Abstract
Several techniques have been employed for deletion of the NKX3.1 gene, resulting in developmental defects of the prostate, including alterations in ductal branching morphogenesis and prostatic secretions as well as epithelial hyperplasia and dysplasia. To investigate whether the CRISPR/Cas9-mediated technique can be applied to study prostate carcinogenesis through exon I deletion of NKX3.1 gene, alterations in the prostatic intraepithelial neoplasia (PIN) and their regulatory mechanism were observed in the prostate of NKX3.1 knockout (KO) mice produced by the CRISPR/Cas9-mediated NKX3.1 mutant gene, at the ages of 16 and 24 weeks. The weight of dorsal-lateral prostate (DLP) and anterior prostate (AP) were observed to be increased in only the 24 weeks KO mice, although morphogenesis was constant in all groups. Obvious PIN 1 and 2 lesions were frequently detected in prostate of the 24 weeks KO mice, as compared with the same age wild type (WT) mice. Ki67, a key indicator for PIN, was densely stained in the epithelium of prostate in the 24 weeks KO mice, while the expression of p53 protein was suppressed in the same group. Also, both the 16 and 24 weeks KO mice reveal inhibition of the PI3K/AKT/mTOR pathway in the prostate. However, prostate specific antigen (PSA) levels and Bax/Bcl-2 expressions were decreased in the prostate of 16 weeks KO mice, and were increased in only the 24 weeks KO mice. Taken together, the results of the present study provide additional evidence that CRISPR/Cas9-mediated exon 1 deletion of the NKX3.1 gene successfully induces PIN lesions, along with significant alterations of Ki67 expression, EGFR signaling pathway, and cancer-regulated proteins.
Keywords: CRISPR/Cas9, prostate, PIN lesion, Ki-67, EGFR signaling pathway
Introduction
NKX3.1 is a well-known prostate tumor suppressor gene which acts as an androgen-regulated transcription factor.1 It is predominantly expressed in the luminal epithelial cells of the prostate and, to a lesser extent, in the testis.2 The NKX3.1 protein regulates branching morphogenesis and epithelial cell differentiation and growth of prostate, and induces differentiation of the prostate gland.3 In addition, NKX3.1 affects the development of prostatic epithelial hyperplasia and dysplasia that progresses to PIN lesions, through the stochastic loss of target gene expression and an extended proliferative phase during normal epithelial turnover.3-5
Function of the NKX3.1 gene has directly been verified by characterization of KO mice generated by gene targeting techniques and Cre-LoxP recombination system.4,5 Three different NKX3.1 KO mice generated by targeted gene disruption display similar phenotypes on the development and carcinogenesis of prostate. Defect in the prostate ductal morphogenesis and secretory protein production, as well as prostatic epithelial hyperplasia and dysplasia resembling PIN lesions, were detected in NKX3.1 heterozygous (+/-) and homozygous (-/-) mice.4,6 The NKX3.1/LacZ null mutants show a significantly defective duct morphogenesis, progressing epithelial hyperplasia, and reduced ductal branching in prostate.7 Significant hyperplasia and defect in the branching morphogenesis of prostate and palatine glands were observed in homozygous mutant mice for NKX3.1.8 Furthermore, conditional NKX3.1-deficient mice produced by the Cre-LoxP-mediated recombination reveal prostatic epithelial hyperplasia and dysplasia at the 20 weeks. In these mice, the expression pattern of some biomarkers that indicate PIN lesions were very similar to human PIN.5 However, no studies have been reported on alterations in the post-birth survival rate of all the above mice. Furthermore, the CRISPR/Cas9 technology has never been applied for the production and tumorigenic phenotype analyses of NKX3.1-deficient mice, although this system has already been successfully applied to target important genes in numerous organisms, including zebrafish, mice, rats, rabbits, monkeys and pigs.9,10
The current study was therefore undertaken to characterize prostate carcinogenesis of NKX3.1 KO mice that have a genetically deleted site of the exon 1 in NKX3.1 gene using the CRISPR/Cas9 system, and to evaluate the applicability of this technique. Our results indicate that the CRISPR/Cas9-mediated NKX3.1-deficient mice show PIN 1 and 2 lesions, accompanied with alterations in the histopathological structure, Ki67 expression, EGFR signaling pathway, anticancer proteins and apoptotic proteins in the prostate at age 24 weeks. We believe this model can be applied as an animal model to study the molecular mechanism for PIN lesions, and evaluate therapeutic drugs for prostate-related diseases.
Materials and Methods
Production and Identification of C57BL/6-NKX3.1em1Hlee/Korl KO Mice
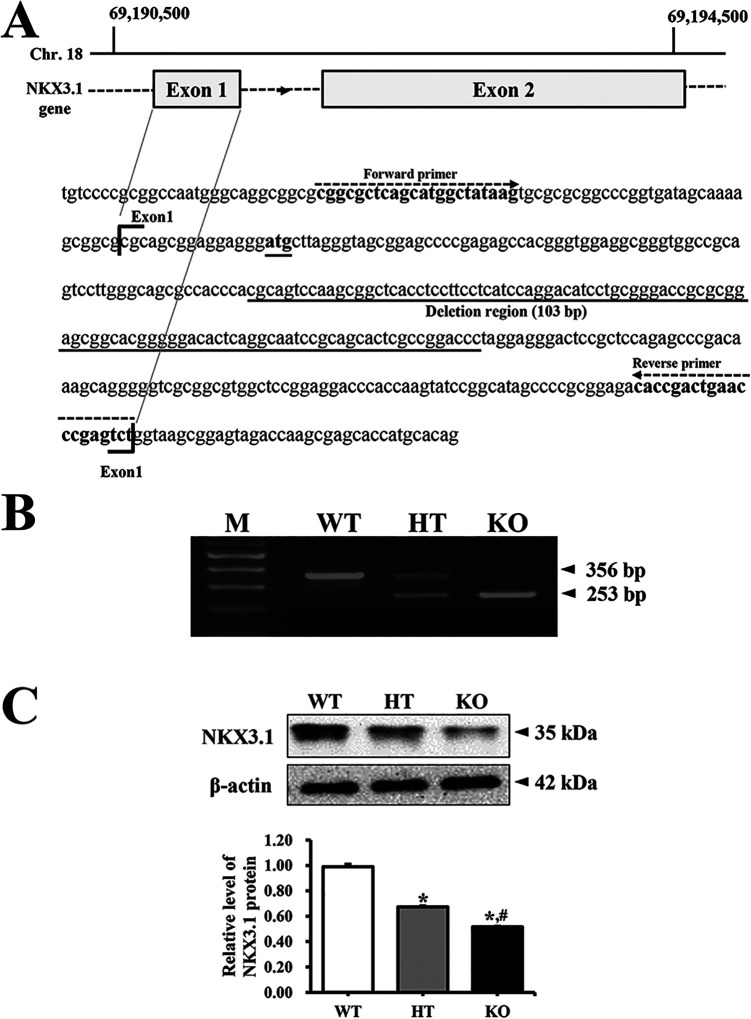
To generate NKX3.1 KO in C57BL/6 mice having indel mutation in exon 1 of NKX3.1 gene, the CRISPR design tool (crispor.tefor.net) was used to select target sequences of small guide RNA (sgRNA). Briefly, a mixture of Cas9 protein (100 ng/μL) (EnGen Cas9 NLS; purchased from NEB) and gRNA (50 ng/μL) was injected into the pronuclei cytoplasm. sgRNAs were generated with the MEGAshotscript T7 kit (Invitrogen, Carlsbad, CA, USA), using sgRNA sequences 5’-AGC CGC TTG GAC TGC GTG GG-3’ and 5’-AGC ACT CGC CGG ACC CTA GGA-3’. Indel mutations of 103 bp in F1 mice were identified by TA cloning and sequencing analyses (Figure 1A).
