Abstract
Many platform chemicals can be produced from renewable biomass by microorganisms, with organic acids making up a large fraction. Intolerance to the resulting low pH growth conditions, however, remains a challenge for the industrial production of organic acids by microorganisms. Issatchenkia orientalis SD108 is a promising host for industrial production because it is tolerant to acidic conditions as low as pH 2.0. With the goal to systematically assess the metabolic capabilities of this non-model yeast, we developed a genome-scale metabolic model for I. orientalis SD108 spanning 850 genes, 1826 reactions, and 1702 metabolites. In order to improve the model’s quantitative predictions, organism-specific macromolecular composition and ATP maintenance requirements were determined experimentally and implemented. We examined its network topology, including essential genes and flux coupling analysis and drew comparisons with the Yeast 8.3 model for Saccharomyces cerevisiae. We explored the carbon substrate utilization and examined the organism’s production potential for the industrially-relevant succinic acid, making use of the OptKnock framework to identify gene knockouts which couple production of the targeted chemical to biomass production. The genome-scale metabolic model iIsor850 is a data-supported curated model which can inform genetic interventions for overproduction.
Keywords: Genome-scale model, Non-model yeast, Constraint-based modeling, Metabolic engineering
Highlights
-
•
Genome-scale metabolic model iIsor850 describes metabolism of I. orientalis SD108.
-
•
Customized biomass reaction highlights differences with S. cerevisiae.
-
•
Chemostat data elucidate growth-associated ATP maintenance.
-
•
Substrate utilization and CRISPR/Cas9 gene knockout phenotypes validate model.
-
•
Model pinpoints candidate gene deletions coupling succinic acid production to growth.
List of abbreviations
- FBA –
Flux Balance Analysis
- FCF –
Flux Coupling Finder
- FVA –
Flux Variability Analysis
- GAM –
Growth-associated Maintenance
- GEM –
Genome-scale model
- GSM –
Genome-scale metabolic
- GPR –
Gene-Protein-Reaction
- NGAM –
Non-growth-associated Maintenance
1. Introduction
Over a decade ago, the US Department of Energy identified twelve building block chemicals that can be produced from sugars, which can be subsequently converted to a number of high-value bio-based chemicals or materials (Werpy and Petersen, 2004). These platform chemicals remain relevant to date and include eight blocks of organic acids which range in length from three to six carbons. The first of these blocks, the 1,4-dicarboxylic acids (i.e., succinic, malic and fumaric acid), has the potential to be a key building block for deriving both commodity and specialty chemicals. For example, succinic acid (formally named butanedioic acid) can readily be converted to polymer precursors such as 1,4-butanediol, N-methyl-2-pyrrolidone, tetrahydrofuran and γ-butyrolactone. Additional acids with multiple applications include 3-hydroxypropionic acid, 2,5-furandicarboxylic acid, itaconic acid, levulinic acid and muconic acid.
Any industrially-viable process relying on a microbial platform to produce this class of compounds will both have to produce efficiently (i.e., with a high yield from sugar) the desired organic acid along with tolerating the low pH associated with a high titer (Abbott et al., 2009). The yeast Issatchenkia orientalis has been proposed to be an excellent candidate since it exhibits both acid and ethanol tolerance (Okuma et al., 1986). I. orientalis is also known as Pichia kudriavzevii, Candida glycerinogenes and Candida krusei (Kurtzman et al., 2008) with a recent study indicating that 32 isolates of P. kudriavzevii and C. krusei are the same species with collinear genomes 99.6% identical in DNA sequence (Douglass et al., 2018). Moreover, strains have been isolated which degrade malic acid (Seo et al., 2007) and grow at a pH of 2.5 on hemicellulosic and cellulosic oligosaccharides obtained by two-step extraction with sulfuric acid from six plant sources (Thalagala et al., 2009). Strains also produce ethanol in media containing 5% sodium sulfate at pH 2.0 (Hisamatsu et al., 2006). Furthermore, recombinant I. orientalis strains can produce 15–20 g/L lactic acid under anaerobic conditions in an unbuffered medium at a pH of 2 (Suominen et al., 2012) and succinic acid with a titer of 11.6 g/L (Xiao et al., 2014). A recent isolate that was subsequently genetically modified and adapted to high lactic acid concentrations was reported to produce as much as 154 g/L d-lactic acid at a pH of 4.7 with the addition of calcium hydroxide (Park et al., 2018).
The strain I. orientalis SD108, which was used as a host for succinic acid overproduction, was found to be tolerant to high levels of succinic acid, itaconic acid, adipic acid, and acetic acid (Xiao et al., 2014), making it an attractive alternative organism for the production of industrially-relevant organic acids. Although it is not as well-studied as the model yeast Saccharomyces cerevisiae, new genetic tools have been developed (Tran et al., 2019; Cao et al., 2020). Advances in domesticating non-model microorganisms, including yeasts, have recently been reviewed (Fatma et al., 2020). As the first eukaryotic genome to be fully sequenced and annotated (Goffeau et al., 1997), S. cerevisiae has a rich history of reconstructed genome-scale metabolic models (Forster et al., 2003; Duarte et al., 2004; Kuepfer et al., 2005; Nookaew et al., 2008; Herrgard et al., 2008; Mo et al., 2009; Dobson et al., 2010; Zomorrodi and Maranas, 2010; Heavner et al., 2012, 2013; Osterlund et al., 2013; Aung et al., 2013) that was recently reviewed (Lopes and Rocha, 2017). Genome-scale metabolic reconstructions are already in place for a growing number of eukaryotic, prokaryotic and archaeal species (O’Brien Edward et al., 2015; Kim et al., 2017) and can be used to evaluate the capabilities of microbial chemical production (Purdy and Reed, 2017). Such curated metabolic reconstructions codify the gene-protein-reaction associations for the biochemical transformations and transport of metabolites that take place in a cell. The resulting genome-scale models globally track mass flows and balance redox reactions through metabolism.
The principles of genome-scale metabolic reconstructions and the mathematical modeling processes have been reviewed extensively before (Feist et al., 2009; Oberhardt et al., 2009; Thiele and Palsson, 2010). Certain steps require particular care. During the process of generating a genome-scale metabolic reconstruction, it is informative to evaluate the network topology for metabolites unreachable under any uptake condition. Optimization tools such as GapFind can automatically identify metabolites which are disconnected from the rest of metabolism and GapFill can generate hypotheses to reconnect them (Satish Kumar et al., 2007). In addition, growth inconsistencies may exist between model predictions and in vivo phenotypes. Substrate utilization prediction inconsistencies can be addressed systematically using optimization tools such as GrowMatch to suggest specific additions or removals of functionality in the model (Kumar and Maranas, 2009). Often, additional experimental pieces of evidence are needed to accept the growth restoring function into the model. These processes should be performed stringently so that gaps are filled only in order to ensure the production of core biomass precursors. Care must be exercised, as non-native functions erroneously appended into the model can adversely affect predictions. Deficiencies in model quality may also arise, in part, because of lack of complete mass balance resulting from non-standardized biomass reactions (Chan et al., 2017). Once a curated genome-scale metabolic reconstruction is in place, optimization tools such as OptKnock (Burgard et al., 2003), OptGene (Patil et al., 2005), OptStrain (Pharkya et al., 2004), GeneForce (Barua et al., 2010) or OptForce (Ranganathan et al., 2010; Ranganathan and Maranas, 2010) can aid in blueprinting successful engineering interventions in microbial strains that lead to the targeted overproduction of desired metabolites (Wang et al., 2017).
Curated genome-scale reconstructions for Candida glabrata (Xu et al., 2013), Pichia pastoris (Chung et al., 2010; Sohn et al., 2010; Caspeta et al., 2012; Tomas-Gamisans et al., 2016), Scheffersomyces stipites (Caspeta et al., 2012; Balagurunathan et al., 2012; Liu et al., 2012), Schizosaccharomyces pombe (Sohn et al., 2012), Yarrowia lipolytica (Loira et al., 2012; Pan and Hua, 2012; Kavscek et al., 2015; Kerkhoven et al., 2016), and Rhodosporidium toruloides (Tiukova et al., 2019; Dinh et al., 2019) are already available. These models offer templates for reconstructing metabolic models for other yeast species. However, no metabolic models for any species of I. orientalis or P. kudriavzevii have been reported in the literature to date. The sequenced genome for I. orientalis SD108 is available (Xiao et al., 2014) and informs the model reconstruction effort presented herein. Filling in this gap with a curated metabolic reconstruction also helps with the KBase effort (Arkin et al., 2018) to catalog and automate the building of fungal models.
In this work, we highlight the development of a genome-scale metabolic (GSM) reconstruction for I. orientalis SD108, referred to hereafter as iIsor850, which contains 850 genes, 1826 reactions, 1702 metabolites, and 874 unique metabolite species distributed between 14 compartments. We used the GSM model Yeast 7.6 of S. cerevisiae (Aung et al., 2013) combined with the KBase database (Arkin et al., 2018) to initiate the draft reconstruction, and reevaluated the model in the light of Yeast 8.3.4 S. cerevisiae model updates. The model was subsequently subjected to network connectivity analysis which suggested refinements in the genes and reactions present, with updated gene-protein-reaction (GPR) associations created for 100 genes, non-homologous to those in the GSM Yeast 8.3.4. A new biomass description was formulated from macromolecular composition measurements performed in this study, and voids in its detailed composition were approximated by the data available in yeast 7.6 (Aung et al., 2013). ATP maintenance requirements were calculated from carbon-limited chemostat data also generated in the current study. We examined the consistency of model predictions of growth phenotypes for carbon and nitrogen substrate utilization as well as those of gene deletions we performed using clustered regularly interspaced short palindromic repeats (CRISPR) Cas9 gene editing tools. After these validations, the model was subsequently used to pinpoint genetic interventions, through the application of OptKnock, that results in the overproduction of succinic acid. Components of these solutions share similarity to approaches implemented in vivo for other organisms, which supports that the model codifies the relevant biochemistry. The model iIsor850 enables the use of metabolic engineering computational tools towards improving I. orientalis SD108 for the production of industrially-relevant organic acids.
2. Materials and methods
2.1. Draft reconstruction from existing fungal genome-scale reconstructions and model refinements
In general, the workflow used followed an established protocol for generating metabolic models utilizing previously built metabolic models for closely related organisms (Mueller et al., 2013). This protocol provides a priority structure for assigning functions to genes using a multiple source annotation workflow. Briefly, the recent genome sequence and gene annotations of I. orientalis SD108 were used for this reconstruction (Xiao et al., 2014). An initial draft reconstruction was assembled by mapping genes and reactions from the S. cerevisiae genome-scale model Yeast 7.6 (Aung et al., 2013), updated with additional information from (Chowdhury et al., 2015). First, homologous genes were determined by bidirectional protein BLAST (Sayers et al., 2020) with an E-value cutoff of 10−5 (Mueller et al., 2013). Next, we evaluated the gene-protein-reaction (GPR) association in Yeast 7.6 using these bidirectional hits and added a reaction to the draft model only if its GPR association was satisfied by the determined gene homologs necessary for a functional protein. In this way, we initially only transferred reactions with sufficient GPR associations. We further extended the draft reconstruction using the KBase “build fungal model” application (Arkin et al., 2018), which extracted homologous genes and associated reactions from a library of fungal genome-scale models using homologous genes identification schematics similar to what we used with the Yeast 7.6 model. We built the initial scaffold using Yeast 7.6 model rather than KBase because the biochemical information in Yeast 7.6 was experimentally verified whenever possible and we wished to draw upon that literature as much as possible. Next, additional reactions and GPRs were manually added using the annotated genome and validated with NCBI’s Conserved Domain Database (Marchler-Bauer et al., 2015). When necessary, missing assignments of reaction compartments were addressed using the protein subcellular localization prediction software DeepLoc (Almagro Armenteros et al., 2017). We followed the BiGG Models knowledgebase (Norsigian et al., 2020) for reaction and metabolite identifiers (BiGG ID) where possible, and adopted a naming scheme for metabolites that indicated compartmentalization.
