetivorans with N2. Finally, to ascertain the ability to study gene regulation using CRISPRi-dCas9, nrpR1, encoding a subunit of the repressor of the nif operon, was targeted. The nrpR1 repression strain grew normally with N2 but had increased nif operon transcript abundance, consistent with NrpR1 acting as a repressor. These results highlight the utility of the system, whereby a single gRNA when expressed with dCas9 can block transcription of targeted genes and operons in M. acetivorans.
IMPORTANCE Genetic tools are needed to understand and manipulate the biology of archaea, which serve critical roles in the biosphere. Methanogenic archaea (methanogens) are essential for the biological production of methane, an intermediate in the global carbon cycle, an important greenhouse gas, and a biofuel. The CRISPRi-dCas9 system in the model methanogen Methanosarcina acetivorans is, to our knowledge, the first Cas9-based CRISPR interference system in archaea. Results demonstrate that the system is remarkably efficient in targeted gene repression and provide new insight into nitrogen fixation by methanogens, the only archaea with nitrogenase. Overall, the CRISPRi-dCas9 system provides a simple, yet powerful, genetic tool to control the expression of target genes and operons in methanogens.
INTRODUCTION
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) system is a primitive adaptive immune system found in prokaryotes (bacteria and archaea) (1, 2). Approximately 40% of bacteria and 90% of archaea contain a CRISPR-Cas system (3). The system is used to combat foreign nucleic acid (e.g., virus) by using previously acquired nucleic acid to generate a guide RNA (gRNA) that targets the foreign nucleic acid for cleavage by a Cas endonuclease (e.g., Cas9). Several types of CRISPR systems have been discovered, and detailed characterization has led to the development of numerous genetic tools (4–6). The type II CRISPR-Cas9 system has garnered the most applications, in part due to DNA targeting by Cas9 being extremely precise with virtually no off-target cleavage (7). For example, systems have been designed to (i) edit genomes by using gRNA-directed Cas9-mediated DNA double-stranded breaks to allow gene deletion and additions using homology-dependent repair (8, 9) and (ii) control gene expression using a nuclease-deficient or dead Cas9 (dCas9) protein that targets specific DNA but does not cut it (10). DNA binding by dCas9 blocks transcription initiation and/or elongation of gRNA-targeted genes. The latter has been termed CRISPR interference (CRISPRi) (11). Gene repression by CRISPRi-dCas9 systems is a powerful gene regulatory tool whereby the expression of multiple gRNAs can allow the repression of several genes at the same time (10). CRISPRi-dCas9 systems have been developed to control gene expression in bacteria (e.g., Escherichia coli) and eukaryotes (e.g., yeast and mammalian cells) but not archaea (11). Archaea play central roles in the cycling of key elements (e.g., carbon, nitrogen, and iron) on Earth, and they serve as important sources for the development of biotechnology, including the production of biofuels and enzymes (12, 13). CRISPRi systems based on endogenous type I and type III CRISPR systems have been developed for the extremophilic archaea Haloferax volcanii and Sulfolobus spp., respectively. The Sulfolobus CRISPRi system is a posttranscriptional gene expression control system that utilizes a CRISPR type III complex to target mRNA for degradation (14–17). The Haloferax CRISPRi system utilizes mutant strains deleted of part or all of the endogenous CRISPR loci and expression of gRNAs that direct the cascade complex to block transcription of a targeted gene or operon (18, 19).
The strictly anaerobic methane-producing archaea (methanogens) are particularly important archaea. Methanogen-produced methane is critical to the global carbon cycle, is a greenhouse gas, and is a biofuel (13). Methanogens are also the only archaea that are capable of dinitrogen (N2) fixation (diazotrophy) (20–22). Methanogens were likely the first diazotrophs and the source of nitrogenase, the unique enzyme that catalyzes the energy-intensive reduction of N2 into usable ammonia (NH3) (23). Advanced genetic tools are necessary to understand the fundamental biology of methanogens and to provide platforms for genetic engineering to facilitate their use in biotechnology. Recently, a CRISPR-Cas9 system was developed to edit the genome of the model methanogen Methanosarcina acetivorans (24). Genome editing by the CRISPR-Cas9 system is driven by native homology-dependent repair (HDR) or by coexpression of the nonhomologous end-joining repair (NHEJ) pathway. Cas9-mediated genome editing has increased the efficiency and speed of genetic analyses of M. acetivorans. However, mutant generation typically requires 4 to 5 weeks (including plasmid construction), and generating strains with multiple mutations can require sequential strain construction (24).
We sought to repurpose the M. acetivorans CRISPR-Cas9 system by replacing Cas9 with dCas9 to generate a system to control gene expression (i.e., CRISPRi-dCas9). To test the CRISPRi-dCas9 system, we selected target genes that are not essential but, when repressed, would give an observable phenotype. M. acetivorans contains the nif operon that encodes the molybdenum (Mo) nitrogenase, as well as operons encoding the alternative vanadium (V) and iron (Fe) nitrogenases (25). Due to substantial ATP consumption required to fix N2, nitrogenase production is highly regulated and occurs only in the absence of a usable nitrogen source (e.g., NH3) (26). All diazotrophs contain Mo nitrogenase, which uses the least amount of ATP (20). The alternative nitrogenases are typically only used when molybdenum is not available (27). Production of functional Mo nitrogenase also requires unique cofactor biogenesis machinery (28). Although M. acetivorans Mo nitrogenase production and nitrogen fixation have not been investigated in detail, that enzyme provides a suitable target for the CRISPRi-dCas9 system since it is only required for growth in the absence of a fixed nitrogen source. Genes encoding the structural, regulatory, and cofactor biogenesis components of Mo nitrogenase were targeted for dCas9-mediated repression. The results demonstrate that the CRISPRi-dCas9 system is remarkably efficient in target gene repression. The M. acetivorans CRISPRi-dCas9 system expands the archaeal genetic toolbox and will specifically expedite the investigation and genetic engineering of methanogens.
RESULTS
Design of a CRISPRi-dCas9 system in M. acetivorans.
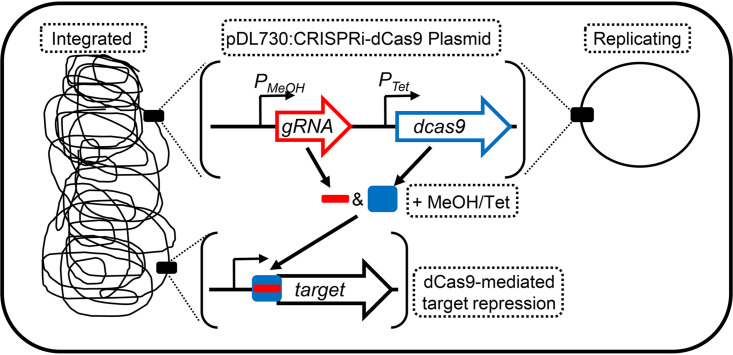
The CRISPR-Cas9 system developed by Nayak and Metcalf (24) was used as the foundation for the development of an M. acetivorans CRISPRi-dCas9 system. In the CRISPR-Cas9 system, one or more gRNAs are expressed from a methanol-inducible promoter (PMeOH) and Cas9 is expressed from a tetracycline-inducible promoter (PTet). Specialized strains (e.g., WWM73) of M. acetivorans are transformed with a plasmid (e.g., pDN203) containing the gRNA(s) and Cas9. Plasmid pDN203 and derivatives can integrate into the chromosome of transformed cells by site-specific recombination or, alternatively, can be converted into a replicating plasmid by cointegration with pAMG40 (24). To convert the CRISPR-Cas9 system into a CRISPRi-dCas9 system, cas9 in pDN203 was replaced with dcas9 from pMJ841 to generate pDL730 (Table 1). Plasmid pDL730 lacks a gRNA and therefore serves as the universal plasmid to incorporate gRNAs. M. acetivorans containing pDL730 was the control strain used in all experiments.
