Abstract
As musculoskeletal (MSK) disorders continue to increase globally, there is an increased need for novel, in vitro models to efficiently study human bone physiology in the context of both healthy and diseased conditions. For these models, the inclusion of innate immune cells is critical. Specifically, signaling factors generated from macrophages play key roles in the pathogenesis of many MSK processes and diseases, including fracture, osteoarthritis, infection etc. In this study, we aim to engineer three-dimensional (3D) and macrophage-encapsulated bone tissues in vitro, to model cell behavior, signaling, and other biological activities in vivo, in comparison to current two-dimensional models. We first investigated and optimized 3D culture conditions for macrophages, and then co-cultured macrophages with mesenchymal stem cells (MSCs), which were induced to undergo osteogenic differentiation to examine the effect of macrophage on new bone formation. Seeded within a 3D hydrogel scaffold fabricated from photocrosslinked methacrylated gelatin, macrophages maintained high viability and were polarized toward an M1 or M2 phenotype. In co-cultures of macrophages and human MSCs, MSCs displayed immunomodulatory activities by suppressing M1 and enhancing M2 macrophage phenotypes. Lastly, addition of macrophages, regardless of polarization state, increased MSC osteogenic differentiation, compared with MSCs alone, with proinflammatory M1 macrophages enhancing new bone formation most effectively. In summary, this study illustrates the important roles that macrophage signaling and inflammation play in bone tissue formation.
Impact statement
There is a lack of in vitro three-dimensional (3D) models that closely recapitulate human musculoskeletal tissues. This study investigates the interactions between primary macrophages (M0, M1, and M2) and mesenchymal stem cells (MSCs). As far as we know, this is also the first in vitro 3D study of a direct cell-to-cell culture of primary human macrophages and MSCs during osteogenesis. Our results suggest that macrophages provide an enhancing role for bone formation, demonstrating the importance of including innate immune cells in 3D in vitro platforms. This model forms the foundation for future efforts to engineer a more physiologically relevant bone in vitro model.
Keywords: human mesenchymal stem cells, macrophages, three-dimensional culture, bone health, microphysiological system, tissue chip
Introduction
Arthritis is a degenerative condition of a joint that causes pain, swelling, and diminished function. Arthritic disorders are the major cause of disability in adults in the United States.1 Between 2013 and 2016, ∼54 million adults in the United States were diagnosed with an arthritic disorder1; it is predicted that by 2040, more than 78 million patients will have one or more joints affected by a type of arthritis (rheumatoid, inflammatory and septic arthritis).2
Until now, there is no definitive cure for most arthritic disorders, and the treatment consists of management protocols focused on pain relief and improvement of joint function. In an effort to better understand the pathology of these diseases, two-dimensional (2D) in vitro studies and animal models have been created; however, they are often costly and limited in scope and scale. While many of these 2D models use human cell lines, they do not accurately simulate the complexity, geometry, and topography found in tissues in vivo. Furthermore, animal cells do not respond to adverse stimuli and drugs in the same way as human cells,3 and animal models are more difficult to use and more costly than in vitro systems. Thus, there is an urgent need for the development of in vitro three-dimensional (3D) models that represent a more plausible human ex vivo platform to allow better recapitulation of native tissues.
The 3D tissue-engineered models have recently received increasing attention due to the growing necessity to simulate the 3D geometry and topography found in vivo compared with 2D in vitro models.4 Cell behavior, cell shape, proliferation, and secretion levels in these 3D tissues differ significantly with respect to the cell characteristics observed in the 2D models.5 These 3D models have been tailored to the particular tissue to be simulated (healthy or diseased), to better recreate the intrinsic properties of the tissue being studied, such as modifications of the extracellular matrix biochemical and mechanical properties, flow rates, co-culture with different cell types, and modular tissue interactions. Moreover, these models facilitate the construction of patient-specific models, opening new doors for personalized medical strategies, innovations, and treatments in a cost-effective manner.
In particular, there is a need for reliable and reproducible models for understanding the biological mechanisms of joint diseases. To address this need, the engineering of 3D musculoskeletal (MSK) tissues, including bone, cartilage, and muscle has evoked increasing interest. Despite these efforts, most of the existing MSK tissue engineering approaches have not considered the crucial role that the innate immune system plays in health and disease, such as in osteoarthritis and rheumatoid arthritis; rather, many of the models have focused on establishing 3D cultures of tissues derived from mesenchymal stem cells (MSCs) only, due to the complexity of culturing immune cells.
Macrophages are dynamic and versatile cells of the innate immune system that are found in practically all MSK tissues, including bone, synovium, muscle, ligament, and tendon. Macrophages are essential in maintaining normal tissue homeostasis and are among the first responders to tissue injury and infection. In general, macrophages can differentiate into two main phenotypes,6 which are mainly characterized by functional parameters. Broadly speaking, undifferentiated (M0) macrophages in the local tissues and those derived from circulating monocytes, can differentiate into M1, or proinflammatory macrophages, which are the initial responders to traumatic events and adverse stimuli. M1 macrophages secrete proinflammatory factors, such as tumor necrosis factor alpha (TNFα), interleukin 1 beta (IL-1β), IL-6, inducible nitric oxide, and others. M2 or anti-inflammatory macrophages are involved in tissue healing, regeneration, and remodeling, and secrete C–C motif chemokine ligand (CCL) 13, CCL17, CCL18, and other factors.7
Macrophages are essential for initiating the physiological sequence of fracture healing. Our group and others have previously shown that macrophages are required for the recruitment and differentiation of MSCs and other bone-forming cells during bone regeneration in 2D systems.8,9 The goal of our study is to develop a 3D system with human primary macrophages and human mesenchymal stem cells (hMSCs) to study inflammation and osteogenesis in a direct co-culture system. For this, we first established the optimal conditions to culture macrophages in 3D scaffolds and then studied the differences between 2D and 3D single cultures of macrophages. In the proposed model, we compared macrophage activation in our 3D system with the standard 2D culture. Then, to the macrophages in 3D scaffolds, we added hMSCs, which were induced to undergo osteogenic differentiation, and assessed osteogenesis after 4 weeks of culture.
To our knowledge, this is the first study that took into consideration human primary macrophages as well as examined direct cell-to-cell interaction between macrophages and hMSCs in 3D culture. This study is the first step in representing in part the complexity of joint tissues in a controlled manner that is not possible in 2D experiments or in animal models. We anticipate that our novel 3D culture system will lead to a better understanding of the pathophysiology and evaluation of potential treatments for arthritic disorders.