Figure 1.
Deletion scheme for NKX3.1 gene and their conformation. (A) After the design and synthesis of 2 highly active CRISPR specific to exon 1 of NKX3.1 gene, small guide mRNA was injected into the cytoplasm of mouse pronuclear-stage embryos to produce NKX3.1 KO mice. (B) DNA-PCR was conducted on genomic DNA isolated from tails of founder mice. After amplification, levels of the 2 products (356 bp and 253 bp) were quantified on 2% agarose gel. Abbreviations: M, Marker; WT, Wild type mice; HT, Heterogenous type mice; KO, Knockout type mice. (C) The expression of NKX3.1 protein was measured by western blot analysis using HRP-labeled anti-rabbit IgG antibody. After intensity of each band was determined using an imaging densitometer, the relative level of the NKX3.1 protein was calculated based on the intensity of actin. Three to 5 mice per group were used to prepare the protein homogenate, and western blot analysis was assayed in duplicate for each sample. Data are reported as the mean ± SD. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to the HT mice.
The deletion of the target gene was identified by DNA-PCR analysis of the genomic DNA isolated from the tails of founder mice. The CRISPR/Cas9-mediated NKX3.1 mutant genes in the C57BL/6 background strains were amplified using primer sets. The WT, heterogeneous type (HT) and KO mice for NKX3.1 gene were identified by sense primer (5′-CGG CGC TCA GCA TGG CTA TAA G-3′) and antisense primer (5′-AGA CTC GGG TTC AGT CGG TG-3′). Thereafter, 10 pmole of sense and antisense primers were added, and the reaction mixture was subjected to 38 cycles of amplification on a Perkin-Elmer Thermal Cycler, as follows: 30 sec, 94°C; 30 sec, 60°C; 1 min, 72°C. After amplification, the final PCR products of 356 bp and 253 bp in tail genomic DNA of WT and KO mice were electrophoresed on 2% agarose gels (Figure 1B).
The levels of NKX3.1 mRNA were measured by RT-PCR analysis. Briefly, total RNA molecules were purified from 9 different tissues including brain, liver, lung, heart, kidney, spleen, thymus, testis, and prostate gland, using RNAzol CS104 (Tel-Test Inc., Friendswood, USA). After synthesis of the complement DNA (cDNA) for 5 µg of total RNA using 200 units of reverse transcriptase (Superscript II, Invitrogen, 200 U/µL), the NKX3.1 transcripts were amplified with specific primers as follows: NKX3.1, sense primer and anti-sense primer; β-actin, sense primer: 5’-TGG AAT CCT GTG GCA TCC ATG AAA C-3’, anti-sense primer: 5’-TAA AAC GCA GCT CAG TAA CAG TCC G-3’. Amplification was conducted in a Perkin-Elmer Thermal Cycler by applying the following cycle: 30 sec at 94°C, 30 sec at 62°C, and 45 sec at 72°C. The final RT-PCR products were separated on 1-2% agarose gel and subsequently visualized by ethidium bromide staining. The densities of specific bands were quantified using the Kodak Electrophoresis Documentation and Analysis System 120 (Eastman Kodak, Rochester, NY). Deletion products (253 bp) of the NKX3.1 transcript were detected in the kidney, spleen, thymus, testis and prostate gland tissues of KO mice, but not in brain, liver, lung and heart (Supplement Figure 1).
Animal Care and Use
The animal protocol used in this study was reviewed and approved by the National Cancer Center Research Institute (NCCRI; Approval Number: NCC-18-391) and the Pusan National University-Institutional Animal Care and Use Committee (PNU-IACUC; Approval Number PNU-2017-1719). C57BL/6 mice were obtained from OrientBio Com. (Seongnam, Korea). Large numbers of WT, HT and KO mice were produced by mating the male and female HT mice. During the experimental period, all mice were handled at the National Cancer Center Research Institute and the Pusan National University Laboratory Animal Resources Center, accredited by the Ministry of Food and Drug Safety (Accredited Unit Number; 00231) and AAALAC International (Accredited Unit Number; 001525). All mice were provided with a standard irradiated chow diet (Purina Mills, Seoungnam, Korea) ad libitum, and were maintained in a specific pathogen-free (SPF) state under a strict light cycle (lights on at 06:00 h and off at 18:00 h) at a temperature of 23 ± 2°C and relative humidity 50 ± 10%.
After harvesting organs from WT, HT and KO mice at 16 (12-17 mice per group) and 24 weeks (5-6 mice per group) mice, morphological alterations in the prostate were directly observed in seminal vesicles (SV), ventral prostate (VP), DLP and AP. The weight of testis, SV, LP, DLP and AP from each mouse was measured by an electronic balance (Mettler Toledo, Greifensee, Switzerland). Total weight of testis, SV, DLP and AP is presented as the whole weight of both lobes, while the weight of VP is a measure of single lobe.
Western Blot Analysis
After isolation of complete form SV, LP, DLP and AP from mice of subset groups (n = 3-5), the same weight of tissue was collected from each lobe and then mixed to use western blot analysis. The mixed lobes were homogenized using a PRO-PREPTM Solution Kit (iNtRON Biotechnology, Sungnam, Korea) supplemented with half tablet of a protein inhibitor cocktail (Roche, Penzberg, Germany), and the mixture was centrifuged at 13,000 rpm for 5 min. The prepared proteins were then electrophoresed on a 10% SDS-PAGE gel, and subsequently transferred onto a nitrocellulose membrane (Amersham Biosciences, Corston, UK) for 2 h at 40 V in transfer buffer (25 mM Trizma-base, 192 mM glycine, and 20% methanol). Efficiency of the transfer and equal protein loading were determined by staining the membrane with Ponceau (Sigma-Aldrich Co., St. Louis, USA), and the gel with Coomassie blue (Sigma-Aldrich Co.). Appropriate dilutions of the following primary antibodies were added to the membranes and allowed to hybridize overnight at 4°C: anti-NKX3.1 (sc-393190, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-AKT (9272S, Cell Signaling Technology, Inc., Danvers, MA, USA), anti-p-AKT (4058S, Cell Signaling Technology, Inc.), anti-PI3K (4292S, Cell Signaling Technology, Inc.), anti-p-PI3K (4228S, Cell Signaling Technology, Inc.), anti-mTOR (2792S, Cell Signaling Technology, Inc.), anti-p-mTOR (2791S, Cell Signaling Technology, Inc.), anti-p53 (Sigma-Aldrich Co.), anti-Bax (ab7977, Abcam, Cambridge, UK), anti-Bcl-2 (ab7973, Abcam) and anti-β-actin (A5316, Sigma–Aldrich Co.). After removing the antibodies, membranes were washed 3 times in a solution composed of 10 mM Trizma-base (Sigma–Aldrich Co) (pH 7.6), 150 mM NaCl (Sigma–Aldrich Co.), and 0.05% Tween-20 (Biosesang, Gyeonggi-do, Korea) for 10 min. The primary antibody conjugated membranes were then incubated with horseradish peroxidase-conjugated anti-secondary antibody (Invitrogen) for 1 h at room temperature. The membrane was washed again as described above, and developed using an enhanced chemiluminescence detection system (Amersham Bioscience). Finally, the results were quantified using the Image Analyzer System (Eastman Kodak 2000MM, NY, USA). All results are expressed as the fold-increase over control value, and were confirmed by 2 independent researchers conducting the experiments at least twice.
