Significance
B cell development and function depend on BCR signaling. Infection of primary B cells by EBV relies on the EBV-encoded protein LMP2A, which is currently thought to mimic BCR signaling. We revise this model based on systematic comparison of BCR and LMP2A signaling at the phosphoproteome and transcriptome levels. Our study identifies LMP2A signaling modules that are not shared by BCR signaling and elucidates their impact on gene expression in primary B cells. We describe a mechanism by which LMP2A interferes with apoptosis and cell-cycle checkpoints to cooperate with oncogenes in promoting cell survival and proliferation. These insights provide a framework in which to understand how EBV promotes malignant transformation of B cells.
Keywords: signal transduction, lymphoma, B cell receptor, Epstein–Barr virus
Abstract
Epstein–Barr virus (EBV) infects human B cells and reprograms them to allow virus replication and persistence. One key viral factor in this process is latent membrane protein 2A (LMP2A), which has been described as a B cell receptor (BCR) mimic promoting malignant transformation. However, how LMP2A signaling contributes to tumorigenesis remains elusive. By comparing LMP2A and BCR signaling in primary human B cells using phosphoproteomics and transcriptome profiling, we identified molecular mechanisms through which LMP2A affects B cell biology. Consistent with the literature, we found that LMP2A mimics a subset of BCR signaling events, including tyrosine phosphorylation of the kinase SYK, the calcium initiation complex consisting of BLNK, BTK, and PLCγ2, and its downstream transcription factor NFAT. However, the majority of LMP2A-induced signaling events markedly differed from those induced by BCR stimulation. These included differential phosphorylation of kinases, phosphatases, adaptor proteins, transcription factors such as nuclear factor κB (NF-κB) and TCF3, as well as widespread changes in the transcriptional output of LMP2A-expressing B cells. LMP2A affected apoptosis and cell-cycle checkpoints by dysregulating the expression of apoptosis regulators such as BCl-xL and the tumor suppressor retinoblastoma-associated protein 1 (RB1). LMP2A cooperated with MYC and mutant cyclin D3, two oncogenic drivers of Burkitt lymphoma, to promote proliferation and survival of primary human B cells by counteracting MYC-induced apoptosis and by inhibiting RB1 function, thereby promoting cell-cycle progression. Our results indicate that LMP2A is not a pure BCR mimic but rather rewires intracellular signaling in EBV-infected B cells that optimizes cell survival and proliferation, setting the stage for oncogenic transformation.
Epstein–Barr virus (EBV) is a member of human herpesviruses and latently infects more than 95% of the adult population. Most primary infections in children are asymptomatic and self-limiting, whereas infections in adolescents can cause infectious mononucleosis (1). The virus persists and remains dormant in memory B cells throughout the lifetime of healthy individuals (2). Importantly, EBV latent infection is associated with several malignancies of both lymphoid and epithelial origins. However, the mechanism by which EBV contributes to malignant transformation is poorly understood.
Four distinct EBV-induced latency gene expression programs have been reported in different cancer types, collectively described as latency programs 0 to III. In latency III, EBV can drive proliferation of newly infected B cells in vivo as well as transform B cells in vitro by expressing all latent gene products. These products include the EBV nuclear antigens (EBNA1, -2, -3, and -LP), latent membrane proteins (LMP1, -2A, and -2B), and EBV-encoded small RNAs as well as viral microRNAs. Expression of latency III program genes can be found in posttransplant lymphoproliferative diseases (PTLDs) affecting immunocompromised individuals. In contrast, immune surveillance in healthy individuals restricts viral gene expression to latency II (also known as the default program), which includes EBNA1, LMP1, LMP2, and noncoding viral RNAs. Latency II B cells can enter the germinal center and further differentiate to memory B cells, where latency I (EBNA1, LMP2A, and viral RNAs) or latency 0 (no viral proteins expressed) is found. Latency II is linked to the pathogenesis of Hodgkin’s lymphoma and latency I is found in subsets of Burkitt lymphoma (1).
Latency programs I to III all share the expression of EBV-encoded LMP2A, a signaling protein that like antigen receptors contains an immunoreceptor tyrosine-based activation motif (ITAM) in its cytoplasmic tail (3). LMP2A is thought to facilitate virus persistence by promoting B cell survival (4–6), and by preventing lytic virus replication (7–9). Functional studies revealed that LMP2A exerts these functions by mimicking B cell receptor (BCR) signaling by for example activating ERK/MAPK and PI-3-kinase pathways (3, 10–14), which are essential for B cell survival and development (4–6, 15). While mimicking BCR signaling, LMP2A is also thought to deprive the BCR signaling complex of its key components such as the tyrosine kinases SYK and LYN, thereby blocking BCR signaling in infected cells to prevent lytic replication (7, 8). This ultimately helps EBV to uncouple B cells from physiological BCR-dependent control mechanisms and supports EBV persistence. However, the molecular mechanisms underlying these two formally contradicting processes remain largely elusive, partly because LMP2A signaling has thus far only been studied at the level of individual proteins or signaling complexes (3, 10–14, 16, 17).
LMP2A expression is found in various B cell malignancies including Hodgkin’s lymphoma, PTLDs, and Burkitt lymphoma (18). By blocking apoptosis and promoting proliferation in premalignant cells, LMP2A is thought to facilitate early transformation during tumorigenesis. However, the molecular role for LMP2A in this process remains unclear. Here, we systematically compared LMP2A and BCR signaling in primary B cells by quantitative phosphoproteomics and gene expression profiling. We found that LMP2A mimics only a small subset of BCR signaling events including the activation of the Ca2+ initiation complex. In contrast, the majority of signaling events were LMP2A-specific and led to transcriptional changes that markedly differed from those triggered by activated BCR. Importantly, we found that one of the key consequences of LMP2A-dependent rewiring of signal transduction is to prime B cells for hyperproliferation and eventually oncogenic transformation by interfering with apoptosis and cell-cycle checkpoints.
Results
Systematic Comparative Analysis of LMP2A and BCR Signaling in Primary B Cells.
To chart LMP2A signaling in B cells and systematically compare it with BCR signaling, we focused on two B cell models: 1) EBV-infected human B cells (lymphoblastoid cell lines, hereinafter LCLs) and 2) B cells derived from tumors of either MYC- or MYC/LMP2A-expressing transgenic mice. LCLs are a bona fide model of latent EBV infection in vitro (19) and were generated by infecting healthy human peripheral blood B cells with either wild-type (EBV-WT) or LMP2A-knockout (LMP2A-KO) EBV. LMP2A and BCR were coexpressed in EBV-WT cells, whereas LMP2A-KO cells lacked LMP2A while retaining the expression of a functional BCR (SI Appendix, Fig. S1A).
We performed a quantitative phosphoproteomic profiling of 1) unstimulated LMP2A-KO cells, 2) 5-min BCR-stimulated LMP2A-KO cells (as a readout for BCR signaling), and 3) unstimulated LMP2A-expressing EBV-WT cells (as a readout for LMP2A signaling) using SILAC (stable isotope labeling by amino acids in cell culture) metabolic labeling combined with mass spectrometry (MS) (Fig. 1A). This approach enabled a site-specific, quantitative phosphoproteome-wide comparison of LMP2A and BCR signaling. For global phosphoproteome (GPome) profiling, phosphopeptides with pS (serine), pT (threonine), and pY (tyrosine) sites were enriched by strong cation exchange (SCX) chromatography in combination with titanium dioxide (TiO2) solid-phase extraction. In addition, as pY sites are underrepresented in GPome analyses, we used pY-specific antibodies to enrich for tyrosine-phosphorylated peptides in a separate set of experiments, thus systematically profiling the tyrosine phosphoproteome (pYome).
