Abstract
T cells expressing the cutaneous lymphocyte antigen (CLA) mediate pathogenic inflammation in atopic dermatitis (AD). The molecular alterations contributing to their dysregulation remain unclear. With the aim to elucidate putative altered pathways in AD we profiled DNA methylation levels and miRNA expression in sorted T cell populations (CD4+, CD4+CD45RA+ naïve, CD4+CLA+, and CD8+) from adult AD patients and healthy controls (HC). Skin homing CD4+CLA+ T cells from AD patients showed significant differences in DNA methylation in 40 genes compared to HC (p < 0.05). Reduced DNA methylation levels in the upstream region of the interleukin-13 gene (IL13) in CD4+CLA+ T cells from AD patients correlated with increased IL13 mRNA expression in these cells. Sixteen miRNAs showed differential expression in CD4+CLA+ T cells from AD patients targeting genes in 202 biological processes (p < 0.05). An integrated network analysis of miRNAs and CpG sites identified two communities of strongly interconnected regulatory elements with strong antagonistic behaviours that recapitulated the differences between AD patients and HC. Functional analysis of the genes linked to these communities revealed their association with key cytokine signaling pathways, MAP kinase signaling and protein ubiquitination. Our findings support that epigenetic mechanisms play a role in the pathogenesis of AD by affecting inflammatory signaling molecules in skin homing CD4+CLA+ T cells and uncover putative molecules participating in AD pathways.
Subject terms: Immunology, Adaptive immunity, Cytokines, Gene regulation in immune cells, Inflammation, Lymphocytes, Immunological disorders, Skin diseases, Epigenetics, Genome, Gene ontology, Gene regulatory networks, Microarrays
Introduction
Atopic dermatitis (AD) is a common chronic inflammatory skin disorder characterized by intense pruritus and xerosis that usually concur with allergic sensitization and elevated plasma immunoglobulin E (IgE) levels1. In recent years, epigenetic modifications are being recognized as key players in the alterations leading to complex inflammatory diseases including AD2–7. By affecting DNA methylation and non-coding RNA levels, these epigenetic modifications determine a complex network of chemical signatures that regulate mRNA expression and are heritable to daughter cells. Therefore, they control the activation or silencing of genes that are critical for cell memory and homeostasis, by regulating which genes are expressed, when, and where8. DNA methylation is an epigenetic modification involving the addition of a methyl group in a cytosine preceding a guanine (CpG site); usually when gene promoters are methylated the gene transcription is suppressed while reduced methylation levels activate genes and promote their expression. When CpG sites have significant differences in DNA methylation between patients and controls, they are named differentially methylated probes (DMPs), and if many of them are close to each other in a gene it is defined as a differentially methylated region (DRM). The contribution of this epigenetic modification in AD has been evaluated in diverse tissues, for instance, DNA methylation in the interleukin 4 receptor gene (IL4R) as detected in cord blood samples was associated with AD at age 12 months9. Decreased methylation in the thymic stromal lymphopoietin (TSLP) promoter resulting in TSLP overexpression has been detected in lesional skin biopsies of AD patients10. DNA methylation differences have also been detected in the epidermis of AD patients11. Altered expression of microRNAs has also shown to play a crucial role in type 2 immunity and in promoting pathogenic mechanisms in AD12–15. However, very few studies have analyzed these epigenetic signatures in purified immune cells from AD patients3.
Circulating T cells can infiltrate human skin as a mechanism of immunosurveillance. They are identified by the expression of the cutaneous lymphocyte-associated antigen (CLA), a cell surface glycoprotein that interacts with E-selectin on the endothelium of postcapillary venules and allows their selective transmigration from the peripheral circulation to the dermis16. AD patients have a higher percentage of circulating CLA+ T cells compared to HC17,18 and the selective expansion of CLA+ T cells in severe AD involves populations with Th2 and Th22 profiles19. Indeed, the frequencies of CLA+ Th2 T cells were similarly expanded across all AD age groups compared with control subjects20,21. CLA+ T cells in the peripheral blood of patients with AD are also functionally disturbed, with increased expression of cytokine receptors that promote their Th2-skewed pattern19. Moreover, they induce IgE production in B cells, enhance eosinophil survival22 and display features of in vivo activation such as the spontaneous release of IL-13 and increased levels of CD25, CD40L and HLA-DR23. Circulating CD4+CLA+ T cells in patients with acute AD furthermore produce significantly higher levels of IL-4, IL-13 and tumor necrosis alpha compared to patients with chronic AD or healthy controls (HC)24. After skin homing, the CLA+ T cells form dermal infiltrates, secrete interferon gamma and become protected from activation-induced cell death25. Recent studies indicate that circulating CLA+ T cells share phenotype, function, and clonotypes with tissue resident CLA+ T cells in the skin26 in agreement with that blood CD4+CLA+ T cells are a reliable surrogate marker of the inflammatory events occurring in the skin16,27. It has also been observed that interaction with external signals from allergens or the skin microbiome can activate CLA+ T cells and promote their pro-inflammatory state28. Indeed, CLA+ T cells are memory cells and antigen-experienced, so they can recognize epitopes in airborne and food allergens29–32 as well as in Staphylococcus aureus18,33.
Even though the alterations in CLA+ T cells from AD patients are well documented34 their extensive molecular characterization, needed to clarify the pathogenesis of AD, is currently missing. Here we compared genome-wide DNA methylation levels and miRNA expression in four sorted blood lymphocyte populations (CD4+, CD4+CD45RA+ naïve, CD4+CLA+ and CD8+) isolated from AD patients and HC. This study shows for the first time that skin-homing CD4+CLA+ memory T cells from AD patients contain dysregulated epigenetic signatures, including differences in DNA methylation in 40 protein-coding genes and 16 differentially expressed miRNAs. An analysis integrating the coordinated response in miRNAs and CpG probes revealed two groups with strong antagonistic behaviors that recapitulated the differences between AD patients and HC. Functional analysis of the genes linked to these groups revealed their association with key cytokine signaling pathways and protein ubiquitination. Our findings support that epigenetic mechanisms play a role in the pathogenesis of AD by affecting inflammatory signaling molecules in skin-homing CD4+CLA+ memory T cells and reveal novel disease candidates.
Results
Purity of isolated T cell populations
Based on our previous observations that sorting by cell-type is crucial for the interpretation of results derived from DNA methylation studies35,36, we isolated four populations of peripheral blood lymphocytes (CD4+, CD4+CD45RA+ naïve, CD4+CLA+ and CD8+ from AD patients with severe phenotype and HC (Table 1). The mean cell surface expression of CD4 and CD8 markers was over 90% in the respective sorted fractions (Supplementary Table S1 online). The mean CD45RA expression was 90% in the sorted naïve cells. The CD45RO+ marker was expressed on 85% of the sorted CD4+CLA+ lymphocytes, confirming their phenotype as memory cells. After cell isolation, we found no significant differences in the mean proportions of sorted positive cells between AD patients and HC (t-test p > 0.05, Supplementary Table S1 online).
Table 1.
Descriptive characteristics of the participants at 450 ml blood donation.
| Healthy controls | AD patients | |
|---|---|---|
| Sample size (n)a | 10 | 10 |
| Age, years (average ± SD) | 37 ± 13.9 | 34.9 ± 14.7 |
| SCORAD (objective)b | n.a | 44 ± 5.5 |
| Total plasma IgE kUA/L, (median, IQR)c | 16.5 (8.2 – 22.5) | 1750 (725 – 3050) |
| Phadiatop ≥ 0.35 kU /L, (n)d | 1/10 | 10/10 |
| Asthma, yes (n)e | 0/10 | 4/10 |
| Rhinitis, yes (n)e | 0/10 | 10/10 |
aGiven the sample size and to avoid confounding effects by gender, only males were analyzed.
bSCORAD: SCORing Atopic Dermatitis. Moderate eczema (score 15 to 40); severe eczema (> 40); n.a = not applicable.
cDetermined by ImmunoCAP (Thermo Fisher, Uppsala, Sweden). Reference value in this population is 122 kUA/L; IQR = interquartile range.
dPhadiatop = analyses of IgE antibodies in plasma to a mix of aeroallergens including birch, timothy, mugwort, mite, cat, dog, horse and mold (Thermo Fisher).
eThe presence of physician-diagnosed asthma and/or rhinitis was obtained by questionnaire at the time of blood sampling.
Comparative analysis of DNA methylation between AD patients and HC
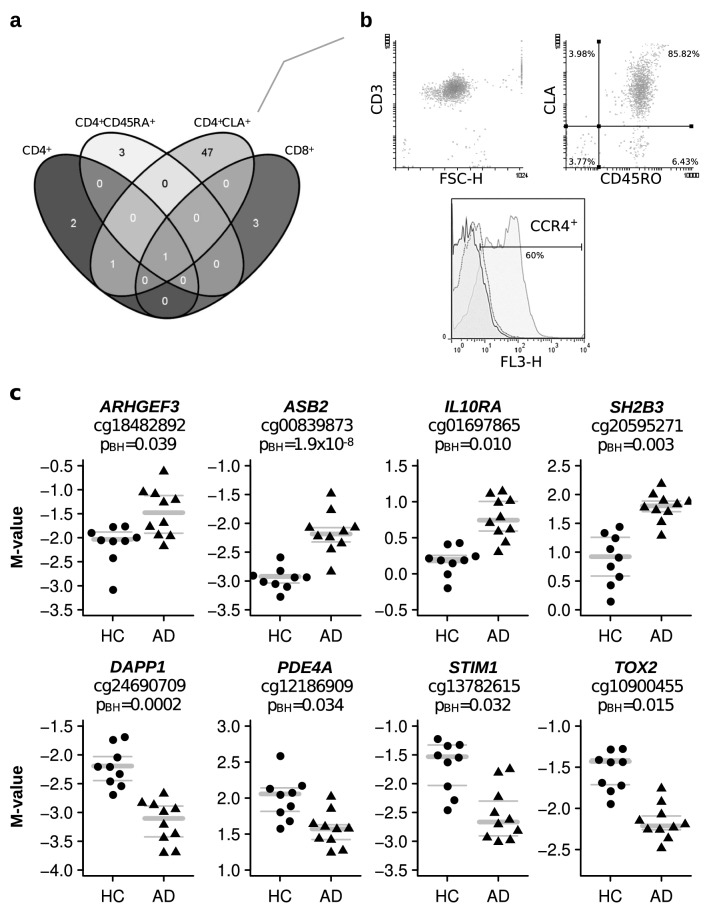
The analysis of the genome-wide DNA methylation levels in the four sorted T cell populations revealed that skin-homing CD4+CLA+ T cells were the subset with the largest numbers of differentially methylated CpG probes (DMPs, n = 49) between AD patients and HC (Fig. 1a). These cells expressed the CD3+ marker, as well as the CLA+ and the CD45RO+ markers, and in about 60% of the population the CCR4 receptor (Fig. 1b). The 49 DMPs in these circulating CD4+CLA+ T cells (BH corrected p value < 0.05) mapped to 35 genes (Table 2). Further analysis of DMRs revealed 5 genes with more than 3 DMPs in the CD4+CLA+ T cells of the AD patients compared to HC (Table 3). These results refined the signals detected in GPR55, MAN1A1 and CDHR3 by using the DMP analysis (Table 2) and uncovered methylation differences in the genes encoding the transcription factor estrogen receptor alpha (ESR1) and the nuclear receptor co-repressor 2 (NCOR2) (Table 3), suggesting that in AD patients the epigenome of circulating CD4+CLA+ T cells is affected in regions encoding transcription factors. The annotated list of 40 differentially methylated genes between AD patients and HC with their cell location and known function are presented in Table 4.