Immunofluorescence (IF) Staining Analysis
For detection of p53 protein via IF staining analysis, prostate tissues of subset groups were fixed in 10% formalin for 48 h, embedded in paraffin block, sliced into 4 µm thick sections, and mounted on a glass slide. Sections were then deparaffinized with xylene, rehydrated with different concentrations of EtOH, and pretreated with blocking buffer containing 10% goat serum in 1× PBS solution, for 30 min at room temperature. The pretreated sections were subsequently incubated with anti-p53 antibody (Sigma-Aldrich Co.), diluted 1:300 in blocking buffer. After thorough washing in 1× PBS solution, the slide sections were incubated with goat FITC-labeled anti-rabbit IgG serum for 1 h, washed thrice in 1× PBS for 3 min each, and mounted with vector shield mounting medium. Finally, the green fluorescence in stained cells was observed at 400× magnification via fluorescence microscopy (Eclipse TX100, Nikon, Tokyo, Japan).
Immunohistochemical (IHC) Staining Analysis
Tissue distribution of Ki67 protein was detected by IHC staining using light microscopy, as previously described.11 Briefly, the prostate tissue samples were fixed in 10% formalin for 12 h, embedded in paraffin, and sliced into 4 µm thick sections. These sections were subsequently deparaffinized with xylene, rehydrated, and pretreated for 30 min at room temperature with PBS blocking buffer containing 10% goat serum (Vector Laboratories, California, USA). The sections were then incubated with primary anti-Ki67 antibody (Abcam), diluted 1:300 in PBS blocking buffer. The antigen-antibody complexes were visualized with biotinylated secondary antibody (goat anti rabbit)-conjugated HRP streptavidin (Histostain-Plus Kit, Zymed, South San Francisco, CA, USA), at a dilution of 1:300 in PBS blocking buffer. Finally, Ki-67 proteins were detected using stable DAB (Invitrogen) and the Leica Application Suite (Leica Microsystems, Wetzlar, Germany).
Histopathological Analysis
To detect alterations in the histopathological structure, prostate tissues were harvested from mice of subset groups, fixed in 10% formalin, embedded in paraffin wax, routinely processed, sectioned into 4 µm thick slices, stained with hematoxylin and eosin (H&E), and examined by light microscopy (Leica Microsystems, Wetzlar, Germany). PIN lesions in the prostate tissue were also identified by Prof. D.V.M. Beum Seok Han at the Department of Pharmaceutical Engineering, Hoseo University, and Dr. D.V.M. Sang Gu Lee at the DD Partner Co., Korea. PIN grades in the histological characteristics of prostate gland were evaluated based on the diagnostic criteria suggested by McNeal and Bostwick research group,12 and Shappell et al. research group.13
Measurement of PSA Concentrations
The level of PSA was quantified using ELISA kits (CSB-E08276m, Cusabio Biotech Co., Ltd., Wuhan, China), according to the manufacturer’s instructions. Briefly, prostate tissue (50 mg) was homogenized in ice-cold 1× PBS (pH 7.2-7.4) using a glass homogenizer (Sigma-Aldrich Co.). The tissue lysates were centrifuged at 1,000 rpm for 5 min at 4°C, and the resultant supernatant was collected for analysis. After addition of the 4 specific hormone antibodies, separately in each well, the supernatant was incubated for 60 min at 37°C, followed by addition of HRP-Streptavidin solution and subsequent incubation for 60 min at 37°C. The TMP OneStep Substrate Reagent was then added, and the mixture was further incubated for 30 min at 37°C. The reaction was terminated following addition of the stop solution. Finally, the absorbance of the reaction mixture was read at 450 nm using a Molecular Devices VERSA max Plate reader (Sunnyvale, CA, USA).
Statistical Analysis
Statistical significance was evaluated using One-way Analysis of Variance (ANOVA) (SPSS for Windows, Release 10.10, Standard Version, Chicago, IL, USA) followed by Tukey post hoc T-test for multiple comparison. All data are expressed as the means ± SD. A p value less than (p < 0.05) is considered statistically significant.
Results
Effects of CRISPR/Cas9-Mediated NKX3.1 Deficiency on the Weight and Morphology of Prostate
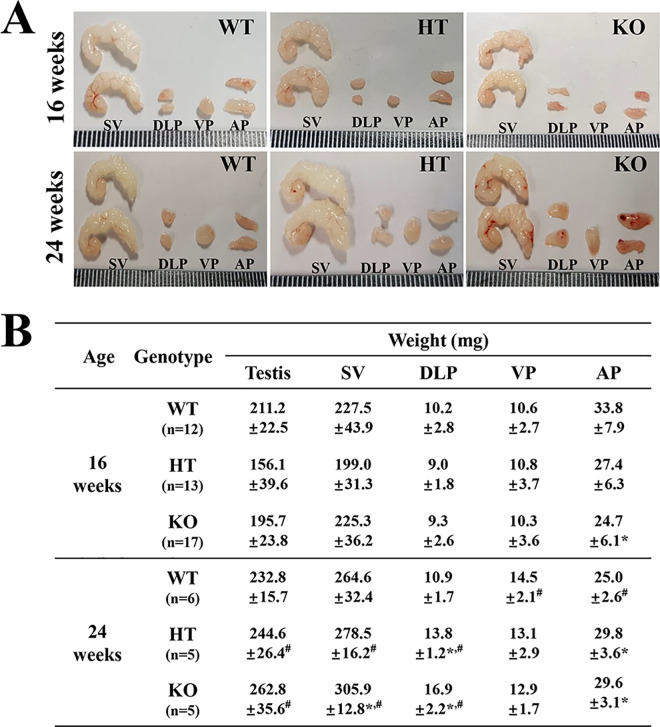
We first verified the successful suppression of NKX3.1 protein and detection of deleted NKX3.1 transcript (253 bp) in the prostate of KO mice, using western blot and RT-PCR analysis (Figure 1B and Supplement Figure 1). Subsequently, the weight and morphological changes in the prostate were compared between WT, HT and KO mice, to determine the effect of CRISPR/Cas9-mediated NKX3.1 deficiency on the morphogenesis of prostate. No significant differences were observed in the morphological features of WT, HT and KO mice, regardless of their age (Figure 2A). However, some significant alterations were detected in the weight of each prostate lobe. In 16 weeks mice, the weight of only AP was decreased in NKX3.1 KO mice compared to WT mice, while the DLP and VP weights remained unchanged. In the 24 weeks mice, the weights of SV, DLP and AP were greater in KO mice as compared to WT mice, but the VP weight remained constant. Moreover, SV, DLP and AP weights of 24 weeks KO mice were significantly increased as compared with those of 16 weeks mice (Figure 2B). These results indicate that CRISPR/Cas9-mediated NKX3.1 deficiency induces an increase in the weights of SV, DLP and AP lobe at age 24 weeks.