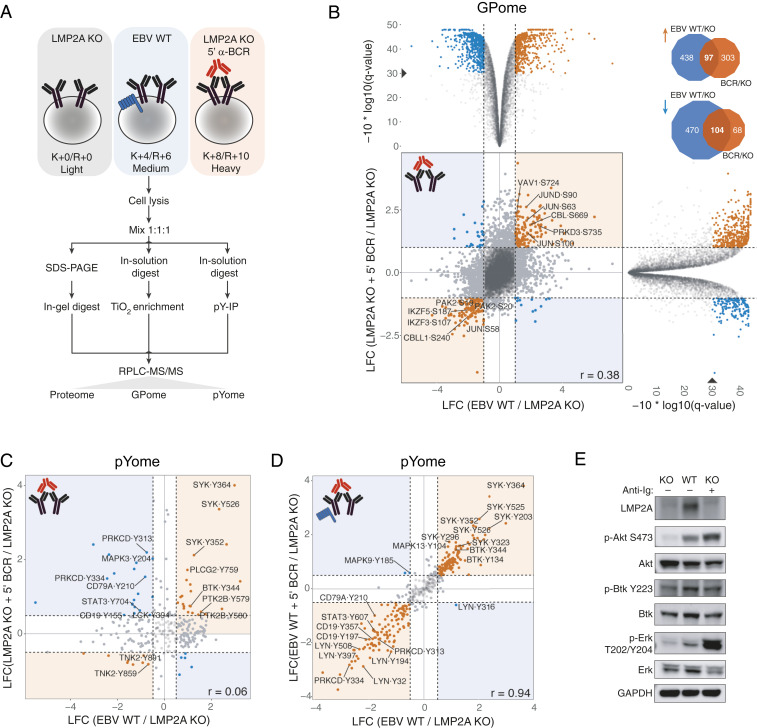
Fig. 1.
Systematic phosphoproteome analysis of LMP2A and BCR signaling in human B cells. (A) Schematic representation of the SILAC-MS approach used for the (phospho)proteomics profiling of LCLs. Cell lines were cultured in the indicated SILAC medium. LMP2A-KO cells were BCR-stimulated for 5 min (red) or left unstimulated (gray). Lysates were mixed in a 1:1:1 ratio and digested with trypsin. Phosphopeptides were enriched by TiO2 chromatography (GPome) or immunoprecipitation of phosphotyrosine (pYome) and analyzed by LC-MS/MS. For total protein analysis (proteome), protein lysates were separated by one-dimensional PAGE, digested with trypsin, and analyzed by LC-MS/MS. (B) Scatterplot of log2 SILAC ratios for the LCL GPome, normalized to unstimulated LMP2A-KO cells. The Spearman’s rank correlation (r) between normalized ratios is shown. Orange and blue dots mark significantly concordantly and discordantly regulated p sites between the two conditions, respectively. Selected protein hits are labeled (gene symbol, residue, and position within the protein are indicated). (B, Top and Right) Volcano plots of individual comparisons are shown. In each plot, normalized log2 SILAC ratios are shown on the x axis while phred-transformed Benjamini–Hochberg adjusted P values (q values) are shown on the y axis. A solid triangle marks a 0.1% false discovery rate level [corresponding to a −10*log10(q) value of 30]. (B, Top Right) Venn diagrams showing the overlap between significantly up- (orange arrow) and down-regulated (blue arrow) p sites in EBV-WT and BCR-stimulated LMP2A-KO conditions. (C) Scatterplot of log2 SILAC ratios for the pYome of BCR-stimulated LMP2A-KO LCLs, normalized to unstimulated LMP2A-KO cells. The Spearman’s rank correlation between normalized ratios is shown. Orange and blue dots mark significantly concordantly and discordantly regulated p sites between the two conditions, respectively. Selected protein hits are labeled (gene symbol, residue, and position within the protein are indicated). (D) Same as C, for BCR-stimulated EBV-WT LCLs. (E) Immunoblots of selected proteins downstream of BCR signaling in the EBV-WT and LMP2A-KO cell lines used in the MS analysis. Anti-Ig indicates BCR stimulation. LCF, log2 fold change.
In the GPome of LCLs, 16,451 class I p sites (p sites with localization probability > 75%) were detected in four biological replicates, of which 7,600 were quantifiable across all three conditions (Dataset S1). These measurements were highly reproducible between replicates (r = 0.75 to 0.93; SI Appendix, Fig. S1B), with 90.5% of the quantified p sites residing on serine, 9.0% on threonine, and 0.5% on tyrosine. A total of 909 class I pY sites were quantified in the corresponding pYome (Dataset S2). A total of 1,109 and 572 significantly regulated p sites (Benjamini–Hochberg adjusted P < 0.001 and absolute log2 fold change ≥ 1) were identified in 734 and 458 proteins in EBV-WT and BCR-stimulated LMP2A-KO cells, respectively. Surprisingly, only 12% of these p sites were shared between LMP2A and BCR signaling (Fig. 1B), suggesting marked differences in their signal transduction pathways. This finding is further supported by a low correlation of phosphorylation changes in both the GPome (r = 0.38; Fig. 1B) and pYome (r = 0.06; Fig. 1C), as well as in a phosphotyrosine Western blot (SI Appendix, Fig. S1C). For instance, core BCR signaling effectors such as the BCR subunits CD79a/b, the BCR coreceptor CD19, and the kinase PRKCD exhibited opposite responses to LMP2A and BCR signaling. LMP2A expression reduced tyrosine phosphorylation of these proteins, which conversely was induced by BCR stimulation (Fig. 1 C and E). In contrast, tyrosine phosphorylation of other BCR signaling effectors including SYK, BTK, and PLCγ2 (Fig. 1C) was observed in response to both LMP2A and BCR signaling, in line with previous reports (11, 12, 15, 20). These results were further supported by a Reactome pathway (21) enrichment analysis of proteins harboring at least one significantly regulated p site in the GPome (SI Appendix, Fig. S1E). This analysis also indicated that LMP2A and BCR signaling differentially affects phosphorylation of proteins involved in cell-cycle checkpoints, chromatin organization, and splicing.
Previous studies showed that LMP2A can block BCR signaling, acting as a dominant-negative and uncoupling B cells from antigen-based stimuli (7–9). However, this observation partly contradicts the notion that LMP2A mimics the BCR (5, 6, 13, 15, 22). To address this issue, we globally profiled tyrosine phosphorylation events in LMP2A-expressing LCLs that were stimulated for 5 min by BCR cross-linking, and compared their response with an unstimulated control. In line with previous studies, the pYome of stimulated and unstimulated LMP2A-expressing LCLs exhibited a nearly perfect correlation (r = 0.94), confirming that LMP2A expression substantially abrogates the BCR signaling response (Fig. 1D and SI Appendix, Fig. S1D). LMP2A expression preserves BCR proximal kinase activation (e.g., increased tyrosine phosphorylation of SYK and BTK) while preventing downstream activation events (such as tyrosine phosphorylation of PRKCD), suggesting that LMP2A largely uncouples BCR downstream effectors from antigen-based external stimuli. Together, our results indicate that LMP2A profoundly rewires BCR signaling in B cells.