We next updated refined the draft model to improve its quality. Specifically, we ensured that every reaction (excluding pseudo and exchange reactions) is mass and charge balanced and as a result we updated metabolite formulae and reaction stoichiometries using standardized metabolite formulae from MetaCyc (Caspi et al., 2018) and ModelSEED (Henry et al., 2010) databases. Furthermore, some GPR associations of S. cerevisiae Yeast 7.6 reactions were recently updated in the yeast model repository (https://github.com/SysBioChalmers/yeast-GEM, version 8.3.4); we examined these updated and modifications were made to the GPR associations. Furthermore, we identified and fixed thermodynamically infeasible cycles, including cycles that allow the unbounded production of ATP. Such cycles were eliminated by simply blocking the reverse direction of the ATP hydrolysis reaction and making the overall reaction irreversible (Fritzemeier et al., 2017). Finally, after all updates described in subsequent sections, we checked the model structure and consistency using the MEMOTE test suite (Lieven et al., 2020) with the model annotations standardized to the MIRIAM namespace (Juty et al., 2012), which is used by MEMOTE. We iteratively addressed flagged issues, and the final version of the model passed all MEMOTE tests.
2.2. Experimental determination of biomass composition
Biomass composition was determined for I. orientalis SD108 cultivated in chemostat using carbon as the limiting nutrient. An overnight stock was prepared in minimal medium that contains yeast nitrogen base without amino acids (YNB, Sigma Y0626) and 20 g/L glucose. It was then inoculated at 1:100 from overnight culture into 250 mL culture to grow in continuous mode in a 500-mL chemostat (Sixfors; Infors AG, Bottmingen, Switzerland) using YNB with 0.8 g/L glucose. The culture was allowed to grow overnight before turning on dilution. Temperature was maintained at 30 °C. Moisturized air was delivered at a flow rate of 20 Nl/h. The culture was stirred at 400 rpm for sufficient oxygen, and was kept at 0.1 h−1 growth rate. After the culture had reached steady state (as determined by pH and cell density which were observed to be stable for at least 12 h), it was harvested for the biomass measurement. The pH in the vessel was not controlled; the starting pH was 5.2, and the final pH was 3.5.
DNA was measured using diphenylamine reagent. 7.5 mL culture was pelleted and washed by 1 mL cold 1 mM HClO4. Serial dilutions of 1 mg/mL Calf thymus DNA (Sigma) were prepared for calibration. Samples were hydrolyzed in 500 μL 1.6 M HClO4 for 30 min at 70 °C, and then reacted with 1 mL diphenylamine reagent (0.5 g diphenylamine in 50 mL glacial acetate, 0.5 mL 98% H2SO4, and 0.125 mL 3.2% acetaldehyde water solution) at 50 °C for 3 h. After centrifugation, the supernatant was taken for OD600 measurement.
RNA was measured by 260 nm absorption. Basically, 2.5 mL culture was pelleted and washed, and digested with 300 μL 0.3M KOH at 37 °C for 60 min. DNA and protein were then precipitated by 100 μL 3M HClO4. Supernatant was taken and precipitate was washed with 600 μL 0.5M HClO4. Absorption of combined supernatant at 260 nm, 900 nm, and 977 nm was measured. RNA concentration was calculated as 5.6·(A260-A260,blank)/(A977-A900) in μg/mL.
Protein was measured using the Biuret method. Briefly, 2.5 mL culture was pelleted, washed and boiled in 100 μL 3M NaOH at 98 °C for 5 min. After cool down, the mixture was reacted with 100 μL CuCO4 for 5 min at room temperature. After centrifugation, supernatant was taken for determination of 555 nm absorption, which was calibrated by serial dilution of Bovine serum albumin (BSA) solutions (Thermo).
Lipid was determined by measuring saponified fatty acids using LC-MS. Briefly, cell pellet from 2.5 mL culture was extracted and saponified in 1 mL 0.3M KOH–MeOH solution at 80 °C for 60 min, then neutralized by 100 μL formic acid, and then extracted by 1 mL hexane 20 nM. 20 nM isotope-labeled fatty acid standards (U–13C–C16:0, U–13C–C18:1, U–13C–C18:2; Cambridge Isotope) were added before saponification as internal standards. Extracted fatty acids were dried under N2 and redissolved in 200 μL acetonitrile:methanol (1:1), and then analyzed by reversed-phase C8 column chromatography coupled to negative-ion mode, full-scan high-resolution LC-MS (Exactive, Thermo).
Carbohydrate was determined by hydrolyzing cell pellet from 2.5 mL culture in 100 μL 2M HCl at 80 °C for 1 h. 0.4 mg U–13C-glucose was added as internal control before hydrolyzation. The lysate was neutralized by 100 μL 2M NH4HCO3, diluted in 1.8 mL 80% MeOH and centrifuged. The supernatant was taken and analyzed by negative-ion mode LC-MS equipped with hydrophilic interaction liquid chromatography (Q Exactive Plus, Thermo). Mass peaks equivalent to C6H12O6 were selected for quantification.
2.3. Generation of biomass reaction
The original Yeast 7.6 biomass reaction was used as the starting point for the biomass reaction. We used the biomass macromolecular composition generated in this study (see above) alongside data from literature to determine the metabolite coefficients in the biomass objective function (specifically for carbon limitation conditions) (see Supplementary Materials 3 for details). We also incorporated other experimentally determined biomass specifications including genome GC content (i.e., 38.33%) (Douglass et al., 2018), and relative abundance of acyl groups and free fatty acids in lipids (this study). We took experimentally determined specifications from S. cerevisiae; specifically amino acid, inorganic compounds (i.e., phosphate, sulfate, and metal ions), and cell wall compositions (Klis et al., 2014), as corresponding data for I. orientalis SD108 were not available. Additional data adopted from the yeast 7.6 model were lipid subspecies composition (e.g., phosphatidylinositol, phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine composition). These data from experiments on S. cerevisiae and Yeast 7.6 were deemed acceptable as I. orientalis and S. cerevisiae are closely related. Without measurements of the soluble metabolite pool, we set the coefficients of twelve cofactors and prosthetic groups to a small number (i.e., to 10−4) so as to impose a biosynthesis requirement on the in silico model; similar measures have been previously adopted for the biomass reactions in other models such as those for S. cerevisiae models (Mo et al., 2009), including Yeast 7.6 (Aung et al., 2013). Metabolite coefficients associated with growth-associated ATP maintenance were also updated (see below). Calculations and detailed listings of metabolites and coefficients in the biomass reaction are provided in Supplementary Materials 3.
2.4. Determination of ATP maintenance requirements
Growth associated ATP maintenance (GAM) and non-growth associated ATP maintenance (NGAM) values were determined using ministat culture data collected as part of this study. The 20 mL culture volume in 55 mL vessel ministat design was adopted from (Miller et al., 2013). We used the same glucose limited conditions as for the chemostat and the same inoculation procedure. Temperature was maintained at 30 °C. Moisturized air was delivered at a flow rate of 7.5Nl/h per vessel. Steady state was assessed with OD and pH, which were observed to be stable for at least 12 h before sampling. We measured the glucose uptake rates over a range of dilution rates (i.e., from 0.11 hr−1 to 0.65 hr−1) by sampling 1 mL culture from a ministat, from which 150 μL supernatant was used to measure glucose by YSI 2900D Biochemistry Analyzer (YSI, Yellow Springs, OH, USA). We used one biological replicate for each dilution rate and performed technical replicates for error analysis. For OD to gDW conversion, we summed the measured biomass components (using the methods described in 4.2) to get gDW for equivalent 1 mL cell having 1 OD. To calculate the ATP maintenance requirement per experimental data point, glucose uptake rate (vglc) was set to the experimentally measured values and growth rate (vbiom) was determined by conversion from the experimentally measured range of dilution rates. We then set the NGAM and GAM demand in the model to zero. Next, ATP maintenance requirement was given by the ATP hydrolysis rate (vatpm) which was found for each dilution rate by constraining the model at the experimental vglc and vbiom values and then maximizing the flux through the reaction ATP + H2O → ADP + H+ + HPO42−. Using linear regression, we estimated the GAM and NGAM as the slope and intercept, respectively, of vatpm as a function of growth rate. We then set the intercept (i.e., NGAM) to a value of 1 as in Yeast 8.3.4 and estimated the GAM as the slope (see Supplementary Materials 4). For these carbon limitation conditions, we measured the compositional profile at the lowest and highest growth rates (data not shown). The results remained relatively constant and in agreement with the composition used during the model biomass reaction construction; thus, no adjustments were made to the biomass reaction whilst determining the GAM value.
2.5. Modeling simulations
Flux balance analysis (FBA) was used throughout the process for model validation and prediction stages (Orth et al., 2010). Model simulations of growth phenotypes were obtained using FBA with the objective of maximizing the biomass reaction (vbiom) whose flux corresponds to the growth rate. During initial testing and model predictions of positive growth on all substrates, the carbon substrate uptake rate was set to a value 3.3 mmol gDW−1 hr−1; we chose this value as a rough estimate for glucose uptake from data in (Xiao et al., 2014) and arbitrarily applied it to each carbon substrate. For subsequent quantitative comparisons with in vivo data, the substrate specific consumption rates (g gDW−1 hr−1), such as glucose (vglc), were set to the values determined experimentally in the current work. For growth predictions involving rich media, supplementary compound uptake rates were set to 0.165 mmol gDW−1 hr−1 (i.e., 5% of default substrate uptake rate of 3.3 mmol gDW−1 hr−1). Simulating rich media components has been described previously (Dinh et al., 2019). In general, the supplementary nutrients present in YNB included thiamine, riboflavin, nicotinate, pyridoxin, folate, (R)-pantothenate, 4-aminobenzoate, and myo-inositol. The undefined composition of yeast extract in Yeast-Peptone-Dextrose (YPD) media was assumed to be that of YNB media plus 20 amino acids and d-glucose. Oxygen and ammonium uptake rates were unconstrained in all simulations unless otherwise noted. Glucose uptake rate was set to 10.0 mmol gDW−1 hr−1 during OptKnock simulations. Oxygen uptake rate was limited to 21.0 mmol gDW−1 hr−1 during OptKnock simulations except during microaerobic conditions, as noted. Gene knockout was translated to the corresponding reaction(s) knockout by examining the Boolean GPR associations, and the reaction was knocked out in the model by setting the corresponding upper and lower flux bounds both to zero. A gene was classified as essential if the maximal growth rate of the corresponding knockout mutant was calculated by FBA to be less than 0.001 hr−1. FBA calculations, Flux Coupling Finder, and OptKnock were implemented using GAMS 24.8.5 using IBM ILOG CPLEX solver on the high-performance computing resource cluster of Pennsylvania State University’s Institute for CyberScience Advanced CyberInfrastructure (ICS-ACI), Roar. Production envelopes were computed using the COBRApy package (version 0.13.4) (Ebrahim et al., 2013) and used IBM ILOG CPLEX solver (version 12.9).
2.6. Batch growth of I. orientalis SD108
YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) was used for routine growth of I. orientalis SD108. Growth rates of I. orientalis SD108 were tested in minimal medium (MM) using different C/N ratios (20 g/L d-glucose, 1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, 0.4–7.1 g/L NH4Cl, C/N = 5:1–90:1, 25 mM Na2HPO4, 150 mM KH2PO4, pH 5.6). Stationary phase I. orientalis SD108 seed cultures were obtained by inoculating single colonies from a YPD agar plate into 25 mL YPD liquid medium in 125 mL baffled flask. For growth, the seed cultures were then used to inoculate into 50 mL minimal medium in 250 mL baffled flask with a starting OD600 of 1. The cells were then grown at 30 °C and 250 rpm. All experimental conditions were performed with four replicates. Growth viability on carbon substrates was tested in minimal media with different carbon substrates as the sole carbon source at 20 g/L unless indicated otherwise. Cell growth was measured by the absorbance at 600 nm using cell density meter. Dry cell weights (CW) were determined as follows. The 1–5 mL of culture samples were collected into pre-weighed tubes and centrifuged at 16,000×g for 5 min. Supernatant was discarded, and pellets were then washed twice with 50 mM phosphate buffered saline. Washed pellets were dried till constant weight at 65 °C for 24–48 h and the tubes were then weighed.
For supernatant composition analysis, I. orientalis wild type or mutant strains were grown in YPD medium containing 2% glucose for overnight at 30 °C and 250 rpm. Next, cells were washed with deionized water and inoculated in 20 mL of respective medium with the initial OD600 of 0.1. For different carbon substrate utilization, YNB medium was supplemented with 2% carbon source (glucose, glycerol, lactic acid, succinic acid, citric acid, ethanol, fructose, xylose) and cell sample was harvested at various time point and supernatant was analyzed for carbon source, organic acid, and alcohol by HPLC (Agilent Technologies 1200 Series, Santa Clara, CA). The HPLC was equipped with a RezexTM ROA-Organic Acid H+ (8%) column (Phenomenex Inc., Torrance, CA) and a refractive index detector (RID). The column was eluted with 0.005 N H2SO4 at a flow rate of 0.6 mL/min at 50 °C (Liu et al., 2019).