Figure 1.
Differentially methylated probes (DMPs) in peripheral blood T cells between AD patients and HC. (a) Venn diagram showing the overlap of DMPs in four different sorted T cell populations. Plotted with the open webtool venny 2.0 (https://bioinfogp.cnb.csic.es/tools/venny/). (b) Representative flow cytometry analysis of CD3, CLA, CD45RO and CCR4 in sorted CD4+CLA+ T cells. Numbers within quadrants represent percentage of cells. FSC-H: forward scatter height; in histogram solid black line: unstained; dotted line: isotype control; gray line: anti CCR4 staining. (c) Eight DMPs in CD4+CLA+ T cells. DNA methylation levels are expressed as M-values, gray bars indicate mean (bold), upper and lower (thin) quartiles. M values above 1 represent that the CpG site is methylated, and M values below − 1 represent that the CpG site is demethylated. Each dot represents an individual, HC (n = 9) and AD patients (n = 10). PBH = Benjamini Hochberg p value.
Table 2.
Annotated list of the 49 differentially methylated probes (DMPs) in CD4+CLA+ cells of AD patients.
| Illumina 450K id | chr | Position (strand) | Gene symbol | Relation to island | Enhancer | DHS | logfc | p value | Benjamini Hochberg p value |
|---|---|---|---|---|---|---|---|---|---|
| Decreased DNA methylation in AD patients | |||||||||
| cg06460587 | chr6 | 31650930 (−) | Island | Yes | −1.38 | 3.75E−06 | 0.035 | ||
| cg24531977 | chr5 | 56204891 (+) | C5orf35 | N_Shore | −1.14 | 2.46E−06 | 0.032 | ||
| cg24690709 | chr4 | 100768712 (+) | DAPP1 | OpenSea | −1.03 | 6.73E−08 | 0.0029 | ||
| cg26077005 | chr6 | 27236793 (+) | OpenSea | −0.99 | 1.85E−07 | 0.0062 | |||
| cg04853218 | chr14 | 55769688 (+) | FBXO34 | OpenSea | Yes | −0.94 | 1.01E−09 | 9.94E−05 | |
| cg08214808 | chr11 | 45922166 (−) | MAPK8IP1 | Island | −0.91 | 2.13E−06 | 0.031 | ||
| cg11770323 | chr13 | 80066032 (+) | NDFIP2 | OpenSea | Yes | −0.90 | 4.18E−08 | 0.0020 | |
| cg21022949 | chr2 | 231809697(+) | OpenSea | −0.88 | 5.38E−07 | 0.011 | |||
| cg07182616 | chr14 | 33409812 (+) | NPAS3 | OpenSea | −0.87 | 3.34E−06 | 0.034 | ||
| cg13782615 | chr11 | 4079556 (+) | STIM1 | OpenSea | Yes | −0.86 | 2.48E−06 | 0.032 | |
| cg00726151 | chr14 | 97881274 (−) | OpenSea | Yes | −0.80 | 3.26E−11 | 6.50E−06 | ||
| cg06854264 | chr1 | 200861254 (+) | C1orf106 | S_Shore | Yes | −0.76 | 3.23E−06 | 0.034 | |
| cg03207915 | chr6 | 119669112 (+) | MAN1A1 | N_Shore | −0.76 | 2.57E−06 | 0.032 | ||
| cg01436550 | chr16 | 24781512 (+) | TNRC6A | OpenSea | Yes | −0.75 | 3.42E−06 | 0.034 | |
| cg12741231 | chr8 | 19321936 (−) | CSGALNACT1 | S_Shelf | −0.73 | 1.14E−10 | 1.51E−05 | ||
| cg07343739 | chrX | 46617524 (+) | SLC9A7 | N_Shore | Yes | −0.72 | 2.37E−06 | 0.032 | |
| cg05313153 | chr8 | 119122430 (+) | EXT1 | N_Shore | −0.71 | 3.92E−07 | 0.0097 | ||
| cg07633835 | chr10 | 5938186 (−) | FBXO18 | OpenSea | Yes | −0.70 | 3.48E−06 | 0.034 | |
| cg25360385 | chr12 | 51786547 (−) | GALNT6 | S_Shore | −0.70 | 3.05E−06 | 0.034 | ||
| cg19722656 | chr6 | 119669372 (−) | MAN1A1 | N_Shore | −0.67 | 1.01E−06 | 0.018 | ||
| cg26780915 | chr7 | 105519144 (+) | S_Shore | Yes | −0.66 | 1.25E−09 | 9.94E−05 | ||
| cg03405260 | chr17 | 77786344 (+) | Island | −0.65 | 5.10E−06 | 0.043 | |||
| cg10900455 | chr20 | 42545099 (−) | TOX2 | Island | Yes | −0.65 | 7.91E−07 | 0.015 | |
| cg08416875 | chr6 | 119669226 (−) | MAN1A1 | N_Shore | −0.64 | 4.10E−06 | 0.037 | ||
| cg13607082 | chr12 | 122652224 (−) | LRRC43 | OpenSea | −0.64 | 3.59E−07 | 0.0095 | ||
| cg15447017 | chr1 | 156095882 (+) | LMNA | OpenSea | −0.63 | 3.49E−07 | 0.0095 | ||
| cg12454975 | chrX | 103356930 (−) | MCART6/ZCCHC18 | N_Shore | Yes | −0.63 | 9.12E−07 | 0.017 | |
| cg08062822 | chrX | 103356845 (−) | MCART6/ZCCHC18 | N_Shore | Yes | −0.62 | 2.49E−06 | 0.032 | |
| cg12186909 | chr19 | 10533016 (−) | PDE4A | S_Shore | −0.57 | 3.10E−06 | 0.034 | ||
| cg12589298 | chr19 | 50828905 (−) | KCNC3 | Island | −0.57 | 2.74E−06 | 0.033 | ||
| cg07910680 | chr18 | 56296449 (+) | ALPK2 | OpenSea | Yes | −0.55 | 5.41E−06 | 0.044 | |
| cg08494390 | chr4 | 87980297 (−) | AFF1 | OpenSea | Yes | −0.54 | 5.90E−06 | 0.047 | |
| cg03403880 | chr2 | 157255372 (−) | N_Shore | −0.52 | 2.66E−07 | 0.0081 | |||
| cg14523284 | chr5 | 131993614 (−) | IL13 | S_Shore | −0.49 | 3.94E−06 | 0.036 | ||
| cg02712553 | chr10 | 64136038 (−) | ZNF365 | S_Shore | −0.49 | 1.90E−06 | 0.029 | ||
| cg17347326 | chr17 | 77779426 (+) | S_Shore | Yes | −0.46 | 3.39E−08 | 0.0019 | ||
| cg05649724 | chr14 | 102415204 (−) | Island | −0.45 | 4.61E−07 | 0.010 | |||
| Increased DNA methylation in AD patients | |||||||||
| cg16312212 | chr8 | 18941574 (+) | OpenSea | Yes | 3.23 | 1.73E−06 | 0.027 | ||
| cg00211609 | chr1 | 1178039 (−) | FAM132A | Island | 1.26 | 5.28E−09 | 0.00035 | ||
| cg04871131 | chr7 | 94954202 (+) | PON1 | S_Shore | Yes | 1.01 | 1.09E−06 | 0.018 | |
| cg20595271 | chr12 | 111889200 (+) | SH2B3 | OpenSea | 0.85 | 8.71E−08 | 0.0034 | ||
| cg00839873 | chr14 | 94421989 (−) | ASB2 | OpenSea | 0.77 | 3.64E−06 | 0.035 | ||
| cg18482892 | chr3 | 56833426 (−) | ARHGEF3 | N_Shelf | 0.73 | 4.46E−06 | 0.039 | ||
| cg01956154 | chr14 | 94423399 (−) | ASB2 | OpenSea | Yes | 0.71 | 5.00E−14 | 1.99E−08 | |
| cg26800893 | chr11 | 67184596 (−) | ATPGD1 | S_Shore | Yes | 0.68 | 1.89E−07 | 0.0062 | |
| cg08943180 | chr1 | 244516022 (+) | C1orf100 | OpenSea | 0.59 | 5.15E−06 | 0.043 | ||
| cg05523877 | chr10 | 72185663 (+) | EIF4EBP2 | OpenSea | 0.53 | 1.53E−06 | 0.025 | ||
| cg01697865 | chr11 | 117856007 (+) | IL10RA | N_Shore | 0.53 | 5.23E−07 | 0.011 | ||
| cg21786381 | chr11 | 75234078 (−) | GDPD5 | N_Shelf | 0.48 | 3.11E−06 | 0.034 | ||
chr = chromosome.
pos = genome coordinate (hg19); DHS = DNAse hypersensitivity site.
logfc = difference between AD patients and HC.
Table 3.
Differentially methylated regions (DMRs) with decreased DNA methylation in CD4+CLA+ T cells from AD patients compared to HC.
| Gene | Chr | DMR Start | Widtha | CpGs | ID | logfc | p value | Benjamini–Hochberg p value |
|---|---|---|---|---|---|---|---|---|
| GPR55 | 2 | 231,790,037 | 776 | 4 | cg16382047 cg14254999 cg19827923 cg13531460 | −0.45 | 5.1 × 10−7 | 0.01 |
| 231,809,610 | 87 | 3 |
cg08840017 cg25013095 cg21022949 |
−0.73 | 1.5 × 10−8 | 0.0006 | ||
| MAN1A1 | 6 | 119,669,112 | 260 | 4 | cg03207915 cg08416875 cg02578070 cg19722656 | −0.68 | 2.5 × 10−8 | 0.0008 |
| ESR1 | 6 | 152,126,895 | 1910 | 23 | See footnoteb | −0.36 | 3.6 × 10−6 | 0.03 |
| CDHR3 | 7 | 105,515,219 | 677 | 2 |
cg03619256 cg20186907 |
−0.55 | 7.8 × 10−7 | 0.01 |
| 105,518,489 | 655 | 2 |
cg00494287 cg26780915 |
−0.68 | 3.6 × 10−10 | 2.4 × 10 − 5 | ||
| NCOR2 | 12 | 124,876,433 | 217 | 3 |
cg16337430 cg16217368 cg11050793 |
−0.53 | 1.8 × 10−6 | 0.02 |
aDMR analysis is based on minfi to collapse connected DNA methylation probes by distance rules; width in base pairs.
bSites in ESR1 cg21157690, cg17264271, cg15543523, cg26089753, cg08884395, cg01715172, cg21608605, cg20627916, cg07671949, cg23164938, cg23165623, cg21614759, cg19411146, cg21950534, cg11813455, cg24900983, cg05171584, cg23467008, cg22839866, cg23009221, cg27316393, cg00655307, cg01777019.
