Abstract
Recombinant adeno-associated viral (rAAV) vector mobilization is a largely theoretical process in which intact AAV vectors spread or “mobilize” from transduced cells and infect additional cells within, or external of, the initial host. This process can be helper virus-independent (vector alone) or helper virus-dependent (de novo rAAV production facilitated by superinfection of both wild-type AAV [wtAAV] and Adenovirus 5 [Ad] helper virus). Herein, rAAV production and mobilization with and without wtAAV were analyzed following plasmid transfection or viral transduction utilizing well-established in vitro conditions and analytical measurements. During in vitro production, wtAAV produced the highest titer with rAAV-luc (4.1 kb), rAAV-IDUA (3.7 kb), and rAAV-Nano-dysferlin (4.9 kb) generating 2.5-, 5.9-, or 10.7-fold lower amounts, respectively. Surprisingly, cotransfection of a wtAAV and an rAAV plasmid resulted in a uniform decrease in production of wtAAV in all instances with a concomitant increase of rAAV such that wtAAV:rAAV titers were at a ratio of 1:1 for all constructs investigated. These results were shown to be independent of the rAAV transgenic sequence, size, transgene, or promoter choice and point to novel aspects of wtAAV complementation that enhance current vector production systems yet to be defined. In a mobilization assay, a sizeable amount of rAAV recovered from infected 293 cell lysate remained intact and competent for a secondary round of infection (termed Ad-independent mobilization). In rAAV-infected cells coinfected with Ad and wtAAV, rAAV particle production was increased >50-fold compared with no Ad conditions. In addition, Ad-dependent rAAV vectors mobilized and resulted in >1,000-fold transduction upon a subsequent second-round infection, highlighting the reality of these theoretical safety concerns that can be manifested under various conditions. Overall, these studies document and signify the need for mobilization-resistant vectors and the opportunity to derive better vector production systems.
Keywords: gene therapy, vector, mobilization, adeno-associated virus, adenovirus
Introduction
Adeno-associated virus (AAV), a Dependovirus of the family Parvoviridae, was first identified as an Adenovirus (Ad) preparation contaminant in 1965 by Atchison et al.1 The linear DNA AAV genome of ∼4.7 kb consists of inverted terminal repeats (ITRs) flanking several open reading frames (ORFs), including rep and cap, which encode proteins involved in genome replication and capsid production, respectively. Although AAV replication is not completely understood, the ITRs serve as the replication origins and work in concert with Rep proteins, the largest of which directly bind the ITR and induce a specific single-strand nick as an initial step in the replication process.2,3
Traditionally, AAV is considered a replication defective virus that requires coinfection of a helper virus, several of which have been identified for completion of its natural life cycle.4 In the absence of a helper virus, little expression of the rep ORFs occurs, and therefore, the AAV genome is minimally replicated and remains latent.5,6 However, AAV replication in the absence of a helper virus has been reported during cellular stress and/or in particular types of cells and/or phases of the cell cycle.7,8
The wild-type AAV2 (wtAAV2) genome was cloned into several plasmid constructs in the 1980s,9–11 and these constructs serve as the parental plasmids of most recombinant AAV (rAAV) vector constructs. In recombinant AAV (rAAV, also termed AAV vectors herein), the ITRs of serotype 2 (ITRs) are the only viral cis elements, flanking transgenic cassettes, as they are required for minimally rAAV genome replication and capsid packaging.12 Currently, AAV vectors are the most promising delivery method for in vivo human gene therapy with successes demonstrated in clinical trials for diverse diseases and a few drugs are FDA-approved and commercialized.2,9,10,12–15 Despite the popularity of AAV-based gene therapies, there remain unanswered questions regarding nearly all aspects of wtAAV and rAAV biology, in addition to the implications of the vector-induced genetic modifications in human patients.16
The rAAV vector itself is replication deficient, as replication requires the Rep proteins (absent from the vector), as well as a helper virus in most reported cases.10,17–23 However, wtAAV, which could supply the Rep and Cap proteins in trans for rAAV genome replication and capsid packaging, is prevalent in the human population.24 Superinfection by other pathogenic viruses, such as herpes simplex virus (HSV) or Ad, which provide “helper functions” for wtAAV or rAAV, is also common in human patient populations. For example, the eye as an external organ offers unique advantages as a gene therapy target,25 some of which also makes it more susceptible to viral infections. Previous reports demonstrated that herpetic keratitis, which is caused by HSV, is the most common corneal infection in the United States with 50,000 new and recurring cases diagnosed annually.26 In addition, several Ad serotypes infect the conjunctiva and cornea and are responsible for 92% of all keratoconjunctivitis.27
rAAV mobilization is a largely theoretical process in which intact AAV vectors spread or “mobilize” from transduced cells and infect additional cells within, or external of, the initial host. This process can be helper virus-independent, in which intracellular intact rAAV particles are released from the cell and infect another cell, or replication-dependent in which there is de novo rAAV production facilitated by superinfection of both wtAAV and a helper virus.28–32 Despite nearly 40 years of rAAV investigations, including broad clinical applications, rAAV mobilization and its potential to induce disease and environmental safety concerns remain an overlooked and understudied problem with only a few publications investigating this phenomenon with no definitive quantification of the events.29,31 In 1980, the rescue and mobilization of the wtAAV genome were first demonstrated in cell cultures.33 In the case of rAAV, Tratschin et al. reported a high frequency of integration and successful rescue from the chromosome by superinfection of wtAAV and Ad, and in some cases, by infection with Ad alone in 293 and Hela cells in 1985.34 Hewitt et al. demonstrated that rAAV vectors utilizing the ITR sequence from AAV5 reduce the risk of mobilization because of the lower frequency of wtAAV5 in human population combined with Rep2's inability to nick the ITR5 sequence.29,35 Due to the lack of an appropriate animal model for studying the mobilization, so far, the only in vivo study to investigate this risk was carried out in nonhuman primates in 1996.31 In that work, the authors demonstrated that rAAV replication and rescue occurred only after a direct administration of a very large dose of wtAAV into the lower respiratory tract before rAAV and Ad administration.31 Whether this condition could ever happen in a natural setting is not clear, but illustrates the early efforts to address this hypothetical concern.
A more likely concern emanates from the risk of rAAV replication-dependent mobilization released into the environment (or sheds) from treated patients following both systemic and local injections of AAV vectors.36–41 As shown in hemophilia clinical trials, AAV vector genomes were detected in the saliva, semen, blood serum, and urine of patients up to 12 weeks postintramuscular42 injections or portal vein infusion.43 In clinical trials using Luxturna (which is locally administered directly to the subretinal space), rAAV shedding was observed in about 40% of the subjects through tears.44 Although it remains to be tested if rAAV infectious particles are also shed from treated patients, these observations underscore the potential of shedding-associated rAAV transduction, with associated off-target transgenic DNA expression in unintended animal and human populations. Complicating this concern are studies, which demonstrate that capsid uncoating is a rate-limiting step in wtAAV and rAAV transduction.45–47 Detection of intact rAAV particles by transmission electron microscopy up to 6 years after administration in the retinas of dogs and primates has been reported.48 It remains possible that a large proportion of AAV vectors that fail to uncoat remain infectious.45,49,50 Whether these vectors could be mobilized in a replication-independent manner and/or shed to mediate transduction in off-target cells, organs, and individuals remains unknown.29
Theoretically, during replication-dependent rAAV mobilization, AAV vector genomes compete with wtAAV genomes for the Rep and Cap proteins, which logically assumes that both wtAAV and rAAV production would decrease in the presence of a helper virus, if Rep or Cap proteins are rate limiting for virion assembly. Herein, this theory is quantitatively assessed under various conditions. The collective results demonstrate that following plasmid transfection: (1) wtAAV production is more efficient than rAAVs, (2) rAAV titers vary significantly in a transgene-dependent manner, (3) following cotransfection of wtAAV and rAAV plasmids, wtAAV demonstrates a modest decrease in the production titer, while rAAV vectors are produced at increased levels, an effect also observed at the level of viral and vector genome replication. Following transduction, Ad coinfection dramatically increased rAAV transduction and helper virus-independent mobilization as high as 10-fold. rAAV virion production was similar to wtAAV upon coinfection in the presence of Ad at a near 1:1 input ratio, and replication-dependent mobilization resulted in ∼1,000-fold higher transduction compared with Ad-independent mobilization. The data generated herein highlight the potential of rAAV vector production in treated patients upon subsequent wtAAV and helper virus infection and raise safety concerns for the treated individual and for the unintended animal and human populations in general.