Materials and Methods
Human monocyte isolation and differentiation to macrophages
Buffy coats from deidentified 20–40-year-old healthy male donors were obtained from the Stanford Blood Center with the approval of the local Ethics Committee. Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats using the Ficoll separation method.10 In brief, the buffy coat was diluted at a 1:1 ratio with 6 nM ethylenediaminetetraacetic acid (EDTA; Sigma-Aldrich, St. Louis, MO) in Dulbecco's phosphate-buffered saline (DPBS; Thermo Fisher Scientific, Waltham, MA) supplemented with 2% heat-inactivated fetal bovine serum (FBS; Life Technologies, Pleasanton, CA), and the solution was carefully pipetted into SepMate-50 PBMC isolation tubes (Stem Cell Technologies, Vancouver, Canada) with 15 mL of Ficoll-Paque PLUS (GE Healthcare, Chicago, IL) in the SepMate insert.
Then the tubes were centrifuged at 300 g for 20 min. The PBMC supernatants were aspirated, and the pellets were washed twice with 1 mM EDTA in DPBS supplemented with 2% FBS and then centrifuged at 300 g for 8 min. The final pellet was resuspended in EasySep Buffer (Stem Cell Technologies), and the monocytes were further purified based on negative selection using the EasySep™ Human Monocyte Isolation Kit (Stem Cell Technologies).
The monocytes were seeded into 10-cm standard tissue culture plates at a density of 1 × 105 cells/cm2 in macrophage-stimulating medium (MSM, Roswell Park Memorial Institute 1640 Medium [RPMI 1640; Life Technologies] supplemented with 5% FBS, 1% Gibco antibiotic/antimycotic solution [Life Technologies], and 100 ng/mL macrophage colony-stimulating factor [M-CSF; PeproTech, Rocky Hill, NJ]) for 6 days to differentiate the adherent monocytes into monocyte-derived macrophages. In detail, after isolating monocytes, the cells were seeded into 10-cm standard tissue culture plates at a density of 1 × 105 cells/cm2. The medium was changed after 3 days of the initial monocyte culture; therefore, the nonadherent monocytes were removed and the medium was replenished to the adherent monocytes.
M-CSF was added to the cell culture medium to differentiate the adherent monocytes to monocyte-derived macrophages (henceforth, we will refer to these cells as macrophages). Only macrophages were activated for this model. Monocytes and macrophages were characterized by fluorescence-activated cell sorting (FACS). Freshly isolated monocytes were washed twice with flow cytometry buffer (1 mM EDTA with 2% FBS in DPBS) and stained with antibodies against CD14 (a monocyte marker; Stem Cell Technologies) and CD45 (common leukocyte antigen; Stem Cell Technologies) according to the manufacturer's protocols.
To characterize macrophage differentiation, the cells were stained with antibodies against CD14, CD71, and CCR5 (BioLegend, San Diego, CA) after culturing in MSM for 6 days according to the manufacturer's protocols. Flow cytometry was performed in a BD LSRII flow cytometry (BD Biosciences, Franklin Lakes, NJ) analyzer at the Stanford Shared FACS Facility. The data collected were analyzed with the FlowJo software (Ashland, OR).
Macrophage culture
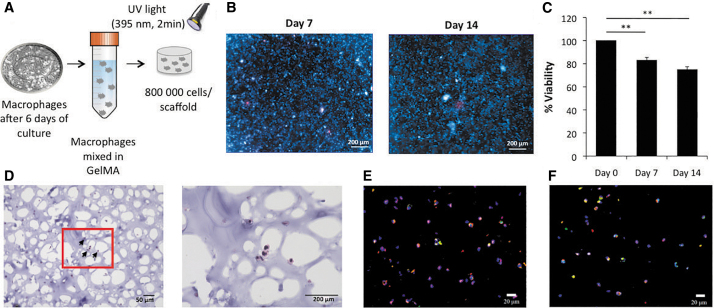
Macrophages were plated in a six-well culture plate at a density of 800,000 cells per well (10 cm2/well) in MSM for the 2D study. Figure 2A shows the 3D macrophage encapsulation protocol. In brief, macrophages were mixed in 50 μL of 15% methacrylated gelatin (GelMA) solution at 800,000 cells/scaffold. The cell/gel solution was added to a silicone mold (5 mm diameter × 2 mm height) and photocrosslinked with UV light (395 nm) for 2 min, forming a 3D scaffold. The scaffolds were placed in a 24-well plate and cultured in MSM, and the medium was refreshed every other day.11 The fabrication method is consistent and the scaffold sizes were found to be comparable among replicates.
FIG. 2.
Macrophages in 3D culture. (A) Procedure for 3D culture of macrophages in GelMA. Monocytes were differentiated into macrophages for 6 days, trypsinized and pelleted. The macrophages were resuspended in GelMA solution and photocrosslinked to fabricate the 3D scaffolds. (B) Live (blue)/Dead (red) staining of constructs at day 7 and 14 (scale bar = 200 μm). (C) Cell viability quantification by PicoGreen DNA assay at days 7 and 14 (normalized to day 0 data). Each value represents the mean ± SD. **p < 0.01, compared with day 0. (D) Hematoxylin and Eosin staining at day 14 (scale bar = 50 μm [left] or 200 μm [right]). Arrows point to macrophages in the scaffold. (E) Immunofluorescence staining of CCR5 (red), CD71 (green), and DAPI nuclear staining (blue). (F) Immunostaining of CCR5 (red), CD68 (green), and DAPI nuclear staining (blue) at day 14 (scale bar = 20 μm). Each value represents the mean ± SD. **p < 0.01 compared with day 0. 3D, three-dimensional; SD, standard deviation; GelMA, methacrylated gelatin; DAPI, 4′,6-diamidino-2-phenylindole. Color images are available online.
Macrophage cell viability
Scaffolds with macrophages were cultured in MSM and collected at days 0, 7, and 14. Cell viability was analyzed using fluorescent live/dead staining (Invitrogen, Carlsbad, CA) and the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). Scaffolds were incubated in live/dead dyes for 12 h and then imaged by an inverted microscope (Observer.Z1; Carl Zeiss, Jena, Germany). For the Quant-iT PicoGreen dsDNA Assay, cellular DNA was released from scaffolds digested by papain (Sigma-Aldrich) in digestion buffer (0.1 M sodium acetate, 0.01 cysteine HCl, 0.05 M EDTA) (1:50) at 60°C overnight. After digestion, the scaffold was stained with PicoGreen according to the manufacturer's protocol and measured at 480/520 nm wavelength in a plate reader (SpectraMax M2e Microplate Reader; Molecular Devices, San Jose, CA). The data from days 7 to 14 were normalized by the DNA amount of the scaffolds at day 0.