TABLE 1.
Plasmids used in this study
| Plasmid | Features | Reference |
|---|---|---|
| pMJ841 | pET-based expression vector containing Spy dcas9 | 48 |
| pDN203 | pJK027A-based plasmid with the native Spy cas9 | 24 |
| pAMG40 | Cointegrates with pJK027A-based plasmids for replication in M. acetivorans | 46 |
| pDL730 | pDN203 with cas9 replaced with dcas9 from pMJ841; gRNA free | This study |
| pDL731 | pDL730 with gRNA-nifD | This study |
| pDL732 | pDL730 with gRNA-Pnif | This study |
| pDL733 | Cointegrate of pDL732 and pAMG40 | This study |
| pDL737 | Cointegrate of pDL730 and pAMG40 | This study |
| pDL739 | pDL730 with gRNA-nrpR1 | This study |
| pDL745 | pDL730 with gRNA-nifB | This study |
CRISPRi-dCas9 strains of M. acetivorans were generated by integrating pDL730 and derivatives into the chromosome of parent strain WWM73. Alternatively, pDL730 could be converted into a replicating plasmid. In all CRISPRi-dCas9 strains, the repression of a target gene was induced by growth with methanol and the addition of tetracycline (Fig. 1). To test the ability of the system to silence or “knock down” transcription of gRNA-targeted genes, we selected the nif operon and related genes for dCas9-mediated repression. These genes should only be expressed when M. acetivorans is not given a usable nitrogen source (e.g., NH4Cl). However, expression of the nif operon in M. acetivorans has not been investigated. Therefore, diazotrophic growth and expression of the nif operon in response to available nitrogen was first determined.
FIG 1.
Design of a CRISPRi-dCas9 system for targeted gene repression in M. acetivorans. The CRISPRi-dCas9 plasmid pDL730 and derivatives can either integrate into the chromosome of M. acetivorans or be converted into a replicating plasmid. Expression of the gRNA and dCas9 is induced by growth with methanol (MeOH) and the addition of tetracycline (Tet).
M. acetivorans expresses Mo nitrogenase (NifHDK) and fixes N2 when grown in the absence of NH4Cl.
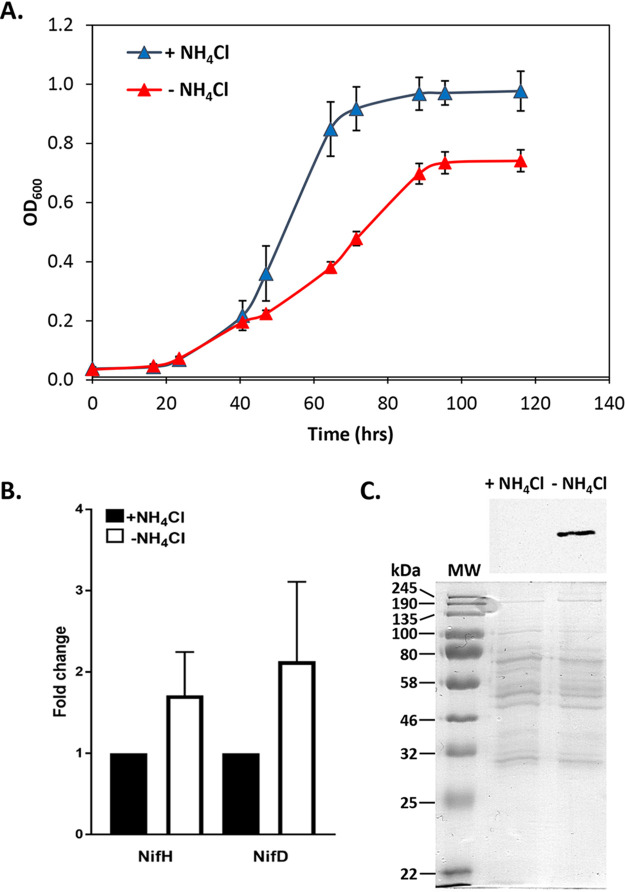
To provide a baseline for CRISPRi-dCas9 experiments, the expression of Mo nitrogenase and diazotrophic growth of M. acetivorans strain WWM73 were examined. Levels of growth of strain WWM73 in sealed tubes with a headspace containing 75% N2 were compared in standard high-salt (HS) medium with or without NH4Cl, the preferred nitrogen source (Fig. 2A). The cell yields and growth rates of cultures lacking NH4Cl were lower than those of cultures containing NH4Cl, consistent with increased energy consumption needed to fix N2. HS medium also contains cysteine, a possible fixed nitrogen source. Recent evidence indicated M. acetivorans can use cysteine as a sole nitrogen source (29). However, extensive experimentation with strain WWM73 and derivatives clearly demonstrates the inability of M. acetivorans to use cysteine as a nitrogen source (see below). Also, M. acetivorans is incapable of growing in cysteine-containing HS medium in tubes with a headspace devoid of N2 (data not shown). Moreover, real-time quantitative PCR (qPCR) analyses revealed increased transcript abundance of both nifH and nifD in cells grown in medium lacking NH4Cl and containing cysteine (Fig. 2B). Finally, NifD was only detected by Western blotting in lysate from cells grown in the absence of NH4Cl (Fig. 2C). These results demonstrate that expression of the nif operon is tightly controlled and that production of Mo nitrogenase occurs only in the absence of a usable nitrogen source (e.g., NH4Cl).
FIG 2.
Effect of NH4Cl on M. acetivorans growth and nif operon expression. (A) Growth in HS medium with or without NH4Cl with a 75% N2 headspace. (B) Fold change in nifH and nifD transcript abundance in cells grown in medium with or without NH4Cl as determined by qPCR. (C, top) Western blot analysis using NifD-specific antibodies on lysates from M. acetivorans cells grown with or without NH4Cl. (C, bottom) SDS-PAGE of lysates used for Western blot. MW, molecular weight standard.
Diazotrophic growth of M. acetivorans is abolished by dCas9-mediated repression of the nif operon.
