Significance
Cells must rapidly and efficiently react to DNA damage to avoid its harmful consequences. Here we report a molecular mechanism that gives rise to a model of how bacterial cells mobilize DNA repair proteins for timely response to genomic stress and initiation of DNA repair upon exposure of single-stranded DNA. We found that bacterial single-stranded DNA binding protein (SSB), a central player in genome metabolism, can undergo dynamic phase separation under physiological conditions. SSB condensates can store a wide array of DNA repair proteins that specifically interact with SSB. However, elevated levels of single-stranded DNA during genomic stress can dissolve SSB condensates, enabling rapid mobilization of SSB and SSB-interacting proteins to sites of DNA damage.
Keywords: SSB, liquid−liquid phase separation, DNA repair, phase transition, membraneless organelle
Abstract
Bacterial single-stranded (ss)DNA-binding proteins (SSB) are essential for the replication and maintenance of the genome. SSBs share a conserved ssDNA-binding domain, a less conserved intrinsically disordered linker (IDL), and a highly conserved C-terminal peptide (CTP) motif that mediates a wide array of protein−protein interactions with DNA-metabolizing proteins. Here we show that the Escherichia coli SSB protein forms liquid−liquid phase-separated condensates in cellular-like conditions through multifaceted interactions involving all structural regions of the protein. SSB, ssDNA, and SSB-interacting molecules are highly concentrated within the condensates, whereas phase separation is overall regulated by the stoichiometry of SSB and ssDNA. Together with recent results on subcellular SSB localization patterns, our results point to a conserved mechanism by which bacterial cells store a pool of SSB and SSB-interacting proteins. Dynamic phase separation enables rapid mobilization of this protein pool to protect exposed ssDNA and repair genomic loci affected by DNA damage.
Single-stranded (ss) DNA-binding proteins (SSB) are essential, ubiquitous, and abundant proteins that are involved in all aspects of genome maintenance in all kingdoms of life (1). Prokaryotes possess a structurally well-conserved SSB protein which forms nucleoprotein filaments with ssDNA to protect it from reannealing and harmful chemical alterations (1, 2). In addition, SSBs recruit essential DNA metabolic enzymes to sites of action via protein−protein interactions (1). In Escherichia coli and most bacteria, SSBs function as homotetramers, but dimeric SSBs are also known (1). Regardless of quaternary structure, functional units of SSBs contain four oligonucleotide/oligosaccharide-binding (OB) folds that assemble into a conserved three-dimensional (3D) structure. Importantly, the vast majority of bacteria encode at least one SSB variant in which the OB fold is followed by a “tail” region which can be subdivided to the intrinsically disordered linker (IDL) and C-terminal peptide (CTP, C-terminal nine amino acids) regions (Fig. 1A) (3–5). The IDL of SSB is one of the few known protein regions with a universally conserved propensity for structural disorder (6); however, its length and amino acid composition are not conserved (3, 7). In contrast, the sequence of the CTP is highly conserved among bacteria, and it mediates essential interactions with at least 18 proteins involved in nucleic acid metabolism (1, 2). Accordingly, CTP mutations weakening protein−protein interactions are lethal (8). The CTP of E. coli SSB is also known to interact with the OB fold, possibly within the tetramer or between adjacent SSB tetramers (9–14).
Fig. 1.
Bioinformatics analysis reveals conserved role of the IDL region of bacterial SSB proteins in phase separation. (A) Crystal structure of the E. coli SSB homotetramer (yellow) bound to two 35mer ssDNA molecules (gray) (Protein Data Bank ID code 1EYG). Each SSB monomer comprises an ssDNA-binding OB fold (oligonucleotide/oligosaccharide-binding domain), an IDL region (yellow line), and the conserved CTP (yellow box; last nine residues indicated). (B–D) The (B) PLAAC (PRD: prion-like domain), (C) PScore, and (D) CatGranule algorithms indicate prion-like features (PLAAC) and a propensity for LLPS for the C-terminal region of E. coli SSB (residues 113 to 177). Predicted LLPS propensities are shown as red lines, whereas the threshold values of the methods are indicated as black dashed lines on each graph. For each method, the residues with predicted values above the corresponding threshold value are the ones predicted with a positive LLPS propensity. For the CatGranule method (D), the threshold value is at y = 0; the dashed line is slightly shifted to enhance its visibility. Note that CatGranule does not provide predicted values for protein termini, resulting in its prediction curve being shorter than the SSB sequence. The algorithms used are validated predictors of LLPS propensity (74). (E) CatGranule-predicted LLPS propensity scores for SSB proteins of representative bacterial strains from 15 major phylogenetic groups of bacteria indicate broad conservation of the LLPS propensity of SSB across the bacterial kingdom. CatGranule provides a profile and a single propensity value for each protein, having a basic minimum threshold of 0.0 and a more stringent threshold of 0.5 for LLPS propensity (see Materials and Methods). For Thermotogae, Aquificae, and Cyanobacteria, less than 50% of SSBs scored positively (0%, 25% and 16.7%, respectively), while, in the other 12 groups, this fraction was above 50%. Overall, LLPS propensity is highly conserved (502 positive hits of 717 SSBs using the stringent threshold). We identified three possible reasons for the low scores of certain SSBs. 1) Thermotogae and Aquificae abound in hyperthermophilic species, wherein structurally disordered regions are generally heavily shortened and affected by adaptive sequence composition changes (75). To assess whether this could be the reason for their low predicted LLPS propensity, we identified hyperthermophilic species [optimum growth temperature ≥ 75 °C based on the Genomes Online (76) and/or BacDive databases (77)] in the whole dataset (respective SSBs highlighted in red). 2) We also identified SSB sequences that appeared to be fragments based on the absence of the CTP motif (and often the IDL too) but could not be excluded due to the lack of orthologs (highlighted in blue). 3) As we do not expect all SSB variants of a strain to drive LLPS, SSBs having an ortholog with higher predicted LLPS propensity retained in the clean dataset were identified and highlighted in green. As expected, the highlighted SSBs tend to have low scores. Cases 1 through 3 together explain ∼35% of CatGranule predictions below the basic threshold, and ∼21.4% of predictions below the stringent threshold.
In the past few years, many eukaryotic and viral proteins were shown to form dynamic liquid condensates termed as membraneless organelles that fulfill crucial functions in normal cellular physiology as well as stress tolerance (15–17). Such condensates include, among others, the nucleolus, stress granules, P bodies, neuronal granules, postsynaptic densities, the heterochromatin, and super enhancers (16). These entities form via liquid−liquid phase separation (LLPS) that is usually driven by multivalent weak protein−protein or protein−nucleic acid interactions, mostly mediated by intrinsically disordered regions (IDRs) of proteins (17). The proteins driving phase separation and their specific interaction partners are highly concentrated in the resulting condensates, while the contents of the condensates can also undergo dynamic exchange with the surroundings through the phase boundary (17, 18). Such phase-separated protein condensates provide diverse functional advantages for cells. For instance, they can serve as activators of reactions due to the high local concentrations of components, as biomolecular filters due to their selectivity, as stress sensors due to their responsiveness, and as reservoirs due to their ability to store macromolecules in an inactive, condensed state (15, 19). Accordingly, mutations affecting the phase separation propensity of proteins are often linked to neurodegeneration and other pathological conditions (20). Whereas LLPS seems to be a heavily exploited process in eukaryotes and viruses (21–24), up until today, only a few bacterial proteins were reported to undergo LLPS. These include Caulobacter crescentus Ribonuclease E (RNase E) that orchestrates RNA degradosomes (25), E. coli FtsZ that forms the division ring during cell divisions (26), and Mycobacterium tuberculosis Rv1747 that is a membrane transporter of cell wall biosynthesis intermediates (27). All in all, the hitherto accumulated observations imply that LLPS is a fundamental mechanism for the effective spatiotemporal organization of cellular space, with emerging but hardly explored functions in bacteria.