LMP2A expression is often found in transformed B cells, for example Burkitt lymphoma cells that are characterized by MYC dysfunction. In this context, LMP2A cooperates with MYC to drive tumor growth in transgenic mouse models (23–25). To test whether our results can be generalized to transformed MYC-dependent B cells, we characterized LMP2A and BCR signaling in an orthogonal model. We subjected mouse B cells derived from MYC- or MYC/LMP2A-expressing tumors to GPome (Dataset S3) and pYome (Dataset S4) profiling by SILAC-MS. We measured quantitative phosphorylation changes in 1) MYC-expressing B cells that were either untreated or stimulated for 5 min by BCR cross-linking (as a readout for BCR signaling) and 2) unstimulated B cells that coexpressed MYC and LMP2A (as a readout for LMP2A signaling). Similar to LCLs, LMP2A and BCR signaling responses in mouse B cells exhibited a low correlation both within the GPome (r = 0.12; SI Appendix, Fig. S2A) and the pYome (r = 0.056; SI Appendix, Fig. S2B). Also, in MYC-transformed mouse B cells, LMP2A led to decreased phosphorylation of BCR signaling proteins including the BCR subunit CD79a and downstream effectors such as PRKCD. In general, the pathways for proteins being either more or less phosphorylated in the presence of LMP2A largely overlapped between LCLs and transformed mouse B cells (SI Appendix, Figs. S1E and S2C). For example, phosphorylation of cell-cycle, BCR, and cytoskeleton regulators was concordantly regulated in both models. These results indicate a high degree of consistency between LCLs and transformed B cells. However, cell type-specific observations were also made. For example, we found LMP2A-dependent MYC phosphorylation at serine 62 in the MYC-transformed B cells only. Notably, increased MYC phosphorylation was not observed upon BCR stimulation. Since pS62 in MYC was shown to be essential for MYC’s oncogenic function in various tumor models (26), our finding might be functionally relevant in the context of the known oncogenic cooperation of LMP2A and MYC.
Rewiring of BCR Signaling Targets Selected Signal Transduction and Effector Nodes.
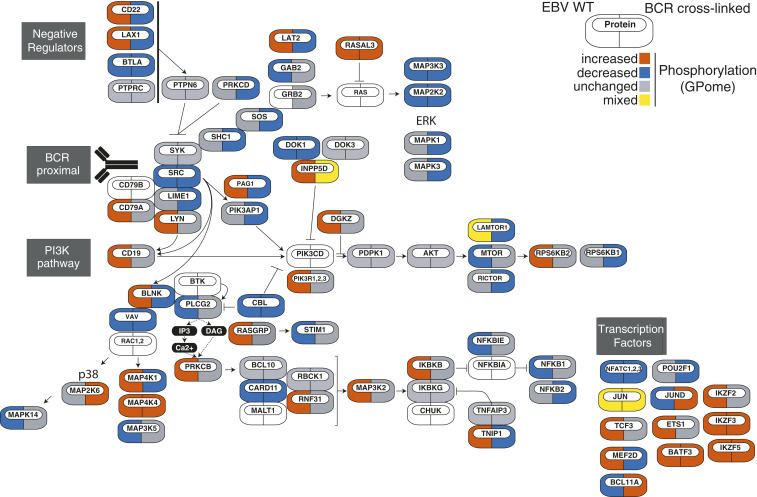
To gain systematic insight into the LMP2A signaling network, we next integrated differential phosphorylation events from the LCL GPome (Fig. 2A) and pYome (hits were defined by absolute log2 fold change ≥ 0.5 and Benjamini–Hochberg adjusted P < 0.01) (SI Appendix, Fig. S3) into a map of known BCR signaling modules and pathways. This analysis highlighted phosphorylation events that are either concordantly or discordantly regulated by LMP2A and BCR signaling. Tyrosine phosphorylation of PI-3-kinase, the BCR-proximal tyrosine kinase SYK, and the Ca2+ initiation complex consisting of BLNK, BTK, and PLCγ2 (SI Appendix, Fig. S3) and serine/threonine phosphorylation of members of the NFAT and Ikaros transcription factor families (Fig. 2) were concordantly regulated. In contrast, the mTOR pathway, components of the MAP kinase signaling cascade, as well as transcriptional regulators including the transcription factors TCF3 and nuclear factor κB (NF-κB) were discordantly regulated (Fig. 2 and SI Appendix, Fig. S3). Furthermore, we found specific differences in the pS/T and/or pY phosphorylation patterns of negative regulators of BCR signaling, including CD22, PTPN6, and INPP5D (SHIP1). LMP2A-directed rewiring of BCR signaling thus primarily targets selected transduction and effector nodes, which likely support EBV persistence in the B cell compartment.
Fig. 2.
Distribution of global phosphorylation events within known BCR signaling modules. The network topology of genes encoding proteins related to BCR signaling is shown. For each protein, significant changes in phosphorylation in the LCL GPome are color-coded as indicated (Left icon: EBV-WT; Right icon: BCR-stimulated LMP2A-KO). Proteins that do not exhibit concordantly increased or decreased phosphorylation across all p sites are referred to as “mixed.”
The B Lymphoid LMP2A Signaling Network.
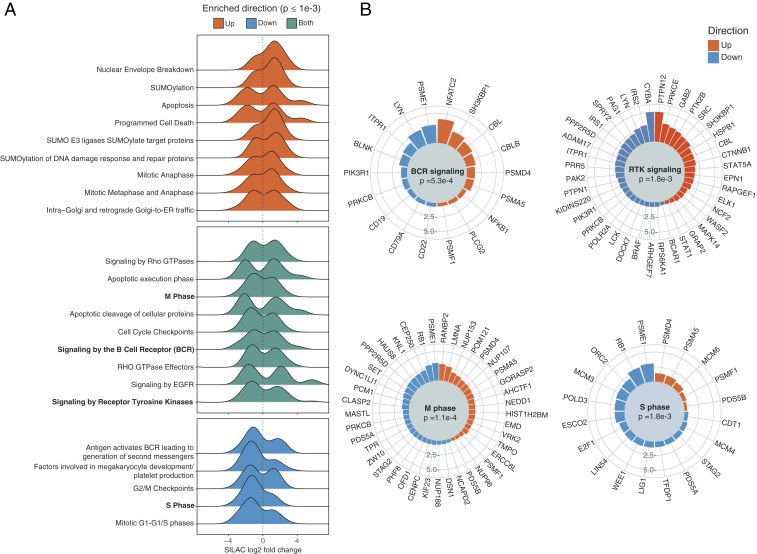
Since our GPome analysis identified >900 differentially phosphorylated sites (in 607 proteins) that were specific to LMP2A expression in LCLs, we next analyzed BCR-independent signaling events induced by LMP2A. To this end, we performed a pathway enrichment analysis using Reactome terms (Fig. 3). Among significantly enriched pathways (Benjamini–Hochberg adjusted P < 0.001), five were identified by LMP2A effectors that were significantly dephosphorylated upon LMP2A expression and that are implicated in cell-cycle regulation and second-messenger production by BCR signaling. This finding agrees with the notion that LMP2A interferes with BCR signaling and regulates cell-cycle progression by reducing phosphorylation of key residues within core cell-cycle regulators such as the tumor suppressor retinoblastoma protein 1 (RB1) (e.g., S612, S788, S795, S807, and S811) and the transcription factor E2F1 (S375). In addition, LMP2A effectors exhibiting increased phosphorylation upon LMP2A expression yielded nine enriched pathways. These included sumoylation of proteins involved in DNA damage repair, a process that has been implicated in primary EBV infection and lytic reactivation (27, 28), as well as apoptosis/programmed cell death, in line with studies showing that LMP2A promotes B cell survival (4–6). Finally, nine pathways were significantly enriched for LMP2A effectors exhibiting discordant phosphorylation patterns upon LMP2A expression. Interestingly, among other signaling processes, signaling by Rho GTPases and Rho GTPase effectors was specifically associated with this class of LMP2A effectors, possibly implicating LMP2A signaling in cytoskeleton regulation.
