Summary
Hepatocellular carcinoma (HCC) is a deadly tumour whose causative agents are generally well known, but whose pathogenesis remains poorly understood. Nevertheless, key genetic alterations are emerging from a heterogeneous molecular landscape, providing information on the tumorigenic process from initiation to progression. Among these molecular alterations, those that affect epigenetic processes are increasingly recognised as contributing to carcinogenesis from preneoplastic stages. The epigenetic machinery regulates gene expression through intertwined and partially characterised circuits involving chromatin remodelers, covalent DNA and histone modifications, and dedicated proteins reading these modifications. In this review, we summarise recent findings on HCC epigenetics, focusing mainly on changes in DNA and histone modifications and their carcinogenic implications. We also discuss the potential drugs that target epigenetic mechanisms for HCC treatment, either alone or in combination with current therapies, including immunotherapies.
Keywords: Hepatocellular carcinoma, Epigenetics, Therapy
Abbreviations: 5acC, 5-acetylcytosine; 5fC, 5-formylcytosine; 5hmC, 5-hydoxymethyl cytosine; 5mC, 5-methylcytosine; Acetyl-CoA, acetyl coenzyme A; α-KG, α-ketoglutarate; BER, base excision repair; BRD, bromodomain; CDA, cytidine deaminase; CGI, CpG island; CIMP, CGI methylator phenotype; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; DNMT, DNA methyltransferase; DNMTi, DNMT inhibitor; FAD, flavin adenine dinucleotide; HAT, histone acetyltransferases; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; HDACi, HDAC inhibitor; HDM, histone demethylase; HMT, histone methyltransferase; KMT, lysine methyltransferase; LSD/KDM, lysine specific demethylases; NAFLD, non-alcoholic fatty liver disease; ncRNAs, non-coding RNAs; NK, natural killer; NPC, nasopharyngeal carcinoma; PD1, programmed cell death protein 1; PD-L1, programmed cell death ligand-1; PHD, plant homeodomain; PTM, post-translational modification; SAM, S-adenosyl-L-methionine; TDG, thymidine-DNA-glycosylase; TERT, telomerase reverse transcriptase; TET, ten-eleven translocation; TME, tumour microenvironment; TSG, tumour suppressor gene; UHRF1, ubiquitin like with PHD and ring finger domains 1; VEGF, vascular endothelial growth factor
Key points.
-
•
The term epigenetics defines somatic heritable differences in the genome not attributable to changes in the primary sequence of DNA. In a multicellular organism, epigenetic mechanisms establish cellular identity out of a common genome.
-
•
Epigenetic mechanisms regulate chromatin conformation, nucleosome positioning and DNA wrapping around nucleosomes, modulating the interaction of the transcriptional machinery with genes, and thus controlling their expression.
-
•
The epigenetic effectors that influence the structure and function of chromatin include chromatin-remodelling complexes, DNA methylation/demethylation enzymes, histone modification enzymes, histone marks readers and non-coding RNAs.
-
•
Chromatin structure, DNA methylation and covalent histone modification patterns are altered in cancer, including hepatocarcinogenesis, and contribute to malignancy from its early stages. Mutations and changes in the expression of epigenetic effectors underlie these alterations.
-
•
DNA methylation and covalent histone modifications are reversible enzymatic reactions amenable to pharmacological intervention with small-molecule inhibitors, epidrugs. Epidrugs are promising therapeutic agents that counteract tumour hallmarks and potentiate the response to chemotherapy, targeted therapy and immunotherapy in hepatocellular carcinoma.
Introduction
Hepatocellular carcinoma (HCC) is the most frequent primary malignancy of the liver, ranking fourth among cancer-related causes of death worldwide.1,2 The World Health Organization projections estimate that by 2030 more than 1 million individuals will die from liver cancer per year.1 The prognosis of HCC remains poor, with a 5-year survival rate of just 18%, highlighting the limitations of available treatments.3 When detected early, HCCs are amenable to locoregional therapies and surgery, however the recurrence rate 5-years post-resection is about 70%.1 Systemic therapies are used for patients diagnosed at more advanced stages. HCC is very resistant to conventional chemotherapies,4 but targeted agents such as the multikinase inhibitors sorafenib, lenvatinib, regorafenib and cabozantinib, as well as monoclonal antibodies like ramucirumab (which targets vascular endothelial growth factor [VEGF] receptor 2), confer some survival benefit.1,3,5,6 Immunotherapy is also being actively tested in HCC, and immune checkpoint inhibitors (ICIs), which are antibodies that block the programmed cell death protein 1 (PD-1)/programmed cell death ligand-1 (PD-L1) pathway, or the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) pathway, have shown clinical activity.3,5,7 Moreover, a recent study showed very promising effects in patients with unresectable HCC that were treated with antibodies targeting PD-L1 and VEGF.8 Despite these advances, the very high mortality rate seen in patients with HCC clearly indicates that the efficacy of systemic therapies needs to be improved. To this end, combination strategies including locoregional approaches, targeted agents and ICIs are of interest, with some combinations being actively investigated.3,9 Nonetheless, when exploring systemic therapies for advanced HCC it is important to bear in mind that this type of tumour usually develops on a background of chronic liver injury, inflammation, fibrosis and cirrhosis, in an organ with impaired metabolic function that renders patients more susceptible to hepatic and systemic toxicities.1,5 Chronic liver injury leading to HCC development is mainly caused by HBV and HCV infections, long-lasting alcohol abuse and non-alcoholic fatty liver disease (NAFLD).1,2,10,11 With the systematic implementation of HBV vaccination and the advent of effective anti-HCV therapies, it is likely that NAFLD will become the dominant cause of HCC in the coming years.2
Development of effective therapies for any cancer relies to a great extent on a deep understanding of the tumour's molecular and cellular biology. Such knowledge may enable the identification (and targeting) of key tumour driver genes, as well as providing biomarkers for prognostic scoring and for the selection of potential responders to molecular therapies.6 Generating a precise molecular portrait and classification of HCC is not an easy task. Factors such as the aetiology of the liver disease, the stage of cancer progression, the molecular heterogeneity between different nodules, and even within the same tumoural mass, have hindered the genomic characterisation of HCC. Nevertheless, over the past decade, next generation sequencing technologies have been used to identify the most frequently recurring mutations, DNA copy number alterations and associated changes in gene expression that contribute to hepatocarcinogenesis.[12], [13], [14], [15], [16], [17], [18], [19] Such studies also enabled the classification of HCCs into different molecular subgroups with associated biological and clinical phenotypes.6,16 The most frequent (95% of tumours) and earliest genetic alteration found in the gradual process of hepatocarcinogenesis is the aberrant expression of telomerase reverse transcriptase (TERT). Upregulation of TERT is due to promoter mutations (40–60%), gene amplification, translocation, and also to viral insertion in HBV-related HCCs.17 The second most frequent mutations activate CTNNB1 (∼30%), the gene coding for β-catenin, or inactivate the tumour protein TP53 (∼30%). Other members of the WNT/β-catenin pathway, such as AXIN1, AXIN2, ZNRF3 and APC are also recurrently mutated and inactivated in HCC.14,16 Mutations in epigenetic modifiers are also frequent in HCC, and altogether they can be found in up to 50% of tumours.12,20 Recurrent inactivating mutations were found in ARID1A and ARID2,21,22 key components of the chromatin remodelling complex SWI/SNF, which has tumour suppressor activity.23 Mutations in histone modifying enzymes were initially described for members of the KMT2 family, such as KMT2D, KMT2B and KMT2C.12,19,22,24,25 However, more recently, the list of epigenetic modifiers mutated in HCC has been significantly expanded.26 DNA copy number alterations were also frequently found, and included deletions of tumour suppressor genes (like PTEN and RB1) and negative cell cycle regulators (such as CDKN2A and CDKN2B),14,19,22,25,27 as well as amplifications in promitogenic genes like FGF19 and CCND1.27,28 This impressive wealth of knowledge has indeed exposed a number of oncogene addiction loops that drive HCC progression.29 However, the most frequently mutated genes identified, such as TERT, CTNNB1 and TP53, are very difficult to target pharmacologically, or are believed to be undruggable.16 While the search for effective inhibitors of key drivers like TERT must continue, exploring new strategies and targets to quell HCC growth is crucial.
One emerging approach for the treatment of solid tumours comes from the field of epigenetics.30,31 Epigenetics refers to heritable traits not attributable to changes in DNA sequence that can control chromatin structure and the accessibility of the transcriptional machinery to DNA, thereby modulating gene expression.32 These mechanisms are thus fundamental for the maintenance of cell identity, but are also heavily implicated in development, stem cell renewal, genome integrity and proliferation.[32], [33], [34] Their deregulation is central to pathogenesis, including tumorigenesis,30 impacting on all hallmarks of cancer.35 Multiple pathways are involved in chromatin dynamics and epigenetic gene regulation, including DNA methylation, ATP-dependent nucleosome remodelling, the introduction of histone variants, post-translational modifications (PTMs) of histones, and non-coding RNAs (ncRNAs).30,32 Contrary to genetic mutations, epigenetic mechanisms such as covalent modifications of DNA and histones are highly flexible and dynamic, involving reversible enzymatic reactions and specific protein-protein interactions, which make them amenable to pharmacological intervention.36 Epigenetic alterations involving the chromatin remodelling machinery and ncRNAs have recently been reviewed elsewhere.[37], [38], [39] Herein, we review basic epigenetic mechanisms and the role of their dysregulation on hepatocarcinogenesis, focusing on DNA methylation and histone PTMs. We also highlight emerging strategies for the molecular targeting of epigenetic mechanisms with so-called “epidrugs” in HCC treatment and prevention.