LLPS is typically driven by multivalent weak interactions, which ensure the dynamic, liquid-like properties of the resulting condensates. Diverse protein sequence compositional features, structural modules, overall architectural properties, and modes of protein−protein interactions are known to favor multivalent weak interactions and thus to promote LLPS. Disordered regions of low sequence complexity, prion-like regions, repeated short linear motifs and their cognate binding domains, nucleic acid-binding domains, and oligomerization domains can all be hallmarks of LLPS propensity based on the properties of the hitherto described cases (28).
The modular architecture of SSB, its ability to homooligomerize and to interact with multiple binding partners, its weakly conserved but compositionally biased IDL region, its conserved short linear CTP motif, and its central integrator role in genome maintenance processes all prompted us to raise the hypothesis that SSB drives the formation of liquid condensates through LLPS. Here we show that SSB is indeed capable of LLPS among physiological conditions, mediated via multifaceted intertetramer interactions involving all SSB protein regions. SSB, ssDNA, and SSB-interacting partners are highly concentrated within the phase-separated droplets, whereas LLPS is overall regulated by the stoichiometry of SSB and ssDNA. Our bioinformatics analysis indicates that the LLPS-forming propensity of the SSB IDL is broadly conserved across all major phylogenetic groups of eubacteria. Together with the recently observed dynamic spot-like subcellular localization patterns of SSB (29), our results suggest that bacterial cells store an abundant pool of SSB and SSB-interacting proteins in phase-separated condensates via a conserved mechanism. The discovered features enable rapid mobilization of SSB to ssDNA regions exposed upon DNA damage or metabolic processes to serve efficient repair, replication, and recombination.
Results
SSB Forms Dynamic LLPS Condensates in Physiologically Relevant Conditions In Vitro.
Due to the fact that SSB shares several features with hitherto described LLPS drivers, we sought to determine whether LLPS could be a yet undiscovered capability of SSB facilitating its multifaceted roles in genome maintenance. We performed a bioinformatics analysis that revealed that E. coli SSB, in particular, its IDL region, shows high propensity for LLPS, according to multiple dedicated sequence-based prediction methods (30–32) (Fig. 1 B–D). This feature of SSB is universally conserved among bacteria, as the majority of analyzed SSBs (72.1%) harbored by representative bacterial species from 15 large phylogenetic groups show similar LLPS propensities (Datasets S1 and S2 and Fig. 1E).
In line with its predicted LLPS propensity, purified E. coli SSB (30 µM; SSB concentrations are expressed as those of tetramer molecules throughout the paper) forms an opaque, turbid solution at low NaCl concentration (50 mM) in vitro at room temperature and also at the native temperature of E. coli cells (37 °C) (Fig. 2 A and B). The process is reversible, as elevation of NaCl concentration to 200 mM leads to immediate loss of turbidity. The total Cl− concentration in E. coli cells depends on their environment (33). Importantly, however, the vast majority of Cl− ions localize to the periplasmatic region, whereas Cl− concentration is undetectably low in the cytoplasm (34, 35) where SSB is located, and the major metabolic anion in the bacterial cytosol is glutamate (Glu; 100 mM) (36). Strikingly, SSB forms a turbid solution even in 200 mM NaGlu (Fig. 2 A and B). (It must be noted that experiments using NaGlu always contained a fixed amount of 20 mM NaCl, for technical reasons.) Investigation of the protein solution by differential interference contrast (DIC) microscopy after diluting SSB into low-salt buffer revealed the formation of regular spherical droplets with diameters in the micrometer range (Fig. 2C). Epifluorescence microscopic experiments obtained using a total internal reflection fluorescence microscope in which solutions of SSB mixed with a fluorescently labeled SSB variant [SSBG26C, allowing for selective labeling with a cysteine-reactive fluorescein dye (37)] were visualized also support the presence of SSB droplets up to 100 mM NaCl or 500 mM NaGlu (Fig. 2D). In addition to Na+ (2 mM to 30 mM), K+ ions are also present in bacterial cells at concentrations up to 300 mM (38). In our epifluorescence microscopic experiments, SSB showed similar behavior in KCl and KGlu (SI Appendix, Fig. S1A) to that seen in NaCl and NaGlu (Fig. 2D): KCl inhibited droplet formation above 100 mM concentration whereas droplets formed even at 500 mM KGlu. Magnesium acetate (MgOAc) had no apparent effect on droplet formation in the investigated concentration range (0 mM to 10 mM) (SI Appendix, Fig. S1B). The apparent diameter of droplets shows a nonnormal (and nonlognormal) distribution (SI Appendix, Fig. S1 C–F) with the median of the apparent droplet diameter slightly, but significantly, decreasing with increasing NaGlu concentration (Fig. 2E) but remaining constant in the investigated MgOAc concentration regime (Fig. 2F). Turbidity measurements also confirmed that, for NaCl, a sharp cutoff concentration around 100 mM exists above which no droplets form, while NaGlu and MgOAc do not influence droplet formation (Fig. 2 G and H). These observations indicate that SSB is able to undergo LLPS under physiologically relevant Na+, K+, Cl−, Mg2+, and glutamate concentrations, even though Cl− ions at higher concentrations inhibit the process in vitro, a property typical of LLPS systems (15, 39).
Fig. 2.
SSB forms phase-separated droplets in physiologically relevant ionic conditions in vitro. (A) Photographs of cuvettes containing 30 µM SSB (SSB concentrations are expressed as of tetramer molecules throughout the paper) and the indicated salt concentrations. Turbid appearance of samples indicates the presence of light-scattering by particles of diameters larger than the wavelength of visible light. (We note that experiments with NaGlu contained a fixed amount of 20 mM NaCl for technical reasons.) (B) Turbidity (measured via optical density at 600 nm [OD600]) of the samples at 25 °C and 37 °C. Turbidity (means ± SEM) decreased with increasing temperature (*: P < 0.01; n.s.: P = 0.076; n = 3, unpaired t test). (C) DIC microscopic images of SSB samples shown in A. (D) Fluorescence microscopic images obtained upon mixing 30 µM SSB and 0.3 µM fluorescein-labeled SSB at the indicated salt concentrations. SI Appendix, Fig. S1A shows MgOAc dependence measurements. (E and F) Salt (E, NaGlu, Inset: NaCl; F, MgOAc) concentration dependence of the apparent droplet diameter (medians and 25/75 percentiles shown as bullets and dashes, respectively), determined from fluorescence microscopic experiments. Lines show linear fits (E, NaGlu: *indicates that the slope is significantly different (P < 0.05) from zero; F, MgOAc: slope not significantly different from zero [n.s., P > 0.05; one-way ANOVA analysis]). Distributions are shown in SI Appendix, Fig. S1 B and C. (G and H) Turbidity of 15 µM SSB samples at the indicated salt concentrations. (I) Example images obtained during FRAP experiments in samples containing 30 µM SSB and 0.3 µM fluorescein-labeled SSB (bleached area diameter: 4,9 µm). (J) After bleaching of fluorophores at the center of droplets (SI Appendix, Fig. S2 G and H), fluorescence intensity recovery (in I) was followed in time. The time course of recovery was fitted by a single exponential decay function (solid line shows best fit). After bleaching, 68 ± 8% of the original fluorescence signal was recovered (recovered fraction) with a half-life (t1/2) of 175 ± 23 s.
During microscopic visualization, droplets sank toward the bottom of the sample chamber, and frequent fusion events were also observed (SI Appendix, Fig. S2A and Movie S1), leading to an increase in apparent droplet diameter in time (SI Appendix, Fig. S2 B and C). This finding supports the liquid nature of SSB droplets. In addition, the turbidity of samples decreased at a rate that showed linear dependence on total SSB concentration, reflecting concentration dependence of droplet fusion frequency (SI Appendix, Fig. S2 D–F). Liquid behavior is also supported by fluorescence recovery after photobleaching (FRAP) experiments (Fig. 2I), where fluorescent molecules at the center of a droplet (SI Appendix, Fig. S2 G and H) were photobleached with high-intensity laser light in a confocal microscope, and then the fluorescence recovery of the photobleached area was followed in time (Fig. 2J). Based on the sizes of the photobleached areas and the measured fluorescence recovery half-life values, the diffusion coefficient of labeled SSB was calculated to be 0.009 ± 0.001 µm2/s (SI Appendix, Table S1). Based on the Einstein−Stokes equation, the theoretical diffusion coefficient of an SSB tetramer [with a Stokes radius of 3.8 nm (40)] in water is ∼50 µm2/s. We note that, while we used the half-life values and the Einstein−Stokes model for simplicity, different diffusion models used for evaluation of FRAP data can lead to diffusion coefficients spreading within about an order of magnitude (41). Our simple estimation indicates that SSB diffuses ∼5,000 times slower in the droplets than it would in water, and thus the apparent dynamic viscosity within the droplets (5,000 mPas) is in the range of that of honey (about 3.6 times that of glycerol).