Macrophage hematoxylin and eosin staining
The scaffolds were collected at day 14, fixed in 4% paraformaldehyde (PFA) for 1 h, washed twice with DPBS, incubated for 24 h in optimal cutting temperature (OCT) compound (Fisher Healthcare, Houston, TX), and flash frozen in liquid nitrogen. The scaffolds were sectioned in 10-μm-thick slices using a LEICA CM 3050-S Cryostat (Wetzlar, Germany); the sections were washed in DPBS for 10 min and stained with Mayer's Hematoxylin (Sigma-Aldrich) solution for 3 min. The slides were washed in running tap water for 5 min, stained with Eosin-Y (Sigma-Aldrich) solution for 30 s and washed with 75%, 95%, 100% EtOH for 1 min each wash, then in Xylene solution, and finally covered by a cover glass with mounting media (Vector Laboratories, Burlingame, CA). The sections were imaged in a standard optical microscope at 40 × and 100 × magnification.
Macrophage immunofluorescence
The scaffold sections were washed in DPBS once and blocked with 5% bovine serum albumin (BSA) in DPBS with 0.3% Triton X-100 for 20 min. Then, the antibodies (CD68, CCR5, CD71) were added directly to the scaffold sections (1:100) in 5% BSA in DPBS with 0.3% Triton X-100 and incubated in the dark for 1 h. The sections were washed in DPBS three times for 5 min. Secondary antibodies were added (Alexa 488, 584) at 1:500 in 5% BSA in DPBS for 1 h. Finally, 4′,6-diamidino-2-phenylindole-containing mounting dye was added to the top of the scaffold sections, and a cover slide was put on top of the sections.
Macrophage polarization from M0 to M1 and M2 types
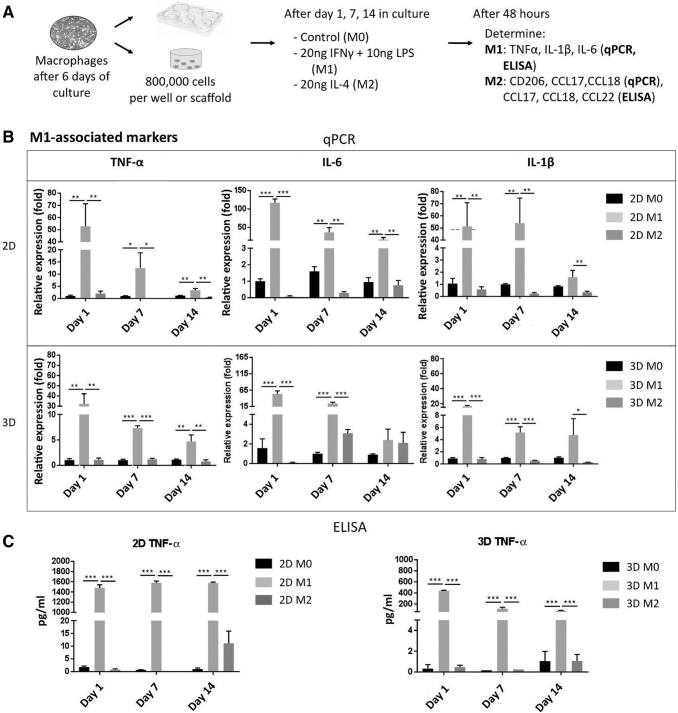
A summary of the experimental steps is described in Figure 3A. Undifferentiated M0 macrophages seeded in the 3D cultures were polarized at days 1, 7, and 14 after initial culture with 20 ng/mL interferon gamma (IFNγ; PeproTech) and 10 ng/mL of lipopolysaccharide (LPS; Sigma-Aldrich) for 48 h for the proinflammatory M1 phenotype; 20 ng/mL IL-4 (PeproTech) was used to polarize M0 to the anti-inflammatory M2 phenotype. RNA and supernatants from the scaffolds were collected for either 2D or 3D cultures after 48 h of polarization, that is, at days 3, 9, and 16 after the initial culture.
FIG. 3.
Macrophage polarization in 2D and 3D cultures to M1. (A) Procedure for macrophage polarization in 2D and 3D cultures. Macrophages in the 3D scaffolds were polarized to either M1 or M2 phenotypes at 1, 7, and 14 days. (B) Expression levels of TNF-α, IL-6, and IL-1β genes in M0, M1, and M2 macrophages to validate macrophage M1 polarization under 2D (top) or 3D (bottom) culture at days 1, 7, and 14. (C) TNFα concentration in the conditioned medium by M0, M1, and M2 on days 1, 7, and 14 under 2D (left) and 3D (right) cultures, which was determined by ELISA. Each value represents the mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001, compared with either 2D M0 or 3D M0 cultures. 2D, two-dimensional; TNFα, tumor necrosis factor alpha; IL, interleukin; ELISA, enzyme-linked immunosorbent assay.
RNA isolation and quantitative reverse transcription/polymerase chain reaction
RNA from the collected scaffolds was obtained by homogenizing them in 550 μL of TRIzol with an RNase-Free Disposable Pellet Pestle (Fisherbrand, Waltham, MA) and vortexed for 5 min. One hundred thirty-seven microliters of chloroform was added, vortexed for 15 s, incubated for 5 min at room temperature, and centrifuged at 12,000 g for 15 min at 4°C. The aqueous phase was separated from the TRIzol layer, and the RNeasy Micro RNA Purification Kit (QIAGEN, Venlo, Limburg, Netherlands) protocol was followed. The RNA concentration and quality were checked by NanoDrop (Thermo Fisher Scientific).
RNA (200 ng) from each sample was reverse transcribed into complementary DNA with the iScript Reverse Transcription Supermix for reverse transcription/quantitative polymerase chain reaction (RT-qPCR; Bio-Rad, Hercules, CA). qPCR was performed with TaqMan (Applied Biosystems, Foster City, CA) chemistry in an ABI7900 HT Sequencing Detection System (Thermo Fisher Scientific) using the probes for genes TNFα, IL-1β, and IL-6 for M1 macrophages, CCL17, CCL18, and CD206 (mannose receptor C type 1) for M2 macrophages, and GAPDH as the internal control. The results were quantified by the −ΔΔCt method to determine gene expression, and the data were normalized with respect to M0 gene expression at the respective day (3, 9, or 16).
Enzyme-linked immunosorbent assay
The conditioned media of the polarization experiments were collected, as indicated above. The secretion of cytokines TNFα, IL-6, and IL-1β by M1 macrophages was confirmed using the respective Enzyme-Linked Immunosorbent Assay (ELISA) Kits. TNFα ELISA Kit was purchased from Invitrogen, and IL-6 and IL-1β ELISA Kits were purchased from R&D Systems (Minneapolis, MN). M2 macrophage cytokine secretion of CCL17 and CCL22 (thymus and activation-regulated chemokine) were also assessed by their correspondent ELISA Kits (R&D Systems). ELISA assays were performed in accordance with the manufacturer's protocols. Finally, the optical absorbance at 450 nm was determined with a plate reader.