Writing, erasing and reading epigenetic marks on chromatin
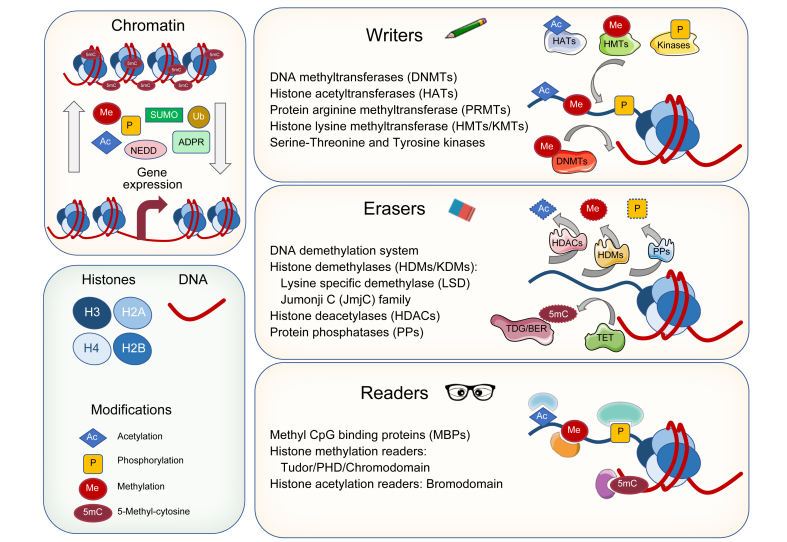
Nuclear DNA needs to be tightly packed, but it also needs to be accessible in a specific and regulated manner to allow for essential processes such as replication, repair and transcription. Efficient packaging of DNA in chromatin is mediated by its interaction with histones and the formation of nucleosomes. In nucleosomes, DNA is wrapped around 2 copies of each of the 4 core histones: H2A, H2B, H3 and H4, and outside the nucleosome the linker histone H1 facilitates further compaction in higher order chromatin structures.40 Nucleosomes are highly dynamic, they can slide along the DNA, fully or partially disassemble and their histone components may be replaced by sequence variants.40 The promoters and enhancers of transcriptionally active genes present reduced nucleosome abundance, which allows the recruitment of the transcriptional machinery and regulatory factors. Compact (repressed) and open (active) chromatin topologies, as found in heterochromatin and euchromatin, respectively, can be dictated by interrelated covalent epigenetic marks deposited on DNA and histones.41,42 DNA methylation mostly occurs on the 5' carbon of cytosine residues (5mC) in CpG dinucleotides. Methylation of CpGs in the so-called CpG islands (CGIs), present in about 70% of human gene promoters, has been widely associated with a closed chromatin conformation and inhibition of transcription initiation (Fig. 1).43 However, it has also been observed that CGI methylation in the transcribed regions of genes, i.e. gene bodies, increases gene expression.44 The list of histone PTMs is extensive and includes acetylation, methylation, phosphorylation, ubiquitinylation, sumoylation, ADP-ribosylation, neddylation, succinylation, crotonylation and butyrylation, among others.45,46 The best characterised modifications take place on amino acids present in the N-terminal tail regions of histones that protrude from the nucleosome surface (Fig. 1). These PTMs can modulate chromatin compaction and control the recruitment of remodelling complexes and transcription factors (TFs).45 In general, histone acetylation is related to gene activation, while methylation, phosphorylation and ubiquitinylation may be stimulatory or inhibitory, and sumoylation has been associated with gene repression.45,46 Interestingly, albeit less studied, PTMs also occur on the lateral surface of the central globular domain of histones, where they may also influence gene expression.47
Fig. 1.
Chromatin conformation and regulation of gene expression by epigenetic effectors acting on DNA and histones: epigenetic writers, erasers and readers.
PHD, plant homeodomain.
Chromatin marks are introduced, removed and recognised by a broad set of proteins called epigenetic modifiers. Epigenetic modifiers can be classified into 3 groups: epigenetic writers, epigenetic erasers and epigenetic readers (Fig. 1). Epigenetic writers are enzymes that add covalent modifications to DNA and histones. They include DNA methyltransferases (DNMTs), which catalyse the transfer of a methyl group from the universal methyl donor S-adenosyl-L-methionine (SAM) to CpG residues. In mammals there are 3 active DNMTs (Table 1): DNMT1, the most abundant DNMT (active at DNA replication foci and considered to be a maintenance DNMT); and DNMT3a and DNMT3b, which are primarily de novo DNMTs. However, these distinctions between maintenance and de novo activities seem not to be absolute.41,48 Histones can be methylated in lysine and arginine residues, and these amino acids can be mono- (me1), di- (me2) or tri-methylated (me3) in their amino and guanidino groups, respectively, making methylation the most complex histone PTM. Histone methylation is carried out by 3 families of enzymes that also use SAM as a cofactor: the SET-domain-containing histone methyltransferases (HMTs), the non-SET-domain-containing HMTs and the protein arginine methyltransferases (Table 1). Depending on the location and the methylation status of the lysine and arginine residues, methylation is associated with transcriptional activation (e.g. H3K4me2,3; H3K9me1; H3K27me1; H3K36me3; H3K79me2,3; H4K20me1; H3R17me2; H4R3me2) or repression (e.g. H3K9me2,3; H3K27me2,3; H4K20me3).45,46,49 Histone acetylation, which is associated with gene transcription, is performed by 3 types of histone acetyltransferases (HATs), belonging to the GNAT, MYST and CBP/p300 families (Table 1). HATs transfer an acetyl group from acetyl coenzyme A (acetyl-CoA) to the amino group of lysine residues.45,49 Histones can also be phosphorylated in serine, threonine and tyrosine residues by a plethora of kinases, a selection of which is indicated in Table 1, with diverse effects on chromatin remodelling and gene expression.50 At this point, it is important to mention that the writing of histone PTMs can be dynamically regulated at different levels, including by extracellular signals, as initially reported for acetylation reactions51 and later profusely analysed for phosphorylation events linked to signalling kinase cascades.50 It is also important to emphasise that an extensive and intricate crosstalk exists between different epigenetic marks in the regulation of gene expression. This was initially shown to occur between DNA CpG methylation and histone deacetylation, leading to chromatin condensation and gene repression.52 This notion has been exponentially extended over the past years, and now we recognise extensive hierarchical relations between DNA methylation and chromatin marks. Just to mention a few, there is an inverse correlation between DNA methylation and H3K4me2/3 and H3K79me3 levels in active gene promoters, while a positive association has been demonstrated between H3K36me3 and DNA methylation in the bodies of actively expressed genes. Conversely, gene expression and the presence of H3K27me3 and H3K9me3 in gene bodies are negatively associated. High H3K9me3 and H4K20me3 levels in gene promoters is also associated with DNA methylation and gene repression.41,42,53 These interactions can operate in both directions and be mutually reinforcing, as described between DNMTs and H3K9 HMTs in gene repression and heterochromatin formation.54
Table 1.
Epigenetic writers, erasers and readers: target residues in DNA and histones, and representative examples.
| Epigenetic modifiers | Major modified/recognised site | Family | Examples |
|---|---|---|---|
| Writers | |||
| DNA methyltransferases (DNMTs) | CpG (Met, 5mC) | DNMT1 DNMT3 | DNMT1A DNMT3A/3B |
| Histone methyltransferases (HMTs): lysine (KMTs) | H3 K4/K9/K27/K36/K79 (Met) H4 K20 (Met) | SUV39 SET1/2 EZH PRDMs | G9a/KMT1C MLL1/KMT2A SETD1A EZH2/KMT6 |
| Histone methyltransferases (HMTs): protein arginine (PRMTs) | H3 R2/R8/R17 (Met) H4 R3 (Met) | PRMT | PRMT1 PRMT4/CARM1 |
| Histone acetyltransferases (HATs) | H3 K9/K14/K56 (Ac) H4 K5/K8/K16 (Ac) H2A K5 (Ac) | GNAT MYST CBP/p300 | GCN5 TIP60 CBP/P300 |
| Serine-Threonine and Tyrosine kinases | Ser (P) Thr (P) Tyr (P) |
Haspin MSK CKII | |
| Erasers | |||
| DNA demethylation system | CpG (Met, 5mC) | TET TDG/BER | |
| Histone demethylases (HDMs/KDMs) | H3 K4/K9/K27/K36/K79 H4 K20 | LSD/KDM1A-B JARID/KDM2-8 | KDM1A/LSD1 KDM4/JMJD2 KDM5/RBP2 |
| Histone deacetylases (HDACs) | H3 K9/K14 H4 K5/K8/K12 |
HDAC I-IV |
HDAC1 Sirtuin |
| Readers | |||
| MBD-containing proteins | Methylated DNA (Methyl-CpG) | MeCP2 MBDs 1–6 SETDB1/2 | |
| Chromo domain- containing proteins | Methylated H3 K4/K9/K27/K36 | CHD1 HP1 | |
| Tudor domain-containing proteins | Methylated H3 K4/K9/K20/K36 | UHRF1 | |
| MBT-containing proteins | Methylated H3 K4/K9/K27/K36 | SFMBT1 MBTD1 | |
| PHD-containing proteins | Acetylated H3 K14 Methylated H3 K4/K9 | TFIID KMT2D | |
| Bromodomain (BRD)-containing proteins | Acetylated H3 K14 Acetylated H4 K5/K8/K16 |
GCN5 BRD4 PCAF (HAT) |
|
| Yeats domain-containing proteins | Acetylated H3 K9 | AF9 |
5mC, 5-methyl-cytosine; Ac, acetylation; MBD, methyl-CpG binding domain; MBT, malignant brain tumour; Me, methylation; PHD, plant homeodomain.
As mentioned, epigenetic marks are reversible and can be removed by a group of dedicated enzymes, collectively named epigenetic erasers (Fig. 1). Regarding DNA demethylation, this process can occur through 2 mechanisms. First, the lack of maintenance methylation during DNA replication, which is mainly carried out by DNMT1 in complex with the UHRF1 adaptor protein,55 can result in the passive dilution of 5mC. More recently, a replication-independent process involving enzymatic conversions has been elucidated. This is performed by a family of dioxygenases known as ten-eleven translocation (TET) enzymes (Table 1), that catalyse the sequential oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) using oxygen and α-ketoglutarate (α-KG) as substrates.56 These oxidized forms of 5mC can also be diluted upon DNA replication. Alternatively, the 5fC and 5caC forms can be removed in a 2-step process involving the sequential action of thymine-DNA-glycosylase (TDG) coupled with base excision repair (BER).56,57 Interestingly, the majority of 5hmC, and also 5fC to a certain extent, are not just short-lived intermediates and can remain in genomic DNA where they may play regulatory roles in gene expression.57,58 Histone demethylation is performed by 2 different families of enzymes with distinct mechanisms of action: the lysine specific demethylases (LSD/KDM1A-B) that use flavin adenine dinucleotide (FAD) as a co-substrate, and the Jumonji (JmjC) domain-containing demethylases (JARID/KDM2-8) which require α-KG and oxygen (Table 1). Interestingly, LSD enzymes can only remove mono- and dimethyl- marks on H3K4 and H3K9, whereas JmjC-containing demethylases can remove all 3 methylation states in all lysine residues.49,59 Depending on the histone and specific lysine residue that is demethylated, KDM activity may contribute to gene repression or activation.59 Acetyl groups from lysine residues are removed by a large set of histone deacetylases (HDACs) which are divided into 4 families: class I, class II, class III and class IV HDACs (Table 1). Class III encompasses the so-called Sirtuins, which present a different mechanism of action to the other 3 classes, requiring NAD+ for their catalytic activity.60 HDAC activity is generally linked to the generation of compact and repressive chromatin. However, the nature of the phosphatases involved in histone dephosphorylation and their regulation remains to be fully characterised.50 From this brief overview of the machinery that establishes the epigenetic marks on chromatin one may grasp its enormous intricacy. Nevertheless, this is just one layer of complexity. Additional aspects that cannot be covered here include the mechanisms involved in the timely recruitment of epigenetic writers and erasers to specific regions of chromatin. Here, TFs, long non-coding RNAs, and the presence of specific protein domains in epigenetic modifiers (i.e. the CXXC un-methylated CpG binding domain) play fundamental roles.54,61,62 Another increasingly recognised and critical regulatory layer emerges from cellular metabolism. As described, most epigenetic writers and erasers utilise metabolites such as oxygen, ATP, SAM, NAD+, acetyl-CoA, FAD and α-KG as cofactors or substrates. Fluctuations in the levels of these metabolites have been shown to impinge on the enzymatic activities of epigenetic modifiers.63 Moreover, elevated levels of other types of cellular metabolites such as the ketone body β-hydroxybutyrate, or the tricarboxylic acid cycle intermediates succinate and fumarate, can inhibit the activities of HDACs and α-KG-dependent enzymes (TETs, KDMs), respectively.64 Therefore, changes in the levels of metabolic substrates and inhibitors may effectively modulate the epigenome, while in turn epigenetic mechanisms control essential metabolic pathways and participate in their alterations in cancer.63,65 This link between metabolism and epigenetics may be particularly relevant for a prominent metabolic organ like the liver.