SSB Drives LLPS at Presumed Cellular Concentrations.
Next we investigated the effect of protein concentration on droplet formation (Fig. 3A) in the presence and absence of 150 mg/mL bovine serum albumin (BSA) used as a molecular crowder to simulate the cellular total protein concentration in E. coli (42). In both conditions, small condensates were detected even at as low as 0.5 µM total SSB concentration; however, at low SSB concentrations, the apparent diameters of SSB droplets were much smaller (Fig. 3B and SI Appendix, Fig. S3), and their small size and rapid diffusion in the solution rendered them difficult to image. The median of the apparent diameter of droplets was dependent on SSB concentration, with a steep increase in the 2.5 µM to 4 µM concentration range, and remained constant above 4 µM independently of the presence of BSA (Fig. 3B). SSB concentration-dependent turbidity measurements also confirmed formation of light-scattering condensates by SSB in the 0.5 µM to 30 µM SSB concentration regime in all conditions except for 200 mM NaCl (Fig. 3C). Recent studies indicate that there are ∼2,000 SSB tetramers present in an E. coli cell (43), in line with an earlier report (44) that also clarified technical issues leading to underestimation of cellular SSB concentrations in preceding studies. Accordingly, it has long been proposed that cells contain considerably more SSB than the amount required for DNA replication under stress-free conditions (44). Thus, the intracellular SSB concentration is about 5 µM, taking into account the average E. coli cell volume (0.65 µm3). Therefore, our results suggest that SSB droplet formation can occur in cells even without the inducing effect of molecular crowding, with droplet sizes being limited by the amount of SSB within the cell. The findings that droplet frequency and size show a marked increase around the physiological SSB concentration (Fig. 3 A and B) are suggestive of the possibility that the LLPS propensity of SSB has been evolutionarily shaped by physiological demands. SSB expression is thought to be under SOS regulation; however, some studies indicate that SSB is expressed constantly in an SOS-independent manner (45–47). Nevertheless, any condition leading to a decrease in cell volume (such as osmotic shock) and/or increased SSB levels could enhance SSB-driven LLPS.
Fig. 3.
SSB forms highly dense droplets at cellular SSB concentrations in vitro. (A) Fluorescence images of samples containing SSB at the indicated concentrations (including 0.3 µM fluorescein-labeled SSB) in the absence and presence of BSA (150 mg/mL). (B) SSB concentration dependence of the apparent diameter of droplets (medians and 25/75 percentiles marked by bullets and dashes, respectively) in the absence and presence of BSA (150 mg/mL). SI Appendix, Fig. S3 shows diameter distributions. (C) SSB concentration dependence of turbidity in the presence of the indicated solution components. (D) Schematics of centrifugation-based concentration determination experiments. (E) Fraction of SSB in the dissolved (black) and droplet (blue) phases. Solid lines show linear fits. Slopes were not significantly different from zero (n.s., P > 0.05; one-way ANOVA analysis). (F) Representative 3D fluorescence image obtained in spinning disk microscopic experiments (15 µM SSB, 0.3 µM fluorescein-labeled SSB). (G) Calculated concentration of SSB in droplets. Solid line shows linear best fit. The slope was not significantly different from zero (n.s, P = 0.804, n = 3, one-way ANOVA analysis). SI Appendix, Fig. S4 shows control experiments revealing the effect of molecular crowders.
Measurements using various concentrations of BSA or polyethylene glycol (PEG20000; a 20,000-Da polymer that is routinely used as a synthetic molecular crowding agent) showed that droplet characteristics are influenced by different crowders (SI Appendix, Fig. S4). Whereas BSA does not influence droplet size distribution, at high polyethylene glycol concentrations, the mixing of SSB with labeled SSB is affected, and thus nonhomogeneous fluorescence distributions and lowered fluorescence intensities were observed in fluorescence microscopic experiments (SI Appendix, Fig. S4 A–F). On the contrary, in DIC experiments, BSA and PEG conditions were indistinguishable (SI Appendix, Fig. S4 A and B). Accordingly, both BSA and PEG altered the turbidity of solutions in a similar fashion (SI Appendix, Fig. S4 G and H) and reduced droplet fusion rates (SI Appendix, Fig. S4 I and J). PEG reduced the diffusion coefficient of labeled SSB in FRAP experiments by 3.9-fold (SI Appendix, Fig. S4 K–M), in line with the altered mixing properties observed in fluorescence experiments (SI Appendix, Fig. S4F).
To calculate the concentration of SSB in the droplets, we separated the condensed fractions of samples of various SSB concentrations using centrifugation, and then determined the protein concentration from the supernatants and back-diluted pellets (resuspended in 1 M NaCl to starting volume) (Fig. 3D). The fraction of SSB molecules localized to the condensed phase increased linearly until 5 µM SSB concentration and remained constant at higher concentrations (up to 30 µM investigated) with an average of 95 ± 3% of SSBs located in the condensed phase in the 5 µM to 30 µM regime (Fig. 3E). Importantly, in the 3 µM to 5 µM SSB concentration regime, 65 to 95% of SSB was in condensed form, further supporting that phase separation can occur at physiologically relevant SSB concentrations, while the size of droplets dynamically increased in the 2.5 µM to 4 µM concentration regime in microscopic experiments (Fig. 3 A and B). Next, we estimated the volume fractionation of droplets and surroundings using 3D images reconstructed from z-stack images prepared using a spinning disk microscope for samples containing 5 µM to 30 µM total SSB concentration, as used in the centrifugation experiments (SSB was mixed with fluorescently labeled SSB in the spinning-disk experiments) (Fig. 3F). Based on the total SSB concentration, the fraction of droplet volume within the sample, and the fraction of SSB molecules localized to the droplets, SSB concentration within droplets was calculated to be 3.9 ± 0.3 mM on average, independent of total SSB concentration (Fig. 3G). Based on the Stokes radius of SSB, the water content of SSB droplets is calculated to be around 46 ± 4 vol/vol%, which is markedly lower than that of the cytoplasm (∼70 vol/vol%).
Multivalent Interactions between SSB IDL Regions Are Required for LLPS, whereas Interactions between the CTP and the OB Fold Enhance LLPS Propensity.
Previous studies suggested that the IDLs of adjacent SSB molecules can interact with each other to enhance the cooperativity of ssDNA binding (12, 13), while the IDL was also proposed to interact with the OB fold (48). Moreover, the central role played by similar low-sequence-complexity, Gln/Gly/Prorich IDRs in previously investigated LLPS systems (49–52) as well as the predicted LLPS propensities suggest a role for the IDL in SSB LLPS (Fig. 1 B–D). To test how the different structural modules of SSB contribute to LLPS, we purified an SSB variant lacking the CTP (SSBdC, comprising amino acids [aa] 1 to 170) and a variant that also lacks the IDL (SSBdIDL, aa 1 to 113) (Fig. 4A and SI Appendix, Fig. S5). Both constructs were previously shown to be able to bind ssDNA (12). Strikingly, in contrast to SSB, SSBdC only forms droplets in the presence of 150 mg/mL BSA, whereas SSBdIDL forms amorphous nonspherical aggregates even in the presence of BSA, as assessed by DIC microscopy (Fig. 4B). In fluorescence experiments, 30 µM SSBdC formed droplets with 0.3 µM fluorescently labeled SSB in both the presence and absence of BSA (Fig. 4C), whereas the formation of SSBdC (30 µM) droplets with 0.3 µM fluorescently labeled SSBdC was only observed in the presence of BSA (Fig. 4D). However, 0.3 µM fluorescently labeled SSBdC formed droplets with 30 µM SSB even in the absence of BSA (Fig. 4D). SSBdIDL was unable to form droplets in any investigated condition, and the aggregated structures had a faint appearance (Fig. 4 B–D). Interestingly, mixing of 30 µM SSBdC with 300 nM fluorescently labeled 79mer homo-deoxythymidine (dT) ssDNA (Cy3-dT79) resulted in droplet formation in both the presence and absence of BSA, whereas experiments using SSBdIDL showed amorphous aggregates (Fig. 4E). Turbidity measurements also support that, in the absence of ssDNA, SSBdC only forms droplets in the presence of BSA (Fig. 4F).