Human mesenchymal stem cell 2D culture and expansion
Human bone marrow-derived MSCs were isolated from the femoral heads of patients undergoing total hip arthroplasty with Institutional Review Board approval (University of Washington and University of Pittsburgh). hMSCs were grown in standard tissue culture plates at a density of 5.7 × 103 cells/cm2 in Gibco Dulbecco's modified Eagle's medium (DMEM) high glucose (Life Technologies) supplemented with 10% hMSC certified FBS (Invitrogen), antibiotic/antimycotic solution and fibroblast growth factor 2 (RayBiotech, Norcross, GA). hMSCs were cultured until reaching 80% confluency, and then the cells were harvested and counted for either passaging or 3D culture. hMSCs used in this study were pooled cells from 10 male and 10 female donors with ages ranging from 30 to 86 years. Cells from passages 4 to 6 were used for the experiments.
Osteogenesis culture
hMSCs (250,000 cells) only were cultured in scaffolds in osteogenic differentiation medium (DMEM, 10% FBS, 1% antibiotic and antimycotic solution, 50 μM l-ascorbic acid [Sigma-Aldrich], 100 nM dexamethasone [Sigma-Aldrich; from day 1 to 14 only], 10 mM β-glycerophosphate [MP Biomedicals], 100 mM Vitamin D3 [Sigma-Aldrich], and 100 ng/mL bone morphogenetic protein 7 [BMP-7; PeproTech]) for 4 weeks.
Osteogenesis co-culture
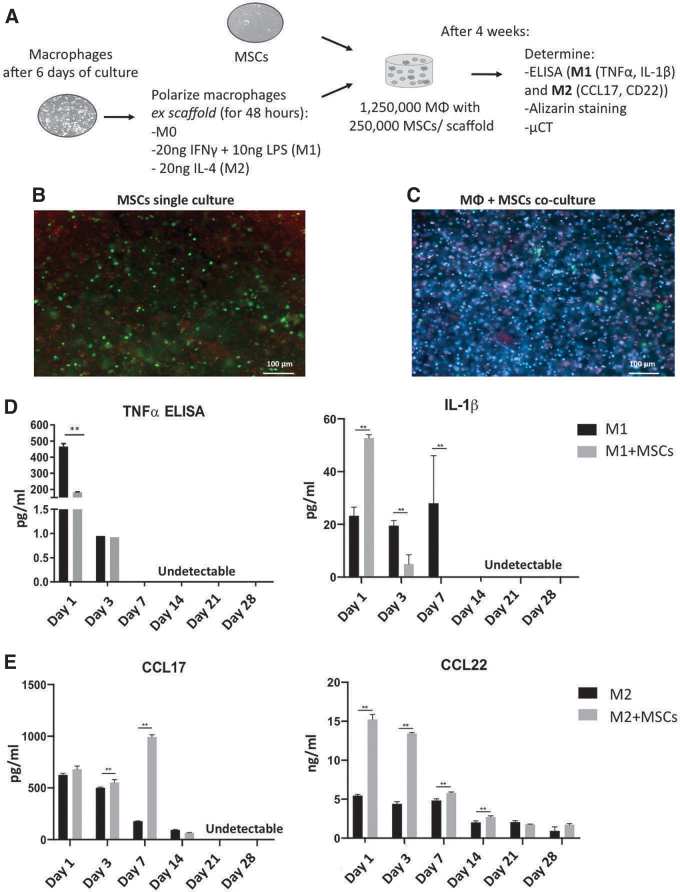
A detailed outline is presented in Figure 5A. Undifferentiated (M0), M1, and M2 macrophages were co-cultured with hMSCs at a ratio of 5:1 (1,250,000 cells:250,000 cells) in GelMA as described above. The 5:1 ratio was found to be the optimal ratio in our previous studies.8 hMSC (250,000 cells) monocultures were seeded in scaffolds as a negative control.
FIG. 5.
Effects of hMSCs on macrophage polarization after 4 weeks of co-culture in 3D scaffolds. (A) hMSCs and macrophage co-culture strategy. Macrophages were first polarized to either M1 or M2, and then mixed with hMSCs in GelMA for the co-culture experiment. No polarization medium was used in the co-culture. (B) Live (green)/Dead (red) staining of hMSCs only culture. (C) Live (blue)/Dead (red) staining of hMSCs (green) co-cultured with M0 macrophages. (D) Secretion profile of M1-associated cytokines TNFα and IL-6 in M1 monocultures and in M1 + hMSC co-cultures and M1 macrophages in osteogenic medium. (E) Secretion profile of M2-associated cytokines CCL22 and CCL17 of M2 monocultures and M2 + hMSC co-cultures in osteogenic medium. hMSCs secretion was depreciable in all the cytokine profiles. Each value represents the mean ± SD. **p < 0.01 compared with either 3D M1 or 3D M2 monocultures. hMSC, human mesenchymal stem cell. Color images are available online.
The scaffolds were cultured in a co-culture medium (DMEM, 10% FBS, 1% antibiotic and antimycotic solution, 50 μM l-ascorbic acid [Sigma-Aldrich], 100 nM dexamethasone [Sigma-Aldrich, added from day 0 to 14], 10 mM β-glycerophosphate [MP Biomedicals], 100 mM Vitamin D3 [Sigma-Aldrich], 100 ng/mL BMP-7 [PeproTech], and 50 ng/mL of M-CSF) for 4 weeks.
Evaluation of osteogenesis
Alkaline phosphatase assay
The co-culture scaffolds were collected after 2 weeks of culture. The scaffolds were homogenized in 200 μL of the alkaline phosphatase (ALP) buffer (ABCAM, Burlingame, CA) with a pestle, and then the mixture (scaffold-buffer) was centrifuged at 10,000 g for 15 min at 4°C. Then, the ALP Kit protocol was followed. The monocultures of hMSCs in growth and co-culture medium were considered as the negative and positive controls, respectively.
Alizarin Red staining
In brief, after 4 weeks of culture, the scaffolds were collected, fixed in 2% PFA for 1 h, and washed twice with DPBS.
Section staining
The scaffolds were incubated for 24 h in OCT compound and frozen in liquid nitrogen. The scaffolds were sectioned into 10-μm-thick slices in a LEICA CM 3050-S Cryostat (Wetzlar, Germany); then the sections were washed in DPBS for 2 min, and stained with 40 mM Alizarin Red (Sigma-Aldrich) solution (with a pH of 4.1–4.3) in deionized water (DI H2O) for 3 min. The slides were washed three times with DI H2O for 5 min each. The sections were imaged in an inverted microscope at 100 × magnification.