In order to translate the epigenetic marks deposited on DNA and histones into functional responses, these modifications must first be recognised. This is mediated by a third group of proteins called epigenetic readers (Table 1), endowed with specialised binding domains for specific covalent modifications. Readers are often found as part of multimeric complexes in association with writers and erasers, enabling the dynamic integration of signals that modulate chromatin conformation.49,66 The best characterised epigenetic readers are those that recognise 5mC, acetyl-lysines and methyl-lysines, while there is less information available for other modifications, such as Ser- and Thr- phosphorylation (Fig. 1).66 The methyl-CpG-binding proteins recognise 5mC through their methyl-CpG binding domain.49 This is a family with 11 members, of which MeCP2 was the first characterised. The presence of MeCP2 is mainly associated with transcriptional inhibition via recruitment of corepressor complexes, like those encompassing HDAC or H3K9 HMT activities, to methylated regions of DNA.52 Interestingly, interactions between MeCP2 and 5mC can also protect these residues from binding and oxidation by TET enzymes.52 Histone methylation readers have been well-characterised, and include a broad variety of proteins that contain different types of methyl-lysine binding domains such as chromodomains, tudor domains, plant homeodomain (PHD) fingers, PWWP motifs, WD40 repeat domains, bromo-adjacent homology domains, or malignant brain tumour domains, among others (Table 1).49 These domains discriminate between specific lysine residues and the degree of their methylation, and can be found on many different protein types. For instance, H3K4me2 and H3K4me3 can be recognised by chromatin remodelers, such as the enzyme CHD1 through its 2 chromodomains, while H3K4me3 is bound by the general transcription factor TFIID through a PHD domain-containing subunit, contributing to enhanced preinitiation complex formation.59 Repressive marks such as H3K9me3 can be recognised by the adaptor protein UHRF1 through a tandem tudor domain, or by a chromodomain present in heterochromatin protein 1 (HP1), leading to chromatin condensation.42,49 Several protein domains can recognise and bind acetylated histones. These domains are classified into 3 major categories: bromodomain (BRD), PHD finger and Yeats domains (Table 1).49 Based on structural similarities, BRD-containing proteins can be subdivided into 8 families, which display different affinities for specific histone acetylation marks, whereas PHD fingers are more flexible, recognising multiple acetylated histones but also methylation marks.67 Proteins containing the Yeats domain recognise several acetylation marks, nonetheless they have recently been shown to efficiently bind crotonylated lysine residues.49 Interestingly, many of the proteins encompassing these histone acetylation reader domains are chromatin remodelling/modifying enzymes, or can recruit these activities to chromatin. Just to name a few, the HAT PCAF encompasses a BRD domain that recognises acetylated H3K14 (H3K14ac) and the HMT KMT2D displays a PHD finger targeting H4K16ac, while the Yeats domain-containing AF9 protein can recruit the HMT DOT1 to chromatin regions displaying the H3K9ac mark.67 Together, these examples illustrate the extremely complex and intertwined nature of epigenetic regulatory mechanisms.
Epigenetic alterations in hepatocarcinogenesis
DNA methylation
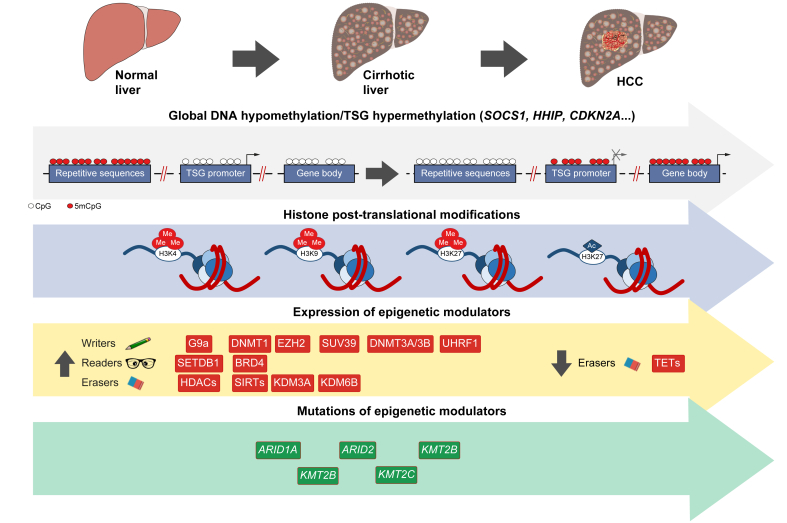
Alterations in DNA methylation, both genome-wide hypomethylation and region-specific hypermethylation, frequently occur in tumours and are among the most consistent epigenetic changes observed during multistage carcinogenesis.68 This also holds true for HCC, where changes in DNA methylation are already observed in the livers of patients with preneoplastic conditions, such as chronic hepatitis and cirrhosis of different aetiologies, including HCV/HBV infection, chronic alcohol consumption and, as more recently described, NAFLD.[69], [70], [71], [72], [73], [74] Furthermore, alterations in DNA methylation markedly increase during the progression from cirrhosis to early and more advanced neoplastic lesions, and many of them are preserved in fully developed HCCs (Fig. 2, Table 2).[70], [71], [72] Importantly, the methylation changes observed in non-tumoural liver tissues from patients with chronic hepatitis and cirrhosis may have prognostic value for HCC development or recurrence.69,71,75 These findings already indicate a causal relationship between epigenetic alterations and liver carcinogenesis. Early studies did not observe significant differences in overall changes to DNA methylation in HCCs from patients with different liver disease aetiologies.76 However, later works implementing more sensitive technologies identified different patterns of altered DNA methylation between HCV, HBV and NAFLD-related HCCs,[70], [71], [72],74 as well as during cirrhotic stages in alcohol or HCV-related carcinogenesis.72 DNA hypomethylation in HCC occurs in repeated DNA sequences, intergenic regions and in CpG sites away from CGIs, the so-called CpG shores, CpG shelfs and “open sea” regions (Fig. 2).77,78 Like in other tumours, DNA hypomethylation was initially associated with genomic instability in HCC.76 More recently, frequent mutations and rearrangements have been confirmed to occur in inactive chromatin regions specifically hypomethylated in HCCs.79 Very interestingly, a genome-wide hypomethylation pattern at transcriptional enhancers has also been reported recently. This was illustrated by the recurrent hypomethylation at the enhancer of C/EBPβ, resulting in the reactivation of its enhancer RNA (eRNA) and gene overexpression, leading to increased tumourigenesis.80 Other examples of genes – with possible tumour-promoting effects – that are hypomethylated and overexpressed in HCC are summarised in Table 2. Together, these observations provide further mechanistic links explaining the role of DNA hypomethylation in hepatocarcinogenesis.
Fig. 2.
Alterations in epigenetic mechanisms during hepatocarcinogenesis.
Ac, acetylation; HCC, hepatocellular carcinoma; Me, methylation; TETs, ten-eleven translocation enzymes; TSG, tumour suppressor gene.
Table 2.
Representative examples of genes with altered CpG methylation and expression in hepatocellular carcinoma.
| Expression | Function | Gene |
|---|---|---|
| Hypermethylation | ||
| Repressed Induced |
Cell cycle and cell growth regulation Cell signalling regulation Gene transcription Metabolic regulation Matrix remodelling Gene transcription |
APC CDH1 CDKN1A CDKN2A CDKN2B PTGS2 DAB2IP DKK3 GNA14 HHIP RASSF1A SFRP2 SOCS1 ESR1 HOXA9 RUNX3 SALL3 TP73 CPS1 FBP1 GSTP1 IGFBP5 MAT1A MMP9 MMP12 WT1 |
| Hypomethylation | ||
| Induced | Gene transcription Metabolic and signalling regulation Chemotaxis and angiogenesis |
C/EBPb IGF2 NOX4 SPINK1 CCL20 ESM1 |
DNA hypermethylation at preneoplastic stages and in HCC tissues occurs predominantly in promoter-associated CGIs (Fig. 2) and cis-regulatory elements, and correlates with reduced gene expression.71,81 Moreover, this epigenetic alteration precedes the appearance of chromosomal instability during hepatocarcinogenesis.68 Hypermethylated and downregulated genes found in hepatocarcinogenesis include well-known tumour suppressor genes (TSGs), regulators of cell signalling, proliferation, survival and metastasis, such as SOCS1, HHIP, SFRP2, APC, RASSF1, CDKN1A, CDKN2B, CDKN2A and CDH1, among others (Fig. 2 and Table 2).39,82 Similar findings were made for metabolic genes like MAT1A, FBP1 and CPS1, involved in SAM synthesis, gluconeogenesis and the urea cycle, respectively, whose repression may contribute to pro-oncogenic metabolic reprogramming.[83], [84], [85] Interestingly, hypermethylation of CGIs located in the gene bodies of bona fide oncogenes has recently been reported in experimental HCC, and this modification was consistently associated with their transcriptional activation.86 Of note, the hypermethylated and transcriptionally active regions in gene bodies seem to be more prone to mutagenic and rearrangement events, at least in HBV-related HCC.79 The strong association between DNA hypermethylation and hepatocarcinogenesis enabled the identification of a DNA-methylation signature in HCC tissues that could predict survival.78 More recently, the existence of a CGI methylator phenotype (CIMP), a biological phenomenon characterised by a subset of concurrently hypermethylated genes, has been identified in HCC tissues. Patients with higher CIMP scores were shown to have worse clinical outcomes, and robust CIMP-based diagnostic and prognostic models were developed based on few CIMP-associated genes.87,88
The mechanisms involved in the dysregulation of DNA methylation in hepatocarcinogenesis are likely multifarious. Oxidative stress,89 as well as HCV and HBV infection, have been shown to alter normal DNA methylation patterns in hepatocytes.16,90,91 The expression of the enzymes involved in DNA methylation and demethylation, DNMTs and TETs, is known to be altered in hepatocarcinogenesis. In this context, DNMT1 and its epigenetic adaptor partner UHRF1 are upregulated in HCC tissues and associated with poor prognosis.[92], [93], [94] Interestingly, the expression of DNMT1 and DNMT3a is already elevated in chronic hepatitis, and DNMT3a and DNMT3b are also overexpressed in HCCs.95 Regarding active DNA demethylation, the levels of 5hmC are consistently reduced in cirrhotic tissues and early HCC stages, remaining low in tumours and correlating with cancer progression.[96], [97], [98] The significant depletion in 5hmC may be attributed in part to the fall in global 5mC levels, but also to the reduced expression and activity of TET enzymes observed in HCC tissues,99 where the concentrations of their substrate α-KG are also markedly diminished.98 Impaired 5hmC turnover and DNA demethylation activity may contribute to the epigenetic repression of TSGs in HCC, as shown for SOCS1.100 However, the functional consequences of 5hmC depletion may still not be fully appreciated. Beyond being an intermediary in the DNA demethylation process, this epigenetic mark is normally enriched in the bodies of actively transcribed genes and enhancer elements; it can modify interactions between DNA and TFs, interfering with maintenance DNA methylation, as the DNMT1-UHRF1 complex does not recognise 5hmC.99