Materials and Methods
Cell culture
The HEK293 cells were maintained in 15-cm-diameter plates in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% heat-inactivated (30 min at 56°C) bovine calf serum (Gibco), 100 U of penicillin per mL, and 100 mg of streptomycin (Gibco) per mL at 37°C in a 5% CO2-air atmosphere.
Plasmids
The plasmids used for transfections were as follows. (1) The rAAV vector plasmids with the AAV2 ITRs9,10: pAAV2-CBA-Luc containing a firefly luciferase reporter cassette,51 pAAV2-EF1α-IDUA harboring a clinical relevant α-l-iduronidase expression construct,52 and pAAV2-C2C27-Nano-dysferlin53 than encodes a truncated human dysferlin protein; (2) the AAV “helper” plasmid pXR2, harboring wtAAV2 rep and AAV2 cap sequence (without ITRs)54; (3) The Ad helper plasmid, pXX680, encoding portions of the Ad genome that assist AAV production55; (4) the plasmid used for wtAAV production that encodes the AAV2 genome: pSSV911; and (5) pcDNA3.1 (originally obtained from Addgene, does not contain ITRs) was used to allow the same total molar mass to be the same for each transfection group. All plasmids will be available upon request from the corresponding author or the Gene Therapy Center at UNC.
AAV DNA replication assay
The AAV DNA replication assay was performed using methods described previously with minor modifications.56–58 Approximately 80% confluent 293 cells in 10-cm-diameter dishes were transfected with plasmids pXX680/pXR2/pAAV-CBA-Luc (to assess rAAV DNA replication), or pXX680/pSSV9/pcDNA3.1 (to assess wtAAV DNA replication), or pXX680/pSSV9/pAAV-CBA-Luc (to assess both rAAV and wtAAV replication), or pAAV-CBA-Luc/pXR2/pcDNA3.1 (serving as negative control). Forty-eight hours post-transfection, Hirt DNA was extracted. In short, the cell pellet from a 10-cm plate was resuspended in 740 μL of Hirt buffer (0.01 M Tris-Hcl Ph 7.5 and 0.01 M EDTA) and lysed by adding 50 μL of 10% sodium dodecyl sulfate (SDS). The cell lysate solution was then mixed with 330 μL of 5 M NaCl, placed overnight at 4°C, and centrifuged at 15,000 rpm at 4°C for 1 h. The supernatant was harvested and mixed with phenol/chloroform/isoamyl alcohol (25:24:1; Sigma) and centrifuged at maximum speed (∼16,000 rpm) for 5 min. The aqueous phase (top layer) was then mixed with an equal volume of chloroform (Sigma). Low-molecular-weight DNA was precipitated by the addition of an equal volume of 100% isopropanol (Fisher), followed by centrifugation in a microcentrifuge tube at ∼16,000 rpm for 15 min at 4°C. The DNA pellet was dissolved in 50 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0) supplemented with 100 μg/mL of RNase A. The Hirt DNA yields were determined by the NanoDrop spectrophotometer, and equivalent amounts of Hirt DNA, with or without prior digestion with DpnI (New England Biolabs), were analyzed on 0.8% agarose gel followed by Southern blotting using a 32P-labeled DNA probe specific for AAV2 rep (for detection of wtAAV2), or CBA (for detection of rAAV2-CBA-Luc), prepared using a random primer labeling kit (Takara).59
AAV production and purification
AAV virions were prepared as previously described,57 with minor modifications. Briefly, triple plasmid transfection of 293 adherent cells was performed with a polyethylenimine (PEI)-MAX (MW, 40,000; Polysciences,) to DNA ratio of 3:1 (a mass-per-mass ratio). Equimolar of total plasmids was used for rAAV and wtAAV for virus production. When packaging wtAAV and rAAV seperately, 0.8 mM (equal to 15 μg) of pXX680 plasmid, 1.6 mM of pXR2 plasmid (equal to 12 μg), and 1.6 mM of the rAAV plasmid were used per 15-cm-diameter dish of 80% confluent 293 cells for rAAV2 vector production. 0.8 mM of pXX680, 1.6 mM of pSSV9, and pcDNA3.1 were used for wtAAV2 production. Under coproduction conditions, 0.8 mM of pXX680, 1.6 mM of pSSV9, and an rAAV plasmid were used for the transfection. pXX680 was cotransfected with pcDNA3.1 as a negative control. Cells and the growth medium were harvested around 65 h following transfection. The cell pellet was collected by centrifugation at 1,500 rpm for 10 min. The virus in the growth medium was recovered by Polyethylene Glycol 8000 (PEG-8000; Fisher Scientific) precipitation. In short, 5 × PEG/NaCl precipitation stock solution (containing 40% PEG-8000 plus 2.5 M NaCl in H2O) was added into the collected medium at a final concentration of 8% PEG followed by centrifugation at 5,000 rpm at 4°C for 30 min. The pellet was then resuspended with PBS and combined with the cell pellet and subjected to disruption by sonication. AAV virions were then purified by CsCl density gradient centrifugation.57 The gradient fractions were then analyzed by alkaline gel electrophoresis and visualized by SYBR Gold Nucleic Acid Gel Stain (Cat# S11494; Thermo Fisher). The peak fraction(s) determined by the alkaline gels were combined, concentrated, and desalted using Amicon Ultra-4 (100 K. MWCO; Millipore, Billerica, MA) with AAV storage solution (1 × PBS with 5% sorbitol and 350 mM NaCl).60 Purified virus was brought to a total volume of 1 mL in low-retention tubes and stored at −80°C.
Adenovirus
The human adenovirus type 5 (Ad5) stock was purchased from ATCC (VR-1516, Manassas, VA) and used as a helper virus for AAV replication.4,55 As a part of our initial characterization, the Ad5 infectious titer (Supplementary Fig. S1) was confirmed in 293 cells. Ad5 stocks were tittered by detection of the Ad hexon protein using the Adeno-X™ Rapid Titer Kit (Cat. No. 632250; Takara, Clontech). The Ad5 cytopathic effect (CPE) was dose dependent, and at an multiplicity of infection (MOI) of 2 to 5, and cells showed obvious CPE 48 h postinfection. Thus, for the experiments herein, an MOI of 5 was chosen unless otherwise indicated.
AAV titer determination
wtAAV2 and rAAV2 particle numbers were determined by a probe-based quantitative PCR (qPCR) analysis following Benzonase treatment to eliminate nonencapsidated DNA.61 Universal probe library (UPL) probe #3 (Roche) and the following primers were used for analysis of wtAAV2: 5′-AGTACCAGCTCCCGTACGTC-3′ and 5′-CATACTGTGGCACCATGAAGAC-3′. UPL probe #29 and primers: forward, 5′-TGAGTACTTGAAATGTCCGTTC-3′ and 5′-GTATTCAGCCCATATCGTTTCAT-3′ were used for AAV2-CBA-Luc; UPL probe #15 and following primers were used for detection of AAV2-EF1α-IDUA: 5′-AAAGGGGGCCAGGTCTAGT-3′ and 5′-ATCTGCTGAGCGACCACCT-3′, UPL probe #46 and primers: forward 5′-CCGACACGCCTACCTGAG-3′ and reverse: 5′-CCGGCACTAAAATCGTCAG-3′ were used for AAV2-C2C27-Nano-dysferlin. Assay parameters with threshold cycle values and calculated quantities for each reaction were exported for further analysis. Plasmid standards were used to determine absolute titers.
Infectious center assay
Infectious center assay was performed following the previously published protocol.57 Briefly, the Hela cells were seeded in 48-well plates at 5.0 × 104 cells/well, and 3 h later, cells were infected with virus. Cells infected with 10-fold dilutions of the wtAAV, in the presence of wtAd5 at a multiplicity of 20 IUs per cell, were utilized as positive controls. While negative controls were Hela cells infected with rAAV or wtAAV alone and HeLa cells with Ad5 alone. The experimental groups were infected with rAAV and wtAd in triplicate. Cells were harvested 42 h postinfection and transferred to membranes using a vacuum device. Membrane were denatured, neutralized, and UV-linked and then hybridized overnight at 65°C with a p32-labeled Rep-specific probe.