2.7. Construction of mutant strains
For construction of mutant strain, we used the CRISPR/Cas9 system developed recently for I. orientalis (Tran et al., 2019). We used the pVT15b plasmid containing improved Cas9 from Streptococcus pyrogens, URA3 expression cassette from I. orientalis; E. coli selection marker and origin of replication; and ARS from S. cerevisiae. The spacer was designed using Benchling (https://benchling.com), and the spacer and HR donor were ordered as gBlocks from Integrated DNA Technologies (IDT, Coralville, IA) and further assembled into CRISPR/Cas9 plasmid using BsaI restriction site by Golden Gate assembly method. Cloned plasmid was further confirmed through the restriction digestion using BsaI and EcoRI restriction enzymes.
For the transformation in I. orientalis, a colony of I. orientalis was inoculated in 2 mL of YPAD for overnight at 30 °C and 250 rpm. Next day culture of I. orientalis was diluted to an initial OD600 of 0.2 and cells were continuously grown until they reached to OD600 of 0.8–1. Cells were collected by centrifugation, washed twice with deionized water, and resuspended in 360 μL of transformation mixture containing 240 μL of 50% w/v PEG3350, 36 μL of 1 M lithium acetate, 50 μL of 2 mg/mL deoxyribonucleic acid from salmon testes (SS-DNA) that was boiled at 100 °C for 5 min and quickly chilled on ice, plasmid (500 ng), and deionized water. After mixing thoroughly, the suspension was subjected to heat shock for 1 h at 42 °C. After the heat shock, cells were collected by gentle centrifugation and spread on the SC-Ura plates. After 3–4 days, when colony appears, deletion of the genes was confirmed through the sequencing or PCR using appropriate primers. A list of strains used in this work are provided in Table 3.
Table 3.
List of strains and plasmids used in this study.
| Strains/Plasmids | Description | Sources |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning host | NEB |
| I. orientalis SD108 | Wild type | Xiao et al., 2014 |
| SD108ΔUra | URA3Δ, host for plasmids in this study | Xiao et al., 2014 |
| SD108Δ2223 | SD108 with disruption of JL09_g2223 | This study |
| SD108Δ850 | SD108 with disruption of JL09_g850 | This study |
| SD108Δ2279 | SD108 with disruption of JL09_g2279 | This study |
| SD108Δ1386 | SD108 with disruption of JL09_g1386 | This study |
| SD108Δ1702 | SD108 with disruption of JL09_g1702 | This study |
| SD108Δ4163 | SD108 with disruption of JL09_g4163 | This study |
| SD108Δ4262 | SD108 with disruption of JL09_g4262 | This study |
| SD108Δ3351 | SD108 with disruption of JL09_g3351 | This study |
| SD108Δ3078 | SD108 with disruption of JL09_g3078 | This study |
| Plasmids | ||
| pVT15b-epi | CRISPR/Cas9 plasmid, containing ScARS, IoURA3, iCas9, RPR1 promoter, and sgRNA scaffold. Used for cloning of guide RNA and homology arm | Tran et al., 2019 |
| pVT15b-XYZ | pVT15b plasmid carrying the homology arm and guide RNA against any one of these genes: g2223, g850, g2279, g1386, g1702, g4163, g4262, g3351, or g3078 |
This study |
3. Results and discussion
3.1. Properties of the model and refinements of the reconstruction
The genome-scale model iIsor850 incorporates relevant yeast biochemistry that has been systematically cataloged into previous genome-scale models for S. cerevisiae (i.e., Yeast 7.6 and subsequent update Yeast 8.3.4 (Aung et al., 2013)) and other KBase fungal models (Arkin et al., 2018)). It augments this knowledgebase with unique metabolic content culled from I. orientalis genome annotation (Xiao et al., 2014) and the literature. iIsor850 is informed by experimental data on growth rates generated in the current work. A majority of the model components, including 88% of genes, 78% of reactions, and 75% of metabolites, are biochemically equivalent to those in S. cerevisiae Yeast 7.6 model. An additional 7% of genes, 5% of reactions, and 6% of metabolites were incorporated from KBase and its “build fungal model” application which contains metabolic reaction information for non-model yeasts. The genome annotation and manual model additions and modifications yielded the remainder of the model (i.e., 5% of the genes and 17% of the reactions). Some of these manual modifications indirectly incorporate reactions included in a different form in the Yeast 7.6 model. For example, variants of diacylglycerol acyltransferase which catalyze reactions involving 32 variants of triacylglycerol in Yeast 7.6 were streamlined into a single reaction in order to more economically reflect the current level of biochemical detail known for I. orientalis. Note that for the sake of convenience 805 out of a total 850 genes in the model are referred to by both the I. orientalis gene locus ID and the corresponding homolog gene name in S. cerevisiae (not all of which are in the Yeast 8.3.4 model) and the remaining 44 absent homologs in S. cerevisiae referred to only by the I. orientalis gene locus ID. By using a more recent version of the consensus yeast model available at https://github.com/SysBioChalmers/yeast-GEM (i.e., Yeast 8.3.4), we updated the GPR assignments for 19 reactions, such as adding an isozyme to the GPR of phosphoglycerate mutase. Unfortunately, we were able to assign GPR associations for only 20% of the transport reactions, which in large part reflects the similar level of lack of knowledge as in the Yeast 8.3.4 model. However, we were able to assign GPR associations to 94% of the metabolic reactions that span all 14 compartments in the model. The statistics of the curated model iIsor850 are summarized in Table 1.
Table 1.
Statistics for genome-scale model iIsor850.
| Properties | Statistics |
|---|---|
| Genesa | 850 |
| % of chromosomal ORFs | 17% |
| without homolog included in Yeast 8.3.4 | 100 |
| without identified S. cerevisiae homolog | 44 |
| Reactions | 1826 |
| Metabolic reactions | 1066 |
| with GPR assigned | 993 |
| Unique metabolic reactions | 1040 |
| Transport reactions | 587 |
| Extracellular transport | 160 |
| with GPR assigned | 71 |
| Intracellular transport | 419 |
| with GPR assigned | 83 |
| Exchange reactions | 170 |
| Metabolites | 1702 |
| Unique metabolites | 874 |
| Compartments | 14 |
The model description for iIsor850 also contains pseudo-genes used to denote spontaneous reactions and those reactions with unknown associated enzymes, which have been excluded from the gene counts.
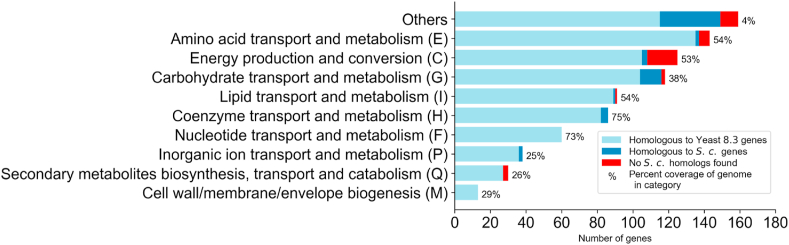
We used eggNOG (Huerta-Cepas et al., 2018), which clusters predicted orthologs (i.e., eukaryotic orthologous groups or KOGs (Koonin et al., 2004)), to classify the genes in the model in order to better highlight those that were or were not homologous to genes in S. cerevisiae and to visualize the metabolic capabilities of the model. Fig. 1 summarizes the final KOG classifications for model iIsor850, as well as the overall coverage of the model of the classifications. Genes that are homologous to those in S. cerevisiae are not evenly distributed in the various functional categories. The categories with the highest fraction of homologous genes include nucleotide transport and metabolism; inorganic ion transport and metabolism; cell wall/membrane/envelope biogenesis; and coenzyme transport and metabolism. The model iIsor850 includes a number of genes without homology to genes in S. cerevisiae which are also dispersed in many categories, including energy production and conversion; carbohydrate transport and metabolism; secondary metabolite biosynthesis, transport and catabolism; and amino acid transport and metabolism. For example, unlike S. cerevisiae, I. orientalis encodes for the complete Complex I NADH dehydrogenase whose 8 subunits are incorporated into iIsor850. In general, these non-homologous genes were included into the model via KBase, the genome annotations and the Conserved Domain Database at NCBI to incorporate pathways, and information on reactions not present in Yeast 8.3.4, such as a heptadecenoyl-CoA synthesis (fatty acid C17:1) reaction. This process was driven by experimental measurements. Unlike S. cerevisiae, I. orientalis contains an ergosterol synthase variant which intrinsically has decreased susceptibility to fluconazole, an anti-fungal drug (Morio et al., 2017; Orozco et al., 1998). The model also uncovered that I. orientalis has genes encoding enzymes that catalyze the entry reactions in biotin formation, which are not present in Yeast 7.6, as many strains of S. cerevisiae do not have corresponding functional genes. We note that 14 genes, including 5 with no homology to genes in S. cerevisiae, were placed in the unassigned category, reflecting the challenge inherent in KOG assignment for non-model organisms. Analysis also revealed as many as 45 reactions for which the number of associated (putative) paralogs is greater in iIsor850 than in Yeast 8.3.4 (Supplementary Materials 1). For instance, 4-aminobutyrate aminotransferase activity is predicted to be encoded by JL09_g1205 or JL09_g1354, whereas Yeast 8.3.4 only has YGR019W associated with this reaction.
Fig. 1.
Classification of I. orientalis SD108 genes contained in iIsor850. Eukaryotic orthologous groups (KOG) annotations (see https://genome.jgi.doe.gov/Tutorial/tutorial/kog.html for details) were generated by analyzing the complete annotated genome of I. orientalis SD108 and then assigned to the genes in the model. Corresponding standard group abbreviations are given in parentheses for each KOG. The group marked Other compiles groups with abbreviations A, B, D, J, K, L, O, S, T, U, V, W, Y, and Z. Genes with multiple KOG assignments were added to all indicated groups. Genes without KOG assignments are not shown. Light blue bars represent genes in iIsor850 that have homologous genes in Yeast 8.3.4, as indicated by the annotations generated during the construction of iIsor850. Similarly, any genes for which the annotations indicated homologous genes in S. cerevisiae but not are contained in Yeast 8.3.4 are shown in dark blue bars. The remaining genes in iIsor850 are indicated in red bars. The percent coverage of iIsor850 of the I. orientalis SD108 genome is indicated by the numbers to the right of each bar. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Biomass composition and experimentally estimated ATP maintenance requirements
We experimentally determined the macromolecular composition of I. orientalis SD108 under carbon limitation (see Methods 4.2). Briefly, protein, carbohydrate, DNA, RNA, and lipid composition were measured for cells growing on glucose in a chemostat at a dilution rate of 0.1 hr−1, as normalized and reported in Table 2 for each category. The corresponding values used in Yeast 7.6 model are listed. Notably, both the protein and lipid fractions in the biomass composition of I. orientalis SD108 are higher than in Yeast 7.6 (i.e., 45.18% vs. 35.71% and 5.00% vs. 0.74%, respectively) whereas the carbohydrate fraction is lower (i.e., 36.80% vs. 52.27%). We also experimentally quantified the relative abundance of acyl groups and free fatty acids in the lipid fraction of the cells (see Methods 4.2). Specifically, these compounds were incorporated into the model replacing the universal free fatty acid metabolite used in Yeast 7.6 for the six free fatty acid compounds found to have an abundance of >1% by weight in the measurement of saponified fatty acids using LC-MS, viz. palmitate (C16:0), palmitoleate (C16:1), 10Z-heptadecenoate (C17:1), oleate (C18:1), linoleate (C18:2) and linolenate (C18:3). The fractional on a per mole basis compositions of these free fatty acids differ substantially from those measured for S. cerevisiae (see Supplementary Materials 2). Three of these free fatty acids (i.e., C17:1, C18:2, and C18:3) have an abundance of <1% in S. cerevisiae. In particular, the inclusion of odd-chain fatty acid C17:1 in the biomass description necessitated the addition of the pathway for producing odd-chain fatty acids, which begins with the use of propionyl-CoA instead of acetyl-CoA used for even-chain fatty acids. Odd-chain fatty acids have an abundance of less than 1% in S. cerevisiae, whereas C17:1 has an abundance of 6% in I. orientalis SD108. Odd-chain fatty acid abundances in iIsor850 are one of the most metabolically unique aspects of I. orientalis. In addition, it uses ubiquinone-7 (Kurtzman et al., 1980) as opposed to ubiquinone-6 used by S. cerevisiae or ubiquinone-9 used by R. toruloides.