Figure 2.
Morphology and weight of prostate and testis in WT, HT and KO mice. (A) Morphological features of SV, DLP, VP and AP were observed after collection from subset groups at 16 and 24 weeks. (B) Weight of each lobe of prostate and testis was measured, as described in the materials and methods. Five to 17 mice per group were used for collecting the prostates, and their weights were assayed in duplicate for each sample. Data are reported as the mean ± SD. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to 16 weeks mice. Abbreviation: SV, Seminal vesicle; DLP, dorsal-lateral prostate; VP, Ventral prostate; AP, Anterior prostate.
Effects of CRISPR/Cas9-Mediated NKX3.1 Deficiency on the Development of PIN Lesions
To observe for development of PIN lesions in the prostate of NKX3.1 KO mice, we observed for alterations in the histopathological features in H&E stained tissue sections of prostate at 16 and 24 weeks. Minor and severe PIN lesions in the H&E stained prostate tissue were clearly observed in only the 24 weeks KO mice, while minor PIN lesions were detected only in the AP of 16 weeks KO mice. Especially, PIN grade 1 (including crowding, stratification and irregular spacing) was significantly increased in VP and AP of KO mice as compared with WT mice at 24 weeks. PIN grade 2 (including tufting, micropapillary, cribriform, flat, nuclear enlargement and less nuclear size variation) first appeared only in the DLP and AP of 24 weeks KO mice (Figure 3 and Table 1).
Figure 3.
Histopathological structures of prostate in WT, HT and KO mice at 16 and 24 weeks. H&E stained sections of each lobe from prostate were observed at 400× magnification using a light microscope. Three to 5 mice per group were used for histological analyses, and their histopathological structure were assayed in duplicate for each sample. Abbreviation: DLP, dorsal-lateral prostate; VP, Ventral prostate; AP, Anterior prostate.
Table 1.
The Grade of Prostatic Intraepithelial Neoplasia (PIN) of NKX3.1 KO Mice in 16 and 24 Weeks.
| Age | Genotype | PIN grade | Detection rate (%) | ||
|---|---|---|---|---|---|
| DLP | VP | AP | |||
| 16 weeks | WT | PIN 1 | - | - | 1.5 |
| PIN 2 | - | - | - | ||
| HT | PIN 1 | - | - | - | |
| PIN 2 | - | - | - | ||
| KO | PIN 1 | - | - | 25 | |
| PIN 2 | - | - | - | ||
| 24 weeks | WT | PIN 1 | 33 | - | 33 |
| PIN 2 | - | - | - | ||
| HT | PIN 1 | 33 | 33 | 33 | |
| PIN 2 | - | - | - | ||
| KO | PIN 1 | - | 66 | 50 | |
| PIN 2 | 66 | - | 50 | ||
Note: PIN 3 was not detected in any tissue samples of prostate.
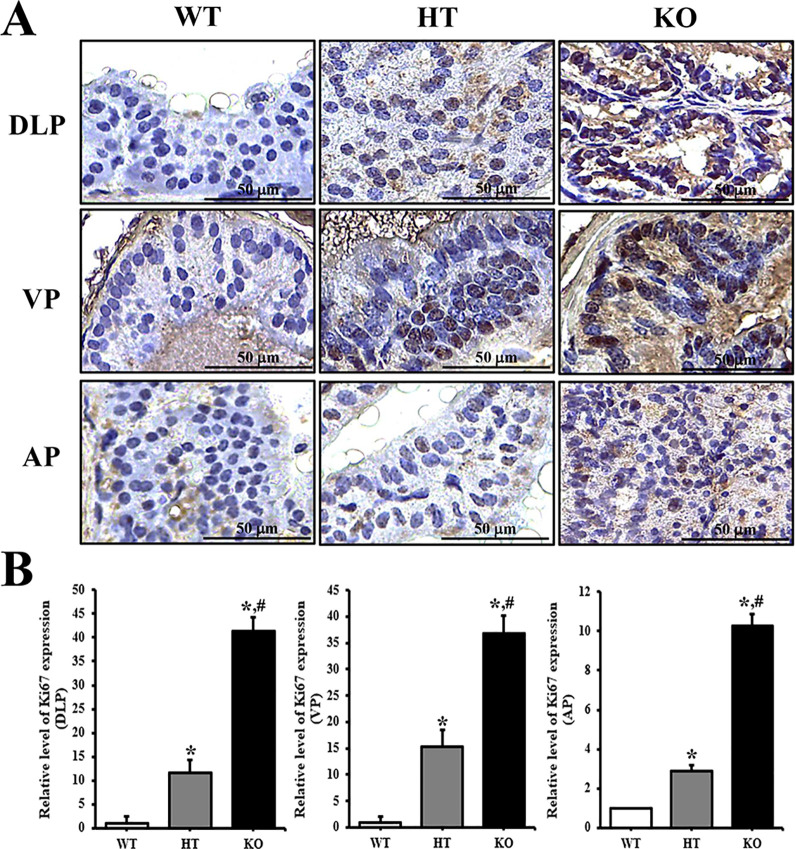
To confirm whether PIN lesions in the prostate of KO mice are accompanied with an alteration of the molecular mechanism, the expression levels of Ki67 protein were measured in the prostate tissue of NKX3.1 KO mice. Significant increase was observed in the tissue distribution of Ki67 protein in DLP, VP and AP lobe of 24 weeks KO mice prostate, although the enhancement rate was greater in VP as compared to other lobes (Figure 4). However, subset groups of 16 weeks mice showed only very limited increase in Ki67 protein level on AP section of KO mice (Supplement Figure 2). Therefore, these results indicate that CRISPR/Cas9-mediated NKX3.1 deficiency successfully induces PIN lesions at 24 weeks.
Figure 4.
Tissue distribution of Ki67 protein in 24 weeks mice. (A) Following staining with Ki67 antibody, the level of cells stained dark brown was examined on the prostate tissue sections of WT, HT and KO mice at 400× magnification. (B) The number of stained cells were counted in specific area and presented on a graph. Three to 5 mice per group were used for preparation of slide section and immunohistochemistry analyses, and the number of stained cells were counted in duplicate for each sample. Data are reported as the mean ± SD. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to 16 weeks mice. Abbreviation: DLP, dorsal-lateral prostate; VP, Ventral prostate; AP, Anterior prostate.