CpG sites indicated in bold were also found as differentially methylated CpG sites in the DMP analysis (see Table 2). Chr: chromosome.
Table 4.
A summary of the 40 differentially methylated genes in CD4+CLA+ T cells of AD patients compared to HC (including genes with DMPs and DMRs).
| Gene symbol | Gene name | Cell location | Function |
|---|---|---|---|
| AFF1 | AF4/FMR2 family, member 1 | Nucleus | Transcription factor |
| ALPK2 | Alpha-kinase 2 | Nucleoplasm | Kinase recognize phosphorylated peptides |
| ARHGEF3 | Rho guanine nucleotide exchange factor (GEF) 3 | Cytosol | Guanine nucleotide exchange factor |
| ASB2 | Ankyrin repeat and SOCS box containing 2 | Cytosol? | Protein polyubiquitination/proteasome degradation |
| ATPGD1 aka. CARNS1 | Carnosine synthase 1 | Unknown | Synthesis of carnosine and homocarnosine |
| C1orf100 | Chromosome 1 open reading frame 100 | Unknown | Uncharacterized protein 147aa 17kda |
| C1orf106 | Chromosome 1 open reading frame 106 | Unknown | Uncharacterized protein 663aa 72.9kda |
| C5orf35 | Chromosome 5 open reading frame 35 | Unknown | Uncharacterized protein 147aa 17kda |
| CDHR3 | Cadherin related family member 3 | Plasma membrane | Calcium-dependent cell adhesion proteins |
| CSGALNACT1 | Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | Golgi | Peptidoglycan glycosyltransferase activity |
| DAPP1 | Dual adaptor of phosphotyrosine and 3-phosphoinositides | Plasma membrane, cytosol | PI3K signaling |
| EIF4EBP2 | Eukaryotic translation initiation factor 4E binding protein 2 | Unknown | Bind eIF4E and inhibit translation initiation |
| ESR1 | Estrogen receptor 1 | Plasma membrane, cytosol and nucleus | Ligand-activated transcription factor |
| EXT1 | Exostosin 1 | Transmembrane glycoprotein er | Catalyzing the synthesis of heparan sulfate |
| C1QTNF12 | Complement C1q tumor necrosis factor-related protein 12 | Secreted protein extracellular | PI3K-Akt signaling pathway |
| FBH1 | F-box protein, helicase, 18 | Nucleus | Unwinds double-stranded DNA in a 3 to 5 direction |
| FBXO34 | F-box protein 34 | Nucleoplasm, cytoplasm | Protein-ubiquitin ligases |
| GALNT6 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) | Golgi | O-linked glycosylation, GalNAc to serine and threonine residues on target proteins |
| GDPD5 | Glycerophosphodiester phosphodiesterase domain containing 5 | Plasma membrane? Cytosol? | Glycerol metabolism, osmotic regulation of cellular glycerophosphocholine |
| GPR55 | G Protein-Coupled Receptor 55 | Plasma membrane | Signaling pathway |
| IL10RA | Interleukin 10 receptor, alpha | Plasma membrane | Akt signalling, IL-10 receptor |
| IL13 | Interleukin 13 | Extracellular space – secreted | Interleukin 13 |
| KCNC3 | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | Plasma membrane | Voltage channel |
| LMNA | Lamin A/C | Nucleus/cytosol | Nuclear assembly, chromatin organization, nuclear membrane |
| LRRC43 | Leucine rich repeat containing protein 43 | Unknown | Unknown |
| MAN1A1 | Mannosidase, alpha, class 1A, member 1 | Golgi membrane/er/secreted? | Maturation of Asn-linked oligosaccharides |
| MAPK8IP1 | Mitogen-activated protein kinase 8 interacting protein 1 | Endoplasmic reticulum/mitocondria | MAPK signaling pathway and Akt Signaling |
| MCART6 aka. SLC25A53 | Solute carrier family 25, member 53 | Unknown | Unknown |
| NCOR2 | Nuclear receptor corepressor 2 | Nucleus | Transcriptional activity of SMAD2/SMAD3-SMAD4 heterotrimer |
| NDFIP2 | Nedd4 family interacting protein 2 | Endosome/golgi/ER | Interacts with HECT domain-containing E3 ubiquitin-protein ligases |
| NPAS3 | Neuronal PAS domain protein 3 | Nucleus | Transcription factor |
| PDE4A | Phosphodiesterase 4A, cAMP-specific | Plasma membrane/cytosol | cellular responses to extracellular signals |
| PON1 | Paraoxonase 1 | Extracellular space – secreted | Aryl-dialkyl phosphatase activity |
| SH2B3 | SH2B adaptor protein 3 | Cytosol | SH2B adaptor family of proteins, signaling activities by growth factor and cytokine receptors |
| SLC9A7 | Solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 | Endosome and Golgi | pH homeostasis in organelles along the secretory and endocytic pathways |
| STIM1 | Stromal interaction molecule 1 | Cytoskeleton, plasma membrane, ER | Mediates Ca2+ influx after depletion of intracellular Ca2+ stores by gating of store-operated Ca2+ influx channels (SOCs) |
| TNRC6A | Trinucleotide repeat containing 6A | Cytosol | Post-transcriptional gene silencing through the RNA interference (RNAi) and microRNA pathways |
| TOX2 | TOX high mobility group box family member 2 | Nucleus | Chromatin remodeling |
| ZCCHC18 | Zinc finger, CCHC domain containing 18 | Nucleus | Nucleic acid binding, TF |
| ZNF365 | Zinc finger protein 365 | Cytoplasm, centrosome | Regulation of mitosis? |
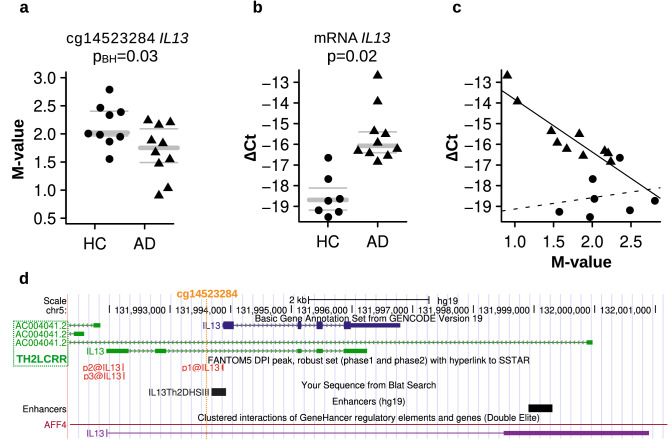
Some of the differentially methylated genes in CD4+CLA+ T cells of AD patients are implicated as key regulators of cytokine signaling pathways and immune inflammation (ARHGEF3, ASB2, DAPP1, IL10RA, PDE4A, SH2B3, STIM1 and TOX2, Fig. 1c). We also found that CD4+CLA+ T cells from AD patients have significantly decreased methylation in the IL13 promoter (Fig. 2). DNA methylation levels at the CpG site cg14523284 in the upstream region of IL13 were significantly lower compared to the levels in HC (Fig. 2a), by contrast, mRNA levels for IL13 were increased in AD patients (Fig. 2b). Spearman correlation tests showed a significant inverse correlation between DNA methylation and IL13 mRNA levels (Spearman rho −0.63, p = 0.006) (Fig. 2c), mainly driven by AD cases. This CpG site with reduced methylation levels in AD patients was at the proximal upstream of the Th2-specific DNA hypersensitive site in the IL13 promoter but within the Th2 locus-control long non-coding RNA37 (Fig. 2d), indicating that this epigenetic modification might functionally explain the augmented capability of CD4+CLA+ T cells of AD patients to produce IL-13. Correlations computed within each group further strengthen the distinct AD vs HC responses, showing a clear trend within the former group (Spearman rho < −0.92, p < 0.0002, FDR < 0.003, n = 10) but no statistically significant relationship in the latter (Spearman rho = 0.39, p > 0.38, FDR > 0.38, n = 7). A linear regression to each of the two datasets shows a statistically significant relationship between delta-Ct and M-value in AD (p < 0.0005, R2 > 0.82) but not in HC (p > 0.65, R2 < 0.05) (Fig. 2c).
Figure 2.
Differential DNA methylation and mRNA levels in the IL13 gene in CD4+CLA+ T cells between AD patients and HC. (a) DNA methylation levels for the DMP located at the IL13 promoter (cg14523284). Each dot represents an individual, HC (n = 9, circle) and AD patients (n = 10, triangle). PBH = Benjamini Hochberg p value. (b) IL13 mRNA levels in CD4+CLA+ T cells between HC (n = 7) and AD patients (n = 10) by qRT-PCR. Gray bars in the panels a and b indicate mean (bold), upper and lower (thin) quartiles. (c) Correlation between IL13 mRNA levels and IL13 DNA methylation levels. Lines of best fit are individually presented for AD (solid line, m < −2.5, p < 0.0005, R2 > 0.82) and HC (dashed line, m = 0.5, p = 0.65, R2 < 0.05). (d) Location of the CpG site cg14523284 at the promoter of IL13 within the T helper type 2 locus control region associated RNA at Chr 5q31.1 (https://genome.ucsc.edu).