Fig. 3.
Pathway enrichment analysis of phosphorylation events induced by LMP2A. (A) Distribution of log2 SILAC ratios for LMP2A effectors exhibiting significantly increased (red) or decreased (blue) phosphorylation in the LCL GPome, for significantly enriched Reactome pathways. Pathways enriched in both protein sets are shown in green. (B) Radial barplot of absolute log2 SILAC ratios for members of selected pathways from A. Proteins are represented by their most regulated p site, defined as the largest absolute SILAC fold change.
Together, our results indicate that LMP2A is a potent signal transducer affecting phosphorylation of a plethora of functionally distinct biological processes such as the cell cycle, apoptosis, cytoskeleton dynamics, and DNA repair.
LMP2A Expression and BCR Signaling Control Distinct Gene Expression Programs.
The multifaceted signaling response observed downstream of LMP2A and its marked difference from the activated BCR prompted us to analyze gene expression changes induced by these two signaling events. To this end, we performed RNA sequencing (RNA-seq) of 1) unstimulated LMP2A-KO cells, 2) LMP2A-KO cells after 3 h of BCR stimulation, and 3) unstimulated EBV-WT cells, measuring messenger RNA (mRNA) expression levels in five biological replicates (Dataset S5) to systematically compare the impact of LMP2A and BCR signaling on gene expression.
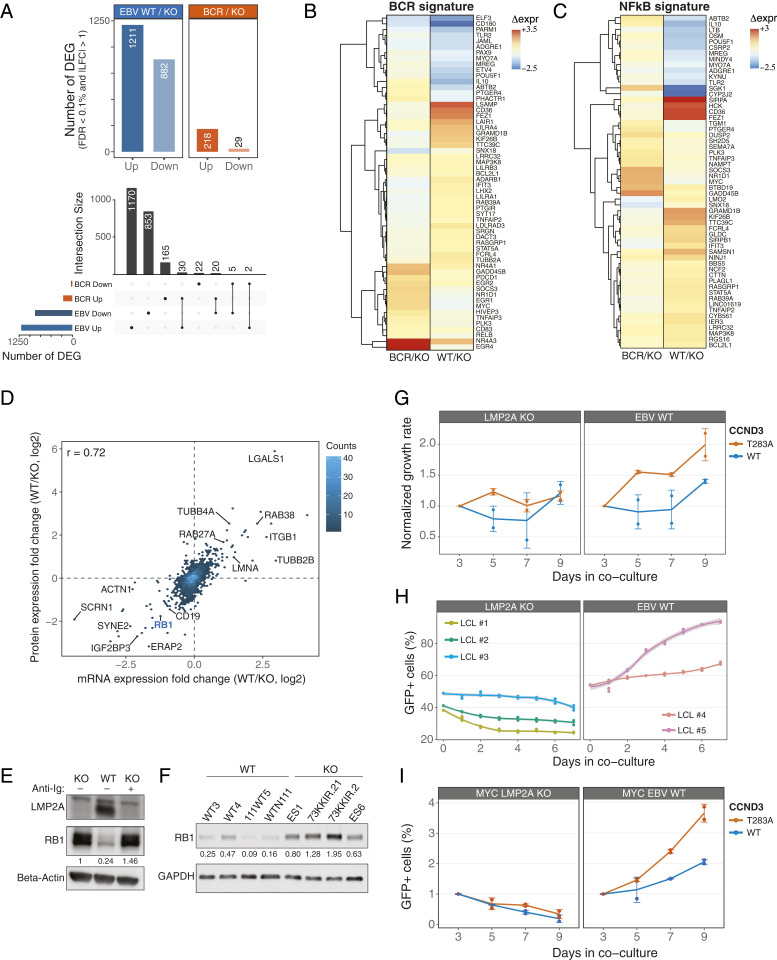
LMP2A signaling induced profound transcriptional changes with 2,093 differentially expressed genes (Benjamini–Hochberg adjusted P < 0.001 and absolute log2 fold change ≥ 1) compared with unstimulated LMP2A-KO cells, whereas BCR stimulation significantly altered the expression of 247 genes (Fig. 4 A, Top). Principal-component analysis and hierarchical clustering of gene expression patterns indicate a clear distinction of BCR- and LMP2A-associated gene expression programs (SI Appendix, Fig. S4 A and B). These results are consistent with a limited overlap in the signaling response elicited by these stimuli. Differentially expressed genes exhibited a marginal yet statistically significant (P < 1e-10, Fisher’s exact test) overlap between LMP2A and activated BCR, with 35 genes (1.7 and 14.2% of significant hits in EBV-WT and BCR-stimulated cells, respectively) concordantly regulated in both conditions (Fig. 4 A, Bottom). Among the concordantly up-regulated genes was the DNA-binding protein inhibitor ID3, which negatively regulates the basic helix–loop–helix transcription factors. It was shown that ID3 is mediating signals from the BCR to cell-cycle progression during the humoral immune response (29) and our data suggest that LMP2A is mimicking this process.
Fig. 4.
LMP2A interferes with the RB1 cell-cycle checkpoint to promote cell proliferation. (A) Total number of differentially expressed genes (DEGs) for the indicated comparisons (Top). The UpSet plot (Bottom) shows the overlap between the indicated sets of DEGs. (B and C) Hierarchical clustering of mRNA expression patterns for BCR (B) and NF-κB (C) gene signatures. Regularized log2 expression values are row − mean subtracted. (D) Density scatterplot of changes in protein (log2 SILAC ratio) and mRNA expression (log2 fold change) levels in EBV-WT relative to LMP2A-KO LCLs. The number of genes in each bin is indicated (color key). The Spearman’s rank correlation coefficient is shown, and selected genes are labeled. (E and F) Immunoblot analyses of RB1 expression in EBV-WT and LMP2A-KO cell lines used in the MS analysis (E) and in additional LCLs (F). (G) Normalized GFP+ expression after transduction of retrovirus expressing GFP-tagged WT or mutant CCND3 in LCLs. Changes in %GFP were normalized to day 3 posttransduction. (H) Percentages of GFP+ LCLs from coculturing transduced GFP-tagged T283A CCND3 cells with parental lines. (I) Same as H, for MYC-overexpressing cells. Graphs represent combined data from two (H and I) or three (G) experiments.