Effects of CRISPR/Cas9-Mediated NKX3.1 Deficiency on the EGFR Signaling Pathway
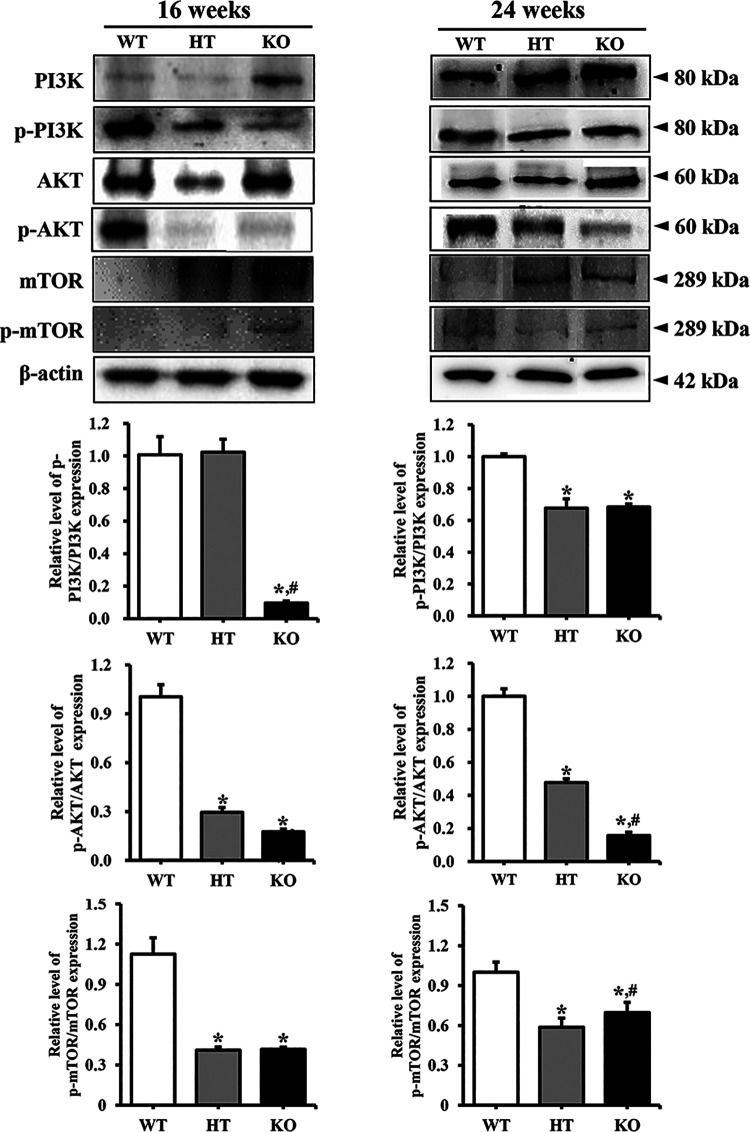
To investigate whether PIN lesions in the prostate of NKX3.1 KO mice are accompanied with alterations in cell survival and proliferation, changes in the members of EGFR downstream signaling pathway were analyzed in the prostate of NKX3.1 KO mice. Similar patterns were observed in the phosphorylation levels of the 3 examined markers, PI3K, AKT and mTOR. These levels were remarkably decreased in the prostate of HT mice or KO mice compared with WT mice at both ages. However, no significant differences were observed in the decrease rate between 16 and 24 weeks KO mice (Figure 5). These results indicate that PIN lesions in the prostate of NKX3.1 KO mice are associated with the suppression of EGFR signaling pathway at 16 and 24 weeks.
Figure 5.
Expression level of various members in the EGFR downstream signaling pathway at 16 and 24 weeks. The expression of several related proteins in the EGFR downstream signaling pathway, including PI3K, p-PI3K, AKT, p-AKT, mTOR and p-mTOR, were measured by western blot analysis using HRP-labeled anti-rabbit IgG antibody. After determining the intensity of each band using an imaging densitometer, the relative levels of proteins were calculated based on the intensity of β-actin. Five to 6 mice per group were used for preparing the protein homogenate, and western blot analyses were assayed in duplicate for each sample. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to the HT mice.
Effects of CRISPR/Cas9-Mediated NKX3.1 Deficiency on the Expression of Cancer-Related Proteins in Prostate
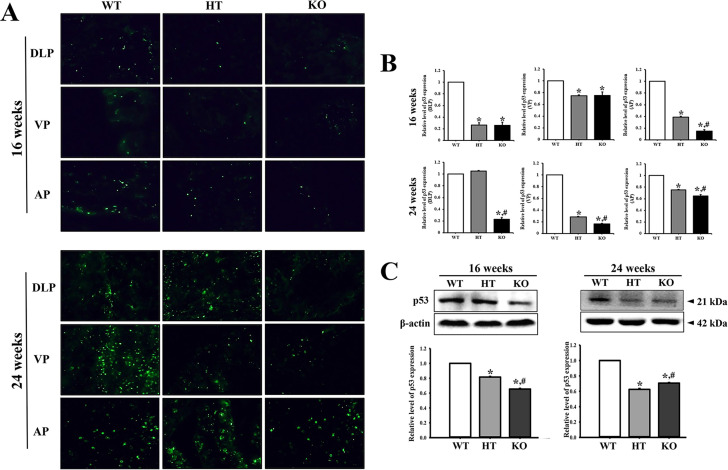
To examine whether PIN lesions in the prostate of CRISPR/Cas9-mediated NKX3.1 KO mice are accompanied with alterations in the levels of cancer-related proteins, we evaluated the expression levels of p53, PSA, Bax and Bcl-2 in the prostate tissue of KO mice. The fluorescence intensity for p53 was lower in the prostates of 16 weeks HT and KO mice than the WT mice. A similar pattern was maintained in the prostate of 24 weeks mice, although the total number of p53-stained fluorescence dots was higher at 24 weeks as compared to 16 weeks. Of the 3 lobes examined, differences on the fluorescence dart for p53 protein were remarkably greater in DLP and VP in the prostate of KO mice as compared to WT mice (Figure 6A and B). Also, western blot results for p53 protein were very similar to fluorescence staining analysis (Figure 6C). Furthermore, increase in the level of PSA was observed to be age dependent; at 24 weeks, the PSA level was remarkably increased in the NKX3.1 HT and KO mice than in WT mice (Figure 7A). Moreover, the expressions of Bax and Bcl-2 were dramatically altered in the prostate of HT and KO mice at both ages. At 16 weeks, the level of Bax/Bcl-2 was decreased in HT and KO mice compared to WT, whereas at 24 weeks, the levels were enhanced in only KO mice as compared to WT and HT mice (Figure 7B). These results therefore indicate that PIN lesions in the prostate of CRISPR/Cas9-mediated NKX3.1 KO mice are related to the suppression of p53 protein, enhancement of PSA levels, and dysregulation of the Bax/Bcl-2 levels.