Scaffold staining
The scaffolds were stained with 40 mM (pH 4.1–4.3) Alizarin Red (Sigma-Aldrich) solution in DI H2O for 15 min and washed five times in DI H2O for 10 min each. Ten percent cetylpyridinium chloride was used to destain the scaffolds, 100 μL aliquots of the destain supernatant of each scaffold were transferred to a 96-well plate, and absorbance was measured at 562 nm.
Micro-computerized tomography
Fixed scaffolds were imaged in a SkyScan 1276 microcomputerized tomography (μCT) machine (Bruker Scientific Instruments, Billerica, MA) at a resolution of 18 μm with a binning of 1008 × 672 and an average of three images per scan. The image analysis was done using CTAN (Billerica, MA) with 210 as the lower threshold and 255 as the upper threshold.
Statistical analysis
One-way analysis of variance with Tukey comparisons and student's t-test were conducted using PRISM 7 (GraphPad Software, San Diego, CA). All experiments were done in triplicates. All the data were graphed as mean ± standard deviation, and the statistical significance threshold was set as *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Characterization of isolated monocytes and macrophages
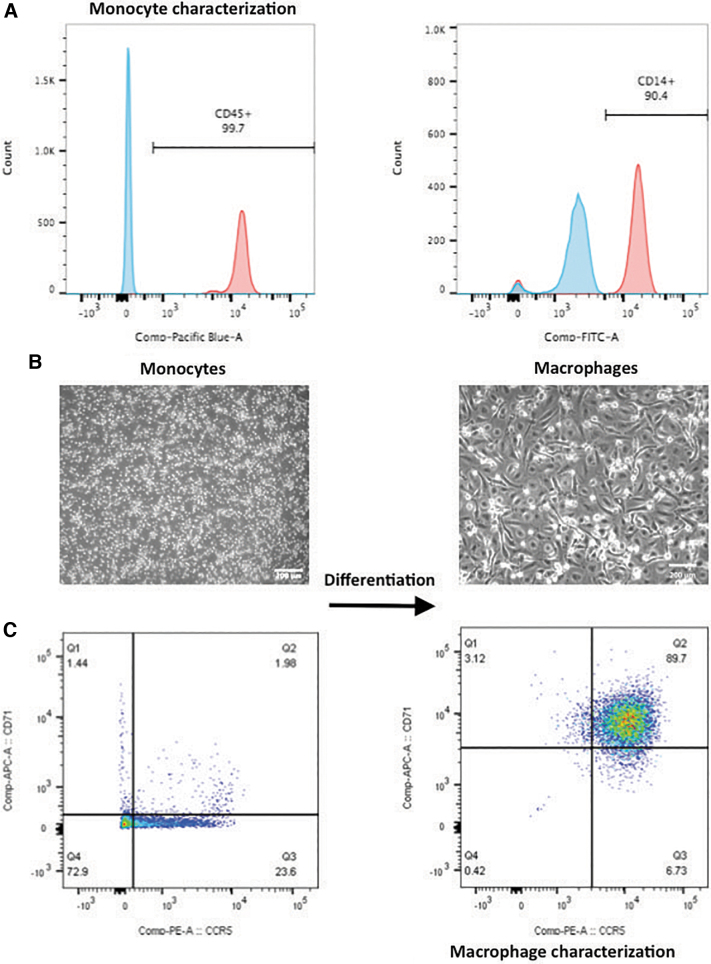
The immunophenotype of the isolated cells was confirmed by FACS (Fig. 1A), with a CD45-positive population of 99.7%, indicating that almost all of the cells were mononuclear leukocytes. In addition, the CD14-positive population represented more than 90% of the total isolated cells, confirming that the isolated cells were mostly monocytes with few leukocytes of other types.
FIG. 1.
Monocyte and macrophage characterization by FACS. (A) Monocyte characterization by FACS. Approximately 99.7% of the isolated cells were leucocytes and 90% of those cells expressed the monocyte-specific marker CD14. (B) (left) Monocyte morphology at day 1 after cell isolation; (right) monocytes cultured in macrophage-stimulating medium after 6 days (scale bar = 200 μm). (C) Monocyte to macrophage differentiation was confirmed by FACS, as indicated by upregulation of macrophage markers CCR5 and CD71 in monocytes after 6 days of culture in macrophage-stimulating medium. FACS, fluorescence-activated cell sorting. Color images are available online.
After 6 days of culture of the monocytes in macrophage-stimulating medium (MSM), changes in cell morphology were observed, passing from round shape to spindle morphology (Fig. 1B). FACS data verified that the monocytes differentiated into mature macrophages, as the expression of CCR5 (MIP-1α receptor) and CD71 (transferrin receptor protein 1) macrophage markers were higher in the monocytes cultured in MSM for 6 days in comparison to the freshly isolated monocyte population (Fig. 1C, left). The overall percentage of CCR5- and CD71-positive cells after 6 days of monocyte culture in MSM was ∼89% (Fig. 1C, right), indicating that 89% of the initial monocyte population differentiated into macrophages.
Macrophage viability decreased slightly after 2 weeks of culture in the 3D scaffolds
Live/Dead staining showed that the majority of the cells were viable at days 7 and 14 (Fig. 2B), indicating that most of the cells in the scaffold survived for at least 2 weeks. A quantitative analysis of the live cell percentage is shown in Figure 2C. We observed a cell survival rate of 82.90% ± 2.53% (with respect to the initial culture) at day 7, and 74.87% ± 2.52% at day 14 of macrophages in 3D culture. These data suggest that macrophages displayed minimal proliferation in the 3D scaffold; however, they were able to secrete the cytokines associated with the macrophage phenotypes (as shown below).
Macrophage morphology and distribution in the scaffold was assessed by hematoxylin and eosin staining (Fig. 2D and Supplementary Fig. S1) and immunofluorescence detection of CCR5 and CD71 (Fig. 2E), as well as CCR5 and CD68 (an intracellular maker for macrophages; Fig. 2F). The staining results showed that macrophages were homogenously distributed throughout the scaffold.
Macrophages were successfully polarized in the 3D scaffolds
Macrophages were polarized to either M1 or M2 phenotype in the 2D and 3D cultures. Their activation was assessed by analyzing the changes in gene expression and secretory profiles in the two cell-culture settings.
The macrophages polarized with LPS + IFNγ were successfully activated to M1 macrophages, as shown by the upregulated gene expression and increased cytokine secretion of M1-associated genes/cytokines (TNFα, IL-6, and IL-β) at days 1, 7, and 14 in 2D and 3D cultures (Fig. 3B), in comparison to the M0 macrophages (p < 0.05). Gene expression increase was higher in the 2D culture than in the 3D culture, and decreased with time in both cases. A similar trend was observed in the ELISA data, where the cytokine secretion profiles reached higher concentrations in the 2D than in 3D cultures, and the concentrations diminished over time (Fig. 3C and Supplementary Fig. S2).