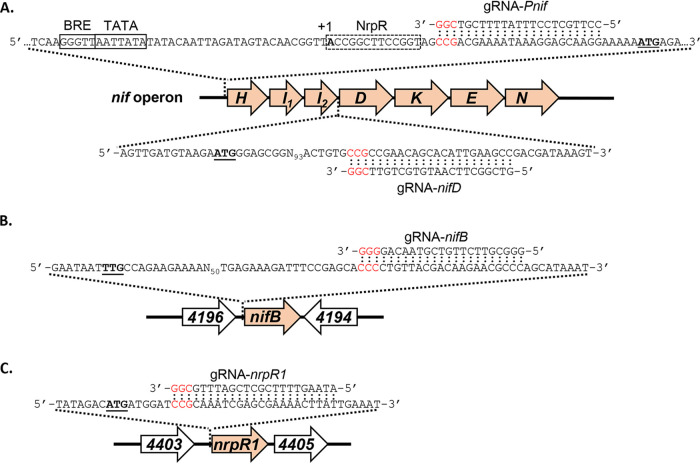
To test the ability of the CRISPRi-dCas9 system to repress expression of Mo nitrogenase, gRNAs were designed to target two separate regions of the nif operon (Table 2 and Fig. 3A). NifH, -D, and -K are the structural components of Mo nitrogenase, NifI1 and -I2 regulate nitrogenase activity, and NifE and -N serve as the M-cluster assembly scaffold (22). NifHDK and NifEN are essential for N2 fixation by Mo nitrogenase (23, 28). A 20-nt gRNA (gRNA-Pnif) was designed to target the sense (nontemplate) strand starting 15 nucleotides downstream of the transcription start site of the nif operon. A second gRNA (gRNA-nifD) targeted the sense strand starting 100 nucleotides downstream of the start codon of nifD. Each gRNA was separately added to pDL730 to generate pDL731 and pDL732 (Table 1). M. acetivorans strain WWM73 was separately transformed with pDL731 or pDL732 to generate strains DJL74 and DJL76, respectively. Control strain DJL72 contained gRNA-free pDL730 (Table 3). To test the effect of each gRNA on diazotrophic growth of M. acetivorans, each strain was grown with methanol in HS medium with or without NH4Cl. Since dCas9 was expressed from a tetracycline-inducible promoter, the effect of the presence and absence of tetracycline on growth was also examined.
TABLE 2.
gRNAs used in this study
| gRNA | Sequencea | Target |
|---|---|---|
| gRNA-Pnif | CCTTGCTCCTTTATTTTCGTCGG | nif operon promoter |
| gRNA-nifD | GTCGGCTTCAATGTGCTGTTCGG | nifD (MA3898) |
| gRNA-nrpR1 | ATAAGTTTTCGCTCGATTTGCGG | nrpRI (MA4404) |
| gRNA-nifB | GGGCGTTCTTGTCGTAACAGGGG | nifB (MA4195) |
PAM sequence is underlined.
FIG 3.
Designed gRNAs for targeted dCas9-mediated gene repression. (A) gRNAs targeting the promoter (gRNA-Pnif) and 5′ end of nifD (gRNA-nifD) of the nif operon. The BRE, TATA, and NrpR operator sequences are boxed. (B) gRNA targeting the 5′ end of nifB. (C) gRNA targeting the 5′ end of nrpR1. Start codons are in bold and underlined. PAM sequences are in red.
TABLE 3.
M. acetivorans strains used in this study
| Strain | Genotype | gRNA | Reference |
|---|---|---|---|
| WWM73 | Δhpt::PmcrB-tetR-ϕC31-int-attP | None | 46 |
| DJL72 | WWM73 att::pDL730 | None | This study |
| DJL74 | WWM73 att::pDL732 | gRNA-Pnif | This study |
| DJL76 | WWM73 att::pDL731 | gRNA-nifD | This study |
| DJL79 | WWM73 att::pDL739 | gRNA-nrpR1 | This study |
| DJL105 | WWM73 att::pDL745 | gRNA-nifB | This study |
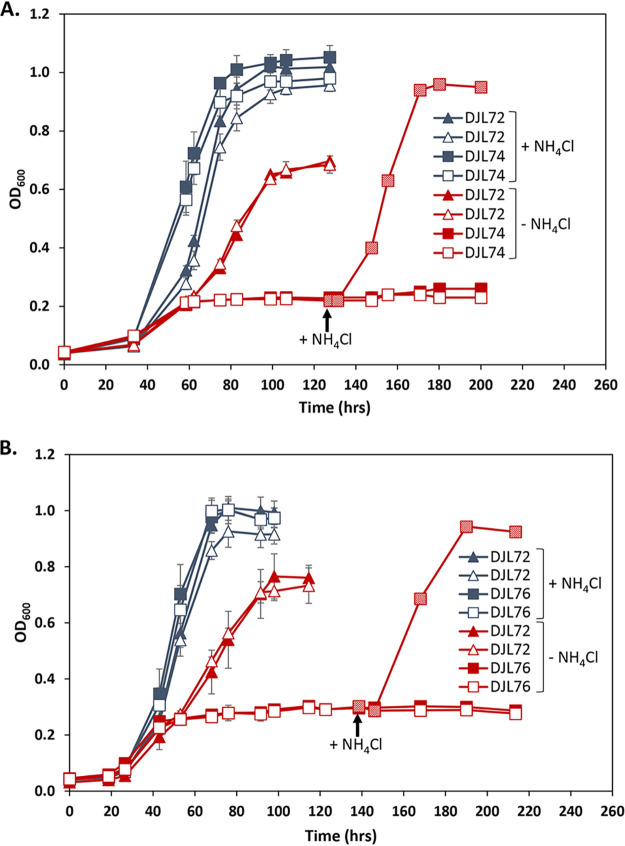
M. acetivorans strain DJL74 grew identically to strain DJL72 in medium with NH4Cl with or without tetracycline (Fig. 4A), indicating that expression of gRNA-Pnif does not impact growth with NH4Cl. However, whereas strain DJL72 was able to grow by fixing N2 in medium lacking NH4Cl, diazotrophic growth of strain DJL74 was abolished (Fig. 4A). Strain DJL74 was only able to grow using initial NH4Cl carried over from the inoculum to reach an optical density at 600 nm (OD600) of ∼0.2 and was unable to switch to diazotrophic growth. The growth profiles of strain DJL74 were identical in the presence and absence of tetracycline, indicating that sufficient dCas9 is produced even in the absence of the inducer. Remarkably, the cessation of growth was absolute; the cells were unable to overcome dCas9-mediated repression of the nif operon, as the OD600 stayed at ∼0.2 for several days. To confirm that the inability of strain DJL74 to grow was due to the lack of fixed nitrogen as a result of dCas9-mediated repression of the nif operon, NH4Cl was added to some of the culture tubes ∼100 h after the cessation of growth. Cells in these cultures immediately started to grow and reached a final OD600 comparable to that of cultures grown initially with NH4Cl (Fig. 4A). An M. acetivorans strain with the replicating version of pDL732 exhibited the same phenotype (see Fig. S1 in the supplemental material), indicating that the location of the CRISPRi-dCas9 plasmid does not impact system function. Expression of gRNA-nifD also abolished diazotrophic growth of strain DJL76 in the absence of NH4Cl (Fig. 4B). The growth profiles of the strains expressing gRNA-Pnif and gRNA-nifD were identical (Fig. 4). Thus, targeting either the nif operon promoter or downstream of the start codon of nifD results in dCas9-mediated repression sufficient to abolish diazotrophic growth of M. acetivorans.
FIG 4.
Comparison of the growth of M. acetivorans strain DJL72 to that of strains DJL74 (A) and DJL76 (B) with methanol and with or without NH4Cl. Strains were grown in HS medium with 125 mM methanol and 18 mM NH4Cl as indicated; 50 μM tetracycline was added to cultures represented by open symbols. Data are the means from four replicates, and error bars represent standard deviations. Arrows denote NH4Cl addition to cultures of DJL74 or DJL76.