Figure 6.
p53 immunofluorescence staining. (A) After staining with specific antibody for p53, the prostate tissue section on glass slide was subsequently reacted with fluorescence-conjugated anti-rabbit IgG antibody. Fluorescence intensity and cellular morphology were observed under a fluorescent microscope (400× magnification). (B) The average number of p53 stained cells was counted in specific area and presented on a graph. Three to 5 mice per group were used for preparation of slide section and immunohistochemistry analyses, and the number of stained cells were counted in duplicate for each sample. (C) The expression of p53 protein was measured by western blot analysis using HRP-labeled anti-rabbit IgG antibody. After determining the intensity of each band using an imaging densitometer, the relative levels were calculated, based on the intensity of β-actin. Five to 6 mice per group were used for preparing the protein homogenate, and western blot analyses were assayed in duplicate for each sample. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to the HT mice. Abbreviation: DLP, dorsal-lateral prostate; VP, Ventral prostate; AP, Anterior prostate.
Figure 7.
Level of PSA and apoptotic proteins. (A) The PSA concentration was measured in the prostate homogenate by enzyme-linked immunosorbent assay. The detection ranges of the kit used is 0.312-20 ng/mL. Five to 6 mice per group were used for preparing the protein lysate, and ELISA were assayed in duplicate for each sample. (B) The expression of Bax and Bcl-2 proteins were measured by western blot analysis using HRP-labeled anti-rabbit IgG antibody. After intensity of each band was determined using an imaging densitometer, the relative levels of the 2 proteins were calculated based on the intensity of β-actin. Five to 6 mice per group were used for preparing the protein homogenate, and western blot analyses were assayed in duplicate for each sample. Data are reported as the mean ± SD. *, p < 0.05 compared to the WT mice. #, p < 0.05 compared to the HT mice.
Discussion
The prostate gland is considered to have paramount importance in human diseases due to the increasing incidence of benign prostatic hyperplasia and carcinoma in aging men; prostate carcinoma now represents the second leading cause of cancer death in American men.14,15 Prostatic cancer is known to be induced by the chromosomal deletion of several genes, including NKX3.1, PTEN, ATBF1 and KLF5.1,16,17 Of the genes known to promote prostate cancer, the NKX3.1 homeobox gene is of particular interest because it maps to the minimal region of the human chromosome 8p211,18 that undergoes loss of heterozygosity in 60%–80% of prostate tumors.19-23 Especially, this protein plays a key role as a homeodomain-containing transcription factor in the suppression of prostate tumorigenesis.24 Therefore, this study investigated prostate carcinogenesis of CRISPR/Cas9-mediated NKX3.1 KO mice at 16 and 24 weeks, by examining whether the NKX3.1 deletion with CRISPR/Cas9-mediated technique contributes to tumor incidence. Our results reveal pronounced PIN lesions with accompanying alterations in cell proliferation and cancer-related proteins in 24 weeks CRISPR/Cas9-mediated NKX3.1 KO mice.
Previous studies have reported that increase of prostate weight can be caused by various experimental conditions in mice. The weight and index of prostate were significantly increased in the testosterone-induced prostatic hyperplasia model and the low dose 17α-ethinyl estradiol (EE) treated model.25,26 Furthermore, prostate weight was enhanced in 8 months CF-1 male mice generated from females administrated low dose of DES during pregnancy.27 The average weight of PSA-Cre;Pten-loxP/loxP prostates increased about 2-fold at 4-5 months, 3-fold at 7-9 months, and 16-fold at 10-14 months, with an obvious enlargement of each prostate lobe observed at the age of 4-5 months.28 Furthermore, a remarkable enhancement of DLP and VP weight were detected at 10, 25, and 80 weeks of age in Pb-PRL transgenic mice expressing prolactin under the control of the prostate-specific rat probasin minimal promoter.29 In the current study, NKX3.1 deletion with CRISPR/Cas9-mediated technique induces the enhancement of DLP and AP weight in 24 weeks KO mice. These alterations observed in the present study are similar to previous studies, although there are few differences in the target lobe with increased weight. Especially, our results show that among prostate lobes, DLP and AP may be more sensitive to NKX3.1 deletion with CRISPR/Cas9-mediated technique. However, the AP of 16 weeks WT mice was heavier than HT and KO mice at the same age. We thought that these results are probably associated with incomplete remove of luminal contents from AP during the experimental process, since clear scientific evidence for significant increase of AP in normal mice has not been found, and these values have large standard deviation.
Several NKX3.1 mutant mice produced in previous studies display a significant alteration in the morphology and histopathology of the prostate, with some showing several defects in the ductal and branching morphogenesis.4,7 Also, severe prostatic epithelial hyperplasia and dysplasia have been observed in homozygous and heterozygous NKX3.1 mutant mice obtained by targeted gene (exon II) disruption in 1 years mice.4 The AP of NKX3.1 HT and KO mice with exon II deletion presented with hyperplasia at age 2 and 10 months, while the DLP of only NKX3.1 KO mice shows severe epithelial hyperplasia at 10 months.7 Furthermore, significant hyperplasia was detected in the epithelial cells of mutant prostate and palatine gland of NKX3.1 homozygous mutant mice with exon II deletion produced by gene targeting technique. After age 6 months, this hyperplasia expanded into the lumen of the ventral and dorsal prostate in NKX3.1 KO mice, although prostate cancer was not detected until 2 years.8 The conditional NKX3.1-deficient mice with exon II deletion showed focal epithelial hyperplasia and PIN lesions resembling human PIN at 28 weeks.5 Furthermore, the PIN lesions of above mice had accompanying expressions of several biomarkers including Ki-67, E-cadherin and high-molecular weight cytokeratins.5 In the current study, we produced novel NKX3.1 KO mice using the CRISPR/Cas9 recombination technique. No significant alterations were observed in the morphogenesis of prostate in both 16 and 24 weeks KO mice. However, PIN grade 1 and 2 lesions were observed in the prostate epithelium of 24 weeks NKX3.1 KO mice, while normal histological features were maintained until 16 weeks. These findings are similar to changes observed in KO mice produced with gene targeting technique and Cre and loxP-mediated recombination system,5 although the targeted gene was different in each study. Especially, a significant increase in PIN grade 1 and 2 lesions was clearly detected in the AP of prostate of 24 weeks NKX3.1 KO mice. Our data shows that PIN lesions of prostate are successfully accelerated under conditions of NKX3.1 exon I deletion induced by the CRISPR/Cas9 system. However, our research has a limitation for detecting the expression of NKX3.1 protein. The western blot band image for the NKX3.1 protein had not disappeared completely in KO mice, as compared to the results obtained by Ouyang et al.30 This result is thought to be due to cross reactivity of antibodies against the NKX3.1 protein, although it was not possible to determine which proteins bind to this antibody.