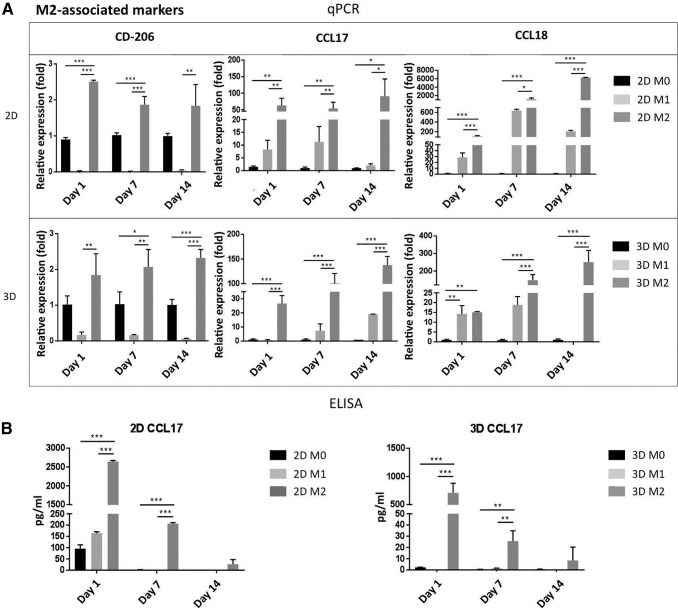
Similarly, the macrophages stimulated with IL-4 showed significantly increased CD206, CCL17, CCL18 gene expression (Fig. 4A) in the 2D than in the 3D culture. However, contrary to the M1 gene expression profile, the expression of the M2 associated genes increased with time in both the 2D and the 3D cultures with respect to M0 macrophages. Secretion of the M2-associated cytokines (CCL17, CCL18, and CCL22) was also higher in the 2D than in the 3D culture (Fig. 4B and Supplementary Fig. S3). The M2 cytokine secretions from the 2D culture were almost double those observed in the 3D culture, and the concentrations decreased during subsequent days. Overall, these results suggest that macrophages embedded in the 3D scaffold were able to polarize into M1 and M2 phenotype after 48 h of stimulation at 1, 7, and 14 days in culture.
FIG. 4.
Macrophage polarization in 2D and 3D cultures to M2. (A) Expression levels of CD-206, CCL17, and CCL18 genes in M0, M1, and M2 macrophages, respectively, to validate macrophage M2 polarization in 2D (top) and 3D (bottom) cultures at days 1, 7, and 14. (B) CCL17 concentration in the conditioned medium by M0, M1, and M2 at days 1, 7, and 14 under 2D (left) and 3D (right) cultures, which was determined by ELISA. Each value represents the mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001, compared with either 2D M0 or 3D M0 cultures. CCL, C–C motif chemokine ligand.
Cell viability in 3D co-culture
Images of live/dead staining of hMSCs in culture alone and in co-culture with macrophages are shown in Figure 5B and C respectively. The majority of cells in both conditions survived after 4 weeks of culture. It is noteworthy that PicoGreen DNA assay-based assessment of cell viability might not reliably differentiate between live hMSCs and live macrophages, because of the different DNA content in each cell type.
hMSCs polarized M1 and M2 macrophages toward an anti-inflammatory phenotype
To determine the effects of hMSCs on macrophages, we compared the cytokine production profile of macrophages monoculture with the macrophage co-culture with hMSCs. The ELISA data presented in Figure 5 showed that a substantial decrease in M1-cytokine secretion (TNFα and IL-1β; Fig. 5D) in the co-cultures compared with the M1 macrophage monoculture (p < 0.05). In addition, when M2 macrophages were co-cultured with hMSCs, the secretion of anti-inflammatory cytokines (CCL17, CCL22; Fig. 5E) significantly increased in comparison to M2 cultures alone.
Macrophages co-cultured with hMSCs enhances hMSC osteogenic differentiation
After 2 weeks in culture, hMSC 3D cultures were harvested for ALP activity assay. The hMSC constructs containing co-cultured M0, M1, or M2 macrophages displayed higher ALP activity than the control group of hMSCs cultured alone (Supplementary Fig. S4). In particular, the cocultures with M1 macrophages showed the highest ALP levels (p < 0.01) of all tested groups.
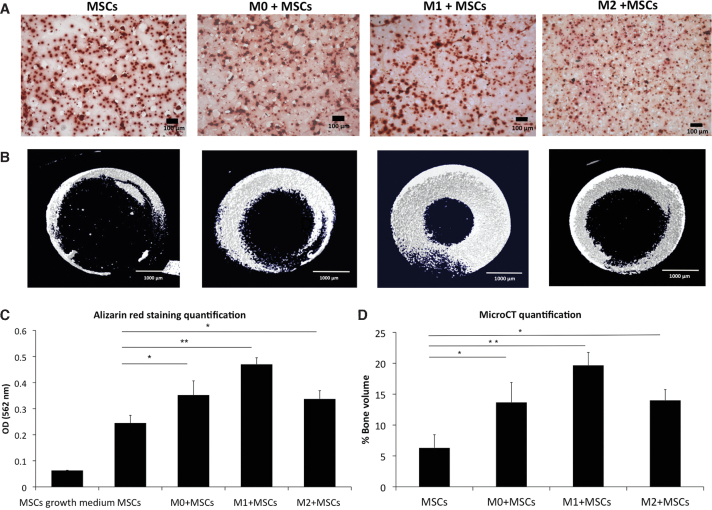
Osteogenesis was further assessed by examining matrix calcification with Alizarin Red staining (Fig. 6A) and μCT (Fig. 6B). The results from these two methods showed similar trends to those from ALP activity assay, namely greater calcium deposition and mineralization was seen in the co-culture constructs than in hMSC monoculture. Moreover, the M1 co-culture scaffolds had the highest calcium deposition and matrix mineralization compared with the other macrophage co-culture phenotypes (Fig. 6C, D), suggesting a potential enhancement effect of macrophages in the M1 cell state on early bone formation.
FIG. 6.
hMSC osteogenesis after 4 weeks of 3D co-culture with macrophages. (A) Alizarin Red staining to assess the calcification in the groups that originally contained hMSC only (MSCs) or hMSC co-cultured with M0 (M0 + MSCs), M1 (M1 + MSCs), or M2 (M2 + MSCs). The red dots in the images show the calcium deposits in the extracellular matrix (scale bar = 100 μm). (B) Representative μCT images of extracellular matrix mineralization in hMSCs monoculture and co-cultures of hMSCs with M0, M1, or M2 macrophages (scale bar = 1000 μm). (C) Quantification of Alizarin Red staining of hMSCs in growth medium, MSCs in osteogenic medium and M0, M1 and M2 co-cultures with MSCs in osteogenic medium in 3D scaffolds. (D) μCT quantification of mineralization in hMSC monocultures and hMSCs + macrophages co-cultures. Each value represents the mean ± SD. *p < 0.05, **p < 0.01, compared with hMSC monocultures in osteogenic medium. μCT, microcomputerized tomography. Color images are available online.