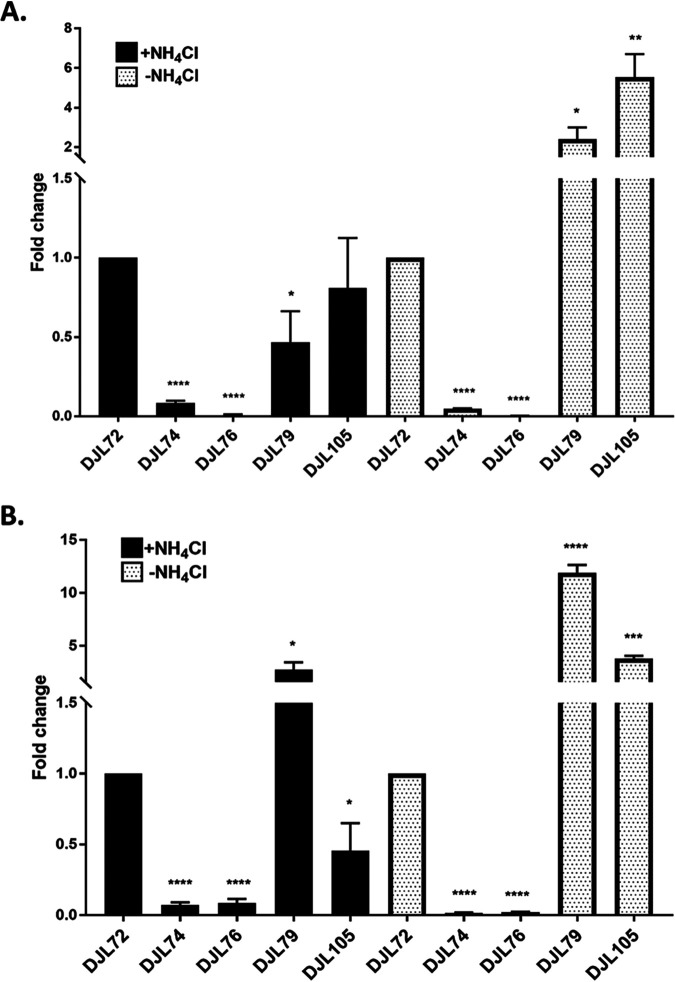
qPCR analysis of the relative transcript abundance of nifH and nifD confirmed significant repression of the nif operon in strains DJL74 and DJL76 compared to that in strain DJL72 (Fig. 5). In both strains, there was a >10-fold (>90%) reduction in both nifH and nifD transcript abundance in cells grown with or without NH4Cl. Thus, induction of the nif operon by the absence of NH4Cl is insufficient to overcome dCas9-mediated repression with either gRNA-Pnif or gRNA-nifD. Interestingly, although gRNA-nifD targets downstream of nifH (Fig. 3), significantly less nifH transcript was detected in cells of strain DJL76. It is possible that blockage of RNAP by dCas9 bound at nifD results in abortive transcripts harboring nifH that are unstable and are subsequently degraded. Consistent with abolished diazotrophic growth, NifD was not detected by Western blotting in lysates from cells of strains DJL74 and DJL76 (Fig. 6). These results reveal that a 20-nucleotide (nt) gRNA is enough to target dCas9 to block transcription of the nif operon, preventing NifHDK production and, subsequently, the ability to grow by fixing N2.
FIG 5.
Relative transcript abundance of nifH (A) and nifD (B) in M. acetivorans CRISPRi-dCas9 strains. Strains were grown in HS medium with 125 mM methanol and with or without NH4Cl. qPCR was performed as described in Materials and Methods. Data are the means from three biological replicates analyzed in duplicate. The relative abundance of nifH and nifD in strain DJL72 (gRNA-less) was set to 1 to determine fold change in the other strains. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FIG 6.
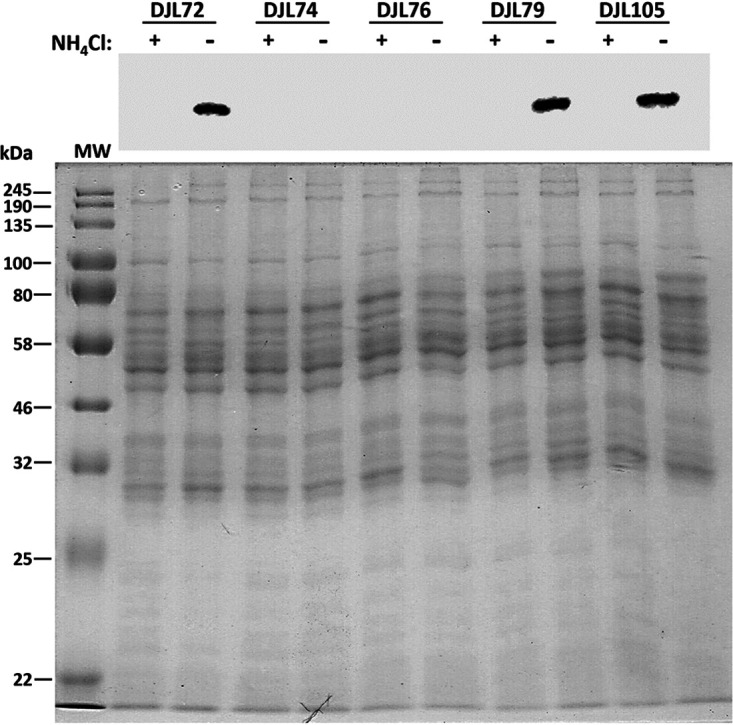
Comparison of NifD abundances in cell lysates from CRISPRi-dCas9 M. acetivorans strains. Strains were grown in HS medium with 125 mM methanol and with or without NH4Cl. (Top) Western blot analysis of lysates was performed with antibodies specific to M. acetivorans NifD. (Bottom) SDS-PAGE of lysates used for Western blot. MW, molecular weight standard.
Repression of nifB by dCas9 inhibits diazotrophic growth of M. acetivorans.