Histone PTMs
The dysregulation of epigenetic modifiers of histones and their role in hepatocarcinogenesis is being actively investigated (Fig. 2). There is some information on the expression of HATs in hepatocarcinogenesis. For instance, a recent study showed that upregulation of hMOF/KAT8 promoted microvascular invasion in HCCs.101 However, many works have reported the overexpression of class I HDACs, like HDAC1 and HDAC2, in HCC tissues and their association with mortality.102,103 Class II and class III HDACs, such as HDAC4, HDAC5, SIRT1, SIRT2 and SIRT7, have also been found upregulated in HCCs, 104,105 and their correlation with tumour progression has been established in some cases.104,106,107 Emerging experimental evidence also supports the involvement of HDACs in NAFLD-related HCC development.108 The mechanisms underlying the dysregulation of HDAC expression in HCC are not fully understood, but a relevant role for specific miRNAs is clearly being elucidated.105,109 A plethora of mechanistic studies have demonstrated the involvement of HDACs in the pathogenesis of HCC. When overexpressed, these epigenetic modifiers display multifaceted pro-oncogenic effects, including the inhibition of TSG expression, activation of cell cycle progression, apoptosis evasion, adaptation to hypoxia and metabolic reprogramming.105,110,111 The molecular mechanisms by which HDACs contribute to carcinogenesis can be quite complex. For instance, HDAC8 upregulation contributes to insulin resistance in NAFLD progression and, in coordination with the HMT KTM6 (EZH2), epigenetically represses the expression of Wnt antagonists, enhancing cell proliferation in HCC.108
Many recent studies demonstrate the altered expression of genes coding for both HMTs and KDMs in HCC tissues. While histone deacetylation is generally associated with repression of gene expression, the intricacies of the histone methylation code make the impact of its dysregulation far more complex. Within the HMT family, one of the best characterised enzymes is EZH2/KMT6, a component of the polycomb repressive complex 2 that mediates gene repression through H3K27 trimethylation.112 Increased EZH2 expression and H3K27me3 levels are found in HCC tissues, correlating with tumour aggressiveness.112 Several mechanisms have been reported to be involved in the pro-tumourigenic effects of EZH2, including the repression of Wnt pathway antagonists, the silencing of tumour suppressor miRNAs or the cooperation with cell cycle-related kinases to enhance androgen receptor signalling.[113], [114], [115] Other HMTs upregulated in HCC and also associated with poor prognosis are SUV39H1/KMT1A, SETDB1/KMT1E and G9a/KMT1C, which target H3K9 and are mostly associated with gene repression.94,[116], [117], [118] The overexpression of these HMTs results in enhanced growth and survival of HCC cells through multiple mechanisms, such as repression of TSG expression in coordination with promoter DNA hypermethylation, the adaptation to hypoxia and pro-oncogenic metabolic reprogramming, as demonstrated for G9a/KMT1C.94,118 Interestingly, a recent study identified the HMTs MLL1/KMT2A and MLL2/KMT2B as transcriptional targets of mutated TP53 (p53R249S), and their pharmacological inhibition abrogated p53R249S-driven HCC cell growth.75 There are also numerous examples of the dysregulation of KDM enzymes in HCC tissues and their association with enhanced tumour progression. For instance, increased KDM3A expression was associated with tumour recurrence after resection,119 while the upregulation of LSD1/KDM1, KDM5B, KDM6B, KDM4B and KDM2A has been related to tumour aggressiveness and poor prognosis.[120], [121], [122], [123], [124] The transcriptional programmes influenced by the overexpression of KDMs are complex to elucidate. The activity of these enzymes may have differential effects on gene expression depending on the specific histone residue on which they act. For example, LSD1/KDM1 and KDM4B, both upregulated in HCC, can remove repressive H3K9 methyl marks, while concomitantly, LSD1/KDM1 and KDM4B can eliminate H3K4 and H3K36 active methyl marks, respectively.59 Nevertheless, several pro-tumorigenic mechanisms triggered by KDMs in HCC have been elucidated, including the promotion of stem cell-like traits by KDM6B and KDM2A,122,124 and the maintenance of glycolytic metabolism, stemness and drug resistance by LSD1/KDM1.125,126
The role of dysregulated histone PTM readers in hepatocarcinogenesis is increasingly being recognised. This is clearly illustrated by BRD-containing protein 4 (BRD4), which was originally shown to be overexpressed in HCC tissues – derived from patients with a poor prognosis – wherein it promoted epithelial-mesenchymal transition.127 BRD4 recognises H3K27ac marks in chromatin, and H3K27ac is highly enriched in large clusters of enhancers, called super-enhancers (SE), that synergistically drive gene expression. Interestingly, many of the H3K27ac-marked SEs in HCC cells were associated with well-known oncogenes, and the presence of BRD4 was necessary for the expression of these SE-driven oncogenes.128
Despite the accumulating data on epigenetic alterations in HCC that we have tried to summarise herein, the epigenetic map of human HCC is far from complete. A recent TCGA-based comprehensive study analysed the mutational status and expression of 90 histone epigenetic readers, writers and erasers in HCC tissues. The authors found that 75% of patients presented at least one somatic mutation in one of the epigenetic modifiers examined, while 20% had more than 5. Regarding gene expression, when epigenetic modifiers were analysed in aggregate, 43% were upregulated and 22% downregulated in HCC tissues vs. non-tumoural tissues.26
Targeting epigenetic mechanisms in HCC
As opposed to gene mutations, the inherent reversibility of epigenetic abnormalities makes them promising targets for small molecules (epidrugs) in cancer treatment. The first FDA-approved epidrugs were DNMT and HDAC inhibitors for the treatment of haematological malignancies.31,129,130 The experimental and clinical experience gathered over the past years has shown us that: i) these specific but at the same time “globally acting” agents can reprogramme cancer stemness through their interaction with multiple genes and pathways, inhibiting cancer initiation and progression;129,131 ii) long-lasting cancer cell reprogramming, and therefore improved activity, can be achieved at low and less toxic doses of epidrugs;129,132 iii) epidrugs can overcome primary resistance and restore sensitivity of cancer cells to targeted agents and conventional chemotherapeutics;129,133,134 iv) epigenetic alterations in cells of the tumour microenvironment (TME), both stromal and immune cells, contribute to carcinogenesis and can be targeted to enhance therapeutic efficacy. Indeed, recent evidence indicates that targeting different epigenetic regulators, including writers (DNMTs and HMTs), readers (BRDs) and erasers (HDACs, KDMs) can increase immune recognition of tumour cells and synergise with immunotherapy.135,136
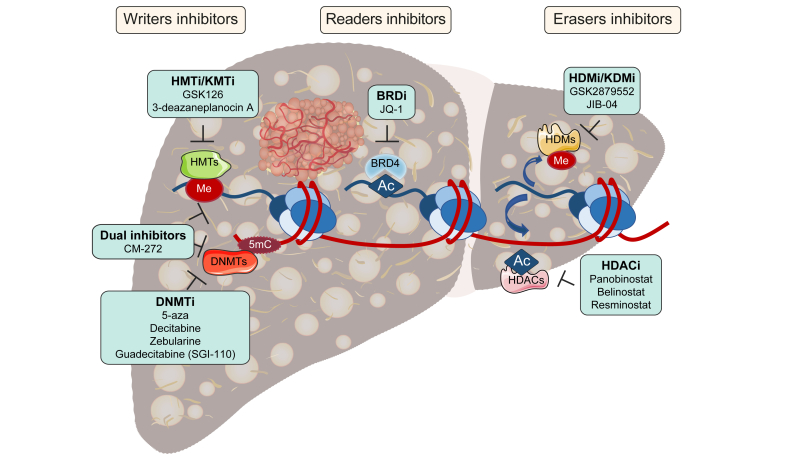
A wide variety of epidrugs have been developed over the past years. Most of them are in use, or are undergoing clinical trials, for the treatment of haematological malignancies (Table 3). [137], [138], [139], [140], [141] However, these compounds are increasingly being experimentally and clinically tested in solid tumours, including HCC (Fig. 3 and Table 3). The effects of DNA methylation inhibitors (DNMTi) on HCC cells have been extensively studied. The first-generation DNMTi included 5-azacytidine (5-aza) and 5-aza-2'-deoxycytidine (decitabine). Both can be incorporated into DNA, bind DNMT1 irreversibly and induce its degradation, leading to DNA demethylation. In addition, significant amounts of 5-aza can also be incorporated into RNA, also altering gene expression.130 In response to these agents TSG expression is reactivated and HCC cells partially recover their hepatocellular differentiation, becoming less tumourigenic and more sensitive to sorafenib.142,143 Decitabine was tested in patients with advanced HCC at low doses, showing favourable toxicity and signs of clinical benefit.144 However, 5-aza and decitabine have very short half-lives in vivo due to degradation by cytidine deaminase (CDA), which is abundant in the liver;39 therefore improved DNMTis have been developed. These include zebularine, an orally available and more stable DNMTi that is potentially less toxic as it does not incorporate into DNA, which has shown preclinical efficacy in a subclass of HCCs with a high degree of CpG methylation.145 Guadecitabine (SGI-110) is another second generation DNMTi consisting of 5-aza-2'-deoxycytidine linked to deoxyguanosine by a phosphodiester bond that is converted in vivo into decitabine; it is resistant to CDA and thus more stable.130 Preclinical studies have shown the inhibitory effects of SGI-110 on HCC growth and its ability to improve the antitumoral actions of sorafenib and oxaliplatin.146,147 Interestingly, besides TSG reinduction, the antitumoral effects of SGI-110 may also be attributable to DNA demethylation in gene body regions and the downregulation of pro-tumorigenic gene expression, including the epigenetic regulator UHRF1 and the HMT KMT6 (EZH2).148 Moreover, SGI-110 robustly reactivated the expression of epigenetically silenced endogenous retroviruses in HCC cells, triggering an innate immune response that can be harnessed to improve ICI sensitivity in vivo.136,148 The results of clinical trials testing SGI-110 administered alone or in combination with other antitumoural or immunotherapeutic agents in patients with advanced HCC are awaited (Table 3).
Table 3.