Table 2.
Summary of iIsor850’s biomass composition.
| Constituentsa | Composition (%) | |||
|---|---|---|---|---|
| iIsor850 | Yeast7.6 | |||
| Proteinb | 45.18 | 35.71 | ||
| l-Alanine | l-Arginine | l-Asparagine | ||
| l-Aspartate | l-Cysteine | l-Glutamine | ||
| l-Glutamate | Glycine | l-Histidine | ||
| l-Isoleucine | l-Leucine | l-Lysine | ||
| l-Methionine | l-Phenylalanine | l-Proline | ||
| l-Serine | l-Threonine | l-Tryptophan | ||
| l-Tyrosine | l-Valine | |||
| Carbohydrate | 36.80 | 52.27 | ||
| 1,3-beta-d-Glucan | 1,6-beta-d-Glucan | Chitin | ||
| N-Glycanc | O-Glycanc | GPI-anchorc | ||
| Lipid | 5.0 | 0.74 | ||
| Episterol | Free fatty acids (6 species)d | Inositol-P-ceramide | ||
| Phosphatidylcholine | Phosphatidylethanolamine | Phosphatidylinositol | ||
| Phosphatidylserine | TAG | |||
| RNAe | 8.63 | 5.85 | ||
| ATP | CTP | GTP | ||
| UTP | ||||
| DNAe | 0.48 | 0.34 | ||
| dATP | dCTP | dGTP | ||
| dTTP | ||||
| Cofactors and prosthetic groups | 0.06 | 0.03 | ||
| S-Adenosyl-L-methionine | Biotin | Coenzyme-A | ||
| FAD | Heme A | NAD | ||
| NADP | Riboflavin | Spermidine | ||
| Tetrahydrofolate | Thiamine diphosphate | |||
| Inorganic ions | 3.85 | 5.06 | ||
| Calcium | Copper | Iron | ||
| Magnesium | Manganese | Phosphate | ||
| Potassium | Sulfate | Zinc | ||
Biomass constituents absent from Yeast 7.6 are underlined. Different representations of Yeast 7.6 biomass constituents are noted below.
Identical to those in yeast 7.6, amino acids in the biomass objective function are in charged-tRNA form, but are listed here as the uncharged amino acids.
The generic mannan (mannose-containing) metabolite in Yeast 7.6 was replaced with three specific essential cell wall components.
Six free fatty acid species were abundant (>1% weight) in growth experiments: palmitate (C16:0), palmitoleate (C16:1), 10Z-heptadecenoate (C17:1), oleate (C18:1), linoleate (C18:2) and linolenate (C18:3).
Monophosphate ribonucleic and deoxyribonucleic acids present in Yeast 7.6 were replaced with the corresponding triphosphate ones in iIsor850.
Overall, a total of 66 metabolites were included in the biomass component list for iIsor850. Overlaps and differences between the model and Yeast 7.6 are summarized in Table 2. The “generic” free fatty acid designation in Yeast 7.6 was replaced with the distinct free fatty acid compounds mentioned above. Eight nucleotide monophosphates were replaced in iIsor850 by the corresponding nucleotide triphosphates and pyrophosphate in order to directly account for DNA and RNA polymerization. Beyond the substitutions detailed above, nine cofactors and prosthetic groups (Coradetti et al., 2018) were added to the list of biomass constituents to bring the model in closer alignment with Yeast 8.3.4. Seven new inorganic ions (i.e., calcium, copper, iron, manganese, magnesium, potassium and zinc) were also included in the biomass reaction following the measurements for S. cerevisiae (Lange and Heijnen, 2001) as these ions are known to be essential (see Supplementary Materials 3). In the absence of direct measurement, the total inorganic ions composition were ported from S. cerevisiae (Lange and Heijnen, 2001), which was the approach earlier considered for another non-model yeast, R. toruloides (Dinh et al., 2019). The cofactor and prosthetic group fractions in the biomass reaction are set to small values (i.e., 10−4 each) which imposes a biosynthesis requirement on the model but keeps their total amount to only 0.06% of the total biomass by weight. We standardized the biomass composition such that the combined coefficient-weighted molecular weight of all constituents is 1 g mmol−1 in order to ensure the consistency of growth yield predictions (Chan et al., 2017). The specifics of the biomass composition used for the model can be found in Supplementary Materials 3. Although the biomass reaction in iIsor850 is customized, most of the list of biomass constituents are not unique to this model – thereby applicable for other yeasts – and could serve as a starting point for other models’ specific biomass reactions by following similar methodologies.
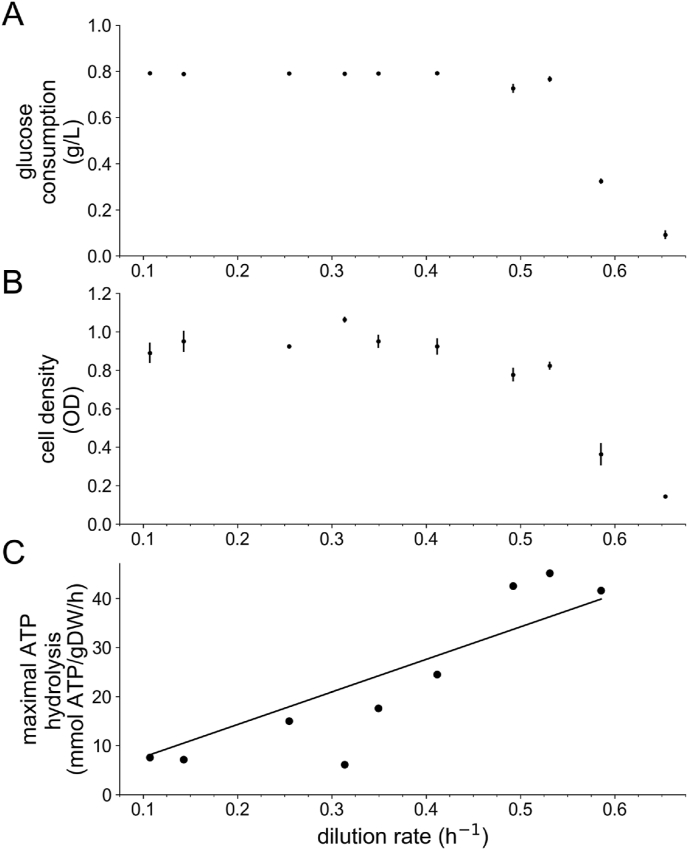
We performed additional chemostat studies under carbon limitation in order to determine the growth-associated ATP maintenance (GAM) and non-growth-associated ATP maintenance (NGAM) for I. orientalis SD108. Correctly assessing both of these maintenance values is vital when quantifying the energetic needs and growth yields properly, with GAM quantifying energetic costs that are not otherwise reflected in the biomass reaction. For this reason, I. orientalis SD108 was cultivated under carbon limitation with dilution rates ranging from 0.02 h−1 to 0.65 h−1; results for the glucose uptake rates and optical density are shown in Fig. 2 Panels A and B, respectively, and listed in Supplementary Materials 4. As observed in Fig. 2A, the glucose consumption in the chemostat is essentially constant with the dilution rate (h−1) over this range except for the highest dilution rates. We then calculated the specific glucose uptake rates (g glucose·g cell mass−1·h−1) and simulated the maximum ATP production for the model for each dilution rate, whilst imposing the corresponding measured biomass production, as shown in Fig. 2C and tabulated in Supplementary Materials 4. Although Yeast 7.6 does not contain an NGAM value, the earlier S. cerevisiae model iMM904 (Mo et al., 2009) uses a value of 1, and this value has recently been adopted into Yeast 8.3.4 as well. Assuming that the intercept (i.e., NGAM) has a value of 1 as in Yeast 8.3.4, we estimated GAM to be 66.37 mmol gDW−1, similar to that in Yeast 7.6 (59.28 mmol gDW−1). Estimated NGAM and GAM values for another non-model yeast, R. toruloides IFO0880, under carbon-limited conditions are 1.01 mmol gDW−1 hr−1 and 140.98 mmol gDW−1, respectively (Dinh et al., 2019). The much larger value for R. toruloides likely reflects that its model has more energy-requiring pathways in metabolism yet to be detailed.
Fig. 2.
ATP maintenance costs. I. orientalis SD108 was grown in a chemostat at various dilution rates and measurements of glucose uptake (A) and cell density (B) were collected. Using these data to set constraints on the glucose uptake rate and the specific growth rate, we used FBA to compute the maximal ATP production rate for each dilution rate using modified version of iIsor850 having a biomass reaction that had the coefficients of the maintenance constants set to zero (C). The resulting slope was used to estimate the growth associated ATP maintenance value in the final curated model.
3.3. Model standardization, annotations, and network analysis
Internal and external consistency are crucial for the quality of genome-scale models (Thiele and Palsson, 2010; Lieven et al., 2020). As mentioned earlier, details such as correctly setting the coefficient-weighted molecular weights in the biomass composition can adversely impact growth yield predictions. We subjected iIsor850 to the MEMOTE suite of tests (Lieven et al., 2020) to evaluate biochemical annotation and network consistency. These tests examined annotations of the model content with regard to standard databases, mass and charge balances, gaps, cycles, among others. After updating iIsor850 to resolve any flagged deficiencies, the model scored an overall 84% on the biochemical annotation and network consistency tests. All metabolites, reactions and genes have at least one annotation to a widely used database including PubChem (Kim et al., 2019), KEGG (Kanehisa et al., 2017), ModelSEED (Henry et al., 2010), ChEBI (Hastings et al., 2016), BioCyc (Karp et al., 2019), UniProt (2019) and NCBI Protein (Sayers et al., 2020), and all conform to MIRIAM guidelines (Novère et al., 2005). The deficiencies in the MEMOTE score lie in the breadth of annotations, as MEMOTE penalizes not using all listed databases (e.g., we do not have annotations to reactome (Jassal et al., 2020), MetaNetX (Moretti et al., 2016), or BRENDA (Jeske et al., 2019)). All items in the model were assigned Systems Biology Ontology (SBO) terms (Courtot et al., 2011). The model was already mass and charge balanced during the initial construction phases, although MEMOTE did assist in detecting 21 metabolites that had inconsistent charges when ignoring compartmentalization. Although reactions were locally charge balanced, in a few edge cases the charges were not identical for the same metabolite species occurring in different compartments. These localized differences arose when combining information from disparate databases, which do not all use the same reference pH value, and were updated. For the purposes of this model, we have used the reference of pH 7.2.
Although as part of the earlier curation we had already removed cycles that led to unbounded fluxes, MEMOTE identified an additional 12 thermodynamically infeasible cycles. Such cycles can adversely affect flux ranges. We resolved these issues by updating the directionality of the reactions and/or transporters, with constraining the latter being the preferred option when available. For example, MEMOTE identified that cycles could arise for 11 amino acids that could transport in and out of the vacuole to the cytosol without any driving force via the corresponding importer, exporter, and efflux transporter, respectively. Such efflux from the vacuole would likely only occur during stress of pathological conditions which trigger autophagy as a survival mechanism (Yang et al., 2006). To address this finding, we made these reactions irreversible in iIsor850 so that each of the corresponding efflux transporters were permitted to only import the amino acid into the vacuole.
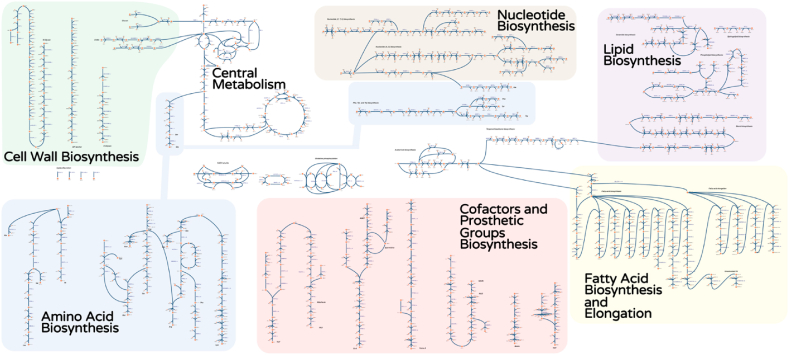
MEMOTE also reported that as many as 639 reactions are blocked, with 81 metabolites that can only be consumed and 102 metabolites that can only be produced. These results are not surprising inasmuch as we were conservative when employing GapFill to bridge network gaps through the addition of reactions, inclusion of transport pathways, and relaxation of irreversibility of reactions already present in the model. Specifically, we only reconnected metabolites with non-gene associated reactions that would ensure that iIsor850 was capable of producing biomass under all appropriate conditions. After correcting for these network gaps which impaired biomass formation, we obtained the model statistics summarized in Table 1. A visualization of the core part of the model using Escher software (King et al., 2015) is shown in Fig. 3.