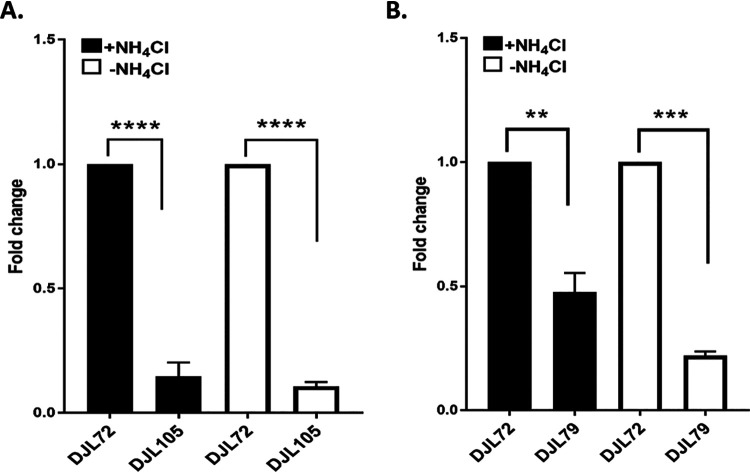
To further ascertain the utility of the CRISPRi-dCas9 system to study gene function in M. acetivorans, we targeted the gene encoding the nitrogenase cofactor biogenesis protein NifB, the radical S-adenosylmethionine (SAM)-dependent enzyme required for the insertion of carbide into the M-cluster of Mo nitrogenase (28). Without the M-cluster, Mo nitrogenase is inactive. Thus, NifB is essential for the ability of diazotrophs to fix N2. M. acetivorans contains a truncated version of NifB, but biochemical evidence revealed it is capable of catalyzing steps leading to synthesis of the M-cluster (30). However, the importance of NifB to diazotrophy by M. acetivorans has not been investigated. A gRNA (gRNA-nifB) was designed to target the sense strand starting ∼100 nucleotides downstream of the start codon of nifB (Table 2 and Fig. 3B). M. acetivorans strain DJL105 was generated, which harbors integrated pDL745 expressing gRNA-nifB (Table 3). Growth of strain DJL105 was identical to that of control strain DJL72 with methanol in HS medium containing NH4Cl (Fig. 7A). However, strain DJL105 exhibited delayed and slower growth in medium lacking NH4Cl. Analysis of nifB transcript abundance by qPCR revealed a 7- to 10-fold (>85%) reduction in strain DJL105 compared to that in strain DJL72 (Fig. 8A). Interestingly, the transcript abundance of nifH and nifD was 4- to 5-fold higher in cells of strain DJL105 than in cells of strain DJL72 grown in the absence of NH4Cl (Fig. 5). However, the level of NifD in lysate from DJL105 cells was similar to the level of NifD in lysate from strain DJL72 cells as determined by Western blotting (Fig. 6). These results are consistent with NifB having a function specific to N2 fixation in M. acetivorans.
FIG 7.
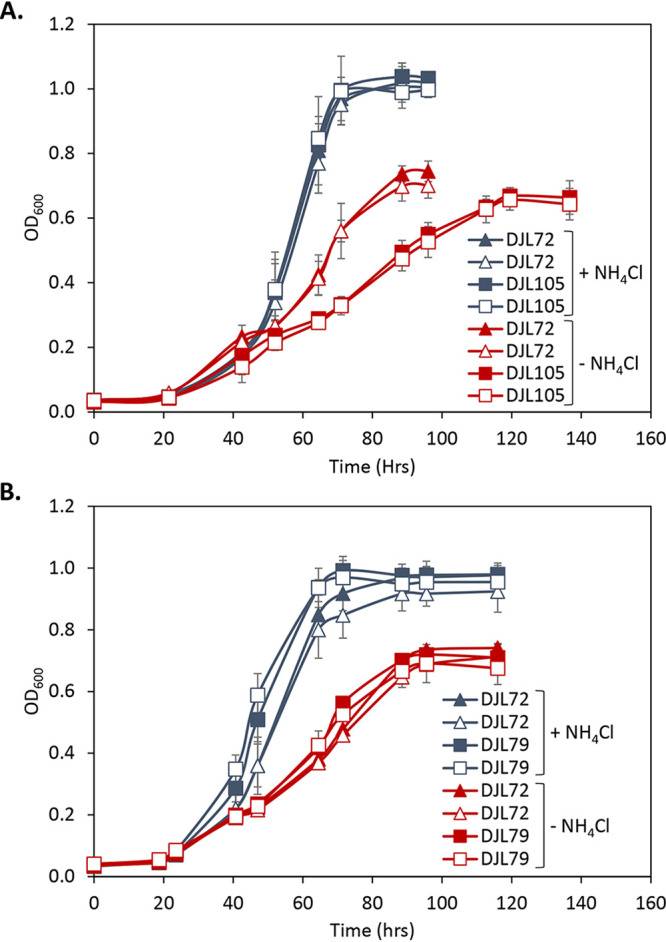
Comparison of the growth of M. acetivorans strain DJL72 to that of strains DJL79 (A) and DJL105 (B) with methanol and with or without NH4Cl. Strains were grown in HS medium with 125 mM methanol and 18 mM NH4Cl as indicated; 50 μM tetracycline was added to cultures represented by open symbols. Data are the means from four replicates, and error bars represent standard deviations.
FIG 8.
Relative transcript abundance of genes targeted for dCas9-mediated repression. (A) Fold change in nifB transcript abundance in strain DJL72 versus DJL105. (B) Fold change in nrpR1 transcript abundance in strain DJL72 versus DJL79. Strains were grown in HS medium with 125 mM methanol and with or without NH4Cl. qPCR was performed as described in Materials and Methods. Data are the means from three biological replicates analyzed in duplicate. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Repression of nrpR1 by dCas9 affects nif operon expression.
Finally, the ability to use the CRISPRi-dCas9 system to study genes involved in regulation of transcription in methanogens was tested. NrpR is the known repressor of nif operon transcription in methanogens (22, 31). Although in vivo analysis of NrpR has not been performed in M. acetivorans, the nif operon contains the identified NrpR operator (Fig. 3A), and recombinant M. acetivorans NrpR is functional in binding DNA (32). Like in other Methanosarcina species, NrpR is encoded by two separate genes in M. acetivorans (32). NrpR1 contains the DNA-binding region, and deletion of only nrpR1 in Methanosarcina mazei results in the loss of the nif operon repression (33). Therefore, only nrpR1 was targeted for dCas9-mediated repression in M. acetivorans. A gRNA (gRNA-nrpR1) was designed to target the sense strand starting six nucleotides downstream of the start codon of nrpR1 (Table 2 and Fig. 3C). M. acetivorans strain DJL79 was generated with integrated pDL739 expressing gRNA-nrpR1 (Table 3). Strain DJL79 and the control strain DJL72 exhibited identical levels of growth in HS medium with or without NH4Cl (Fig. 7B). However, the transcript abundance of nrpR1 was 2-fold (52%) and 5-fold (78%) reduced in DJL79 cells grown with and without NH4Cl, respectively (Fig. 8). Moreover, DJL79 had altered nif operon expression (Fig. 5). The transcript abundances of both nifH and nifD were significantly higher in DJL79 cells than in DJL72 control cells grown in the absence of NH4Cl (nif-inducing conditions). Cells of DJL79 grown in the presence of NH4Cl had more nifD detected, but less nifH, than the control cells. NifD was not detected in lysate from cells grown with NH4Cl (Fig. 6), despite the increased transcript abundance. The lower levels of nifH detected in strain DJL79 and the lack of NifD protein under nif-repressing conditions (+NH4Cl) are consistent with significant posttranscriptional regulation of the nif operon (34–36). Similar levels of NifD were observed in cells of strains DJL72 and DJL79 when grown under nif-inducing conditions (−NH4Cl) (Fig. 6). These results demonstrate that dCas9-mediated repression of NrpR1 alters transcription of the nif operon, consistent with NrpR as the repressor of the nif operon.