miRNA deregulation in CD4+CLA+ T cells of AD patients
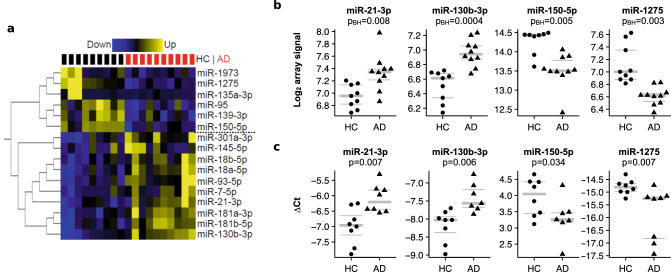
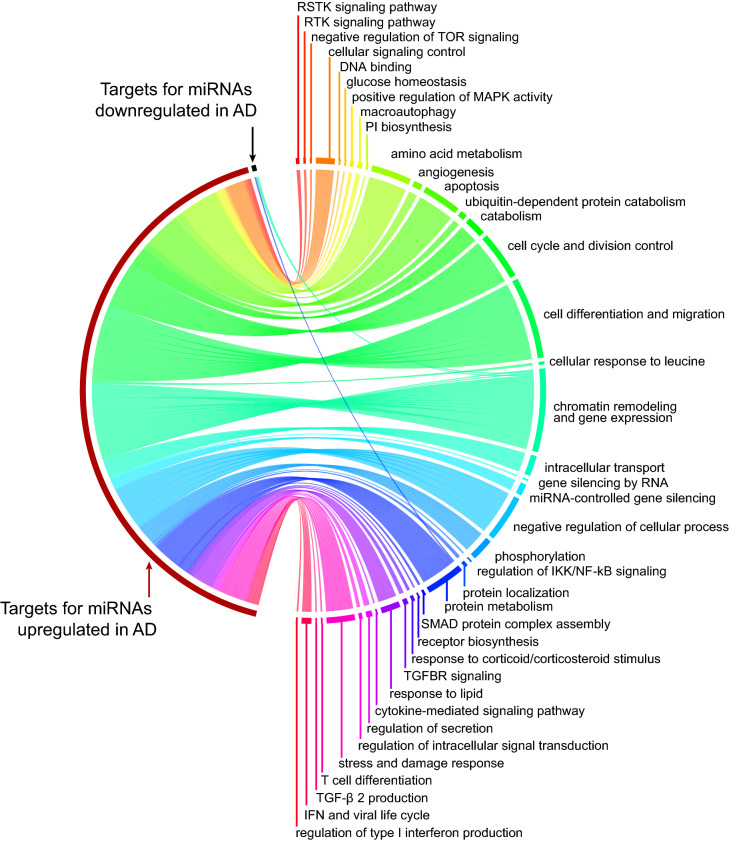
The analysis of global miRNA expression levels in the four T cell populations revealed that only the CD4+CLA+ T cells contain differentially expressed miRNAs (n = 16) between AD patients and HC (BH corrected p value < 0.05). In AD patients, 10 miRNAs were up-regulated, and 6 miRNAs were down-regulated, allowing a clear distinction between AD patients and HC (Fig. 3a). We selected 8 differentially expressed miRNAs from the microarray analysis (miR-7-5p, miR-21-3p, miR-93-5p, miR-130b-3p, miR-145-5p, miR-150-5p, miR-181b-5p and miR-1275) for technical validation by qPCR. Significant differences between AD patients and HC could be confirmed by qPCR for four of them, miR-21-3p, miR-130b-3p, miR-150-5p and miR-1275 (Fig. 3b,c). Next, we performed gene set enrichment analysis on the predicted miRNA targets of upregulated and downregulated miRNAs in AD (Fig. 4) and found 202 biological processes associated with the targets of the miRNAs dysregulated in AD (Supplementary Table S2 online). The top pathways (FDR < 1.1 × 10−5) included cell differentiation and migration, apoptosis ubiquitin-dependent protein catabolic process, transforming growth factor beta receptor signaling pathway and positive regulation of MAP kinase activity. We found that ESR1, NDFIP2, ASB2 and TNRC6A genes which were differentially methylated in AD patients (Table 4) were also targeted by upregulated miRNAs in AD patients (Supplementary Table S2 online), suggesting complex interactions between these epigenetic layers.
Figure 3.
Differentially expressed miRNAs in CD4+CLA+ T cells between AD patients and HC. (a) Differential miRNA expression by miRNA microarray between HC (n = 9) and AD patients (n = 10). Fold expression of 16 miRNAs with significant differences between AD patients and HC (Benjamini Hochberg corrected p value < 0.05). Blue indicates downregulation and yellow indicates upregulation. Each row corresponds to a miRNA and each column to 1 sample. Black and red squares on the top indicate HC and AD samples, respectively. Six down-regulated and 10 up-regulated and miRNAs in AD patients are indicated to the right of the heatmap. Software used Glucore Omics Explorer (https://www.qlucore.com/). (b) Log2 miRNA levels from the microarray analysis between HC and AD patients. The array level indicates the amount of miRNA based on the fluorescence signal in the Cy3 channel. PBH = Benjamini Hochberg p value. (c) ∆-Ct miRNA levels confirmed with qPCR between HC and AD patients. Gray bars in the panels B and C indicate mean (bold), upper and lower quartiles; each dot represents an individual.
Figure 4.
Functional enrichment analysis on the targets of AD perturbed miRNAs (FDR < 0.05). On the right hand key biological processes are summarized by their similarity. The lines are proportional to the number of biological processes associated with targets of miRNAs that are upregulated (red) or downregulated (black) in AD patients. The complete list of 202 processes is presented in Supplementary Table S2 online. This circular layout was created with the free R package circlize (https://cran.r-project.org/web/packages/circlize/index.html).
Network analysis of coordinated epigenetic responses reveals potentially perturbed pathways in circulating CD4+CLA+ T cells from AD patients
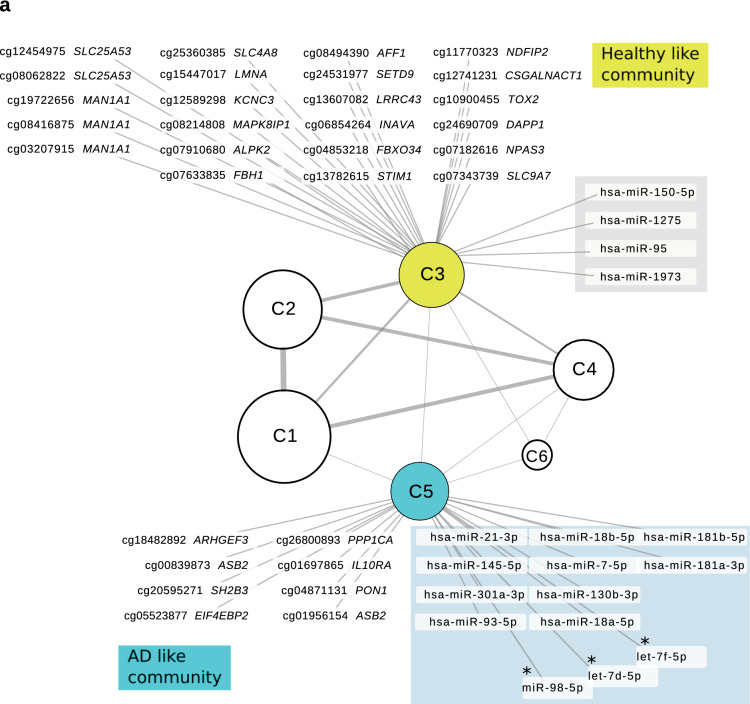
To further explore the coordinated epigenetic relationships, we performed network analysis using the full set of miRNAs, as well as the top ~ 1% CpG sites showing the largest differences between AD and HC. An association analysis (absolute Spearman ρ > 0.75, FDR < 0.001) was used to identify strongly interconnected communities of miRNA/CpG sites. Communities are sets of miRNA/CpG sites that tend to display globally coordinated expression patterns, thus highlighting potentially harmonized regulatory effectors38. Importantly, the set of miRNA/CpG sites within a community are more strongly associated with their respective community than between communities. Our analyses highlighted six communities based on the global coordinated associations between miRNA and CpG sites. Interestingly, we identified one miRNA/CpG community (C5) that recapitulated the genes showing significant differences in the AD group (Fig. 5a). This community has 640 elements: 122 miRNAs and 518 CpG sites that included 8 out of 12 CpG sites with increased DNA methylation in AD patients (Table 2) but none with decreased methylation. This community also contained all 10 miRNAs that had been identified as upregulated in the AD group by the differential expression analysis (Fig. 3a), suggesting that our approach was powered enough to capture AD associated features and its neighbors including three miRNAs targeting IL13 (hsa-miR-98-5p, hsa-let-7d-5p, hsa-let-7f.-5p) (Fig. 5a). On the other hand, community C3 has 765 elements: 15 miRNAs and 765 CpG sites including CpG probes with increased DNA methylation in HC (DAPP1, STIM1, PDEA4 and TOX2) and 4 miRNAs with increased expression in HC (Fig. 1b & Fig. 3). Overall, communities C3 and C5 reflect the antagonistic behavior that we had observed in HC and AD patients (Fig. 1b). Detailed information on the CpG sites and miRNAs detected in each community is presented in Supplementary Table S3 online.
Figure 5.
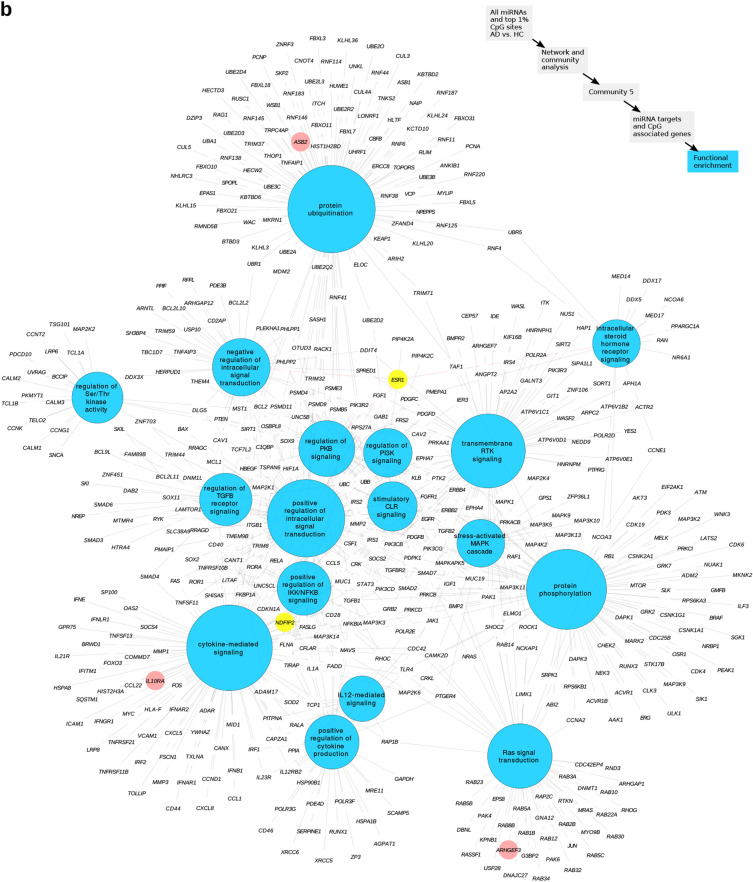
Integrated network analysis of miRNAs and CpG sites (CpGs) highlights coordinated epigenetic changes in CD4+CLA+ T cells. (a) Based on the set of associated miRNAs and CpG sites (absolute Spearman ρ > 0.75, FDR < 0.001) we identified 6 communities (C1–C6) of highly interconnected miRNAs/CpGs. These shows coordinated expression patterns, with strong interconnections within but not between communities. Note that communities 3 (yellow) and 5 (blue) show many of the differential miRNAs/CpGs detected in HC and AD patients: CpG sites (black), miRNAs (shadow), and miRNAs targeting IL13 (with asterisk). The node size for each community is proportional to the number of elements in each community. Line thickness connecting the communities is proportional to the number of connections between them. Detailed information on these communities is presented in Supplementary Table S3 online. (b) Functional enrichment analysis on putative regulated targets revealed 17 key pathways associated with AD (community C5) and are indicated as blue circles. Genes highlighted in pink and yellow were also detected as differentially methylated in the comparison between AD patients and HC (Table 2). The networks were created in Cytoscape 3.7.2 (https://cytoscape.org/).