Examples of epigenetic drugs undergoing clinical trials.
| Drug | Disease | Phase | Reference/Clinical trial number |
|---|---|---|---|
| DNMTi | |||
| Azacitidine | CMML, AML, MDS | Clinical practice | (137) |
| Decitabine | CMML, AML, MDS | Clinical practice | (137) |
| Decitabine + Chemo- or immunotherapy | HCC | Phase I/II | NCT01799083 |
| Guadecitabine (SGI-110) | AML | Phase III | NCT02920008 |
| Guadecitabine (SGI-110) + sorafenib + oxaliplatin | HCC | Phase II | NCT01752933 |
| Guadecitabine (SGI-110) + durvalumab | HCC and biliopancreatic tumours | Phase I | NCT03257761 |
| TdCyd (4'-thio-2'- deoxycytidine) | Advanced solid tumours | Phase I | NCT02423057 |
| HDACi | |||
| Belinostat (PXD-101) | Relapsed or refractory PTCL | Clinical practice | (138) |
| Belinostat (PXD-101) | HCC | Phase I/II | NCT00321594 |
| Chidamide | PTCL | Phase II | NCT02944812 |
| Panobinostat | Multiple myeloma | Clinical practice | (139) |
| Quisinostat | Ovarian cancer | Phase II | NCT02948075 |
| Resminostat + sorafenib | HCC | Phase I/II | NCT00943449 |
| Romidepsin | CTCL | Clinical practice | (140) |
| Vorinostat | CTCL | Clinical practice | (141) |
| HMTi | |||
| MAK683 | DLBCL, NPC and other advanced solid tumours | Phase I/II | NCT02900651 |
| Tazemetostat (EPZ-6438) | Refractory B cell (NHL) with EZH2 gene mutation | Phase I/II | NCT03456726 |
| HDMi | |||
| GSK2879552 | Relapsed or refractory SCLC | Phase II | NCT02034123 |
| INCB059872 | Advanced solid tumours and hematologic malignancies | Phase I/II | NCT02712905 |
| BETi | |||
| BMS-986158 | Advanced solid tumours and hematologic malignancies | Phase I/II | NCT02419417 |
| GS-5829 | Solid tumours, lymphoma | Phase I | NCT02392611 |
| INCB057643 | Advanced solid tumours and hematologic malignancies | Phase I/II | NCT02711137 |
| PF-06821497 | SCLC, DLBCL, CRPC, and FL | Phase I | NCT03460977 |
| ZEN003694 | Triple negative breast cancer | Phase II | NCT03901469 |
AML, acute myeloid leukaemia; BETi, bromodomain and extra-terminal domain inhibitors; CMML, chronic myelomonocytic leukaemia; CRPC, castration-resistant prostate cancer; CTCL, cutaneous T cell lymphoma; DLBCL, diffuse large-B cell lymphoma; DNMTi, DNA methyltransferase inhibitor; FL, follicular lymphoma; HCC, hepatocellular carcinoma; HDACi, histone deacetylase inhibitor; HDMi, histone demethylase inhibitor; HMTi, histone methyltransferase inhibitor; MDS, myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; NPC, nasopharyngeal carcinoma; PTCL, peripheral T cell lymphoma; SCLC, small cell lung cancer.
Fig. 3.
Drugs targeting epigenetic writers, erasers and readers in HCC therapy.
5mC, 5-methyl-cytosine; Ac, acetylation; BRDi, bromodomain inhibitor; DNMT, DNA methyltransferase; DNMTi, DNMT inhibitor; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; HDACi, HDAC inhibitor; HDM, histone demethylase; HDMi, HDM inhibitor; HMT, histone methyltransferase; HMTi, HMT inhibitor; KMT, lysine methyltransferase; KMTi, KMT inhibitor; KDMi, lysine specific demethylase inhibitor; Me, methylation.
HDAC inhibitors (HDACi) have also been approved and clinically evaluated, mostly for the treatment of haematological malignancies (Table 3).31,130 Their efficacy in experimental HCC has been demonstrated. For instance, the non-selective pan-HDACi panobinostat was found to inhibit HCC proliferation, induce apoptosis, reprogramme cancer cell metabolism and reduce tumour angiogenesis.39 Another pan-HDACi, belinostat, also showed experimental efficacy and its clinical performance in patients with unresectable HCC suggests some value as a second-line treatment.39,105 Interestingly, the combination of belinostat with ICIs increased their efficacy in an experimental model of HCC.149 Resminostat, an oral pan-HDACi, was tested in patients with advanced HCC who had previously progressed on sorafenib. The combined administration of resminostat with sorafenib showed clinical efficacy, indicating that this HDACi may restore sensitivity to sorafenib.150 Mechanistically, the ability of resminostat to induce the reversion of HCC cells' stem-like properties may underlie the increased cytotoxic effects of the multikinase inhibitor.151 A recent study also provided a rationale for SIRT7 inhibition to increase the efficacy of immunotherapy (anti PD-L1) in mouse models, however potent and specific SIRT7 small molecule inhibitors still need to be synthesised.152
The development of HMT and HDM inhibitors is also being actively pursued, with some compounds being clinically tested in haematological malignancies (Table 3).49,153 In the context of HCC, the KMT6/EZH2 inhibitor GSK126 has recently been shown to enhance natural killer (NK) cell-mediated HCC cell death through its ability to re-induce the expression of NK cell ligands in HCC cells.154 Another recent study demonstrated that combined administration of 5-aza and 3-deazaneplanocin A (a non-specific EZH2 inhibitor) enhanced intratumor T cell trafficking and improved the antineoplasic effects of ICIs in a model of subcutaneously implanted HCC cells.155 However, another recent report found that administration of decitabine and GSK126 resulted in an impaired antitumorigenic T cell response and increased growth of orthotopically implanted HCC cells.156 Although, targeting G9a/KMT1C with different selective inhibitors reactivated TSG expression in HCC cells and demonstrated antitumoral effects in both in vitro and in vivo HCC xenograft models.118
Regarding KDM (HDM) targeting, inhibition of LSD-1/KDM1 with GSK2879552 could revert stem cell-like properties and re-sensitise HCC cells to sorafenib in vivo.157 More recently the JmjC KDM family inhibitor JIB-04 was shown to exert potent antitumoral effects in an immunocompetent mouse model of inflammation and fibrosis-associated HCC.26
As mentioned, epigenetic mechanisms act in concert in the normal regulation of gene expression, but can also become dysregulated in a “coordinated” manner to drive disease progression.41 This notion has led to the development of a novel set of drugs that combine dual inhibitory activities by acting on different epigenetic targets, or against an epigenetic and a non-epigenetic enzyme.36 One of these agents is CM-272,158 a new class of dual inhibitors of G9a/KMTC1 and DNMT1, enzymes that coordinate to mediate TSG silencing and foster cancer growth.159 CM-272 showed anti-HCC efficacy in vitro and in vivo, being able to restore the differentiated phenotype of HCC cells and to abate the pro-tumorigenic effects of the fibrogenic stroma.94 CM-272 has been shown to potentiate the efficacy of ICIs in other solid tumours;160 thus, evaluation of this combination in immunocompetent HCC models is warranted.
Pharmacological targeting of epigenetic readers is also an area of active research, and studies on haematological malignancies are also pioneering the field (Table 3).161 As mentioned, BRD4 and H3K27ac marks are known to be increased in HCC.128,162 Targeting the H3K27ac reader BRD4 with the inhibitor JQ-1 reduced HCC cell growth and survival,26,128 and most interestingly it prevented non-alcoholic steatohepatitis-associated experimental HCC development.162 Besides disrupting tumour-intrinsic oncogenic pathways, interference with BRD4 in myeloid-derived suppressor cells inhibits the development of fibrosis-associated experimental HCC, and enhances ICI efficacy.163 Together, these findings highlight the potential of preventing/treating HCC by targeting epigenetic mechanisms in the fibrotic stroma. Indeed, fibrogenesis is a key contributor to HCC development, and dysregulation of epigenetic circuits is central to liver fibrogenic cell activation and HCC progression.164,165 Pharmacological interference with such circuits is therefore emerging as an alternative strategy to halt liver fibrosis, and ultimately HCC development, as shown for BRD4 inhibitors 166 and more recently for the dual G9a/DNMT inhibitor CM-272.167
Conclusions
Our understanding of epigenetics has increased significantly over the last 2 decades. We are now aware of its importance for the regulation of cell function and its involvement in cancer development. However, we have also learned that the complexity of epigenetic regulatory circuits is enormous, and we have barely begun to scratch the surface of the iceberg. Although not covered here because of space limitations, epigenetic modifiers such as HMTs, KDMs and HDACs may also act on multiple protein targets, besides histones, with important regulatory consequences.30,129 Moreover, a given epigenetic modifier, such as G9a/KMT1C, can behave as a coactivator or corepressor of the transcription of different target genes in the same cell,168 while others like EZH2/KMT6 may act as gene repressors or as transcriptional activators depending on the tumour type where they are overexpressed.30 It is therefore critical to unravel all the dimensions of the epigenetic machinery to fully understand their pathophysiological implications. Given the magnitude of the task, the implementation of artificial intelligence tools for the molecular analyses of clinical samples and relevant experimental models appears essential.
Despite our still limited knowledge, current evidence suggests that epigenetic therapies also hold promise for the treatment of HCC, particularly in combination with other chemotherapeutics or with ICIs. However, there are still important general issues that need to be addressed when epigenetic drugs are considered as combination partners in cancer therapy. One relevant aspect is the potential systemic toxicity of these compounds. In general, combinations with chemotherapeutic agents or ICIs have been well tolerated.169 Nevertheless, adverse reactions to epidrugs may emerge due to their pleiotropic effects on gene expression, their activity on non-histone targets, or their interaction with non-tumoural cells.31,170 The specificity issues of epigenetic drugs are difficult to harness, as the diversity of their biological effects depends not only on the quality of their chemical design, but mostly on the complex functional interactions among their targets, as previously discussed. Accumulating evidence indicates that epidrug toxicity may be averted or attenuated by modifying the administration strategies. Importantly, it should be considered that dosing epigenetic drugs below their maximum tolerated dose can still result in full pharmacodynamic effects.171,172 Additionally, in combination regimens, selection of treatment sequence and schedule (i.e. concomitant administration, sequential administration, intermittent dosing, epigenetic priming, etc.) may not only reduce toxicity but also leverage synergies and overcome intrinsic and acquired resistance.169 The selection of adequate dosages and schedules may be particularly important in combinations between epidrugs and ICIs, as pro-tumorigenic effects have been recorded for both types of agents in experimental and clinical settings.156,173 Therefore, administration strategies still need to be thoroughly addressed in carefully designed clinical studies. Another determinant for the optimal application of epigenetic therapies is the availability of epigenetic biomarkers. Ideally, molecular biomarkers would enable patient selection and stratification, as well as the prediction of therapeutic response. However, the diversity and dynamism of epigenetic marks make the development of biomarkers for heterogeneous tumours such as HCC particularly challenging. So far, the identification of mutations in epigenetic genes, and the detection of CpG methylation at specific loci constitute the most successful epigenetic biomarkers in cancer.31 All in all, extracting the full potential of epidrugs for HCC treatment will require a precision-medicine approach, involving multidisciplinary cooperation and ad hoc trial designs. Nevertheless, the potential reward is clearly worth the effort.