AAV mobilization detection
Ad-independent rAAV mobilization (defined as vectors failing to uncoat and shedding into nontargeted cells) and replication-dependent mobilization of rAAV2 (defined as the rAAV vector genome being replicated and encapsidated in the presence of wtAAV2 and Ad5) were mimicked in vitro by successive infection of 293 cells. Briefly, 293 cells were seeded at 70% confluence in 24-well plates. The cultures were infected with rAAV2-CBA-Luc at 100, 1,000, or 10,000 vg/cell, with or without the presence of wtAAV2 (1,000 vg/cell) and Ad5. Forty-eight hours after addition of Ad, the medium was removed, cells were harvested and washed with PBS. The cell pellets were then resuspended in 200 μL of PBS and subjected to three cycles of freeze/thaw lysis in a dry ice/ethanol bath. The cell debris was then removed by centrifugation, and the lysate supernatants were either used for qPCR detection of Benzonase-resistant AAV genomes or for a second-round infection. For the qPCR detection, an equal volume of the lysate supernatant was digested with Benzonase for 1 h, heated at 95°C for 15 min, diluted in H2O (due to the qPCR being sensitive to the inhibitors released from the crude lysate, the supernatant was diluted for at least 50-fold), and then directly used as a template for qPCR using UPL probes. The lower limit of detection (defined as the lowest titer detected within the range of the linear standard curve) of qPCR was determined according to the standard curve created by serial dilutions of plasmids. To rule out preexisting latent infection of wtAAV in the producer line, the 293 cells were tested by qPCR and confirmed negative for wtAAV (Supplementary Fig. S2). Also, the plasmid preparations used for transfection were all negative for wtAAV2 detection. For the second-round infection, an equal volume of the lysate supernatant was added to fresh 293 cells with or without the addition of Ad (MOI = 5). The transduction was evaluated ∼42 h postinfection by reading the luciferase activity. In short, cells were harvested, extensively washed, and directly lysed using 1 × passive lysis buffer (Promega). Upon addition of substrate (E1483; Promega), light emission was measured using a Perkin Elmer plate reader (machine model: VICTOR3 1420 Multilabel Plate Reader), and the relative luminescence unit was presented as the mean ± SD. Several controls were used to exclude the possibility that the luciferase activity detected in the second round of infection was not a carryover of the luciferase protein from the first-round cell lysate supernatant. First, as mentioned above, at the end of the second-round infection, the medium, containing the lysate supernatant from the first round of cell lysates, was removed and the cells were extensively washed. In addition, when the lysate supernatant from luciferase plasmid-transfected 293 cells was applied to fresh 293 cells, no luciferase activity was observed. Last, we further distinguished the luciferase activity by addition of Ad5 to all groups in the second round of infection. Theoretically, the Ad5 would increase the rAAV transduction, but would be unlikely to increase the activity of preexisted luciferase protein carryover. The experiment was repeated on three separate occasions, and each experimental group was performed in triplicate.
Statistics
Statistical analyses were conducted with GraphPad Prism software version 8 (San Diego, CA). Student's t-test, ordinary one-way ANOVA, and two-way ANOVA were used for data comparisons. Differences were considered significant when p < 0.05. Data are shown as mean ± SD.
Results
Viral genome replication following plasmid transfection
To investigate an initial step during wtAAV and rAAV virion production, viral DNA replication was investigated following plasmid transfections in the absence or presence of the Ad helper plasmid pXX680.55 Equal amounts of Hirt DNA isolated 48 h post-transfection were treated with DpnI endonuclease to differentiate the input plasmids from newly replicated DNA, and then loaded for the agarose gel electrophoresis followed by Southern blot analysis. The wtAAV2 genome, the rAAV vector map, and the probes used for radioactive detection of the wtAAV or rAAV genomes are depicted in Fig. 1A. A representative Southern blot revealed bands of the 4.7 kb monomer duplexes and the 9.4 kb dimer duplexes of wtAAV genomes, which indicated that the wtAAV2 genome was efficiently replicated following plasmid transfection with the presence of pXX680 (Fig. 1B, lane “WT”). Under nonreplicating conditions (without pXX680), wtAAV replication was not detected as expected (Fig. 1B, lane “Ctrl”). In the presence of an rAAV plasmid, the wtAAV genome was also replicated, however, to a lesser extent (Fig. 1B lane “WT” vs. lane “WT+rAAV”). As shown in Fig. 1C, both monomeric and dimeric rAAV-Luc replicative forms were observed in the rAAV-only group (Fig. 1C, lane rAAV), and in the rAAV and wtAAV cotransfection group (Fig. 1C, lane WT+rAAV). Interestingly, the rAAV DNA was observed to replicate slightly more efficiently than in the absence of the wtAAV plasmid (Fig. 1C, lane “rAAV” vs. “WT+rAAV”).
Figure 1.
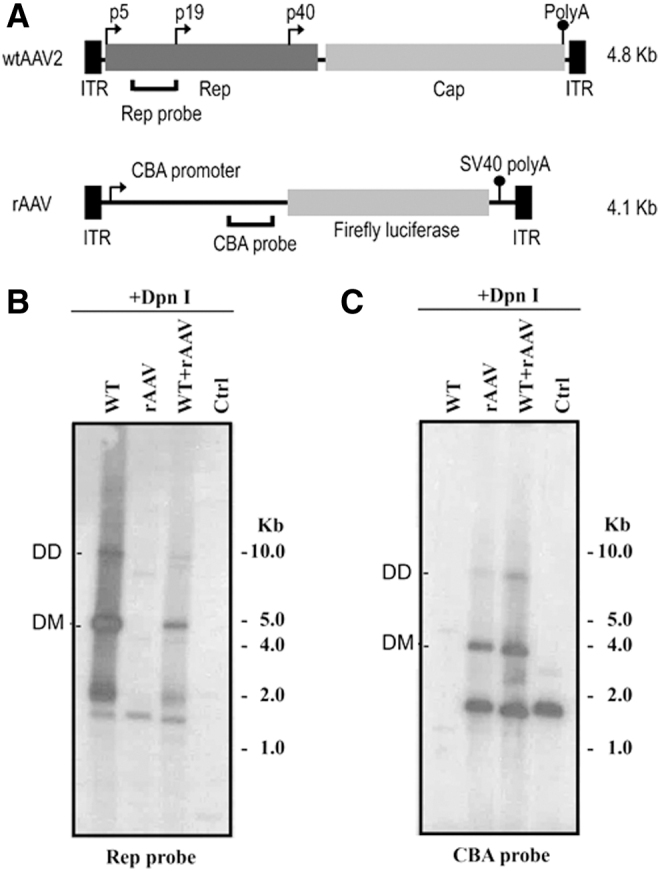
Replication assay of wtAAV and rAAV following cotransfection of wt and rAAV plasmids detected by Southern blot analysis. (A) Schematic maps of the wtAAV2 and rAAV-CBA-Luc, including the probes used for detection. Southern blot analysis of wtAAV2 (B) or rAAV DNA replication (C) using the indicated probes. Equimolar of total plasmids transfected into 293 cells. To assess wtAAV DNA replication, cells were transfected with pxx680/pSSV9/pcDNA3.1; for rAAV, cells were transfected with pxx680/pXR2/pAAV-CBA-Luc; for wtAAV+rAAV group, cells were transfected with pxx680/pSSV9/pAAV-CBA-Luc; for control, cells were transfected with pAAV-CBA-Luc/pXR2/pcDNA3.1. Hirt DNA was recovered at 48 h post-transfection. Equal amounts of DNA was digested with Dpn I, separated on 0.8% agarose gel, and subjected to Southern blot analysis using 32p-labeled rep or CBA probe. CBA, Cytomegalovirus enhancer/chicken β-actin promoter; ITR, inverted terminal repeats;. rAAV, recombinant adeno-associated viral; wtAAV, wild-type AAV.