The list of miRNAs targets together with the genes annotated to contain CpG sites associated with each community were further functionally characterized (Supplementary Table S4 online). Community 5 (C5) showed many processes related to cell signaling. Indeed, top pathways (FDR < 10−8) associated with community C5 revealed 17 involved in protein ubiquitination, positive regulation of intracellular signal transduction, protein phosphorylation, positive regulation of IKK/NFkB signaling, Ras protein signal transduction, stress-activated MAPK cascade and cytokine-mediated signaling (Fig. 5b). Overall, this suggests that epigenetic alterations in CD4+CLA+ T cells of AD patients may be affecting receptors and adaptors that are crucial for the regulation of cytokine signaling. Genes that showed increased DNA methylation in AD were represented in these pathways, and included ASB2 in protein ubiquitination, IL10RA in cytokine mediated signaling and ESR1 as a common gene in several C5 pathways (Fig. 5b).
Discussion
Several studies have shown disturbed biology in skin homing CLA+ T cells in AD patients but the underlying mechanisms explaining the alterations in this cell population remain unclear. We here analyzed the combined profiles of DNA methylation and miRNA expression in sorted peripheral blood T cell populations from AD patients compared to HC. This study revealed for the first-time significant differences in the DNA methylation levels of several key immune genes in skin homing CD4+CLA+ T cells from AD patients. The most significant differences among the 40 differentially methylated genes were found in ASB2, DAPP1, FBXO34 and NDFIP2 (Table 2). We also found significant DNA methylation differences in genes known to be genetically associated with AD predisposition including IL1339,40, IL10RA41, ZNF36542 and STIM143.
Our results revealed for the first time a significant inverse correlation between reduced DNA methylation in the IL13 promoter and increased IL13 mRNA expression in CD4+CLA+ T cells of AD patients (Fig. 2c), providing insights into the molecular events that might lead to the remarkable ability of CLA+ T cells from AD patients to secrete IL-1322,24,44. The relationship between reduced DNA methylation in the IL13 promoter of AD patients and increased IL-13 expression support that this cytokine is a central pathogenic mediator in AD45 and therapies targeting this molecule or its receptors (e.g. Dupilumab) might be beneficial in patients with severe atopic phenotypes46. The increased DNA methylation levels in the IL13 promoter observed in HC could explain why the IL13 mRNA expression was significantly reduced in this group (Fig. 2). DNA methylation differences in the IL13 gene (cg04303330) has been also described by Boorgula et al., in whole blood samples from patients with the phenotype of AD with eczema herpeticum47.
We also found reduced methylation in the gene PDE4A in AD patients, a molecule that promotes downstream inflammatory pathways and is amenable to be targeted with phosphodiesterase inhibitors (Fig. 1b)48. Since CLA+ T cells recirculate between skin and blood and mediate pathogenic inflammation, antibodies targeting adhesion molecules used by CLA+ T cells to mediate their transendothelial migration49 or anti-CLA antibodies are being attempted and proposed as promising therapeutic options in AD50. However, more studies are needed because CLA+ T cells are involved in immunosurveillance and preventing their migration to skin produces CLA+ leukocytosis and possibly alterations in other tissues.
We discovered 16 miRNAs to be differently expressed in CD4+CLA+ T cells from AD patients (Fig. 3), all except for miR-21 previously undescribed in AD12. The magnitude of the expression differences between patients and controls was small, but the effect sizes for miRNA variation are unknown. The up-regulation of miR-21 and miR-145 may reflect the pro-inflammatory status of CD4+ CLA+ T cells in AD patients. Indeed, miR-21 is involved in the polarization of adaptive immune responses and has been found upregulated in lesional skin biopsies of AD patients and in lesional skin of patients with contact dermatitis after challenge with diphenylcyclopropenone51. The altered miRNA signatures in AD may differ depending on the cell type or tissue studied. We did not find any differences in the expression of miR-146a or miR-155 between AD and HC, previously reported to be up-regulated in lesional skin biopsies of AD patients52,53 consistent with that in those studies the significant differences were attributed to the keratinocytes. Moreover, we discovered 202 biological processes significantly enriched in targets of AD-associated miRNAs of which many were implicated in cell signaling, transforming growth factor beta production and interferon responses (Fig. 4). These analyses highlighted ESR1 as a target of several AD upregulated miRNAs, involved in several processes such as cell differentiation and migration, transforming growth factor beta receptor signaling pathway and T cell differentiation (Supplementary Table S2 online); suggesting for the first time that this transcription factor is a dysregulated mediator of several putative disturbed pathways in CLA+ T cells from AD patients. These observations are in line with recent studies showing that estrogen-responsive genes may influence IL-13 production in patients with eosinophilic esophagitis54. We also evaluated putative targets of the dysregulated miRNAs in CD4+CLA+ T cells, not only for genes expressed in CD4+ T cells but using the entire miRTarBase targets, since these altered miRNAs may be released from the T cell and exert their effects on skin cells, endothelium or other immune cells.
The integrative network analysis of coordinated changes of DNA methylation and miRNA in CD4+CLA+ T cells revealed several pathways and new candidate dysregulated genes for AD (Fig. 5b). These included ASB2 a gene encoding the ankyrin-repeat suppressor of cytokine signaling (SOCS) box-containing protein 2, an E3 ubiquitin ligase that promotes protein degradation by coupling SOCS proteins with the elongin BC complex and mediate the ubiquitination of Notch targets such as E2A and Janus kinase (Jak) 255 and Jak356. ASB2 is known to be expressed in T helper 2 cells (Th2), is regulated by GATA3 and promotes Th2 phenotype in vivo57. Further studies are needed to elucidate the role of ASB2 in CLA+ T cells from AD patients. In contrast, HC showed increased methylation in NDFIP2 a gene encoding for the Nedd4 Family Interacting Protein 2 which has been described to limit the cytokine signaling and expansion of effector Th2 cells58 by promoting degradation of JAK1, probably by ITCH- and NEDD4L-mediated ubiquitination59. Two other genes from the F-box family (FBXO18 and FBXO24) implicated in protein ubiquitination were found differentially methylated in AD patients (Table 2), altogether suggesting that genes implicated in protein ubiquitination are epigenetically altered in CD4+CLA+ T cells from AD patients, and may promote inflammation by altering signaling. Ubiquitin ligases also mediate inhibition of activation of induced cell death (AICD) and contribute to lymphocyte accumulation60, therefore, altered epigenetic signatures in genes involved in protein ubiquitination may contribute to the resistance to apoptosis that acquires the CLA+ T cells upon skin infiltration25.
The coordinated epigenetic changes highlight several genes and pathways involved in intracellular signaling and were consistently associated with AD features in the CD4+CLA+ T cells from patients. These included cytokine-mediated signaling associated with IL10RA and its intracellular adaptor SH2B3 which constrains cytokine signals and might influence inflammatory immune responses in peripheral lymphoid tissues61. Also, the Ras signaling transduction pathway implicating ARHGEF3; as well as positive regulation of IKK/NFKB signaling implicating NDFIP2 (Fig. 5b). Among genes in community C3, TOX2 showed increased methylation in HC (Fig. 1b & Fig. 5a); this transcription factor is being recognized as a regulator of T cell differentiation but further studies are needed to evaluate its role in CLA+ T cells62,63.
We analyzed CD4+CD45RA+ naïve T cells since we hypothesized that epigenetic signatures predisposing to T cell dysregulation in AD patients may already be present at this cell stage before the effector or memory T helper cell differentiation. In agreement with Han et al.64, we found almost no differences in DNA methylation levels between CD4+ naïve T cells from AD patients compared to HC (Fig. 1a); neither in the unfractionated CD4+ T cells as described by Rodriguez et al.11, nor in CD8+ T cells supporting that epigenetic alterations are principally detected in CD4+CLA+ T cells. The CD8+CLA+ T cells also have an important role in AD65,66, although recent studies highlighted their pathogenic role in psoriasis67. Further studies are needed to evaluate epigenetic signatures in the CD8+CLA+ T cell populations in AD patients.
Provided that T cell turnover between skin and blood involves active de-homing from skin and migration through the lymph nodes and peripheral circulation26, it is conceivable that CD4+CLA+ T cells isolated from blood could be informative on pathogenic processes occurring in skin. It remains to be determined at which developmental state the CD4+CLA+ T cells acquired these altered epigenetic signatures in AD patients. Most probably by signals received after antigen priming or during their recirculation within skin, lymph nodes and the peripheral blood. Exposure to environmental signals from the skin microbiome or allergens could also be involved. We do not know the TCR specificity of the CD4+CLA+ T cells analyzed here but previous studies have shown that CLA+ T cells in AD preferentially respond to allergens and Staphylococcal enterotoxin B68.
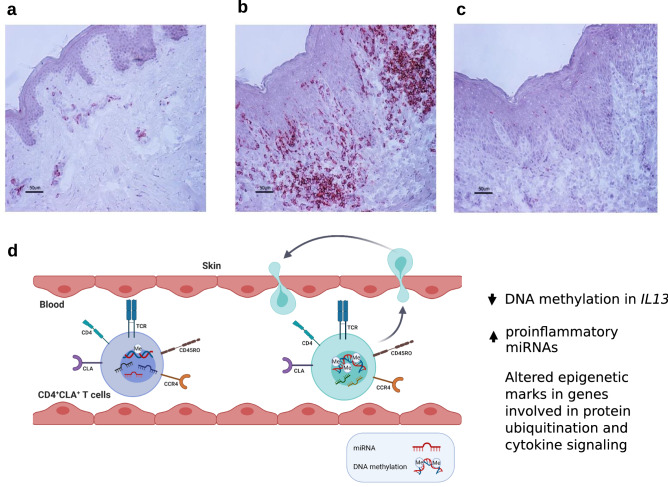
We did not compare frequencies of CLA+ T cells between AD patients and HC in peripheral circulation, however, in agreement with previous reports25, we found abundant infiltrates of CLA+ cells in the lesional skin biopsy specimens of the AD patients compared to HC (Fig. 6a-c), which were also dominated by CD3+ and CD4+ cells in consecutive sections (data not shown). Given the larger numbers of these epigenetically altered CD4+CLA+ T cells in the skin of patients (Fig. 6b) and the recirculation of these cells, we hypothesize that the epigenetic alterations detected in CLA+ T cells from peripheral blood promote pro-inflammatory functions in skin of patients and contribute to AD immunopathology (Fig. 6d). It should be noted that AD is a heterogeneous disease with different immunophenotypes in the cellular infiltrates, including not only Th2 inflammation but also other cell populations69–71. We here included adult patients with a phenotype of severe AD (objective SCORAD above 40) and allergen specific IgE sensitization. Therefore, the epigenetic signatures detected in this study and their related genes may be associated with this phenotype of patients, and more research is needed to evaluate their implications in other AD cohorts.