Financial support
Work in the authors' laboratory is supported by: CIBERehd; grants PI16/01126 and PI19/00613 from Instituto de Salud Carlos III (ISCIII) co-financed by “Fondo Europeo de Desarrollo Regional” (FEDER) “Una manera de hacer Europa”; grants 58/17 and 2018-055 from Gobierno de Navarra; grants SAF2014-54191-R, SAF2017-88933-R, PID2019-104878R-100/AEI/10.13039/501100011033 and PID2019-104265RB-100/AEI/10.13039/501100011033 from FEDER/Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación; HEPACARE Project from Fundación La Caixa. Gobierno de Navarra fellowship to LC; AECC post-doctoral fellowship to MA and Ramón y Cajal Program contract RYC2018-024475-1 to MGFB.
Authors' contributions
All authors systematically revised the literature and revised the manuscript. CB and MAA supervised, drafted and revised the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2020.100167.
Contributor Information
Carmen Berasain, Email: cberasain@unav.es.
Matias A. Avila, Email: maavila@unav.es.
Supplementary data
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn K.A., Petrick J.L., El-Serag H.B. Epidemiology of hepatocellular carcinoma. Hepatology. 2020 doi: 10.1002/hep.31288. hep.31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarchoan M., Agarwal P., Villanueva A., Rao S., Dawson L.A., Llovet J.M. Recent developments and therapeutic strategies against hepatocellular carcinoma. Cancer Res. 2019;79:4326–4330. doi: 10.1158/0008-5472.CAN-19-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin J.J.G., Herraez E., Lozano E., Macias R.I.R., Briz O. Models for understanding resistance to chemotherapy in liver cancer. Cancers (Basel) 2019;11:1677. doi: 10.3390/cancers11111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerbes A., Zoulim F., Tilg H., Dufour J.-F., Bruix J., Paradis V. Gut roundtable meeting paper: selected recent advances in hepatocellular carcinoma. Gut. 2018;67:380–388. doi: 10.1136/gutjnl-2017-315068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet J.M., Montal R., Sia D., Finn R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greten T.F., Sangro B. Targets for immunotherapy of liver cancer. J Hepatol. 2018;68:157–166. doi: 10.1016/j.jhep.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 9.Cheng A.L., Hsu C., Chan S.L., Choo S.P., Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72:307–319. doi: 10.1016/j.jhep.2019.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara N., Friedman S.L., Goossens N., Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526–549. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avila M.A., Dufour J.F., Gerbes A.L., Zoulim F., Bataller R., Burra P. Recent advances in alcohol-related liver disease (ALD): summary of a Gut round table meeting. Gut. 2020;69:764–780. doi: 10.1136/gutjnl-2019-319720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 13.Ally A., Balasundaram M., Carlsen R., Chuah E., Clarke A., Dhalla N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze K., Nault J.C., Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031–1042. doi: 10.1016/j.jhep.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 16.Dhanasekaran R., Nault J.C., Roberts L.R., Zucman-Rossi J. Genomic medicine and implications for hepatocellular carcinoma prevention and therapy. Gastroenterology. 2019;156:492–509. doi: 10.1053/j.gastro.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebouissou S., Nault J.C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72:215–229. doi: 10.1016/j.jhep.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–577.e22. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 19.Totoki Y., Tatsuno K., Covington K.R., Ueda H., Creighton C.J., Kato M. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 20.Schulze K., Imbeaud S., Letouzé E., Alexandrov L.B., Calderaro J., Rebouissou S. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Zhao H., Zhang X., Wood L.D., Anders R.A., Choti M.A. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad, Ben I. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittal P., Roberts C.W.M. The SWI/SNF complex in cancer — biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17:435–448. doi: 10.1038/s41571-020-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleary S.P., Jeck W.R., Zhao X., Chen K., Selitsky S.R., Savich G.L. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58:1693–1702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kan Z., Zheng H., Liu X., Li S., Barber T.D., Gong Z. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayo J., Fiore E.J., Dominguez L.M., Real A., Malvicini M., Rizzo M. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J Hepatol. 2019;71:78–90. doi: 10.1016/j.jhep.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Ahn S.M., Jang S.J., Shim J.H., Kim D., Hong S.M., Sung C.O. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 28.Sawey E.T., Chanrion M., Cai C., Wu G., Zhang J., Zender L. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zucman-Rossi J., Villanueva A., Nault J.-C., Llovet J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 30.Pfister S.X., Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 31.Berdasco M., Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 32.Brien G.L., Valerio D.G., Armstrong S.A. Exploiting the epigenome to control cancer-promoting gene-expression programs. Cancer Cell. 2016;29:464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macchi F., Sadler K.C. Unraveling the epigenetic basis of liver development, regeneration and disease. Trends Genet. 2020;36:587–597. doi: 10.1016/j.tig.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Arechederra M., Berasain C., Avila M.A., Fernández-Barrena M.G. Chromatin dynamics during liver regeneration. Semin Cell Dev Biol. 2020;97:38–46. doi: 10.1016/j.semcdb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Flavahan W.A., Gaskell E., Bernstein B.E. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357 doi: 10.1126/science.aal2380. eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cossío F.P., Esteller M., Berdasco M. Towards a more precise therapy in cancer: exploring epigenetic complexity. Curr Opin Chem Biol. 2020;57:41–49. doi: 10.1016/j.cbpa.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Hu B., Lin J.Z., Yang X.B., Sang X.T. The roles of mutated SWI/SNF complexes in the initiation and development of hepatocellular carcinoma and its regulatory effect on the immune system: a review. Cell Prolif. 2020;53:e12791. doi: 10.1111/cpr.12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manna D., Sarkar D. Non-coding RNAs: regulating disease progression and therapy resistance in hepatocellular carcinoma. Cancers (Basel) 2020;12:1243. doi: 10.3390/cancers12051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toh T.B., Lim J.J., Chow E.K.-H. Epigenetics of hepatocellular carcinoma. Clin Transl Med. 2019;8:13. doi: 10.1186/s40169-019-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai W.K.M., Pugh B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. 2017;18:548–562. doi: 10.1038/nrm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du J., Johnson L.M., Jacobsen S.E., Patel D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeltsch A., Broche J., Bashtrykov P. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes (Basel) 2018;9:566. doi: 10.3390/genes9110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 44.Yang X., Han H., DeCarvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Z., Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaur J., Daoud A., Eblen S.T. Targeting chromatin remodeling for cancer therapy. Curr Mol Pharmacol. 2019;12:215–229. doi: 10.2174/1874467212666190215112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence M., Daujat S., Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 49.Biswas S., Rao C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018;837:8–24. doi: 10.1016/j.ejphar.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 50.Kumar R., Deivendran S., Santhoshkumar T.R., Pillai M.R. Signaling coupled epigenomic regulation of gene expression. Oncogene. 2017;36:5917–5926. doi: 10.1038/onc.2017.201. [DOI] [PubMed] [Google Scholar]
- 51.Latasa M.U., Boukaba A., García-Trevijano E.R., Torres L., Rodríguez J.L., Caballería J. Hepatocyte growth factor induces MAT2A expression and histone acetylation in rat hepatocytes: role in liver regeneration 1. FASEB J. 2001;15:1248–1250. doi: 10.1096/fj.00-0556fjev1. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt A., Zhang H., Cardoso M.C. MeCP2 and chromatin compartmentalization. Cells. 2020;9:878. doi: 10.3390/cells9040878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noh K.M., Allis C.D., Li H. Reading between the lines: “aDD”-ing histone and DNA methylation marks toward a new epigenetic “sum”. ACS Chem Biol. 2016;11:554–563. doi: 10.1021/acschembio.5b00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soshnev A.A., Josefowicz S.Z., Allis C.D. Greater than the sum of parts: complexity of the dynamic epigenome. Mol Cell. 2016;62:681–694. doi: 10.1016/j.molcel.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferry L., Fournier A., Tsusaka T., Adelmant G., Shimazu T., Matano S. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell. 2017;67:550–565.e5. doi: 10.1016/j.molcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Wu X., Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 57.Rasmussen K.D., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfeifer G.P., Szabó P.E., Song J. Protein interactions at oxidized 5-methylcytosine bases. J Mol Biol. 2020;432:1718–1730. doi: 10.1016/j.jmb.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milazzo G., Mercatelli D., Di Muzio G., Triboli L., De Rosa P., Perini G. Histone deacetylases (HDACs): evolution, specificity, role in transcriptional complexes, and pharmacological actionability. Genes. 2020;11:556. doi: 10.3390/genes11050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laisné M., Gupta N., Kirsh O., Pradhan S., Defossez P.A. Mechanisms of DNA methyltransferase recruitment in mammals. Genes (Basel) 2018;9:617. doi: 10.3390/genes9120617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Begolli R., Sideris N., Giakountis A. LncRNAs as chromatin regulators in cancer: From molecular function to clinical potential. Cancers (Basel) 2019;11:1524. doi: 10.3390/cancers11101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ly C.H., Lynch G.S., Ryall J.G. A metabolic roadmap for somatic stem cell fate. Cell Metab. 2020;31:1–16. doi: 10.1016/j.cmet.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y., Dong C., Zhou B.P. Metabolic reprogram associated with epithelial-mesenchymal transition in tumor progression and metastasis. Genes Dis. 2020;7:172–184. doi: 10.1016/j.gendis.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang T., Gong Y., Meng H., Li C., Xue L. Symphony of epigenetic and metabolic regulation - interaction between the histone methyltransferase EZH2 and metabolism of tumor. Clin Epigenetics. 2020;12:72. doi: 10.1186/s13148-020-00862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mio C., Bulotta S., Russo D., Damante G. Reading cancer: chromatin readers as druggable targets for cancer treatment. Cancers (Basel) 2019;11:61. doi: 10.3390/cancers11010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu H., Wei T., Cai Y., Jin J. Small molecules targeting the specific domains of histone-mark readers in cancer therapy. Molecules. 2020;25:578. doi: 10.3390/molecules25030578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanai Y., Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 69.Nagashio R., Arai E., Ojima H., Kosuge T., Kondo Y., Kanai Y. Carcinogenetic risk estimation based on quantification of DNA methylation levels in liver tissue at the precancerous stage. Int J Cancer. 2011;129:1170–1179. doi: 10.1002/ijc.26061. [DOI] [PubMed] [Google Scholar]