AAV virion production following plasmid transfection
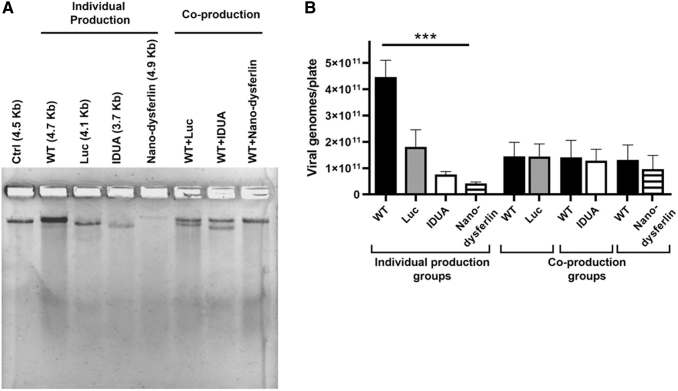
First, the wtAAV or rAAV particle yields were determined individually following a triple transfection protocol in 293 cells54,55 using 15-cm-diameter plates. As described in the Materials and Methods section, for rAAV production, an rAAV plasmid {pITR2-CBA-luc (4.1 kb), or pITR2-EF1α-opt-IDUA (3.7 kb),62 or pITR2-C2C27-Nano-dysferlin (4.9 kb)53} was used along with pXR2 (rep2cap2)55 and the Ad helper plasmid pXX680.55 To produce wtAAV, pSSV911 was used along with pXX680 and an additional plasmid, pcDNA3.1 (to maintain consistent molar amount of total plasmids). Sixty-five hours post-transfection, virions were purified by CsCl gradient centrifugation. Alkaline gel electrophoresis followed by SYBR gold staining was used to visualize the packaged genome integrity and relative viral titer by loading the same recovered volume of the preparation (Fig. 2A), and qPCR was utilized to determine the absolute titers (Fig. 2B). When packaging the wtAAV and rAAV separately, wtAAV2 preparations consistently demonstrated the highest titer at about 4.5e11 viral genome/plate (vg/plate), which was 3–10-fold greater than all rAAV2 preparations (Fig. 2B, individual production groups). Among the three different rAAV2 preparations, significant differences in production titers were noted that varied approximately three- to four-fold, with AAV2-C2C27-Nano-dysferlin (4.9 kb) having the lowest titer of 4.2e10 vg/plate, AAV2-EF1α-opt-IDUA (3.7 kb) being intermediate at 7.6e10 vg/plate, while AAV2-CBA-luc produced the most rAAV2 particles at 1.8e11 vg/plate (Fig. 2B, coproduction groups).
Figure 2.
AAV virion production following cotransfection of wt and rAAV plasmids. (A) Viral genome integrity and relative viral titer visualized by alkaline agarose gel electrophoresis. A reference vector was loaded at 5e10 vg (Ctrl). (B) Virus titer determined by probe-based qPCR presented as the total vg/plate (mean ± SD). ***p < 0.001 (p = 0.0006), Kruskal–Wallis test. qPCR, quantitative PCR.
In addition, possible wtAAV genome (partial or full length) contamination in the three different rAAV preparations was investigated. The results of qPCR using a probe specific to cap2 sequence demonstrate that when producing rAAV vectors with a triple plasmid transfection protocol in 293 cells, one of the most commonly used methods,55 all rAAV preparations contained Benzonase-resistant wtAAV cap sequence contamination, the amount of which ranged from 0.8% to 1.7% of the intended encapsidated rAAV genomes, depending on the transgenic sequence and/or size (Supplementary Fig. S1A). To check if the detected wtAAV sequence contamination is replication-competent, an infectious center assay was performed (Supplementary Fig. S3). wtAAV replication was not detected in the vector preparations under the conditions tested (specified in the Materials and Methods section).
Next, rAAV and wtAAV virion production was assessed following plasmid cotransfections of wtAAV and rAAV plasmids. These experiments relied on the cotransfection of equal moles of pSSV9 and pITR2-transgenic, along with pXX680. In contrast to the production of wtAAV and rAAV separately, wtAAV2 virion production during coproduction setting was decreased approximately threefold with titers not significantly different than the production of rAAV2 (Fig. 2B, coproduction groups). This result was shown to be independent of the transgenic sequence of the rAAV vectors used herein (Fig. 2B, coproduction groups).
Significant Ad-independent rAAV mobilization occurs
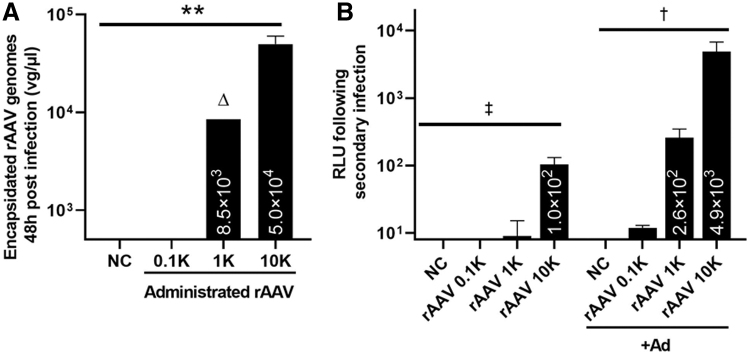
Ad-independent mobilization events following primary cell transduction was investigated by a secondary infection. Briefly, 293 cells were infected with rAAV2-CBA-Luc at doses of 100, 1,000, or 10,000 vg/cell. Forty-eight hours postinfection, the medium was removed; cells were harvested and extensively washed with PBS. The cell pellets were then resuspended in 200 μL of PBS and subjected to freeze/thaw. Then the cell debris was removed by centrifugation, and the supernatant was used for qPCR detection of rAAV genomes and for their ability to mediate subsequent transduction in fresh cells (Fig. 3). As described in the Materials and Methods section, for qPCR detection, the harvested supernatant was digested by Benzonase to remove nucleic acids that are not encapsidated, and then heated at 95°C to denature the AAV capsid and to release the viral genomes. Forty-eight hours postinfection, intact rAAV remained at detectable levels depending on the infectious dose (Fig. 3A). To check whether these intracellularly released rAAV virions were competent for replication-independent mobilization, the lysate supernatants from individual wells were added to fresh 293 cells for a second round of infection with or without coinfection by Ad5. Approximately 42 h postaddition, these second-round cells were lysed and subjected to luciferase activity detection to measure transduction. As shown in Fig. 3B and Supplementary Fig. S4, dose-dependent rAAV mobilization occurred resulting in luciferase activity. In the presence of Ad5, rAAV secondary transduction increased >10-fold compared with the no Ad5 groups (Fig. 3B). It should be noted that several controls were performed as outlined in the Materials and Methods section to demonstrate that these results are not a carryover of luciferase protein from the first-round lysate and therefore represent productive replication-independent rAAV mobilization that is significantly enhanced by Ad5 following serial infection.
Figure 3.
Ad-independent mobilization. (A) Quantitative analysis of the encapsidated rAAV using probe-based qPCR at 48 h postinfection. Cells were infected with rAAV at indicated doses. Forty-eight hours postinfection, cells were harvested and subjected to extensive wash and three rounds of freeze/thaw. Lysate debris was removed by centrifugation, and the supernatant was treated with Benzonase for 1 h at 37°C, heated at 95°C for 15 min, diluted for at least 50-fold in H2O, and then used as a template for qPCR. The rAAV was detected using UPL probe/primers specific to the luciferase coding sequence. Data are presented as mean ± SD (vg/μL). **p < 0.01 (p = 0.0028), 10K versus NC, one-way ANOVA test, n = 3. Δ, only one repeat was right above the lower detection limit, and the other repeats were detectable, but did not fall in the linear range of the standard curve. (B) Luciferase activity from the second round of infection. Individual wells from the first-round infection were subjected to extensive wash and then resuspended in the PBS, followed by freeze/thaw. The cell debris was removed by centrifugation, and the supernatants were added to fresh 293 cells with or without the presence of Ad for the second-round infection. Forty-two hours postinfection, the cells were harvested for the luciferase activity measurement. Data are shown as mean ± SD. ‡p < 0.0001, one-way ANONVA. †p = 0.0006, one-way ANONVA. NC, negative control without addition of AAV; rAAV, rAAV2-CBA-Luc; 0.1K, 1K, and 10K represent rAAV administered at 100, 1,000, and 10,000 vg/cell, respectively.