DISCUSSION
CRISPR-mediated transcriptional regulation is an extremely powerful tool for targeted and tunable control of gene expression. In bacteria such as E. coli, a single gRNA along with dCas9 is sufficient to elicit up to 99% repression of a synthetic target gene (10). A gRNA targeting the sense (nontemplate) strand near the promoter or ribosome binding site (RBS) effectively silences transcription initiation and elongation. However, using dCas9 with a single gRNA is less efficient in silencing transcription in eukaryotes (e.g., Saccharomyces cerevisiae) (37, 38). The repression of target genes is improved by fusing additional regulatory domains to dCas9. An initial study showed that fusing the mammalian repressor Mxi1 to dCas9 greatly enhanced targeted gene repression over that by dCas9 alone (39). This difference may be due to the increased complexity of transcription initiation and elongation in eukaryotes. Although archaea have compact genomes, like bacteria, with genes lacking introns and arranged in operons, the archaeal transcription machinery is homologous to eukaryotic machinery (e.g., RNA polymerase [RNAP]) and shares similar mechanisms of transcription initiation and elongation (40). However, like that seen in bacteria, results in this study with M. acetivorans reveal that expression of dCas9 with a single gRNA is sufficient to silence gene expression in archaea. Separate gRNAs targeting the sense strand of the nif operon promoter and near the start codon of nifD repressed nif operon transcription, eliminated NifD production, and subsequently abolished growth with N2. The repression of the nif operon was efficient and stable; strains expressing either gRNA-Pnif or gRNA-nifD were never able to overcome dCas9-mediated repression and grow with N2. Remarkably, M. acetivorans could not relieve repression to an extent that would allow production of sufficient nitrogenase to support growth, despite the strongest selective pressure (i.e., to grow). In fact, the cells remained in a viable but nongrowing state for at least 2 weeks. The gRNAs targeting the promoter and the start codon worked equally well, indicating that dCas9 is capable of blocking transcription initiation and elongation in archaea. Importantly, based on the significant repression of genes targeted by gRNA-nifD, gRNA-nifB, and gRNA-nrpR1, in theory, any M. acetivorans gene could be silenced by expression of a single gRNA targeting the sense strand after the start codon.
dCas9-mediated repression of putative nitrogen fixation genes in M. acetivorans provides new insight, as well as confirms existing knowledge, about the expression, regulation, and maturation of nitrogenase in methanogens. These results confirm that the nif operon encodes Mo nitrogenase, which is required for diazotrophic growth in medium containing molybdenum. M. acetivorans can also fix nitrogen using V and/or Fe nitrogenases in molybdenum-deplete medium (our unpublished results). The regulation of the alternative nitrogenases has not been investigated in methanogens. Interestingly, M. acetivorans did not compensate for the loss of Mo nitrogenase by producing an alternative nitrogenase to fix N2, at least under the conditions tested, which has been seen in some diazotrophic bacteria (41). These results indicate that molybdenum is likely the critical effector molecule that controls expression of V and Fe nitrogenases in M. acetivorans.
NifB is essential for the activity of all nitrogenases (42). However, dCas9-mediated repression of nifB did not abolish diazotrophic growth of strain DJL105, unlike the repression of the nif operon. NifB converts [4Fe-4S] clusters into NifB-co, an [8Fe-9S-C] cluster that is the precursor to the active site cluster (M-cluster, [7Fe-9S-C-Mo-homocitrate]) in NifDK (28). The genome of M. acetivorans encodes a single NifB that lacks a NifX-like domain found in some bacteria. Recombinant M. acetivorans NifB supports the in vitro synthesis of the M-cluster in NifDK (30). Unlike the structural components (NifDK) of Mo nitrogenase, which are needed in high abundance (43), only a small amount of NifB is likely required for maturation of nitrogenase. Therefore, the impaired growth of strain DJL105 in the absence of NH4Cl could be due to minimal production of NifB. Alternatively, an unknown protein could compensate for the loss of NifB production. Additional experimentation is required to confirm that NifB encoded by MA4195 is solely required for the maturation of nitrogenase. Interestingly, a significant increase in the transcript abundance of nifH and nifD was observed in strain DJL105 cells grown without NH4Cl (Fig. 5). NifB limitation likely decreases the level of functional Mo nitrogenase. To compensate, the cells increase nif operon transcription to attempt to produce more Mo nitrogenase. These results provide new insight into the relationship between nitrogenase maturation and expression in methanogens.
NrpR is a transcriptional repressor of the nitrogen fixation and assimilation genes in methanogens and has been extensively studied in Methanococcus maripaludis and M. mazei but not M. acetivorans (22, 31–33, 44). M. acetivorans strain DJL79 had higher transcript abundance under nitrogen-sufficient conditions (+NH4Cl) than the control, revealing that NrpR is also the repressor of the nif operon in this methanogen. Increased transcription of the nif operon in strain DJL79 did not result in increased NifD production, consistent with additional posttranscriptional regulation, and underscores the complexity of nitrogenase regulation in diazotrophs (34, 36). Interestingly, nifH and nifD transcript abundance was highest in strain DJL79 under nitrogen limitation, indicating that NrpR may primarily function to titrate the level of transcription during nitrogen limitation (−NH4Cl).
Compared to the endogenous CRISPRi systems in Haloferax and Sulfolobus for silencing genes in archaea, the exogenous dCas9-based CRISPRi system in Methanosarcina is simpler and more efficient. The Haloferax CRISPRi system requires the use of mutant strains deleted of all or parts of the endogenous type I CRISPR components (18, 19). Thus, use of the system is restricted to specialized strains, and since the endogenous system is used, it is unclear if these mutations cause unknown effects. Moreover, the Haloferax system only blocks transcription initiation, not elongation, because only gRNAs that target the template strand near the transcriptional start site result in significant repression (18, 19). In contrast to the Methanosarcina CRISPRi-dCas9 and Haloferax CRISPRi systems, the Sulfolobus CRISPRi system degrades existing mRNA using endogenous type III CRISPR system components. Greater than 90% reduction in mRNA and subsequent protein activity can be achieved by the Sulfolobus system (17). However, optimal mRNA reduction requires multiple gRNAs and/or increased gRNA dosage (15, 17). In contrast, the M. acetivorans CRISPRi-dCas9 system requires only two components, dCas9 and a single gRNA, to efficiently block both transcription initiation and elongation. Although specialized strains of M. acetivorans are needed to integrate pDL730 and derivatives into the chromosome, these strains are not required if the plasmid is first converted into the replicating version (Fig. 1). Therefore, any existing M. acetivorans strain could be transformed with replicating pDL730 and derivatives, allowing targeted gene repression in existing mutants. Although not demonstrated here, we expect that expression of multiple gRNAs targeting different genes/operons in a single strain will result in simultaneous repression, and we are currently testing this possibility.
There are limitations to the current CRISPRi-dCas9 system. Like the Haloferax and Sulfolobus CRISPRi systems, there is a lack of control. It is not possible to turn on/off repression of a target gene/operon. Like the M. acetivorans CRISPR-Cas9 system, where tetracycline was not required to induce genome editing (24), tetracycline had no effect on dCas9-mediated repression of target genes in the CRISPRi-dCas9 strains. This limits the ability to study essential or critical genes, since the system is stuck in the “on” state. We are currently attempting to engineer additional control in the system.
In summary, the results in this study demonstrate that the CRISPRi-dCas9 system is a simple yet powerful new tool to understand and control the biology of M. acetivorans. Importantly, since no additional processing of transformants is required, CRISPRi-dCas9 strains can be generated in as little as 2 weeks (including plasmid construction). The system will expedite understanding gene function, regulation, and interaction in methanogens and can likely be adapted for use in other nonextremophilic archaea as well.