To gain insight into functional categories of differentially regulated genes, we performed a gene signature enrichment analysis using gene signatures previously defined in BCR-dependent lymphoma models (30) and a lenient gene expression cutoff (absolute fold change ≥ 1.5). This analysis identified 29 (19 positively associated and 10 negatively associated) and 28 (all positively associated) signatures that were enriched in LMP2A-expressing cells and BCR-stimulated LMP2A-KO cells, respectively, with an overlap of 4 signatures between the two conditions (Dataset S6). Notably, the latter were related to BCR activation and stimulation of NF-κB, a transcription factor activated by a number of stimuli including BCR signaling, and were enriched in both conditions (Fig. 4 B and C). However, the genes driving the enrichment of these signatures were largely stimulus-specific, in line with the differential phosphorylation events mapping to these pathways (Fig. 4 B and C) and highlighting largely distinct gene expression programs. In addition, genes involved in the regulation of apoptosis were significantly overrepresented (P < 1e-6, one-sided Fisher’s exact test) among LMP2A-specific differentially expressed genes. LMP2A led to a significant down-regulation of proapoptotic genes such as BIM and BNIP3L, whereas antiapoptotic genes such as BCL2L10 were up-regulated. This result indicates that LMP2A induces a prosurvival program in B cells.
Because gene and protein expression levels are not necessarily highly correlated (31), we profiled proteomic changes induced by LMP2A by measuring total protein expression by MS. This analysis quantified the expression of 4,080 proteins across all five biological replicates (Dataset S7), and identified 273 significantly regulated hits (Benjamini–Hochberg adjusted P < 0.001 and absolute log2 fold change ≥ 1). Protein and mRNA expression changes in EBV-WT LCLs relative to LMP2A-KO cells were highly correlated (r = 0.72; Fig. 4D). In line with previous studies and our gene expression analysis, regulators of apoptosis and cell-cycle checkpoints were among the most regulated proteins. We validated LMP2A-induced down-regulation of the proapoptotic factor BIM, as well as up-regulation of the antiapoptotic effectors BCL2L10 and BCL-xL (SI Appendix, Fig. S5A).
Interestingly, the cell-cycle regulator and tumor suppressor RB1 was significantly down-regulated in LMP2A-expressing cells at both the mRNA and protein levels (Fig. 4 D and E), a finding that was further validated in four independent LMP2A-expressing and LMP2A-negative LCL pairs (Fig. 4F).
LMP2A Promotes Cell Proliferation by Interfering with the RB1 Cell-Cycle Checkpoint.
Because LMP2A signaling influences the expression and activity of cell-cycle and survival regulators and because of its expression in specific lymphoma types, we hypothesized that LMP2A cooperates with oncogenic drivers in EBV-infected B cells. To test this hypothesis, we retrovirally overexpressed MYC and a cyclin D3 mutant (CCND3 T283A) (either alone or in combination) together with green fluorescent protein (GFP) in LMP2A-KO and LMP2A-expressing EBV-WT LCLs. Besides MYC dysregulation, CCND3 T283A is one of the most recurrent oncogenic events in Burkitt lymphoma cells (32). This mutation is thought to stabilize cyclin D3 expression, thereby inhibiting RB1 function via CDK4/6 (32). To test if LMP2A cooperates with wild-type and/or mutant CCND3 in vitro, we performed a competitive proliferation assay by coculturing nontransduced parental LCLs with those expressing GFP-tagged cyclin D3 WT or mutant versions. Interestingly, neither WT CCND3 nor the mutant variant provided a competitive advantage to LMP2A-KO cells. However, the expression of mutant CCND3 in LMP2A-expressing LCLs resulted in a significant outgrowth of these cells (Fig. 4G). The effect was specific for mutant CCND3 because transduction of GFP-tagged WT CCND3 did not change the relative percentage of GFP+ EBV-WT cells. This result was validated in three additional LMP2A-KO and two EBV-WT LCLs (Fig. 4H). More strikingly, MYC-overexpressing cells only survived in the presence of LMP2A and the combination of LMP2A, MYC, and mutant CCND3 showed the strongest outgrowth in our assay, suggesting an additive effect in promoting cell survival and proliferation (Fig. 4I). These results indicate that LMP2A, MYC, and CCND3 T283A jointly induce survival and hyperproliferation of B cells, most likely by interfering with the RB1 cell-cycle control checkpoint and by induction of apoptosis inhibitors such as BCL-xL.
Discussion
One of the main features of EBV is its ability to persist throughout the life of the host, where it remains dormant in long-lived memory B cells (2). EBV can reach this cell subset by adapting as B lymphocytes differentiate from naïve to memory cells without reducing their fitness. Normal B cell differentiation and survival are tightly controlled by BCR signaling (33, 34). Hence, EBV has developed strategies to hijack BCR signaling in order to take over control of B cell fate decisions. Previous studies from our group (4) and others (5, 6, 35) demonstrated that EBV utilizes LMP2A as a functional BCR mimic to promote B cell survival and differentiation, while simultaneously blocking physiological B cell responses. However, the molecular pathways engaged by LMP2A to control B cell fate decisions, as well as its role in lymphomagenesis, remained elusive.
Identification of the ITAM signaling motif in LMP2A as well as its interaction with LYN and SYK have further supported a model where LMP2A acts as a BCR mimic. However, LMP2A and BCR are structurally distinct signal transducers and to what extent LMP2A mimics BCR signaling at the molecular level is not known. Here, we elucidated the LMP2A signaling network in primary B cells and systematically compared it with the BCR signaling response by using an integrated multiomics approach. Consistent with previous studies (11, 12, 17), LMP2A utilized BCR signaling effectors, such as SYK, BLNK, BTK, and PLCγ2 (Figs. 1 and 2). Surprisingly, however, the majority of LMP2A signaling events at the phosphoproteome level strongly differed from those induced by the BCR, underscoring that LMP2A precisely rewires BCR signaling rather than mimicking it. This rewiring process is also reflected by profound LMP2A-dependent changes in the transcriptome that differ from those induced by BCR stimulation (Fig. 4). Many changes in the proteome driven by LMP2A are a consequence of a fundamental rewiring of the transcriptome. While LMP2A affects pathways in common with direct BCR stimulation, LMP2A regulates distinct subsets of genes within these pathways. LMP2A expression, in general, is not as powerful as BCR cross-linking for inducing immediate-early, BCR, or NF-κB–associated signaling genes. Instead, LMP2A expression elicits a unique interferon response, as compared with BCR stimulation, perhaps as an unavoidable consequence of viral transformation. Moreover, LMP2A specifically induces BCL-xL and BCL2L10, antiapoptotic proteins which likely help ensure the survival of infected cells. Our results uncovered LMP2A as a unique signal transducer and suggest that LMP2A has a wider impact on many cellular pathways.