Figure 6.
Identification of CLA+ cells in skin biopsies and epigenetic changes detected in circulating CD4+CLA+ T cells from AD patients that might contribute to skin inflammation. A representative immunohistochemistry staining of the distribution of CLA+ cells in skin biopsies from (a) a healthy control, (b) lesional skin from an AD patient, and (c), rat IgM used as isotype control. Scale bars represent 50 µm. (d) Circulating CD4+CLA+ T cells from AD patients (light blue cell) show significant differences in DNA methylation and miRNA levels compared to CD4+CLA+ T cells from HC (purple cell). The main differences were detected in the reduced DNA methylation of the IL13 gene, the increased expression of proinflammatory miRNAs and coordinated epigenetic changes in genes involved in protein ubiquitination and cytokine signaling in AD patients. Since these CD4+CLA+ T cells can recirculate between skin and blood25, these altered epigenetic marks might contribute to AD immunopathology.
Several methodological aspects add strength to this study. To avoid the confounding effects that cell heterogeneity has on the interpretation of epigenetic and expression studies, we analyzed sorted T cell populations in peripheral blood. In addition, we used a robust methodology that interrogated DNA methylation levels in ~ 450 000 CpG sites through the genome and global miRNA expression in ~ 2000 miRNAs. At several loci, the magnitude of the epigenetic differences between AD and HC were relatively small, yet statistically significant after correction by multiple testing. Several of the loci detected in the comparison between AD patients and HC (Fig. 1b & Table 2) showed coordinated changes when analyzed with a different algorithm for network analysis (Fig. 5a). Our study has also some limitations, to avoid the confounding effects of gender, we included only male AD patients that even though they were selected to be as much homogeneous as possible had some differences in their total serum IgE levels, AD severity and the presence of comorbidities such as asthma (Table 1). It is worth noting that a larger sample size would assist in identifying important biological signals in the etiology of AD. Due to the limitations in the amount of cells that were sorted per individual, we could only measure mRNA expression in few genes by TaqMan. We detected significant differences in IL13 expression, but we could not interrogate how the coordinated changes in miRNA and CpG methylation affects global gene expression. Nevertheless, this study revealed new genes and biological processes that deserve to be further validated at mRNA and protein levels in other cohorts of AD patients and functionally evaluated at the cellular level to dissect their role in AD pathogenesis.
In summary, we here discovered putative altered molecular pathways in circulating CD4+CLA+ T cells from AD patients, involving disease-associated signatures in DNA methylation and miRNA levels. The identified loci highlight new candidates in AD, including genes mediating intracellular cell signaling and adaptor molecules of the IL-10/IL-13 interleukin signaling pathway as well as genes involved in protein ubiquitination. Our findings support epigenetic profiling as a valuable tool to uncover putative molecules participating in disease pathways. Further studies are needed to define the downstream effects of these epigenetic alterations in AD immunopathology and evaluate if environmental signals at the target organ (e.g., skin microbiota) induce the detected epigenetic differences in circulating CD4+CLA+ T cells from AD patients.
Methods
Study population and samples
Twenty age-matched adult male individuals (10 AD patients and 10 HC) were selected from a Swedish eczema study72. All participants were asked to visit the Dermatology Unit at the Karolinska University Hospital in Solna and were examined by a dermatologist for their general physical conditions, in case of AD patients, also for the severity of the eczema. Afterwards, all provided a 450 ml blood donation (see below). The cases fulfilled the following inclusion criteria: (1) a physician-diagnosis of moderate or severe AD as determined by the objective Scoring Atopic Dermatitis index (SCORAD) (2) elevated levels of total plasma IgE > 122 kUA/L (ImmunoCAP, Thermo Fisher Scientific, Uppsala, Sweden), (3) positive allergen-specific IgE > 0.35 kUA/L as determined by Phadiatop (Thermo Fisher, Uppsala, Sweden), and (4) besides AD, a clinical history of asthma and/or rhinitis. All AD patients had used topical corticosteroids at least 30 days before blood sampling. Controls were healthy individuals without clinical history of eczema, asthma or other allergic diseases and plasma IgE levels below 122 kUA/L. On the same day as the 450 ml blood donation, skin biopsies were obtained from AD patients (n = 5) and from HC (n = 5). Punch biopsies (4 mm) were taken under local anesthesia (Lidocaine 5 mg/ml with epinephrine 5 µg/mL, Astra, Södertälje, Sweden) from eczema lesions in the AD patients located in the popliteal fossa, upper/mid back or shoulders and healthy skin of HC from the low-back (lumbar) region. The biopsies were snap-frozen on dry ice and stored at −80 °C until immunohistochemistry analysis (see below). This study was conducted in accordance with the Helsinki Declaration ethical principles for medical research and was approved by the Regional Ethical Review Board in Stockholm (Dnrs 04–593/1, 2008/1122–32, 2010/754–32, and 2011/1051–31). All participants gave their written informed consent.
Isolation of T cell populations
A 450 ml blood donation was obtained at the Blood Transfusion Center Karolinska University Hospital and processed immediately for cell isolation. Peripheral blood mononuclear cells (PBMCs) were separated by density centrifugation on Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden) and then labeled for magnetic associated cell sorting (MACS) to obtain CD4+ T cells and CD8+ T cells, CD4+CD45RA+ naïve T cells and skin-homing CD4+CLA+ T cells (Miltenyi Biotec, Gladbach, Germany). Cell viability after sorting was > 90% in all populations (as determined by trypan blue exclusion) and the purity was examined by flow cytometry (see below). Two aliquots were stored at −80 °C: one containing 5 × 106 cells for DNA extraction as described previously36 and another containing a maximum of 10 × 106 cells homogenized in QIAzol lysis reagent (Qiagen, Hilden, Germany) for total RNA extraction.
Flow cytometry
Cells were re-suspended in FACS buffer (0.1% BSA in PBS) at a final concentration of 0.1 × 106 cells per tube. Fc-receptors were blocked with 1 µl of normal mouse serum (Dako A/S, Glostrup, Denmark) for 10 min at 4 °C. Optimized panels of fluorochrome-conjugated monoclonal antibodies (Supplementary Table S1 online) were added to the cells, in a final volume of 100 µl and incubated for 30 min at 4 °C. Every staining included the unstained sample and the corresponding panels of isotype controls to set the gates for positive and negative populations. In addition to the markers described in Supplementary Table S1 online, which were measured in all the sorted samples, we also analyzed the cell surface expression of CCR4 (HC = 1, AD = 1) to better define the immunophenotype of the CD4+CLA+ T cells during validation of the cell sorting protocol (PerCP Anti-human CD194 (CCR4), clone L291H4, mIgG1 κ. Biolegend). Data were acquired using a FACS Calibur (BD Biosciences, San Jose, CA, USA), to at least 5000 events per population and analyzed by FlowJo vX.0.7 (FlowJo, LLC, Ashland, OR, USA).
DNA methylation analysis
Genomic DNA was extracted using the QIAamp DNA Mini kit (Qiagen) and DNA concentrations were assessed by Qubit and Nanodrop. DNA purity was evaluated by the A260/A280 ratio by Nanodrop. After diluting to normalize the concentrations the DNA samples were bisulfite treated using the EZ-96 DNA Methylation kit (Zymo Research Corp., Irvine, CA, USA) according to manufacturer’s instructions. Denatured bisulfite-treated DNA was amplified, fragmented and hybridized onto the Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA, USA), following manufacturer’s instructions, at the Mutation Analysis Core Facility (Karolinska Institutet, Stockholm, Sweden). The .idat files for each sample obtained from Genome Studio were imported in R and read in using the minfi package73 with the genome annotation based on GRCh37/hg19. A total of 77 samples were analyzed: 20 CD4+(HC = 10, AD = 10), 18 CD4+CD45RA+ naïve (HC = 8, AD = 10), 19 CD4+CLA+ cells (HC = 9, AD = 10), and 20 CD8+ (HC = 10, AD = 10). The data were normalized using the subset-quantile within array normalization (Swan) method74. The probes overlapping with known SNPs were removed, leaving 398 494 CpG sites for further analysis. Differentially methylated CpG sites between AD patients and controls were obtained using linear models (y ~ group + subject + age) and pairwise comparisons with empirical Bayes as implemented in limma75. DMRs were analyzed with the minfi package73. All the CpG sites presented as having differential methylation between AD patients and HC had a p value < 0.05 after Benjamini-Hochberg (BH) correction for multiple testing.
RNA isolation and miRNA measurements
mRNA and miRNA were obtained by phenol/chloroform extraction. RNA integrity was evaluated using the Nanochip kits in Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) resulting in RIN values (mean ± SD) of 9.31 ± 0.38. miRNA levels were measured for 4774 probes (representing 2006 human miRNAs, miRBase database release 19) using the SurePrint G3 (8 × 60 K) miRNA array (Agilent Technologies). The raw .txt files obtained from the Agilent feature extraction software were imported in R and analyzed with limma75. After quality check 71 samples remained for the analysis of differential miRNA expression: 17 CD4+ (HC = 8, AD = 9), 15 CD4+CD45RA+ naïve (HC = 6, AD = 9), 19 CD4+CLA+ (HC = 9, AD = 10) and 20 CD8+ (HC = 10, AD = 10). To ensure homogeneity of analytical strategy with the DNA methylation, the same statistical approach described above was also used to find differentially expressed miRNAs. Experimentally validated human miRNA targets were determined through Ingenuity Pathway Analysis (IPA) (https://www.ingenuity.com) and miRTarBase76.
Network analysis
Association analyses were performed after removing features with null variance and based on the entire set of quantified miRNAs and a subset of DNA methylation probes as follows. The full DNA methylation set comprising > 398,000 probes was filtered by first determining CpG probe UCSC classifications for the Illumina 450 K manifest through the FDb.InfiniumMethylation.hg19 R package in R 3.6.1, and excluded if no gene association was identified (e.g. intergenic probes). Secondly, we selected the top varying CpG probes (FDR < 0.1, 1.44% top probes) based on the limma comparison of AD patients and HC. Association analyses were then performed on the resulting set of 455 miRNA and 4369 CpG probes by Spearman correlations. P values were corrected for multiple hypothesis testing considering Benjamin-Hochberg false discovery rates as indicated throughout. The resulting set of positively correlated interactions were processed through igraph (https://igraph.org/) in Python 3.7. Node communities were computed through the Leiden algorithm through modularity optimization38 and antagonistic feature relationships were identified as described by Benfeitas et al77.