Discussion
Macrophages play a critical role not only in normal bone tissue homeostasis and bone regeneration, but also in various pathological conditions impacting bone and surrounding tissues. To this end, we have established, using a photocrosslinked GelMA hydrogel scaffold, an in vitro 3D culture model of macrophages, as well as co-cultured macrophages and hMSCs, to investigate the biological interactions between the innate immune system and MSCs undergoing osteogenic differentiation.
We first analyzed the effect of 3D culturing on macrophage behavior. Macrophages displayed good survival to at least 2 weeks in the 3D culture, and maintained surface marker expression as well as the ability to polarize to M1 and M2 phenotypes. However, lower levels of both pro- and anti-inflammatory gene expression and cytokine secretion were seen in 3D culture compared with 2D cultures.
Some research groups have tried to expound upon these 2D versus 3D cytokine secretion differences in cell behavior using other cell types and materials. Donaldson et al. and other groups demonstrated a similar trend of lower proinflammatory gene expression and cytokine secretion of monocytes grown in 3D than in 2D cultures using GelMA gels.12–14 Their data showed that a percentage of proinflammatory stimuli, such as LPS, as well as secreted cytokines (TNFα, IL-1β, and IL-6), were trapped by the gel itself.
It is thought that the gelatin amino acid structure enhances cytokine (particularly proinflammatory cytokines) binding to GelMA12 but more studies need to be done to better understand the interaction between cytokines and gelatin. This “mop-up” or sequestering effect limits the availability of the stimuli factors for cells to respond to, causing a decrease in gene expression and cytokine secretion, while also reducing the total detected protein secretion in the supernatant as compared with the 2D culture. Of note, this might also be the case in vivo, where various extracellular matrix components could bind cytokines and other signaling molecules, limiting their immediate effects.
Another possible explanation of the differences observed between our 2D and 3D macrophage systems might be attributed to the intrinsic mechanical and chemical properties of the 3D system itself, such as porosity, stiffness, and adhesion sites. For example, podosomes are specialized contact cell structures between the cell and the extracellular matrix,15 and the distribution of podosomes in macrophages differs in 2D versus 3D.16
In 2D cultures, macrophages' podosomes are distributed throughout the entire cell surface that is in contact with the culture plate. However, the development of cell podosomes in 3D cultures is contingent on the availability of cell adhesion sites throughout the extracellular matrix, which also dictates changes in cell morphology for different geometries.17 This heterogeneity in the substrate will also impede cell–cell contacts between macrophages, as fewer podosomes form per cell and the decreased intercellular interaction will result in an attenuated pro- or anti-inflammatory response, since macrophages depend on cell–cell communication.18
Interestingly, we observed marked crosstalk between hMSCs and macrophages in 3D culture. While the secretion of proinflammatory cytokines by M1 macrophages was reduced in the presence of hMSCs, the secretion of anti-inflammatory cytokines increased in M2 macrophage coculture with hMSCs. These immunomodulatory effects of hMSCs on macrophages are consistent with previous reports in vitro and in vivo.19–21
Moreover, we demonstrated that the co-culture of M0, M1, or M2 macrophages with hMSCs enhanced osteogenesis in 3D gels. The ALP, μCT, and Alizarin Red results showed that matrix mineralization increased significantly more in hMSCs co-cultured with macrophages than hMSCs monocultures. Specifically, the co-culture of hMSCs with M1 phenotype exhibited the highest calcium deposition compared with hMSCs only. These results suggest that the stage of acute inflammation (M1 polarization) is necessary to “prime” the local milieu for optimal recovery and bone healing, as observed in a previous study.8 However, our previous work used murine cells in a 2D format; the current study continues to expand upon these paradigms and uses human primary cells in a 3D format to further approximate in vivo physiology.
Our results highlight the importance of crosstalk between macrophages and MSCs during osteogenesis in 3D cultures. There have been attempts by other groups to incorporate monocytes or macrophages into 3D tissue systems.22–27 Nevertheless, the results are controversial; while some groups have shown a similar synergistic effect in bone formation when hMSCs are co-cultured with macrophages,25–27 others have observed the opposite effect of macrophages on bone formation.22–24 It is important to acknowledge that these co-culture studies differ from the current one in cell species type (murine vs. human), monocyte or macrophage type, hMSC origin, and scaffold material.
Because of the complexity of the immune cell culture system, most of the innate immune cells and MSCs co-culture studies have used established cell lines of monocytes or macrophages (Raw 274.6, a murine macrophage cell line or THP-1, a human monocyte cell line), or systems with no direct cell–cell contact of macrophages and MSCs in the cultures, that is, macrophages placed on top of the gel instead of mixed together with MSCs in the 3D hydrogel, as we have shown in the present study. While established cell lines, such as THP-1 cells, avoid the cellular variability observed among patients, the secretory gene, and proliferation profiles differ significantly from those of the average healthy human primary macrophages.28
Furthermore, the use of human primary cells allows the study of specific genders, ages, or health states of donors and, with this, the potential of future translational applications. Investigation of the different 3D in vitro models is limited, and further studies are needed to establish an ideal platform for engineering bone tissue.
There are some limitations in our study that will need to be assessed. GelMA was selected as the scaffold of choice for our engineered bone tissue. This gelatin-derived material was selected because its structure offers more control of its physical (e.g., stiffness) and chemical properties compared with other natural scaffolds such as collagen or gelatin itself, making it a useful surrogate for bone tissue engineering.29,30 However, Donaldson and others12 observed that the viability of macrophages in 15% GelMA decreased with time, which could be related to the density of the matrix.
Further studies are needed to optimize the characteristics of the scaffold so that it is able to support macrophage proliferation without limiting bone formation. Importantly, it is noteworthy that 3D scaffolds allow greater potential to manipulate matrix geometries and architectures to achieve a more physiologically relevant microenvironment that mimics that of tissues, such as bone, in vivo.
In conclusion, we have described and tested a novel 3D model for investigating the interactions between macrophages (M0, M1, and M2) and hMSCs during osteogenesis. Our results provide evidence that macrophages enhance MSC-mediated bone formation in 3D GelMA matrices, especially under the proinflammatory phenotype. This observation has direct significance in bone tissue engineering and in the development of novel cell-based bone graft substitutes. The present study forms the foundation for our future efforts to engineer a physiologically relevant in vitro model to simulate healthy and diseased MSK tissues.