MATERIALS AND METHODS
M. acetivorans strains and growth.
M. acetivorans strains used are listed in Table 3. M. acetivorans strain WWM73 served as the parent strain for all experiments. HS medium was prepared as previously described (45), except NH4Cl was omitted. Methanol, sulfide, tetracycline, puromycin, and NH4Cl were added from anaerobic sterile stocks using sterile syringes prior to inoculation. All M. acetivorans strains were grown at 35°C in Balch tubes containing 10 ml HS medium with 125 mM methanol and 0.025% sulfide. Headspace gas composition was 75% N2, 20% CO2, and 5% H2. Tetracycline (50 μM), puromycin (2 μg/ml), and NH4Cl (18 mM) were added where indicated. Growth was measured by monitoring optical density at 600 nm (OD600) using a spectrophotometer.
Construction of M. acetivorans CRISPRi-dCas9 plasmids and strains.
All primers (Table 4) and synthetic DNA fragments (gblocks) were designed using Geneious Prime and purchased from Integrated DNA Technologies (IDT). To construct a universal CRISPRi-dCas9 plasmid, cas9 was replaced with dcas9 in pDN203 (gift from the Metcalf lab) (24). First, dCas9 was PCR amplified using Q5 high-fidelity DNA polymerase (New England Biolabs) with specific primers (Table 4) and pMJ841 as a template. The dcas9 PCR amplicon was purified using a Wizard SV Gel and PCR Clean-up kit (Promega) and digested with AscI and HindIII. pDN203 was similarly digested with AscI and HindIII, and the plasmid lacking cas9 was purified from an agarose gel using the Wizard SV Gel and PCR Clean-up kit. Digested dcas9 was ligated into pDN203 using T4 DNA ligase followed by transformation of E. coli DH5α competent cells (New England Biolabs). Transformants were screened by PCR, and the final universal CRISPRi-dCas9 plasmid (pDL730) containing dcas9 was verified by sequencing (Eurofins Genomics LLC, USA). All other plasmids used to target specific genes and operons (Table 1) for dCas9-mediated repression were constructed by introducing a gblock that contained a specific gRNA into AscI-digested pDL730 using a Gibson Assembly Ultra kit (catalog number GA1200-10; Synthetic Genomics, Inc.) according to the manufacturer’s instructions. CRISPRi-dCas9 plasmids were converted into M. acetivorans replicating plasmids by retrofitting with pAMG40 (46) using a Gateway BP Clonase II kit (Invitrogen) according to the manufacturer’s instructions. M. acetivorans strain WWM73 was separately transformed with replicating and nonreplicating plasmids using a liposome-mediated transformation protocol as described previously (47). Transformants were selected by anaerobic growth on agar plates containing 125 mM methanol and 2 μg/ml puromycin. Colonies were screened by PCR, and a positive colony was selected as the new CRISPRi-dCas9 strain (Table 3). Strains were maintained in HS medium with 125 mM methanol and 2 μg/ml puromycin.
TABLE 4.
Primers used in this study
| Primer | Sequence (5′–3′) | Use |
|---|---|---|
| dCas9.F | ATGGATAAGAAATACTCAATAGGC | Cloning dcas9 |
| dCas9.HindIII | GTATAGCATACATTATACGAAGTTATCAAGAAGCTTTTAGTCACCTCCTAGCTG | |
| CasHdrSeq.F | CAGCTAGCTAACGCGTATTAAAGG | Confirmation of Gibson assembly |
| Cas9NHEJ.R | GCCATTTTTCTTCTCACCGGGGAGCTGAGC | |
| RepA.F | CTGAGTTCTGATTTTAGTTCTAGACC | Confirmation of replicating plasmid |
| RepA.R | TCAATCACCCCCAAACATCATG | |
| C2Achr1 | GAAGCTTCCCCTTGCCAAT | Confirmation of integrated plasmid |
| C2Achr2 | TTGATTCGGATA CCCTGAGC | |
| Plscreen3 | GCAAAGAAAAGCCAGTATGGA | |
| Plscreen4 | TTTTTCGTCTCAGCCAATCC | |
| 16sRNA.F | GGTACGGGTTGTGAGAGCAA | qPCR: MA1618 |
| 16sRNA.R | CTCGGTGTCCCCTTATCACG | |
| NifH.F | AATCTTACTGCGGCCCTCTC | qPCR: MA3895 |
| NifH.R | GTGTCAAGTACGGTCTTCTGGT | |
| NifD.F | CGCCCGCTGTGAAGGATATA | qPCR: MA3898 |
| NifD.R | TTATGTCAAAGGGAGTGGGGTC | |
| NrpRI.F | TCCGCTACCACCTGAGGAT | qPCR: MA4404 |
| NrpRI.R | GAGGGCATCATTCATCTCGCT | |
| NifB.F | ACTCTGCCAGGATCTGCTTTG | qPCR: MA4195 |
| NifB.R | TCCGGGAATTTGACTGCGTAA |
Gene expression analysis.
M. acetivorans cells (4 ml of culture) were harvested at mid-log phase (OD600 of 0.2 to 0.4) by centrifugation at 5,000 × g inside an anaerobic chamber (Coy Laboratories). The cell pellets were resuspended in 1 ml TRIzol (catalog number 15596-026; Life Technologies) and stored at −80°C until use. Total RNA was purified from cells using a Direct-zol RNA Miniprep kit (catalog number R2050; Zymo Research) according to the manufacturer’s instructions followed by an additional DNase treatment using a DNA-free DNA removal kit (catalog number AM1906; Thermo Fisher Scientific). RNA concentrations were determined using a Thermo Scientific NanoDrop 2000 spectrophotometer. To determine transcript abundance by real-time quantitative PCR (qPCR), cDNA was generated with 300 ng of total RNA using an iScript Select cDNA synthesis kit (catalog number 1708897; Bio-Rad) with the following conditions: 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min. cDNA was stored at −20°C until use. Each qPCR mixture (10 μl) contained 1× SsoAdvanced universal SYBR green supermix (catalog number 1725271; Bio-Rad), 300 nM forward and reverse primers (Table 4), and 300-fold-diluted cDNA. The qPCRs were performed using a CFX96 real-time PCR detection system (Bio-Rad) for all samples under the following conditions: 95°C for 1 min followed by 37 cycles of 95°C for 20 s and 60°C for 20 s; this was followed by melting curve analysis to ensure the specificity of amplification. qPCR data were analyzed using the comparative threshold cycle (ΔΔCq) method with 16S RNA used as an internal control. To determine the transcript abundance of a specific gene, ΔCq values of the gene in the samples were calibrated to the average of its ΔCq values in the control group (DJL72 cells) unless indicated otherwise. Duplicate qPCRs were conducted for three biological replicates for each strain.
Western blot analysis.
M. acetivorans cells were harvested at mid-log phase (OD600 of 0.2 to 0.4) by centrifugation at 5,000 × g inside an anaerobic chamber (Coy Laboratories). Cell pellets were resuspended in buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Tween 20, 1 mM benzamidine hydrochloride hydrate, and 2 mM EDTA), sonicated 2 to 3 times in an ice bath using a Branson Digital Sonifier 450 (Branson Ultrasonics), and centrifuged at 14,500 × g for 3 min. The total protein concentrations in the cell lysates were measured by the Bradford method using Bio-Rad protein assay dye reagent concentrate (catalog number 5000006) according to the manufacturer’s instructions. Protein (5 μg) samples were resolved in a 10% SDS-PAGE gel and transferred to an Amersham Protran NC membrane (catalog number 10600005; Fisher Scientific). The membrane was blocked with blocking buffer (5% skim milk in Tris-buffered saline with 0.1% Tween [TBST]) for 15 min at room temperature (RT) and probed with a custom anti-NifD antibody (GenScript) overnight. The membrane was washed at least three times with TBST for 5 min each and then incubated with the secondary antibody (goat anti-rabbit IgG) for 1 h at RT with slow shaking. After three washes, the membrane was developed with an enhanced chemiluminescence (ECL) reagent (Amersham ECL Prime Western blotting detection reagent, catalog number RPN2232; GE Healthcare, UK) and scanned with a FluorChem 8900 imaging system (Alpha Innotech, San Leandro, CA). Western blot analyses of lysates from all strains were performed with at least three biological replicates.