LMP2A expression in various B cell lymphomas is thought to contribute to lymphomagenesis through largely unknown mechanisms. Previous studies in a murine MYC-driven lymphoma model (25, 36) showed that LMP2A interferes with the G1 checkpoint during cellular transformation. Here we demonstrated that LMP2A down-regulates cell-cycle checkpoint genes like CDKN1B (p27) and CHEK1 and the tumor suppressor RB1. RB1 expression was repressed at both the mRNA and protein levels by LMP2A in human B cells. Lower levels of RB1 provided an advantageous platform for cellular outgrowth when combined with an oncogenic mutant form of CCND3 that further suppresses RB1 protein expression (Fig. 4). Since recurrent CCND3 mutations have been reported in endemic as well as sporadic Burkitt lymphoma (32, 37), the observed cooperative effect might influence proliferation and potentially other characteristics of these tumors. Moreover, alteration of the apoptotic threshold by up-regulation of BCL-xL and dysregulation of other survival signals further contributes to the functional cooperation of LMP2A and oncogenic drivers like MYC. Our data indicate that LMP2A is versatile in coupling with different oncogenes to prime B cells for abnormal proliferation and eventually malignant transformation.
Taken together, this study elucidates the signaling pathways that are regulated by LMP2A to influence B cell differentiation and survival. While we identified some commonalities in signaling output between LMP2A and BCR cross-linking, this study revealed many more profound differences between LMP2A and BCR signaling, in terms of changes in the phosphoproteome, the total proteome, and the transcriptome. Finally, with regard to LMP2A’s effect on lymphomagenesis, LMP2A clearly interferes with cell-cycle checkpoint proteins, in particular RB1, and, moreover, it cooperated with MYC by inhibiting its proapoptotic effects. Our comprehensive map of EBV LMP2A signaling in human and mouse B cells may guide further genetic, biochemical, and functional studies that focus on the interplay between EBV and B cells and may help to identify novel cellular targets in EBV-associated cancers.
Materials and Methods
Cell Culture and BCR Stimulation.
EBV-transformed LCLs were generated as described previously (7). Mouse lymphoma cell lines were derived from primary tumor cells from λ-MYC and LMP2A/λ-MYC transgenic mice (23). All cell lines were cultured in RPMI (Corning) supplemented with 10% fetal bovine serum (Corning) and penicillin/streptomycin (Sigma) at 37 °C and 5% CO2. SILAC labeling of cell lines was performed as previously described (38). Briefly, cells were cultured in SILAC-RPMI medium 1640 without arginine and lysine (Thermo Fisher Scientific) supplemented with 10% dialyzed fetal calf serum (Sigma) and SILAC amino acids (Cambridge Isotopes). “Light” SILAC medium contained [12C614N4]-l-arginine and [12C614N2]-l-lysine; “medium” SILAC medium contained [13C614N4]-l-arginine and l-lysine-4,4,5,5-D4; and “heavy” SILAC medium contained [13C615N4]-l-arginine and [13C615N2]-l-lysine. For BCR stimulation, cells were first starved in RPMI without supplements for 20 min. In LCLs, the BCR was subsequently stimulated for 5 min (for all proteome analyses) or 3 h (for RNA-seq) at 37 °C with 20 µg/mL F(ab)2 anti-human pan-immunoglobulin (Ig) antibody (Southern Biotech). Mouse B cells were stimulated with 10 µg/mL F(ab)2 anti-mouse IgM antibody (Jackson ImmunoResearch). For proteome and global phosphoproteome analyses, SILAC-labeled cells were lysed in 1% Nonidet P-40 lysis buffer containing 50 mM Tris⋅HCl (pH 7.8), 150 mM NaCl, 0.5 mM ethylenediaminetetraacetate (EDTA), 2 mM Na3VO4, and 1 mM NaF, supplemented with a protease inhibitor mixture (Roche). Cell lysis for proteomic profiling of tyrosine phosphorylation was performed according to the manufacturer’s instructions by using a urea-based lysis buffer (PTMScan Kit; Cell Signaling Technology).
Reagents.
Antibodies used for immunoblots were BLNK, p-BLNK (Y96), ERK, p-ERK, p-SYK, BTK, p-BTK, CD79a, p-CD79a, p-CD19, RB, BCL2L10, and BCL-xL (Cell Signaling Technology), SYK and CD19 (Santa Cruz), p-Tyr (Millipore), p-BLNK Y84 (BD Biosciences), actin (Sigma), and BIM and GAPDH (Abcam). The antibody against LMP2A clone 14B7 was previously described (9). Tyrosine-phosphorylated peptides were enriched by the PTMScan Phospho-Tyrosine Rabbit mAb Kit (P-Tyr-1000; Cell Signaling Technology).
Retroviral plasmids expressing GFP-tagged WT or T283A CCND3 (32) and c-MYC were contributed by the L.M.S. laboratory. All viral vectors were packaged by cotransfecting pMSCV gag/pol and pHitg (kindly provided by Eva Gottwein, Northwestern University, Chicago, IL) in 293T cells using TransIT-LT1 Transfection Reagent (Mirus). The virus supernatants were concentrated using a Lenti-X concentrator (Clontech) according to the manufacturer’s protocol.
Competitive Assay and Flow Cytometry.
LCLs were plated at 6 × 105 cells and transduced with GFP-tagged WT or T283A CCND3 retroviruses. Percentages of GFP+ cells were then measured every 48 h after transduction using FACSCanto II (BD Biosciences). For competitive assays, LCLs transduced with T283A CCND3 were sorted using the BD FACSAria SORP System. GFP+ LCLs were subsequently mixed with the respective parental lines at a 1:1 ratio and the change in GFP+ cells was measured by flow cytometry. Surface BCR expression was determined by staining LCLs with goat anti-human Ig-Alexa Fluor 647 (Southern Biotech).
Global Proteome Analysis.
For protein expression analysis, cell lysates of light-labeled unstimulated LMP2A-KO, medium-labeled unstimulated LMP2A-WT, and heavy-labeled LMP2A-KO after 3 h of BCR stimulation were mixed in a 1:1:1 ratio. Extracted proteins were then separated by sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE) using precast Bis-Tris minigels (NuPAGE Novex 4 to 12%; Life Technologies) and visualized by staining with Coomassie brilliant blue (Serva). Each lane was cut into 23 slices, reduced with dithiothreitol (Sigma-Aldrich), alkylated with iodoacetamide (IAM; Sigma-Aldrich), in gel-digested with trypsin (Promega), extracted, and analyzed by mass spectrometry.
Protein Digestion and Phosphopeptide Enrichment for Global Phosphoproteome Analysis.
For phosphoproteome analysis, Halt Protease and Phosphatase Inhibitor Mixture (100×) (Thermo Fisher Scientific) and 0.5 M EDTA were added to SILAC-labeled cell lysates. These were then sonicated using a Bioruptor and the supernatant was carefully collected after centrifugation at 14,000 × g for 10 min. Protein concentration was measured by Bradford assay, and equal amounts of SILAC-labeled cell lysates were mixed, treated with 10 mM Tris(2-carboxyethyl)phosphine (Thermo Fisher Scientific) and 15 mM IAM for 1 h at 37 °C in the dark for reduction and alkylation, and precipitated with acetone overnight. The precipitate was redissolved in 8 M urea (Sigma-Aldrich) and 25 mM ammonium bicarbonate (Sigma-Aldrich) and then diluted to a urea concentration <1 M. Proteins were then digested with sequencing-grade trypsin (Promega) at an enzyme/protein ratio of 1:50 (weight [wt]/wt) at 37 °C overnight. Digestion was stopped by adding trifluoroacetic acid (TFA; Roth) to a final concentration of 0.5%.