Significant Ad-dependent rAAV mobilization occurs
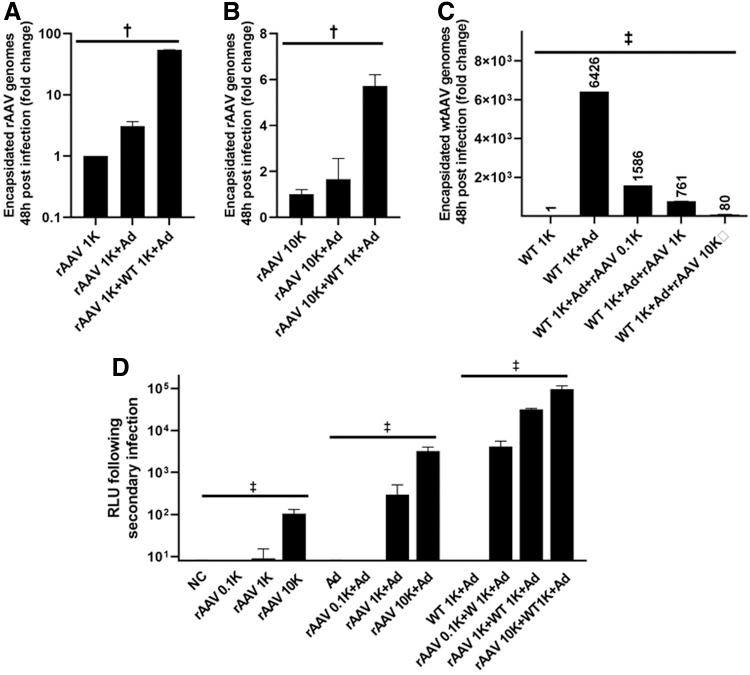
Next, rAAV Ad-dependent mobilization in the presence and absence of wtAAV and a helper virus was investigated similarly by two rounds of infection. For the first infection, 293 cells were infected with rAAV at increasing doses and/or wtAAV (fixed dose of 1,000 vg/cell). Ad5 (MOI = 5) or PBS was added to the wells 4 h later. The cells were harvested 48 h post-AAV addition and used for three different measurements: (1) luciferase activity following the primary infection; (2) qPCR detection of encapsidated genomes; and (3) the second-round transduction efficiency mediated by the rAAV released from primary infected cells at 42 h postinfection.
The luciferase activity of the first infection was measured at 48 h postinfection as described in the Materials and Methods section. As expected, the rAAV transduction efficiency was dose dependent with dramatic enhancement in the presence of Ad5 (>10-fold increase) consistent with Fig. 3B and previous reports (Supplementary Fig. S4).4,63,64 However, when rAAV and wtAAV were coadministered to cells followed by Ad5 infection, luciferase activity in this setting decreased compared with the levels observed in a similar setting without wtAAV (Supplementary Fig. S2).
To examine rAAV production in the presence of Ad5 and/or wtAAV in a transduction setting, qPCR was used to quantitatively assess the Benzonase-resistant rAAV vector titer 48 h post-transduction. Interesting, in cells administered 1,000 or 10,000 rAAV vg/cell and Ad, without intentional wtAAV inoculation, the recovered rAAV titer increased approximately two- to five-fold in a dose-dependent manner (Fig. 4A, B, rAAV+Ad vs. rAAV). In cells given both rAAV+wtAAV (each at 1,000 vg/cell) and then superinfected by Ad5 (MOI = 5), the encapsidated rAAV titer increased ∼100-fold compared with transduction by rAAV alone (Fig. 4A, rAAV 1K+WT 1K+Ad vs. rAAV 1K) 48 h postinfection, suggesting de novo rAAV production (Fig. 4A). The magnitude of this effect was observed to be dose dependent, in that when 10-fold more rAAV was used, only a 7-fold increase in rAAV titer was observed (Fig. 4B, rAAV 10K+WT 1K+Ad vs. rAAV 10K). When analyzing intact wtAAV production following transduction in cells superinfected with Ad5, a > 6,000-fold increase in wtAAV virion titer was observed 48 h postinfection (Fig. 4C, WT 1K+Ad vs. WT 1K). Interestingly, in cells coinfected with wtAAV and rAAV in the presence of Ad, wtAAV production was dramatically inhibited by rAAV in a manner that directly correlated with the administered rAAV dose (Fig. 4C and Supplementary Fig. S5). When the original input of rAAV was 10-fold higher than wtAAV, the total production of rAAV outcompeted the wtAAV (Supplementary Fig. S5).
Figure 4.
Quantitative analysis of Ad-dependent production and mobilization with the presence of wtAAV and/or Ad. (A, B) Quantitative analysis of the rAAV vector genomes using probe-based qPCR. Cells were infected with rAAV at indicated doses, in the presence and absence of wtAAV (1,000 v/cell) and/or Ad (MOI = 5). Forty-eight hours post-rAAV infection, cells were harvested, extensively washed, and digested with Benzonase. Encapsidated rAAV virions were detected by probe-based qPCR. Data are presented as mean ± SD (vg/μL). †p < 0.001 (p = 0.0001), one-way ANONVA. (C) Quantitative analysis of the wtAAV vector genomes using probe-based qPCR. Cells were infected with wtAAV (1,000 vg/cell), in the presence or absence of Ad (MOI = 5) and/or rAAV2-CBA-Luc (at indicated doses). Forty-eight hours postinfection, cells were harvested, extensively washed, and digested with Benzonase. Encapsidated wtAAV virions were detected by qPCR. Data are presented as mean ± SD (vg/μL). ‡p < 0.0001, One-way ANOVA test, n = 3; (D) Luciferase activity from the second round of infection. Cells from the first-round infection were subjected to extensive wash and then resuspended in PBS, followed by freeze/thaw. The cell debris was removed by centrifugation, and the supernatants were added to fresh 293 cells for the second-round infection. Forty-two hours later, cells from the second-round infection were harvested for the luciferase activity measurement. Data are presented as the mean ± SD. ‡p < 0.0001, two-way ANOVA. NC, negative control without addition of AAV; WT, wtAAV2; rAAV, rAAV2-CBA-Luc; 0.1K, 1K, and 10K represent AAV administered at 100, 1,000, and10,000 vg/cell, respectively. RLU, relative luminescence unit.
Next, to determine if Ad-dependent mobilization occurs, the infected cell lysate was administered to fresh cells, and rAAV transduction was assayed by luciferase activity 42 h later. Significant enhancement of secondary infection was observed in primary cells administered rAAV and Ad, with no intentional wtAAV administration (Fig. 4D, rAAV+Ad vs. rAAV). In primary cells coadministered rAAV and wtAAV in the presence of Ad, mobilized rAAV resulted in ∼1,000-fold enhancement of luciferase activity following secondary cell transduction depending on the original dose (Fig. 4D, rAAV+WT 1K+Ad vs. rAAV).
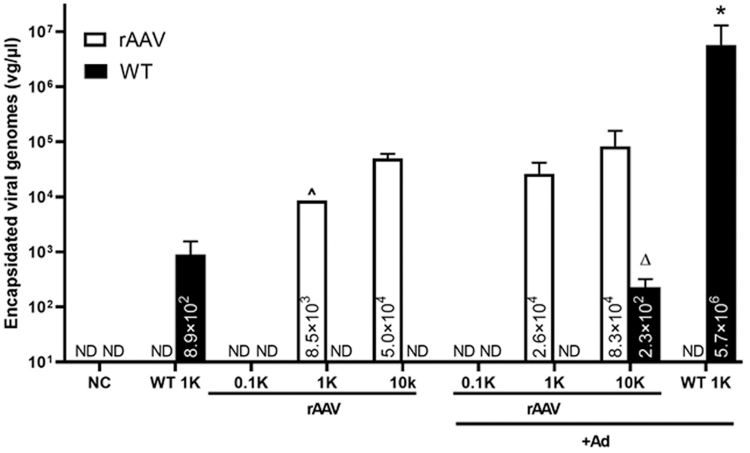
wtAAV genome sequence contamination in the rAAV preparations may facilitate the replication-dependent replication
wtAAV genome sequence contamination of all rAAV preparations used herein was observed, which is consistent with observations of multiple other groups65,66 when using the triple transfection production protocol (Supplementary Fig. S1A). We next examined whether these contaminants could supply the required Rep and Cap for replication-dependent mobilization. The wtAAV and rAAV titer was examined 48 h postaddition of Ad5 (MOI = 5). As shown in Fig. 5, the wtAAV production was increased >1,000-fold in the presence of Ad5 compared with the no-Ad5 group. Interestingly, although the wtAAV sequence was not detectable in the rAAV-Luc-alone groups, following superinfection of Ad5, detection of the wtAAV genome became evident in the 10,000 vg/cell dose, suggesting Ad-dependent replication and encapsidation of wtAAV-contaminant particles in the rAAV preparation. This observation highly suggests that the contaminating wtAAV-like particles detected in the rAAV preparations were replication competent (Fig. 5). In addition, the rAAV-Luc was slightly increased in both doses of 1,000 and 10,000 vg/cell, further suggesting that this replication-competent wtAAV might supply the Rep and Cap for the production of rAAV-Luc.