Functional annotation
Gene set enrichment analysis was performed for Gene Ontology Biological Processes (2018) and KEGG 2019 pathways through the Enrichr78 API in Python 3.7 and considering the entire set of possible miRNA target genes and CpG-associated genes as background, where applicable. GO terms were further filtered for redundancy based on their similarity through Revigo79. Associations with miRNA targets were plotted using circus through the R circlize package80 and Cytoscape 3.7.2 (https://cytoscape.org/).
qPCR validation of miRNA and mRNA expression in CD4+CLA+ T cells
To validate differentially expressed miRNAs, cDNA was prepared from the total RNA of CD4+CLA+ T cells (15 ng) from 10 AD patients and 9 HC (the same samples as used in the Agilent array) using the miRCURY LNA Universal RT microRNA PCR kit (Exiqon A/S, Vedbaek, Denmark) according to the manufacturer’s protocol. miRNAs were amplified using the miRCURY LNA SYBR Green master mix and specific PCR primer sets for 8 miRNAs (Exiqon) according to the manufacturer’s instructions. SNORD44 (snRNA) was used as endogenous control as described previously81. To validate mRNA targets, cDNA was prepared using the high capacity RNA to cDNA kit (Life Technologies) starting from 80 ng of total RNA from 8 HC and 10 AD patients. Gene expression levels (mRNA) were measured using validated Taqman probes (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s instructions using beta-2 microglobulin (B2M) as the reference gene. Quantitative real-time PCR was performed in the Biorad CFX96 system (BioRad Inc., Hercules, CA, USA) with each sample in duplicate, and the gene expression data were exported from the DataAssist Software v3.01. Differential expression between AD patients and HC was calculated using the comparative Ct method. In brief, the average Ct value for each miRNA was subtracted from the Ct value of SNORD44 to obtain the delta-Ct. The comparisons of delta-Ct between AD patients and HC were calculated by unpaired t-tests and a p value < 0.05 was considered significant.
Immunohistochemistry
The frozen skin biopsy specimens were embedded in optimal cutting temperature compound and six µm thin sections were prepared in a microtome-cryostat, two sections per glass slide. The tissue architecture and degree of cell infiltrates were evaluated by hematoxylin and eosin staining. For the evaluation of T cell markers, the sections were acetone fixed and stained using the avidin–biotin complex method (Vectastain Elite ABC-kit, Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions with the following primary antibodies: anti-CLA (rat IgM, clone: HECA-452), anti-CD4 (mouse IgG1, clone: SK3), anti-CD3 (mouse IgG1, clone: SK7), all from BD Pharmingen (San Diego, CA, USA). Rat IgM (clone R4-22, BD Pharmingen) and mouse IgG1 (clone MG1-45, Biolegend) were included in each staining as isotype controls. Biotinylated antibodies targeting rat IgM (clone G53-238, BD Pharmingen) and mouse IgG1 (BA-2001, Vector Laboratories) were used as secondary antibodies. The sections were counterstained with hematoxylin.
Statistical analysis
Statistical approaches were chosen according to the diverse data types in this study and explained in their respective methods sections. Hypothesis testing was performed by considering the null hypothesis of the absence of an association between the compared variables. The associations were tested according to the nature of the data: continuous vs continuous (Spearman rank correlation test); continuous vs categorical (t-test or Mann Whitney test according to data distribution). p values were corrected by the Benjamini–Hochberg procedure for multiple testing and a value < 0.05 was considered statistically significant.
Supplementary information
Acknowledgments
We thank the patients and healthy controls that kindly participated in the study, Catharina Johansson for the coordination of donor recruitment, the personnel from the Transfusion Center at the Karolinska University Hospital Solna for help with the blood collection. Helena Persson for advice on miRNA extractions and Lovisa E. Reinius for insightful discussions. The Mutation Analysis Core Facility (MAF) for the laboratory work with the Illumina 450K arrays, the Bioinformatic and Expression Core Facility (BEA) for the laboratory work with the miRNA arrays and the National Bioinformatics Infrastructure Sweden (NBIS) for bioinformatic support. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC 2019/8-236. This study was supported by grants from the Swedish Research Foundation of Strategic Research (RBc08-0027), the Swedish Research Council, through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and the Karolinska Institutet and the Karolinska University Hospital, the Cancer and Allergy Fund, and the Hesselman’s Foundation.
Author contributions
A.S. was responsible for the recruitment of the AD patients and HC. G.W. under supervision of L.L. examined all individuals. Conceived and planned the experiments: N.A., C.S., J.K., D.G., and A.S. N.A. and A.A. performed the cell sorting and the immunohistochemistry experiments. N.A. performed the flow cytometry analysis and the DNA/RNA isolations. C.S. and J.K. supervised the molecular studies. D.G. performed the pre-processing of microarray data and the statistical comparisons between patients and controls. N.A. and S.B. assisted D.G. with the bioinformatic analysis of microarray data. S.B. performed the qPCR validation of differentially expressed miRNAs and analyzed the data together with S.K. N.A. interpreted the results and performed the functional annotation of AD-associated signatures together with R.B. and J.L. N.A., S.K. and R.B. prepared the figures. N.A. wrote the manuscript together with AS. All authors participated in revising the manuscript and approved the final version.
Funding
Open Access funding provided by Karolinska Institutet.
Data availability
The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files. DNA methylation and miRNA levels are available from the corresponding authors on reasonable request.
Competing interests
A. Scheynius is a member in the Joint Steering Committee for the Human Translational Microbiome Program at Karolinska Institutet together with Ferring Pharmaceuticals, Switzerland. The rest of the authors declare that they have no relevant conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathalie Acevedo, Email: nacevedoc@unicartagena.edu.co.
Annika Scheynius, Email: annika.scheynius@ki.se.
Supplementary information
is available for this paper at 10.1038/s41598-020-74798-z.
References
- 1.Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat. Rev. Dis. Primers. 2018;4:1. doi: 10.1038/s41572-018-0001-z. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, et al. Demethylation of the FCER1G promoter leads to FcepsilonRI overexpression on monocytes of patients with atopic dermatitis. Allergy. 2012;67:424–430. doi: 10.1111/j.1398-9995.2011.02760.x. [DOI] [PubMed] [Google Scholar]
- 3.Liang Y, et al. Thymic stromal lymphopoietin epigenetically upregulates Fc receptor gamma subunit-related receptors on antigen-presenting cells and induces TH2/TH17 polarization through dectin-2. J. Allergy Clin. Immunol. 2019;144:1025–1035. doi: 10.1016/j.jaci.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Cho HJ, et al. Prenatal 25-hydroxyvitamin D deficiency affects development of atopic dermatitis via DNA methylation. J. Allergy Clin. Immunol. 2019;143:1215–1218. doi: 10.1016/j.jaci.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Kumar D, et al. A functional SNP associated with atopic dermatitis controls cell type-specific methylation of the VSTM1 gene locus. Genome Med. 2017;9:18. doi: 10.1186/s13073-017-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tost J. A translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 2018;142:715–726. doi: 10.1016/j.jaci.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Martin MJ, et al. Genetics and Epigenetics of Atopic Dermatitis: An Updated Systematic Review. Genes (Basel) 2020 doi: 10.3390/genes11040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potaczek DP, et al. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9:539–571. doi: 10.2217/epi-2016-0162. [DOI] [PubMed] [Google Scholar]
- 9.Barton SJ, et al. DNA methylation of Th2 lineage determination genes at birth is associated with allergic outcomes in childhood. Clin. Exp. Allergy. 2017;47:1599–1608. doi: 10.1111/cea.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Zhou B, Zhao M, Tang J, Lu Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin. Exp. Dermatol. 2014;39:48–53. doi: 10.1111/ced.12206. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez E, et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J. Invest. Dermatol. 2014;134:1873–1883. doi: 10.1038/jid.2014.87. [DOI] [PubMed] [Google Scholar]
- 12.Guidi R, Wedeles CJ, Wilson MS. ncRNAs in Type-2 immunity. Noncoding RNA. 2020 doi: 10.3390/ncrna6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baskara-Yhuellou I, Tost J. The impact of microRNAs on alterations of gene regulatory networks in allergic diseases. Adv. Protein Chem. Struct. Biol. 2020;120:237–312. doi: 10.1016/bs.apcsb.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Liew WC, et al. Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Vaher H, et al. miR-10a-5p is increased in atopic dermatitis and has capacity to inhibit keratinocyte proliferation. Allergy. 2019;74:2146–2156. doi: 10.1111/all.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czarnowicki T, Santamaria-Babi LF, Guttman-Yassky E. Circulating CLA(+) T cells in atopic dermatitis and their possible role as peripheral biomarkers. Allergy. 2017;72:366–372. doi: 10.1111/all.13080. [DOI] [PubMed] [Google Scholar]
- 17.Teraki Y, Hotta T, Shiohara T. Increased circulating skin-homing cutaneous lymphocyte-associated antigen (CLA)+ type 2 cytokine-producing cells, and decreased CLA+ type 1 cytokine-producing cells in atopic dermatitis. Br. J. Dermatol. 2000;143:373–378. doi: 10.1046/j.1365-2133.2000.03665.x. [DOI] [PubMed] [Google Scholar]
- 18.Torres MJ, et al. Circulating CLA+ lymphocytes from children with atopic dermatitis contain an increased percentage of cells bearing staphylococcal-related T-cell receptor variable segments. Clin. Exp. Allergy. 1998;28:1264–1272. doi: 10.1046/j.1365-2222.1998.00397.x. [DOI] [PubMed] [Google Scholar]
- 19.Czarnowicki T, et al. Severe atopic dermatitis is characterized by selective expansion of circulating TH2/TC2 and TH22/TC22, but not TH17/TC17, cells within the skin-homing T-cell population. J. Allergy Clin. Immunol. 2015;136:104–115. doi: 10.1016/j.jaci.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Czarnowicki T, et al. Evolution of pathologic T-cell subsets in patients with atopic dermatitis from infancy to adulthood. J. Allergy Clin. Immunol. 2020;145:215–228. doi: 10.1016/j.jaci.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czarnowicki T, et al. Early pediatric atopic dermatitis shows only a cutaneous lymphocyte antigen (CLA)(+) TH2/TH1 cell imbalance, whereas adults acquire CLA(+) TH22/TC22 cell subsets. J. Allergy Clin. Immunol. 2015;136:941–951. doi: 10.1016/j.jaci.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akdis CA, Akdis M, Simon HU, Blaser K. Regulation of allergic inflammation by skin-homing T cells in allergic eczema. Int. Arch. Allergy Immunol. 1999;118:140–144. doi: 10.1159/000024051. [DOI] [PubMed] [Google Scholar]