Fig. 4.
Multifaceted interactions of SSB structural regions are required for efficient LLPS. (A) Schematic domain structure of SSB constructs (see also Fig. 1A). Numbers indicate amino acid positions at boundaries of structural regions. SSBdC lacks the CTP region, whereas SSBdIDL lacks the IDL and CTP regions. For site-specific fluorescent labeling, SSB variants harboring the G26C substitution were used (SI Appendix, Fig. S5) (37). In addition to SSB constructs, we also generated chimeric proteins of eGFP and the SSB C-terminal regions to test the role of the IDL and CTP regions in the absence of the OB fold (SI Appendix, Fig. S6). (B) DIC images of 30 µM SSB constructs in the presence and absence of 150 mg/mL BSA. (C–E) Fluorescence images of 30 µM unlabeled SSB constructs mixed with 0.3 µM (C) fluorescein-labeled SSB, (D) 0.3 µM Alexa555-labeled SSBdC, or (E) 0.3 µM cyanine 3 (Cy3) labeled dT79 ssDNA (79-nucleotide-long homopolymer deoxythymidine) in the presence and absence of 150 mg/mL BSA. (F) SSBdC concentration dependence of sample turbidity in the presence and absence of 150 mg/mL BSA (in 50 mM NaGlu). SSB data from Fig. 3C are shown for comparison. (G) Model of LLPS-driving intertetramer SSB interactions based on the structure of SSB, the SSB-CTP interaction, and the revealed roles of structural regions in LLPS.
To test the role of the SSB OB fold in LLPS, we generated two chimeric proteins. We fused the IDL plus CTP regions of SSB C terminally to monomeric enhanced green fluorescent protein (eGFP) (SI Appendix, Fig. S6 A and B). We also generated an eGFP fusion variant wherein a short flexible (Gly-Gly-Ser)4 linker connected the CTP to eGFP instead of the IDL (SI Appendix, Fig. S6 A and B). Neither of these constructs (including eGFP alone) was able to preferentially partition into SSB droplets regardless of the presence of BSA (SI Appendix, Fig. S6 C–E). However, when using PEG, the eGFP fusion proteins, but not eGFP alone, were able to enrich in SSB droplets with similar efficiency (SI Appendix, Fig. S6 C–E) and showed diffusion characteristics similar to those of SSB in the absence of PEG (SI Appendix, Fig. S6 F–H). This finding indicates that, under PEG-induced crowding conditions, the interaction between the eGFP-fused CTP and SSB’s OB fold enables the partitioning of eGFP into phase-separated SSB droplets, with the IDL having no apparent effect. Taken together, the above results highlight that all structural modules of SSB contribute to efficient phase separation. The tetramerization of OB folds brings about multivalency and enhances the local concentration of IDL regions, whereas weak IDL−IDL (and/or IDL−OB fold) interactions drive LLPS and bring about the liquid nature of the condensates (similarly to the IDRs of eukaryotic LLPS driver proteins), while the specific CTP−OB fold interaction enhances this LLPS propensity (Fig. 4G).
ssDNA Binding to SSB Inhibits Phase Separation due to Competition between ssDNA and CTP Binding to the OB Fold.
A major cellular function of SSB is to cover ssDNA. Accordingly, we observed that fluorescently labeled ssDNA (Cy3-dT79) is enriched in SSB droplets compared to the surroundings, when the DNA concentration was low compared to that of SSB (Figs. 4B and 5A). We also found that ssDNA can diffuse within the droplets 2.3 ± 0.5 times faster than SSB, possibly due to its rapid dynamic interaction with adjacent SSB tetramers (SI Appendix, Fig. S7 A–C) (53). Intriguingly, a labeled ssRNA molecule behaved similarly under these conditions (Fig. 5A). SSB was earlier shown to bind ssRNA, albeit with lower affinity than that for ssDNA (40, 54, 55). We also find that the interaction of SSB with ssRNA is weaker than that with ssDNA (SI Appendix, Fig. S7 D and E), but this interaction is still sufficient to drive RNA enrichment in SSB droplets due to the high concentration of SSB in this phase (Figs. 3G and 5A), raising the possibility of yet undiscovered roles for SSB in RNA metabolism. Indeed, recently SSB was found to enhance the function of RNase HI in R-loop removal (56).
Fig. 5.
SsDNA regulates SSB phase separation by competing with the SSB CTP for OB fold binding. (A) Fluorescence microscopic images of 30 µM SSB, and 0.3 µM fluorescein-labeled SSB (green channel) in the presence and absence of 0.1 µM Cy3-labeled dT79 or ssRNA (41 nt, nonhomopolymeric) (red channel). SI Appendix, Fig. S7 A–E shows diffusion characteristics of Cy3-dT79 in droplets and binding characteristics of SSB to Cy3-dT79 and Cy3-ssRNA. (B) The ssDNA dependence of SSB droplet formation, as observed in fluorescence microscopic images of samples containing 30 µM SSB, 0.3 µM fluorescein-labeled SSB, and unlabeled dT79 at the indicated concentrations. SI Appendix, Fig. S7F shows additional data recorded in the presence of molecular crowding agents (BSA and polyethylene glycol [PEG20000]), using longer ssDNA (poly-dT with average length between 600 nt and 1,000 nt), and controls for ssDNA dependence measurements. (C) The dT79 concentration dependence of apparent droplet diameters (medians and 25/75 percentiles marked by bullets and dashes, respectively) in the presence and absence of 150 mg/mL BSA. See SI Appendix, Fig. S7 H–K for additional experiments and distributions of apparent diameters. Lines show linear best fits. One-way ANOVA analysis indicates that the slopes are not significantly different from zero (n.s., P > 0.05). (D) The ssDNA concentration dependence of turbidity of samples containing SSB at the indicated concentrations. Solid lines show fits using a quadratic binding equation (SI Appendix, Eq. S2; best-fit parameters are shown in SI Appendix, Table S2). (Inset) Apparent binding site size of SSB at saturating ssDNA concentrations (means ± SEM). Values at different SSB concentrations are not significantly different (n.s., P = 0.979, n =3, one-way ANOVA). SI Appendix, Fig. S8 shows additional ssDNA concentration-dependent turbidity measurements at higher NaGlu concentration, in the presence of molecular crowders or using dT36 or poly-dT. (E) Fluorescence anisotropy-based experiments monitoring SSB binding to 15 nM fluorescein-labeled, isolated SSB CTP in the presence and absence of 50 µM dT79. Solid line shows best fit based on the Hill equation (SI Appendix, Eq. S1). Fit results are shown in SI Appendix, Table S3. (F) Schematic model for the LLPS-inhibiting effect of ssDNA (black line). SSB tetramers and structural regions are shown as in Fig. 4G.
In the above experiments, substoichiometric amounts of ssDNA (or ssRNA) over SSB DNA binding sites were used. Therefore, we also examined the effect of higher DNA concentrations on SSB-driven LLPS. Strikingly, droplets disappeared with increasing concentrations of unlabeled dT79 (Fig. 5B and Movies S2 and S3) or poly-dT (homo-deoxythymidine ssDNA with average length 600 nt to 1,000 nt), regardless of the presence of BSA or PEG (SI Appendix, Fig. S7 F and G). The apparent diameter of droplets was not influenced by the presence of ssDNA in either the absence or the presence of BSA (Fig. 5C and SI Appendix, Fig. S7 H–K). However, the number of detected droplets decreased with increasing ssDNA concentration in all studied conditions (Fig. 5B and SI Appendix, Fig. S7 F and G) along with the measured turbidity of the samples (Fig. 5D and SI Appendix, Fig. S8). Strikingly, in the presence of PEG, a shallower ssDNA dependence of turbidity was detected compared to other conditions, indicating a decreased sensitivity of droplet disruption by ssDNA (SI Appendix, Fig. S8E).