Fig. 3.
Escher map of core pathways in iIsor850. An Escher map consists of metabolites (circles) connected via reactions (lines) which indicate directionality of allowable fluxes (arrows). The core components of iIsor850 were drawn and placed according to relevant classifications. For clarity, not all reactions for the metabolites are shown, and some metabolites occur more than once (i.e., cofactors). Amino acid biosynthesis is indicated in blue, nucleotide biosynthesis is brown, lipid biosynthesis is purple, cell wall biosynthesis is green, fatty acid biosynthesis and elongation is yellow, cofactors and prosthetic groups biosynthesis is red. Central metabolism is uncolored in the middle-top of the figure. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Under aerobic glucose minimal media conditions, model simulations indicate that there are 237 essential genes in the model (i.e., 28% of the included genes). These essential genes correspond to 374 essential reactions in the model (i.e., 21.2% of the reactions). The proportion of genes and reactions classified as computationally essential for Yeast 8.3.4 under the same growth conditions is lower (i.e., 13%). Unlike S. cerevisiae, however, extensive and comprehensive gene deletion studies and in vivo essentiality data do not exist for I. orientalis, and so classification into true and false positives and negatives cannot currently be determined for the essentiality predictions. We did however perform some selected gene knockouts during model validation (see below).
Using constraints corresponding to aerobic glucose minimal media conditions, we examined coupled reaction sets using the Flux Coupling Finder (FCF) (Burgard et al., 2004). For any two fluxes in a fully coupled reaction set, a non-zero flux for a given one implies a fixed non-zero value for the other member, and vice versa. Reaction flux coupling information can be helpful when designing strain optimization strategies (Reimers et al., 2015). We found that there were 81 fully coupled reaction sets spanning 352 reactions. The majority (i.e., 44) were sets that only had two members, and the bulk of these two-member sets were adjacent reactions in a linear pathway. The largest fully coupled set contained 94 reactions, including biomass. The second largest set of 28 reactions included reactions such as acetyl-CoA C-acyltransferase (octanoyl-CoA) involved in fatty acid metabolism. Other sets included three of size 9, 8, and 6 that contained stretches of linear subpathways involved in histidine metabolism, ergosterol biosynthesis, and chorismite metabolism, respectively. The remainder of the fully coupled sets (i.e., 32) had fewer than 5 reactions each. Only 7 partially coupled reaction sets occur, for which a non-zero flux for one member implies a non-zero, though variable, flux for another, and vice versa. These sets were all small, with 3 containing 3 members and 5 containing only 2 members. The reactions in these sets were involved in pathways such as fatty acid degradation in the peroxisome, pyrimidine metabolism in the cytosol, and glycerophospholipid biosynthesis in the endoplasmic reticulum membrane. The complete list of all fully, partially, and directionally coupled reactions can be found in Supplementary Materials 7. We use this reaction coupling information to streamline subsequent analyses, including designing metabolic engineering interventions to select reactions for elimination, as described below.
3.4. Substrate utilization and growth phenotype predictions for gene knockouts
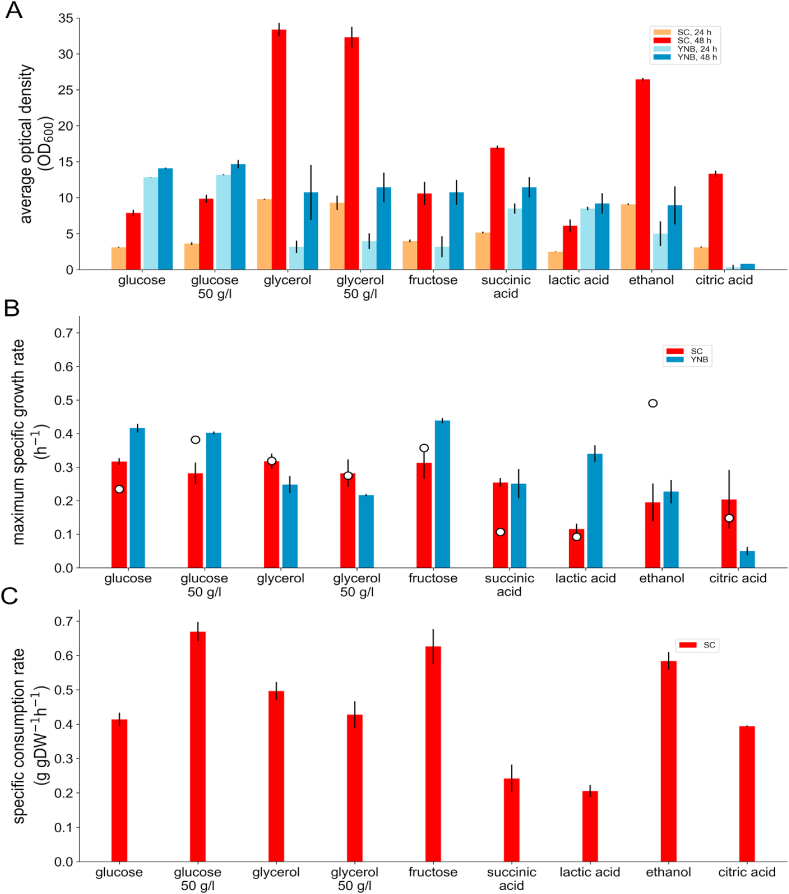
Substrate utilization data are somewhat scarce for I. orientalis, especially for substrates other than glucose (Abbott et al., 2009) or ethanol (Okuma et al., 1986). Growth/no growth have previously been tested on 34 carbon substrates and glucose with 5 nitrogen substrates (Hisamatsu et al., 2006) as well as a separate study of 26 carbon-only substrates (Seo et al., 2007). However, different strains could have differences in substrate utilization preferences (as evidenced by the differences between the results from (Hisamatsu et al., 2006) and (Seo et al., 2007)). In the former study (Hisamatsu et al., 2006), seven of the carbon substrates scored positive (i.e., glucose, lactose, glycerol, lactate, succinate, citrate, and ethanol), four were delayed (i.e., xylose, sucrose, xylitol, and glucuono-1,5-lactone), and three were weak (i.e., raffinose, melezitose, and propane 1,2-diol). In the latter study (Seo et al., 2007), six scored positive (i.e., same as the positives in study (Hisamatsu et al., 2006), except for lactose). Quantitative growth rates are more informative in assessing the model predicted substrate-dependent biomass yields. Therefore, we performed shake flask experiments for growth on 15 carbon substrates (see Supplementary Materials 5), using both the earlier qualitative I. orientalis data (Seo et al., 2007; Hisamatsu et al., 2006) and the predictions of iIsor850 as guides. Fig. 4A shows summary results for each substrate that exhibited growth (see Supplementary Materials 5 for data) at two of the time points for two of the basal growth media used. We approximated the maximal growth rate on each carbon substrate for the two growth basal media, shown in Fig. 4B, as the mid-log exponential growth rate. We compared these values to those generated by the model using the corresponding substrate uptake rate estimates shown in Figure 4C. The model correctly predicted positive growth for the same seven carbon substrates that could be used for growth in vivo (i.e., glycerol, ethanol, succinic acid, citric acid, fructose, glucose, and lactic acid), and correctly predicted no growth for the remaining substrates tested other than d-xylose. Interestingly, fructose was not tested in the previous studies but both the model and experiments demonstrated growth. The test of lactose scored negative for growth, which is in agreement with previously reported data (Seo et al., 2007) (data not shown). Quantitatively, the model reasonably matched the growth rate for five of these seven carbon substrates with notable exceptions of underpredicting the growth on ethanol and overpredicting succinic acid (see Figure 4B). The model was also unable to explain the low growth on citrate in YNB medium. We furthermore found that I. orientalis SD108 was unable to grow on d-xylose as a sole carbon source, despite the model’s prediction of growth which were consistent with the presence in its genome of all genes needed for d-xylose conversion to d-xylose-5-P and funneling to pentose phosphate pathway (same as in the draft genome of P. kudriavzevii M12 (Chan et al., 2012)). However, we found that when using glucose and xylose together as the carbon source, after complete utilization of glucose I. orientalis SD108 starts utilizing xylose but at a very slow rate; approximately 10 mol% of the xylose consumed was converted into xylitol found in the media (see Supplementary Materials 5 for time-course media concentrations and OD measurements). We note that P. kudriavzevii VTT-C-75010 can consume d-xylose, enabling its conversion into d-xylonic acid upon the recombinant expression of d-xylose dehydrogenase and initial growth on glucose to maintain biomass production (Toivari et al., 2013).
Fig. 4.
Carbon substrate utilization. In panel A, growth of I. orientalis SD108 in batch culture are shown for two different base media: a synthetic defined medium (SC) show in red bars and rich medium (YNB), shown in blue bars each supplemented with different carbon sources as indicated, using optical density as the measurement for growth. Data shown here are for the two time points used to score overall growth, with the lighter bars indicating 24 h after inoculation and darker bars indicating 48 h. Higher resolution time-course batch cultures (see Supplemental Materials 4) were used to estimate the specific growth rate for each base medium and carbon substrate combination (panel B), with SC medium in red bars and YNB medium in blue. Model predictions for growth rate on SC for the different carbon substrates are indicated as circles in panel B, using experimentally determined specific consumption rates (panel C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As mentioned when presenting network connectivity results above, genome-wide gene essentiality data are not currently available for I. orientalis. In lieu of such data, we identified differences in predictions in I. orientalis and corresponding homologs in S. cerevisiae. In particular, two genes that are predicted by iIsor850 to be essential in I. orientalis have homologs in S. cerevisiae that are non-essential in vivo and according to the model Yeast 8.3.4. Both of these genes, JL09_g1956 (YLR342W; 1,3-beta-glucan synthase) and JL09_g1840 (YBR161W; mannosylinositol phosphorylceramide (MIPC) synthase) have isozymes/paralogs present in S. cerevisiae that were not found in I. orientalis (i.e., YGR032W and YPL057C, respectively), thus resulting in the essentiality of these genes. An additional 11 genes are essential in I. orientalis, but without homologs in S. cerevisiae with which to compare. An additional six genes in I. orientalis have essential homologs in S. cerevisiae but are predicted by iIsor850 to be non-essential in I. orientalis. For four of these (i.e., JL09_g5079 (YKL001C; adenylyl-sulfate kinase), JL09_g4689 (YNL220W; adenylosuccinate synthase), JL09_g4696 (YOR303W; carbamoyl-phosphate synthase (glutamine-hydrolyzing)), and JL09_g5138 (YNR050C; saccharopine dehydrogenase (NADP, l-glutamate forming)) involved with the synthesis of methionine, adenine, arginine and lysine, respectively, two nearly identical isozymes (i.e., sequence identity > 99%) were classified by BLAST searches during the model construction. I. orientalis SD108 is diploid (Xiao et al., 2014), suggesting that these putative isozymes could in fact be alleles of the same gene on the homologous chromosomes. Examining sequences before and after the ORFs (i.e., JL09_g2650 and JL09_g5079, JL09_g4686 and JL09_g4689, JL09_g4693 and JL09_g4696, and JL09_g5105 and JL09_g5138, respectively) revealed similar high sequence identity, thus lending credence to this hypothesis. We attempted to knock out adenylyl-sulfate kinase in I. orientalis SD108 but were unable to target only one of the sequences, and the simultaneous knockout of both was auxotrophic for methionine (data not shown), thus confirming the essential nature of the associated reaction. For the fifth case of YER026C (phosphatidylserine synthase), the protein sequences of the putative isozymes (i.e., JL09_g964 and JL09_g5170) were 83% identical. We were able to target independently the sequence with the higher identity to YER026C in S. cerevisiae, but the secondary isozyme was unable to rescue this knock out. It is possible that the isozyme is not expressed under the tested conditions or perhaps typically catalyzes a different reaction and its secondary catalytic activity as phosphatidylserine synthase is insufficient for growth. For the sixth gene JL09_g4419 (YFL017C; N-acetylglucosamine-6-phosphate synthase) which is involved in chitin formation, iIsor850 predicted that it is non-essential if N-acetylglucosamine-6-phosphate deacetylase is reversible. We were unable to knock out JL09_g4419 suggesting that the deacetylase is probably not reversible under the examined conditions. Indeed, we note that the reverse direction of N-acetylglucosamine-6-phosphate deacetylase purified from E. coli is reported to be kinetically unfavorable (Souza et al., 1997).