- 70.Um T.H., Kim H., Oh B.K., Kim M.S., Kim K.S., Jung G. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54:939–947. doi: 10.1016/j.jhep.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 71.Nishida N., Kudo M., Nagasaka T., Ikai I., Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56:994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- 72.Hlady R.A., Tiedemann R.L., Puszyk W., Zendejas I., Roberts L.R., Choi J.H. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget. 2014;5:9425–9443. doi: 10.18632/oncotarget.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wijetunga N.A., Pascual M., Tozour J., Delahaye F., Alani M., Adeyeye M. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene. 2017;36:2030–2044. doi: 10.1038/onc.2016.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuramoto J., Arai E., Tian Y., Funahashi N., Hiramoto M., Nammo T. Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis. 2017;38:261–270. doi: 10.1093/carcin/bgx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding X., He M., Chan A.W.H., Song Q.X., Sze S.C., Chen H. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology. 2020;157:1630–1645.e6. doi: 10.1053/j.gastro.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Calvisi D.F., Ladu S., Gorden A., Farina M., Lee J.S., Conner E.A. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen J., Wang S., Zhang Y.J., Wu H.C., Kibriya M.G., Jasmine F. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34–43. doi: 10.4161/epi.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villanueva A., Portela A., Sayols S., Battiston C., Hoshida Y., Méndez-González J. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956. doi: 10.1002/hep.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hama N., Totoki Y., Miura F., Tatsuno K., Saito-Adachi M., Nakamura H. Epigenetic landscape influences the liver cancer genome architecture. Nat Commun. 2018;9:1643. doi: 10.1038/s41467-018-03999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiong L., Wu F., Wu Q., Xu L., Cheung O.K., Kang W. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat Commun. 2019;10:335. doi: 10.1038/s41467-018-08245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Revill K., Wang T., Lachenmayer A., Kojima K., Harrington A., Li J. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424–1435. doi: 10.1053/j.gastro.2013.08.055. e1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishida N., Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma:A systematic review. Curr Genomics. 2011;12:130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Avila M.A., Berasain C., Torres L., Martín-Duce A., Corrales F.J., Yang H. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 84.Hirata H., Sugimachi K., Komatsu H., Ueda M., Masuda T., Uchi R. Decreased expression of fructose-1,6-bisphosphatase associates with glucose metabolism and tumor progression in hepatocellular carcinoma. Cancer Res. 2016;76:3265–3276. doi: 10.1158/0008-5472.CAN-15-2601. [DOI] [PubMed] [Google Scholar]
- 85.Liu H., Dong H., Robertson K., Liu C. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. Am J Pathol. 2011;178:652–661. doi: 10.1016/j.ajpath.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arechederra M., Daian F., Yim A., Bazai S.K., Richelme S., Dono R. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat Commun. 2018;9:3164. doi: 10.1038/s41467-018-05550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng J., Wei D., Ji Y., Chen L., Yang L., Li G. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10:42. doi: 10.1186/s13073-018-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G., Xu W., Zhang L., Liu T., Jin G., Song J. Development and validation of a CIMP-associated prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;47:128–141. doi: 10.1016/j.ebiom.2019.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim S.O., Gu J.M., Kim M.S., Kim H.S., Park Y.N., Park C.K. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140. doi: 10.1053/j.gastro.2008.07.027. 2140.e1-8. [DOI] [PubMed] [Google Scholar]
- 90.Park I.Y., Sohn B.H., Yu E., Suh D.J., Chung Y.H., Lee J.H. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 91.Okamoto Y., Shinjo K., Shimizu Y., Sano T., Yamao K., Gao W. Hepatitis virus infection affects DNA methylation in mice with humanized livers. Gastroenterology. 2014;146:562–572. doi: 10.1053/j.gastro.2013.10.056. [DOI] [PubMed] [Google Scholar]
- 92.Saito Y., Kanai Y., Nakagawa T., Sakamoto M., Saito H., Ishi H. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 93.Mudbhary R., Hoshida Y., Chernyavskaya Y., Jacob V., Villanueva A., Fiel M.I. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bárcena-Varela M., Caruso S., Llerena S., Álvarez-Sola G., Uriarte I., Latasa M.U. Dual targeting of histone methyltransferase G9a and DNA-methyltransferase 1 for the treatment of experimental hepatocellular carcinoma. Hepatology. 2019;69:587–603. doi: 10.1002/hep.30168. [DOI] [PubMed] [Google Scholar]
- 95.Saito Y., Kanai Y., Sakamoto M., Saito H., Ishii H., Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 96.Udali S., Guarini P., Moruzzi S., Ruzzenente A., Tammen S.A., Guglielmi A. Global DNA methylation and hydroxymethylation differ in hepatocellular carcinoma and cholangiocarcinoma and relate to survival rate. Hepatology. 2015;62:496–504. doi: 10.1002/hep.27823. [DOI] [PubMed] [Google Scholar]
- 97.Hlady R.A., Sathyanarayan A., Thompson J.J., Zhou D., Wu Q., Pham K. Integrating the epigenome to identify drivers of hepatocellular carcinoma. Hepatology. 2019;69:639–652. doi: 10.1002/hep.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J., Jiang J., Mo J., Liu D., Cao D., Wang H. Global DNA 5-hydroxymethylcytosine and 5-formylcytosine contents are decreased in the early stage of hepatocellular carcinoma. Hepatology. 2019;69:196–208. doi: 10.1002/hep.30146. [DOI] [PubMed] [Google Scholar]
- 99.Wang P., Yan Y., Yu W., Zhang H. Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma. Cell Prolif. 2019;52:e12626. doi: 10.1111/cpr.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Q., Yin D., Zhang Y., Yu L., Li X.D., Zhou Z.J. MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis. Cell Death Dis. 2017;8:e2906. doi: 10.1038/cddis.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poté N., Cros J., Laouirem S., Raffenne J., Negrão M., Albuquerque M. The histone acetyltransferase hMOF promotes vascular invasion in hepatocellular carcinoma. Liver Int. 2020;40:956–967. doi: 10.1111/liv.14381. [DOI] [PubMed] [Google Scholar]
- 102.Quint K., Agaimy A., Di Fazio P., Montalbano R., Steindorf C., Jung R. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011;459:129–139. doi: 10.1007/s00428-011-1103-0. [DOI] [PubMed] [Google Scholar]
- 103.Ler S.Y., Leung C.H.W., Khin L.W., Lu G.D., Salto-Tellez M., Hartman M. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast Asian population. Oncol Rep. 2015;34:2238–2250. doi: 10.3892/or.2015.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim J.K., Noh J.H., Jung K.H., Eun J.W., Bae H.J., Kim M.G. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 105.Zhao J., Gray S.G., Greene C.M., Lawless M.W. Unmasking the pathological and therapeutic potential of histone deacetylases for liver cancer. Expert Rev Gastroenterol Hepatol. 2019;13:247–256. doi: 10.1080/17474124.2019.1568870. [DOI] [PubMed] [Google Scholar]
- 106.Jiang H., Zhang X., Tao Y., Shan L., Jiang Q., Yu Y. Prognostic and clinicopathologic significance of SIRT1 expression in hepatocellular carcinoma. Oncotarget. 2017;8:52357–52365. doi: 10.18632/oncotarget.14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen J., Chan A.W.H., To K.F., Chen W., Zhang Z., Ren J. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology. 2013;57:2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 108.Tian Y., Wong V.W.S., Wong G.L.H., Yang W., Sun H., Shen J. Histone deacetylase HDAC8 promotes insulin resistance and β-catenin activation in NAFLD-associated hepatocellular carcinoma. Cancer Res. 2015;75:4803–4816. doi: 10.1158/0008-5472.CAN-14-3786. [DOI] [PubMed] [Google Scholar]
- 109.Kim H.S., Shen Q., Nam S.W. Histone deacetylases and their regulatory microRNAs in hepatocarcinogenesis. J Korean Med Sci. 2015;30:1375–1380. doi: 10.3346/jkms.2015.30.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee Y.H., Seo D., Choi K.J., Andersen J.B., Won M.A., Kitade M. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res. 2014;74:4752–4761. doi: 10.1158/0008-5472.CAN-13-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang J., Jin X., Yan Y., Shao Y., Pan Y., Roberts L.R. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci Rep. 2017;7:43864. doi: 10.1038/srep43864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Au S.L.K., Ng I.O.L., Wong C.M. Epigenetic dysregulation in hepatocellular carcinoma: focus on polycomb group proteins. Front Med. 2013;7:231–241. doi: 10.1007/s11684-013-0253-7. [DOI] [PubMed] [Google Scholar]
- 113.Cheng A.S.L., Lau S.S., Chen Y., Kondo Y., Li M.S., Feng H. EZH2-mediated concordant repression of Wnt antagonists promotes β-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–4039. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 114.Au S.L.K., Wong C.C.L., Lee J.M.F., Fan D.N.Y., Tsang F.H., Ng I.O.L. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–631. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- 115.Feng H., Yu Z., Tian Y., Lee Y.Y., Li M.S., Go M.Y.Y. A CCRK-EZH2 epigenetic circuitry drives hepatocarcinogenesis and associates with tumor recurrence and poor survival of patients. J Hepatol. 2015;62:1100–1111. doi: 10.1016/j.jhep.2014.11.040. [DOI] [PubMed] [Google Scholar]
- 116.Ngo-Yin Fan D., Ho-Ching Tsang F., Hoi-Kam Tam A., Leung-Kuen Au S., Chak-Lui Wong C., Wei L. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57:637–647. doi: 10.1002/hep.26083. [DOI] [PubMed] [Google Scholar]
- 117.Wong C.M., Wei L., Law C.T., Ho D.W.H., Tsang F.H.C., Au S.L.K. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 118.Wei L., Chiu D.K.C., Tsang F.H.C., Law C.T., Cheng C.L.H., Au S.L.K. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol. 2017;67:758–769. doi: 10.1016/j.jhep.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 119.Yamada D., Kobayashi S., Yamamoto H., Tomimaru Y., Noda T., Uemura M. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann Surg Oncol. 2012;19:S355–S364. doi: 10.1245/s10434-011-1797-x. [DOI] [PubMed] [Google Scholar]
- 120.Zhao Z.K., Yu H.F., Wang D.R., Dong P., Chen L., Wu W.G. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol. 2012;18:6651–6656. doi: 10.3748/wjg.v18.i45.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shigekawa Y., Hayami S., Ueno M., Miyamoto A., Suzaki N., Kawai M. Overexpression of KDM5B/JARID1B is associated with poor prognosis in hepatocellular carcinoma. Oncotarget. 2018;9:34320–34335. doi: 10.18632/oncotarget.26144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang B., Qi G., Tang F., Yuan S., Wang Z., Liang X. Aberrant JMJD3 expression upregulates slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res. 2016;76:6520–6532. doi: 10.1158/0008-5472.CAN-15-3029. [DOI] [PubMed] [Google Scholar]