We further investigated the expression level of biomarkers for PIN lesions in the prostate of CRISPR/Cas9-mediated NKX3.1 KO mice. Immunohistochemistry analysis revealed increasing numbers of Ki67-stained cells during the deficiency of the NKX3.1 gene (Figure 4). A similar pattern was observed for the expression of E-cadherin in the prostate of other NKX3.1 KO mice. Development of PIN lesions is also reported to show increased expression of several biomarkers which affect the proliferative potential of dysplastic cells.31 Especially, significant enhancement of Ki67 and E-cadherin expressions have been observed in the prostate of Cre- and loxP-mediated NKX3.1 KO mice.5 These findings of previous studies are in complete agreement with the results of our study. However, to gain better understanding of the underlying molecular events involved in the development of PIN lesions, further researches using multiple biomarkers of PIN lesions in the prostate of NKX3.1 KO mice are required.
EGF is an important factor in maintaining homeostasis of the prostate epithelium, although it is also a well-known key factor expressed during late stages of inflammation and wound healing.32,33 Furthermore, after binding to their ligand, EGFR mediates certain signaling pathways in various cancers to promote fundamental cellular events, including survival, proliferation and migration.34,35 The expression level of this receptor increases in primary and metastatic prostate cancers.36,37 Of the downstream members of EGFR, the PI3K/AKT pathway participates in various cellular metabolisms, tumor progression (including development, growth, proliferation and metastases), and cytoskeleton reorganization.38 This pathway is considered one of the most prominent alternate pathways in aggressive prostate cancers. Aberration of the PI3K/AKT pathway has been detected in early (40%) and advanced stages (70-100%) of prostate cancer.39,40 Especially, PI3K was activated in several prostate cancer models generated with PTEN gene disruption, although this alteration was not investigated in NKX3.1 KO mice.41,42 The expression level of p-AKT is reported to be remarkably increased in the NKX3.1−/−/Pten+/− mice, while NKX3.1−/−/Pten+/+ mice show no significant changes.6 In the current study, we first analyzed the PI3K signaling pathway as one of the EGFR downstream signaling pathways in prostate of CRISPR/Cas9-mediated NKX3.1 KO mice. We observed significant inhibition of the PI3K/AKT/mTOR pathway, regardless of age. These results for PI3K in NKX3.1 KO mice differ from previous studies, wherein the PTEN deficiency was associated with the activation of PI3K. These differences can be attributed to the properties and types of receptors, although PI3K is common in their downstream signaling pathway. Moreover, in the present study, there are few discordances between cell proliferation on the histological structure (Figure 3) and suppression of EGFR downstream signaling pathway (Figure 5) at age 16 weeks. These differences are due to the fact that molecular alterations induced by NKX3.1 deficiency may not be completely reflected into histological and morphological changes of prostate tissue at 16 weeks, although they are fully reflected at 24 weeks.
p53 has an important role as a nuclear transcription factor, and stimulates the transcription of numerous target genes involved in the regulation of cell cycle arrest and apoptosis.43,44 This protein can be stabilized by NKX3.1 through regulation of MDM2 activity in the nucleus.45,30 Also, the expression level of p53 significantly decreases in the prostate of NKX3.1 KO mice.46 In this study, tissue distribution of the p53 protein was compared in 3 lobes of the prostate in WT, HT and KO mice. A significant decrease of p53 was observed in KO mice, as compared with WT mice. These results are similar to previous studies, although different animals were examined. Our results provide additional evidence that NKX3.1 regulates the expression level of p53 proteins to balance cell proliferation, survival and differentiation.
To summarize, the present study investigated the effects of CRISPR/Cas9-mediated NKX3.1 exon I deletion on the prostate carcinogenesis of C57BL/6 mice. Taken together, our results indicate that deficiency of NKX3.1 created with the CRISPR/Cas9-mediated recombination induces the development of PIN lesions, accompanied with aberration of histological structure, Ki-67 expression, EGFR downstream signaling pathway, PSA level and apoptotic protein levels. However, further studies investigating the onset point and detection of prolonged clinical symptoms are required before these models can be applied as efficacy tests for prostate cancer drugs.
Supplemental Material
Supplement_figure_1_(TCRT-19-0256) for Deletion of NKX3.1 via CRISPR/Cas9 Induces Prostatic Intraepithelial Neoplasia in C57BL/6 Mice by Jin Ju Park, Ji Eun Kim, Yoon Jeon, Mi Rim Lee, Jun Young Choi, Bo Ram Song, Ji Won Park, Mi Ju Kang, Hyeon Jun Choi, Su Ji Bae, Ho Lee, Byeong Cheol Kang and Dae Youn Hwang in Technology in Cancer Research & Treatment
Supplement_figure_2_(TCRT-19-0256) for Deletion of NKX3.1 via CRISPR/Cas9 Induces Prostatic Intraepithelial Neoplasia in C57BL/6 Mice by Jin Ju Park, Ji Eun Kim, Yoon Jeon, Mi Rim Lee, Jun Young Choi, Bo Ram Song, Ji Won Park, Mi Ju Kang, Hyeon Jun Choi, Su Ji Bae, Ho Lee, Byeong Cheol Kang and Dae Youn Hwang in Technology in Cancer Research & Treatment
Abbreviations
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9
- KO
knockout type
- HT
heterogeneous type
- WT
wild type
- DLP
dorsal-lateral prostate
- VP
ventral prostate
- AP
anterior prostate
- SV
seminal vesicle
- PIN
mouse prostatic intraepithelial neoplasia
- ELISA
Enzyme-linked immunosorbent assay
- FITC
Fluorescein isothiocyanate
- H&E
Hematoxylin and Eosin
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- I.V.
intravenous
- PCR
polymerase chain reaction
- Bax
BCL2 associated X
- Bcl-2
B-cell lymphoma 2
- EGFR
epithelial growth factor receptor
- PI3K
phosphoinositide 3-kinase.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This present study was supported by a grant from BIOREIN (Laboratory Animal Bio Resources Initiative) from the Ministry of Food and Drug Safety.
ORCID iD: Dae Youn Hwang  https://orcid.org/0000-0002-5144-1725
https://orcid.org/0000-0002-5144-1725
Supplemental Material: Supplemental material for this article is available online.