Data availability.
The raw data from the growth studies and qPCR will be available upon request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dipti Nayak and Bill Metcalf for providing M. acetivorans strains and plasmids.
This work was supported in part by DOE Biosciences grant number DE-SC0019226 (D.J.L.), NSF grant number MCB1817819 (D.J.L.), and the Arkansas Biosciences Institute (D.J.L.), the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 3.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charpentier E, Doudna JA. 2013. Biotechnology: rewriting a genome. Nature 495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 6.Sampson TR, Weiss DS. 2014. Exploiting CRISPR/Cas systems for biotechnology. Bioessays 36:34–38. doi: 10.1002/bies.201300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Zhang L, Huang X. 2014. Genome modification by CRISPR/Cas9. FEBS J 281:5186–5193. doi: 10.1111/febs.13110. [DOI] [PubMed] [Google Scholar]
- 10.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrera MA, Blamey JM. 2018. Biotechnological applications of archaeal enzymes from extreme environments. Biol Res 51:37. doi: 10.1186/s40659-018-0186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 14.Peng W, Feng M, Feng X, Liang YX, She Q. 2015. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zink IA, Pfeifer K, Wimmer E, Sleytr UB, Schuster B, Schleper C. 2019. CRISPR-mediated gene silencing reveals involvement of the archaeal S-layer in cell division and virus infection. Nat Commun 10:4797. doi: 10.1038/s41467-019-12745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zebec Z, Manica A, Zhang J, White MF, Schleper C. 2014. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 42:5280–5288. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zebec Z, Zink IA, Kerou M, Schleper C. 2016. Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 (Bethesda) 6:3161–3168. doi: 10.1534/g3.116.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stachler AE, Schwarz TS, Schreiber S, Marchfelder A. 2020. CRISPRi as an efficient tool for gene repression in archaea. Methods 172:76–85. doi: 10.1016/j.ymeth.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Stachler AE, Marchfelder A. 2016. Gene repression in haloarchaea using the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas I-B system. J Biol Chem 291:15226–15242. doi: 10.1074/jbc.M116.724062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leigh JA. 2000. Nitrogen fixation in methanogens: the archaeal perspective. Curr Issues Mol Biol 2:125–131. [PubMed] [Google Scholar]
- 22.Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol 61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Ribbe MW. 2015. Nitrogenase and homologs. J Biol Inorg Chem 20:435–445. doi: 10.1007/s00775-014-1225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayak DD, Metcalf WW. 2017. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc Natl Acad Sci U S A 114:2976–2981. doi: 10.1073/pnas.1618596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, et al. . 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters JW, Fisher K, Dean DR. 1995. Nitrogenase structure and function: a biochemical-genetic perspective. Annu Rev Microbiol 49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 27.Mus F, Alleman AB, Pence N, Seefeldt LC, Peters JW. 2018. Exploring the alternatives of biological nitrogen fixation. Metallomics 10:523–538. doi: 10.1039/C8MT00038G. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Ribbe MW. 2016. Biosynthesis of the metalloclusters of nitrogenases. Annu Rev Biochem 85:455–483. doi: 10.1146/annurev-biochem-060614-034108. [DOI] [PubMed] [Google Scholar]
- 29.Nayak DD, Metcalf WW. 2019. Methylamine-specific methyltransferase paralogs in Methanosarcina are functionally distinct despite frequent gene conversion. ISME J 13:2173–2182. doi: 10.1038/s41396-019-0428-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fay AW, Wiig JA, Lee CC, Hu Y. 2015. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc Natl Acad Sci U S A 112:14829–14833. doi: 10.1073/pnas.1510409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lie TJ, Leigh JA. 2003. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol 47:235–246. doi: 10.1046/j.1365-2958.2003.03293.x. [DOI] [PubMed] [Google Scholar]
- 32.Lie TJ, Dodsworth JA, Nickle DC, Leigh JA. 2007. Diverse homologues of the archaeal repressor NrpR function similarly in nitrogen regulation. FEMS Microbiol Lett 271:281–288. doi: 10.1111/j.1574-6968.2007.00726.x. [DOI] [PubMed] [Google Scholar]
- 33.Weidenbach K, Ehlers C, Kock J, Ehrenreich A, Schmitz RA. 2008. Insights into the NrpR regulon in Methanosarcina mazei Go1. Arch Microbiol 190:319–332. doi: 10.1007/s00203-008-0369-3. [DOI] [PubMed] [Google Scholar]
- 34.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 35.Halbleib CM, Ludden PW. 2000. Regulation of biological nitrogen fixation. J Nutr 130:1081–1084. doi: 10.1093/jn/130.5.1081. [DOI] [PubMed] [Google Scholar]
- 36.Prasse D, Forstner KU, Jager D, Backofen R, Schmitz RA. 2017. sRNA154 a newly identified regulator of nitrogen fixation in Methanosarcina mazei strain Go1. RNA Biol 14:1544–1558. doi: 10.1080/15476286.2017.1306170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farzadfard F, Perli SD, Lu TK. 2013. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanegas KG, Lehka BJ, Mortensen UH. 2017. SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb Cell Fact 16:25. doi: 10.1186/s12934-017-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blombach F, Matelska D, Fouqueau T, Cackett G, Werner F. 2019. Key concepts and challenges in archaeal transcription. J Mol Biol 431:4184–4201. doi: 10.1016/j.jmb.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Oda Y, Samanta SK, Rey FE, Wu L, Liu X, Yan T, Zhou J, Harwood CS. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J Bacteriol 187:7784–7794. doi: 10.1128/JB.187.22.7784-7794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arragain S, Jimenez-Vicente E, Scandurra AA, Buren S, Rubio LM, Echavarri-Erasun C. 2017. Diversity and functional analysis of the FeMo-cofactor maturase NifB. Front Plant Sci 8:1947. doi: 10.3389/fpls.2017.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dingler C, Kuhla J, Wassink H, Oelze J. 1988. Levels and activities of nitrogenase proteins in Azotobacter vinelandii grown at different dissolved oxygen concentrations. J Bacteriol 170:2148–2152. doi: 10.1128/JB.170.5.2148-2152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidenbach K, Ehlers C, Kock J, Schmitz RA. 2010. NrpRII mediates contacts between NrpRI and general transcription factors in the archaeon Methanosarcina mazei Go1. FEBS J 277:4398–4411. doi: 10.1111/j.1742-4658.2010.07821.x. [DOI] [PubMed] [Google Scholar]
- 45.Sowers KR, Boone JE, Gunsalus RP. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol 59:3832–3839. doi: 10.1128/AEM.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from the growth studies and qPCR will be available upon request.