Figure 5.
Quantitative analysis of replication-competent wtAAV. Cells were infected with wtAAV (1,000 vg/cell) or rAAV at indicated doses, with or without Ad (MOI = 5). Forty-eight hours postinfection, cells were harvested and subjected to extensive wash and three rounds of freeze/thaw. The lysate debris was removed by centrifugation and the lysate supernatant was treated with Benzonase for 1 h at 37°C, heated at 95°C for 15 min, diluted in molecular-grade water, and used for qPCR using UPL probes, the rAAV genome was detected with probe/primers specific to luciferase coding sequence, and the wtAAV genome was detected using probe/primers specific to Cap gene. Data are presented as the mean ± SD (vg/μL). *p < 0.05, WT 1K+Ad significant difference from WT 1K, n = 3, T test; ▵statistics were impossible to be calculated due to the wtAAV genome in the rAAV-alone groups being below the detection limit. ^Only one in three repeats was right above the detection limit. MOI, multiplicity of infection.
Discussion
rAAV has become one of the most utilized delivery formats for human gene therapy with hundreds of clinical trials addressing numerous diverse diseases. In fact, currently, three rAAV drugs have been approved by the European and the U.S. Food and Drug Administration and estimations by the FDA have predicted that 10–20 new gene therapy drugs will be approved by 2025. Generally, positive therapeutic results have been obtained from clinical applications, and overall, AAV gene therapy has garnered a strong safety profile. However, newly characterized concerns regarding the therapeutic use of AAV vectors remain, including the following: (1) capsid and transgenic product immunogenicity, (2) demonstrations of chromosomal integration, (3) the potential for oncogenesis, and (4) potential complications related to off-target transduction in the treated subjects,67–72 including nonpatient bystanders resulting from rAAV shedding.37,38,40 Surprisingly, rAAV mobilization, another largely theoretical concern associated with rAAV vectors, has been underappreciated in the AAV research community with only sparse reports over several decades.10,29,31,33 wtAAV is reported to have better yields than rAAV during production in the laboratory,73 yet, in a gene therapy setting, when a cell is transduced by rAAV and co- or superinfected by wtAAV and a helper virus, it is unclear whether wtAAV retains its advantage for replication and/or capsid packaging thereby potentially inhibiting de novo rAAV production in patients. In the current study, the reality of these concerns has been investigated in a quantitative manner in both transfection and transduction contexts in cell culture with several novel findings: (1) Ad-independent rAAV mobilization resulting in serial transduction is substantial (Figs. 3 and 6); (2) with the presence of both wtAAV and rAAV, there is no obvious bias between wtAAV and rAAV for production; however, a dose-dependent effect exists (Fig. 4C and Supplementary Fig. S5); and (3) replication-dependent mobilization results in >1,000-fold higher serial transduction compared with Ad-independent mobilization (Figs. 4D and 6). The data generated herein expose the potential for AAV gene therapy-treated patients to produce and disseminate rAAV, and thereby highlight the need for development of better vector production protocols that completely eliminate Benzonase-resistant wtAAV sequence contamination and perhaps safer mobilization-resistant AAV vectors. In addition, a yet to be defined aspect that increases rAAV production in the presence of a wtAAV plasmid is reported (Fig. 2).
Figure 6.
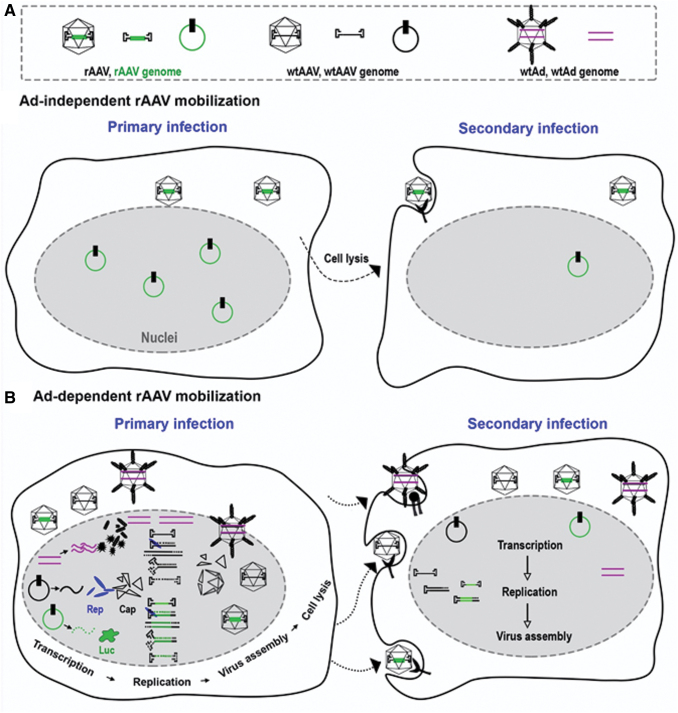
Model of Ad-independent and -dependent rAAV mobilization. (A) Ad-independent mobilization. During the primary infection, some of the AAV vectors remain intact while others uncoat and persist in the nucleus as double-strand circular episomes that mediate transgene expression. Upon cell lysis, intact particles mobilize to other cells and mediate secondary cell transduction. (B) Ad-dependent mobilization. In the presence of wtAAV and a helper virus (Ad depicted), Ad provides helper function to the wtAAV to allow Rep and Cap gene expression. Rep proteins have the capacity to replicate both WT and transgenic AAV genomes and package them into AAV capsids resulting in de novo production of rAAV (and wtAAV) post-transduction. Upon cell lysis, the AAV virions mediate a second round of infection resulting in serial transduction by both previously uncoated and newly produced rAAV virion. Color images are available online.
One obstacle for the characterization of rAAV mobilization is the lack of an ideal in vivo model to represent a natural infection situation in humans. In a lot of instances, Ad (or a different helper virus) and wtAAV present as natural coinfections.74 It has long been the assumption that attributes of the wtAAV genome, such as the size and sequence, make it a more attractive substrate for Rep-mediated replication and capsid packaging compared with an rAAV transgenic genome.73 This hypothetical preference for the wtAAV was observed previously73 and consistently herein, with wtAAV producing at a 3–10-fold higher titer compared with rAAV depending on the transgene (Fig. 2B). However, following cotransfection of wtAAV and rAAV plasmids, rAAV vector DNA tends to replicate more efficiently in the presence of wtAAV (Fig. 1C), and there is little to no bias observed toward wtAAV production (Fig. 2A, B). This may be attributed to the equal efficiency of wtAAV and rAAV genome encapsidation,75 yet the improved efficiency of the rAAV vector yield may be a result of when the rep and cap genes are supplied by the wtAAV genome, which is assumed to have the optimal rep/cap expression ratios during production in the laboratory setting.56 Although still unknown, this result may be related to the following: (1) inherent ITR transcriptional activity that may fine-tune expression of known or currently undescribed AAV ORFs76 and/or perhaps (2) the ability of the wtAAV helper plasmid to replicate, an activity shown to increase rAAV production.56 Since the rAAV vector production via cotransfection in the laboratory mimics the production of wtAAV and rAAV with regard to replication substrates, these observations strongly suggest the likelihood of wtAAV and rAAV particle production at similar efficiencies after cotransduction (Fig. 2).
In the studies herein, it was also found that rAAV preparations produced by a common triple transfection protocol51 are contaminated with the Benzonase-resistant wtAAV capsid sequence when examined by qPCR (Supplementary Fig. S1A). This finding is consistent with several other reports demonstrating wtAAV sequence contamination in rAAV preparations at a range of 0.01–10%.37,65,66,77 However, qPCR only detects a region of the wtAAV recognized by the chosen probe/primer set and it remains unknown if full-length wtAAV genomes were present as preparation contaminants. If these wtAAV sequences proved to be replication competent in cis, trans, or perhaps following recombination events during replication, it may decrease the stringency required for AAV vector mobilization, potentially by eliminating the requirement for subsequent (or prior) wtAAV transduction of an rAAV genome harboring cell (since it was provided as an rAAV preparation contaminant). To further investigate this possibility, an infectious center assay was performed and the results (Supplementary Fig. S3) failed to detect replication-competent rAAV under the tested conditions (Supplementary Fig. S3). As such, the potential for wtAAV contamination in rAAV preparations to induce rAAV production following transduction remains unanswered and requires further investigations at clinical doses in a relevant animal model.