SSB can bind ssDNA in two major binding modes that can reversibly interconvert (2, 57). Therefore, we tested how binding modes affect droplet formation. At low salt concentrations (depending largely on salt type) and high SSB:ssDNA concentration ratios, the binding mode equilibrium is shifted toward the so-called 35-nt mode in which, on average, two monomers of the SSB tetramer bind ssDNA with an apparent binding site size of 35 nt (2, 57, 58). High salt concentrations and low SSB:ssDNA concentration ratios favor the 65-nt binding mode in which ssDNA is wrapped around all four SSB tetramers (2, 57). The analysis of ssDNA-dependent turbidity curves revealed that, in all conditions, except for PEG, turbidity linearly decreases with ssDNA concentration (as expected for a stoichiometric binding reaction; Fig. 5D and SI Appendix, Fig. S8 A–D and Table S2) until the point at which all SSB tetramers become ssDNA bound and therefore droplet formation is completely inhibited, regardless of the binding mode (Fig. 5D and SI Appendix, Fig. S8). Previous studies using NMR spectroscopy showed that the CTP binds to the same region on the OB fold as does ssDNA (10) and it was shown that the IDL, and especially the CTP, influence ssDNA binding (2, 9, 12, 14). In line with these observations, we found that, when SSB is bound to ssDNA, the binding of the isolated CTP peptide to SSB is significantly weaker than that for DNA-free SSB (Fig. 5E). This observation, together with the LLPS-promoting effect of the CTP (Fig. 4 and SI Appendix, Fig. S6), suggests that droplet formation is regulated by the competition between the CTP and ssDNA for the OB fold (Fig. 5F).
Even Weak Interaction Partners of SSB Enrich in Phase-Separated SSB Condensates.
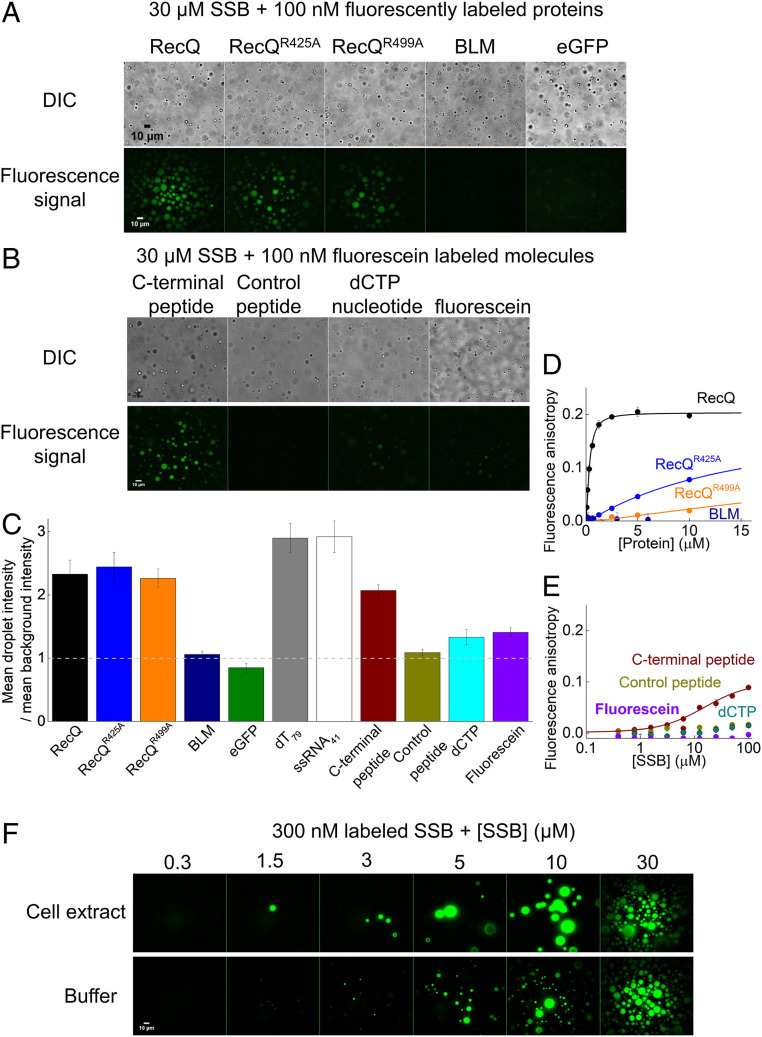
The above results demonstrate that specific interactions with SSB allow enrichment of interactors in SSB droplets. Thus, we tested how the interaction strength and the molecular size of interactors influence their ability to partition into phase-separated SSB condensates (Fig. 6 A and B and SI Appendix, Fig. S9A). E. coli RecQ helicase, a 67-kDa protein that was shown to interact with SSB dominantly via the CTP (59, 60), readily enriches in SSB droplets (Fig. 6 A, C, and D). Interestingly, RecQ point mutants with a markedly weakened affinity to the CTP (61) also enrich in the droplets with similar efficiency (Fig. 6 A, C, and D and SI Appendix, Table S3). In contrast, BLM helicase (a human RecQ homolog) and eGFP are both unable to enrich in SSB droplets (Fig. 6 A, C, and D and SI Appendix, Table S3), highlighting that specific interactions with SSB are required for proteins to partition into SSB condensates. Probably due to the high SSB concentration within the droplets (Fig. 3G), even weak interactions (Kd in the range of tens of micromolars) appear to be sufficient for enrichment in the SSB condensates. In line with this proposition, the isolated SSB CTP peptide became enriched in the droplets, but a control peptide that does not interact with SSB was not enriched (Fig. 6 B, C, and E and SI Appendix, Table S3). In addition, small molecules, such as deoxycytidine triphosphate (dCTP) or fluorescein, are slightly enriched in droplets, but not as effectively as SSB interactors, indicating that small molecules are able to enter the separated phase, and the droplet environment has a slight concentrating effect (Fig. 6 B and C). As expected for interactors, both RecQ and the isolated CTP peptide can diffuse within the droplets, and their rate of diffusion is apparently dictated by their size (SI Appendix, Fig. S9 B–D).
Fig. 6.
Specific interactions drive the enrichment of SSB-interacting partners in phase-separated SSB droplets. (A) DIC and fluorescence microscopic images obtained upon mixing 30 µM SSB and 0.1 µM of various fluorescently labeled proteins (Alexa488-labeled RecQ helicase (wild type or variants harboring the R425A [RecQR425A] or R499A [RecQR499A] substitutions), fluorescein-labeled human BLM (Bloom’s syndrome) helicase, or eGFP). See SI Appendix, Fig. S9 for additional data on protein constructs. (B) DIC and fluorescence microscopic images obtained upon mixing 30 µM SSB and 0.1 µM fluorescein-labeled isolated SSB CTP, fluorescein-labeled control peptide (12mer), fluorescein-labeled dCTP or fluorescein. (C) Enrichment of various molecular components in SSB droplets, inferred from the ratio of the mean signal intensity within droplets and the mean background intensity, determined from background-uncorrected fluorescence images recorded for the indicated fluorescent molecules. (D and E) Fluorescence anisotropy titrations of (D) 15 nM fluorescein-labeled isolated SSB CTP with unlabeled proteins used in A (except eGFP) and (E) titrations of 15 nM fluorescein-labeled CTP, control peptide, fluorescein labeled dCTP, or fluorescein with unlabeled SSB. Solid lines show best fits based on the Hill equation (SI Appendix, Eq. S1). Best-fit parameters are shown in SI Appendix, Table S3. (F) Fluorescence images obtained in E. coli cell extract and in a buffer solution (20 mM Tris-OAc pH 7.5, 5 mM MgOAc, 50 mM NaGlu) upon addition of 300 nM fluorescein-labeled SSB plus unlabeled SSB to reach the indicated total SSB concentrations. Images were background corrected.