We also examined predicted non-essential genes whose deletion in iIsor850 leads to an impact on biomass yield. Upon inspection, we found that the deletion of most of these genes only had a minor effect (i.e., max growth rate was predicted to be within 98% that of the wild type). However, for four enzymatic activities the model predicted a substantial reduction, but not an elimination, of growth upon their individual deletion. These enzymes were cytochrome reductase, ATP synthase, complex I NADH dehydrogenase, and phosphoglycerate kinase. The first three are multi-subunit enzyme complexes with 8, 14, and 8 gene products, respectively, predicted to be subunits. The associated genes for these four enzymatic activities are given within Supplementary Materials 7. We individually targeted three subunits of cytochrome reductase (i.e., JL09_g850, JL09_g2223, and JL09_g2279) for deletion in vivo but only saw a reduction of cell growth rate to as low as 86% that of wild-type for the three mutants, as opposed to the 23% of the wild-type growth rate predicted by the model. When we targeted JL09_g1386 encoding one of the subunits of ATP synthase, we saw a substantial reduction in growth rate in vivo to 35% that of wild-type, which compared favorably to the 40% reduction predicted by the model. When examining the impact on NADH dehydrogenase, we targeted in vivo JL09_g1702 and JL09_g4151. We observed a reduction in growth to 87% that of wild-type for the mutants, which was only slightly higher than the 76% predicted by the model. We were unsuccessful in achieving a viable deletion of JL09_g220 which encodes phosphoglycerate kinase, despite the fact that the predicted reduction in growth rate was only slight, to 91%. This result is consistent with observations for S. cerevisiae, in that the enzyme is essential in vivo, yet Yeast 8.3.4 also predicts viability of the knockout.
Transketolase (TKL) carries out both reactions transketolase 1 (TKT1) and transketolase 2 (TKT2). Unlike S. cerevisiae, which has two paralogs TKL1 (YPR074C) and TKL2 (YBR117C), the only homolog we identified in I. orientalis SD108 is locus JL09_g3308. When knocking out JL09_g3308, FBA results indicated no growth on glucose, which is consistent with the results in S. cerevisiae for the tkl1 tkl2 double null mutant that is auxotrophic for aromatic amino acids. Using the succinic acid producing mutant strain described in (Xiao et al., 2014), we attempted the construction of knockout mutant for JL09_g4514 (GND), however we were unable to create the null GND mutant in vivo. In S. cerevisiae, unlike I. orientalis, there are paralogs of this gene, GND1 (YHR183W) and GND2 (YGR256W). Although in S. cerevisiae gnd1 or gnd2 null mutants are viable, the double mutant gnd1 gnd2 is not viable, possibly because of the accumulation of 6-O-phosphono-D-glucono-1,5-lactone. We were also unable to create an ENO deletion mutant by knocking out JL09_g3824. As with GND, in S. cerevisiae, there are paralogs ENO1 and ENO2 for enolase which are absent in I. orientalis. Although in S. cerevisiae eno1 or eno2 null mutants are viable, the double mutant eno1 eno2 is inviable (i.e., synthetic lethal). FBA on the model results indicated no growth on glucose for both the GND and ENO eliminations. We note that Yeast 8.3.4 also incorrectly predicts viability of the GND knockout as well as tolerance of all four lower glycolysis reaction elimination, including ENO knockout. We were able to construct a malate dehydrogenase knockout (JL09_g238, YKL085W) in I. orientalis SD108 and confirmed its viable growth in YPD medium.
The model also predicted that I. orientalis SD108 may contain a complete pathway to produce biotin (vitamin H), unlike many strains of S. cerevisiae, including the reference genome strain S288c. Specifically, we found loci that correspond to BIO1 and BIO6 (i.e., JL09_g2082 and JL09_g2083, respectively). Both genes were found to be expressed by qPCR and I. orientalis SD108 was able to grow on modified YPD medium without biotin supplementation with no observed differences in medium supplemented with or without biotin (see Supplementary Materials 6). We were unable to knockout either gene and rescue with biotin supplementation.
3.5. Model-guided metabolic engineering strategies for succinic acid production
We evaluated the applicability of the model for guiding metabolic engineering strategies for the industrial production of organic acids, by using the OptKnock framework (Burgard et al., 2003). OptKnock identifies reaction deletions, upon which gene deletions can be suggested based on model’s GPR associations, that lead to the targeted overproduction of a desired metabolite by ensuring that the target metabolite production is coupled to growth. Specifically, as proof of concept we applied the OptKnock framework using iIsor850 for growth on glucose with the target of succinic acid production. For all OptKnock analyses in this section, we excluded transporters and reactions without assigned GPR associations from being selected as targets, and also excluded all but one representative member of each fully coupled reaction set described above. Reactions are located in the cytoplasm unless indicated otherwise with a compartment denotation appended (e.g., with _m for mitochondrion).
As found by FBA, the minimal oxygen uptake that does not impact the maximum growth rate is 18.18 mol on a 10 mol glucose basis, and so we set a constraint that limits oxygen uptake to no greater than this amount, which we refer to hereafter as aerobic conditions. Initially, under aerobic growth we identified as many as 105 distinct solutions involving combinations of 1–3 of 36 sets of reactions, as listed in Supplementary Materials 6, that enforce coupling of growth with succinic acid production (i.e., production occurs to maximize biomass production). The growth coupling requirement of succinic acid production eliminated an additional 11 solutions that had a lower bound of zero for succinic acid production at maximum biomass flux (identified using flux variability analysis (FVA) (Mahadevan and Schilling, 2003)). Notably, reaction succinate dehydrogenase (ubiquinone-7) (SUCDq7_m) occurred in every solution. Succinate dehydrogenase is the electron transport chain known as Complex II and occurs in the mitochondria via the involvement of multiple subunits. The GPR association for this reaction in iIsor850 is “SDH1 and SDH2 and SMI1 and (TIM18 or SDH4) and (SDH5 or SDH8)”. In the only single reaction (i.e., SUCDq7_m) elimination solution identified by OptKnock, at least 0.15 mol succinic acid are produced using a basis of 10.0 mol glucose with a corresponding reduction in growth rate of only 2% compared to wild-type. Upon deleting of both JL09_g265 (SDH1) and JL09_g3504 (SDH2) in vivo we observed a slight increase in succinic acid production consistent with the OptKnock predictions. The knockout in the corresponding succinate dehydrogenase enzyme complex in S. cerevisiae (i.e., a sdh3 null mutant) has been shown to have improved both succinic acid titer and yield (Otero et al., 2013).
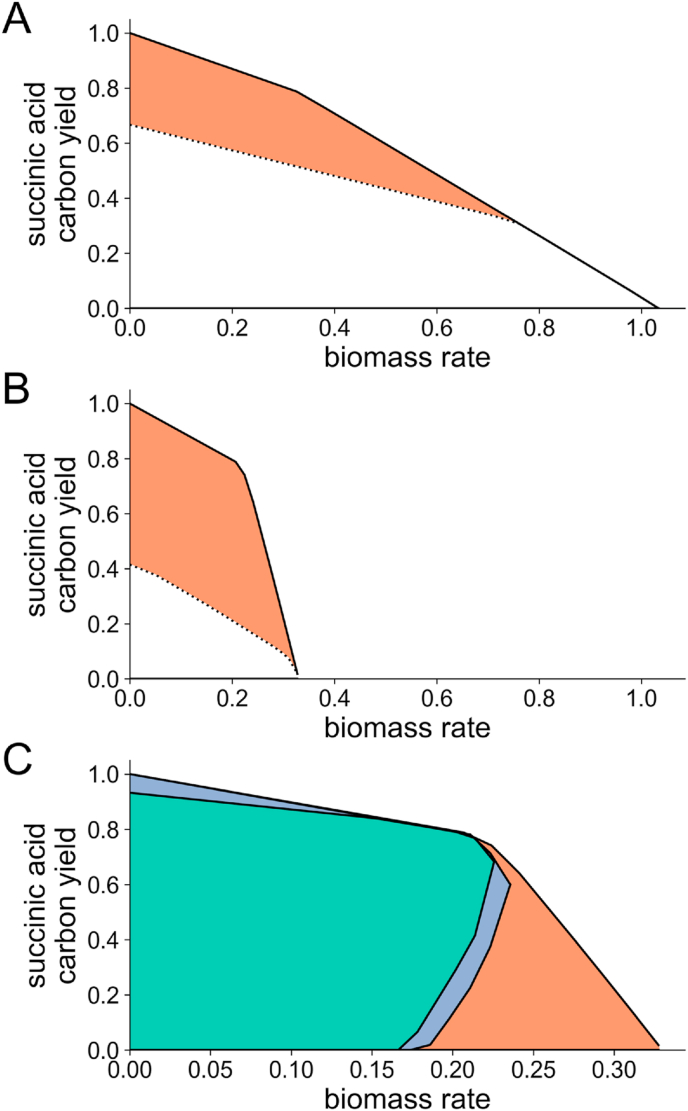
Two concerns with the industrial applicability arose of the solutions found for the wild type strain under aerobic glucose growth. The first is the ability to implement the solution in vivo. Because OptKnock operates on the reaction level, we examined the GPR associations for these reactions and evaluated the feasibility of the associated gene knockouts. Filtering out all reaction eliminations from the solution sets reported to be inviable in vivo in S. cerevisiae reduces the number of solutions to 35 that have exactly three reaction eliminations in each. The second is that most these solutions (i.e., 30) had just greater than 25% carbon yield of succinic acid, and the top remaining solution involved a guaranteed succinic acid yield of just 38.7% carbon yield and 62% of biomass yield compared to wild-type. In fact, flux variability analysis (FVA) revealed that the maximal conversion of glucose into succinic acid for the wild type network is 1:1 and occurs when cells are not growing (i.e., no glucose is converted into biomass). This condition, however, still causes the loss of 2 of the 6 carbons in glucose, resulting in a maximum of 66.7% carbon yield for succinic acid. Heterologous expression of a fumarate reductase (frd) gene codon optimized for S. cerevisiae has been shown in vivo to result in increased succinic acid productivity (Xiao et al., 2014). By adding the reaction fumarate reductase to the network, along with a reaction to regenerate its co-factor, FVA indicated that all 6 carbons of glucose could be routed into succinic acid for the maximum theoretical yield of 15.0 mol succinic acid using a basis of 10.0 mol glucose (i.e., a carbon yield of 100%). Production envelopes for succinic acid for both the wild-type and +frd are shown in Fig. 5A.
Fig. 5.
Succinic acid production envelopes for growth on glucose. A) Aerobic conditions for both wild type (dotted) and with heterologous expression of fumarate reductase (frd) (solid). The increased production capacity enabled by frd expression is shaded. B) Microaerobic conditions for both wild type (dotted) and with heterologous expression of fumarate reductase (frd) (solid). The increased production capacity enabled by frd expression is shaded. For both the maximum growth rate is lower than aerobic conditions (cf. panel A), whereas the absolute maximum succinic acid conversion possible is lower for only the wild type (cf. panel A). C) Microaerobic production of succinic acid from glucose with heterologous frd expression with a set of 5 OptKnock suggested simultaneous reaction eliminations (green shaded), and with a subset of 3 OptKnock suggested simultaneous reaction eliminations (purple shaded), indicating the impact of successive eliminations. The same conditions without the reaction eliminations is shown for comparison (orange shaded). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
FVA further uncovered that high oxygen uptake rates would in principle allow the cell to divert most glucose to biomass rather than succinic acid, as indicated by the rightmost parts of Fig. 5A, thus reducing carbon yield of succinic acid. We examined the impact of setting an additional constraint to simulate microaerobic condition as a model-testable strategy to improve succinic acid production. Introducing microaerobic conditions during the shift from aerobic (growth phase) to anaerobic (production phase) conditions has been observed in vivo to improve succinic acid production in E. coli (Martinez et al., 2010). Also, an engineered Corynebacterium glutamicum exhibited increased production of acetate and succinic acid under microaerobic conditions (Yamauchi et al., 2014). Using the glucose uptake basis used previously (i.e. 10 mol glucose), we found that a minimal amount of 3.17 mol oxygen is required for the maximal theoretical succinic acid yield. Such uptake amount was used in our simulation for microaerobic conditions, and the resulting maximum growth rate is 31.7% of that in aerobic conditions. Production envelopes showing carbon yield for succinic acid under these microaerobic conditions is shown in Fig. 5B. We note that even under minimal oxygen uptake levels (e.g., 0.057 mol, which permits 10% of the maximum aerobic biomass flux rate) the maximum carbon yield for succinic acid is 83.1%. These results underscore the impact operational conditions can have on production.