In the transduction analysis, rAAV transduction efficiency was substantially increased by the presence of Ad consistent with published studies.63 This enhancement remains uncharacterized and may be related to the Ad-induced CPE effect of Ad, which may alter gene expression (viral and host) and therefore vector load. Interestingly, the Ad enhancement appeared to be inhibited by the addition of wtAAV, since rAAV transduction in the presence of Ad and wtAAV was decreased when compared with the rAAV+Ad groups (Supplementary Fig. S2); this phenomenon is not understood and requires additional investigation.78,79
Notably, rAAV particle production, following cell transduction, increased up to 100-fold in cells also infected with wtAAV and Ad (Fig. 4A). These replication-dependent rAAV vectors mobilized and resulted in a 1,000-fold increase in transduction efficiency in subsequently infected cells (Figs. 4D and 6). The extent of rAAV production directly correlated to the original input of materials, and the increased production of rAAV decreased the magnitude of the wtAAV production (Fig. 4C and Supplementary Fig. S5). Although we did look at ratios of wtAAV to rAAV in the above experiment, it should be noted that wtAAV contamination in clinical rAAV preparations is likely several magnitudes less than the intended AAV vector (Fig. 2), thereby conferring a production advantage to the rAAV in treated patient cells. This may lead to transgene-specific toxicity as particular transgene products used in clinic could have detrimental effects in off-target cells and/or healthy bystanders with the observations that tissue-specific promoter restriction and regulatory mRNA targets engineered into transgenic cassettes are incomplete.
Regarding the overall safety concern of rAAV mobilization, usually neutralizing antibodies to the AAV capsid inhibit superinfection of the same AAV serotype (dependent upon dose, route of administration, etc.). The level of the antibody response to date from clinical trials has prevented readministration and typically prevents all subsequent serotype administration due to cross antigen presentation. Although this immune response is extremely robust based on current vector doses, there is a window of vulnerability, namely: (1) wtAAV infection post-AAV vector administration, yet before the onset of capsid neutralizing antibody production (an ∼2 week window80), (2) superinfection of a wtAAV serotype that can evade the neutralizing antibodies induced by the therapeutic vector, (3) local injection of rAAV that results in minimal to no neutralizing antibody generation,44,62 and (4) replication-competent wtAAV as a contaminant of clinical rAAV preparations that is coinjected into the patient at the same time as the rAAV therapeutic and activated by the helper virus or various stress conditions.37,66,77 Concerns of interhuman spread of replication-independent and/or replication-dependent mobilized rAAV represent a formal safety concern for most mammals in general, and is theoretically dependent upon many factors, including preexisting capsid neutralizing antibodies, route of transmission, and helper virus and/or wtAAV infection in the mammalian bystander. However, the reality of this situation remains to be formally demonstrated.
In conclusion, the collective data emphasize the pragmatic risk of rAAV mobilization. Furthermore, evidence of a diminished capsid neutralizing antibody response over time in patients rekindles the risk of rAAV mobilization as well.81 As the field progresses to more efficient AAV vectors for targeting, tropism, and ability to treat more broad diseases, the data herein provided a proof-of-concept for safety concerns and highlight the need for mobilization-resistant AAV vectors. One methodology is to alter the rAAV ITR sequence in a manner that allows efficient AAV vector production only in a laboratory setting. This notion was partially supported by a previous report in which vector genomes utilizing the rarer ITR5 sequence, resistant to replication initiated by common Rep 2 or several other AAV serotypes, were generated and tested, although wtAAV5 still exists in human populations.29 Ultimately, rationally designed ITRs that are resistant to replication/mobilization by all naturally occurring Rep proteins, yet allow high titer production by a novel laboratory-restricted Rep-like protein, would overcome rAAV mobilization concerns highlighted herein are required and such reagents are already under active investigation.35
Supplementary Material
Acknowledgments
This study was supported by grants from the NIH RO1AI072176-06A1 (M.L.H.) and the Pfizer-NCBiotech Distinguished postdoctoral Fellowship (L.S.). The authors are grateful to Drs. Chengwen Li and Chuan-An Zhang for their excellent technical support. They also thank Dr. Jacquelyn J. Bower, Prabhakar Bastola, and Malik Moncalvo for their critical review of the article.
Author Disclosure
M.L.H. is a cofounder of Bedrock Therapeutics and RainBIO, Inc. M.L.H. has other unrelated technology licensed to Asklepios BioPharmaceutical for which he has received royalties. R.J.S. is the founder and a shareholder at Asklepios BioPharmaceutical and Bamboo Therapeutics, Inc. He holds patents that have been licensed by UNC to Asklepios Biopharmaceutical, for which he receives royalties. He has consulted for Baxter Healthcare and has received payment for speaking.
Funding Information
This study was supported by grants from the National Institutes of Health (NIH) RO1AI072176-06A1 (M.L.H.) and Pfizer-NCBiotech Distinguished Postdoctoral Fellowship (L.S.).
Supplementary Material
REFERENCES
- 1. Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science 1965;149:754–756 [DOI] [PubMed] [Google Scholar]
- 2. Berns KI. The unusual properties of the AAV inverted terminal repeat. Hum Gene Ther 2020;31:518–523 [DOI] [PubMed] [Google Scholar]
- 3. Srivastava A, Lusby EW, Berns KI. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol 1983;45:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferrari FK, Samulski T, Shenk T, et al. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol 1996;70:3227–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laughlin CA, Cardellichio CB, Coon HC. Latent infection of KB cells with adeno-associated virus type 2. J Virol 1986;60:515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Philpott NJ, Gomos J, Berns KI, et al. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc Natl Acad Sci U S A 2002;99:12381–12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol 1987;61:972–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang XS, Srivastava A. Rescue and autonomous replication of adeno-associated virus type 2 genomes containing Rep-binding site mutations in the viral p5 promoter. J Virol 1998;72:4811–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laughlin CA, Tratschin JD, Coon H, et al. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 1983;23:65–73 [DOI] [PubMed] [Google Scholar]
- 10. Samulski RJ, Berns KI, Tan M, et al. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A 1982;79:2077–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samulski RJ, Chang LS, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol 1987;61:3096–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilmott P, Lisowski L, Alexander IE, et al. A user's guide to the inverted terminal repeats of adeno-associated virus. Hum Gene Ther Methods 2019;30:206–213 [DOI] [PubMed] [Google Scholar]
- 13. Lusby E, Fife KH, Berns KI. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol 1980;34:402–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 2020;21:255–272 [DOI] [PubMed] [Google Scholar]
- 15. Keeler AM, Flotte TR. Recombinant adeno-associated virus gene therapy in light of Luxturna (and Zolgensma and Glybera): where are we, and how did we get here? Annu Rev Virol 2019;6:601–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berns KI, Muzyczka N. AAV: an overview of unanswered questions. Hum Gene Ther 2017;28:308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang XS, Srivastava A. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J Virol 1997;71:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang XS, Qing K, Ponnazhagan S, et al. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J Virol 1997;71:3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 1990;61:447–457 [DOI] [PubMed] [Google Scholar]
- 20. McCarty DM, Ryan JH, Zolotukhin S, et al. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol 1994;68:4998–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brister JR, Muzyczka N. Mechanism of Rep-mediated adeno-associated virus origin nicking. J Virol 2000;74:7762–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao X, Xiao W, Li J, et al. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol 1997;71:941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pereira DJ, McCarty DM, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol 1997;71:1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hüser D, Khalid D, Lutter T, et al. High prevalence of infectious adeno-associated virus (AAV) in human peripheral blood mononuclear cells indicative of T lymphocytes as sites of AAV persistence. J Virol 2017;91:e02137-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon K, Del Medico A, Sander I, et al. Gene therapies in ophthalmic disease. Nat Rev Drug Discov 2019;18:415–416 [DOI] [PubMed] [Google Scholar]
- 26. Farooq AV, Shukla D. Herpes simplex epithelial and stromal keratitis: an epidemiologic update. Surv Ophthalmol 2012;57:448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bialasiewicz A. Adenoviral keratoconjunctivitis. Sultan Qaboos Univ Med J 2007;7:15–23 [PMC free article] [PubMed] [Google Scholar]
- 28. Rademaker HJ, El Hassan MAA, Versteeg GA, et al. Efficient mobilization of E1-deleted adenovirus type 5 vectors by wild-type adenoviruses of other serotypes. J Gen Virol 2002;83:1311–1314 [DOI] [PubMed] [Google Scholar]