SSB Condensates Form also in Cytosolic Extracts, Supporting the Occurrence of LLPS In Vivo.
To further assess the possibility that SSB phase separation occurs in the cellular environment, we investigated condensate formation in E. coli cell extracts using epifluorescence microscopy (see SI Appendix, Supplementary Materials and Methods for more specifications). Condensate formation was not observed upon addition of 0.3 µM fluorescein-labeled SSB alone to the extracts. However, droplets formed when unlabeled SSB was also added to at least 1.5 µM total SSB concentration (Fig. 6F). Under identical conditions, eGFP was not able to form condensates in control experiments (SI Appendix, Fig. S10A). Moreover, SSB condensates disappeared upon the addition of NaCl or dT79 (SI Appendix, Fig. S10 B and C) similar to what was observed in measurements performed in simple buffer solutions (Figs. 2D and 5B). Additional experiments suggested that the requirement for added SSB (at a minimum of 1.5 µM) for droplet formation originated from the presence of significant amounts of ssDNA generated during cell lysis (SI Appendix, Fig. S10D and corresponding legend). Taken together, these measurements clearly demonstrate that SSB-driven LLPS can indeed occur in the cytosolic molecular environment.
Bioinformatics Analysis Indicates Conservation of LLPS Propensity in Human SSB Homologs.
Besides Replication Protein A, the long-known main eukaryotic ssDNA-binding protein, recent studies identified human SSB homologs hSSB1/SOSB1 and hSSB2/SOSB2 that are similar in size to bacterial SSBs and similarly possess C-terminal IDL regions (62, 63). Using the bioinformatics tools applied in Fig. 1 B–D, we assessed whether the LLPS propensity of bacterial SSB proteins is conserved in these human homologs. The algorithms indicated similar degrees of LLPS propensity for the IDL regions of both human proteins to that for E. coli SSB, indicating a potential role for LLPS in eukaryotic DNA metabolic processes (SI Appendix, Fig. S11).
Discussion
Taken together, here we show that E. coli SSB undergoes LLPS and forms viscous, liquid-state protein droplets under physiologically relevant ionic conditions and protein concentrations, in both the presence and absence of molecular crowders. Efficient phase separation requires all structural modules of SSB and is regulated by the specific interaction between the CTP and the OB fold as well as the stoichiometry of available SSB and ssDNA. Saturation of SSB binding sites by ssDNA, independent of SSB’s DNA-binding mode, prevents LLPS, because ssDNA and the CTP compete for the same binding site on the OB folds. We also observed an LLPS inhibitory effect specifically for Cl− ions, which were shown to be able to interact with the OB fold, likely through lysine side chains that were previously shown to interact with phosphate groups of ssDNA (64). As the CTP interaction site of the OB fold is located at its ssDNA binding site, it is possible that Cl− ions compete with the CTP for binding to the OB, as does ssDNA. In line with this proposition, in NMR studies, increasing NaCl concentrations inhibited the OB fold–CTP interaction, whereas even 300 mM NaGlu had no effect on the interaction (11). While other effects of Cl− cannot be excluded, these results highlight the importance of the OB fold−CTP interaction in LLPS. Nevertheless, the Cl− concentration in the E. coli cytosol is much lower than 100 mM (34, 35), underscoring that the LLPS-inhibiting effect of Cl− is not relevant under cellular conditions.
Given that an E. coli cell may contain around 2,000 SSB tetramers and that we found the concentration of SSB to be steady around 3 mM within the droplets, the theoretical maximum for intracellular SSB droplet size is ∼117 nm in diameter that could be achieved if the entire pool of cellular SSB molecules would form a single droplet. Although particles of this size range can be visualized by superresolution microscopy, the compelling in vivo demonstration of LLPS by SSB is highly challenging.
Previous studies using widefield fluorescence microscopy on cells expressing SSB proteins labeled on their C terminus with fluorescent proteins showed the presence of SSB foci dominantly at replication forks (65, 66). These experiments required the simultaneous expression of wild-type SSB, as the fusion of fluorescent proteins to the C terminal inhibits the essential functions of SSB. This limitation has been recently overcome by a novel SSB fluorescent protein toolbox in which different fluorescent proteins are inserted into the IDL region (67). This approach revealed the same widefield fluorescence localization pattern for SSB as that in the aforementioned experiments (65, 66). Nevertheless, the effect of fluorescent tags on the phase separation properties of SSB are yet unknown. Importantly, a recent study using structured illumination superresolution microscopy combined with C-terminal GFP-labeled SSB showed that, in E. coli cells, SSB not only localizes to DNA replication forks but also forms multiple DNA-free protein foci near the inner cell membrane (29). Membrane localization is explained by specific binding of SSB to lipid membrane components. Upon DNA damage, membrane-localized SSB were shown to disperse, and, concomitantly, SSB relocalized to sites on the bacterial chromosome, likely to those containing exposed ssDNA (29). Importantly, the ability of SSB to undergo ssDNA concentration-dependent LLPS, discovered in our current study, can explain the observed localization patterns. We propose that, when only a small amount of ssDNA is exposed, the majority of cellular SSB molecules (together with interacting partner proteins) will localize to (multiple) membrane-proximal condensates, with dynamic exchange between condensed and noncondensed SSB pools. An increase in exposed ssDNA sites (e.g., upon DNA damage) can trigger very rapid dispersion of condensates as noncondensed cytosolic SSB molecules bind to ssDNA, leading to relocalization of SSB to sites along the genome (Fig. 7). Considering the observation of multiple SSB foci (29), the diameter of individual SSB droplets must be smaller than 100 nm. Based on the diffusion coefficient of phase-separated SSB estimated in our study (SI Appendix, Table S1), the contents of an intracellular SSB droplet with a 100-nm diameter can be released with a half-time of 70 ms even in the absence of additional signaling components. Relocalization is expected to occur even more rapidly for smaller droplets. We note that our estimation for the diffusion coefficient results from a simple model used to evaluate the FRAP results, and different models could yield values within a margin of one order of magnitude (41). However, even 10 times slower SSB dynamics would still allow a very rapid cellular response.
Fig. 7.
Proposed model for the in vivo role of SSB LLPS. The model is based on data presented here and earlier in vivo imaging results of Zhao et al. (29).
Our current finding that even weak interaction partners of SSB are specifically enriched in the condensates is in line with previous findings showing that SSB interaction partners can form complexes with SSB even in the absence of ssDNA in vivo (68). Importantly, our findings highlight that the view that weak protein−protein interactions are functionally irrelevant needs to be revisited in the light of recently discovered properties of phase-separated protein condensates.
Previously, it was indicated that the amount of cellular SSB is considerably higher than that needed to cover ssDNA-containing sites during replication (44), but the role and localization of this “excess” SSB content was unknown. Based on the above findings, we propose that bacterial cells constitutively store a fraction of the SSB pool and SSB-interacting proteins in a condensed form at the inner membrane (Fig. 7). Supporting this proposition, SSB interacting partners RecG and PriA have been shown to localize to membrane-associated foci in vivo (68). The condensate-localized SSB pool can be rapidly mobilized for rapid response to genomic stress and initiation of DNA repair. Interestingly, it was previously shown that bacterial cells expressing an IDL-deleted, but CTP-containing, SSB variant in place of wild-type SSB were viable in a stress-free laboratory environment (69), likely due to the ability of SSB to efficiently bind to replication sites independent of its LLPS capability. However, another study showed that, when the IDL of E. coli SSB is removed or its composition is altered, the sensitivity of cells to ultraviolet (UV) damage is increased (12). The finding that the LLPS propensity of the IDL is highly conserved among bacteria (Fig. 1E) suggests that, while the ability of SSB to drive LLPS is not necessarily essential for cell viability under stress-free laboratory conditions, it confers important adaptive advantages in free-living bacteria, such as the adaptability to environmental stress and related DNA damage. In line with our conclusions, a recent work using a novel, ClpB disaggregase-based genetic test showed that a segment of the Campylobacter hominis SSB IDR can form prion-like condensates in live E. coli cells (70).