We thus decided to explore the ramifications of modifying both the network and operational conditions. We subsequently reapplied OptKnock using 1) the network augmented with fumarate reductase, 2) an additional constraint on oxygen uptake to not exceed the microaerobic value, 3) coupled-sets of reactions to reduce the set of reactions allowed to be selected, and 4) the prevention of selecting reactions associated with inviable in vivo mutants in S. cerevisiae. The results were not simply a recapitulation of the previous aerobic results. We identified as many as 336 different solutions that enforce coupling of growth with succinic acid production (i.e., non-zero required production of succinic acid for a biomass yield of at least 10% of the microaerobic maximum value. These solutions contained between 1 and 6 from a list of 78 different reactions. Unlike under aerobic conditions, succinate dehydrogenase (ubiquinone-7) (SUCDq7_m) only occurred in a few the solutions (i.e., less than 10%). Examination of solutions containing SUCDq7_m revealed that these were a subset of the aerobic solutions and were below the median microaerobic solution; the best only guaranteed a 18.8% carbon yield, accentuating the difference that operating conditions can make. Furthermore, no single reaction occurred in every solution, and the triple knockout solutions were not all supersets of the double knockout solutions. Fig. 5C illustrates the production envelope of one of the top solutions, which involves the simultaneous elimination of 1) pyruvate decarboxylase, 2) aspartate transaminase, 3) valine transaminase, 4) glucose 6-phosphate dehydrogenase, and 5) homoserine dehydrogenase. This solution results in 66% carbon yield of succinic acid at maximal growth, which is 69% of that for the +frd strain with no reaction eliminations under microaerobic conditions. A subset of this solution, consisting of the first 3 reaction eliminations (i.e., pyruvate decarboxylase, aspartate transaminase, and valine transaminase) was also identified as a potential solution, albeit with an inferior predicted conversion, as indicated in Fig. 5C. Among the reactions present in some solutions we found threonine aldolase, GLY1 (YEL046C), which has been identified elsewhere as a potential knockout target, based on gene expression data for an evolved strain of S. cerevisiae that had deletions in both succinate dehydrogenase and 3-phosphoglycerate dehydrogenase and exhibited improved succinic acid titers (Otero et al., 2013). We also found PGL (6-phosphogluconolactonase), the elimination of which results in no flux through the pentose phosphate pathway (PPP). We note that a strategy for increasing succinic acid production by disrupting PPP via a G6PD (glucose-6-phosphate dehydrogenase) knockout was described in S. cerevisiae (Patil et al., 2005).
4. Conclusion
In this work, we introduced the genome-scale metabolic model iIsor850 which encapsulates the metabolic capabilities of I. orientalis SD108. To improve the predictive capabilities of the model we derived its biomass composition based in part on our biomass composition measurements and estimated ATP maintenance requirements using chemostats growth data collected for the present study. We generated in vivo carbon substrate utilization data and captured in silico all seven of the 15 which enabled positive growth phenotypes. By predicting and testing in vivo gene essentiality and growth defects resulting from gene deletions, we were able to validate and improve the model GPR association assignments. In general, the modeling and validation stages revealed some of the complexities that can occur when working with non-model organisms, especially those that are not monoploid. We envision that iIsor850 will facilitate the metabolic engineering of I. orientalis for organic acid production.
Previous studies have examined the impact of extracellular pH has on S. cerevisiae and found increased activity of the plasma membrane ATPase, at the expense of ATP (Pampulha and Loureirodias, 1989). In particular, incubating S. cerevisiae in acetic acid dissipates the proton motive force across the plasma membrane (Guldfeldt and Arneborg, 1998). Such effects might impact I. orientalis SD108, especially at higher titers of organic acid production and could necessitate reevaluating the ATP maintenance costs associated with the production environment.
Looking to the future, we note that subsequent iterations of metabolic reconstructions of I. orientalis can incorporate additional biochemical information in order to address aspects of iIsor850, such as reactions without associated genes or reconnecting blocked metabolites. For instance, the remaining blocked metabolites identified by MEMOTE are targets for further verification (for instance by probing the occurrence of these metabolites in vivo) or hypotheses generation for possible reaction functionalities involving these metabolites which could be experimentally examined, thereby resulting in subsequent updates to the model. In addition, genome-wide single gene deletion data is necessary to improve the GPR association assignments (Thiele and Palsson, 2010); such experiments for I. orientalis could prove insightful for refining iIsor850. Additionally, continued efforts on genetic tools in I. orientalis will streamline efforts to perform interventions predicted to achieve metabolic engineering targets. Although the phylum to which I. orientalis belongs (i.e., Ascomycota) contains other genome-scale network reconstructions (Monk et al., 2014), iIsor850 provides a more exact metabolic reconstruction for simulation and expands the coverage of fungal genomes.
Availability of data and material
The model is provided in Supplementary Materials 8 in JSON and SBML (version 3, level 1) versions. A list of putative paralogs is in Supplementary Materials 1. The data for biomass composition for free fatty acids are provided in Supplementary Materials 2. The formulation of the biomass reaction is available in Supplementary Materials 3. The data for glucose uptake rates under carbon limited conditions in a chemostat are in Supplementary Materials 4. The carbon substrate growth data and growth phenotype data for the gene deletion strains are provided in Supplementary Materials 5. qtPCR data for biotin experiments are in Supplementary Materials 6. The Flux Coupling Finder results and OptKnock strain redesign suggestions are provided in Supplementary Materials 7.
CRediT authorship contribution statement
Patrick F. Suthers: Methodology, Software, Investigation, Data curation, Writing - original draft, Visualization. Hoang V. Dinh: Methodology, Software, Data curation, Writing - review & editing, Visualization. Zia Fatma: Methodology, Investigation, Writing - review & editing. Yihui Shen: Methodology, Investigation, Writing - review & editing. Siu Hung Joshua Chan: Methodology, Software, Writing - review & editing. Joshua D. Rabinowitz: Supervision, Resources, Writing - review & editing. Huimin Zhao: Conceptualization, Supervision, Resources, Writing - review & editing, Project administration. Costas D. Maranas: Conceptualization, Supervision, Resources, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Computations for this research were performed on the Pennsylvania State University’s Institute for Computational and Data Sciences’ Roar supercomputer.
This work was supported by U. S. Department of Energy Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018260.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00148.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abbott D.A., Zelle R.M., Pronk J.T., van Maris A.J. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9(8):1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018;36(7):566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung H.W., Henry S.A., Walker L.P. Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism. Ind. Biotechnol. 2013;9(4):215–228. doi: 10.1089/ind.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagurunathan B., Jonnalagadda S., Tan L., Srinivasan R. Reconstruction and analysis of a genome-scale metabolic model for Scheffersomyces stipitis. Microb. Cell Factories. 2012;11:27. doi: 10.1186/1475-2859-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua D., Kim J., Reed J.L. An automated phenotype-driven approach (GeneForce) for refining metabolic and regulatory models. PLoS Comput. Biol. 2010;6(10) doi: 10.1371/journal.pcbi.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Nikolaev E.V., Schilling C.H., Maranas C.D. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14(2):301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Fatma Z., Song X., Hsieh P.-H., Tran V.G., Lyon W.L., Sayadi M., Shao Z., Yoshikuni Y., Zhao H. A genetic toolbox for metabolic engineering of Issatchenkia orientalis. Metab. Eng. 2020;59:87–97. doi: 10.1016/j.ymben.2020.01.005. [DOI] [PubMed] [Google Scholar]
- Caspeta L., Shoaie S., Agren R., Nookaew I., Nielsen J. Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials. BMC Syst. Biol. 2012;6:24. doi: 10.1186/1752-0509-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.F., Gan H.M., Ling H.L., Rashid N.A. Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot. Cell. 2012;11(10):1300–1301. doi: 10.1128/EC.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.H.J., Cai J., Wang L., Simons-Senftle M.N., Maranas C.D. Standardizing biomass reactions and ensuring complete mass balance in genome-scale metabolic models. Bioinformatics. 2017;33(22):3603–3609. doi: 10.1093/bioinformatics/btx453. [DOI] [PubMed] [Google Scholar]
- Chowdhury R., Chowdhury A., Maranas C.D. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models. Metabolites. 2015;5(4):536–570. doi: 10.3390/metabo5040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.K., Selvarasu S., Andrea C., Ryu J., Lee H., Ahn J., Lee H., Lee D.Y. Genome-scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement. Microb. Cell Factories. 2010;9:50. doi: 10.1186/1475-2859-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti S.T., Pinel D., Geiselman G.M., Ito M., Mondo S.J., Reilly M.C., Cheng Y.-F., Bauer S., Grigoriev I.V., Gladden J.M., Simmons B.A., Brem R.B., Arkin A.P., Skerker J.M. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. Elife. 2018;7 doi: 10.7554/eLife.32110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M., Juty N., Knupfer C., Waltemath D., Zhukova A., Drager A., Dumontier M., Finney A., Golebiewski M., Hastings J. Controlled vocabularies and semantics in systems biology. Mol. Syst. Biol. 2011;7:543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh H.V., Suthers P.F., Chan S.H.J., Shen Y., Xiao T., Deewan A., Jagtap S.S., Zhao H., Rao C.V., Rabinowitz J.D., Maranas C.D. A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data. Metab Eng Commun. 2019;9 doi: 10.1016/j.mec.2019.e00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson P.D., Smallbone K., Jameson D., Simeonidis E., Lanthaler K., Pir P., Lu C., Swainston N., Dunn W.B., Fisher P. Further developments towards a genome-scale metabolic model of yeast. BMC Syst. Biol. 2010;4:145. doi: 10.1186/1752-0509-4-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass A.P., Offei B., Braun-Galleani S., Coughlan A.Y., Martos A.A.R., Ortiz-Merino R.A., Byrne K.P., Wolfe K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLoS Pathog. 2018;14(7) doi: 10.1371/journal.ppat.1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte N.C., Herrgard M.J., Palsson B.O. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14(7):1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A., Lerman J.A., Palsson B.O. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatma Z., Schultz J.C., Zhao H. Recent advances in domesticating non-model microorganisms. Biotechnol. Prog. 2020 doi: 10.1002/btpr.3008. [DOI] [PubMed] [Google Scholar]
- Feist A.M., Herrgard M.J., Thiele I., Reed J.L., Palsson B.O. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 2009;7(2):129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J., Famili I., Fu P., Palsson B.O., Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13(2):244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzemeier C.J., Hartleb D., Szappanos B., Papp B., Lercher M.J. Erroneous energy-generating cycles in published genome scale metabolic networks: identification and removal. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A.E.A., Aert R., Agostini-Carbone M.L., Ahmed A., Aigle M., Alberghina L., Albermann K., Albers M., Aldea M., Alexandraki D. The yeast genome directory. Nature. 1997;387(6632):5–6. [Google Scholar]