Digested peptides were collected by centrifugation at 14,000 × g for 15 min and desalted on Waters Oasis HLB solid-phase extraction columns. The eluted cleared peptides were then evaporated to dryness on a SpeedVac concentrator (Thermo Fisher Scientific). Phosphopeptides were enriched directly from desalted peptides of mixed cell lysates before high-pH C18 prefractionation. In brief, digested dried peptides were redissolved in incubation buffer (80% [volume (vol)/vol] acetonitrile [ACN], 5% [vol/vol] TFA, 5% [vol/vol] glycerol), and then incubated with TiO2 beads (10 μm; GL Sciences) at a 1:8 peptide:bead ratio (wt/wt) with end-over-end rotation at room temperature for 20 min. The peptide concentration was maintained at around 2 to 5 mg/mL during the incubation. After incubation, all TiO2 beads were loaded onto an empty spin column (5-μm frit; Hoefer) and washed three times each with incubation buffer, 80% (vol/vol) ACN, 5% (vol/vol) TFA, and 60% (vol/vol) ACN, 0.1% (vol/vol) TFA. Phosphopeptides were then eluted with 1% NH4OH (pH ≥ 10.5) and acidified immediately with 10% (vol/vol) TFA to adjust the pH to less than 3. The eluate was desalted on a C18 spin column and fractionated using basic reverse-phase chromatography on an Agilent 1100 Series Capillary LC System. Enriched phosphopeptides were resuspended in 1 μL mobile phase B (10 mM ammonium hydroxide in 80% ACN, pH 10) and 49 μL mobile phase A (10 mM ammonium hydroxide in water, pH 10), injected onto an XBridge C18 column (3.5 μm, 150 × 1.0 mm; Waters), and eluted using mobile phases A and B with a gradient (2 to 5% B, 0 to 7 min; 5 to 15% B, 7 to 27 min; 15 to 30% B, 27 to 42 min; 30 to 50% B, 42 to 50 min; 50 to 90% B, 50 to 51 min; 90 to 90% B, 51 to 56.5 min; 90 to 2% B, 56.5 to 57 min; 2 to 2% B, 57 to 64 min) at a flow rate of 60 μL/min. Peptides were detected at 214 nm; 50 fractions were collected along with the liquid chromatography (LC) separation in a time-based mode from 0 to 64 min and then concatenated into 14 fractions by combining fractions 6, 18, 30, and 42; 7, 19, 31, and 43; and so forth (especially 0 to 5 min as fraction 5 and 54 to 64 min as fraction 14). The combined fractions were then dried on a SpeedVac concentrator and stored at −80 °C.
Phosphopeptide Enrichment for pYome Analysis.
Antibody-based enrichment for tyrosine-phosphorylated peptides was performed with the Phospho-Tyrosine Rabbit mAb Kit (P-Tyr-1000; Cell Signaling Technology). Cell lysates from SILAC-labeled unstimulated LMP2A-KO, unstimulated LMP2A-WT, and LMP2A-KO after 3 h of BCR stimulation were mixed in a 1:1:1 ratio according to protein concentration and prepared according to the manufacturer’s instructions (Cell Signaling Technology).
Liquid Chromatography-Tandem Mass Spectrometry Analysis and Data Processing.
All samples were measured on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) coupled to an UltiMate 3000 Rapid Separation LC System (Dionex). Samples were preconcentrated and desalted on a trap column (5 mm length, 30 μm inner diameter; Thermo Fisher Scientific) at 10 μL/min in loading buffer (2% [vol/vol] ACN, 0.1% formic acid [FA]). Peptides were separated on a self-made capillary column (ReproSil-Pur 120 C18-AQ, 1.9 μm, 300 × 0.075 mm; Dr. Maisch) using buffer A (0.1% FA in water) and buffer B (0.1% FA in 80% [vol/vol] acetonitrile) with a 90-min linear gradient (2 to 2% B, 0 to 3 min; 2 to 40% B, 3 to 73 min; 40 to 60% B, 73 to 76 min; 60 to 90% B, 76 to 76.1 min; 90 to 90% B, 76.1 to 82 min; 90 to 2% B, 82 to 82.1 min; 2 to 2% B, 82.1 to 90 min) at a constant flow rate of 300 nL/min. The mass spectrometer was operated in data-dependent acquisition mode using a top-20 method with a survey scan resolution setting of 120,000 full width at half maximum (FWHM) and a tandem mass spectrometry (MS/MS) resolution setting of 30,000 FWHM at 200 m/z. Higher-energy collisional dissociation was performed with an normalized collision energy setting of 28% and an isolation width of 1.4 m/z. Automatic gain control target values and maximum ion injection time for MS and MS/MS were set at 1 × 106 in 40 ms and 1 × 105 in 128 ms, respectively.
Raw files were processed using MaxQuant (v1.5.5.1; Max Planck Institute of Biochemistry) (39). MS/MS spectra were searched against a UniProtKB human database containing 92,954 protein entries (downloaded February 2017) supplemented with 245 frequently observed contaminants via the Andromeda search engine (40). For phosphoproteome analysis, precursor and fragment ion mass tolerances were set to 6 and 20 parts per million after initial recalibration, respectively. STY phosphorylation, protein N-terminal acetylation, and methionine oxidation were allowed as variable modifications. Cysteine carbamidomethylation was set as a fixed modification. Enzyme specificity was set to trypsin allowing N-terminal cleavage to proline. Minimum peptide length was set to seven amino acids, with a maximum of two missed cleavages. The false discovery rate (FDR) was set to 1% on peptide, modification site, and protein level using a forward-and-reverse concatenated decoy database approach.
For SILAC quantitation, multiplicity was set to 3 for triple (Lys+0/Arg+0, Lys+4/Arg+6, Lys+8/Arg+10) labeling. At least two ratio counts were required for peptide quantitation. Both the “match between runs” and “requantify” options of MaxQuant were enabled. For proteome analysis, variable modifications did not consist of STY phosphorylation and all of the other parameter settings were identical to the phosphoproteome analysis.
RNA-Seq.
RNA was extracted from five replicates (107 cells each) for each condition using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. DNase digestion and RNA cleanup were done following the instructions in appendix E of the RNeasy Mini Kit (Qiagen) manual. RNA concentrations were determined using the Qubit RNA HS Assay Kit and a Qubit 3 fluorometer (both Thermo Fisher Scientific). RNA quality control was done using a Bioanalyzer 2100 and RNA Nano Chips (Agilent Technologies); RNA integrity number values for the individual samples ranged between 8.8 and 10. Fifty-base pair single-read RNA sequencing was done at the Genomics and Proteomics Core Facility at the German Cancer Research Center (DKFZ) on a HiSeq 2000 v4 (Illumina).
Bioinformatic Data Analysis.
Proteome data analysis.
Data postprocessing was performed using Perseus (v1.6.0.2). After removing all decoy hits and potential contaminant entries, identified phosphosites (p sites) with localization probability <0.75 were discarded.