Besides membrane-associated SSB condensate formation, our results raise the possibility that some form of condensation could take place also at DNA replication and repair sites. Our results show that LLPS does occur at low ssDNA:SSB molar ratios, and DNA is embedded in the SSB droplets (Fig. 5A). Thus, when the free SSB pool markedly exceeds the DNA-bound fraction, condensate formation could occur even at genomic DNA sites. Moreover, the same forces that drive LLPS may also drive the IDL-dependent non-nearest-neighbor cooperative interactions of DNA-bound SSB tetramers (12, 13, 71). An earlier single-molecule imaging study showed that high acetate and glutamate concentrations lead to hypercompaction of the SSB−ssDNA filament that can be explained based on similar interactions (72).
All in all, our findings align well with recent discoveries suggesting that the formation of liquid condensates through LLPS is a universally exploited fundamental mechanism that provides efficient means for optimizing the spatiotemporal organization of cellular space according to functional needs (73). Our results imply that, among other key cellular processes such as RNA degradation (25), cell division (26), and membrane transport (27), bacterial DNA replication and repair mechanisms also benefit from the advantages provided by dynamic liquid condensates. Importantly, the architectural similarities of human SSB homologs hSSB1/SOSB1 and hSSB2/SOSB2 to bacterial SSBs along with their predicted LLPS propensity (SI Appendix, Fig. S11) suggest that SSB phase separation may be a broadly conserved feature also playing a role in eukaryotic DNA metabolic processes.
Materials and Methods
Detailed descriptions of materials and methods and equations used in fits are provided in SI Appendix, Supplementary Materials and Methods.
Reagents and Experimental Conditions.
DNA concentrations are expressed as those of oligonucleotide or polynucleotide molecules (as opposed to those of constituent nucleotide units) unless otherwise stated. SSB concentrations are expressed as those of tetramer molecules.
Measurements were performed at 25 °C in LLPS buffer containing 20 mM Tris-acetate (Tris-OAc) pH 7.5, 5 mM MgOAc, 50 mM NaGlu, 20 mM NaCl unless otherwise indicated. Due to the NaCl content of SSB stock solutions, all NaGlu experiments contained a constant concentration of 20 mM NaCl. Experiments monitoring the effect of NaCl were performed in the absence of NaGlu. Cloning, expression, and purification of proteins used in the study and peptides are described in SI Appendix, Supplementary Materials and Methods.
Bioinformatics Analysis.
We used the web servers of three dedicated prediction methods to obtain the LLPS propensity profile of E. coli and human SSBs (hSSB1/SOSB1 [also known as NABP2] and hSSB2/SOSB2 [also known as NABP1]). The prion-like amino acid composition (PLAAC) method (31) predicts prion-like domains based on sequence composition properties. It was used with parameters core length = 30 and α = 100. The PScore method (default parameters used) estimates the ability of proteins to undergo LLPS solely based on their propensity for long-range planar pi−pi contacts (32). The CatGranule method (30) (default parameters used) is a machine learning approach that was trained on a set of granule-forming proteins and predicts LLPS propensity based on RNA binding, structural disorder, and amino acid composition features.
To predict the LLPS propensity of the clean set of 717 SSBs (SI Appendix, Materials and Methods), the PLAAC and CatGranule web servers were used.
Although these methods are preliminary and only take into account a restricted set of features indicative of LLPS, they—especially in combination—are highly useful for exploratory assessment of LLPS propensity (74).
Fluorescence Anisotropy Titrations.
Fifteen nanomolars fluorescein-labeled molecules were titrated with increasing concentrations of SSB in LLPS buffer at 25 °C.
Turbidity Measurements.
Turbidity of 40-µL samples (light depletion at 600-nm wavelength) was measured in a Tecan Infinite Nano+ plate reader instrument at 25 °C or 37 °C.
Electrophoretic Mobility Shift Assay.
The 200 nM 5′-Cy3-labeled, 41-nt ssRNA (5′-Cy3-UAAGAGCAAGAUGUUCUAUAAAAGAUGUCCUAGCAAGGCAC-3′) or 41-nt ssDNA (5′ Cy3 - dT41) was titrated with increasing concentrations of SSB (0 nM to 500 nM) in LLPS buffer at 25 °C. Molecular species were separated on a 1 wt/vol% agarose gel and detected using a Typhoon Trio+ Variable Mode Imager.
Centrifugation-Based Concentration Determination.
SSB in various concentrations was incubated in LLPS buffer (20 mM Tris-OAc pH 7.5, 20 mM NaCl, 50 mM NaGlu and 5 mM MgOAc) at 25 °C. Samples were centrifuged using a tabletop Eppendorf centrifuge (13,400 rpm [12,000 × g], 30 min). Protein concentrations of the supernatant and resuspended pellet were determined with UV spectrophotometry.
Microscopy and Image Analysis.
DIC images were captured using a Zeiss AxioImager M2 upright microscope. Raw, unprocessed DIC images are shown in the paper.
Epifluorescence images were captured with a Nikon Eclipse Ti-E TIRF microscope in epifluorescent mode. Raw, unprocessed images were analyzed using the Fiji (ImageJ) software. For illustration purposes, images were background corrected (SI Appendix, Supplementary Materials and Methods). Methods of determination of size distribution and fluorescent partitioning are described in SI Appendix, Supplementary Materials and Methods.
FRAP experiments were performed at 23 °C using an inverted LSM800 confocal microscope (Zeiss).
For evaluation of droplet volumes, microscopic images were taken on a Zeiss Spinning Disk system at 24 °C. Image analysis methods are described in SI Appendix, Supplementary Materials and Methods.
Data Analysis.
Mean ± SEM (SE of mean) values are reported in the paper unless otherwise specified. For apparent droplet size distributions, mean ± SD values and medians are shown. Sample sizes (n) are given for the number of observed particles in fluorescence microscopic experiments or the number of ensemble in vitro measurements performed (n = 3 unless otherwise specified).
Supplementary Material
Acknowledgments
This work was supported by the Human Frontier Science Program (Grant RGY0072/2010 to M.K.), the “Momentum” Program of the Hungarian Academy of Sciences (Grant LP2011-006/2011 to M.K.), Grants ELTE (Eötvös Loránd University) KMOP‐4.2.1/B‐10‐2011‐0002, NKFIH (National Research, Development and Innovation Office) K-116072, NKFIH K-123989, and NKFIH ERC_HU 117680 to M.K. and Grant NKFIH FK-128133 to R.P. Both G.M.H. and R.P. are supported by the Premium Postdoctoral Program of the Hungarian Academy of Sciences (Grants PREMIUM-2017-17 to G.M.H. and PREMIUM-2017-48 to R.P.). The project was supported by the NKFIH (Grant VEKOP-2.3.3-15-2016-00007 to ELTE) grant. Z.J.K. and J.P. are supported by the New National Excellence Program of the Ministry for Innovation and Technology (Grants ÚNKP-19-3 to Z.J.K. and ÚNKP-19-2 to J.P.). This work was completed in the ELTE Thematic Excellence Programme supported by the Hungarian Ministry for Innovation and Technology. We are grateful to Dr. Tibor Kovács for the help with DIC measurements, Gábor Szegvári for the custom-written script for volume estimation of droplets observed in spinning disc fluorescence microscopic experiments, and Anna Budai for the help in the cloning of expression vectors encoding GFP fusion proteins.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. I.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000761117/-/DCSupplemental.
Data Availability.
All data generated or analyzed during this study are included in this published article and Datasets S1 and S2. Source code of the script used for analyses of spinning disk microscopy data are freely available at GitHub, https://github.com/SzegaX/VolumeOfSmearedSpheres.