Supplementary Material
Acknowledgments
The authors would like to thank Yusuke Kohno, MD, PhD, Masaya Ueno, MD, PhD, Takeshi Utsunomiya, MD, PhD, and Cristobal Franco-de-Leon, PhD. They also thank Timothy Doyle, PhD for his support in the MicroCT at the Stanford Small Animal Imaging Service Center and to the NIH on the S10 grant-funded Bruker Skyscan 1276 MicroCT.
Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the National Institutes of Health grant UG3TR002136.
Supplementary Material
References
- 1. Barbour K.E., Helmick C.G., Boring M., and Brady T.J.. Vital signs: prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation—United States, 2013–2015. MMWR Morb Mortal Wkly Rep 66, 246, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hootman J.M., Helmick C.G., Barbour K.E., Theis K.A., and Boring M.A.. Updated projected prevalence of self-reported doctor-diagnosed arthritis and arthritis-attributable activity limitation among US adults, 2015–2040. Arthritis Rheumatol 68, 1582, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fang Y., and Eglen R.M.. Three-dimensional cell cultures in drug discovery and development. SLAS Discov 22, 456, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duval K., Grover H., Han L.-H., et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology 32, 266, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 5. Eglen R.M., and Klein J.-L.. Three-dimensional cell culture: a rapidly emerging approach to cellular science and drug discovery. SLAS Discov 22, 453, 2017 [DOI] [PubMed] [Google Scholar]
- 6. Murray P.J., Allen J.E., Biswas S.K., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murray P.J. Macrophage polarization. Annu Rev Physiol 79, 541, 2017 [DOI] [PubMed] [Google Scholar]
- 8. Lu L.Y., Loi F., Nathan K., et al. Pro-inflammatory M1 macrophages promote Osteogenesis by mesenchymal stem cells via the COX-2-prostaglandin E2 pathway. J Orthopaed Res 35, 2378, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loi F., Córdova L.A., Zhang R., et al. The effects of immunomodulation by macrophage subsets on osteogenesis in vitro. Stem Cell Res Ther 7, 15, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennett W.E., and Cohn Z.A.. The isolation and selected properties of blood monocytes. J Exp Med 123, 145, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin H., Cheng A.W.-M., Alexander P.G., Beck A.M., and Tuan R.S.. Cartilage tissue engineering application of injectable gelatin hydrogel with in situ visible-light-activated gelation capability in both air and aqueous solution. Tissue Eng Part A 20, 2402, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donaldson A.R., Tanase C.E., Awuah D., et al. Photocrosslinkable gelatin hydrogels modulate the production of the major pro-inflammatory cytokine, TNF-α, by human mononuclear cells. Front Bioeng Biotechnol 6, 116, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benton J.A., DeForest C.A., Vivekanandan V., and Anseth K.S.. Photocrosslinking of gelatin macromers to synthesize porous hydrogels that promote valvular interstitial cell function. Tissue Eng Part A 15, 3221, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frasca G., Cardile V., Puglia C., Bonina C., and Bonina F.. Gelatin tannate reduces the proinflammatory effects of lipopolysaccharide in human intestinal epithelial cells. Clin Exp Gastroenterol 5, 61, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Linder S., and Kopp P.. Podosomes at a glance. J Cell Sci 118, 2079, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Wiesner C., Le-Cabec V., El Azzouzi K., Maridonneau-Parini I., and Linder S.. Podosomes in space. Cell Adhes Migr 8, 179, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen S., Jones J.A., Xu Y., Low H.-Y., Anderson J.M., and Leong K.W.. Characterization of topographical effects on macrophage behavior in a foreign body response model. Biomaterials 31, 3479, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fortes F.S.A., Pecora I.L., Persechini P.M., et al. Modulation of intercellular communication in macrophages: possible interactions between GAP junctions and P2 receptors. J Cell Sci 117, 4717, 2004 [DOI] [PubMed] [Google Scholar]
- 19. Kim J., and Hematti P.. Mesenchymal stem cell-educated macrophages: a novel type of alternatively activated macrophages. Exp Hematol 37, 1445, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eggenhofer E., and Hoogduijn M.J.. Mesenchymal stem cell-educated macrophages. Transplant Res 1, 12, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vasandan A.B., Jahnavi S., Shashank C., Prasad P., Kumar A., and Prasanna S.J.. Human mesenchymal stem cells program macrophage plasticity by altering their metabolic status via a PGE2-dependent mechanism. Sci Rep 6, 38308, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang H., Husch J.F.A., Zhang Y., Jansen J.A., Yang F., and van den Beucken J.J.J.P.. Coculture with monocytes/macrophages modulates osteogenic differentiation of adipose-derived mesenchymal stromal cells on poly(lactic-co-glycolic) acid/polycaprolactone scaffolds. J Tissue Eng Regen Med 13, 785, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang H., Zhang Y., Jansen J.A., and van den Beucken J.J.J.P.. Effect of monocytes/macrophages on the osteogenic differentiation of adipose-derived mesenchymal stromal cells in 3D co-culture spheroids. Tissue Cell 49, 461, 2017 [DOI] [PubMed] [Google Scholar]
- 24. Jeon O.H., Panicker L.M., Lu Q., Chae J.J., Feldman R.A., and Elisseeff J.H.. Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci Rep 6, 26761, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Böse T., Unger R.E., Jansen J.A., Kirkpatrick C.J., and van den Beucken J.J.J.P.. Macrophage type modulates osteogenic differentiation of adipose tissue MSCs. Cell Tissue Res 369, 273, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guihard P., Danger Y., Brounais B., et al. Induction of osteogenesis in mesenchymal stem cells by activated monocytes/macrophages depends on oncostatin M signaling. Stem Cells 30, 762, 2012 [DOI] [PubMed] [Google Scholar]
- 27. Pirraco R.P., Reis R.L., and Marques A.P.. Effect of monocytes/macrophages on the early osteogenic differentiation of hBMSCs. J Tissue Eng Regen Med 7, 392, 2013 [DOI] [PubMed] [Google Scholar]
- 28. Heil T.L., Volkmann K.R., Wataha J.C., and Lockwood P.E.. Human peripheral blood monocytes versus THP-1 monocytes for in vitro biocompatibility testing of dental material components. J Oral Rehabil 29, 401, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Dong Z., Yuan Q., Huang K., Xu W., Liu G., and Gu Z.. Gelatin methacryloyl (GelMA)-based biomaterials for bone regeneration. RSC Adv 9, 17737, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Celikkin N., Mastrogiacomo S., Jaroszewicz J., Walboomers X.F., and Swieszkowski W.. Gelatin methacrylate scaffold for bone tissue engineering: the influence of polymer concentration. J Biomed Mater Res Part A 106, 201, 2018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.