References
- 1. He WW, Sciavolino PJ, Wing J, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43(1):69–77. [DOI] [PubMed] [Google Scholar]
- 2. Bieberich CJ, Fujita K, He WW, Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J Biol Chem. 1996;271(50):31779–31782. [DOI] [PubMed] [Google Scholar]
- 3. Abdulkadir SA. Mechanisms of prostate tumorigenesis: roles for transcription factors NKX3.1 and Egr1. Ann N Y Acad Sci. 2005;1059:33–40. [DOI] [PubMed] [Google Scholar]
- 4. Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13(8):966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdulkadir SA, Magee JA, Peters TJ, et al. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22(5):1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim MJ, Cardiff RD, Desai N, et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99(5):2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider A, Brand T, Zweigerdt R, Arnold H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech Dev. 2000;95(1-2):163–174. [DOI] [PubMed] [Google Scholar]
- 8. Tanaka M, Komuro I, Inagaki H, Jenkins NA, Copeland NG, Izumo S. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev Dyn. 2000; 219: 248–260. [DOI] [PubMed] [Google Scholar]
- 9. Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–843. [DOI] [PubMed] [Google Scholar]
- 10. Yang D, Xu J, Zhu T, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim JE, Park JW, Kang MJ, et al. Laxative effect of Spicatoside A by cholinergic regulation of enteric nerve in Loperamide-induced constipation: ICR mice model. Molecules. 2019;24(5):896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McNeal JE, Bostwick DG. Intraductal dysplasia: a premalignant lesion of the prostate. Hum Pathol. 1986;17(1):64–71. [DOI] [PubMed] [Google Scholar]
- 13. Shappell SB, Thomas GV, Roberts RL, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64(6):2270–2305. [DOI] [PubMed] [Google Scholar]
- 14. Coffey RJ, Richards JS, Remmert CS, LeRoy SS, Schoville RR, Baldwin PJ. An introduction to critical paths. Qual Manag Health Care. 1992;14(1):45–54. [PubMed] [Google Scholar]
- 15. Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics. CA Cancer J Clin. 1998;48(1):6–29. [DOI] [PubMed] [Google Scholar]
- 16. Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. [DOI] [PubMed] [Google Scholar]
- 17. Sun H, Lesche R, Li DM, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96(11):6199–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voeller HJ, Augustus M, Madike V, Bova GS, Carter KC, Gelmann EP. Coding region of NKX3.1, a prostate-specific homeobox gene on 8p21, is not mutated in human prostate cancers. Cancer Res. 1997;57(20):4455–4459. [PubMed] [Google Scholar]
- 19. Bergerheim US, Kunimi K, Collins VP, Ekman P. Deletion mapping of chromosomes 8, 10, and 16 in human prostatic carcinoma. Genes Chromosomes Cancer. 1991;3(3):215–220. [DOI] [PubMed] [Google Scholar]
- 20. Bova GS, Carter BS, Bussemakers MJG, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 1993;53(17):3869–3873. [PubMed] [Google Scholar]
- 21. Trapman J, Sleddens HF, van der Weiden MM, et al. Loss of heterozygosity of chromosome 8 microsatellite loci implicates a candidate tumor suppressor gene between the loci D8S87 and D8S133 in human prostate cancer. Cancer Res. 1994;54(23):6061–6064. [PubMed] [Google Scholar]
- 22. Cher ML, Bova GS, Moore DH, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56(13):3091–3102. [PubMed] [Google Scholar]
- 23. Vocke CD, Pozzatti RO, Bostwick DG, et al. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 1996;56(10):2411–2416. [PubMed] [Google Scholar]
- 24. Logan M, Anderson PD, Saab ST, Hameed O, Abdulkadir SA. RAMP1 is a direct NKX3.1 target gene up-regulated in prostate cancer that promotes tumorigenesis. Am J Pathol. 2013;183(3):951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Basha SZ, Mohamed GA, Abdel-Naim AB, Hasan A, Abdel-Lateff A. Cucurbitacin E glucoside from Citrullus colocynthis inhibits testosterone-induced benign prostatic hyperplasia in mice. Drug Chem Toxicol. 2019:1–11. doi:10.1080/01480545.2019.1635149 [DOI] [PubMed] [Google Scholar]
- 26. Thayer KA, Ruhlen RL, Howdeshell KL, et al. Altered prostate growth and daily sperm production in male mice exposed prenatally to subclinical doses of 17α-ethinyl oestradiol. Hum Reprod. 2001;16(5):988–996. [DOI] [PubMed] [Google Scholar]
- 27. vom Saal FS, Timms BG, Montano MM, et al. Prostate enlargement in mice due to fetal exposure to low doses of estradiol or diethylstilbestrol and opposite effects at high doses. Proc Natl Acad Sci U S A. 1997;94(5):2056–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma X, Ziel-van der Made AC, Autar B, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65(13):5730–5739. [DOI] [PubMed] [Google Scholar]
- 29. Kindblom J, Dillner K, Sahlin L, et al. Prostate hyperplasia in a transgenic mouse with prostate-specific expression of prolactin. Endocrinology. 2003;144(6):2269–2278. [DOI] [PubMed] [Google Scholar]
- 30. Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65(15):6773–6779. [DOI] [PubMed] [Google Scholar]
- 31. Myers RB, Grizzle WE. Biomarker expression in prostatic intraepithelial neoplasia. Eur Urol. 1996;30(2):153–166. [DOI] [PubMed] [Google Scholar]
- 32. Fowler JE, Jr, Lau JL, Ghosh L, Mills SE, Mounzer A. Epidermal growth factor and prostatic carcinoma: an immunohistochemical study. J Urol. 1988;139(4):857–861. [DOI] [PubMed] [Google Scholar]
- 33. Bodnar RJ. Epidermal growth factor and epidermal growth factor receptor: The Yin and Yang in the treatment of cutaneous wounds and cancer. Adv Wound Care (New Rochelle). 2013;2(1):24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31(6):637–643. [DOI] [PubMed] [Google Scholar]
- 36. Di Lorenzo G, Tortora G, D’Armiento FP, et al. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin Cancer Res. 2002;8(11):3438–3444. [PubMed] [Google Scholar]
- 37. Hofer DR, Sherwood ER, Bromberg WD, Mendelsohn J, Lee C, Kozlowski JM. Autonomous growth of androgen-independent human prostatic carcinoma cells: role of transforming growth factor α. Cancer Res. 1991; 51: 2780–2785. [PubMed] [Google Scholar]
- 38. Toren P, Zoubeidi A. Targeting the PI3K/Akt pathway in prostate cancer: challenges and opportunities (review). Int J Oncol. 2014;45(5):1793–1801. [DOI] [PubMed] [Google Scholar]
- 39. Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu L, Dong X. Complex impacts of PI3K/AKT inhibitors to androgen receptor gene expression in prostate cancer cells. PLoS One. 2014;9:e108780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–221. [DOI] [PubMed] [Google Scholar]
- 43. Sionov RV, Haupt Y. The cellular response to p53: The decision between life and death. Oncogene. 1999;18(45):6145–6157. [DOI] [PubMed] [Google Scholar]
- 44. Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13(2):293–325. [DOI] [PubMed] [Google Scholar]
- 45. Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lei Q, Jiao J, Xin L, et al. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9(5):367–378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement_figure_1_(TCRT-19-0256) for Deletion of NKX3.1 via CRISPR/Cas9 Induces Prostatic Intraepithelial Neoplasia in C57BL/6 Mice by Jin Ju Park, Ji Eun Kim, Yoon Jeon, Mi Rim Lee, Jun Young Choi, Bo Ram Song, Ji Won Park, Mi Ju Kang, Hyeon Jun Choi, Su Ji Bae, Ho Lee, Byeong Cheol Kang and Dae Youn Hwang in Technology in Cancer Research & Treatment
Supplement_figure_2_(TCRT-19-0256) for Deletion of NKX3.1 via CRISPR/Cas9 Induces Prostatic Intraepithelial Neoplasia in C57BL/6 Mice by Jin Ju Park, Ji Eun Kim, Yoon Jeon, Mi Rim Lee, Jun Young Choi, Bo Ram Song, Ji Won Park, Mi Ju Kang, Hyeon Jun Choi, Su Ji Bae, Ho Lee, Byeong Cheol Kang and Dae Youn Hwang in Technology in Cancer Research & Treatment