- Guldfeldt L.U., Arneborg N. Measurement of the effects of acetic acid and extracellular pH on intracellular pH of nonfermenting, individual Saccharomyces cerevisiae cells by fluorescence microscopy. Appl. Environ. Microbiol. 1998;64(2):530–534. doi: 10.1128/aem.64.2.530-534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44(D1):D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner B.D., Smallbone K., Barker B., Mendes P., Walker L.P. Yeast 5 - an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst. Biol. 2012;6:55. doi: 10.1186/1752-0509-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner B.D., Smallbone K., Price N.D., Walker L.P. Version 6 of the consensus yeast metabolic network refines biochemical coverage and improves model performance. Database. 2013;2013:bat059. doi: 10.1093/database/bat059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28(9):977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- Herrgard M.J., Swainston N., Dobson P., Dunn W.B., Arga K.Y., Arvas M., Bluthgen N., Borger S., Costenoble R., Heinemann M. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 2008;26(10):1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu M., Furubayashi T., Karita S., Mishima T., Isono N. Isolation and identification of a novel yeast fermenting ethanol under acidic conditions. J. Appl. Glycosci. 2006;53(2):111–113. [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen Lars J. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske L., Placzek S., Schomburg I., Chang A., Schomburg D. BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 2019;47(D1):D542–D549. doi: 10.1093/nar/gky1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juty N., Le Novere N., Laibe C. Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res. 2012;40:D580–D586. doi: 10.1093/nar/gkr1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q. The BioCyc collection of microbial genomes and metabolic pathways. Briefings Bioinf. 2019;20(4):1085–1093. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavscek M., Bhutada G., Madl T., Natter K. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst. Biol. 2015;9:72. doi: 10.1186/s12918-015-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven E.J., Pomraning K.R., Baker S.E., Nielsen J. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst Biol Appl. 2016;2:16005. doi: 10.1038/npjsba.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.J., Kim H.U., Lee S.Y. Current state and applications of microbial genome-scale metabolic models. Curr. Opin. Struct. Biol. 2017;2:10–18. [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z.A., Drager A., Ebrahim A., Sonnenschein N., Lewis N.E., Palsson B.O., Escher A web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput. Biol. 2015;11(8) doi: 10.1371/journal.pcbi.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F.M., de Koster C.G., Brul S. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell. 2014;13:2–9. doi: 10.1128/EC.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B.S. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuepfer L., Sauer U., Blank L.M. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005;15(10):1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.S., Maranas C.D. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput. Biol. 2009;5(3) doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C.P., Smiley M.J., Johnson C.J. Emendation of the genus Issatchenkia kudriavzev and comparison of species by deoxyribonucleic acid reassociation, mating reaction, and ascospore ultrastructure. Int. J. Syst. Evol. Microbiol. 1980;30(2):503–513. [Google Scholar]
- Kurtzman C.P., Robnett C.J., Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 2008;8(6):939–954. doi: 10.1111/j.1567-1364.2008.00419.x. [DOI] [PubMed] [Google Scholar]
- Lange H.C., Heijnen J.J. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001;75(3):334–344. doi: 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., Babaei P., Bartell J.A., Blank L.M., Chauhan S., Correia K. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020;38(3):272–276. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zou W., Liu L., Chen J. A constraint-based model of Scheffersomyces stipitis for improved ethanol production. Biotechnol. Biofuels. 2012;5(1):72. doi: 10.1186/1754-6834-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-J., Zhang G.-C., Kwak S., Oh E.J., Yun E.J., Chomvong K., Cate J.H.D., Jin Y.-S. Overcoming the thermodynamic equilibrium of an isomerization reaction through oxidoreductive reactions for biotransformation. Nat. Commun. 2019;10(1):1356. doi: 10.1038/s41467-019-09288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loira N., Dulermo T., Nicaud J.M., Sherman D.J. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst. Biol. 2012;6:35. doi: 10.1186/1752-0509-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes H., Rocha I. Genome-scale modeling of yeast: chronology, applications and critical perspectives. FEMS Yeast Res. 2017;17(5) doi: 10.1093/femsyr/fox050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003;5(4):264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Bennett G.N., San K.Y. Metabolic impact of the level of aeration during cell growth on anaerobic succinate production by an engineered Escherichia coli strain. Metab. Eng. 2010;12(6):499–509. doi: 10.1016/j.ymben.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Miller A.W., Befort C., Kerr E.O., Dunham M.J. Design and use of multiplexed chemostat arrays. JoVE. 2013;(72) doi: 10.3791/50262. e50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M.L., Palsson B.O., Herrgard M.J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J., Nogales J., Palsson B.O. Optimizing genome-scale network reconstructions. Nat. Biotechnol. 2014;32(5):447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- Moretti S., Martin O., Van Du Tran T., Bridge A., Morgat A., Pagni M. MetaNetX/MNXref-reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016;44(D1):D523–D526. doi: 10.1093/nar/gkv1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morio F., Jensen R.H., Le Pape P., Arendrup M.C. Molecular basis of antifungal drug resistance in yeasts. Int. J. Antimicrob. Agents. 2017;50(5):599–606. doi: 10.1016/j.ijantimicag.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Mueller T.J., Berla B.M., Pakrasi H.B., Maranas C.D. Rapid construction of metabolic models for a family of Cyanobacteria using a multiple source annotation workflow. BMC Syst. Biol. 2013;7:142. doi: 10.1186/1752-0509-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookaew I., Jewett M.C., Meechai A., Thammarongtham C., Laoteng K., Cheevadhanarak S., Nielsen J., Bhumiratana S. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst. Biol. 2008;2:71. doi: 10.1186/1752-0509-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norsigian C.J., Pusarla N., McConn J.L., Yurkovich J.T., Drager A., Palsson B.O., King Z. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020;48(D1):D402–D406. doi: 10.1093/nar/gkz1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novère N.L., Finney A., Hucka M., Bhalla U.S., Campagne F., Collado-Vides J., Crampin E.J., Halstead M., Klipp E., Mendes P. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat. Biotechnol. 2005;23(12):1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Oberhardt M.A., Palsson B.O., Papin J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma Y., Endo A., Iwasaki H., Ito Y., Goto S. Isolation and properties of ethanol-using yeasts with acid and ethanol tolerance. J. Ferment. Technol. 1986;64:379–382. [Google Scholar]
- Orozco A.S., Higginbotham L.M., Hitchcock C.A., Parkinson T., Falconer D., Ibrahim A.S., Ghannoum M.A., Filler S.G. Mechanism of fluconazole resistance in Candida krusei. Antimicrob. Agents Chemother. 1998;42(10):2645–2649. doi: 10.1128/aac.42.10.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund T., Nookaew I., Bordel S., Nielsen J. Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling. BMC Syst. Biol. 2013;7:36. doi: 10.1186/1752-0509-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero J.M., Cimini D., Patil K.R., Poulsen S.G., Olsson L., Nielsen J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PloS One. 2013;8(1) doi: 10.1371/journal.pone.0054144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien Edward J., Monk Jonathan M., Palsson Bernhard O. Using genome-scale models to predict biological capabilities. Cell. 2015;161(5):971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampulha M.E., Loureirodias M.C. Combined effect of acetic-acid, ph and ethanol on intracellular ph of fermenting yeast. Appl. Microbiol. Biotechnol. 1989;31(5–6):547–550. [Google Scholar]
- Pan P., Hua Q. Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PloS One. 2012;7(12) doi: 10.1371/journal.pone.0051535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Bae J.H., Ko H.J., Lee S.H., Sung B.H., Han J.I., Sohn J.H. Low-pH production of d-lactic acid using newly isolated acid tolerant yeast Pichia kudriavzevii NG7. Biotechnol. Bioeng. 2018;115(9):2232–2242. doi: 10.1002/bit.26745. [DOI] [PubMed] [Google Scholar]
- Patil K.R., Rocha I., Forster J., Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinf. 2005;6 doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharkya P., Burgard A.P., Maranas C.D. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14(11):2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy H.M., Reed J.L. Evaluating the capabilities of microbial chemical production using genome-scale metabolic models. Curr. Opin. Struct. Biol. 2017;2:91–97. [Google Scholar]
- Ranganathan S., Maranas C.D. Microbial 1-butanol production: identification of non-native production routes and in silico engineering interventions. Biotechnol. J. 2010;5(7):716–725. doi: 10.1002/biot.201000171. [DOI] [PubMed] [Google Scholar]
- Ranganathan S., Suthers P.F., Maranas C.D. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010;6(4) doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers A.C., Goldstein Y., Bockmayr A. Generic flux coupling analysis. Math. Biosci. 2015;262:28–35. doi: 10.1016/j.mbs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Satish Kumar V., Dasika M.S., Maranas C.D. Optimization based automated curation of metabolic reconstructions. BMC Bioinf. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2020;48(D1):D9–D16. doi: 10.1093/nar/gkz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.H., Rhee C.H., Park H.D. Degradation of malic acid by Issatchenkia orientalis KMBL 5774, an acidophilic yeast strain isolated from Korean grape wine pomace. J. Microbiol. 2007;45(6):521–527. [PubMed] [Google Scholar]
- Sohn S.B., Graf A.B., Kim T.Y., Gasser B., Maurer M., Ferrer P., Mattanovich D., Lee S.Y. Genome-scale metabolic model of methylotrophic yeast Pichia pastoris and its use for in silico analysis of heterologous protein production. Biotechnol. J. 2010;5(7):705–715. doi: 10.1002/biot.201000078. [DOI] [PubMed] [Google Scholar]
- Sohn S.B., Kim T.Y., Lee J.H., Lee S.Y. Genome-scale metabolic model of the fission yeast Schizosaccharomyces pombe and the reconciliation of in silico/in vivo mutant growth. BMC Syst. Biol. 2012;6:49. doi: 10.1186/1752-0509-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza J.M., Plumbridge J.A., Calcagno M.L. N-acetylglucosamine-6-phosphate deacetylase from Escherichia coli: purification and molecular and kinetic characterization. Arch. Biochem. Biophys. 1997;340(2):338–346. doi: 10.1006/abbi.1997.9780. [DOI] [PubMed] [Google Scholar]
- Suominen P., Aristidou A., Pentilla M., Ilmen M., Ruohonen L., Koivuranta K., Roberg-Perez K. US Patent; 2012. Genetically modified yeast of the species Issatchenkia orientalis and closely relates species, and fermentation processes using same. [Google Scholar]
- Thalagala T.A.T.P., Kodama S., Mishima T., Isono N., Furujyo A., Kawasaki Y., Hisamatsu M. Study on ethanol fermentation using D-glucose rich fractions obtained from lignocelluloses by a two-step extraction with sulfuric acid and Issatchenkia orientalis MF 121. J. Appl. Glycosci. 2009;56(1):7–11. [Google Scholar]
- Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiukova I.A.P.S., Sandgren M., Nielsen J., Kerkhoven E.J. Genome-scale Model of Rhodotorula Toruloides Metabolism. Biotechnol. Bioeng. 2019;116:3396–3408. doi: 10.1002/bit.27162. [DOI] [PubMed] [Google Scholar]
- Toivari M., Vehkomaki M.L., Nygard Y., Penttila M., Ruohonen L., Wiebe M.G. Low pH D-xylonate production with Pichia kudriavzevii. Bioresour. Technol. 2013;133:555–562. doi: 10.1016/j.biortech.2013.01.157. [DOI] [PubMed] [Google Scholar]
- Tomas-Gamisans M., Ferrer P., Albiol J. Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PloS One. 2016;11(1) doi: 10.1371/journal.pone.0148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V.G., Cao M., Fatma Z., Song X., Zhao H. Development of a CRISPR/Cas9-Based tool for gene deletion in Issatchenkia orientalis. mSphere. 2019;4(3) doi: 10.1128/mSphere.00345-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dash S., Ng C.Y., Maranas C.D. A review of computational tools for design and reconstruction of metabolic pathways. Synth Syst Biotechnol. 2017;2(4):243–252. doi: 10.1016/j.synbio.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werpy T., Petersen G. vol I. 2004. Top Value Added Chemicals from Biomass. (Results of Screening for Potential Candidates from Sugars and Synthesis Gas). [Google Scholar]
- Xiao H., Shao Z., Jiang Y., Dole S., Zhao H. Exploiting Issatchenkia orientalis SD108 for succinic acid production. Microb. Cell Factories. 2014;13:121. doi: 10.1186/s12934-014-0121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Liu L., Zou W., Liu J., Hua Q., Chen J. Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata. Mol. Biosyst. 2013;9(2):205–216. doi: 10.1039/c2mb25311a. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Hirasawa T., Nishii M., Furusawa C., Shimizu H. Enhanced acetic acid and succinic acid production under microaerobic conditions by Corynebacterium glutamicum harboring Escherichia coli transhydrogenase gene pntAB. J. Gen. Appl. Microbiol. 2014;60(3):112–118. doi: 10.2323/jgam.60.112. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D.J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17(12):5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomorrodi A.R., Maranas C.D. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data. BMC Syst. Biol. 2010;4:178. doi: 10.1186/1752-0509-4-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The model is provided in Supplementary Materials 8 in JSON and SBML (version 3, level 1) versions. A list of putative paralogs is in Supplementary Materials 1. The data for biomass composition for free fatty acids are provided in Supplementary Materials 2. The formulation of the biomass reaction is available in Supplementary Materials 3. The data for glucose uptake rates under carbon limited conditions in a chemostat are in Supplementary Materials 4. The carbon substrate growth data and growth phenotype data for the gene deletion strains are provided in Supplementary Materials 5. qtPCR data for biotin experiments are in Supplementary Materials 6. The Flux Coupling Finder results and OptKnock strain redesign suggestions are provided in Supplementary Materials 7.