- 123.Lu J.W., Ho Y.J., Lin L.I., Huang Y.C., Yeh K.T., Lin Y.H. JMJD2B as a potential diagnostic immunohistochemical marker for hepatocellular carcinoma: a tissue microarray-based study. Acta Histochem. 2015;117:14–19. doi: 10.1016/j.acthis.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 124.Lin Q., Wu Z., Yue X., Yu X., Wang Z., Song X. ZHX2 restricts hepatocellular carcinoma by suppressing stem cell-like traits through KDM2A-mediated H3K36 demethylation. EBioMedicine. 2020;53:102676. doi: 10.1016/j.ebiom.2020.102676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sakamoto A., Hino S., Nagaoka K., Anan K., Takase R., Matsumori H. Lysine demethylase LSD1 coordinates glycolytic and mitochondrial metabolism in hepatocellular carcinoma cells. Cancer Res. 2015;75:1445–1456. doi: 10.1158/0008-5472.CAN-14-1560. [DOI] [PubMed] [Google Scholar]
- 126.Lei Z.J., Wang J., Xiao H.L., Guo Y., Wang T., Li Q. Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5 + liver cancer initiating cells by suppressing negative regulators of β-catenin signaling. Oncogene. 2015;34:3188–3198. doi: 10.1038/onc.2015.129. [DOI] [PubMed] [Google Scholar]
- 127.Zhang P., Dong Z., Cai J., Zhang C., Shen Z., Ke A. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Immunopathol Pharmacol. 2015;28:36–44. doi: 10.1177/0394632015572070. [DOI] [PubMed] [Google Scholar]
- 128.Tsang F.H., Law C., Tang T.C., Cheng C.L., Chin D.W., Tam W.V. Aberrant super-enhancer landscape in human hepatocellular carcinoma. Hepatology. 2019;69:2502–2517. doi: 10.1002/hep.30544. [DOI] [PubMed] [Google Scholar]
- 129.Azad N., Zahnow C.A., Rudin C.M., Baylin S.B. The future of epigenetic therapy in solid tumours - lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bennett R.L., Licht J.D. Targeting epigenetics in cancer. Annu Rev Pharmacol Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tsui Y.M., Chan L.K., Ng I.O.L. Cancer stemness in hepatocellular carcinoma: mechanisms and translational potential. Br J Cancer. 2020;122:1428–1440. doi: 10.1038/s41416-020-0823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tsai H.C., Li H., Van Neste L., Cai Y., Robert C., Rassool F.V. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oronsky B.T., Oronsky A.L., Lybeck M., Oronsky N.C., Scicinski J.J., Carter C. Episensitization: defying time's arrow. Front Oncol. 2015;5:134. doi: 10.3389/fonc.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ravindran Menon D., Hammerlindl H., Torrano J., Schaider H., Fujita M. Epigenetics and metabolism at the crossroads of stress-induced plasticity, stemness and therapeutic resistance in cancer. Theranostics. 2020;10:6261–6277. doi: 10.7150/thno.42523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu Y., Chan Y.T., Tan H.Y., Li S., Wang N., Feng Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19:79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Topper M.J., Vaz M., Marrone K.A., Brahmer J.R., Baylin S.B. The emerging role of epigenetic therapeutics in immuno-oncology. Nat Rev Clin Oncol. 2020;17:75–90. doi: 10.1038/s41571-019-0266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Diesch J., Zwick A., Garz A.K., Palau A., Buschbeck M., Götze K.S. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin Epigenetics. 2016;8:71. doi: 10.1186/s13148-016-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sawas A., O'connor O.A., Radeski D. Belinostat in patients with refractory or relapsed peripheral T-cell lymphoma: a perspective review. Ther Adv Hematol. 2015;6:202–208. doi: 10.1177/2040620715592567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cavenagh J.D., Popat R. Optimal management of histone deacetylase inhibitor-related adverse events in patients with multiple myeloma: a focus on panobinostat. Clin lymphoma Myeloma Leuk. 2018;18:501–507. doi: 10.1016/j.clml.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 140.Reddy S.A. Romidepsin for the treatment of relapsed/refractory cutaneous T-cell lymphoma (mycosis fungoides/Sézary syndrome): use in a community setting. Crit Rev Oncol Hematol. 2016;106:99–107. doi: 10.1016/j.critrevonc.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 141.Bubna A.K. Vorinostat-An overview. Indian J Dermatol. 2015;60:419. doi: 10.4103/0019-5154.160511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu J., Liu Y., Meng L., Liu K., Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. 2017;38:899–907. doi: 10.3892/or.2017.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gailhouste L., Liew L.C., Yasukawa K., Hatada I., Tanaka Y., Nakagama H. Differentiation therapy by epigenetic reconditioning exerts antitumor effects on liver cancer cells. Mol Ther. 2018;26:1840–1854. doi: 10.1016/j.ymthe.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mei Q., Chen M., Lu X., Li X., Duan F., Wang M. An open-label, single-arm, phase I/II study of lowerdose decitabine based therapy in patients with advanced hepatocellular carcinoma. Oncotarget. 2015;6:16698–16711. doi: 10.18632/oncotarget.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andersen J.B., Factor V.M., JU Marquardt, Raggi C., Lee Y.-H., Seo D. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci Transl Med. 2010;2:54ra77. doi: 10.1126/scitranslmed.3001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kuang Y., El-Khoueiry A., Taverna P., Ljungman M., Neamati N. Guadecitabine (SGI-110) priming sensitizes hepatocellular carcinoma cells to oxaliplatin. Mol Oncol. 2015;9:1799–1814. doi: 10.1016/j.molonc.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jueliger S., Lyons J., Cannito S., Pata I., Pata P., Shkolnaya M. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics. 2016;11:709–720. doi: 10.1080/15592294.2016.1214781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu M., Zhang L., Li H., Hinoue T., Zhou W., Ohtani H. Integrative epigenetic analysis reveals therapeutic targets to the DNA methyltransferase inhibitor guadecitabine (SGI-110) in hepatocellular carcinoma. Hepatology. 2018;68:1412–1428. doi: 10.1002/hep.30091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Llopiz D., Ruiz M., Villanueva L., Iglesias T., Silva L., Egea J. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol Immunother. 2019;68:379–393. doi: 10.1007/s00262-018-2283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bitzer M., Horger M., Giannini E.G., Ganten T.M., Wörns M.A., Siveke J.T. Resminostat plus sorafenib as second-line therapy of advanced hepatocellular carcinoma – the SHELTER study. J Hepatol. 2016;65:280–288. doi: 10.1016/j.jhep.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 151.Soukupova J., Bertran E., Peñuelas-Haro I., Urdiroz-Urricelqui U., Borgman M., Kohlhof H. Resminostat induces changes in epithelial plasticity of hepatocellular carcinoma cells and sensitizes them to sorafenibinduced apoptosis. Oncotarget. 2017;8:110367–110379. doi: 10.18632/oncotarget.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xiang J., Zhang N., Sun H., Su L., Zhang C., Xu H. Disruption of SIRT7 increases the efficacy of checkpoint inhibitor via MEF2D regulation of programmed cell death 1 ligand 1 in hepatocellular carcinoma cells. Gastroenterology. 2020;158:664–678.e24. doi: 10.1053/j.gastro.2019.10.025. [DOI] [PubMed] [Google Scholar]
- 153.Rugo H.S., Jacobs I., Sharma S., Scappaticci F., Paul T.A., Jensen-Pergakes K. The promise for histone methyltransferase inhibitors for epigenetic therapy in clinical oncology: a narrative review. Adv Ther. 2020;37:3059–3082. doi: 10.1007/s12325-020-01379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bugide S., Green M.R., Wajapeyee N. Inhibition of Enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc Natl Acad Sci U S A. 2018;115:E3509–E3518. doi: 10.1073/pnas.1802691115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hong Y.K., Li Y., Pandit H., Li S.P., Pulliam Z., Zheng Q. Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell Immunol. 2019;336:66–74. doi: 10.1016/j.cellimm.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 156.Wei Y., Lao X.M., Xiao X., Wang X.Y., Wu Z.J., Zeng Q.H. Plasma cell polarization to the immunoglobulin G phenotype in hepatocellular carcinomas involves epigenetic alterations and promotes hepatoma progression in mice. Gastroenterology. 2019;156:1890–1904.e16. doi: 10.1053/j.gastro.2019.01.250. [DOI] [PubMed] [Google Scholar]
- 157.Huang M., Chen C., Geng J., Han D., Wang T., Xie T. Targeting KDM1A attenuates Wnt/β-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 2017;398:12–21. doi: 10.1016/j.canlet.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 158.San José-Enériz E., Agirre X., Rabal O., Vilas-Zornoza A., Sanchez-Arias J.A., Miranda E. Discovery of first-in-class reversible dual small molecule inhibitors against G9a and DNMTs in hematological malignancies. Nat Commun. 2017;8:15424. doi: 10.1038/ncomms15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wozniak R.J., Klimecki W.T., Lau S.S., Feinstein Y., Futscher B.W. 5-Aza-2'-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 160.Segovia C., San José-Enériz E., Munera-Maravilla E., Martínez-Fernández M., Garate L., Miranda E. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat Med. 2019;25:1073–1081. doi: 10.1038/s41591-019-0499-y. [DOI] [PubMed] [Google Scholar]
- 161.Stathis A., Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8:24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 162.Jühling F., Hamdane N., Crouchet E., Li S., El Saghire H., Mukherji A. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut. 2020 doi: 10.1136/gutjnl-2019-318918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu M., Zhou J., Liu X., Feng Y., Yang W., Wu F. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365–379. doi: 10.1136/gutjnl-2018-317257. [DOI] [PubMed] [Google Scholar]
- 164.Moran-Salvador E., Mann J. Epigenetics and liver fibrosis. Cell Mol Gastroenterol Hepatol. 2017;4:125–134. doi: 10.1016/j.jcmgh.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Barcena-Varela M., Colyn L., Fernandez-Barrena M.G. Epigenetic mechanisms in hepatic stellate cell activation during liver fibrosis and carcinogenesis. Int J Mol Sci. 2019;20:2507. doi: 10.3390/ijms20102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ding N., Hah N., Yu R.T., Sherman M.H., Benner C., Leblanc M. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci U S A. 2015;112:15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Barcena-Varela M., Paish H., Alvarez L., Uriarte I., Latasa M.U., Santamaria E. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut. 2020 doi: 10.1136/gutjnl-2019-320205. [DOI] [PubMed] [Google Scholar]
- 168.Stallcup M.R., Poulard C. Gene-specific actions of transcriptional coregulators facilitate physiological plasticity: evidence for a physiological coregulator code. Trends Biochem Sci. 2020;45:497–510. doi: 10.1016/j.tibs.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morel D., Jeffery D., Aspeslagh S., Almouzni G., Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours — past lessons and future promise. Nat Rev Clin Oncol. 2020;17:91–107. doi: 10.1038/s41571-019-0267-4. [DOI] [PubMed] [Google Scholar]
- 170.Ganesan A., Arimondo P.B., Rots M.G., Jeronimo C., Berdasco M. The timeline of epigenetic drug discovery: from reality to dreams. Clin Epigenetics. 2019;11:174. doi: 10.1186/s13148-019-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Choy E., Flamand Y., Balasubramanian S., Butrynski J.E., Harmon D.C., George S. Phase 1 study of oral abexinostat, a histone deacetylase inhibitor, in combination with doxorubicin in patients with metastatic sarcoma. Cancer. 2015;121:1223–1230. doi: 10.1002/cncr.29175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Italiano A., Soria J.C., Toulmonde M., Michot J.M., Lucchesi C., Varga A. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 173.Grecea M., Marabelle A., Ammari S., Massard C., Champiat S. Managing hyperprogressive disease in the era of programmed cell death protein 1/programmed death-ligand 1 blockade: a case discussion and review of the literature. Oncologist. 2020;25:369–374. doi: 10.1634/theoncologist.2019-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.