All subsequent analyses were carried out using R v3.6.0 (41) and Bioconductor (42) using log2-transformed SILAC ratios for proteins/p sites. For the total proteome and GPome, only proteins/p sites detected across all biological replicates were considered. For the LCL pYome, p-site detection in at least four replicates was required. Differential phosphorylation/protein expression analyses were performed using the limma package (v3.42.0). For the total proteome and GPome, statistically significant hits were defined by an absolute log2 fold change ≥1 and a Benjamini–Hochberg adjusted P ≤ 1e-3. For the pYome, an absolute log2 fold change ≥0.5 and a Benjamini–Hochberg adjusted P ≤ 1e-2 were used as a cutoff to define significant hits. Pathway enrichment analysis was performed with the ReactomePA package (v1.30.0) (43), using Entrez gene identifiers and Reactome database terms. A Benjamini–Hochberg adjusted P ≤ 0.01 was used to define significantly enriched pathways.
RNA-seq data analysis.
Raw data were subjected to quality control with FastQC v0.11.4. Short reads were aligned to the human reference genome (Ensembl GRCh38 release 90) using STAR v2.4.2a. Gene count tables were generated while mapping, using Gencode gene annotation release 27 (GRCh38.p10). Exploratory analyses and differential gene expression analysis were carried out with DESeq2 v1.20.0 (44). For sample clustering and principal-component analysis, genes with zero counts across all samples were removed from the analysis. For differential expression analysis, independent filtering and a 0.1% FDR level (Benjamini–Hochberg adjusted P ≤ 1e-3) were used. A Wald test was used for pairwise comparisons, whereas a likelihood ratio test was used to extract significant differences across three conditions. Hits with an absolute log2 fold change ≥1 were retained.
Supplementary Material
Acknowledgments
We thank members of the T.O. and R.L. laboratories for help in completion of this study. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, Deutsche Krebshilfe (DKH 70113536) (to T.O.), and NIH (R01 CA073507) (to R.L., K.F., M.I., and S.J.S.). Flow cytometry and cell sorting were performed at the Northwestern University Flow Cytometry Core Facility (supported by Cancer Center Support Grant NCI CA060553) and Interdepartmental Immunobiology Flow Cytometry Core Facility.
Footnotes
Competing interest statement: F.C. is a co-founder of enGene Statistics GmbH. T.O. reported grants from Merck KGaA, and Gilead, and personal fees from Merck KGaA, Roche, Kronos Bio, all outside the submitted work.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007946117/-/DCSupplemental.
Data Availability.
Mass spectrometry and RNA-sequencing data reported in this paper have been deposited in the PRIDE Proteomics Identifications Database (PRIDE Project: PXD018566). RNA-sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) with BioProject accession number PRJNA622883.
References
- 1.Longnecker R., Kieff E., Cohen J. I., “Epstein-Barr virus” in Fields Virology, Fields B. N., Knipe D. M., Howley P. M., Eds. (Lippincott Williams & Wilkins, Philadelphia, ed. 6, 2013), Vol. 2, pp. 1898–1959. [Google Scholar]
- 2.Babcock G. J., Decker L. L., Volk M., Thorley-Lawson D. A., EBV persistence in memory B cells in vivo. Immunity 9, 395–404 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Fruehling S., Longnecker R., The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 235, 241–251 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Caldwell R. G., Wilson J. B., Anderson S. J., Longnecker R., Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9, 405–411 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Casola S. et al., B cell receptor signal strength determines B cell fate. Nat. Immunol. 5, 317–327 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Mancao C., Hammerschmidt W., Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 110, 3715–3721 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller C. L., Lee J. H., Kieff E., Longnecker R., An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. U.S.A. 91, 772–776 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller C. L. et al., Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2, 155–166 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Fruehling S. et al., Identification of latent membrane protein 2A (LMP2A) domains essential for the LMP2A dominant-negative effect on B-lymphocyte surface immunoglobulin signal transduction. J. Virol. 70, 6216–6226 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fruehling S., Swart R., Dolwick K. M., Kremmer E., Longnecker R., Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J. Virol. 72, 7796–7806 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merchant M., Longnecker R., LMP2A survival and developmental signals are transmitted through Btk-dependent and Btk-independent pathways. Virology 291, 46–54 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Engels N. et al., Epstein-Barr virus latent membrane protein 2A (LMP2A) employs the SLP-65 signaling module. J. Exp. Med. 194, 255–264 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portis T., Longnecker R., Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23, 8619–8628 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Swart R., Ruf I. K., Sample J., Longnecker R., Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 74, 10838–10845 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamitani T. et al., Evasion of affinity-based selection in germinal centers by Epstein-Barr virus LMP2A. Proc. Natl. Acad. Sci. U.S.A. 112, 11612–11617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M., Ikeda A., Longan L. C., Longnecker R., The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 268, 178–191 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Rovedo M., Longnecker R., Epstein-Barr virus latent membrane protein 2A preferentially signals through the Src family kinase Lyn. J. Virol. 82, 8520–8528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cen O., Longnecker R., Latent membrane protein 2 (LMP2). Curr. Top. Microbiol. Immunol. 391, 151–180 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Thorley-Lawson D. A., Allday M. J., The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat. Rev. Microbiol. 6, 913–924 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Miller C. L. et al., Epstein-Barr virus protein LMP2A regulates reactivation from latency by negatively regulating tyrosine kinases involved in sIg-mediated signal transduction. Infect. Agents Dis. 3, 128–136 (1994). [PubMed] [Google Scholar]
- 21.Jassal B. et al., The Reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson-Mungerson M. A., Caldwell R. G., Bultema R., Longnecker R., Epstein-Barr virus LMP2A alters in vivo and in vitro models of B-cell anergy, but not deletion, in response to autoantigen. J. Virol. 79, 7355–7362 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bieging K. T., Amick A. C., Longnecker R., Epstein-Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 17945–17950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bultema R., Longnecker R., Swanson-Mungerson M., Epstein-Barr virus LMP2A accelerates MYC-induced lymphomagenesis. Oncogene 28, 1471–1476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fish K., Sora R. P., Schaller S. J., Longnecker R., Ikeda M., EBV latent membrane protein 2A orchestrates p27kip1 degradation via Cks1 to accelerate MYC-driven lymphoma in mice. Blood 130, 2516–2526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X. et al., Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 71, 925–936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenney S. C., Mertz J. E., Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 26, 60–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hau P. M., Tsao S. W., Epstein-Barr virus hijacks DNA damage response transducers to orchestrate its life cycle. Viruses 9, E341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan L., Sato S., Frederick J. P., Sun X. H., Zhuang Y., Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol. Cell. Biol. 19, 5969–5980 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelan J. D. et al., A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 560, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwanhäusser B. et al., Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Schmitz R. et al., Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melchers F., Checkpoints that control B cell development. J. Clin. Invest. 125, 2203–2210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burger J. A., Wiestner A., Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 18, 148–167 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Mancao C., Altmann M., Jungnickel B., Hammerschmidt W., Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood 106, 4339–4344 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fish K., Chen J., Longnecker R., Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood 123, 530–540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grande B. M. et al., Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 133, 1313–1324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corso J. et al., Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc. Natl. Acad. Sci. U.S.A. 113, 5688–5693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Cox J. et al., Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 41.R Core Team , R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, Vienna, 2015). [Google Scholar]
- 42.Huber W. et al., Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., He Q. Y., ReactomePA: An R/Bioconductor package for Reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry and RNA-sequencing data reported in this paper have been deposited in the PRIDE Proteomics Identifications Database (PRIDE Project: PXD018566). RNA-sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) with BioProject accession number PRJNA622883.