- 29. Hewitt FC, Li C, Gray SJ, et al. Reducing the risk of adeno-associated virus (AAV) vector mobilization with AAV type 5 vectors. J Virol 2009;83:3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans JT, Garcia JV. Lentivirus vector mobilization and spread by human immunodeficiency virus. Hum Gene Ther 2000;11:2331–2339 [DOI] [PubMed] [Google Scholar]
- 31. Afione SA, Conrad CK, Kearns WG, et al. In vivo model of adeno-associated virus vector persistence and rescue. J Virol 1996;70:3235–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turner AM, De La Cruz J, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol Ther 2009;17:360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheung AK, Hoggan MD, Hauswirth WW, et al. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol 1980;33:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tratschin JD, Miller IL, Smith MG, et al. Adeno-associated virus vector for high-frequency integration, expression, and rescue of genes in mammalian cells. Mol Cell Biol 1985;5:3251–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hewitt FC, Samulski RJ. Creating a novel origin of replication through modulating DNA-protein interfaces. PLoS One 2010;5:e8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferla R, Alliegro M, Marteau JB, et al. Non-clinical safety and efficacy of an AAV2/8 vector administered intravenously for treatment of mucopolysaccharidosis type VI. Mol Ther Methods Clin Dev 2017;6:143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tenenbaum L, Lehtonen E, Monahan PE. Evaluation of risks related to the use of adeno-associated virus-based vectors. Curr Gene Ther 2003;3:545–565 [DOI] [PubMed] [Google Scholar]
- 38. Barnard AR, Rudenko AN, MacLaren RE. Vector shedding and immunogenicity sampling for retinal gene therapy. Methods Mol Biol 2018;1715:359–371 [DOI] [PubMed] [Google Scholar]
- 39. Seitz IP, Michalakis S, Wilhelm B, et al. Superior retinal gene transfer and biodistribution profile of subretinal versus intravitreal delivery of AAV8 in nonhuman primates. Invest Ophthalmol Vis Sci 2017;58:5792–5801 [DOI] [PubMed] [Google Scholar]
- 40. Schenk-Braat EA, van Mierlo MM, Wagemaker G, et al. An inventory of shedding data from clinical gene therapy trials. J Gene Med 2007;9:910–921 [DOI] [PubMed] [Google Scholar]
- 41. Song L, Llanga T, Conatser LM, et al. Serotype survey of AAV gene delivery via subconjunctival injection in mice. Gene Ther 2018;25:402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manno CS, Chew AJ, Hutchison S, et al. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood 2003;101:2963–2972 [DOI] [PubMed] [Google Scholar]
- 43. Manno CS, Pierce GF, Arruda VR, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 2006;12:342–347 [DOI] [PubMed] [Google Scholar]
- 44. Russell S, Bennett J, Wellman JA, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 2017;390:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol 2000;74:2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas CE, Storm TA, Huang Z, et al. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol 2004;78:3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rossi A, Dupaty L, Aillot L, et al. Vector uncoating limits adeno-associated viral vector-mediated transduction of human dendritic cells and vector immunogenicity. Sci Rep 2019;9:3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stieger K, Schroeder J, Provost N, et al. Detection of intact rAAV particles up to 6 years after successful gene transfer in the retina of dogs and primates. Mol Ther 2009;17:516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao PJ, Samulski RJ. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol 2012;86:10462–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson JS, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol 2009;83:2632–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song L, Li X, Jayandharan GR, et al. High-efficiency transduction of primary human hematopoietic stem cells and erythroid lineage-restricted expression by optimized AAV6 serotype vectors in vitro and in a murine xenograft model in vivo. PLoS One 2013;8:e58757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vance M, Llanga T, Bennett W, et al. AAV gene therapy for MPS1-associated corneal blindness. Sci Rep 2016;6:22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Llanga T, Nagy N, Conatser L, et al. Structure-based designed nano-dysferlin significantly improves dysferlinopathy in BLA/J mice. Mol Ther 2017;25:2150–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grieger JC, Soltys SM, Samulski RJ. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol Ther 2016;24:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 1998;72:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li C, Samulski RJ. Serotype-specific replicating AAV helper constructs increase recombinant AAV type 2 vector production. Virology 2005;335:10–21 [DOI] [PubMed] [Google Scholar]
- 57. Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc 2006;1:1412–1428 [DOI] [PubMed] [Google Scholar]
- 58. Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 1967;26:365–369 [DOI] [PubMed] [Google Scholar]
- 59. Song L, Bower JJ, Hirsch ML. Preparation and administration of adeno-associated virus vectors for corneal gene delivery. Methods Mol Biol 2020;2145:77–102 [DOI] [PubMed] [Google Scholar]
- 60. Wang Y, Ling C, Song L, et al. Limitations of encapsidation of recombinant self-complementary adeno-associated viral genomes in different serotype capsids and their quantitation. Hum Gene Ther Methods 2012;23:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song L, Kauss MA, Kopin E, et al. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy 2013;15:986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyadera K, Conatser L, Llanga TA, et al. Intrastromal gene therapy prevents and reverses advanced corneal clouding in a canine model of mucopolysaccharidosis I. Mol Ther 2020;28:1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xiao W, Warrington KH, Hearing P, et al. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J Virol 2002;76:11505–11517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Teramoto S, Bartlett JS, McCarty D, et al. Factors influencing adeno-associated virus-mediated gene transfer to human cystic fibrosis airway epithelial cells: comparison with adenovirus vectors. J Virol 1998;72:8904–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Allen JM, Debelak DJ, Reynolds TC, et al. Identification and elimination of replication-competent adeno-associated virus (AAV) that can arise by nonhomologous recombination during AAV vector production. J Virol 1997;71:6816–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang XS, Khuntirat B, Qing K, et al. Characterization of wild-type adeno-associated virus type 2-like particles generated during recombinant viral vector production and strategies for their elimination. J Virol 1998;72:5472–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chandler RJ, Sands MS, Venditti CP. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum Gene Ther 2017;28:314–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grossman Z, Berns KI, Winocour E. Structure of simian virus 40-adeno-associated virus recombinant genomes. J Virol 1985;56:457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ulusoy A, Sahin G, Björklund T, et al. Dose optimization for long-term rAAV-mediated RNA interference in the nigrostriatal projection neurons. Mol Ther 2009;17:1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hirsch ML, Fagan BM, Dumitru R, et al. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS One 2011;6:e27520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ertl HCJ, High KA. Impact of AAV capsid-specific T-cell responses on design and outcome of clinical gene transfer trials with recombinant adeno-associated viral vectors: an evolving controversy. Hum Gene Ther 2017;28:328–337 [DOI] [PubMed] [Google Scholar]
- 72. Botquin V, Cid-Arregui A, Schlehofer JR. Adeno-associated virus type 2 interferes with early development of mouse embryos. J Gen Virol 1994;75 (Pt 10):2655–2662 [DOI] [PubMed] [Google Scholar]
- 73. Zeltner N, Kohlbrenner E, Clément N, et al. Near-perfect infectivity of wild-type AAV as benchmark for infectivity of recombinant AAV vectors. Gene Ther 2010;17:872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Blacklow NR, Hoggan MD, Rowe WP. Isolation of adenovirus-associated viruses from man. Proc Natl Acad Sci U S A 1967;58:1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Timpe J, Bevington J, Casper J, et al. Mechanisms of adeno-associated virus genome encapsidation. Curr Gene Ther 2005;5:273–284 [DOI] [PubMed] [Google Scholar]
- 76. Earley LF, Conatser LM, Lue VM, et al. Adeno-associated virus serotype-specific inverted terminal repeat sequence role in vector transgene expression. Hum Gene Ther 2020;31:151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nony P, Chadeuf G, Tessier J, et al. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J Virol 2003;77:776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Labow MA, Graf LH, Berns KI. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol 1987;7:1320–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hörer M, Weger S, Butz K, et al. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol 1995;69:5485–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li C, Narkbunnam N, Samulski RJ, et al. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther 2012;19:288–294 [DOI] [PubMed] [Google Scholar]
- 81. Chirmule N, Xiao W, Truneh A, et al. Humoral immunity to adeno-associated virus type 2 vectors following administration to murine and nonhuman primate muscle. J Virol 2000;74:2420–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.