References
- 1.Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L., SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antony E., Lohman T. M., Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 86, 102–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianco P. R., The tale of SSB. Prog. Biophys. Mol. Biol. 127, 111–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghunathan S., Ricard C. S., Lohman T. M., Waksman G., Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 94, 6652–6657 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savvides S. N. et al., The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 13, 1942–1947 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pancsa R., Tompa P., Essential functions linked with structural disorder in organisms of minimal genome. Biol. Direct 11, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paradžik T., Filić Ž., Vujaklija D., Variations in amino acid composition in bacterial single stranded DNA–binding proteins correlate with GC content. Period. Biol. 118, 385–397 (2017). [Google Scholar]
- 8.Antony E. et al., Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozlov A. G., Cox M. M., Lohman T. M., Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 285, 17246–17252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shishmarev D. et al., Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy. Nucleic Acids Res. 42, 2750–2757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su X.-C. et al., Bound or free: Interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry 53, 1925–1934 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Kozlov A. G. et al., Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J. Mol. Biol. 427, 763–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlov A. G., Shinn M. K., Weiland E. A., Lohman T. M., Glutamate promotes SSB protein-protein interactions via intrinsically disordered regions. J. Mol. Biol. 429, 2790–2801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlov A. G., Jezewska M. J., Bujalowski W., Lohman T. M., Binding specificity of Escherichia coli single-stranded DNA binding protein for the chi subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry 49, 3555–3566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Banani S. F. et al., Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancsa R., Schad E., Tantos A., Tompa P., Emergent functions of proteins in non-stoichiometric supramolecular assemblies. Biochim. Biophys. Acta. Proteins Proteomics 1867, 970–979 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Heinrich B. S., Maliga Z., Stein D. A., Hyman A. A., Whelan S. P. J., Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio 9, e02290-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolic J. et al., Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 8, 58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolic J., Lagaudrière-Gesbert C., Scrima N., Blondel D., Gaudin Y., Rabies virus factories are formed by liquid-liquid phase separation. Med. Sci. 34, 203–205 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y., Su J. M., Samuel C. E., Ma D., Measles virus forms inclusion bodies with properties of liquid organelles. J. Virol. 93, e00948-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Husini N., Tomares D. T., Bitar O., Childers W. S., Schrader J. M., α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71, 1027–1039.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monterroso B. et al., Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 20, e45946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinkel F. et al., Phase separation and clustering of an ABC transporter in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 116, 16326–16331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittag T., Parker R., Multiple modes of protein-protein interactions promote RNP granule assembly. J. Mol. Biol. 430, 4636–4649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao T. et al., Super-resolution imaging reveals changes in Escherichia coli SSB localization in response to DNA damage. Genes Cells 24, 814–826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolognesi B. et al., A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vernon R. M. et al., Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz S. G., Wilson N. L., Epstein W., Cation transport in Escherichia coli. II. Intracellular chloride concentration. J. Gen. Physiol. 46, 159–166 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock J. B., Rauch B., Roseman S., Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 252, 7850–7861 (1977). [PubMed] [Google Scholar]
- 35.Stull F., Hipp H., Stockbridge R. B., Bardwell J. C. A., In vivo chloride concentrations surge to proteotoxic levels during acid stress. Nat. Chem. Biol. 14, 1051–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett B. D. et al., Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillingham M. S. et al., Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys. J. 95, 3330–3339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richey B. et al., Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J. Biol. Chem. 262, 7157–7164 (1987). [PubMed] [Google Scholar]
- 39.Lin Y.-H., Forman-Kay J. D., Chan H. S., Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry 57, 2499–2508 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Meyer R. R., Laine P. S., The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 54, 342–380 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor N. O., Wei M.-T., Stone H. A., Brangwynne C. P., Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J. 117, 1285–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuznetsova I. M., Turoverov K. K., Uversky V. N., What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 15, 23090–23140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt A. et al., The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bobst E. V., Bobst A. M., Perrino F. W., Meyer R. R., Rein D. C., Variability in the nucleic acid binding site size and the amount of single-stranded DNA-binding protein in Escherichia coli. FEBS Lett. 181, 133–137 (1985). [DOI] [PubMed] [Google Scholar]
- 45.Villani G., Pierre A., Salles B., Quantification of SSB protein in E. coli and its variation during RECA protein induction. Biochimie 66, 471–476 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Perrino F. W., Rein D. C., Bobst A. M., Meyer R. R., The relative rate of synthesis and levels of single-stranded DNA binding protein during induction of SOS repair in Escherichia coli. Mol. Gen. Genet. 209, 612–614 (1987). [DOI] [PubMed] [Google Scholar]
- 47.Courcelle J., Khodursky A., Peter B., Brown P. O., Hanawalt P. C., Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158, 41–64 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bianco P. R. et al., The IDL of E. coli SSB links ssDNA and protein binding by mediating protein-protein interactions. Protein Sci. 26, 227–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakthavachalu B. et al., RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Mannen T., Yamashita S., Tomita K., Goshima N., Hirose T., The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J. Cell Biol. 214, 45–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milovanovic D., Wu Y., Bian X., De Camilli P., A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabari B. R. et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov A. G., Lohman T. M., Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry 41, 11611–11627 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Molineux I. J., Pauli A., Gefter M. L., Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Res. 2, 1821–1837 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Overman L. B., Bujalowski W., Lohman T. M., Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry 27, 456–471 (1988). [DOI] [PubMed] [Google Scholar]
- 56.Wolak C. et al., Interaction with single-stranded DNA-binding protein localizes ribonuclease HI to DNA replication forks and facilitates R-loop removal. Mol. Microbiol. 114, 495–509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lohman T. M., Ferrari M. E., Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Bujalowski W., Lohman T. M., Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 25, 7799–7802 (1986). [DOI] [PubMed] [Google Scholar]
- 59.Mills M. et al., RecQ helicase triggers a binding mode change in the SSB-DNA complex to efficiently initiate DNA unwinding. Nucleic Acids Res. 45, 11878–11890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shereda R. D., Bernstein D. A., Keck J. L., A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 282, 19247–19258 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Shereda R. D., Reiter N. J., Butcher S. E., Keck J. L., Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB’s C terminus. J. Mol. Biol. 386, 612–625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashton N. W., Bolderson E., Cubeddu L., O’Byrne K. J., Richard D. J., Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BMC Mol. Biol. 14, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawson T. et al., A structural perspective on the regulation of human single-stranded DNA binding protein 1 (hSSB1, OBFC2B) function in DNA repair. Comput. Struct. Biotechnol. J. 17, 441–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozlov A. G., Lohman T. M., Effects of monovalent anions on a temperature-dependent heat capacity change for Escherichia coli SSB tetramer binding to single-stranded DNA. Biochemistry 45, 5190–5205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D. J., Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spenkelink L. M. et al., Recycling of single-stranded DNA-binding protein by the bacterial replisome. Nucleic Acids Res. 47, 4111–4123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dubiel K. et al., Development of a single-stranded DNA-binding protein fluorescent fusion toolbox. Nucleic Acids Res. 48, 6053–6067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu C. et al., SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells 21, 163–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curth U., Genschel J., Urbanke C., Greipel J., In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 24, 2706–2711 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleming E., Yuan A. H., Heller D. M., Hochschild A., A bacteria-based genetic assay detects prion formation. Proc. Natl. Acad. Sci. U.S.A. 116, 4605–4610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kozlov A. G., Shinn M. K., Lohman T. M., Regulation of nearest-neighbor cooperative binding of E. coli SSB protein to DNA. Biophys. J. 117, 2120–2140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell J. C., Liu B., Kowalczykowski S. C., Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. eLife 4, e08646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mészáros B. et al., PhaSePro: The database of proteins driving liquid–liquid phase separation. Nucleic Acids Res. 48, D360–D367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vernon R. M., Forman-Kay J. D., First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 58, 88–96 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Pancsa R., Kovacs D., Tompa P., Misprediction of structural disorder in halophiles. Molecules 24, 479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukherjee S. et al., Genomes OnLine database (GOLD) v.6: Data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reimer L. C. et al., BacDive in 2019: Bacterial phenotypic data for high-throughput biodiversity analysis. Nucleic Acids Res. 47, D631–D636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and Datasets S1 and S2. Source code of the script used for analyses of spinning disk microscopy data are freely available at GitHub, https://github.com/SzegaX/VolumeOfSmearedSpheres.