- 23.Akdis M, Akdis CA, Weigl L, Disch R, Blaser K. Skin-homing, CLA+ memory T cells are activated in atopic dermatitis and regulate IgE by an IL-13-dominated cytokine pattern: IgG4 counter-regulation by CLA- memory T cells. J. Immunol. 1997;159:4611–4619. [PubMed] [Google Scholar]
- 24.Antunez C, et al. Different cytokine production and activation marker profiles in circulating cutaneous-lymphocyte-associated antigen T cells from patients with acute or chronic atopic dermatitis. Clin. Exp. Allergy. 2004;34:559–566. doi: 10.1111/j.1365-2222.2004.1933.x. [DOI] [PubMed] [Google Scholar]
- 25.Akdis M, Trautmann A, Klunker S, Blaser K, Akdis CA. Cytokine network and dysregulated apoptosis in atopic dermatitis. Acta Odontol. Scand. 2001;59:178–182. doi: 10.1080/000163501750266783. [DOI] [PubMed] [Google Scholar]
- 26.Klicznik MM, et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 2019 doi: 10.1126/sciimmunol.aav8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czarnowicki T, et al. Blood endotyping distinguishes the profile of vitiligo from that of other inflammatory and autoimmune skin diseases. J. Allergy Clin. Immunol. 2019;143:2095–2107. doi: 10.1016/j.jaci.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 28.De Jesus-Gil C, et al. CLA(+) T cell response to microbes in psoriasis. Front Immunol. 2018;9:1488. doi: 10.3389/fimmu.2018.01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abernathy-Carver KJ, Sampson HA, Picker LJ, Leung DY. Milk-induced eczema is associated with the expansion of T cells expressing cutaneous lymphocyte antigen. J. Clin. Invest. 1995;95:913–918. doi: 10.1172/JCI117743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campana R, et al. Epicutaneous allergen application preferentially boosts specific T cell responses in sensitized patients. Sci. Rep. 2017;7:11657. doi: 10.1038/s41598-017-10278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SM, et al. Cutaneous lymphocyte antigen and alpha4beta7 T-lymphocyte responses are associated with peanut allergy and tolerance in children. Allergy. 2012;67:336–342. doi: 10.1111/j.1398-9995.2011.02765.x. [DOI] [PubMed] [Google Scholar]
- 32.Jung T, Schulz S, Zachmann K, Neumann C. Expansion and proliferation of skin-homing T cells in atopic dermatitis as assessed at the single cell level. Int. Arch. Allergy Immunol. 2003;130:143–149. doi: 10.1159/000069010. [DOI] [PubMed] [Google Scholar]
- 33.Strickland I, Hauk PJ, Trumble AE, Picker LJ, Leung DY. Evidence for superantigen involvement in skin homing of T cells in atopic dermatitis. J. Invest. Dermatol. 1999;112:249–253. doi: 10.1046/j.1523-1747.1999.00502.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferran M, Santamaria-Babi LF. Pathological mechanisms of skin homing T cells in atopic dermatitis. World Allergy Organ. J. 2010;3:44–47. doi: 10.1097/WOX.0b013e3181d675f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinius LE, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spurlock CF, 3rd, et al. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mucha S, et al. Protein-coding variants contribute to the risk of atopic dermatitis and skin-specific gene expression. J. Allergy Clin. Immunol. 2019 doi: 10.1016/j.jaci.2019.10.030. [DOI] [PubMed] [Google Scholar]
- 40.Weidinger S, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum. Mol. Genet. 2013;22:4841–4856. doi: 10.1093/hmg/ddt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohara Y, et al. A major determinant quantitative-trait locus responsible for atopic dermatitis-like skin lesions in NC/Nga mice is located on Chromosome 9. Immunogenetics. 2001;53:15–21. doi: 10.1007/s002510000286. [DOI] [PubMed] [Google Scholar]
- 42.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 43.Fuchs S, et al. Antiviral and regulatory T cell immunity in a patient with stromal interaction molecule 1 deficiency. J. Immunol. 2012;188:1523–1533. doi: 10.4049/jimmunol.1102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antunez C, et al. Calcitonin gene-related peptide modulates interleukin-13 in circulating cutaneous lymphocyte-associated antigen-positive T cells in patients with atopic dermatitis. Br. J. Dermatol. 2009;161:547–553. doi: 10.1111/j.1365-2133.2009.09318.x. [DOI] [PubMed] [Google Scholar]
- 45.Bieber T. Interleukin-13: Targeting an underestimated cytokine in atopic dermatitis. Allergy. 2020;75:54–62. doi: 10.1111/all.13954. [DOI] [PubMed] [Google Scholar]
- 46.Moyle M, Cevikbas F, Harden JL, Guttman-Yassky E. Understanding the immune landscape in atopic dermatitis: the era of biologics and emerging therapeutic approaches. Exp. Dermatol. 2019;28:756–768. doi: 10.1111/exd.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boorgula MP, et al. Replicated methylation changes associated with eczema herpeticum and allergic response. Clin. Epigenet. 2019;11:122. doi: 10.1186/s13148-019-0714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H, et al. Application of topical phosphodiesterase 4 inhibitors in mild to moderate atopic dermatitis: a systematic review and meta-analysis. JAMA Dermatol. 2019;155:585–593. doi: 10.1001/jamadermatol.2019.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harper EG, et al. Efalizumab therapy for atopic dermatitis causes marked increases in circulating effector memory CD4+ T cells that express cutaneous lymphocyte antigen. J. Invest. Dermatol. 2008;128:1173–1181. doi: 10.1038/sj.jid.5701169. [DOI] [PubMed] [Google Scholar]
- 50.Sernicola A, Russo I, Silic-Benussi M, Ciminale V, Alaibac M. Targeting the cutaneous lymphocyte antigen (CLA) in inflammatory and neoplastic skin conditions. Expert Opin. Biol. Ther. 2020;20:275–282. doi: 10.1080/14712598.2020.1715937. [DOI] [PubMed] [Google Scholar]
- 51.Vennegaard MT, et al. Allergic contact dermatitis induces upregulation of identical microRNAs in humans and mice. Contact Dermat. 2012;67:298–305. doi: 10.1111/j.1600-0536.2012.02083.x. [DOI] [PubMed] [Google Scholar]
- 52.Sonkoly E, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 2010;126(581–589):e581–e520. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 53.Rebane A, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin. Immunol. 2014;134:836–847. doi: 10.1016/j.jaci.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler JC, et al. 17beta-Estradiol protects the esophageal epithelium from IL-13-induced barrier dysfunction and remodeling. J. Allergy Clin. Immunol. 2019;143:2131–2146. doi: 10.1016/j.jaci.2018.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nie L, et al. Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell Res. 2011;21:754–769. doi: 10.1038/cr.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W, Sun XH. A mechanism underlying NOTCH-induced and ubiquitin-mediated JAK3 degradation. J. Biol. Chem. 2011;286:41153–41162. doi: 10.1074/jbc.M111.273755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spinner CA, et al. The E3 Ubiquitin Ligase Asb2alpha in T Helper 2 Cells Negatively Regulates Antitumor Immunity in Colorectal Cancer. Cancer Immunol Res. 2019;7:1332–1344. doi: 10.1158/2326-6066.CIR-18-0562. [DOI] [PubMed] [Google Scholar]
- 58.O'Leary CE, et al. Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4+ effector T cells. Nat. Commun. 2016;7:11226. doi: 10.1038/ncomms11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trimpert C, et al. NDFIP allows NEDD4/NEDD4L-induced AQP2 ubiquitination and degradation. PLoS ONE. 2017;12:e0183774. doi: 10.1371/journal.pone.0183774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salva KA, Reeder MJ, Lloyd R, Wood GS. c-CBL E3 Ubiquitin ligase expression increases across the spectrum of benign and malignant T-cell skin diseases. Am. J. Dermatopathol. 2017;39:731–737. doi: 10.1097/DAD.0000000000000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mori T, et al. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-gamma-producing T cells. J. Immunol. 2014;193:1728–1736. doi: 10.4049/jimmunol.1303243. [DOI] [PubMed] [Google Scholar]
- 62.Xu W, et al. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 2019;51:826–839. doi: 10.1016/j.immuni.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc. Natl. Acad. Sci. USA. 2019;116:12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han J, et al. The characteristics of genome-wide DNA methylation in naive CD4+ T cells of patients with psoriasis or atopic dermatitis. Biochem. Biophys. Res. Commun. 2012;422:157–163. doi: 10.1016/j.bbrc.2012.04.128. [DOI] [PubMed] [Google Scholar]
- 65.Zhang BX, et al. Attenuation of peripheral regulatory T-cell suppression of skin-homing CD8(+)T cells in atopic dermatitis. Yonsei Med. J. 2015;56:196–203. doi: 10.3349/ymj.2015.56.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seneviratne SL, et al. Interleukin-4 promotes human CD8 T cell expression of CCR7. Immunology. 2007;120:66–72. doi: 10.1111/j.1365-2567.2006.02478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casciano F, et al. CCR4(+) skin-tropic phenotype as a feature of central memory CD8(+) T cells in healthy subjects and psoriasis patients. Front Immunol. 2020;11:529. doi: 10.3389/fimmu.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferran M, et al. Circulating CLA+ T lymphocytes as peripheral cell biomarkers in T-cell-mediated skin diseases. Exp. Dermatol. 2013;22:439–442. doi: 10.1111/exd.12154. [DOI] [PubMed] [Google Scholar]
- 69.Simon D, Wollenberg A, Renz H, Simon HU. Atopic dermatitis: collegium internationale allergologicum (CIA) update 2019. Int. Arch. Allergy Immunol. 2019;178:207–218. doi: 10.1159/000497383. [DOI] [PubMed] [Google Scholar]
- 70.Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol. 2019;143:1–11. doi: 10.1016/j.jaci.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 71.Sugita K, Akdis CA. Recent developments and advances in atopic dermatitis and food allergy. Allergol. Int. 2020;69:204–214. doi: 10.1016/j.alit.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 72.Mittermann I, et al. IgE sensitization profiles differ between adult patients with severe and moderate atopic dermatitis. PLoS ONE. 2016;11:e0156077. doi: 10.1371/journal.pone.0156077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou CH, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benfeitas R, et al. Characterization of heterogeneous redox responses in hepatocellular carcinoma patients using network analysis. EBioMedicine. 2019;40:471–487. doi: 10.1016/j.ebiom.2018.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 81.Chavali S, et al. MicroRNAs act complementarily to regulate disease-related mRNA modules in human diseases. RNA. 2013;19:1552–1562. doi: 10.1261/rna.038414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper and its supplementary information files. DNA methylation and miRNA levels are available from the corresponding authors on reasonable request.